
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 21 July 2020
Sec. Comparative Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01362
This article is part of the Research Topic Vaccines and Immunostimulants for Finfish View all 21 articles
 Biswajit Maiti1*
Biswajit Maiti1* Saurabh Dubey2
Saurabh Dubey2 Hetron Mweemba Munang'andu2*
Hetron Mweemba Munang'andu2* Iddya Karunasagar3
Iddya Karunasagar3 Indrani Karunasagar1,3
Indrani Karunasagar1,3 Øystein Evensen2*
Øystein Evensen2*Aquaculture is one of the fastest-growing food-producing sectors in the world. However, its growth is hampered by various disease problems due to infectious microorganisms, including Gram-negative bacteria in finfish aquaculture. Disease control in aquaculture by use of antibiotics is not recommended as it leads to antibiotic residues in the final product, selection, and spread of antibiotic resistance in the environment. Therefore, focus is on disease prevention by vaccination. All Gram-negative bacteria possess surface-associated outer membrane proteins (OMPs), some of which have long been recognized as potential vaccine candidates. OMPs are essential for maintaining the integrity and selective permeability of the bacterial membrane and play a key role in adaptive responses of bacteria such as solute and ion uptake, iron acquisition, antimicrobial resistance, serum resistance, and bile salt resistance and some adhesins have virulence attributes. Antigenic diversity among bacterial strains even within the same bacterial species has constrained vaccine developments, but OMPs that are conserved across serotypes could be used as potential candidates in vaccine development, and several studies have demonstrated their efficacy and potential as vaccine candidates. In this review, we will look into the application of OMPs for the design of vaccines based on recombinant proteins, subunit vaccines, chimeric proteins, and DNA vaccines as new-generation vaccine candidates for major bacterial pathogens of fish for sustainable aquaculture.
Asia accounts for more than 80% of the global aquaculture production of which India is the third largest producer (1, 2). Indian aquaculture has enormous potential and contributes significantly to the country's economy and its foreign exchange earnings. Finfishes are the most cultured species in the world, and India is no exception, contributing to 68% of world food fish aquaculture production with the other groups being molluskan shellfish (oyster, clam, mussel, and scallop), crustaceans (shrimps and prawns), and other fishes (1). A half of global food fish consumption comes from aquaculture. However, there is a need to improve management practices in order to reduce the disease burden and usage of drugs for disease treatment.
Although aquaculture is fast growing in Asian countries such as India, the largest proportion of fish farming is done by low-resource farmers in earthen ponds and floating cages in rivers and lakes whose environmental conditions support the survival of opportunistic pathogenic bacteria that cause disease in fish. Implementation of biosecurity measures is mostly low while the use of antibiotics is high, posing the danger of drug resistance (3–5). As such, vaccination is considered to be the most effective environmentally friendly disease control strategy. However, the most prevalent diseases infecting the top farmed fish species in each country guide the choice and priority of vaccine development. India being a country mostly producing carp, pathogens infecting top farmed carp species would be a priority for fish vaccines. Another confounding factor is the choice of vaccine delivery system as to whether vaccines should be administered by injection, orally, or immersion, which is guided by factors such the cost of vaccination, labor input, stress on fish, and other factors. Hence, the objective of this review is to bring into perspective the major fish species farmed in India together with major pathogens that need vaccine development. We also wanted to highlight the shortcomings of whole cell inactivated (WCI) and attenuated live vaccines used elsewhere that have paved way to research on the use of outer membrane protein (OMP) vaccines in Indian aquaculture. Herein, we provide an up-to-date status of ongoing research on OMP vaccines being developed against major pathogens infecting the top-farmed fish species in Indian aquaculture.
Food and nutritional security are being addressed through aquaculture due to stagnation of capture fisheries. In 2016, total global production of fish, crustaceans, mollusks, and other aquatic animals reached 170.9 million tons (MT) in which a large volume (>88%) was utilized for human consumption (1, 2). Global aquaculture is a fast-growing vital sector for the production of high-protein food, having an average annual growth rate of 5.8% during 2001–2016 (6). In India, aquaculture is a rapidly growing fisheries sector with an annual growth rate of over 7% of which freshwater aquaculture contributed 95% of the total annual production of 5.77 mt (MT) by 2017 (7). As discussed by Jayasankar (8), advances in carp breeding technologies and traditional polyculture system have contributed to increased production of the three India major carp species, namely, catla (Catla catla), rohu (Labeo rohita), and mrigal (Cirrhinus mrigala), accounting for 70–75% of total freshwater production. This is followed by the culture of three exotic carp species comprising of common carp (Cyprinus carpio), silver carp (Hypophthalmichthys molitrix), and grass carp (Ctenopharyngodon idella) that account for 20–30% of freshwater fish species. The increase in stocking density brought about by intensified farming systems led to increased output from 500 to 600 kg/ha to 3,000 kg/ha, resulting in fish farmers achieving higher production levels of 6–8 t/ha/year, while national average output increased from 50 kg/ha/year in 1974–1975 to about 2,135 kg/ha/year in 1994–1995 and 2,270 kg/ha/year in 2003–2004 (9, 10). Due to the contribution of public and private hatcheries in the production of about 40 billion fry in 2017, it is projected that by 2020, total carp production would exceed 15 mt due to intensive farming systems supported by high stocking densities (8). This increasing trend in stocking density could be contributing to the increase in disease outbreaks as a result of the increase in the disease transmission index as well as induction of stress predisposing fish to various infections.
The major diseases of finfish in Indian aquaculture are caused by bacterial infections (7, 11). Viral pathogens like tilapia lake virus (TiLV) (12), nodavirus (13), Koi herpesvirus virus (KHV) (14), and red sea bream iridovirus (RSBIV) (15, 16) are not pathogens of top farmed fish species in India. The major parasite-causing disease in fish in India is Ichthyophthirius mulifiliis (11) whose impact is not severe compared to bacterial pathogens. Overall, viral and parasitic diseases cause less economic losses unlike bacterial diseases that cause an adverse economic impact, calling for the urgent need of protective vaccines (17). The most prevalent bacterial pathogens in Indian aquaculture belong to the genera Aeromonas, Edwardsiella, Vibrio, and Flavobacterium, infecting the top farmed fish species (11, 18). Table 1 shows fish species infected by these bacteria and their occurrence during different stages of the fish production cycles. Other pathogenic bacterial genera that are associated with fish diseases in India include Streptococcus, Pseudomonas, and Mycobacterium.
Aeromonads belong to the family Aeromonadaceae, and the most common species associated with fish diseases in India are Aeromonas hydrophila (24, 25), Aeromonas sobria (26, 27), Aeromonas caviae (28), and Aeromonas veronii (29). They are natural inhabitants of aquatic environments such as freshwater, estuarine, and infrequently marine waters (25). These pathogens cause hemorrhagic septicemia, tail-rot (or fin-rot), red sore, ulcerative disease, dropsy, asymptomatic septicemia, exophthalmos, and ulceration in different fish species (30).
Vibriosis is one of the most critical fish diseases caused by members of the genus Vibrio that are ubiquitous in aquatic environments. The disease affects both cold-water and warmwater fish species, including sea bass, carp, catfish, salmon, flounder, and eel across the world. In India, the Vibrio species known to cause diseases include Vibrio anguillarum, Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio ordalii, and Vibrio vulnificus of which classical vibriosis is mostly caused by V. anguillarum (20, 31).
Another important genus is Edwardsiella, which is ubiquitous in aquatic environments and is responsible for high mortality in several commercial fish species including carp, catfish, and tilapia in India. Previous studies have shown that the most common Edwardsiella species infecting fish in India was Edwardsiella tarda (22) as the causative agent of septicemia in warmwater fish species, especially catfish. It also causes fish gangrene, emphysematous putrefactive disease, red disease, and enteric septicemia in carp, catfish, and several other fish species (32, 33). However, in a recent study, we showed that piscine Edwardsiella isolates from 10 fish species in India belonged to Edwardsiella piscicida and Edwardsiella anguillarum (34). Therefore, it is likely that all fish isolates previously classified as E. tarda were either E. piscicida or E. anguillarum.
Among the Flavobacterium, Flavobacterium columnare is the most common isolate and often associated with columnaris in farmed catfish (Clarias batrachus), carp (C. carpio), rohu (L. rohita), catla (C. catla), and other fish species (35, 36). Other Flavobacterium species reported to cause disease in fish in India include Flavobacterium aquaticum, Flavobacterium granuli, Flavobacterium hercynium, and Flavobacterium terrae (35).
The common denominator for all these bacteria species is that they ubiquitously live in water and are able to survive under different environmental conditions (25, 37–40) becoming pathogenic as fish become vulnerable to infection when predisposing factors such as high stocking densities that stress fish leading to immunosuppression favor infection establishment. Therefore, the increase in stocking density aimed at increasing productivity in Indian aquaculture discussed in the section Fish Aquaculture: Present Status could be contributing to the increase in disease outbreaks caused by these bacteria species.
