
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 16 July 2020
Sec. Cytokines and Soluble Mediators in Immunity
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01424
This article is part of the Research Topic Modulating Cytokines as Treatment for Autoimmune Diseases and Cancer View all 14 articles
Cytokines are small signaling proteins that have central roles in inflammation and cell survival. In the half-century since the discovery of the first cytokines, the interferons, over fifty cytokines have been identified. Amongst these is interleukin (IL)-6, the first and prototypical member of the IL-6 family of cytokines, nearly all of which utilize the common signaling receptor, gp130. In the last decade, there have been numerous advances in our understanding of the structural mechanisms of IL-6 family signaling, particularly for IL-6 itself. However, our understanding of the detailed structural mechanisms underlying signaling by most IL-6 family members remains limited. With the emergence of new roles for IL-6 family cytokines in disease and, in particular, roles of IL-11 in cardiovascular disease, lung disease, and cancer, there is an emerging need to develop therapeutics that can progress to clinical use. Here we outline our current knowledge of the structural mechanism of signaling by the IL-6 family of cytokines. We discuss how this knowledge allows us to understand the mechanism of action of currently available inhibitors targeting IL-6 family cytokine signaling, and most importantly how it allows for improved opportunities to pharmacologically disrupt cytokine signaling. We focus specifically on the need to develop and understand inhibitors that disrupt IL-11 signaling.
In 1957, interferons were the first cytokines to be identified as secreted protein products induced following virus infection (1). In the subsequent decades, similar proteins, including the colony stimulating factors (CSFs) (2–4), Interleukin (IL)-2 (5, 6), and IL-3 (7, 8) were identified as secreted molecules able to support the growth of various hematopoietic cell linages in vitro. In 1974, the broad term “cytokine” was introduced (9) and in 1979 the term “interleukin” was introduced to standardize the names of the proteins now known as IL-1 and IL-2 (10). Over the next decade, radiolabelling studies revealed that cytokines bound distinct and unique receptors on the cell surface (11). It was also revealed that some cytokines, such as granulocyte-macrophage CSF (GM-CSF), IL-5 and IL-3 compete for a low-affinity receptor (12, 13), foreshadowing the identification of the β common receptor.
Following the discovery of the first cytokines, the mechanisms of intracellular signal transduction by cytokines remained elusive. The first transcriptional activator to be well-characterized was interferon-stimulated gene factor 3 (ISGF3), a multi-component protein complex consisting of what is now known as signal transducer and activator of transcription (STAT)1 and STAT2 (14, 15). Subsequently, related STAT proteins were identified as being activated via cytokine stimulation (16, 17). It was also shown that these factors were tyrosine phosphorylated (18, 19) on cytokine activation. The kinases responsible for this phosphorylation, the Janus kinases (JAKs) were first identified through a PCR screen of a murine hematopoietic cell line (20, 21). Their significance was unclear until the early 1990s, when they were shown to be activated as a result of cytokine binding and to phosphorylate the transcription factors that were already identified as key for interferon signal transduction (22). Subsequently, different members of the JAK family were found to be responsible for signal transduction by numerous cytokines (23–25). In 1997, the negative feedback regulators of the pathway, the suppressors of cytokine signaling (SOCS) proteins were identified (26–28). The key components of cytokine signaling using the JAK-STAT pathway were thus understood by the late 1990s, although many of the detailed molecular mechanisms are still unknown and remain under intense investigation today.
IL-6 family cytokines belong to a large group that signal via the JAK-STAT pathway, are characterized by a four α-helical bundle structure, and share receptors with similar structures consisting of several fibronectin type III (Fn3) and immunoglobulin-like (Ig-like) domains (29–31). Other cytokines, such as the IL-1/IL-18 family and the TNF-α family are structurally distinct from the four-α helical bundle family (32), utilize different signaling mechanisms, and are thus beyond the scope of this review. Conversely, several protein hormones, such as leptin, growth hormone (GH), prolactin and erythropoietin (EPO) utilize similar signal transduction mechanisms, are structurally related to the four-α helical bundle cytokines, and are thus best categorized alongside them (30, 33). The discovery of GH and EPO predate that of the interferons by several decades (34–37), but they were not recognized as related until they were cloned, sequenced, and significant sequence homology was noted between the receptors, GHR and EPOR (38, 39).
The four-α helical bundle cytokine family is the largest cytokine family. Both class I cytokines (e.g., GH, IL-6, IL-11) and class II cytokines (e.g., IFN-α, IL-10) utilize receptors that are broadly similar in structure and initiate similar intracellular signaling mechanisms (29). Cytokines from both classes are characterized by a compact α-helical bundle formed by four anti-parallel α-helices, arranged in an up-up-down-down topology (29, 31). This arrangement of helices necessitates long loops joining the helices (Figure 1A). Secondary structure in the loops is common, for example, the loop joining the C and D helices in IL-6 (the CD loop) contains a short α-helix (45), and in IL-4 (46) and GM-CSF (41), the AB and CD loops form a small anti-parallel β-sheet on the same face of the cytokine (Figure 1A). The topology of the four-α helical bundle fold provides a large surface area for the cytokine to bind its receptors.
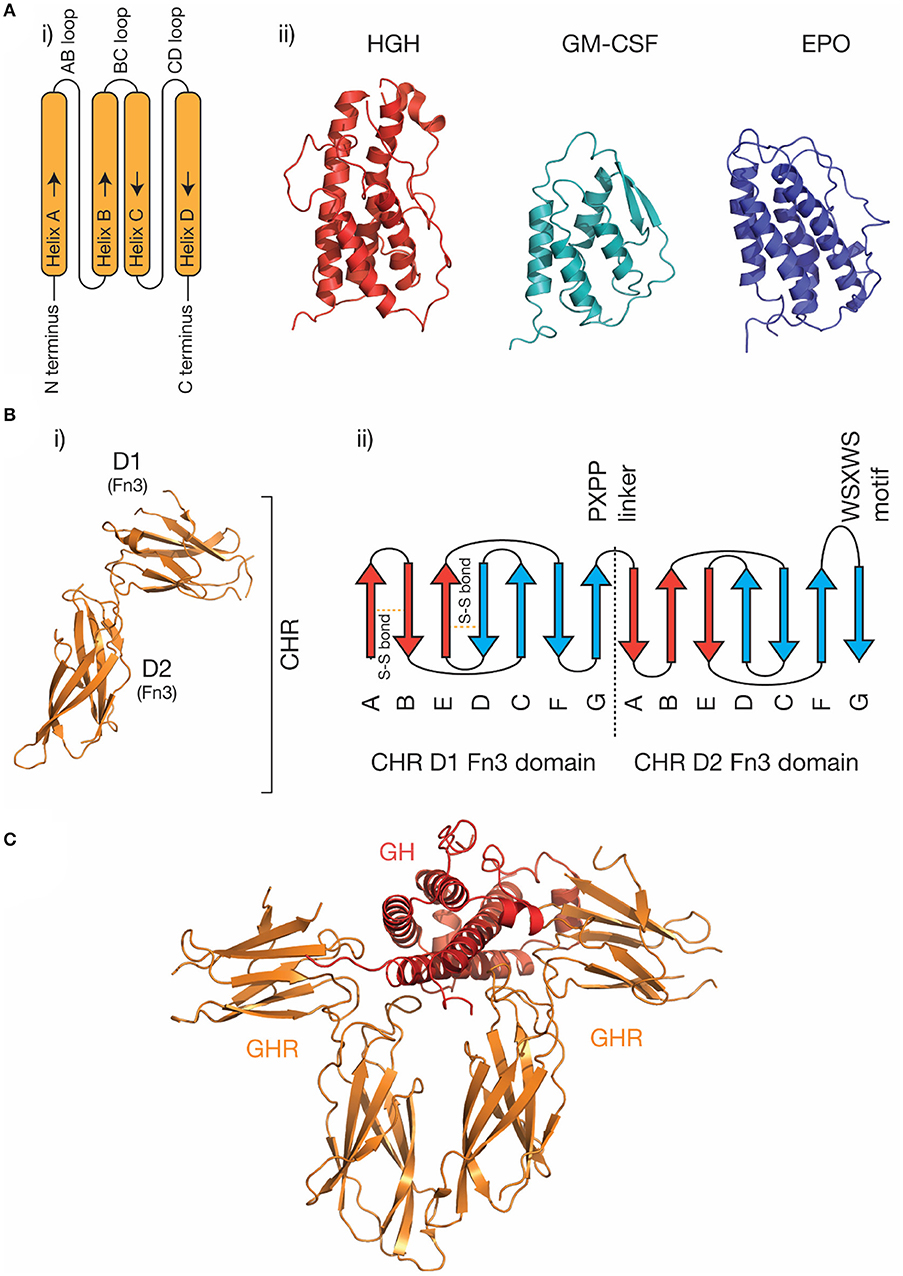
Figure 1. The structure of cytokines and receptors. (A) (i) A schematic of the four-α helical bundle topology of hematopoietic cytokines, (ii) cartoon representations of the structures of several representative cytokines; human growth hormone [PDB ID: 1HGU (40)], GM-CSF [PDB ID: 1CSG (41)], and erythropoietin [PDB ID: 1BUY (42)]. (B) The structure of the growth hormone receptor [PDB ID: 2AEW (43)]. The two Fn3 domains that make up the CHR are indicated, and a typical topology (30) for the two Fn3 domains in the CHR is shown in (ii). The conserved disulfide bonds in the N-terminal domain, the linker sequence, and the conserved WSXWS motif are indicated. (C) The structure of the growth hormone/growth hormone receptor complex [PDB ID: 3HHR (44)].
Cytokine receptors are generally modular, single-pass transmembrane proteins, with a large extracellular region consisting of multiple all-β Ig-like domains and Fn3 domains (33). Both domains possess a β-sandwich structure, with two anti-parallel β sheets (Figure 1B). The exception are the IL-2Rα/IL-15Rα receptors, which consist of two all-β sushi domains, unrelated to the Ig and Fn3 domains comprising other cytokine receptors (33, 47, 48). The cytokine binding domains of the receptors consist of two Fn3 domains at approximately a 90° angle, forming the cytokine binding homology region (CHR) (30). Cytokines bind at the junction of these two domains. Each of the two domains of the CHR possess conserved features, the N-terminal domain of the CHR has two conserved disulphide bonds, and in class I cytokine receptors of the C-terminal domain of the CHR has a highly conserved Trp-Ser-X-Trp-Ser motif (WSXWS) motif (30). The WSXWS motif generally forms a “ladder” consisting of cation-π interactions between the tryptophan and arginine side chains. The precise structural role of the WSXWS motif is still unclear. It may stabilize the receptor, since mutations in the WSXWS motif result in a non-functional receptor (49, 50), and a rare genetic disease results from a mutation in the WSXWS motif of GHR (51). In IL-21Rα, the first Trp of the WSXWS motif is C-mannosylated and this modified Trp forms stabilizing interactions with other glycans and amino acid residues in the structure (52). The extensive glycosylation, both Trp C-mannosylation, and N-linked glycosylation gives IL-21Rα the structure of an “A-frame,” with a glycan chain forming a bridge between the two domains in the receptor. Similar Trp C-mannosylation has been detected in the p40 subunit of IL-12 by mass spectrometry (53), but has not been observed in crystal structures which include p40 (54–56), possibly reflecting incomplete incorporation of the modification in recombinant protein. Recent studies have suggested that, in addition to being a stabilizing structural element, the WSXWS motif undergoes a conformational change on cytokine binding, suggesting it has a role in receptor activation (57).
Beyond the CHR, many cytokine receptors have additional extracellular domains. These domains have varied roles, for example in correctly orienting the receptor to allow the activation of intracellular kinases (58), to facilitate cytokine binding (59), or to modulate intracellular trafficking to the membrane (60). While, most cytokine receptors are single-pass transmembrane proteins, an exception is the ciliary neurotrophic factor (CNTF) receptor, which is lipid anchored (61). The structures of cytokine receptor transmembrane domains have been solved, generally by nuclear magnetic resonance (NMR) spectroscopy (62–64). Single-pass transmembrane cytokine receptors also possess an intracellular domain that is assumed to be highly dynamic (65, 66). In the case of signal-transducing cytokine receptors, the intracellular domain binds signal transducing molecules, such as the JAKs, STATs, and the SOCS proteins.
Understanding the molecular details of cytokine engagement requires detailed structural knowledge of the complexes formed by cytokines and receptors. The first cytokine/receptor complex structure solved was the GH:GHR complex in 1992 (Figure 1C), which revealed GH bound to a dimer of GHR (44). The most surprising feature of the structure was the observation that two chemically distinct binding sites on GH bind similar epitopes on GHR. Following the GH:GHR structure, more complex structures followed, such as the tetrameric viral IL-6 (67) complex, the hexameric IL-6 (68) complex, and the dodecameric GM-CSF (69) complex, providing a more thorough understanding of cytokine/receptor engagement from several cytokine families. To date, no high-resolution structures have been solved that include the transmembrane or intracellular regions of cytokine receptors, although low-resolution negative-stain electron microscopy studies have captured the overall organization of these complexes (65, 70, 71).
The use of shared signal transducing receptors by cytokines is common. For example, three cytokines utilize the common β chain (βc), IL-3, IL-5, and GM-CSF (72), six cytokines utilize the common γ chain (γc), IL-2, IL-7, IL-9, IL-13, IL-15, and IL-21 (73), and more than ten cytokines utilize glycoprotein (gp)130, including IL-6, IL-11, leukemia inhibitory factor (LIF), CNTF and oncostatin M (OSM) (74, 75). As structures have now been solved of several representative cytokines from these families, the mechanisms of shared receptor use have begun to be understood. For example, the γc receptor has a large binding surface in the CHR, allowing it to bind structurally diverse cytokines (48, 73), in contrast, gp130 has a structurally rigid, chemically diverse binding surface at the CHR, with different gp130-binding cytokines interacting with different but overlapping regions of the surface (76). In shared receptor systems, cytokine-specific receptors with restricted expression, such as IL-6Rα or IL-15Rα, serve to limit the activity of cytokines to specific target cells despite their utilization of similar intracellular signaling pathways.
The JAK-STAT pathway is the most well-studied pathway activated in response to cytokines (Figure 2A). The major components of the pathway are cytokine, cytokine receptor, kinase (i.e., JAK), signal transducer (i.e., STAT), and negative feedback regulators (i.e., SOCS). JAKs are associated with the cytoplasmic domains of signal-transducing cytokine receptors and consist of four domains, a kinase domain, pseudokinase domain, 4.1 ezrin radixin moesin (FERM) domain, and Src homology 2 (SH2) phosphotyrosine-binding domain. The pseudokinase domain regulates the kinase domain (77), with the term “Janus kinase” referring to the presence of two kinase domains, real and pseudo, named for the two-faced Roman god (21). The FERM/SH2 domains form a single structural unit (78, 79), and are responsible for interacting with the cytokine receptor, through defined motifs on the receptor, termed Box 1 and Box 2 (80). Cytokine binding results in the activation and phosphorylation of the kinases, which then phosphorylate the cytokine receptor at STAT binding sites, serving to recruit STATs. Bound STATs are themselves phosphorylated, resulting in the activation of the STAT dimer, its translocation to the nucleus, and the expression of cytokine responsive genes. Importantly, different kinases are associated with different cytokine receptors—for example, the IFNα/β receptor primarily uses tyrosine kinase 2 (TYK2) (22) and βc primarily uses JAK2 (81). Furthermore, different receptor-kinase complexes result in activation of different STAT proteins—for example, STAT1/2 for IFNα/βR (22), STAT5 for βc (81), leading to different gene expression programs in response to signaling.
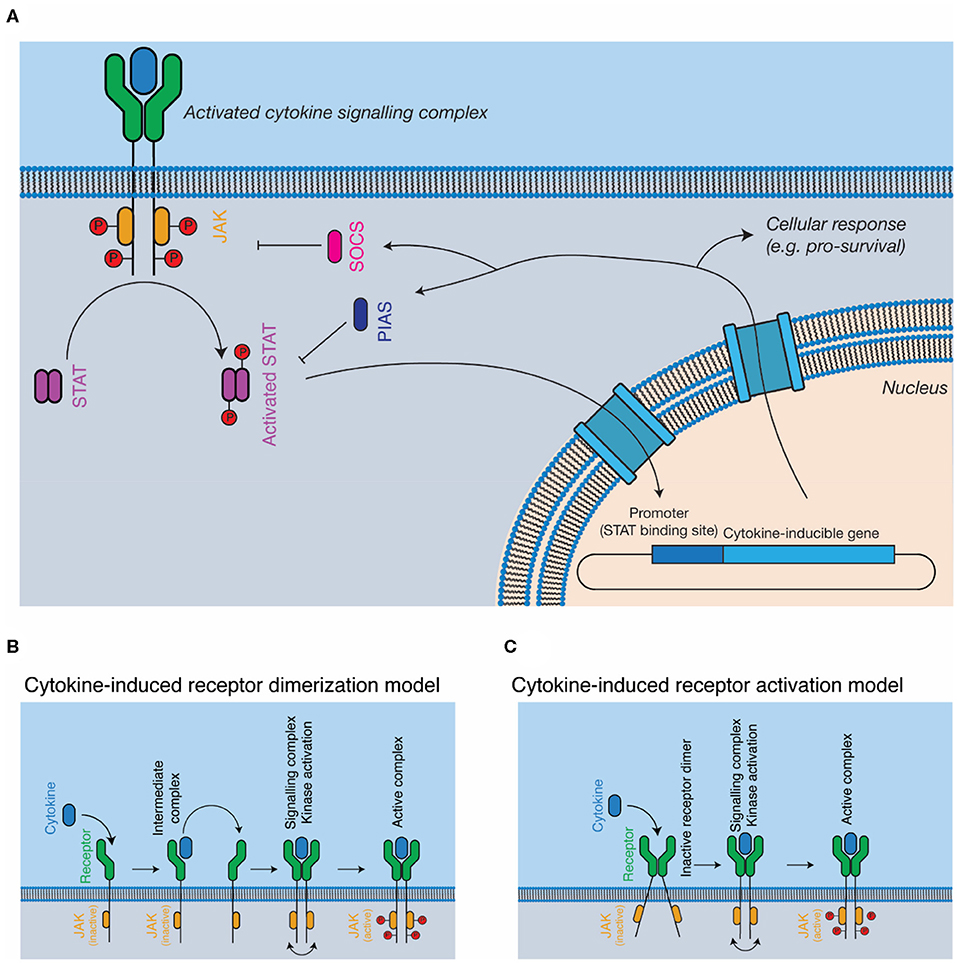
Figure 2. Cytokine signal transduction. (A) General schematic of the JAK-STAT pathway. Cytokine binding results in the activation of intracellular kinases (JAKs) that phosphorylate and activate STATs, which subsequently translocate to the nucleus, resulting in altered gene expression, and negative feedback on the pathway through the SOCS proteins. (B,C) Models for complex activation. Cytokines are thought to either, (B) dimerise receptors on the cell surface, resulting in kinase autophosphorylation and activation or (C) bind to pre-dimerised receptors on the cell surface, resulting in receptor activation through conformational alterations of the receptor dimer.
