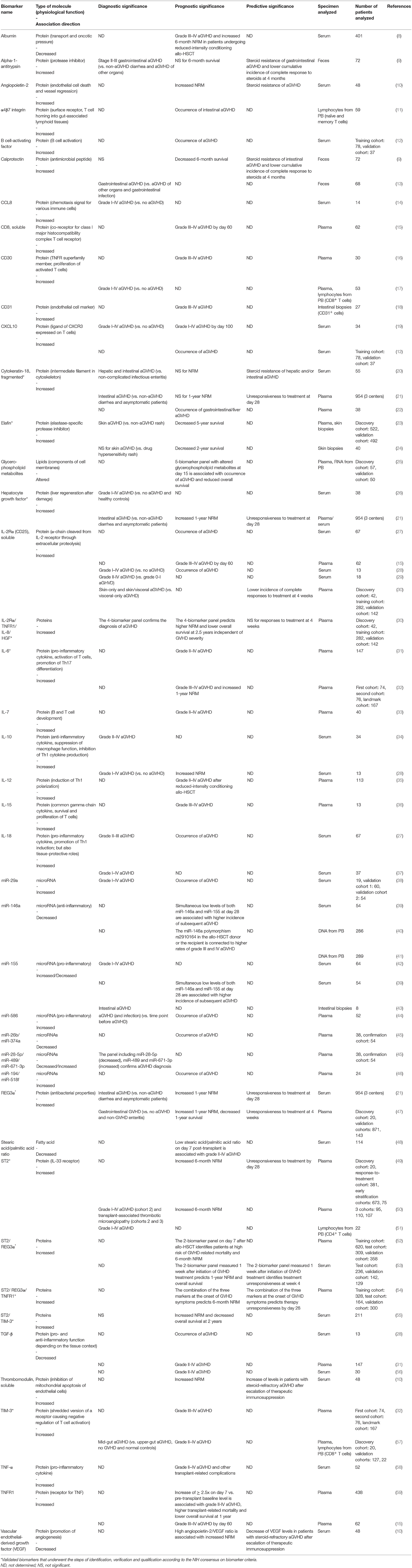
- 1Department of Immunology, Memorial Sloan Kettering Cancer Center, Sloan Kettering Institute, New York, NY, United States
- 2Department of Medicine I, Faculty of Medicine, Medical Center, University of Freiburg, Freiburg, Germany
- 3German Cancer Consortium (DKTK), Partner Site Freiburg, Freiburg, Germany
- 4Signalling Research Centres BIOSS and CIBSS – Centre for Integrative Biological Signalling Studies, University of Freiburg, Freiburg, Germany
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a well-established curative treatment for various malignant hematological diseases. However, its clinical success is substantially limited by major complications including graft-vs.-host disease (GVHD) and relapse of the underlying disease. Although these complications are known to lead to significant morbidity and mortality, standardized pathways for risk stratification of patients undergoing allo-HSCT are lacking. Recent advances in the development of diagnostic and prognostic tools have allowed the identification of biomarkers in order to predict outcome after allo-HSCT. This review will provide a summary of clinically relevant biomarkers that have been studied to predict the development of acute GVHD, the responsiveness of affected patients to immunosuppressive treatment and the risk of non-relapse mortality. Furthermore, biomarkers associated with increased risk of relapse and subsequent mortality will be discussed.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is the only curative treatment for a variety of malignant hematological diseases. A major complication after allo-HSCT consists of acute graft-vs.-host disease (aGVHD), which occurs when immunocompetent T cells of the allo-HSCT donor recognize antigens on recipient cells as foreign and attack recipient tissue, mainly the skin, gastrointestinal tract and liver (1), but as shown more recently, also the central nervous system (2). Several immunosuppressive agents are used for the treatment of aGVHD (3). While aGVHD leads to significant morbidity and mortality, donor T cell effector functions are necessary for the elimination of remaining malignant cells after allo-HSCT. This phenomenon, termed graft-vs.-leukemia (GVL) effect, is crucial for reducing the risk of relapse of the underlying disease, a complication occurring in a large portion of patients and causing substantially reduced survival after allo-HSCT (4, 5). In order to improve outcome after allo-HSCT, it would be desirable to predict which patients are at a high risk to develop aGVHD, how they respond to corticosteroids and what their risk of non-relapse mortality (NRM) as well as relapse is. To address these questions, multiple candidate biomarkers have been determined and correlated with clinical outcome, with some having been validated in large patient cohorts.
Biomarkers for Acute Graft-vs.-Host Disease and Non-relapse Mortality
Even when patients are cured of their underlying disease after allo-HSCT, their life expectancy remains inferior to that of age-matched general population due to NRM (6). Major risk factors of NRM include acute and chronic GVHD, infections, organ failure and second cancers (7). This review will focus on candidate and validated biomarkers that have been investigated in transplanted patients in order to predict the risk of aGVHD and the response to immunosuppressive therapy (Table 1).
A biomarker is defined as a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes or pharmacologic responses to a therapeutic intervention (60). The Biomarker Working Group of the National Institutes of Health (NIH) Consensus Development Project on Criteria for Clinical Trials in Chronic GVHD as well as the North-American and European Consortium distinguished four categories of GVHD biomarkers (61, 62): (1) diagnostic biomarkers, which identify GVHD patients at the onset of clinical disease, (2) prognostic biomarkers, which categorize patients by degree of risk for GVHD occurrence, progression or resolution before the onset of clinical disease, (3) predictive biomarkers, which categorize patients by their likelihood of response or outcome to a particular treatment before initiation of the treatment, and (4) response-to-treatment biomarkers, which monitor patients' response to GVHD treatment after initiation of therapy and which can substitute for a clinical efficacy endpoint.
Before being considered for standard clinical use, the development of biomarkers has to undergo a multi-step process consisting of (61): (1) identification of potential biomarker candidates in a small experiment of well-matched cases and controls selected from the populations in which the biomarker is intended for use, (2) verification by confirming the analytical validity and practicality of the test in an independent patient cohort, and (3) qualification by testing the impact on patient outcomes.
Immune Cell-Derived Biomarkers
Early approaches to identify biomarkers for aGVHD mainly focused on the detection of inflammatory cytokines involved in the pathogenesis of the disorder. Increased levels of interleukin (IL)-12 and IL-18, two cytokines known to promote T cell differentiation into T helper (Th) 1 cells with subsequent interferon-γ production, have been shown to correlate with severity of aGVHD (27, 35, 37). High levels of the key pro-inflammatory cytokine tumor necrosis factor (TNF)-α, mainly produced by macrophages, as well as elevated serum levels of its receptor TNFR1 were also found to be associated with severe aGVHD (15, 58, 59). Studies on another pro-inflammatory cytokine, IL-6, validated that increased levels at the time period before or at the onset of GVHD symptoms predicted development of severe GVHD (31, 32). Several studies described an association between levels of soluble IL-2 receptor α (IL-2Rα) and the occurrence of aGVHD (15, 27–29). Furthermore, IL-2Rα levels at GVHD onset were associated with complete responses to treatment at 4 weeks (30). B cell-activating factor (BAFF) as an indicator of B cell activation was also found to be increased pre-transplant and on day 14 in aGVHD patients (12).
Not only have increased levels of various pro-inflammatory cytokines (depicted in Figure 1) been identified as potential biomarkers for aGVHD, but also cytokines with anti-inflammatory effects and their dysregulation have been investigated. Decreased levels of transforming growth factor β (TGF-β), which is involved in the generation of regulatory T cells (Tregs) and inhibition of lymphocyte activation, have been associated with GVHD incidence and severity (28, 31, 56). Interestingly, IL-10, which is known to suppress macrophage functions and inhibit expression of Th1 cytokines, was demonstrated to be increased in aGVHD patients (28, 34). The authors hypothesize that high levels of IL-10 during GVHD are produced in response to the existing inflammation in order to inhibit further production of pro-inflammatory cytokines.
