- 1NIHR Health Protection Research Unit in Emerging and Zoonotic Infections, Centre for Global Vaccine Research, Institute of Infection and Global Health, University of Liverpool, Liverpool, United Kingdom
- 2Division of Vaccine Discovery, La Jolla Institute for Immunology, La Jolla, CA, United States
- 3Tropical and Infectious Disease Unit, Liverpool University Hospitals, Liverpool, United Kingdom
Dengue virus (DENV, family Flaviviridae, genus Flavivirus) exists as four distinct serotypes. Generally, immunity after infection with one serotype is protective and lifelong, though exceptions have been described. However, secondary infection with a different serotype can result in more severe disease for a minority of patients. Host responses to the first DENV infection involve the development of both cross-reactive antibody and T cell responses, which, depending upon their precise balance, may mediate protection or enhance disease upon secondary infection with a different serotype. Abundant evidence now exists that responses elicited by DENV infection can cross-react with other members of the genus Flavivirus, particularly Zika virus (ZIKV). Cohort studies have shown that prior DENV immunity is associated with protection against Zika. Cross-reactive antibody responses may enhance infection with flaviviruses, which likely accounts for the cases of severe disease seen during secondary DENV infections. Data for T cell responses are contradictory, and even though cross-reactive T cell responses exist, their clinical significance is uncertain. Recent mouse experiments, however, show that cross-reactive T cells are capable of mediating protection against ZIKV. In this review, we summarize and discuss the evidence that T cell responses may, at least in part, explain the cross-protection seen against ZIKV from DENV infection, and that T cell antigens should therefore be included in putative Zika vaccines.
Introduction
During the last two decades, the rate of infections with flaviviruses, particularly dengue virus (DENV) and Zika virus (ZIKV), has risen significantly (Figure 1). At present, half of the world's population is considered at risk for DENV and cases of ZIKV continue to be reported globally, including the first local cases in southern Europe (1, 2). DENV and ZIKV are spread via the bite of infected mosquitoes, Aedes spp., whose expanding ecological niches beyond the tropical and sub-tropical regions pose a major public health threat (3). Infection with DENV can present with a spectrum of clinical manifestations ranging from an asymptomatic illness to an acute fever/arthralgia/rash (dengue fever, DF) that usually is self-limiting, to more severe disease (dengue hemorrhagic fever/dengue shock syndrome, DHF/DSS) that is characterized by vascular leakage and/or hemorrhage (4). ZIKV causes a similar febrile illness that is often mild, with the exception of rare cases of neurological disease, such as Guillain-Barré syndrome (5). In pregnancy, ZIKV infection is associated with adverse fetal/neonatal outcomes such as congenital Zika syndrome (CZS) (6). Given that these viruses share similar geographic distributions and high sequence homology, immunological cross-reactivity between DENV and ZIKV is a well-recognized and unsurprising phenomenon. In this review, we will focus on the role of T cells during DENV and ZIKV infections in humans and in animal models, summarizing the major findings, discussing how cross-reactivity might impact immunity, and providing evidence why incorporating T cell epitopes into vaccine design is favorable.
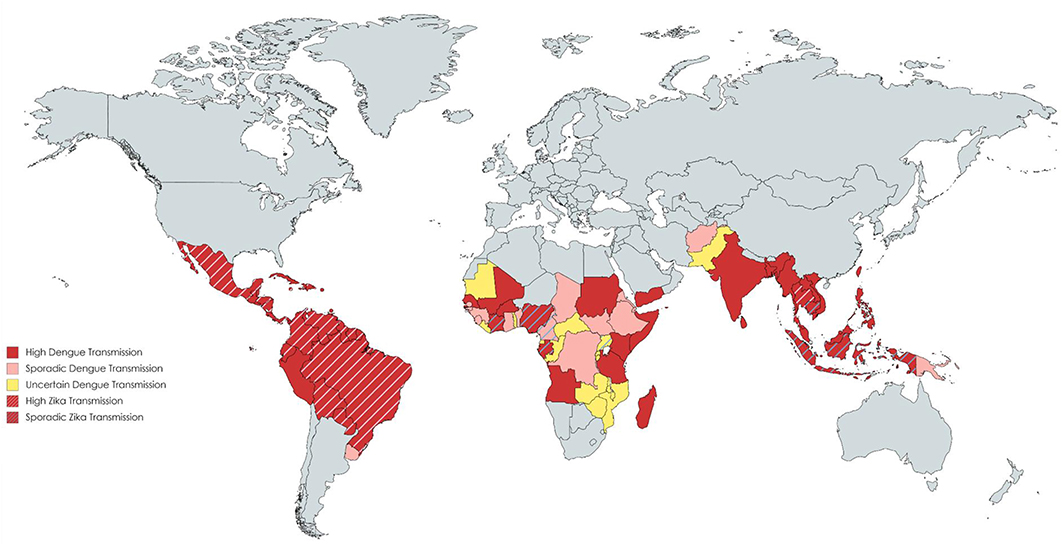
Figure 1. Distribution of countries with Dengue and Zika transmission. Data compiled using the CDC yellow book and HealthMap©. Each color reflects a varying transmission rate from high dengue transmission (in red) to uncertain dengue transmission (in yellow). Countries with cross-hatched lines reflect those with high or sporadic Zika transmission.
Virology
There are four dengue viruses, DENV1-4, which are antigenically distinct (hence called serotypes) and possibly represent four distinct introductions into humans from the sylvatic cycle in non-human primates (7). On occasion several serotypes can circulate concomitantly within endemic areas, or as individual serotypes in sequence (8). A primary dengue infection generally results in lifelong immunity against the same serotype, although homotypic DENV re-infections have also been described (9). DENV infections can generate cross-reactive, poorly neutralizing antibodies that bind the other serotypes (10). Upon secondary infection with a heterologous DENV serotype, there is then a risk of severe disease, thought to be mediated via a mechanism called antibody-mediated enhancement (ADE) (11, 12). ADE arises when antibodies against one serotype can bind to, but not fully neutralize, another DENV serotype. These virus-antibody complexes can bind to the Fcγ receptors on the surface of mononuclear phagocytes enhancing viral entry and facilitating viral replication (13).
ZIKV has three genotypes, East African, West African and Asian (14). Recent Zika outbreaks have indicated a role for pre-existing immunity against DENV to modulate ZIKV infection (15). Data from a large pediatric cohort in Nicaragua found that prior DENV infection reduced the risk of symptomatic ZIKV infection by about one third (16) and in a Brazilian cohort high pre-existing antibody titers to DENV were associated with reduced risk of ZIKV infection and symptoms (17). Furthermore, protection against congenital Zika syndrome was shown to be associated with prior DENV immunity (18).
Viral Structure
The flavivirus virion is enveloped, and contains a single-stranded, positive-sense RNA that is ~11 kb size. The viral genome encodes three structural proteins [capsid, precursor membrane (prM) and envelope (E)] involved in virion assembly and seven non-structural (NS) proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) that function in the viral life cycle (19). The canonical role of NS proteins is in viral replication where with host factors they function in the assembly of the membrane-bound multi-protein replication complex (RC). NS proteins are also the target of most of the flavivirus CD8 T cell epitopes (20–22). A mature flavivirus particle has a well-organized outer glycoprotein shell with an icosahedral T = 3 symmetry, a host derived lipid bilayer membrane and a poorly defined inner nucelocapsid core (23). Flavivirus particles can assume various morphologies (immature, mosaic-like, and mature) that vary between flaviviruses and have important implications on antibody binding specifically regarding the availability and accessibility of epitopes (24) [reviewed in (25)].
Antibody-Mediated Immunity
Neutralizing antibody plays a crucial role in immunity to flaviviruses. Animal models show that robustly neutralizing monoclonal antibodies are sufficient for protection against many flaviviruses (24, 26, 27). However, antibody responses against flaviviruses can also be notoriously cross-reactive and the neutralization potential of these antibodies can vary considerably. The neutralizing antibody response is directed against the E protein, but responses against other proteins such as prM and NS1 also form a significant fraction of the response after both DENV and ZIKV infections (25, 28–30).
In recent years, much has been learned about anti-flavivirus antibody responses by cloning antibodies from infected or previously infected humans. These studies have demonstrated that, while many different classes of antibody are made, those which most potently neutralize frequently recognize quaternary epitopes on the viral surface and bind across multiple Envelope proteins (24, 31–33). The probable mechanism of neutralization by antibodies against quaternary epitopes is through interfering with viral fusion by locking the particle in a non-fusogenic form (24). Although some of these antibody classes can neutralize all four DENV serotypes and ZIKV (34, 35), they may not be durable in humans and their effectiveness may wane with time (36). In fact, a period of cross-protection exists even after a primary DENV infection, as observed by Sabin who found that subjects re-challenged with heterotypic DENV infection were protected if the re-challenge occurred 2–3 months after the original infection (37).
A major epitope recognized by the human antibody response is the viral fusion-loop of E domain II (25). Fusion loop antibodies are highly cross-reactive and strongly binding, but weakly neutralizing, not able to cross-neutralize other viruses, and can mediate ADE (38). During secondary flavivirus infections, the resulting antibody response can be predominantly focused upon the earlier infecting virus, a phenomenon known as “original antigenic sin (OAS);” and thus may be poorly neutralizing against the current infection (39). It may be challenging for new vaccines (such as those against ZIKV), when introduced in areas of intense flavivirus transmission, to protect if the balance of enhancing and neutralizing antibody is not optimal, or the development of new antibody responses is impaired (38). In addition, the development of congenital Zika syndrome has been linked to ADE (40) and could be due to the presence of cross-reactive fusion loop antibodies. Prior DENV immunity can protect against ZIKV, and, in the cases where inefficient antibody responses arise, possibly due to OAS, it might be that a cross-reactive CD8+ T cell response contributes to protection.
