
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 18 August 2020
Sec. Mucosal Immunity
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01747
This article is part of the Research Topic Immune Responses of the Mucosal Epithelium in Chronic Lung Diseases View all 15 articles
 Bilal Alashkar Alhamwe1,2,3
Bilal Alashkar Alhamwe1,2,3 Sarah Miethe1,4
Sarah Miethe1,4 Elke Pogge von Strandmann3
Elke Pogge von Strandmann3 Daniel P. Potaczek1,5†
Daniel P. Potaczek1,5† Holger Garn1,4*†
Holger Garn1,4*†Asthma is a chronic inflammatory disease of the respiratory tract characterized by recurrent breathing problems resulting from airway obstruction and hyperresponsiveness. Human airway epithelium plays an important role in the initiation and control of the immune responses to different types of environmental factors contributing to asthma pathogenesis. Using pattern recognition receptors airway epithelium senses external stimuli, such as allergens, microbes, or pollutants, and subsequently secretes endogenous danger signaling molecules alarming and activating dendritic cells. Hence, airway epithelial cells not only mediate innate immune responses but also bridge them with adaptive immune responses involving T and B cells that play a crucial role in the pathogenesis of asthma. The effects of environmental factors on the development of asthma are mediated, at least in part, by epigenetic mechanisms. Those comprise classical epigenetics including DNA methylation and histone modifications affecting transcription, as well as microRNAs influencing translation. The common feature of such mechanisms is that they regulate gene expression without affecting the nucleotide sequence of the genomic DNA. Epigenetic mechanisms play a pivotal role in the regulation of different cell populations involved in asthma pathogenesis, with the remarkable example of T cells. Recently, however, there is increasing evidence that epigenetic mechanisms are also crucial for the regulation of airway epithelial cells, especially in the context of epigenetic transfer of environmental effects contributing to asthma pathogenesis. In this review, we summarize the accumulating evidence for this very important aspect of airway epithelial cell pathobiology.
Asthma is a chronic inflammatory disease of the airways, in which airway obstruction and hyperresponsiveness underlie recurrent breathing problems, with symptoms being especially pronounced during disease exacerbations (1, 2). Respiratory tract epithelium plays an important role in asthma by initiating and controlling immune responses to different types of pathogenic environmental factors, including allergens, viruses, pollutants, and others. The biology of the airway epithelium in health and its pathobiology in asthma are regulated by epigenetic mechanisms forming the intercellular homeostatic system responding to internal as well as external changing conditions on the level of transcriptional and posttranscriptional regulation of gene expression (3, 4).
The airway epithelium is the first structure of the body getting into contact with inhaled air with all its containing environmental components. Initially, it was thought to just constitute a mechanical barrier to enable the bidirectional transfer of air to and from the gas-exchanging alveolar structures. Over the last years, it turned out, however, that the airway epithelium in general and the bronchial epithelium as a major part of it in particular represent a much more complex tissue fulfilling a variety of additional functions such as retrograde transport of inhaled particles, establishment of a biochemical barrier system, and initiation and regulation of innate and adaptive immune mechanisms by release of various cytokines and chemokines. By this, it represents an integrative part of the innate immune system, the coordinated activity of which is essential for maintaining the local tissue and even systemic body integrity (5). To exert these diverse functions, the bronchial epithelium is composed of multiple structurally and/or functionally differing cell types, such as ciliated cells (mucociliary transport), goblet cells (mucus secretion), tuft and M cells (luminal signal sampling and antigen presentation), ionocytes (water regulation), and club cells (mucus and surfactant protein production) (6). All these cell types develop from local stem cell precursors, called basal cells (7). It is quite obvious that the continuous development of the different cell types from such precursors, as well as their concerted action under healthy conditions, requires a high level of control and regulation (8). In asthma, the underlying control mechanisms are disturbed by both external (environmental factors such as allergens, pollen, bacteria, viruses) and internal (i.e., cytokines, chemokines, low-molecular-weight mediators produced by innate and adaptive immune cells) influences, resulting in dysregulated activities of the bronchial epithelium (9). This includes hypersecretion of mucus, release of epithelial-derived cytokines called alarmins [e.g., interleukin 25 (IL-25), IL-33, thymic stromal lymphopoietin], chemokines, and antimicrobial peptides, as well as uncontrolled proliferation and differentiation processes, altogether leading to functional [e.g., airway hyperresponsiveness(AHR)] and structural (e.g., airway remodeling) changes that represent characteristic features of asthma pathology (10). Not unexpectedly, because of the close relation to environmental influences and their changes, epigenetic regulation processes are crucially involved in the appropriate development, maintenance, and functionality of the different components of the airway epithelium (11). Chronic inflammatory processes such as in asthma are expected to interfere with these well-balanced epigenetic mechanisms in the epithelium of the airways. This may happen at the level of the aforementioned finally differentiated cell types and by changing related gene expression patterns that influence their functional behavior. It is also conceivable that epigenetic changes occur already at the level of the basal cells, which would then be inherited to all kinds of cells developing from the affected precursors with multiple functional consequences (12). It needs to be considered that these mechanisms may either lead to further perpetuation of the disease process or, alternatively, represent repair activities initiated to get the complex system back to steady state, that is, healthy conditions.
