- 1Clinical Medicine College, Weifang Medical University, Weifang, China
- 2Department of Rheumatology, The Affiliated Hospital of Weifang Medical University, Weifang, China
- 3Department of Gastrointestinal and Anal Diseases Surgery, The Affiliated Hospital of Weifang Medical University, Weifang, China
- 4Functional Laboratory, Clinical Medicine College of Weifang Medical University, Weifang, China
- 5Department of Operating Room, Zhucheng People's Hospital, Zhucheng, China
Rheumatoid arthritis is a common systemic and autoimmune disease characterized by symmetrical and inflammatory destruction of distal joints. Its primary pathological characters are synovitis and vasculitis. Accumulating studies have implicated the critical role of non-coding RNAs (ncRNAs) in inflammation and autoimmune regulation, primarily including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA). NcRNAs are significant regulators in distinct physiological and pathophysiological processes. Many validated non-coding RNAs have been identified as promising biomarkers for the diagnosis and treatment of RA. This review will shed some light on RA pathogenesis and be helpful for identifying potential ncRNA biomarkers for RA.
Introduction
Rheumatoid arthritis (RA) is a type of chronic autoimmune disease, characterized by synovitis and vasculitis in pathology. It is a highly disabling disease due to joint deformity and loss of function (1). The main clinical features of RA typically are symmetrical polyarthritis with distal joint redness, swelling, and pain, especially the small joints of hands and feet (2). Approximately 1% of the population is affected with RA worldwide, with a higher prevalence in Europeans and Asians (3). Studies have implicated the significant and complex roles of genetic factor and environmental factor in the etiology of RA (4, 5). It has been well-documented that inflammatory response and immunological disorders critically contribute to RA. However, the precise pathogenesis and etiology of RA remain to be completely elucidated (6). To the best of our knowledge, common laboratory tests used for RA generally include erythrocyte sedimentation rate (ESR), c-reactive protein (CRP), rheumatoid factor (RF), and anti-cyclic peptide containing citrulline (anti-CCP) antibodies (7). Nevertheless, they lack specificity and have low priority. As a result, identification of novel and promising biomarkers for RA is essential for its early diagnosis and treatment.
In human, non-protein coding genes occupy ~70% of the genome. Accumulating data have suggested non-coding RNAs (ncRNAs) play important roles in regulating autoimmunity and inflammation (8). Due to increasing development of microarray sequencing techniques and bioinformatics analysis, many ncRNAs have been identified and validated in many kinds of diseases (9–12). They can be regarded as promising biomarkers predicting the occurrence and progression of cancer, cardiovascular disease and autoimmune disease, and so on (9–12). Different autoimmune disease has different ncRNA expression profile in diverse cells and tissues. In addition, there are still some ncRNAs dysregulated in several kinds of inflammatory or autoimmune diseases with similarities. Accumulating studies have suggested some ncRNAs are specifically expressed in RA, mainly including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) (7, 13, 14). Previously, we have identified the specific profile of miRNAs and lncRNAs differentially expressed in RA, which can serve as promising markers for RA diagnosis and treatment (15–17). Nonetheless, the modifying effects and molecular mechanism of those specifically expressed ncRNAs in RA pathogenesis have not been fully elucidated up to date.
In the present study, some functional ncRNAs have been listed in Table 1. The potential targets and mechanisms of them are also summarized. We aim to focus on the current knowledge of ncRNAs in RA, primarily including miRNAs, lncRNAs, and circRNAs by reviewing all currently published studies. Clarification of the expression and molecular mechanism of dysregulated ncRNAs in inflammation and autoimmunity will help to understand the pathogenesis of RA. Most importantly, identifying the targeted genes of those aberrantly expressed ncRNAs in RA will be useful for investigating promising biomarkers for its early diagnosis and efficient treatment.
MiRNAs
MiRNAs are evolutionarily conserved and usually have a length of 18–25 nucleotides, which regulate the expression of targeted genes at the post-transcriptional level by promoting the degradation of mRNA or repressing its translation (7). Accumulated studies have suggested the critical role of miRNAs in several kinds of autoimmune diseases, such as systemic lupus erythematosus (SLE), RA and Sjögren's syndrome (35). However, the expression and function of those aberrantly expressed miRNAs may be different in diverse autoimmune diseases. MiRNAs play a pivotal role in the regulation of multiple physiological and pathological processes, including cell cycle, stem cell maintenance, organ development, angiogenesis, and carcinogenesis (36). A number of well-established miRNAs have been regarded as candidate biomarkers for RA due to their critical role in regulating inflammation and autoimmunity (37). They are widely expressed in various cells, tissues, or microsomes and contribute to the pathogenesis of RA (37). Besides, some miRNAs are differentially expressed in response to TNF inhibitor treatment and other conventional therapies (38). Accordingly, miRNA can serve as predictive factor for the clinical response to biological therapies among RA patients.
