
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 04 August 2020
Sec. Comparative Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01596
This article is part of the Research Topic Novel Insights into Insect Antiviral Immunity View all 16 articles
In nature, plant viruses are mostly transmitted by hemipteran insects, such as aphids, leafhoppers, and whiteflies. However, the molecular mechanisms underlying the interactions between virus and insect vector are poorly known. Here, we investigate the proteomic interactions between tomato yellow leaf curl virus (TYLCV, genus Begomovirus, family Geminiviridae), a plant virus, and its vector whitefly (Bemisia tabaci) species complex. First, using a yeast two-hybrid system, we identified 15 candidate whitefly proteins interacting with the coat protein of TYLCV. GO and KEGG pathway analysis implicated that these 15 whitefly proteins are of different biological functions/processes mainly including metabolic process, cell motility, signal transduction, and response to stimulus. We then found that the whitefly protein tumorous imaginal discs (Tid), one of the 15 whitefly proteins identified, had a stable interaction with TYLCV CP in vitro, and the DnaJ_C domain of Tid301−499aa may be the viral binding site. During viral retention, the expression of whitefly protein Tid was observed to increase at the protein level, and feeding whiteflies with dsRNA or antibody against Tid resulted in a higher quantity of TYLCV in the whitefly body, suggesting the role of Tid in antiviral infection. Our data indicate that the induction of Tid following viral acquisition is likely a whitefly immune response to TYLCV infection.
Many plant viruses, such as species of the Luteoviridae, Geminiviridae, and Nanoviridae families, are transmitted by hemipteran insects in a persistent, circulative manner (1). During the long-term virus-vector interactions, insect vectors have developed two inevitable physical barriers to virus movement: midgut and salivary glands (1, 2). Initially, the vector ingests virions from virus-infected plants; then, virions enter the insect midgut lumen and subsequently cross through the midgut epithelial cells to be released into the hemolymph. Afterwards, virions move along with the hemolymph and reach the salivary glands from which they are injected into plants together with whitefly saliva secretion (1, 2). During this circulative journey, viruses need to interact with the insect vector in a coordinated manner for successful transmission to occur; at the same time, viral infection may activate immune reactions from its vector (3, 4).
Begomoviruses (genus Begomovirus, family Geminiviridae) are a group of single-stranded circular DNA viruses, which are transmitted by whiteflies of the Bemisia tabaci species complex in a circulative manner (5, 6). Some begomoviruses are serious viral disease agents of many crops worldwide. For example, tomato yellow leaf curl virus (TYLCV) is transmitted by a notorious invading species of whitefly, provisionally named as Middle East–Asia Minor 1 (MEAM1), of the B. tabaci species complex and has caused enormous damage to the production of tomato and some other crops in many countries/regions in the last three decades (7–9). Similar to other begomoviruses and other circulatively transmitted viruses, ingested TYLCV moves along the path of stylet-midgut-hemolymph-salivary glands in whitefly vectors. During the movement, TYLCV depends on clathrin-mediated endocytosis to enter the midgut cells and then accumulates in intracellular vesicle-like structures (10–12). At the same time, a viral infection activates the whitefly autophagy pathway, which plays an important role in the antiviral response (13, 14).
Up to now, the coat protein (CP) is the only structural protein of begomoviruses known to be involved in viral movement in the vector (15). CP gene replacement results in dramatic changes in characteristics of viral acquisition and transmission by whitefly vector (16–19). However, so far, only a few whitefly proteins have been reported to interact with the viral CP. The heat shock protein 70 (HSP70) and vesicle-associated membrane protein–associated protein B (VAPB) show inhibitory roles in virus transmission (20, 21), and a peptidoglycan recognition protein BtPGRP acts in whitefly immunity (22). In contrast, GroEL produced by secondary endosymbionts Hamiltonella or Arsenophonus may protect the virus from degradation in vector hemolymph (23, 24), and the midgut protein, cyclophilin B and collagen protein may assist in viral transmission (25–27). Vitellogenin may enable transovarial transmission of virus to the next generation of whitefly (28). The putative roles of BtHSP16, thioredoxin-like protein (TLP) and protein BtR242 produced by Rickettsia in the viral transmission are yet unclear (29–31). Despite this progress, the functions of some of the abovementioned proteins require further validation, and many more vector components remain to be discovered to achieve an adequate understanding of begomovirus-whitefly interactions.
In this study, first, using the yeast two hybrid (Y2H) system, we identified 15 candidate whitefly proteins interacting with TYLCV CP, including the evolutionarily highly conserved protein tumorous imaginal discs (Tid). As the mammalian homolog of whitefly Tid has been implicated for its role in a variety of signaling pathways and autophagy (32, 33), we then conducted a series of molecular experiments and bioassays to examine in vitro interaction between whitefly Tid and TYLCV CP. Following viral infection, increase of whitefly Tid at the protein level exerted constraints on viral retention. Our data provide novel insights into begomovirus-whitefly interactions, indicating the negative impact of Tid on viral retention.
TYLCV clone isolate SH2 (GenBank accession number: AM282874.1) was agro-inoculated into tomato plants (Solanum lycopersicon L. cv. Hezuo903) at the 3–4 true leaf stage. The tomato plants were then cultivated to the 7–8 true leaf stage, and plants showing typical symptoms were taken for use in experiments. Cotton plants (Gossypium hirsutum L. cv. Zhemian, 1793) were cultivated to the 6–7 true leaf stage for whitefly culture maintenance and experiments. A stock culture of MEAM1 whitefly was maintained in insect-proof cages on cotton plants at 26 ± 1°C, 60% relative humidity and 14 h light/10 h darkness.
The Y2H assay based on the matchmaker gold yeast two-hybrid system (Cat. No. 630489; Clontech) was used to explore the interactions between whitefly proteins and TYLCV CP. The cDNA library of whitefly was constructed in the prey plasmid of SfiI-digested pGADT7. The full-length of TYLCV CP gene was cloned into the bait plasmid of pGBKT7 after Nde I and EcoR I restriction. Primers used for cloning are listed in Supplementary Table 1. We used the following procedure for the Y2H assay: (1) transform the recombinant plasmid pGBKT7-TYLCV CP into the Y2H Gold yeast strain; (2) select the yeast strain on synthetic defined minimal medium lacking tryptophan (S.D./-Trp); (3) extract the yeast protein by yeast total protein extraction kit (Cat. No.C500013; Sangon Biotech) and confirm the expression of TYLCV CP in yeast in a Western blot by anti-TYLCV CP antibody (provided by Professor Jian-Xiang Wu); (4) conduct the auto-activation detection; (5) transform the cDNA library of whitefly into the Y2HGold yeast strain containing the bait plasmid pGBKT7-TYLCV CP; (6) observe the growth of yeast strain on the double dropout medium (DDO: S.D./-Leu/-Trp) and triple dropout medium (TDO: S.D./-His/-Leu/-Trp) with 40 μg/ml X-alpha-Gal and 125 ng/ml aureobasidin A (AbA) (TDO/X/A), select the positive clones on TDO/X/A; (7) restreak these positive clones on quadruple dropout medium (QDO: S.D./Ade/-His/-Leu/-Trp) with 40 μg/ml X-alpha-Gal and 125 ng/ml AbA (QDO/X/A) to eliminate the false positives; (8) recover the prey plasmids from the positive clones and transform them into Escherichia coli strain DH5α, sequence, and identify their interactions with TYLCV CP again. The different fragments screened from the whitefly cDNA library were used in a BLAST search of the NCBI database (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi), and the sequences of these fragments screening in the Y2H assay were deposited in GenBank.