T Cell Responses to DENV in Humans
Early work on cellular immunity to DENV demonstrated that T cell responses were readily detectable, and serotype cross-reactive responses of both CD4+ and CD8+ T cells were described (20–22, 41–46). The existence of serotype cross-reactivity at the level of individual T cell epitopes was found in both subjects given an experimental DENV vaccine (47, 48) and after natural exposure (49). In fact, a single DENV infection can elicit a cross-reactive T cell response against several serotypes (50), and the same T cell receptor (TCR) can recognize epitopes from multiple serotypes (51–53). Although variant epitopes may be recognized by the same TCR, the degree of overall serotype cross-reactivity is also likely to be influenced by the targeting of immunodominant responses, for example non-structural (NS) proteins are more highly conserved than structural proteins (Table 1). Responses biased toward sequences that are conserved between serotypes (possibly in NS proteins) give rise to higher potential for serotype cross-reactivity (55). The factors that determine whether responses are focused on conserved or variant epitopes are not known. However, interestingly, the pattern of conserved/serotype specific epitope recognition was remarkably similar in two different populations studied (Figure 2), implying that the factors underlying the phenomenon are not constrained to specific populations (55). The immunodominant targets of the T-cell response can vary between CD4+ and CD8+ T cells (Figure 3), and also between DENV serotypes (22, 55), as well as with exposure to other flaviviruses. Interestingly, the stimulus for the most cross-reactive T cell responses of all appears to be the tetravalent live attenuated DENV vaccine TV003, which includes the NS protein from DENV serotypes 1, 3, and 4, where the vast majority of the response is directed against NS proteins (Figure 4) (56). CD8+ T cells from TV003 vaccines can also cross-recognize ZIKV peptides, suggesting that the tetravalent DENV vaccination can induce T cell cross-reactivity across DENV serotypes and the closely related ZIKV (54).
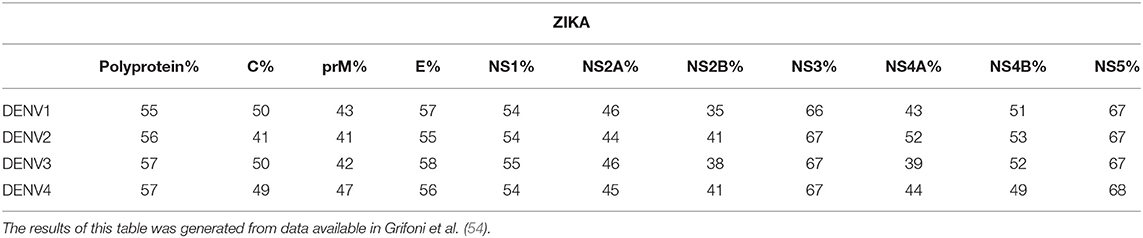
Table 1. Percent homology across structural and non-structural proteins between Zika and DENV serotypes 1–4.
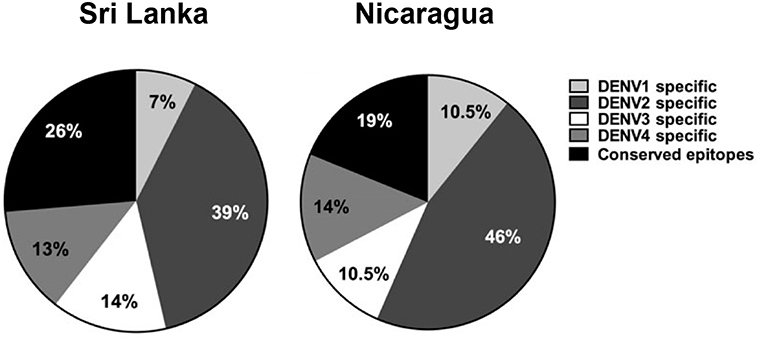
Figure 2. Distribution of CD8+ T cell epitopes across the four DENV serotypes in two clinically characterized cohorts from Sri Lanka and Nicaragua. Adapted from results in Gordon et al. (16), Weiskopf et al. (55).
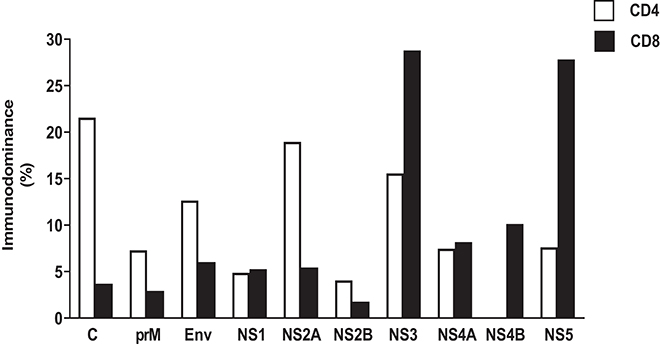
Figure 3. Immunodominant protein pattern of DENV-specific CD4 and CD8 T cell response based on IEDB data. HLA class I and class II restricted epitopes for CD4 and CD8 DENV-specific T cell responses, respectively, have been derived querying IEDB database (www.IEDB.org) on 02-Aug-2019. Epitope list was filtered using Immunobrowser, selecting epitope lists tested in at least 10 donors. Protein immunodominance was calculated as percentage of epitopes per protein per HLA restriction.
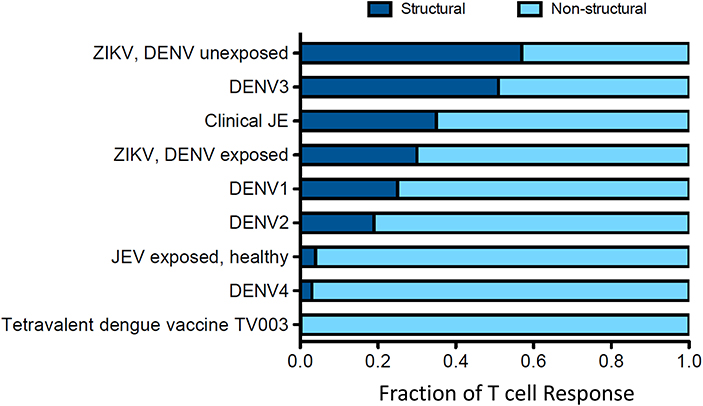
Figure 4. Distribution of the T cell response to structural and non-structural components is influenced by prior clinical history. Data for this figure was compiled from the following studies: Turtle et al. (20), Weiskopf et al. (21, 56), Grifoni et al. (54), and Delgado et al. (57). T cell responses are from subjects with natural infections or vaccine recipients. Responses were identified by ELISPOT and intracellular cytokine staining (ICS) assays.
Demonstrating a clear role for DENV specific T cell responses in protection or disease has been more challenging. Initially, it was thought that the T cell response against DENV was pathological to the host. Some variant DENV epitopes may function as inefficient TCR agonists (58), and during acute disease CD8+ T cell responses can be more focused on variant epitopes from a previous infection (original antigenic sin) (51), possibly leading to less efficient responses. Moreover, many of these DENV specific T cells were found to be apoptotic (51). Some studies have demonstrated that ex-vivo T cell cytokine responses are greater with more severe disease, when tested shortly after disease onset (59, 60), suggesting a role for T cells in mediating excessive inflammation. In addition, degranulation of T cells [a surrogate marker for cytotoxicity (61)] was not greater in DHF (60, 62), implying that it is in fact the balance between cytokine production (pathological?) and killing (protective?) that may influence disease phenotype.
However, not all studies have shown a relationship between acute responses and disease phenotype, with some authors pointing out that the appearance of DENV specific T cells occurs after resolution of clinical outcomes in severe dengue (63). Furthermore, not all studies show a relationship between higher cytokine production and disease severity in DHF (64). In fact, when sampled early in the disease course, or as in one prospective study before disease onset, T cell cytokine production correlated with lower viremia and less severe disease (65, 66), implying a protective role for T cells. One study of HLA association with disease severity of dengue showed certain alleles to be protective whilst others were detrimental, potentially explaining why studies of T cells and disease severity give discrepant results (21, 67, 68).
DENV/ZIKV T Cell Cross-Reactivity
Given the potential for T cell responses to DENV to be protective, at least in some circumstances, it is therefore possible that T cells primed by DENV could recognize ZIKV and be protective. Grifoni et al. found that in DENV exposed individuals, the T cell response to ZIKV is earlier, larger and exhibits greater cytotoxic capacity (54). In the same study it was shown that in two large cohorts from Sri Lanka and Nicaragua (54), the imprint of previous DENV exposure is clearly detectable, and that the resulting T cell response to Asian ZIKV was biased toward the non-structural proteins (Figure 4). Similarly, evidence of pre-existing flavivirus immunity has been shown to result in enhanced T cell responses directed to NS3 of DENV and African ZIKV (69). Whether such responses are protective is unknown, but two studies have demonstrated that short-term T cell cultures of flavivirus specific T cells are capable of killing targets pulsed with peptides that are found in ZIKV, indicating that they likely have anti-viral function (Figure 5) (20, 70). With DENV infection appearing to confer partial protection against Zika illness (16, 18), cross-reactive T cell responses may be one such mechanism by which this protection is mediated. Additionally, transcriptomic profiles of ZIKV-specific CD8+ T cells in DENV naïve or pre-exposed patients showed no qualitative differences in ZIKV- specific CD8+ T cell responses supporting the fact that cross-reactive T cell responses share the same protective phenotype observed after single flavivirus exposure (71). This phenomenon may also not be confined to ZIKV. There is some evidence that partial protection against Japanese Encephalitis (JE) appears to be conferred by prior DENV infection, with the number of JE cases lower than expected in areas with DENV outbreaks (72). Also it has been shown that the severity of JE is reduced by previous flavivirus infection (73).
