- 1School of Pharmacy and Biomolecular Sciences and Tissue Engineering Research Group, Royal College of Surgeons in Ireland, Dublin, Ireland
- 2Department of Anatomy and Regenerative Medicine and Tissue Engineering Research Group, Royal College of Surgeons in Ireland, Dublin, Ireland
The circadian cycle allows organisms to track external time of day and predict/respond to changes in the external environment. In higher order organisms, circadian rhythmicity is a central feature of innate and adaptive immunity. We focus on the role of the molecular clock and circadian rhythmicity specifically in monocytes and macrophages of the innate immune system. These cells display rhythmicity in their internal functions, such as metabolism and inflammatory mediator production as well as their external functions in pathogen sensing, phagocytosis, and migration. These inflammatory mediators are of clinical interest as many are therapeutic targets in inflammatory disease such as cardiovascular disease, diabetes, and rheumatoid arthritis. Moreover, circadian rhythm disruption is closely linked with increased prevalence of these conditions. Therefore, understanding the mechanisms by which circadian disruption affects monocyte/macrophage function will provide insights into novel therapeutic opportunities for these chronic inflammatory diseases.
Introduction
Circadian rhythms are oscillations in physiology and behavior with a 24-h periodicity. This rhythmicity first arose at the cellular level, ~2.5 billion years ago. Organisms evolved this strategy as an adaptation to rhythmic changes in oxidative stress caused by the rotation of the earth on its axis (1). A common hypothesis is that rhythmic cycles of peroxiredoxins conferred a selective advantage on photosynthetic bacteria, allowing them to detoxify reactive oxygen species (ROS) derived from daily oxygen consumption. Today, mammalian circadian rhythms are more complex and molecular clocks throughout the body can synchronize physiological and behavioral activities to appropriate times of the 24-h day, thus maximizing energy efficiency (2–4).
The term “circadian” was coined by Franz Halberg in 1959. It was Halberg who carried out a seminal study showing that survival rates in mice were dependent on the time-of-day when Escherichia coli (E. Coli) endotoxin was injected (5). Interestingly, the response to endotoxin relies heavily on cells of the innate immune system, the branch of immunity which provides the first line of defense against infection and damage. Monocytes and macrophages are central to innate immunity (6) and their molecular clocks have been implicated in multiple inflammatory disorders (7). Monocytes are short-lived, motile cells found in blood, bone marrow, and spleen (6). They quickly respond and migrate to sites of infection. They are often considered a systemic reservoir of myeloid precursors, important in the renewal of tissue macrophages and dendritic cells. Macrophages, on the other hand, are long-lived tissue-specific cells with roles ranging from tissue homeostasis to immune response generation against pathogens (6). In this review, we will discuss our understanding of the molecular mechanisms governing circadian control of monocytes/macrophages and their potential impact on chronic inflammatory disease.
The Molecular Clock
Virtually all cell types have an internal molecular clock (8, 9). However, the master-clock resides in the suprachiasmatic nucleus (SCN) of the hypothalamus. The SCN processes external light signals, generating rhythmic signals via the hypothalamic-pituitary-adrenal (HPA) axis and autonomic nervous system, which synchronize peripheral clocks in tissues (10, 11). The molecular clock is regulated by a series of interlocking transcription-translation feedback loops (TTFLs), powered by the heterodimeric pairing of BMAL1 and CLOCK (Figure 1A). BMAL1 is the master clock regulator and its deletion ablates most rhythmic activity (12). BMAL1:CLOCK heterodimers bind E-box sites on DNA and facilitate the transcription of clock-controlled genes (CCG). Included in CCGs are the clock's negative regulators, period (PER) and cryptochrome (CRY), which translocate to the nucleus, disrupt the BMAL1:CLOCK heterodimer and inhibit their own expression (13–15). Other regulators within this circuitry include RAR-related orphan receptor alpha (RORα) and the nuclear receptor REV-ERBα, which promote and suppress Bmal1 transcription, respectively (16, 17). Comprehensive details of the molecular circadian circuitry have been reviewed elsewhere (18, 19).
