
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 08 May 2020
Sec. Viral Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.00939
This article is part of the Research TopicCoronavirus Disease (COVID-19): Pathophysiology, Epidemiology, Clinical Management and Public Health ResponseView all 400 articles
Zoonotic infections are an imminent threat to human health. Pangolins were recently identified as carriers and intermediate hosts of coronaviruses. Previous research has shown that infection with coronaviruses activates an innate immune response upon sensing of viral RNA by interferon-induced with helicase C domain 1 (IFIH1), also known as MDA5. Here, we performed a comparative genomics study of RNA sensor genes in three species of pangolins. DDX58/RIG-I, a sensor of cytoplasmic viral RNA and toll-like receptors (TLR) 3, 7, and 8, which bind RNA in endosomes, are conserved in pangolins. By contrast, IFIH1 a sensor of intracellular double-stranded RNA, has been inactivated by mutations in pangolins. Likewise, Z-DNA-binding protein (ZBP1), which senses both Z-DNA and Z-RNA, has been lost during the evolution of pangolins. These results suggest that the innate immune response to viruses differs significantly between pangolins and other mammals, including humans. We put forward the hypothesis that loss of IFIH1 and ZBP1 provided an evolutionary advantage by reducing inflammation-induced damage to host tissues and thereby contributed to a switch from resistance to tolerance of viral infections in pangolins.
Emerging infectious diseases represent a major challenge to public health. The transmission of pathogens from other vertebrate animals to humans is of particular concern because the resulting diseases, known as zoonoses, have caused major epidemics in the past and continue to pose enormous threats to the human population, as exemplified by the recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak (1, 2). In a broader sense, viral and bacterial pathogens are among the strongest drivers of evolutionary change and the genomes of vertebrate species have been shaped, to a large extent, by adaptations to pathogens.
To cope with viral infections, vertebrate species have evolved response strategies which can be classified into resistance and tolerance (3). Resistance depends on the efficient sensing of the infection and mounting of antiviral responses that involve programmed death of infected cells, suppression of viral replication, inflammation and the establishment of adaptive immunity. However, pathogens can also trigger overreactions of the immune system which cause more harm to the individual than the infectious agent itself (4, 5). Therefore, tolerance to infections has evolved as an alternative response of many hosts to specific pathogens (6, 7). In this scenario, the pathogens are not efficiently eliminated but the pathogen or defense-induced damage to the host is reduced. Tolerance does not depend on, or is even impeded by, the early sensing of pathogen-associated patterns (PAMPs) and its mechanisms of protection are not yet fully understood (6, 8, 9). Species that tolerate infections can carry a high burden of infectious agents, and therefore may be important reservoirs for transmissions to other species. This notion is supported by the finding that bats tolerate many viral infections some of which have spread to humans causing zoonoses such as Ebola, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) (7).
Pangolins have been identified, besides bats, as a possible source of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease 2019 (COVID-19) (10–14). Eight species of pangolins form the mammalian order Pholidota which is most closely related to Carnivora (cat-like and dog-like carnivorans). They are insectivorous and toothless animals whose body is largely covered by keratinous scales. The immune defense of pangolins has not been characterized yet except for reports on the deficiencies of TLR5, a receptor of bacterial flagellin (15) and interferon-ε, an antiviral cytokine of epithelia (16, 17). The receptor of SARS-CoV-2, i.e., angiotensin I converting enzyme 2 (ACE2) is conserved in pangolins (18) and coronaviruses isolated from pangolins have a receptor binding domain in their spike protein that is uniquely similar to that of SARS-CoV-2 (10, 19).
Antiviral defense of vertebrates is initiated by sensors of viral nucleic acids. Infections with RNA viruses, such as coronaviruses, influenza viruses and Ebolavirus activate sensors of extracellular or endosomal RNA, such as TLR3, TLR7, and TLR8 (20), and sensors of intracellular RNA, such as IFIH1/MDA5, ZBP1, and DDX58/RIG-I (21–28). These sensors are specific for different subtypes of RNAs that constitute the viral genome or appear during viral replication or gene expression and they activate distinct cellular and organismal responses, such as necroptotic cell death, interferon signaling and inflammation (27, 29).