The SOCS proteins, which are expressed as a consequence of cytokine activation, negatively regulate the pathway (27). The SOCS proteins recruit the E3 ligase, Cullin5, resulting in the degradation of the receptor complex in the proteasome (82, 83). Two SOCS proteins, SOCS1 (84) and SOCS3 (85), also directly inhibit the kinase activity of the JAKs. The protein inhibitor of activated STAT (PIAS) proteins inhibit the activity of STAT through mechanisms that include directly blocking STAT interaction with nuclear DNA (86, 87). Several phosphatases act as negative regulators of signaling, such as the SH2-domain containing phosphatases, SHP1 and SHP2 (88, 89) and protein-tyrosine phosphatase (PTP) 1B (90). The lymphocyte adaptor protein, Lnk, serves as an additional negative regulator of signaling by several cytokines that signal using JAK2 (91).
The exact mechanisms by which cytokine engagement triggers signal transduction remain unclear and are the subject of active investigation. In the classical model of cytokine signaling, dimerization of signal transducing receptors simply brings the associated JAKs close enough in proximity to phosphorylate each other in trans (44, 92) (Figure 2B). However, several cytokine receptors, including GHR (43, 93), EPOR (94), and gp130 (95, 96) have been shown to exist as preformed dimers at the cell membrane (Figure 2C). Investigations of GHR suggest that cytokine binding results in a rearrangement of the transmembrane α-helices of the receptor, a conformational change that lifts pseudokinase domain mediated inhibition of the JAKs (43, 93). Determining the universality of such a mechanism will require the study of additional cytokine receptors, particularly those that signal through more complex hetero-dimeric or larger signaling complexes.
In addition to the JAK-STAT pathway, cytokines can utilize alternative signaling pathways, including the mitogen-activated protein kinase (MAPK) pathway, and the phosphoinositide 3-kinase (PI3K) pathway (81). The multi-adaptor protein SH2 domain containing tyrosine phosphatase (SHP2) interacts with several cytokine receptors and provides the link between the receptors and the MAPK pathway (97). Signaling through these pathways is generally less well understood than the JAK-STAT pathway.
The IL-6 family of cytokines is one of the largest cytokine families (Figure 3). These cytokines are unified by the near-universal use of the shared signal transducing receptor, gp130. The exception is IL-31, which uses the related receptor IL-31Rα, also known as gp130-like receptor (GPL) (102, 103). The distinct biological activity of IL-6 family cytokines is controlled by the restricted expression of the cytokine-specific receptors, such as IL-6Rα and IL-11Rα by a limited subset of cell types (104). Several cytokines can bind IL-6Rα in addition to IL-6, including CNTF (105), the IL-27 subunit IL-27p28 (also known as IL-30) (106), a IL-27p28 fusion with cytokine-like factor (107), and human herpes virus 8 IL-6 (vIL-6) (108), a viral analog of IL-6 with ~25% sequence identity to mammalian IL-6 (109). Receptor promiscuity is thus a common feature of the IL-6 family.
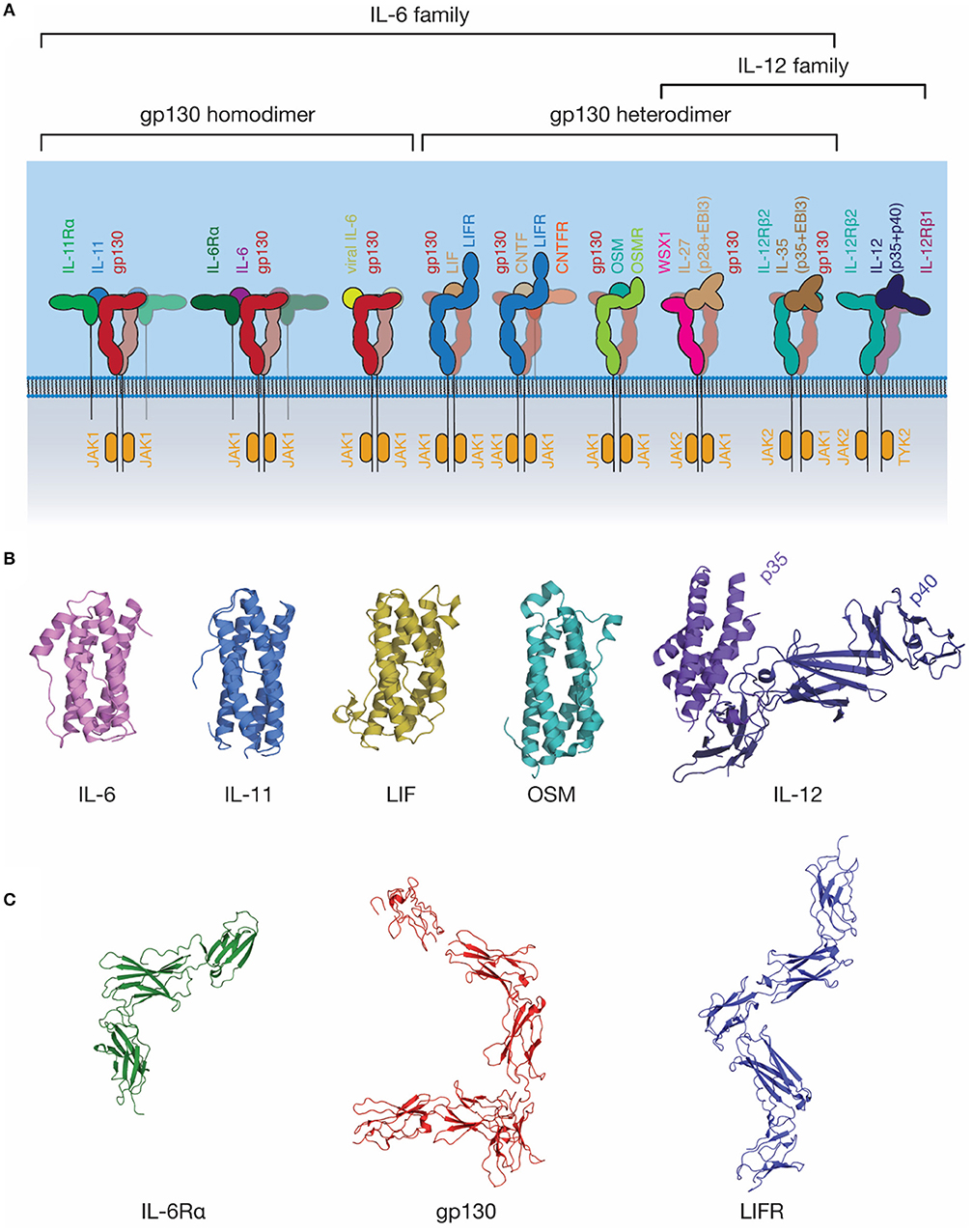
Figure 3. The IL-6 family of cytokines. (A) A schematic representation of selected IL-6 and IL-12 family cytokine-receptor complexes, illustrating the diversity in the stoichiometry of signaling complexes employed. Indicative JAK family members utilized by each signal transducing receptor are shown. (B) The structures of several IL-6 and IL-12 family cytokines: IL-6 [PDB ID: 1ALU (45)]; IL-11, [PDB ID: 4MHL (98)]; LIF [PDB ID: 1LKI (99)]; OSM [PDB ID: 1EVS (100)]; IL-12 [PDB ID: 1F45 (54)]. (C) The structures of extracellular domains of IL-6 family cytokine receptors: IL-6Rα [PDB ID: 1N26 (101)], the common signal transducing receptor, gp130 [PDB ID: 3L5H (58)] and the receptor for LIF and several other IL-6 family cytokines, LIFR [PDB ID: 3E0G (65)].
IL-6 was initially identified under several names in the 1980s (110, 111) as a protein involved in B-cell differentiation (112), a plasmacytoma growth factor (113), and a protein involved in the induction of acute phase proteins in the liver (114). Subsequent cloning of these proteins showed that they were all identical, thus they were given a common name, IL-6. IL-6 is the most well-characterized member of this family structurally, with crystal structures of IL-6 solved in 1997 (45, 115), the structure of IL-6Rα solved in 2002 (101), and the structure of the IL-6 signaling complex solved in 2003 (68) (Figures 3B,C, 4A). IL-6 is a typical four-α helical bundle cytokine, with the expected up-up-down-down arrangement of α-helices, with an additional, short α-helix in the CD loop (Figure 3B). The extracellular region of IL-6Rα consists of three domains (101), an N-terminal Ig-like domain, and two Fn3 domains, which form the IL-6 binding CHR (Figure 3C). The N-terminal Ig domain adopts a distorted Ig-like fold, and is dispensable for cytokine binding and biological activity (60, 68), although there is some evidence that it is required for correct trafficking of the receptor (60). IL-6 binds the surface formed by the two Fn3 domains, D2 and D3, comprising the CHR (68). C-terminal of the structured extracellular domains (D1-D3), there is a long linker region (52 residues), predicted to be disordered, that appears to function as a spacer in the signaling complex between the structured extracellular domains and the membrane (116–118).
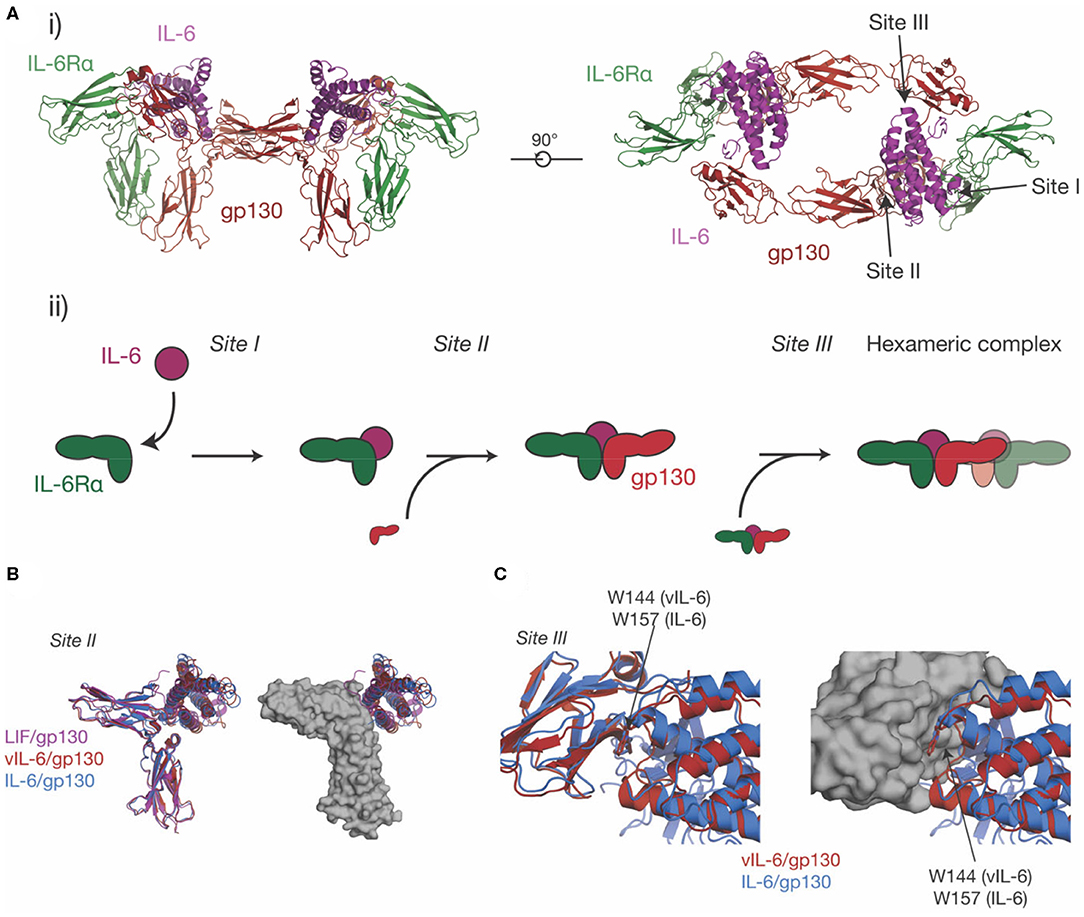
Figure 4. The structure of the IL-6 signaling complex. (A) (i) Two views of the structure of the hexameric IL-6/ IL-6Rα/gp130 complex [PDB ID: 1P9M (68)]. The three binding sites on the cytokine (site-I, binding IL-6Rα, site-II, binding gp130, site-III binding gp130), are indicated in the figure. The stepwise assembly of the complex is shown in (ii), with the interactions mediated by each of the binding sites indicated. (B) The binding of three IL-6 family cytokines to the CHR of gp130, IL-6, vIL-6 [PDB ID: 1I1R (67)] and LIF [PDB ID: 1PVH (76)]. The three cytokines do not induce significant rearrangements in the CHR of gp130 but adopt a different pose on the CHR and bind different regions in the surface of gp130. (C) The binding of IL-6 and vIL-6 to gp130 D1, the interaction that forms the hexameric complex. The two cytokines engage gp130 D1 in an analogous way. The key tryptophan “hot-spot” residue in site-III is indicated.
Gp130 is the common signal transducing molecule for nearly all IL-6 family cytokines, and some cytokines in the closely related IL-12 family. It was first identified in 1989 (119) as the component of the IL-6 signaling complex involved in signal transduction, and subsequently cloned in 1990 (120). Following this, gp130 was recognized as being a common component of the IL-11 (121), OSM (122), LIF and CNTF (123) signaling complexes. Structures of the CHR domains of gp130 became available in 1998 (124), and the full extracellular region of gp130 in 2010 (58) (Figure 3C). The extracellular domains of gp130 are those of a typical “tall” cytokine receptor, consisting of six domains, an N-terminal Ig-like domain, and five Fn3 domains (58). The first three, membrane-distal domains (D1-D3) are involved in cytokine recognition and complex formation, and are sufficient to bind cytokines and form a complex in solution (68, 76). The membrane-distal domains are also directly involved in gp130 activation, with oncogenic mutations that result in cytokine-independent activation of gp130 clustering in D2 (125). These mutations are thought to act by disrupting the D2/D3 interdomain linker, allowing the receptor to adopt an active conformation in the absence of ligand (126).
The three membrane proximal domains of gp130 (D4-D6) are not directly involved in binding the cytokine, but are required for signal transduction, as deletion of any of the domains results in an inactive receptor (127). Electron microscopy shows that the membrane-proximal domains are involved in the correct orientation of the intracellular kinases for signal transduction (65, 70, 71, 128). In addition to the extracellular domains, gp130 contains a large intracellular domain, which is involved in binding molecules required for signal transduction. Structurally, little is known about the intracellular domain of gp130, although NMR studies have shown that the isolated intracellular domain is disordered (65). JAK1, which mediates intracellular signaling, has been shown to bind gp130 at the Box 1 motif in the intracellular domain of gp130 (80). STAT3 (17, 129) and STAT1 (130) bind at C-terminal phosphotyrosine residues in the intracellular domain of gp130 (131). Specifically STAT3 utilizes Tyr767, Tyr814, Tyr905, and Tyr915, while STAT1 utilizes Tyr905 and Tyr915 (132). SHP2 is also recruited by gp130 at the intracellular domain (133), interacting with Tyr759 providing the link between gp130 and the MAPK pathway (134). The same Tyr759 allows for SOCS3 regulation of cytokine signaling (27, 85).
Prior to the determination of the structure of IL-6 in complex with the cytokine binding domains of its receptors (Figure 4Ai) (68), there was extensive evidence from analytical ultracentrifugation and electrophoresis that the complex was hexameric, comprising two copies each of IL-6, IL-6Rα, and gp130 (135–137). Concurrently, mutagenic studies identified three binding sites on IL-6 (136), which were later confirmed in the structure of the complex (68). Site-I is responsible for binding IL-6Rα, site-II is responsible for binding the first molecule of gp130, and site-III is responsible for binding the second molecule of gp130, resulting in the formation of the hexameric complex (Figures 4Ai, ii). Site-I and site-II are positioned on the cytokine in a broadly analogous manner to GH and form a similar trimeric complex, with IL-6 binding the CHRs of IL-6Rα and gp130 (33) (Figure 4Ai). The distinct cytokine:Ig domain interaction between the cytokine and D1 of gp130 is unique to IL-6 family cytokines (138). This interaction is formed by site-III on the cytokine. The complex is formed by ten interdependent interfaces between IL-6 and the two receptors, and between the receptors, with the earlier binding events creating composite binding surfaces to enable subsequent receptor recruitment. The structure of the IL-6 signaling complex has aided drug design studies (139, 140), showing its value in the design of novel therapeutics.
The site-II/CHR region of gp130 is involved in the binding of all gp130-binding cytokines. Alongside the structure of the IL-6 signaling complex, structures were solved of vIL-6 in complex with gp130 (67) and LIF in complex with gp130 (76). All three cytokines engage the CHR of gp130 via the site-II region of the cytokine (Figure 4B). The structures showed that vIL-6, IL-6, and LIF engage different but overlapping binding regions in the CHR of gp130, with the three cytokines adopting different poses. A key residue in site-II of gp130, Phe169, forms important interactions with IL-6, vIL-6, and LIF. Surprisingly, the cytokine binding surface of gp130 is relatively rigid, and does not significantly change conformation in response to the binding of different cytokines (76). The CHR of gp130 presents a large, chemically diverse binding surface and the different regions engaged by IL-6, vIL-6, and LIF result in each cytokine/gp130 interaction displaying different thermodynamic properties (76). The size and “thermodynamic plasticity” (76) of the CHR of gp130 is thought to result in its promiscuous binding to multiple cytokines (33, 76).
IL-6 and vIL-6 interact with the Ig-like domain D1 of gp130 through site-III on the cytokine. The interactions between IL-6/gp130 D1 and vIL-6/gp130 D1 are broadly analogous (Figure 4C). In both complexes, a conserved tryptophan is the key hydrophobic “hot spot” residue (Trp157 in human IL-6, Trp144 in vIL-6), providing ~25% of the buried surface area at site-III. Likewise, the N-terminus of gp130 forms a short mainchain-mainchain interaction with the AB loop of the cytokine (67, 68). The site-III interface on gp130 D1 is otherwise relatively chemically and structurally featureless (33), providing a low-affinity binding surface that is reliant on prior interactions with other receptors for stable complex assembly. An interaction similar to the gp130-D1 interaction is formed by LIF with the Ig-D3 and Fn3-D4 of LIFR, although this interaction buries more surface area and forms more polar interactions (59).
No structural data are available for the gp130 binding epitopes of any IL-6 family cytokines other than vIL-6, IL-6, and LIF. Mutagenesis of gp130 shows that IL-11 and IL-6 both require D1 of gp130 for signaling, and bind a similar epitope in the CHR (141). Monoclonal antibodies against gp130 have been developed that antagonize signaling through specific cytokines, including IL-11 and IL-6-specific neutralizing antibodies, suggesting that each cytokine engages gp130 using a structurally different mechanism (142); however, the structural basis of this specificity is currently unknown.
No high-resolution structures are available of the complete extracellular regions of any IL-6 family cytokine complex. All complexes described above comprise heavily truncated forms of the receptors to facilitate crystallization. Electron microscopy (both cryogenic and negative stain) has been used to study several complexes, including the IL-6 complex (70, 71), the LIF complex (65), and the IL-11 complex (128). The resolution in these studies is insufficient to resolve structural detail of the complex, although they reveal a common “doughnut-shaped” architecture, with the “legs” of the tall cytokine receptors, LIFR and gp130, bent to create a complex with a hole in the middle. The details of any contacts between the membrane proximal domains of the receptors in these complexes remain to be elucidated and will require the determination of high-resolution structures of the complete extracellular regions of the complexes.