Epigenetics comprises molecular mechanisms of inheritable but reversible phenotypic changes that lead to modified gene expression without alterations at the level of the DNA sequence (13). In the human genome, 80% of the DNA is packed into nucleosomes, and the rest forms linkers between nucleosomes. The nucleosomes are further packed into dense three-dimensional structures called chromosomes (14). The core components of the nucleosome are histone proteins, which are accessible to different types of posttranslational modifications (PTMs), including acetylation, methylation, phosphorylation, sumoylation, and ubiquitination. Posttranslational modifications, especially if occurring at important regulatory genomic regions such as enhancers or promoters, are able to change the accessibility of the DNA to the transcriptional machinery, which is associated with active, poised, or silenced status of transcriptional activity. For example, histone acetylations, the changes introduced by histone acetyltransferases (HATs) and removed by histone deacetylases (HDACs), are usually associated with transcriptional activation of the gene (15, 16). DNA methylation, in which a methyl group is enzymatically added to the cytosine ring of DNA, is another type of the epigenetic modification. While the methylation reaction is catalyzed by DNA methyltransferases, ten-eleven translocation (TET) methylcytosine dioxygenase family proteins mediate DNA demethylation. DNA methylation is typically associated with gene repression (3, 17). In addition to the classical epigenetic modifications mentioned above, different types of the non-coding RNAs such as microRNAs (miRNAs) and others, for instance, piwi-interacting RNAs or small nucleolar RNAs, are involved in the epigenetic regulation of gene expression. Briefly, miRNAs exert their silencing effects through the binding to the mature mRNA molecules in the cytosol that leads to mRNA degradation or reduction in the translational efficiency of the ribosomes (18, 19) (Figure 1).
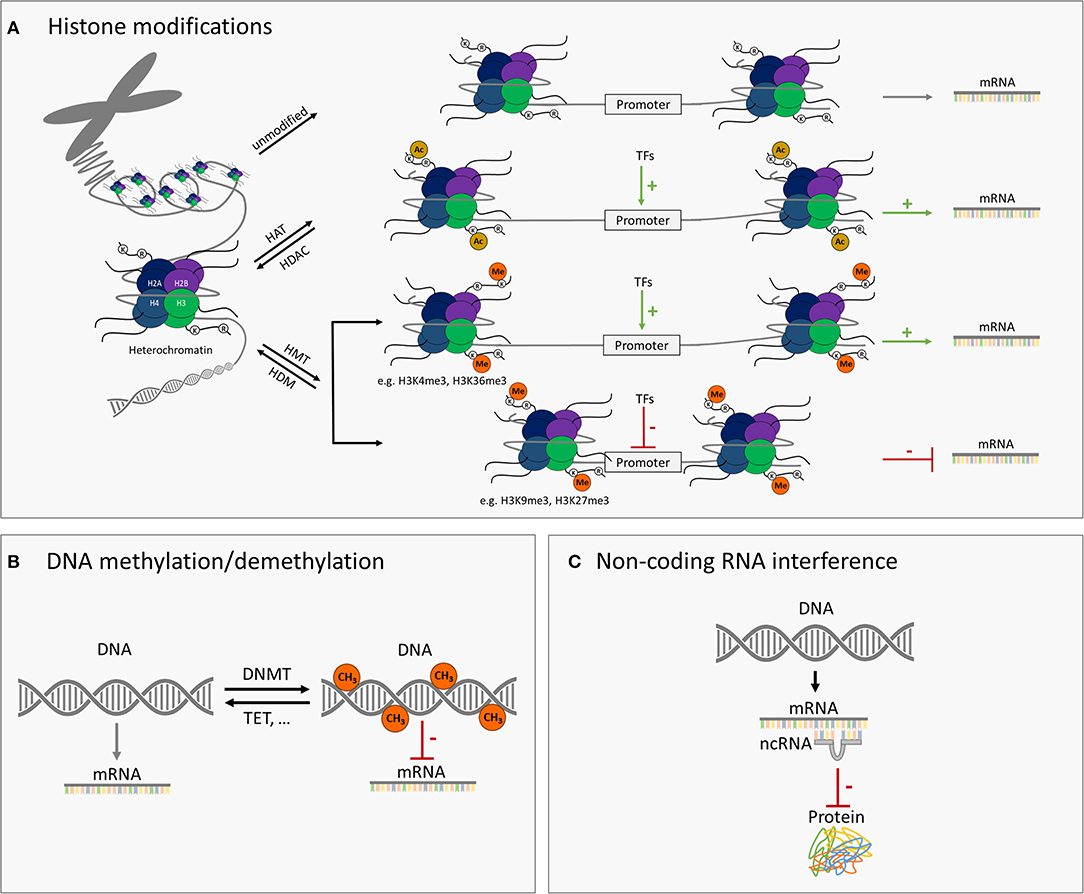
Figure 1. Schematic illustration of major epigenetic modifications. (A) Modification of histones such as histone acetylation/deacetylation via histone acetyltransferases (HATs)/histone deacetylases (HDAC) and methylation/demethylation via histone methyltransferases (HMT)/histone demethylases (HDM) can either activate or repress the target gene transcription. Histone acetylation is typically associated with higher expression of the gene. Histone methylation can be related to either higher or lower transcriptional activity, depending on the amino acid residue modified and the number of methyl groups added. (B) DNA methylation or demethylation of genomic DNA through DNA methyltransferases (DNMT) or ten-eleven translocation (TET) enzymes and others, respectively. Higher level of DNA methylation is typically associated with lower transcriptional activity of the respective gene. (C) MicroRNAs (miRNAs) and further small non-coding RNAs can interfere with gene expression through base pairing with messenger RNAs and thus inhibiting their translation into the encoded protein.
