
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 10 June 2020
Sec. B Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01025
This article is part of the Research Topic The Germinal Center Reaction and Antibody Diversification Mechanisms in B Cell Neoplasia and Autoimmune Disease View all 7 articles
Germinal center (GC) B cell differentiation is critical for the production of affinity-matured pathogen-specific antibodies, the dysregulation of which may lead to humoral immunodeficiency or autoimmunity. The development of an in vivo screening system for factors regulating GC B cell differentiation has been a challenge. Here we describe a small-scale in vivo screening system with NP-specific B1-8hi cells and a retroviral shRNA library targeting 78 candidate genes to search for B cell-intrinsic factors that specifically regulate GC B cell differentiation. Zdhhc2, a gene encoding palmitoyltransferase ZDHHC2 and highly expressed in GC B cells, is identified as a strong positive regulator of GC B cell differentiation. B1-8hi cells transduced with Zdhhc2-shRNA are severely compromised in differentiating into GC B cells. A further analysis of in vitro differentiated B cells transduced with Zdhhc2-shRNA shows that Zdhhc2 is critical for the proliferation and the survival of B cells stimulated by CD40L, BAFF, and IL-21 and consequently impacts on their differentiation into GC B cells and post-GC B cells. These studies not only identify Zdhhc2 as a novel regulator of GC B cell differentiation but also represent a proof of concept of in vivo screen for regulators of GC B cell differentiation.
The germinal center (GC) B cell response is a fundamental process in humoral immunity that enables the rapid evolution and selection of B cells, so B cells that express pathogen-specific affinity-matured B cell receptors (BCRs) with reduced autoreactivity can be selected for antibody production (1, 2). GC B cell differentiation is a T cell-dependent process in which antigen-specific B cells present antigens to cognate T cells, which in turn provide reciprocal activating signals for the differentiation for both GC B cells and follicular T helper (TFH) cells. Antigen-specific B/T cell interactions result in the activation and the proliferation of both B and T cells and their chemokine-dependent migration to the B cell follicles to form the GCs (3–7). In the dark zone of GCs, B cells proliferate rapidly and undergo extensive activation-induced cytidine deaminase-mediated somatic hypermutation, which leads to BCR diversification (8). The expanded and BCR-diversified B cells migrate to the light zone of GCs and compete for capturing antigens and T cell help (6, 9–13). It has been proposed that high-affinity B cells can capture and present more antigens to TFH cells and consequently acquire sufficient signals from TFH cells for their further differentiation into plasmablasts and memory B cells (14–16). At the same time, autoreactive B cells are counter-selected to ensure the production of pathogen-specific affinity-matured BCRs with reduced autoreactivity (2). The integrity of this process is critical for humoral immunity and requires careful regulation (17). A deficiency in the key signals involved in this process, such as CD40L/CD40 and ICOSL/ICOS signals, can lead to either humoral immune deficiency or autoimmune diseases, or sometimes both, such as lupus (18–21). Therefore, it is crucial to understand the molecular mechanism that regulates GC B cell differentiation.
Previously, a number of factors critical for GC B cell differentiation have been identified (22–27), but none of them was initially identified through in vivo screen, as far as we know. For instance, BCL6 (encoded by Bcl6), a key transcription factor essential for GC B cell differentiation, was noticed because of its mutations in GC B cell-derived diffuse large B cell lymphoma (22). Bcl6-deficient mice have normal primary follicles but lack GCs after immunization (22, 28). MYC, another important transcription factor essential for GC formation and maintenance (29), was initially noticed due to its upregulated expression in B cell-derived Burkitt lymphoma (30). IRF4, critical for GC formation and antibody production (24, 25), was initially cloned from a translocation breakpoint in myeloma (31). More recently, several factors (such as ephrin B1 and Cbl ubiquitin ligases) involved in regulating GC B cell differentiation were identified due to their distinct expression patterns in GC B cells (32, 33). The development of in vivo screening systems for B cell-intrinsic factors regulating GC B cell differentiation has been a challenge, which has hindered the discovery of new genes implicated in GC B cell differentiation.
In vivo screens in mouse models have been mainly applied in the context of tumorigenesis based on either spontaneous or site-directed mutagenesis approaches, such as mutation-inducing chemicals, shRNA, and CRISPR/Cas9 systems (34–47). These screens are based on the principles that either gain-of-function mutations in oncogenes or loss-of-function mutations in tumor-suppressive genes can promote tumorigenesis in various tumor models, including tumors derived from B- and T-lineage cells, breast cancer, and glioblastoma (34, 35, 37, 44). A similar strategy has also been exploited to screen genes that regulate B cell differentiation in the bone marrow, where both positive and negative selections take place (48). In a screen for microRNA that regulates B cell tolerance, miR-148a was identified as a critical regulator of B cell tolerance and autoimmunity that can promote the survival of autoreactive immature B cells (48). In another screen for genes that regulate T cell differentiation during lymphocytic choriomeningitis virus infection, Ccnt1 was identified to promote both CD4 and CD8 T cell differentiation (49).
Since the in vivo screen depends on genetic manipulation and selection, we reasoned that these two factors could be achieved by retroviral transduction in antigen-specific B cells and the selection of these B cells in GC responses. Here we show that retrovirally transduced antigen-specific B cells can be used to screen regulators for GC B cell differentiation in vivo and identify Zdhhc2 as a novel positive regulator.
B1-8hi (B6.129P2-PtrpcaIghtm1Mnz/J) mice were purchased from the Jackson laboratory. Wild-type C57BL/6 mice were purchased from Shanghai SLAC Laboratory Animal Company. All mice were maintained in a specific-pathogen-free animal facility at Shanghai Jiao Tong University School of Medicine (SJTUSM).
The shRNA sequences were either designed by the Broad Institute GPP Web Portal or reported previously (50). The retroviral shRNA library was constructed by inserting the mix of shRNA double-strand fragments with 5′-BamHI and 3′-EcoRI sticky ends into the pSIREN-RetroQ_mCherry retroviral vector, in which the puromycin-resistant gene of pSIREN-RetroQ (Clontech) was replaced by the mCherry sequence from the mCherry-pBAD vector (Addgene).
For the study of Bcl6–shRNA and Zdhhc2-shRNA, Bcl6–shRNA (5′- GATCCGCTGTCAAAGAGAAG
GCTTTATTCAAGAGATAAAGCCTTCTCTTTGACAGCTTTTTTGATATCG-3′) was inserted into the retroviral pSIREN-RetroQ_GFP vector, in which the puromycin-resistant gene of pSIREN-RetroQ was replaced by the green fluorescent protein (GFP) gene sequence from the PMKO.1-GFP vector (Addgene), Zdhhc2-shRNA-2 (5′-GATCCGTGACAGATGCCAACTTATAATTCAAGAGATTATAAGTTGGCATCTGTCA
CTTTTTTGATATCG-3′) and Zdhhc2-shRNA-4 (5′-GATCCGCTACTCCTGCGGGACTAAATTTTCAAGAGA
AATTTAGTCCCGCAGGAGTAGCTTTTTTGATATCG-3′) were inserted into the retroviral pSIREN-RetroQ_mCherry vector, and a scramble shRNA (5′- GATCCGTGCGTTGCTAGTACCAACCTATTCAAGAGA
TAGGTTGGTACTAGCAACGCACTTTTTTGATATCG-3′) was inserted into the retroviral pSIREN-RetroQ_mCherry vector as controls.
