- 1George S. Wise Faculty of Life Sciences, School of Molecular Cell Biology and Biotechnology, Tel Aviv University, Tel Aviv, Israel
- 2Department of Pharmaceutical Chemistry, The University of Kansas, Lawrence, KS, United States
- 3Department of Chemical and Petroleum Engineering, The University of Kansas, Lawrence, KS, United States
Monoclonal antibodies (mAbs) are a crucial asset for human health and modern medicine, however, the repeated administration of mAbs can be highly immunogenic. Drug immunogenicity manifests in the generation of anti-drug antibodies (ADAs), and some mAbs show immunogenicity in up to 70% of patients. ADAs can alter a drug’s pharmacokinetic and pharmacodynamic properties, reducing drug efficacy. In more severe cases, ADAs can neutralize the drug’s therapeutic effects or cause severe adverse events to the patient. While some contributing factors to ADA formation are known, the molecular mechanisms of how therapeutic mAbs elicit ADAs are not completely clear. Accurate ADA detection is necessary to provide clinicians with sufficient information for patient monitoring and clinical intervention. However, ADA assays present unique challenges because both the analyte and antigen are antibodies, so most assays are cumbersome, costly, time consuming, and lack standardization. This review will discuss aspects related to ADA formation following mAb drug administration. First, we will provide an overview of the prevalence of ADA formation and the available diagnostic tools for their detection. Next, we will review studies that support possible molecular mechanisms causing the formation of ADA. Finally, we will summarize recent approaches used to decrease the propensity of mAbs to induce ADAs.
Introduction
In the last three decades, the pharmaceutical industry experienced a massive shift toward the use of protein drugs, often referred to as “biologics.” Biologics offer higher specificity and better characterized mechanisms of action compared to small molecule drugs, and their use has revolutionized the treatment of a wide range of diseases and disorders. In general, monoclonal antibodies (mAbs) are the most widely used class of biologics (1).
Monoclonal antibodies account for a growing number of blockbuster drugs with their US sales reaching over $24 billion (2), and will maintain a dominant position in the pharmaceutical market that exceeds $125 billion by the end of 2020 (3).
To date, over 73 mAbs have been approved by the United States Food and Drug Administration (FDA) and the European Medicines Agency (EMA). Hundreds more mAbs are in different stages of clinical developmental. mAbs are used for various clinical indications including cancer, chronic autoimmune and inflammatory diseases, allergies, infections, transplantations, and cardiovascular diseases (4).
The mechanism of action (MOA) of mAbs can vary across different use cases. For example, the anti-CD20 rituximab induces cell death by binding to surface receptors, resulting in a signaling cascade that leads to apoptosis (5). Other mAbs, including the anti-HER-2 trastuzumab, block receptor-ligand interactions to achieve a desired effect, either by blocking the receptor domain to inhibit an activation signal by removing a soluble ligand entirely from circulation (6). mAbs can also induce fragment crystallizable (Fc)-dependent effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC), which are important for the anti-CD20 drug obinutuzumab that is used for the treatment of lymphoproliferative disorders (7). Other mAbs target specific proteins involved in pathogenesis of disease, such as anti-TNFα mAbs infliximab and adalimumab that are used to treat inflammatory bowel disease (IBD) and rheumatoid arthritis (RA) (8). Other mAbs in this category are omalizumab, an anti-IgE mAb that is used to treat patients with allergic asthma (9), palivizumab which targets an epitope in the A antigenic site of the F protein of the respiratory syncytial virus (RSV) (10), and bezlotoxumab which binds and neutralizes Clostridium difficile toxin B (11). Some mAbs, such as cetuximab and panitumumab (12), target the epidermal growth factor receptor (EGFR) which is overexpressed in a number of cancers. In recent years, checkpoint inhibitor mAbs were also developed to manipulate anti-tumor T-cell responses, like the anti-PD-1 nivolumab that is used to treat melanoma and non-small cell lung cancer (13).
The tremendous progress in mAb discovery began in 1975, when Köhler and Milstein reported in vitro screening and production of murine mAbs from hybridomas (14). In the late 1980s, murine mAbs were in rapid clinical development, but had significant drawbacks as they were often induced allergic reactions and the formation of human anti-mouse antibodies (HAMA). Examples include T101 used to treat chronic lymphocytic leukemia (CLL) and cutaneous T cell lymphoma (CTCL), and 9.2.27 to treat melanoma (15). Additionally, murine mAbs exhibited a relatively short half-life in humans, possibly due to low affinity toward the human neonatal Fc receptor (FcRn) (16), and were relatively poor recruiters of effector functions, crucial for some mAb efficacy (17).
To overcome the immunogenicity and reduced effector function of murine mAbs, chimeric antibodies (mouse–human) were next developed by fusing the antigen-specific variable domain of a murine mAb with the constant domains of a human mAb. This resulted in chimeric mAbs of approximately 65% human origin by amino acid content (18). Human gene sequences were mostly taken from the κ light chain and the IgG1 heavy chain, as IgG1 has the highest efficiency in activating complement and cytotoxic effector cells, and the κ light chain is more common in human serum antibodies (19, 20). The development of chimeric mAbs indeed reduced immunogenicity and increased efficacy. For example, metastatic colorectal carcinoma patients who received the chimeric mAb 17-1A did not show any toxic or allergic reactions, and the chimeric antibody was significantly less immunogenic than its parental murine antibody (21).
Chimeric mAbs exhibited an extended half-life and reduced immunogenicity, but they still presented a considerably high propensity for ADA induction (22). Aiming to further reduce mAb immunogenicity, humanized mAbs were developed by grafting the murine complementarity determining regions (CDR) onto framework regions (FR) of the human mAb heavy and light chain variable domains (VH and VL, respectively), for mAbs that are approximately 95% human (23). mAb humanization often significantly reduces immunogenicity and ADA formation (24).
Technological advances of phage display technology (25, 26) based on human single chain Fv (scFv) libraries (27) next enabled the discovery of antibodies comprised entirely of human genes. These human mAbs were additionally aided by the more recent development of transgenic mouse strains expressing human antibody variable domains (28–30).
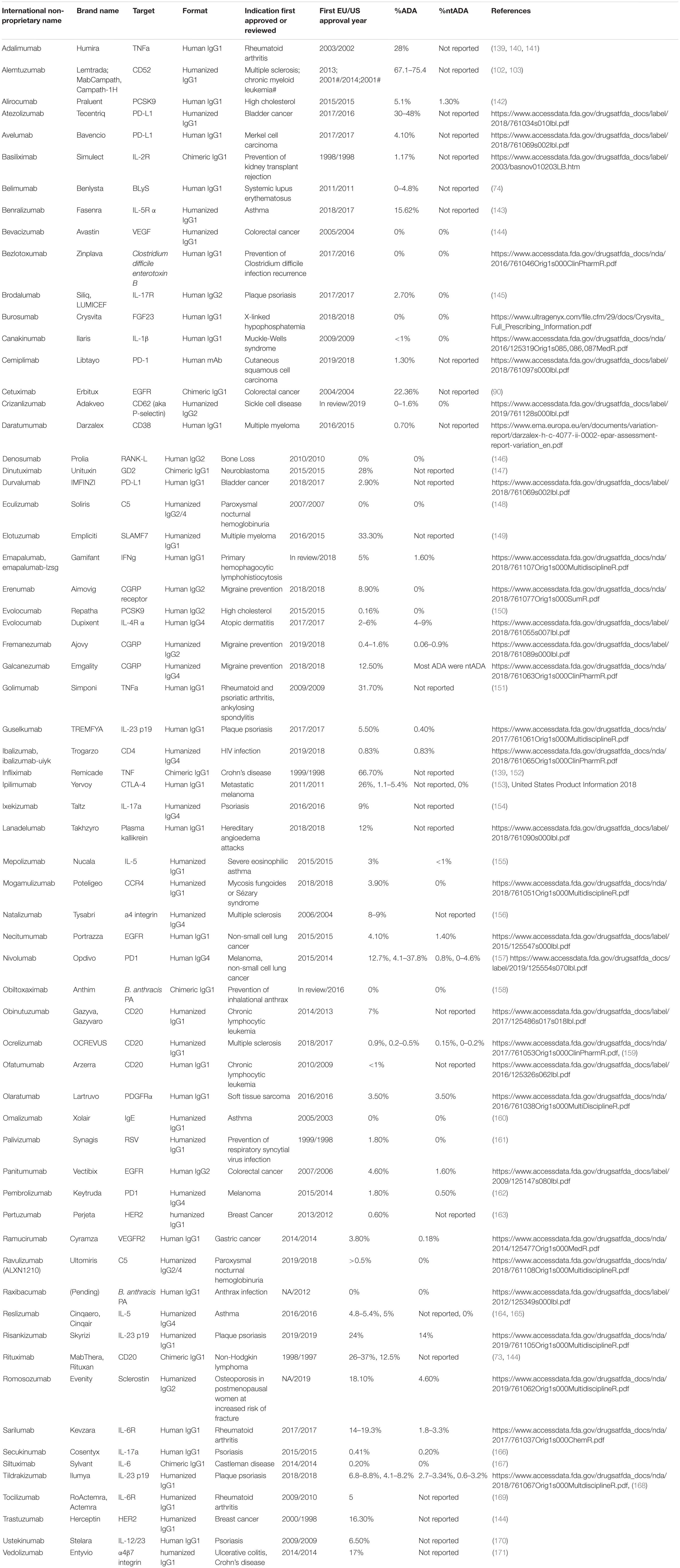
While both humanized and fully human mAbs reduce immunogenic potential and show properties similar to human endogenous IgGs, they fail to completely eliminate mAb immunogenicity and ADA formation (31). Table 1 summarizes mAbs that are currently approved in the US and EU, along with their reported immunogenicity rates.
In the past decade, next-generation sequencing (NGS) technologies enabled a rapid increase in the capacity to sequence human and animal genomes (32). Like many other areas of modern biology, NGS is now frequently used in basic and applied immunology. NGS is often applied for sequencing the VH and VL antibody domains (33–36), as well as T-cell receptors (37, 38) and antibody derivative [e.g., scFv, F(ab)] libraries screened using display systems (39–41). NGS analysis of B cells can elucidate the features of antibody immune responses at a molecular level, and has been further exploited for advanced mAb discovery and engineering (42–44).
In addition to NGS of bulk populations, single-cell sequencing comprises an important group of technologies for antibody discovery, as single cell data is necessary to reveal the native VH and VL pairing. Previous studies were able to obtain VH and VL chain pairing from isolated plasmablasts (PB) in immunized mice (34, 45, 46) and antigen-specific PB from tetanus-vaccinated human patients (33).