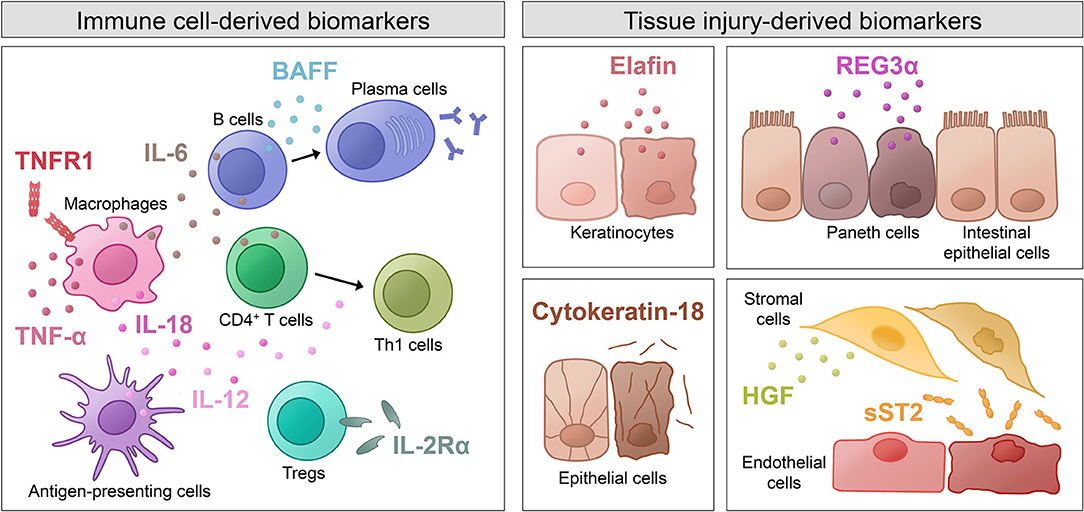
Figure 1. Shown are immune cell-derived molecules and tissue injury-derived molecules as well as the cells that they originate from. The molecules have various physiological functions and were described as biomarkers for acute GVHD. BAFF, B cell-activating factor; HGF, hepatocyte growth factor; IL, interleukin; REG3α, regenerating islet-derived protein 3α; sST2, soluble isoform of suppression of tumorigenicity 2; Th1 cells, T helper 1 cells; TNF-α, tumor necrosis factor α; TNFR1, tumor necrosis factor receptor 1; Tregs, regulatory T cells.
Other molecules found in the plasma that are related to immune cell activation and that were investigated as potential biomarkers in aGVHD include chemokines, such as CXCL10 and CXCL11 as mediators of leukocyte chemotaxis (12), the soluble extracellular domain of T cell immunoglobulin and mucin domain 3 (TIM-3) (32, 57) and α4β7 integrin, a surface molecule involved in lymphocyte trafficking to intestinal lymphoid tissue (11).
Tissue Injury-Derived Biomarkers
Novel advances in proteomic analyses have allowed screening of large numbers of patient samples and identification of novel biomarker candidates. Some of these potential biomarkers are not directly involved in the pathogenesis of aGVHD, but rather indicate end-organ tissue injury caused by the inflammatory processes in aGVHD (depicted in Figure 1). Since certain molecules are released from particular cell types, some biomarkers have diagnostic value for specific GVHD target organs. For instance, elafin, an elastase-specific protease inhibitor, was identified as a diagnostic and prognostic biomarker for skin GVHD, which is associated with higher incidence and lower overall survival (23, 24). Regenerating islet-derived protein 3α (REG3α), a C-type lectin secreted by Paneth cells, was validated as a prognostic marker for aGVHD of the intestinal tract (47). When epithelial cell death occurs, the intermediate filament cytokeratin-18 is cleaved, and the fragments released into the serum were found to be elevated in patients with intestinal and liver GVHD (20–22). Hepatocyte growth factor (HGF), a molecule involved in tissue repair, was shown to be elevated in liver GVHD patients, probably due to increased release from the target organ as a physiologic response to GVHD tissue damage (21, 26). A marker that indicates tissue damage especially in endothelial and stromal cells is the soluble form of suppression of tumorigenicity 2 (ST2). ST2 is a member of the IL-1 receptor family with a transmembrane isoform and a soluble (sST2) isoform. Latter acts as a decoy receptor for IL-33 and was shown to correlate with the risk of therapy-resistant aGVHD and 6-month NRM (49).
Plasma Biomarker Panels
A large number of molecules in the plasma have been identified as potential biomarkers, but changes observed in single candidates mostly lacked sufficient specificity and sensitivity to be introduced into routine clinical use. A first 4-biomarker panel consisting of IL-2Rα, TNFR1, IL-8, and HGF was validated for confirmation of aGVHD diagnosis and prediction of survival independent of GVHD severity (30). A combination algorithm using the concentrations of ST2, REG3α, and TNFR1 measured at the onset of aGVHD symptoms was developed to assess therapy responsiveness within 28 days and the probability of 6-month NRM (54). The combination of ST2 and REG3α measured 7 days after allo-HSCT was shown to be connected to increased aGVHD-related death risk (52). The same algorithm using high levels of ST2 and REG3α applied 1 week after the initiation of GVHD treatment was able to identify treatment unresponsiveness at week 4 (53).
Metabolic Biomarkers
Given that the type of saturated fatty acid present in the diet can significantly affect lymphocyte functions (63), an untargeted metabolomics study demonstrated that patients with lower serum stearic acid/palmitic acid ratios on day 7 after transplantation were more likely to develop aGVHD, while no differences in NRM were observed (48). Another study reported significant variation in microbiota-derived metabolites at the onset of aGVHD, especially in aryl hydrocarbon receptor ligands, bile acids and plasmalogens (64). A recent integrated metabolomics and transcriptomics study uncovered an altered glycerophospholipid (GPL) metabolism signature of aGVHD, which was used to develop a biomarker panel with prognostic value using five GPL metabolites (25).
MicroRNAs as Biomarkers
Besides soluble factors in the blood of the GVHD patients, microRNAs (miRs), which determine the transcription of multiple target genes, were evaluated after allo-HSCT [reviewed in (1)]. MiRs are potent regulators of multiple pro-inflammatory target genes and readily measurable in patient serum. Multiple miRs in the serum were strongly connected to aGVHD risk (46, 65), in particular miR-155 and miR-146a (39, 42). MiRs, such as miR-155, can also be found in intestinal biopsies of patients with aGVHD (43). Several miRs were studied in mouse models of GVHD and were shown to promote or inhibit GVHD, including miR-155 (43, 66), miR-146a (40, 41), and miR-100 (18). MiR-155 was found to be essential for CXCR4-dependent donor T cell migration during GVHD (43) and NLRP3 inflammasome activation in dendritic cells (66). The miR-146a polymorphism rs2910164 in either the allo-HSCT donor or recipient was connected to higher rates of grade III and IV aGVHD (40, 41).
Microbiome-Associated Changes as Biomarkers
Major shifts in the composition of the intestinal flora have been observed during allo-HSCT as well as GVHD (67). Different studies showed that loss of intestinal microbiota diversity and predominance of a single bacterial genus, e.g., Enterococcus, were associated with occurrence of intestinal GVHD as well as overall mortality after engraftment (67, 68). On the other hand, harboring increased amounts of bacteria belonging to the genus Blautia was associated with reduced GVHD mortality in two independent cohorts (69). Another study identified increases in Lactobacillales and decreases in Clostridiales at GVHD onset (70). These shifts in species abundance and measures of diversity [reviewed in (71)] could potentially serve as biomarkers for outcome after allo-HSCT.