As shown in Table 1, a variety of miRNAs are differentially expressed and dysregulated in RA, which can negatively regulate targeted genes, such as those genes encoding cytokines, chemokines, and inflammation-related signaling molecules, and thus participate in the pathogenesis of RA (22, 39, 40). Moreover, it has been well-established some nanovesicles-delivered miRNAs specially expressed in RA and exert modifying effects on inflammation and autoimmunity, such as exosomes-encapsulated miRNAs (15, 16). Exosomes are cell-derived vesicles encapsulating functional molecules such as RNAs, DNAs, proteins, and lipids (41, 42). Exosomes usually mediate intracellular communication by delivering functional RNAs from donor to receipted cells, including ncRNAs of miRNAs, lncRNAs as well as circRNAs (Figure 1). Mounting data have implicated exosomes and their encapsulated functional ncRNAs have been recognized as potential biomarkers for RA, especially exosome-encapsulated miRNAs (16, 18, 43).
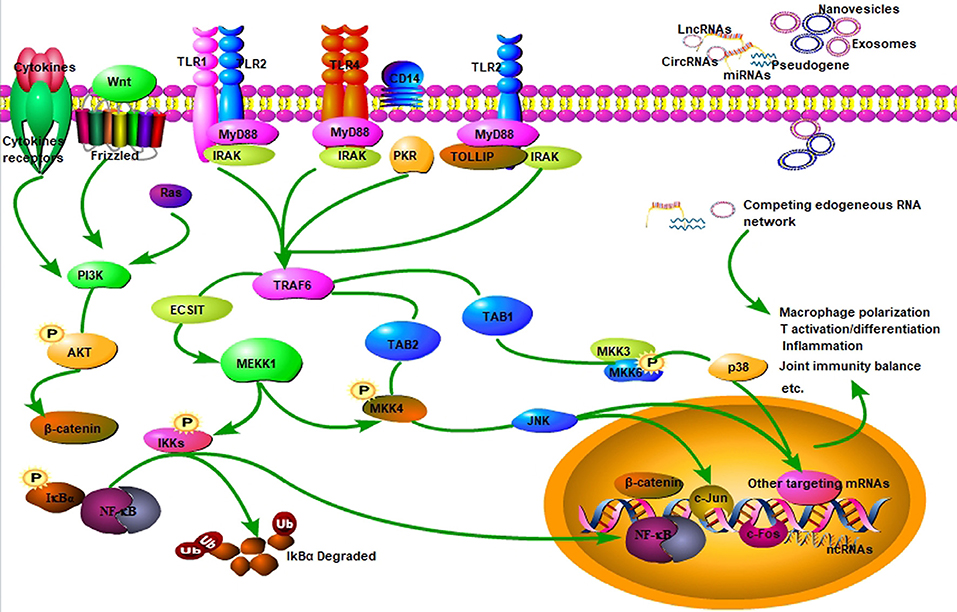
Figure 1. Signaling pathway of ncRNAs in RA. NcRNAs (miRNA, lncRNA, circRNA) are involved in regulating inflammation and autoimmunity, such as immune cell activation, differentiation, and polarization. Some ncRNAs are encapsulated in nanovesicles and exert critical effects on inflammatory and immune cells, and some can function as ceRNA by sponging miRNAs in RA. NcRNAs participate in RA inflammation and autoimmune disorders primarily through TLR4/NF-κB, MAPK/PI3K, and Wnt3a/β-catenin signaling pathways, and so on.
Growing data have revealed that many free miRNAs and exosome-delivered miRNAs are closely associated with RA (44, 45). The molecular mechanism of differentially expressed miRNAs in RA has been widely investigated by many published studies, particularly regarding their altering effects on inflammation and autoimmunity (15, 46–48). Toll-like receptors (TLRs), such as TLR2 and TLR4, are vital pattern recognition receptors (PRRs) functioning as a bridge linking immunomodulation and inflammatory response in many autoimmune diseases, including RA (35, 49, 50). Mechanisms of different TLRs in immune and inflammatory cells have been extensively investigated (Figure 1). Our previous study has demonstrated that miR-6089 inhibits inflammatory response via targeting TLR4 (16). It has been well-documented signaling pathways of TLRs/NF-κB, cytokines, and chemokines as well as Wnt signal play vital roles in regulating inflammatory response and immunological reaction that are involved in RA pathogenesis (Figure 1) (15, 46–48). The study by Guo et al. has shown that the proliferation, apoptosis and migration of fibroblast-like synoviocytes in RA can be affected by miR-338-5p via targeting NFAT5 (19). MiR-708-5p can promote the apoptosis of fibroblast-like synoviocytes and alleviate RA through Wnt3a/β-catenin pathway (20). MAPK signaling is also well-documented in regulating miRNAs in RA (Figure 1) (21, 46, 51). Our previous study has demonstrated that exosome-delivered miR-548a-3p regulates macrophages-mediated inflammation through TLR4/NF-κB signaling pathway in RA (15). Therefore, miR-548a-3p may serve as a promising marker for RA, because it can alleviate inflammation in RA. The miR-548a-3p/TLR4/NF-κB axis will offer new therapeutic strategies for RA. Taken together, the differentially expressed miRNAs in peripheral circulation or extracellular vesicles or synovium tissues in RA would be identified as important biological targets for the diagnosis and treatment of RA patients. Nevertheless, more pre-clinical or clinical experiments are warranted for more investigations.