For in vivo screen, the retroviruses of shRNA library including 78 candidate genes were packaged in Phoenix cells; B1-8hi splenic cells were stimulated with anti-CD180 (0.25 μg/ml, clone RP/14, BD Bioscience) for 24 h, then spin-infected at 2,000 g for 1.5 h with retroviruses in the presence of polybrene (8 μg/ml) (TR-1003-G, Millipore), and cultured overnight before transferring into eight wild-type C57BL/6 mice by tail vein injection (5~10 × 106 cells per mouse). The recipients were immunized intraperitoneally with 100 μg of NP49-CGG (Biosearch Technologies, N-5055E) in Alum (Pierce, 77,161) per mouse the day after transfer. The GC B cells and the non-GC B cells were MACS-sorted [according to (51)] from splenic cells pooled from eight recipients at 10 days later. The total genomic DNA was extracted from sorted GC B cells and non-GC B cells, and each template was amplified five times in parallel. The shRNA fragments were amplified by nested PCR and subjected to next-generation sequencing. The primers used for the nested PCR are 5′-GAAGAGGGCCTATTTCCCATGATTC-3′ and 5′-ACTTCCATTTGTCACGTCCTGCAC-3′ for the 1st round of PCR, 5′-GGACTATCATATGCTTACCGTAACTTGA-3′ and 5′-TGGATGTGGAATGTGTGCGA-3′ for the 2nd round of PCR. The shRNA fragments were subjected to next-generation sequencing (Illumina Hiseq X Ten). Two independent screens were performed.
For the study of Bcl6–shRNA and Zdhhc2-shRNA, the retroviruses containing these gene-specific shRNAs and their scramble shRNA or empty vector controls were packaged in Phoenix cells. The B1-8hi cells were cultured as described above and separately transduced with retroviruses containing these gene-specific shRNAs and scramble shRNAs tagged with mCherry reporter for Zdhhc2-shRNA or empty vectors tagged with GFP reporter for Bcl6–shRNA. The B1-8hi cells transduced with the gene-specific or matched scramble shRNAs or empty vectors were mixed at a ratio of 1:1, with the B1-8hi cells transduced with the empty vectors tagged with the alternative fluorescent reporter and adoptively transferred into wild-type C57BL/6 recipient mice (5~10 × 106 cells per mouse), which were immunized intraperitoneally with 100 μg of NP49-CGG/Alum on the day after transfer, and analyzed by flow cytometry 10 days after immunization.
To analyze the abundance of each shRNA fragment in the next-generation sequencing data, AWK (https://www.gnu.org/software/gawk/manual/gawk.html) and Python scripts were used to count the occurrence of each shRNA segment of the ~400 shRNA vectors in the five parallel data sets of each GC and non-GC B cell sample. The percentages of each shRNA fragment in the five parallel data sets of each sample were calculated, and the median value was used to represent its abundance in the sample. The shRNAs with median percentage values exceeding 0.025% (10 times less than the average abundance of 400 shRNA) in either non-GC or GC B cells were further analyzed to screen genes that specifically impact on GC B cells. The fold change was calculated as the GC/non-GC ratio for the abundance of each shRNA construct. The fold change values, expressed as binary logarithm (log2), and the reciprocals of their standard deviations were used to identify shRNA constructs with large and consistent (large reciprocal values) impacts on GC B cell differentiation.
The flow cytometry analysis was performed using the BD LSRFortessa™ X-20 analyzer (BD Biosciences). Mouse splenic cells were stained with anti-B220 (RA3-6B2, BioLegend), CD38 (90, eBioscience), CD95 (Jo2, BD), CD45.1 (A20, BioLegend), and CD45.2 (104, BioLegend) for the in vivo study of B1-8hi cell differentiation or with anti-B220, CD95, GL7 (GL7, BioLegend), CD138 (281-2, BioLegend), biotin anti-mouse IgG1 (RMG1-1, BioLegend), and BV785 streptavidin (BioLegend) for the in vitro analysis of GC B cell and plasma cell differentiation in the co-culture system or with anti-B220, annexin V (BD), and 7AAD (BD) for the analysis of cell apoptosis. The transduced A20 cell lines (kindly provided by Dr. Jeffrey Ravetch, Rockefeller University) were stained with anti-mouse BCL6 (K112-91, BD) for the knockdown efficiency of Bcl6-specific shRNA.
The sorted GC B cells were lysed in 10 mM Tris-HCl, pH 8.0, and 0.1 mM ethylenediaminetetraacetic acid (EDTA), proteinase K (B600452, Sangon Biotech), was added at a concentration of 0.5 mg/ml; the mixture was incubated at 50°C for 2.5 h and then at 95°C for 10 min. The sorted non-GC B cells were lysed in genomic DNA buffer without sodium dodecyl sulfate (SDS; 100 mM NaCl, 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA), mixed with an equal volume of genomic DNA buffer with 1% SDS and 0.2 mg/ml proteinase K, and incubated for 4 h at 37°C on a shaking heating block (Allsheng). Isopropanol was added to precipitate genomic DNA, which was further washed with 70% ethanol and resolved in ddH2O. Both GC B cell lysates and purified non-GC B cell genomic DNA were used as templates to amplify the shRNA fragments.
RNA was prepared with TRIzol (Invitrogen) from the GC B cells and the non-GC B cells of Peyer's patches by fluorescence-activated cell sorting. Quantitative real-time PCR was performed with One-Step SYBR PrimeScript RT-PCR Kit II (Perfect Real Time) (RR086A, Takara) and a 7,500 Fast Real-Time PCR System (Applied Biosystems). Gene expression was normalized to that of β-actin, and data were presented as a fold difference of the normalized values relative to that of the control samples by ‘2−ΔΔCT’ (change in cycling threshold). The following primer sets were used:
β-actin forward: 5′-CGCCACCAGTTCGCCATGGA-3′,
β-actin reverse: 5′-TACAGCCCGGGGAGCATCGT-3′,
Zdhhc2 forward: 5′-TGGTCTGCCTGATACTCAAGCCAAG-3′,
Zdhhc2 reverse: 5′- CTGAAACCCAAGCTGAATCCGTTC-3′.
Wild-type (WT) C57BL/6 splenic single cells were prepared and cultured in RPMI-1640 medium supplemented with 10% FBS (Gibco), 2 mM L-glutamine (Gibco), 1% Pen/Strep (100 units/ml penicillin, 100 μg/ml streptomycin, Hyclone), 10 mM HEPES (pH 7.2–7.5, Gibco), 1 mM sodium pyruvate (Gibco), 1% non-essential amino acids (Gibco), and 50 μM 2-mercaptoethanol. Splenocytes (1 × 105 cells per well) were co-cultured overnight with NB-21.2D9 feeder cells (irradiated with 8 Gy X-ray, 1.5 × 104 cells per well) in a 24-well plate (Corning), together with 2 ng/ml mIL-4 (Sino Biological). These cells were then spin-infected with retroviruses containing Zdhhc2- or scramble shRNAs at 2,000 g for 1.5 h and further cultured at the same condition. B cell differentiation and apoptosis were analyzed by flow cytometry at 48 h after transduction. To analyze cell proliferation, the transduced B cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) (Invitrogen) at 48 h after transduction and further co-cultured with irradiated NB-21.2D9 feeder cells in the presence of 2 ng/ml mIL-4 for 2 days before the flow cytometry analysis.
All statistical tests were performed using the GraphPad Prism 6.0 software. The specific statistical test for each experiment is described in the figure legends. Unpaired t-test with a two-tailed 95% confidence interval was employed for the comparison of two groups. One-way ANOVA with Dunnett's test or two-way ANOVA with Sidak's test was applied to multiple comparisons. A P value <0.05 was considered as statistically significant.
To establish an in vivo screening system for B cell-intrinsic factors that regulate GC B cell differentiation, we hypothesized to exploit the clonal expansion feature of antigen-specific GC B cells and a retroviral shRNA system that has been previously described (52). To validate the system, we first confirmed that B1-8hi cells, an established NP-specific B cell response model (21), can differentiate into GC B cells in vivo, both before and after in vitro retroviral transduction and adoptive cell transfer. As shown in Supplemental Figure 1, fresh B1-8hi cells expanded from 0.13% in the non-GC B cell compartment to ~48% in the GC B cell compartment when adoptively transferred into WT recipient mice and challenged with NP-CGG/Alum, suggesting that fresh B1-8hi cells can efficiently differentiate into GC B cells. At the same time, retrovirally transduced B1-8hi cells could also differentiate into GC B cells, although at a lower efficiency, as shown by an expansion from 0.79% in the non-GC B cell compartment to 2.96% in the GC B cell compartment.
