
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 28 August 2020
Sec. Viral Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01637
This article is part of the Research Topic Sensing DNA in Antiviral Innate Immunity View all 14 articles
 Yue Lu1,2,3,4
Yue Lu1,2,3,4 Leiliang Zhang1,2,3,4,5*
Leiliang Zhang1,2,3,4,5*As pattern recognition receptors, cytosolic DNA sensors quickly induce an effective innate immune response. Poxvirus, a large DNA virus, is capable of evading the host antiviral innate immune response. In this review, we summarize the latest studies on how poxvirus is sensed by the host innate immune system and how poxvirus-encoded proteins antagonize DNA sensors. A comprehensive understanding of the interplay between poxvirus and DNA-sensing antiviral immune responses of the host will contribute to the development of new antiviral therapies and vaccines in the future.
Poxvirus is a double-stranded DNA (dsDNA) virus (1, 2) that replicates completely in the cytoplasm. Members of the Poxviridae family include variola virus (VARV), vaccinia virus (VACV), ectromelia virus (ECTV), and monkeypox virus (MPXV). VACV is a prototype member of the Orthopoxvirus genus of the Poxviridae family and has been used as a live vaccine for smallpox eradication (3). Interest in VACV persists because it is an excellent model for studying host pathogen interactions and cell biology (4). ECTV is a mouse-specific pathogen that has been used as a model to study the pathogenesis and immunobiology of Orthopoxvirus infection (5, 6).
The innate immune response against viruses is not only the first line of defense against viral infection but is also important for the establishment of adaptive immunity against viruses. The recognition of the viral DNA genome by DNA sensors, including cyclic GMP–AMP synthase (cGAS), DNA-dependent protein kinase (DNA-PK), and IFN-γ inducible protein 16 (IFI16), is the first step in the innate immune response (7, 8). Next, innate immune signal transduction is initiated by activating adaptor proteins, such as stimulator of interferon genes (STING), resulting in the production of a large number of defense molecules in the host, including interferons (IFNs) and pro-inflammatory cytokines and chemokines (9–11).
The evolutionary arms race between the virus and the host leads to the virus-mediated antagonism of antiviral immunity. Viruses hide their DNA from cellular sensing systems and/or inactivate sensors and downstream signal transduction pathways. These viral strategies include separation or modification of viral nucleic acids, interfering with specific post-translational modifications of pattern recognition receptors (PRRs) or their adaptors, degradation or cleavage of PRRs or adaptors, and separating or repositioning PRRs. For instance, there are a variety of proteins that inhibit the activation of the transcription factors interferon regulatory factor 3 (IRF3) and nuclear factor kappa B (NF-κB) or the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway (2).
Poxvirus encodes the largest number of immune antagonistic virus proteins, thereby showing the most diverse immune escape strategies (4). During infection, these immunomodulatory proteins are delivered to the cytoplasm of the host cell to combat the innate immune response (12, 13). The conserved central region of the poxvirus genome encodes the open reading frames (ORFs) essential for virus replication. The other ORFs are non-essential for viral replication in cell culture (14), with most associated with targeting the innate immune system (15, 16).
In this review, we summarize the DNA-sensing signal pathways in poxvirus-infected cells. In particular, we focus on DNA sensors (cGAS/DNA-PK/IFI16), an adaptor protein (STING), and host defense molecules (IFNs/cytokines). We also describe how poxvirus targets DNA sensors to abrogate the antiviral immune response. Understanding antiviral immunity and poxvirus-mediated antagonism mechanisms may guide the development of live attenuated vaccines and antiviral therapies.
In 2013, Sun et al. discovered a new DNA sensor, cGAS, which advanced our understanding of innate DNA sensing (17). cGAS, an enzyme belonging to the ancient oligoadenylate synthase (OAS) protein family (18), is a universal cytoplasmic DNA sensor upstream of STING. cGAS recognizes a large number of cytoplasmic DNA viruses (HSV-1, KSHV, and VACV) and retroviruses (HIV-1, HIV-2) (19–24). cGAS is activated upon binding to DNA, which catalyzes the production of 2'3'-cGAMP from ATP and GTP, resulting in the binding of second messenger cyclic GMP–AMP (cGAMP) to STING (17, 25–29). As an adaptor protein, STING recruits TBK1, which phosphorylates IRF3. Then, IRF3 is relocated to the nucleus to induce IFN and thus establishes an antiviral state (19, 30–33). NF-κB is also activated by STING (32).
The cGAS–STING pathway is very important for sensing ECTV infection, inducing type I IFN production and controlling ECTV replication (34). In the lymph nodes of mice infected with ECTV, inflammatory monocytes (IMOs) are the main cells producing type I IFN in draining lymph nodes (DLNs). To induce the expression of IFN and pro-inflammatory cytokines, IMOs require STING–IRF7 and STING–NF-κB (10).