Here we report a unique degeneration of the innate immune response against RNA viruses in pangolins.
The following genome sequences of pangolin species were analyzed: Malayan pangolin (Manis javanica), Assembly: ManJav1.0 (GCA_001685135.1), submitted by The International Pangolin Research Consortium (16); Chinese pangolin (Manis pentadactyla), Assembly: M_pentadactyla-1.1.1 (GCA_000738955.1), submitted by Washington University; Tree pangolin (Manis tricuspis), Assembly: ManTri_v1_BIUU (GCA_004765945.1), submitted by Broad Institute. Gene annotations were available in GenBank only for M. javanica (NCBI Manis javanica Annotation Release 100).
Shared order of gene arrangement (synteny) in the Malayan pangolin (M. javanica), cat, dog, cattle, mouse, and human was assessed by comparison of gene loci that were downloaded from GenBank at https://www.ncbi.nlm.nih.gov/gene/ (last accessed on 27 March, 2020). In addition, Basic Local Alignment Search Tool (BLAST) was used to find regions of local similarity between sequences (30). Amino acid and nucleotide sequence were aligned with the Multalin software (31). Divergence times of evolutionary lineages were obtained from the Timetree website (www.timetree.org) (32).
IFIH1, also known as melanoma differentiation-associated protein 5 (MDA5), binds to double-stranded RNA in the cytosol and signals through mitochondrial antiviral-signaling protein (MAVS) to activate expression of interferons and to induce inflammation (33). IFIH1 senses cytoplasmic RNA of coronaviruses and other viruses (27, 34, 35). Comparison of the IFIH1 gene locus showed conservation of the arrangement of IFIH1 relative to the neighboring genes in mammals (Figure 1A). In the Malayan pangolin, IFIH1 is inactivated by more than 10 frameshift and in-frame stop mutations. In silico translation of the pangolin IFIH1 pseudogene (GenBank gene ID: 108398082) and alignment of the resulting amino acid sequence to that of human IFIH1 showed numerous disruptive mutations (Figure S1A). An open reading frame in exon 1 of the Malayan pangolin encodes a theoretical protein that lacks essential domains and has only 100 amino acid residues whereas functional IFIH1 proteins consist of more than 1,000 amino acid residues (Figure S2). Detailed comparative analysis of exon 1 showed the presence of multiple frameshift mutations and in-frame stop codons in the IFIH1 genes of Malayan, Chinese and tree pangolins (Figure 1B). One of the frameshift mutations and one premature stop mutation are shared by all three species, suggesting that these mutations have already been present in their last common ancestor that lived more than 20 million years ago (32).