To validate an in vivo screening system based on retroviral shRNA knockdown, a shRNA vector targeting Bcl6 was tested. The Bcl6–shRNA was confirmed to efficiently knockdown BCL6 expression (Figure 1A). As illustrated in Figure 1B, B1-8hi cells transduced with the Bcl6–shRNA vector containing a GFP reporter were mixed with B1-8hi cells transduced with empty vector containing a mCherry reporter, adoptively transferred into WT recipient mice, and evaluated for their expansion in the GC B cell compartment 10 days after NP-CGG/Alum immunization. As shown in Figure 1C, while the empty vector-transduced cells were present in both non-GC and GC B cell compartments, the Bcl6–shRNA transduced cells were almost completely absent from the GC B cell compartment, but not in the non-GC B cell compartment, which led to a significantly reduced GC/non-GC ratio for Bcl6–shRNA transduced cells (Figure 1D). While these results are consistent with the previous report that Bcl6 is essential for GC B cell differentiation (28), they also provide the basis for the use of retroviral shRNA transduced B1-8hi B cells for in vivo screen.
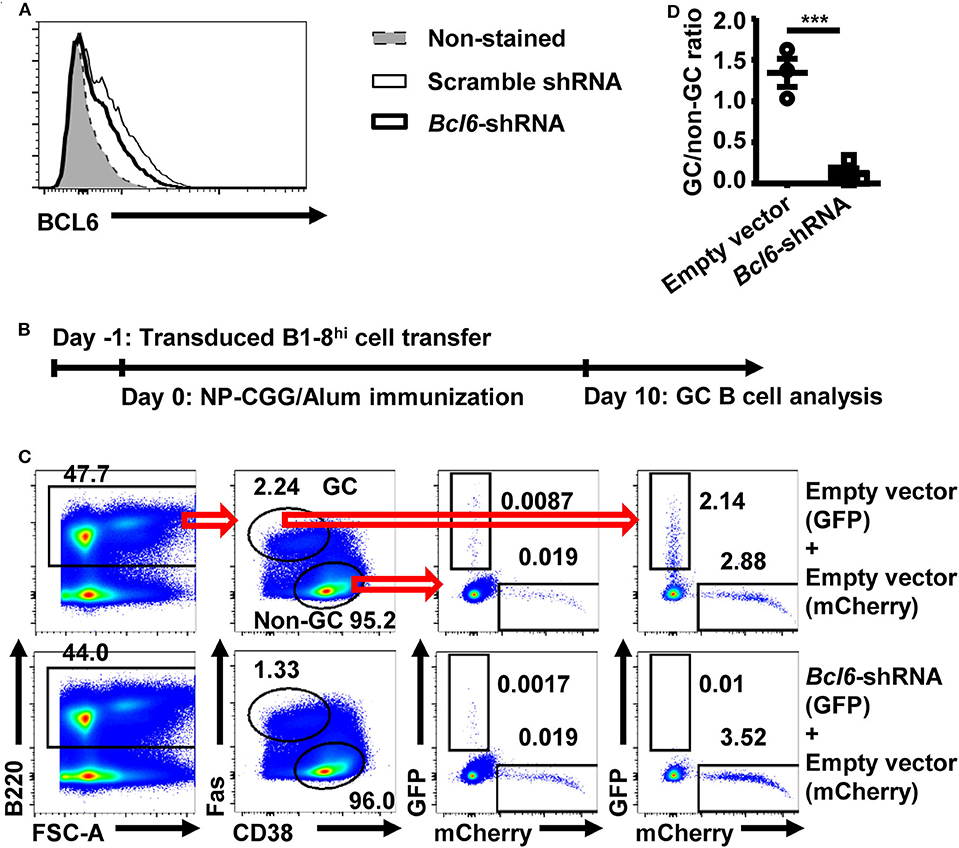
Figure 1. Retrovirally transduced B1-8hi cells can be used to study the shRNA function. (A) Representative flow cytometry profile showing the intranuclear BCL6 levels in untransduced A20 cells or transduced A20 cells with scramble shRNA or Bcl6–shRNA retroviral vectors. Murine B cell lymphoma A20 cells were either untransduced or transduced with the indicated retroviral vectors and analyzed for intranuclear BCL6 levels by flow cytometry 3 days later. (B) B1-8hi cell transfer model for evaluating germinal center (GC) B cell differentiation. Fresh or retrovirally transduced B1-8hi cells were adoptively transferred to wild-type (WT) recipients on day−1, immunized with NP-CGG/Alum on day 0, and analyzed for GC B cell differentiation on day 10. (C,D) Representative flow cytometry profiles (C) showing the percentages of empty vector and Bcl6–shRNA transduced B1-8hi cells (expressing a GFP reporter) as well as the co-transferred empty vector transduced B1-8hi cells (expressing a mCherry reporter) in non-GC B cells and GC B cells, and representative graph (D) showing the GC/non-GC ratios of percentages of transduced B1-8hi cells (expressing GFP) normalized to empty vector transduced cells (expressing mCherry). The empty vector or Bcl6–shRNA transduced B1-8hi cells (expressing GFP) were mixed 1:1 with empty vector transduced B1-8hi cells (expressing mCherry) and adoptively transferred into WT recipient mice and evaluated for their percentages among non-GC B cells and GC B cells (C) 10 days after NP-CGG/Alum immunization. The ratio of the percentage of empty vector or Bcl6–shRNA transduced cells (expressing GFP) in GC B cells to that in the matched non-GC B cells was calculated and normalized to the ratio of empty vector transduced cells (expressing mCherry), expressed as GC/non-GC ratio (D). Each symbol in (D) represents data from an individual mouse. The bars represent mean ± SEM; ***p ≤ 0.001, unpaired two-tailed t-test. A representative of two independent experiments is shown.
To test whether retroviral shRNA transduced B1-8hi cells can be used for in vivo screen, a small shRNA library of ~400 shRNA retroviral constructs targeting 78 candidate genes (five shRNA constructs for each gene, Supplemental Table 1) was constructed and confirmed by next-generation sequencing, with ~95% constructs comprising more than 0.025% of all sequences (Supplemental Figure 2). These genes are differentially expressed in GC and follicular B cells based on the Immunological Genome Project database (http://rstats.immgen.org/PopulationComparison/index.html) and based on the scarcity of information regarding their impacts on GC B cell differentiation. As illustrated in Figure 2A, B1-8hi B cells transduced with the shRNA library were transferred into WT recipient mice and challenged with NP-CGG/Alum immunization. The transduction efficiency of B1-8hi cells with the library is ~ 24% as analyzed 24 hours or 43~72% as analyzed 48 hours after transduction (Supplemental Figure 3). The GC B cells and the non-GC B cells were sorted 10 days later and analyzed for the abundance of these shRNA constructs in these cells. We reasoned that shRNA constructs that target the critical positive regulators of GC B cell differentiation will be less abundant and vice versa for the critical negative regulators (Figure 2B).