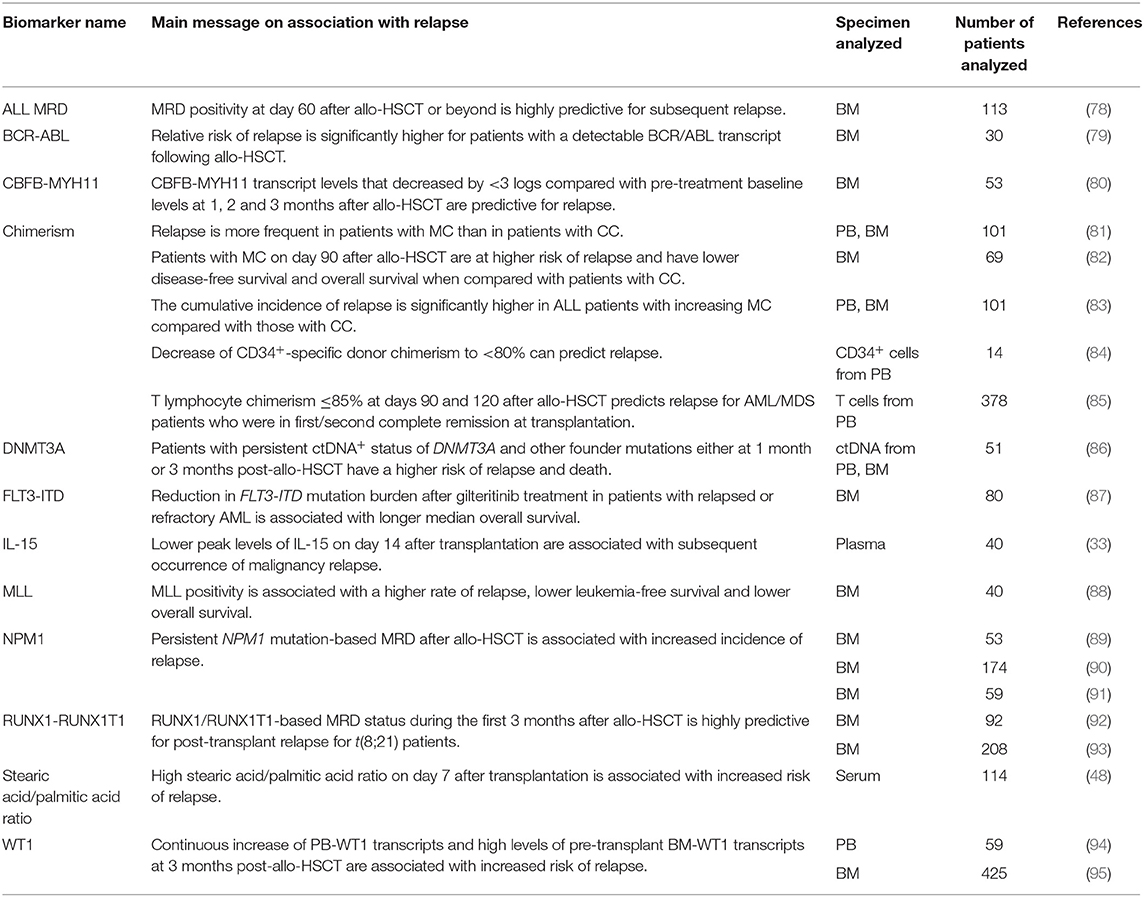
Biomarkers for Relapse
Relapse of the underlying disease is the main cause of death in the first years after allo-HSCT (72, 73). Leukemia cells use various mechanisms to escape the allogeneic immune system, such as loss of human leukocyte antigen (HLA) molecules (74), downregulation of HLA expression (75), upregulation of immune checkpoint ligands (76) and others [reviewed in (77)]. A summary of various biomarkers that have been evaluated for prediction of relapse can be found in Table 2.
Measurable Residual Disease
Measurable residual disease (MRD, also referred to as minimal residual disease) can be used to identify remaining leukemic cells that are below the limit of detection of morphological assessment (96). MRD monitoring can thus help to identify patients with increased risk of relapse after allo-HSCT. However, not all patients with MRD positivity will relapse clinically, and some patients will relapse despite negative MRD results. The following paragraphs will focus on MRD detection in acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL), which, taken together, account for a large portion of indications for allo-HSCT (97).
Given the molecular diversity of acute leukemia, different methods are applied for MRD detection. Multiparameter flow cytometry and real-time quantitative polymerase chain reaction (PCR) are widely used, while newer technologies are emerging, e.g., droplet digital PCR (ddPCR) and next-generation sequencing (NGS) (98).
Overexpression of Wilms tumor 1 (WT1) is found in most AML patients and can be measured in peripheral blood (PB) or bone marrow (BM) (99, 100). Patients who displayed increased WT1 transcripts in the PB after allo-HSCT or who failed to clear their high levels of pre-transplant WT1 transcripts in the BM at 3 months post-allo-HSCT were shown to be at increased risk of relapse (94, 95). Mutation in nucleophosmin 1 (NPM1) is present in around one-third of adult AML patients (101). Several studies showed an association between persistent NPM1 mutation-based MRD after allo-HSCT and increased incidence of relapse (89–91). Core binding factor (CBF) AML is characterized by the presence of the chromosomal rearrangements t(8;21) and inv(16), causing production of the fusion transcripts RUNX1/RUNX1T1 and CBFB-MYH11, respectively (102). RUNX1/RUNX1T1-based MRD status in t(8;21) AML patients during the first 3 months after allo-HSCT was found to be highly predictive for post-transplant relapse (92). Similarly, CBFB-MYH11 transcript levels that decreased by <3 logs compared with pre-treatment baseline levels at 1, 2, and 3 months after allo-HSCT were demonstrated to be predictive for relapse (80). Interestingly, low levels of CBF fusion transcripts were observed to persist in long-term transplant survivors (103). The mixed leukemia lineage (MLL) gene (also termed KMT2A), is frequently disrupted in AML by different chromosomal rearrangements involving other partner chromosomes (104). MLL positivity was shown to be associated with a higher rate of relapse, lower leukemia-free survival and lower overall survival (88). The detection of driver mutations associated with clonal hematopoiesis of indeterminate potential (CHIP), such as mutations in DNMT3A, TET2, and ASXL, is complex because these mutations might be derived from the allo-HSCT donor (105). Some studies indicate that residual allelic burdens associated with CHIP were not suitable for MRD testing in remission to predict relapse rate (106, 107). However, in a study utilizing personalized ddPCR, patients with persistent ctDNA+ status of DNMT3A and other driver mutations either at 1 or 3 months post-allo-HSCT had a significantly higher risk of relapse and death compared with those with negative status (86). Additionally, increasing ctDNA levels between 1 and 3 months post-allo-HSCT was a precise predictor of relapse (86). Mutations in the fms-like tyrosine kinase 3 (FLT3) gene producing internal tandem duplications (FLT3-ITD) are common in AML and are known to be associated with poor prognosis (108). A novel NGS-based MRD assay detecting FLT3-ITD showed that reduction in mutation burden after treatment with gilteritinib, a FLT3 inhibitor, in patients with relapsed or refractory AML (NCT02014558) was linked to longer median overall survival (87). Also, RAS mutations (NRAS and KRAS) can be detected after allo-HSCT, and a link of KRAS downstream signaling with NLRP3 inflammasome activation was recently reported (109), showing a potential pro-inflammatory activity of certain oncogenic mutations.