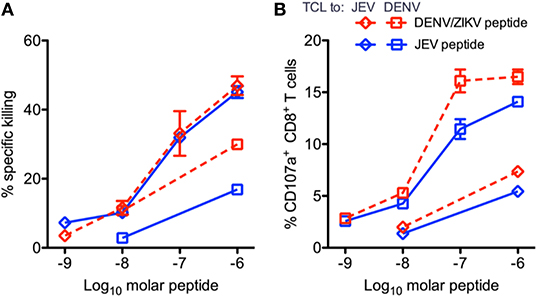
Figure 5. Peptide-pulsed, CFSE-labeled, HLA-matched targets were incubated with CD8+ T cell line effector cells, and the percent specific killing was measured by flow cytometry in response to JEV and DENV/ZIKV peptides. Percent killing (A) and the percent of CD107+ CD8+ T cells (B) are shown. Diamonds indicate T cell lines expanded with JEV peptide, and squares indicate lines expanded with DENV/ZIKV peptide. Assays were performed in duplicate for each T cell line/peptide pair. Error bars represent standard error of the mean. This figure was adapted from Turtle et al. (20).
These effects may be mediated by T cell responses (20). The degree to which T cell responses are targeted to structural vs. non-structural flavivirus proteins may vary according to previous exposure. For example, in the case of ZIKV, being affected by prior DENV infection biases the response toward the non-structural proteins (Figure 4). Responses against non-structural proteins tend to be more cross-reactive, meaning that previous dengue infection has the potential to bias the T cell response to ZIKV toward more cross-reactive epitopes. In an environment where multiple flavivirus exposure occurs, such epitopes, are likely to receive a great number of re-stimulations and may rise to the top of a “hierarchy of immunodominance.” Including such epitopes in vaccines, therefore, has merit in that such a vaccine may be made more effective if there is a degree of pre-existing immunity in the population, lowering the threshold for vaccine responses to be generated. Although further studies are required to unequivocally show that DENV-primed T cell responses can mediate protection against ZIKV in humans, mouse studies provide convincing evidence that T cells can mediate cross-protection.
T Cell Responses in Mice and Non-Human Primates (NHP)
Mice are not natural hosts for flaviviruses as the murine type I IFN system provides a very effective defense, which thwarts viral dissemination and thus prevents them being useful models of severe disease phenotypes (74). As such, to establish a model of productive viral infection, which can be used to examine T cell function and test potential vaccine candidates, multiple strains of immunocompromised mice have been generated (74). These strains include mice deficient in either type I or type II or both IFN receptors, mice with STAT2 knocked-out, mice with mouse STAT2 replaced by human STAT2, and more nuanced models where type I IFN receptors are absent in specific cells or tissues (74). In combination with either mouse-adapted or human viral strains, this has established an infection model that closely, but not perfectly, mimics human disease. For DENV and ZIKV, strains that are commonly used either lack type I IFN receptor (IFNAR−/− and A129) or both type I and II receptors (AG129 mice) (74). Human Leucocyte Antigen (HLA)-transgenic mice have also been used to model CD8+ and/or CD4+ T cell responses in flavivirus infection. Work in HLA-transgenic mice show a broad epitope repertoire, whilst some HLA variants, such as HLA-B*0702, have been specifically studied due to a known association with high T cell response frequency and magnitude in humans, as well as decreased susceptibility to severe dengue disease (75, 76).
Control of primary DENV infection in mice requires CD8+ T cells to a greater extent than CD4+ cells. In IFNAR−/− mice, depletion of CD8+ cells was directly associated with increased viral burden in tissues that was not ameliorated by the transfer of serum or B cells (77). Protection in the study was mediated by increased cytotoxic activity in DENV-specific CD8+ T cells; activity which was further enhanced when mice received a peptide vaccination (77). However, challenge experiments in other mouse models (HEPG2-grafted SCID) find that DENV-specific CD8+ T cell responses were associated with reduced mortality, which suggests that T cells may contribute to disease severity in some instances, and prevent mortality in others (78). Crossing IFNAR−/− mice with HLA transgenic mice showed that protective CD8+ T cell responses tended to be polyfunctional and principally targeted non-structural proteins such as NS3 and NS5, similar to that in humans (75). Responses targeted against NS proteins have also shown their protective potential in homotypic secondary DENV infections where wildtype mice primed with a non-lethal DENV2 strain ACS46 were challenged with a lethal encephalitic homotypic strain JHA1 (79). In this model, protective immunity was reduced when both CD4+ and CD8+ T cells were depleted. In most challenge models CD4+ T cells play an accessory and non-essential role in which they contribute to viral clearance when induced by immunization (80).
Models of heterologous DENV infection also demonstrate the importance of T cells during the anti-DENV response. Collectively these studies show that CD8+ responses can protect against heterologous DENV challenge in non-lethal (81) and lethal models (82). CD8+ T cell responses contribute to protection during heterotypic reinfection, whereas homotypic reinfection can be contained by neutralizing antibodies against the infecting serotype (81), as is believed to be the case in humans. Comparison of specific and cross-reactive T cell responses in IFNAR−/− mice reveal that, despite their relatively low magnitude and avidity, cross-reactive CD8+ T cells from prior DENV exposure reduce viral load and exhibit a polyfunctional response in a manner comparable to that of serotype-specific cells (46). However, in a model of secondary DENV infection resulting in severe disease, cross-reactive CD4+ and CD8+ T cells were found to be pathogenic in wildtype mice infected with a non-mouse adapted DENV strain (83). In summary, the contribution of T cells to disease and protection in dengue mouse models is still not fully understood. The variability in current data are likely shaped by factors such as differences in mouse immune function, infection methods, strain difference, and experimental end-points.
Similar to observations for DENV, experiments in mice in which T cell responses are lacking, or on the other hand are enhanced (e.g., through peptide immunization or adoptive cell transfer), demonstrate a protective role for CD8+ T cells against ZIKV (84, 85). CD8+ T cell responses in primary ZIKV infection appear to be essential for immunity. In LysMCre+ IFNARfl/fl (type I IFN receptor absent only in myeloid cells) and IFNAR−/− mice, ZIKV-immune CD8+ T cells protect against infection through cytotoxic, polyfunctional cellular responses (46, 86). However, in some instances, the resulting cytotoxicity may damage the host, in a tissue specific manner. For example, in IFNAR−/− mice ZIKV infection of astrocytes results in a breakdown of the blood-brain barrier, allowing an influx of CD8+ T cells into the central nervous system (CNS) where they mediate apoptosis of ZIKV-infected neurons, but also results in severe neuropathology (87). Similarly, CD8+ cellular infiltration was also found in the CNS following ZIKV infection in C57/BL6 neonatal mice who developed hind limb collapse, cerebellar degeneration (88) and in the case of adult wildtype C57BL/6 mice, encephalitis (89). Whilst the CD8+ T cell response may be detrimental in the CNS, in IFNAR−/− pregnant mice cross-reactive DENV-specific CD8+ cells are protective against ZIKV infection of the fetus, including the fetal central nervous system, and are associated with increased fetal growth and viability (90). The CNS may represent a special case, where infection in the absence of CD8+ T cells results in severe viral pathology, and in the presence of CD8+ T cells in immunopathology, with little difference in survival in either case, as is seen in Japanese encephalitis (91).
Responses to sequential DENV-ZIKV infection (summarized in Figure 6) share similarities with secondary heterotypic DENV infection. Firstly, DENV-immune CD8+ T cell responses—either from prior exposure, peptide immunization or transfer of DENV-immune CD8+ T cells—can protect against ZIKV infection (90, 92, 93). This is an important result, which corroborates human studies that demonstrate prior DENV immunity can reduce the risk of Zika infection (16). Likewise, prior exposure to DENV provided IFNAR−/− mice protection against maternal and fetal ZIKV infection as compared with non-immune controls (90). As in heterotypic DENV, in mice as well as humans—the immunodominance pattern of the CD8+ T cell response to ZIKV infection was altered by prior DENV immunity and focused on conserved cross-reactive epitopes (93). ZIKV/DENV cross-reactive T cells performed comparably to ZIKV-specific T cells in viral load reduction in the serum and brain of knockout mice, which have key implications for ZIKV vaccine development.