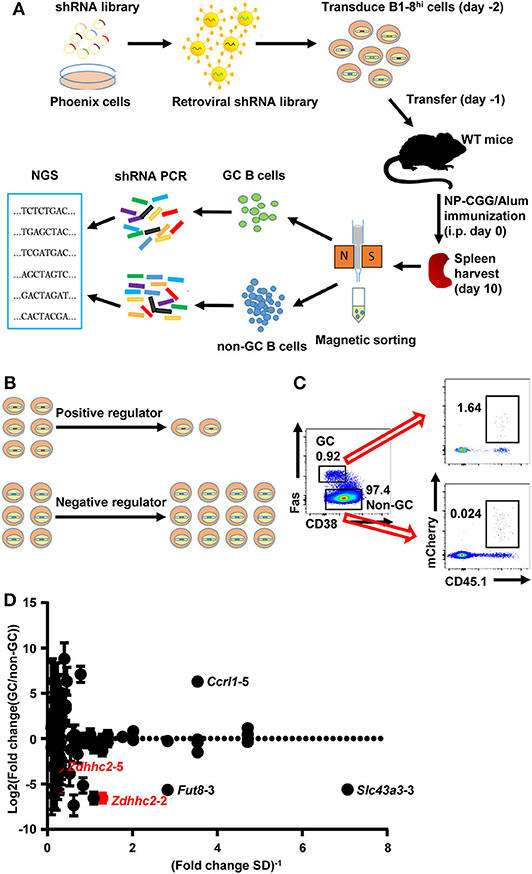
Figure 2. Overview of the shRNA in vivo screening system for regulators of germinal center (GC) B cell differentiation. (A) A schematic view of the in vivo screening system for regulators of GC B cell differentiation. The shRNA retroviral library was produced in Phoenix cells and used to transduce B1-8hi cells on day−2, which were adoptively transferred into eight wild-type (WT) recipient mice on day−1. These mice were immunized with NP-CGG/Alum on day 0, sacrificed on day 10, and subjected to splenic GC and non-GC B cell isolation by magnetic sorting. The shRNA sequences were amplified by PCR and determined by next-generation sequencing to evaluate the abundance of each shRNA construct. (B) Illustration of the screening strategy: shRNA constructs that target the positive regulators of GC B cell differentiation are less abundant and vice versa for negative regulators. (C) Representative flow cytometry profiles showing the percentages of retroviral shRNA library transduced cells (expressing mCherry) in GC and non-GC B cells in mice treated as described in (A). (D) Plot of the binary logarithm of shRNA's fold change values (calculated as the ratio of each shRNA's abundance in GC B cells to that in the corresponding non-GC B cells) against the reciprocals of their standard deviations [(fold change SD)−1]. Two independent screens were performed.
As shown in Figure 2C, a small percentage of GC B cells expressing fluorescent reporter genes (mCherry) was observed, suggesting that the retroviral constructs had successfully transduced these cells. The total number of retrovirally transduced B cells is ~10 times more than that of the shRNA constructs, providing the basis for a further screen. The shRNA fragments were amplified from these cells and subjected to next-generation sequencing. To minimize the variation, each sample was amplified and sequenced five times in parallel, and the median percentage values of each shRNA fragment were used to represent its abundance. To screen for genes that specifically impact on GC B cell differentiation, we focused on shRNA constructs that consistently recovered in either the non-GC or the GC B cell compartment. Based on two independent experiments, 85 shRNA constructs (targeting 52 out of 78 candidate genes) fall into this category (Supplemental Table 2). The fold change, defined as the abundance of each shRNA construct in GC B cells relative to the corresponding non-GC B cells, was plotted against the reciprocal of its standard deviation to help in the identification of shRNA constructs with large and consistent impacts on GC B cell differentiation. As shown in Figure 2D, B1-8hi cells transduced with several shRNAs (Zdhhc2-2, Fut8-3, Slc43a3-3, etc.) had a consistently reduced differentiation to GC B cells relative to non-GC B cells, suggesting that they target potential positive regulators. In contrast, B1-8hi cells transduced with some shRNA constructs (Ccrl1-5, etc.) that transduced cells were observed to be relatively more abundant in GC B cells than in non-GC B cells, suggesting their potential roles in negatively regulating GC B cell differentiation. Other shRNAs for these genes (Zdhhc2, Fut8, Slc43a3, and Ccrl1) are either lost in both screens (Zdhhc2-1, Fut8-2,5, Slc43a3-1,5, and Ccrl1-1,2,3,4) or only recovered in one experiment (Zdhhc2-3,4, Fut8-1,4, and Slc43a3-2,4), except for Zdhhc2-5, which displayed variable levels of inhibition of GC B cell differentiation (Figure 2D).
To verify the in vivo screening results, Zdhhc2 shRNA was further studied, given its strong activity as observed in the screen. The knockdown effect of Zdhhc2-shRNA-2 was confirmed in in vitro differentiated B cells with GC B cell phenotype (53), where Zdhhc2-shRNA-2 transduced cells displayed significantly reduced Zdhhc2 mRNA levels (Figure 3A), consistent with the results of previous studies on this shRNA (50). Zdhhc2-shRNA-2 transduced B1-8hi cells (mCherry+) were cotransferred with empty vector transduced B1-8hi cells (GFP+) into WT recipient mice and evaluated for their expansion in the GC B cell compartment 10 days after NP-CGG/Alum immunization (Figure 1B). While comparable levels of Zdhhc2-shRNA-2 and empty vector transduced cells were observed in non-GC B cells, the Zdhhc2-shRNA-2 transduced B1-8hi cells were essentially absent in GC B cells (Figure 3B) and had significantly reduced GC/non-GC ratios (Figure 3C). To further validate that Zdhhc2 is a positive regulator, Zdhhc2-shRNA-4, another Zdhhc2-shRNA described previously (50), was also tested. As shown in Figures 3B,C, Zdhhc2-shRNA-4 could also significantly reduce the differentiation of B1-8hi cells into GC B cells. These data suggest that Zdhhc2 is a positive regulator of GC B cell differentiation.
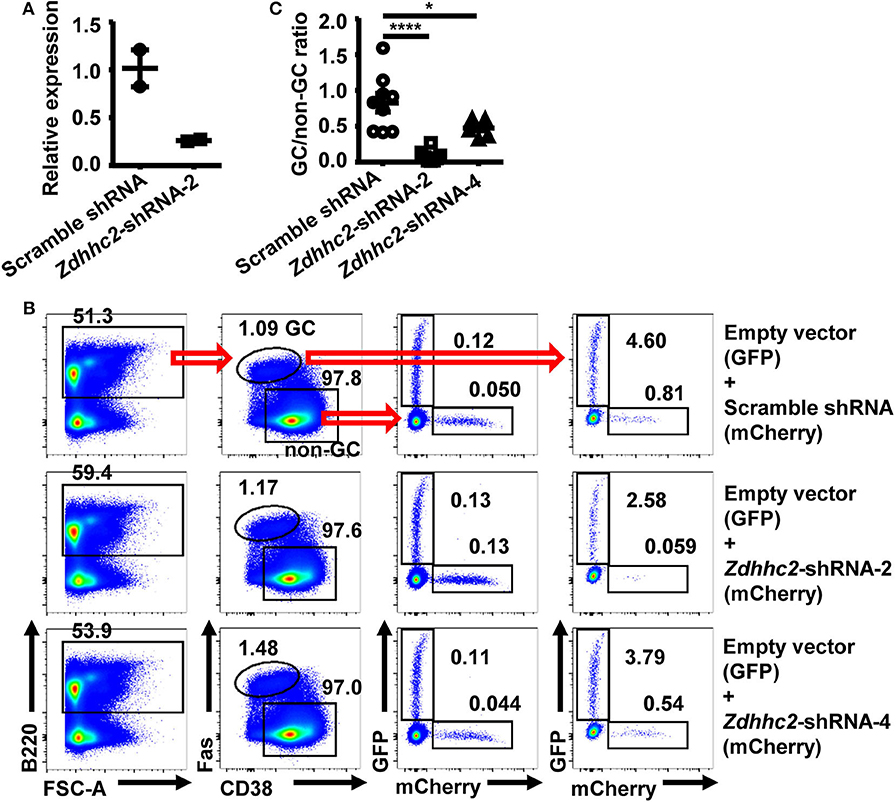
Figure 3. Reduced in vivo differentiation of Zdhhc2-shRNA transduced B cells into germinal center (GC) B cells. (A) Graph showing the relative mRNA levels in scramble shRNA and Zdhhc2-shRNA transduced B cells. Each symbol represents an independent culture. (B,C) Representative flow cytometry profiles (B) showing the precentages of scramble shRNA or Zdhhc2-shRNA transduced B1-8hi cells (expressing mCherry) as well as the co-transferred empty vector transduced B1-8hi cells (expressing GFP) in non-GC B cells and GC B cells, and representative graph (C) showing the GC/non-GC ratio of percentages of scramble shRNA or Zdhhc2-shRNA transduced B1-8hi cells (expressing mCherry) normalized to empty vector transduced cells (expressing GFP), analyzed as described in Figures 1C,D. Each symbol in (C) represents data from an individual mouse. The bars represent mean ± SEM; *p ≤ 0.05, ****p ≤ 0.0001, one-way ANOVA with Dunnett's multiple-comparisons test. A representative of two independent experiments is shown.