Intensive aquaculture systems where single or multiple fish species are cultured at high densities facilitate high transmission of pathogens between individual fish. Although biosecurity measures that include quarantine, sanitation, and disinfection as well as the use of probiotics, disease-free brood stock, immunostimulants, and quality feed have been shown to reduce disease transmission, these measures do not always ensure total elimination of infectious agents. On the other hand, use of antibiotics poses the risk of selection of drug resistance in pathogens, making the treatment ineffective, spread of resistance determinants to other bacteria (41), and antibiotic residues in food (42). To prevent the recurrence of disease outbreaks and widespread use of antibiotics in aquatic environments in India, the most environment-friendly practical approach would be vaccination. For example, vaccination of Atlantic salmon (Salmo salar L.) against pathogens such as Aeromonas salmonicida and Vibrio salmonicida for more than 30 years contributed to a significant reduction of antibiotics use in Norway from nearly 50,000 kg of antibiotics in 1987 to <1,000–2,000 kg in 1997 (43).
Vaccines in aquaculture are either administered by injection, oral route, or immersion. Advantages of oral and immersion vaccine delivery systems are that they are less labor intensive, uses the natural route of pathogen exposure, and less stressful on fish, while vaccination by injection is labor intensive, bypasses the natural route of pathogen exposure, and is stressful on fish. However, vaccination by injection guarantees delivery of the same antigen dose to all fish, while immersion and oral vaccine delivery do not (44). Vaccines administered by injection require high labor costs for individual handling, which is expensive for the majority of low-resource fish farmers in India. On the contrary, vaccine delivery by immersion or oral does not require high labor costs because fish are vaccinated in bulk at the same time orally through feed or by immersion in vaccine-containing water. The major predicament with vaccine delivery by immersion is that practically it cannot be done in open water in ponds or cages floating in rivers and lakes. On the other hand, the major drawback with oral vaccination is that vaccines administered by ingestion are degraded in the acidic environment of the stomach/foregut before they reach the intestine where they are potentially taken up by cells of the innate immune system for local antigen presentation or transport to major immune organs (kidney/spleen) (45). There are few oral vaccines licensed to date (46), and new approaches using new technologies such as poly D, L lactic-co-glycolic acid (PLGA) nanoparticle vaccines that can protect antigens against low pH degradation in the stomach/gut are considered better alternatives.
Traditionally, fish vaccines are made of live-attenuated or WCI vaccines (47, 48). WCI bacterial vaccines are prepared by chemical or heat inactivation of bacteria, and they account for the largest proportion of commercial vaccines used in aquaculture worldwide (49). They are safe because they are not infectious (“killed”) but have the disadvantage of being less immunogenic, needing adjuvants to produce long-term protective immunity (49). They elicit humoral immune responses that to a lesser extent confer protection against intracellular replicating bacteria such as A. hydrophila, Edwardsiella spp., or Piscirickettsia sp. because they do not induce cell-mediated immune (CMI) responses needed to eliminate intracellularly replicating bacteria (50). In contrast, live attenuated vaccines are highly immunogenic and have the ability to evoke both humoral and CMI responses needed to eliminate extra- and intracellular bacteria, but they pose the danger of reversion to virulence (50). DNA vaccines meet challenges of genetically modified organism (GMO) regulations; although the vaccines by themselves are not considered GMOs, vaccinated fish are considered GMOs under certain conditions. In the case of Indian aquaculture where intracellular replicating bacteria such as A. hydrophila and Edwardsiella spp. account for a large proportion of pathogens infecting top farmed fish species, there is a need for vaccines able to evoke both humoral and CMI responses. Currently, there are no licensed attenuated live vaccines against diseases caused by these pathogens. Hence, the use of genetically engineered vaccines using immunogenic proteins such as OMPs encoded in carrier vectors able to evoke both humoral and CMI responses is considered to be a better alternative.
Several studies show that bacterial OMPs have the potential to serve as vaccine candidates for immunization against bacteria infecting fish (49). OMPs are the essential component of outer membranes and are found in many prokaryotes (bacteria) as well as in specific organelles like mitochondria, chloroplasts (51) of eukaryotic cells, possibly even in archaea (52). In general, about 2–3% of the total bacterial genes encode OMPs in Gram-negative bacteria (53). They are made of β-barrel structures that contain 8–22 β-strands, which are antiparallel to each other and tilted strongly on the barrel axis (54). They are shaped in different forms such as monomers, homo-dimers, and/or homo-trimers in the outer membrane of which more than a dozen OMP structures have been resolved. As shown in Figure 1, the structural layout of OMPs shows that the C and N-terminal ends of OMPs are directed toward the periplasm, while surface loops (marked as L1, L2, L3, and so on) are located on the outermost exterior where they are exposed to the outside environment. Several studies have shown that the surface periplasmic that turns together with surface loops of OMPs have more sequence variations than the β-sheet strands, which are conserved in most bacteria species (51, 55). For example, Braun and Cole (56) found a low amino acid sequence similarity of the periplasmic turns and surface loops (54%) while β-sheets similarity was higher (74%) among OmpA proteins of Serratia marcescens. The strategic location of surface loops being exposed to the exterior surface render them ideal for interaction with host cells while their sequence differences could account for antigenic diversity within bacterial species (57). As such, OMPs are considered potential vaccine candidates since they are (i) highly immunogenic due to their exposed epitopes on the bacterial outer cell surface and (ii) highly conserved among different serovars and within Gram-negative bacteria (58–63). Suffice to point out that some OMPs also work as adhesins facilitating the attachment and penetration of bacteria into the host cells, thereby contributing to virulence (58–63). In addition, OMPs carry pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS) recognized by pathogen recognition receptors (PRRs) found on host cells such as monocytes, macrophages, neutrophils, and dendritic cells involved in antigen uptake, processing, and presentation to cells of the adaptive immune system for induction of long-term protective immunity. Overall, this supports the use of OMPs as ideal vaccine candidates for both intra- and extracellular bacteria that are endemic in Indian aquaculture. As shown in Table 1, various OMPs have been used for vaccine development against various pathogens infecting different fish species in India.
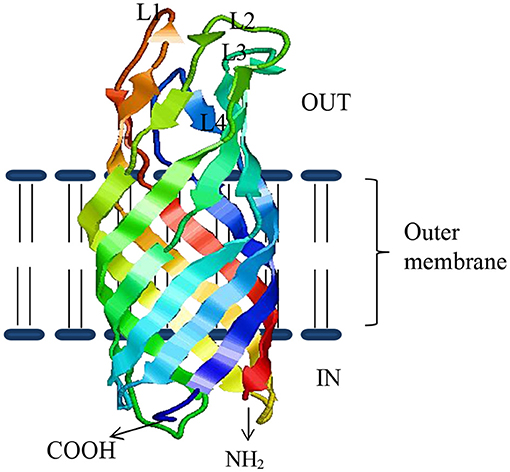
Figure 1. The β-sheeted architecture of OmpW protein of A. hydrophila. Four outside exposed loops of the protein are indicated as L1–L4, respectively.
Epitope mapping of antigenic proteins recognized by B and T cells is crucial for optimal vaccine design. One approach suggested by Rappuoli (64) is to use reverse vaccinology in which several molecules are screened using in silico analysis to identify potential vaccine candidates (65). Figure 2 illustrates the use of reverse vaccinology in vaccine design. In India, various studies have been conducted aimed at identifying bacterial antigenic proteins using in silico analysis as shown in Table 2. Nucleotide or genome sequence of several OMPs can be retrieved from the databases for in silico analysis. There are several bioinformatics tools available used to identify open reading frames (ORFs) encoding putative omp genes while the basic local alignment search tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of the National Center for Biotechnology Information (NCBI) can be used for sequence verification. After predicting ORFs encoding the putative omp genes, the next step is to apply a battery of algorithms designed to extract as much information about the ORF as possible, including tentative molecular weight, pI, and hydrophobic nature of the protein. In general, OMPs include signal peptide required for translocation from the cytoplasm to the outer membrane of cells. The SignalP 5.0 server (http://www.cbs.dtu.dk/services/SignalP/) (83) offers a platform based on a combination of several artificial neural networks that can predict the presence of signal peptides and identify the cleavage sites in proteins. The number of domains and motifs can be found with the help of a domain finder and motif finder. Further, beta-barrel OMPs can be predicted and two-dimensional topology can be analyzed using the online software PRED-TMBB (http://bioinformatics.biol.uoa.gr/PRED-TMBB) (84). The degree of immunogenicity associated with specific OMPs can be predicted using various tools such as the EMBOSS server (http://bioinfo.nhri.org.tw/gui/) (85), which is one of the popular online sites used for the determination of antigenic sites present in the protein. The presence of B- and T-cell epitopes can be identified in OMP sequences using different software. For example, the locations of linear B-cell epitopes in the OMP sequence can be identified by the BepiPred server (http://www.cbs.dtu.dk/services/BepiPred/) (86) that uses a combination of hidden Markov model and propensity scale methods. The commonly used T-cell epitope predicting tool like NetCTL (http://www.cbs.dtu.dk/services/NetCTL/) (87) identifies protein sequences using major histocompatibility complex (MHC) class I binding prediction of 12 MHC supertypes including the supertypes A26 and B39. Peptide–MHC class I binding can be predicted using NetMHC server (http://www.cbs.dtu.dk/services/NetMHC/) (88) that works on artificial neural networks (ANNs) and weight matrices.