LncRNAs
LncRNA is a newly identified non-coding RNA widely expressed in various tissues of the human body, which consists of more than 200 nucleotides in length (17). According to the structure and function of lncRNA, lncRNAs can be divided into five categories: sense, antisense, bidirectional, intronic, and intergenic (52). Some lncRNAs exert oncogenic properties in cancer (53), while some can inhibit the development and progression of malignancies due to distinct expression and biological effects in cancer cells (54). Accumulating studies have implicated that a variety of lncRNAs are found to be differentially expressed and confer effects on immune cells in several kinds of autoimmune diseases, including RA (55–58). Different autoimmune diseases have specific lncRNA expression profiles, which may be also specifically expressed in different cells and tissues. Besides, the lncRNA expression profile in RA can be influenced by different therapy strategies demonstrated by Guo et al. (59). Furthermore, it has been reported that a number of lncRNAs are dysregulated and associated with organ damage in systemic lupus erythematosus (SLE) compared with RA (60), which suggests a critical role of ncRNA in regulating specific organ damage in autoimmune diseases. LncRNA H19, Hotair, lincRNA-p21, C5T1, LOC100652951, and LOC100506036 have been verified to be dysregulated in T cells, peripheral blood mononuclear cells (PBMCs), exosomes, and synovial cells in RA, which are associated with inflammation and immune reaction in RA (Table 1) (24–27, 61). The lncRNA expression profile in RA is different in diverse types of immune cell, such as B cells, nature killer (NK) cells, and T cells, which suggests immune cell-type specificity of lncRNA expression (62). Identification of aberrantly expressed lncRNAs in RA and exploration of the underlying molecular mechanisms will offer a new direction to understand the pathogenesis of RA.
The regulatory mechanism of lncRNAs is complicated and needs to be investigated by more functional and mechanical experiments. T lymphocytes-mediated autoimmune response plays an important role in the development of RA (63, 64). Moreover, the abnormally expressed lncRNAs in T cells can influence their function and facilitate or suppress immune and inflammatory reactions in RA, such as lncRNA FAM66C, LOC100652951, and LOC100506036 (27, 64, 65). PBMC and exosome-derived Hotair are demonstrated to affect the migration of activated macrophages and the expression of MMP-2 and MMP-13 in RA (61). An lncRNA NTT/PBOV1 axis has been elucidated by a published study, which is capable of regulating monocyte differentiation in RA (28). LncRNA HOTAIR is documented to alleviate RA by targeting miR-138 and inhibit the activation of NF-κB pathway in LPS-treated chondrocytes, suggesting an lncRNA-miRNA interaction in RA pathogenesis (29). In our previous study, five lncRNAs are reported to be significantly up-regulated in serum samples of RA patients, including RNA143598, RNA143596, HIX0032090, IGHCγl, and XLOC-002730 (17) (Table 1). Some of these aberrantly expressed lncRNAs are associated with the disease course, anti-CCP antibody level and disease activity of RA (17). The bioinformatics analysis indicates that classic signaling pathways of TLRs, cytokines, NF-κB, and IRF3/IRF7 that are most likely involved in RA with regard to lncRNAs regulation (17). More interestingly, HIX0032090 has been demonstrated to participate in RA pathogenesis by functioning as a competitive endogenous RNA (ceRNA) for miRNA in our recently published study (23). Nevertheless, more future studies are warranted to elucidate the molecular mechanism of those dysregulated lncRNAs in RA initiation and progression. Taken together, these available data have suggested the immune cell specificity of lncRNA expressed in RA.
Mounting evidence has suggested lncRNA, the same as pseudogenes, circRNAs and competing mRNAs, can function as ceRNA based on a lncRNA-miRNA-mRNA network in autoimmune disease, vascular disease, cancer, and so on (Table 1 and Figure 2) (66–70). LncRNA may facilitate the expression and function of the targeted mRNA by sponging miRNA, and thus participates in regulating immune cell activity and function (71). Jiang et al. have found that three lncRNAs, namely S5645.1, XR_006437.1 and J01878, can serve as promising biomarkers for RA via ceRNA network (30). It has also been demonstrated that lncRNA GAPLINC enhances cell proliferation, migration, and generation of proinflammatory cytokines by sponging miR-382-5p and miR-575 in fibroblast-like synoviocytes (31). Similarly, ZFAS1, a newly identified lncRNA in RA, is shown to modulate fibroblast-like synoviocytes migration and invasion by targeting miR-27a as a sponge (32). As mentioned above, an lncRNA HOTAIR-miR-138-NF-κB axis has also been established in chondrocytes in RA (29). Accordingly, lncRNA may function through ceRNA mechanism by sponging one or more miRNAs in immune cell or parenchymal cell, such as chondrocytes (Figure 2). Identification of lncRNA-miRNA-mRNA ceRNA network provides new insight into the pathogenesis of RA. Key molecules and signaling pathway in this network will serve as ideal diagnostic and therapeutic targets for RA.