In addition to “classic” IL-6 signaling through membrane-bound IL-6Rα and gp130, IL-6 can also bind a soluble form of IL-6Rα (sIL-6Rα). The IL-6/sIL-6Rα complex can then engage membrane-bound gp130, allowing the stimulation of cells that do not express IL-6Rα, a process known as trans-signaling (119, 143) (Figure 5). IL-6 trans-signaling is implicated in IL-6 mediated inflammation (143). sIL-6Rα is generated through alternative splicing (144) and through cleavage of the intact receptor by the membrane-bound metalloproteases, ADAM10 and ADAM17, resulting in shedding of the extracellular receptor domains (143). The physiological antagonist of trans-signaling is soluble gp130 (sgp130), which can bind to the sIL-6Rα/IL-6 complex extracellularly, thereby neutralizing its cellular activity (145).
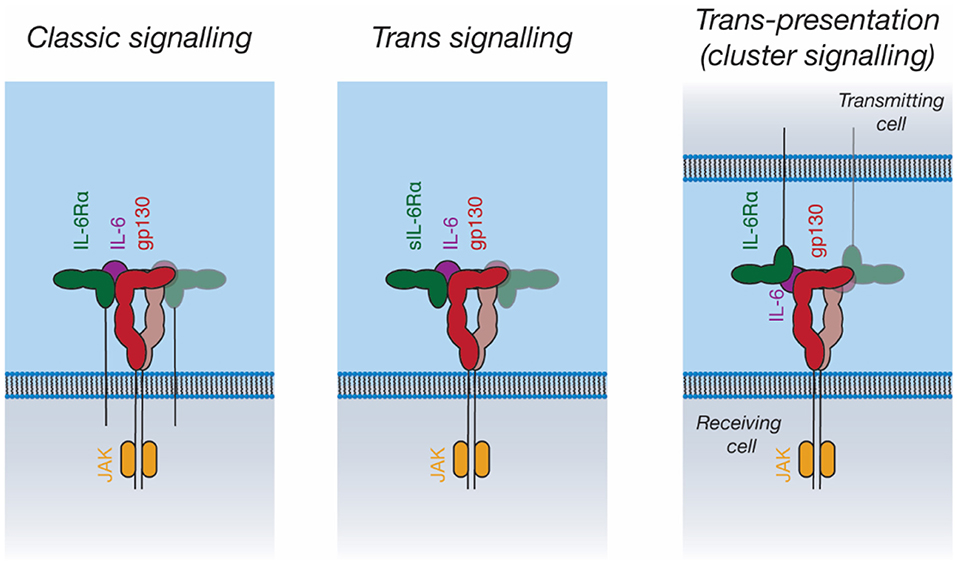
Figure 5. Signal transduction by IL-6. IL-6 can activate intracellular pathways in three ways: “Classic” signaling, in which IL-6 binds to membrane-bound IL-6Rα, and subsequently binds to membrane-bound gp130 on the same cell; “trans-signaling”, in which IL-6 binds to soluble IL-6Rα, subsequently binding membrane-bound gp130; and “trans-presentation,” in which IL-6 binds membrane-bound IL-6Rα on a “transmitting cell” and subsequently engages gp130 on a neighboring (“receiving”) cell, activating intracellular signaling pathways in the receiving cell.
IL-11 trans-signaling has recently been identified (146). The membrane metalloprotease ADAM10 can cleave IL-11Rα to produce sIL-11Rα, which can engage IL-11 and gp130 in an analogous manner to IL-6/sIL-6Rα (146). To date, no clear biological role has been ascribed to IL-11 trans-signaling. In diseases shown to be driven by classic IL-11 signaling, for example gastrointestinal cancers, it has been shown that there is no role for IL-11 trans-signaling (147). Likewise, the loss of classic IL-11 signaling is associated with defects in embryo implantation; however, the inhibition of IL-11 trans-signaling in mice does not result in infertility (148). Fusion proteins of IL-6 with IL-6Rα and IL-11 with IL-11Rα (“hyper-IL-6 and hyper-IL-11”) are used to mimic trans-signaling experimentally (149, 150).
Recent studies have proposed a third IL-6 signaling mechanism, trans-presentation, whereby IL-6 binds IL-6Rα on a “transmitting cell,” which then presents the IL-6/IL-6Rα complex to gp130-expressing cells (Figure 5) (151, 152). This was shown to be critical for the differentiation of TH17 T helper cells, where IL-6/IL-6Rα is presented in trans by dendritic cells (151). Trans-presentation has also been shown to be possible for IL-11Rα, however a defined biological role for this has not been identified (152). Trans-presentation of IL-6 family cytokines has not yet been characterized structurally; such a signaling mode would require large rearrangements of the IL-6 signaling complex components. Other cytokines such as IL-2 (153) and IL-15 (154) can utilize similar trans-presentation mechanisms, where dendritic cells present the cytokine in trans to antigen-specific T-cells (48, 155).
The IL-12 family of cytokines is closely related structurally to the IL-6 family of cytokines, indeed, it has been suggested that a clear distinction between the two families is almost impossible to define (156). In contrast to the majority of the IL-6 family, all IL-12 family cytokines consist of two subunits, a smaller four-α helical subunit, and a larger all-β protein cytokine receptor subunit, which is analogous to the α-receptors for IL-6 and IL-11. For example, IL-12 consists of two subunits, p35, analogous to a four-α helical bundle cytokine, and p40, which resembles a class I cytokine receptor (Figure 3B) (54).
IL-27 and IL-35 are two IL-12 family cytokines that utilize gp130 as a signal transducing molecule and, thus, are also grouped as members of the IL-6 family (Figure 3A) (157, 158). IL-27 consists of a complex of IL-27p28 and Epstein–Barr virus-induced gene 3 (EBI3) that signals through a heterodimer of WSX1 and gp130 (Figure 3A) (157). In addition to this complex, IL-27p28 may utilize IL-6Rα as the cytokine-receptor subunit to signal through a gp130 dimer (106). IL-27p28 was also shown to antagonize IL-6 and IL-27 signaling through gp130, but not OSM signaling, suggesting that IL-27p28 may compete with cytokines that bind D1 of gp130 (159). IL-35 can signal using a heterodimer of IL-12Rβ2 and gp130, or homodimers of either IL-12Rβ2 or gp130; however, the molecular mechanisms underpinning this promiscuity are currently unclear (158). Broadly, these findings suggest an evolutionary relationship between the IL-6 and IL-12 families of cytokines and underscore the promiscuity of cytokine receptors in the IL-6/IL-12 superfamily.
A distant homolog of gp130 has been identified in Drosophila melanogaster, the receptor domeless (dome) (160), which is the likely evolutionary ancestor to all IL-6 family cytokine receptors (161). Dome shares a similar domain structure to gp130 and LIFR, and has a putative CHR, albeit with low sequence identity to the CHR of gp130. A putative ligand for Dome, Unpaired-3 (Upd3) (162) has also been identified, alongside JAK kinases (Hopscotch) and STAT transcription factors (Marelle) (163). The Dome-Hopscotch pathway has been shown to have several roles in Drosophila physiology, including in responding to bacterial infection (164), in oogenesis (164), in hemocyte proliferation (165), and in tissue development (166, 167), showing that cytokine pleiotropy is a common feature in metazoans. Neither dome or Upd3 have been studied structurally, although recombinant Upd3 has been produced, and has been shown by circular dichroism spectroscopy to have a predominately α-helical secondary structure (168). Zebrafish possess a mammalian-like cohort of cytokines, with relatives of all extant mammalian cytokine families present, suggesting that an increase in diversity of cytokines and receptors occurred with the evolution of the adaptive immune system in vertebrates (169, 170).
IL-11 was first identified in 1990, following the discovery of a protein factor that stimulated a murine plasmacytoma cell line previously thought to be IL-6 dependent (171). The following year, IL-11 was also identified as a factor secreted from a bone marrow derived cell line culture, which inhibited adipogenesis in preadipocytes (172, 173), thus the pleiotropic nature of IL-11 signaling was appreciated early. While there was a flurry of activity surrounding IL-11 in the 1990s, there has been less research activity since. However, in the last decade there has been a renewed interest in IL-11 following its emerging role in numerous diseases.
In contrast to IL-6, LIF and other IL-6 family cytokines, little was previously known about the structure of IL-11 or IL-11Rα. We reported the first crystal structure of IL-11 in 2014 (98) and have recently reported a higher-resolution structure of the cytokine (Figure 6A) (174). Our structures show that IL-11 is ~5 Å longer than IL-6, suggesting differences in binding mode and geometry within the signaling complex. Likewise, the IL-11Rα binding site (site-I) and the first gp130 binding site (site-II) of IL-11, previously identified through mutagenesis (175, 176), are different in chemical character to IL-6, with site-I more hydrophobic (Figure 6B). Our recent structure of IL-11Rα (Figure 6C) (174) revealed that the cytokine binding site of the receptor is more hydrophobic in character than IL-6Rα, consistent with the corresponding site of IL-11 and suggesting distinct mechanisms of cytokine engagement.
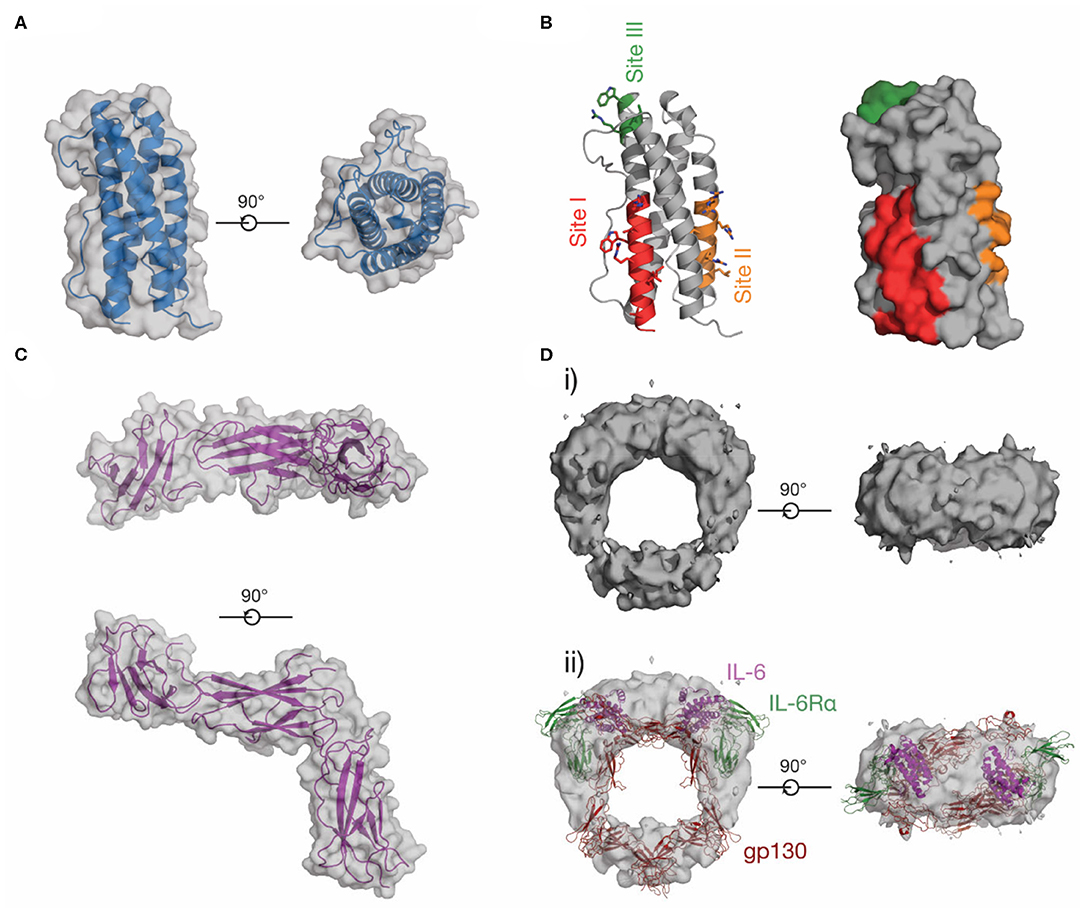
Figure 6. The structure of IL-11. (A) Two views of the structure of IL-11 [PDB ID: 6O4O (174)]. (B) Receptor-binding sites on IL-11, which have been identified through mutagenic studies on human and mouse IL-11. (C) Two views of the structure of the extracellular domains of IL-11Rα [PDB ID: 6O4P (174)]. (D) A low-resolution EM map of the extracellular IL-11 signaling complex [EMD-1223 (128)] (i), additionally shown overlaid with a model of the extracellular IL-6 signaling complex, (ii) [generated from PDB IDs: 3L5H (58), 1P9M (68)].
No high-resolution structural data for the IL-11 signaling complex are currently available in the literature, although sequence analysis (121, 177) and our structural data (174) show that IL-11Rα and IL-6Rα are structurally similar. The IL-11 signaling complex, like the IL-6 signaling complex, is thought to be hexameric, as shown by immunoprecipitation and electrophoresis (178). Contemporaneous mutagenic studies (175, 176, 179) also identified site-I, II, and III on IL-11 (Figure 6B), suggesting that IL-6 and IL-11 form an active signaling complex using a broadly similar mechanism. A low-resolution (~30 Å) cryoEM density map of the IL-11 signaling complex extracellular domains (128) (Figure 6D) shows that the overall arrangement of the complex is broadly similar to the IL-6 signaling complex (Figure 6Dii), although the details of complex formation were not clear at this resolution. We have recently solved structures of the IL-11 signaling complex that provide high resolution detail of the assembled complex (unpublished).
Early studies of IL-11 revealed that it was a potent hematopoietic factor, acting synergistically in culture with other cytokines, such as IL-3 (180, 181) and IL-4 (182). In particular, IL-11 was found to have a role in megakaryocytopoiesis, causing the maturation of megakaryocytes, large cells which form platelets (181). In mice, IL-11 alone is a potent hematopoietic stimulator following radiation therapy and chemotherapy, and markedly increases platelet counts (183). Recombinant IL-11 is approved by the FDA to treat thrombocytopenia following radiation treatment in humans (184), and is commonly prescribed to breast cancer patients. In addition to its well-characterized role in megakaryocytopoiesis, IL-11 has other roles in hematopoiesis (185), for example, in lymphopoiesis (186), in erythropoiesis (187), and in myelopoiesis (188).
IL-11 signaling has been shown to promote osteoblast differentiation, and thus bone formation, with IL-11Rα knockout mice showing abnormal craniofacial features (189–191). In humans, mutations in the genes for IL-11 and IL-11Rα are associated with a reduction in height (192, 193), suggesting that IL-11 signaling has a role in regulating growth. Likewise, a genetic variant in the gene for IL-11, resulting in a substitution mutation (R112H), is associated with osteoarthritis and a reduction in height (192, 194). Biochemical characterization of the mutant cytokine has shown that it does not alter the biological activity of IL-11, but compromises the stability of the protein (195).
Over the past decade, a number of studies have identified mutations in the gene for IL-11Rα, which result in a genetic disease associated with craniosynostosis (196–198). Craniosynostosis is a condition in which bone plates in the skull fuse too early, resulting in facial abnormalities and an abnormally shaped skull. The disease is rare, and has been found in families with diverse geographic origins (196). Generally, the disease occurs as a result of point substitution mutations in the extracellular domains of IL-11Rα (196, 199), and many of these mutations are situated in regions distant from the putative cytokine or receptor binding sites. Several of the mutations have been shown to impair correct processing and surface expression of the receptor (199). Molecular dynamics simulations using our IL-11Rα structure indicate that some mutations destabilize the receptor and may have indirect effects on the cytokine binding region (174).
IL-11 is highly expressed as a consequence of viral induced asthma (200), and overexpression of IL-11 in the airways of mice results in remodeling of the airways, inflammation and asthma-like symptoms (201). Subsequent studies have shown that IL-11 signaling is critical for a TH2-mediated inflammatory response in the lung (202), and that inhibition of IL-11 signaling in the lung alleviates inflammation, implying that IL-11 signaling is a therapeutic target in asthma (203). Similarly, IL-11 has been shown to drive lung inflammation in a murine model of Mycobacterium tuberculosis infection (204).
Female knock-out mice lacking the gene for IL-11Rα are infertile, and cannot undergo the uterine transformations required for embryo survival (205). Likewise, IL-11 and IL-11Rα have been localized to reproductive tissues during early pregnancy in primates, suggesting a role in placentation and decidualization (206). Related to this, inhibition of IL-11 signaling impairs decidualization and prevents pregnancy in mice, suggesting that therapeutic inhibition of IL-11 may be a potent contraceptive (207). Defects in the production of IL-11 have also been associated with anembryonic pregnancy, a cause of miscarriage (208). IL-11 signaling inhibits and regulates invasion of extravillous trophoblasts, cells which are key in placentation for the formation of blood vessels (209–211). Thus, elevated IL-11 is associated with preeclampsia, a disease where placentation is impaired, resulting in hypertension (211). Together, these studies suggest that IL-11 has key roles in driving the tissue transformations that occur as a result of pregnancy.
IL-11 has been implicated in fibrosis of the heart (212), liver (213), and lung (214, 215). Fibrosis is the generation of excess connective tissue, and is a hallmark of several diseases, including late-stage cardiovascular disease, and liver diseases such as non-alcoholic liver disease. In the heart, IL-11 has recently been identified as a key fibrotic factor, acting downstream of the main fibrotic factor TGFβ1, driving fibrotic protein synthesis in an autocrine manner (212). IL-11 has a similar role in driving inflammation and fibrosis of the liver (213). Interestingly, in both cases, the effect has been shown to be driven by non-canonical signaling via the MAPK/ERK pathway, rather than via the JAK-STAT pathway. Surprisingly, canonical IL-11 signaling via STAT3 has previously been ascribed a cardioprotective role, inhibiting cardiovascular fibrosis and preventing cardiovascular remodeling following myocardial infarction (216). These contradictory observations may be a consequence of the source of IL-11 used in either study, as it was shown that human IL-11, previously used to show that IL-11 is cardioprotective, does not activate mouse cardiac fibroblasts, while murine IL-11 strongly activates murine cardiac fibroblasts (212). Alternatively, it may suggest different roles for IL-11 in response to different cardiovascular stresses. More broadly, this may reflect an inadequate understanding of the species-specific effects of IL-11, or differences in signaling in humans as compared to mice.
IL-11 signaling drives several cancer hallmarks (217, 218) including cell survival, metastasis, and invasion (219–221). IL-11 levels are significantly higher in a murine model of gastric cancer (222), and IL-11 is the major factor that drives STAT3 activation and corresponding inflammation in murine gastric and colon cancer models, as well as human cell line xenograph models of these cancers (221). A role for IL-11 signaling in breast cancer has been less well-described, but elevated levels of IL-11 and IL-11Rα are associated with poor patient outcomes (223, 224) and both IL-11 and IL-6 are associated with breast cancer metastasis into bone (225). IL-11 is also associated with endometrial cancer, and is associated with increasing tumor grade (226). Elevated levels of IL-11 are found in several other types of cancer, including pancreatic cancer (227), skin cancer (228), and bone cancer (229), although a precise role for IL-11 signaling in many of these cancers remains to be defined.