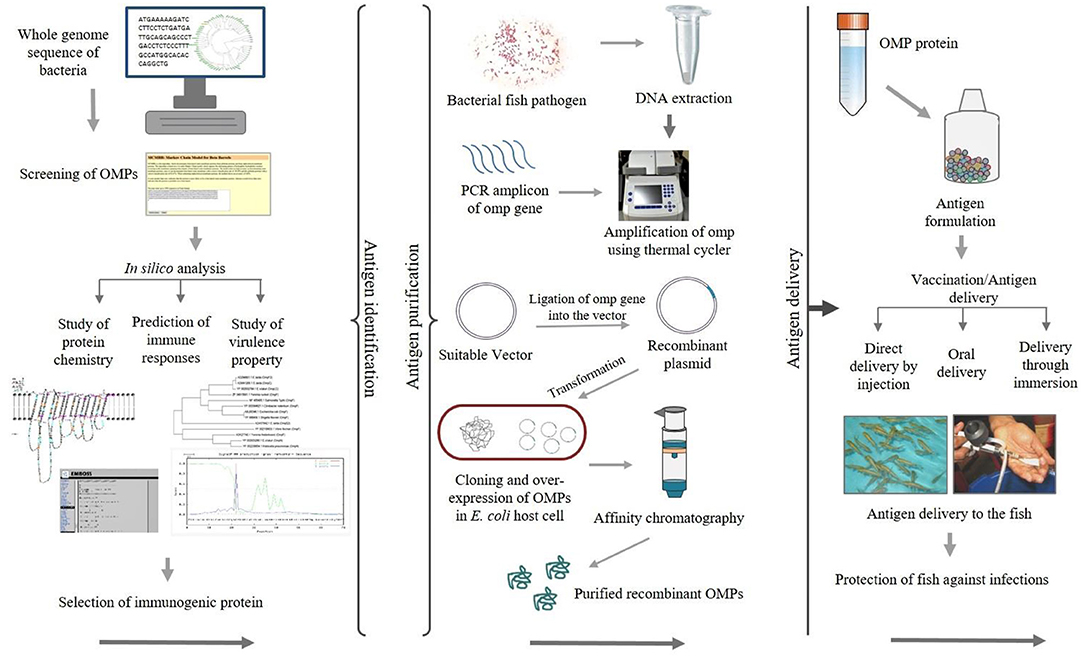
Figure 2. Schematic diagram showing the application of recombinant outer membrane proteins (OMPs) through a reverse vaccinology approach.
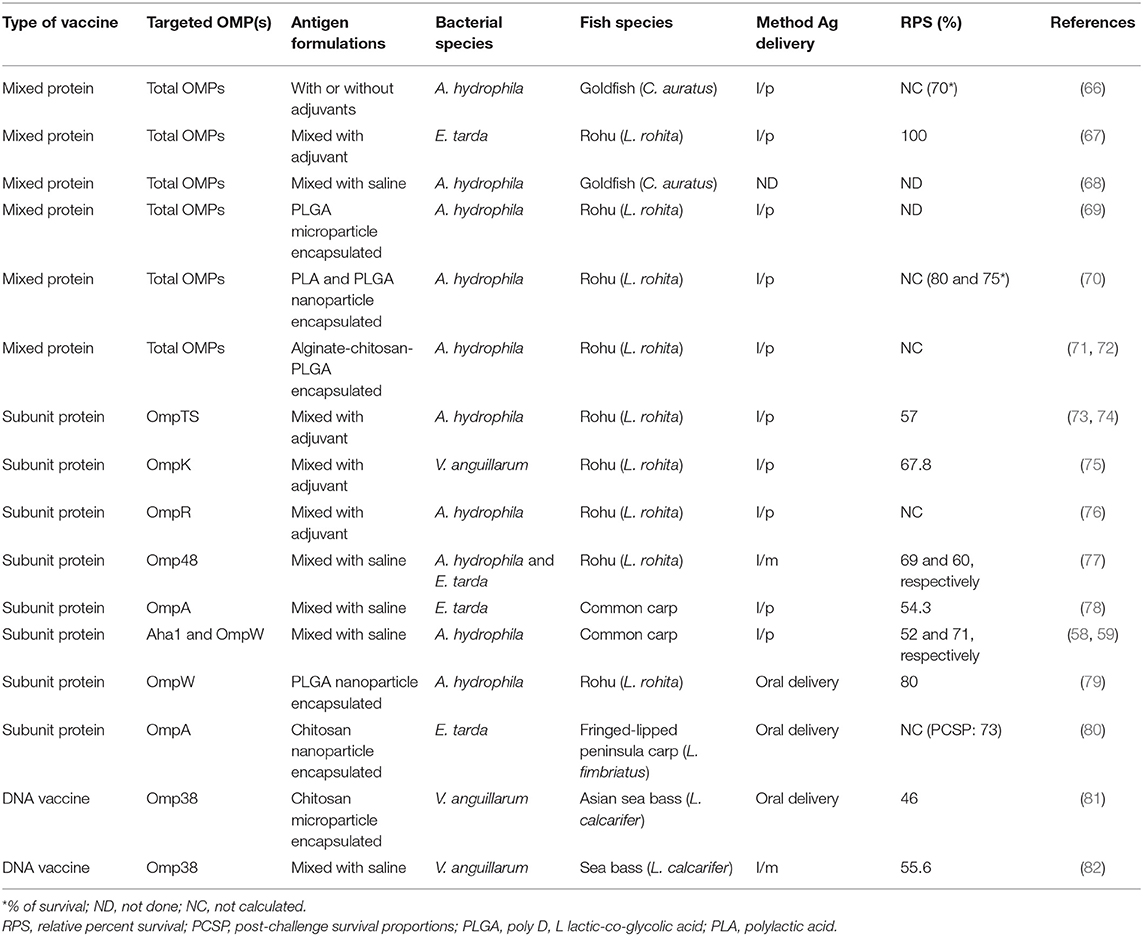
Table 2. Outer membrane protein (OMP)-based vaccination studies conducted in India against major bacterial fish pathogens.
Another approach used for epitope mapping that has gained precedent in recent years is whole-genome sequencing (WGS) of pathogens used to identify new antigens. Together with recombinant DNA technology, WGS has contributed to improving OMP vaccine design, while protein sequence comparison has proved to be a powerful tool used to identify immunogenic proteins that are broadly protective against variant pathogen strains. For example, Dubey et al. (34) used protein sequence comparison and phylogenetic analysis to show that the OmpW of E. piscicida and E. anguillarum had high similarity, suggesting that a common antigen can be used against isolates from different fish species and geographical areas in Asia.
Genetically engineered vaccines are made of purified recombinant proteins or subunit of proteins expressed in heterologous vectors (89). The main advantage of recombinant vaccines is safety because they only contain the antigenic protein and not the entire pathogens. Moreover, genetically engineered vaccines help remove undesired harmful antigens or cleave out epitopes that stimulate T-suppressor cells. The common genetically engineered vaccines used for delivery of OMPs and other antigens in aquaculture are subunit and DNA vaccines.
In recent years, recombinant OMPs are widely tested as subunit vaccines for various pathogens in different fish species since they are highly immunogenic and they are considered safe because they only contain the antigen proteins and not the entire pathogen (59, 65, 71, 77–80, 90–105). Subunit vaccines either (i) are made of specific targeted epitopes identified from total OMPs using technologies such in silico analysis or mass spectrometry (Table 2) or (ii) use total OMP expressed in recombinant expression vectors (Table 3).
In India, Maji et al. (68) fractionated the A. hydrophila OMP using gel permeation and ion-exchange chromatography and generated 10 fractions of which two of the fractionated antigens made of 23-kDa and 57-kDa polypeptides had higher sero-reactivity than the crude OMP. In another study, Kumar et al. (67) used isoelectric focusing (IEF) followed by two-dimensional polyacrylamide gel electrophoresis (PAGE) and mass spectrophotometry to identify two immunogenic proteins (OMP assembly factor YaeT and GroEL) from E. tarda OMP that produced 100% protection after challenge in vaccinated rohu. Sharma and Dixit (106) used in silico analysis to eliminate nonspecific binding epitopes and selectively identified four immunodominant B-cell epitopes of the A. hydrophila OmpF that were highly immunogenic. They showed that the region harboring 66–80 aa residues of the OmpF had the highest reactivity in ELISA, clearly indicating that the OmpF epitope66−80 was the most potent vaccine candidate against A. hydrophila. In another study, Sharma and Dixit (108) used a bioinformatic algorithm to show that the linear B-cell epitopes covering 143–175 aa of A. hydrophila OmpC had the highest cross reactivity with the parent OmpC protein. Antibody isotyping, cytokine ELISA, and cytokine array analysis revealed a Th2 skewed immune response. Mahendran et al. (109) used in silico immunoinformatics to identify T-cell epitopes with binding interaction between E. tarda TolC and F. columnare FCOLo peptides of OMPs with MHC-I alleles. Altogether, these studies show that specific immunogenic proteins can be identified targeting B- and T-cell epitopes from total/crude OMPs for use in vaccine design.