To further investigate the function of Zdhhc2 in GC B cell differentiation, a culture system in which mouse naïve B cells undergo extensive proliferation and differentiation into GC-phenotype B cells and plasma cells was exploited (53). In this system, the naïve B cells were co-cultured with NB21.2D9 feeder cell lines that express CD40L, BAFF, and IL-21. At 24 h after co-culture, the splenic B cells were transduced with Zdhhc2-shRNA or scramble shRNA retroviral vectors and analyzed for their differentiation (Figure 4A). Consistent with the previous report, the splenic B cells can differentiate into GC phenotype B cells (B220+Fas+GL7+, referred to as “iGC”), which are most abundant on day 4, and then to plasma cells (B220+CD138+IgG1+, referred to as “PC”) (Supplemental Figure 4). Importantly, the percentage and the number of Zdhhc2-shRNA-2 transduced iGC B cells were significantly reduced as compared to those of scramble shRNA transduced iGC B cells from as early as day 4 (Figures 4B,D,E). The reduction peaked at days 6 and 7 and is not due to the retroviral transduction process as the percentages of scramble shRNA transduced iGC B cells and plasma cells are almost maintained constant (Figures 4D,F). A quantification of the percentage and the number of plasma cells also supports that Zdhhc2-shRNA-2 transduced B cells have reduced differentiation into plasma cells (Figures 4C,F,G).
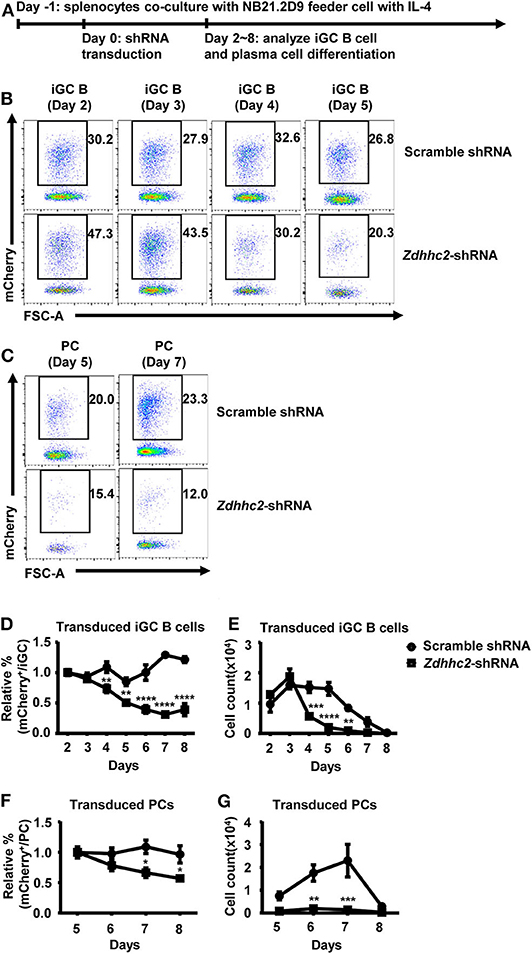
Figure 4. Reduced in vitro differentiation of Zdhhc2-shRNA transduced B cells into germinal center (GC) B cells and plasma cells. (A) The in vitro model for analyzing iGC B cell and plasma cell differentiation. The wild-type (WT) splenic cells were co-cultured with irradiated NB-21.2D9 feeder cells (expressing CD40L, BAFF, and IL21) in the presence of 2 ng/ml of IL-4 on day−1, transduced with scramble shRNA or Zdhhc2-shRNA-2 retroviral vectors (expressing mCherry) on day 0, and analyzed for iGC B cell and plasma cell differentiation from day 2 to day 8. (B,C) Representative flow cytometry profiles showing the percentages of scramble shRNA or Zdhhc2-shRNA transduced cells (mCherry+) among iGC B cells (B220+Fas+GL7+) as cultured in (A) on days 2–5 (B) and plasma cells (B220+CD138+IgG1+ or PC) on day 5 and day 7 (C). (D–G) Representative graphs showing the relative percentage (D,F) or the absolute number (E,G) of scramble shRNA or Zdhhc2-shRNA transduced cells among iGC B cells (normalized to the values on day 2) (D,E) or plasma cells (normalized to the values on day 5) (F,G) at the indicated time points (days 2–8 for iGC B cells, days 5–8 for PC) in the cultures described as in (A). The bars represent mean ± SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, two-way ANOVA with Sidak's multiple-comparisons test. A representative of two independent experiments is shown.
To further explore the role of Zdhhc2 in GC B cell differentiation, its expression was analyzed. As shown in Figure 5A, the mRNA levels of Zdhhc2 in GC B cells, quantified by qPCR, are about 20-fold higher than those in follicular B cells. Zdhhc2 expression is also induced in in vitro differentiated iGC B cells (Figure 5B). These results are consistent with previously published microarray and RNA-seq data (Supplemental Figure 5A) and data from the Immunological Genome Project (Supplemental Figures 5B,C) (54, 55).
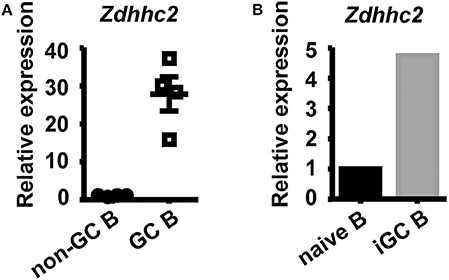
Figure 5. High expression levels of Zdhhc2 in germinal center (GC) B cells. (A,B) Graphs showing the relative Zdhhc2 mRNA levels, quantified by qRT-PCR, in non-GC B and GC B cells sorted by fluorescence-activated cell sorting (FACS) from Peyer's patches of wild-type mice (A), FACS-sorted splenic naïve B cells, and induced GC B cells (iGC B) cultured as in Figure 4A. Each symbol in (A) represents the sample from an individual mouse. N = 1 in (B).
To further investigate how Zdhhc2 knockdown impacts on GC B cell differentiation, we analyzed its impact on B cell proliferation and apoptosis. We noticed that the percentage and the number of Zdhhc2-shRNA transduced B-lineage cells were lower than those of scramble shRNA transduced cells in the in vitro culture system (Figures 6A–C), suggesting that Zdhhc2-shRNA may have an impact on B cell proliferation and survival. To investigate this possibility, Zdhhc2-shRNA transduced iGC B cells were labeled with CFSE and analyzed 2 days later. As shown in Figures 6D,E, Zdhhc2-shRNA transduced cells have significantly higher CFSE levels as compared to scramble shRNA transduced cells. In contrast, the co-cultured non-transduced cells have comparable CFSE levels (Figures 6D,E). These results suggest that Zdhhc2-shRNA transduction inhibits B cell proliferation in response to CD40L/BAFF/IL-21 stimulation. At the same time, increased percentages of apoptotic cells (annexin V+7AAD−), which was only observed from day 7, and dead cells (annexin V+7AAD+), from as early as day 5, were also observed among Zdhhc2-shRNA transduced cells as compared to the scramble shRNA transduced cells (Figures 6F–H). Collectively, these results suggest that Zdhhc2 can regulate GC B cell differentiation by regulating both B cell proliferation and survival.