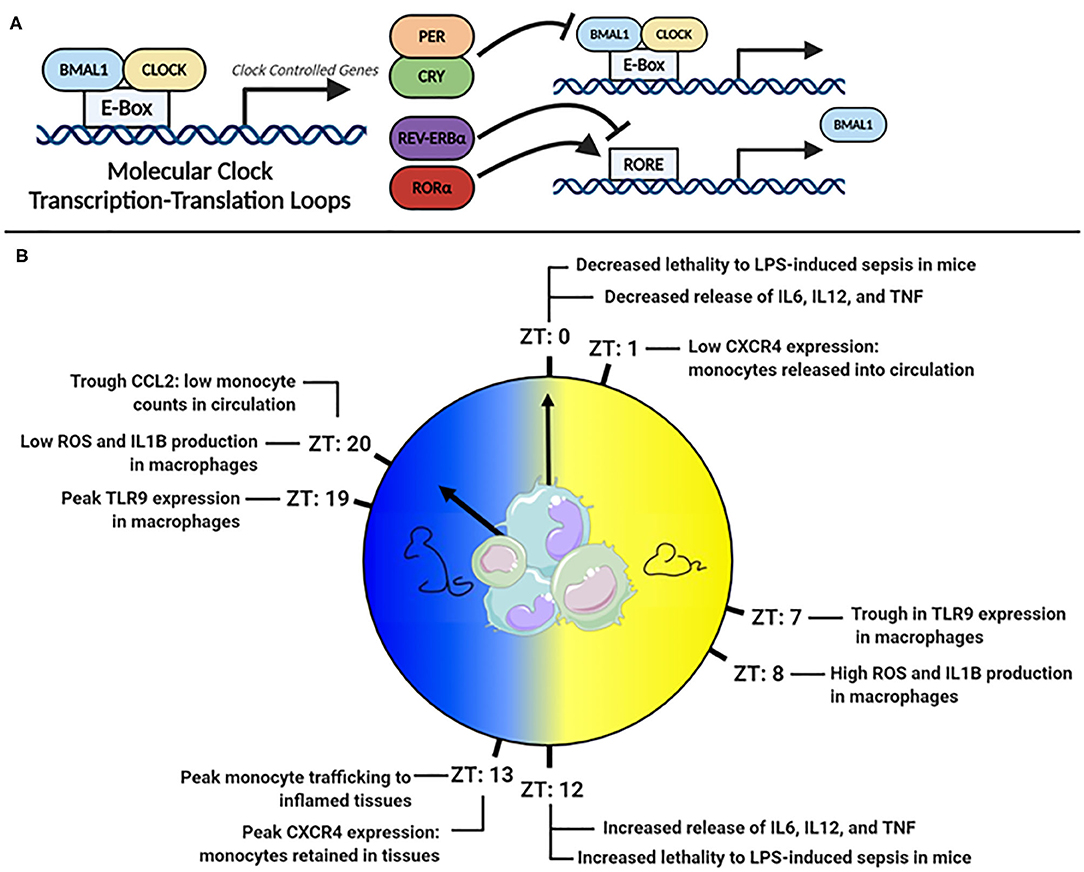
Figure 1. (A) The molecular clock transcription factor feedback loop. (B) Peaks troughs in murine monocyte and macrophage inflammatory mediators across the 24 h day.
A powerful example of the clocks influence is that 43% of murine protein-coding genes across 12 organs display circadian cycling, in an organ-dependent manner (8). In baboons, a closer cousin of humans, an astonishing 80% of protein-coding genes across 64 tissues displayed rhythmicity (9). Monocytes and macrophages also express a robust molecular clock (20–22) and at least 8% of transcripts in murine peritoneal macrophages are circadian (23). Many of these cycling transcripts are involved in key innate-immune functions, such as antigen presentation, immune regulation, and phagocytosis (23). Given that their primary role is to sense and respond to challenges from pathogens, which would be driven by rhythms in feeding and activity, it is unsurprising that significant rhythmicity has been documented in monocyte and macrophage function (Figure 1B).
Pattern Recognition Receptors
Monocytes and macrophages use membrane-bound pattern recognition receptors (PRRs), to sense the external environment for infection and damage. PRRs are capable of recognizing pathogen-associated or damage-associated molecular patterns (PAMPs/DAMPs) facilitating responses to harmful substances (24). PAMPs include non-self-molecules, such as bacterial deoxyribonucleic acids and lipoproteins, which represent infection (25). DAMPs are endogenous molecules that represent deviation from homeostasis to the body, e.g., adenosine triphosphate (ATP) and DNA, which are released from cells with cell-death or damage. Perhaps the most well-studied group of PRRs are the toll-like receptors (TLRs), which induce a series of signaling cascades that convert monocytes and macrophages from quiescence to immunologically active (26).
TLR4, a surface bound receptor, senses the endotoxin LPS found on gram-negative bacteria such as E. coli and Salmonella (27). Halberg's observation that E. coli endotoxin-induced death varied with time-of-day of injection suggested that TLR4 receptor signaling was under circadian control (5). A more recent study found that mice injected with LPS at Zeitgeber time (ZT) 0 (lights on and beginning of rest phase) were less likely to succumb to disease than mice injected at ZT12 [(lights off and beginning of active phase (28)] (Figure 1B). Deletion of Bmal1 in myeloid cells, which includes monocytes and macrophages, resulted in loss of time-of-day protection. However, circadian oscillation in Tlr4 expression has not been observed in macrophages (29), but many genes downstream of TLR4 were cycling (23) such as IkBα, which negatively regulates NF-κB, and Adam17, a metalloproteinase involved in TNFα release (30). Direct circadian impact on other TLRs, such as TLR9, an intracellular receptor that senses bacterial and viral DNA, has been observed (29). In a TLR9-dependent model of sepsis, greater lethality was observed at the time-of-day coinciding with highest TLR9 expression in splenic macrophages and B cells. In splenic macrophages, rhythms have been also observed in the mRNA expression of Tlr2 and Tlr6, peaking at ZT14 (31). Therefore, circadian control in some TLRs and downstream signaling pathways appears as an important mechanism directing circadian inflammation. However, circadian TLR expression differs amongst immune cells (31), and an explanation of this, and the functional consequences of it, are still largely unknown.
Immunometabolism
Immune cell activation following PRR stimulation requires significant amounts of energy. Immunometabolism is an emerging field seeking to understand how cellular metabolism impacts immunity (32, 33). Multiple relationships exist between clock function and metabolism in liver (34–37) and muscle (38–40). However, the specific relationships between the molecular clock, metabolism, and immunity have yet to be fully determined (41, 42). Many immunometabolism investigations have been on macrophages, whose activity covers a spectrum of phenotypes. LPS stimulation promotes a pro-inflammatory M1-like phenotype characterized by increased glycolysis and decreased oxidative phosphorylation (43). In contrast, M2-like stimuli, such as IL-4, decrease glycolysis, promote oxidative phosphorylation, and generating an anti-inflammatory state (44). These metabolic shifts can directly impact outcomes of certain pathologies, such as sepsis (45). Interestingly, BMAL1 suppresses sepsis through its impact upon glycolytic metabolism (46). BMAL1 transcriptionally targets the glycolytic enzyme Pkm2, negatively regulating glycolysis, lactate production, and the immune checkpoint protein PD-L1. Deletion of Bmal1 in macrophages diminished their ability to control glycolysis and increased downstream PD-L1/T-cell mediated septic shock, revealing the importance of macrophage clock function on metabolic control of inflammation.
