- 1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, OH, United States
- 2Department of Zoonoses, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt
- 3Division of Medical Laboratory Science, School of Health and Rehabilitation Sciences, The Ohio State University, Columbus, OH, United States
- 4The James Comprehensive Cancer Center, Pelotonia Institute for Immuno-Oncology, The Ohio State University, Columbus, OH, United States
- 5Division of Pulmonary Medicine, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States
- 6Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 7Marker Therapeutics Inc., Houston, TX, United States
- 8Department of Veterinary Biosciences, College of Veterinary Medicine, Ohio State University, Columbus, OH, United States
- 9Infectious Diseases Institute, The Ohio State University, Columbus, OH, United States
The new pandemic virus SARS-CoV-2 emerged in China and spread around the world in <3 months, infecting millions of people, and causing countries to shut down public life and businesses. Nearly all nations were unprepared for this pandemic with healthcare systems stretched to their limits due to the lack of an effective vaccine and treatment. Infection with SARS-CoV-2 can lead to Coronavirus disease 2019 (COVID-19). COVID-19 is respiratory disease that can result in a cytokine storm with stark differences in morbidity and mortality between younger and older patient populations. Details regarding mechanisms of viral entry via the respiratory system and immune system correlates of protection or pathogenesis have not been fully elucidated. Here, we provide an overview of the innate immune responses in the lung to the coronaviruses MERS-CoV, SARS-CoV, and SARS-CoV-2. This review provides insight into key innate immune mechanisms that will aid in the development of therapeutics and preventive vaccines for SARS-CoV-2 infection.
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) reportedly emerged at a live animal market in the Chinese city of Wuhan is currently causing a pandemic and negatively affecting global health (1–3). There are more than 11 million confirmed SARS-CoV-2 infections with an mortality rate that widely varies by country and region (4). Even in industrialized countries, SARS-CoV-2 led healthcare systems approach the brink of collapse by overwhelming their capacity and straining resources. Governments and local leaders ordered the shutdown of cities, regions, countries leading to massive disruptions in the local and global economy. Unlike previous Coronavirus (CoV) infections, the rapid global spread, high transmission rate, longer incubation time, and disease severity of SARS-CoV-2 requires a better understanding of the epidemiology and immunopathogenesis of this viral outbreak in order to learn from this experience and to manage future pandemics.
SARS-CoV-2 is a highly pathogenic CoV (5) (case-fatality rate of 3.6–3.8%) (4, 6) that is related to Severe Acute Respiratory Syndrome CoV (SARS-CoV) (case-fatality rate of 14–15%) and the Middle East Respiratory Syndrome CoV (MERS-CoV) (case-fatality rate of 34.4%), see also Table 1 (138, 139). SARS-CoV, SARS-CoV-2 and MERS-CoV target the lower respiratory system, causing respiratory illnesses, including severe pneumonia, acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) (140, 141). SARS-CoV-2 infection results in higher hospitalization rates in the elderly (>65) and persons with pre-existing conditions including hypertension, diabetes and obesity compared to rates among younger populations without pre-existing conditions (Table 1) (142, 143). In addition to an age disparity, males with COVID-19 appear to have higher risk for worse outcomes and death (143, 144). Epidemiological research of the SARS and MERS infections also showed that males had a higher mortality rate than females (144–146).
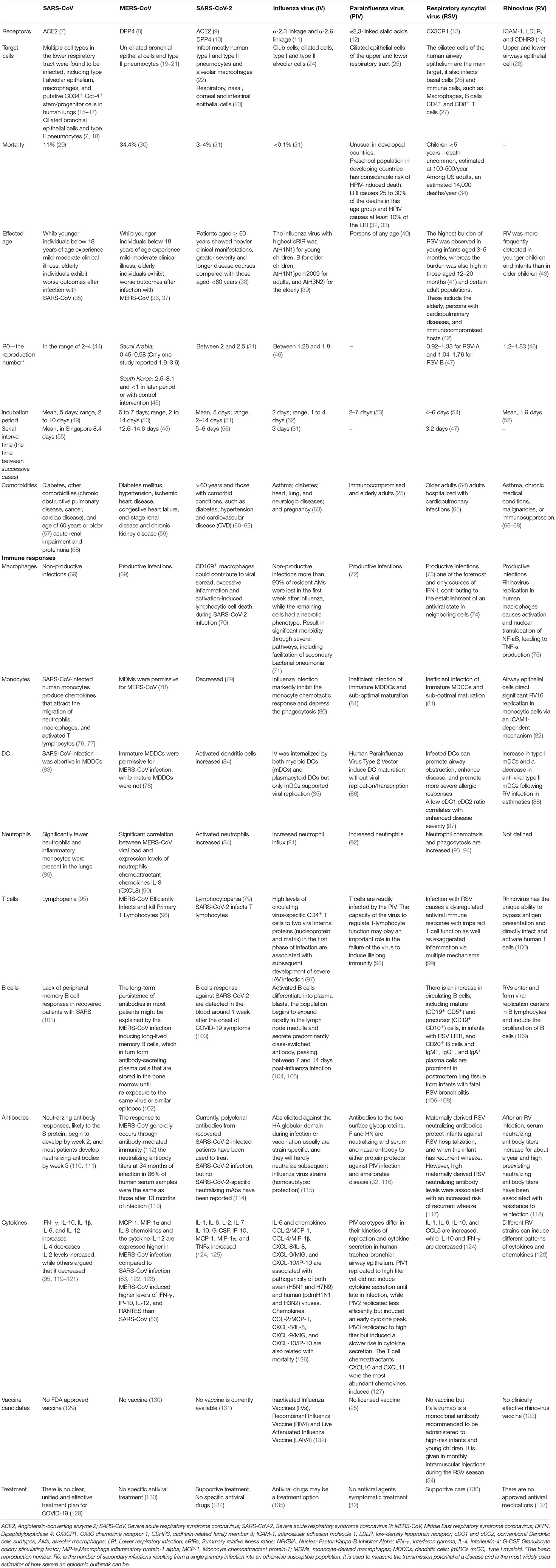
Table 1. Comparison of Immune pathogenesis between highly pathogenic coronaviruses and other significant respiratory viral infections.
While SARS-CoV-2 is a novel coronavirus, several important insights have already been made about its basic mode of transmission. Virus particles are inhaled in respiratory droplets expelled from the airways of infected individuals. Angiotensin-converting enzyme 2 (ACE2), expressed on the ciliated bronchial cells, endothelial cells, and type I and II alveolar cells, is the host receptor for cell entry into the respiratory tract by both SARS-CoV-2 and SARS-CoV (Table 1) (147–150). The spike protein (S) of CoV is responsible for the entry of the virus into the target cell (Figure 1) (147, 151). ACE2 is a type I transmembrane metallocarboxypeptidase that plays a vital role in the Renin-Angiotensin System (RAS) (152, 153), which in turn is critical in maintaining blood pressure homeostasis as well as fluid and salt balance in mammals. ACE2 is found in vascular endothelial cells, in the renal tubular epithelium, and in Leydig cells of the testes (154). Studies have shown that ACE2 is expressed in gastrointestinal (GI) tissues, making it a potential site for harboring SARS-CoV (155). This may be one of the reasons for GI pathology reported in some patients with COVID-19 and viral shedding in stool. In contrast, MERS-CoV uses dipeptidyl-peptidase 4 (DPP4) as an entry receptor, which is expressed on endothelial cells and some epithelial tissues (Table 1) (19, 156).
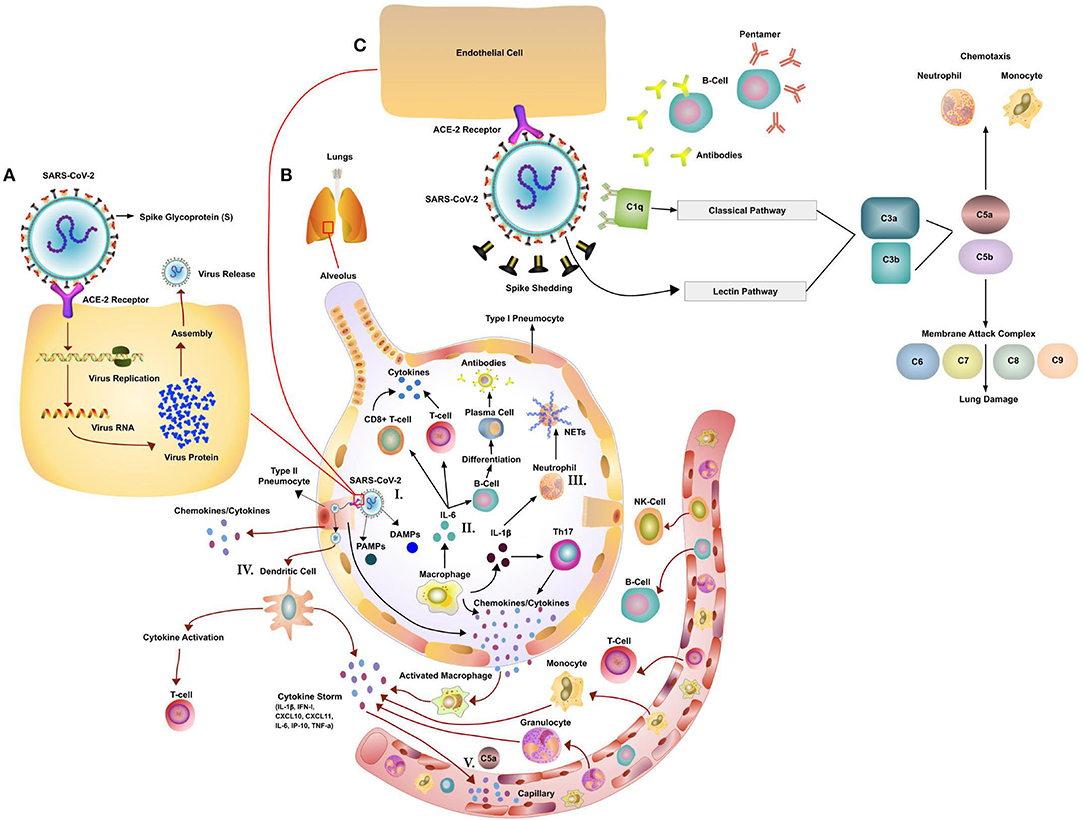
Figure 1. Potential Immune Pathogenesis of SARS-Cov-2. (A) Replication cycle of SARS-CoV-2: Spike protein on the SARS-CoV-2 binds to angiotensin converting enzyme 2 (ACE2), a cell-surface protein. The virion releases its RNA. Some RNA is translated into proteins by the host cell's machinery. Proteins and RNA are assembled into a new virion in the Golgi and released. (B) The innate and adaptive immune responses to Coronavirus (CoV) infection. (I). Initiation of immune response via PAMPs/DAMPS. The host innate immune system detects CoV infections by using pattern recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (PAMPs) and Damage (Danger)-Associated Molecular Patterns (DAMPs). (II) Activation of T-cells and B-cells via cytokines and activation of the complement system. CoV infection leads to macrophages activation and release of inflammatory cytokines. This in turn activates T and B cells and promotes differentiation. Multiple different T cell subsets (i.e., Th1 and Th17) are involved, releasing cytokines for immune response amplification. (III) Activation of Neutrophils (NET formation) Neutrophils, attracted by chemokines/cytokines swarm to the site of infection. Subsequently activated neutrophils undergo degranulation and NET formation releasing intracellular DAMPs, DNA, histones, neutrophil elastase that activate the PRRs of surrounding immune and non-immune cells to induce cytokine secretion. Neutrophils and neutrophil extracellular traps (NETs) drive necroinflammation in COVID-19. The extracellular DNA released by NETs activates platelets and aggregated NETs provide a scaffold for binding of erythrocytes and activated platelets that promote thrombus formation. (IV) Dendritic Cell mediated activation of T-cells. DCs present viral antigens to T-cells inducing activation. (V) Cytokine and C5a led to influx of immune cells. Secrete chemokines, cytokines and complement C5a attract immune cells. (C) Effects of CoV-mediated complement activation. SARS-CoV-2 has been shown to activate the lectin (MBL) complement pathway. Antibodies (early stage IgM or at a later stage IgG) to the virus can activate the classic complement pathway. Both pathways converge at C3. C3 can be converted into C3a and C3b. C3b mediates pathogen opsonization and activates the conversion of C5 into C5a and C5b. C5b mediates the formation of the membrane attack complex, which leads to cell lysis. C3a and C5a promote immune cell recruitment to the site of infection.