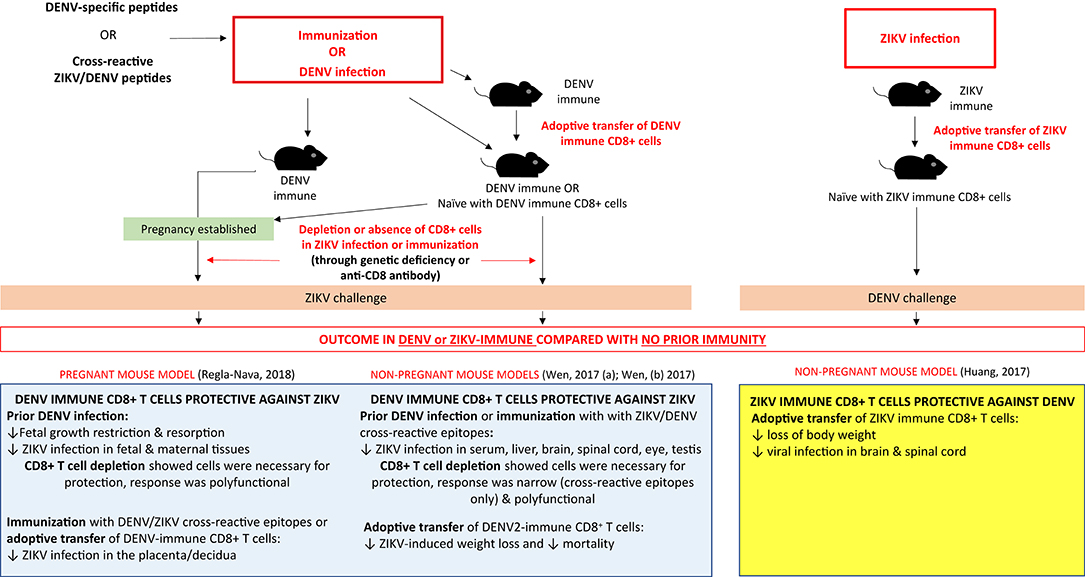
Figure 6. The CD8+ T cell response and impact of prior flavivirus immunity in secondary flavivirus infection in mice. Summary of experiments (infections, immunizations, adoptive cell transfer, and depletions) demonstrating the protective effect of prior DENV immunity on ZIKV challenge and prior ZIKV immunity on DENV challenge in pregnant and non-pregnant mouse models.
Contrary to mice, non-human primates (NHP) are natural hosts for DENV and ZIKV. Several different primate models have been employed, including rhesus and cynomolgus macaques. In line with epidemiological observations in humans, experiments in NHP demonstrate that dengue immunity may curb Zika replication and potentially symptoms (94). Performing the order of viral challenge the other way around gave a similar result, where macaques with prior ZIKV immunity mounted strong humoral and cellular responses against DENV (95). Furthermore, this model found that a longer convalescence between ZIKV and DENV challenge was associated with higher and more durable antibody and T cell responses suggesting that ZIKV immune memory can contribute to protection against DENV (95).
Dengue and ZIKA Vaccines in Development
There are currently three dengue vaccines that have either been tested in, or are currently in, phase III trials. At the time of writing the only licensed dengue vaccine is Sanofi-Pasteur's Dengvaxia® (CYD-TDV), a live-attenuated, chimeric, tetravalent vaccine, in which the genes encoding prM and E of the four DENV serotypes have been inserted into YFV-17D (Figure 7) (96). The vaccine was developed to produce DENV neutralizing antibodies in human subjects, but protection against disease is incomplete despite high levels of seroconversion (97, 98), and one trial found no efficacy at all against DENV2 (99). During longer follow up of clinical trial participants, it was observed that young children in the vaccinated group had excess hospital admissions due to dengue compared with the placebo group (100). Dengvaxia is most effective in individuals who are DENV-seropositive at the time of immunization, while in seronegative subjects the vaccine is not protective and increases the risk of severe disease (101). One hypothesis for failure to protect DENV-naïve subjects may be that the T cell response generated was directed against the yellow fever NS proteins present within the vaccine, rather than DENV NS proteins (68, 102). These observations suggest the need to determine the optimal T cell antigens and incorporate them into new vaccines. The other two dengue vaccines in development are based on full length DENV. One [Takeda, tetravalent dengue vaccine (TAK-003)] uses an attenuated strain of DENV2, with the prM and E genes of the other serotypes inserted into it (103). Regardless of dengue serostatus, TAK-003 elicited strong humoral responses against all four DENV serotypes (104), and generated a polyfunctional CD8+ T cell response to the non-structural proteins of DENV2, which cross-reacted against DENV1, DENV3, and DENV4 (105). Preliminary findings of a phase III trial show TAK-003 to have ~81% efficacy against symptomatic dengue (106), though protection against DENV3 was slightly lower. Therefore, a vaccine that induces better—and potentially more cross-reactive T cell responses also seems to have higher efficacy. However, these vaccines have not been compared directly and TAK-003 may still enhance disease in DENV naïve people, and could protect through a mechanism not involving CD8+ T cells. The other dengue vaccine in phase III trials is TV003, which is a tetravalent formulation of DENV1-3 with an additional chimeric DENV4 with the DENV2 prM and E genes inserted (Figure 7). Administration of TV003 induced a T cell response which predominantly targeted conserved epitopes of NS3 and NS5 (55) and was found to be immunogenic in subjects with prior flavivirus exposure (107). Field efficacy data are not yet available for TV003, but the vaccine protects against rash and viremia in a dengue human challenge model (108).
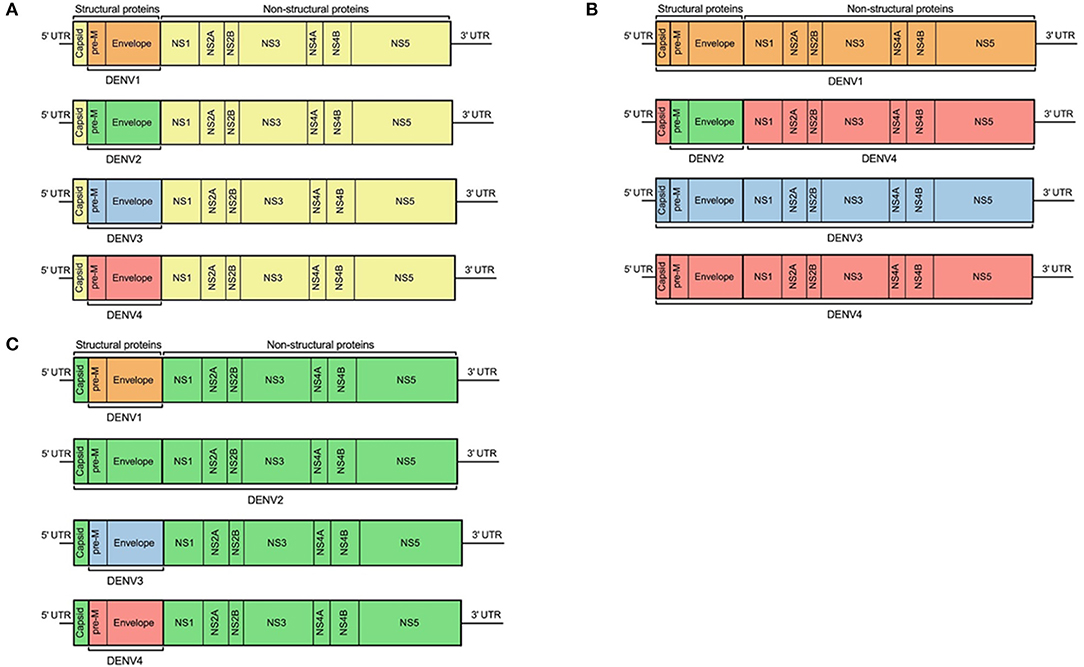
Figure 7. Design schematic of three dengue vaccines. (A) represents Sanofi's licensed vaccine, Dengvaxia®, (B) represents the NIH live attenuated tetravalent dengue vaccine candidate (TV003), and (C) represents Takeda's tetravalent dengue vaccine (TDV). Each color represents regions from different flaviviruses: yellow, YFV; orange, DENV1; green, DENV2; blue, DENV3; and red, DENV4.
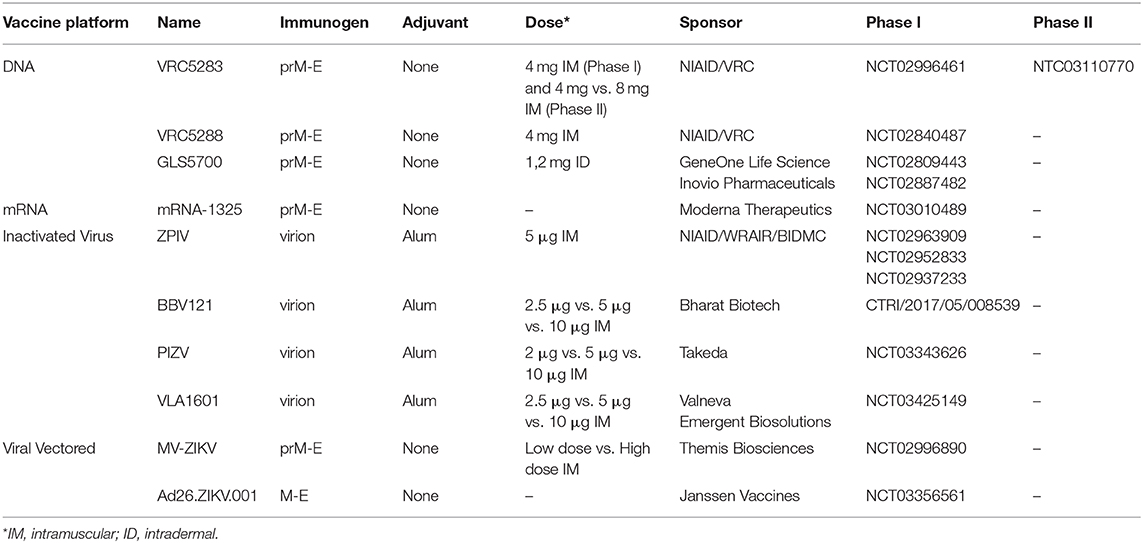
The majority of Zika vaccines are still in phase I/II trials (Table 2). These vaccines include DNA/mRNA, purified inactivated ZIKV, and recombinant virus-vectored vaccines; with most vaccine constructs containing the prM and E antigens as the main immunogen, as these proteins are targets for neutralizing antibodies (109). Three DNA-based vaccines (GLS-5700, VRC5283, and VRC5288) that have entered human testing show promising results, with one, VRC5283 advancing into phase II. VRC5283 and VRC5288 are both chimeric vaccines that utilize a JE virus (JEV) prM signal sequence followed by either the full-length E protein from wildtype ZIKV (VRC5283) or a modified E region in which the terminal 98 amino acids are exchanged with the analogous JEV sequence (VRC5288) (110). Both vaccines were shown to be immunogenic in mouse and NHP models (110) and a phase I trial found that vaccination with VRC5283 elicited neutralizing antibodies and cellular responses in all of the participants (111). A randomized placebo-controlled phase II study of VRC5283 is currently underway. A limitation of VRC5283 is whether the incorporation of a sequence from a different flavivirus (JEV prM) will provide protection against congenital Zika infection (111). To address this point, the vaccine was tested in a non-human primate pregnancy model. Vaccinated animals displayed fewer fetal losses and had reduced placental and fetal pathology; vaccine protection correlated with serum neutralizing antibody and antiviral T cell responses (112). However, these results may have to be considered cautiously as the model does not reflect early gestational exposure to ZIKV (112).