Mitochondria have extended their reach beyond energy production and are now considered central hubs of immunity (47, 48). They achieve this through production of metabolites (47) and ROS (49), and by altering their morphology, which can affect metabolism and signal transduction pathways (48). Mitochondrial morphology describes the elongation (fusion) or segmentation (fission) of mitochondria within the cell (50–53). Rhythms in mitochondrial morphology and membrane potential have been observed in synchronized peritoneal macrophages in vitro (54). However, whether mitochondrial morphology is under circadian control in innate immunity is still unknown. Thus, determining circadian immunometabolism of the innate immune system will provide new insights into a range of diseases and pathologies.
Inflammatory Mediators
While classifying macrophages into M1 vs. M2 is convenient, the reality is that macrophages in vivo are highly plastic cells existing across a spectrum of activation states (55, 56). Nonetheless, macrophages isolated from mice lacking the clock genes Per1 and Per2 preferentially display an M1-like pro-inflammatory phenotype. However, this phenotype is attenuated following overexpression of PPARγ (57), a critical regulator of M2-like macrophage polarization (58). The circadian hormone melatonin promotes an M2-like phenotype, acting through the clock component RORα and through metabolite-signaling dependent mechanisms (59, 60). When mice were exposed to a shifted light-dark cycle and fed a high-fat diet, more M1-like macrophage polarization was observed compared to normal light-dark cycles controls (61). IL-6 expression in peritoneal macrophages isolated from chronically clock-disrupted mice was increased following exposure to LPS (62). The response of individual macrophages to LPS has been shown to be dependent on the circadian genes Nfil3 and Dbp (63). These two genes are in antiphase to each other and have opposite effects on LPS induced inflammation. NFIL3 and DBP competitively bind to the promoter of Il12b repressing and enhancing its expression, respectively. The oscillations of these circadian proteins provide variation in the response to LPS across the circadian day. This study highlights the molecular clock as a potential mechanism by which genetically identical cells of the same lineage may respond differently to the same stimuli. Taken together, data from these studies illustrate the importance of circadian rhythmicity and the molecular clock on macrophage polarization.
Our current understanding of clock control of macrophage/monocyte expression of chemokines and cytokines is summarized in Figures 2A,B, respectively. Serum levels of IL-6, IL-12, CCL5, CXCL1, and CCL2 were shown to be higher in WT mice injected with LPS at the time of transition to dark phase. Similarly, serum levels of CCL2, IL-1β, IL-6, and IFN-γ were higher in mice infected with Listeria at ZT8 vs. ZT0 (22) and deletion of Bmal1 in the myeloid lineage of mice also increased these cytokines (22). BMAL1 directly induces the master antioxidant transcription factor NRF2, which diurnally regulates ROS in myeloid cells, limiting HIF-1α induced IL-1β (64). Others have demonstrated increased Hif1a and Il1b expression in macrophages with deletion of Bmal1 (65). This was due to a loss of BMAL1's epigenetic role in down regulating TLR4 responsive enhancer RNA (eRNA) expression (65). CLOCK enhances gene expression of Il-6, Il1b, Tnfa, Cxcl1, Ifnb, and Ccl2 (66). CLOCK also boosts NF-kB activity in mouse endothelial fibroblasts (MEFs), while BMAL1 works to sequester CLOCK from the NF-κB subunit p65 in MEFs (67). Whether this mechanism exists in macrophages is unknown but warrants investigation. However, Bmal1 deletion in myeloid cells is known to increase expression of p65, mediated by miR-155, with a subsequent increase in TNFα. TLR4 activation by LPS also results in increased levels of miR-155, which targets Bmal1 for degradation, potentiating inflammation (28). This evidence suggests that BMAL1 suppresses, while CLOCK potentiates, the inflammatory response in macrophages. Further work is needed to clarify the direct and indirect regulatory mechanisms impacted by these circadian transcription factors.
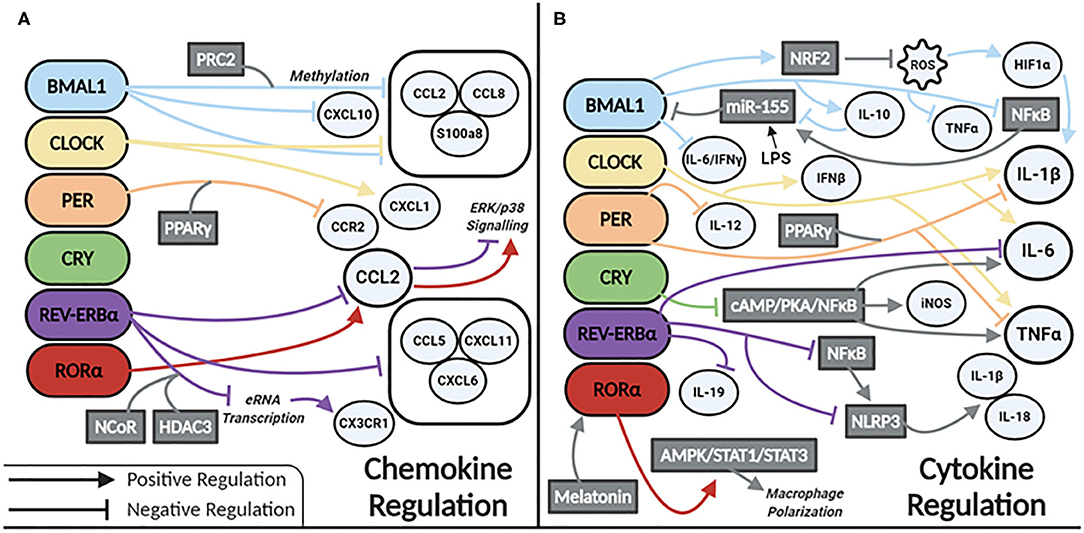
Figure 2. Circadian clock proteins mediated molecular regulation of (A) chemokines and (B) cytokines.