DNA methylation is probably the best studied epigenetic modification in general but also in relation to asthma. Although studies conducted so far on the involvement of DNA methylation in asthma have mostly used already available DNA samples and/or DNA extracted from easily available tissues, also lower airway epithelial cells (AECs) have been investigated (Figure 2). Stefanowicz et al. (20) performed a comparative DNA methylation analysis of 807 genes in bronchial AECs and peripheral blood mononuclear cells (PBMCs) obtained from atopics, atopic asthmatics, non-atopic asthmatics, or healthy controls. They identified signature sets of CpG sites differentially methylated between AECs and PBMCs, which were either independent of the disease phenotype or specific to healthy controls, atopics, or asthmatics. Although no differences in the DNA methylation status were found between disease phenotypes in PBMCs, they were observed between asthmatics and atopics in AECs (20). Kim et al. (21) comparatively analyzed genome-wide DNA methylation levels in bronchial mucosa tissues obtained from atopic and non-atopic asthmatics and healthy controls. Although the methylation levels were similar between asthmatics and controls, a set of loci has been identified with significant differences in DNA methylation between atopic and non-atopic asthmatics (21). Clifford et al. (22) investigated in turn the effects of experimental respiratory tract exposure to allergen, diesel exhaust, or both as a coexposure, always observing only minimal resulting changes in the bronchial epithelial DNA methylome of the participating individuals. They found, however, that if any of the two insults occurs in advance of the other (crossover exposure with a 4-week interval), the initial one primes the bronchial epithelial DNA methylome for the second, resulting in cumulative epigenetic changes with potential biological relevance (22).
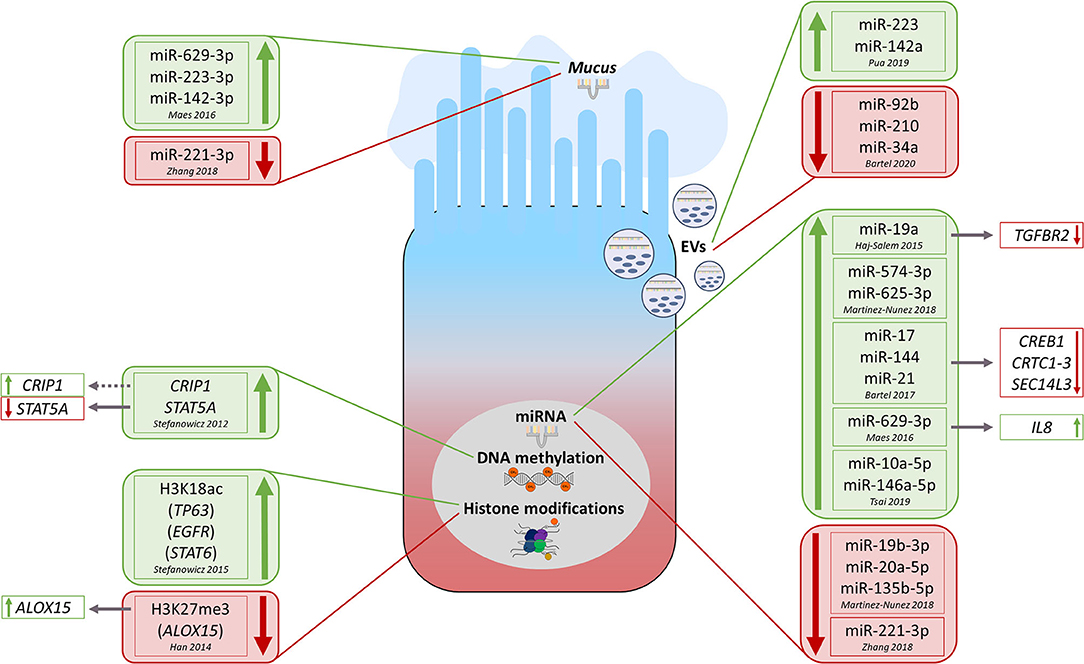
Figure 2. Overview of currently known key epigenetic modifications observed in lower airway epithelial cells from asthma/allergic airway inflammation conditions and—if known—associated functional consequences. The green color always indicates upregulation of the respective modification in asthmatics vs. healthy while red color identifies opposite regulation. EVs, extracellular vesicles; miRNA, microRNA; H3K18ac, histone H3K18 acetylation; H3K27me3, histone H3K27me3 trimethylation.
In most cases, however, DNA methylation studies conducted in airway tissue in the context of asthma have not been performed in bronchial or lung ACEs but rather in nasal epithelial cells (NECs) due to easier accessibility (23–28). Cardenas et al. (23) conducted an epigenome-wide study on DNA methylation using nasal swabs collected in a large group of early teenagers deriving from a birth cohort. They identified multiple DNA methylation loci associated with asthma, allergies, and related clinical or laboratory parameters (23). In another cohort of adolescents, Forno et al. (24) performed in turn an epigenome-wide analysis of DNA methylation in nasal epithelium. The major findings of this study, replicated in two independent cohorts, comprised the identification of specific DNA methylation profiles associated with atopy and atopic asthma and a nasal methylation panel that could classify children by atopy or atopic asthma (24). Reese et al. (25) sought to identify differential DNA methylation related to pediatric asthma in blood from newborns and school-aged children. Interestingly, they were able to replicate in eosinophils or nasal respiratory epithelium most of the asthma-related differential methylation signatures initially detected in blood (25). Brugha et al. (29) comparatively analyzed DNA methylation in airway and surrogate tissues. They found that the methylation profile in nasal epithelium was most representative of that in the airway epithelium, whereas the profile in buccal cells was moderately and that in blood was least similar (29). In our view, these results clearly suggest that DNA methylation studies performed in the context of asthma as an airway disease should preferentially be conducted using AECs or NECs. Although beyond the scope of this review, we would like to mention that, in our view, sorted specific white blood cell populations would be highly valuable to study systemic adaptive immunity DNA methylation patterns underlying asthma. However, how well those signatures correspond to local lung DNA methylation patterns would need to be assessed in separate studies. Back to NECs, Xiao et al. (27) showed that nasal DNA methylation at the promoter of the vanin 1 gene (VNN1) might be a clinically useful biomarker of corticosteroid treatment response in asthmatic children. Another study from the same group demonstrated in turn that DNA methylation at the TET methylcytosine dioxygenase 1 gene (TET1) contributes to traffic-related air pollution and asthma (26).