Whitefly proteins identified from the Y2H assay system were categorized according to their gene ontology (GO) annotation using the Blast2GO software and then performed using the OmicShare tools, a free online platform for data analysis (http://www.omicshare.com/tools). The metabolic pathway analysis of these proteins was conducted according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation (https://www.kegg.jp/blastkoala/). Network diagrams were created using the database search tool for the retrieval of interacting genes/proteins (STRING 9.1; http://stringdb.org). All of these analyses were conducted by the full length of amino acid sequences.
Quantitative (q) PCR was performed on CFX connect real-time PCR system (Bio-Rad, USA) using the SYBR Premix Ex Taq II (Cat. No. RR820A, Takara). β-Actin was used as an internal reference, and relative abundance of TYLCV or transcript was calculated by 2−ΔΔCt. Primers used for real-time PCR are listed in Supplementary Table 1.
DNA templates with a T7 promoter at both ends of selected genes were used to synthesize dsRNA following the manufacturer's instruction of the T7 high-yield transcription kit (Cat. No.TR101-02; Vazyme). Then, dsRNA was purified using phenol: chloroform extraction, isopropanol precipitation, and resuspended in nuclease-free water. The size and quality of the dsRNA were confirmed by 1% agarose gel electrophoresis, and its quantity was measured using Nanodrop (Thermo Scientific, USA). DsGFP was used as a control. Primers used for DNA template synthesis are listed in Supplementary Table 1.
Whitefly adults within 7 days post-emergence were collected from cotton plants. A group of 250 adults were released into a glass tube 1.5 cm in diameter and 10 cm in length. According to Pan et al. (10), for dsRNA silencing, whiteflies were fed on 15% sucrose solution containing 200 ng/μl dsRNA for 48 h, and 15% sucrose solution with the same amount dsGFP was used as control. For antibody feeding, Tid polyclonal antibody (PcAb) was mixed with 15% sucrose solution with a dilution rate of 1:50 for 24 h, and 15% sucrose solution with the same dilution of rabbit pre-immune serum was set as control.
For viral acquisition after dsTid or dsUBR7 feeding (dsGFP was used as control), whiteflies were caged with leaves from the same branch of TYLCV-infected tomato plants for 6, 12, or 24 h, respectively, and then transferred to feed on cotton for 48 h for viral retention. Female adults were collected in groups of 10 each and homogenized in 100 μL lysis buffer for relative viral quantity analysis (10). Three biological replicates were conducted for relative viral quantity analysis by real-time PCR. For the subsequent experiments of membrane feeding of dsRNA or antibody against Tid, whiteflies were caged with leaves of two symmetrical leaves of the same height on the same branch of TYLCV-infected tomato plants for 12 h and then transferred to feed on cotton for 48 h for viral retention. Three to five biological replicates were conducted for relative viral quantity analysis by real-time PCR.
The amino acid sequence of Tid fragment screened from the Y2H assay (Tid-S, GenBank: MT505751) was aligned with the Tid full-length (Tid-FL, GenBank: MT505750) using Clustal X (2.0). Phylogenetic reconstruction was conducted using the maximum likelihood (ML) method and the global transvers time (GTR) model implemented in the MEGA v.6 program (34). Support for the internal nodes of the trees was evaluated using the bootstrap method with 10,000 replicates. The protein domain, transmembrane region, and signal peptide predictions were conducted using the NCBI conserved domain database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/), respectively. The 3-D structure of protein Tid was predicted using swissmodel (http://swissmodel.expasy.org).
The ORF of Tid-FL (GenBank: MT505750) was amplified from the whitefly cDNA using PrimerSTAR max DNA polymerase (Cat. No. R045A; Takara) and then cloned into pET28a plasmid for fusion with His tag. His-Tid-FL was expressed in inclusion bodies of E. coli strain Rosetta, and following renaturation and purification of inclusion body protein, His-Tid-FL was used to immunize rabbits to obtain a Tid-specific PcAb by HuaBio (China). Primers used in this experiment are listed in Supplementary Table 1. The specificity of Tid rabbit PcAb is shown in Supplementary Figure 1.
Tid-S, Tid76−138aa (226-414 bp of Tid-FL, DnaJ domain), Tid239−299aa [715-897 bp of Tid-FL, four repeats of a CXXCXGX(G) motif], and Tid301−419aa (901-1,257 bp of Tid-FL, DnaJ_C domain) were cloned into pMAL-c5X for fusion with MBP tag, accordingly. TYLCV CP was cloned into pGEX-6p-1 for fusion with GST tag. These recombinant proteins were expressed in E. coli strain Rosetta and purified. GST-TYLCV CP was bound to glutathione agarose beads (Cat. No.17-5132-01; GE Healthcare) for 2–4 h at 4°C. Then the mixtures were centrifuged for 5 min at 1,000 rpm, and the supernatants were discarded. Agarose beads were washed five times with 1 × phosphate-buffer saline (PBS). Different purified and desalinated MBP-tag fusion proteins or the native whitefly proteins extracted by cytoplasmic extraction buffer (Cat. No.SC-003; Invent) were added to the beads, respectively, and incubated for 4 h at 4°C. These mixtures were centrifuged and washed five times with 1 × PBS, and the bead-bound proteins were eluted by boiling in PAGE buffer (Cat. No. FD 002; FDbio) for 10 min. Finally, these proteins were separated by SDS/PAGE gel electrophoresis and detected by Western blot using anti-MBP antibody (Cat. No. ab49923; Abcam) or anti-Tid antibody. Primers used are listed in Supplementary Table 1.
Whitefly adults within 7 days post-emergence from cotton were transferred to TYLCV-infected tomato for 12 h and then transferred to feed on cotton for 48 h. Un-infected tomato was used as a control. Whitefly adults (three biological replicates) were collected as groups of 30 adults for analyzing gene expression of Tid at the transcriptional level. Total RNA of whitefly was isolated with TRIzol (Cat. No. 15596-026; Invitrogen), and reverse transcription was done using the PrimeScript RT reagent kit (Cat. No. DRR037A; Takara). For translational-level analysis, 100 whitefly adults were collected as one sample for protein extraction by RIPA (Cat. No. P0013B; Beyotime). Then, we used the BCA protein assay (Cat. No. 23250; Thermo Scientific) to determine and unify the concentration of protein samples. Western blot analysis was conducted by anti-Tid antibody, using anti-β-actin antibody (Cat. No. E021020-01; Earthox) as a control. The translational-level analysis was repeated three times, and ImageJ was used to quantify the relative protein level, Following dsTid membrane feeding, 12 h viral acquisition, and 48 h viral retention, the expressions of Tid at transcriptional and translational levels were analyzed as described above.