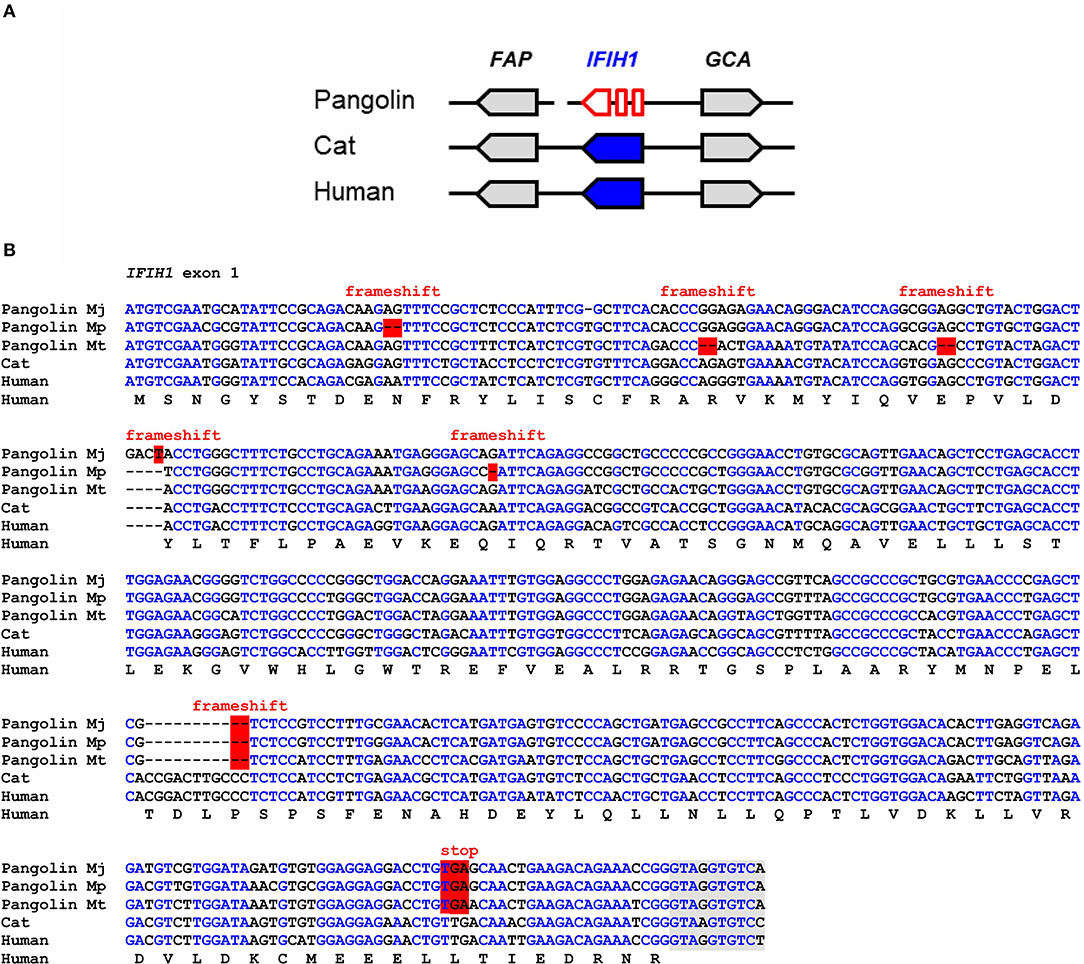
Figure 1. IFIH1 is a pseudogene in pangolins. (A) Gene locus of IFIH1 in the pangolin (M. javanica), cat, and human. Genes are represented by arrows pointing in the direction of transcription. A sequence gap is located between FAP and IFIH1 in the pangolin. (B) Inactivating mutations in exon 1 of IFIH1 in three species of pangolins. Nucleotide sequences of pangolins, cat and human were aligned. The coding sequence of human IFIH1 was translated and the amino acid sequence is shown below the nucleotide sequences. Frameshift mutations and in-frame stop codons are highlighted by red shading. Nucleotides conserved in more than 50% of the sequences are indicated by blue fonts. Nucleotides in the flanking region of the first intron are shown with gray shading. Nucleotide sequence accession numbers (GenBank): Human (NC_000002.12, nucl. 162317845-162318307, compl.), cat (NC_018730.3, nucl. 154125204-154125666, compl.), Malayan pangolin (NW_016533891.1, nucl. 53417-53871, compl.), Chinese pangolin (JPTV01003556.1, nucl. 39028-39476, compl.), tree pangolin (SOZM010146646.1, nucl. 741-1188, compl.). Abbreviations: compl., complementary; nucl., nucleotide numbers; Mj, Manis javanica; Mp, Manis pentadactyla; Mt, Manis tricuspis.
ZBP1 binds to left-handed double helix structures of DNA and RNA (Z-DNA and Z-RNA) and thereupon triggers necroptosis and inflammation through interactions with receptor-interacting serine/threonine-protein kinase 3 (RIPK3) (36). Influenza virus and other viruses induce ZBP1-mediated innate immune responses in humans and mice (24, 25). Comparison of the ZBP1 gene locus showed conservation of the arrangement of ZBP1 relative to the neighboring genes in mammals (Figure 2A). In the Malayan pangolin, ZBP1 is inactivated by multiple in-frame stop codons. In silico translation of the pangolin ZBP1 pseudogene (GenBank gene ID: 108390931) and alignment of the resulting amino acid sequence to that of human ZBP1 showed premature termination of the translation product and lack of the carboxy-terminal half of the protein (Figure S1B). Mutations that prevent the production of a functional protein were found in all segments of the ZBP1 pseudogene of the Malayan pangolin. The nucleotide sequence alignment of ZBP1 exon 4 shows the presence of in-frame stop codons in three species of pangolins (M. javanica, M. pentadactyla, M. tricuspis) (Figure 2B).