By using cGAS-deficient mice, researchers showed that type I IFN is not produced during VACV infection (35, 36). In addition, cGAMP, produced by cGAS in virus-infected cells, can be transferred to uninfected neighboring cells through gap junctions, where it promotes STING activation and antiviral immunity reactions independent of type I IFN (37).
Interferon-induced oligoadenylate synthetase-like (OASL) binds specifically to cGAS and inhibits cGAS enzyme activity in the process of DNA virus infection, which inhibits IFN induction and promotes DNA virus replication through the cGAS–STING DNA sensing pathway (38). Deletion of human OASL and mouse OASL2 can inhibit DNA virus infection. OASL1 and OASL2 are negative feedback regulators of cGAS and inhibit cGAS-mediated type I IFN induction (38).
The modified VACV Ankara strain (MVA) has been designed as a vaccine vector (39–41), and it can effectively prevent VARV and MPXV infection (42, 43). IFN in MVA-infected conventional dendritic cells (cDCs) is produced independently of the RNA-sensing pathway mediated by MDA5, MAVS, TLR3, or TRIF and is not affected by the absence of TLR9/MyD88 in the DNA sensing pathway in vivo. The cGAS/STING-mediated DNA-sensing pathway plays a key role in MVA-induced IFN production in CDCs. MVA infection of cDCs triggers the phosphorylation of TBK1 and IRF3, which is abolished in the absence of cGAS and STING. Similar results were also observed in mouse models (44).
Of the 10 TLRs found in humans, TLR9 is the only known DNA sensor. TLR9 specifically recognizes the unmethylated CpG motif in dsDNA (CpG DNA), which is common in bacterial and viral genomes (32, 45–47). TLR9 recruits the adaptor protein MyD88 and then recruits tumor necrosis factor receptor associated factor 6 (TRAF6) and IκB kinase (IKK) complexes; the former leads to the activation of IRF7 and ultimately induces the production of type I IFN (48, 49), and the latter leads to the activation of NF-κB, resulting in the induction of inflammatory cytokines (50).
TLR9/MyD88 sensing increased the expression of the NKG2D ligand in virus-infected migratory dendritic cells (mDCs), and induced production of IFN-γ in classical NK cells and innate lymphoid cells (ILCs). IFN-γ induces CXCL9 in uninfected IMOs and induces the recruitment of protective NK cells to DLNs (51). In CD11c+ cells, MyD88–IRF7 recruit IMOs to DLNs, and although the TLR9–MyD88–IRF7 signaling pathway is necessary for IMOs recruitment to DLNs, it is not directly necessary for type I IFN production. The induction of type I IFN in DLNs during ECTV infection is due to the indirect recognition of the virus by the TLR9–MyD88–IRF7 and STING–IRF7/NF-κB pathways (52). Compared with wild-type mice, mice lacking TLR9 and MyD88 showed higher viral loads, more severe pathological liver and spleen conditions, and increased susceptibility to ECTV infection (53–55). C57BL/6 mice lacking IRF7 and NF-κB, which are downstream targets of TLR9–MyD88 and STING, are highly susceptible to ECTV infection (52).
Absent in melanoma 2 (AIM2), a member of the PYHIN protein family, is a receptor of cytoplasmic DNA. AIM2 senses viral DNA and can activate the inflammasome pathway (56, 57), which plays an important role in the production of pro-inflammatory cytokines and the clearance of infected cells through pyroptosis (58). After AIM2 binds to DNA through its HIN200 domain, caspase-1 is recruited and activated, leading to the production of inflammatory cytokines, including IL-1β and IL-18. Disabling AIM2 inhibits caspase-1 activation by cytoplasmic dsDNA and VACV infection (59, 60). More importantly, AIM2-deficient cells have a defective innate immune response to VACV (61).
IFI16, a member of the PYHIN protein family, recognizes the DNA virus genome in the nucleus and activates antiviral gene expression and the inflammasome-mediated immune response. IFI16 is mainly located in the nucleus but can also shuttle between the cytoplasm and nucleus in different types of cells (62). IFI16 could bind to the dsDNA fragment of 70 bp from the VACV genome (48). It can also interact with STING to induce the TBK1-dependent IFN-β response. The nuclear induction of IFI16 upon cell exposure to viral DNA activates the inflammasome pathway through ASC and caspase-1, resulting in the production of IL-1β and IL-18 (63).
Both IFI16 and cGAS are necessary for the activation of STING, which is induced by cGAMP. They interact with STING to promote its phosphorylation and translocation. IFI16 is the main nuclear DNA receptor, while cGAS plays an auxiliary role. For example, upon the stabilization of IFI16 to initiate or prolong signal enhancement, the synergistic effect of IFI16 and cGAS can induce immune signaling in response to exogenous DNA in the nucleus (64, 65).
DNA-PK is a protein kinase that binds to cytoplasmic DNA. It is composed of Ku70, Ku80, and catalytic subunit DNA-PKCs. In the case of VACV infection, DNA-PK relies on STING, TBK1, and IRF3 to induce cytokine production (32, 45–47). PRR detection of DNA triggers the production of type I IFN, cytokines, and chemokines through the STING pathway (51).