Comparison of the relative abundance of virus in whitefly and expression levels of genes were performed using an independent t-test with P < 0.05 as the threshold of significant difference (*P < 0.05, **P < 0.01, and ***P < 0.001). All the statistical analyses were performed using SPSS 20.0 (SPSS Inc., USA).
As shown in Supplementary Figure 2A, the Y2H system was used to examine the interactions between whitefly proteins and TYLCV CP. The titer of the primary whitefly cDNA library was ~5.0 × 106 cfu with an average insert size of 1 kb, meeting the requirements of a standard cDNA library (Supplementary Figure 2B). The fusion expression of TYLCV CP with GAL4 DNA-BD in the yeast (≈46 kDa) was verified using Western blot analysis (Supplementary Figure 2C). The auto-activation detection showed that the bait plasmid pGBKT7-TYLCV CP could be used in this Y2H system (Supplementary Figure 2D). After Y2H screening, 26 positive clones were isolated, and 15 unique whitefly proteins were identified (Table 1). To identify the one-to-one interaction between bait and prey protein, the interactions between these 15 screened whitefly proteins and TYLCV CP were validated using the Y2H assay (Figure 1A), combined with reported interactions between MEAM1 whitefly and TYLCV, and a protein interaction network was generated, including the predicted interactions among whitefly proteins (Figure 1B).
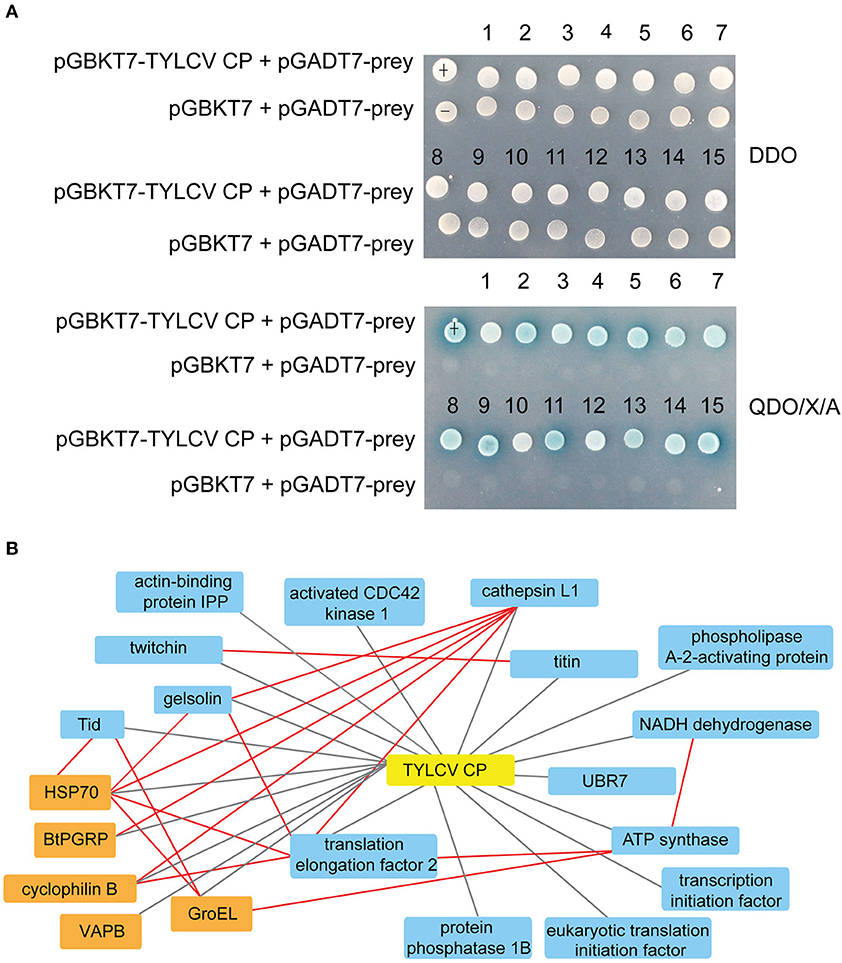
Figure 1. Interactions between whitefly proteins and TYLCV CP. (A) Confirmed interactions between TYLCV CP and screened proteins of B. tabaci using Y2H assay. TYLCV CP and 15 respective prey proteins were used to cotransform yeast for growth on DDO and QDO/X/A selective medium. pGBKT7-p53 and pGADT7-LargeT were used as positive controls (+); pGBKT7-p53 and pGADT7 were used as negative controls (–). (B) Protein interaction network was constructed using TYLCV CP and 20 whitefly protein homologs of Drosophila melanogaster; 20 whitefly proteins include 15 candidate whitefly proteins (blue) obtained in this study and five other whitefly proteins (orange) available in the literature related to MEAM1 whitefly-TYLCV interactions. The red line means the interaction predicted from the database search tool for the retrieval of interacting genes/proteins (STRING 9.1; http://stringdb.org), the black line stands for the interaction supported by experiments.
According to GO and KEGG analyses (Figures 2, 3), the 15 interactors from the Y2H assay (Table 1) were classified into different groups, mainly including metabolic process, cell motility, signal transduction, and response to stimulus. The GO analysis suggests that the 15 proteins may be responsible for 17 different biological processes, mainly involved in cellular and metabolic processes with different distributions inside and outside of cells; most of them shared the binding activity, and about half of them possess catalytic activity (Figure 2). The KEGG pathway analysis suggests that the 15 proteins can be classified into 7 groups (Figure 3). For example, gelsolin-like isoform X2 belongs to the pathway of cell motility; protein phosphatase 1B as a member of the MAPK signaling pathway belongs to the group of signal transduction. The whitefly autophagy pathway and ubiquitin-proteasome system have been shown to play a role in antiviral response (13, 14, 35). Among the 15 whitefly proteins, there is a ubiquitin-protein ligase (UBR7) and a protein Tid related to macro-autophagy (33). Both of these two proteins belong to the biological process of response to stimulus (GO: 0050896).