A recently introduced technology combines proteomic analyses of antibodies in blood or secretions with NGS analysis of antibody-encoding B cells. Proteomics thus provides invaluable information about the molecular, monoclonal properties of human serum antibodies in health and disease (46–48). All of the above recently developed technologies have expedited mAb discovery and revolutionized our understanding about the nature of the immune responses, including in the formation of ADAs following immunization and administration of mAbs.
Monoclonal antibodies immunogenicity is mainly manifested in ADA generation (49). The formation of ADAs alters a drug’s bioavailability and pharmacokinetic and pharmacodynamic properties, and most often reduces drug efficacy (50, 51). ADAs have a significant impact on mAb drug safety, as they can lead to serious adverse immune reactions in the clinic (52). Patients with ADAs can be stratified by their effect on the clinical treatment course. Patients are designated as having primary loss of response (LOR) when the administrated mAb fails to show any efficacy within several weeks following treatment initiation, or secondary LOR when patients show significant side effects or the drug loses effectiveness over time despite an initial therapeutic response (53–55).
For multiple decades, many studies focused on possible mechanisms that govern ADA formation, development of improved assays for ADA detection, and advancement of tools for immunogenicity and prediction of ADA formation. This review provides an overview on these topics, underlining the challenges and potential solutions for this important research field. While this review focuses on ADA as an important outcome of mAb immunogenicity, there are other immunogenicity outcomes such as allergic reactions, cytopenia, and anaphylaxis that are widely reviewed elsewhere (56).
The Molecular Mechanisms That Lead to ADA Formation
Anti-drug antibodies can be generated by a T-cell dependent or independent B cell activation pathway. In the T-cell dependent pathway, mAbs act as antigens and are internalized by antigen presenting cells (APCs), processed, and presented to T cells via the cognate interaction between the MHC class II molecules and T-cell receptor. Depending on the cytokine milieu during this interaction, several different immune responses can occur (57). In the T-cell dependent pathway, ADAs are generated when a T helper cell (Th) differentiates into a Th1 or Th2 phenotype and, following their cognate interactions with B cells, induces the proliferation of plasma cells (PC) that secrete ADAs. Previous studies showed that a Th2 response mostly induce ADA production of the IgG4 isotype, in comparison to the Th1 response, that in the case of anti-factor VIII elicits the generation of IgG1 and IgG2 ADA (58, 59).
For example, infliximab-specific Th2 cells can be detected in circulation after infliximab infusion, and these cells were correlated with the presence of infliximab-specific ADA (60). Interestingly, this cellular response was observed mostly in patients with hypersensitivity reactions, rather than in the LOR group. In another study, T cell epitopes of infliximab and rituximab were identified by isolating antibody-specific T cells after repeated rounds of antibody-loaded dendritic cells (DCs) in co-culture (61). These T cells were specific to peptides derived from VH and VL and encompassed CDRs and FRs, reflecting the immunogenicity of the chimeric part of these antibodies. Importantly, these peptides were also eluted from antibody-loaded DCs, highlighting the importance of MHC Class II antigen presentation in the ADA formation process.
In contrast, for the T cell independent pathway mAbs with multiple epitopes can crosslink B cell receptors (BCRs) and stimulate B cells to differentiate into PC to produce ADAs (62–66). It was previously demonstrated that impurities and aggregates of the mAbs may increase the number of adjacent epitopes on the mAb, potentially steering the immune response toward a T-cell independent pathway by B cell crosslinking (67–70).
Drug and Patient Characteristics Contributing to ADA Formation
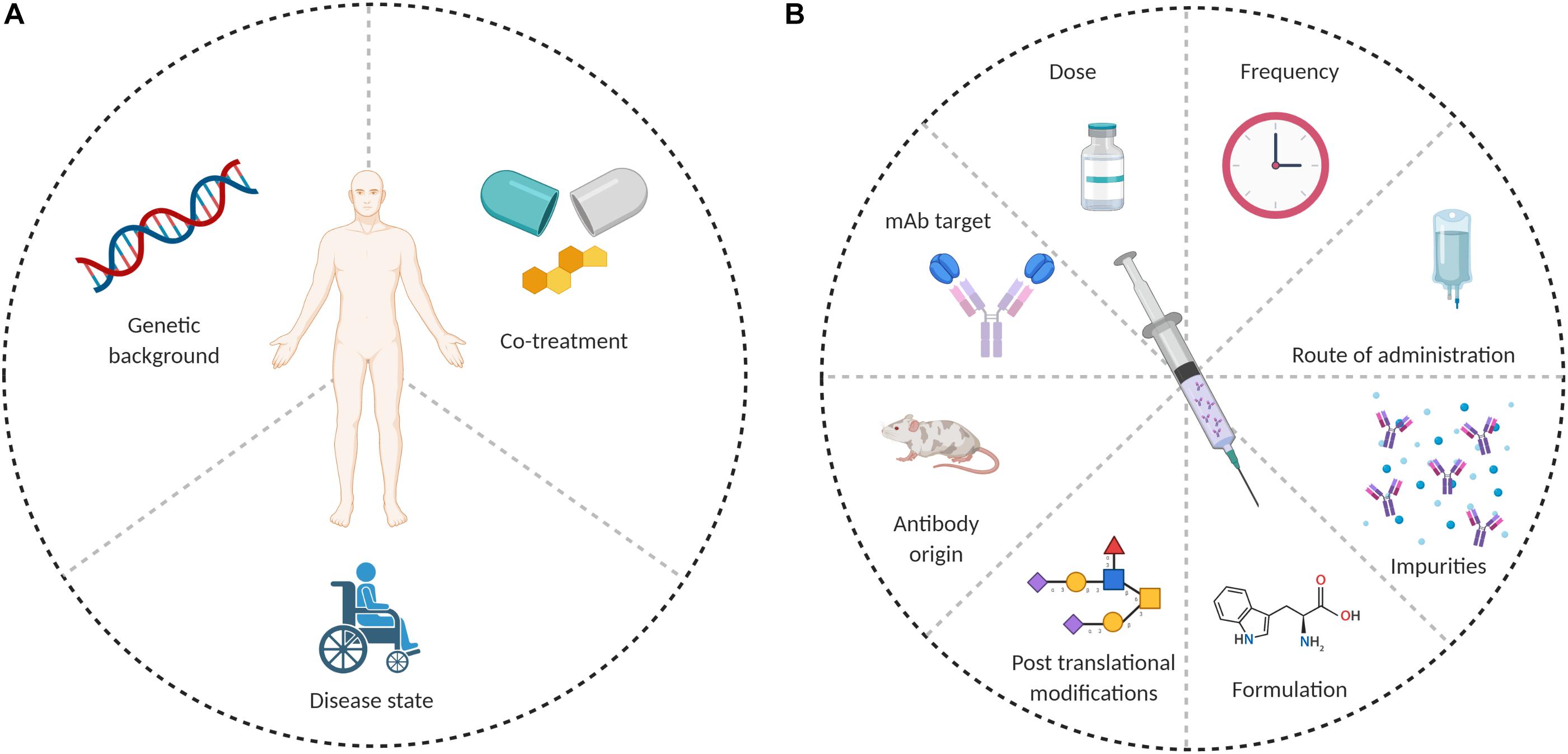
Anti-drug antibodies formation depends on the interplay between several factors, which can be patient-related or drug-related. Possible causes for ADA formation are summarized in Figure 1.
Patient-Related Factors
The study of why and how ADAs are generated is complicated by the fact that some patients develop ADAs and some, with the same clinical indication and receiving the same therapeutic mAb, do not. The extent of immunogenicity thus differs among patients receiving the same mAb, which could be related to the immune pathways underlying the pathogenesis of the disease (71). For example, RA patients have a higher likelihood of developing ADAs toward a mAb drug than spondyloarthritis patients (57). When examining a specific disease or immune target, different mAbs may have a varying effect on the induction of ADAs. RA patients develop higher ADA levels when treated with two different mAbs (72). In multiple sclerosis (MS) patients, treatment with rituximab (chimeric anti-CD20 mAb) generated an unwanted immune response in up to 37% of patients (73). On the contrary, belimumab (a fully human anti B-cell activating factor (BAFF) mAb), which is used to treat systemic lupus erythematosus (SLE) patients, showed low rates induction ADA (74). Of note, in autoimmune diseases the hyperactivation of both the innate and adaptive immune responses may further complicate the study of mAb immunogenicity (57, 75). On the other hand, when administering mAbs to cancer patients, ADA formation often depends on the stage of the cancer. ADA levels tend to be higher in early stages of the disease than in later stages (76).
Much of the variability in the propensity of administrated mAb to induce ADA formation may result from different immune contexts; Principally, disease status and HLA alleles, which could promote or inhibit an ADA response. The idea that ADA formation is often derived from a T-dependent response has recently led to studies focusing on how ADA formation correlates with HLA polymorphism in the population. Although limited by sample size, Benucci et al. showed that patients with the HLA-DRβ-11, HLA-DQ-03, and HLA-DQ-05 alleles were at a higher risk to develop ADA responses after treatment with an anti-TNF mAb (5 different mAbs were included in this study) (77). Another report revealed that a G1m1 allotype in the IgG1 created a protease cleavage site in the CH3 domain of the antibody Fc and enabled presentation of a CH315–29 peptide epitope (78). The CH315–29 peptide epitope was tolerated in patients with a G1m1 allotype. However, donors homozygous for nG1m1 did not natively display the G1m1 MHC-II peptide and developed T cell CD4+ responses against antibody therapeutics containing the G1m1 allotype sequence; these ADA were also correlated with HLA-DRB1∗07 allele. Some therapeutic mAbs (including trastuzumab) do not harbor this allotype, which could partially explain differences in immunogenicity across different mAb drugs (78, 79). This allotype difference could impact future development of antibody products, since ∼40% of the Caucasian population is homozygous for nG1m1, and thus may be at a greater risk for ADA generation (80). In two recent studies, ADA formation against infliximab and adalimumab was correlated with the HLADQA1∗05A > G genotype in IBD patients (81, 82). One detailed recent study examined the immune response to natalizumab, a humanized monoclonal IgG4 antibody to α4 integrins that is used to treat patients with MS, and that induces ADA formation in ∼6% of the patients. The immune response was found to be polyclonal and targeted different epitopes of the natalizumab idiotype, with a single immunodominant T cell epitope spanning the FR2-CDR2 region of the VL (83). Generation of a T cell-dependent ADA response is also a multifactorial process, depending not only on the existence of a potential MHC-II peptide epitope in the mAb, but also on the ability of that epitope to be processed, presented and recognized by T cells. The influence of HLA allotypes on the probability of ADA responses should be considered during the design of immunogenicity studies and clinical trials for mAb development. Conclusions from studies that rely on smaller cohorts might not have general applicability for ADA predictions if the study population has substantially different MHC-II gene backgrounds from a larger treatment population.
