Corrigendum: Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases
- Department of Microbiology, Faculty of Medicine, Shimane University, Shimane, Japan
Epstein-Barr virus (EBV) is an oncogenic human herpes virus that was discovered in 1964. Viral non-coding RNAs, such as BamHI-A rightward fragment-derived microRNAs (BART miRNAs) or BamHI-H rightward fragment 1-derived miRNAs (BHRF1 miRNA) in EBV-infected cells have been recently reported. Host miRNAs are also upregulated upon EBV infection. Viral and host miRNAs are important in maintaining viral infection and evasion of host immunity. Although miRNAs in EBV-infected cells often promote cell proliferation by targeting apoptosis or cell cycle, this review focuses on the regulation of the recognition of the host immune system. This review firstly describes the location and organization of two clusters of viral miRNAs, then describes evasion from host immune surveillance systems by modulating viral gene expression or inhibiting innate and acquired immunity by viral miRNAs as well as host miRNAs. Another topic is the enigmatic depletion of viral miRNAs in several types of EBV-infected tumor cells. Finally, this review introduces the strong correlation of nasopharyngeal cancer cases with a newly identified single nucleotide polymorphism that enhances BART miRNA promoter activity.
Introduction
Epstein-Barr virus (EBV) is a double-stranded DNA virus that belongs to the Gammaherpesvirus subfamily and was discovered in a Burkitt's lymphoma (BL) cell (1). EBV primarily infects B cells via the high-affinity receptor CD21; it also infects CD21-negative T cells, natural killer (NK) cells, and epithelial cells using low-affinity receptors (2). EBV causes the primary acute disease “infectious mononucleosis” in adolescents (3). Following a primary infection in B lymphocytes or epithelial cells, EBV establishes a chronic infection known as latent infection.
The two infection cycles that enable successful propagation of the EBV progeny viruses are lytic and latent infection. During lytic infection, all the viral genes are expressed and the viral genome is rapidly replicated. In contrast, latent infection involves the restricted expression of a number of viral genes. Here, EBV evades host immune surveillance and the copy number of DNA in the viral daughter cells are maintained by synchronous duplication of viral and host genomes. A small subset of viral genes and microRNAs (miRNAs) expressed during the latent infection maintain viral episomes and stimulate host cell proliferation. EBV propagates viral genomes together with host cells during latent infection.
Host cell proliferation associated with latent EBV infection induces malignancies, such as BL, Hodgkin's lymphoma (HL), EBV-positive diffuse large B-cell lymphoma (DLBCL), extranodal NK/T-cell lymphoma-nasal type (ENKL), nasopharyngeal carcinoma (NPC), and EBV-associated gastric carcinoma. EBV also causes the severe infectious disease called chronic active EBV infection (3–6).
A miRNA is a non-coding single-stranded RNA comprising 20–22 bases that regulates post-transcriptional gene expression. More than 60% of protein-coding genes are regulated by miRNAs in mammals (7). miRNAs are present in both eukaryotic and viral genomes, such as the EBV genome (8). Viral miRNAs are incorporated into the RNA-induced silencing complex and this miRNA complex interacts with the 3′ untranslated region of host and viral mRNAs. This suppresses the expression of target gene(s) via translational repression or mRNA degradation (9). Viral miRNAs suppress target genes in the EBV and host genomes to maintain latent EBV infection, evade the host immune surveillance system, and promote tumorigenic growth of infected cells among other functions (10).
Here we discuss the role of EBV-encoded miRNAs in maintaining latent and lytic infection along with the function of host and viral miRNAs in regulating immune responses in EBV-associated diseases.