Finally, allelic differences in DNA methylation and thus gene expression in AECs can mediate the effects of certain genetic variants known to be associated with susceptibility to childhood asthma, such as those in chromosome 17q21 (30, 31).
In addition to DNA methylation, also histone modifications participate in epithelial (patho-) mechanisms related to asthma (Figure 2). Stefanowicz et al. (32) compared global and gene-specific alveolar epithelial cells histone acetylation and methylation status between asthmatics and healthy subjects. Generally, they observed higher global H3K18ac and H3K9me3 levels in asthmatic subjects. In more detail, they found in asthmatics a higher association of H3K18ac (but not H3K9me3) around the transcription start sites of TP63 (tumor protein p63, ΔNp63 isoform), EGFR (epidermal growth factor receptor), and STAT6 (signal transducer and activator of transcription 6) genes. Finally, they detected a non-significant increase in protein expression of those three genes in AECs treated with trichostatin A, an HDAC inhibitor (HDACi) (32). In another work, the same group comparatively analyzed the expression of 82 epigenetic modifying enzymes in AECs and bronchial fibroblasts obtained from asthmatics and healthy controls (33). Thirty-nine enzymes were differentially expressed between AECs and bronchial fibroblasts, 24 of which passed the correction for multiple testing. Six histone modifiers turned out to be differentially expressed in AECs between asthmatics and non-asthmatics, however, mostly not significantly when corrected for multiple testing (33).
Beneficial effects of HDACi have been observed in murine models of allergic airway inflammation (AAI) mimicking features of human allergic asthma (34, 35). Application of HDACi in an ovalbumin (OVA)–based model reduced airway inflammation, remodeling, and AHR. In addition, HDACi treatment was associated with lower expression of transforming growth factor β1 (TGF-β1) in AECs and diminished synthesis of contractile proteins by airway smooth muscle cells (34). HDACi treatment in mice subjected to a house dust mite (HDM)–based model was in turn able to prevent them from developing AHR and AAI. Moreover, HDACi restored the integrity of the ex vivo–cultured NECs isolated from AR patients (35). Significantly lower H3K27me3 levels at the promoter of the arachidonate 15-lipoxygenase (ALOX15) gene (ALOX15) were observed in human lung epithelial A549 cells after the treatment with IL-4, which coincided with higher ALOX15 mRNA levels (36).
Targeting histone modification—related mechanisms turned out to be effective also in a cockroach allergen extract–induced mouse model of mixed granulocytic (eosinophilic and neutrophilic), TH2/TH17-driven asthma (37). Specifically, whereas a bromo- and extraterminal (BET) inhibitor was already alone able to abolish TH17-driven neutrophilic inflammation, in combination with dexamethasone it completely blocked both TH2- and TH17-driven immune responses in the lung, which was associated with reductions in lung eosinophilia and neutrophilia, and mucin secretion. Furthermore, BET inhibition improved cockroach allergen extract– or IL-17A–induced increase in markers of glucocorticoid insensitivity [i.e., decrease in HDAC2 expression (38)] in murine or human AECs, respectively (37). In another study, Hdac2+/− mice subjected to an HDM-induced AAI model demonstrated stronger inflammatory infiltration as well as higher expression of type 2 cytokines and IL-17A in the lung tissue compared to wild-type animals. Additional IL-17A depletion was able to reverse these HDAC2 impairment-induced effects (39). In turn, HDM and IL-17A synergistically reduced HDAC2 expression in human bronchial epithelial cells (BECs) in vitro. Besides, silencing the HDAC2-encoding gene further enhanced HDM- and/or IL-17A–induced inflammatory cytokines in human BECs, whereas HDAC2 overexpression or knockdown of the gene encoding IL-17A was able to reduce the release of such inflammatory cytokines (39). Taken together, original findings by Zijlstra et al. (38), who first discovered IL-17A–induced steroid resistance mediated by a reduction of HDAC2 activity, have thus been corroborated and expanded.