Accumulating data suggest that the innate anti-viral response and adaptive immunity may contribute to a cytokine storm leading to systemic hyper inflammation and exacerbation of the disease in patients with (a) co-morbidities (b) older than 65 years of age (c) of the male sex. The exact role of the innate immune system in disease pathogenesis and prevention between the sexes and the impact of age is not fully elucidated. In addition, the potential dysregulation of the innate immune response by SARS-CoV and SARS-CoV-2 is not completely understood which warrants further research.
Respiratory Epithelial Cell and Resident Innate Immune Cell Responses to Respiratory Viral Infections
The cells of the airway epithelium are the first line of defense, providing a mechanical barrier (mucociliary escalator) that expels particles and pathogens via cilia, mucus, and induced coughing (157, 158). This barrier includes cells of the pulmonary epithelium and tissue-resident macrophages and dendritic cells (DCs). The macrophages and DCs express pattern recognition receptors (PRRs) that can detect molecules from pathogens (Pathogen-Associated Molecular Patterns—PAMPs) or molecules released from damaged cells (Damage or Danger-Associated Molecular Patterns—DAMPs) (158–160). In the lung, there are two populations of macrophages, alveolar and interstitial macrophages (161). In addition to these macrophages, DCs play a vital role in facilitating the host defenses against respiratory diseases (162–164). DCs can be divided into plasmacytoid (pDC) and myeloid types (mDC) (165–167). Macrophages and the two DC subtypes trigger antiviral responses by generating a substantial amount of type I interferon and these cells play important roles in immune surveillance in the airways and the distal lung (74, 168–172). During steady state, the DC density in lung associated tissue declines from the trachea to the alveoli (173) while the representation of cells in macrophage compartment seems more complex (174). If a virus infects airway epithelial cells, the viral RNA would be sensed via intrinsic innate receptors, including RIG-1, MDA5, NLRP3 inflammasome, and the RNA sensing TLRs 7 and 8. In the case of influenza A infection, triggering the PRRs causes a strong induction of type 1 interferon (IFN) in epithelial cells (175, 176). In other viral infections, such as Respiratory Syncytial Virus (RSV), alveolar macrophages are the predominant source of type 1 IFNs (161). Furthermore, respiratory epithelial cells and lung macrophages are capable of secreting a broad range of chemokines like IL-8, Macrophage inflammatory protein-1 (MIP-1), RANTES and cytokines including TNF-α, IL-6, IL-1β that influence the types of immune cells being recruited to the area in response to acute viral infections (177, 178).
Macrophages, depending on their polarization status of either M1 or M2, and in a similar way as airway epithelial cells, can further elicit a Th1 or Th2 response (158, 161, 178). In the case of influenza virus infection, the magnitude of epithelial cell response can be proportional to the amount of virus which result in paracrine induction of IFN λ (175). Not only can airway epithelial cells produce a large array of cytokines/chemokines in response to an acute viral infection, but, depending on the magnitude of PRR engagement and the combination of PAMPs and DAMPs triggered, these epithelial cells can modulate the type of chemokines/cytokines produced and influence the influx of innate and adaptive immune cells (158, 160). The response to different viral infections is generally similar, however, the response can be tailored in timing, magnitude and the induced gene signatures in response to each virus (179). Unlike RSV and MERS-CoV, which both productively infect alveolar macrophages (73, 180), seasonal influenza and SARS-CoV usually lead to non-productive infections in these cells (181). In addition, SARS-CoV infection of primary monocytes yielded little virus, likely due to the suppressive effects of IFN-α (182). Thus, the initial cell type(s) involved in propagating a viral infection intensifies the complexity of the immune response.
Another key factor that determines the magnitude of the immune response is the induction and rate of cell death. Although related, MERS-CoV induces widespread cell death when compared to SARS-CoV in human bronchial epithelial cell cultures (183). However, the SARS-CoV open reading frame 3a (ORF3a) protein can induced necrotic cell death in a variety of cell lines (184). The same ORF3a protein can activate the NLRP3 inflammasome, leading to activation of NF-κB and elevated secretion of active IL-1β in cell culture (185).
Cytokine profiles of macrophages activated by SARS-CoV and MERS-CoV are different (180). Nonetheless, both MERS-CoV and SARS-CoV, in human epithelial cell and fibroblast culture, show a delay (24–30 h post-infection) in the induction of proinflammatory cytokines (186), with slightly different cytokine/chemokine profiles. This delay in cytokine induction was confirmed in another study using the same epithelial cell lines (187) as well as in human alveolar type II cells (18). In both cell lines and primary alveolar type II cells, SARS-CoV induced IFN-β, IFN-λ, CXCL10, CXCL11, IL-6, IP-10, and TNF-α (18, 187). MERS-CoV did not induce IFN-β but induced higher level of IL-8 transcript in cell culture. However, no difference in IL-8 production was observed between SARS-CoV and MERS-CoV at 48 h post-infection (186). This was confirmed in-vivo using a non-human primate model comparing the immune responses to SARS-CoV infection between young adult cynomolgus macaques (3–5 yrs) and older macaques (10–19 yrs) (188).
Interestingly, the high induction of IL-8 was observed on a transcript level in the older animals, while the younger once showed higher levels of IFN-β transcript (188). In all animals, the expression of IFN-β was inversely correlated with the pathology score, supporting the role of IFN-β in controlling disease severity (188) and introducing a potential area of research to define age disparity observed in patients infected with SARS-CoV-2. Both older age and male sex are important factors associated with high mortality of SARS-CoV and SARS-CoV-2 infection (189, 190). Many viruses have evolved to disrupt and subvert the immune responses. A common virus that is well-known to affect the lower airway and counteract the immune response is RSV (178, 191). The RSV genome encodes non-structural proteins NS1 and NS2 that can block type 1 IFN production and signaling in cell cultures (191). Similar to RSV, the Measle virus V protein blocks IFN-α and β signaling by inhibiting Stat1 and Stat2 signaling in cell lines (192), whereas MERS-CoV M protein also suppresses type 1 IFN by inactivating IRF3 (193), leading to the low expression of IFN-β.
In contrast to reports in epithelial cell lines or primary alveolar type II cell culture and observations in non-human primates, SARS-CoV nucleocapsid (N) protein and membrane (M) protein, as well as nsp1, can suppress IFN response via various mechanisms in cell lines (194–196). To bridge the dichotomy of inhibition of IFN signaling in cell lines, and the IFN expression in vivo, cells recruited by the infection need to be considered as a potential source. As previously discussed, infected epithelial cells via paracrine signaling to neighboring cells and resident macrophages, secrete chemokines and cytokines to attract other immune cells.
In general, monocytes/macrophages are recruited by CCL3 (MIP-1a), CCL2 (MCP-1), and neutrophils are recruited by IL-8 (CXCL8), CXCL2, and CXCL5 chemokines, all of which can be secreted by airway epithelial cells (178, 197, 198). Both monocytes and neutrophils are also recruited by complement fragment (anaphylatoxin) C5a (Figure 1). Both Influenza and SARS virus can induce acute lung injury (ALI) which is accompanied by high levels of C5a, leading to the influx and activation of innate immune cells (199) (Figure 1).
Serum samples and lung tissue of SARS patients showed high-level expression of CXCL10 (IP-10), which is also found to be induced by SARS-CoV in the epithelial cell line Calu-3 (200). Significant neutrophil, macrophage and CD8 T-cell infiltration can be found in the lung of SARS patients by immunohistochemistry (76, 77, 201). In addition to post-mortem lung histology analysis in patients with SARS-CoV, experiments using Rhesus macaques infected with SARS-CoV found monocyte and macrophage recruitment. The accumulation of pathogenic inflammatory monocyte-macrophages (IMMs) was also observed in a SARS-CoV mouse model. The accumulation of IMMs resulted in heightened lung inflammatory cytokine/chemokine levels, extensive vascular leakage, and impaired virus-specific T cell responses (202).
A strong infiltration of CD68 and Mac387 positive monocytes/macrophages were found in the human and animals lung samples (203, 204). Macrophages further stimulate fibroblasts to deposit extracellular matrix leading to pulmonary fibrosis (205), which was also observed in patients who recovered from SARS (206, 207). Autopsy samples acquired from patients with SARS-CoV-2 patients contained viral nucleocapsid protein (NP) positive CD68+ macrophages in the capillaries of the spleen and in lymph nodes, indicating that SARS-CoV-2 might migrate into the spleen and lymph nodes through macrophages. This study also found that CD169+ macrophages appear to mediate SARS-CoV-2 into these tissue sites, contributing to virus dissemination (208). Similar to SARS-CoV-2, SARS viral particles and genomic sequences were detected in monocytes, macrophages as well as within different organs of SARS patients (15). SARS-CoV was shown to infect both immature and mature human monocyte-derived DCs by electron microscopy and immunofluorescence. The detection of negative strands of SARS-CoV RNA in DCs suggests viral replication, but no increase in viral RNA was observed (209). As mentioned above, there was no perceivable increase to SARS-CoV replication in primary monocytes (182). Another study looked at SARS-CoV and MERS-CoV replication in human macrophages, human monocyte-derived macrophages, and dendritic cells (MDDCs) and found that both viruses replicated poorly. MERS-CoV induced IFN-λ1, CXCL10, and MxA mRNAs in both macrophages and MDDCs, however, SARS-CoV was unable to induce such responses (69). Interestingly, depletion of inflammatory monocyte-macrophages in the mouse model partially protected from lethal SARS infection (210). These data suggest that monocytes, macrophages and dendritic cells have essential roles in CoV infection. The severity of disease and the response to these viruses seems to be dependent on the induced cytokine/chemokine profiles and the amplification of the immune response by the recruited cells. Growing evidence of dysregulation of an innate anti-viral response originates from studies using clinical samples (211) and murine models (202, 212, 213).