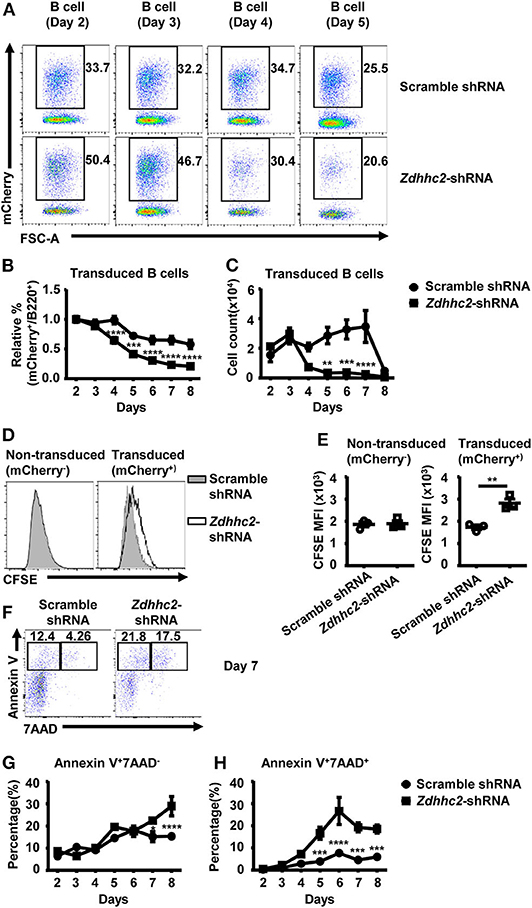
Figure 6. The reduced proliferation and the increased cell death in Zdhhc2-shRNA transduced B cells. (A) Representative flow cytometry profiles showing the percentages of scramble shRNA or Zdhhc2-shRNA transduced cells (mCherry+) among B cells (B220+) cultured as in Figure 4 on days 2–5. (B,C) Representative graphs showing the relative percentage (normalized to the values on day 2) (B) or the absolute number (C) of scramble shRNA or Zdhhc2-shRNA transduced cells among B cells at the indicated time points in the culture described as in Figure 4. (D,E) Representative flow cytometry profiles showing the carboxyfluorescein succinimidyl ester (CFSE) levels (D) and graphs showing the CFSE MFI levels (E) in scramble shRNA or Zdhhc2-shRNA transduced B cells (mCherry+) and non-transduced B cells (mCherry−). The splenic cells were cultured and transduced as described in Figure 4A on day −1 and day 0, then labeled with CFSE on day 2 and further co-cultured with irradiated NB-21.2D9 feeder cells, and analyzed for CFSE levels on day 4. (F,G,H) Representative flow cytometry profiles (F) and graphs showing the percentages of apoptotic cells (Annexin-V+7AAD−) (G) and dead cells (Annexin-V+7AAD+) (H) in scramble shRNA or Zdhhc2-shRNA transduced B cells (mCherry+). Each symbol represents an independent culture. The bars represent mean ± SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, two-way ANOVA with Sidak's multiple-comparisons test (B,C,G,H) or unpaired two-tailed t-test (E). The data are representative of two independent experiments.
Here we show the proof of concept of a functional in vivo screen for genes that regulate GC B cell differentiation using retroviral shRNA vector transduced antigen-specific B cells. Despite that this approach only allows the screen of relatively small library given that only a small fraction of retrovirally transduced cells is present in the GC B cell compartment (~4,000 retrovirally transduced B1-8hi cells were recovered in each screen, which is ~10 times more than that of the shRNA constructs). Because of this limitation, loss of some shRNAs in the screen is expected. At the same time, some shRNA constructs may be lost due to their biological activity, which could not be distinguished at this point. Regardless, some novel strong regulators of GC B cell differentiation can be identified. We noticed that the knockdown of Bcl6 results in a more substantial reduction in GC B cells as compared to the reduction observed in heterozygous Bcl6 mice (vs. WT mice), where the percentage of GC B cells is reduced to ~50% (56). We reasoned that in our system the Bcl6–shRNA transduced B1-8hi cells need to compete with Bcl6-sufficient B1-8hi cells in the same mice, where the selection disadvantage of Bcl6–shRNA transduced B1-8hi cells might become more evident under such competition pressure, a phenomenon that we have previously observed (57). Therefore, one advantage of our system is to provide the sensitivity of competition models. This system provides a new way to discover genes that regulate GC B cell differentiation and perhaps post-GC B cell differentiation, such as memory and plasma cell differentiation.
The in vivo screening system may be further optimized in the future for the higher-throughput screen. It is noted that while the retroviral transduction efficiency can reach ~50%, as assessed by flow cytometry at 2 days after transduction, the efficiency of these retrovirally transduced B1-8hi cells is reduced as compared to that of fresh B1-8hi cells without the retroviral transduction procedure, suggesting that the stimulation during the retroviral transduction procedure is detrimental, to some extent, for GC B cell differentiation. It is possible that moving the retroviral transduction to earlier B cell development stages may help to optimize the system, which is the subject of a future study.
The identification of Zdhhc2 in our screen suggests that Zdhhc2 is a strong regulator of GC B cell differentiation. As far as we know, no study of Zdhhc2 in B cells has been described. As a member of DHHC-palmitoyl transferases, Zdhhc2 has been mainly studied in the field of cancer and neuroscience (58, 59). In humans, aberrant ZDHHC2 expression or translocation has been described in several cancers, including acute myeloid leukemia and hepatocellular carcinoma (59). Palmitoylation mediated by DHHC-palmitoyl transferases has been proposed to regulate oncogenic RAS signaling and tumor suppressor localization to the plasma membrane (59). ZDHHC2 has also been implicated in mediating the palmitoylation of PSD-95 and AKAP79/150 required for neuronal activity (60, 61). The reported substrates of ZDHHC2 also includes CD9 and CD151 in HEK293 cells (62), Lck in T cells (63), and CKAP4 in hepatocellular carcinoma and interstitial cystitis (64–66).
The function of Zdhhc2 in B cell is, however, poorly understood. Palmitoylation of CD81 has been found to be necessary for the raft-stabilizing function of the CD19/CD21/CD81 complex to facilitate BCR signaling (67). The B lymphocyte-specific immune regulators CD20 and CD23 have been identified as novel palmitoylated proteins in human B cells (68). Since the reversible cycles of palmitoylation and depalmitoylation have been proposed to regulate the precise membrane localization of proteins, membrane association, protein stability and traffic, and cell signaling processes (58, 69–75), we speculate that Zdhhc2 may regulate GC B cell differentiation through modulating the palmitoylation of unknown targets associated with cell proliferation and survival in B cells. The underlying molecular mechanism has remained to be revealed.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
The animal study was reviewed and approved by SJTUSM Institutional Animal Care and Use Committee.
FL and RZ designed the study and analyzed the results and wrote the paper. RZ performed the experiments. HZ, YZ, and CH provided technical supports. DL provided next-generation sequencing data processing supports. All authors reviewed and approved the manuscript.
This work was supported by the Chinese Mega Project on Infectious Diseases Grant 2018ZX10302301, the National Natural Science Foundation of China (NNSFC) Project 31422020, 973 Program 2014CB943600, and NNSFC Projects 31600704 and 31800778. FL was supported by the “Shu Guang” project from Shanghai Municipal Education Commission and Shanghai Education Development Foundation. HZ was supported by the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning. HZ, YZ, and FL were also supported by the innovative research team of high-level local universities in Shanghai (SSMU-2DCX20180100).