MRD monitoring in B- or T-lymphoid malignancies includes detection of a leukemia-associated immunophenotype (LAIP) by flow cytometry as well as detection of disease-specific T cell receptor or immunoglobulin gene rearrangements by PCR (110, 111). Several studies in the pediatric setting of ALL have shown that patients with detectable MRD after allo-HSCT were more likely to experience relapse (78, 112, 113). In adult patients with Philadelphia chromosome-positive ALL, MRD positivity in terms of detectable BCR/ABL transcript was found to be associated with increased risk of relapse (79).
Chimerism
Studies on different hematological malignancies showed the relevance of chimerism and its kinetics for the prediction of relapse (110). For instance, the cumulative incidence of relapse was found to be significantly higher in patients with AML, myelodysplastic syndrome (MDS), chronic myeloid leukemia (CML) and ALL with increasing mixed chimerism (MC) than in those with complete chimerism (CC) (81–83). Lineage-specific chimerism analysis may increase the specificity in predicting relapse (114). A prospective study found that the decrease of CD34+-specific donor chimerism to <80% had 100% sensitivity and 86% accuracy in predicting relapse (84). T lymphocyte chimerism ≤ 85% at days 90 and 120 after allo-HSCT was shown to predict relapse for patients who were in first/second complete remission at transplantation (85).
Plasma Biomarkers
Levels of ST2 and REG3α were previously used to develop an algorithm that predicts the risk of severe GVHD and NRM. The authors used this same algorithm to show that low levels of ST2 and REG3α on day 28 after allo-HSCT in patients who had not developed GVHD were associated with higher risk of relapse than severe GVHD and NRM (115). This observation suggests that the patients who are at low risk of developing severe GVHD, but who remain at an increased risk of relapse, might benefit from early taper of prophylactic immunosuppression in order to enhance GVL effects. Low peak levels at day 14 of another candidate biomarker connected to aGVHD, IL-15, were shown to be associated with subsequent occurrence of malignancy relapse (33).
A recent study aimed to develop a plasma signature to identify GVL effects without GVHD by conducting plasma proteomics and systems biology analyses of patients in relapse after allo-HSCT who were treated with allogeneic donor lymphocyte infusions (116). A unique 61-protein signature was identified in patients with GVL without GVHD, of which 43 genes were further confirmed using single-cell RNA sequencing analysis in activated T cells. Novel markers, such as RPL23, ILF2, CD58, and CRTAM, were identified and will need further validation in other cohorts.
Metabolic Biomarkers
An untargeted metabolomic study showed that in a patient cohort with AML, ALL and Non-Hodgkin lymphoma, a high ratio between serum stearic acid and palmitic acid on day 7 after transplantation was associated with increased risk of relapse, suggesting that the measurement of this ratio may improve risk stratification after allo-HSCT (48).
Conclusion
Acute GVHD and relapse of the underlying disease form the two major complications after allo-HSCT, leading to significant morbidity and mortality. Recent advances in proteomic analyses allowed the identification of numerous candidate biomarkers for aGVHD. Of note, the discovery of these candidate biomarkers was mostly based on evaluation at a single center and only a limited number of studies met the criteria of verifying and qualifying these candidates as actual biomarkers according to the NIH consensus. Those and possibly other yet to be discovered biomarkers hold promise to better predict the risk of aGVHD and aGVHD-related mortality, which could lead to a more individualized GVHD prophylaxis approach. Monitoring of MRD and chimerism is the most commonly used tool to detect relapse after allo-HSCT. The ultimate significance of MRD monitoring, in particular, remains to be further investigated. MRD detection techniques are constantly improving. However, clinical trials will be necessary to define standardized pathways for MRD testing and MRD-directed therapy intervention in clinical practice.
Author Contributions
SC and RZ wrote the manuscript together. All authors contributed to the article and approved the submitted version.
Funding
SC was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft, Germany. RZ was supported by the Deutsche Forschungsgemeinschaft via SFB TRR167, SFB 850, ERC Consolidator grant (681012 GvHDCure to RZ), by the Germany's Excellence Strategy (CIBSS – EXC-2189 – Project ID 390939984).
Conflict of Interest
RZ received speaker fees from Novartis, Incyte, and Mallinckrodt. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms I. Calmbach for help with the references.
References
1. Zeiser R, Blazar BR. Acute graft-versus-host disease - biologic process, prevention, and therapy. N Eng J Med. (2017) 377:2167–79. doi: 10.1056/NEJMra1609337
2. Mathew NR, Vinnakota JM, Apostolova P, Erny D, Hamarsheh S, Andrieux G, et al. Graft-versus-host disease of the CNS is mediated by TNF upregulation in microglia. J Clin Invest. (2020) 130:1315–29. doi: 10.1172/JCI130272
3. Zeiser R, von Bubnoff N, Butler J, Mohty M, Niederwieser D, Or R. Ruxolitinib for glucocorticoid-refractory acute graft-versus-host disease. N Eng J Med. (2020) 382:1800–10. doi: 10.1056/NEJMoa1917635
4. Dickinson AM, Norden J, Li S, Hromadnikova I, Schmid C, Schmetzer H, et al. Graft-versus-leukemia effect following hematopoietic stem cell transplantation for leukemia. Front Immunol. (2017) 8:496. doi: 10.3389/fimmu.2017.00496
5. Mathew NR, Baumgartner F, Braun L, David O'Sullivan Thomas S, Waterhouse M, Müller TA, et al. Sorafenib promotes graft-versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD mutant leukemia cells. Nat Med. (2018) 24:282–91.