Drug-Related Factors
The molecular mechanisms that lead to induction of ADAs were initially related to the murine origin of the first mAbs, which were recognized as “non-self” by the human immune system. Unfortunately, even the use of complete human antibody genes has not completely eliminated immunogenicity and the associated induction of ADA (84). Fully human mAbs contain new epitopes in the CDRs that can steer the immune response through an idiotype/anti-idiotype interaction (85, 86). As discussed above, mAb-derived peptides presented by MHC-II are necessary for T cell-dependent ADA formation. Efforts to remove T cell epitopes during mAb engineering are used consistently, but the high genetic variability of human populations greatly complicates efforts to remove all MHC-II-binding peptides from human mAbs (87, 88).
Changes in Fc glycosylation may also affect ADA induction. The removal of N-linked glycosylation of the Fc was shown to reduce immunogenicity (89). Fully human mAbs lacking Fc functions were also shown to be immunogenic and have direct effects on the ability to recruit macrophages and activate complement. For example, galactose-α-1,3-galactose, which is a foreign glycan not found in humans, is present on the antigen-binding (Fab) portion of the cetuximab VH (a chimeric mAb used in cancer therapy targeting the EGF receptor). This glycan was shown to induce ADA formation of the IgE isotype, and was responsible for anaphylactic reactions in patients (90, 91). On the other hand, immunogenicity is sometimes linked to impurities in the formulation process, and not necessarily due to glycosylation differences. A review of the differences between 18 biosimilars and mAbs originators concluded that the differences between them are mainly in glycosylation patterns, and do not impact immunogenicity (92).
Other drug related factors that play a role in mAb immunogenicity are “danger signals” that are released by tissues undergoing stress, damage or abnormal death. The danger model was first suggested in 1994, were it was first postulated that the immune system responds to substances that cause damage, rather than to those that are simply foreign (93, 94). In the case of therapeutic antibodies, process related impurities (such as aggregates and residual DNA or proteins from the mAb expression system) can influence immunogenicity (95).
The mAb target may also have high importance for the MOA of ADA formation. We recently found that repeated administration of infliximab (a TNFα antagonist) results in a vaccine-like response, where ADA formation is governed by the extrafollicular T cell-independent immune response (96). The administration of infliximab blocks TNFα and shifts the immune response toward the marginal zone (MZ) instead of the germinal center (GC), as observed in TNFα knockout mice (97). Another possible explanation is that a strong T cell-independent immune response in the MZ may be induced by a drug/ADA/TNFα immunocomplex (IC). As a trimer, TNFα may form “super complexes” upon engagement with TNFα antagonistic antibodies (98–100).
Another example of mAb target importance is alemtuzumab, a mAb specific to the CD52 lymphocyte cell surface glycoprotein. Alemtuzumab is used to treat MS (101) and induces ADAs in about 85% of patients, of which around 92% develop neutralizing ADAs (102). Alemtuzumab’s high frequency of ADA induction may be related to CD52 expression patterns. Alemtuzumab targets APCs, which include DCs, monocytes, and memory B cells, based on their CD52 expression. When monocytes repopulate, they encounter the circulating mAb that rapidly presents antigen to the antigen-specific lymphocytes (103, 104). Memory B cells often exhibit homeostatic expansion following treatment with alemtuzumab (105), which could complement ADA generation.
mAb dosage and schedule are other possible factors influencing ADA formation rates. Increased numbers of injections and higher mAb doses are associated with higher ADA risk, although some cases of chronic treatment and higher doses have lower immunogenicity (92, 106). For example, rituximab, a chimeric mAb anti-CD20, targets surface antigens on pre-B cells and B cells before their differentiation into PCs. As rituximab selectively depletes CD20 positive B cells, it does not affect mature PCs and does not have a propensity to elicit ADAs (107).
Assays for Immunogenicity Assessment and Tools for Immunogenicity Reduction
Pre-clinical Setting
Due to the growing importance of mAb immunogenicity, there has been a growing need for tools to assess immunogenicity and reduce the propensity of mAbs to induce ADAs. Great efforts in tools such as in silico prediction algorithms and cell based experimental assays are facilitating immunogenicity assessment, especially during the initial development phases of the mAb (108).
In silico CD4+ T cell epitope prediction models are often used to identify potentially immunogenic MHC-II peptide epitopes. These algorithms are based on the affinity of mAb-derived peptides to MHC-II (109–111).
With recent advances in proteomics and sequencing, several MHC-II peptide epitope databases have been constructed that provide a library of MHC-II binding data to enable immunogenicity prediction (112). Most algorithms that predict the immunogenic sequences recognized by T cells are later confirmed by assessing peptide binding to MHC molecules (88, 113). For example, a strong correlation was found between in silico evaluation of T cell epitopes from a recombinant Fc fusion protein, and the immunogenicity rate when administered to patients in a clinical trial (114). While such predictive algorithms are common used, they capture only a fraction of the system’s complexity. Most CD4 + T cell epitope prediction algorithms are based on binding affinity and stability to MHC-II molecules (88, 110), but fail to consider other essential factors in the recognition of T cell epitopes. Among these factors are protease cleavage sites (115), T cell precursor frequency (116), and peptide and T cell competition (117).
Experimental tools are also used to make pre-clinical predictions about mAb immunogenicity risk. These include HLA binding assays, DC related assays, T cell stimulation assays, peripheral blood mononuclear cell (PBMC) stimulation assays, and various animal models (115). HLA binding and DC antigen presentation assays can evaluate potential T cell epitopes derived from the mAb, while T cell and PBMC stimulation assays examine whether a mAb can activate immune cells in vitro and ex vivo in terms of cell proliferation and cytokine release. For example, T cell epitopes in the variable regions of infliximab and rituximab were able to stimulate peripheral blood mononuclear cells (PBMCs) to secrete a variety of cytokines (61). In another study, the immunogenicity of secukinumab, an anti- interleukin-17A mAb used to treat plaque psoriasis, was assessed by examining T-cell proliferation (118).
Each of these experimental tools has limitations in assessing and predicting immunogenicity. While considered reliable and straightforward, most of the experimental assays are labor intensive and are impractical to implement with a large number of mAb candidates. These assays are often performed with cells derived from a naïve population, where the frequency of antigen-specific cells is relatively low and precludes a clear positive result due to low signal-to-noise ratios (88).
Other advancements are being made in the development of mAbs to which patients will be more tolerant. A previous study identified a set of naturally occurring human regulatory T cell epitopes (“Tregitopes”), present in the Fc and Fab domains of IgG, that induce tolerance when co-administered with other proteins (119). When incubated with PBMCs in vitro, Tregitopes activated CD4+ T cells and increased expression of regulatory cytokines, chemokines, and CD25/Foxp3. When were administered in vivo with protein antigens, Tregitopes inhibited T cell proliferation, reduced effector cytokine expression, and induced antigen-specific adaptive tolerance. Co-administration of Tregitopes along with mAbs may be a useful tool for tolerization of mAbs.
Clinical Settings
Early and accurate ADA detection is extremely important for patients treated with biologics, especially for mAbs (120). ADA detection is required to provide the clinicians with sufficient information to monitor treatment and determine optimal intervention strategies (121). Detection of ADA against therapeutic mAbs is highly challenging since both the drug and the analyte are antibodies. Moreover, immunoassays are prone to biases due to the presence of the drug and immune-complexes in patients’ serum. Historically, studies of the response following mAb administration and ADA prevalence have been inconsistent, partly due to the various assay formats used to monitor immunogenicity in clinical trials (122). Each available format has its limitations that can reduce the assay’s utility in clinical and research settings, and also complicate interpretation of the data. Some assays have poor dynamic range and may generate false-negative results because of interfering interactions with the active drug, or false-positive results due to other antibodies like rheumatoid factor (123). Figure 2 shows the competing factors which affect accurate measurement of ADAs.
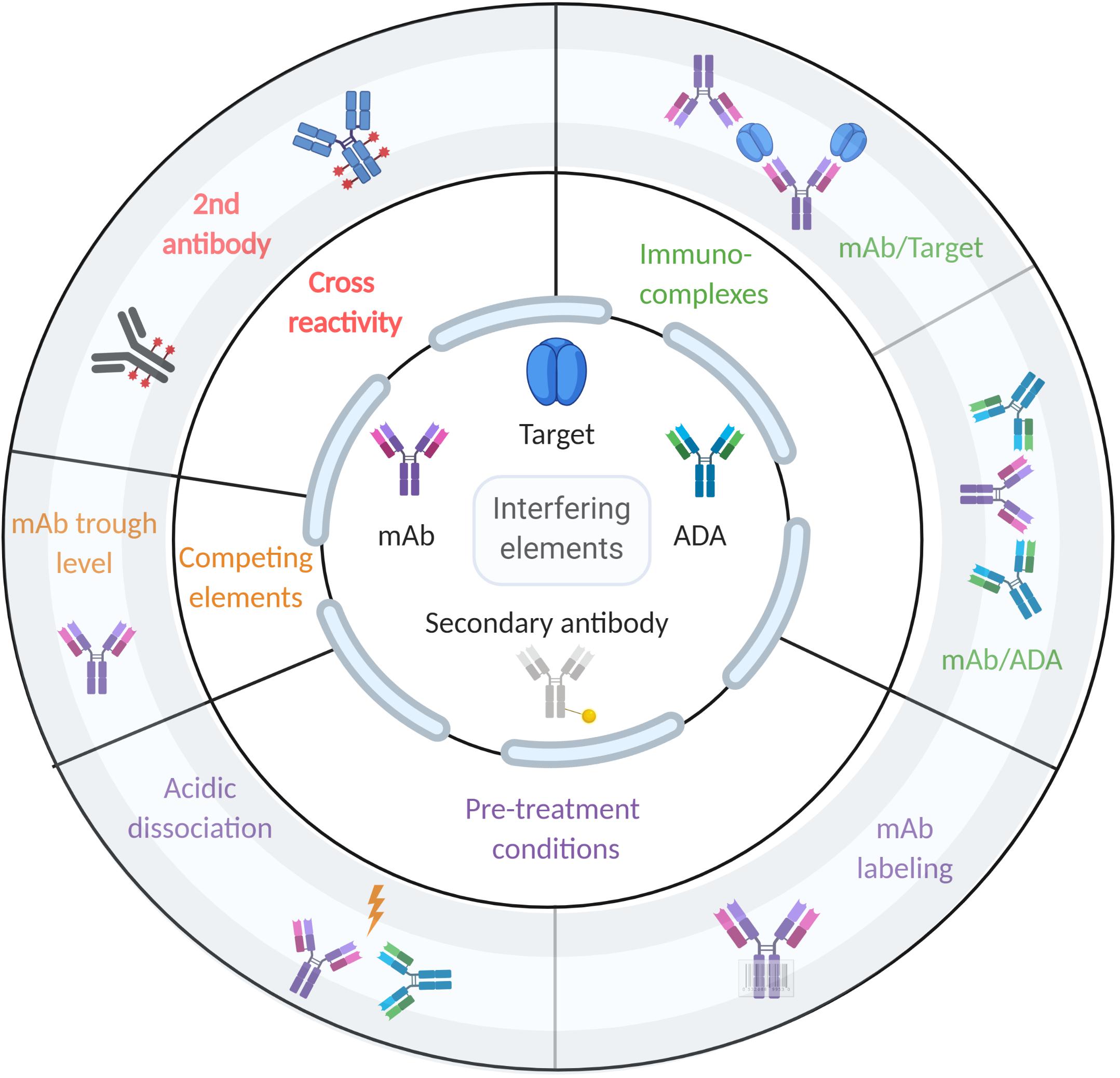
Figure 2. Factors that affect ADA detection in immunoassays. The center of the figure designates the components that could interfere with ADA detection (i.e., mAb, target, ADA, and secondary antibody). The middle circle designates the type of interference, while the outer circle provides examples of such interferences.