Given the role of cytokine signaling in numerous pathological conditions there is broad interest in the development of therapeutic agents that block their activity. Generally, inhibition can occur at several points in the cytokine signaling pathway—either by preventing the protein-protein interactions on the cell surface, or by targeting components of the signal transduction machinery within the cell. Conversely, recombinant cytokines can also be used to therapeutically boost cytokine signaling. Here we provide an overview of several approaches to therapeutically modulate cytokine signaling that are in development, as well as those currently used in the clinic. We focus our discussion on how advances in these areas may inform the design of IL-11 signaling inhibitors suitable for clinical use.
JAK inhibitors are widely used, orally bioavailable, small molecules for the treatment of blood cancers and inflammatory diseases (230) (Figure 7). Six JAK inhibitors are used clinically, with several in development. For example, the JAK1/2 selective inhibitor ruxolitinib (231) is used to treat a group of rare blood cancers associated with an activating mutation in JAK2. Similarly, tofacitinib (non-selective) and baricitinib (selective for JAK1/2) are JAK inhibitors used to treat the inflammatory disease rheumatoid arthritis (232, 233). JAK inhibitors are now undergoing clinical trials for a broader array of inflammatory diseases (234). Challenges with developing JAK inhibitors are largely a consequence of the inherently non-specific nature of the drugs. Moreover, JAK inhibition may be associated with severe side effects, including opportunistic viral infections, likely a consequence of inhibition of interferon-mediated protective antiviral signaling (235). Similarly, due to the central roles of cytokine driven JAK activation in hematopoiesis, JAK inhibitors have been noted to cause mild anemia and neutropenia (236, 237). Despite this, JAK inhibitors are widely used, and efforts to develop novel JAK inhibitors, particularly inhibitors that are selective for a specific kinase, are ongoing.
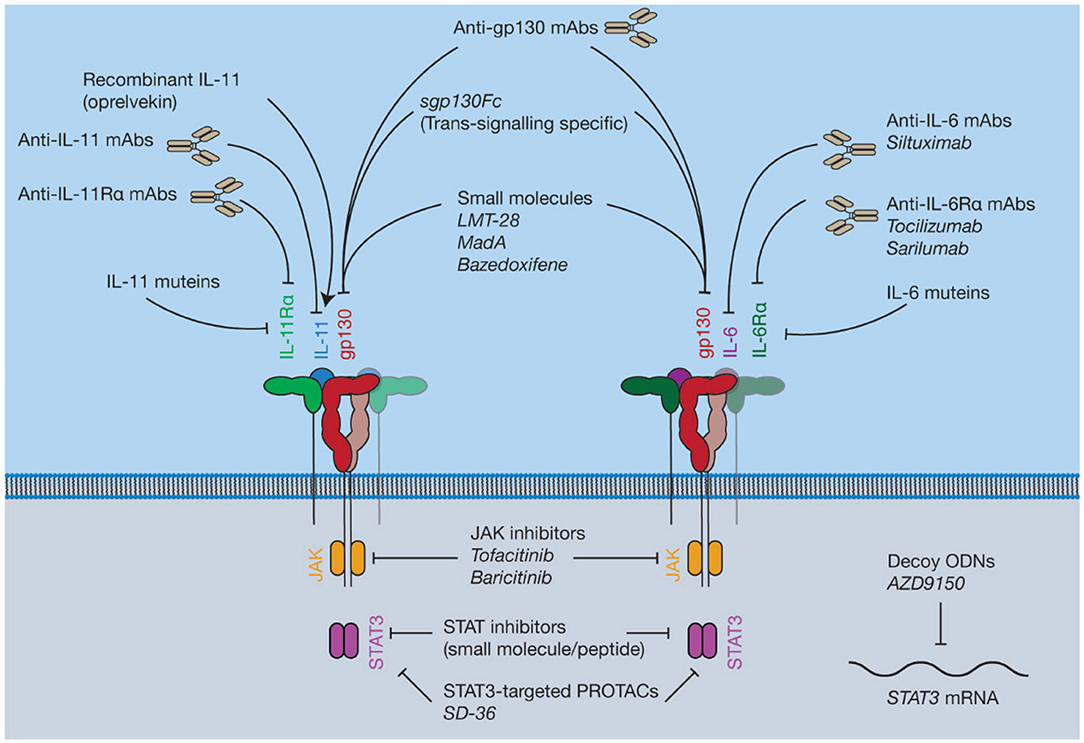
Figure 7. Pharmacological approaches to target IL-6 and IL-11 signaling. Current inhibitors of IL-6 and IL-11 signaling include protein antagonists such as cytokine mutants and antibodies, small molecule protein-protein interaction (PPI) inhibitors targeting gp130, recombinant IL-11, small molecule inhibitors of proteins in the intracellular JAK-STAT pathway, and decoy oligodeoxynucleotides (ODNs) targeting the STAT3 mRNA.
Inhibitors of STAT activity are in various stages of development (238). Phase I and II trials have been conducted on several drug candidates targeting STAT3, although the results are pending (239, 240). These inhibitors are generally peptides or small molecules designed to prevent STAT dimerization (241, 242), or decoy oligodeoxynucleotides (ODNs) designed to target expression of the STAT gene directly (243). Recently, a small-molecule proteolysis targeting chimera (PROTAC), SD-36 (244), which selectively targets STAT3 over other STAT family members, has been described. Direct inhibition of activated STATs is at a less advanced stage compared to kinase inhibitors, or drugs targeting the cytokine/receptor interaction directly, with current inhibitors having low potency and poor pharmacokinetic properties (245). For example, curcumin, an extract of the turmeric plant, Curcuma longa, has been used in traditional medicine for centuries for its anti-inflammatory properties (246). Mass spectrometric and computational docking studies have shown that curcumin directly interacts with STAT3 to inhibit phospho-STAT3 dimerization (247). Several in vitro studies demonstrate that curcumin is an inhibitor of STAT3 signaling (247, 248). However, the use of curcumin as a drug candidate or treatment is controversial (246, 249). Generally, direct targeting of STATs may not have clear benefits over existing therapeutic strategies, such as JAK inhibitors, which may hinder clinical uptake of STAT inhibitors.
Several small molecules have been described that are believed to bind to gp130 and inhibit the protein-protein interactions (PPIs) that result in complex formation (Figure 7). Despite the challenges of targeting PPIs, as they present large flat binding surfaces (250), small molecule modulation of PPIs is potentially invaluable therapeutically. Small molecule inhibitors could be more specific for the inhibition of signaling through individual cytokines compared to JAK inhibitors, which modulate the signaling of numerous cytokines. Moreover, PPI-inhibitors would likely be cheaper, orally bioavailable, and have a shorter half-life compared to biologic therapies, which is beneficial in the event of serious adverse events (251).
Madindoline A (MadA), a natural product isolated from Streptomyces nitrosporeus culture, is a small molecule shown to specifically inhibit the activity of IL-6 and IL-11 in vitro (252). MadA has subsequently been shown to inhibit the action of IL-6/IL-11, but not LIF, in bone resorption and macrophage differentiation (253). Additional studies have shown that MadA binds specifically to gp130, with a low affinity (254). Chemical synthesis of MadA is difficult (255) and it is produced in low yields by bacterial fermentation, limiting its potential as a drug candidate for large scale production.
The small molecule gp130 inhibitor SC144 was identified serendipitously from efforts to design a human immunodeficiency virus (HIV) integrase inhibitor, which would be a potential anti-HIV drug (256, 257). Several candidate HIV integrase inhibitors were highly cytotoxic (258). A library was built from these cytotoxic molecules (256) and further screening and lead optimization resulted in the synthesis of SC144 (257), which was effective against a variety of cancer models (259). Subsequently, it was shown that SC144 binds gp130 and inhibits the activity of IL-6 and LIF, likely through binding the CHR of gp130, resulting in suppression of cancer growth in human ovarian cancer xenographs (260). Following this initial description of its activity, SC144 has been used by various groups as an experimental tool to block IL-6 signaling through gp130 [see for example (261–263)].
Another small molecule inhibitor that has been shown to bind to gp130, LMT-28, was identified by screening a library of ~1,000 compounds (264). Computational docking suggested that LMT-28 binds to D1 of gp130, and the putative binding region in D1 of gp130 was supported using site-directed mutagenesis (265). Likewise, SPR showed that LMT-28 specifically bound gp130, with a dissociation constant (KD) of 7.4 μM, and LMT-28 was able to out-compete IL-6/IL-6Rα for gp130 binding (264). LMT-28 has been shown to specifically inhibit IL-6/IL-11 driven cell proliferation, and block IL-6-driven inflammation in vivo (264). In contrast, LMT-28 does not inhibit OSM/LIF activity, consistent with a binding site in D1 of gp130 (264).
Bazedoxifene is an FDA-approved selective estrogen receptor modulator used clinically in combination with other drugs to treat osteoporosis in elderly women (266). It was recently shown that bazedoxifene inhibited gp130 signaling, following an in silico screen on the IL-6/gp130 site-III interface (139). Bazedoxifene has been shown to suppress STAT3 activation through IL-6, inhibit tumor growth in a murine model of rhabdomyosarcoma, a soft-tissue sarcoma (267), and inhibit the proliferation of IL-6 dependent cell lines (268). Bazedoxifene has also been shown to block STAT3 activation by IL-11 in human cancer cell lines, and reduce the tumor burden in murine models of gastric cancer (140). Bazedoxifene was also shown to inhibit IL-6 signaling in triple negative breast cancer cell lines (269), and in murine models of the inflammatory cardiovascular disease abdominal aortic aneurysm (270). Recently, more efficacious analogs of bazedoxifene have been synthesized (271). Given that bazedoxifene is already used clinically, and thus has an established safety profile, it represents a potential small molecule inhibitor of both IL-11 and IL-6 signaling that could be used therapeutically.
Generally, with some exceptions, recombinant cytokines have not seen wide use therapeutically. Although rare, long-term treatment with recombinant cytokines can result in the generation of endogenous antibodies against the cytokine (272). More generally, the pleiotropic nature of many cytokines may result in unpredictable and intolerable inflammation-associated side-effects, which could limit the use of recombinant cytokines in the clinic (273, 274).
Recombinant human IL-11 (oprelvekin) was FDA-approved in 1998 (184, 275, 276) for the treatment of thrombocytopenia (low platelet levels) in myelosuppressive chemotherapy, as a substitute for platelet transfusions. Oprelvekin has also undergone a clinical trial for use thrombocytopenia in myelodysplastic syndrome, in which the bone marrow fails to properly mature blood cells (277). Oprelvekin is, however, not widely used, both for reasons of cost (278) and due to toxicity associated with mild anemia, periostitis, edema and in some cases neuropathy (279, 280). This toxicity can be managed by limiting the dose of oprelvekin (281). IL-11 also has anti-inflammatory properties, and oprelvekin has also undergone small clinical trials in inflammatory bowel disease (282) and rheumatoid arthritis (283). Both trials were inconclusive, and no further trials for either of these indications have been published.
Numerous monoclonal antibodies (mAbs) are used clinically to target IL-6 signaling (284), for example, the anti-IL-6Rα mAbs tocilizumab (285) and sarilumab (286), and the anti-IL-6 mAb siltuximab (287) are used to treat several diseases including rheumatoid arthritis and kidney cancer (Figure 7). Antibodies targeting IL-6 signaling are generally well-tolerated but have been noted to result in adverse events. For example, long-term clinical trials have noted that tocilizumab treatment can result in opportunistic infection, neutropenia and gastrointestinal disorders (288, 289), likewise infection, fatigue and neutropenia have been noted as potential adverse effects of siltuximab (290). The anti-IL-6 mAb olokizumab is currently undergoing a phase III clinical trial for rheumatoid arthritis (ClinicalTrials.gov identifier NCT02760368). Structures show that the olokizumab Fab blocks site-III of IL-6, preventing formation of the IL-6 hexameric complex (291). Structures have also been solved of two anti-IL-6 Fabs, which bind site-I, mimicking the IL-6/IL-6Rα interaction (292). No structures are available of the FDA-approved anti-IL-6 signaling antibodies in complex with their antigen.
Viral infections, including influenza (293), and severe acute respiratory syndrome (SARS) (294, 295), caused by SARS-coronavirus (CoV), can induce cytokine release syndrome (often referred to as “cytokine storm”), a severe immune reaction frequently associated with elevated serum IL-6 (296, 297). Severe coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2 (298), is associated with elevated serum IL-6 and cytokine release syndrome (299–301). Thus, IL-6 signaling inhibition may be a strategy for managing severe and critical COVID-19 (302). Accordingly, tocilizumab is currently undergoing several expedited clinical trials in severe and critical COVID-19 patients (for example, ChiCTR ID: ChiCTR2000029765, ChiCTR2000030894; ClinicalTrials.gov ID: NCT04315480, NCT04317092, NCT04372186, NCT04320615) (303). Tocilizumab appears to reduce mortality in severe and critical COVID-19 patients (300, 304–307), however in some cases poor outcomes have been noted (308).
Antibodies against IL-11 (214, 309) and IL-11Rα (213, 310, 311) that inhibit IL-11 signaling have been described and patented, although none are clinically available. The mechanisms of action of these antibodies have not been described in the literature.
Antibodies against gp130 have been described (142) that specifically antagonize signaling through a specific cytokine or cytokines, although they are not used in the clinic. The structural basis of this specificity is currently unknown, although epitope mapping studies have been conducted on the antibodies (142, 312), which show that the IL-11-specific mAb, B-P4, binds the membrane proximal region (D4-D6) of gp130 and not at the CHR. The OSM/LIF-specific mAb (B-K5), CNTF-specific mAb (B-P8) and broadly neutralizing mAb (B-R3) bind at the CHR of gp130, presumably sterically interfering with cytokine binding (142, 312).
Many of the harmful, pro-inflammatory effects of IL-6 signaling are believed to be caused by trans IL-6 signaling (143). Soluble gp130 (sgp130) is an antagonist of trans IL-6 signaling (145). Sgp130 fused to an IgG Fc fragment (sgp130Fc, olamkicept) is currently under development as an IL-6 trans-signaling specific inhibitor (313). The effect of sgp130Fc treatment has been studied in animal models of a number of inflammatory diseases including several cancers (314, 315), arthritis (316, 317), inflammatory bowel disease (318, 319), and pancreatitis-associated lung inflammation (320). The side effects of existing treatments targeting IL-6 signaling in humans are believed to result from a blockade of classic signaling, resulting in an increased susceptibility to infections, due to the key role of IL-6 signaling in responding to infection (313, 321). In animal models, blockade of IL-6 trans-signaling does not alter the IL-6 dependent response to infection (321). Sgp130Fc is currently undergoing phase II clinical trials for colitis (313) (ClinicalTrials.gov ID: NCT03235752; DRKS-ID: DRKS00010101). An anti-trans-signaling nanobody has also been developed (322) which specifically recognizes an epitope formed between IL-6 and IL-6Rα, although the structural mechanism behind inhibition has not been described. IL-11 trans-signaling has not been ascribed the same biological significance as IL-6 trans-signaling, regardless, sgp130Fc is used as a tool to study IL-11 trans-signaling (146), and may be a useful therapy in the case that IL-11 trans-signaling is found to be pathologically important.
In the past decades, systematic mutagenesis or phage display was used to generate antagonistic variants of IL-6, IL-11, and LIF by altering affinity to IL-6Rα, IL-11Rα, LIFR, or gp130 (203, 323, 324). These antagonists generally function by selectively increasing affinity to one cytokine receptor, and decreasing affinity to a second cytokine receptor, allowing the non-signaling competent mutant to compete with endogenous cytokine for its receptor. For example, a LIF mutein (324) was developed using phage display to increase the affinity for LIFR, while incorporating mutations that reduced the affinity for gp130. This enables the LIF mutein to compete with endogenous LIF for LIFR binding, while the LIF mutein has reduced capacity to form signaling complex with gp130, resulting in inhibition of signaling by LIF. A similar approach was used to design an IL-11 mutein (203). The mutein incorporates two sets of mutations, a mutation in site-III to reduce binding to gp130, and mutations in the AB loop intended to increase affinity to IL-11Rα allowing the IL-11 mutein to compete with endogenous IL-11 for IL-11Rα, and reduce signaling through IL-11.
Recently, a novel CNTF signaling agonist, IC7, was designed (325) by substituting site-III on IL-6 with site-III on CNTF (which binds LIFR), resulting in a cytokine that signals through a gp130/LIFR heterodimer, while being dependent on IL-6Rα, a signaling mode which is not used by any known IL-6 family cytokine (325). Recombinant CNTF has undergone clinical trials previously to treat type-2 diabetes (326), however the trials were halted due the potential immunogenicity of recombinant CNTF. IC7 provides a therapeutic benefit in animal models of diet-induced obesity, and was not observed to have any severe inflammatory or immunogenic side-effects, suggesting that IC7 holds promise as a novel cytokine treatment for diabetes (325).
An additional approach to develop cytokine signaling modulators is the use of computationally de novo designed proteins. A notable recent example of the use of protein design is in the development of IL-2 signaling modulators (327). De novo designed proteins, which have low sequence identity to endogenous cytokines, can reduce the risk of immunogenicity when using recombinant cytokines or cytokine mutants as drugs. The use of de novo protein design may allow the development of IL-11 agonists or antagonists with low immunogenicity that are more potent than existing therapies.
As new roles for cytokines in disease are discovered, the development of therapeutics to inhibit their action invariably follows. Our rapidly increasing understanding of the importance of IL-11 signaling in disease underscores its potential as a therapeutic target. However, the development and appropriate characterization of inhibitors of IL-11 signaling has not matured at the same pace. Detailed biophysical and structural information obtained in parallel with pre-clinical testing can greatly facilitate design, specificity, and potency of new cytokine inhibitors, ensuring that the best therapeutics are entered into clinical trials. Thus, improved structural and molecular understanding of the IL-11 signaling complex and current generation inhibitors will be of great benefit for therapeutic development programs targeting IL-11.
RM wrote the manuscript and prepared the figures. TP wrote the manuscript. MG wrote the manuscript and supervised the studies. All authors contributed to the article and approved the submitted version.
This work was supported by the National Health & Medical Research Council of Australia (APP1147621 and APP1080498) and Worldwide Cancer Research (14-1197). RM was the recipient of an Australian Government Research Training Program Scholarship. TP was the recipient of a Victorian Cancer Agency Fellowship (MCRF16009), a Dyson Bequest WEHI Centenary Fellowship, and a Viertel Senior Medical Research Fellowship. MG was the recipient of an Australian Research Council Future Fellowship (project number FT140100544). Funding from the Victorian Government Operational Infrastructure Support Scheme was acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Isaacs J, Lindemann A. Virus interference: I. The interferon. Proc R Soc London Ser B Biol Sci. (1957) 147:258–67. doi: 10.1098/rspb.1957.0048
2. Bradley TR, Metcalf D. The growth of mouse bone marrow cells in vitro. Aust J Exp Biol Med Sci. (1966) 44:287–99. doi: 10.1038/icb.1966.28
3. Burgess AW, Camakaris J, Metcalf D. Purification and properties of colony-stimulating from mouse lung-conditioned medium. J Biol Chem. (1977) 252:1998–2003.
4. Sparrow LG, Metcalf D, Hunkapiller MW, Hood LE, Burgess AW. Purification and partial amino acid sequence of asialo murine granulocyte-macrophage colony stimulating factor. Proc Natl Acad Sci USA. (1985) 82:292–6. doi: 10.1073/pnas.82.2.292
5. Morgan DA, Ruscetti FW, Gallo R. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science. (1976) 193:1007–8. doi: 10.1126/science.181845
6. Gillis S, Ferm MM, Ou W, Smith KA. T cell growth factor: parameters of production and a quantitative microassay for activity. J Immunol. (1978) 120:2027–32.