In addition to dysregulated cellular responses, the complement system may play an important role in SARS-CoV-2 infection (Figure 1). Evidence comes from SARS infected patients who had lower levels of mannan binding lectin (MBL) in serum compared to healthy controls (214). The SARS patients with a higher frequency of MBL gene polymorphisms were associated with lower serum levels or deficiency of MBL (214). It is still unknown if this is also true for COVID-19 patients, which requires further investigation. In cell culture experiments SARS-CoV was able to bind and activate the complement cascade and block viral infection (214). Preliminary findings in a limited number COVID-19 patients found significant deposits of the membrane attack complex (MAC), C4d and MBL-associated serine protease (MASP)2 in the microvasculature indicating sustained, systemic activation (215). The SARS-CoV-2 spike protein was co-localized with C4d and MAC (215). In a non-peer reviewed publication by Gao et al., MERS-CoV, SARS-CoV and SARS-CoV-2 N protein are able to bind to MASP-2 leading to enhanced complement activation (216) (Figure 1). In a later phase of the infection, the complement system might be also triggered via antibodies bound to the virus (Classic activation pathway, Figure 1). This excessive complement activation might play a role in multi organ damage in severe COVID-19 cases (217). In a MERS-CoV mouse model the blockade of the C5a-C5aR axis alleviated not only lung damage but also spleen damage (218). Mice treated with a monoclonal antibody to C5a showed reduced lung infiltration of CD68+ cells and significantly lower cytokine levels of IL-1 β, TNF-α, INF-γ and IL-12 (218). Complement blockade might be an important way to curb part of the immune dysregulation associated with COVID-19. Overall, we need to look closer at the role of the complement system, the recruited innate immune cells and their combined role in pathogenesis, viral clearance and the eventual resolution of the infection.
The Role of Neutrophils
The most abundant leukocytes, neutrophils, play a critical role in clearing viral infections. Neutrophils, attracted by chemokines/cytokines released by tissue-resident macrophages and DCs, swarm to the site of infection. They recognize and release bioactive compounds, including cytokines, chemokines and ROS, as well as NOS in the very early phase of the infection (219, 220). Neutrophils modulate other innate and adaptive immune responses via cytokine/chemokine release and cell death and, therefore, can ameliorate or exacerbate disease progression. Neutrophils infiltrate tissues infected by CoV, including SARS-CoV, Rat coronavirus (rCoV), and Mouse Hepatitis Virus (MHV). A high neutrophil count in the blood of SARS patients at the time of hospital admission is associated with a poor prognosis (221, 222). Gao et al. suggested that patients with SARS-CoV-2 pneumonia can be stratified by neutrophil to lymphocyte ratio (NLR) and age (216). Patients older than 50 years of age and having an NLR ≥ 3.13 had more severe illness, so rapid access to the intensive care unit is required (79, 223). Experiments in mice showed that SARS-CoV disease severity in older mice correlated with increased pulmonary inflammation and influx of neutrophils (224, 225). Infection of rats with rCoV could lead to neutrophil infiltrating into the respiratory tract early after inoculation, followed by the recruitment of macrophages and lymphocytes (226). Infection of mice with a neurotropic murine CoV (MHV-JHM) showed infiltration of neutrophils into the brain as early as the first day after inoculation, which then promoted the recruitment of other types of inflammatory cells into the brain, likely through the loss of the blood-brain barrier integrity (227). Gene expression analysis in experimentally infected rhesus macaques with MERS-CoV revealed the recruitment of neutrophils into infected lung tissue (228, 229).
Angiotensin-converting enzyme inhibitors (ACE-Is) could serve as a potential risk for fatal COVID-19 through the up-regulation of ACE2 (230) and may provide a direct linkage to neutrophils and disease progression. Investigators found that ACE2 modulates IL-17-mediated neutrophil influx by impacting STAT3 activity (231). Animal models used to study the pathogenesis of SARS-CoV-2 have revealed important roles of neutrophils in infection and confirmed findings observed in patients. A new aspect in SARS-CoV-2 infection is the potential role of neutrophil extracellular traps (NETs). The process of NET formation is a specific type of cell death that can be triggered under inflammatory conditions (232, 233), such as GM-CSF+C5a, IL-8, IFN-α+C5a or other TLR response mediators; all conditions present in severe SARS-CoV-2 infection (232, 233). The NET formation has been observed in COVID-19 patients and may contribute to thrombotic complications in COVID-19 patients (234, 235). Microvascular injury and thrombosis have been reported in COVID-19 patients, increasing the likelihood that neutrophil NET formation might play a role (215, 236, 237). NET formation was reported to be involved in clot formation and thrombosis and can further increase inflammation (232, 233). Therefore, neutrophils can attract a second wave of immune cells to the site of infection by cytokine/chemokine secretion as well as via NETosis (238, 239), which included monocytes and natural killer cells. On the other hand aggregated NETs were reported to limit inflammation by degrading cytokines and chemokines and disrupting neutrophil recruitment and activation (240). Despite the presence of neutrophils in SARS-CoV-2-infected tissues, their role in the clearance and/or immunopathology of the viral infection remains unclear. Future studies on the responses of neutrophils to SARS-CoV-2-infection or infected cells in vitro may elucidate the role of neutrophils in the pathogenesis of respiratory CoV infections.
The Potential Role of Natural Killer Cells
Natural Killer (NK) cells are a heterogenic immune cell subset that acts promptly to combat viral infections. They produce significant amounts of IFN-γ, kill virus-infected cells, provide direct support to other innate immune cells, and aid in the adaptive immune response to counter viruses. NK cells express activating receptors that detect viral antigens, enabling the destruction of infected cells (241–244). Lectin-like receptor CD94 and killer immunoglobulin-like receptors, such as CD158b, regulate the function of NK cells. A study of 221 patients with SARS explored the relationship of the number of NK cells and the expression level of their immunoglobulin-like receptor CD158b in the peripheral blood to the severity of SARS (245). The overall count of NK cells and CD158+ NK cells and the percentage of CD158+ NK cells in patients with SARS were significantly lower than counts in healthy subjects (245). A separate study that analyzed lymphocytes and lymphocyte subsets in a cohort of 38 patients with SARS observed reduced NK cell counts in 21 patients (55%) (246). Clinical reports reveal that children appear to have a milder form of SARS-CoV-2, with peripheral blood lymphocyte levels remaining in the normal range, suggesting less immune dysfunction following the disease (247). This could be attributed to healthy children expressing lymphocytes, especially NK cells, in a greater quantity compared to healthy adults (248). Interestingly, previous studies found rapid and significant restoration of lymphocyte subsets including, NK cells, in peripheral blood in patients recovering from the initial stages of SARS infection (249). Although the primary mechanism for the decrease in NK cells and other subsets during disease onset remains unknown, their contribution to SARS-CoV-2 needs further study especially from a treatment perspective.
The Contribution by the Innate Lymphocytes
Innate lymphoid cells (ILCs) are a family of innate immune cells that include ILC1, ILC2 and ILC3. Although ILC2 facilitates lung repair after injury, the role of ILCs during respiratory viral infection is not clearly defined (250). Evidence for the potential involvement of ILC2 cells in the lung during viral infection was reported in a murine model (251). This study found a rapid accumulation of ILC2 cells in the lung after an influenza virus infection, however their initial contribution to exacerbation of the disease was limited (251). A recent study identified an interaction between ACE2-expressing SARS-CoV-2 target cells and ILCs in the colon (252). Thus, elucidating the role of ILC subsets will be important in understanding the pathogenesis of SARS, SARS-CoV-2 and MERS infections.
Roles of Interferons in Coronavirus Infection
There is distinct evidence indicating an important role of IFNs in SARS and other CoV infections (201, 253). The sera of patients with SARS revealed the presence of high levels of IL-1, IL-6, INFγ, CCL2, CXCL10, and IL-8 and products of interferon stimulated genes (254, 255). High expression levels of ISGs such as CD58, IFNAR1, and IFNGR1 and IFN-stimulated chemokines CXCL10 and CCL2 were observed in another cohort of SARS patients and were correlated with the severity of pathogenesis (256). Significant upregulation of CXCL10 gene expression was observed in the severe phase of patients who died from SARS. This data is corroborated by studies in patients with MERS that found upregulation of CXCL10 in the serum of patients who developed pneumonia (254). CXCL10 and INFα were also correlated with severity of disease (255).
The importance of IFN signaling in response to CoV infection has been well-demonstrated in several knockout mouse models. Type I, II, and III IFN signaling deficient mice have increased susceptibility to mouse-adapted SARS-CoV strains (257, 258). Studies using mice lacking the IFNAR1 and IFNLR1 or STAT1 identified higher SARS-CoV replication in the lungs and delayed virus clearance (259, 260). Another study with MERS-CoV in mice expressing the human DPP4 receptor showed a role for the IFNAR1 in viral clearance and lung inflammation (112). These mouse models suggest an important role of IFN response for CoV clearance. This quick expanding medical literature is very suggestive of an important role of IFN responses for CoV control and clearance.
Innate Immune Evasion Strategy of Human Coronaviruses
Many viruses have evolved to disrupt and subvert the immune response. RSV counteracts the immune response (178, 191); as discussed earlier, the RSV genome encodes non-structural proteins (NS1 and NS2) that are able to block type 1 IFN production and signaling in cell cultures (191). Similar to RSV, the Measle virus V protein blocks IFN-α and β signaling by inhibiting Stat1 and Stat2 signaling in cell culture lines (192). CoVs have developed several ways to escape from innate immune pressure. MERS-CoV M protein suppresses type 1 IFN by blocking the IRF3 activation (193), explaining the low expression of IFN-β. In various cell lines, SARS-CoV nucleocapsid (N) protein, membrane (M) protein, as well as nsp1, were reported to suppress IFN response (194, 196, 261). The nucleocapsid protein (N) of SARS-CoV interferes with the function of IRF3. Although it does not form a complex with RIG-I or MDA5, RNA binding activity at the initial recognition stage of viral RNA potentially contributes to immune evasion (261, 262).
Aside from the HCoV, structural proteins, accessory, and non-structural proteins (nsp) are involved in innate immunity modulation. In both SARS-CoV and MERS-CoV, host mRNA endonucleolytic cleavage is promoted by nsp1 protein, which modifies capped non-viral RNAs (263, 264). Nsp1 in SARS-CoV prevents host mRNA translation by interacting with the 40S subunit of the ribosome; in turn, transcription and translation of viral RNA is given preference over the host mRNA (263). Another study found that additional SARS-CoV nsp1 residues interfered with IFN-dependent signaling (265). IFN production is affected by nsp3 proteins in SARS-CoV and MERS-CoV. These proteins have both papain-like protease (PLpro) and a PLP2 domain, and the PLpro domains in both SARS-CoV and MERS-CoV downregulate mRNA levels of CCL5, INFβ, CXCL10, and other pro-inflammatory cytokines (266). The suppression of IFN responses by SARS-CoV PLpro is due to the inhibition of phosphorylation of IFN-regulatory factor 3 (IRF3) and its subsequent translocation to the nucleus where it enhances IFN gene transcription (267). MERS-CoV PLpro also suppresses RIG-I and MDA5 and antagonizes IFN induction (266, 268). In HCoV-229E and SARS-CoV suppression of IFN responses, the key molecule is a ADP-ribose-1-monophosphatase macrodomain encoded within nsp3 (269). Accessory proteins are not key in viral replication; however, in human CoV, this group of proteins are involved in diverse cellular signaling, including cell proliferation, apoptosis, and IFN signaling (270). By downregulating phosphorylation and nuclear translocation of IRF3, Open Reading Frame ORF3b and -6 interfere with IFNβ synthesis and prevent IFNβ-induced activation of IFN-stimulated response element (ISRE) in the promoter of ISG in SARS-CoV (262). In cells transfected with ORF4a, -4b, and -5 of MERS-CoV, IFNβ promoter-driven luciferase activity is significantly reduced, and it may follow a similar pattern of suppression of IRF3 nuclear translocation (141). Therefore, the suppression of signaling events in infected immune and airway epithelial cells, as well as the magnitude of suppression due to elevated expression levels of these accessory proteins, needs to be further elucidated to understand delayed or hyperimmune responses and cytokine storm that occurs in CoV infection.