EBV-Encoded miRNAs (EBV miRNAs)
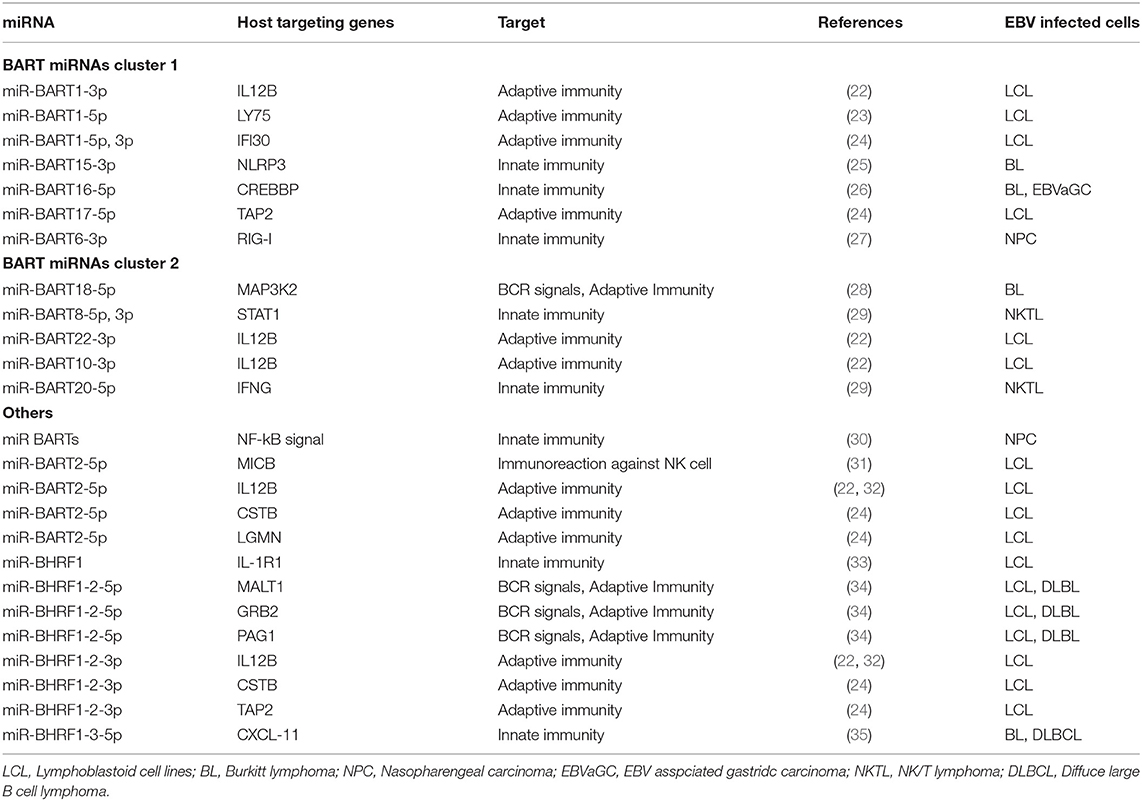
EBV-encoded BamH I-A rightward transcripts (BARTs) are alternatively spliced non-coding RNAs abundantly expressed during latent infection (11). B95-8 is a representative EBV strain with a deletion in a major portion of BART. This strain can transform B lymphocytes and produce progeny viral particles in abundance (12). Because previous EBV studies have mostly based on the in vitro immortalizing assay of primary B lymphocytes, the role of BART in the viral life cycle could only be studied after the discovery of BART miRNAs.
Wild-type EBV contains 44 BART miRNAs that are separated by an intron resulting in BART miRNA clusters 1 and 2 (13). Double-stranded RNAs transcribed from the EBV genome are processed by the host miRNA machinery to produce viral miRNAs (9). BART miRNA cluster 1 contains primary transcripts for eight miRNA (pri-miRNAs), namely pri-miR-BART1, 3–6, and 15–17. BART miRNA cluster 2 encodes 13 pri-miRNAs, including pri-miR-BART21, 18, 7, 8, 9, 22, 10, 11, 12, 19, 20, 13, and 14. The deletion in B95-8 encompasses pri-miR-BART15 to the 13 pri-miRNAs in cluster 2 (13) (Figure 1A).