6. Wingard JR, Majhail NS, Brazauskas R, Wang Z, Sobocinski KA, Jacobsohn D, et al. Long-term survival and late deaths after allogeneic hematopoietic cell transplantation. J Clin Oncol. (2011) 29:2230–9. doi: 10.1200/JCO.2010.33.7212
7. Styczynski J, Tridello G, Koster L, Iacobelli S, van Biezen A, van der Werf S, et al. Death after hematopoietic stem cell transplantation: changes over calendar year time, infections and associated factors. Bone Marrow Transplant. (2020) 55:126–36. doi: 10.1038/s41409-019-0624-z
8. Rezvani AR, Storer BE, Storb RF, Mielcarek M, Maloney DG, Sandmaier BM, et al. Decreased serum albumin as a biomarker for severe acute graft-versus-host disease after reduced-intensity allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. (2011) 17:1594–601. doi: 10.1016/j.bbmt.2011.07.021
9. Rodriguez-Otero P, Porcher R, Peffault de Latour R, Contreras M, Bouhnik Y, Xhaard A, et al. Fecal calprotectin and alpha-1 antitrypsin predict severity and response to corticosteroids in gastrointestinal graft-versus-host disease. Blood. (2012) 119:5909–17. doi: 10.1182/blood-2011-12-397968
10. Luft T, Dietrich S, Falk C, Conzelmann M, Hess M, Benner A, et al. Steroid-refractory GVHD: T-cell attack within a vulnerable endothelial system. Blood. (2011) 118:1685–92. doi: 10.1182/blood-2011-02-334821
11. Chen YB, Kim HT, McDonough S, Odze RD, Yao X, Lazo-Kallanian S, et al. Up-Regulation of alpha4beta7 integrin on peripheral T cell subsets correlates with the development of acute intestinal graft-versus-host disease following allogeneic stem cell transplantation. Biol Blood Marrow Transplant. (2009) 15:1066–76. doi: 10.1016/j.bbmt.2009.05.003
12. Ahmed SS, Wang XN, Norden J, Pearce K, El-Gezawy E, Atarod S, et al. Identification and validation of biomarkers associated with acute and chronic graft versus host disease. Bone Marrow Transplant. (2015) 50:1563–71. doi: 10.1038/bmt.2015.191
13. Chiusolo P, Metafuni E, Giammarco S, Bellesi S, Piccirillo N, Fanali C, et al. Role of fecal calprotectin as biomarker of gastrointestinal GVHD after allogeneic stem cell transplantation. Blood. (2012) 120:4443–4. doi: 10.1182/blood-2012-08-447326
14. Hori T, Naishiro Y, Sohma H, Suzuki N, Hatakeyama N, Yamamoto M, et al. CCL8 is a potential molecular candidate for the diagnosis of graft-versus-host disease. Blood. (2008) 1111:4403–12. doi: 10.1182/blood-2007-06-097287
15. August KJ, Chiang KY, Bostick RM, Flanders WD, Waller EK, Langston A, et al. Biomarkers of immune activation to screen for severe, acute GVHD. Bone Marrow Transplant. (2011) 64:601–4. doi: 10.1038/bmt.2010.165
16. Hubel K, Cremer B, Heuser E, von Strandmann EP, Hallek M, Hansen HP. A prospective study of serum soluble CD30 in allogeneic hematopoietic stem cell transplantation. Transpl Immunol. (2010) 23:215–9. doi: 10.1016/j.trim.2010.07.001
17. Chen YB, McDonough S, Hasserjian R, Chen H, Coughlin E, Illiano C, et al. Expression of CD30 in patients with acute graft-versus-host disease. Blood. (2012) 120:691–6. doi: 10.1182/blood-2012-03-415422
18. Leonhardt F, Grundmann S, Behe M, Bluhm F, Dumont RA, Braun F, et al. Inflammatory neovascularization during graft-versus-host disease is regulated by αv integrin and miR-100. Blood. (2013) 121:3307–18. doi: 10.1182/blood-2012-07-442665
19. Piper KP, Horlock C, Curnow SJ, Arrazi J, Nicholls S, Mahendra P, et al. CXCL10-CXCR3 interactions play an important role in the pathogenesis of acute graft-versus-host disease in the skin following allogeneic stem-cell transplantation. Blood. (2007) 110:3827–32. doi: 10.1182/blood-2006-12-061408
20. Luft T, Conzelmann M, Benner A, Rieger M, Hess M, Strohhaecker U, et al. Serum cytokeratin-18 fragments as quantitative markers of epithelial apoptosis in liver and intestinal graft-versus-host disease. Blood. (2007) 110:4535–42. doi: 10.1182/blood-2006-10-049817
21. Harris AC, Ferrara JL, Braun TM, Holler E, Teshima T, Levine JE, et al. Plasma biomarkers of lower gastrointestinal and liver acute GVHD. Blood. (2012) 119:2960–3. doi: 10.1182/blood-2011-10-387357
22. Waterhouse M, Samek E, Torres M, Bertz H, Finke J. Diagnostic utility of a soluble cytokeratin 18 assay for gastrointestinal graft-vs-host disease detection. Clin Chem Lab Med. (2011) 49:1695–7. doi: 10.1515/CCLM.2011.644
23. Paczesny S, Braun TM, Levine JE, Hogan J, Crawford J, Coffing B, et al. Elafin is a biomarker of graft-versus-host disease of the skin. Sci Transl Med. (2010) 2:13ra2. doi: 10.1126/scitranslmed.3000406
24. Brüggen M, Petzelbauer P, Greinix H, Contassot E, Jankovic D, French L, et al. Epidermal elafin expression is an indicator of poor prognosis in cutaneous graft-versus-host disease. J Invest Dermatol. (2015) 135:999–1006. doi: 10.1038/jid.2014.489
25. Liu Y, Huang A, Chen Q, Chen X, Fei Y, Zhao X, et al. A distinct glycerophospholipid metabolism signature of acute graft versus host disease with predictive value. JCI Insight. (2019) 5:129494. doi: 10.1172/jci.insight.129494
26. Okamoto T, Takatsuka H, Fujimori Y, Wada H, Iwasaki T, Kakishita E. Increased hepatocyte growth factor in serum in acute graft-versus-host disease. Bone Marrow Transplant. (2001) 28:197–200. doi: 10.1038/sj.bmt.1703095
27. Shaiegan M, Iravani M, Babaee G, Ghavamzadeh A. Effect of IL-18 and sIL2R on aGVHD occurrence after hematopoietic stem cell transplantation in some Iranian patients. Transpl Immunol. (2006) 15:223–7. doi: 10.1016/j.trim.2005.10.002
28. Visentainer JEL, Lieber SR, Persoli LBL, Vigorito AC, Aranha FJP, de Brito Eid KA, et al. Serum cytokine levels and acute graft-versus-host disease after HLA-identical hematopoietic stem cell transplantation. Exp Hematol. (2003) 31:1044–50. doi: 10.1016/S0301-472X(03)00264-9
29. Nakamura H, Komatsu K, Ayaki M, Kawamoto S, Murakami M, Uoshima N, et al. Serum levels of soluble IL-2 receptor, IL-12, IL-18, and IFN-gamma in patients with acute graft-versus-host disease after allogeneic bone marrow transplantation. J Allergy Clin Immunol. (2000) 106:45–50. doi: 10.1067/mai.2000.106774
30. Paczesny S, Krijanovski OI, Braun TM, Choi SW, Clouthier SG, Kuick R, et al. A biomarker panel for acute graft-versus-host disease. Blood. (2009) 113:273–8. doi: 10.1182/blood-2008-07-167098
31. Malone FR, Leisenring WM, Storer BE, Lawler R, Stern JM, Aker SN, et al. Prolonged anorexia and elevated plasma cytokine levels following myeloablative allogeneic hematopoietic cell transplant. Bone Marrow Transplant. (2007) 40:765–72. doi: 10.1038/sj.bmt.1705816
32. McDonald GB, Tabellini L, Storer BE, Lawler RL, Martin PJ, Hansen JA. Plasma biomarkers of acute GVHD and nonrelapse mortality: predictive value of measurements before GVHD onset and treatment. Blood. (2015) 126:113–20. doi: 10.1182/blood-2015-03-636753