The majority of OMP subunit vaccines are made of entire ORFs of total OMPs expressed and purified from heterologous vectors (Table 3). Bader et al. (111) showed that total OMP extracted from Edwardsiella ictaluri had low protection in channel catfish vaccinated with 3.13 or 6.25 μg of OMP, but a higher dose of OMP (12.5 μg) produced higher protection [relative percent survival (RPS) = 67.5]. Khushiramani et al. (73, 74) showed that the A. hydrophila OmpTS produced 57% survival in rohu after challenge. Similarly, Wang et al. (112) compared the protective ability of a 20-kDa protein of A. hydrophila OmpW with a kDa adhesin protein (Aha1) of A. hydrophila in common carp (C. carpio) and showed that the rOmpW (RPS = 71%) had superior protection over the Aha1 (RPS = 52%) after challenge (59). In another study, Khushiramani et al. (77) showed that the rOmp48 produced high protection in rohu against multiple fish pathogens viz A. hydrophila (RPS = 69%) and E. tarda (RPS = 60%), indicating that Omp48 could be used against multiple pathogens. A study by Maiti et al. (78) reported RPS = 54.3% in common carp using vaccinated rOmpA after challenge with E. tarda. Similarly, Hamod et al. (75) showed high protection (RPS = 67.8%) in adult rohu vaccinated with a rOmpK subunit vaccine after challenge with Vibrio anguillarum. Dash et al. (76) showed that a rOmpR vaccine adjuvanted with mineral oil produced 54 and 90% survival in rohu after challenge with A. hydrophila at 56 and 140 days post vaccination, respectively. In the same study, Dash et al. (76) used the same rOmpR vaccine with a modified adjuvant of mineral oil mixed with phosphate buffered saline (PBS) at equal volumes (1:1 ratio) and showed protection of 67 and 87% after challenge with A. hydrophila at 56 and 140 days post vaccination, respectively. Put together, these studies show that OMP vaccines are being developed against major fish pathogens such as A. hydrophila, E. tarda, and V. anguillarum (Table 1) and that vaccine efficacy trials are mostly done in fish species such as rohu, common carp, and channel catfish that are among the top farmed species in the Indian aquaculture. In addition, these studies also show that different OMPs such as OmpA, OmpK, OmpR, OmpW, OmpTS, and Omp48 are being used in the design of subunit vaccines in India.
Another important vaccination approach used for the delivery of OMP antigens is the use of plasmid vectors to produce DNA vaccines able to transcribe and translate the immunogenic OMP genes intracellularly (113). DNA vaccines possess several advantages over WCI vaccines such as the stimulation of both humoral and CMI responses (50, 114, 115) unlike WCI vaccines that only stimulate humoral responses (49). Moreover, DNA vaccines do not require the potentiation effect of adjuvants unlike WCI vaccines that have been shown to have severe side effects caused by adjuvants incorporated in vaccine formulations (116). In addition, DNA vaccines do not pose the danger of reversion to virulence unlike live attenuated vaccine that pose the risk of reverting to virulence. However, there are some drawbacks associated with DNA vaccination of which the most important is the possibility of integration of plasmid DNA into the host genome, which pose the danger of being transferred to other aquatic organisms and humans (117). Other difficulties include the cost of preparation and method of administration. In India, a porin gene encoding 38-kDa major OMP (Omp38) of V. anguillarum was used to construct a DNA vaccine for immunization of sea bass (Lates calcarifer) administered by intramuscular injection by Kumar et al. (82). After challenge with V. anguillarum, vaccinated sea bass was protected (RPS = 55.6%) unlike the control group that had high mortality. In another trial, Asian sea bass vaccinated using a DNA vaccine showed moderate protection (RPS = 46%) after challenge with V. anguillarum (81). Overall, there are few studies on OMP-based DNA vaccine compared to those with subunit vaccines for fish carried out in India so far.
PLGA, polylactic acid (PLA), and chitosan are polymers, commonly used for vaccine delivery as nanoparticles because of their biodegradable nontoxic properties (118–120). Moreover, they are easy to produce and are relatively affordable. They are attractive for oral vaccination because they easily adsorb to epithelial cells and penetrate the mucosal barrier where they are taken up by antigen-presenting cells (APCs). And as such, they can be bioengineered to enhance their adsorption on mucosal cells. Cellular uptake of PLGA nanoparticles is widely documented as shown that they are easily engulfed by various phagocytic cells such as monocytes, macrophages, neutrophils, and dendritic cells that serve as APCs leading to activation of cells of the adaptive immune system for induction of long-term protective immunity (121–126). They protect the vaccines from degradation, and they have been shown to have some potentiation effect able to enhance their uptake and enable slow release of antigens at deposition sites (127–129). Put together, these attributes render use of biodegradable nanoparticles as a better option for oral delivery of OMP vaccines than feed-coated oral vaccines.
Behera et al. (69) used PLGA microparticle for delivery A. hydrophila OMPs in rohu in which they observed an increase in several innate immune parameters such as respiratory burst, lysozyme, and complement activity alongside an increase in long-term expression of antibody responses against A. hydrophila in rohu. In another study, Behera et al. (72) showed 90% survival in rohu vaccinated with A. hydrophila OMP PLGA microsphere vaccine than in control fish vaccinated with OMPs that had 100% mortality after challenge with A. hydrophila. Similarly, Rauta and Nayak (70) showed high antibody responses and survival in rohu vaccinated with PLA-OMP (80%) and PLGA-OMP (75%) nanoparticles after challenge with A. hydrophila. Dubey et al. (79) immunized rohu using OmpW encapsulated in PLGA nanoparticles by oral vaccination and showed a dose-dependent protective immunity in which fish vaccinated with a low antigen dose had 48.3% survival while fish vaccinated with a high antigen dose had 73.3 % after challenge with A. hydrophila. In another study, Dubey et al. (80) showed that the OmpA encapsulated in chitosan nanoparticles (73.3%) had superior protection over WCI vaccine (48.3%) in Labeo fimbriatus after challenge with E. tarda. In general, studies on biodegradable nanoparticle fish vaccines are increasing in India because of safety and ease of administration orally through feed and absence of side effects. On the other hand, WCI vaccine formulations with adjuvants such as mineral oils have been linked to side effects in fish (116).
While OMPs have proved to be protective antigens ideal for vaccine development, there are several factors that make the design of fish vaccines using OMPs a challenge. For example, the surface loops that encode epitopes for B-cell binding have been shown to be highly divergent for some bacterial species, making it difficult to choose antigens with a broad protective ability against variant strains for use in different ecosystems. One of the challenges in bioengineering of OMP vaccines is LPS detoxification. LPS activates the innate immune system via Toll-like receptor (TLR)4 of which excessive TLR4 activation causes endotoxicity, leading to excessive inflammatory cytokine expression (130). While Zollinger et al. (131) described detoxification of LPS using a detergent extraction process, other scientists have used bioengineering techniques for LPS detoxification (132, 133). Leitner et al. (134) showed that genetic modification of LPS lipid A of Vibrio cholerae detoxified the LPS activity and elicited the production of highly protective antibodies, while Watkins et al. (135) detoxified LPS by producing truncated LPS containing lipid IVa instead of full LPS. Endotoxicity of LPS encoded in OMPs used for fish vaccine design has not been determined, and this poses a threat in the safety of OMPs used for fish vaccines. Another challenge in OMP-based vaccines is selecting epitopes able to evoke both humoral and CMI responses. OMP surface antigens encode epitopes specific for B-cell binding (136), while luminal antigens shielded inside β-sheets have been shown to be skewed toward CMI responses (137). The challenge is to identify luminal peptides suitable for producing T-cell vaccines. While OMPs are, by themselves, potent adjuvants able to activate the innate immune system through interaction between their PAMPs and host TLRs, they require additional conventional adjuvants to sustain long-term activation of the innate immune system of which the choice of adjuvant can be a challenge especially for oral vaccines (138, 139).
OMPs are essential molecules of Gram-negative bacteria as they play various roles including adaptation, immunogenicity, and pathogenesis of bacterium. They possess epitopes essential for binding to B and T lymphocytes, rendering them to be ideal vaccine candidates for both extra- and intra-cellular replicating bacteria. And as shown herein, they have been widely used in vaccine development for the Indian aquaculture, which has a high prevalence of both extra- and intracellular replicating bacterial pathogens such as Edwardsiella spp., Aeromonads, Vibrio spp., and Flavobacterium spp. The quest to develop safe vaccines that do not pose the danger of reversion to virulence, such as live attenuated vaccines, coupled with the need for vaccines able to evoke both humoral and CMI responses, unlike WCI vaccines, has extended the search for protective vaccines to include OMPs in vaccine development. Evidence obtained through work carried out by several groups in India reveals that OMPs are potent immunogenic molecules able to provide significant protection in fish when delivered as subunit, DNA, or PLGA/chitosan nanoparticle vaccines. Despite so, there is a need for optimization of several factors such as the choice of antigen delivery systems whether to use intra- or extra-cellular delivery as well as whether to use oral, immersion, or injectable vaccine delivery systems and to develop prime-boost vaccination regimes that confer the highest protection throughout the fish production cycle. Nonetheless, this review shows that OMP subunit, DNA, and PLGA/chitosan nanoparticle vaccines could form a large proportion of future vaccines for fish bacterial diseases in India.
BM conceptualized the initial draft of the manuscript. SD and HM conceived and reviewed the manuscript. ØE, IdK, and InK revised the manuscript. All authors read and approved publication of the manuscript.
This study was supported by the Department of Science and Technology (DST), Government of India, through the Indo-Norway joint project (INT/NOR/RCN/BIO/P-01/2018) and through the project ‘‘Biotechnology applied for controlling diseases in aquaculture in Norway and India'' funded by the Research Council of Norway (grant 283566).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
2. FAO. The State of World Fisheries and Aquaculture 2018-Meeting the Sustainable Development Goals (2018).
3. John N, Abdulla MH. Prevalence, distribution and drug resistance of motile aeromonads in freshwater ornamental fishes. Indian J Fish. (2012) 59:161–4.