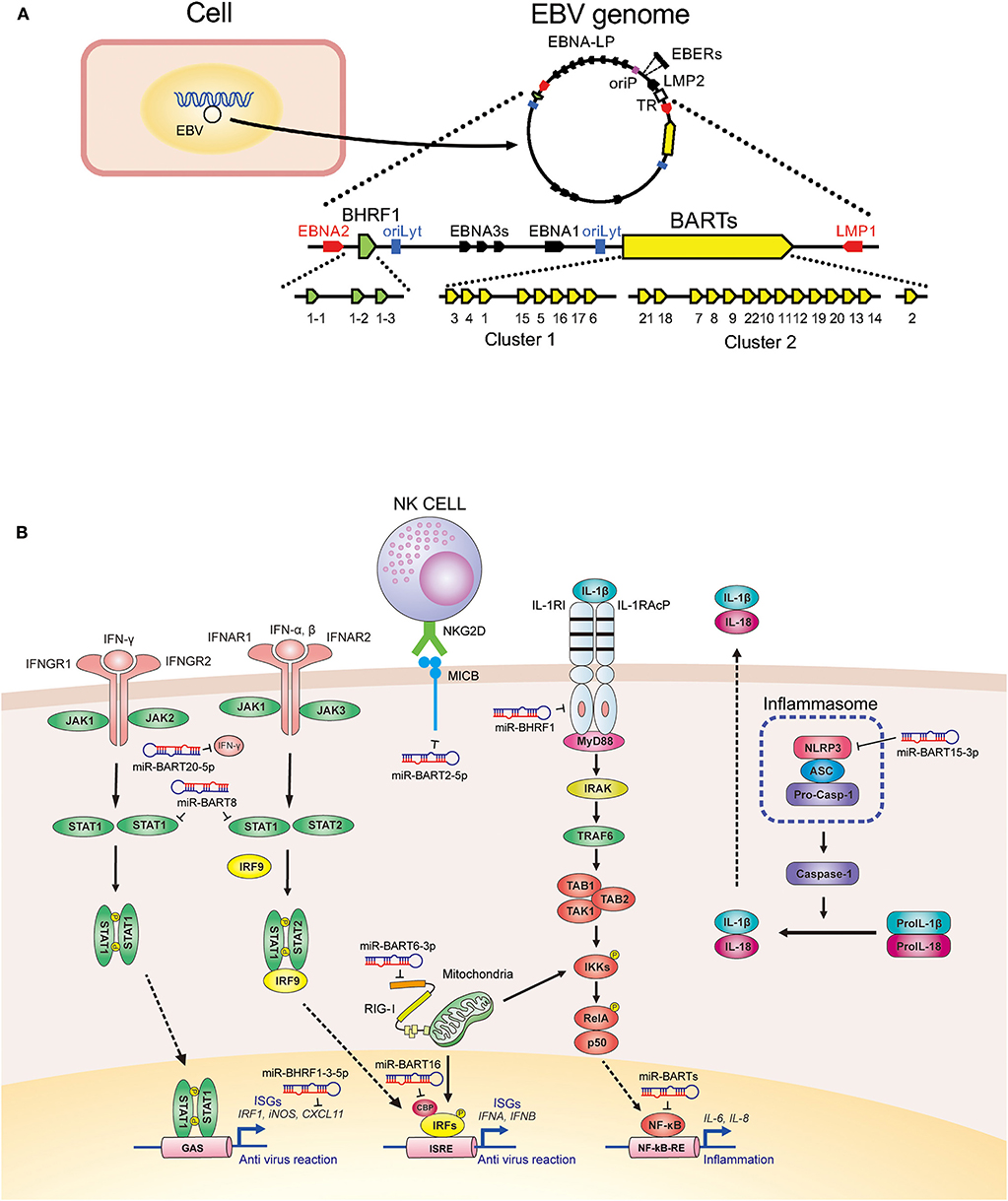
Figure 1. Expression and function of Epstein-Barr virus-derived microRNAs (EBV-derived miRNAs). (A) Genomic organization of EBV miRNAs. (B) Signaling cascades involved in EBV miRNA-mediated repression of host innate immunity.
The gene for BamH I-H right fragment 1 (BHRF1) encodes for three pri-miRNAs called pri-miR-BHRF1-1, -BHRF1-2, and -BHRF1-3. BHRF1 miRNAs are expressed during lytic infection, inhibit apoptosis, and favor proliferation of infected cells to enable the early phase of viral propagation (14) (Figure 1A).
Since viruses infect eukaryotic organisms to proliferate, viral miRNAs regulate host cell function and viral life cycle, including viral infection and development of viral progeny (9). EBV miRNAs are more strongly expressed in ENKL and NPC/EBV-associated gastric carcinoma as compared to B cell lymphomas (15).