REV-ERBα also has a role in regulating inflammation in macrophages. Global Rev-Erbα deletion ablates time-of-day gating of peritoneal macrophage IL-6 production. Alveolar macrophages isolated from these mice have heightened inflammation in terms of Il6, Ccl2, and Ccl5 expression (68). Conversely, a REV-ERBα agonist suppresses expression of Il6, Il19, Cxcl6, Cxcl11, and Ccl2 in LPS-stimulated human monocyte-derived macrophages (69). REV-ERBα directly binds to the promoter of Ccl2 (70) attenuating its expression. However, RORα binding promotes Ccl2 expression (70). REV-ERBα has additional anti-inflammatory function via recruitment of the NCoR-HDAC corepressor complex, inhibiting eRNA transcription, and subsequent downstream mRNA transcription of the inflammatory genes Cx3cr1 and Mmp9 (71). In terms of neuroinflammation, REV-ERBα negatively regulates microglial expression of Il1b, Il6, and Ccl2 (72). In a murine model of DSS-induced colitis NF-κB signaling is also increased with Rev-Erbα deletion (72, 73), and interestingly this was shown to promote indirect activation of the NLRP3 inflammasome. A model of fulminant hepatitis (74) demonstrated direct negative regulation of Nlrp3 mRNA by REV-ERBα (74). Thus, a wealth of evidence is emerging that REV-ERBα through various mechanisms, is a suppressor of inflammation in macrophages.
Per1/Per2 also impacts macrophage inflammatory responses. Mice lacking these genes have increased expression of Il1b and Tnfa basally, as well as in response to LPS (57). The repressive effects of the PER complex are mediated through PPARγ. PER1 and PPARγ bind the Ccr2 promoter to inhibit its expression. Deletion of Per1 increases expression of Ccr2 and migratory activity in macrophages (75). Peritoneal macrophages lacking Per2 have heightened responses to TLR9 activation displaying heightened TNFα and IL-12 production (29). CRY also suppresses the inflammatory response in macrophages via negative regulation of the cAMP-PKA-NF-κB pathway (76). Loss of CRY results in constitutive upregulation of Il6, Tnfa, and inos. Thus, the PER/CRY complex is another mechanism of clock-related suppression of inflammatory mediators in macrophages.
Thus, the activation state and regulation of cytokines, chemokines, ROS, miRNAs, and eRNAs in monocytes and macrophages are directly and indirectly targeted by components of the circadian machinery. Circadian regulation of these mediators ensures a closely controlled and appropriately timed macrophage/monocyte response to challenge and infection.
Phagocytosis
A crucial function of macrophages is the ingestion of pathogens via phagocytosis. Diurnal regulation of phagocytosis has been demonstrated in ex vivo peritoneal macrophages (20). Synchronized peritoneal macrophages in vitro, suggested circadian rhythmicity in phagocytic activity (54). Another study showed similar time-of-day variation in peritoneal macrophage phagocytosis ex vivo, but found that this pattern was lost in vivo (77). A recent study demonstrated deletion of Bmal1 creates a more phagocytic, motile, and ultimately antimicrobial macrophage via a RhoA-dependent mechanism, which impacted on Streptococcus pneumoniae lung infection. This is an extremely interesting development, however, whether BMAL1 and other clock components play this role in response to the full range of pathogens and foreign bodies that macrophages can phagocytose, is yet unknown.
Circadian Migration
Appropriate cell migration of immune cells into tissues is critical for protective immunity. Monocyte migration is highly rhythmic and is tightly controlled by autocrine monocyte signaling as well as from the destination tissue. In mice, total-blood leukocyte numbers peak during the behavioral rest phase (ZT5), whereas their migration into bone-marrow and organ tissues peaks during the behavioral active phase (ZT13) (78). Central to regulating monocyte trafficking into tissues is the chemokine receptor CXCR4, whose expression on monocytes peaks at ZT13 (79). Decreased expression of surface CXCR4 abrogates rhythmic diurnal oscillations in monocytes, and has also been shown to affect their homing ability to peripheral organs such as the liver and lung (80). Myeloid deletion of Bmal1 ablates the rhythmic trafficking of monocytes between bone-marrow, blood and peripheral organs, highlighting the importance of intrinsic monocyte clocks in this process (22). Inflammatory monocyte chemotaxis is also dependent on chemokine signaling through the CCL2:CCR2 axis (81). BMAL1 has been shown to control circadian monocyte trafficking by inhibiting the transcription of the chemokines Ccl2 and Ccl8 through recruitment of the polycomb repressive complex 2 (PRC2) (22). Monocyte release into circulation, as well as their eventual infiltration into organ tissues, is clearly dependent on the circadian expression of receptors and chemokines (82).