DNA viruses usually release genomic DNA into the nucleus of host cells after entry. Heterogeneous nuclear ribonucleoprotein A2B1 (HnRNPA2B1) recognizes viral DNA, undergoes homodimerization, and is demethylated by arginine demethylase JMJD6 at Arg226. This modification results in hnRNPA2B1 translocation to the cytoplasm and activation of the TBK1–IRF3 pathway, which enhances IFN-α/β production. In addition, hnRNPA2B1 promotes the modification of N6-methyladenosine (m6A) and the nuclear and cytoplasmic transport of cGAS, IFI16, and spiny mRNA. These factors mediate the amplified activation of the cytoplasmic TBK1–IRF3 pathway. Therefore, nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses (52).
RNA polymerase III is a new type of dsDNA cytoplasmic DNA sensor, and RIG-I is pivotal in sensing viral RNA. AT-rich dsDNA serves as a template for this DNA sensor, RNA polymerase III converts poly(dA:dT) to poly(A:U)-rich dsRNA, which, in turn, serves as a RIG-I agonist. Then, activation of RIG-I by this dsRNA induces the production of type I IFN and activation of the transcription factor NF-κB (53–55).
Poxvirus inhibits innate immunity through diverse mechanisms that involve multiple players including sensors, adaptors, and effectors. In this review, we focus on sensors and the most recent studies on adaptors and effectors. Therefore, only a small number of poxvirus immune antagonistic proteins are discussed. More poxvirus immune evasion mechanisms have been summarized in previous studies (4, 66–69).
cGAS is the main sensor that mediates IRF activation and ISG response to VACV lacking F17 (44, 70). The poxvirus F17 protein hijacks the mammalian target of rapamycin (mTOR) regulatory factors Raptor and Rictor, leading to an mTOR imbalance. Excess mTOR accumulates in the Golgi apparatus and causes mTOR-dependent cGAS degradation, thus inactivating the cGAS–STING pathway (71). In contrast, when VACV lacking F17 infects the cells, cGAS activates STING. Then, STING is phosphorylated, dimerized, and translocated from the endoplasmic reticulum (ER) to the perinuclear region, where it mediates the activation of IRF3 (72).
DNA-PK can be antagonized by VACV proteins C16 and C4. C16 and C4 bind to Ku and block the binding of Ku to DNA (73), resulting in the reduced production of cytokines and chemokines, decreased recruitment of inflammatory cells, and inhibition of IRF3 signaling. The response to VACV infection is weakened in cells and mice lacking DNA-PK components (49). A model infected with C16-knockout VACV show fewer signs of disease and upregulated cytokine synthesis (73, 74). C4 inhibits NF-κB signaling (75) and cytokine production in vitro and in vivo. The loss of C4 enhances the recruitment and activation of cells involved in innate and acquired immunity.
Georgana et al. studied the activation of innate immune signals by four different VACV prototypes. They found that the virulent Copenhagen and Western Reserve VACV strains inhibited STING dimerization and phosphorylation during infection and in response to transfected DNA and cGAMP, thus effectively inhibiting DNA sensing and the activation of IRF3. However, an attenuated MVA strain showed the opposite result, and IRF3 was activated by cGAS and STING after infection (70). Georgana et al. found that virus-encoded protein C16 is a viral DNA sensing inhibitor that acts upstream of STING and has the ability to block STING activation (70).
The mutation of serine to alanine in the IκBα-like motif of A49 prevented β-TrCP binding, stabilized p-IκBα and inhibited the activation of NF-κB (76). B14 targets IKK complex and inhibits the activation of NF-κB in response to TNF-α, IL-1β, Poly(I:C), and PMA (77). The intracellular immunomodulatory proteins K1L, N1L, and A52R can inhibit the NF-κB signaling pathway (44, 78, 79).
VACV virulence factor N1 is a 14 kDa cytoplasmic protein that facilitates an increase in virulence (80, 81) and plays an inhibitory role in the cGAS–STING–IRF3-dependent cytoplasmic DNA-sensing pathway and in IFN-β gene induction (82).
Poxvirus protein serine protease inhibitor 2 (SPI-2) and cytokine response modifier (CrmA) are involved in a variety of poxvirus immune escape strategies. SPI-2 and CrmA target caspase-1 to prevent apoptosis and cytokine activation. The ectopic expression of SPI-2 or CrmA inhibits the induction of IFN-β and its downstream genes. SPI-2 and CrmA can also bind to TBK1 and IKKε to disrupt the STING-TBK1/IKK ε-IRF3 complex, which is a newly discovered mechanism of the SPI-2/CrmA–mediated immune escape of poxvirus (83).
VACV expresses many proteins that antagonize the IFN system. C6 is a multifunctional IFN inhibitor expressed prior to viral genome replication and resides in the cytoplasm and nucleus. It can reduce IFN production and inhibit IFN-induced signal transduction, thus inhibiting ISG expression (84). C6 inhibits the activation of IRF3 by binding to TBK1 in the cytoplasm, thus blocking the induction of IFN by IRF3 (84).