An ELISA-based bridging assay is one of the most commonly used assays for ADA screening, where the mAb drug is used to first capture ADA present in the patient sera, and the latter are detected by adding additional labeled mAb as a secondary probe. Bridging ELISA assays are used for ADA detection of a large variety of mAbs, and some include an acidic step to dissociate ADA from the mAb. The excess mAb is then captured or removed, and free ADA can be detected. These assays often have significantly higher background and suffer from low sensitivity due to the disassociation of antibodies. Bridging assays can also result in false-negatives, as they are more likely to “miss” low affinity IgM ADAs present in early stages of the immune response (124). Most ELISA-based bridging assays are also sensitive to the mAbs’ trough levels (levels of circulating mAb at sampling time). ADA and mAbs tend to form high molecular weight immune-complexes, making ADA detection more challenging (125). To overcome this challenge, several drug-tolerant assays have been developed to measure ADA levels in the presence of high mAb concentrations (126). Most of these assays also use an acidic treatment step. Several other techniques have been reported to evaluate serum ADA levels. These assays include radio-immunoassays (127), Biotin-drug Extraction with Acid Dissociation (BEAD) (128), Precipitation and Acid dissociation (PANDA) (129), Affinity Capture Elution ELISA (ACE) (130), and Homogenous Mobility Shift Assay (HMSA) (131); these assays have been reviewed in detail elsewhere (126). While these assays presumably detect all serum ADA, they primarily provide qualitative measures to assist healthcare providers deciding on appropriate patient interventions, and many (if not all) studies underestimate actual ADA levels. These assays also lack standardization that could enable comparisons of ADA levels across health centers. The great diversity in these assays poses tremendous difficulty in studying ADA levels between different mAbs, across studies of the same mAb, and across different assays.
In a clinical context, it important both to assess ADA levels in patient serum, and also to assess the presence of neutralizing antibodies that interfere with biological and clinical activity of the mAb. The neutralizing effect of ADAs can be assayed by testing whether ADAs in serum inhibit binding of the mAb to its target (132). Several cell-based assays were developed to detect ntADA in patients’ serum. One of these assays is a functional ADA cell-based assay that was developed to quantify the activity of TNFα antagonists. This assay assesses both drug activity and ntADA levels (133), but correlations between the clinical outcome and assay results were not thoroughly tested. Another assay developed for ntADA detection is the reporter gene assay, which is based on excretion of IL8 by HT29 cells due to TNFα stimulation (77). When the assay was applied to sera samples with low-level ADA, it detected ntADA even prior to clinical LOR to the mAb, which allows the prediction of clinical LOR with high probability.
While these assays are accurate and sensitive, they require an active cell line, which complicates assay implementation. We recently reported on a newly developed quantitative bio-immunoassay for quantifying ADA specific to TNFα antagonists. The bio-immunoassay was further modified to easily assess the neutralization capacity of ADA using an in vitro assay (96). This assay can be readily used in a clinical setting that performs routine ADA measurements.
Other clinical approaches to reduce immunogenicity include active interference of the T cell responses to mAbs, thereby inducing individual tolerance of the immune system (“tolerization”).
For example, administration of methotrexate (MTX) with infliximab reduced ADA formation in RA patients (134). MTX also reversed high ADA levels in infantile Pompe disease patients treated with rituximab, when administered alongside bortezomib, a proteasome activity inhibitor that leads to cell death (135). Azathioprine is also an immunosuppressive drug that can be given in combination with infliximab or adalimumab to improve treatment and reduce immunogenicity and ADA formation (136–138). However, such non-specific immunosuppressive approaches have potentially harmful side effects that must be balanced with the patient’s overall treatment plan.
Concluding Remarks
Monoclonal antibodies have the potential to treat a wide range of diseases and disorders, but they can be highly immunogenic and induce undesirable ADA responses. ADAs can reduce mAb drug efficacy by altering its bioavailability and/or accelerating clearance from circulation. While the molecular mechanisms of ADA generation are not fully understood, it is dependent on both patient and drug characteristics. While early ADAs were related to the murine origin of the first mAb therapeutics, ADAs also occur against fully human mAbs. Indeed, complete humanization cannot completely abrogate mAb immunogenicity and ADA formation. The questions of why and how ADA are generated also depend on variability of the reported immunogenicity rates, which emphasizes the need for standardized clinical assays for ADA detection. Understanding the mechanisms of ADA generation and the major factors that influence immunogenicity of mAbs will help us design safer mAbs with lower drug rejection rates. Recent and ongoing efforts to study mAb immunogenicity at the molecular level is augmenting our understanding of these mechanisms that lead to ADA formation, which may help provide new guidelines to improve the safety and efficacy of mAb therapeutics.
Author Contributions
AV-M, MG-G, BD, and YW wrote the sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
Funding for this work was provided by the United States-Israel Binational Science Foundation #2017359, and by US NIH grants R21AI143407, R21AI144408, and DP5OD023118.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Colette Worcester for assistance with manuscript preparation.
References
1. Chan, JCN, Chan, ATC. Biologics and biosimilars: what, why and how? ESMO Open. (2017) 2:e000180. doi: 10.1136/esmoopen-2017-000180
2. Aggarwal, RS. What’s fueling the biotech engine-2012 to 2013. Nat Biotechnol. (2014) 32:32–9. doi: 10.1038/nbt.2794
3. Ecker, DM, Jones, SD, Levine, HL. The therapeutic monoclonal antibody market. MAbs. (2015) 7:9–14. doi: 10.4161/19420862.2015.989042
4. Grilo, AL, Mantalaris, A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. (2019) 37:9–16. doi: 10.1016/j.tibtech.2018.05.014
5. Alas, S, Bonavida, B. Rituximab inactivates signal transducer and activation of transcription 3 (STAT3) activity in B-non-hodgkin’s lymphoma through inhibition of the interleukin 10 autocrine / paracrine loop and results in down-regulation of Bcl-2 and sensitization to cytot. Cancer Res. (2001) 61:5137–44.
6. Nahta, R, Esteva, FJ. Herceptin: mechanisms of action and resistance. Cancer Lett. (2006) 232:123–38. doi: 10.1016/j.canlet.2005.01.041
7. Said, R, Tsimberidou, AM. Obinutuzumab for the treatment of chronic lymphocytic leukemia and other B-cell lymphoproliferative disorders. Expert Opin Biol Ther. (2017) 17:1463–70.
8. Lim, H, Lee, SH, Lee, HT, Lee, JU, Son, JY, Shin, W, et al. Structural biology of the TNFalpha antagonists used in the treatment of rheumatoid arthritis. Int J Mol Sci. (2018) 19:768. doi: 10.3390/ijms19030768
9. Schulman, ES. Development of a monoclonal anti-immunoglobulin E antibody (omalizumab) for the treatment of allergic respiratory disorders. Am J Respir Crit Care Med. (2001) 164:S6–11.
10. Rogovik, AL, Carleton, B, Solimano, A, Goldman, RD. Palivizumab for the prevention of respiratory syncytial virus infection. Can Fam Physician. (2010) 56:769–72.
11. Wilcox, MH, Gerding, DN, Poxton, IR, Kelly, C, Nathan, R, Birch, T, et al. Bezlotoxumab for prevention of recurrent clostridium difficile infection. N Engl J Med. (2017) 376:305–17.
12. García-Foncillas, J, Sunakawa, Y, Aderka, D, Wainberg, Z, Ronga, P, Witzler, P, et al. Distinguishing features of cetuximab and panitumumab in colorectal cancer and other solid tumors. Front Oncol. (2019) 9:849. doi: 10.3389/fonc.2019.00849
13. Rajan, A, Kim, C, Heery, CR, Guha, U, Gulley, JL. Nivolumab, anti-programmed death-1 (PD-1) monoclonal antibody immunotherapy: role in advanced cancers. Hum Vaccin Immunother. (2016) 12:2219–31. doi: 10.1080/21645515.2016.1175694
14. Kohler, G, Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. (1975) 256:495–7. doi: 10.1038/256495a0
15. Schroff, RW, Foon, KA, Beatty, SM, Oldham, RK, Morgan, AC Jr. Human anti-murine immunoglobulin responses in patients receiving monoclonal antibody therapy. Cancer Res. (1985) 45:879–85.
16. Pyzik, M, Rath, T, Lencer, WI, Baker, K, Blumberg, RS. FcRn: the architect behind the immune and nonimmune functions of IgG and albumin. J Immunol. (2015) 194:4595–603. doi: 10.4049/jimmunol.1403014
17. Ober, RJ, Radu, CG, Ghetie, V, Ward, ES. Differences in promiscuity for antibody-FcRn interactions across species: implications for therapeutic antibodies. Int Immunol. (2001) 13:1551–9. doi: 10.1093/intimm/13.12.1551
18. Morrison, SL, Johnson, MJ, Herzenberg, LA, Oi, VT. Chimeric human antibody molecules: mouse antigen-binding domains with human constant region domains. Proc Natl Acad Sci USA. (1984) 81:6851–5. doi: 10.1073/pnas.81.21.6851
19. MacLennan, IC, Howard, A, Gotch, FM, Quie, PG. Effector activating determinants on IgG. I. The distribution and factors influencing the display of complement, neutrophil and cytotoxic B-cell determinants on human IgG sub-classes. Immunology. (1973) 25:459–69.