The transition of inflammatory monocyte from blood into peripheral tissues is also dependent on the leukocyte adhesion cascade. This process involves direct interactions between endothelial vascular cells and infiltrating leukocytes (83). At the start of this process, chemokines are released into the blood from tissue-resident cells. These activate receptors on circulating monocytes that boost the expression of adhesion molecules to facilitate trans-endothelial migration. The molecular clock in monocytes is crucial in this regulation. Deletion of Bmal1 in monocytes results in increased expression of CD18 integrin (a transmembrane receptor facilitating extracellular matrix adhesion), decreased chemokine receptor CCR2, and loss of rhythm in gene expression of L-selectin (a homing receptor that aids binding to endothelial cells). The ultimate consequence of these changes in adhesion and chemokine receptors, is disruption of monocyte trafficking to target tissues (80). Adrenergic nerves of the autonomic nervous system are also important in governing leukocyte recruitment to tissues in a circadian manner through adrenoreceptor signaling in endothelial cells, which leads to increased expression of ICAM1 (an adhesion molecule that aids in transmigration of immune cells) (78). Circadian expression of endothelial ICAM1 and VCAM1 in the lungs and liver, which peak at ZT13, coincides with maximum leukocyte recruitment, and this peak at ZT13 in endothelial ICAM1 and VCAM1 is ablated with Bmal1 deletion (80). Deletion of Bmal1 in endothelial cells ablates the rhythmic migration of all leukocyte subsets, including inflammatory monocytes. Unlike adrenergic signaling, glucocorticoid signaling by the adrenal gland is not required for circadian trafficking of monocytes to organs such as the spleen (23).
Diurnal monocyte trafficking corresponds to the immune response to Listeria. Intraperitoneal infection at ZT8 results in lower counts of colony forming units (CFU), but increased lethality compared to infection at ZT0 (22). Myeloid deletion of Bmal1 results in greater lethality to Listeria infection. This indicates that BMAL1 prevents an overactive and lethal immune response to Listeria by dampening monocyte trafficking. Indeed, myeloid Bmal1 deletion also results in greater numbers of inflammatory monocytes in circulation and in the spleen (22). Collectively, these data demonstrate the central role of the molecular clock in controlling monocyte migration via chemokines, their receptors, and adhesion molecules, which is highly relevant to the immune response and to disease susceptibility and progression.
Conclusions
It is evident that the molecular clock exerts significant control over key functions of innate immunity, in particular monocytes and macrophages. However, our modern 24/7 lifestyles are at odds with this closely regulated endogenous 24-h system of control. This is leading to a global increase in the prevalence of circadian disruption in human health (84). An estimated 20% of the work force are shift workers who, due to their schedule, are at increased risk for obesity, diabetes, cardiovascular disease, and cancer (85, 86). Furthermore, at least 80% of us experience social jet lag (84), defined as misalignment between our body clocks and social behaviors (87). Shift workers have been shown to have altered numbers of immune cells such as monocytes (85) and altered rhythms in cytokine output (88)—and similar effects are likely to be present in all of us who experience significant social jetlag. It is conceivable that these circadian disruptions are an important link between lifestyle, behavior, and disease. Most of the studies discussed in this review used animal models of circadian disruption via jet lag/shift work models, or circadian gene deletion, to study their effects on inflammatory functions of monocytes and macrophages. These will help us to understand and explain key mechanisms in the pathogenesis of many human inflammatory disorders such as cardiovascular disease (89), asthma (90), rheumatoid arthritis (91), and potentially many others. Understanding the underlying molecular mechanisms by which the molecular clock controls metabolism, phagocytosis, pattern recognition, and inflammatory mediator production in monocytes and macrophages will help us to develop new tools and therapies for chronic disease. These will help us to manage the conflicting pressures of modern lifestyles, that have developed in recent decades, with the tightly controlled internal circadian system that has evolved over a much longer timescale.
Author Contributions
GT and JO'S contributed equally in the research, writing, and creating of diagrams for this manuscript. OK aided in writing and critically evaluated the manuscript. AC provided ideas for and aided in writing and critically evaluated the manuscript. JE provided the layout for the manuscript, wrote and performed a critical evaluation of the manuscript, and provided final checks of the manuscript's quality. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the following awards: a Science Foundation Ireland [Career Development Award (17/CDA/4699)] and Marie Sklodowska-Curie Actions (MSCA) award to OK, and a Science Foundation Ireland [Career Development Award (17/CDA/4688)] and an Irish Research Council Laureate Award (IRCLA/2017/110) to AC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. (2012) 485:459–64. doi: 10.1038/nature11088