7. Johnson GR. Colony formation in agar by adult bone marrow multipotential hemopoietic cells. J Cell Physiol. (1980) 103:371–83. doi: 10.1002/jcp.1041030302
8. Ihle JN, Keller J, Oroszlan S, Henderson LE, Copeland TD, Fitch F, et al. Biologic properties of homogeneous interleukin 3. I. Demonstration of WEHI-3 growth factor activity, mast cell growth factor activity, p cell-stimulating factor activity, colony-stimulating factor activity, and histamine-producing cell-stimulating factor. J Immunol. (1983) 131:282–7.
9. Cohen S, Bigazzi PE, Yoshida T. Similarities of T cell function in cell-mediated immunity and antibody production. Cell Immunol. (1974) 12:150–9. doi: 10.1016/0008-8749(74)90066-5
10. Arden LA, Brunner TK, Cerottini J-C, Dayer J-M, De Weck AL, Dinarello CA, et al. Revised nomenclature for antigen-nonspecific T cell proliferation and helper factors. J Immunol. (1979) 123:2928–9.
11. Branca AA, Baglioni C. Evidence that types I and II interferons have different receptors. Nature. (1981) 294:768–70. doi: 10.1038/294768a0
12. Walker F, Burgess AW. Specific binding of radioiodinated granulocyte-macrophage colony-stimulating factor to hemopoietic cells. EMBO J. (1985) 4:933–9. doi: 10.1002/j.1460-2075.1985.tb03721.x
13. Walker F, Nicola NA, Metcalf D, Burgess AW. Hierarchical down-modulation of hemopoietic growth factor receptors. Cell. (1985) 43:269–76. doi: 10.1016/0092-8674(85)90032-7
14. Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci USA. (1990) 87:8555–9. doi: 10.1073/pnas.87.21.8555
15. Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA. (1992) 89:7836–9. doi: 10.1073/pnas.89.16.7836
16. Zhong Z, Wen Z, Darnell JE. Stat3 and Stat4: Members of the family of signal transducers and activators of transcription. Proc Natl Acad Sci USA. (1994) 91:4806–10. doi: 10.1073/pnas.91.11.4806
17. Zhong Z, Wen Z, Darnell JE Jr. Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. (1994) 264:95–8. doi: 10.1126/science.8140422
18. Schindler C, Shuai K, Prezioso VR, Darnell JE. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. (1992) 257:809–13. doi: 10.1126/science.1496401
19. Shuai K, Stark GR, Kerr M, Darnell JE Jr. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science. (1993) 261:1744–6. doi: 10.1126/science.7690989
20. Wilks AF. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci USA. (1989) 86:1603–7. doi: 10.1073/pnas.86.5.1603
21. Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zürcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol. (1991) 11:2057–65. doi: 10.1128/MCB.11.4.2057
22. Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon α/β signaling pathway. Cell. (1992) 70:313–22. doi: 10.1016/0092-8674(92)90105-L
23. Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature. (1993) 366:0–129. doi: 10.1038/366129a0
24. Johnston JA, Kawamura M, Kirken RA, Chen YQ, Blake TB, Shibuya K, et al. Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature. (1994) 370:151–3. doi: 10.1038/370151a0
25. Nicholson SE, Oates AC, Harpur AG, Ziemiecki A, Wilks AF, Layton JE. Tyrosine kinase JAK1 is associated with the granulocyte-colony-stimulating factor receptor and both become tyrosine-phosphorylated after receptor activation. Proc Natl Acad Sci USA. (1994) 91:2985–8. doi: 10.1073/pnas.91.8.2985
26. Kajita T, Kishimoto T, Naka T, Narazaki M, Hirata M, Matsumoto T, et al. Structure and function of a new STAT-induced STAT inhibitor. Nature. (1997) 387:924–9. doi: 10.1038/43219
27. Starr R, Willson TA, Viney EM, Murry LJL, Rayner JR, Jenkins BJ, et al. A family of cytokine-inducible inhibitors of signalling. Nature. (1997) 387:917–21. doi: 10.1038/43206
28. Yoshimura A, Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. (1997) 387:921–4. doi: 10.1038/43213
29. Bazan JF. Haemopoietic receptors and helical cytokines. Immunol Today. (1990) 11:350–4. doi: 10.1016/0167-5699(90)90139-Z
30. Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA. (1990) 87:6934–8. doi: 10.1073/pnas.87.18.6934
31. Sprang SR, Bazan JF. Cytokine structural taxonomy and mechanisms of receptor engagement. Curr Opin Struct Biol. (1993) 3:815–27. doi: 10.1016/0959-440X(93)90144-A
32. Morris R, Kershaw NJ, Babon JJ. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. (2018) 27:1984–2009. doi: 10.1002/pro.3519
33. Boulanger MJ, Garcia KC. Shared cytokine signaling receptors: structural insights from the GP130 system. Adv Protein Chem. (2004) 68:107–46. doi: 10.1016/S0065-3233(04)68004-1
34. Li CH, Evans HM. The isolation of pituitary growth hormone. Science. (1944) 99:183–4. doi: 10.1126/science.99.2566.183
35. Bonsdorff E, Jalavisto E. A humoral mechanism in anoxic erythrocytosis. Acta Physiol Scand. (1948) 16:150–70. doi: 10.1111/j.1748-1716.1948.tb00535.x
36. Lindholm J. Growth hormone: historical notes. Pituitary. (2006) 9:5–10. doi: 10.1007/s11102-006-7557-4
37. Jelkmann W. Erythropoietin after a century of research: younger than ever. Eur J Haematol. (2007) 78:183–205. doi: 10.1111/j.1600-0609.2007.00818.x
38. D'andrea AD, Fasman GD, Lodish HF. Erythropoietin receptor and interleukin-2 receptor β chain: a new receptor family. Cell. (1989) 58:1023–4. doi: 10.1016/0092-8674(89)90499-6
39. Cosman D, Lyman SD, Idzerda RL, Beckmann MP, Park LS, Goodwin RG, et al. A new cytokine receptor superfamily. Trends Biochem. (1993) 15:265–70. doi: 10.1016/0968-0004(90)90051-C
40. Chantalat L, Jones ND, Korber F, Navaza J, Pavlovsky AG. The crystal structure of wild-type growth-hormone at 2.5 angstrom resolution. Protein Peptide Lett. (1995) 2:333–40.
41. Walter MR, Cook WJ, Ealick SE, Nagabhushan TL, Trotta PP, Bugg CE. Three-dimensional structure of recombinant human granulocyte-macrophage colony-stimulating factor. J Mol Biol. (1992) 224:1075–85. doi: 10.1016/0022-2836(92)90470-5
42. Cheetham JC, Smith DM, Aoki KH, Stevenson JL, Hoeffel TJ, Syed RS, et al. NMR structure of human erythropoietin and a comparison with its receptor bound conformation. Nat Struct Biol. (1998) 5:861–6. doi: 10.1038/2302
43. Brown RJ, Adams JJ, Pelekanos RA, Wan Y, Mckinstry WJ, Palethorpe K, et al. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol. (2005) 12:814–21. doi: 10.1038/nsmb977
44. De Vos A, Ultsch M, Kossiakoff A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. (1992) 255:306–12. doi: 10.1126/science.1549776
45. Somers W, Stahl M, Seehra JS. 1.9 Å crystal structure of interleukin 6: Implications for a novel mode of receptor dimerization and signaling. EMBO J. (1997) 16:989–97. doi: 10.1093/emboj/16.5.989
46. Redfield C, Smith LJ, Boyd J, Lawrence GM, Edwards RG, Gershater CJ, et al. Analysis of the solution structure of human interleukin-4 determined by heteronuclear three-dimensional nuclear magnetic resonance techniques. J Mol Biol. (1994) 238:23–41. doi: 10.2210/pdb1itm/pdb
47. Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. The structure of interleukin-2 complexed with its alpha receptor. Science. (2005) 308:1477–80. doi: 10.1126/science.1109745
48. Ring AM, Lin J-X, Feng D, Mitra S, Rickert M, Bowman GR, et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat Immunol. (2012) 13:1187–95. doi: 10.1038/ni.2449
49. Yoshimura A, Zimmers T, Neumann D, Longmore G, Yoshimura Y, Lodish HF. Mutations in the Trp-Ser-X-Trp-Ser motif of the erythropoietin receptor abolish processing, ligand binding, and activation of the receptor. J Biol Chem. (1992) 267:11619–25.
50. Hilton DJ, Watowich SS, Katz L, Lodish HF. Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J Biol Chem. (1996) 271:4699–708. doi: 10.1074/jbc.271.9.4699
51. Duriez B, Sobrier ML, Duquesnoy P, Tixier-Boichard M, Decuypere E, Coquerelle G, et al. A naturally occurring growth hormone receptor mutation: in vivo and in vitro evidence for the functional importance of the WS motif common to all members of the cytokine receptor superfamily. Mol Endocrinol. (1993) 7:806–14. doi: 10.1210/mend.7.6.8361502
52. Hamming OJ, Kang L, Svensson A, Karlsen JL, Rahbek-Nielsen H, Paludan SR, et al. Crystal structure of interleukin-21 receptor (IL-21R) bound to IL-21 reveals that sugar chain interacting with WSXWS motif is integral part of IL-21R. J Biol Chem. (2012) 287:9454–60. doi: 10.1074/jbc.M111.311084
53. Doucey MA, Hess D, Blommers MJJ, Hofsteenge J. Recombinant human interleukin-12 is the second example of a C-mannosylated protein. Glycobiology. (1999) 9:435–41. doi: 10.1093/glycob/9.5.435
54. Yoon C, Johnston SC, Tang J, Stahl M, Tobin JF, Somers WS. Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12. EMBO J. (2000) 19:3530–41. doi: 10.1093/emboj/19.14.3530
55. Beyer BM, Ingram R, Ramanathan L, Reichert P, Le HV, Madison V, et al. Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody. J Mol Biol. (2008) 382:942–55. doi: 10.1016/j.jmb.2008.08.001
56. Lupardus PJ, Garcia KC. The structure of interleukin-23 reveals the molecular basis of p40 subunit sharing with interleukin-12. J Mol Biol. (2008) 382:931–41. doi: 10.1016/j.jmb.2008.07.051
57. Dagil R, Knudsen MJ, Olsen JG, O'shea C, Franzmann M, Goffin V, et al. The WSXWS motif in cytokine receptors is a molecular switch involved in receptor activation: insight from structures of the prolactin receptor. Structure. (2012) 20:270–82. doi: 10.1016/j.str.2011.12.010
58. Xu Y, Kershaw NJ, Luo CS, Soo P, Pocock MJ, Czabotar PE, et al. Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J Biol Chem. (2010) 285:21214–8. doi: 10.1074/jbc.C110.129502
59. Huyton T, Zhang J-G, Luo CS, Lou M-Z, Hilton DJ, Nicola N, et al. An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. Proc Natl Acad Sci USA. (2007) 104:12737–42. doi: 10.1073/pnas.0705577104
60. Vollmer P, Oppmann B, Voltz N, Fischer M, Rose-John S. A role for the immunoglobulin-like domain of the human IL-6 receptor: Intracellular protein transport and shedding. Eur J Biochem. (1999) 263:438–46. doi: 10.1046/j.1432-1327.1999.00511.x
61. Davis S, Aldrich TH, Valenzuela DM, Wong V, Furth ME, Squinto SP, et al. The receptor for ciliary neurotrophic factor. Science. (1991) 253:59–63. doi: 10.1126/science.1648265
62. Bugge K, Papaleo E, Haxholm GW, Hopper JTS, Robinson CV, Olsen JG, et al. A combined computational and structural model of the full-length human prolactin receptor. Nat Commun. (2016) 7:1–11. doi: 10.1038/ncomms11578
63. Schmidt T, Ye F, Situ AJ, An W, Ginsberg MH, Ulmer TS. A conserved ectodomain-transmembrane domain linker motif tunes the allosteric regulation of cell surface receptors. J Biol Chem. (2016) 291:17536–46. doi: 10.1074/jbc.M116.733683
64. Bocharov EV, Lesovoy DM, Bocharova OV, Urban AS, Pavlov KV, Volynsky PE, et al. Structural basis of the signal transduction via transmembrane domain of the human growth hormone receptor. Biochim Biophys Acta General Subj. (2018) 1862:1410–20. doi: 10.1016/j.bbagen.2018.03.022
65. Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol Cell. (2008) 31:737–48. doi: 10.1016/j.molcel.2008.08.011
66. Haxholm GW, Nikolajsen LF, Olsen JG, Fredsted J, Larsen FH, Goffin V, et al. Intrinsically disordered cytoplasmic domains of two cytokine receptors mediate conserved interactions with membranes. Biochem J. (2015) 468:495–506. doi: 10.1042/BJ20141243
67. Chow D-C, He X, Snow AL, Rose-John S, Garcia K. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. (2001) 291:2150–5. doi: 10.1126/science.1058308
68. Boulanger MJ, Chow D-C, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. (2003) 300:2101–4. doi: 10.1126/science.1083901
69. Hansen G, Hercus TR, Mcclure BJ, Stomski FC, Dottore M, Powell J, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. (2008) 134:496–507. doi: 10.1016/j.cell.2008.05.053
70. Skiniotis G, Boulanger MJ, Garcia KC, Walz T. Signaling conformations of the tall cytokine receptor gp130 when in complex with IL-6 and IL-6 receptor. Nat Struct Mol Biol. (2005) 12:545–51. doi: 10.1038/nsmb941
71. Lupardus PJ, Skiniotis G, Rice AJ, Thomas C, Fischer S, Walz T, et al. Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Rα cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure. (2011) 19:45–55. doi: 10.1016/j.str.2010.10.010
72. Broughton SE, Nero TL, Dhagat U, Kan WL, Hercus TR, Tvorogov D, et al. The βc receptor family - Structural insights and their functional implications. Cytokine. (2015) 74:247–58. doi: 10.1016/j.cyto.2015.02.005
73. Laporte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. (2008) 132:259–72. doi: 10.1016/j.cell.2007.12.030
74. Rose TM, Bruce AG. Oncostatin M is a member of a cytokine family that includes leukemia-inhibitory factor, granulocyte colony-stimulating factor, and interleukin 6. Proc Natl Acad Sci USA. (1991) 88:8641–5. doi: 10.1073/pnas.88.19.8641
75. Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. (1997) 15:797–819. doi: 10.1146/annurev.immunol.15.1.797
76. Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol Cell. (2003) 12:577–89. doi: 10.1016/S1097-2765(03)00365-4
77. Lupardus PJ, Ultsch M, Wallweber H, Bir Kohli P, Johnson AR, Eigenbrot C. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci USA. (2014) 111:8025–30. doi: 10.1073/pnas.1401180111
78. Wallweber HJA, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-α receptor by tyrosine kinase 2. Nat Struct Mol Biol. (2014) 21:443–8. doi: 10.1038/nsmb.2807
79. Ferrao R, Wallweber HJA, Ho H, Tam C, Franke Y, Quinn J, et al. The structural basis for class II cytokine receptor recognition by JAK1. Structure. (2016) 24:897–905. doi: 10.1016/j.str.2016.03.023
80. Tanner JW, Chen W, Young RL, Longmore GD, Shaw AS. The conserved box 1 motif of cytokine receptors is required for association with JAK kinases. J Biol Chem. (1995) 270:6523–30. doi: 10.1074/jbc.270.12.6523
81. Hercus TR, Dhagat U, Kan WLT, Broughton SE, Nero TL, Perugini M, et al. Signalling by the βc family of cytokines. Cytokine Growth Factor Rev. (2013) 24:189–201. doi: 10.1016/j.cytogfr.2013.03.002
82. Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS box encodes a hierarchy of affinities for Cullin5: implications for ubiquitin ligase formation and cytokine signalling suppression. J Mol Biol. (2009) 387:162–74. doi: 10.1016/j.jmb.2009.01.024
83. Kershaw NJ, Laktyushin A, Nicola NA, Babon JJ. Reconstruction of an active SOCS3-based E3 ubiquitin ligase complex in vitro: identification of the active components and JAK2 and gp130 as substrates. Growth Factors. (2014) 32:1–10. doi: 10.3109/08977194.2013.877005
84. Liau NPD, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E. The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun. (2018) 9:1–14. doi: 10.1038/s41467-018-04013-1
85. Kershaw NJ, Murphy JM, Liau NPD, Varghese LN, Laktyushin A, Whitlock EL, et al. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol. (2013) 20:469–76. doi: 10.1038/nsmb.2519
86. Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. (1997) 278:1803–5. doi: 10.1126/science.278.5344.1803
87. Shuai K, Liu B. Regulation of gene-activation pathways by pias proteins in the immune system. Nat Rev Immunol. (2005) 5:593–605. doi: 10.1038/nri1667
88. David M, Chen HE, Goelz S, Larner AC, Neel BG. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. (1995) 15:7050–8. doi: 10.1128/MCB.15.12.7050
89. Alicea-Velázquez NL, Jakoncic J, Boggon TJ. Structure-guided studies of the SHP-1/JAK1 interaction provide new insights into phosphatase catalytic domain substrate recognition. J Struct Biol. (2013) 181:243–51. doi: 10.1016/j.jsb.2012.12.009
90. Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. (2001) 276:47771–4. doi: 10.1074/jbc.C100583200
91. Tong W, Zhang J, Lodish HF. Lnk inhibits erythropoiesis and Epo-dependent JAK2 activation and downstream signaling pathways. Blood. (2005) 105:4604–12. doi: 10.1182/blood-2004-10-4093
92. Goeddel V, Wells JA, Fuh G, Cunningham BC, Fukunaga R, Nagata S, et al. Rational design of potent antagonists to the human growth hormone receptor. Science. (1992) 256:1677–80. doi: 10.1126/science.256.5064.1677
93. Brooks A, Dai W, O'mara M, Abankwa D, Chhabra Y, Pelekanos R, et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. (2014) 344:703–4. doi: 10.1126/science.1249783
94. Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. (1999) 283:987–90. doi: 10.1126/science.283.5404.987
95. Giese B, Roderburg C, Sommerauer M, Wortmann SB, Metz S, Heinrich PC, et al. Dimerization of the cytokine receptors gp130 and LIFR analysed in single cells. J Cell Sci. (2005) 118:5129–40. doi: 10.1242/jcs.02628
96. Tenhumberg S, Schuster B, Zhu L, Kovaleva M, Scheller J, Kallen KJ, et al. gp130 dimerization in the absence of ligand: preformed cytokine receptor complexes. Biochem Biophys Res Commun. (2006) 346:649–57. doi: 10.1016/j.bbrc.2006.05.173