33. Thiant S, Yakoub-Agha I, Magro L, Trauet J, Coiteux V, Jouet JP, et al. Plasma levels of IL-7 and IL-15 in the first month after myeloablative BMT are predictive biomarkers of both acute GVHD and relapse. Bone Marrow Transplant. (2010) 45:1546–52. doi: 10.1038/bmt.2010.13
34. Liem LM, van Houwelingen HC, Goulmy E, Mattsson J. Serum cytokine levels after HLA-identical bone marrow transplantation. Transplantation. (1998) 66:863–71. doi: 10.1097/00007890-199810150-00009
35. Mohty M, Blaise D, Faucher C, Vey N, Bouabdallah R, Stoppa A-M, et al. Inflammatory cytokines and acute graft-versus-host disease after reduced-intensity conditioning allogeneic stem cell transplantation. Blood. (2005) 106:4407–11. doi: 10.1182/blood-2005-07-2919
36. Sakata N, Yasui M, Okamura T, Inoue M, Yumura-Yagi K, Kawa K. Kinetics of plasma cytokines after hematopoietic stem cell transplantation from unrelated donors: the ratio of plasma IL-10/sTNFR level as a potential prognostic marker in severe acute graft-versus-host disease. Bone Marrow Transplant. (2001) 27:1153–61. doi: 10.1038/sj.bmt.1703060
37. Fujimori Y, Takatsuka H, Takemoto Y, Hara H, Okamura H, Nakanishi K, et al. Elevated interleukin (IL)-18 levels during acute graft-versus-host disease after allogeneic bone marrow transplantation. Brit J Haematol. (2000) 109:4373–80. doi: 10.1046/j.1365-2141.2000.02095.x
38. Ranganathan P, Ngankeu A, Zitzer NC, Leoncini P, Yu X, Casadei L, et al. Serum miR-29a is upregulated in acute graft-versus-host disease and activates dendritic cells through TLR binding. J Immunol. (2017) 198:2500–12. doi: 10.4049/jimmunol.1601778
39. Atarod S, Ahmed MM, Lendrem C, Pearce KF, Cope W, Norden J, et al. miR-146a and miR-155 expression levels in acute graft-versus-host disease incidence. Front Immunol. (2016) 7:56–61. doi: 10.3389/fimmu.2016.00056
40. Stickel N, Prinz G, Pfeifer D, Hasselblatt P, Schmitt-Graeff A, Follo M, et al. MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GvHD. Blood. (2014) 124:2586–95. doi: 10.1182/blood-2014-04-569046
41. Stickel N, Hanke K, Marschner D, Prinz G, Köhler M, Melchinger W, et al. MicroRNA-146a reduces MHC-II expression via targeting JAK/STAT-signaling in dendritic cells after stem cell transplantation. Leukemia. (2017) 31:2732–41. doi: 10.1038/leu.2017.137
42. Xie LN, Zhou F, Liu XM, Fang Y, Yu Z, Song NX, et al. Serum microRNA155 is increased in patients with acute graft-versus-host disease. Clin Transplant. (2014) 28:314–23. doi: 10.1111/ctr.12314
43. Ranganathan P, Heaphy CE, Costinean S, Stauffer N, Na C, Hamadani M, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. (2012) 119:4786–97. doi: 10.1182/blood-2011-10-387522
44. Wang Y, Zhao X, Ye X, Luo H, Zhao T, Diao Y, et al. Plasma microRNA-586 is a new biomarker for acute graft-versus-host disease. Ann Hematol. (2015) 94:1505–14. doi: 10.1007/s00277-015-2414-z
45. Zhang C, Bai N, Huang W, Zhang P, Luo Y, Men S, et al. The predictive value of selected serum microRNAs for acute GVHD by TaqMan MicroRNA arrays. Ann Hematol. (2016) 95:1833–43. doi: 10.1007/s00277-016-2781-0
46. Gimondi S, Dugo M, Vendramin A, Bermema A, Biancon G, Cavané A, et al. Circulating miRNA panel for prediction of acute graft-versus-host disease in lymphoma patients undergoing matched unrelated hematopoietic stem cell transplantation. Exp Hematol. (2016) 44:624–634. doi: 10.1016/j.exphem.2016.03.005
47. Ferrara JL, Harris AC, Greenson JK, Braun TM, Holler E, Teshima T, et al. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood. (2011) 118:6702–8. doi: 10.1182/blood-2011-08-375006
48. Wu X, Xie Y, Wang C, Han Y, Bao X, Ma S, et al. Prediction of acute GVHD and relapse by metabolic biomarkers after allogeneic hematopoietic stem cell transplantation. JCI Insight. (2018) 3:99672. doi: 10.1172/jci.insight.99672
49. Vander Lugt BMT, Hanash TM, Ritz S, Ho J, Antin VT, Zhang JH, et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. N Engl J Med. (2013) 369:529–39. doi: 10.1056/NEJMoa1213299
50. Rotz SJ, Dandoy CE, Davies SM. ST2 and endothelial injury as a link between GVHD and microangiopathy. N Eng J Med. (2017) 376:1189–90. doi: 10.1056/NEJMc1700185
51. Reichenbach DK, Schwarze V, Matta BM, Tkachev V, Lieberknecht E, Liu Q, et al. The IL-33/ST2 axis augments effector T cell responses during acute GVHD. Blood. (2015) 125:3183–92. doi: 10.1182/blood-2014-10-606830
52. Hartwell MJ, Özbek U, Holler E, Renteria AS, Major-Monfried H, Reddy P, et al. An early-biomarker algorithm predicts lethal graft-versus-host disease and survival. JCI Insight. (2017) 2:e89798. doi: 10.1172/jci.insight.89798
53. Major-Monfried H, Renteria AS, Pawarode A, Reddy P, Ayuk F, Holler E, et al. MAGIC biomarkers predict long-term outcomes for steroid-resistant acute GVHD. Blood. (2018) 131:2846–55. doi: 10.1182/blood-2018-01-822957
54. Levine JE, Braun TM, Harris AC, Holler E, Taylor A, Miller H, et al. A prognostic score for acute graft-versus-host disease based on biomarkers: a multicentre study. Lancet Haematol. (2015) 2:e21–9. doi: 10.1016/S2352-3026(14)00035-0
55. Abu Zaid M, Wu J, Wu C, Logan BR, Yu J, Cutler C, et al. Plasma biomarkers of risk for death in a multicenter phase 3 trial with uniform transplant characteristics post-allogeneic HCT. Blood. (2017) 129:162–70. doi: 10.1182/blood-2016-08-735324
56. Remberger M, Jaksch M, Uzunel M, Mattsson J. Serum levels of cytokines correlate to donor chimerism and acute graft-vs-host disease after haematopoietic stem cell transplantation. Eur J Haematol. (2003) 70:384–91. doi: 10.1034/j.1600-0609.2003.00078.x
57. Hansen JA, Hanash SM, Tabellini L, Baik C, Lawler RL, Grogan BM, et al. A novel soluble form of Tim-3 associated with severe graft-versus-host disease. Biol Blood Marrow Transplant. (2016) 19:1323–30. doi: 10.1016/j.bbmt.2013.06.011
58. Holler E, Kolb HJ, Moller A, Kempeni J, Liesenfeld S, Pechumer H, et al. Increased serum levels of tumor necrosis factor alpha precede major complications of bone marrow transplantation. Blood. (1990) 75:1011–6. doi: 10.1182/blood.V75.4.1011.1011
59. Choi SW, Kitko CL, Braun T, Paczesny S, Yanik G, Mineishi S, et al. Change in plasma tumor necrosis factor receptor 1 levels in the first week after myeloablative allogeneic transplantation correlates with severity and incidence of GVHD and survival. Blood. (2008) 1539–42. doi: 10.1182/blood-2008-02-138867
60. BDW. Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. (2011) 69:89–95. doi: 10.1067/mcp.2001.113989
61. Paczesny S, Hakim FT, Pidala J, Cooke KR, Lathrop J, Griffith LM, et al. National institutes of health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: III. The 2014 biomarker working group report. Biol Blood Marrow Transplant. (2015) 21:780–92. doi: 10.1016/j.bbmt.2015.01.003
62. Wolff D, Greinix H, Lee SJ, Gooley T, Paczesny S, Pavletic S, et al. Biomarkers in chronic graft-versus-host disease: quo vadis? Bone Marrow Transplant. (2018) 53:832–7. doi: 10.1038/s41409-018-0092-x