2. Man K, Loudon A, Chawla A. Immunity around the clock. Science. (2016) 354:999–1003. doi: 10.1126/science.aah4966
3. Riede SJ, van der Vinne V, Hut RA. The flexible clock: predictive and reactive homeostasis, energy balance and the circadian regulation of sleep-wake timing. J Exp Biol. (2017) 220:738–49. doi: 10.1242/jeb.130757
4. Wang GZ, Hickey SL, Shi L, Huang HC, Nakashe P, Koike N, et al. Cycling transcriptional networks optimize energy utilization on a genome scale. Cell Rep. (2015) 13:1868–80. doi: 10.1016/j.celrep.2015.10.043
5. Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med. (1960) 103:142–4. doi: 10.3181/00379727-103-25439
6. Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. (2014) 5:514. doi: 10.3389/fimmu.2014.00514
7. Carter SJ, Durrington HJ, Gibbs JE, Blaikley J, Loudon AS, Ray DW, et al. A matter of time: study of circadian clocks and their role in inflammation. J Leukoc Biol. (2016) 99:549–60. doi: 10.1189/jlb.3RU1015-451R
8. Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. (2014) 111:16219–24. doi: 10.1073/pnas.1408886111
9. Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. (2018) 359:eaao0318. doi: 10.1126/science.aao0318
10. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. (2010) 72:517–49. doi: 10.1146/annurev-physiol-021909-135821
11. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. (2012) 35:445–62. doi: 10.1146/annurev-neuro-060909-153128
12. Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 Is an essential component of the master circadian pacemaker in mammals. Cell. (2000) 103:1009–17. doi: 10.1016/S0092-8674(00)00205-1
13. Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. (1998) 280:1564–9. doi: 10.1126/science.280.5369.1564
14. Langmesser S, Tallone T, Bordon A, Rusconi S, Albrecht U. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol Biol. (2008) 9:41. doi: 10.1186/1471-2199-9-41
15. Yoo S-H, Ko CH, Lowrey PL, Buhr ED, Song E-j, Chang S, et al. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A. (2005) 102:2608–13. doi: 10.1073/pnas.0409763102
16. Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. (2005) 12:441–8. doi: 10.1038/nsmb925
17. Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. (2005) 20:391–403. doi: 10.1177/0748730405277232
18. Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. (2006) 15:R271–R7. doi: 10.1093/hmg/ddl207
19. Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. (2014) 24:90–9. doi: 10.1016/j.tcb.2013.07.002
20. Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. (2007) 30:621–6. doi: 10.1248/bpb.30.621
21. Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun. (2012) 26:407–13. doi: 10.1016/j.bbi.2011.10.001
22. Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. (2013) 341:1483–8. doi: 10.1126/science.1240636
23. Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. (2009) 106:21407–12. doi: 10.1073/pnas.0906361106
24. Medzhitov R, Janeway CA Jr. Decoding the patterns of self and nonself by the innate immune system. Science. (2002) 296:298–300. doi: 10.1126/science.1068883
25. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. (2010) 140:805–20. doi: 10.1016/j.cell.2010.01.022
26. Vidya MK, Kumar VG, Sejian V, Bagath M, Krishnan G, Bhatta R. Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int Rev Immunol. (2018) 37:20–36. doi: 10.1080/08830185.2017.1380200
27. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
28. Curtis AM, Fagundes CT, Yang G, Palsson-McDermott EM, Wochal P, McGettrick AF, et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A. (2015) 112:7231–6. doi: 10.1073/pnas.1501327112
29. Silver Adam C, Arjona A, Walker Wendy E, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. (2012) 36:251–61. doi: 10.1016/j.immuni.2011.12.017
30. Bell JH, Herrera AH, Li Y, Walcheck B. Role of ADAM17 in the ectodomain shedding of TNF-α and its receptors by neutrophils and macrophages. J Leukoc Biol. (2007) 82:173–6. doi: 10.1189/jlb.0307193
31. Silver AC, Buckley SM, Hughes ME, Hastings AK, Nitabach MN, Fikrig E. Daily oscillations in expression and responsiveness of Toll-like receptors in splenic immune cells. Heliyon. (2018) 4:e00579. doi: 10.1016/j.heliyon.2018.e00579
32. O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. (2016) 16:553–65. doi: 10.1038/nri.2016.70
33. Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic instruction of immunity. Cell. (2017) 169:570–86. doi: 10.1016/j.cell.2017.04.004
34. Robles MS, Humphrey SJ, Mann M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. (2017) 25:118–27. doi: 10.1016/j.cmet.2016.10.004
35. Reinke H, Asher G. Circadian clock control of liver metabolic functions. Gastroenterology. (2016) 150:574–80. doi: 10.1053/j.gastro.2015.11.043
36. Wang Y, Song L, Liu M, Ge R, Zhou Q, Liu W, et al. A proteomics landscape of circadian clock in mouse liver. Nat Commun. (2018) 9:1553. doi: 10.1038/s41467-018-03898-2
37. Jacobi D, Liu S, Burkewitz K, Kory N, Knudsen Nelson H, Alexander Ryan K, et al. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 22:709–20. doi: 10.1016/j.cmet.2015.08.006
38. Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, et al. Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. (2017) 25:86–92. doi: 10.1016/j.cmet.2016.09.010