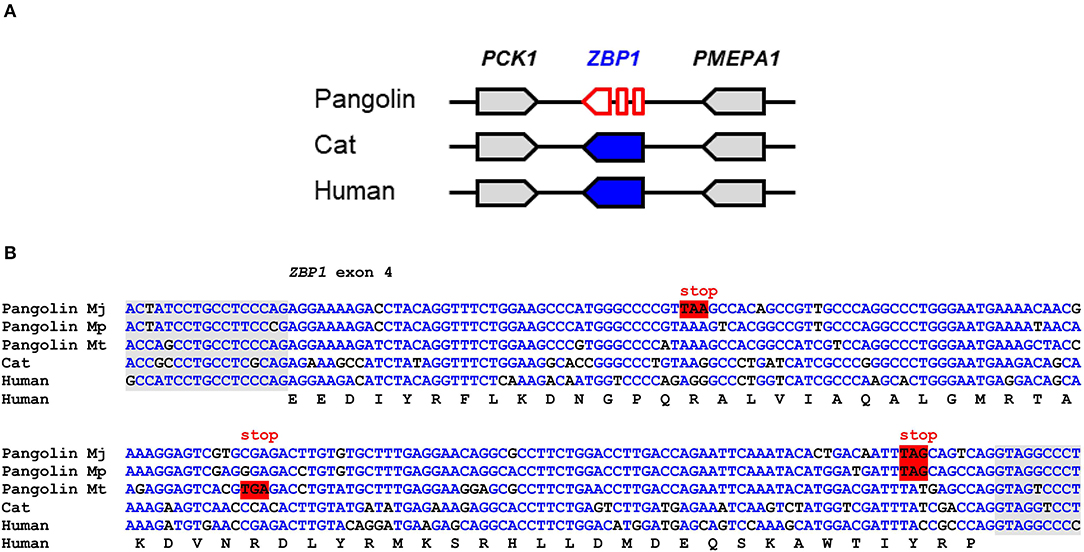
Figure 2. ZBP1 is a pseudogene in pangolins. (A) Gene locus of ZBP1 in the pangolin (M. javanica), cat, and human. Genes are represented by arrows pointing in the direction of transcription. (B) Inactivating mutations in exon 4 of ZBP1 in three species of pangolins. Nucleotide sequences of pangolins, cat and human were aligned. The coding sequence of human ZBP1 was translated and the amino acid sequence is shown below the nucleotide sequences. In-frame stop codons are highlighted by red shading. Nucleotides conserved in more than 50% of the sequences are indicated by blue fonts. Nucleotides in the flanking region of the introns are shown with gray shading. Nucleotide sequence accession numbers (GenBank): Human (NC_000020.11, nucl. 57614878.0.57615077, compl.), cat (NC_018725.3, nucl. 5721658-5721857), Malayan pangolin (NW_016529116.1, nucl. 156452-156651, compl.), Chinese pangolin (JPTV01006633.1, nucl. 23295.0.23494), tree pangolin (SOZM010101098.1, nucl. 532-731). Abbreviations: compl., complementary; nucl., nucleotide numbers; Mj, Manis javanica; Mp, Manis pentadactyla; Mt, Manis tricuspis.
In contrast to IFIH1 and ZBP1, the genes encoding the intracellular RNA sensor RIG-I, i.e., DExD/H-box helicase 58 (DDX58), and TLR3, TLR7, and TLR8 which control the sensing of RNA in endosomes and a series of other genes involved in antiviral signaling and defense, such as MAVS, RIPK3, MLKL, SKIV2L, OAS2, RNASEL, and EIF2AK2 (PKR) do not contain disruptive mutations and therefore appear to be intact in the Malayan pangolin (M. javanica) (Table S1). DDX58 contains in-frame stop codons and frameshift mutations in the tree pangolin (M. tricuspis) but not in the Chinese pangolin (M. pentadactyla) (Figure S3), suggesting that the tree pangolin lacks functional DDX58/RIG-I in addition to the two intracellular RNA sensors (IFIH1 and ZBP1) absent in all pangolins.