Poxvirus encodes several soluble IFN receptors. For instance, VACV B8 interacts with IFN-γ and prevents it from binding to IFN-γ receptors (85–87). VACV B18 binds to type I IFN and blocks the signal transduction of IFNAR (88–91).
In this review, we discussed the interplay between poxvirus and host antiviral innate immune factors, particularly focusing on the STING pathway (Figure 1). The sensor proteins upstream of STING are cGAS, DNA-PK, and IFI16. There are two pathways of downstream STING effectors: TBK1-IRF3 and IKK-NF-κB. These two signaling pathways induce the production of IFNs and cytokines. In addition, we also described other signaling pathways that trigger the innate immune response.
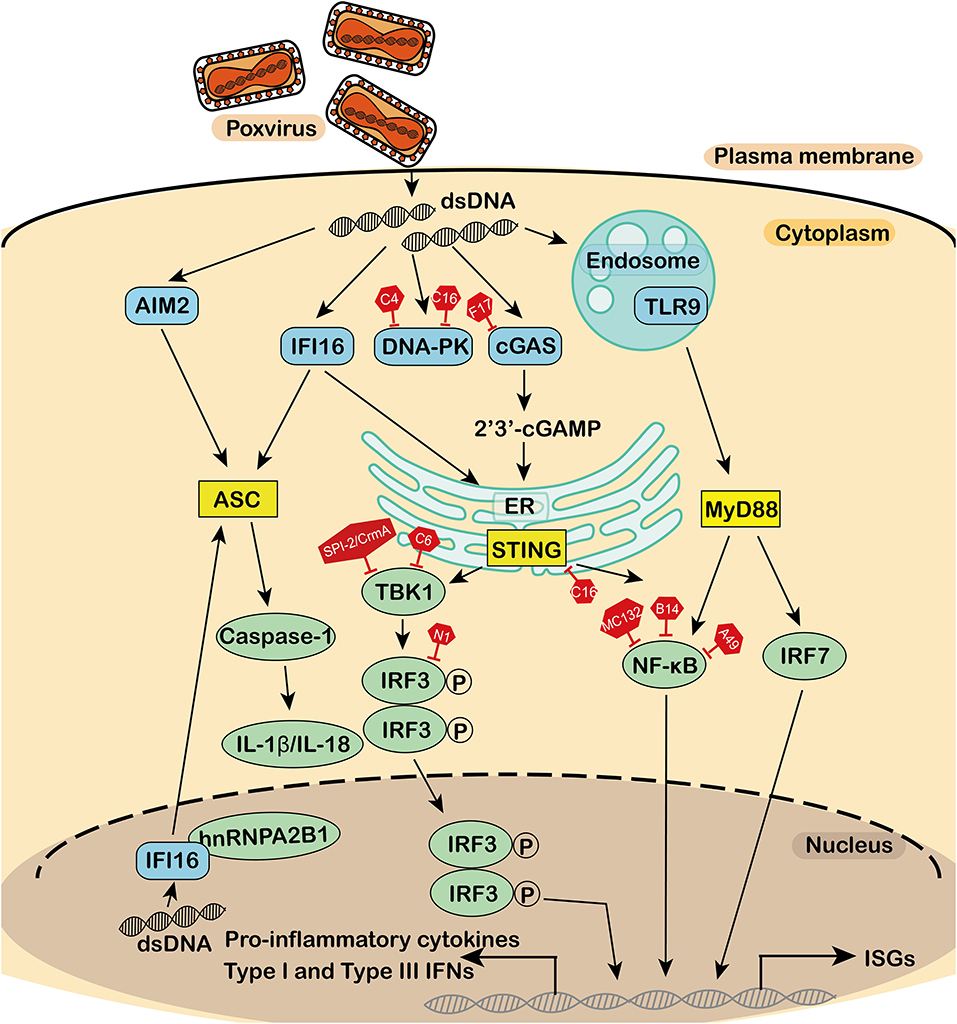
Figure 1. Antagonism of the DNA sensor by poxvirus. During poxvirus infection, the cytosolic DNA sensor activates the adaptor, which in turn activates a series of downstream effectors to produce interferons, cytokines, and interleukins for an antiviral immune response. DNA sensors, adaptors, effectors, and virus-encoded inhibitors are in blue, yellow, green, and red, respectively. dsDNA, double-stranded DNA; TLR9, toll-like receptor 9; IFI16, interferon-γ inducible protein 16; cGAS, cyclic guanosine monophosphate-adenosine monophosphate synthase; DNA-PK, DNA-dependent protein kinase; AIM2, absent in melanoma 2; MyD88, myeloid differentiation factor 88; STING, stimulator of interferon genes; ASC, apoptosis-associated speck-like protein containing a CARD; IFN, interferons; CK, cytokines; NF-κB, nuclear factor κB; TBK1, TANK-binding kinase 1; IRF3, interferon regulatory factor 3; P, phosphorylation; IRF7, interferon regulatory factor 7; IL, interleukin; HnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; ER, endoplasmic reticulum; ISGs, IFN stimulating genes.