4. Sahoo P, Swaminathan TR, Abraham TJ, Kumar R, Pattanayak S, Mohapatra A, et al. Detection of goldfish haematopoietic necrosis herpes virus (Cyprinid herpesvirus-2) with multi-drug resistant Aeromonas hydrophila infection in goldfish: first evidence of any viral disease outbreak in ornamental freshwater aquaculture farms in India. Acta Tropica. (2016) 161:8–17. doi: 10.1016/j.actatropica.2016.05.004
5. Singh AK, Rathore G, Singh V, Mani I, Singh RK, Mishra SK, et al. Bacterial resistance to oxytetracycline in different life stages of Indian freshwater carp aquaculture system. Int J Microbiol. (2009) 1:25–34. doi: 10.9735/0975-5276.1.1.25-34
6. Fisheries F. World Aquaculture 2010: Food and Agriculture Organization of the United Nations (2011).
7. Novotny L, Dvorska L, Lorencova A, Beran V, Pavlik I. Fish: a potential source of bacterial pathogens for human beings. A review. J Vet Med. (2004) 49:343–58. doi: 10.17221/5715-VETMED
8. Jayasankar P. Present status of freshwater aquaculture in India-A review. Indian J Fish. (2018) 65:157–65. doi: 10.21077/ijf.2018.65.4.81300-20
9. Katiha PK, Jena J, Pillai N, Chakraborty C, Dey MM. Inland aquaculture in India: past trend, present status and future prospects. Aquacult Econ Manag. (2005) 9:237–64. doi: 10.1080/13657300590961573
10. Katiha PK editor. Freshwater aquaculture in India: Status, potential and constraints. In: National Centre for Agricultural Economics and Policy Research, Workshop Proceedings. New Delhi (2000). p. 98–108.
11. Mishra SS, Rakesh D, Dhiman M, Choudhary P, Debbarma J, Sahoo S, et al. Present status of fish disease management in freshwater aquaculture in India: state-of-the-art-review. J Aquacult Fish. (2017) 1:003. doi: 10.24966/AAF-5523/100003
12. Behera B, Pradhan P, Swaminathan T, Sood N, Paria P, Das A, et al. Emergence of tilapia lake virus associated with mortalities of farmed Nile tilapia Oreochromis niloticus (Linnaeus 1758) in India. Aquaculture. (2018) 484:168–74. doi: 10.1016/j.aquaculture.2017.11.025
13. Azad I, Shekhar M, Thirunavukkarasu A, Poornima M, Kailasam M, Rajan J, et al. Nodavirus infection causes mortalities in hatchery produced larvae of Lates calcarifer: first report from India. Dis Aquat Organ. (2005) 63:113–8. doi: 10.3354/dao063113
14. Swaminathan TR, Kumar R, Dharmaratnam A, Basheer V, Sood N, Pradhan P, et al. Emergence of carp edema virus in cultured ornamental koi carp, Cyprinus carpio koi, in India. J Gen Virol. (2016) 97:3392–9. doi: 10.1099/jgv.0.000649
15. Girisha S, Puneeth T, Nithin M, Kumar BN, Ajay S, Vinay T, et al. Red sea bream iridovirus disease (RSIVD) outbreak in Asian seabass (Lates calcarifer) cultured in open estuarine cages along the west coast of India: First report. Aquaculture. (2020) 520:734712. doi: 10.1016/j.aquaculture.2019.734712
16. Sivasankar P, John KR, George MR, Mageshkumar P, Manzoor MM, Jeyaseelan MJP. Characterization of a virulent ranavirus isolated from marine ornamental fish in India. Virusdisease. (2017) 28:373–82. doi: 10.1007/s13337-017-0408-2
17. Nayak SK. Current status of Aeromonas hydrophila vaccine development in fish: an Indian perspective. Fish Shellfish Immunol. (2020) 100:283–99. doi: 10.1016/j.fsi.2020.01.064
18. Das MK, Das RK. A review of the fish disease epizootic ulcerative syndrome in India. Environ Ecol. (1993) 11:134–45.
19. Karunasagar I, Rosalind GM, Gopal Rao K. Aeromonas hydrophila septicaemia of Indian major carps in some commercial fish farms of West Godavari District, Andhra Pradesh. Curr Sci (Bangalore). (1989) 58:1044–5.
20. Mukherjee SC. Fish Diseases in India, Their Causes and Control Measures - Winter School on Recent Advances in Diagnosis and Management of Diseases in Mariculture, 7th to 27th November 2002, Course Manual. Central Marine Fisheries Research Institute, Cochin, India (2002).
21. Das BK. Bacterial Diseases of fish. Fish Health management, National Training Programme. Central Institute of Freshwater Aquaculture, Bhubaneswar, India (2011).
22. Kumar P, Adikesavalu H, Abraham TJ. Prevalence of Edwardsiella tarda in commercially important finfish and shellfish of Bihar and West Bengal, India. J Coast Life Med. (2016) 4:30–5. doi: 10.12980/jclm.4.2016apjtd-2014-0184
23. Surendraraj A, Farvin KS, Yathavamoorthi R, Thampuran N. Enteric bacteria associated with farmed freshwater fish and its culture environment in Kerala, India. Res J Microbiol. (2009) 4:334–44. doi: 10.3923/jm.2009.334.344
24. Shome R, Shome B, Ram N. Study of virulence factors of Aeromonas hydrophila isolates causing acute abdominal dropsy and ulcerative diseases in Indian major carps. Indian J Fish. (1999) 46:133–40.
25. Janda JM, Abbott SL. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin Infect Dis. (1998) 27:332–44. doi: 10.1086/514652
26. Dar G, Kamili A, Chishti M, Dar S, Tantry T, Ahmad F. Characterization of Aeromonas sobria isolated from Fish Rohu (Labeo rohita) collected from polluted pond. J Bacteriol Parasitol. (2016) 7:3. doi: 10.4172/2155-9597.1000273
27. Das A, Rathore A, Janani C, Hemanth C, Balakrishnan R. Diagnosis of motile Aeromonas sobria from cat fishes with septicemia by PCR. IOSR J Appl Phys. (2013) 2:87–91. doi: 10.9790/2380-0268791
28. Thomas J, Madan N, Nambi K, Majeed SA, Basha AN, Hameed ASS. Studies on ulcerative disease caused by Aeromonas caviae-like bacterium in Indian catfish, Clarias batrachus (Linn). Aquaculture. (2013) 376:146–50. doi: 10.1016/j.aquaculture.2012.11.015
29. Sreedharan K, Philip R, Singh ISB. Characterization and virulence potential of phenotypically diverse Aeromonas veronii isolates recovered from moribund freshwater ornamental fishes of Kerala, India. Antonie Van Leeuwenhoek. (2013) 103:53–67. doi: 10.1007/s10482-012-9786-z
30. Karunasagar I, Rosalind GM, Gopal Rao K. Aeromonas hydrophila septicaemia of Indian major carps in some commercial fish farms of West Godavari District, Andhra Pradesh. Curr Sci (Bangalore). (1989) 58:1044–5.
31. Das B. Bacterial Diseases of Fish. Fish Health management, National Training Programme. Central Institute of Freshwater Aquaculture, Bhubaneswar, India. (2011).
32. Egusa S. Some bacterial diseases of freshwater fishes in Japan. Fish Pathol. (1976) 10:103–14. doi: 10.3147/jsfp.10.103
33. Meyer F, Bullock G. Edwardsiella tarda, a new pathogen of channel catfish (Ictalurus punctatus). Appl Microbiol. (1973) 25:155. doi: 10.1128/AEM.25.1.155-156.1973
34. Dubey S, Maiti B, Kim SH, Sivadasan SM, Kannimuthu D, Pandey PK, et al. Genotypic and phenotypic characterization of Edwardsiella isolates from different fish species and geographical areas in Asia: implications for vaccine development. J Fish Dis. (2019) 42:835–50. doi: 10.1111/jfd.12984
35. Verma DK, Rathore G. New host record of five Flavobacterium species associated with tropical fresh water farmed fishes from North India. Braz J Microbiol. (2015) 46:969–76. doi: 10.1590/S1517-838246420131081
36. Prasad Y, Kumar D, Sharma AK. Lytic bacteriophages specific to Flavobacterium columnare rescue catfish, Clarias batrachus (Linn.) from columnaris disease. J Environ Biol. (2011) 32:161–8.
37. Gao X, Pi D, Chen N, Li X, Liu X, Yang H, et al. Survival, virulent characteristics, and transcriptomic analyses of the pathogenic Vibrio anguillarum under starvation stress. Front Cell Infect Microbiol. (2018) 8:389. doi: 10.3389/fcimb.2018.00389
38. Parveen S, Islam MS, Huq A. Abundance of Aeromonas spp. in river and lake waters in and around Dhaka, Bangladesh. J Diarrhoeal Dis Res. (1995) 13:183–6.
39. Holmes P, Niccolls LM, Sartory DP. The ecology of mesophilic Aeromonas in the aquatic environment. In: Austin B, Altwegg M, Gosling PJ, Joseph S, editors. The Genus Aeromonas. West Sussex: Wiley (1996). p. 127–50.