We screened the genomes of mammals from diverse phylogenetic lineages for functional copies (devoid of frameshift mutations and premature in-frame stop codons) of ZBP1, IFIH1 and other RNA sensor genes. Mapping the presence or absence of these genes onto the phylogenetic tree suggested that loss of both ZBP1 and IFIH1 occurred in the pangolin lineages soon after divergence from the lineage leading to Carnivora (represented by cat, dog and bear in Figure 3A). Other genes implicated in anti-RNA-viral defense are conserved in the selected set of species (Figure 3A; Table S2).
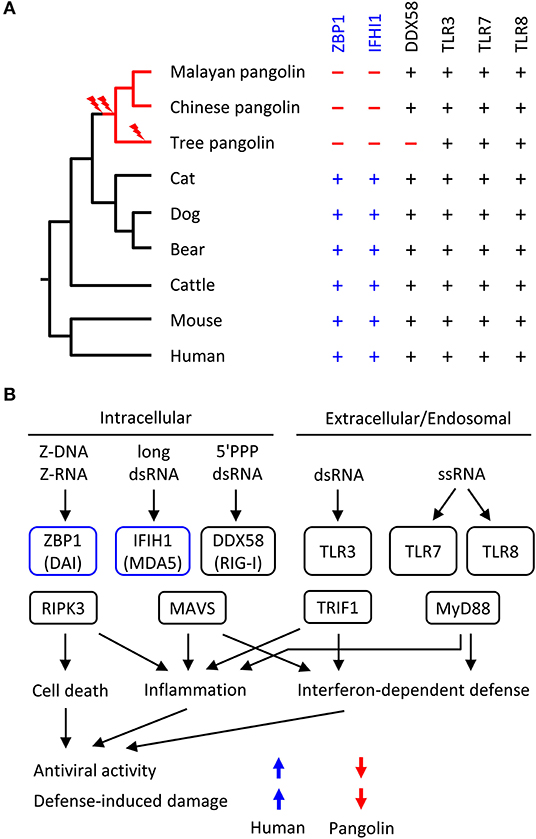
Figure 3. Evolution of RNA sensor genes and possible implications on antiviral responses in pangolins. (A) Phylogenetic tree of mammals and comparison of presence (+) or absence (–) of RNA sensor genes. Evolutionary gene loss (indicated by lightning bolt symbols) was inferred from the species distribution of the genes. Species: Malayan pangolin (Manis javanica), Chinese pangolin (Manis pentadactyla), tree pangolin (Manis tricuspis), cat (Felis catus), dog (Canis lupus familiaris), bear (Ursus arctos horribilis), cattle (Bos taurus), mouse (Mus musculus), human (Homo sapiens). (B) Schematic overview of innate immune sensors of viral RNA and signaling in mammals. Only RNA sensors investigated in this study are shown. The schematic includes the hypothesis about IFIH1 and ZBP1-dependent differences in the antiviral activity and defense-induced damage to the host. The directions of the colored arrows indicate the effects of the presence or absence of RNA sensors. 5'PPP, triphosphorylated at the 5'-end; ds, double-stranded; ss, single-stranded.
Based on the known target specificities of mammalian RNA sensors (Figure 3B), the loss of ZBP1 and IFIH1 suggests that the response to Z-RNA and long double-stranded RNA is diminished in pangolins. Accordingly, the resistance to RNA viruses that depend on cytoplasmic Z-RNA and long double-stranded RNA for replication has likely decreased in the evolution of pangolins. We put forward the hypothesis that strong antiviral defense was harmful and loss of ZBP1 and IFIH1 provided an evolutionary advantage by increasing tolerance to infections by certain RNA viruses, including coronaviruses.