DL was employed by the company Boston Consulting Group.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Zhaoyuan Hou for providing retroviral pSIREN-RetroQ vector, Dr. Jeffrey Ravetch (The Rockefeller University) for providing A20 cells. We also thank the help of the staff of the flow cytometry core facility of Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine. We acknowledge the assistance of staff in the Department of Laboratory Animal Science, Shanghai Jiao Tong University School of Medicine.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01025/full#supplementary-material
1. Burnett DL, Langley DB, Schofield P, Hermes JR, Chan TD, Jackson J, et al. Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination. Science. (2018) 360:223–6. doi: 10.1126/science.aao3859
2. Brink R, Phan TG. Self-reactive B cells in the germinal center reaction. Annu Rev Immunol. (2018) 36:339–57. doi: 10.1146/annurev-immunol-051116-052510
3. Allen CD, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol. (2004) 5:943–52. doi: 10.1038/ni1100
4. Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. (2007) 315:528–31. doi: 10.1126/science.1136736
5. Caron G, Le Gallou S, Lamy T, Tarte K, Fest T. CXCR4 expression functionally discriminates centroblasts versus centrocytes within human germinal center B cells. J Immunol. (2009) 182:7595–602. doi: 10.4049/jimmunol.0804272
6. Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. (2010) 143:592–605. doi: 10.1016/j.cell.2010.10.032
7. Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. (2007) 179:5099–108. doi: 10.4049/jimmunol.179.8.5099
8. Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. (2011) 110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3
9. Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. (2012) 30:429–57. doi: 10.1146/annurev-immunol-020711-075032
10. Meyer-Hermann ME, Maini PK, Iber D. An analysis of B cell selection mechanisms in germinal centers. Math Med Biol. (2006) 23:255–77. doi: 10.1093/imammb/dql012
11. Rolf J, Bell SE, Kovesdi D, Janas ML, Soond DR, Webb LM, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. (2010) 185:4042–52. doi: 10.4049/jimmunol.1001730
12. Mandel TE, Phipps RP, Abbot A, Tew JG. The follicular dendritic cell: long term antigen retention during immunity. Immunol Rev. (1980) 53:29–59. doi: 10.1111/j.1600-065X.1980.tb01039.x
13. Mandel T, Phipps R, Abbot A, Tew J. Long-term antigen retention by dendritic cells in the popliteal lymph node of immunized mice. Immunology. (1981) 43:353.
14. Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. (2005) 5:230–42. doi: 10.1038/nri1572
15. Gray D. Immunological memory. Annu Rev Immunol. (1993) 11:49–77. doi: 10.1146/annurev.iy.11.040193.000405
16. Weisel FJ, Zuccarino-Catania GV, Chikina M, Shlomchik MJ. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity. (2016) 44:116–30. doi: 10.1016/j.immuni.2015.12.004
17. De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. (2015) 15:137. doi: 10.1038/nri3804
18. Grammer AC, Slota R, Fischer R, Gur H, Girschick H, Yarboro C, et al. Abnormal germinal center reactions in systemic lupus erythematosus demonstrated by blockade of CD154-CD40 interactions. J Clin Invest. (2003) 112:1506–20. doi: 10.1172/JCI200319301
19. Lougaris V, Badolato R, Ferrari S, Plebani A. Hyper immunoglobulin M syndrome due to CD40 deficiency: clinical, molecular, and immunological features. Immunol Rev. (2005) 203:48–66. doi: 10.1111/j.0105-2896.2005.00229.x
20. Warnatz K, Bossaller L, Salzer U, Skrabl-Baumgartner A, Schwinger W, van der Burg M, et al. Human ICOS deficiency abrogates the germinal center reaction and provides a monogenic model for common variable immunodeficiency. Blood. (2006) 107:3045–52. doi: 10.1182/blood-2005-07-2955
21. Zhang W, Zhang H, Liu S, Xia F, Kang Z, Zhang Y, et al. Excessive CD11c(+)Tbet(+) B cells promote aberrant TFH differentiation and affinity-based germinal center selection in lupus. Proc Natl Acad Sci USA. (2019) 116:18550–60. doi: 10.1073/pnas.1901340116
22. Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. (1997) 276:589–92. doi: 10.1126/science.276.5312.589
23. Cutrona G, Ulivi M, Fais F, Roncella S, Ferrarini M. Transfection of the c-myc oncogene into normal Epstein-Barr virus-harboring B cells results in new phenotypic and functional features resembling those of Burkitt lymphoma cells and normal centroblasts. J Exp Med. (1995) 181:699–711. doi: 10.1084/jem.181.2.699
24. Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. (1997) 275:540–3. doi: 10.1126/science.275.5299.540
25. Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, Shi W, et al. Transcription factor IRF4 regulates germinal center cell formation through a B cell–intrinsic mechanism. J Immunol. (2014) 192:3200–6. doi: 10.4049/jimmunol.1303216
26. Vikstrom I, Carotta S, Luthje K, Peperzak V, Jost PJ, Glaser S, et al. Mcl-1 is essential for germinal center formation and B cell memory. Science. (2010) 330:1095–9. doi: 10.1126/science.1191793
27. Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. (1999) 274:18470–6. doi: 10.1074/jbc.274.26.18470
28. Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. (1997) 186:439–48. doi: 10.1084/jem.186.3.439
29. Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. (2012) 13:1092–100. doi: 10.1038/ni.2418
30. Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. (2008) 8:976–90. doi: 10.1038/nrc2231
31. De Silva NS, Simonetti G, Heise N, Klein U. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol Rev. (2012) 247:73–92. doi: 10.1111/j.1600-065X.2012.01113.x
32. Lu P, Shih C, Qi H. Ephrin B1-mediated repulsion and signaling control germinal center T cell territoriality and function. Science. (2017) 356:eaai9264. doi: 10.1126/science.aai9264
33. Li X, Gadzinsky A, Gong L, Tong H, Calderon V, Li Y, et al. Cbl ubiquitin ligases control B cell exit from the germinal-center reaction. Immunity. (2018) 48:530–41. doi: 10.1016/j.immuni.2018.03.006
34. Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. (2009) 16:324–35. doi: 10.1016/j.ccr.2009.08.015
35. Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. (2010) 12:372–9. doi: 10.1038/ncb2037
36. Adams BD, Guo S, Bai H, Guo Y, Megyola CM, Cheng J, et al. An in vivo functional screen uncovers miR-150-mediated regulation of hematopoietic injury response. Cell Rep. (2012) 2:1048–60. doi: 10.1016/j.celrep.2012.09.014
37. Murugaesu N, Iravani M, van Weverwijk A, Ivetic A, Johnson DA, Antonopoulos A, et al. An in vivo functional screen identifies ST6GalNAc2 sialyltransferase as a breast cancer metastasis suppressor. Cancer Discov. (2014) 4:304–17. doi: 10.1158/2159-8290.CD-13-0287
38. Ashenden M, van Weverwijk A, Murugaesu N, Fearns A, Campbell J, Gao Q, et al. An in vivo functional screen identifies JNK signaling as a modulator of chemotherapeutic response in breast cancer. Mol Cancer Ther. (2017) 16:1967–78. doi: 10.1158/1535-7163.MCT-16-0731
39. Malina A, Mills JR, Cencic R, Yan Y, Fraser J, Schippers LM, et al. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. (2013) 27:2602–14. doi: 10.1101/gad.227132.113