Several recent studies have highlighted the importance of miRNAs in the regulation of epithelial pathobiology in asthma (Figure 2). Bartel et al. (40) combined different approaches such as in vivo studies in mice with OVA- or HDM-induced AAI, ex vivo/in vitro experiments including luciferase reporter assay and stimulation–expression analyses, miRNA/mRNA microarrays, and in silico approaches. This composed strategy enabled the authors to identify the transcription factor cAMP-responsive element binding protein (Creb1) and its transcriptional coactivators (Crtc1-3) as targets for miR-17, miR-144, and miR-21, all three deregulated in lungs of mice with AAI. Moreover, they observed downregulation of Sec14-like 3 (Sec14l3), a putative target of Creb1, in both AAI models and in primary normal human BECs upon IL-13 treatment suggesting that miRNA-regulated Crtc1-3 and Sec14l3 play a role in early epithelial responses to type 2 stimuli (40). Microarray analysis of miRNA expression in bronchoscopy-isolated human BECs showed in turn an upregulation of miR-19a in samples obtained from severe asthmatic subjects compared to those from mild asthmatics and healthy controls (41). Furthermore, luciferase reporter assay- and Western blot–based functional studies demonstrated miR-19a to enhance proliferation of BECs in severe asthma through targeting TGF-β receptor 2 gene (TGFBR2) mRNA (41). Using subcellular fractionation and RNA sequencing (Frac-seq) in human primary BECs from healthy controls and severe asthmatics, Martinez-Nunez et al. (42) assessed paired genome-wide expression of miRNAs along with cytoplasmic (total) and polyribosome-bound (translational) mRNA levels. They identified a hub of six dysregulated miRNAs, displaying preference for polyribosome-bound mRNAs, which accounted for ~90% of whole miRNA targeting. Interestingly, transfection of such miRNAs into BECs obtained from healthy subjects turned them into cells mimicking features of those obtained from severe asthmatics (42).
Recently, extracellular vesicles (EVs) transferring miRNAs between cells have been identified as a novel mechanism of intercellular communication (3, 43, 44). Of note, the composition of the extracellular miRNA pool in the lung of mice was very similar to that of the airway epithelium, and ~80% of the detected EVs were of epithelial origin (45). However, following the induction of AAI, the presence of miRNAs preferentially expressed by immune cells, such as miR-223 and miR-142a, and hematopoietic cell–derived EVs increased also substantially, indicating an importance of the extracellular miRNA pool for the development of local allergic inflammatory processes (45). Gupta et al. (46) focused on EVs secreted by two kinds of human airway cell cultures, that is, primary tracheobronchial cells and a cultured AEC line (Calu-3). Their data suggest that cellular information can be transferred between AECs via miRNA-containing EVs, which may thereby contribute to epithelial biology and remodeling (46). Another study profiled the expression of miRNAs in EVs secreted from the apical and basal sides by normal human BECs treated with IL-13 in order to induce an asthma-like response (47). Significant candidates were then confirmed in EVs isolated from nasal lavages obtained from children with mild to moderate or severe asthma and healthy control subjects. Interestingly, levels of miR-92b, miR-210, and miR-34a turned out to correlate with lung function measures (47).
Two studies investigated miRNAs in asthmatic sputum (48, 49). In two independent cohorts, Maes et al. (48) found a significant upregulation of miR-629-3p, miR-223-3p, and miR-142-3p in sputum of severe asthmatics compared to healthy controls, with the highest levels in patients with neutrophilic asthma. Of those three miRNAs associated with sputum neutrophilia and airway obstruction, miR-629-3p was expressed in BECs. Interestingly, transfection of human BECs with a miR-629-3p mimic induced expression of IL-8, the sputum levels of which were significantly increased and positively correlated with sputum neutrophilia in severe asthmatics (48). Zhang et al. (49) found in turn that epithelial, sputum, and plasma miR-221-3p levels were significantly lower in asthmatics compared to healthy controls. In addition, levels of epithelial and sputum miR-221-3p inversely correlated with airway eosinophilia (49).
Finally, Tsai et al. (50) sought to find the common miRNA-related effects in BECs obtained from subjects with asthma and chronic obstructive pulmonary disease (COPD). First detected with next-generation sequencing, the upregulation of miR-10a-5p and miR-146a-5p in BECs obtained from both asthma and COPD patients was subsequently confirmed by quantitative polymerase chain reaction. Moreover, compared to healthy controls, also serum miR-146a-5p levels were higher in asthma and COPD subjects (50). Further research will establish whether miRNAs mediating intercellular communication can be used for clinical applications as biomarkers or therapeutic targets.
Airborne viruses, for instance, human rhinoviruses (HRVs), stimulate asthma exacerbations. In addition, repeated early life infections with such viruses can lead to the development of a persistent asthma phenotype, especially in children with atopic susceptibility (3, 51). Interestingly, some studies suggest that the effects of respiratory viral infections are at least partly mediated by epigenetic changes in airway epithelial cells. It has been demonstrated that ex vivo experimental HRV infection of NECs obtained from asthmatic children significantly changes patterns of DNA methylation and mRNA expression (52). Moreover, HRV infection in young children has been associated with changes in the airway secretory miRNome, characterized by a highly specific additional appearance of miR-155 in nasal secretion EVs (53). In turn, BECs obtained from asthmatics have been shown to be characterized by dysregulated miR-22 expression after experimental ex vivo infection with influenza A virus (IAV) (54). Other epigenetic modifications, specifically histone methylations, also seem to be involved in the regulation of epithelial antiviral responses (55).
Dysregulated epithelial–mesenchymal transition (EMT) is the process driven mostly by TGF-β1, which strongly contributes to the establishment of the persistent asthma phenotype, that is, to disease chronification (56). Epigenetic mechanisms seem to play an important role in EMT. It has been demonstrated in mouse models mimicking allergic asthma that miR-448-5p can inhibit TGF-β1–induced EMT and pulmonary fibrosis (57). Applying an epigenome-wide approach in a human study, McErlean et al. (58) identified in turn multiple loci showing differential H3K27ac enrichment in asthma, which clustered at genes associated with type 2–driven asthma and EMT.