97. Salmond RJ, Alexander DR. SHP2 forecast for the immune system: fog gradually clearing. Trends Immunol. (2006) 27:154–60. doi: 10.1016/j.it.2006.01.007
98. Putoczki TL, Dobson RCJ, Griffin MDW. The structure of human interleukin-11 reveals receptor-binding site features and structural differences from interleukin-6. Acta Crystallograph Sect D Biol Crystallogr. (2014) 3:2277–85. doi: 10.1107/S1399004714012267
99. Robinson RC, Grey LM, Staunton D, Vankelecom H, Vernallis AB, Moreau JF, et al. The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding. Cell. (1994) 77:1101–16. doi: 10.1016/0092-8674(94)90449-9
100. Deller MC, Hudson KR, Ikemizu S, Bravo J, Jones EY, Heath JK. Crystal structure and functional dissection of the cytostatic cytokine oncostatin M. Structure. (2000) 8:863–74. doi: 10.1016/S0969-2126(00)00176-3
101. Varghese J, Moritz R, Lou M, Van Donkelaar A, Ji H, Ivancic N, et al. Structure of the extracellular domains of the human interleukin-6 receptor α-chain. Proc Natl Acad Sci USA. (2002) 99:15959–64. doi: 10.1073/pnas.232432399
102. Diveu C, Lelièvre E, Perret D, Lak-Hal AHL, Froger J, Guillet C, et al. GPL, a novel cytokine receptor related to GP130 and leukemia inhibitory factor receptor. J Biol Chem. (2003) 278:49850–9. doi: 10.1074/jbc.M307286200
103. Dillon SR, Sprecher C, Hammond A, Bilsborough J, Rosenfeld-Franklin M, Presnell SR, et al. Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat Immunol. (2004) 5:752–60. doi: 10.1038/ni1084
104. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. (2014) 13:397–406. doi: 10.1074/mcp.M113.035600
105. Schuster B, Kovaleva M, Sun Y, Regenhard P, Matthews V, Grötzinger J, et al. Signaling of human ciliary neurotrophic factor (CNTF) revisited: the interleukin-6 receptor can serve as an α-receptor for CNTF. J Biol Chem. (2003) 278:9528–35. doi: 10.1074/jbc.M210044200
106. Garbers C, Spudy B, Aparicio-Siegmund S, Waetzig GH, Sommer J, Hölscher C, et al. An lnterleukin-6 receptor-dependent molecular switch mediates signal transduction of the IL-27 cytokine subunit p28 (IL-30) via a gp130 protein receptor homodimer. J Biol Chem. (2013) 288:4346–54. doi: 10.1074/jbc.M112.432955
107. Crabé S, Guay-Giroux A, Tormo AJ, Duluc D, Lissilaa R, Guilhot F, et al. The IL-27 p28 subunit binds cytokine-like factor 1 to form a cytokine regulating NK and T cell activities requiring IL-6R for signaling. J Immunol. (2009) 183:7692–702. doi: 10.4049/jimmunol.0901464
108. Boulanger MJ, Chow DC, Brevnova E, Martick M, Sandford G, Nicholas J, et al. Molecular mechanisms for viral mimicry of a human cytokine: activation of gp130 by HHV-8 interleukin-6. J Mol Biol. (2004) 335:641–54. doi: 10.1016/j.jmb.2003.10.070
109. Neipel F, Albrecht J-C, Ensser A, Fleckenstein B, Huang Y-QI, Friedman-Kien AE. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. (1997) 71:839–42. doi: 10.1128/JVI.71.1.839-842.1997
111. Kang S, Narazaki M, Metwally H, Kishimoto T. Historical overview of the interleukin-6 family cytokine. J Exp Med. (2020) 217:e20190347. doi: 10.1084/jem.20190347
112. Hirano T, Yasukawa K, Harada H, Taga T, Watanabe Y, Matsuda T, et al. Complementary DNA for a novel human interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature. (1986) 324:73–6. doi: 10.1038/324073a0
113. Van Damme J, Opdenakker G, Simpson RJ, Rubira MR, Cayphas S, Vink A, et al. Hybridoma / plasmacytoma growth factor induced by interleukin 1 and tumor necrosis factor. J Exp Med. (1987) 165:914–9. doi: 10.1084/jem.165.3.914
114. Gauldie J, Richards C, Harnish D, Lansdorp P, Baumann H. Interferon beta 2/B-cell stimulatory factor type 2 shares identity with monocyte-derived hepatocyte-stimulating factor and regulates the major acute phase protein response in liver cells. Proc Natl Acad Sci USA. (1987) 84:7251–5. doi: 10.1073/pnas.84.20.7251
115. Xu GY, Yu H, Hong J, Stahl M, Mcdonagh T, Kay LE, et al. Solution structure of recombinant human interleukin-6. J Mol Biol. (1997) 268:468–81. doi: 10.1006/jmbi.1997.0933
116. Baran P, Nitz R, Groẗzinger J, Scheller J, Garbers C. Minimal interleukin 6 (IL-6) receptor stalk composition for IL-6 receptor shedding and IL-6 classic signaling. J Biol Chem. (2013) 288:14756–68. doi: 10.1074/jbc.M113.466169
117. Nitz R, Lokau J, Aparicio-Siegmund S, Scheller J, Garbers C. Modular organization of interleukin-6 and interleukin-11 α-receptors. Biochimie. (2015) 119:175–82. doi: 10.1016/j.biochi.2015.11.005
118. Lokau J, Garbers C. The length of the interleukin-11 receptor stalk determines its capacity for classic signaling. J Biol Chem. (2018) 293:6398–409. doi: 10.1074/jbc.RA118.001879
119. Taga T, Hibi M, Hirata Y, Yamasaki K, Yasukawa K, Matsuda T, et al. Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell. (1989) 58:573–81. doi: 10.1016/0092-8674(89)90438-8
120. Hibi M, Murakami M, Saito M, Hirano T, Taga T, Kishimoto T. Molecular cloning and expression of an IL-6 signal transducer, gp130. Cell. (1990) 63:1149–57. doi: 10.1016/0092-8674(90)90411-7
121. Hilton DJ, Hilton AA, Raicevic A, Rakar S, Harrison-Smith M, Gough NM, et al. Cloning of a murine IL-11 receptor alpha-chain; requirement for gp130 for high affinity binding and signal transduction. EMBO J. (1994) 13:4765–75. doi: 10.1002/j.1460-2075.1994.tb06802.x
122. Gimpel SD, Thut CJ, Mcgourty J, Brasher KK, King JA, Gillis S, et al. The IL-6 signal transducer, gpl30: an oncostatin M receptor and affinity converter for the LIF receptor. Science. (1992) 255:1434–7. doi: 10.1126/science.1542794
123. Ip NY, Nye SH, Boulton TG, Davis S, Taga T, Li Y, et al. CNTF and LIF act on neuronal cells via shared signaling pathways that involve the IL-6 signal transducing receptor component gp130. Cell. (1992) 69:1121–32. doi: 10.1016/0092-8674(92)90634-O
124. Bravo J, Staunton D, Heath JK, Yvonne Jones E. Crystal structure of a cytokine-binding region of gp130. EMBO J. (1998) 17:1665–74. doi: 10.1093/emboj/17.6.1665
125. Spannbauer MM, Trautwein C. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumors. Nature. (2009) 49:1387–9. doi: 10.1002/hep.22902
126. Schütt A, Zacharias M, Schneider N, Horn S, Grötzinger J, Rose-John S, et al. gp130 activation is regulated by D2-D3 interdomain connectivity. Biochem J. (2013) 450:487–96. doi: 10.1042/BJ20121660
127. Timmermann A, Küster A, Kurth I, Heinrich PC, Müller-Newen G. A functional role of the membrane-proximal extracellular domains of the signal transducer gp130 in heterodimerization with the leukemia inhibitory factor receptor. Eur J Biochem. (2002) 269:2716–26. doi: 10.1046/j.1432-1033.2002.02941.x
128. Matadeen R, Hon WC, Heath JK, Jones EY, Fuller S. The dynamics of signal triggering in a gp130-receptor complex. Structure. (2007) 15:441–8. doi: 10.1016/j.str.2007.02.006
129. Lütticken C, Wegenka UM, Yuan J, Buschmann J, Schindler C, Ziemiecki A, et al. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science. (1994) 263:89–92. doi: 10.1126/science.8272872
130. Hemmann U, Gerhartz C, Heesel B, Sasse J, Kurapkat G, Grötzinger J, et al. Differential activation of acute phase response factor/Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. J Biol Chem. (2002) 271:12999–3007. doi: 10.1074/jbc.271.22.12999
131. Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE, Yancopoulos GD. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. (1995) 267:1349–53. doi: 10.1126/science.7871433
132. Gerhartz C, Hessel B, Sasse J, Hemman U, Landgraf C, Schneider-Mergener J, et al. Differential activation of acute phase response factor/Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130: I. Definition of a novel phosphotyrosine motif mediating STAT1 activation. J Biol Chem. (1996) 271:12991–8. doi: 10.1074/jbc.271.22.12991
133. Anhuf D, Weissenbach M, Schmitz J, Sobota R, Hermanns HM, Radtke S, et al. Signal transduction of IL-6, leukemia-inhibitory factor, and oncostatin M: structural receptor requirements for signal attenuation. J Immunol. (2000) 165:2535–43. doi: 10.4049/jimmunol.165.5.2535
134. Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. (2003) 374:1–20. doi: 10.1042/bj20030407
135. Ward LD, Howlett GJ, Discolo G, Yasukawa K, Hammacher A, Moritz RL, et al. High affinity interleukin-6 receptor is a hexameric complex consisting of two molecules each of interleukin-6, interleukin-6 receptor, and gp-130. J Biol Chem. (1994) 269:23286–9. doi: 10.1016/1043-4666(94)90186-4
136. Paonessa G, Graziani R, De Serio A, Savino R, Ciapponi L, Lahm A, et al. Two distinct and independent sites on IL-6 trigger gp 130 dimer formation and signalling. EMBO J. (1995) 14:1942–51. doi: 10.1002/j.1460-2075.1995.tb07186.x
137. Chow D-C, Ho J, Nguyen Pham TL, Rose-John S, Garcia KC. In vitro reconstitution of recognition and activation complexes between interleukin-6 and gp130. Biochemistry. (2001) 40:7593–603. doi: 10.1021/bi010192q
138. Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. (2009) 27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616
139. Li H, Xiao H, Lin L, Jou D, Kumari V, Lin J, et al. Drug design targeting protein-protein interactions (PPIs) using multiple ligand simultaneous docking (MLSD) and drug repositioning: discovery of raloxifene and bazedoxifene as novel inhibitors of IL-6/GP130 interface. J Med Chem. (2014) 57:632–41. doi: 10.1021/jm401144z
140. Thilakasiri P, Huynh J, Poh AR, Tan CW, Nero TL, Tran K, et al. Repurposing the selective estrogen receptor modulator bazedoxifene to suppress gastrointestinal cancer growth. EMBO Mol Med. (2019) 11:e9539. doi: 10.15252/emmm.201809539
141. Kurth I, Horsten U, Pflanz S, Dahmen H, Küster A, Grötzinger J, et al. Activation of the signal transducer glycoprotein 130 by both IL-6 and IL-11 requires two distinct binding epitopes. J Immunol. (1999) 162:1480–7.
142. Wijdenes J, Heinrich PC, Müller-Newen G, Roche C, Gu Z.-J, Cldment C, et al. Interleukin-6 signal transducer gp130 has specific binding sites for different cytokines as determined by antagonistic and agonistic anti-gp130 monoclonal antibodies. Eur J Immunol. (1995) 25:3474–81. doi: 10.1002/eji.1830251240
143. Chalaris A, Garbers C, Rabe B, Rose-John S, Scheller J. The soluble Interleukin 6 receptor: generation and role in inflammation and cancer. Eur J Cell Biol. (2011) 90:484–94. doi: 10.1016/j.ejcb.2010.10.007
144. Lust JA, Donovan KA, Kline MP, Greipp PR, Kyle RA, Maihle NJ. Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine. (1992) 4:96–100. doi: 10.1016/1043-4666(92)90043-Q
145. Jostock T, Müllberg J, Özbek S, Atreya R, Blinn G, Voltz N, et al. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur J Biochem. (2001) 268:160–7. doi: 10.1046/j.1432-1327.2001.01867.x
146. Lokau J, Nitz R, Agthe M, Monhasery N, Aparicio-Siegmund S, Schumacher N, et al. Proteolytic cleavage governs interleukin-11 trans-signaling. Cell Rep. (2016) 14:1761–73. doi: 10.1016/j.celrep.2016.01.053
147. Balic JJ, Garbers C, Rose-John S, Yu L, Jenkins BJ. Interleukin-11-driven gastric tumourigenesis is independent of trans-signalling. Cytokine. (2017) 92:118–23. doi: 10.1016/j.cyto.2017.01.015
148. Agthe M, Garbers Y, Putoczki T, Garbers C. Interleukin-11 classic but not trans-signaling is essential for fertility in mice. Placenta. (2017) 57:13–6. doi: 10.1016/j.placenta.2017.05.015
149. Fischer M, Goldschmitt J, Peschel C, Brakenhoff JPG, Kallen K-J, Wollmer A, et al. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol. (1997) 15:142–5. doi: 10.1038/nbt0297-142
150. Dams-Kozlowska HH, Gryska KK, Kwiatkowska-Borowczyk EE, Izycki DD, Rose-John SS, Mackiewicz A. A designer hyper interleukin 11 (H11) is a biologically active cytokine. BMC Biotechnol. (2012) 12:8. doi: 10.1186/1472-6750-12-8
151. Heink S, Yogev N, Garbers C, Herwerth M, Aly L, Gasperi C, et al. Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells. Nat Immunol. (2016) 18:74–85. doi: 10.1038/ni.3632
152. Lamertz L, Rummel F, Polz R, Baran P, Hansen S, Waetzig GH, et al. Soluble gp130 prevents interleukin-6 and interleukin-11 cluster signaling but not intracellular autocrine responses. Sci Signal. (2018) 11:eaar7388. doi: 10.1126/scisignal.aar7388
153. Wuest SC, Edwan JH, Martin JF, Han S, Perry JSA, Cartagena CM, et al. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat Med. (2011) 17:604–9. doi: 10.1038/nm.2365
154. Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Rα recycles and presents IL-15 in trans to neighboring cells. Immunity. (2002) 17:537–47. doi: 10.1016/S1074-7613(02)00429-6
155. Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. (2005) 310:1159–63. doi: 10.1126/science.1117893
156. Rose-John S, Scheller J, Schaper F. “Family reunion” - a structured view on the composition of the receptor complexes of interleukin-6-type and interleukin-12-type cytokines. Cytokine Growth Factor Rev. (2015) 26:471–4. doi: 10.1016/j.cytogfr.2015.07.011
157. Pflanz S, Hibbert L, Mattson J, Rosales R, Vaisberg E, Bazan JF, et al. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J Immunol. (2004) 172:2225–31. doi: 10.4049/jimmunol.172.4.2225
158. Collison LW, Delgoffe GM, Guy CS, Vignali KM, Chaturvedi V, Fairweather D, et al. The composition and signaling of the IL-35 receptor are unconventional. Nat Immunol. (2012) 13:290–9. doi: 10.1038/ni.2227
159. Stumhofer JS, Tait ED, Iii WJQ, Hosken N, Spudy B, Goenka R, et al. A role for IL-27p28 as an antagonist of gp130-mediated signaling. Nat Immunol. (2010) 11:1119–26. doi: 10.1038/ni.1957
160. Brown S, Hu N, Hombría JCG. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. (2001) 11:1700–5. doi: 10.1016/S0960-9822(01)00524-3
161. Jones SA, Jenkins BJ. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol. (2018) 18:773–89. doi: 10.1038/s41577-018-0066-7
162. Harrison DA, Mccoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. (1998) 12:3252–63. doi: 10.1101/gad.12.20.3252
163. Zeidler MP, Bausek N. The Drosophila JAK-STAT pathway. Jak Stat. (2013) 2:e25353. doi: 10.4161/jkst.25353
164. Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. (2003) 5:441–50. doi: 10.1016/S1534-5807(03)00244-2
165. Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. (1995) 14:1412–20. doi: 10.1002/j.1460-2075.1995.tb07127.x
166. Perrimon N, Mahowald AP. l(1)hopscotch, a larval-pupal zygotic lethal with a specific maternal effect on segmentation in Drosophila. Dev Biol. (1986) 118:28–41. doi: 10.1016/0012-1606(86)90070-9
167. Wang H, Chen X, He T, Zhou Y, Luo H. Evidence for tissue-specific JAK/STAT target genes in Drosophila optic lobe development. Genetics. (2013) 195:1291–306. doi: 10.1534/genetics.113.155945
168. Oldefest M, Nowinski J, Hung CW, Neelsen D, Trad A, Tholey A, et al. Upd3 - an ancestor of the four-helix bundle cytokines. Biochem Biophys Res Commun. (2013) 436:66–72. doi: 10.1016/j.bbrc.2013.04.107
169. Liongue C, Ward AC. Evolution of class I cytokine receptors. BMC Evolut Biol. (2007) 7:120. doi: 10.1186/1471-2148-7-120
170. Liongue C, Sertori R, Ward AC. Evolution of cytokine receptor signaling. J Immunol. (2016) 197:11–8. doi: 10.4049/jimmunol.1600372
171. Paul SR, Bennett F, Calvetti J, Kelleher K, Wood CR, O'hara RM, et al. Molecular cloning of a cDNA encoding interleukin 11, a stromal cell-derived lymphopoietic and hematopoietic cytokine. Proc Natl Acad Sci USA. (1990) 87:7512–6. doi: 10.1073/pnas.87.19.7512
172. Kawashima I, Ohsumi J, Mita-Honjo K, Shimoda-Takano K, Ishikawa H, Sakakibara S, et al. Molecular cloning of cDNA encoding adipogenesis inhibitory factor and identity with interleukin-11. FEBS J. (1991) 283:199–202. doi: 10.1016/0014-5793(91)80587-S
173. Keller DC, Du XX, Srour EF, Hoffman R, Williams DA. Interleukin-11 inhibits adipogenesis and stimulates myelopoiesis in human long-term marrow cultures. Blood. (1993) 82:1428–35. doi: 10.1182/blood.V82.5.1428.1428
174. Metcalfe RD, Aizel K, Zlatic CO, Nguyen PM, Morton CJ, Lio, et al. The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement. J Biol Chem. (2020) 295:8285–301. doi: 10.1074/jbc.RA119.012351
175. Barton V, Hudson KR, Heath JK. Identification of three distinct receptor binding sites of murine interleukin-11. J Biol Chem. (1999) 274:5755–61. doi: 10.1074/jbc.274.9.5755
176. Tacken I, Dahmen H, Boisteau O, Minvielle S, Jacques Y, Grötzinger J, et al. Definition of receptor binding sites on human interleukin-11 by molecular modeling-guided mutagenesis. Eur J Biochem. (1999) 265:645–55. doi: 10.1046/j.1432-1327.1999.00755.x