Viruses are potent drivers of evolutionary adaptations in their hosts. Both insufficient and overshooting responses to viral infections have deleterious effects, leading to strong selection for resistant or tolerant host genotypes (37, 38). Bats have retained functional RNA sensor genes (Table S3) but exert only dampened antiviral responses, indicating that they have adapted to the evolutionary pressure from viruses by decreasing inflammatory responses and by enhancing tolerance to viral replication (39–42). The results of the present study suggest that pangolins are another group of mammals with evolutionarily downregulated defense against a subset of viruses, namely those sensed by IFIH1/MDA5 or ZBP1 in other species. Our data urge to study the virus burden of pangolins, their antiviral immune response and their ability to act as reservoirs for viruses with zoonotic potential, especially coronaviruses. While genetic suppression of IFIH1/MDA5 and ZBP1-dependent pathways had neutral or beneficial effects in the evolution of pangolins, pharmaceutical suppression of IFIH1/MDA5 and ZBP1-dependent signaling may be beneficial for human patients with overreactions to viral nucleic acids.
All datasets presented in this study are included in the article/Supplementary Material.
HF and LE designed the study and performed bioinformatic analyses. HF, ET, and LE wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00939/full#supplementary-material
1. Plowright RK, Parrish CR, McCallum H, Hudson PJ, Ko AI, Graham AL, et al. Pathways to zoonotic spillover. Nat Rev Microbiol. (2017) 15:502–10. doi: 10.1038/nrmicro.2017.45
2. Fauci AS, Lane HC, Redfield RR. Covid-19 - navigating the uncharted. N Engl J Med. (2020) 382:1268–9. doi: 10.1056/NEJMe2002387
3. Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. (2008) 8:889–95. doi: 10.1038/nri2432
4. Oldstone MB. Lessons learned and concepts formed from study of the pathogenesis of the two negative-strand viruses lymphocytic choriomeningitis and influenza. Proc Natl Acad Sci USA. (2013) 110:4180–3. doi: 10.1073/pnas.1222025110
5. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. (2020) 395:1033–4. doi: 10.1016/S0140-6736(20)30628-0
6. Soares MP, Teixeira L, Moita LF. Disease tolerance and immunity in host protection against infection. Nat Rev Immunol. (2017) 17:83–96. doi: 10.1038/nri.2016.136
7. Hayman DTS. Bat tolerance to viral infections. Nat Microbiol. (2019) 4:728–9. doi: 10.1038/s41564-019-0430-9
8. Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. (2012) 335:936–41. doi: 10.1126/science.1214935
9. Luan HH, Wang A, Hilliard BK, Carvalho F, Rosen CE, Ahasic AM, et al. GDF15 is an inflammation-induced central mediator of tissue tolerance. Cell. (2019) 178:1231–44. doi: 10.1016/j.cell.2019.07.033
10. Lam TT, Shum MH, Zhu H, Tong Y, Ni X, Liao Y, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. (2020). doi: 10.1038/s41586-020-2169-0. [Epub ahead of print].
11. Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. (2020) 30:1346–51. doi: 10.1016/j.cub.2020.03.022
12. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. (2020) 26:450–2. doi: 10.1038/s41591-020-0820-9
13. Zhang Y, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. (2020) 181:223–7. doi: 10.1016/j.cell.2020.03.035
14. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
15. Sharma V, Walther F, Hecker N, Stuckas H, Hiller M. Convergent losses of TLR5 suggest altered extracellular flagellin detection in four mammalian lineages. Mol Biol Evol. (2020) msaa058. doi: 10.1093/molbev/msaa058
16. Choo SW, Rayko M, Tan TK, Hari R, Komissarov A, Wee WY, et al. Pangolin genomes and the evolution of mammalian scales and immunity. Genome Res. (2016) 26:1312–22. doi: 10.1101/gr.203521.115
17. Fung KY, Mangan NE, Cumming H, Horvat JC, Mayall JR, Stifter SA, et al. Interferon-ε protects the female reproductive tract from viral and bacterial infection. Science. (2013) 339:1088–92. doi: 10.1126/science.1233321
18. Luan J, Lu Y, Jin X, Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun. (2020) 526:165–69. doi: 10.1016/j.bbrc.2020.03.047
19. Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J Proteome Res. (2020) 19:1351–60. doi: 10.1021/acs.jproteome.0c00129
20. Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. (2006) 7:131–7. doi: 10.1038/ni1303
21. Maelfait J, Liverpool L, Bridgeman A, Ragan KB, Upton JW, Rehwinkel J. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. (2017) 36:2529–43. doi: 10.15252/embj.201796476
22. Roth-Cross JK, Bender SJ, Weiss SR. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol. (2008) 82:9829–38. doi: 10.1128/JVI.01199-08
23. Li J, Liu Y, Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J Virol. (2010) 84:6472–82. doi: 10.1128/JVI.00016-10
24. Kuriakose T, Man SM, Malireddi RK, Karki R, Kesavardhana S, Place DE, et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. (2016) 1:aag2045. doi: 10.1126/sciimmunol.aag2045
25. Zhang T, Yin C, Boyd DF, Quarato G, Ingram JP, Shubina M, et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell. (2020) 180:1115–29. doi: 10.1016/j.cell.2020.02.050
26. Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci USA. (2017) 114:E4251–60. doi: 10.1073/pnas.1618310114
27. Lee HC, Chathuranga K, Lee JS. Intracellular sensing of viral genomes and viral evasion. Exp Mol Med. (2019) 51:1–13. doi: 10.1038/s12276-019-0299-y
28. Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci USA. (2020) 117:8094–103. doi: 10.1073/pnas.1921485117
29. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. (2006) 441:101–5. doi: 10.1038/nature04734
30. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
31. Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. (1988) 16:10881–90. doi: 10.1093/nar/16.22.10881
32. Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. (2015) 32:835–45. doi: 10.1093/molbev/msv037
33. Chow KT, Gale M Jr., Loo YM. RIG-I and other RNA sensors in antiviral immunity. Annu Rev Immunol. (2018) 36:667–94. doi: 10.1146/annurev-immunol-042617-053309
34. Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. (2011) 12:137–43. doi: 10.1038/ni.1979
35. Zalinger ZB, Elliott R, Rose KM, Weiss SR. MDA5 is critical to host defense during infection with murine coronavirus. J Virol. (2015) 89:12330–40. doi: 10.1128/JVI.01470-15
36. Kuriakose T, Kanneganti TD. ZBP1: Innate sensor regulating cell death and inflammation. Trends Immunol. (2018) 39:123–34. doi: 10.1016/j.it.2017.11.002
37. Koonin EV. Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci. (2016) 371:20150442. doi: 10.1098/rstb.2015.0442
38. Berezovskaya F, Karev GP, Katsnelson MI, Wolf YI, Koonin EV. Stable coevolutionary regimes for genetic parasites and their hosts: you must differ to coevolve. Biol Direct. (2018) 13:27. doi: 10.1186/s13062-018-0230-9
39. Pavlovich SS, Lovett SP, Koroleva G, Guito JC, Arnold CE, Nagle ER, et al. The Egyptian Rousette genome reveals unexpected features of bat antiviral immunity. Cell. (2018) 173:1098–110. doi: 10.1016/j.cell.2018.03.070
40. Ahn M, Anderson DE, Zhang Q, Tan CW, Lim BL, Luko K, et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol. (2019) 4:789–99. doi: 10.1038/s41564-019-0371-3
41. Banerjee A, Baker ML, Kulcsar K, Misra V, Plowright R, Mossman K. Novel insights into immune systems of bats. Front Immunol. (2020) 11:26. doi: 10.3389/fimmu.2020.00026
Keywords: zoonosis, pangolin, coronavirus, RNA sensor, innate immunity, inflammation, tolerance, gene loss
Citation: Fischer H, Tschachler E and Eckhart L (2020) Pangolins Lack IFIH1/MDA5, a Cytoplasmic RNA Sensor That Initiates Innate Immune Defense Upon Coronavirus Infection. Front. Immunol. 11:939. doi: 10.3389/fimmu.2020.00939
Received: 31 March 2020; Accepted: 22 April 2020;
Published: 08 May 2020.
Edited by:
Shen-Ying Zhang, The Rockefeller University, United StatesReviewed by:
Marina Cella, Washington University School of Medicine in St. Louis, United StatesCopyright © 2020 Fischer, Tschachler and Eckhart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leopold Eckhart, bGVvcG9sZC5lY2toYXJ0QG1lZHVuaXdpZW4uYWMuYXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.