Modulation of Viral Gene Expression by EBV miRNAs
EBV miRNAs regulate viral antigen expression (10). miR-BART1-5p, miR-BART16-5p, miR-BART17-5p, and miR-BART9-5p suppress the increase in expression of the highly immunogenic viral latent membrane protein 1 (LMP1) (16, 17). miR-BART22 inhibits the expression of the immunogenic latent membrane protein 2A (18). miR-BART20-5p represses the synthesis of viral transcription factors BamH I-Z leftward reading frame 1 (BZLF1) and BamH I-R leftward reading frame 1 that enable switching between latent and lytic EBV infection (19). miR-BART2-5p hinders the production of viral DNA polymerase BamH I-A leftward reading frame 5 during the latent phase to prevent the transition to lytic replication (13, 20). Therefore, viral strains deficient in all BART miRNAs cannot maintain latent infection since they strongly express BZLF1 that allows the switch to lytic replication (21).
Regulation of Host Immunity by EBV miRNAs
Suppression of Host Innate Immunity by EBV miRNAs
EBV miRNAs target viral and host genes involved in innate immunity (Figure 1B and Table 1) (10).
During lytic infection, miR-BHRF1-2-5p targets the 3′ untranslated region of the interleukin-1 receptor 1 (IL-1R1) and suppresses IL-1 signaling (33). miR-BHRF1-3-5p in EBV-infected B cells downregulates C-X-C motif chemokine 11 (CXCL-11) that is a downstream effector in interferon gamma (IFN-γ) signaling (35).
miR-BART6-3p targets the retinoic acid-inducible gene-I (RIG-I) (an intracellular receptor for double-stranded RNA), thereby suppressing host innate immune responses (27). miR-BART20-5p and miR-BART8 target IFN-γ and the signal transducer and activator of transcription 1 (STAT1), respectively, ultimately suppressing cellular immunity against tumor cells (29). miR-BART16 targets the cAMP response element-binding protein-binding protein (CBP) (a transcriptional coactivator for type I IFN signaling) in EBV-infected B cells and epithelial cells to inhibit IFN signaling (26). miR-BART15-3p targets the NLR family pyrin domain-containing protein 3 (NLRP3; a member of the inflammasome) and inhibits the synthesis of IL-1β and IL-18 (25, 36). miR-BART2-5p maintains tumor cell survival by downregulating the major histocompatibility complex (MHC) class I polypeptide-related sequence B (MICB) recognized by the natural killer group 2 member D receptor present on NK cells (31, 37).
The BART miRNA coding sequence from the Akata strain was inserted into the B95-8 strain to restore the deleted region (30). As compared to the parental B95-8 strain, the restored B95-8 strain showed a decrease in the activity of nuclear factor kappa light chain enhancer of activated B cells (NF-κB) (30).
Inhibition of Host Adaptive Immunity by EBV miRNAs
EBV miRNAs also suppress host adaptive immunity (Table 1) (10). BART miRNAs regulate adaptive immunity during latent and lytic infection. In comparison, BHRF1 miRNAs regulate adaptive immunity only during lytic infection. miR-BART1-3p, miR-BART2-5p, miR-BART10-3p, miR-BART22-3p, and miR-BHRF1-2-3p suppress the expression of IL-12B in infected cells. There is a significant decrease in the levels of IL-12 in EBV-infected B lymphocytes that impairs the differentiation of CD4+ T cells into T helper 1 (Th1) cells, thereby abrogating host immune response. Thus, there is a reduction in cytotoxic T cells specific for the EBV antigens (22, 32, 38).
miR-BHRF1-3-5p and miR-BART17-5p target transporter associated with antigen processing 2 (TAP2) that transports antigenic peptides to MHC class I molecules, thus, viral antigen presentation is impaired in CD8+ T cells (24). EBV miRNAs also target genes involved in antigen processing, such as cystatin-B (CSTB), asparagine endopeptidase (LGMN), and gamma-interferon-inducible lysosomal thiol reductase (IFI30). Thus, antigen presentation is reduced in EBV-infected cells. Similarly, immunodeficient mice transplanted with human hematopoietic stem cells and infected with EBV possess proliferating EBV-infected B lymphocytes owing to reduced immune recognition by the human CD8+ T cells (39).