Epigenetic mechanisms play a very important role in the epithelial pathobiology of asthma. While histone modifications seem to be especially interesting as possible therapeutic targets, DNA methylation and miRNAs, also from the easily accessible nasal epithelium, show a substantial diagnostic potential. Although the data gathered by now (for overview, see also Supplementary Table 1) already strongly suggest a usefulness of epigenetics in the asthma management, further studies, especially those considering the complex interplay of different epigenetic mechanisms and those focusing on a single-cell type or investigations on the single cell level, are needed.
BA: draft writing and figure drafts. SM: draft writing and final figures. ES: editing and reviewing. DP: conceptualization, draft writing, and reviewing. HG: conceptualization, coordination, draft writing, and reviewing. All authors contributed to the article and approved the submitted version.
BA was supported by the German Academic Exchange Service (DAAD; Personal Reference no. 91559386). Parts of the study were funded by grants GRK 2573/1 and KFO325 Project A3 to ES.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01747/full#supplementary-material
1. Miethe S, Guarino M, Alhamdan F, Simon H-U, Renz H, Dufour J-F, et al. Effects of obesity on asthma: immunometabolic links. Polish Arch Internal Med. (2018) 128:469–77. doi: 10.20452/pamw.4304
2. Koczulla AR, Vogelmeier CF, Garn H, Renz H. New concepts in asthma: clinical phenotypes and pathophysiological mechanisms. Drug Discov Today. (2017) 22:388–96. doi: 10.1016/j.drudis.2016.11.008
3. Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. (2017) 9:539–71. doi: 10.2217/epi-2016-0162
4. Pepper AN, Renz H, Casale TB, Garn H. Biologic therapy and novel molecular targets of severe asthma. J Allergy Clin Immunol Pract. (2017) 5:909–16. doi: 10.1016/j.jaip.2017.04.038
5. Potaczek DP, Miethe S, Schindler V, Alhamdan F, Garn H. Role of airway epithelial cells in the development of different asthma phenotypes. Cell Signal. (2020) 69:109523. doi: 10.1016/j.cellsig.2019.109523
6. Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. (2018) 560:377–81. doi: 10.1038/s41586-018-0394-6
7. Watson JK, Rulands S, Wilkinson AC, Wuidart A, Ousset M, van Keymeulen A, et al. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. (2015) 12:90–101. doi: 10.1016/j.celrep.2015.06.011
8. Loxham M, Davies DE. Phenotypic and genetic aspects of epithelial barrier function in asthmatic patients. J Allergy Clin Immunol. (2017) 139:1736–51. doi: 10.1016/j.jaci.2017.04.005
9. Holgate ST. The sentinel role of the airway epithelium in asthma pathogenesis. Immunol Rev. (2011) 242:205–19. doi: 10.1111/j.1600-065X.2011.01030.x
10. Lloyd CM, Saglani S. Epithelial cytokines and pulmonary allergic inflammation. Curr Opin Immunol. (2015) 34:52–8. doi: 10.1016/j.coi.2015.02.001
11. Wawrzyniak P, Wawrzyniak M, Wanke K, Sokolowska M, Bendelja K, Rückert B, et al. Regulation of bronchial epithelial barrier integrity by type 2 cytokines and histone deacetylases in asthmatic patients. J Allergy Clin Immunol. (2017) 139:93–103. doi: 10.1016/j.jaci.2016.03.050
12. Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. (2018) 560:649–54. doi: 10.1038/s41586-018-0449-8
13. Lacal I, Ventura R. Epigenetic inheritance: concepts, mechanisms and perspectives. Front Mol Neurosci. (2018) 11:292. doi: 10.3389/fnmol.2018.00292
14. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell: Chromosomal DNA and Its Packaging in the Chromatin Fiber. 4th ed. New York, NY: Garland Science (2002).
15. Harb H, Alashkar Alhamwe B, Garn H, Renz H, Potaczek DP. Recent developments in epigenetics of pediatric asthma. Curr Opin Pediatr. (2016) 28:754–63. doi: 10.1097/MOP.0000000000000424
16. Alaskhar Alhamwe B, Khalaila R, Wolf J, Bülow V, von Harb H, Alhamdan F, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. (2018) 14:39. doi: 10.1186/s13223-018-0259-4
17. Alashkar Alhamwe B, Alhamdan F, Ruhl A, Potaczek DP, Renz H. The role of epigenetics in allergy and asthma development. Curr Opin Allergy Clin Immunol. (2020) 20:48–55. doi: 10.1097/ACI.0000000000000598
18. Hu G, Niu F, Humburg BA, Liao K, Bendi S, Callen S, et al. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget. (2018) 9:18648–63. doi: 10.18632/oncotarget.24307
19. Karlsson O, Baccarelli AA. Environmental health and long non-coding RNAs. Curr Environ Health Rep. (2016) 3:178–87. doi: 10.1007/s40572-016-0092-1
20. Stefanowicz D, Hackett T-L, Garmaroudi FS, Günther OP, Neumann S, Sutanto EN, et al. DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children. PLoS ONE. (2012) 7:e44213. doi: 10.1371/journal.pone.0044213