40. Welker TL, Shoemaker CA, Arias CR, Klesius PH. Transmission and detection of Flavobacterium columnare in channel catfish Ictalurus punctatus. Dis Aquat Organ. (2005) 63:129–38. doi: 10.3354/dao063129
41. Karunasagar I, Pai R, Malathi G, Karunasagar I. Mass mortality of Penaeus monodon larvae due to antibiotic-resistant Vibrio harveyi infection. Aquaculture. (1994) 128:203–9. doi: 10.1016/0044-8486(94)90309-3
42. Bower C, Daeschel M. Resistance responses of microorganisms in food environments. J Int J Food Microbiol. (1999) 50:33–44. doi: 10.1016/S0168-1605(99)00075-6
43. Sommerset I, Krossøy B, Biering E, Frost P. Vaccines for fish in aquaculture. Expert Rev Vaccines. (2005) 4:89–101. doi: 10.1586/14760584.4.1.89
44. Munang'andu HM, Evensen Ø. Correlates of protective immunity for fish vaccines. Fish Shellfish Immunol. (2019) 85:132–40. doi: 10.1016/j.fsi.2018.03.060
45. Martín-Martín A, Simón R, Abós B, Díaz-Rosales P, Tafalla C. Rainbow trout mount a robust specific immune response upon anal administration of thymus-independent antigens. Dev Comp Immunol. (2020) 109:103715. doi: 10.1016/j.dci.2020.103715
46. Embregts CW, Forlenza M. Oral vaccination of fish: lessons from humans and veterinary species. Dev Comp Immunol. (2016) 64:118–37. doi: 10.1016/j.dci.2016.03.024
47. Hirst ID, Ellis AE. Iron-regulated outer membrane proteins of Aeromonas salmonicida are important protective antigens in Atlantic salmon against furunculosis. Fish Shellfish Immunol. (1994) 4:29–45. doi: 10.1006/fsim.1994.1004
48. Karunasagar I, Ali A, Otta S. Immunization with bacterial antigens: infections with motile aeromonads. Dev Biol Stand. (1997) 90:135–41.
49. Munang'andu HM, Mutoloki S, Evensen Ø. Non-replicating vaccines. In: Gudding R, Lillehaug A, Evensen Ø, editors. Fish Vaccination. West Sussex: Wiley Blackwell (2014). p. 22–32.
50. Munang'andu HM. Intracellular bacterial infections: a challenge for developing cellular mediated immunity vaccines for farmed fish. Microorganisms. (2018) 6:33. doi: 10.3390/microorganisms6020033
51. Schulz GE. β-barrel membrane proteins. Curr Opin Struct Biol. (2000) 10:443–7. doi: 10.1016/S0959-440X(00)00120-2
52. Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. (2003) 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003
53. Wimley WC. The versatile β-barrel membrane protein. Curr Opin Struct Biol. (2003) 13:404–11. doi: 10.1016/S0959-440X(03)00099-X
54. Bond PJ, Sansom MS. The simulation approach to bacterial outer membrane proteins. Mol Membr Biol. (2004) 21:151–61. doi: 10.1080/0968760410001699169
55. Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. (2002) 1565:308–17. doi: 10.1016/S0005-2736(02)00577-1
56. Braun G, Cole ST. DNA sequence analysis of the Serratia marcescens ompA gene: implications for the organisation of an enterobacterial outer membrane protein. Mol Gen Genet. (1984) 195:321–8. doi: 10.1007/BF00332766
57. Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. (2000) 37:239–53. doi: 10.1046/j.1365-2958.2000.01983.x
58. Maiti B, Raghunath P, Karunasagar I, Karunasagar I. Cloning and expression of an outer membrane protein OmpW of Aeromonas hydrophila and study of its distribution in Aeromonas spp. J Appl Microbiol. (2009) 107:1157–67. doi: 10.1111/j.1365-2672.2009.04296.x
59. Maiti B, Shetty M, Shekar M, Karunasagar I, Karunasagar I. Evaluation of two outer membrane proteins, Aha1 and OmpW of Aeromonas hydrophila as vaccine candidate for common carp. Vet Immunol Immunopathol. (2012) 149:298–301. doi: 10.1016/j.vetimm.2012.07.013
60. Vàzquez-Juárez RC, Romero MJ, Ascencio F. Adhesive properties of a LamB-like outer-membrane protein and its contribution to Aeromonas veronii adhesion. J Appl Microbiol. (2004) 96:700–8. doi: 10.1111/j.1365-2672.2004.02177.x
61. Vazquez-Juarez RC, Gomez-Chiarri M, Barrera-Saldaña H, Hernandez-Saavedra N, Dumas S, Ascencio F. Evaluation of DNA vaccination of spotted sand bass (Paralabrax maculatofasciatus) with two major outer-membrane protein-encoding genes from Aeromonas veronii. Fish Shellfish Immunol. (2005) 19:153–63. doi: 10.1016/j.fsi.2004.12.007
62. Ebanks RO, Goguen M, McKinnon S, Pinto DM, Ross NW. Identification of the major outer membrane proteins of Aeromonas salmonicida. Dis Aquat Organ. (2005) 68:29–38. doi: 10.3354/dao068029
63. Neema M, Karunasagar I. In silico homology modeling and epitope prediction of outer membrane protein OMP W, a potential vaccine candidate against Edwardsiella tarda. Int J Curr Microbiol Appl Sci. (2018) 41:1349–58. doi: 10.20546/ijcmas.2018.703.319
64. Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. (2000) 3:445–50. doi: 10.1016/S1369-5274(00)00119-3
65. Baliga P, Shekar M, Venugopal MN. Potential outer membrane protein candidates for vaccine development against the pathogen Vibrio anguillarum: a reverse vaccinology based identification. J Curr Microbiol. (2018) 75:368–77. doi: 10.1007/s00284-017-1390-z
66. Viji VT, Deepa K, Velmurugan S, Donio MBS, Jenifer JA, Babu MM, et al. Vaccination strategies to protect goldfish Carassius auratus against Aeromonas hydrophila infection. J Dis Aquat Organ. (2013) 104:45–57. doi: 10.3354/dao02581
67. Kumar G, Rathore G, El-Matbouli M. Outer membrane protein assembly factor YaeT (omp85) and GroEL proteins of Edwardsiella tarda are immunogenic antigens for Labeo rohita (Hamilton). J Fish Dis. (2014) 37:1055–9. doi: 10.1111/jfd.12205
68. Maji S, Mali P, Joardar SN. Immunoreactive antigens of the outer membrane protein of Aeromonas hydrophila, isolated from goldfish, Carassius auratus (Linn.). Fish Shellfish Immunol. (2006) 20:462–73. doi: 10.1016/j.fsi.2005.06.003
69. Behera T, Nanda P, Mohanty C, Mohapatra D, Swain P, Das B, et al. Parenteral immunization of fish, Labeo rohita with Poly D, L-lactide-co-glycolic acid (PLGA) encapsulated antigen microparticles promotes innate and adaptive immune responses. Fish Shellfish Immunol. (2010) 28:320–5. doi: 10.1016/j.fsi.2009.11.009
70. Rauta PR, Nayak B. Parenteral immunization of PLA/PLGA nanoparticle encapsulating outer membrane protein (Omp) from Aeromonas hydrophila: evaluation of immunostimulatory action in Labeo rohita (rohu). Fish Shellfish Immunol. (2015) 44:287–94. doi: 10.1016/j.fsi.2015.02.007
71. Zhang Y-q, Zhang T-t, Li J-n, Liu X-l, Li L. Design and evaluation of a tandemly arranged outer membrane protein U (OmpU) multi-epitope as a potential vaccine antigen against Vibrio mimicus in grass carps (Ctenopharyngodon idella). Vet Immunol Immunopathol. (2014) 160:61–9. doi: 10.1016/j.vetimm.2014.03.016
72. Behera T, Swain P. Alginate-chitosan-PLGA composite microspheres induce both innate and adaptive immune response through parenteral immunization in fish. Fish Shellfish Immunol. (2013) 35:785–91. doi: 10.1016/j.fsi.2013.06.012
73. Khushiramani R, Girisha SK, Karunasagar I, Karunasagar I. Cloning and expression of an outer membrane protein ompTS of Aeromonas hydrophila and study of immunogenicity in fish. Protein Expr Purif. (2007) 51:303–7. doi: 10.1016/j.pep.2006.07.021
74. Khushiramani R, Girisha SK, Karunasagar I, Karunasagar I. Protective efficacy of recombinant OmpTS protein of Aeromonas hydrophila in Indian major carp. Vaccine. (2007) 25:1157–8. doi: 10.1016/j.vaccine.2006.10.032
75. Hamod MA, Nithin MS, Shukur YN, Karunasagar I, Karunasagar I. Outer membrane protein K as a subunit vaccine against V. anguillarum. Aquaculture. (2012) 354-355:107–10. doi: 10.1016/j.aquaculture.2012.03.033
76. Dash P, Sahoo P, Gupta P, Garg L, Dixit AJF. Immune responses and protective efficacy of recombinant outer membrane protein R (rOmpR)-based vaccine of Aeromonas hydrophila with a modified adjuvant formulation in rohu (Labeo rohita). Fish Shellfish Immunol. (2014) 39:512–23. doi: 10.1016/j.fsi.2014.06.007
77. Khushiramani RM, Maiti B, Shekar M, Girisha SK, Akash N, Deepanjali A, et al. Recombinant Aeromonas hydrophila outer membrane protein 48 (Omp48) induces a protective immune response against Aeromonas hydrophila and Edwardsiella tarda. Res Microbiol. (2012) 163:286–91. doi: 10.1016/j.resmic.2012.03.001