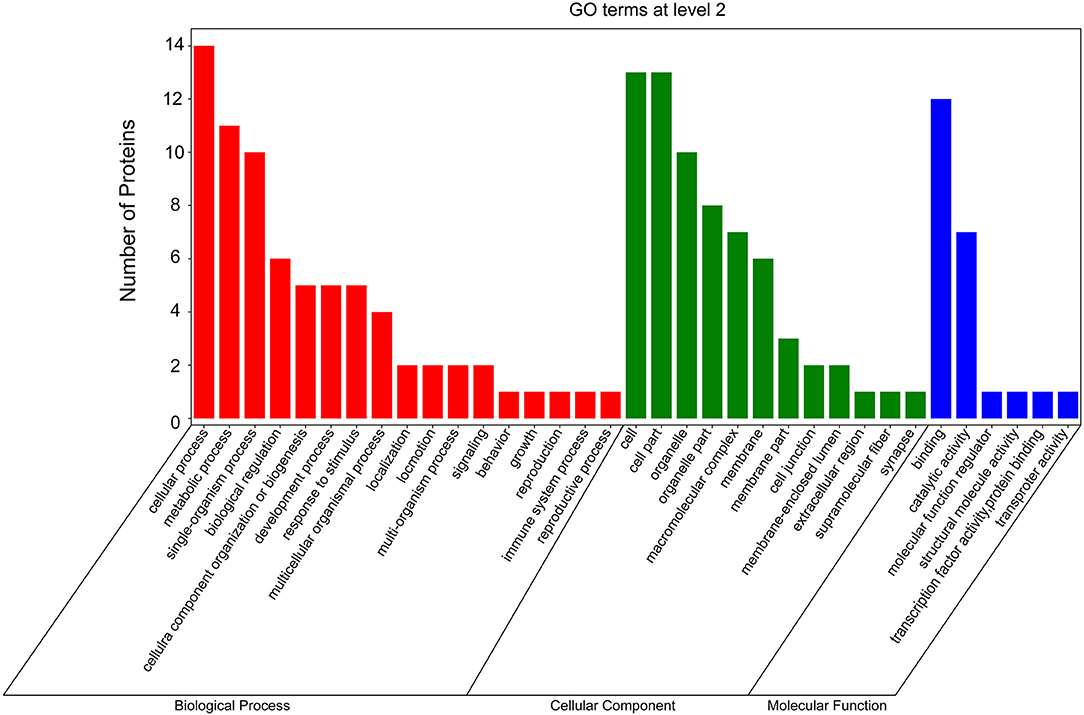
Figure 2. GO analysis of the 15 putative interactors inferred via screening in the Y2H system. Different colors represent different GO categories. GO annotation was conducted by the Blast2GO software, and the figure was generated using the OmicShare tools, a free online platform for data analysis (http://www.omicshare.com/tools).
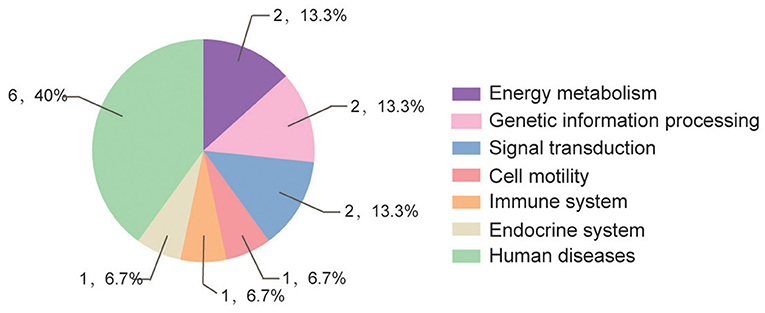
Figure 3. Pathway distribution of the 15 putative interactors inferred via screening in the Y2H system. The metabolic pathway analysis of these proteins was conducted according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation (https://www.kegg.jp/blastkoala/); in total, 15 pathways were identified for the 15 prey proteins.
To examine the role of proteins Tid and UBR7 on virus retention, whiteflies that had received dsRNA interference treatment were transferred to feed on a TYLCV-infected tomato for 6, 12, or 24 h, respectively, and then transferred to feed on cotton for 48 h. At the end of each of the three time points, after dsTid interference, the relative viral quantity in whiteflies significantly increased compared to the control (Figures 4A–C) although, for UBR7 dsRNA interference, following a viral acquisition access period for 12 h, the defense ability of the whitefly against TYLCV retention significantly decreased (Figure 4B). When the intervals of the viral acquisition access period lasted 6 or 24 h, the defense ability of the whitefly against TYLCV retention had non-significant decrease (Figures 4A,C). Based on these results, we selected Tid for the following experiments.
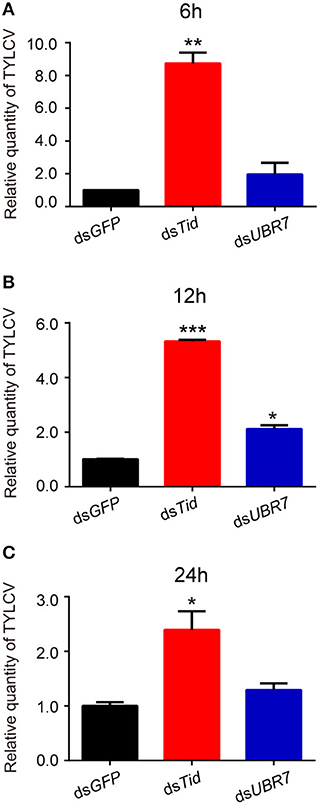
Figure 4. Effects of dsRNA interference of Tid and UBR7 on TYLCV retention. After dsRNA feeding, whiteflies of the interference treatments were caged to feed on leaves of the same branch of a TYLCV-infected tomato plant for 6 h (A), 12 h (B), or 24 h (C), and then transferred to feed on cotton for 48 h to test the viral retention ability of the whitefly by qPCR. GFP was used as a control. Whitefly females were collected in groups of 10 each and homogenized in 100 μL lysis buffer for relative viral quantity analysis. In (A), dsTid: n = 3, t = −11.716, P = 0.0072; dsUBR7: n = 3, t = −1.327, P = 0.3158; in (B), dsTid: n = 3, t = −66.018, P < 0.0001; dsUBR7: n = 3, t = −7.855, P = 0.0138; and in (C), dsTid: n = 3, t = −4.035, P = 0.0157; dsUBR7: n = 3, t = −2.058, P = 0.1087. Independent t-test was used here and the differences between treatments were considered significant when *P < 0.05; **P < 0.01, ***P < 0.001.
After sequencing the Tid prey plasmid screened from Y2H, we obtained an 855 bp long (285 aa) Tid-S sequence (GenBank: MT505751), having a 60% coverage (164–448aa) of Tid-FL. Tid-FL (GenBank: MT505750, ≈52 kDa) has a DnaJ domain (N-terminal, 76–138aa), a DnaJ_C domain (C-terminal, 301–419aa), and four repeats of a CXXCXGX(G) motif (239–299aa). Tid-S contains only the CXXCXGX(G) motifs and DnaJ_C domain (Figure 5A). Tid-FL has no transmembrane domain or signal peptide, and its 3-D structure model is shown in Figure 5B. Phylogenetic analysis of B. tabaci Tid and 16 other insect Tid proteins showed that B. tabaci Tid forms a monophyletic lineage with species of Hymenoptera and appears closely related to the genus Drosophila (Figure 5C). This DnaJ domain-containing protein is evolutionarily highly conserved; Tid in mammals and that of Drosophila show 54.9% identity in amino acid sequences (36), and the Tid of whitefly and that of Drosophila melanogaster show 54.0% identity in amino acid sequences.