20. Reff, ME, Carner, K, Chambers, KS, Chinn, PC, Leonard, JE, Raab, R, et al. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. (1994) 83:435–45. doi: 10.1182/blood.v83.2.435.bloodjournal832435
21. LoBuglio, AF, Wheeler, RH, Trang, J, Haynes, A, Rogers, K, Harvey, EB, et al. Mouse/human chimeric monoclonal antibody in man: kinetics and immune response. Proc Natl Acad Sci USA. (1989) 86:4220–4. doi: 10.1073/pnas.86.11.4220
22. Presta, LG. Engineering of therapeutic antibodies to minimize immunogenicity and optimize function. Adv Drug Deliv Rev. (2006) 58:640–56. doi: 10.1016/j.addr.2006.01.026
23. Jones, PT, Dear, PH, Foote, J, Neuberger, MS, Winter, G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. (1986) 321:522–5. doi: 10.1038/321522a0
24. Hwang, WY, Foote, J. Immunogenicity of engineered antibodies. Methods. (2005) 36:3–10. doi: 10.1016/j.ymeth.2005.01.001
25. Winter, G, Griffiths, AD, Hawkins, RE, Hoogenboom, HR. Making antibodies by phage display technology. Annu Rev Immunol. (1994) 12:433–55. doi: 10.1146/annurev.iy.12.040194.002245
26. Vaughan, TJ, Williams, AJ, Pritchard, K, Osbourn, JK, Pope, AR, Earnshaw, JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. (1996) 14:309–14. doi: 10.1038/nbt0396-309
27. Azriel-Rosenfeld, R, Valensi, M, Benhar, I. A human synthetic combinatorial library of arrayable single-chain antibodies based on shuffling in vivo formed CDRs into general framework regions. J Mol Biol. (2004) 335:177–92. doi: 10.1016/j.jmb.2003.10.053
28. Green, LL, Hardy, MC, Maynard-Currie, CE, Tsuda, H, Louie, DM, Mendez, MJ, et al. Antigen-specific human monoclonal antibodies from mice engineered with human Ig heavy and light chain YACs. Nat Genet. (1994) 7:13–21. doi: 10.1038/ng0594-13
29. Cohenuram, M, Saif, MW. Panitumumab the first fully human monoclonal antibody: from the bench to the clinic. Anticancer Drugs. (2007) 18:7–15. doi: 10.1097/cad.0b013e32800feecb
30. Rau, R. Adalimumab (a fully human anti-tumour necrosis factor alpha monoclonal antibody) in the treatment of active rheumatoid arthritis: the initial results of five trials. Ann Rheum Dis. (2002) 61(Suppl. 2):ii70–3.
31. Baker, MP, Reynolds, HM, Lumicisi, B, Bryson, CJ. Immunogenicity of protein therapeutics: the key causes, consequences and challenges. Self Nonself. (2010) 1:314–22. doi: 10.4161/self.1.4.13904
32. Luciani, F, Bull, RA, Lloyd, AR. Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol. (2012) 30:443–52. doi: 10.1016/j.tibtech.2012.05.005
33. DeKosky, BJ, Ippolito, GC, Deschner, RP, Lavinder, JJ, Wine, Y, Rawlings, BM, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. (2013) 31:166–9. doi: 10.1038/nbt.2492
34. DeKosky, BJ, Kojima, T, Rodin, A, Charab, W, Ippolito, GC, Ellington, AD, et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. (2015) 21:86–91. doi: 10.1038/nm.3743
35. Doria-Rose, NA, Schramm, CA, Gorman, J, Moore, PL, Bhiman, JN, DeKosky, BJ, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. (2014) 509:55–62.
36. Wu, X, Zhou, T, Zhu, J, Zhang, B, Georgiev, I, Wang, C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. (2011) 333:1593–602.
37. Robins, HS, Campregher, PV, Srivastava, SK, Wacher, A, Turtle, CJ, Kahsai, O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. (2009) 114:4099–107. doi: 10.1182/blood-2009-04-217604
38. Rosati, E, Dowds, CM, Liaskou, E, Henriksen, EKK, Karlsen, TH, Franke, A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol. (2017) 17:61. doi: 10.1186/s12896-017-0379-9
39. Glanville, J, Zhai, W, Berka, J, Telman, D, Huerta, G, Mehta, GR, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci USA. (2009) 106:20216–21. doi: 10.1073/pnas.0909775106
40. Vaisman-Mentesh, A, Wine, Y. Monitoring phage biopanning by next-generation sequencing. Methods Mol Biol. (2018) 1701:463–73. doi: 10.1007/978-1-4939-7447-4_26
41. Wang, B, DeKosky, BJ, Timm, MR, Lee, J, Normandin, E, Misasi, J, et al. Functional interrogation and mining of natively paired human VH:VL antibody repertoires. Nat Biotechnol. (2018) 36:152–5. doi: 10.1038/nbt.4052
42. Georgiou, G, Ippolito, GC, Beausang, J, Busse, CE, Wardemann, H, Quake, SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol. (2014) 32:158–68. doi: 10.1038/nbt.2782
43. Reddy, ST, Ge, X, Miklos, AE, Hughes, RA, Kang, SH, Hoi, KH, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. (2010) 28:965–9. doi: 10.1038/nbt.1673
44. Saggy, I, Wine, Y, Shefet-Carasso, L, Nahary, L, Georgiou, G, Benhar, I. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng Des Sel. (2012) 25:539–49. doi: 10.1093/protein/gzs060
45. Lee, J, Boutz, DR, Chromikova, V, Joyce, MG, Vollmers, C, Leung, K, et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med. (2016) 22:1456–64. doi: 10.1038/nm.4224
46. Lavinder, JJ, Wine, Y, Giesecke, C, Ippolito, GC, Horton, AP, Lungu, OI, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA. (2014) 111:2259–64. doi: 10.1073/pnas.1317793111
47. Boutz, DR, Horton, AP, Wine, Y, Lavinder, JJ, Georgiou, G, Marcotte, EM. Proteomic identification of monoclonal antibodies from serum. Anal Chem. (2014) 86:4758–66. doi: 10.1021/ac4037679
48. Wine, Y, Boutz, DR, Lavinder, JJ, Miklos, AE, Hughes, RA, Hoi, KH, et al. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci USA. (2013) 110:2993–8. doi: 10.1073/pnas.1213737110
49. van Schouwenburg, PA, Rispens, T, Wolbink, GJ. Immunogenicity of anti-TNF biologic therapies for rheumatoid arthritis. Nat Rev Rheumatol. (2013) 9:164–72. doi: 10.1038/nrrheum.2013.4
50. Atzeni, F, Talotta, R, Salaffi, F, Cassinotti, A, Varisco, V, Battellino, M, et al. Immunogenicity and autoimmunity during anti-TNF therapy. Autoimmun Rev. (2013) 12:703–8. doi: 10.1016/j.autrev.2012.10.021
51. De Groot, AS, Scott, DW. Immunogenicity of protein therapeutics. Trends Immunol. (2007) 28:482–90. doi: 10.1016/j.it.2007.07.011
52. Hansel, TT, Kropshofer, H, Singer, T, Mitchell, JA, George, AJ. The safety and side effects of monoclonal antibodies. Nat Rev Drug Discov. (2010) 9:325–38. doi: 10.1038/nrd3003
53. de Vries, MK, Wolbink, GJ, Stapel, SO, de Groot, ER, Dijkmans, BA, Aarden, LA, et al. Inefficacy of infliximab in ankylosing spondylitis is correlated with antibody formation. Ann Rheum Dis. (2007) 66:133–4. doi: 10.1136/ard.2006.057745
54. Yanai, H, Hanauer, SB. Assessing response and loss of response to biological therapies in IBD. Am J Gastroenterol. (2011) 106:685–98. doi: 10.1038/ajg.2011.103
55. Ben-Horin, S, Chowers, Y. Review article: loss of response to anti-TNF treatments in Crohn’s disease. Aliment Pharmacol Ther. (2011) 33:987–95. doi: 10.1111/j.1365-2036.2011.04612.x
56. Baldo, BA. Adverse events to monoclonal antibodies used for cancer therapy: focus on hypersensitivity responses. Oncoimmunology. (2013) 2:e26333. doi: 10.4161/onci.26333
57. Talotta, R, Rucci, F, Canti, G, Scaglione, F. Pros and cons of the immunogenicity of monoclonal antibodies in cancer treatment: a lesson from autoimmune diseases. Immunotherapy. (2019) 11:241–54. doi: 10.2217/imt-2018-0081
58. Reding, MT, Lei, S, Lei, H, Green, D, Gill, J, Conti-Fine, BM. Distribution of Th1- and Th2-induced anti-factor VIII IgG subclasses in congenital and acquired hemophilia patients. Thromb Haemost. (2002) 88:568–75. doi: 10.1055/s-0037-1613257