40. Weber J, Ollinger R, Friedrich M, Ehmer U, Barenboim M, Steiger K, et al. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc Natl Acad Sci USA. (2015) 112:13982–7. doi: 10.1073/pnas.1512392112
41. Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. (2015) 160:1246–60. doi: 10.1016/j.cell.2015.02.038
42. Maresch R, Mueller S, Veltkamp C, Ollinger R, Friedrich M, Heid I, et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun. (2016) 7:10770. doi: 10.1038/ncomms10770
43. Katigbak A, Cencic R, Robert F, Senecha P, Scuoppo C, Pelletier J. A CRISPR/cas9 functional screen identifies rare tumor suppressors. Sci Rep. (2016) 6:38968. doi: 10.1038/srep38968
44. Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci. (2017) 20:1329–41. doi: 10.1038/nn.4620
45. Xu C, Qi X, Du X, Zou H, Gao F, Feng T, et al. PiggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc Natl Acad Sci USA. (2017) 114:722–7. doi: 10.1073/pnas.1615735114
46. Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. (2017) 547:413–8. doi: 10.1038/nature23270
47. Song CQ, Li Y, Mou H, Moore J, Park A, Pomyen Y, et al. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology. (2017) 152:1161–73. doi: 10.1053/j.gastro.2016.12.002
48. Gonzalez-Martin A, Adams BD, Lai M, Shepherd J, Salvador-Bernaldez M, Salvador JM, et al. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat Immunol. (2016) 17:433–40. doi: 10.1038/ni.3385
49. Chen R, Bélanger S, Frederick Megan A, Li B, Johnston Robert J, Xiao N, et al. In vivo RNA interference screens identify regulators of antiviral CD4+ and CD8+ T cell differentiation. Immunity. (2014) 41:325–38. doi: 10.1016/j.immuni.2014.08.002
50. Brichta L, Shin W, Jackson-Lewis V, Blesa J, Yap EL, Walker Z, et al. Identification of neurodegenerative factors using translatome-regulatory network analysis. Nat Neurosci. (2015) 18:1325–33. doi: 10.1038/nn.4070
51. Cato MH, Yau IW, Rickert RC. Magnetic-based purification of untouched mouse germinal center B cells for ex vivo manipulation and biochemical analysis. Nat Protoc. (2011) 6:953–60. doi: 10.1038/nprot.2011.344
52. Muppidi JR, Schmitz R, Green JA, Xiao W, Larsen AB, Braun SE, et al. Loss of signalling via Galpha13 in germinal centre B-cell-derived lymphoma. Nature. (2014) 516:254–8. doi: 10.1038/nature13765
53. Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. (2011) 2:465. doi: 10.1038/ncomms1475
54. Shi W, Liao Y, Willis SN, Taubenheim N, Inouye M, Tarlinton DM, et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. (2015) 16:663–73. doi: 10.1038/ni.3154
55. Heng TS, Painter MW, Immunological Genome Project C. The immunological genome project: networks of gene expression in immune cells. Nat Immunol. (2008) 9:1091–4. doi: 10.1038/ni1008-1091
56. Huang C, Geng H, Boss I, Wang L, Melnick A. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B-cell differentiation. Blood. (2014) 123:1012–20. doi: 10.1182/blood-2013-07-518605
57. Li M, Lazorchak AS, Ouyang X, Zhang H, Liu H, Arojo OA, et al. Sin1/mTORC2 regulate B cell growth and metabolism by activating mTORC1 and Myc. Cell Mol Immunol. (2019) 16:757–69. doi: 10.1038/s41423-018-0185-x
58. Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neurosci. (2010) 11:161–75. doi: 10.1038/nrn2788
59. Ko PJ, Dixon SJ. Protein palmitoylation and cancer. EMBO Rep. (2018) 19:e46666. doi: 10.15252/embr.201846666
60. Noritake J, Fukata Y, Iwanaga T, Hosomi N, Tsutsumi R, Matsuda N, et al. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J Cell Biol. (2009) 186:147–60. doi: 10.1083/jcb.200903101
61. Woolfrey KM, Sanderson JL, Dell'Acqua ML. The palmitoyl acyltransferase DHHC2 regulates recycling endosome exocytosis and synaptic potentiation through palmitoylation of AKAP79/150. J Neurosci. (2015) 35:442–56. doi: 10.1523/JNEUROSCI.2243-14.2015
62. Sharma C, Yang XH, Hemler ME. DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol Biol Cell. (2008) 19:3415–25. doi: 10.1091/mbc.e07-11-1164
63. Zeidman R, Buckland G, Cebecauer M, Eissmann P, Davis DM, Magee AI. DHHC2 is a protein S-acyltransferase for Lck. Mol Membr Biol. (2011) 28:473–86. doi: 10.3109/09687688.2011.630682
64. Planey SL, Keay SK, Zhang CO, Zacharias DA. Palmitoylation of cytoskeleton associated protein 4 by DHHC2 regulates antiproliferative factor-mediated signaling. Mol Biol Cell. (2009) 20:1454–63. doi: 10.1091/mbc.e08-08-0849
65. Zhang J, Planey SL, Ceballos C, Stevens SM, Jr., Keay SK, et al. Identification of CKAP4/p63 as a major substrate of the palmitoyl acyltransferase DHHC2, a putative tumor suppressor, using a novel proteomics method. Mol Cell Proteomics. (2008) 7:1378–88. doi: 10.1074/mcp.M800069-MCP200
66. Li SX, Tang GS, Zhou DX, Pan YF, Tan YX, Zhang J, et al. Prognostic significance of cytoskeleton-associated membrane protein 4 and its palmitoyl acyltransferase DHHC2 in hepatocellular carcinoma. Cancer. (2014) 120:1520–31. doi: 10.1002/cncr.28593
67. Cherukuri A, Carter RH, Brooks S, Bornmann W, Finn R, Dowd CS, et al. B cell signaling is regulated by induced palmitoylation of CD81. J Biol Chem. (2004) 279:31973–82. doi: 10.1074/jbc.M404410200
68. Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, et al. Proteomic analysis of S-acylated proteins in human B cells reveals palmitoylation of the immune regulators CD20 and CD23. PLoS ONE. (2012) 7:e37187. doi: 10.1371/journal.pone.0037187
69. Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. (2007) 8:74–84. doi: 10.1038/nrm2084
70. Guan X, Fierke CA. Understanding protein palmitoylation: biological significance and enzymology. Sci China Chem. (2011) 54:1888–97. doi: 10.1007/s11426-011-4428-2
71. El-Husseini Ael D, Bredt DS. Protein palmitoylation: a regulator of neuronal development and function. Nat Rev Neurosci. (2002) 3:791–802. doi: 10.1038/nrn940
72. Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. (2010) 191:23–9. doi: 10.1083/jcb.200911143
73. Sanders SS, De Simone FI, Thomas GM. mTORC1 signaling is palmitoylation-dependent in hippocampal neurons and non-neuronal cells and involves dynamic palmitoylation of LAMTOR1 and mTOR. Front Cell Neurosci. (2019) 13:115. doi: 10.3389/fncel.2019.00115
74. Zingler P, Sarchen V, Glatter T, Caning L, Saggau C, Kathayat RS, et al. Palmitoylation is required for TNF-R1 signaling. Cell Commun Signal. (2019) 17:90. doi: 10.1186/s12964-019-0405-8
Keywords: germinal center B cell, in vivo screen, shRNA, GC selection, Zdhhc2
Citation: Zhao R, Zhang H, Zhang Y, Li D, Huang C and Li F (2020) In vivo Screen Identifies Zdhhc2 as a Critical Regulator of Germinal Center B Cell Differentiation. Front. Immunol. 11:1025. doi: 10.3389/fimmu.2020.01025
Received: 11 March 2020; Accepted: 28 April 2020;
Published: 10 June 2020.
Edited by:
Jeroen E. J. Guikema, Amsterdam University Medical Center (UMC), NetherlandsReviewed by:
Agnieszka Dzikiewicz-Krawczyk, Institute of Human Genetics (PAN), PolandCopyright © 2020 Zhao, Zhang, Zhang, Li, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fubin Li, ZnViaW4ubGlAc2p0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.