Summary
In addition to revealing our unpreparedness of handling a worldwide pandemic by a viral infection, COVID-19 exposed our lack of understanding of the pathogenesis of diseases as well as the host immunity. The interaction of the host innate immune system and other factors including age, sex, and pre-existing conditions need further investigation regarding disease severity and morbidity of SARS/MERS and COVID-19. Disease severity and its related progression are further associated with dysregulation of multiple components of both innate and adaptive immune responses leading to a cytokine storm and severe pathology. For the development of a therapeutic intervention, it is vital to elucidate the interplay among the different layers of the innate immune response and their relation to the clinical factors associated with increased morbidity and mortality in CoV infection. Investments in basic science research are needed to help elucidate the roles of different immune cells, and their contribution to disease severity; it will pave the way to prevent or abrogate CoV outbreaks and potentially new viruses.
Author Contributions
NL, TD, DH, ZL, and NF performed the literature search, analyzed the literature, and wrote the manuscript. HA-H, BS, YW, and RS performed the literature search and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
TD is employed by company Marker Therapeutics Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. (2020) 395:470–3. doi: 10.1016/S0140-6736(20)30185-9
2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
3. Ryu S, Chun BC, Korean Society of Epidemiology -nCo VTFT. An interim review of the epidemiological characteristics of 2019 novel coronavirus. Epidemiol Health. (2020) 42:e2020006. doi: 10.4178/epih.e2020006
4. Khafaie MA, Rahim F. Cross-country comparison of case fatality rates of COVID-19/SARS-COV-2. Osong Public Health Res Perspect. (2020) 11:74–80. doi: 10.24171/j.phrp.2020.11.2.03
5. Sun P, Lu X, Xu C, Sun W, Pan B. Understanding of COVID-19 based on current evidence. J Med Virol. (2020) 92:548–51. doi: 10.1002/jmv.25722
6. Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, et al. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel Coronavirus Disease 2019. (COVID-19). J Microbiol Biotechnol. (2020). 30:313–24. doi: 10.4014/jmb.2003.03011
7. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. (2003) 426:450–4. doi: 10.1038/nature02145
8. Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. (2013) 23:986–93. doi: 10.1038/cr.2013.92
9. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. (2020) 367:1444–8. doi: 10.1126/science.abb2762
10. Li Y, Zhang Z, Yang L, Lian X, Xie Y, Li S, et al. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. iScience. (2020) 23:101160. doi: 10.1016/j.isci.2020.101160
11. Shinya K, Kawaoka Y. [Influenza virus receptors in the human airway]. Uirusu. (2006) 56:85–9. doi: 10.2222/jsv.56.85
12. Tong J, Fu Y, Meng F, Kruger N, Valentin-Weigand P, Herrler G. The sialic acid binding activity of human parainfluenza virus 3 and mumps virus glycoproteins enhances the adherence of group b streptococci to HEp-2 Cells. Front Cell Infect Microbiol. (2018) 8:280. doi: 10.3389/fcimb.2018.00280
13. Anderson CS, Chu CY, Wang Q, Mereness JA, Ren Y, Donlon K, et al. CX3CR1 as a respiratory syncytial virus receptor in pediatric human lung. Pediatr Res. (2020) 87:862–7. doi: 10.1038/s41390-019-0677-0
14. Basnet S, Palmenberg AC, Gern JE. Rhinoviruses and their receptors. Chest. (2019) 155:1018–25. doi: 10.1016/j.chest.2018.12.012
15. Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. (2005) 202:415–24. doi: 10.1084/jem.20050828
16. Chen Y, Chan VS, Zheng B, Chan KY, Xu X, To LY, et al. A novel subset of putative stem/progenitor CD34+Oct-4+ cells is the major target for SARS coronavirus in human lung. J Exp Med. (2007) 204:2529–36. doi: 10.1084/jem.20070462
17. Nicholls JM, Butany J, Poon LL, Chan KH, Beh SL, Poutanen S, et al. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. (2006) 3:e27. doi: 10.1371/journal.pmed.0030027
18. Qian Z, Travanty EA, Oko L, Edeen K, Berglund A, Wang J, et al. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am J Respir Cell Mol Biol. (2013) 48:742–8. doi: 10.1165/rcmb.2012-0339OC
19. Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. (2013) 495:251–4. doi: 10.1038/nature12005
20. Abeloos J, Rolly G, Timperman J, Watson AA. Anaesthetic and medico-legal problems in patients intoxicated by alcohol. Med Sci Law. (1985) 25:131–5. doi: 10.1177/002580248502500210
21. Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA. (2013) 110:16157–62. doi: 10.1073/pnas.1311542110
22. Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. (2020). doi: 10.1093/cid/ciaa410. [Epub ahead of print].
23. Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. (2020) 26:681–7. doi: 10.1038/s41591-020-0868-6
24. Fiege JK, Langlois RA. Investigating influenza A virus infection: tools to track infection and limit tropism. J Virol. (2015) 89:6167–70. doi: 10.1128/JVI.00462-15
25. Branche AR, Falsey AR. Parainfluenza virus infection. Semin Respir Crit Care Med. (2016) 37:538–54. doi: 10.1055/s-0036-1584798
26. Persson BD, Jaffe AB, Fearns R, Danahay H. Respiratory syncytial virus can infect basal cells and alter human airway epithelial differentiation. PLoS ONE. (2014) 9:e102368. doi: 10.1371/journal.pone.0102368
27. Raiden S, Sananez I, Remes-Lenicov F, Pandolfi J, Romero C, De Lillo L, et al. Respiratory Syncytial Virus (RSV) infects CD4+ T cells: frequency of circulating CD4+ RSV+ T cells as a marker of disease severity in young children. J Infect Dis. (2017) 215:1049–58. doi: 10.1093/infdis/jix070
28. Ljubin-Sternak S, Mestrovic T, Ivkovic-Jurekovic I, Kolaric B, Slovic A, Forcic D, et al. The emerging role of rhinoviruses in lower respiratory tract infections in children - clinical and molecular epidemiological study from Croatia, 2017-2019. Front Microbiol. (2019) 10:2737. doi: 10.3389/fmicb.2019.02737
29. WHO. Summary Table of SARS Cases by Country, 1 November 2002. - 7 August Report. (2003). Available online at: https://www.who.int/csr/sars/country/2003_08_15/en/
30. WHO. Middle East Respiratory Syndrome Coronavirus (MERS-CoV). (2019). Available online at: https://www.who.int/emergencies/mers-cov/en/
31. WHO. Q&A: Influenza and COVID-19 - Similarities and Differences. (2020). Available online at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/q-a-similarities-and-differences-covid-19-and-influenza?gclid=EAIaIQobChMIy-WTm6GY6gIVi8qyCh3iXwFrEAAYASAAEgIDBvD_BwE
32. Henrickson KJ. Parainfluenza VIruses. Clin Microbiol Rev. (2003) 16:242–64. doi: 10.1128/CMR.16.2.242-264.2003
33. Berman S. Epidemiology of acute respiratory infections in children of developing countries. Rev Infect Dis. (1991) 13(Suppl. 6):S454–62. doi: 10.1093/clinids/13.Supplement_6.S454
34. CDC. Respiratory Syncytial Virus-Associated Mortality (RSV-Associated Mortality). 2019 Case Definition. (2019). Available online at: https://wwwn.cdc.gov/nndss/conditions/respiratory-syncytial-virus-associated-mortality/case-definition/2019/
35. Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. (2003) 361:1319–25. doi: 10.1016/S0140-6736(03)13077-2
36. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. (2013) 13:752–61. doi: 10.1016/S1473-3099(13)70204-4
37. Saad M, Omrani AS, Baig K, Bahloul A, Elzein F, Matin MA, et al. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int J Infect Dis. (2014) 29:301–6. doi: 10.1016/j.ijid.2014.09.003
38. Liu Y, Mao B, Liang S, Yang JW, Lu HW, Chai YH, et al. Association between age and clinical characteristics and outcomes of COVID-19. Eur Respir J. (2020) 55:2001112. doi: 10.1183/13993003.01112-2020
39. Caini S, Spreeuwenberg P, Kusznierz GF, Rudi JM, Owen R, Pennington K, et al. Distribution of influenza virus types by age using case-based global surveillance data from twenty-nine countries, 1999-2014. BMC Infect Dis. (2018) 18:269. doi: 10.1186/s12879-018-3181-y
40. Lau SK, To WK, Tse PW, Chan AK, Woo PC, Tsoi HW, et al. Human parainfluenza virus 4 outbreak and the role of diagnostic tests. J Clin Microbiol. (2005) 43:4515–21. doi: 10.1128/JCM.43.9.4515-4521.2005
41. Ueno F, Tamaki R, Saito M, Okamoto M, Saito-Obata M, Kamigaki T, et al. Age-specific incidence rates and risk factors for respiratory syncytial virus-associated lower respiratory tract illness in cohort children under 5 years old in the Philippines. Influenza Other Respir Viruses. (2019) 13:339–53. doi: 10.1111/irv.12639
42. Falsey AR, Walsh EE. Respiratory syncytial virus infection in adults. Clin Microbiol Rev. (2000) 13:371–84. doi: 10.1128/CMR.13.3.371
43. Linder JE, Kraft DC, Mohamed Y, Lu Z, Heil L, Tollefson S, et al. Human rhinovirus C: Age, season, and lower respiratory illness over the past 3 decades. J Allergy Clin Immunol. (2013) 131:69–77.e1–6. doi: 10.1016/j.jaci.2012.09.033
44. WHO. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS) WHO. Contract No.: WHO/CDS/CSR/GAR/2003.11. (2003). Available online at: https://www.who.int/csr/sars/WHOconsensus.pdf?ua=1
45. Park JE, Jung S, Kim A, Park JE. MERS transmission and risk factors: a systematic review. BMC Public Health. (2018) 18:574. doi: 10.1186/s12889-018-5484-8
46. Biggerstaff M, Cauchemez S, Reed C, Gambhir M, Finelli L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis. (2014) 14:480. doi: 10.1186/1471-2334-14-480
47. Otomaru H, Kamigaki T, Tamaki R, Okamoto M, Alday PP, Tan AG, et al. Transmission of respiratory syncytial virus among children under 5 years in households of rural communities, the Philippines. Open Forum Infect Dis. (2019) 6:ofz045. doi: 10.1093/ofid/ofz045
48. Scully EJ, Basnet S, Wrangham RW, Muller MN, Otali E, Hyeroba D, et al. Lethal respiratory disease associated with human rhinovirus C in wild chimpanzees, Uganda, (2013). Emerg Infect Dis. (2018) 24:267–74. doi: 10.3201/eid2402.170778
49. Varia M, Wilson S, Sarwal S, McGeer A, Gournis E, Galanis E, et al. Investigation of a nosocomial outbreak of severe acute respiratory syndrome (SARS) in Toronto, Canada. CMAJ. (2003) 169:285–92.