59. Reding, MT. Immunological aspects of inhibitor development. Haemophilia. (2006) 12(Suppl. 6):30–5; discussion 35–6.
60. Vultaggio, A, Petroni, G, Pratesi, S, Nencini, F, Cammelli, D, Milla, M, et al. Circulating T cells to infliximab are detectable mainly in treated patients developing anti-drug antibodies and hypersensitivity reactions. Clin Exp Immunol. (2016) 186:364–72. doi: 10.1111/cei.12858
61. Hamze, M, Meunier, S, Karle, A, Gdoura, A, Goudet, A, Szely, N, et al. Characterization of CD4 T cell epitopes of infliximab and rituximab identified from healthy donors. Front Immunol. (2017) 8:500. doi: 10.3389/fimmu.2017.00500
62. Vos, Q, Lees, A, Wu, ZQ, Snapper, CM, Mond, JJ. B-cell activation by T-cell-independent type 2 antigens as an integral part of the humoral immune response to pathogenic microorganisms. Immunol Rev. (2000) 176:154–70. doi: 10.1034/j.1600-065x.2000.00607.x
63. Alugupalli, KR, Leong, JM, Woodland, RT, Muramatsu, M, Honjo, T, Gerstein, RM. B1b lymphocytes confer T cell-independent long-lasting immunity. Immunity. (2004) 21:379–90. doi: 10.1016/j.immuni.2004.06.019
64. Obukhanych, TV, Nussenzweig, MC. T-independent type II immune responses generate memory B cells. J Exp Med. (2006) 203:305–10. doi: 10.1084/jem.20052036
65. El Shikh, ME, El Sayed, RM, Szakal, AK, Tew, JG. T-independent antibody responses to T-dependent antigens: a novel follicular dendritic cell-dependent activity. J Immunol. (2009) 182:3482–91. doi: 10.4049/jimmunol.0802317
66. Taillardet, M, Haffar, G, Mondière, P, Asensio, MJ, Gheit, H, Burdin, N, et al. The thymus-independent immunity conferred by a pneumococcal polysaccharide is mediated by long-lived plasma cells. Blood. (2009) 114:4432–40. doi: 10.1182/blood-2009-01-200014
67. Yin, L, Chen, X, Vicini, P, Rup, B, Hickling, TP. Therapeutic outcomes, assessments, risk factors and mitigation efforts of immunogenicity of therapeutic protein products. Cell Immunol. (2015) 295:118–26. doi: 10.1016/j.cellimm.2015.03.002
68. Batista, FD, Harwood, NE. The who, how and where of antigen presentation to B cells. Nat Rev Immunol. (2009) 9:15–27. doi: 10.1038/nri2454
69. Kumar, S, Singh, SK, Wang, X, Rup, B, Gill, D. Coupling of aggregation and immunogenicity in biotherapeutics: T- and B-cell immune epitopes may contain aggregation-prone regions. Pharm Res. (2011) 28:949–61. doi: 10.1007/s11095-011-0414-9
70. Fehr, T, Bachmann, MF, Bucher, E, Kalinke, U, Di Padova, FE, Lang, AB, et al. Role of repetitive antigen patterns for induction of antibodies against antibodies. J Exp Med. (1997) 185:1785–92. doi: 10.1084/jem.185.10.1785
71. Ben-Horin, S, Heap, GA, Ahmad, T, Kim, H, Kwon, T, Chowers, Y. The immunogenicity of biosimilar infliximab: can we extrapolate the data across indications? Expert Rev Gastroenterol Hepatol. (2015) 9(Suppl. 1):27–34. doi: 10.1586/17474124.2015.1091307
72. Spinelli, FR, Valesini, G. Immunogenicity of anti-tumour necrosis factor drugs in rheumatic diseases. Clin Exp Rheumatol. (2013) 31:954–63.
73. Dunn, N, Juto, A, Ryner, M, Manouchehrinia, A, Piccoli, L, Fink, K, et al. Rituximab in multiple sclerosis: frequency and clinical relevance of anti-drug antibodies. Mult Scler. (2018) 24:1224–33. doi: 10.1177/1352458517720044
74. Blair, HA, Duggan, ST. Belimumab: a review in systemic lupus erythematosus. Drugs. (2018) 78:355–66. doi: 10.1007/s40265-018-0872-z
75. Tovey, MG, Lallemand, C. Immunogenicity and other problems associated with the use of biopharmaceuticals. Ther Adv Drug Saf. (2011) 2:113–28. doi: 10.1177/2042098611406318
76. van Brummelen, EM, Ros, W, Wolbink, G, Beijnen, JH, Schellens, JH. Antidrug antibody formation in oncology: clinical relevance and challenges. Oncologist. (2016) 21:1260–8. doi: 10.1634/theoncologist.2016-0061
77. Benucci, M, Damiani, A, Li Gobbi, F, Bandinelli, F, Infantino, M, Grossi, V, et al. Correlation between HLA haplotypes and the development of antidrug antibodies in a cohort of patients with rheumatic diseases. Biologics. (2018) 12:37–41. doi: 10.2147/btt.s145941
78. Stickler, MM, Reddy, A, Xiong, JM, Hinton, PR, DuBridge, R, Harding, FA. The human G1m1 allotype associates with CD4+ T-cell responsiveness to a highly conserved IgG1 constant region peptide and confers an asparaginyl endopeptidase cleavage site. Genes Immun. (2011) 12:213–21. doi: 10.1038/gene.2010.68
79. Carter, P, Presta, L, Gorman, CM, Ridgway, JB, Henner, D, Wong, WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. (1992) 89:4285–9. doi: 10.1073/pnas.89.10.4285
80. Johnson, MJ, de Lange, G, Cavalli-Sforza, LL. Ig gamma restriction fragment length polymorphisms indicate an ancient separation of Caucasian haplotypes. Am J Hum Genet. (1986) 38:617–40.
81. Sazonovs, A, Kennedy, NA, Moutsianas, L, Heap, GA, Rice, DL, Reppell, M, et al. HLA-DQA1∗05 carriage associated with development of anti-drug antibodies to infliximab and adalimumab in patients with Crohn’s disease. Gastroenterology. (2020) 158:189–99.
82. Wilson, A, Peel, C, Wang, Q, Pananos, AD, Kim, RB. HLADQA1∗05 genotype predicts anti-drug antibody formation and loss of response during infliximab therapy for inflammatory bowel disease. Aliment Pharmacol Ther. (2020) 51:356–63. doi: 10.1111/apt.15563
83. Cassotta, A, Mikol, V, Bertrand, T, Pouzieux, S, Le Parc, J, Ferrari, P, et al. A single T cell epitope drives the neutralizing anti-drug antibody response to natalizumab in multiple sclerosis patients. Nat Med. (2019) 25:1402–7. doi: 10.1038/s41591-019-0568-2
84. Pecoraro, V, De Santis, E, Melegari, A, Trenti, T. The impact of immunogenicity of TNFalpha inhibitors in autoimmune inflammatory disease. A systematic review and meta-analysis. Autoimmun Rev. (2017) 16:564–75. doi: 10.1016/j.autrev.2017.04.002
85. Harding, FA, Stickler, MM, Razo, J, DuBridge, RB. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. MAbs. (2010) 2:256–65. doi: 10.4161/mabs.2.3.11641
86. Jerne, NK. The generative grammar of the immune system. EMBO J. (1985) 4:847–52. doi: 10.1002/j.1460-2075.1985.tb03709.x
87. Griswold, KE, Bailey-Kellogg, C. Design and engineering of deimmunized biotherapeutics. Curr Opin Struct Biol. (2016) 39:79–88. doi: 10.1016/j.sbi.2016.06.003
88. Jawa, V, Cousens, LP, Awwad, M, Wakshull, E, Kropshofer, H, De Groot, AS. T-cell dependent immunogenicity of protein therapeutics: preclinical assessment and mitigation. Clin Immunol. (2013) 149:534–55. doi: 10.1016/j.clim.2013.09.006
89. Zhou, Q, Qiu, H. The mechanistic impact of N-glycosylation on stability, pharmacokinetics, and immunogenicity of therapeutic proteins. J Pharm Sci. (2019) 108:1366–77. doi: 10.1016/j.xphs.2018.11.029
90. Chung, CH, Mirakhur, B, Chan, E, Le, QT, Berlin, J, Morse, M, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med. (2008) 358:1109–17.
91. Berg, EA, Platts-Mills, TA, Commins, SP. Drug allergens and food–the cetuximab and galactose-alpha-1,3-galactose story. Ann Allergy Asthma Immunol. (2014) 112:97–101. doi: 10.1016/j.anai.2013.11.014
92. Doevendans, E, Schellekens, H. Immunogenicity of innovative and biosimilar monoclonal antibodies. Antibodies. (2019) 8:21. doi: 10.3390/antib8010021
93. Gallucci, S, Matzinger, P. Danger signals: SOS to the immune system. Curr Opin Immunol. (2001) 13:114–9. doi: 10.1016/s0952-7915(00)00191-6
94. Matzinger, P. Tolerance, danger, and the extended family. Annu Rev Immunol. (1994) 12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015
95. Niazi, S. Biosimilarity: The FDA Perspective. Boca Raton, FL: CRC Press. (2018). doi: 10.1146/annurev.iy.12.040194.005015
96. Vaisman-Mentesh, A, Rosenstein, S, Yavzori, M, Dror, Y, Fudim, E, Ungar, B, et al. Molecular landscape of anti-drug antibodies reveals the mechanism of the immune response following treatment with TNFα antagonists. Front Immunol. (2019) 10:2921. doi: 10.3389/fimmu.2019.02921
97. Pasparakis, M, Alexopoulou, L, Episkopou, V, Kollias, G. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J Exp Med. (1996) 184:1397–411. doi: 10.1084/jem.184.4.1397
98. van Schie, KA, Kruithof, S, Ooijevaar-de Heer, P, Derksen, NIL, van de Bovenkamp, FS, Saris, A, et al. Restricted immune activation and internalisation of anti-idiotype complexes between drug and antidrug antibodies. Ann Rheum Dis. (2018) 77:1471–9. doi: 10.1136/annrheumdis-2018-213299
99. Arnoult, C, Brachet, G, Cadena Castaneda, D, Azzopardi, N, Passot, C, Desvignes, C, et al. Crucial role for immune complexes but not FcRn in immunization against Anti-TNF-alpha antibodies after a single injection in mice. J Immunol. (2017) 199:418–24. doi: 10.4049/jimmunol.1601246
100. Bar-Yoseph, H, Pressman, S, Blatt, A, Vainberg, SG, Maimon, N, Starosvetsky, E, et al. Infliximab-tumor necrosis factor complexes elicit formation of anti-drug antibodies. Gastroenterology. (2019) 157:1338–51.e8.
101. Cohen, JA, Coles, AJ, Arnold, DL, Confavreux, C, Fox, EJ, Hartung, HP, et al. Alemtuzumab versus interferon beta 1a as first-line treatment for patients with relapsing-remitting multiple sclerosis: a randomised controlled phase 3 trial. Lancet. (2012) 380:1819–28.