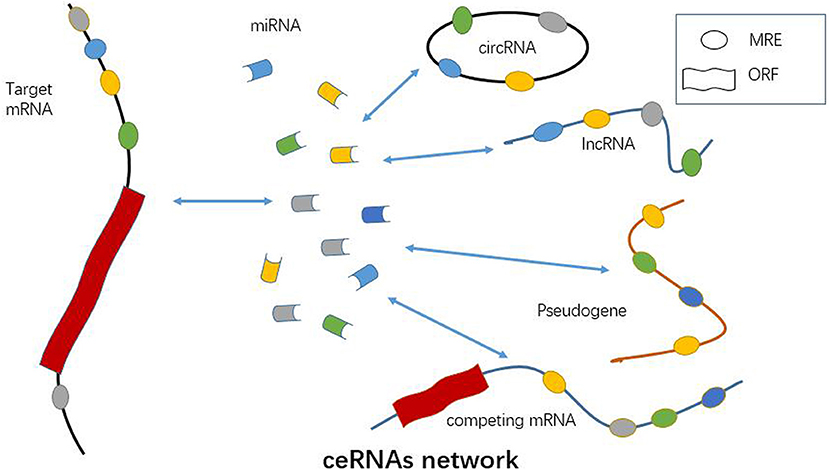
Figure 2. Functional role of ncRNAs as ceRNA. A number of miRNAs, lncRNAs, circRNAs, pseudogenes, and competing mRNAs can act as ceRNA and promote the expression of targeted mRNAs via sponging miRNAs. CeRNA network is a crucial mechanism of ncRNAs involved in RA pathogenesis.
CircRNAs
Circular RNA (circRNA) is an endogenous non-coding RNA, the most representative characteristic of which is the covalently closed RNA circle without 5′ end caps or 3′ poly (1) tails (33, 72). This circular structure is usually stable with the half-life larger than 48 h (73). CircRNAs are primarily divided into three types of circRNA including exonic circRNAs (ecircRNAs), circular intronic RNAs (ciRNAs), and exon-intron circRNAs (EIciRNAs) (74). The production of circRNAs in cells is usually attributed to exon skipping and circularization driven by intron pairing or RNA binding protein (74). Apart from mammals, numerous circRNAs have been demonstrated to be expressed in fungi, plants, and protists (75–78). Most importantly, the expression of circRNAs is in a tissue-specific manner (79). Usually, circRNAs can be found in peripheral blood, exosomes, and tissues. Similar to lncRNA, circRNA can also serve as miRNA sponge, which can combine with miRNAs and thereby insulate them from the natural mRNAs (70, 80–83) (Figure 2). Available data have revealed ecircRNA confers critical effects on several pathological and physiological processes mainly through ceRNA mechanism in cytoplasm (74). However, circRNAs of ciRNAs and EIciRNAs usually regulate the targeted genes in nucleus (84–86). CircRNAs serve as miRNAs sponge and facilitate the expression of targeted mRNAs by inhibiting the effect of miRNA (82, 87). CeRNA is also an essential way for circRNA in regulations of autoimmunity and inflammation (88, 89) (Figure 2). However, there are few publications elucidating the ceRNA regulatory mechanism of circRNA in RA up till now.
CircRNAs are suggested in regulating diverse immune disorders due to their various forms of epigenetic modification, for instance, miRNA sponge and miRNA reservoir (74, 79, 83). Accumulated data have implicated the vital role of circRNAs in multiple kinds of diseases, such as cancer, neurologic disorders and cardiovascular diseases (74, 90–92). The critical role of circRNAs in antiviral immunity has been well-documented, which offers potential therapeutic strategies for antiviral therapy by targeting circRNAs (93–95). The study by Ma et al. shows the evidence that circARSP91 promotes cancer immune surveillance by regulating NK cells in liver cancer, suggesting a critical role of circRNA in tumor immunity (96). In addition, circRNA Malat-1 has been suggested as a key regulator in alloimmune rejection by promoting dendritic cells to induce T cell exhaustion and regulatory T cell generation, which implicates the pivotal role of circRNA in adaptive immunity (97). Taken together, circRNA plays critical roles not only in innate immunity but adaptive immunity.
During the past few years, the role of circRNAs in RA has drawn more and more attention. There is specific circRNA expression profile in RA as demonstrated by microarray chip analysis (14, 33, 98). As shown in Table 1, many circRNAs have been documented to be aberrantly expressed in RA, such as circRNA_092516, circRNA_003524, circRNA_103047, and circRNA_101873. CircRNAs can be up-regulated or down-regulated in peripheral blood or tissues in RA. Interaction between miRNA and circRNA is also revealed in RA, which implicates the circRNA-miRNA network in autoimmune regulation (99). It has been shown that has-circ-0001859 is identified in synovial tissues, which regulates synovial inflammation via sponging miR-204/211 and targeting ATF1 (34). Accordingly, circRNAs can regulate RA through ceRNA network (Figure 2). Nevertheless, little is known about the downstream signaling pathway of circRNA in regulating autoimmunity and inflammation. More studies are warranted to elucidate this issue in future. It is also prospective to investigate novel diagnostic and therapeutic strategies for RA by targeting circRNAs.