Subcellular compartments are involved in the spatiotemporal interplay between poxviruses and DNA sensing molecules. TLR9 is located in endosomes, while STING is located in the ER. Yip1 Domain Family Member 5 (YIPF5) is recycled between the ER and the Golgi, involving the maintenance of the Golgi structure. YIPF5 recruits STING to COPII vesicles and facilitates STING trafficking from the ER to the Golgi apparatus, triggering type I IFN production (92). Interestingly, cGAS and IFI16 are located in the nucleus and cytoplasm. Acetylation of nuclear localization signal sequences targets IFI16 to the cytoplasm, thus fine-tuning the subcellular distribution of IFI16. Endogenous cGAS seems to be uniformly distributed in the cytoplasm and nucleus (93). Although poxvirus replicates in the cytoplasm, many viral proteins are located in the nucleus. cGAS and IFI16 are partially localized to the nucleus; however, no nuclear poxvirus proteins are reported to antagonize cGAS or IFI16 in the nucleus.
To successfully survive, the poxvirus genome encodes a number of immunomodulatory proteins to escape the innate immune response. The key challenge is to translate the viral evasion mechanism into useful applications for the development of new vaccines and antiviral drugs. Knockouts of immunomodulatory proteins or the depletion of specific viral PRR antagonistic mechanisms may lead to changes in virulence and/or the immune response, which may effectively induce long-lasting immune antiviral responses and may improve the immunogenicity of viral vectors.
Through these recent achievements, we have gained a richer understanding of viral evasion mechanisms in host cells. However, there are gaps that need to be investigated further. Firstly, how the interplay between poxvirus and innate immune response affects human viral diseases is unknown. Secondly, what are the relative contributions of the many DNA sensors required for poxvirus sensing? There is no definite answer to date. Finally, what might be the unique viral ligands that activate distinct DNA sensors? Are these DNA sensors involved in different cell types? Determining the molecular mechanism of poxvirus evasion will not only greatly contribute to important insights for the development of antiviral drugs and vaccines but will also provide a viral model for the future study of viral antagonism to host immunity.
LZ conceived the work and modified the manuscript. YL drafted the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from National Natural Science Foundation of China (81871663 and 81672035) and Academic Promotion Programme of Shandong First Medical University (2019LJ001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Moss B. (2015). Poxvirus membrane biogenesis. Virology. 479–80:619–26. doi: 10.1016/j.virol.2015.02.003
2. Smith GL, Talbot-Cooper C, Lu Y. How does vaccinia virus interfere with interferon? Adv Virus Res. (2018) 100:355–78. doi: 10.1016/bs.aivir.2018.01.003
3. Andzhaparidze OG, Slepushkin AN. The 30th anniversary of the adoption by the World Health Organization of the program for smallpox eradication throughout the world. Vopr Virusol. (1988) 33:517–23.
4. Smith GL, Benfield CTO, Maluquer de Motes C, Mazzon M, Ember SWJ, Ferguson BJ, et al. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol. (2013) 94:2367–92. doi: 10.1099/vir.0.055921-0
5. Garver J, Weber L, Vela EM, Anderson M, Warren R, Merchlinsky M, et al. Ectromelia virus disease characterization in the BALB/c mouse: a surrogate model for assessment of smallpox medical countermeasures. Viruses. (2016) 8:70203. doi: 10.3390/v8070203
6. Usme-Ciro JA, Paredes A, Walteros DM, Tolosa-Perez EN, Laiton-Donato K, Pinzon MD, et al. Detection and molecular characterization of zoonotic poxviruses circulating in the Amazon Region of Colombia, 2014. Emerg Infect Dis. (2017) 23:649–53. doi: 10.3201/eid2304.161041
7. Luecke S, Paludan SR. Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine. (2017) 98:4–14. doi: 10.1016/j.cyto.2016.10.003
8. Ma Z, Ni G, Damania B. Innate sensing of DNA virus genomes. Annu Rev Virol. (2018) 5:341–62. doi: 10.1146/annurev-virology-092917-043244
9. Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci USA. (2013) 110:E4492–501. doi: 10.1073/pnas.1316194110
10. Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. (2014) 32:461–88. doi: 10.1146/annurev-immunol-032713-120156
11. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. (2015) 33:257–90. doi: 10.1146/annurev-immunol-032414-112240