102. Baker, D, Herrod, SS, Alvarez-Gonzalez, C, Giovannoni, G, Schmierer, K. Interpreting lymphocyte reconstitution data from the pivotal phase 3 trials of alemtuzumab. JAMA Neurol. (2017) 74:961–9. doi: 10.1001/jamaneurol.2017.0676
103. Li, Z, Richards, S, Surks, HK, Jacobs, A, Panzara, MA. Clinical pharmacology of alemtuzumab, an anti-CD52 immunomodulator, in multiple sclerosis. Clin Exp Immunol. (2018) 194:295–314. doi: 10.1111/cei.13208
104. Thomas, K, Eisele, J, Rodriguez-Leal, FA, Hainke, U, Ziemssen, T. Acute effects of alemtuzumab infusion in patients with active relapsing-remitting MS. Neurol Neuroimmunol Neuroinflamm. (2016) 3:e228. doi: 10.1212/nxi.0000000000000228
105. Cox, AL, Thompson, SA, Jones, JL, Robertson, VH, Hale, G, Waldmann, H, et al. Lymphocyte homeostasis following therapeutic lymphocyte depletion in multiple sclerosis. Eur J Immunol. (2005) 35:3332–42. doi: 10.1002/eji.200535075
106. Herskovitz, J, Ryman, J, Thway, T, Lee, S, Zhou, L, Chirmule, N, et al. Immune suppression during preclinical drug development mitigates immunogenicity-mediated impact on therapeutic exposure. AAPS J. (2017) 19:447–55. doi: 10.1208/s12248-016-0026-8
107. Cohen, SB, Emery, P, Greenwald, MW, Dougados, M, Furie, RA, Genovese, MC, et al. Rituximab for rheumatoid arthritis refractory to anti-tumor necrosis factor therapy: Results of a multicenter, randomized, double-blind, placebo-controlled, phase III trial evaluating primary efficacy and safety at twenty-four weeks. Arthritis Rheum. (2006) 54:2793–806. doi: 10.1002/art.22025
108. Gokemeijer, J, Jawa, V, Mitra-Kaushik, S. How close are we to profiling immunogenicity risk using in silico algorithms and in vitro methods? An industry perspective. AAPS J. (2017) 19:1587–92. doi: 10.1208/s12248-017-0143-z
109. Messitt, TJ, Terry, F, Moise, L, Martin, W, De Groot, AS. A comparison of two methods for T cell epitope mapping: “cell free” in vitro versus immunoinformatics. Immunome Res. (2011) 7:e6.
110. Yin, L, Calvo-Calle, JM, Dominguez-Amorocho, O, Stern, LJ, HLA-Dm constrains epitope selection in the human CD4 T cell response to vaccinia virus by favoring the presentation of peptides with longer HLA-DM-mediated half-lives. J Immunol. (2012) 189:3983–94. doi: 10.4049/jimmunol.1200626
111. Sacks, D, Baxter, B, Campbell, BCV, Carpenter, JS, Cognard, C, Dippel, D, et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke. (2018) 13:612–32.
112. Schlessinger, A, Ofran, Y, Yachdav, G, Rost, B. Epitome: database of structure-inferred antigenic epitopes. Nucleic Acids Res. (2006) 34:D777–80.
113. Fleri, W, Paul, S, Dhanda, SK, Mahajan, S, Xu, X, Peters, B, et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. (2017) 8:278. doi: 10.3389/fimmu.2017.00278
114. Koren, E, De Groot, AS, Jawa, V, Beck, KD, Boone, T, Rivera, D, et al. Clinical validation of the “in silico” prediction of immunogenicity of a human recombinant therapeutic protein. Clin Immunol. (2007) 124:26–32. doi: 10.1016/j.clim.2007.03.544
115. Manoury, B, Mazzeo, D, Fugger, L, Viner, N, Ponsford, M, Streeter, H, et al. Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat Immunol. (2002) 3:169–74. doi: 10.1038/ni754
116. Harrington, CJ, Paez, A, Hunkapiller, T, Mannikko, V, Brabb, T, Ahearn, M, et al. Differential tolerance is induced in T cells recognizing distinct epitopes of myelin basic protein. Immunity. (1998) 8:571–80. doi: 10.1016/s1074-7613(00)80562-2
117. Kedl, RM, Kappler, JW, Marrack, P. Epitope dominance, competition and T cell affinity maturation. Curr Opin Immunol. (2003) 15:120–7. doi: 10.1016/s0952-7915(02)00009-2
118. Karle, A, Spindeldreher, S, Kolbinger, F. Secukinumab, a novel anti-IL-17A antibody, shows low immunogenicity potential in human in vitro assays comparable to other marketed biotherapeutics with low clinical immunogenicity. MAbs. (2016) 8:536–50. doi: 10.1080/19420862.2015.1136761
119. De Groot, AS, Moise, L, McMurry, JA, Wambre, E, Van Overtvelt, L, Moingeon, P, et al. Activation of natural regulatory T cells by IgG Fc-derived peptide “Tregitopes”. Blood. (2008) 112:3303–11. doi: 10.1182/blood-2008-02-138073
120. Krieckaert, C, Rispens, T, Wolbink, G. Immunogenicity of biological therapeutics: from assay to patient. Curr Opin Rheumatol. (2012) 24:306–11. doi: 10.1097/bor.0b013e3283521c4e
121. Hart, MH, de Vrieze, H, Wouters, D, Wolbink, GJ, Killestein, J, de Groot, ER, et al. Differential effect of drug interference in immunogenicity assays. J Immunol Methods. (2011) 372:196–203. doi: 10.1016/j.jim.2011.07.019
122. Vincent, FB, Morand, EF, Murphy, K, Mackay, F, Mariette, X, Marcelli, C. Antidrug antibodies (ADAb) to tumour necrosis factor (TNF)-specific neutralising agents in chronic inflammatory diseases: a real issue, a clinical perspective. Ann Rheum Dis. (2013) 72:165–78. doi: 10.1136/annrheumdis-2012-202545
123. Tatarewicz, S, Miller, JM, Swanson, SJ, Moxness, MS. Rheumatoid factor interference in immunogenicity assays for human monoclonal antibody therapeutics. J Immunol Methods. (2010) 357:10–6. doi: 10.1016/j.jim.2010.03.012
124. Collet-Brose, J, Couble, PJ, Deehan, MR, Nelson, RJ, Ferlin, WG, Lory, S. Evaluation of multiple immunoassay technology platforms to select the anti-drug antibody assay exhibiting the most appropriate drug and target tolerance. J Immunol Res. (2016) 2016:5069678.
125. Wang, YM, Fang, L, Zhou, L, Wang, J, Ahn, HY. A survey of applications of biological products for drug interference of immunogenicity assays. Pharm Res. (2012) 29:3384–92. doi: 10.1007/s11095-012-0833-2
126. Bloem, K, Hernandez-Breijo, B, Martinez-Feito, A, Rispens, T. Immunogenicity of therapeutic antibodies: monitoring antidrug antibodies in a clinical context. Ther Drug Monit. (2017) 39:327–32. doi: 10.1097/ftd.0000000000000404
127. Svenson, M, Geborek, P, Saxne, T, Bendtzen, K. Monitoring patients treated with anti-TNF-α biopharmaceuticals: assessing serum infliximab and anti-infliximab antibodies. Rheumatology. (2007) 46:1828–34. doi: 10.1093/rheumatology/kem261
128. Lofgren, JA, Wala, I, Koren, E, Swanson, SJ, Jing, S. Detection of neutralizing anti-therapeutic protein antibodies in serum or plasma samples containing high levels of the therapeutic protein. J Immunol Methods. (2006) 20:101–8. doi: 10.1016/j.jim.2005.10.007
129. Zoghbi, J, Xu, Y, Grabert, R, Theobald, V, Richards, S. A breakthrough novel method to resolve the drug and target interference problem in immunogenicity assays. J Immunol Methods. (2015) 426:62–9. doi: 10.1016/j.jim.2015.08.002
130. Schmidt, E, Hennig, K, Mengede, C, Zillikens, D, Kromminga, A. Immunogenicity of rituximab in patients with severe pemphigus. Clin Immunol. (2009) 132:334–41. doi: 10.1016/j.clim.2009.05.007
131. Hernandez-Breijo, B, Chaparro, M, Cano-Martinez, D, Guerra, I, Iborra, M, Cabriada, JL, et al. Standardization of the homogeneous mobility shift assay protocol for evaluation of anti-infliximab antibodies. Application of the method to Crohn’s disease patients treated with infliximab. Biochem Pharmacol. (2016) 122:33–41. doi: 10.1016/j.bcp.2016.09.019
132. Gupta, S, Indelicato, SR, Jethwa, V, Kawabata, T, Kelley, M, Mire-Sluis, AR, et al. Recommendations for the design, optimization, and qualification of cell-based assays used for the detection of neutralizing antibody responses elicited to biological therapeutics. J Immunol Methods. (2007) 321:1–18. doi: 10.1016/j.jim.2006.12.004
133. Lallemand, C, Kavrochorianou, N, Steenholdt, C, Bendtzen, K, Ainsworth, MA, Meritet, JF, et al. Reporter gene assay for the quantification of the activity and neutralizing antibody response to TNFα antagonists. J Immunol Methods. (2011) 373:229–39. doi: 10.1016/j.jim.2011.08.022
134. Maini, RN, Breedveld, FC, Kalden, JR, Smolen, JS, Davis, D, Macfarlane, JD, et al. Therapeutic efficacy of multiple intravenous infusions of anti-tumor necrosis factor alpha monoclonal antibody combined with low-dose weekly methotrexate in rheumatoid arthritis. Arthritis Rheum. (1998) 41:1552–63. doi: 10.1002/1529-0131(199809)41:9<1552::aid-art5>3.0.co;2-w
135. Banugaria, SG, Prater, SN, McGann, JK, Feldman, JD, Tannenbaum, JA, Bailey, C, et al. Bortezomib in the rapid reduction of high sustained antibody titers in disorders treated with therapeutic protein: lessons learned from Pompe disease. Genet Med. (2013) 15:123–31. doi: 10.1038/gim.2012.110
136. Ruffolo, C, Scarpa, M, Bassi, N. Infliximab, azathioprine, or combination therapy for Crohn’s disease. N Engl J Med. (2010) 363:1086–7; author reply 1087–8.