Conclusions and Future Directions
In the last few years, ncRNAs have been regarded as hot points in many scientific fields worldwide. Role of ncRNAs in regulating inflammation and autoimmunity has drawn widely attention. Although specific expression profiles of miRNAs, lncRNAs and circRNAs in RA have been well-documented in many currently published studies, the molecular mechanism behind ncRNAs regulation in RA is not very clear yet. Those aberrantly expressed ncRNAs participate in the pathogenesis of RA primarily by regulating autoimmunity and inflammation. Up to now, Wnt3a/β-catenin, TLR/NF-κB, and MAPK signaling pathways have been well-established in regulating the differentially expressed ncRNAs in RA. Most interestingly, elucidation of the lncRNA/circRNA-miRNA-mRNA ceRNA network sheds light on the pathogenesis of RA. Researchers are encouraged to investigate novel strategies for the early diagnosis and treatment of RA by targeting ncRNAs and relevant key signaling pathways in the future.
Author Contributions
JW, SY, HL, and JY carried out literature research and reviewed all articles. JW, SY, and HL wrote the paper. ZW and DX edited the article.
Funding
This work was supported by grants from the National Natural Science Foundation, China (81601408), Natural Science Foundation for Young Scholars, Shandong, China (ZR2019QH012 and ZR2016HQ12), and Science and Technology Development Program, Weifang, China (2019YX020, 2019GX031, and 2017YX019).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. (2010) 376:1094–108. doi: 10.1016/S0140-6736(10)60826-4
2. Davis JM III, Matteson EL. My treatment approach to rheumatoid arthritis. Mayo Clin Proc. (2012) 87:659–73. doi: 10.1016/j.mayocp.2012.03.011
3. Lawrence RC, Felson DT, Helmick CG, Arnold LM, Choi H, Deyo RA, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. (2008) 58:26–35. doi: 10.1002/art.23176
4. MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. (2000) 43:30–7. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B
5. Padyukov L, Silva C, Stolt P, Alfredsson L, Klareskog L. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. (2004) 50:3085–92. doi: 10.1002/art.20553
6. Ursini F, Russo E, Letizia Hribal M, Mauro D, Savarino F, Bruno C, et al. Abatacept improves whole-body insulin sensitivity in rheumatoid arthritis: an observational study. Medicine. (2015) 94:e888. doi: 10.1097/MD.0000000000000888
7. Tavasolian F, Abdollahi E, Rezaei R, Momtazi-Borojeni AA, Henrotin Y, Sahebkar A. Altered expression of microRNAs in rheumatoid arthritis. J Cell Biochem. (2018) 119:478–87. doi: 10.1002/jcb.26205
8. Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. (2012) 22:1646–57. doi: 10.1101/gr.134767.111
9. Leimena C, Qiu H. Non-coding RNA in the pathogenesis, progression and treatment of hypertension. Int J Mol Sci. (2018) 19:E927. doi: 10.3390/ijms19040927
10. Wang B, Yao Q, Xu D, Zhang JA. MicroRNA-22–3p as a novel regulator and therapeutic target for autoimmune diseases. Int Rev Immunol. (2017) 36:176–81. doi: 10.1080/08830185.2017.1281272
11. Wang Y, Xu D, Wang B, Hou X. Could microRNAs be regulators of gout pathogenesis? Cell Physiol Biochem. (2015) 36:2085–92. doi: 10.1159/000430176
12. Ramon YCS, Segura MF, Hummer S. Interplay between ncRNAs and cellular communication: a proposal for understanding cell-specific signaling pathways. Front Genet. (2019) 10:281. doi: 10.3389/fgene.2019.00281
13. Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. (2018) 415:11–9. doi: 10.1016/j.canlet.2017.11.018
14. Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep. (2017) 16:8029–36. doi: 10.3892/mmr.2017.7638
15. Wang Y, Zheng F, Gao G, Yan S, Zhang L, Wang L, et al. MiR-548a-3p regulates inflammatory response via TLR4/NF-κB signaling pathway in rheumatoid arthritis. J Cell Biochem. (2018) 120:1133–40. doi: 10.1002/jcb.26659
16. Xu D, Song M, Chai C, Wang J, Jin C, Wang X, et al. Exosome-encapsulated miR-6089 regulates inflammatory response via targeting TLR4. J Cell Physiol. (2019) 234:1502–11. doi: 10.1002/jcp.27014
17. Xu D, Jiang Y, Yang L, Hou X, Wang J, Gu W, et al. Long noncoding RNAs expression profile and functional networks in rheumatoid arthritis. Oncotarget. (2017) 8:95280–92. doi: 10.18632/oncotarget.20036
18. Chen Z, Wang H, Xia Y, Yan F, Lu Y. Therapeutic potential of mesenchymal cell-derived miRNA-150–5p-expressing exosomes in rheumatoid arthritis mediated by the modulation of MMP14 and VEGF. J Immunol. (2018) 201:2472–82. doi: 10.4049/jimmunol.1800304