12. Hwang S, Kim KS, Flano E, Wu TT, Tong LM, Park AN, et al. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe. (2009) 5:166–78. doi: 10.1016/j.chom.2008.12.013
13. Schmidt FI, Bleck CK, Reh L, Novy K, Wollscheid B, Helenius A, et al. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep. (2013) 4:464–76. doi: 10.1016/j.celrep.2013.06.028
14. Gubser C, Hue S, Kellam P, Smith GL. Poxvirus genomes: a phylogenetic analysis. J Gen Virol. (2004) 85:105–17. doi: 10.1099/vir.0.19565-0
15. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. (2008) 8:911–22. doi: 10.1038/nri2436
16. Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, et al. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. (2012) 150:831–41. doi: 10.1016/j.cell.2012.05.049
17. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. (2013) 339:786–91. doi: 10.1126/science.1232458
18. Kranzusch PJ, Wilson SC, Lee AS, Berger JM, Doudna JA, Vance RE. Ancient origin of cGAS-STING reveals mechanism of universal 2',3' cGAMP signaling. Mol Cell. (2015) 59:891–903. doi: 10.1016/j.molcel.2015.07.022
19. Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. (2009) 106:8653–8. doi: 10.1073/pnas.0900850106
20. Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. (2013) 341:903–6. doi: 10.1126/science.1240933
21. Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. (2013) 39:1132–42. doi: 10.1016/j.immuni.2013.11.002
22. Gao J, Tao J, Liang W, Jiang Z. Cyclic (di)nucleotides: the common language shared by microbe and host. Curr Opin Microbiol. (2016) 30:79–87. doi: 10.1016/j.mib.2015.12.005
23. Tao J, Zhou X, Jiang Z. cGAS-cGAMP-STING: the three musketeers of cytosolic DNA sensing and signaling. IUBMB Life. (2016) 68:858–70. doi: 10.1002/iub.1566
24. Kieser KJ, Kagan JC. Multi-receptor detection of individual bacterial products by the innate immune system. Nat Rev Immunol. (2017) 17:376–90. doi: 10.1038/nri.2017.25
25. Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. (2013) 498:380–4. doi: 10.1038/nature12306
26. Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. (2013) 3:1355–61. doi: 10.1016/j.celrep.2013.05.009
27. Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, et al. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. (2013) 153:1094–107. doi: 10.1016/j.cell.2013.04.046
28. Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. (2013) 339:826–30. doi: 10.1126/science.1229963
29. Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. (2013) 51:226–35. doi: 10.1016/j.molcel.2013.05.022
30. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. (2008) 455:674–8. doi: 10.1038/nature07317
31. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. (2008) 29:538–50. doi: 10.1016/j.immuni.2008.09.003
32. Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. (2012) 5:ra20. doi: 10.1126/scisignal.2002521
33. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. (2015) 347:aaa2630. doi: 10.1126/science.aaa2630
34. Cheng WY, He XB, Jia HJ, Chen GH, Jin QW, Long ZL, et al. The cGas-sting signaling pathway is required for the innate immune response against ectromelia virus. Front Immunol. (2018) 9:1297. doi: 10.3389/fimmu.2018.01297
35. Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. (2013) 341:1390–4. doi: 10.1126/science.1244040
36. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. (2014) 505:691–5. doi: 10.1038/nature12862
37. Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. (2013) 503:530–4. doi: 10.1038/nature12640
38. Ghosh A, Shao L, Sampath P, Zhao B, Patel NV, Zhu J, et al. Oligoadenylate-synthetase-family protein OASL inhibits activity of the DNA sensor cGAS during DNA virus infection to limit interferon production. Immunity. (2019) 50:51–63 e55. doi: 10.1016/j.immuni.2018.12.013
39. Meyer H, Sutter G, Mayr A. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J Gen Virol. (1991) 72:1031–8. doi: 10.1099/0022-1317-72-5-1031
40. McCurdy LH, Larkin BD, Martin JE, Graham BS. Modified vaccinia Ankara: potential as an alternative smallpox vaccine. Clin Infect Dis. (2004) 38:1749–53. doi: 10.1086/421266
41. Vollmar J, Arndtz N, Eckl KM, Thomsen T, Petzold B, Mateo L, et al. Safety and immunogenicity of IMVAMUNE, a promising candidate as a third generation smallpox vaccine. Vaccine. (2006) 24:2065–70. doi: 10.1016/j.vaccine.2005.11.022
42. Sutter G, Staib C. Vaccinia vectors as candidate vaccines: the development of modified vaccinia virus Ankara for antigen delivery. Curr Drug Targets Infect Disord. (2003) 3:263–71. doi: 10.2174/1568005033481123
43. Gomez CE, Najera JL, Krupa M, Esteban M. The poxvirus vectors MVA and NYVAC as gene delivery systems for vaccination against infectious diseases and cancer. Curr Gene Ther. (2008) 8:97–120. doi: 10.2174/156652308784049363
44. Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, et al. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog. (2014) 10:e1003989. doi: 10.1371/journal.ppat.1003989
45. Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. (2008) 451:725–9. doi: 10.1038/nature06537
46. Zyzak J, Mitkiewicz M, Leszczyńska E, Reniewicz P, Moynagh PN, Siednienko J. HSV-1/TLR9-mediated IFNβ and TNFα induction is mal-dependent in macrophages. J Innate Immun. (2019) 18:1–12. doi: 10.1159/000504542
47. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. (2009) 461:788–92. doi: 10.1038/nature08476
48. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. (2010) 11:997–1004. doi: 10.1038/ni.1932
49. Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. (2012) 1:e00047. doi: 10.7554/eLife.00047.012
50. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. (2011) 34:637–50. doi: 10.1016/j.immuni.2011.05.006
51. Wong E, Xu RH, Rubio D, Lev A, Stotesbury C, Fang M, et al. Migratory dendritic cells, group 1 innate lymphoid cells, and inflammatory monocytes collaborate to recruit NK cells to the virus-infected lymph node. Cell Rep. (2018) 24:142–54. doi: 10.1016/j.celrep.2018.06.004
52. Xu RH, Wong EB, Rubio D, Roscoe F, Ma X, Nair S, et al. Sequential activation of two pathogen-sensing pathways required for type i interferon expression and resistance to an acute DNA virus infection. Immunity. (2015) 43:1148–59. doi: 10.1016/j.immuni.2015.11.015
53. Samuelsson C, Hausmann J, Lauterbach H, Schmidt M, Akira S, Wagner H, et al. Survival of lethal poxvirus infection in mice depends on TLR9, and therapeutic vaccination provides protection. J Clin Invest. (2008) 118:1776–84. doi: 10.1172/JCI33940
54. Sutherland DB, Ranasinghe C, Regner M, Phipps S, Matthaei KI, Day SL, et al. Evaluating vaccinia virus cytokine co-expression in TLR GKO mice. Immunol Cell Biol. (2011) 89:706–15. doi: 10.1038/icb.2010.157
55. Rubio D, Xu RH, Remakus S, Krouse TE, Truckenmiller ME, Thapa RJ, et al. Crosstalk between the type 1 interferon and nuclear factor kappa B pathways confers resistance to a lethal virus infection. Cell Host Microbe. (2013) 13:701–10. doi: 10.1016/j.chom.2013.04.015
56. Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. (2009) 10:266–72. doi: 10.1038/ni.1702
57. Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. (2009) 323:1057–60. doi: 10.1126/science.1169841
58. Kanneganti TD. Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol. (2010) 10:688–98. doi: 10.1038/nri2851
59. Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. (2009) 458:509–13. doi: 10.1038/nature07710
60. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. (2009) 458:514–8. doi: 10.1038/nature07725
61. Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. (2010) 11:395–402. doi: 10.1038/ni.1864
62. Dell'Oste V, Gatti D, Giorgio AG, Gariglio M, Landolfo S, De Andrea M. The interferon-inducible DNA-sensor protein IFI16: a key player in the antiviral response. New Microbiol. (2015) 38:5–20.
63. Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. (2011) 9:363–75. doi: 10.1016/j.chom.2011.04.008
64. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. (2000) 408:740–5. doi: 10.1038/35047123
65. Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. (2004) 5:190–8. doi: 10.1038/ni1028
66. Rahman MM, McFadden G. Modulation of NF-κB signalling by microbial pathogens. Nat Rev Microbiol. (2011) 9:291–306. doi: 10.1038/nrmicro2539
67. Brady G, Bowie AG. Innate immune activation of NFκB and its antagonism by poxviruses. Cytokine Growth Factor Rev. (2014) 25:611–20. doi: 10.1016/j.cytogfr.2014.07.004
68. Bidgood SR, Mercer J. Cloak and dagger: alternative immune evasion and modulation strategies of poxviruses. Viruses. (2015) 7:4800–25. doi: 10.3390/v7082844
69. Shisler JL. Immune evasion strategies of molluscum contagiosum virus. Adv Virus Res. (2015) 92:201–52. doi: 10.1016/bs.aivir.2014.11.004
70. Georgana I, Sumner RP, Towers GJ, Maluquer de Motes C. Virulent poxviruses inhibit DNA sensing by preventing STING activation. J Virol. (2018) 92:17. doi: 10.1128/JVI.02145-17
71. Meade N, King M, Munger J, Walsh D. mTOR dysregulation by vaccinia virus F17 controls multiple processes with varying roles in infection. J Virol. (2019) 93:19. doi: 10.1128/JVI.00784-19
72. Meade N, Furey C, Li H, Verma R, Chai Q, Rollins MG, et al. Poxviruses evade cytosolic sensing through disruption of an mTORC1-mTORC2 regulatory circuit. Cell. (2018) 174:1143–57 e1117. doi: 10.1016/j.cell.2018.06.053