50. Virlogeux V, Fang VJ, Park M, Wu JT, Cowling BJ. Comparison of incubation period distribution of human infections with MERS-CoV in South Korea and Saudi Arabia. Sci Rep. (2016) 6:35839. doi: 10.1038/srep35839
51. Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung SM, et al. Incubation period and other epidemiological characteristics of 2019. Novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med. (2020) 9:538. doi: 10.3390/jcm9020538
52. Lessler J, Reich NG, Brookmeyer R, Perl TM, Nelson KE, Cummings DA. Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect Dis. (2009) 9:291–300. doi: 10.1016/S1473-3099(09)70069-6
53. CDC. Human Parainfluenza Viruses (HPIVs). (2019). Available online at: https://www.cdc.gov/parainfluenza/hcp/clinical.html
54. CDC. Respiratory Syncytial Virus Infection (RSV). (2018). Available online at: https://www.cdc.gov/rsv/clinical/index.html
55. Lipsitch M, Cohen T, Cooper B, Robins JM, Ma S, James L, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. (2003) 300:1966–70. doi: 10.1126/science.1086616
56. WHO. Q&As on COVID-19 and Related Health Topics. (2019). Available online at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub
57. Booth CM, Matukas LM, Tomlinson GA, Rachlis AR, Rose DB, Dwosh HA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. (2003) 289:2801–9. doi: 10.1001/jama.289.21.JOC30885
58. Chu KH, Tsang WK, Tang CS, Lam MF, Lai FM, To KF, et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. (2005) 67:698–705. doi: 10.1111/j.1523-1755.2005.67130.x
59. Alqahtani FY, Aleanizy FS, Ali El Hadi Mohamed R, Alanazi MS, Mohamed N, Alrasheed MM, et al. Prevalence of comorbidities in cases of Middle East respiratory syndrome coronavirus: a retrospective study. Epidemiol Infect. (2018) 147:1–5. doi: 10.1017/S0950268818002923
60. Ganguly R, Waldman RH. Respiratory tract cell-mediated immunity. Bull Eur Physiopathol Respir. (1977) 13:95–102.
61. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
62. Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. (2020) 94:91–5. doi: 10.1016/j.ijid.2020.03.017
63. Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, et al. Hospitalized patients with 2009. H1N1 influenza in the United States, April-June. 2009. N Engl J Med. (2009) 361:1935–44. doi: 10.1056/NEJMoa0906695
64. Ackerson B, Tseng HF, Sy LS, Solano Z, Slezak J, Luo Y, et al. Severe morbidity and mortality associated with respiratory syncytial virus versus influenza infection in hospitalized older adults. Clin Infect Dis. (2019) 69:197–203. doi: 10.1093/cid/ciy991
65. Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. (2005) 352:1749–59. doi: 10.1056/NEJMoa043951
66. Iroh Tam PY, Zhang L, Cohen Z. Clinical characteristics and outcomes of human rhinovirus positivity in hospitalized children. Ann Thorac Med. (2018) 13:230–6. doi: 10.4103/atm.ATM_291_17
67. Costa C, Bergallo M, Astegiano S, Sidoti F, Terlizzi ME, Gambarino S, et al. Detection of human rhinoviruses in the lower respiratory tract of lung transplant recipients. Arch Virol. (2011) 156:1439–43. doi: 10.1007/s00705-011-0986-z
68. Parody R, Rabella N, Martino R, Otegui M, del Cuerpo M, Coll P, et al. Upper and lower respiratory tract infections by human enterovirus and rhinovirus in adult patients with hematological malignancies. Am J Hematol. (2007) 82:807–11. doi: 10.1002/ajh.20974
69. Tynell J, Westenius V, Ronkko E, Munster VJ, Melen K, Osterlund P, et al. Middle East respiratory syndrome coronavirus shows poor replication but significant induction of antiviral responses in human monocyte-derived macrophages and dendritic cells. J Gen Virol. (2016) 97:344–55. doi: 10.1099/jgv.0.000351
70. Park MD. Macrophages: a Trojan horse in COVID-19? Nat Rev Immunol. (2020) 20:351. doi: 10.1038/s41577-020-0317-2
71. Ghoneim HE, Thomas PG, McCullers JA. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol. (2013) 191:1250–9. doi: 10.4049/jimmunol.1300014
72. Briggs CM, Holder RC, Reid SD, Parks GD. Activation of human macrophages by bacterial components relieves the restriction on replication of an interferon-inducing parainfluenza virus 5 (PIV5) P/V mutant. Microbes Infect. (2011) 13:359–68. doi: 10.1016/j.micinf.2010.12.005
73. Panuska JR, Cirino NM, Midulla F, Despot JE, McFadden ER Jr, Huang YT. Productive infection of isolated human alveolar macrophages by respiratory syncytial virus. J Clin Invest. (1990) 86:113–9. doi: 10.1172/JCI114672
74. Goritzka M, Makris S, Kausar F, Durant LR, Pereira C, Kumagai Y, et al. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J Exp Med. (2015) 212:699–714. doi: 10.1084/jem.20140825
75. Laza-Stanca V, Stanciu LA, Message SD, Edwards MR, Gern JE, Johnston SL. Rhinovirus replication in human macrophages induces NF-kappaB-dependent tumor necrosis factor alpha production. J Virol. (2006) 80:8248–58. doi: 10.1128/JVI.00162-06
76. Hu W, Yen YT, Singh S, Kao CL, Wu-Hsieh BA. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol. (2012) 25:277–88. doi: 10.1089/vim.2011.0099
77. Yen YT, Liao F, Hsiao CH, Kao CL, Chen YC, Wu-Hsieh BA. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J Virol. (2006) 80:2684–93. doi: 10.1128/JVI.80.6.2684-2693.2006
78. Cong Y, Hart BJ, Gross R, Zhou H, Frieman M, Bollinger L, et al. MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS ONE. (2018) 13:e0194868. doi: 10.1371/journal.pone.0194868
79. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. (2020) 71:762–8. doi: 10.2139/ssrn.3541136
80. Gardner ID, Lawton JW. Depressed human monocyte function after influenza infection in vitro. J Reticuloendothel Soc. (1982) 32:443–8.
81. Le Nouen C, Munir S, Losq S, Winter CC, McCarty T, Stephany DA, et al. Infection and maturation of monocyte-derived human dendritic cells by human respiratory syncytial virus, human metapneumovirus, and human parainfluenza virus type 3. Virology. (2009) 385:169–82. doi: 10.1016/j.virol.2008.11.043
82. Zhou X, Zhu L, Lizarraga R, Chen Y. Human airway epithelial cells direct significant rhinovirus replication in monocytic cells by enhancing ICAM1 Expression. Am J Respir Cell Mol Biol. (2017) 57:216–25. doi: 10.1165/rcmb.2016-0271OC
83. Chu H, Zhou J, Wong BH, Li C, Cheng ZS, Lin X, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. (2014) 454–455:197–205. doi: 10.1016/j.virol.2014.02.018
84. Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. (2020) 27:883–90. e2. doi: 10.1016/j.chom.2020.04.017
85. Smed-Sorensen A, Chalouni C, Chatterjee B, Cohn L, Blattmann P, Nakamura N, et al. Influenza A virus infection of human primary dendritic cells impairs their ability to cross-present antigen to CD8 T cells. PLoS Pathog. (2012) 8:e1002572. doi: 10.1371/journal.ppat.1002572
86. Hara K, Fukumura M, Ohtsuka J, Kawano M, Nosaka T. Human parainfluenza virus type 2 vector induces dendritic cell maturation without viral RNA replication/transcription. Hum Gene Ther. (2013) 24:683–91. doi: 10.1089/hum.2013.024
87. Andrade CA, Pacheco GA, Galvez NMS, Soto JA, Bueno SM, Kalergis AM. Innate immune components that regulate the pathogenesis and resolution of hRSV and hMPV infections. Viruses. (2020) 12:637. doi: 10.3390/v12060637
88. Kirchberger S, Majdic O, Steinberger P, Bluml S, Pfistershammer K, Zlabinger G, et al. Human rhinoviruses inhibit the accessory function of dendritic cells by inducing sialoadhesin and B7-H1 expression. J Immunol. (2005) 175:1145–52. doi: 10.4049/jimmunol.175.2.1145
89. Gralinski LE, Sheahan TP, Morrison TE, Menachery VD, Jensen K, Leist SR, et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. (2018) 9:e01753–18. doi: 10.1128/mBio.01753-18
90. Alosaimi B, Hamed ME, Naeem A, Alsharef AA, AlQahtani SY, AlDosari KM, et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. (2020) 126:154895. doi: 10.1016/j.cyto.2019.154895
91. Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. (2014) 14:315–28. doi: 10.1038/nri3665
92. Pickles RJ, DeVincenzo JP. Respiratory syncytial virus (RSV) and its propensity for causing bronchiolitis. J Pathol. (2015) 235:266–76. doi: 10.1002/path.4462
93. Noah TL, Ivins SS, Murphy P, Kazachkova I, Moats-Staats B, Henderson FW. Chemokines and inflammation in the nasal passages of infants with respiratory syncytial virus bronchiolitis. Clin Immunol. (2002) 104:86–95. doi: 10.1006/clim.2002.5248
94. Sheeran P, Jafri H, Carubelli C, Saavedra J, Johnson C, Krisher K, et al. Elevated cytokine concentrations in the nasopharyngeal and tracheal secretions of children with respiratory syncytial virus disease. Pediatr Infect Dis J. (1999) 18:115–22. doi: 10.1097/00006454-199902000-00007
95. Wong RS, Wu A, To KF, Lee N, Lam CW, Wong CK, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. (2003) 326:1358–62. doi: 10.1136/bmj.326.7403.1358
96. Chu H, Zhou J, Wong BH, Li C, Chan JF, Cheng ZS, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. (2016) 213:904–14. doi: 10.1093/infdis/jiv380
97. Zhao Y, Zhang YH, Denney L, Young D, Powell TJ, Peng YC, et al. High levels of virus-specific CD4+ T cells predict severe pandemic influenza A virus infection. Am J Respir Crit Care Med. (2012) 186:1292–7. doi: 10.1164/rccm.201207-1245OC
98. Sieg S, Muro-Cacho C, Robertson S, Huang Y, Kaplan D. Infection and immunoregulation of T lymphocytes by parainfluenza virus type 3. Proc Natl Acad Sci USA. (1994) 91:6293–7. doi: 10.1073/pnas.91.14.6293
99. Openshaw PJ, Chiu C. Protective and dysregulated T cell immunity in RSV infection. Curr Opin Virol. (2013) 3:468–74. doi: 10.1016/j.coviro.2013.05.005
100. Ilarraza R, Wu Y, Skappak CD, Ajamian F, Proud D, Adamko DJ. Rhinovirus has the unique ability to directly activate human T cells in vitro. J Allergy Clin Immunol. (2013) 131:395–404. doi: 10.1016/j.jaci.2012.11.041
101. Tang F, Quan Y, Xin ZT, Wrammert J, Ma MJ, Lv H, et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. (2011) 186:7264–8. doi: 10.4049/jimmunol.0903490
102. Mubarak A, Alturaiki W, Hemida MG. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J Immunol Res. (2019) 2019:6491738. doi: 10.1155/2019/6491738
103. Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. (2020) 26:453–5. doi: 10.1038/s41591-020-0819-2
104. Kavaler J, Caton AJ, Staudt LM, Gerhard W. A B cell population that dominates the primary response to influenza virus hemagglutinin does not participate in the memory response. Eur J Immunol. (1991) 21:2687–95. doi: 10.1002/eji.1830211107
105. Rothaeusler K, Baumgarth N. B-cell fate decisions following influenza virus infection. Eur J Immunol. (2010) 40:366–77. doi: 10.1002/eji.200939798