63. Jeffery NM, Sanderson P, Newsholme EA, Calder PC. Effects of varying the type of saturated fatty acid in the rat diet upon serum lipid levels and spleen lymphocyte functions. Biochim Biophys Acta. (1997) 134:223–36. doi: 10.1016/S0005-2760(96)00174-9
64. Michonneau D, Latis E, Curis E, Dubouchet L, Ramamoorthy S, Ingram B, et al. Metabolomics analysis of human acute graft-versus-host disease reveals changes in host and microbiota-derived metabolites. Nat Commun. (2019) 10:5695. doi: 10.1038/s41467-019-13498-3
65. Xiao B, Wang Y, Li W, Baker M, Guo J, Corbet K, et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood. (2013) 122:3365–75. doi: 10.1182/blood-2013-06-510586
66. Chen S, Smith BA, Iype J, Prestipino A, Pfeifer D, Grundmann S, et al. MicroRNA-155-deficient dendritic cells cause less severe GVHD through reduced migration and defective inflammasome activation. Blood. (2015) 126:103–12. doi: 10.1182/blood-2014-12-617258
67. Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. (2014) 124:1174–82. doi: 10.1182/blood-2014-02-554725
68. Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. (2014) 20:640–5. doi: 10.1016/j.bbmt.2014.01.030
69. Jenq R, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. (2015) 21:1373–83. doi: 10.1016/j.bbmt.2015.04.016
70. Jenq R, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. (2012) 209:903–11. doi: 10.1084/jem.20112408
71. Andermann TM, Peled JU, Ho C, Reddy P, Riches M, Storb R, et al. The microbiome and hematopoietic cell transplantation: past, present, and future. Biol Blood Marrow Transplant. (2018) 24:1322–40. doi: 10.1016/j.bbmt.2018.02.009
72. Horowitz M, Schreiber H, Elder A, Heidenreich O, Vormoor J, Toffalori C, et al. Epidemiology and biology of relapse after stem cell transplantation. Bone Marrow Transplant. (2018) 53:1379–89. doi: 10.1038/s41409-018-0171-z
73. Zeiser R, Beelen DW, Bethge W, Bornhäuser M, Bug G, Burchert A, et al. Biology-driven approaches to prevent and treat relapse of myeloid neoplasia after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2019) 25:128–40. doi: 10.1016/j.bbmt.2019.01.016
74. Vago L, Perna SK, Zanussi M, Mazzi B, Barlassina C, Stanghellini MT, et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Eng J Med. (2009) 361:478–88. doi: 10.1056/NEJMoa0811036
75. Christopher M, Petti AA, Miller CA, Rettig MP, Duncavage EJ, Klco JM, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Eng J Med. 379:2330–41. doi: 10.1056/NEJMoa1808777
76. Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med. (2019) 25:603–11. doi: 10.1038/s41591-019-0400-z
77. Zeiser R, Vago L. Mechanisms of immune escape after allogeneic hematopoietic cell transplantation. Blood. (2019) 133:1290–97. doi: 10.1182/blood-2018-10-846824
78. Bader P, von Stackelberg A, Eckert C, Salzmann-Manrique E, Meisel R, Poetschger U, et al. Monitoring of minimal residual disease after allogeneic stem-cell transplantation in relapsed childhood acute lymphoblastic leukemia allows for the identification of impending relapse: results of the ALL-BFM-SCT 2003 trial. J Clin Oncol. (2015) 33:1275–84. doi: 10.1200/JCO.2014.58.4631
79. Radich J, Gehly G, Lee A, Avery R, Bryant E, Edmands S, et al. Detection of bcr-abl transcripts in Philadelphia chromosome-positive acute lymphoblastic leukemia after marrow transplantation. Blood. (1997) 89:2602–9. doi: 10.1182/blood.V89.7.2602
80. Tang F, Xu LP, Zhang XH, Chen H, Chen YH, Han W, et al. Monitoring of post-transplant CBFB-MYH11 as minimal residual disease, rather than KIT mutations, can predict relapse after allogeneic haematopoietic cell transplantation in adults with inv(16) acute myeloid leukaemia. Br J Haematol. (2018) 180:448–51. doi: 10.1111/bjh.14340
81. Wäsch R, Bertz H, Kunzmann R, Finke J. Incidence of mixed chimaerism and clinical outcome in 101 patients after myeloablative conditioning regimens and allogeneic stem cell transplantation. Br J Haematol. (2000) 109:743–50. doi: 10.1046/j.1365-2141.2000.02110.x
82. Lamba R, Abella E, Kukuruga D, Klein J, Savasan S, Abidi MH, et al. Mixed hematopoietic chimerism at day 90 following allogenic myeloablative stem cell transplantation is a predictor of relapse and survival. Leukemia. (2004) 18:1681–6. doi: 10.1038/sj.leu.2403468
83. Terwey TH, Hemmati PG, Nagy M, Pfeifer H, Gökbuget N, Brüggemann M, et al. Comparison of chimerism and minimal residual disease monitoring for relapse prediction after allogeneic stem cell transplantation for adult acute lymphoblastic leukemia. Biol Blood Marrow Transplant. (2014) 20:1522–9. doi: 10.1016/j.bbmt.2014.05.026
84. Unnikrishnan A, Meacham AM, Goldstein SS, Ta M, Leather HL, Cogle CR, et al. CD34+ chimerism analysis for minimal residual disease monitoring after allogeneic hematopoietic cell transplantation. Leuk Lymphoma. (2018) 74:110–2. doi: 10.1016/j.leukres.2018.10.007
85. Lee HC, Saliba RM, Rondon G, Chen J, Charafeddine Y, Medeiros LJ, et al. Mixed T lymphocyte chimerism after allogeneic hematopoietic transplantation is predictive for relapse of acute myeloid leukemia and myelodysplastic syndromes. Biol Blood Marrow Transplant. (2015) 21:1948–51. doi: 10.1016/j.bbmt.2015.07.005
86. Nakamura S, Yokoyama K, Shimizu E, Yusa N, Kondoh K, Ogawa M, et al. Prognostic impact of circulating tumor DNA status post-allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood. (2019) 133:2682–95. doi: 10.1182/blood-2018-10-880690
87. Levis MJ, Perl AE, Altman JK, Gocke CD, Bahceci E, Hill J, et al. A next-generation sequencing-based assay for minimal residual disease assessment in AML patients with FLT3-ITD mutations. Blood Adv. (2018) 2:825–31. doi: 10.1182/bloodadvances.2018015925
88. Liu J, Wang Y, Xu LP, Liu DH, Qin YZ, Chang YJ, et al. Monitoring mixed lineage leukemia expression may help identify patients with mixed lineage leukemia–rearranged acute leukemia who are at high risk of relapse after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2014) 20:929–36. doi: 10.1016/j.bbmt.2014.03.008
89. Schnittger S, Kern W, Tschulik C, Weiss T, Dicker F, Falini B, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. (2009) 114:2220–31. doi: 10.1182/blood-2009-03-213389
90. Shayegi N, Kramer M, Bornhäuser M, Schaich M, Schetelig J, Platzbecker U, et al. The level of residual disease based on mutant NPM1 is an independent prognostic factor for relapse and survival in AML. Blood. (2013) 122:83–92 (2013). doi: 10.1182/blood-2012-10-461749
91. Zhou Y, Othus M, Walter RB, Estey EH, Wu D, Wood BL. Deep NPM1 sequencing following allogeneic hematopoietic cell transplantation improves risk assessment in adults with NPM1-mutated AML. Biol Blood Marrow Transplant. (2018) 24:1615–20. doi: 10.1016/j.bbmt.2018.04.017