The B cell receptor (BCR) that mediates adaptive immunity as well as lytic infection in EBV-infected B lymphocytes is inhibited by miR-BHRF1-2-5p and miR-BART2-5p (34). miR-BART18-5p targets mitogen-activated protein kinase kinase kinase 2 (MAP3K2) that is a downstream effector in BCR signaling (28). The miR-BHRF1 cluster is considered to suppress constitutive lytic infection and adaptive immunity.
Lymphocyte antigen 75 (LY75) is a membrane protein that is expressed on dendritic cells and induces differentiation of Th0 to Th1 cells. miR-BART1-5p (transferred by exosomes) targets LY75 in dendritic cells suppressing Th1 cell differentiation (23).
The roles of EBV miRNAs in suppressing innate and adaptive immunity has been summarized in Figure 1B.
Host miRNA-Mediated Evasion of the Immune System by EBV-Infected Cells
EBV exploits host miRNAs to escape from the immune system. EBNA2 is a viral protein that expressed during type III latency and upregulates miR-21, that subsequently downregulates myeloid differentiation factor 88 (MyD88) and IL-1 receptor-associated kinase 1 (IRAK1) (40). The miR-17-92 cluster, which is essential for the differentiation of immune cells, is highly expressed in EBV-positive tumors, such as NPC (41) and DLBCL (42). High expression of miR-17-92 in B cells, T cells, NK cells, macrophages, and dendritic cells is known to inhibit cellular differentiation and function (43).
In EBV-infected B lymphocytes, viral LMP1 activates NF-κB signaling and host miR-155. But miR-155 attenuates NF-κB signaling to stabilize persistent infection (44). The miR-155 also targets suppressor of cytokine signaling 1 (SOCS1), a suppressor of the JAK-STAT signal (45). Though miR-155 is upregulated, strong expression of SOCS1 can be observed in EBV-infected cells (46). Simultaneous upregulation of SOCS1 and miR-155 has become an important controversy for researchers who study herpesviruses (47). It might be possible that miR-155 may target another gene expressed higher than SOCS1 in NPC cells.
Depletion of Viral miRNAs in EBV-Associated Tumors
In EBV-infected epithelial tumor cells, BART miRNAs are highly expressed and help in evading immune recognition (10). However, the BART miRNA clusters are frequently depleted in virus causing chronic active EBV infection, ENKL, and DLBCL (48, 49). BART miRNA were found lacking in 71% of DLBCL cases (49). On the other hand, DLBCL patients with high BART miRNA expression in the blood showed worse prognosis than patients with low expression (50). Although high expression of BART miRNAs is possibly important for malignant transformation of lymphoma, it may be disadvantageous for lymphoma cells survival by escaping immune surveillance.
Similarly, LMP1 is expressed in all the early NPC tumor cells and contributes to pleiotropy in NPCs (51). However, once NF-κB signaling is sufficiently active in NPC tumor cells, LMP1 is frequently downregulated (52).
As mutations and/or promoter methylation accumulate in the host genome, the presence of the viral genome may no longer be required for the growth of the tumor cell. In such a situation, carrying large EBV genomes may be a burden for host cells; thus, cells harboring the defective, but oncogenic, EBV genome may proliferate faster than cells infected with EBV having the complete genome. Alternatively, the increased levels of BART miRNAs may repress the expression of genes important for survival of EBV-positive cells. Therefore, further investigation is necessary to discern the physiological significance of BART miRNAs in EBV-positive tumor cells.
Single Nucleotide Polymorphisms (SNPs) in the Viral miRNA Promoters
BART miRNAs are important in evading the immune system and inhibiting apoptosis. However, multiple BART miRNAs frequently target the same gene to induce a high level of repression (16, 17). This hinders the development of efficacious drugs that must target each BART miRNA in EBV-associated malignancies. Thus, blocking the BART miRNA promoters could be a better strategy to target all the necessary miRNAs (53, 54). We have recently reported a characteristic SNP in the promoter of BART that increases BART promoter activity. This SNP is frequently detected in EBV-associated NPC with an odds ratio of 5.7 (55). Therefore, studying the promoter of BART and the SNPs associated with it can help develop strong candidates that suppress BART transcription.