78. Maiti B, Shetty M, Shekar M, Karunasagar I, Karunasagar I. Recombinant outer membrane protein A (OmpA) of Edwardsiella tarda, a potential vaccine candidate for fish, common carp. Microbiol Res. (2011) 167:1–7. doi: 10.1016/j.micres.2011.02.002
79. Dubey S, Avadhani K, Mutalik S, Sivadasan S, Maiti B, Paul J, et al. Aeromonas hydrophila OmpW PLGA nanoparticle oral vaccine shows a dose-dependent protective immunity in rohu (Labeo rohita). Vaccines. (2016) 4:21. doi: 10.3390/vaccines4020021
80. Dubey S, Avadhani K, Mutalik S, Sivadasan S, Maiti B, Girisha S, et al. Edwardsiella tarda OmpA encapsulated in chitosan nanoparticles shows superior protection over inactivated whole cell vaccine in orally vaccinated fringed-lipped peninsula carp (Labeo fimbriatus). Vaccines. (2016) 4:40. doi: 10.3390/vaccines4040040
81. Kumar SR, Ahmed VI, Parameswaran V, Sudhakaran R, Babu VS, Hameed ASS. Potential use of chitosan nanoparticles for oral delivery of DNA vaccine in Asian sea bass (Lates calcarifer) to protect from Vibrio (Listonella) anguillarum. Fish Shellfish Immunol. (2008) 25:47–56. doi: 10.1016/j.fsi.2007.12.004
82. Kumar SR, Parameswaran V, Ahmed VPI, Musthaq SS, Hameed ASS. Protective efficiency of DNA vaccination in Asian seabass (Lates calcarifer) against Vibrio anguillarum. Fish Shellfish Immunol. (2007) 23:316–26. doi: 10.1016/j.fsi.2006.11.005
83. Nielsen H, Tsirigos KD, Brunak S, von Heijne G. A brief history of protein sorting prediction. Protein J. (2019) 38:200–16. doi: 10.1007/s10930-019-09838-3
84. Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of β-barrel outer membrane proteins. Nucleic Acids Res. (2004) 32:W400–W4. doi: 10.1093/nar/gkh417
85. Kolaskar A, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. (1990) 276:172–4. doi: 10.1016/0014-5793(90)80535-Q
86. Larsen JEP, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. (2006) 2:2. doi: 10.1186/1745-7580-2-2
87. Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. (2007) 8:424. doi: 10.1186/1471-2105-8-424
88. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. (2016) 32:511–7. doi: 10.1093/bioinformatics/btv639
89. Hansson M, Nygren PA, Ståhl S. Design and production of recombinant subunit vaccines. Biotechnol Appl Biochem. (2000) 32:95–107. doi: 10.1042/BA20000034
90. SongLin G, PanPan L, JianJun F, JinPing Z, Peng L, LiHua D. A novel recombinant bivalent outer membrane protein of Vibrio vulnificus and Aeromonas hydrophila as a vaccine antigen of American eel (Anguilla rostrata). Fish Shellfish Immunol. (2015) 43:477–84. doi: 10.1016/j.fsi.2015.01.017
91. Luo Z, Fu J, Li N, Liu Z, Qin T, Zhang X, et al. Immunogenic proteins and their vaccine development potential evaluation in outer membrane proteins (OMPs) of Flavobacterium columnare. J Aquacult Fish. (2016) 1:1–8. doi: 10.1016/j.aaf.2016.10.002
92. Nguyen HT, Nguyen TTT, Chen YC, Vu-Khac H, Wang PC, Chen SC. Enhanced immune responses and effectiveness of refined outer membrane protein vaccines against Vibrio harveyi in orange-spotted grouper (Epinephelus coioides). J Fish Dis. (2018) 41:1349–58. doi: 10.1111/jfd.12828
93. Wang E, Qin Z, Yu Z, Ai X, Wang K, Yang Q, et al. Molecular characterization, phylogenetic, expression, and protective immunity analysis of OmpF, a promising candidate immunogen against Yersinia ruckeri infection in channel catfish. J Front Immunol. (2018) 9:2003. doi: 10.3389/fimmu.2018.02003
94. Ningqiu L, Junjie B, Shuqin W, Xiaozhe F, Haihua L, Xing Y, et al. An outer membrane protein, OmpK, is an effective vaccine candidate for Vibrio harveyi in Orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. (2008) 25:829–33. doi: 10.1016/j.fsi.2008.09.007
95. Cai S-H, Yaol S-Y, Lu Y-S, Wu Z-H, Jian J-C, Wang B. Immune response in Lutjanus erythropterus induced by the major outer membrane protein (OmpU) of Vibrio alginolyticus. Dis Aquat Organ. (2010) 90:63–8. doi: 10.3354/dao02206
96. Cai SH, Lu YS, Wu ZH, Jian JC. Cloning, expression of Vibrio alginolyticus outer membrane protein-OmpU gene and its potential application as vaccine in crimson snapper, Lutjanus erythropterus Bloch. J Fish Dis. (2013) 36:695–702. doi: 10.1111/jfd.12036
97. Wang Q, Chen J, Liu R, Jia J. Identification and evaluation of an outer membrane protein OmpU from a pathogenic Vibrio harveyi isolate as vaccine candidate in turbot (Scophthalmus maximus). Lett Appl Microbiol. (2011) 53:22–9. doi: 10.1111/j.1472-765X.2011.03062.x
98. Lun J, Xia C, Yuan C, Zhang Y, Zhong M, Huang T, et al. The outer membrane protein, LamB (maltoporin), is a versatile vaccine candidate among the Vibrio species. Vaccine. (2014) 32:809–15. doi: 10.1016/j.vaccine.2013.12.035
99. Liu F, Tang X, Sheng X, Xing J, Zhan W. Edwardsiella tarda outer membrane protein C: an immunogenic protein induces highly protective effects in flounder (Paralichthys olivaceus) against edwardsiellosis. Int J Mol Sci. (2016) 17:1117. doi: 10.3390/ijms17071117
100. Yang Q, Pan Y-L, Wang K-Y, Wang J, He Y, Wang E-L, et al. OmpN, outer membrane proteins of Edwardsiella ictaluri are potential vaccine candidates for channel catfish (Ictalurus punctatus). Mol Immunol. (2016) 78:1–8. doi: 10.1016/j.molimm.2016.08.011
101. Abdelhamed H, Ibrahim I, Nho SW, Banes MM, Wills RW, Karsi A, et al. Evaluation of three recombinant outer membrane proteins, OmpA1, Tdr, and TbpA, as potential vaccine antigens against virulent Aeromonas hydrophila infection in channel catfish (Ictalurus punctatus). Fish Shellfish Immunol. (2017) 66:480–6. doi: 10.1016/j.fsi.2017.05.043
102. Yadav SK, Sahoo PK, Dixit A. Characterization of immune response elicited by the recombinant outer membrane protein OmpF of Aeromonas hydrophila, a potential vaccine candidate in murine model. Mol Biol Rep. (2014) 41:1837–48. doi: 10.1007/s11033-014-3033-9
103. Le H, LiHua D, JianJun F, Peng L, SongLin G. Immunogenicity study of an expressed outer membrane protein U of Vibrio vulnificus in Japanese eel (Anguilla japonica). J Appl Microbiol. (2018) 125:1642–54. doi: 10.1111/jam.14068
104. He L, Wu L, Lin P, Zhai S, Guo S, Xiao Y, et al. First expression and immunogenicity study of a novel trivalent outer membrane protein (OmpII-UA) from Aeromonas hydrophila, Vibrio vulnificus and Edwardsiella anguillarum. Aquaculture. (2020) 519:734932. doi: 10.1016/j.aquaculture.2020.734932
105. Guo S, He L, Wu L, Xiao Y, Zhai S, Yan Q. Immunization of a novel bivalent outer membrane protein simultaneously resisting Aeromonas hydrophila, Edwardsiella anguillarum and Vibrio vulnificus infection in European eels (Angullia angullia). Fish Shellfish Immunol. (2020) 97:46–57. doi: 10.1016/j.fsi.2019.12.044
106. Sharma M, Dixit A. Identification and immunogenic potential of B cell epitopes of outer membrane protein OmpF of Aeromonas hydrophila in translational fusion with a carrier protein. Appl Microbiol Biotechnol. (2015) 99:6277–91. doi: 10.1007/s00253-015-6398-3
107. Upadhyaya T, Singh RK, Upadhyaya T, Singh RK, Dixit A. Molecular cloning and sequence analysis of lamB encoding outer membrane maltose-inducible porin of Aeromonas hydrophila: full length research paper. DNA Seq. (2007) 18:302–6. doi: 10.1080/10425170701248608
108. Sharma M, Dixit A. Immune response characterization and vaccine potential of a recombinant chimera comprising B-cell epitope of Aeromonas hydrophila outer membrane protein C and LTB. Vaccine. (2016) 34:6259–66. doi: 10.1016/j.vaccine.2016.10.064
109. Mahendran R, Jeyabaskar S, Sitharaman G, Michael RD, Paul AV. Computer-aided vaccine designing approach against fish pathogens Edwardsiella tarda and Flavobacterium columnare using bioinformatics softwares. Drug Des Dev Ther. (2016) 10:1703. doi: 10.2147/DDDT.S95691
110. Neema M, Karunasagar I, Karunasagar I. Structural and functional characterization of outer membrane protein N in Edwardsiella ictaluri: a bioinformatic approach. Intl J Pharm Sci Res. (2011) 2:13–26.