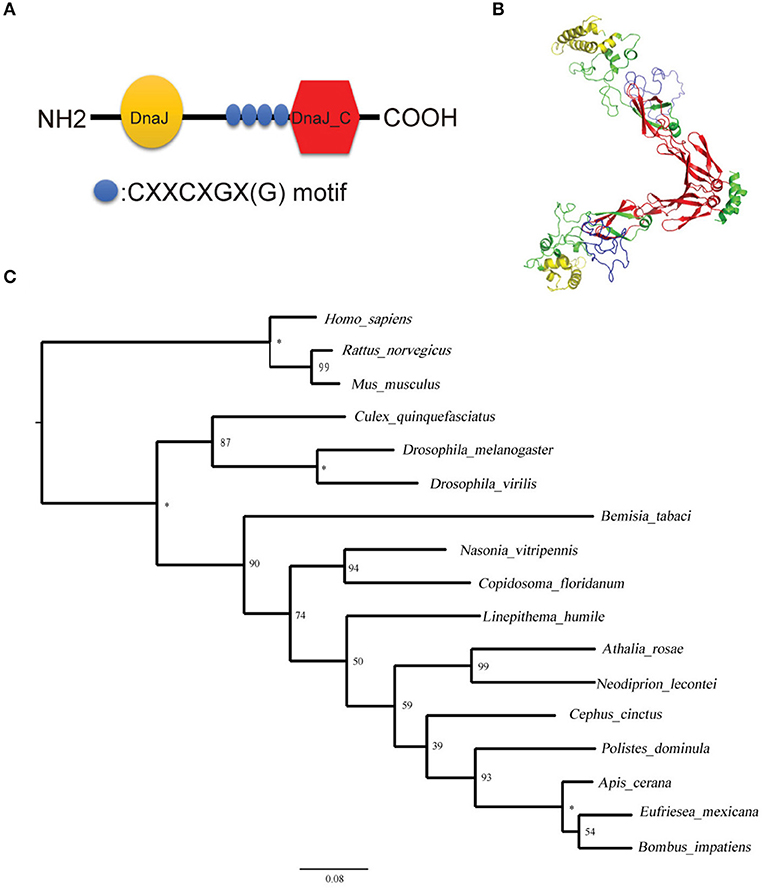
Figure 5. Structural and phylogenetic analysis of the protein Tid. (A) Graphic presentation of the Tid structure. (B) 3-D structure of protein Tid. Asterisk indicates that the nodes were 100% supported. (C) Phylogenetic tree of B. tabaci-Tid and other arthropods and mammals were constructed using MEGA v.6 with the maximum likelihood (ML) method. Numbers next to the branches indicated bootstrap value of each internal branch in the phylogenetic tree nodes from 10000 replicates. Tid sequences include Bemisia tabaci (MT505750), Homo sapiens (NM 001286516), Rattus norvegicus (NM 001038596), Mus musculus (NM 001135112), Culex quinquefasciatus (XM 001848856), Drosophila melanogaster (NM 001259554), Drosophila virilis (XM 002059276), Bemisia tabaci (XP 018917603), Nasonia vitripennis (XM 016983982), Copidosoma floridanum (XM 014350678), Linepithema humile (XM 012379048), Athalia rosae (XM 012406140), Neodiprion lecontei (XM 015659368), Cephus cinctus (XM 015737810), Polistes dominula (XM 015322978), Apis cerana (XM 017059752), Eufriesea mexicana (XM 017897896), and Bombus impatiens (XM 012384798).
TYLCV CP fused with GST and Tid-S tagged with MBP were used to verify their interaction through GST pull-down analysis (Figure 6A). Using the fusion protein GST-TYLCV CP as a bait protein and native whitefly proteins extracted by cytoplasmic extraction buffer (Cat. No.SC-003; Invent) as prey proteins, whitefly endogenous Tid could co-elute with GST-fused TYLCV CP but not with GST (Figure 6B). Further, we tested the interaction between TYLCV CP and different regions of Tid-FL mentioned above: Tid76−138aa (DnaJ domain), Tid239−299aa (four repeats of a CXXCXGX(G) motif), and Tid301−419aa (DnaJ_C domain). The results showed that Tid76−138aa and Tid239−299aa show no binding activity with TYLCV CP (Figure 6C); the binding site of TYLCV CP may be located in the C terminal of Tid-FL (Figure 6D).
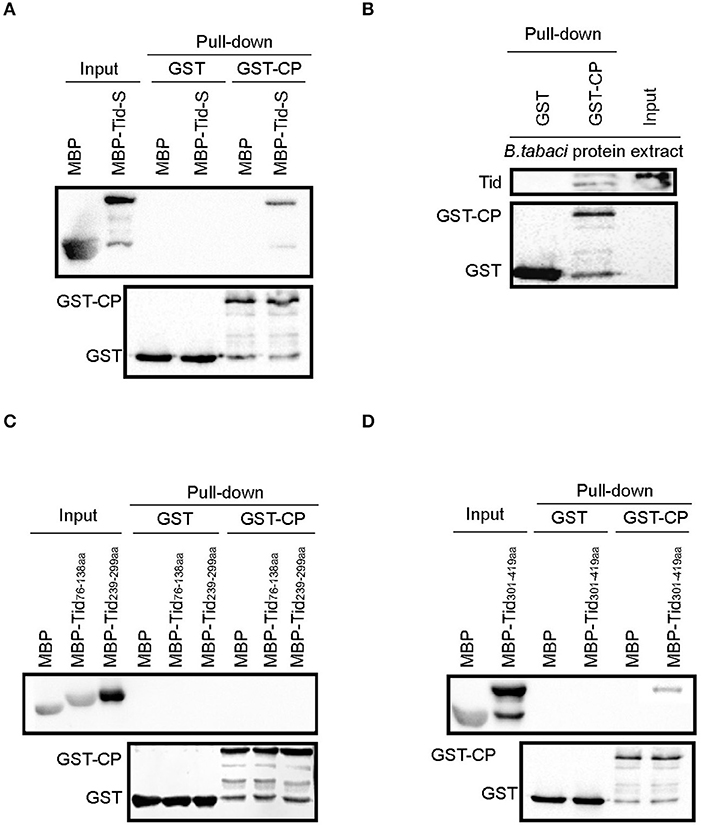
Figure 6. Interaction analysis between Tid and TYLCV CP. GST-TYLCV CP was used as bait protein, (A) MBP-Tid-S was used as prey protein, confirmation of the interaction between GST-TYLCV CP and MBP-Tid-S using GST pull-down; (B) native whitefly proteins extracted by cytoplasmic extraction buffer (Cat. No.SC-003; Invent) were used as prey proteins, identification of the interaction between whitefly endogenous Tid and GST-TYLCV CP by GST pull-down; (C) MBP-Tid76−138aa and MBP-Tid239−299aa were, respectively, used as prey protein, the interactions between these two Tid regions and GST-TYLCV CP were conducted via GST pull-down. (D) The C terminal of Tid301−419aa was used as prey protein, identification of the interaction between Tid301−419aa and GST-TYLCV CP via GST pull-down.
Following viral infection, the expression of Tid at both transcriptional and translational levels was tested. Data demonstrates that there was no significant change of the expression of Tid at transcriptional level (Figure 7A). However, Western blot analysis showed TYLCV infection could significantly increase the expression of Tid at protein level (Figure 7B).