177. Chérel M, Sorel M, Lebeau B, Dubois S, Moreau JF, Bataille R, et al. Molecular cloning of two isoforms of a receptor for the human hematopoietic cytokine interleukin-11. Blood. (1995) 86:2534–40. doi: 10.1182/blood.V86.7.2534.bloodjournal8672534
178. Barton V, Hall M, Hudson KR, Heath JK. Interleukin-11 signals through the formation of a hexameric receptor complex. J Biol Chem. (2000) 275:36197–203. doi: 10.1074/jbc.M004648200
179. Czupryn M, Bennett F, Dube J, Grant K, Scoble H, Sookdeo H, et al. Alanine-scanning mutagenesis of human interleukin-11: identification of regions important for biological activity. Ann NY Acad Sci. (1995) 762:152–64. doi: 10.1111/j.1749-6632.1995.tb32323.x
180. Musashi M, Yang YC, Paul SR, Clark SC, Sudo T, Ogawa M. Direct and synergistic effects of interleukin 11 on murine hemopoiesis in culture. Proc Natl Acad Sci USA. (1991) 88:765–9. doi: 10.1073/pnas.88.3.765
181. Teramura M, Kobayashi S, Hoshino S, Oshimi K, Mizoguchi H. Interleukin-11 enhances human megakaryocytopoiesis in vitro. Blood. (1992) 79:327–31. doi: 10.1182/blood.V79.2.327.327
182. Musashi M, Clark SC, Sudo T, Urdal DL, Ogawa M. Synergistic interactions between interleukin-11 and interleukin-4 in support of proliferation of primitive hematopoietic progenitors of mice. Blood. (1991) 78:1448–51. doi: 10.1182/blood.V78.6.1448.1448
183. Leonard JP, Quinto CM, Kozitza MK, Neben TY, Goldman SJ. Recombinant human interleukin-11 stimulates multilineage hematopoietic recovery in mice after a myelosuppressive regimen of sublethal irradiation and carboplatin. Blood. (1994) 83:1499–506. doi: 10.1182/blood.V83.6.1499.bloodjournal8361499
184. Wilde MI, Faulds D. Oprelvekin: a review of its pharmacology and therapeutic potential in chemotherapy-induced thrombocytopenia. BioDrugs. (1998) 10:159–71. doi: 10.2165/00063030-199810020-00006
185. Du X, Williams D. Interleukin-11: review of molecular, cell biology, and clinical use. Blood. (1997) 89:3897–908. doi: 10.1182/blood.V89.11.3897
186. Anderson KC, Morimoto C, Paul SR, Chauhan D, Williams D, Cochran M, et al. Interleukin-11 promotes accessory cell-dependent B-cell differentiation in humans. Blood. (1992) 80:2797–804. doi: 10.1182/blood.V80.11.2797.bloodjournal80112797
187. Quesniaux VFJ, Clark SC, Turner K, Fagg B. Interleukin-11 stimulates multiple phases of erythropoiesis in vitro. Blood. (1992) 80:1218–23. doi: 10.1182/blood.V80.5.1218.1218
188. Cairo MS, Plunkett JM, Nguyen A, Schendel P, Van De Ven C. Effect of interleukin-11 with and without granulocyte colony-stimulating factor on in vivo neonatal rat hematopoiesis: induction of neonatal thrombocytosis by interleukin-11 and synergistic enhancement of neutrophilia by interleukin-11 + granulocyte colony-stimulating factor. Pediatr Res. (1993) 34:56–61. doi: 10.1203/00006450-199307000-00014
189. Takeuchi Y, Watanabe S, Ishii G, Takeda S, Nakayama K, Fukumoto S, et al. Interleukin-11 as a stimulatory factor for bone formation prevents bone loss with advancing age in mice. J Biol Chem. (2002) 277:49011–8. doi: 10.1074/jbc.M207804200
190. Sims NA, Jenkins BJ, Nakamura A, Quinn JMW, Li R, Gillespie MT, et al. Interleukin-11 receptor signaling is required for normal bone remodeling. J Bone Mineral Res. (2005) 20:1093–102. doi: 10.1359/JBMR.050209
191. Johnson RW, Mcgregor NE, Brennan HJ, Crimeen-Irwin B, Poulton IJ, Martin TJ, et al. Glycoprotein130 (Gp130)/interleukin-6 (IL-6) signalling in osteoclasts promotes bone formation in periosteal and trabecular bone. Bone. (2015) 81:343–51. doi: 10.1016/j.bone.2015.08.005
192. Lanktree MB, Guo Y, Murtaza M, Glessner JT, Bailey SD, Onland-Moret NC, et al. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am J Hum Genet. (2011) 88:6–18. doi: 10.1016/j.ajhg.2010.11.007
193. Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, et al. Rare and low-frequency coding variants alter human adult height. Nature. (2017) 542:186–90. doi: 10.1038/nature21039
194. Styrkarsdottir U, Lund SH, Thorleifsson G, Zink F, Stefansson OA, Sigurdsson JK, et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat Genet. (2018) 50:1681–7. doi: 10.1038/s41588-018-0247-0
195. Lokau J, Göttert S, Arnold P, Düsterhöft S, López DM, Grötzinger J, et al. The SNP rs4252548 (R112H) which is associated with reduced human height compromises the stability of IL-11. Biochim Biophys Acta Mol Cell Res. (2017) 1865:496–506. doi: 10.1016/j.bbamcr.2017.12.003
196. Nieminen P, Morgan NV, Fenwick AL, Parmanen S, Veistinen L, Mikkola ML, et al. Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am J Hum Genet. (2011) 89:67–81. doi: 10.1016/j.ajhg.2011.05.024
197. Neveling K, Elcioglu N, Konas E, Akarsu NA, Müller-Newen G, Richardson R, et al. Mutations in the interleukin receptor IL11RA cause autosomal recessive Crouzon-like craniosynostosis. Mol Genet Genomic Med. (2013) 1:223–37. doi: 10.1002/mgg3.28
198. Brischoux-Boucher E, Trimouille A, Baujat G, Goldenberg A, Schaefer E, Guichard B, et al. IL11RA-related Crouzon-like autosomal recessive craniosynostosis in 10 new patients: resemblances and differences. Clin Genet. (2018) 94:373–80. doi: 10.1111/cge.13409
199. Agthe M, Brügge J, Garbers Y, Wandel M, Kespohl B, Arnold P, et al. Mutations in craniosynostosis patients cause defective interleukin-11 receptor maturation and drive craniosynostosis-like disease in mice. Cell Rep. (2018) 25:10–8. doi: 10.1016/j.celrep.2018.09.005
200. Einarsson O, Geba GP, Zhu Z, Landry M, Elias JA. Interleukin-11: stimulation in vivo and in vitro by respiratory viruses and induction of airways hyperresponsiveness. J Clin Investig. (1996) 97:915–24. doi: 10.1172/JCI118514
201. Tang W, Geba GP, Zheng T, Ray P, Homer RJ, Kuhn C, et al. Targeted expression of IL-11 in the murine airway causes lymphocytic inflammation, bronchial remodeling, and airways obstruction. J Clin Investig. (1996) 98:2845–53. doi: 10.1172/JCI119113
202. Chen Q, Rabach L, Noble P, Zheng T, Lee CG, Homer RJ, et al. IL-11 receptor α in the pathogenesis of IL-13-induced inflammation and remodeling. J Immunol. (2005) 174:2305–13. doi: 10.4049/jimmunol.174.4.2305
203. Chun GL, Hartl D, Matsuura H, Dunlop FM, Scotney PD, Fabri LJ, et al. Endogenous IL-11 signaling is essential in Th2- and IL-13-induced inflammation and mucus production. Am J Respir Cell Mol Biol. (2008) 39:739–46. doi: 10.1165/rcmb.2008-0053OC
204. Kapina MA, Shepelkova GS, Avdeenko VG, Guseva AN, Kondratieva TK, Evstifeev VV, et al. Interleukin-11 drives early lung inflammation during Mycobacterium tuberculosis infection in genetically susceptible mice. PLoS ONE. (2011) 6:e21878. doi: 10.1371/journal.pone.0021878
205. Robb L, Li R, Hartley L, Nandurkar HH, Koentgen F, Begley CG. Infertility in female mice lacking the receptor for interleukin 11 is due to a defective uterine response to implantation. Nat Med. (1998) 4:303–8. doi: 10.1038/nm0398-303
206. Dimitriadis E, Robb L, Liu YX, Enders AC, Martin H, Stoikos C, et al. IL-11 and IL-11Rα immunolocalisation at primate implantation sites suports a role for IL-11 in placentation and fetal development. Reprod Biol Endocrinol. (2003) 1:1–10. doi: 10.1186/1477-7827-1-34
207. Menkhorst E, Salamonsen L, Robb L, Dimitriadis E. IL11 Antagonist inhibits uterine stromal differentiation, causing pregnancy failure in mice. Biol Reprod. (2009) 80:920–7. doi: 10.1095/biolreprod.108.073601
208. Chen HF, Lin CY, Chao KH, Wu MY, Yang YS, Ho NN. Defective production of interleukin-11 by decidua and chorionic villi in human anembryonic pregnancy. J Clin Endocrinol Metab. (2002) 87:2320–8. doi: 10.1210/jcem.87.5.8478
209. Paiva P, Salamonsen LA, Manuelpillai U, Walker C, Tapia A, Wallace EM, et al. Interleukin-11 promotes migration, but not proliferation, of human trophoblast cells, implying a role in placentation. Endocrinology. (2007) 148:5566–72. doi: 10.1210/en.2007-0517
210. Paiva P, Salamonsen LA, Manuelpillai U, Dimitriadis E. Interleukin 11 inhibits human trophoblast Invasion indicating a likely role in the decidual restraint of trophoblast invasion during placentation. Biol Reprod. (2008) 80:302–10. doi: 10.1095/biolreprod.108.071415
211. Winship AL, Koga K, Menkhorst E, Van Sinderen M, Rainczuk K, Nagai M, et al. Interleukin-11 alters placentation and causes preeclampsia features in mice. Proc Natl Acad Sci USA. (2015) 112:15928–33. doi: 10.1073/pnas.1515076112
212. Schafer S, Viswanathan S, Widjaja AA, Lim WW, Moreno-Moral A, Delaughter DM, et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. (2017) 552:110–5. doi: 10.1038/nature24676
213. Widjaja AA, Singh BK, Adami E, Viswanathan S, Dong J, D'agostino GA, et al. Inhibiting interleukin 11 signaling reduces hepatocyte death and liver fibrosis, inflammation, and steatosis in mouse models of nonalcoholic steatohepatitis. Gastroenterology. (2019) 157:777–92.e714. doi: 10.1053/j.gastro.2019.05.002
214. Ng B, Dong J, Viswanathan S, D'agostino GA, Widjaja AA, Lim, et al. IL-11 is a therapeutic target in idiopathic pulmonary fibrosis. Sci Translat Med. (2019) 11:eaaw1237. doi: 10.1126/scitranslmed.aaw1237
215. Strikoudis A, Cieślak A, Loffredo L, Chen YW, Patel N, Saqi A, et al. Modeling of fibrotic lung disease using 3D organoids derived from human pluripotent stem cells. Cell Rep. (2019) 3709–23. doi: 10.1016/j.celrep.2019.05.077
216. Obana M, Maeda M, Takeda K, Hayama A, Mohri T, Yamashita T, et al. Therapeutic activation of signal transducer and activator of transcription 3 by interleukin-11 ameliorates cardiac fibrosis after myocardial infarction. Circulation. (2010) 121:684–91. doi: 10.1161/CIRCULATIONAHA.109.893677
217. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. (2000) 100:57–70. doi: 10.1016/S0092-8674(00)81683-9
218. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
219. Sotiriou C, Lacroix M, Lespagnard L, Larsimont D, Paesmans M, Body JJ. Interleukins-6 and−11 expression in primary breast cancer and subsequent development of bone metastases. Cancer Lett. (2001) 169:87–95. doi: 10.1016/S0304-3835(01)00524-9
220. Calon A, Espinet E, Palomo-Ponce S, Tauriello DVF, Iglesias M, Céspedes MV, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. (2012) 22:571–84. doi: 10.1016/j.ccr.2012.08.013
221. Putoczki T, Thiem S, Loving A, Busuttil R, Wilson N, Ziegler P, et al. Interleukin-11 is the dominant Il-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell. (2013) 24:257–71. doi: 10.1016/j.ccr.2013.06.017
222. Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Investig. (2008) 118:1727–38. doi: 10.1172/JCI34944
223. Hanavadi S, Martin TA, Watkins G, Mansel RE, Jiang WG. Expression of interleukin 11 and its receptor and their prognostic value in human breast cancer. Ann Surg Oncol. (2006) 13:802–8. doi: 10.1245/ASO.2006.05.028
224. Johnstone CN, Chand A, Putoczki TL, Ernst M. Emerging roles for IL-11 signaling in cancer development and progression: focus on breast cancer. Cytokine Growth Factor Rev. (2015) 26:489–98. doi: 10.1016/j.cytogfr.2015.07.015
225. Ren L, Wang X, Dong Z, Liu J, Zhang S. Bone metastasis from breast cancer involves elevated IL-11 expression and the gp130/STAT3 pathway. Med Oncol. (2013) 30:634. doi: 10.1007/s12032-013-0634-4
226. Yap J, Salamonsen LA, Jobling T, Nicholls PK, Dimitriadis E. Interleukin 11 is upregulated in uterine lavage and endometrial cancer cells in women with endometrial carcinoma. Reprod Biol Endocrinol. (2010) 8:1–11. doi: 10.1186/1477-7827-8-63
227. Bellone G, Smirne C, Mauri FA, Tonel E, Carbone A, Buffolino A, et al. Cytokine expression profile in human pancreatic carcinoma cells and in surgical specimens: implications for survival. Cancer Immunol Immunother. (2006) 55:684–98. doi: 10.1007/s00262-005-0047-0
228. Paglia D, Oran A, Sauder DN, Mckenzie RC, Kerbel RS. Expression of leukemia inhibitory factor and interleukin-11 by human melanoma cell lines: LIF, IL-6, and IL-11 are not coregulated. J Interferon Cytokine Res. (1995) 15:455–60. doi: 10.1089/jir.1995.15.455
229. Elias JA, Tang W, Horowitz MC. Cytokine and hormonal stimulation of human osteosarcoma interleukin-11 production. Endocrinology. (1995) 136:489–98. doi: 10.1210/endo.136.2.7835281
230. Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O'shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov. (2017) 16:843–62. doi: 10.1038/nrd.2017.201
231. Mesa RA, Yasothan U, Kirkpatrick P. Ruxolitinib. Nat Rev Drug Discov. (2012) 11:103–4. doi: 10.1038/nrd3652
232. Fleischmann R, Kremer J, Cush J, Schulze-Koops H, Connell CA, Bradley JD, et al. Placebo-controlled trial of tofacitinib monotherapy in rheumatoid arthritis. N Engl J Med. (2012) 367:495–507. doi: 10.1056/NEJMoa1109071
233. Mogul A, Corsi K, Mcauliffe L. Baricitinib: the second FDA-approved JAK inhibitor for the treatment of rheumatoid arthritis. Ann Pharmacother. (2019) 53:943–53. doi: 10.1177/1060028019839650
234. Fragoulis GE, Mcinnes IB, Siebert S. JAK-inhibitors. New players in the field of immune-mediated diseases, beyond rheumatoid arthritis. Rheumatology. (2019) 58:i43–i54. doi: 10.1093/rheumatology/key276
235. Strand V, Ahadieh S, French J, Geier J, Krishnaswami S, Menon S, et al. Systematic review and meta-analysis of serious infections with tofacitinib and biologic disease-modifying antirheumatic drug treatment in rheumatoid arthritis clinical trials. Arthr Res Ther. (2015) 17:1–9. doi: 10.1186/s13075-015-0880-2
236. Vincenti F, Tedesco Silva H, Busque S, O'connell P, Friedewald J, Cibrik D, et al. Randomized phase 2b trial of tofacitinib (CP-690,550) in de novo kidney transplant patients: Efficacy, renal function and safety at 1 year. Am J Transplant. (2012) 12:2446–56. doi: 10.1111/j.1600-6143.2012.04127.x
237. Wollenhaupt J, Silverfield J, Lee EB, Curtis JR, Wood SP, Soma K, et al. Safety and efficacy of tofacitinib, an oral janus kinase inhibitor, for the treatment of rheumatoid arthritis in open-Label, longterm extension studies. J Rheumatol. (2014) 41:837–52. doi: 10.3899/jrheum.130683
238. Miklossy G, Hilliard TS, Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat Rev Drug Discov. (2013) 12:611–29. doi: 10.1038/nrd4088
239. Wong AL, Soo RA, Tan DS, Lee SC, Lim JS, Marban PC, et al. Phase I and biomarker study of OPB-51602, a novel signal transducer and activator of transcription (STAT) 3 inhibitor, in patients with refractory solid malignancies. Ann Oncol. (2015) 26:998–1005. doi: 10.1093/annonc/mdv026
240. Reilley MJ, Mccoon P, Cook C, Lyne P, Kurzrock R, Kim Y, et al. STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: results of a phase 1b trial. J Immunother Cancer. (2018) 6:119. doi: 10.1186/s40425-018-0436-5
241. Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. (2001) 276:45443–55. doi: 10.1074/jbc.M107527200
242. Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA. (2005) 102:4700–5. doi: 10.1073/pnas.0409894102
243. Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Translat Med. (2015) 7:314ra185. doi: 10.1126/scitranslmed.aac5272
244. Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, Mceachern D, et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell. (2019) 36:498–511.e417. doi: 10.1016/j.ccell.2019.10.002
245. Wong ALA, Hirpara JL, Pervaiz S, Eu JQ, Sethi G, Goh BC. Do STAT3 inhibitors have potential in the future for cancer therapy? Expert Opin Investig Drugs. (2017) 26:883–7. doi: 10.1080/13543784.2017.1351941
246. Nelson KM, Dahlin JL, Bisson J, Graham J, Pauli GF, Walters MA. The essential medicinal chemistry of curcumin. J Med Chem. (2017) 60:1620–37. doi: 10.1021/acs.jmedchem.6b00975
247. Hahn YI, Kim SJ, Choi BY, Cho KC, Bandu R, Kim KP, et al. Curcumin interacts directly with the Cysteine 259 residue of STAT3 and induces apoptosis in H-Ras transformed human mammary epithelial cells. Sci Rep. (2018) 8:1–14. doi: 10.1038/s41598-018-23840-2
248. Glienke W, Maute L, Wicht J, Bergmann L. Curcumin inhibits constitutive STAT3 phosphorylation in human pancreatic cancer cell lines and downregulation of Survivin/BIRC5 gene expression. Cancer Investig. (2010) 28:166–71. doi: 10.3109/07357900903287006
249. Burgos-Morón E, Calderón-Montaño JM, Salvador J, Robles A, López-Lázaro M. The dark side of curcumin. Int J Cancer. (2010) 126:1771–5. doi: 10.1002/ijc.24967
250. Nero TL, Morton CJ, Holien JK, Wielens J, Parker MW. Oncogenic protein interfaces: small molecules, big challenges. Nat Rev Cancer. (2014) 14:248–62. doi: 10.1038/nrc3690
251. Wilson CGM, Arkin MR. Small-molecule inhibitors of IL-2/IL-2R: lessons learned and applied. In: Vassilev L, Fry D, editors. Small-Molecule Inhibitors of Protein-Protein Interactions. Berlin: Springer Berlin Heidelberg (2011). p. 25–59.