21. Kim Y-J, Park S-W, Kim T-H, Park J-S, Cheong HS, Shin HD, et al. Genome-wide methylation profiling of the bronchial mucosa of asthmatics: relationship to atopy. BMC Med Genet. (2013) 14:39. doi: 10.1186/1471-2350-14-39
22. Clifford RL, Jones MJ, MacIsaac JL, McEwen LM, Goodman SJ, Mostafavi S, et al. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J Allergy Clin Immunol. (2017) 139:112–21. doi: 10.1016/j.jaci.2016.03.046
23. Cardenas A, Sordillo JE, Rifas-Shiman SL, Chung W, Liang L, Coull BA, et al. The nasal methylome as a biomarker of asthma and airway inflammation in children. Nat Commun. (2019) 10:3095. doi: 10.1038/s41467-019-11058-3
24. Forno E, Wang T, Qi C, Yan Q, Xu C-J, Boutaoui N, et al. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: a genome-wide study. Lancet Respir Med. (2019) 7:336–46. doi: 10.1016/S2213-2600(18)30466-1
25. Reese SE, Xu C-J, den Dekker HT, Lee MK, Sikdar S, Ruiz-Arenas C, et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol. (2019) 143:2062–74. doi: 10.1016/j.jaci.2018.11.043
26. Somineni HK, Zhang X, Biagini Myers JM, Kovacic MB, Ulm A, Jurcak N, et al. Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J Allergy Clin Immunol. (2016) 137:797–805.e5. doi: 10.1016/j.jaci.2015.10.021
27. Xiao C, Biagini Myers JM, Ji H, Metz K, Martin LJ, Lindsey M, et al. Vanin-1 expression and methylation discriminate pediatric asthma corticosteroid treatment response. J Allergy Clin Immunol. (2015) 136:923–31.e3. doi: 10.1016/j.jaci.2015.01.045
28. Zhang X, Biagini Myers JM, Burleson JD, Ulm A, Bryan KS, Chen X, et al. Nasal DNA methylation is associated with childhood asthma. Epigenomics. (2018) 10:629–41. doi: 10.2217/epi-2017-0127
29. Brugha R, Lowe R, Henderson AJ, Holloway JW, Rakyan V, Wozniak E, et al. DNA methylation profiles between airway epithelium and proxy tissues in children. Acta Paediatr. (2017) 106:2011–6. doi: 10.1111/apa.14027
30. Moussette S, Al Tuwaijri A, Kohan-Ghadr H-R, Elzein S, Farias R, Bérubé J, et al. Role of DNA methylation in expression control of the IKZF3-GSDMA region in human epithelial cells. PLoS ONE. (2017) 12:e0172707. doi: 10.1371/journal.pone.0172707
31. Toncheva AA, Potaczek DP, Schedel M, Gersting SW, Michel S, Krajnov N, et al. Childhood asthma is associated with mutations and gene expression differences of ORMDL genes that can interact. Allergy. (2015) 70:1288–99. doi: 10.1111/all.12652
32. Stefanowicz D, Lee JY, Lee K, Shaheen F, Koo H-K, Booth S, et al. Elevated H3K18 acetylation in airway epithelial cells of asthmatic subjects. Respir Res. (2015) 16:95. doi: 10.1186/s12931-015-0254-y
33. Stefanowicz D, Ullah J, Lee K, Shaheen F, Olumese E, Fishbane N, et al. Epigenetic modifying enzyme expression in asthmatic airway epithelial cells and fibroblasts. BMC Pulm Med. (2017) 17:24. doi: 10.1186/s12890-017-0371-0
34. Ren Y, Su X, Kong L, Li M, Zhao X, Yu N, et al. Therapeutic effects of histone deacetylase inhibitors in a murine asthma model. Inflamm Res. (2016) 65:995–1008. doi: 10.1007/s00011-016-0984-4
35. Steelant B, Wawrzyniak P, Martens K, Jonckheere A-C, Pugin B, Schrijvers R, et al. Blocking histone deacetylase activity as a novel target for epithelial barrier defects in patients with allergic rhinitis. J Allergy Clin Immunol. (2019) 144:1242–53.e7. doi: 10.1016/j.jaci.2019.04.027
36. Han H, Xu D, Liu C, Claesson H-E, Björkholm M, Sjöberg J. Interleukin-4-mediated 15-lipoxygenase-1 trans-activation requires UTX recruitment and H3K27me3 demethylation at the promoter in A549 cells. PLoS ONE. (2014) 9:e85085. doi: 10.1371/journal.pone.0085085
37. Nadeem A, Ahmad SF, Al-Harbi NO, Siddiqui N, Ibrahim KE, Attia SM. Inhibition of BET bromodomains restores corticosteroid responsiveness in a mixed granulocytic mouse model of asthma. Biochem Pharmacol. (2018) 154:222–33. doi: 10.1016/j.bcp.2018.05.011
38. Zijlstra GJ, Hacken NHT, ten Hoffmann RF, van Oosterhout AJM, Heijink IH. Interleukin-17A induces glucocorticoid insensitivity in human bronchial epithelial cells. Eur Respir J. (2012) 39:439–45. doi: 10.1183/09031936.00017911
39. Lai T, Wu M, Zhang C, Che L, Xu F, Wang Y, et al. HDAC2 attenuates airway inflammation by suppressing IL-17A production in HDM-challenged mice. Am J Physiol Lung Cell Mol Physiol. (2019) 316:L269–79. doi: 10.1152/ajplung.00143.2018
40. Bartel S, Schulz N, Alessandrini F, Schamberger AC, Pagel P, Theis FJ, et al. Pulmonary microRNA profiles identify involvement of Creb1 and Sec14l3 in bronchial epithelial changes in allergic asthma. Sci Rep. (2017) 7:46026. doi: 10.1038/srep46026