106. Roman M, Calhoun WJ, Hinton KL, Avendano LF, Simon V, Escobar AM, et al. Respiratory syncytial virus infection in infants is associated with predominant Th-2-like response. Am J Respir Crit Care Med. (1997) 156:190–5. doi: 10.1164/ajrccm.156.1.9611050
107. Reed JL, Welliver TP, Sims GP, McKinney L, Velozo L, Avendano L, et al. Innate immune signals modulate antiviral and polyreactive antibody responses during severe respiratory syncytial virus infection. J Infect Dis. (2009) 199:1128–38. doi: 10.1086/597386
108. Raes M, Peeters V, Alliet P, Gillis P, Kortleven J, Magerman K, et al. Peripheral blood T and B lymphocyte subpopulations in infants with acute respiratory syncytial virus brochiolitis. Pediatr Allergy Immunol. (1997) 8:97–102. doi: 10.1111/j.1399-3038.1997.tb00151.x
109. Aab A, Wirz O, van de Veen W, Sollner S, Stanic B, Ruckert B, et al. Human rhinoviruses enter and induce proliferation of B lymphocytes. Allergy. (2017) 72:232–43. doi: 10.1111/all.12931
110. Nie Y, Wang G, Shi X, Zhang H, Qiu Y, He Z, et al. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. (2004) 190:1119–26. doi: 10.1086/423286
111. Temperton NJ, Chan PK, Simmons G, Zambon MC, Tedder RS, Takeuchi Y, et al. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg Infect Dis. (2005) 11:411–6. doi: 10.3201/eid1103.040906
112. Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. (2014) 111:4970–5. doi: 10.1073/pnas.1323279111
113. Payne DC, Iblan I, Rha B, Alqasrawi S, Haddadin A, Al Nsour M, et al. Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg Infect Dis. (2016) 22:1824–6. doi: 10.3201/eid2210.160706
114. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. (2020) 41:355–9. doi: 10.1016/j.it.2020.03.007
115. Padilla-Quirarte HO, Lopez-Guerrero DV, Gutierrez-Xicotencatl L, Esquivel-Guadarrama F. Protective antibodies against influenza proteins. Front Immunol. (2019) 10:1677. doi: 10.3389/fimmu.2019.01677
116. Smith CB, Purcell RH, Bellanti JA, Chanock RM. Protective effect of antibody to parainfluenza type 1 virus. N Engl J Med. (1966) 275:1145–52. doi: 10.1056/NEJM196611242752101
117. Stensballe LG, Ravn H, Kristensen K, Agerskov K, Meakins T, Aaby P, et al. Respiratory syncytial virus neutralizing antibodies in cord blood, respiratory syncytial virus hospitalization, and recurrent wheeze. J Allergy Clin Immunol. (2009) 123:398–403. doi: 10.1016/j.jaci.2008.10.043
118. Barclay WS, al-Nakib W, Higgins PG, Tyrrell DA. The time course of the humoral immune response to rhinovirus infection. Epidemiol Infect. (1989) 103:659–69. doi: 10.1017/S095026880003106X
119. Beijing Group of National Research Project for SARS. Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chin Med J. (2003) 116:1283–7.
120. Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. (2004) 136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x
121. Li Z, Guo X, Hao W, Wu Y, Ji Y, Zhao Y, et al. The relationship between serum interleukins and T-lymphocyte subsets in patients with severe acute respiratory syndrome. Chin Med J. (2003) 116:981–4.
122. Zhou J, Chu H, Li C, Wong BH, Cheng ZS, Poon VK, et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. (2014) 209:1331–42. doi: 10.1093/infdis/jit504
123. Cheung CY, Poon LL, Ng IH, Luk W, Sia SF, Wu MH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. (2005) 79:7819–26. doi: 10.1128/JVI.79.12.7819-7826.2005
124. Bohmwald K, Galvez NMS, Canedo-Marroquin G, Pizarro-Ortega MS, Andrade-Parra C, Gomez-Santander F, et al. Contribution of cytokines to tissue damage during human respiratory syncytial virus infection. Front Immunol. (2019) 10:452. doi: 10.3389/fimmu.2019.00452
125. Conti P, Ronconi G, Caraffa A, Gallenga CE, Ross R, Frydas I, et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J Biol Regul Homeost Agents. (2020) 34:1. doi: 10.23812/CONTI-E
126. Betakova T, Kostrabova A, Lachova V, Turianova L. Cytokines induced during influenza virus infection. Curr Pharm Des. (2017) 23:2616–22. doi: 10.2174/1381612823666170316123736
127. Schaap-Nutt A, Liesman R, Bartlett EJ, Scull MA, Collins PL, Pickles RJ, et al. Human parainfluenza virus serotypes differ in their kinetics of replication and cytokine secretion in human tracheobronchial airway epithelium. Virology. (2012) 433:320–8. doi: 10.1016/j.virol.2012.08.027
128. Rajan D, McCracken CE, Kopleman HB, Kyu SY, Lee FE, Lu X, et al. Human rhinovirus induced cytokine/chemokine responses in human airway epithelial and immune cells. PLoS ONE. (2014) 9:e114322. doi: 10.1371/journal.pone.0114322
129. Yang Y, Peng F, Wang R, Yange M, Guan K, Jiang T, et al. The deadly coronaviruses: the 2003. SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. (2020) 109:102434. doi: 10.1016/j.jaut.2020.102434
130. CDC. Middle East Respiratory Syndrome (MERS). (2019). Available online at: https://www.cdc.gov/coronavirus/mers/about/prevention.html
131. Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, Evaluation and Treatment Coronavirus (COVID-19). Treasure Island, FL: StatPearls (2020).
132. CDC. Influenza Vaccines — United States, 2019–20 Influenza Season*. (2019). Available online at: https://www.cdc.gov/flu/professionals/vaccines.htm
133. Papi A, Contoli M. Rhinovirus vaccination: the case against. Eur Respir J. (2011) 37:5–7. doi: 10.1183/09031936.00145710
134. Jiang F, Deng L, Zhang L, Cai Y, Cheung CW, Xia Z. Review of the clinical characteristics of Coronavirus Disease 2019. (COVID-19). J Gen Intern Med. (2020). 35:1545–9. doi: 10.1007/s11606-020-05762-w
135. CDC. Influenza (Flu). (2019). Available online at: https://www.cdc.gov/flu/treatment/index.html
136. Eiland LS. Respiratory syncytial virus: diagnosis, treatment and prevention. J Pediatr Pharmacol Ther. (2009) 14:75–85. doi: 10.5863/1551-6776-14.2.75
137. Greenberg SB. Update on human rhinovirus and coronavirus infections. Semin Respir Crit Care Med. (2016) 37:555–71. doi: 10.1055/s-0036-1584797
138. Heugel J, Martin ET, Kuypers J, Englund JA. Coronavirus-associated pneumonia in previously healthy children. Pediatr Infect Dis J. (2007) 26:753–5. doi: 10.1097/INF.0b013e318054e31b
139. Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. (2007) 119:e70–6. doi: 10.1542/peds.2006-1406
140. Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. (2003) 348:1967–76. doi: 10.1056/NEJMoa030747
141. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. (2012) 367:1814–20. doi: 10.1056/NEJMoa1211721
142. Garg S, Kim L, Whitaker M, O'Halloran A, Cummings C, Holstein R, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed Coronavirus Disease 2019. - COVID-NET, 14 States, March 1-30, 2020. MMWR Morb Mortal Wkly Rep. (2020) 69:458–64. doi: 10.15585/mmwr.mm6915e3
143. Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. (2020) 323:2052–9. doi: 10.1001/jama.2020.6775
144. Jin JM, Bai P, He W, Wu F, Liu XF, Han DM, et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. (2020) 8:152. doi: 10.3389/fpubh.2020.00152
145. Karlberg J, Chong DS, Lai WY. Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am J Epidemiol. (2004) 159:229–31. doi: 10.1093/aje/kwh056
146. Alghamdi IG, Hussain, II, Almalki SS, Alghamdi MS, Alghamdi MM, et al. The pattern of Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive epidemiological analysis of data from the Saudi Ministry of Health. Int J Gen Med. (2014) 7:417–23. doi: 10.2147/IJGM.S67061
147. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271–80.e8. doi: 10.1016/j.cell.2020.02.052
148. Tseng CT, Huang C, Newman P, Wang N, Narayanan K, Watts DM, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol. (2007) 81:1162–73. doi: 10.1128/JVI.01702-06
149. Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA. (2005) 102:7988–93. doi: 10.1073/pnas.0409465102
150. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. (2004) 203:631–7. doi: 10.1002/path.1570
151. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. (2016) 3:237–61. doi: 10.1146/annurev-virology-110615-042301
152. Riordan JF. Angiotensin-I-converting enzyme and its relatives. Genome Biol. (2003) 4:225. doi: 10.1186/gb-2003-4-8-225
153. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. (2020) 94. doi: 10.1128/JVI.00127-20
154. Kuba K, Imai Y, Ohto-Nakanishi T, Penninger JM. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol Ther. (2010) 128:119–28. doi: 10.1016/j.pharmthera.2010.06.003
155. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. (2003) 348:1953–66. doi: 10.1056/NEJMoa030781
156. Lambeir AM, Durinx C, Scharpe S, De Meester I. Dipeptidyl-peptidase IV from bench to bedside: an update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit Rev Clin Lab Sci. (2003) 40:209–94. doi: 10.1080/713609354
157. Whitsett JA. Airway Epithelial Differentiation and Mucociliary Clearance. Ann Am Thorac Soc. (2018) 15(Suppl. 3):S143–8. doi: 10.1513/AnnalsATS.201802-128AW
158. Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. (2015) 16:27–35. doi: 10.1038/ni.3045
159. Grassin-Delyle S, Abrial C, Salvator H, Brollo M, Naline E, Devillier P. The role of toll-like receptors in the production of cytokines by human lung macrophages. J Innate Immun. (2020) 12:63–73. doi: 10.1159/000494463
160. Leiva-Juarez MM, Kolls JK, Evans SE. Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol. (2018) 11:21–34. doi: 10.1038/mi.2017.71
161. Byrne AJ, Mathie SA, Gregory LG, Lloyd CM. Pulmonary macrophages: key players in the innate defence of the airways. Thorax. (2015) 70:1189–96. doi: 10.1136/thoraxjnl-2015-207020
162. Kopf M, Schneider C, Nobs SP. The development and function of lung-resident macrophages and dendritic cells. Nat Immunol. (2015) 16:36–44. doi: 10.1038/ni.3052
163. Jung HE, Kim TH, Lee HK. Contribution of dendritic cells in protective immunity against respiratory syncytial virus infection. Viruses. (2020) 12:102. doi: 10.3390/v12010102
164. Duan M, Hibbs ML, Chen W. The contributions of lung macrophage and monocyte heterogeneity to influenza pathogenesis. Immunol Cell Biol. (2017) 95:225–35. doi: 10.1038/icb.2016.97
165. Reizis B. Plasmacytoid dendritic cells: development, regulation, and function. Immunity. (2019) 50:37–50. doi: 10.1016/j.immuni.2018.12.027
166. Collin M, Bigley V. Human dendritic cell subsets: an update. Immunology. (2018) 154:3–20. doi: 10.1111/imm.12888
167. Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. (2015) 15:471–85. doi: 10.1038/nri3865
168. Lau YL, Peiris JS, Law HK. Role of dendritic cells in SARS coronavirus infection. Hong Kong Med J. (2012) 18(Suppl. 3):28–30.