111. Bader JA, Shoemaker CA, Klesius PH. Immune response induced by N-lauroylsarcosine extracted outer-membrane proteins of an isolate of Edwardsiella ictaluri in channel catfish. Fish Shellfish Immunol. (2004) 16:415–28. doi: 10.1016/j.fsi.2003.07.003
112. Wang N, Yang Z, Zang M, Liu Y, Lu C. Identification of Omp38 by immunoproteomic analysis and evaluation as a potential vaccine antigen against Aeromonas hydrophila in Chinese breams. Fish Shellfish Immunol. (2013) 34:74–81. doi: 10.1016/j.fsi.2012.10.003
113. Liu MA. DNA vaccines: a review. J Intern Med. (2003) 253:402–10. doi: 10.1046/j.1365-2796.2003.01140.x
114. Munang'andu HM, Evensen Ø. A review of intra-and extracellular antigen delivery systems for virus vaccines of finfish. J Immunol Res. (2015) 2015:960859. doi: 10.1155/2015/960859
115. Yamasaki M, Araki K, Nakanishi T, Nakayasu C, Yamamoto A. Role of CD4+ and CD8α+ T cells in protective immunity against Edwardsiella tarda infection of ginbuna Crucian carp, Carassius auratus langsdorfii. Fish Shellfish Immunol. (2014) 36:299–304. doi: 10.1016/j.fsi.2013.11.016
116. Mutoloki S, Alexandersen S, Evensen Ø. Sequential study of antigen persistence and concomitant inflammatory reactions relative to side-effects and growth of Atlantic salmon (Salmo salar L.) following intraperitoneal injection with oil-adjuvanted vaccines. Fish Shellfish Immunol. (2004) 16:633–44. doi: 10.1016/j.fsi.2003.10.002
117. Rogan D, Babiuk L. Novel vaccines from biotechnology. Rev Sci Tech. (2005) 24:159. doi: 10.20506/rst.24.1.1561
118. Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers. (2011) 3:1377–97. doi: 10.3390/polym3031377
119. Akagi T, Baba M, Akashi M. Biodegradable nanoparticles as vaccine adjuvants and delivery systems: regulation of immune responses by nanoparticle-based vaccine. Polym Nanomed. (2011) 247:31–64. doi: 10.1007/12_2011_150
120. Borchard G, Esmaeili F, Heuking S. Chitosan-based delivery systems for mucosal vaccination. In: Sarmento B, Das Nevas J, editors. Chitosan-Based Systems for Biopharmaceuticals: Delivery, Targeting Polymer Therapeutics. John Wiley & Sons, Ltd. (2012). p. 211–24.
121. Newman KD, Elamanchili P, Kwon GS, Samuel J. Uptake of poly(D,L-lactic-co-glycolic acid) microspheres by antigen-presenting cells in vivo. J Biomed Mater Res. (2002) 60:480–6. doi: 10.1002/jbm.10019
122. Prior S, Gander B, Blarer N, Merkle HP, Subirá MaL, Irache JM, et al. In vitro phagocytosis and monocyte-macrophage activation with poly(lactide) and poly(lactide-co-glycolide) microspheres. Eur J Pharm Sci. (2002) 15:197–207. doi: 10.1016/S0928-0987(01)00218-4
123. Diwan M, Elamanchili P, Lane H, Gainer A, Samuel J. Biodegradable nanoparticle mediated antigen delivery to human cord blood derived dendritic cells for induction of primary T cell responses. J Drug Target. (2003) 11:495–507. doi: 10.1080/10611860410001670026
124. Elamanchili P, Lutsiak CME, Hamdy S, Diwan M, Samuel J. “Pathogen-mimicking” nanoparticles for vaccine delivery to dendritic cells. J Immunother. (2007) 30:378–95. doi: 10.1097/CJI.0b013e31802cf3e3
125. Waeckerle-Men Y, Groettrup M. PLGA microspheres for improved antigen delivery to dendritic cells as cellular vaccines. Adv Drug Deliv Rev. (2005) 57:475–82. doi: 10.1016/j.addr.2004.09.007
126. Desai MP, Labhasetwar V, Walter E, Levy RJ, Amidon GL. The Mechanism of uptake of biodegradable microparticles in caco-2 cells is size dependent. Pharm Res. (1997) 14:1568–73. doi: 10.1023/A:1012126301290
127. Calvo P, Remuñan-López C, Vila-Jato JL, Alonso MJ. Chitosan and chitosan/ethylene oxide-propylene oxide block copolymer nanoparticles as novel carriers for proteins and vaccines. Pharm Res. (1997) 14:1431–6. doi: 10.1023/A:1012128907225
128. Panyam J, Dali MM, Sahoo SK, Ma W, Chakravarthi SS, Amidon GL, et al. Polymer degradation and in vitro release of a model protein from poly (D, L-lactide-co-glycolide) nano-and microparticles. J Control Release. (2003) 92:173–87. doi: 10.1016/S0168-3659(03)00328-6
129. Sahana D, Mittal G, Bhardwaj V, Kumar MR. PLGA nanoparticles for oral delivery of hydrophobic drugs: influence of organic solvent on nanoparticle formation and release behavior in vitro and in vivo using estradiol as a model drug. J Pharm Sci. (2008) 97:1530–42. doi: 10.1002/jps.21158
130. Fitzgerald KA, Rowe DC, Golenbock DT. Endotoxin recognition and signal transduction by the TLR4/MD2-complex. Microbes Infect. (2004) 6:1361–7. doi: 10.1016/j.micinf.2004.08.015
131. Zollinger W, Mandrell R, Altieri P, Berman S, Lowenthal J, Artenstein MS. Safety and immunogenicity of a Neisseria meningitidis type 2 protein vaccine in animals and humans. J Infect Dis. (1978) 137:728–39. doi: 10.1093/infdis/137.6.728
132. Zariri A, Pupo E, Van Riet E, Van Putten JP, van der Ley P. Modulating endotoxin activity by combinatorial bioengineering of meningococcal lipopolysaccharide. Sci Rep. (2016) 6:36575. doi: 10.1038/srep36575
133. Zariri A, van der Ley P. Biosynthetically engineered lipopolysaccharide as vaccine adjuvant. Expert Rev Vaccines. (2015) 14:861–76. doi: 10.1586/14760584.2015.1026808
134. Leitner DR, Feichter S, Schild-Prüfert K, Rechberger GN, Reidl J, Schild S, et al. Lipopolysaccharide modifications of a cholera vaccine candidate based on outer membrane vesicles reduce endotoxicity and reveal the major protective antigen. Infect Immun. (2013) 81:2379–93. doi: 10.1128/IAI.01382-12
135. Watkins HC, Rappazzo CG, Higgins JS, Sun X, Brock N, Chau A, et al. Safe recombinant outer membrane vesicles that display M2e elicit heterologous influenza protection. Mol Ther. (2017) 25:989–1002. doi: 10.1016/j.ymthe.2017.01.010
136. Gerritzen MJ, Martens DE, Wijffels RH, van der Pol L, Stork M. Bioengineering bacterial outer membrane vesicles as vaccine platform. Biotechnol Adv. (2017) 35:565–74. doi: 10.1016/j.biotechadv.2017.05.003
137. Galen JE, Curtiss R 3rd. The delicate balance in genetically engineering live vaccines. Vaccine. (2014) 32:4376–85. doi: 10.1016/j.vaccine.2013.12.026
138. Apostólico JdS, Lunardelli VAS, Coirada FC, Boscardin SB, Rosa DS. Adjuvants: classification, modus operandi, and licensing. J Immunol Res. (2016) 2016:1459394. doi: 10.1155/2016/1459394
Keywords: outer membrane proteins (OMPs), vaccination, fish, aquaculture, fish pathogens
Citation: Maiti B, Dubey S, Munang'andu HM, Karunasagar I, Karunasagar I and Evensen Ø (2020) Application of Outer Membrane Protein-Based Vaccines Against Major Bacterial Fish Pathogens in India. Front. Immunol. 11:1362. doi: 10.3389/fimmu.2020.01362
Received: 29 February 2020; Accepted: 28 May 2020;
Published: 21 July 2020.
Edited by:
Geert Wiegertjes, Wageningen University and Research, NetherlandsReviewed by:
Bo Peng, Sun Yat-sen University, ChinaCopyright © 2020 Maiti, Dubey, Munang'andu, Karunasagar, Karunasagar and Evensen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biswajit Maiti, bWFpdGkuYkBuaXR0ZS5lZHUuaW4=; Hetron Mweemba Munang'andu, aGV0cm9uZXkubXdlZW1iYS5tdW5hbmdhbmR1QG5tYnUubm8=; Øystein Evensen, b3lzdGVpbi5ldmVuc2VuQG5tYnUubm8=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.