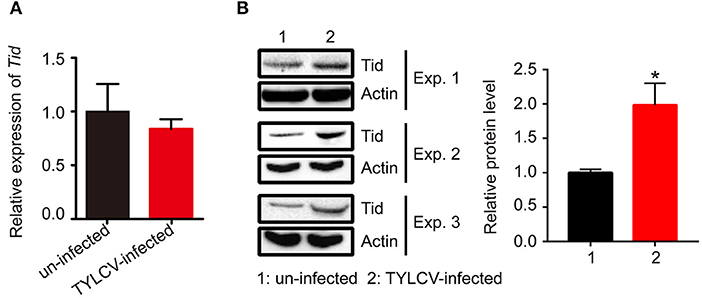
Figure 7. The expression of Tid following TYLCV acquisition. (A) The change of the Tid expression following viral acquisition at transcriptional level was analyzed by qPCR analysis (n = 3, t = 0.594, P = 0.5842). (B) The expression of Tid following viral acquisition at protein level was analyzed by Western blot analysis; 100 whitefly adults were collected as one sample for protein extraction and BCA protein assay was used to determine and unify the concentration of protein samples. Three biological replicates were set, and the results were quantified by ImageJ, t = −3.077, P = 0.0370. Independent t-test was used here and the differences between treatments were considered significant when *P < 0.05.
Following dsRNA feeding, the adults were transferred to feed on TYLCV-infected tomato plants for 12 h for virus acquisition and then were transferred to feed on cotton for 48 h for observation on virus retention. Data showed that the expression of Tid in whiteflies was effectively knocked down via dsRNA interference (Figures 8A,B), and knockdown of Tid expression resulted in significant increases of relative virus quantity in whiteflies (Figure 8C). In addition, blocking Tid function by anti-Tid antibody likewise resulted in significantly higher relative virus quantity in whiteflies during virus retention (Figure 8D).
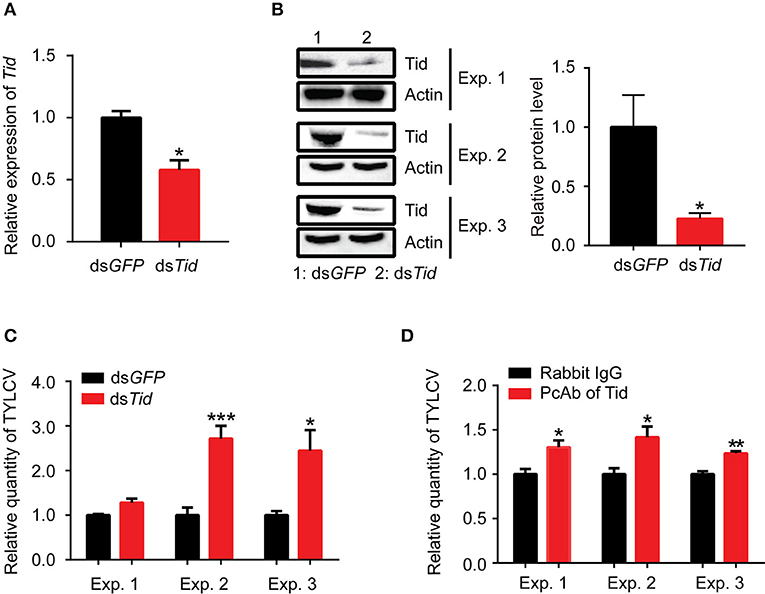
Figure 8. Tid restricts viral retention in whiteflies. After feeding with dsRNA, (A) Tid mRNA levels after were analyzed by qPCR analysis. Whitefly adults were collected as groups of 30 adults for RNA isolation and cDNA synthesis (n = 3, t = 4.46, P = 0.0111); (B) Tid protein levels were analyzed by Western blot analysis; 100 whitefly adults were collected as one sample for protein extraction and BCA protein assay was used to determine and unify the concentration of protein samples. Three biological replicates were set and the results were quantified by ImageJ, t = 2.82, P = 0.0478. (C) After dsTid interference and viral acquisition, TYLCV levels in whitefly whole body was analyzed by qPCR analysis. Whitefly females were collected in groups of 10 each and homogenized in 100 μL lysis buffer for relative viral quantity analysis (Exp. 1, n = 4–5, t = −2.85, P = 0.0565; Exp. 2, n = 5, t = −5.12, P = 0.0009; Exp. 3, n = 3, t = −3.06, P = 0.0375). (D) Effect of feeding whiteflies with an antibody against Tid: quantity of virus in the whole body (Exp. 1, n = 5, t = −3.11, P = 0.0145; Exp. 2, n = 4, t = −2.96, P = 0.0252; Exp. 3, n = 3–4, t = −5.32, P = 0.0031). Independent t-test was used here and the differences between treatments were considered significant when *P < 0.05; **P < 0.01, ***P < 0.001.
Investigation of the interactions between begomoviruses and whitefly proteins can provide new knowledge of the virus transportation journey in vector. In this study, 15 candidate whitefly proteins of various categories were detected that may interact with TYLCV CP. In further tests of Tid and UBR7, two of the 15 candidate proteins detected showed that both proteins posed an adverse effect on viral retention, and Tid had a stronger effect than UBR7. A stable interaction between whitefly Tid and TYLCV CP was then observed, and the C-terminal of Tid was observed to be the likely binding site. Viral infection could increase the expression of whitefly Tid at the protein level; feeding whiteflies with dsRNA or antibody against Tid resulted in a significantly higher quantity of TYLCV in the body of whiteflies following viral acquisition. Altogether, these data reveal one novel whitefly protein that may function in antiviral response.
The insect innate immune system incurs physical, cellular, and humoral responses to invaders (37), and it is common for insect vectors to take advantage of their immunity to fight against viral infection. Wang et al. (22) demonstrate that whitefly protein BtPGRP with antibacterial activity acts in multiple immune-response functions. Wang et al. (38) show that insect vectors could operate the c-Jun N-terminal kinase (JNK) signaling pathway for controlling viral transmission, causing a significant reduction in virus accumulation and transmission. The studies of Luan et al. (13) and Wang et al. (14) indicate that autophagy is involved in whitefly repression of begomovirus infection and triggers complex interactions between virus and insect vector. A previous study reported a mammalian homolog of whitefly Tid, which acted as a key regulator in mediating autophagy independently of HSP70 (33). Data available to date indicate that both Tid and HSP 70 play a role in repressing virus infection [(20); this study]; however, the relationships among whitefly autophagy, Tid, HSP70, and TYLCV CP remain unclear. Molecular mechanisms underlying the activation of autophagy pathway by TYLCV-infection in whiteflies warrant further investigation. Our findings provide clues for future studies on these issues.