169. Fitzgerald-Bocarsly P, Dai J, Singh S. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. (2008) 19:3–19. doi: 10.1016/j.cytogfr.2007.10.006
170. Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. (2013) 31:563–604. doi: 10.1146/annurev-immunol-020711-074950
171. Sang Y, Miller LC, Blecha F. Macrophage polarization in virus-host interactions. J Clin Cell Immunol. (2015) 6:311. doi: 10.4172/2155-9899.1000311
172. Franken L, Schiwon M, Kurts C. Macrophages: sentinels and regulators of the immune system. Cell Microbiol. (2016) 18:475–87. doi: 10.1111/cmi.12580
173. Condon TV, Sawyer RT, Fenton MJ, Riches DW. Lung dendritic cells at the innate-adaptive immune interface. J Leukoc Biol. (2011) 90:883–95. doi: 10.1189/jlb.0311134
174. Schyns J, Bureau F, Marichal T. Lung interstitial macrophages: past, present, and future. J Immunol Res. (2018) 2018:5160794. doi: 10.1155/2018/5160794
175. Ramos I, Smith G, Ruf-Zamojski F, Martinez-Romero C, Fribourg M, Carbajal EA, et al. Innate immune response to influenza virus at single-cell resolution in human epithelial cells revealed paracrine induction of interferon lambda 1. J Virol. (2019) 93:e00559-19. doi: 10.1128/JVI.00559-19
176. Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. (2009) 30:556–65. doi: 10.1016/j.immuni.2009.02.005
177. Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol. (2011) 45:189–201. doi: 10.1165/rcmb.2011-0011RT
178. Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev. (2011) 24:210–29. doi: 10.1128/CMR.00014-10
179. VanLeuven JT, Ridenhour BJ, Gonzalez AJ, Miller CR, Miura TA. Lung epithelial cells have virus-specific and shared gene expression responses to infection by diverse respiratory viruses. PLoS ONE. (2017) 12:e0178408. doi: 10.1371/journal.pone.0178408
180. Thistle J, Martinon D, Weers PM. Helix 1 tryptophan variants in Galleria mellonella apolipophorin III. Chem Phys Lipids. (2015) 193:18–23. doi: 10.1016/j.chemphyslip.2015.10.002
181. Dong S, Zhang X, He Y, Xu F, Li D, Xu W, et al. Synergy of IL-27 and TNF-alpha in regulating CXCL10 expression in lung fibroblasts. Am J Respir Cell Mol Biol. (2013) 48:518–30. doi: 10.1165/rcmb.2012-0340OC
182. Yilla M, Harcourt BH, Hickman CJ, McGrew M, Tamin A, Goldsmith CS, et al. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. (2005) 107:93–101. doi: 10.1016/j.virusres.2004.09.004
183. Zhou J, Chu H, Chan JF, Yuen KY. Middle East respiratory syndrome coronavirus infection: virus-host cell interactions and implications on pathogenesis. Virol J. (2015) 12:218. doi: 10.1186/s12985-015-0446-6
184. Bonora M, Wieckowsk MR, Chinopoulos C, Kepp O, Kroemer G, Galluzzi L, et al. Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene. (2015) 34:1608. doi: 10.1038/onc.2014.462
185. Siu KL, Yuen KS, Castano-Rodriguez C, Ye ZW, Yeung ML, Fung SY, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. (2019) 33:8865–77. doi: 10.1096/fj.201802418R
186. Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. (2013) 94(Pt. 12):2679–90. doi: 10.1099/vir.0.055533-0
187. Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS ONE. (2010) 5:e8729. doi: 10.1371/journal.pone.0008729
188. Smits SL, de Lang A, van den Brand JM, Leijten LM, van IWF, Eijkemans MJ, et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. (2010) 6:e1000756. doi: 10.1371/journal.ppat.1000756
189. Korean Society of Infectious Diseases and Korea Centers for Disease Control and Prevention. Analysis on 54 mortality cases of Coronavirus Disease 2019 in the Republic of Korea from January 19 to March 10, 2020. J Korean Med Sci. (2020) 35:e132. doi: 10.3346/jkms.2020.35.e132
190. Leung GM, Ho LM, Lam TH, Hedley AJ, Peiris JS. Prevalence of SARS-CoV antibody in all Hong Kong patient contacts. Hong Kong Med J. (2009) 15(Suppl. 9):27–9.
191. Pierangeli A, Scagnolari C, Antonelli G. Respiratory syncytial virus. Minerva Pediatr. (2018) 70:553–65. doi: 10.23736/S0026-4946.18.05312-4
192. Takeuchi K, Kadota SI, Takeda M, Miyajima N, Nagata K. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. (2003) 545:177–82. doi: 10.1016/S0014-5793(03)00528-3
193. Lui PY, Wong LY, Fung CL, Siu KL, Yeung ML, Yuen KS, et al. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg Microbes Infect. (2016) 5:e39. doi: 10.1038/emi.2016.33
194. Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. (2008) 82:4471–9. doi: 10.1128/JVI.02472-07
195. Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. (2009) 284:16202–9. doi: 10.1074/jbc.M109.008227
196. Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, et al. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol. (2017) 91:e02143-16. doi: 10.1128/JVI.02143-16
197. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. (2013) 13:159–75. doi: 10.1038/nri3399
198. Miura TA, Holmes KV. Host-pathogen interactions during coronavirus infection of primary alveolar epithelial cells. J Leukoc Biol. (2009) 86:1145–51. doi: 10.1189/jlb.0209078
199. Wang R, Xiao H, Guo R, Li Y, Shen B. The role of C5a in acute lung injury induced by highly pathogenic viral infections. Emerg Microbes Infect. (2015) 4:e28. doi: 10.1038/emi.2015.28
200. Jiang Y, Xu J, Zhou C, Wu Z, Zhong S, Liu J, et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. (2005) 171:850–7. doi: 10.1164/rccm.200407-857OC
201. Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. (2003) 361:1773–8. doi: 10.1016/S0140-6736(03)13413-7
202. Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. (2016) 19:181–93. doi: 10.1016/j.chom.2016.01.007
203. Tse GM, To KF, Chan PK, Lo AW, Ng KC, Wu A, et al. Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS). J Clin Pathol. (2004) 57:260–5. doi: 10.1136/jcp.2003.013276
204. Liu L, Wei Q, Nishiura K, Peng J, Wang H, Midkiff C, et al. Spatiotemporal interplay of severe acute respiratory syndrome coronavirus and respiratory mucosal cells drives viral dissemination in rhesus macaques. Mucosal Immunol. (2016) 9:1089–101. doi: 10.1038/mi.2015.127
205. Zhou X, Moore BB. Location or origin? What is critical for macrophage propagation of lung fibrosis? Eur Respir J. (2018) 51:1800103. doi: 10.1183/13993003.00103-2018
206. Antonio GE, Wong KT, Hui DS, Wu A, Lee N, Yuen EH, et al. Thin-section CT in patients with severe acute respiratory syndrome following hospital discharge: preliminary experience. Radiology. (2003) 228:810–5. doi: 10.1148/radiol.2283030726
207. Lau YL, Peiris JS. Pathogenesis of severe acute respiratory syndrome. Curr Opin Immunol. (2005) 17:404–10. doi: 10.1016/j.coi.2005.05.009
208. Feng Z, Diao B, Wang R, Wang G, Wang C, Tan Y, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv [Preprint]. (2020). doi: 10.1101/2020.03.27.20045427
209. Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. (2005) 106:2366–74. doi: 10.1182/blood-2004-10-4166
210. Channappanavar R, Fett C, Mack M, Ten Eyck PP, Meyerholz DK, Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J Immunol. (2017) 198:4046–53. doi: 10.4049/jimmunol.1601896
211. Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. (2007) 81:8692–706. doi: 10.1128/JVI.00527-07
212. Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. (2020) 583:830–3. doi: 10.1101/2020.02.07.939389
213. Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. (2020) 182:50–8.e8. doi: 10.1016/j.cell.2020.05.027
214. Ip WK, Chan KH, Law HK, Tso GH, Kong EK, Wong WH, et al. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J Infect Dis. (2005) 191:1697–704. doi: 10.1086/429631
215. Magro C, Mulvey JJ, Berlin D, Nuovo G, Salvatore S, Harp J, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl Res. (2020) 220:1–13. doi: 10.1016/j.trsl.2020.04.007
216. Gao T, Hu M, Zhang X, Li H, Zhu L, Liu H, et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv [Preprint]. (2020) doi: 10.1101/2020.03.29.20041962
217. Noris M, Benigni A, Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. (2020) 98:314–22. doi: 10.1016/j.kint.2020.05.013
218. Jiang Y, Zhao G, Song N, Li P, Chen Y, Guo Y, et al. Blockade of the C5a-C5aR axis alleviates lung damage in hDPP4-transgenic mice infected with MERS-CoV. Emerg Microbes Infect. (2018) 7:77. doi: 10.1038/s41426-018-0063-8
219. Galani IE, Andreakos E. Neutrophils in viral infections: current concepts and caveats. J Leukoc Biol. (2015) 98:557–64. doi: 10.1189/jlb.4VMR1114-555R
220. Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol. (2017) 8:550. doi: 10.3389/fimmu.2017.00550
221. Manocha S, Walley KR, Russell JA. Severe acute respiratory distress syndrome (SARS): a critical care perspective. Crit Care Med. (2003) 31:2684–92. doi: 10.1097/01.CCM.0000091929.51288.5F
222. Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. (2003) 348:1986–94. doi: 10.1056/NEJMoa030685
223. Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, et al. Neutrophil-to-lymphocyte ratio predicts severe illness patients with 2019. Novel coronavirus in the early stage. J Transl Med. (2020) 18:206. doi: 10.1186/s12967-020-02374-0
224. Nagata N, Iwata N, Hasegawa H, Fukushi S, Harashima A, Sato Y, et al. Mouse-passaged severe acute respiratory syndrome-associated coronavirus leads to lethal pulmonary edema and diffuse alveolar damage in adult but not young mice. Am J Pathol. (2008) 172:1625–37. doi: 10.2353/ajpath.2008.071060