73. Peters NE, Ferguson BJ, Mazzon M, Fahy AS, Krysztofinska E, Arribas-Bosacoma R, et al. A mechanism for the inhibition of DNA-PK-mediated DNA sensing by a virus. PLoS Pathog. (2013) 9:e1003649. doi: 10.1371/journal.ppat.1003649
74. Fahy AS, Clark RH, Glyde EF, Smith GL. Vaccinia virus protein C16 acts intracellularly to modulate the host response and promote virulence. J Gen Virol. (2008) 89:2377–87. doi: 10.1099/vir.0.2008/004895-0
75. Ember SWJ, Ren H, Ferguson BJ, Smith GL. Vaccinia virus protein C4 inhibits NF-kappaB activation and promotes virus virulence. J Gen Virol. (2012) 93:2098–108. doi: 10.1099/vir.0.045070-0
76. Mansur DS, Maluquer de Motes C, Unterholzner L, Sumner RP, Ferguson BJ, Ren H, et al. Poxvirus targeting of E3 ligase beta-TrCP by molecular mimicry: a mechanism to inhibit NF-kappaB activation and promote immune evasion and virulence. PLoS Pathog. (2013) 9:e1003183. doi: 10.1371/journal.ppat.1003183
77. Chen RA, Ryzhakov G, Cooray S, Randow F, Smith GL. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog. (2008) 4:e22. doi: 10.1371/journal.ppat.0040022
78. Lynch HE, Ray CA, Oie KL, Pollara JJ, Petty IT, Sadler AJ. Modified vaccinia virus Ankara can activate NF-kappaB transcription factors through a double-stranded RNA-activated protein kinase (PKR)-dependent pathway during the early phase of virus replication. Virology. (2009) 391:177–86. doi: 10.1016/j.virol.2009.06.012
79. Willis KL, Patel S, Xiang Y, Shisler JL. The effect of the vaccinia K1 protein on the PKR-eIF2alpha pathway in RK13 and HeLa cells. Virology. (2009) 394:73–81. doi: 10.1016/j.virol.2009.08.020
80. Kotwal GJ, Hugin AW, Moss B. Mapping and insertional mutagenesis of a vaccinia virus gene encoding a 13,800-Da secreted protein. Virology. (1989) 171:579–87. doi: 10.1016/0042-6822(89)90627-2
81. Bartlett N, Symons JA, Tscharke DC, Smith GL. The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J Gen Virol. (2002) 83:1965–76. doi: 10.1099/0022-1317-83-8-1965
82. DiPerna G, Stack J, Bowie AG, Boyd A, Kotwal G, Zhang Z, et al. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by toll-like receptors. J Biol Chem. (2004) 279:36570–8. doi: 10.1074/jbc.M400567200
83. Qin Y, Li M, Zhou SL, Yin W, Bian Z, Shu HB. SPI-2/CrmA inhibits IFN-beta induction by targeting TBK1/IKKepsilon. Sci Rep. (2017) 7:10495. doi: 10.1038/s41598-017-11016-3
84. Unterholzner L, Sumner RP, Baran M, Ren H, Mansur DS, Bourke NM, et al. Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog. (2011) 7:e1002247. doi: 10.1371/journal.ppat.1002247
85. Alcami A, Smith GL. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol. (1995) 69:4633–9. doi: 10.1128/JVI.69.8.4633-4639.1995
86. Mossman K, Upton C, Buller RM, McFadden G. Species specificity of ectromelia virus and vaccinia virus interferon-gamma binding proteins. Virology. (1995) 208:762–9. doi: 10.1006/viro.1995.1208
87. Symons JA, Tscharke DC, Price N, Smith GL. A study of the vaccinia virus interferon-gamma receptor and its contribution to virus virulence. J Gen Virol. (2002) 83:1953–64. doi: 10.1099/0022-1317-83-8-1953
88. Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem. (1995) 270:15974–8. doi: 10.1074/jbc.270.27.15974
89. Symons JA, Alcami A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. (1995) 81:551–60. doi: 10.1016/0092-8674(95)90076-4
90. Alcami A, Symons JA, Smith GL. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol. (2000) 74:11230–9. doi: 10.1128/JVI.74.23.11230-11239.2000
91. Montanuy I, Alejo A, Alcami A. Glycosaminoglycans mediate retention of the poxvirus type I interferon binding protein at the cell surface to locally block interferon antiviral responses. FASEB J. (2011) 25:1960–71. doi: 10.1096/fj.10-177188
92. Ran Y, Xiong MG, Xu ZS, Luo WW, Wang SY, Wang YY. YIPF5 is essential for innate immunity to DNA virus and facilitates COPII-dependent STING trafficking. J Immunol. (2019) 203:1560–70. doi: 10.4049/jimmunol.1900387
Keywords: poxvirus, cGAS, DNA-PK, IFI16, STING
Citation: Lu Y and Zhang L (2020) DNA-Sensing Antiviral Innate Immunity in Poxvirus Infection. Front. Immunol. 11:1637. doi: 10.3389/fimmu.2020.01637
Received: 15 April 2020; Accepted: 18 June 2020;
Published: 28 August 2020.
Edited by:
Chunfu Zheng, Fujian Medical University, ChinaReviewed by:
Kai Huang, University of Texas Medical Branch at Galveston, United StatesCopyright © 2020 Lu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leiliang Zhang, YXJtemhhbmdAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.