Additionally, the roles of other candidate proteins detected in this study are also worth exploring. Gelsolin is a key regulator of actin filament assembly and disassembly (39), and actin has been shown to interact with several viral proteins and plays important roles in viral transmission. For example, interactions between non-structural protein Pns10 of rice dwarf virus and the cytoplasmic actin of leafhoppers is correlated with insect vector specificity (40); the non-structural protein P7-1 of reovirus southern rice black-streaked dwarf virus generates tubules and this tubules associate with the actin cytoskeleton in insect vector (Sogatella furcifera) cells (41, 42). In addition, MAPK signaling pathway is known to be activated by a diverse group of viruses and has important roles in viral replication (43), such as supporting assembly and maturation of West Nile virus and dengue virus (44, 45), regulating multiple steps of influenza A virus replication (46) and so on. In view of the potential role of protein phosphatase in regulating the life cycle of Simian Virus 40 (47), a study of the relationship of protein phosphatase 1B (a member of the MAPK signaling pathway) with TYLCV infection may be worthwhile. These investigations may lead to a comprehensive recognition of whitefly binding partners of viral CP and better understanding of the complex interactions between begomoviruses and their whitefly vectors.
All datasets generated for this study are included in the article/Supplementary Material.
JZ and TG designed this study and conducted most experiments as well as data analysis. JZ drafted and revised the manuscript. TL did the qPCR analysis and participated in statistical analysis. J-CZ made bioinformatic analysis. FW participated in Y2H screening. X-WW participated in manuscript preparation. S-SL provided supervision for the study and participated in manuscript preparation and revision. All authors read and approved the final version of the manuscript.
This study was financially supported by the National Natural Science Foundation of China (Project Nos. 31930092 and 31925033).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JN-C declared a past co-authorship with several of the authors SS-L and XW-W to the handling editor.
We thank Professor Jianxiang Wu, Institute of Biotechnology, Zhejiang University, for providing monoclonal antibodies of TYLCV CP. We also thank our lab engineer Gen-Hong Yan for his effort in plant growing and maintenance and Yuenan Zhou and Wenqiang Xia of our institute for their valuable discussions during this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01596/full#supplementary-material
1. Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol. (2008) 46:327–59. doi: 10.1146/annurev.phyto.022508.092135
2. Gray S, Cilia M, Ghanim M. Circulative, “nonpropagative” virus transmission: an orchestra of virus-, insect-, and plant-derived instruments. Adv Virus Res. (2014) 89:141–99. doi: 10.1016/B978-0-12-800172-1.00004-5
3. Gray SM, Banerjee N. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol Mol Biol Rev. (1999) 63:128–48. doi: 10.1128/MMBR.63.1.128-148.1999
4. Power AG. Insect transmission of plant viruses: a constraint on virus variability. Curr Opin Plant Biol. (2000) 3:336–40. doi: 10.1016/S1369-5266(00)00090-X
5. Harrison B, Robinson D. Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (begomoviruses). Annu Rev Phytophathol. (1999) 37:369–98. doi: 10.1146/annurev.phyto.37.1.369
6. Ran R, Kanakala S, Kliot A, Pakkianathan BC, Farich BA, Santana-Magal N, et al. Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr Opin Virol. (2015) 15:1–8. doi: 10.1016/j.coviro.2015.06.008
7. Li M, Hu J, Xu F, Liu SS. Transmission of Tomato yellow leaf curl virus by two invasive biotypes and a Chinese indigenous biotype of the whitefly Bemisia tabaci. Int J Pest Manag. (2010) 56:275–80. doi: 10.1080/09670871003743428
8. Moriones E, Navas-Castillo J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. (2000) 71:123–34. doi: 10.1016/S0168-1702(00)00193-3
9. Rojas MR, Macedo M, Maliano MR, Soto-Aguilar M, Souza JO, et al. World management of geminiviruses. Annu Rev Phytopathol. (2018) 56:637–77. doi: 10.1146/annurev-phyto-080615-100327
10. Pan LL, Chen QF, Zhao JJ, Guo T, Wang XW, Hariton-Shalev A, et al. Clathrin-mediated endocytosis is involved in Tomato yellow leaf curl virus transport across the midgut barrier of its whitefly vector. Virology. (2017) 502:152–9. doi: 10.1016/j.virol.2016.12.029
11. Uchibori M, Hirata A, Suzuki M, Ugaki M. Tomato yellow leaf curl virus accumulates in vesicle-like structures in descending and ascending midgut epithelial cells of the vector whitefly, Bemisia tabaci, but not in those of nonvector whitefly Trialeurodes vaporariorum. J Gen Plant Pathol. (2013) 79:115–22. doi: 10.1007/s10327-012-0426-2
12. Xia WQ, Liang Y, Chi Y, Pan LL, Zhao J, Liu SS, et al. Intracellular trafficking of begomoviruses in the midgut cells of their insect vector. PLoS Pathog. (2018) 14:e1006866. doi: 10.1371/journal.ppat.1006866
13. Luan JB, Li JM, Varela N, Wang YL, Li FF, Bao YY, et al. Global analysis of the transcriptional response of whitefly to Tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J Virol. (2011) 85:3330–40. doi: 10.1128/JVI.02507-10
14. Wang LL, Wang XR, Wei XM, Huang H, Wu JX, Chen XX, et al. The autophagy pathway participates in resistance to Tomato yellow leaf curl virus infection in whiteflies. Autophagy. (2016). 12:1560–74. doi: 10.1080/15548627.2016.1192749
15. Harrison BD, Swanson MM, Fargette D. Begomovirus coat protein: serology, variation and functions. Mol Plant Pathol. (2002) 60:257–71. doi: 10.1006/pmpp.2002.0404
16. Höfer P, Bedford ID, Markham PG, Jeske H, Frischmuth T. Coat protein gene replacement results in whitefly transmission of an insect nontransmissible geminivirus isolate. Virology. (1997) 236:288–95. doi: 10.1006/viro.1997.8751
17. Höhnle M, Höfer P, Bedford ID, Briddon RW, Markham PG, Frischmuth T. Exchange of three amino acids in the coat protein results in efficient whitefly transmission of a nontransmissible abutilon mosaic virus isolate. Virology. (2001) 290:164–71. doi: 10.1006/viro.2001.1140
18. Wei J, Zhao JJ, Zhang T, Li FF, Ghanim M, Zhou XP, et al. Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of begomoviruses. J Virol. (2014) 88:13460–8. doi: 10.1128/JVI.02179-14
19. Guo T, Zhao J, Pan LL, Geng L, Lei T, Wang XW, et al. The level of midgut penetration of two begomoviruses affects their acquisition and transmission by two species of Bemisia tabaci. Virology. (2018) 515:66–73. doi: 10.1016/j.virol.2017.12.004