137. Anderson, PJ. Tumor necrosis factor inhibitors : clinical implications of their different immunogenicity profiles. Semin Arthritis Rheum. (2005) 34:19–22. doi: 10.1016/j.semarthrit.2005.01.005
138. Garces, S, Demengeot, J, Benito-Garcia, E. The immunogenicity of anti-TNF therapy in immune-mediated inflammatory diseases: a systematic review of the literature with a meta-analysis. Ann Rheum Dis. (2013) 72:1947–55. doi: 10.1136/annrheumdis-2012-202220
139. Quistrebert, J, Hassler, S, Bachelet, D, Mbogning, C, Musters, A, Tak, PP, et al. Incidence and risk factors for adalimumab and infliximab anti-drug antibodies in rheumatoid arthritis: a European retrospective multicohort analysis. Semin Arthritis Rheum (2019) 48:967–75. doi: 10.1016/j.semarthrit.2018.10.006
140. Paul, S, Moreau, AC, Del Tedesco, E, Rinaudo, M, Phelip, JM, Genin, C, et al. Pharmacokinetics of adalimumab in inflammatory bowel diseases: a systematic review and meta-analysis. Inflamm Bowel Dis. (2014) 20:1288–95. doi: 10.1097/mib.0000000000000037
141. Bartelds, GM, Krieckaert, CL, Nurmohamed, MT, van Schouwenburg, PA, Lems, WF, Twisk, JW, et al. Development of antidrug antibodies against adalimumab and association with disease activity and treatment failure during long-term follow-up. JAMA. (2011) 305:1460–8. doi: 10.1001/jama.2011.406
142. Roth, EM, Goldberg, AC, Catapano, AL, Torri, A, Yancopoulos, GD, Stahl, N, et al. Antidrug antibodies in patients treated with alirocumab. N Engl J Med. (2017) 376:1589–90. doi: 10.1056/nejmc1616623
143. Yan, L, Wang, B, Chia, YL, Roskos, LK. Population pharmacokinetic modeling of benralizumab in adult and adolescent patients with asthma. Clin Pharmacokinet. (2019) 58:943–58. doi: 10.1007/s40262-019-00738-4
144. Saffari, F, Jafarzadeh, A, Kalantari Khandani, B, Saffari, F, Soleimanyamoli, S, Mohammadi, M. Immunogenicity of rituximab, trastuzumab, and bevacizumab monoclonal antibodies in patients with malignant diseases. Int J Cancer Manag. (2018) 11:e64983.
145. Bagel, J, Lebwohl, M, Israel, RJ, Jacobson, A. Immunogenicity and skin clearance recapture in clinical studies of brodalumab. J Am Acad Dermatol. (2020) 82:344–51. doi: 10.1016/j.jaad.2019.05.094
146. Chen, Q, Hu, C, Liu, Y, Song, R, Zhu, W, Zhao, H, et al. Pharmacokinetics, pharmacodynamics, safety, and tolerability of single-dose denosumab in healthy Chinese volunteers: a randomized, single-blind, placebo-controlled study. PLoS One. (2018) 13:e0197984. doi: 10.1371/journal.pone.0197984
147. Ozkaynak, MF, Gilman, AL, London, WB, Naranjo, A, Diccianni, MB, Tenney, SC, et al. Trial of chimeric antibody 14.18 with GM-CSF, IL-2, and isotretinoin in high-risk neuroblastoma patients following myeloablative therapy: children’s oncology group study ANBL0931. Front Immunol. (2018) 9:1355. doi: 10.3389/fimmu.2018.01355
148. Hillmen, P, Muus, P, Szer, J, Hill, A, Hochsmann, B, Kulasekararaj, A, et al. Assessment of human antihuman antibodies to eculizumab after long-term treatment in patients with paroxysmal nocturnal hemoglobinuria. Am J Hematol. (2016) 91:E16–7.
149. Passey, C, Mora, J, Dodge, R, Gibiansky, L, Sheng, J, Roy, A, et al. An integrated assessment of the effects of immunogenicity on the pharmacokinetics, safety, and efficacy of elotuzumab. AAPS J. (2017) 19:557–67. doi: 10.1208/s12248-016-0033-9
150. Koren, MJ, Sabatine, MS, Giugliano, RP, Langslet, G, Wiviott, SD, Ruzza, A, et al. Long-term efficacy and safety of evolocumab in patients with hypercholesterolemia. J Am Coll Cardiol. (2019) 74:2132–46. doi: 10.1016/j.jacc.2019.08.1024
151. Leu, JH, Adedokun, OJ, Gargano, C, Hsia, EC, Xu, Z, Shankar, G. Immunogenicity of golimumab and its clinical relevance in patients with rheumatoid arthritis, psoriatic arthritis and ankylosing spondylitis. Rheumatology. (2019) 58:441–6. doi: 10.1093/rheumatology/key309
152. Ben-Horin, S, Yavzori, M, Katz, L, Kopylov, U, Picard, O, Fudim, E, et al. The immunogenic part of infliximab is the F(ab’)2, but measuring antibodies to the intact infliximab molecule is more clinically useful. Gut. (2010) 60:41–8. doi: 10.1136/gut.2009.201533
153. Kverneland, AH, Enevold, C, Donia, M, Bastholt, L, Svane, IM, Nielsen, CH. Development of anti-drug antibodies is associated with shortened survival in patients with metastatic melanoma treated with ipilimumab. Oncoimmunology. (2018) 7:e1424674. doi: 10.1080/2162402x.2018.1424674
154. Gordon, KB, Blauvelt, A, Papp, KA, Langley, RG, Luger, T, Ohtsuki, M, et al. Phase 3 trials of ixekizumab in moderate-to-severe plaque psoriasis. N Engl J Med. (2016) 375:345–56.
155. Ortega, H, Lemiere, C, Llanos, JP, Forshag, M, Price, R, Albers, F, et al. Outcomes following mepolizumab treatment discontinuation: real-world experience from an open-label trial. Allergy Asthma Clin Immunol. (2019) 15:37.
156. Sandborn, WJ, Colombel, JF, Enns, R, Feagan, BG, Hanauer, SB, Lawrance, IC, et al. Natalizumab induction and maintenance therapy for Crohn’s disease. N Engl J Med. (2005) 353:1912–25.
157. Agrawal, S, Statkevich, P, Bajaj, G, Feng, Y, Saeger, S, Desai, DD, et al. Evaluation of immunogenicity of nivolumab monotherapy and its clinical relevance in patients with metastatic solid tumors. J Clin Pharmacol. (2017) 57:394–400. doi: 10.1002/jcph.818
158. Nagy, CF, Leach, TS, King, A, Guttendorf, R. Safety, pharmacokinetics, and immunogenicity of obiltoxaximab after intramuscular administration to healthy humans. Clin Pharmacol Drug Dev. (2018) 7:652–60. doi: 10.1002/cpdd.410
159. Song A, Hendricks R, Chung S, Wang Q, Chin P, Garren, H. Immunogenicity with repeated dosing of ocrelizumab in patients with multiple sclerosis (P2.087). Neurology (2016) 86:P2.087. doi: 10.1002/cpdd.410
160. Somerville, L, Bardelas, J, Viegas, A, D’Andrea, P, Blogg, M, Peachey, G. Immunogenicity and safety of omalizumab in pre-filled syringes in patients with allergic (IgE-mediated) asthma. Curr Med Res Opin. (2014) 30:59–66. doi: 10.1185/03007995.2013.844115
161. Null, D Jr., Pollara, B, Dennehy, PH, Steichen, J, Sanchez, PJ, Givner, LB, et al. Safety and immunogenicity of palivizumab (Synagis) administered for two seasons. Pediatr Infect Dis J. (2005) 24:1021–3. doi: 10.1097/01.inf.0000183938.33484.bd
162. van Vugt, MJH, Stone, JA, De Greef, R, Snyder, ES, Lipka, L, Turner, DC, et al. Immunogenicity of pembrolizumab in patients with advanced tumors. J Immunother Cancer. (2019) 7:212.
163. Kirschbrown, WP, Wang, B, Nijem, I, Ohtsu, A, Hoff, PM, Shah, MA, et al. Pharmacokinetic and exposure-response analysis of pertuzumab in patients with HER2-positive metastatic gastric or gastroesophageal junction cancer. Cancer Chemother Pharmacol. (2019) 84:539–50. doi: 10.1007/s00280-019-03871-w
164. Zou, L, Pukac, L, Shalit, Y, Hickey, L, Garin, M, Liu, P. Immunogenicity assessment of intravenous administration of reslizumab in patients with asthma in phase 3 clinical studies. Eur Respir J. (2018) 52:A1032.
165. Maspero, J. Reslizumab in the treatment of inadequately controlled asthma in adults and adolescents with elevated blood eosinophils: clinical trial evidence and future prospects. Ther Adv Respir Dis. (2017) 11:311–25. doi: 10.1177/1753465817717134
166. Mrowietz, U, Leonardi, CL, Girolomoni, G, Toth, D, Morita, A, Balki, SA, et al. Secukinumab retreatment-as-needed versus fixed-interval maintenance regimen for moderate to severe plaque psoriasis: a randomized, double-blind, noninferiority trial (SCULPTURE). J Am Acad Dermatol. (2015) 73:27–36.e1. doi: 10.1016/j.jaad.2015.04.011
167. Deisseroth, A, Ko, CW, Nie, L, Zirkelbach, JF, Zhao, L, Bullock, J, et al. FDA approval: siltuximab for the treatment of patients with multicentric Castleman disease. Clin Cancer Res. (2015) 21:950–4. doi: 10.1158/1078-0432.ccr-14-1678
168. Kimball AB, Kerbusch T, van Aarle F, Kulkarni P, Li Q, Blauvelt A, et al. Assessment of the effects of immunogenicity on the pharmacokinetics, efficacy and safety of tildrakizumab. Br J Dermatol. (2020) 182:180–189. doi: 10.1158/1078-0432.ccr-14-1678
169. Burmester, GR, Choy, E, Kivitz, A, Ogata, A, Bao, M, Nomura, A, et al. Low immunogenicity of tocilizumab in patients with rheumatoid arthritis. Ann Rheum Dis. (2017) 76:1078–85. doi: 10.1136/annrheumdis-2016-210297
170. Chiu, H-Y, Chu, TW, Cheng, Y-P, Tsai, T-F. The association between clinical response to ustekinumab and immunogenicity to ustekinumab and prior adalimumab. PLoS One. (2015) 10:e0142930. doi: 10.1371/journal.pone.0142930
Keywords: monoclonal antibodies, anti-drug antibodies, immune response, immunogenicity, neutralizing antibodies
Citation: Vaisman-Mentesh A, Gutierrez-Gonzalez M, DeKosky BJ and Wine Y (2020) The Molecular Mechanisms That Underlie the Immune Biology of Anti-drug Antibody Formation Following Treatment With Monoclonal Antibodies. Front. Immunol. 11:1951. doi: 10.3389/fimmu.2020.01951
Received: 18 March 2020; Accepted: 20 July 2020;
Published: 18 August 2020.
Edited by:
Susan Richards, Sanofi Genzyme, United StatesReviewed by:
Marc Pallardy, Université Paris-Sud, FranceAnnie Searls De Groot, EpiVax, Inc., United States
Copyright © 2020 Vaisman-Mentesh, Gutierrez-Gonzalez, DeKosky and Wine. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yariv Wine, eWFyaXYud2luZUBnbWFpbC5jb20=
 Anna Vaisman-Mentesh
Anna Vaisman-Mentesh Matias Gutierrez-Gonzalez
Matias Gutierrez-Gonzalez Brandon J. DeKosky
Brandon J. DeKosky Yariv Wine
Yariv Wine