252. Õmura S, Hayashi M, Tomoda H. Recent progress of the research on novel microbial metabolites. Pure Appl Chem. (1999) 71:1673–81. doi: 10.1351/pac199971091673
253. Hayashi M, Rho M-C, Enomoto A, Fukami A, Kim Y-P, Kikuchi Y, et al. Suppression of bone resorption by madindoline A, a novel nonpeptide antagonist to gp130. Proc Natl Acad Sci USA. (2002) 99:14728–33. doi: 10.1073/pnas.232562799
254. Saleh AZM, Greenman KL, Billings S, Van Vranken DL, Krolewski JJ. Binding of madindoline A to the extracellular domain of gp130. Biochemistry. (2005) 44:10822–7. doi: 10.1021/bi050439+
255. Sunazuka T, Hirose T, Shirahata T, Harigaya Y, Hayashi M, Komiyama K, et al. Total synthesis of (+)-madindoline A and (-)-madindoline B, potent, selective inhibitors of interleukin 6. Determination of the relative and absolute configurations. J Am Chem Soc. (2000) 122:2122–3. doi: 10.1021/ja9938074
256. Plasencia C, Dayam R, Wang Q, Pinski J, Burke TR, Quinn DI, et al. Discovery and preclinical evaluation of a novel class of small-molecule compounds in hormone-dependent and -independent cancer cell lines. Mol Cancer Ther. (2005) 4:1105–13. doi: 10.1158/1535-7163.MCT-04-0288
257. Grande F, Aiello F, Grazia OD, Brizzi A, Garofalo A, Neamati N. Synthesis and antitumor activities of a series of novel quinoxalinhydrazides. Bioorgan Med Chem. (2007) 15:288–94. doi: 10.1016/j.bmc.2006.09.073
258. Neamati N, Hong H, Owen JM, Sunder S, Winslow HE, Christensen JL, et al. Salicylhydrazine-containing inhibitors of HIV-1 integrase: implication for a selective chelation in the integrase active site. J Med Chem. (1998) 41:3202–9. doi: 10.1021/jm9801760
259. Plasencia C, Grande F, Oshima T, Cao X, Yamada R, Sanchez T, et al. Discovery of a novel quinoxalinhydrazide with a broad-spectrum anticancer activity. Cancer Biol Ther. (2009) 8:458–65. doi: 10.4161/cbt.8.5.7741
260. Xu S, Grande F, Garofalo A, Neamati N. Discovery of a novel orally active small-molecule gp130 inhibitor for the treatment of ovarian cancer. Mol Cancer Ther. (2013) 12:937–49. doi: 10.1158/1535-7163.MCT-12-1082
261. Sun S, Wang R, Song J, Guan M, Li N, Zhang X, et al. Blocking gp130 signaling suppresses autotoxin expression in adipocytes and improves insulin sensitivity in diet-induced obesity. J Lipid Res. (2017) 58:2102–13. doi: 10.1194/jlr.M075655
262. Ham S, Lima LG, Chai EPZ, Muller A, Lobb RJ, Krumeich S, et al. Breast cancer-derived exosomes alter macrophage polarization via gp130/STAT3 signaling. Front Immunol. (2018) 9:1–10. doi: 10.3389/fimmu.2018.00871
263. Gao M, Yin D, Chen J, Qu X. Activating the interleukin-6-Gp130-STAT3 pathway ameliorates ventricular electrical stability in myocardial infarction rats by modulating neurotransmitters in the paraventricular nucleus. BMC Cardiovasc Disord. (2020) 20:60. doi: 10.1186/s12872-020-01363-x
264. Hong S-S, Choi JH, Lee SY, Park Y-H, Park K-Y, Lee JY, et al. A novel small-molecule inhibitor targeting the IL-6 receptor β subunit, glycoprotein 130. J Immunol. (2015) 195:237–45. doi: 10.4049/jimmunol.1402908
265. Singh S, Gajulapati V, Gajulapati K, Goo JI, Park YH, Jung HY, et al. Structure-activity relationship study of a series of novel oxazolidinone derivatives as IL-6 signaling blockers. Bioorgan Med Chem Lett. (2016) 26:1282–6. doi: 10.1016/j.bmcl.2016.01.016
266. Biskobing DM. Update on bazedoxifene: a novel selective estrogen receptor modulator. Clin. Intervent Aging. (2007) 2:299–303.
267. Xiao H, Bid HK, Chen X, Wu X, Wei J, Bian Y, et al. Repositioning Bazedoxifene as a novel IL-6/GP130 signaling antagonist for human rhabdomyosarcoma therapy. PLoS ONE. (2017) 12:1–19. doi: 10.1371/journal.pone.0180297
268. Yadav A, Kumar B. Bazedoxifene enhances the anti-tumor effects of cisplatin and radiation treatment by blocking IL-6 signaling in head and neck cancer. Oncotarget. (2016) 8:66912–24. doi: 10.18632/oncotarget.11464
269. Tian J, Chen X, Fu S, Zhang R, Pan L, Cao Y, et al. Bazedoxifene is a novel IL-6/GP130 inhibitor for treating triple-negative breast cancer. Breast Cancer Res Treat. (2019) 175:553–66. doi: 10.1007/s10549-019-05183-2
270. Yan D, Ma H, Shi W, Luo P, Liu T, Guo J, et al. Bazedoxifene attenuates abdominal aortic aneurysm formation via downregulation of interleukin-6/glycoprotein 130/signal transducer and activator of transcription 3 signaling pathway in apolipoprotein E–knockout mice. Front Pharmacol. (2020) 11:392. doi: 10.3389/fphar.2020.00392
271. Song D, Yu W, Ren Y, Zhu J, Wan C, Cai G, et al. Discovery of bazedoxifene analogues targeting glycoprotein 130. Eur J Med Chem. (2020) 199:112375. doi: 10.1016/j.ejmech.2020.112375
272. De Groot AS, Scott DW. Immunogenicity of protein therapeutics. Trends Immunol. (2007) 28:482–90. doi: 10.1016/j.it.2007.07.011
273. Dinarello CA. Historical insights into cytokines. Eur J Immunol. (2007) 37:34–45. doi: 10.1002/eji.200737772
274. Baldo BA. Side effects of cytokines approved for therapy. Drug Safety. (2014) 37:921–43. doi: 10.1007/s40264-014-0226-z
275. Tepler I, Elias L, Smith JW, Hussein M, Rosen G, Chang AY, et al. A randomized placebo-controlled trial of recombinant human interleukin-11 in cancer patients with severe thrombocytopenia due to chemotherapy. Blood. (1996) 87:3607–14.
276. Kaye JA. FDA licensure of neumega to prevent severe chemotherapy-induced thrombocytopenia. Stem Cells. (1998) 16:207–23. doi: 10.1002/stem.5530160724
277. Montero AJ, Estrov Z, Freireich EJ, Khouri IF, Koller CA, Kurzrock R. Phase II study of low-dose interleukin-11 in patients with myelodysplastic syndrome. Leukemia Lymphoma. (2006) 47:2049–54. doi: 10.1080/10428190600758058
278. Cantor SB, Elting LS, Hudson DV, Rubenstein EB. Pharmacoeconomic analysis of oprelvekin (recombinant human interleukin-11) for secondary prophylaxis of thrombocytopenia in solid tumor patients receiving chemotherapy. Cancer. (2003) 97:3099–106. doi: 10.1002/cncr.11447
279. Bussel JB, Mukherjee R, Stone AJ. A pilot study of rhuIL-11 treatment of refractory ITP. Am J Hematol. (2001) 66:172–7. doi: 10.1002/1096-8652(200103)66:3<172::AID-AJH1041>3.0.CO;2-Q
280. Milman E, Berdon WE, Garvin JH, Cairo MS, Bessmertny O, Ruzal-Shapiro C. Periostitis secondary to interleukin-11 (Oprelvekin, Neumega). Treatment for thrombocytopenia in pediatric patients. Pediatric Radiol. (2003) 33:450–2. doi: 10.1007/s00247-003-0893-x
281. Tsimberidou AM, Giles FJ, Khouri I, Bueso-Ramos C, Pilat S, Thomas DA, et al. Low-dose interleukin-11 in patients with bone marrow failure: update of the M. D Anderson Cancer Center experience. Ann Oncol. (2005) 16:139–45. doi: 10.1093/annonc/mdi007
282. Sands BE, Bank S, Sninsky CA, Robinson M, Katz S, Singleton JW, et al. Preliminary evaluation of safety and activity of recombinant human interleukin 11 in patients with active Crohn's disease. Gastroenterology. (1999) 117:58–64. doi: 10.1016/S0016-5085(99)70550-0
283. Moreland L, Gugliotti R, King K, Chase W, Weisman M, Greco T, et al. Results of a phase-I/II randomized, masked, placebo-controlled trial of recombinant human interleukin-11 (rhIL-11) in the treatment of subjects with active rheumatoid arthritis. Arthr Res. (2001) 3:247–52. doi: 10.1186/ar309
284. Kang S, Tanaka T, Narazaki M, Kishimoto T. Targeting interleukin-6 signaling in clinic. Immunity. (2019) 50:1007–23. doi: 10.1016/j.immuni.2019.03.026
285. Burmester GR, Rigby WF, Van Vollenhoven RF, Kay J, Rubbert-Roth A, Kelman A, et al. Tocilizumab in early progressive rheumatoid arthritis: FUNCTION, a randomised controlled trial. Ann Rheum Dis. (2016) 75:1081–91. doi: 10.1136/annrheumdis-2015-207628
286. Lamb YN, Deeks ED. Sarilumab: a review in moderate to severe rheumatoid arthritis. Drugs. (2018) 78:929–40. doi: 10.1007/s40265-018-0929-z
287. Rossi JF, Négrier S, James ND, Kocak I, Hawkins R, Davis H, et al. A phase I/II study of siltuximab (CNTO 328), an anti-interleukin-6 monoclonal antibody, in metastatic renal cell cancer. Br J Cancer. (2010) 103:1154–62. doi: 10.1038/sj.bjc.6605872
288. Nishimoto N, Miyasaka N, Yamamoto K, Kawai S, Takeuchi T, Azuma J. Long-term safety and efficacy of tocilizumab, an anti-IL-6 receptor monoclonal antibody, in monotherapy, in patients with rheumatoid arthritis (the STREAM study): evidence of safety and efficacy in a 5-year extension study. Ann Rheum Dis. (2009) 68:1580–4. doi: 10.1136/ard.2008.092866
289. Jones G, Sebba A, Gu J, Lowenstein MB, Calvo A, Gomez-Reino JJ, et al. Comparison of tocilizumab monotherapy versus methotrexate monotherapy in patients with moderate to severe rheumatoid arthritis: the AMBITION study. Ann Rheum Dise. (2010) 69:88–96. doi: 10.1136/ard.2008.105197
290. Rhee FV, Casper C, Voorhees PM, Fayad LE, Gibson D, Kanhai K, et al. Long-term safety of siltuximab in patients with idiopathic multicentric Castleman disease: a prespecified, open-label, extension analysis of two trials. Lancet Haematol. (2020) 3026:1–9. doi: 10.1016/S2352-3026(19)30257-1
291. Shaw S, Bourne T, Meier C, Carrington B, Gelinas R, Henry A, et al. Discovery and characterization of olokizumab. mAbs. (2014) 6:773–81. doi: 10.4161/mabs.28612
292. Blanchetot C, De Jonge N, Desmyter A, Ongenae N, Hofman E, Klarenbeek A, et al. Structural mimicry of receptor interaction by antagonistic interleukin-6 (IL-6) antibodies. J Biol Chem. (2016) 291:13846–54. doi: 10.1074/jbc.M115.695528
293. Liu Q, Zhou YH, Yang ZQ. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol. (2016) 13:3–10. doi: 10.1038/cmi.2015.74
294. Zhang Y, Li J, Zhan Y, Wu L, Yu X, Zhang W, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immunity. (2004) 72:4410–5. doi: 10.1128/IAI.72.8.4410-4415.2004
295. Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J General Virol. (2013) 94:2679–90. doi: 10.1099/vir.0.055533-0
296. Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. (2013) 368:1509–18. doi: 10.1056/NEJMoa1215134
297. Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect Biol. (2014) 6:a016295. doi: 10.1101/cshperspect.a016295
298. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
299. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Investig. (2020) 130:2620–9. doi: 10.1172/JCI137244
300. Xu X, Han M, Li T, Sun W, Wang D, Fu B, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. (2020) 117:10970–5. doi: 10.1073/pnas.2005615117
301. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
302. Mehta P, Mcauley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. (2020) 395:1033–4. doi: 10.1016/S0140-6736(20)30628-0
303. Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun. (2020) 111:102452. doi: 10.1016/j.jaut.2020.102452
304. Alattar R, Ibrahim TBH, Shaar SH, Abdalla S, Shukri K, Daghfal JN, et al. Tocilizumab for the Treatment of Severe COVID-19. J Med Virol. (2020). doi: 10.1002/jmv.25964. [Epub ahead of print].
305. Fontana F, Alfano G, Mori G, Amurri A, Lorenzo T, Ballestri M, et al. Covid-19 pneumonia in a kidney transplant recipient successfully treated with Tocilizumab and Hydroxychloroquine. Am J Transplant. (2020) 20:1902–6. doi: 10.1111/ajt.15935
306. Luo P, Liu Y, Qiu L, Liu X, Liu D, Li J. Tocilizumab treatment in COVID-19: a single center experience. J Med Virol. (2020) 92:814–8. doi: 10.1002/jmv.25801
307. Zhang X, Song K, Tong F, Fei M, Guo H, Lu Z, et al. First case of COVID-19 in a patient with multiple myeloma successfully treated with tocilizumab. Blood Adv. (2020) 4:1307–10. doi: 10.1182/bloodadvances.2020001907
308. Radbel J, Narayanan N, Bhatt PJ. Use of tocilizumab for COVID-19 infection-induced cytokine release syndrome: a cautionary case report. Chest. (2020). doi: 10.1016/j.chest.2020.04.024. [Epub ahead of print].
309. Deguchi Y, Nishina T, Asano K, Ohmuraya M, Nakagawa Y, Nakagata N, et al. Generation of and characterization of anti-IL-11 antibodies using newly established Il11-deficient mice. Biochem Biophys Res Commun. (2018) 505:453–9. doi: 10.1016/j.bbrc.2018.09.128
310. Blanc C, Vusio P, Schleinkofer K, Boisteau O, Pflanz S, Grotzinger J, et al. Monoclonal antibodies against the human interleukin-11 receptor alpha-chain (IL-11Ra) and their use in studies of human mononuclear cells. J Immunol Methods. (2000) 241:43–59. doi: 10.1016/S0022-1759(00)00194-0
311. Winship AL, Van Sinderen M, Donoghue J, Rainczuk K, Dimitriadis E. Targeting interleukin-11 receptor-α impairs human endometrial cancer cell proliferation and invasion in vitro and reduces tumor growth and metastasis in vivo. Mol Cancer Ther. (2016) 15:720–30. doi: 10.1158/1535-7163.MCT-15-0677
312. Müller-Newen G, Küster A, Wijdenes J, Schaper F, Heinrich PC. Studies on the interleukin-6-type cytokine signal transducer gp130 reveal a novel mechanism of receptor activation by monoclonal antibodies. J Biol Chem. (2000) 275:4579–86. doi: 10.1074/jbc.275.7.4579
313. Garbers C, Heink S, Korn T, Rose-John S. Interleukin-6: designing specific therapeutics for a complex cytokine. Nat Rev Drug Discov. (2018) 17:395–412. doi: 10.1038/nrd.2018.45
314. Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, et al. Stat3/Socs3 activation by iL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. (2011) 19:456–69. doi: 10.1016/j.ccr.2011.03.009
315. Bergmann J, Müller M, Baumann N, Reichert M, Heneweer C, Bolik J, et al. IL-6 trans-signaling is essential for the development of hepatocellular carcinoma in mice. Hepatology. (2017) 65:89–103. doi: 10.1002/hep.28874
316. Nowell MA, Richards PJ, Horiuchi S, Yamamoto N, Rose-John S, Topley N, et al. Soluble IL-6 receptor governs IL-6 activity in experimental arthritis: blockade of arthritis severity by soluble glycoprotein 130. J Immunol. (2003) 171:3202–9. doi: 10.4049/jimmunol.171.6.3202
317. Richards PJ, Nowell MA, Horiuchi S, Mcloughlin RM, Fielding CA, Grau S, et al. Functional characterization of a soluble gp130 isoform and its therapeutic capacity in an experimental model of inflammatory arthritis. Arthr Rheum. (2006) 54:1662–72. doi: 10.1002/art.21818
318. Atreya R, Mudter J, Finotto S, Müllberg J, Jostock T, Wirtz S, et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in Crohn disease and experimental colitis in vivo. Nat Med. (2000) 6:583–8. doi: 10.1038/75068
319. Mitsuyama K, Matsumoto S, Rose-John S, Suzuki A, Hara T, Tomiyasu N, et al. STAT3 activation via interleukin 6 trans-signalling contributes to ileitis in SAMP1/Yit mice. Gut. (2006) 55:1263–9. doi: 10.1136/gut.2005.079343
320. Zhang H, Neuhöfer P, Song L, Rabe B, Lesina M, Kurkowski MU, et al. IL-6 trans-signaling promotes pancreatitis- associated lung injury and lethality. J Clin Investig. (2013) 123:1019–31. doi: 10.1172/JCI64931
321. Hoge J, Yan I, Jänner N, Schumacher V, Chalaris A, Steinmetz OM, et al. IL-6 controls the innate immune response against listeria monocytogenes via classical IL-6 signaling. J Immunol. (2013) 190:703–11. doi: 10.4049/jimmunol.1201044
322. Adams R, Burnley RJ, Valenzano CR, Qureshi O, Doyle C, Lumb S, et al. Discovery of a junctional epitope antibody that stabilizes IL-6 and gp80 protein:protein interaction and modulates its downstream signaling. Sci Rep. (2017) 7:37716. doi: 10.1038/srep37716
323. De Hon FD, Ehlers M, Rose-John S, Ebeling SB, Bos HK, Aarden LA, et al. Development of an interleukin (IL) 6 receptor antagonist that inhibits IL-6-dependent growth of human myeloma cells. J Exp Med. (1994) 180:2395–400. doi: 10.1084/jem.180.6.2395
324. Fairlie WD, Uboldi AD, Mccoubrie JE, Wang CC, Lee EF, Yao S, et al. Affinity maturation of leukemia inhibitory factor and conversion to potent antagonists of signaling. J Biol Chem. (2004) 279:2125–34. doi: 10.1074/jbc.M310103200
325. Findeisen M, Allen TL, Henstridge DC, Kammoun H, Brandon AE, Baggio LL, et al. Treatment of type 2 diabetes with the designer cytokine IC7Fc. Nature. (2019) 574:63–8. doi: 10.1038/s41586-019-1601-9
326. Ettinger MP, Littlejohn TW, Schwartz SL, Weiss SR, Mcilwain HH, Heymsfield SB, et al. Recombinant variant of ciliary neurotrophic factor for weight loss in obese adults: a randomized, dose-ranging study. J Am Med Assoc. (2003) 289:1826–32. doi: 10.1001/jama.289.14.1826
Keywords: cytokine, interleukin, IL-11, IL-6, JAK, STAT, structural biology, drug development
Citation: Metcalfe RD, Putoczki TL and Griffin MDW (2020) Structural Understanding of Interleukin 6 Family Cytokine Signaling and Targeted Therapies: Focus on Interleukin 11. Front. Immunol. 11:1424. doi: 10.3389/fimmu.2020.01424
Received: 03 March 2020; Accepted: 02 June 2020;
Published: 16 July 2020.
Edited by:
Erwan Mortier, INSERM U1232 Centre de Recherche en Cancérologie et Immunologie Nantes Angers (CRCINA), FranceReviewed by:
Gareth Wyn Jones, Cardiff University School of Medicine, United KingdomCopyright © 2020 Metcalfe, Putoczki and Griffin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tracy L. Putoczki, cHV0b2N6a2kudEB3ZWhpLmVkdS5hdQ==; Michael D. W. Griffin, bWdyaWZmaW5AdW5pbWVsYi5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.