Conclusion
EBV uses miRNAs to switch between lytic and latent infection. This helps maintain EBV infection and evade recognition of EBV by the host immune system by reducing viral gene (antigenic) expression. EBV miRNAs also target and suppress genes involved with host immunity. This oncogenic virus also exploits miRNAs for malignant transformation. Exosomes secreted from EBV-infected B lymphocytes contain a large amount of host and viral miRNAs that are transferred to epithelial cells (56). Therefore, miRNAs derived from EBV-infected cells may affect infected and uninfected host cells. Finally, future research may help treat EBV-associated malignancies by developing anti-tumor drugs that inhibit BART promoter activity.
Author Contributions
HI wrote the manuscript. HK, AK, and YK prepared the table and figures. HY complied the manuscript.
Funding
This review was supported by KAKENHI (Grant-in-Aid for Scientific Research) from the Ministry of Education, Culture, Sports, Science and Technology (16H05843, 18K07147, 18K07148, and 18K15168), the Health and Labour Sciences Research Grant from the Ministry of Health, Labour and Welfare, Japan, and the Kobayashi Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Ms. Sayuri Hamada for preparing the figures.
References
1. Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet. (1964) 1:702–3. doi: 10.1016/S0140-6736(64)91524-7
2. van Zyl DG, Mautner J, Delecluse HJ. Progress in EBV vaccines. Front Oncol. (2019) 9:104. doi: 10.3389/fonc.2019.00104
3. Young LS, Murray PG. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene. (2003) 22:5108-21. doi: 10.1038/sj.onc.1206556
4. Harabuchi Y, Namanaka N, Kataura A, Imai S, Kinoshita T, Mizuno F, et al. Epstein-Barr virus in nasal T-cell lymphomas in patients with lethal midline granuloma. Lancet. (1990) 335:128-30. doi: 10.1016/0140-6736(90)90002-M
5. Burke AP, Yen TS, Shekitka KM, Sobin LH. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol. (1990) 3:377-80.
6. Kikuta H, Taguchi Y, Tomizawa K, Kojima K, Kawamura N, Ishizaka A, et al. Epstein-Barr virus genome-positive T lymphocytes in a boy with chronic active EBV infection associated with Kawasaki-like disease. Nature. (1988) 333:455-7. doi: 10.1038/333455a0
7. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92-105. doi: 10.1101/gr.082701.108
8. Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science. (2004) 30:734-6. doi: 10.1126/science.1096781
9. Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. (2009) 457:421-5. doi: 10.1038/nature07757
10. Kim H, Iizasa H, Kanehiro Y, Fekadu S, Yoshiyama H. Herpesviral microRNAs in cellular metabolism and immune responses. Front Microbiol. (2017) 8:1318. doi: 10.3389/fmicb.2017.01318
11. Edwards RH, Marquitz AR, Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol. (2008) 82:9094-106. doi: 10.1128/JVI.00785-08
12. Miller G, Robinson J, Heston L, Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci USA. (1974) 71:4006-10. doi: 10.1073/pnas.71.10.4006
13. Cai X, Schäfer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. (2006) 2:e23. doi: 10.1371/journal.ppat.0020023
14. Seto E, Moosmann A, Grömminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. (2010) 6:e1001063. doi: 10.1371/journal.ppat.1001063
15. Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J, et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. (2011) 7:e1002193. doi: 10.1371/journal.ppat.1002193
16. Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. (2007) 104:16164-9. doi: 10.1073/pnas.0702896104
17. Zhang Y, Zhang W, Liu W, Liu H, Zhang Y, Luo B. Epstein-Barr virus miRNA-BART16 modulates cell proliferation by targeting LMP1. Virus Res. (2018) 256:38-44. doi: 10.1016/j.virusres.2018.08.001
18. Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL, et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. (2009) 11:1174-84. doi: 10.1593/neo.09888
19. Jung YJ, Choi H, Kim H, Lee SK. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J Virol. (2014) 88:9027-37. doi: 10.1128/JVI.00721-14
20. Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. (2008) 36:666-75. doi: 10.1093/nar/gkm1080
21. Lin X, Tsai MH, Shumilov A, Poirey R, Bannert H, Middeldorp JM, et al. The Epstein-Barr virus BART miRNA cluster of the M81 strain modulates multiple functions in primary B cells. PLoS Pathog. (2015) 11:e1005344. doi: 10.1371/journal.ppat.1005344
22. Tagawa T, Albanese M, Bouvet M, Moosmann A, Mautner J, Heissmeyer V, et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J Exp Med. (2016) 213:2065-80. doi: 10.1084/jem.20160248
23. Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. (2012) 8:e1002484. doi: 10.1371/journal.ppat.1002484
24. Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J, et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc Natl Acad Sci USA. (2016) 113:E6467-75. doi: 10.1073/pnas.1605884113
25. Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, et al. miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J Immunol. (2012) 189:3795-9. doi: 10.4049/jimmunol.1200312
26. Ho oykaas MJG, van Gent M, Soppe JA, Kruse E, Boer IGJ, van Leenen D, et al. EBV microRNA BART16 suppresses type I IFN signaling. J Immunol. (2017) 198:4062-73. doi: 10.4049/jimmunol.1501605
27. Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X, et al. Epstein-Barr virus miR-BART6-3p inhibits the RIG-I pathway. J Innate Immun. (2017) 9:574-86. doi: 10.1159/000479749
28. Qiu J, Thorley-Lawson DA. EBV microRNA BART18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci USA. (2014) 111:11157-62. doi: 10.1073/pnas.1406136111
29. Huang WT, Lin CW. EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-γ-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am J Pathol. (2014) 184:1185-97. doi: 10.1016/j.ajpath.2013.12.024
30. Yajima M, Miyata M, Ikuta K, Hasegawa Y, Oneyama C, Kanda T. Efficient Epstein-Barr virus progeny production mediated by cancer-derived LMP1 and virally-encoded microRNAs. Microorganisms. (2019) 7:E119. doi: 10.3390/microorganisms7050119
31. Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. (2009) 5:376-85. doi: 10.1016/j.chom.2009.03.003
32. Schultze JL, Michalak S, Lowne J, Wong A, Gilleece MH, Gribben JG, et al. Human non-germinal center B cell interleukin (IL)-12 production is primarily regulated by T cell signals CD40 ligand, interferon gamma, and IL-10: role of B cells in the maintenance of T cell responses. J Exp Med. (1999) 189:1-12. doi: 10.1084/jem.189.1.1
33. Skinner CM, Ivanov NS, Barr SA, Chen Y, Skalsky RL. An Epstein-Barr virus microRNA blocks interleukin-1 (IL-1) signaling by targeting IL-1 receptor 1. J Virol. (2017) 91:e00530-17. doi: 10.1128/JVI.00530-17
34. Chen Y, Fachko D, Ivanov NS, Skinner CM, Skalsky RL. Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog. (2019) 15:e1007535. doi: 10.1371/journal.ppat.1007535
35. Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. (2008) 68:1436-42. doi: 10.1158/0008-5472.CAN-07-5126
36. Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. (2002) 10:417-26. doi: 10.1016/S1097-2765(02)00599-3
37. Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. (1998) 279:1737-40. doi: 10.1126/science.279.5357.1737
38. Athie-Morales V, Smits HH, Cantrell DA, Hilkens CM. Sustained IL-12 signaling is required for Th1 development. J Immunol. (2004) 172:61-9. doi: 10.4049/jimmunol.172.1.61
39. Murer A, Rühl J, Zbinden A, Capaul R, Hammerschmidt W, Chijioke O, et al. MicroRNAs of Epstein-Barr virus attenuate T-cell-mediated immune control in vivo. MBio. (2019) 10:e01941-18. doi: 10.1128/mBio.01482-19