41. Haj-Salem I, Fakhfakh R, Bérubé J-C, Jacques E, Plante S, Simard MJ, et al. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFβR2 gene in severe asthma. Allergy. (2015) 70:212–9. doi: 10.1111/all.12551
42. Martinez-Nunez RT, Rupani H, Platé M, Niranjan M, Chambers RC, Howarth PH, et al. Genome-wide posttranscriptional dysregulation by MicroRNAs in human asthma as revealed by Frac-seq. J Immunol. (2018) 201:251–63. doi: 10.4049/jimmunol.1701798
43. Tost J. A translational perspective on epigenetics in allergic diseases. J Allergy Clin Immunol. (2018) 142:715–26. doi: 10.1016/j.jaci.2018.07.009
44. Guiot J, Struman I, Louis E, Louis R, Malaise M, Njock M-S. Exosomal miRNAs in lung diseases: from biologic function to therapeutic targets. J Clin Med. (2019) 8:1345. doi: 10.3390/jcm8091345
45. Pua HH, Happ HC, Gray CJ, Mar DJ, Chiou N-T, Hesse LE, et al. Increased hematopoietic extracellular RNAs and vesicles in the lung during allergic airway responses. Cell Rep. (2019) 26:933–44.e4. doi: 10.1016/j.celrep.2019.01.002
46. Gupta R, Radicioni G, Abdelwahab S, Dang H, Carpenter J, Chua M, et al. Intercellular communication between airway epithelial cells is mediated by exosome-like vesicles. Am J Respir Cell Mol Biol. (2019) 60:209–20. doi: 10.1165/rcmb.2018-0156OC
47. Bartel S, La Grutta S, Cilluffo G, Perconti G, Bongiovanni A, Giallongo A, et al. Human airway epithelial extracellular vesicle miRNA signature is altered upon asthma development. Allergy. (2020) 75:346–56. doi: 10.1111/all.14008
48. Maes T, Cobos FA, Schleich F, Sorbello V, Henket M, Preter K, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol. (2016) 137:1433–46. doi: 10.1016/j.jaci.2016.02.018
49. Zhang K, Liang Y, Feng Y, Wu W, Zhang H, He J, et al. Decreased epithelial and sputum miR-221-3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. Am J Physiol Lung Cell Mol Physiol. (2018) 315:L253–L264. doi: 10.1152/ajplung.00567.2017
50. Tsai M-J, Tsai Y-C, Chang W-A, Lin Y-S, Tsai P-H, Sheu C-C, et al. Deducting microRNA-mediated changes common in bronchial epithelial cells of asthma and chronic obstructive pulmonary disease-a next-generation sequencing-guided bioinformatic approach. Int J Mol Sci. (2019) 20:553. doi: 10.3390/ijms20030553
51. Potaczek DP, Unger SD, Zhang N, Taka S, Michel S, Akdag N, et al. Development and characterization of DNAzyme candidates demonstrating significant efficiency against human rhinoviruses. J Allergy Clin Immunol. (2019) 143:1403–15. doi: 10.1016/j.jaci.2018.07.026
52. Pech M, Weckmann M, König IR, Franke A, Heinsen F-A, Oliver B, et al. Rhinovirus infections change DNA methylation and mRNA expression in children with asthma. PLoS ONE. (2018) 13:e0205275. doi: 10.1371/journal.pone.0205275
53. Gutierrez MJ, Gomez JL, Perez GF, Pancham K, Val S, Pillai DK, et al. Airway secretory microRNAome changes during rhinovirus infection in early childhood. PLoS ONE. (2016) 11:e0162244. doi: 10.1371/journal.pone.0162244
54. Moheimani F, Koops J, Williams T, Reid AT, Hansbro PM, Wark PA, et al. Influenza A virus infection dysregulates the expression of microRNA-22 and its targets; CD147 and HDAC4, in epithelium of asthmatics. Respir Res. (2018) 19:145. doi: 10.1186/s12931-018-0851-7
55. Spalluto CM, Singhania A, Cellura D, Woelk CH, Sanchez-Elsner T, Staples KJ, et al. IFN-γ influences epithelial antiviral responses via histone methylation of the RIG-I promoter. Am J Respir Cell Mol Biol. (2017) 57:428–38. doi: 10.1165/rcmb.2016-0392OC
56. Rout-Pitt N, Farrow N, Parsons D, Donnelley M. Epithelial mesenchymal transition (EMT): a universal process in lung diseases with implications for cystic fibrosis pathophysiology. Respir Res. (2018) 19:136. doi: 10.1186/s12931-018-0834-8
57. Yang Z-C, Qu Z-H, Yi M-J, Shan Y-C, Ran N, Xu L, et al. MiR-448-5p inhibits TGF-β1-induced epithelial-mesenchymal transition and pulmonary fibrosis by targeting Six1 in asthma. J Cell Physiol. (2019) 234:8804–14. doi: 10.1002/jcp.27540
Keywords: airway, allergy, asthma, epigenetic, epithelium, histone, methylation, microRNA (miRNA)
Citation: Alashkar Alhamwe B, Miethe S, Pogge von Strandmann E, Potaczek DP and Garn H (2020) Epigenetic Regulation of Airway Epithelium Immune Functions in Asthma. Front. Immunol. 11:1747. doi: 10.3389/fimmu.2020.01747
Received: 27 February 2020; Accepted: 30 June 2020;
Published: 18 August 2020.
Edited by:
Christian Herr, Saarland University Hospital, GermanyReviewed by:
Yogesh Singh, Tübingen University Hospital, GermanyCopyright © 2020 Alashkar Alhamwe, Miethe, Pogge von Strandmann, Potaczek and Garn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Holger Garn, Z2FybkBzdGFmZi51bmktbWFyYnVyZy5kZQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.