19. Guo T, Ding H, Jiang H, Bao N, Zhou L, Zhao J. miR-338–5p Regulates the viability, proliferation, apoptosis and migration of rheumatoid arthritis fibroblast-like synoviocytes by targeting NFAT5. Cell Physiol Biochem. (2018) 49:899–910. doi: 10.1159/000493222
20. Wu J, Fan W, Ma L, Geng X. miR-708–5p promotes fibroblast-like synoviocytes' cell apoptosis and ameliorates rheumatoid arthritis by the inhibition of Wnt3a/beta-catenin pathway. Drug Des Devel Ther. (2018) 12:3439–47. doi: 10.2147/DDDT.S177128
21. Yang Z, Wang J, Pan Z, Zhang Y. miR-143–3p regulates cell proliferation and apoptosis by targeting IGF1R and IGFBP5 and regulating the Ras/p38 MAPK signaling pathway in rheumatoid arthritis. Exp Ther Med. (2018) 15:3781–90. doi: 10.3892/etm.2018.5907
22. Churov AV, Oleinik EK, Knip M. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun Rev. (2015) 14:1029–37. doi: 10.1016/j.autrev.2015.07.005
23. Yan S, Wang P, Wang J, Yang J, Lu H, Jin C, et al. Long Non-coding RNA HIX003209 promotes inflammation by sponging miR-6089 via TLR4/NF-κB signaling pathway in rheumatoid arthritis. Front Immunol. (2019) 10:2218. doi: 10.3389/fimmu.2019.02218
24. Stuhlmuller B, Kunisch E, Franz J, Martinez-Gamboa L, Hernandez MM, Pruss A, et al. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am J Pathol. (2003) 163:901–11. doi: 10.1016/S0002-9440(10)63450-5
25. Spurlock CF III, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. (2014) 66:2947–57. doi: 10.1002/art.38805
26. Messemaker TC, Frank-Bertoncelj M, Marques RB, Adriaans A, Bakker AM, Daha N, et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. (2016) 17:85–92. doi: 10.1038/gene.2015.54
27. Lu MC, Yu HC, Yu CL, Huang HB, Koo M, Tung CH, et al. Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol Res. (2016) 64:576–83. doi: 10.1007/s12026-015-8756-8
28. Yang CA, Li JP, Yen JC, Lai IL, Ho YC, Chen YC, et al. lncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int J Mol Sci. (2018) 19:2806. doi: 10.3390/ijms19092806
29. Zhang HJ, Wei QF, Wang SJ, Zhang XY, Geng Q, Cui YH, et al. LncRNA HOTAIR alleviates rheumatoid arthritis by targeting miR-138 and inactivating NF-κB pathway. Int Immunopharmacol. (2017) 50:283–90. doi: 10.1016/j.intimp.2017.06.021
30. Jiang H, Ma R, Zou S, Wang Y, Li Z, Li W. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol Biosyst. (2017) 13:1182–92. doi: 10.1039/C7MB00094D
31. Mo BY, Guo XH, Yang MR, Liu F, Bi X, Liu Y, et al. Long non-coding RNA GAPLINC promotes tumor-like biologic behaviors of fibroblast-like synoviocytes as microRNA sponging in rheumatoid arthritis patients. Front Immunol. (2018) 9:702. doi: 10.1136/annrheumdis-2018-eular.3584
32. Ye Y, Gao X, Yang N. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Hum Cell. (2018) 31:14–21. doi: 10.1007/s13577-017-0179-5
33. Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, et al. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem. (2017) 42:651–9. doi: 10.1159/000477883
34. Li B, Li N, Zhang L, Li K, Xie Y, Xue M, et al. Hsa_circ_0001859 Regulates ATF2 expression by functioning as an MiR-204/211 sponge in human rheumatoid arthritis. J Immunol Res. (2018) 2018:9412387. doi: 10.1155/2018/9412387
35. Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. (2016) 15:1171–80. doi: 10.1016/j.autrev.2016.09.003
36. Yang N, Zhu S, Lv X, Qiao Y, Liu YJ, Chen J. MicroRNAs: pleiotropic regulators in the tumor microenvironment. Front Immunol. (2018) 9:2491. doi: 10.3389/fimmu.2018.02491
37. Chen XM, Huang QC, Yang SL, Chu YL, Yan YH, Han L, et al. Role of micro RNAs in the pathogenesis of rheumatoid arthritis: novel perspectives based on review of the literature. Medicine. (2015) 94:e1326. doi: 10.1097/MD.0000000000001326
38. Liu Y, Han Y, Qu H, Fang J, Ye M, Yin W. Correlation of microRNA expression profile with clinical response to tumor necrosis factor inhibitor in treating rheumatoid arthritis patients: a prospective cohort study. J Clin Lab Anal. (2019) 33:e22953. doi: 10.1002/jcla.22953
39. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
40. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. (2013) 12:847–65. doi: 10.1038/nrd4140
41. Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. (2006) 107:102–8. doi: 10.1016/j.imlet.2006.09.005
42. van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. (2012) 64:676–705. doi: 10.1124/pr.112.005983
43. Withrow J, Murphy C, Liu Y, Hunter M, Fulzele S, Hamrick MW. Extracellular vesicles in the pathogenesis of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. (2016) 18:286. doi: 10.1186/s13075-016-1178-8
44. Zakeri Z, Salmaninejad A, Hosseini N, Shahbakhsh Y, Fadaee E, Shahrzad MK, et al. MicroRNA and exosome: key players in rheumatoid arthritis. J Cell Biochem. (2019) 120:10930–44. doi: 10.1002/jcb.28499
45. Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. (2008) 10:R101. doi: 10.1186/ar2493
46. Sujitha S, Rasool M. MicroRNAs and bioactive compounds on TLR/MAPK signaling in rheumatoid arthritis. Clin Chim Acta. (2017) 473:106–15. doi: 10.1016/j.cca.2017.08.021
47. Sharma AR, Sharma G, Lee SS, Chakraborty C. miRNA-regulated key components of cytokine signaling pathways and inflammation in rheumatoid arthritis. Med Res Rev. (2016) 36:425–39. doi: 10.1002/med.21384
48. Miao CG, Yang YY, He X, Li XF, Huang C, Huang Y, et al. Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cell Signal. (2013) 25:2069–78. doi: 10.1016/j.cellsig.2013.04.002
49. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
50. Xu D, Yan S, Wang H, Gu B, Sun K, Yang X, et al. IL-29 Enhances LPS/TLR4-mediated inflammation in rheumatoid arthritis. Cell Physiol Biochem. (2015) 37:27–34. doi: 10.1159/000430330
51. Moran-Moguel MC, Petarra-Del Rio S, Mayorquin-Galvan EE, Zavala-Cerna MG. Rheumatoid arthritis and miRNAs: a critical review through a functional view. J Immunol Res. (2018) 2018:2474529. doi: 10.1155/2018/2474529
52. Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, et al. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS ONE. (2014) 9:e104044. doi: 10.1371/journal.pone.0104044
53. Jin Y, Wu P, Zhao W, Wang X, Yang J, Huo X, et al. Long noncoding RNA LINC00165-induced by STAT3 exerts oncogenic properties via interaction with polycomb repressive complex 2 to promote EMT in gastric cancer. Biochem Biophys Res Commun. (2018) 507:223–30. doi: 10.1016/j.bbrc.2018.11.012
54. Zeng H, Wang J, Chen T, Zhang K, Chen J, Wang L, et al. Downregulation of lncRNA OIP5-AS1 inhibits breast cancer progression by targeting SOX2 via miRNA-129–5p upregulation. Cancer Sci. (2019) 110:289–302. doi: 10.1111/cas.13879
55. Scaria V. Joining the long shots: emerging evidence on the role of long noncoding RNAs in rheumatoid arthritis. Int J Rheum Dis. (2014) 17:831–3. doi: 10.1111/1756-185X.12570
56. Sigdel KR, Cheng A, Wang Y, Duan L, Zhang Y. The emerging functions of long noncoding RNA in immune cells: autoimmune diseases. J Immunol Res. (2015) 2015:848790. doi: 10.1155/2015/848790
57. Liang J, Chen W, Lin J. LncRNA: An all-rounder in rheumatoid arthritis. J Transl Int Med. (2019) 7:3–9. doi: 10.2478/jtim-2019-0002
58. Hur K, Kim SH, Kim JM. Potential implications of long noncoding RNAs in autoimmune diseases. Immune Netw. (2019) 19:e4. doi: 10.4110/in.2019.19.e4
59. Guo S, Liu J, Jiang T, Lee D, Wang R, Zhou X, et al. (5R)-5-Hydroxytriptolide (LLDT-8) induces substantial epigenetic mediated immune response network changes in fibroblast-like synoviocytes from rheumatoid arthritis patients. Sci Rep. (2019) 9:11155. doi: 10.1038/s41598-019-47411-1
60. Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, et al. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. (2015) 17:131. doi: 10.1186/s13075-015-0632-3
61. Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. (2015) 15:121–6. doi: 10.1007/s10238-013-0271-4
62. Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. (2014) 6:88. doi: 10.1186/s13073-014-0088-0
63. Mori M, Hashimoto M, Matsuo T, Fujii T, Furu M, Ito H, et al. Cell-contact-dependent activation of CD4(+) T cells by adhesion molecules on synovial fibroblasts. Mod Rheumatol. (2017) 27:448–56. doi: 10.1080/14397595.2016.1220353
64. Houtman M, Shchetynsky K, Chemin K, Hensvold AH, Ramskold D, Tandre K, et al. T cells are influenced by a long non-coding RNA in the autoimmune associated PTPN2 locus. J Autoimmun. (2018) 90:28–38. doi: 10.1016/j.jaut.2018.01.003
65. Aterido A, Palacio C, Marsal S, Avila G, Julia A. Novel insights into the regulatory architecture of CD4+ T cells in rheumatoid arthritis. PLoS ONE. (2014) 9:e100690. doi: 10.1371/journal.pone.0100690
66. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. (2018) 19:E1310. doi: 10.3390/ijms19051310
67. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. (2016) 17:272–83. doi: 10.1038/nrg.2016.20
68. Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. Sci World J. (2014) 2014:896206. doi: 10.1155/2014/896206
69. Fan Z, Gao S, Chen Y, Xu B, Yu C, Yue M, et al. Integrative analysis of competing endogenous RNA networks reveals the functional lncRNAs in heart failure. J Cell Mol Med. (2018) 22:4818–29. doi: 10.1111/jcmm.13739
70. Li LJ, Zhao W, Tao SS, Leng RX, Fan YG, Pan HF, et al. Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Expert Opin Ther Targets. (2017) 21:639–48. doi: 10.1080/14728222.2017.1319938