In addition to DNA-based vaccines, a ZIKV purified inactivated vaccine (ZPIV) based on the Puerto Rican strain PRVABC59 was found to be immunogenic at phase I with an acceptable safety profile (113). Vaccination with ZPIV elicited neutralizing antibody responses in the majority of tested individuals (113). One vaccine recipient from the trial produced cross-neutralizing antibodies to both ZIKV and DENV; responses which were linked to the individual's prior flavivirus exposure (114). These antibodies were shown to target the E domain I/III linker and could protect IFNAR−/− mice challenged either with ZIKV or DENV-2 (114). The durability of these protective responses were only evaluated up to 8 weeks post-vaccination (114) and longer follow up is still needed to fully demonstrate the longevity of these responses.
Enhanced immunogenicity associated with viral vectors (115) make these attractive candidates for Zika antigen delivery. Of the vaccines in clinical testing, two use viral vectors: one an attenuated measles strain (116) and the other a replication-incompetent human adenovirus serotype 26 (Ad26) (117). A single immunization with the Ad26 construct containing the Zika prM and E (Ad26.ZIKV.M-Env) antigens was able to elicit protective humoral and cellular responses in mice and NHP (117). Ad26.ZIKV.M-Env also protected IFNAR−/− dams and fetuses from ZIKV in a pregnancy model (118).
As discussed previously, all of vaccine constructs in clinical testing target structural proteins and may therefore not be optimal in their ability to induce cellular responses or boost pre-existing responses. Therefore, vaccines that include non-structural proteins should be considered, given that vaccination with non-structural proteins can induce cytotoxic T cell and polyfunctional helper T cell responses (119). Furthermore, given that non-structural proteins can elicit cross-reactive T cell responses, vaccines that incorporate these proteins may provide suitable priming for the development of memory responses during secondary flavivirus challenge (74, 94, 95, 109, 110, 115, 120–140).
Conclusion
There still remains a need to develop Zika vaccines, and also to better understand the cross-reactivity of flavivirus immune responses so that this can be harnessed for the emergent flaviviruses of the future. Dengue infection appears to be protective against ZIKV through mechanisms mediated by cross-reactive T cell responses against NS proteins. Given that in general, non-structural proteins elicit the most cross-reactive responses, and that these responses are likely to be protective against other viruses beyond dengue, there is a strong argument for including non-structural proteins to act as CD8 T cell antigens in novel flavivirus vaccines. As the majority of the population in flavivirus endemic areas are repeatedly exposed, vaccines incorporating non-structural antigens may be more efficient and may require fewer doses due to the constant boosting of an existing T cell response. Finally, the degree of cross-reactivity seen with human CD8+ T cell responses to flaviviruses raises the possibility of engineering a single component containing T cell antigens that could be used in multiple vaccines, or even in a multivalent vaccine, provided suitable B cell antigens can be found.
Author Contributions
KS contributed to the conceptualization of the article, wrote the first draft, and reviewed the manuscript. SL and LG contributed to conceptualization of the article and wrote the first draft of sections of the article. AG contributed viral homology measures and also to conceptualization of the article. DW and LT contributed to conceptualization of the article, contributed to early drafts, and reviewed the article. All authors reviewed the final draft.
Funding
This article is an independent research funded by the National Institute for Health Research (NIHR, www.nihr.ac.uk) Health Protection Research Unit in Emerging and Zoonotic Infections (grant No. IS-HPU-1112-10117), Global Health Research Group on Brain Infections (No. 17/63/110) and National Institutes of Health contracts HHSN27220140045C and HHSN75N9301900065. LT is a Wellcome clinical career development fellow, supported by grant number 205228/Z/16/Z. SL, LT, and DW were supported by the European Union Zika Preparedness Latin American Network consortium (ZikaPLAN). ZikaPLAN has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no 734584.
Disclaimer
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or Public Health England. This work was conducted independently of influence from the NIHR.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. (2012) 6:e1760. doi: 10.1371/journal.pntd.0001760
2. Brady OJ, Hay SI. The first local cases of Zika virus in Europe. Lancet. (2019) 394:1991–2. doi: 10.1016/S0140-6736(19)32790-4
3. Reinhold JM, Lazzari CR, Lahondere C. Effects of the environmental temperature on aedes aegypti and aedes albopictus mosquitoes: a review. Insects. (2018) 9:E158. doi: 10.3390/insects9040158
4. Hasan S, Jamdar SF, Alalowi M, Al Ageel Al Beaiji SM. Dengue virus: a global human threat: review of literature. J Int Soc Prev Community Dent. (2016) 6:1–6. doi: 10.4103/2231-0762.175416
5. Reid S, Rimmer K, Thakur K. Zika virus and neurologic disease. Neurol Clin. (2018) 36:767–87. doi: 10.1016/j.ncl.2018.06.003
6. Lima GP, Rozenbaum D, Pimentel C, Frota ACC, Vivacqua D, Machado ES, et al. Factors associated with the development of Congenital Zika Syndrome: a case-control study. BMC Infect Dis. (2019) 19:277. doi: 10.1186/s12879-019-3908-4
7. Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol. (2011) 9:532–41. doi: 10.1038/nrmicro2595
8. Chen R, Vasilakis N. Dengue–quo tu et quo vadis? Viruses. (2011) 3:1562–608. doi: 10.3390/v3091562
9. Waggoner JJ, Balmaseda A, Gresh L, Sahoo MK, Montoya M, Wang C, et al. Homotypic dengue virus reinfections in nicaraguan children. J Infect Dis. (2016) 214:986–93. doi: 10.1093/infdis/jiw099
10. Smith SA, de Alwis AR, Kose N, Harris E, Ibarra KD, Kahle KM, et al. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. MBio. (2013) 4:e00873–13. doi: 10.1128/mBio.00873-13
12. Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science. (2017) 358:929–32. doi: 10.1126/science.aan6836
13. Flipse J, Smit JM. The complexity of a dengue vaccine: a review of the human antibody response. PLoS Negl Trop Dis. (2015) 9:e0003749. doi: 10.1371/journal.pntd.0003749
14. Poland GA, Kennedy RB, Ovsyannikova IG, Palacios R, Ho PL, Kalil J. Development of vaccines against Zika virus. Lancet Infect Dis. (2018) 18:e211–9. doi: 10.1016/S1473-3099(18)30063-X
15. Gunawardana SA, Shaw RH. Cross-reactive dengue virus-derived monoclonal antibodies to Zika virus envelope protein: panacea or pandora's box? BMC Infect Dis. (2018) 18:641. doi: 10.1186/s12879-018-3572-0
16. Gordon A, Gresh L, Ojeda S, Katzelnick LC, Sanchez N, Mercado JC, et al. Prior dengue virus infection and risk of Zika: a pediatric cohort in Nicaragua. PLoS Med. (2019) 16:e1002726. doi: 10.1371/journal.pmed.1002726
17. Rodriguez-Barraquer I, Costa F, Nascimento EJM, Nery NJ, Castanha PMS, Sacramento GA, et al. Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science. (2019) 363:607–10. doi: 10.1126/science.aav6618
18. Pedroso C, Fischer C, Feldmann M, Sarno M, Luz E, Moreira-Soto A, et al. Cross-protection of dengue virus infection against congenital zika syndrome, Northeastern Brazil. Emerg Infect Dis. (2019) 25:1485–93. doi: 10.3201/eid2508.190113