92. Wang Y, Wu DP, Liu QF, Qin YZ, Wang JB, Xu LP, et al. In adults with t(8;21)AML, posttransplant RUNX1/RUNX1T1-based MRD monitoring, rather than c-KIT mutations, allows further risk stratification. Blood. (2014) 124:1880–6. doi: 10.1182/blood-2014-03-563403
93. Qin YZ, Wang Y, Xu LP, Zhang XH, Chen H, Han W, et al. The dynamics of RUNX1-RUNX1T1 transcript levels after allogeneic hematopoietic stem cell transplantation predict relapse in patients with t(8;21) acute myeloid leukemia. J Hematol Oncol. (2017) 10:44. doi: 10.1186/s13045-017-0414-2
94. Rautenberg C, Pechtel S, Hildebrandt B, Betz B, Dienst A, Nachtkamp K, et al. Wilms' tumor 1 gene expression using a standardized european leukemianet-certified assay compared to other methods for detection of minimal residual disease in myelodysplastic syndrome and acute myelogenous leukemia after allogeneic blood stem cell transplantation. Biol Blood Marrow Transplant. (2018) 24:2337–43. doi: 10.1016/j.bbmt.2018.05.011
95. Cho BS, Min GJ, Park SS, Shin SH, Yahng SA, Jeon YW, et al. WT1 measurable residual disease assay in patients with acute myeloid leukemia who underwent allogeneic hematopoietic stem cell transplantation: optimal time points, thresholds, and candidates. Biol Blood Marrow Transplant. (2019) 10:1925–32. doi: 10.1016/j.bbmt.2019.05.033
96. Hourigan CS, Karp JE. Minimal residual disease in acute myeloid leukaemia. Nat Rev Clin Oncol. (2013) 10:460–71. doi: 10.1038/nrclinonc.2013.100
97. Passweg J, Baldomero H, Basak GW, Chabannon C, Corbacioglu S, Duarte R, et al. The EBMT activity survey report 2017: a focus on allogeneic HCT for nonmalignant indications and on the use of non-HCT cell therapies. Bone Marrow Transplant. (2019) 54:1575–85. doi: 10.1038/s41409-019-0465-9
98. Schuurhuis GJ, Heuser M, Freeman S, Béné MC, Buccisano F, Cloos J, et al. Minimal/measurable residual disease in AML: a consensus document from the european leukemianet MRD working party. Blood. (2018) 131:1275–91. doi: 10.1182/blood-2017-09-801498
99. Inoue K, Sugiyama H, Ogawa H, Nakagawa M, Yamagami T, Miwa H, et al. WT1 as a new prognostic factor and a new marker for the detection of minimal residual disease in acute leukemia. Blood. (1994) 84:3071–9. doi: 10.1182/blood.V84.9.3071.3071
100. Bergmann L, Miething C, Maurer U, Brieger J, Karakas T, Weidmann E, et al. High levels of Wilms' tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. (1997) 90:1217–25. doi: 10.1182/blood.V90.3.1217
101. Rau R, Brown P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity. Hematol Oncol. (2009) 27:171–81. doi: 10.1002/hon.904
102. Sangle NA, Perkins SL. Core-binding factor acute myeloid leukemia. Arch Pathol Lab Med. (2011) 135:1504–9. doi: 10.5858/arpa.2010-0482-RS
103. Jurlander J, Caligiuri MA, Ruutu T, Baer MR, Strout MP, Oberkircher AR, et al. Persistence of the AML1/ETO fusion transcript in patients treated with allogeneic bone marrow transplantation for t(8;21) leukemia. Blood. (1996) 88:2183–91. doi: 10.1182/blood.V88.6.2183.bloodjournal8862183
104. Muñoz L, Nomdedéu JF, Villamor N, Guardia R, Colomer D, Ribera JM, et al. Acute myeloid leukemia with MLL rearrangements: clinicobiological features, prognostic impact and value of flow cytometry in the detection of residual leukemic cells. Leukemia. (2003) 1:76–82. doi: 10.1038/sj.leu.2402708
105. Frick M, Chan W, Arends CM, Hablesreiter R, Halik A, Heuser M, et al. Role of donor clonal hematopoiesis in allogeneic hematopoietic stem-cell transplantation. J Clin Oncol. (2019) 37:375–85. doi: 10.1200/JCO.2018.79.2184
106. Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Al Hinai A, Zeilemaker A, et al. Molecular minimal residual disease in acute myeloid leukemia. N Eng J Med. (2018) 378:1189–99. doi: 10.1056/NEJMoa1716863
107. Gaidzik V, Weber D, Paschka P, Kaumanns A, Krieger S, Corbacioglu A, et al. DNMT3A mutant transcript levels persist in remission and do not predict outcome in patients with acute myeloid leukemia. Leukemia. (2018) 32:30–37. doi: 10.1038/leu.2017.200
108. Patel J, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Eng J Med. (2012) 366:1079–89. doi: 10.1056/NEJMoa1112304
109. Hamarsheh S, Osswald L, Saller BS, Unger S, De Feo D, Vinnakota JM, et al. Oncogenic KrasG12D causes myeloproliferation via NLRP3 inflammasome activation. Nat Commun. (2020) 11:1659. doi: 10.1038/s41467-020-15497-1
110. Kröger N, Miyamura K, Bishop MR. Minimal residual disease following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2016) 17:94–100. doi: 10.1016/j.bbmt.2010.10.031
111. Craddock C, Hoelzer D, Komanduri KV. Current status and future clinical directions in the prevention and treatment of relapse following hematopoietic transplantation for acute myeloid and lymphoblastic leukemia. Bone Marrow Transplant. (2019) 54:6–16. doi: 10.1038/s41409-018-0203-8
112. Dominietto A, Pozzi S, Miglino M, Albarracin F, Piaggio G, Bertolotti F, et al. Donor lymphocyte infusions for the treatment of minimal residual disease in acute leukemia. Blood. (2007) 109:5063–4. doi: 10.1182/blood-2007-02-072470
113. Elorza I, Palacio C, Dapena JL, Gallur L, Sánchez de Toledo J, Díaz de Heredia C. Relationship between minimal residual disease measured by multiparametric flow cytometry prior to allogeneic hematopoietic stem cell transplantation and outcome in children with acute lymphoblastic leukemia. Haematologica. (2010) 95:936–41. doi: 10.3324/haematol.2009.010843
114. Zeiser R, Spyridonidis A, Wasch R, Ihorst G, Grullich C, Bertz H, et al. Evaluation of immunomodulatory treatment based on conventional and lineage-specific chimerism analysis in patients with myeloid malignancies after myeloablative allogeneic hematopoietic cell transplantation. Leukemia. (2005) 19:814–21. doi: 10.1038/sj.leu.2403719
115. Aziz MD, Shah J, Kapoor U, Dimopoulos C, Anand S, Augustine A, et al. Disease risk and GVHD biomarkers can stratify patients for risk of relapse and nonrelapse mortality post hematopoietic cell transplant. Leukemia. (2020) 34:1898–906. doi: 10.1038/s41375-020-0726-z
Keywords: biomarker, GVHD, steroid-refractory graft-vs.-host disease, immune cells, relapse, minimal residual disease
Citation: Chen S and Zeiser R (2020) Novel Biomarkers for Outcome After Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 11:1854. doi: 10.3389/fimmu.2020.01854
Received: 10 May 2020; Accepted: 09 July 2020;
Published: 18 August 2020.
Edited by:
John Andrew Snowden, Sheffield Teaching Hospitals NHS Foundation Trust, United KingdomReviewed by:
Hildegard Theresia Greinix, Medical University of Graz, AustriaBipin N. Savani, Vanderbilt University, United States
Copyright © 2020 Chen and Zeiser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert Zeiser, cm9iZXJ0LnplaXNlckB1bmlrbGluaWstZnJlaWJ1cmcuZGU=
 Sophia Chen
Sophia Chen Robert Zeiser
Robert Zeiser