39. Dirk van M, Jan H, Bas H, Frank AJLS, Johanna AJ, Joris H, et al. Demonstration of a day-night rhythm in human skeletal muscle oxidative capacity. Mol Metab. (2016) 5:635–45. doi: 10.1016/j.molmet.2016.06.012
40. McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. (2007) 31:86–95. doi: 10.1152/physiolgenomics.00066.2007
41. Carroll RG, Timmons GA, Cervantes-Silva MP, Kennedy OD, Curtis AM. Immunometabolism around the clock. Trends Mol Med. (2019) 25:612–25. doi: 10.1016/j.molmed.2019.04.013
42. Early JO, Curtis AM. Immunometabolism: is it under the eye of the clock? Semin Immunol. (2016) 28:478–90. doi: 10.1016/j.smim.2016.10.006
43. Kelly B, O'Neill LAJ. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. (2015) 25:771–84. doi: 10.1038/cr.2015.68
44. Huang SC, Smith AM, Everts B, Colonna M, Pearce EL, Schilling JD, et al. Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis is essential for macrophage alternative activation. Immunity. (2016) 45:817–30. doi: 10.1016/j.immuni.2016.09.016
45. Arts RJ, Gresnigt MS, Joosten LA, Netea MG. Cellular metabolism of myeloid cells in sepsis. J Leukoc Biol. (2017) 101:151–64. doi: 10.1189/jlb.4MR0216-066R
46. Deng W, Zhu S, Zeng L, Liu J, Kang R, Yang M, et al. The circadian clock controls immune checkpoint pathway in sepsis. Cell Rep. (2018) 24:366–78. doi: 10.1016/j.celrep.2018.06.026
47. Mills EL, Kelly B, O'Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol. (2017) 18:488. doi: 10.1038/ni.3704
48. Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. (2015) 42:406–17. doi: 10.1016/j.immuni.2015.02.002
49. Chen Y, Zhou Z, Min W. Mitochondria, oxidative stress and innate immunity. Front Physiol. (2018) 9:1487. doi: 10.3389/fphys.2018.01487
50. Burté F, Carelli V, Chinnery PF, Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat Rev Neurol. (2015) 11:11–24. doi: 10.1038/nrneurol.2014.228
51. Dorn II GW. Mitochondrial dynamism and heart disease: changing shape and shaping change. EMBO Mol Med. (2015) 7:865–77. doi: 10.15252/emmm.201404575
52. Wang L, Ishihara T, Ibayashi Y, Tatsushima K, Setoyama D, Hanada Y, et al. Disruption of mitochondrial fission in the liver protects mice from diet-induced obesity and metabolic deterioration. Diabetologia. (2015) 58:2371–80. doi: 10.1007/s00125-015-3704-7
53. Touvier T, De Palma C, Rigamonti E, Scagliola A, Incerti E, Mazelin L, et al. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. (2015) 6:e1663. doi: 10.1038/cddis.2014.595
54. Oliva-Ramirez J, Moreno-Altamirano MM, Pineda-Olvera B, Cauich-Sanchez P, Sanchez-Garcia FJ. Crosstalk between circadian rhythmicity, mitochondrial dynamics and macrophage bactericidal activity. Immunology. (2014) 143:490–7. doi: 10.1111/imm.12329
55. Ruytinx P, Proost P, Van Damme J, Struyf S. Chemokine-induced macrophage polarization in inflammatory conditions. Front Immunol. (2018) 9:1930. doi: 10.3389/fimmu.2018.01930
56. Bosco MC. Macrophage polarization: reaching across the aisle? J Allergy Clin Immunol. (2019) 143:1348–50. doi: 10.1016/j.jaci.2018.12.995
57. Xu H, Li H, Woo SL, Kim SM, Shende VR, Neuendorff N, et al. Myeloid cell-specific disruption of Period1 and Period2 exacerbates diet-induced inflammation and insulin resistance. J Biol Chem. (2014) 289:16374–88. doi: 10.1074/jbc.M113.539601
58. Nelson VL, Nguyen HCB, Garcia-Canaveras JC, Briggs ER, Ho WY, DiSpirito JR, et al. PPARgamma is a nexus controlling alternative activation of macrophages via glutamine metabolism. Genes Dev. (2018) 32:1035–44. doi: 10.1101/gad.312355.118
59. Liu Z, Gan L, Zhang T, Ren Q, Sun C. Melatonin alleviates adipose inflammation through elevating alpha-ketoglutarate and diverting adipose-derived exosomes to macrophages in mice. J Pineal Res. (2018) 64:e12455. doi: 10.1111/jpi.12455
60. Ding S, Lin N, Sheng X, Zhao Y, Su Y, Xu L, et al. Melatonin stabilizes rupture-prone vulnerable plaques via regulating macrophage polarization in a nuclear circadian receptor RORα-dependent manner. J Pineal Res. (2019) 67:e12581. doi: 10.1111/jpi.12581
61. Kim SM, Neuendorff N, Alaniz RC, Sun Y, Chapkin RS, Earnest DJ. Shift work cycle-induced alterations of circadian rhythms potentiate the effects of high-fat diet on inflammation and metabolism. FASEB J. (2018) 32:3085–95. doi: 10.1096/fj.201700784R
62. Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, et al. Dysregulation of inflammatory responses by chronic circadian disruption. J Immunol. (2010) 185:5796–805. doi: 10.4049/jimmunol.1001026
63. Allen NC, Philip NH, Hui L, Zhou X, Franklin RA, Kong Y, et al. Desynchronization of the molecular clock contributes to the heterogeneity of the inflammatory response. Sci Signal. (2019) 12:eaau1851. doi: 10.1126/scisignal.aau1851
64. Early JO, Menon D, Wyse CA, Cervantes-Silva MP, Zaslona Z, Carroll RG, et al. Circadian clock protein BMAL1 regulates IL-1beta in macrophages via NRF2. Proc Natl Acad Sci U S A. (2018) 115:E8460–e8. doi: 10.1073/pnas.1800431115
65. Oishi Y, Hayashi S, Isagawa T, Oshima M, Iwama A, Shimba S, et al. Bmal1 regulates inflammatory responses in macrophages by modulating enhancer RNA transcription. Sci Rep. (2017) 7:7086. doi: 10.1038/s41598-017-07100-3
66. Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, et al. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci U S A. (2013) 110:9897–902. doi: 10.1073/pnas.1120636110
67. Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, et al. Core circadian protein CLOCK is a positive regulator of NF-κB-mediated transcription. Proc Natl Acad Sci U S A. (2012) 109:E2457–E65. doi: 10.1073/pnas.1206274109
68. Pariollaud M, Gibbs JE, Hopwood TW, Brown S, Begley N, Vonslow R, et al. Circadian clock component REV-ERBα controls homeostatic regulation of pulmonary inflammation. J Clin Invest. (2018) 128:2281–96. doi: 10.1172/JCI93910
69. Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. (2012) 109:582–7. doi: 10.1073/pnas.1106750109
70. Sato S, Sakurai T, Ogasawara J, Takahashi M, Izawa T, Imaizumi K, et al. A circadian clock gene, Rev-erbα, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J Immunol. (2014) 192:407–17. doi: 10.4049/jimmunol.1301982
71. Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. (2013) 498:511–5. doi: 10.1038/nature12209
72. Griffin P, Dimitry JM, Sheehan PW, Lananna BV, Guo C, Robinette ML, et al. Circadian clock protein Rev-erbα regulates neuroinflammation. Proc Natl Acad Sci U S A. (2019) 116:5102–7. doi: 10.1073/pnas.1812405116
73. Wang S, Lin Y, Yuan X, Li F, Guo L, Wu B. REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat Commun. (2018) 9:4246. doi: 10.1038/s41467-018-06568-5
74. Pourcet B, Zecchin M, Ferri L, Beauchamp J, Sitaula S, Billon C, et al. Nuclear receptor subfamily 1 group D member 1 regulates circadian activity of NLRP3 inflammasome to reduce the severity of fulminant hepatitis in mice. Gastroenterology. (2018) 154:1449–64.e20. doi: 10.1053/j.gastro.2017.12.019
75. Wang T, Wang Z, Yang P, Xia L, Zhou M, Wang S, et al. PER1 prevents excessive innate immune response during endotoxin-induced liver injury through regulation of macrophage recruitment in mice. Cell Death Dis. (2016) 7:e2176. doi: 10.1038/cddis.2016.9
76. Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci U S A. (2012) 109:12662–7. doi: 10.1073/pnas.1209965109
77. Geiger SS, Curtis AM, O'Neill LA, Siegel RM. Daily variation in macrophage phagocytosis is clock-independent and dispensable for cytokine production. Immunology. (2019) 157:122–36. doi: 10.1111/imm.13053
78. Scheiermann C, Kunisaki Y, Lucas D, Chow A, Jang JE, Zhang D, et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. (2012) 37:290–301. doi: 10.1016/j.immuni.2012.05.021
79. Chong SZ, Evrard M, Devi S, Chen J, Lim JY, See P, et al. CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J Exp Med. (2016) 213:2293–314. doi: 10.1084/jem.20160800
80. He W, Holtkamp S, Hergenhan SM, Kraus K, de Juan A, Weber J, et al. Circadian expression of migratory factors establishes lineage-specific signatures that guide the homing of leukocyte subsets to tissues. Immunity. (2018) 49:1175–90.e7. doi: 10.1016/j.immuni.2018.10.007
81. Volpe S, Cameroni E, Moepps B, Thelen S, Apuzzo T, Thelen M. CCR2 Acts as scavenger for CCL2 during monocyte chemotaxis. PLoS One. (2012) 7:e37208. doi: 10.1371/journal.pone.0037208
82. Pick R, He W, Chen C-S, Scheiermann C. Time-of-day-dependent trafficking and function of leukocyte subsets. Trends Immunol. (2019) 40:524–37. doi: 10.1016/j.it.2019.03.010
83. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. (2007) 7:678–89. doi: 10.1038/nri2156
84. Sulli G, Lam MTY, Panda S. Interplay between circadian clock and cancer: new frontiers for cancer treatment. Trends Cancer. (2019) 5:475–94. doi: 10.1016/j.trecan.2019.07.002
85. Loef B, Nanlohy NM, Jacobi RHJ, van de Ven C, Mariman R, van der Beek AJ, et al. Immunological effects of shift work in healthcare workers. Sci Rep. (2019) 9:18220. doi: 10.1038/s41598-019-54816-5
86. Wang XS, Armstrong ME, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: the epidemiological evidence. Occup Med. (2011) 61:78–89. doi: 10.1093/occmed/kqr001
87. Wittmann M, Dinich J, Merrow M, Roenneberg T. Social jetlag: misalignment of biological and social time. Chronobiol Int. (2006) 23:497–509. doi: 10.1080/07420520500545979
88. Cuesta M, Boudreau P, Dubeau-Laramée G, Cermakian N, Boivin DB. Simulated night shift disrupts circadian rhythms of immune functions in humans. J Immunol. (2016) 196:2466–75. doi: 10.4049/jimmunol.1502422
89. McAlpine CS, Swirski FK. Circadian influence on metabolism and inflammation in atherosclerosis. Circ Res. (2016) 119:131–41. doi: 10.1161/CIRCRESAHA.116.308034
90. Durrington HJ, Farrow SN, Loudon AS, Ray DW. The circadian clock and asthma. Thorax. (2014) 69:90–2. doi: 10.1136/thoraxjnl-2013-203482
Keywords: circadian, macrophage, monocyte, molecular clock, inflammation, cell migration, immunometabolism, phagocytosis
Citation: Timmons GA, O'Siorain JR, Kennedy OD, Curtis AM and Early JO (2020) Innate Rhythms: Clocks at the Center of Monocyte and Macrophage Function. Front. Immunol. 11:1743. doi: 10.3389/fimmu.2020.01743
Received: 24 April 2020; Accepted: 30 June 2020;
Published: 04 August 2020.
Edited by:
Christoph Scheiermann, Ludwig Maximilian University of Munich, GermanyReviewed by:
Bert Maier, Charité—University Medicine Berlin, GermanyAdam C. Silver, University of Hartford, United States
Copyright © 2020 Timmons, O'Siorain, Kennedy, Curtis and Early. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Annie M. Curtis, YW5uaWVjdXJ0aXNAcmNzaS5jb20=; James O. Early, amFtZXNlYXJseUByY3NpLmNvbQ==
†These authors have contributed equally to this work
 George A. Timmons
George A. Timmons James R. O'Siorain1
James R. O'Siorain1 Oran D. Kennedy
Oran D. Kennedy Annie M. Curtis
Annie M. Curtis James O. Early
James O. Early