19. Perera R, Kuhn RJ. Structural proteomics of dengue virus. Curr Opin Microbiol. (2008) 11:369–77. doi: 10.1016/j.mib.2008.06.004
20. Turtle L, Bali T, Buxton G, Chib S, Chan S, Soni M, et al. Human T cell responses to Japanese encephalitis virus in health and disease. J Exp Med. (2016) 213:1331–52. doi: 10.1084/jem.20151517
21. Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci USA. (2013) 110:e2046–53. doi: 10.1073/pnas.1305227110
22. Rivino L, Kumaran EA, Jovanovic V, Nadua K, Teo EW, Pang SW, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol. (2013) 87:2693–706. doi: 10.1128/JVI.02675-12
23. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. (2002) 108:717–25. doi: 10.1016/s0092-8674(02)00660-8
24. Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. (2015) 349:88–91. doi: 10.1126/science.aaa8651
25. Slon Campos JL, Mongkolsapaya J, Screaton GR. The immune response against flaviviruses. Nat Immunol. (2018) 19:1189–98. doi: 10.1038/s41590-018-0210-3
26. Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. (2005) 11:522–30. doi: 10.1038/nm1240
27. Fernandez E, Dejnirattisai W, Cao B, Scheaffer SM, Supasa P, Wongwiwat W, et al. Human antibodies to the dengue virus E-dimer epitope have therapeutic activity against Zika virus infection. Nat Immunol. (2017) 18:1261–09. doi: 10.1038/ni.3849
28. Culshaw A, Mongkolsapaya J, Screaton G. The immunology of Zika Virus. F1000Res. (2018) 7:203. doi: 10.12688/f1000research.12271.1
29. Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. (2010) 328:745–8. doi: 10.1126/science.1185181
30. Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. (2016) 353:823–6. doi: 10.1126/science.aaf8505
31. Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. (2010) 8:271–83. doi: 10.1016/j.chom.2010.08.007
32. Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, Crowe JE Jr. Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol. (2014) 88:12233–41. doi: 10.1128/JVI.00247-14
33. Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. (2015) 520:109–13. doi: 10.1038/nature14130
34. Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinsky A, et al. Corrigendum: a new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. (2015) 16:785. doi: 10.1038/ni0715-785a
35. Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. (2016) 536:48–53. doi: 10.1038/nature18938
36. Collins MH, McGowan E, Jadi R, Young E, Lopez CA, Baric RS, et al. Lack of durable cross-neutralizing antibodies against zika virus from dengue virus infection. Emerg Infect Dis. (2017) 23:773–81. doi: 10.3201/eid2305.161630
37. Sabin AB. Research on dengue during world war II. Am J Trop Med Hyg. (1952) 1:30–50. doi: 10.4269/ajtmh.1952.1.30
38. Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. (2016) 17:1102–8. doi: 10.1038/ni.3515
39. Halstead SB, Rojanasuphot S, Sangkawibha N. Origin0061l antigenic sin in dengue. Am J Trop Med Hyg. (1983) 32:154–6. doi: 10.4269/ajtmh.1983.32.154
40. Robbiani DF, Olsen PC, Costa F, Wang Q, Oliveira TY, Nery N Jr, et al. Risk of Zika microcephaly correlates with features of maternal antibodies. J Exp Med. (2019) 216:2302–15. doi: 10.1084/jem.20191061
41. Kurane I, Innis BL, Nisalak A, Hoke C, Nimmannitya S, Meager A, et al. Human T cell responses to dengue virus antigens. Proliferative responses and interferon gamma production. J Clin Invest. (1989) 83:506–13. doi: 10.1172/JCI113911
42. Kurane I, Meager A, Ennis FA. Dengue virus-specific human T cell clones. Serotype crossreactive proliferation, interferon gamma production, and cytotoxic activity. J Exp Med. (1989) 170:763–75. doi: 10.1084/jem.170.3.763
43. Bukowski JF, Kurane I, Lai CJ, Bray M, Falgout B, Ennis FA. Dengue virus-specific cross-reactive CD8+ human cytotoxic T lymphocytes. J Virol. (1989) 63:5086–91. doi: 10.1128/JVI.63.12.5086-5091.1989
44. Shrestha B, Diamond MS. Role of CD8+ T cells in control of West Nile virus infection. J Virol. (2004) 78:8312–21. doi: 10.1128/JVI.78.15.8312-8321.2004
45. St John AL, Rathore APS. Adaptive immune responses to primary and secondary dengue virus infections. Nat Rev Immunol. (2019) 19:218–30. doi: 10.1038/s41577-019-0123-x
46. Elong Ngono A, Vizcarra EA, Tang WW, Sheets N, Joo Y, Kim K, et al. Mapping and role of the CD8(+) T cell response during primary zika virus infection in mice. Cell Host Microbe. (2017) 21:35–46. doi: 10.1016/j.chom.2016.12.010
47. Zivny J, Kurane I, Leporati AM, Ibe M, Takiguchi M, Zeng LL, et al. A single nine-amino acid peptide induces virus-specific, CD8+ human cytotoxic T lymphocyte clones of heterogeneous serotype specificities. J Exp Med. (1995) 182:853–63. doi: 10.1084/jem.182.3.853
48. Livingston PG, Kurane I, Dai LC, Okamoto Y, Lai CJ, Men R, et al. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J Immunol. (1995) 154:1287–95.
49. Mathew A, Kurane I, Green S, Stephens HA, Vaughn DW, Kalayanarooj S, et al. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J Virol. (1998) 72:3999–4004. doi: 10.1128/JVI.72.5.3999-4004.1998
50. Friberg H, Burns L, Woda M, Kalayanarooj S, Endy TP, Stephens HA, et al. Memory CD8+ T cells from naturally acquired primary dengue virus infection are highly cross-reactive. Immunol Cell Biol. (2011) 89:122–9. doi: 10.1038/icb.2010.61
51. Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. (2003) 9:921–7. doi: 10.1038/nm887
52. Dong T, Moran E, Vinh Chau N, Simmons C, Luhn K, Peng Y, et al. High pro-inflammatory cytokine secretion and loss of high avidity cross-reactive cytotoxic T-cells during the course of secondary dengue virus infection. PLoS ONE. (2007) 2:e1192. doi: 10.1371/journal.pone.0001192
53. Imrie A, Meeks J, Gurary A, Sukhbataar M, Kitsutani P, Effler P, et al. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J Virol. (2007) 81:10081–91. doi: 10.1128/JVI.00330-07
54. Grifoni A, Pham J, Sidney J, O'Rourke PH, Paul S, Peters B, et al. Prior Dengue virus exposure shapes T cell immunity to Zika virus in humans. J Virol. (2017)91:e01469–17. doi: 10.1128/JVI.01469-17
55. Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, et al. Human CD8+ T-cell responses against the 4 dengue virus serotypes are associated with distinct patterns of protein targets. J Infect Dis. (2015) 212:1743–51. doi: 10.1093/infdis/jiv289
56. Weiskopf D, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, et al. The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes. J Virol. (2015) 89:120–8. doi: 10.1128/JVI.02129-14
57. Delgado FG, Torres KI, Castellanos JE, Romero-Sanchez C, Simon-Loriere E, Sakuntabhai A, et al. Improved immune responses against zika virus after sequential dengue and zika virus infection in humans. Viruses. (2018) 10:E480. doi: 10.3390/v10090480
58. Zivny J, DeFronzo M, Jarry W, Jameson J, Cruz J, Ennis FA, et al. Partial agonist effect influences the CTL response to a heterologous dengue virus serotype. J Immunol. (1999) 163:2754–60.
59. Appanna R, Huat TL, See LL, Tan PL, Vadivelu J, Devi S. Cross-reactive T-cell responses to the nonstructural regions of dengue viruses among dengue fever and dengue hemorrhagic fever patients in Malaysia. Clin Vaccine Immunol. (2007) 14:969–77. doi: 10.1128/CVI.00069-07
60. Duangchinda T, Dejnirattisai W, Vasanawathana S, Limpitikul W, Tangthawornchaikul N, Malasit P, et al. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc Natl Acad Sci USA. (2010) 107:16922–7. doi: 10.1073/pnas.1010867107
61. Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. (2003) 281:65–78. doi: 10.1016/S0022-1759(03)00265-5
62. Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. (2006) 176:3821–9. doi: 10.4049/jimmunol.176.6.3821
63. Dung NT, Duyen HT, Thuy NT, Ngoc TV, Chau NV, Hien TT, et al. Timing of CD8+ T cell responses in relation to commencement of capillary leakage in children with dengue. J Immunol. (2010) 184:7281–7. doi: 10.4049/jimmunol.0903262
64. Simmons CP, Dong T, Chau NV, Dung NT, Chau TN, Thao le TT, et al. Early T-cell responses to dengue virus epitopes in vietnamese adults with secondary dengue virus infections. J Virol. (2005) 79:5665–75. doi: 10.1128/JVI.79.9.5665-5675.2005
65. Wijeratne DT, Fernando S, Gomes L, Jeewandara C, Ginneliya A, Samarasekara S, et al. Quantification of dengue virus specific T cell responses and correlation with viral load and clinical disease severity in acute dengue infection. PLoS Negl Trop Dis. (2018) 12:e0006540. doi: 10.1371/journal.pntd.0006540
66. Hatch S, Endy TP, Thomas S, Mathew A, Potts J, Pazoles P, et al. Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J Infect Dis. (2011) 203:1282–91. doi: 10.1093/infdis/jir012
67. Loke H, Bethell DB, Phuong CX, Dung M, Schneider J, White NJ, et al. Strong HLA class I–restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J Infect Dis. (2001) 184:1369–73. doi: 10.1086/324320
68. Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol. (2015) 15:745–59. doi: 10.1038/nri3916
69. Herrera BB, Tsai WY, Chang CA, Hamel DJ, Wang WK, Lu Y, et al. Sustained Specific and cross-reactive T cell responses to zika and dengue virus NS3 in West Africa. J Virol. (2018) 92:e01992–17. doi: 10.1128/JVI.01992-17
70. Lim MQ, Kumaran EAP, Tan HC, Lye DC, Leo YS, Ooi EE, et al. Cross-Reactivity and anti-viral function of dengue capsid and NS3-specific memory T cells toward zika virus. Front Immunol. (2018) 9:2225. doi: 10.3389/fimmu.2018.02225
71. Grifoni A, Costa-Ramos P, Pham J, Tian Y, Rosales SL, Seumois G, et al. Cutting edge: transcriptional profiling reveals multifunctional and cytotoxic antiviral responses of zika virus-specific CD8(+) T cells. J Immunol. (2018) 201:3487–91. doi: 10.4049/jimmunol.1801090
72. Grossman RA, Edelman R, Gould DJ. Study of Japanese encephalitis virus in Chiangmia Valley, Thailand. VI. Summary and conclusions. Am J Epidemiol. (1974) 100:69–76. doi: 10.1093/oxfordjournals.aje.a112010
73. Libraty DH, Nisalak A, Endy TP, Suntayakorn S, Vaughn DW, Innis BL. Clinical and immunological risk factors for severe disease in Japanese encephalitis. Trans R Soc Trop Med Hyg. (2002) 96:173–8. doi: 10.1016/S0035-9203(02)90294-4
74. Alves Dos Santos E, Fink K. Animal models for Dengue and Zika vaccine development. Adv Exp Med Biol. (2018) 1062:215–39. doi: 10.1007/978-981-10-8727-1_16
75. Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, et al. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J Immunol. (2011) 187:4268–79. doi: 10.4049/jimmunol.1101970
76. Weiskopf D, Angelo MA, Sidney J, Peters B, Shresta S, Sette A. Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J Virol. (2014) 88:11383–94. doi: 10.1128/JVI.01108-14
77. Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, et al. A protective role for dengue virus-specific CD8+ T cells. J Immunol. (2009) 182:4865–73. doi: 10.4049/jimmunol.0801974
78. An J, Zhou DS, Zhang JL, Morida H, Wang JL, Yasui K. Dengue-specific CD8+ T cells have both protective and pathogenic roles in dengue virus infection. Immunol Lett. (2004) 95:167–74. doi: 10.1016/j.imlet.2004.07.006
79. Amorim JH, dos Santos Alves RP, Bizerra R, Araujo Pereira S, Ramos Pereira L, Nascimento Fabris DL, et al. Antibodies are not required to a protective immune response against dengue virus elicited in a mouse encephalitis model. Virology. (2016) 487:41–9. doi: 10.1016/j.virol.2015.10.006
80. Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, et al. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol. (2010) 185:5405–16. doi: 10.4049/jimmunol.1001709
81. Zellweger RM, Tang WW, Eddy WE, King K, Sanchez MC, Shresta S. CD8+ T cells can mediate short-term protection against heterotypic dengue virus reinfection in mice. J Virol. (2015) 89:6494–505. doi: 10.1128/JVI.00036-15
82. Zompi S, Santich BH, Beatty PR, Harris E. Protection from secondary dengue virus infection in a mouse model reveals the role of serotype cross-reactive B and T cells. J Immunol. (2012) 188:404–16. doi: 10.4049/jimmunol.1102124
83. Talarico LB, Batalle JP, Byrne AB, Brahamian JM, Ferretti A, Garcia AG, et al. The role of heterotypic DENV-specific CD8(+)T lymphocytes in an immunocompetent mouse model of secondary dengue virus infection. EBioMedicine. (2017) 20:202–16. doi: 10.1016/j.ebiom.2017.04.033
84. Roth C, Delgado FG, Simon-Loriere E, Sakuntabhai A. Immune responses to dengue and zika viruses-guidance for T cell vaccine development. Int J Environ Res Public Health. (2018) 15:E385. doi: 10.3390/ijerph15020385
85. Huang H, Li S, Zhang Y, Han X, Jia B, Liu H, et al. CD8(+) T cell immune response in immunocompetent mice during zika virus infection. J Virol. (2017) 91:e00900–17. doi: 10.1128/JVI.00900-17
86. Hassert M, Brien JD, Pinto AK. Mouse models of heterologous flavivirus immunity: a role for cross-reactive T cells. Front Immunol. (2019) 10:1045. doi: 10.3389/fimmu.2019.01045
87. Jurado KA, Yockey LJ, Wong PW, Lee S, Huttner AJ, Iwasaki A. Antiviral CD8 T cells induce Zika-virus-associated paralysis in mice. Nat Microbiol. (2018) 3:141–7. doi: 10.1038/s41564-017-0060-z
88. Manangeeswaran M, Ireland DD, Verthelyi D. Zika (PRVABC59) infection is associated with T cell infiltration and neurodegeneration in CNS of immunocompetent neonatal C57Bl/6 mice. PLoS Pathog. (2016) 12:e1006004. doi: 10.1371/journal.ppat.1006004
89. Hayashida E, Ling ZL, Ashhurst TM, Viengkhou B, Jung SR, Songkhunawej P, et al. Zika virus encephalitis in immunocompetent mice is dominated by innate immune cells and does not require T or B cells. J Neuroinflammation. (2019) 16:177. doi: 10.1186/s12974-019-1566-5
90. Regla-Nava JA, Elong Ngono A, Viramontes KM, Huynh AT, Wang YT, Nguyen AT, et al. cross-reactive dengue virus-specific CD8(+) T cells protect against Zika virus during pregnancy. Nat Commun. (2018) 9:3042. doi: 10.1038/s41467-018-05458-0
91. Larena M, Regner M, Lee E, Lobigs M. Pivotal role of antibody and subsidiary contribution of CD8+ T cells to recovery from infection in a murine model of Japanese encephalitis. J Virol. (2011) 85:5446–55. doi: 10.1128/JVI.02611-10
92. Wen J, Elong Ngono A, Regla-Nava JA, Kim K, Gorman MJ, Diamond MS, et al. Dengue virus-reactive CD8(+) T cells mediate cross-protection against subsequent Zika virus challenge. Nat Commun. (2017) 8:1459. doi: 10.1038/s41467-017-01669-z
93. Wen J, Tang WW, Sheets N, Ellison J, Sette A, Kim K, et al. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8(+) T cells. Nat Microbiol. (2017) 2:17036. doi: 10.1038/nmicrobiol.2017.36
94. Pantoja P, Perez-Guzman EX, Rodriguez IV, White LJ, Gonzalez O, Serrano C, et al. Zika virus pathogenesis in rhesus macaques is unaffected by pre-existing immunity to dengue virus. Nat Commun. (2017) 8:15674. doi: 10.1038/ncomms15674
95. Perez-Guzman EX, Pantoja P, Serrano-Collazo C, Hassert MA, Ortiz-Rosa A, Rodriguez IV, et al. Time elapsed between Zika and dengue virus infections affects antibody and T cell responses. Nat Commun. (2019) 10:4316. doi: 10.1038/s41467-019-12295-2
96. Prompetchara E, Ketloy C, Thomas SJ, Ruxrungtham K. Dengue vaccine: global development update. Asian Pac J Allergy Immunol. (2019). doi: 10.12932/AP-100518-0309. [Epub ahead of print].
97. Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. (2014) 384:1358–65. doi: 10.1016/S0140-6736(14)61060-6
98. Villar L, Dayan GH, Arredondo-Garcia JL, Rivera DM, Cunha R, Deseda C, et al. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med. (2015) 372:113–23. doi: 10.1056/NEJMoa1411037
99. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. (2012) 380:1559–67. doi: 10.1016/S0140-6736(12)61428-7
100. Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, et al. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med. (2015) 373:1195–206. doi: 10.1056/NEJMoa1506223
101. Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, et al. Effect of dengue serostatus on dengue vaccine safety and efficacy. N Engl J Med. (2018) 379:327–40. doi: 10.1056/NEJMoa1800820
102. Halstead SB. Which dengue vaccine approach is the most promising, and should we be concerned about enhanced disease after vaccination? there is only one true winner. Cold Spring Harb Perspect Biol. (2018) 10:a030700. doi: 10.1101/cshperspect.a030700
103. Osorio JE, Wallace D, Stinchcomb DT. A recombinant, chimeric tetravalent dengue vaccine candidate based on a dengue virus serotype 2 backbone. Expert Rev Vaccines. (2016) 15:497–508. doi: 10.1586/14760584.2016.1128328
104. Saez-Llorens X, Tricou V, Yu D, Rivera L, Jimeno J, Villarreal AC, et al. Immunogenicity and safety of one versus two doses of tetravalent dengue vaccine in healthy children aged 2-17 years in Asia and Latin America: 18-month interim data from a phase 2, randomised, placebo-controlled study. Lancet Infect Dis. (2018) 18:162–70. doi: 10.1016/S1473-3099(17)30632-1
105. Chu H, George SL, Stinchcomb DT, Osorio JE, Partidos CD. CD8+ T-cell responses in flavivirus-naive individuals following immunization with a live-attenuated tetravalent dengue vaccine candidate. J Infect Dis. (2015) 212:1618–28. doi: 10.1093/infdis/jiv258
106. Biswal S, Reynales H, Saez-Llorens X, Lopez P, Borja-Tabora C, Kosalaraksa P, et al. Efficacy of a tetravalent dengue vaccine in healthy children and adolescents. N Engl J Med. (2019) 381:2009–19. doi: 10.1056/NEJMoa1903869
107. Whitehead SS, Durbin AP, Pierce KK, Elwood D, McElvany BD, Fraser EA, et al. In a randomized trial, the live attenuated tetravalent dengue vaccine TV003 is well-tolerated and highly immunogenic in subjects with flavivirus exposure prior to vaccination. PLoS Negl Trop Dis. (2017) 11:e0005584. doi: 10.1371/journal.pntd.0005584
108. Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med. (2016) 8:330ra36. doi: 10.1126/scitranslmed.aaf1517
109. Lin HH, Yip BS, Huang LM, Wu SC. Zika virus structural biology and progress in vaccine development. Biotechnol Adv. (2018) 36:47–53. doi: 10.1016/j.biotechadv.2017.09.004
110. Dowd KA, Ko SY, Morabito KM, Yang ES, Pelc RS, DeMaso CR, et al. Rapid development of a DNA vaccine for Zika virus. Science. (2016) 354:237–40. doi: 10.1126/science.aai9137
111. Gaudinski MR, Houser KV, Morabito KM, Hu Z, Yamshchikov G, Rothwell RS, et al. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet. (2018) 391:552–62. doi: 10.1016/S0140-6736(17)33105-7
112. Van Rompay KKA, Keesler RI, Ardeshir A, Watanabe J, Usachenko J, Singapuri A, et al. DNA vaccination before conception protects Zika virus-exposed pregnant macaques against prolonged viremia and improves fetal outcomes. Sci Transl Med. (2019) 11:eaay2736. doi: 10.1126/scitranslmed.aay2736
113. Modjarrad K, Lin L, George SL, Stephenson KE, Eckels KH, De La Barrera RA, et al. Preliminary aggregate safety and immunogenicity results from three trials of a purified inactivated Zika virus vaccine candidate: phase 1, randomised, double-blind, placebo-controlled clinical trials. Lancet. (2018) 391:563–71. doi: 10.1016/S0140-6736(17)33106-9
114. Dussupt V, Sankhala RS, Gromowski GD, Donofrio G, De La Barrera RA, Larocca RA, et al. Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor. Nat Med. (2020) 26:228–35. doi: 10.1038/s41591-019-0746-2
115. Garg H, Mehmetoglu-Gurbuz T, Joshi A. Recent advances in Zika virus vaccines. Viruses. (2018) 10:e631. doi: 10.3390/v10110631
116. Nurnberger C, Bodmer BS, Fiedler AH, Gabriel G, Muhlebach MD. A measles virus-based vaccine candidate mediates protection against zika virus in an allogeneic mouse pregnancy model. J Virol. (2019) 93:e01485–18. doi: 10.1128/JVI.01485-18
117. Cox F, van der Fits L, Abbink P, Larocca RA, van Huizen E, Saeland E, et al. Adenoviral vector type 26 encoding Zika virus (ZIKV) M-Env antigen induces humoral and cellular immune responses and protects mice and nonhuman primates against ZIKV challenge. PLoS ONE. (2018) 13:e0202820. doi: 10.1371/journal.pone.0202820
118. Larocca RA, Mendes EA, Abbink P, Peterson RL, Martinot AJ, Iampietro MJ, et al. Adenovirus vector-based vaccines confer maternal-fetal protection against Zika virus challenge in pregnant IFN-alphabetaR(-/-) mice. Cell Host Microbe. (2019) 26:591–600 e4. doi: 10.1016/j.chom.2019.10.001
119. Grubor-Bauk B, Wijesundara DK, Masavuli M, Abbink P, Peterson RL, Prow NA, et al. NS1 DNA vaccination protects against Zika infection through T cell-mediated immunity in immunocompetent mice. Sci Adv. (2019) 5:eaax2388. doi: 10.1126/sciadv.aax2388
120. Osorio JE, Brewoo JN, Silengo SJ, Arguello J, Moldovan IR, Tary-Lehmann M, et al. Efficacy of a tetravalent chimeric dengue vaccine (DENVax) in Cynomolgus macaques. Am J Trop Med Hyg. (2011) 84:978–87. doi: 10.4269/ajtmh.2011.10-0592
121. Sumathy K, Kulkarni B, Gondu RK, Ponnuru SK, Bonguram N, Eligeti R, et al. Protective efficacy of Zika vaccine in AG129 mouse model. Sci Rep. (2017) 7:46375. doi: 10.1038/srep46375
122. Shan C, Muruato AE, Nunes BTD, Luo H, Xie X, Medeiros DBA, et al. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat Med. (2017) 23:763–7. doi: 10.1038/nm.4322
123. Wang R, Liao X, Fan D, Wang L, Song J, Feng K, et al. Maternal immunization with a DNA vaccine candidate elicits specific passive protection against post-natal Zika virus infection in immunocompetent BALB/c mice. Vaccine. (2018) 36:3522–32. doi: 10.1016/j.vaccine.2018.04.051
124. Wang R, Gao N, Li Y, Fan D, Zhen Z, Feng K, et al. Cross-protection against four serotypes of dengue virus in mice conferred by a Zika DNA vaccine. Front Cell Infect Microbiol. (2019) 9:147. doi: 10.3389/fcimb.2019.00147
125. Griffin BD, Muthumani K, Warner BM, Majer A, Hagan M, Audet J, et al. DNA vaccination protects mice against Zika virus-induced damage to the testes. Nat Commun. (2017) 8:15743. doi: 10.1038/ncomms15743
126. Li A, Yu J, Lu M, Ma Y, Attia Z, Shan C, et al. A Zika virus vaccine expressing premembrane-envelope-NS1 polyprotein. Nat Commun. (2018) 9:3067. doi: 10.1038/s41467-018-05276-4
127. Brault AC, Domi A, McDonald EM, Talmi-Frank D, McCurley N, Basu R, et al. A Zika vaccine targeting NS1 protein protects immunocompetent adult mice in a lethal challenge model. Sci Rep. (2017) 7:14769. doi: 10.1038/s41598-017-15039-8
128. Sariol CA, White LJ. Utility, limitations, and future of non-human primates for dengue research and vaccine development. Front Immunol. (2014) 5:452. doi: 10.3389/fimmu.2014.00452
129. Ambuel Y, Young G, Brewoo JN, Paykel J, Weisgrau KL, Rakasz EG, et al. A rapid immunization strategy with a live-attenuated tetravalent dengue vaccine elicits protective neutralizing antibody responses in non-human primates. Front Immunol. (2014) 5:263. doi: 10.3389/fimmu.2014.00263
130. Sims S, Willberg C, Klenerman P. MHC-peptide tetramers for the analysis of antigen-specific T cells. Expert Rev Vaccines. (2010) 9:765–74. doi: 10.1586/erv.10.66
131. Kanthaswamy S, Ng J, Satkoski Trask J, George DA, Kou AJ, Hoffman LN, et al. The genetic composition of populations of cynomolgus macaques (Macaca fascicularis) used in biomedical research. J Med Primatol. (2013) 42:120–31. doi: 10.1111/jmp.12043
132. Osuna CE, Whitney JB. Nonhuman Primate Models of Zika Virus Infection, Immunity, and Therapeutic Development. J Infect Dis. (2017) 216:S928–34. doi: 10.1093/infdis/jix540
133. Dudley DM, Aliota MT, Mohr EL, Weiler AM, Lehrer-Brey G, Weisgrau KL, et al. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun. (2016) 7:12204. doi: 10.1038/ncomms12204
134. Abbink P, Larocca RA, De La Barrera RA, Bricault CA, Moseley ET, Boyd M, et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. (2016) 353:1129–32. doi: 10.1126/science.aah6157
135. Muthumani K, Griffin BD, Agarwal S, Kudchodkar SB, Reuschel EL, Choi H, et al. In vivo protection against ZIKV infection and pathogenesis through passive antibody transfer and active immunisation with a prMEnv DNA vaccine. NPJ Vaccines. (2016) 1:16021. doi: 10.1038/npjvaccines.2016.21
136. Brewoo JN, Kinney RM, Powell TD, Arguello JJ, Silengo SJ, Partidos CD, et al. Immunogenicity and efficacy of chimeric dengue vaccine (DENVax) formulations in interferon-deficient AG129 mice. Vaccine. (2012) 30:1513–20. doi: 10.1016/j.vaccine.2011.11.072
137. Zust R, Dong H, Li XF, Chang DC, Zhang B, Balakrishnan T, et al. Rational design of a live attenuated dengue vaccine: 2'-o-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog. (2013) 9:e1003521. doi: 10.1371/journal.ppat.1003521
138. Zellweger RM, Shresta S. Mouse models to study dengue virus immunology and pathogenesis. Front Immunol. (2014) 5:151. doi: 10.3389/fimmu.2014.00151
139. Bente DA, Melkus MW, Garcia JV, Rico-Hesse R. Dengue fever in humanized NOD/SCID mice. J Virol. (2005) 79:13797–9. doi: 10.1128/JVI.79.21.13797-13799.2005
Keywords: dengue, Zika, T-cells, cross-reactivity, vaccine, epitope, animal models
Citation: Subramaniam KS, Lant S, Goodwin L, Grifoni A, Weiskopf D and Turtle L (2020) Two Is Better Than One: Evidence for T-Cell Cross-Protection Between Dengue and Zika and Implications on Vaccine Design. Front. Immunol. 11:517. doi: 10.3389/fimmu.2020.00517
Received: 15 October 2019; Accepted: 06 March 2020;
Published: 25 March 2020.
Edited by:
Allan Randrup Thomsen, University of Copenhagen, DenmarkReviewed by:
James J. Kobie, University of Alabama at Birmingham, United StatesKatja Fink, HuMabs BioMed SA, Switzerland
Copyright © 2020 Subramaniam, Lant, Goodwin, Grifoni, Weiskopf and Turtle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lance Turtle, bGFuY2UudHVydGxlQGxpdmVycG9vbC5hYy51aw==
 Krishanthi S. Subramaniam
Krishanthi S. Subramaniam Suzannah Lant
Suzannah Lant Lynsey Goodwin
Lynsey Goodwin Alba Grifoni
Alba Grifoni Daniela Weiskopf
Daniela Weiskopf Lance Turtle
Lance Turtle