20. Götz M, Popovski S, Kollenberg M, Gorovits R, Brown JK, Cicero JM, et al. Implication of Bemisia tabaci heat shock protein 70 in begomovirus-whitefly interactions. J Virol. (2012) 86:13241–52. doi: 10.1128/JVI.00880-12
21. Zhao J, Chi Y, Zhang XJ, Wang XW, Liu SS. Implication of whitefly vesicle associated membrane protein-associated protein B in the transmission of Tomato yellow leaf curl virus. Virology. (2019) 535:210–7. doi: 10.1016/j.virol.2019.07.007
22. Wang ZZ, Shi M, Huang YC, Wang XW, Stanley D, Chen XX. A peptidoglycan recognition protein acts in whitefly (Bemisia tabaci) immunity and involves in begomovirus acquisition. Sci Rep. (2016) 6:37806. doi: 10.1038/srep37806
23. Gottlieb Y, Zchorifein E, Mozesdaube N, Kontsedalov S, Skaljac M, Brumin M, et al. The transmission efficiency of Tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J Virol. (2010) 84:9310–7. doi: 10.1128/JVI.00423-10
24. Rana VS, Sing ST, Priya NG, Kumar J, Rajagopal R. Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary gland of whitefly B. tabaci PLoS ONE. (2012) 7:e42168. doi: 10.1371/journal.pone.0042168
25. Rana VS, Popli S, Saurav GK, Raina HS, Chaubey R, Ramamurthy VV, et al. A Bemisia tabaci midgut protein interacts with begomoviruses and plays a role in virus transmission. Cell Microbiol. (2015) 18:663–78. doi: 10.1111/cmi.12538
26. Kanakala S, Ghanim M. Implication of the whitefly Bemisia tabaci cyclophilin B protein in the transmission of Tomato yellow leaf curl virus. Front Plant Sci. (2016) 7:1702. doi: 10.3389/fpls.2016.01702
27. Rana VS, Popli S, Saurav GK, Raina HS, Jamwal R, Chaubey R, et al. Implication of the whitefly, Bemisia tabaci, collagen protein in begomoviruses acquisition and transmission. Phytopathology. (2019) 109:1481–93. doi: 10.1094/PHYTO-03-18-0082-R
28. Wei J, He YZ, Guo Q, Guo T, Liu YQ, Zhou XP, et al. Vector development and vitellogenin determine the transovarial transmission of begomoviruses. Proc Natl Acad Sci USA. (2017) 114:6747–51. doi: 10.1073/pnas.1701720114
29. Ohnesorge S, Bejarano ER. Begomovirus coat protein interacts with a small heat-shock protein of its transmission vector (Bemisia tabaci). Insect Mol Biol. (2010) 18:693–703. doi: 10.1111/j.1365-2583.2009.00906.x
30. Saurav GK, Rana VS, Popli S, Daimei G, Rajagopal R. A thioredoxin-like protein of Bemisia tabaci interacts with coat protein of begomoviruses. Virus Genes. (2019) 55:356–67. doi: 10.1007/s11262-019-01657-z
31. Lei T, Zhao J, Wang WL, Liu YQ, Liu S. Impact of a novel Rickettsia symbiont on the life history and virus transmission capacity of its host whitefly (Bemisia tabaci). Insect Sci. (2020). doi: 10.1111/1744-7917.12797. [Epub ahead of print].
32. Gaestel M. Molecular chaperones in signal transduction. Handb Exp Pharmacol. (2006) 172:93–109. doi: 10.1007/3-540-29717-0_4
33. Niu G, Zhang H, Liu D, Chen L, Belani C, Wang HG, et al. Tid1, the mammalian homologue of drosophila tumor suppressor tid56, mediates macroautophagy by interacting with beclin1-containing autophagy protein complex. J Biol Chem. (2015) 290:18102–10. doi: 10.1074/jbc.M115.665950
34. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–29. doi: 10.1093/molbev/mst197
35. Xia WQ, Liang Y, Liu YQ, Liu SS, Wang XW. Effects of ubiquitin-proteasome system on Tomato yellow leaf curl virus in whitefly Bemisia tabaci (Hemiptera: Aleyrodidae). Acta Entomologica Sinica. (2017) 60:1411–9. In Chinese with English abstract. doi: 10.16380/j.kcxb.2017.12.007
36. Schilling B, De-Medina T, Syken J, Vidal M, Münger K. A novel human dnaj protein, hTid-1, a homolog of the drosophila tumor suppressor protein Tid56, can interact with the human papillomavirus type 16 E7 oncoprotein. Virology. (1998) 247:74–85. doi: 10.1006/viro.1998.9220
37. Strand MR. In insect immunology. In: Beckage NE, editor. Insect Immunity. Amsterdam: Elsevier Academic Press (2008), p. 25–47.
38. Wang W, Zhao W, Li J, Luo L, Cui F. The c-jun n-terminal kinase pathway of a vector insect is activated by virus capsid protein and promotes viral replication. eLife. (2017) 6:e26591. doi: 10.7554/eLife.26591
39. Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. (1999) 274:33179. doi: 10.1074/jbc.274.47.33179
40. Chen Q, Wang HT, Ren TY, Xie LH, Wei TY. Interaction between non-structural protein Pns10 of rice dwarf virus and cytoplasmic actin of leafhoppers is correlated with insect vector specificity. J. Gen. Virol. (2015) 96:933–8. doi: 10.1099/jgv.0.000022
41. Liu Y, Jia DS, Che H, Chen Q, Xie LH, et al. The P7-1 protein of southern rice black-streaked dwarf virus, a fifijivirus, induces the formation of tubular structures in insect cells. Arch Virol. (2011) 156:1729–36. doi: 10.1007/s00705-011-1041-9
42. Jia DS, Mao QZ, Chen HY, Wang AM, Liu YY, Wang HT, et al. Virus-induced tubule: a vehicle for rapid spread of virions through basal lamina from midgut epithelium in the insect vector. J Virol. (2014) 88:10488–500. doi: 10.1128/JVI.01261-14
43. Kumar R, Khandelwal N, Thachamvally R, Tripathi BN, Barua S, Kashyap S, et al. Role of MAPK/MNK1 signaling in virus replication. Virus Res. (2018) 253:48–61. doi: 10.1016/j.virusres.2018.05.028
44. Hirsch AJ, Medigeshi GR, Meyers HL, DeFilippis V, Fruh K, Briese T, et al. The Src family kinase c-Yes is required for maturation of West Nile virus particles. J Virol. (2005) 79:11943–51. doi: 10.1128/JVI.79.18.11943-11951.2005
45. Chu JJ, Yang PL. C-src protein kinase inhibitors block assembly and maturation of dengue virus. Proc Natl Acad Sci USA. (2007) 104:3520–5. doi: 10.1073/pnas.0611681104
46. Kumar N, Liang Y, Parslow TG, Liang Y. Receptor tyrosine kinase inhibitors block multiple steps of influenza A virus replication. J Virol. (2011) 85:2818–27. doi: 10.1128/JVI.01969-10
Keywords: whitefly, TYLCV, interaction, Tid, antiviral infection
Citation: Zhao J, Guo T, Lei T, Zhu J-C, Wang F, Wang X-W and Liu S-S (2020) Proteomic Analyses of Whitefly-Begomovirus Interactions Reveal the Inhibitory Role of Tumorous Imaginal Discs in Viral Retention. Front. Immunol. 11:1596. doi: 10.3389/fimmu.2020.01596
Received: 10 February 2020; Accepted: 16 June 2020;
Published: 04 August 2020.
Edited by:
Xiao-Qiang Yu, University of Missouri–Kansas City, United StatesReviewed by:
Jesús Navas-Castillo, Institute of Subtropical and Mediterranean Horticulture La Mayora, SpainCopyright © 2020 Zhao, Guo, Lei, Zhu, Wang, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu-Sheng Liu, c2hzaGxpdUB6anUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.