40. Rosato P, Anastasiadou E, Garg N, Lenze D, Boccellato F, Vincenti S, et al. Differential regulation of miR-21 and miR-146a by Epstein-Barr virus-encoded EBNA2. Leukemia. (2012) 26:2343-52. doi: 10.1038/leu.2012.108
41. Luo Z, Dai Y, Zhang L, Jiang C, Li Z, Yang J, et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis. (2013) 34:415-25. doi: 10.1093/carcin/bgs329
42. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. (2005) 435:828-33. doi: 10.1038/nature03552
43. Kuo G, Wu CY, Yang HY. MiR-17-92 cluster and immunity. J Formos Med Assoc. (2019) 118:2-6. doi: 10.1016/j.jfma.2018.04.013
44. Lu F, Weidmer A, Liu CG, Volinia S, Croce CM, Lieberman PM. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. (2008) 82:10436-43. doi: 10.1128/JVI.00752-08
45. Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. (2010) 70:3119-27. doi: 10.1158/0008-5472.CAN-09-4250
46. Delgado-Ortega M, Marc D, Dupont J, Trapp S, Berri M, Meurens F. SOCS proteins in infectious diseases of mammals. Vet Immunol Immunopathol. (2013) 151:1-19. doi: 10.1016/j.vetimm.2012.11.008
47. Alston CI, Dix RD. SOCS and herpesviruses, with emphasis on cytomegalovirus retinitis. Front Immunol. (2019) 10:732. doi: 10.3389/fimmu.2019.00732
48. Peng RJ, Han BW, Cai QQ, Zuo XY, Xia T, Chen JR, et al. Genomic and transcriptomic landscapes of Epstein-Barr virus in extranodal natural killer T-cell lymphoma. Leukemia. (2019) 33:1451-62. doi: 10.1038/s41375-018-0324-5
49. Okuno Y, Murata T, Sato Y, Muramatsu H, Ito Y, Watanabe T, et al. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat Microbiol. (2019) 4:544. doi: 10.1038/s41564-019-0387-8
50. Higuchi H, Yamakawa N, Imadome KI, Yahata T, Kotaki R, Ogata J, et al. Role of exosomes as a proinflammatory mediator in the development of EBV-associated lymphoma. Blood. (2018) 131:2552-67. doi: 10.1182/blood-2017-07-794529
51. Pathmanathan R, Prasad U, Sadler R, Flynn K, Raab-Traub N. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. (1995) 333:693-8. doi: 10.1056/NEJM199509143331103
52. Tsao SW, Tsang CM, Lo KW. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci. (2017) 372:20160270. doi: 10.1098/rstb.2016.0270
53. de Jesus O, Smith PR, Spender LC, Elgueta Karstegl C, Niller HH, Huang D, et al. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J Gen Virol. (2003) 84:1443-50. doi: 10.1099/vir.0.19054-0
54. Chen H, Huang J, Wu FY, Liao G, Hutt-Fletcher L, Hayward SD. Regulation of expression of the Epstein-Barr virus BamHI-A rightward transcripts. J Virol. (2005) 79:1724-33. doi: 10.1128/JVI.79.3.1724-1733.2005
55. Kim H, Burassakarn A, Kang Y, Iizasa H, Yoshiyama H. A single nucleotide polymorphism in the BART promoter region of Epstein-Barr virus isolated from nasopharyngeal cancer cells. Biochem Biophys Res Commun. (2019) 520:373-8. doi: 10.1016/j.bbrc.2019.10.028
Keywords: microRNAs, herpes virus, immune evasion, BART miRNA, BHRF miRNA
Citation: Iizasa H, Kim H, Kartika AV, Kanehiro Y and Yoshiyama H (2020) Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases. Front. Immunol. 11:367. doi: 10.3389/fimmu.2020.00367
Received: 02 October 2019; Accepted: 14 February 2020;
Published: 03 March 2020.
Edited by:
Afsar Raza Naqvi, University of Illinois at Chicago, United StatesReviewed by:
Elena Martinelli, Population Council, United StatesFrançois J. M. A. Meurens, UMR INRA-Oniris 1300 Oniris-Nantes Atlantic National College of Veterinary Medicine, France
Shashank Tripathi, Indian Institute of Science (IISc), India
Copyright © 2020 Iizasa, Kim, Kartika, Kanehiro and Yoshiyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hironori Yoshiyama, eW9zaXlhbWFAbWVkLnNoaW1hbmUtdS5hYy5qcA==
 Hisashi Iizasa
Hisashi Iizasa Hyoji Kim
Hyoji Kim Andy Visi Kartika
Andy Visi Kartika Yuichi Kanehiro
Yuichi Kanehiro Hironori Yoshiyama
Hironori Yoshiyama