225. Raj GD, Savage CE, Jones RC. Effect of heterophil depletion by 5-fluorouracil on infectious bronchitis virus infection in chickens. Avian Pathol. (1997) 26:427–32. doi: 10.1080/03079459708419224
226. Haick AK, Rzepka JP, Brandon E, Balemba OB, Miura TA. Neutrophils are needed for an effective immune response against pulmonary rat coronavirus infection, but also contribute to pathology. J Gen Virol. (2014) 95(Pt. 3):578–90. doi: 10.1099/vir.0.061986-0
227. Zhou J, Stohlman SA, Hinton DR, Marten NW. Neutrophils promote mononuclear cell infiltration during viral-induced encephalitis. J Immunol. (2003) 170:3331–6. doi: 10.4049/jimmunol.170.6.3331
228. de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci USA. (2013) 110:16598–603. doi: 10.1073/pnas.1310744110
229. Guery B, Poissy J, el Mansouf L, Sejourne C, Ettahar N, Lemaire X, et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. (2013) 381:2265–72. doi: 10.1016/S0140-6736(13)60982-4
230. Kuster GM, Pfister O, Burkard T, Zhou Q, Twerenbold R, Haaf P, et al. SARS-CoV2: should inhibitors of the renin-angiotensin system be withdrawn in patients with COVID-19? Eur Heart J. (2020) 41:1801–3. doi: 10.1093/eurheartj/ehaa235
231. Sodhi CP, Nguyen J, Yamaguchi Y, Werts AD, Lu P, Ladd MR, et al. A dynamic variation of pulmonary ACE2 is required to modulate neutrophilic inflammation in response to Pseudomonas aeruginosa lung infection in mice. J Immunol. (2019) 203:3000–12. doi: 10.4049/jimmunol.1900579
232. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. (2018) 18:134–47. doi: 10.1038/nri.2017.105
233. Zawrotniak M, Rapala-Kozik M. Neutrophil extracellular traps (NETs) - formation and implications. Acta Biochim Pol. (2013) 60:277–84. doi: 10.18388/abp.2013_1983
234. Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. (2020) 217:e20200652. doi: 10.1084/jem.20200652
235. Mozzini C, Girelli D. The role of neutrophil extracellular traps in Covid-19: only an hypothesis or a potential new field of research? Thromb Res. (2020) 191:26–7. doi: 10.1016/j.thromres.2020.04.031
236. Wichmann D, Sperhake JP, Lutgehetmann M, Steurer S, Edler C, Heinemann A, et al. Autopsy findings and venous thromboembolism in patients with COVID-19. Ann Intern Med. (2020) M20–2003. doi: 10.7326/M20-2003
237. Levi M, Thachil J, Iba T, Levy JH. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. (2020) 7:e438–40. doi: 10.1016/S2352-3026(20)30145-9
238. Soehnlein O, Lindbom L, Weber C. Mechanisms underlying neutrophil-mediated monocyte recruitment. Blood. (2009) 114:4613–23. doi: 10.1182/blood-2009-06-221630
239. Prame Kumar K, Nicholls AJ, Wong CHY. Partners in crime: neutrophils and monocytes/macrophages in inflammation and disease. Cell Tissue Res. (2018) 371:551–65. doi: 10.1007/s00441-017-2753-2
240. Schauer C, Janko C, Munoz LE, Zhao Y, Kienhofer D, Frey B, et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med. (2014) 20:511–7. doi: 10.1038/nm.3547
241. Norris S, Doherty DG, Curry M, McEntee G, Traynor O, Hegarty JE, et al. Selective reduction of natural killer cells and T cells expressing inhibitory receptors for MHC class I in the livers of patients with hepatic malignancy. Cancer Immunol Immunother. (2003) 52:53–8. doi: 10.1007/s00262-002-0331-1
242. Dukers DF, Vermeer MH, Jaspars LH, Sander CA, Flaig MJ, Vos W, et al. Expression of killer cell inhibitory receptors is restricted to true NK cell lymphomas and a subset of intestinal enteropathy-type T cell lymphomas with a cytotoxic phenotype. J Clin Pathol. (2001) 54:224–8. doi: 10.1136/jcp.54.3.224
243. Ahmad R, Sindhu ST, Tran P, Toma E, Morisset R, Menezes J, et al. Modulation of expression of the MHC class I-binding natural killer cell receptors, and NK activity in relation to viral load in HIV-infected/AIDS patients. J Med Virol. (2001) 65:431–40. doi: 10.1002/jmv.2053
244. Husain Z, Levitan E, Larsen CE, Mirza NM, Younes S, Yunis EJ, et al. HLA-Cw7 zygosity affects the size of a subset of CD158b+ natural killer cells. J Clin Immunol. (2002) 22:28–36. doi: 10.1023/A:1014204519468
245. National Research Project for SARS, Beijing Group. The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am J Clin Pathol. (2004) 121:507–11. doi: 10.1309/WPK7Y2XKNF4CBF3R
246. Cui W, Fan Y, Wu W, Zhang F, Wang JY, Ni AP. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis. (2003) 37:857–9. doi: 10.1086/378587
247. Cao Q, Chen YC, Chen CL, Chiu CH. SARS-CoV-2 infection in children: transmission dynamics and clinical characteristics. J Formos Med Assoc. (2020) 119:670–3. doi: 10.1016/j.jfma.2020.02.009
248. Cristiani L, Mancino E, Matera L, Nenna R, Pierangeli A, Scagnolari C, et al. Will children reveal their secret? The coronavirus dilemma. Eur Respir J. (2020). doi: 10.1183/13993003.00749-2020. [Epub ahead of print].
249. Li T, Qiu Z, Zhang L, Han Y, He W, Liu Z, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. (2004) 189:648–51. doi: 10.1086/381535
250. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. (2011) 12:1045–54. doi: 10.1038/ni.2131
251. Li BWS, de Bruijn MJW, Lukkes M, van Nimwegen M, Bergen IM, KleinJan A, et al. T cells and ILC2s are major effector cells in influenza-induced exacerbation of allergic airway inflammation in mice. Eur J Immunol. (2019) 49:144–56. doi: 10.1002/eji.201747421
252. Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. (2020) 526:135–40. doi: 10.1016/j.bbrc.2020.03.044
253. Lo AW, Tang NL, To KF. How the SARS coronavirus causes disease: host or organism? J Pathol. (2006) 208:142–51. doi: 10.1002/path.1897
254. Min CK, Cheon S, Ha NY, Sohn KM, Kim Y, Aigerim A, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep. (2016) 6:25359. doi: 10.1038/srep25359
255. Kim ES, Choe PG, Park WB, Oh HS, Kim EJ, Nam EY, et al. Clinical progression and cytokine profiles of Middle East respiratory syndrome coronavirus infection. J Korean Med Sci. (2016) 31:1717–25. doi: 10.3346/jkms.2016.31.11.1717
256. Moran JV, Zimmerly S, Eskes R, Kennell JC, Lambowitz AM, Butow RA, et al. Mobile group II introns of yeast mitochondrial DNA are novel site-specific retroelements. Mol Cell Biol. (1995) 15:2828–38. doi: 10.1128/MCB.15.5.2828
257. Hogan RJ, Gao G, Rowe T, Bell P, Flieder D, Paragas J, et al. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol. (2004) 78:11416–21. doi: 10.1128/JVI.78.20.11416-11421.2004
258. Frieman MB, Chen J, Morrison TE, Whitmore A, Funkhouser W, Ward JM, et al. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. (2010) 6:e1000849. doi: 10.1371/journal.ppat.1000849
259. Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. (2010) 84:5670–7. doi: 10.1128/JVI.00272-10
260. Mahlakoiv T, Ritz D, Mordstein M, DeDiego ML, Enjuanes L, Muller MA, et al. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J Gen Virol. (2012) 93(Pt. 12):2601–5. doi: 10.1099/vir.0.046284-0
261. Lu X, Pan J, Tao J, Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. (2011) 42:37–45. doi: 10.1007/s11262-010-0544-x
262. Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. (2007) 81:548–57. doi: 10.1128/JVI.01782-06
263. Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. (2011) 7:e1002433. doi: 10.1371/journal.ppat.1002433
264. Lokugamage KG, Narayanan K, Nakagawa K, Terasaki K, Ramirez SI, Tseng CT, et al. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J Virol. (2015) 89:10970–81. doi: 10.1128/JVI.01352-15
265. Jauregui AR, Savalia D, Lowry VK, Farrell CM, Wathelet MG. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS ONE. (2013) 8:e62416. doi: 10.1371/journal.pone.0062416
266. Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. (2014) 450–451:64–70. doi: 10.1016/j.virol.2013.11.040
267. Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. (2007) 282:32208–21. doi: 10.1074/jbc.M704870200
268. Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, et al. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J Gen Virol. (2014) 95(Pt. 3):614–26. doi: 10.1099/vir.0.059014-0
269. Kuri T, Eriksson KK, Putics A, Zust R, Snijder EJ, Davidson AD, et al. The ADP-ribose-1”-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J Gen Virol. (2011) 92(Pt. 8):1899–905. doi: 10.1099/vir.0.031856-0
Keywords: SARS-CoV-2, COVID-19, innate immune responses, Coronavirus (CoV), Coronavirus (2019-nCoV) outbreak
Citation: Ahmed-Hassan H, Sisson B, Shukla RK, Wijewantha Y, Funderburg NT, Li Z, Hayes D Jr, Demberg T and Liyanage NPM (2020) Innate Immune Responses to Highly Pathogenic Coronaviruses and Other Significant Respiratory Viral Infections. Front. Immunol. 11:1979. doi: 10.3389/fimmu.2020.01979
Received: 04 May 2020; Accepted: 22 July 2020;
Published: 18 August 2020.
Edited by:
Donald Sodora, Seattle Children's Research Institute, United StatesReviewed by:
Jianzhong Zhu, Yangzhou University, ChinaLaura Fantuzzi, Istituto Superiore di Sanità (ISS), Italy
Copyright © 2020 Ahmed-Hassan, Sisson, Shukla, Wijewantha, Funderburg, Li, Hayes, Demberg and Liyanage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Namal P. M. Liyanage, bmFtYWwubGl5YW5hZ2VAb3N1bWMuZWR1
†These authors have contributed equally to this work
 Hanaa Ahmed-Hassan
Hanaa Ahmed-Hassan Brianna Sisson1†
Brianna Sisson1† Rajni Kant Shukla
Rajni Kant Shukla Yasasvi Wijewantha
Yasasvi Wijewantha Thorsten Demberg
Thorsten Demberg Namal P. M. Liyanage
Namal P. M. Liyanage