71. Mousavi MJ, Jamshidi A, Chopra A, Aslani S, Akhlaghi M, Mahmoudi M. Implications of the noncoding RNAs in rheumatoid arthritis pathogenesis. J Cell Physiol. (2018) 234:335–47. doi: 10.1002/jcp.26911
72. Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. (2016) 32:309–16. doi: 10.1016/j.tig.2016.03.002
73. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
74. Yang L, Fu J, Zhou Y. Circular RNAs and their emerging roles in immune regulation. Front Immunol. (2018) 9:2977. doi: 10.3389/fimmu.2018.02977
75. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. (2015) 4:e07540. doi: 10.7554/eLife.07540
76. Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, Sabeti PC. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics. (2015) 16:454. doi: 10.1186/s12864-015-1603-4
77. Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. (2015) 21:2076–87. doi: 10.1261/rna.052282.115
78. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. (2014) 9:e90859. doi: 10.1371/journal.pone.0090859
79. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, et al. Circular RNAs in immune responses and immune diseases. Theranostics. (2019) 9:588–607. doi: 10.7150/thno.29678
80. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
81. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
82. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. (2016) 238:42–51. doi: 10.1016/j.jbiotec.2016.09.011
83. Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. (2017) 8:73271–81. doi: 10.18632/oncotarget.19154
84. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
85. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
86. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Corrigendum: exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2017) 24:194. doi: 10.1038/nsmb0217-194a
87. Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol. (2015) 36:479–88. doi: 10.1007/s13277-015-3093-z
88. Su Q, Lv X. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis. Genomics. (2019). doi: 10.1016/j.ygeno.2019.10.006. [Epub ahead of print].
89. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. (2018) 17:79. doi: 10.1186/s12943-018-0827-8
90. Sheng JQ, Liu L, Wang MR, Li PY. Circular RNAs in digestive system cancer: potential biomarkers and therapeutic targets. Am J Cancer Res. (2018) 8:1142–56.
91. Ojha R, Nandani R, Chatterjee N, Prajapati VK. Emerging role of circular rnas as potential biomarkers for the diagnosis of human diseases. Adv Exp Med Biol. (2018) 1087:141–57. doi: 10.1007/978-981-13-1426-1_12
92. Yang J, Chen M, Cao RY, Li Q, Zhu F. The role of circular RNAs in cerebral ischemic diseases: ischemic stroke and cerebral ischemia/reperfusion injury. Adv Exp Med Biol. (2018) 1087:309–25. doi: 10.1007/978-981-13-1426-1_25
93. Xia X, Tang X, Wang S. Roles of CircRNAs in autoimmune diseases. Front Immunol. (2019) 10:639. doi: 10.3389/fimmu.2019.00639
94. Wang M, Yu F, Wu W, Zhang Y, Chang W, Ponnusamy M, et al. Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. (2017) 13:1497–506. doi: 10.7150/ijbs.22531
95. Cadena C, Hur S. Antiviral immunity and circular RNA: no end in sight. Mol Cell. (2017) 67:163–4. doi: 10.1016/j.molcel.2017.07.005
96. Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. (2019) 17:388–97. doi: 10.3892/ol.2018.9606
97. Zhang Y, Zhang G, Liu Y, Chen R, Zhao D, McAlister V, et al. GDF15 Regulates malat-1 circular RNA and inactivates NFκB signaling leading to immune tolerogenic DCs for preventing alloimmune rejection in heart transplantation. Front Immunol. (2018) 9:2407. doi: 10.3389/fimmu.2018.02407
98. Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing C, et al. Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin Exp Immunol. (2018) 194:118–24. doi: 10.1111/cei.13181
Keywords: circRNA, exosome, lncRNA, microRNA, non-coding RNA, rheumatoid arthritis
Citation: Wang J, Yan S, Yang J, Lu H, Xu D and Wang Z (2020) Non-coding RNAs in Rheumatoid Arthritis: From Bench to Bedside. Front. Immunol. 10:3129. doi: 10.3389/fimmu.2019.03129
Received: 10 June 2019; Accepted: 23 December 2019;
Published: 28 January 2020.
Edited by:
Pier Luigi Meroni, Italian Auxological Institute (IRCCS), ItalyReviewed by:
Tatsuya Atsumi, Hokkaido University, JapanChary Lopez-Pedrera, Maimonides Biomedical Research Institute of Cordoba (IMIBIC), Spain
Copyright © 2020 Wang, Yan, Yang, Lu, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengyan Wang, MjE2NTA1NjQ2OUBxcS5jb20=; Donghua Xu, Zmxvd2VyMzIyQDE2My5jb20=
†These authors share first authorship
 Jinghua Wang
Jinghua Wang Shushan Yan
Shushan Yan Jinghan Yang
Jinghan Yang Hongying Lu4†
Hongying Lu4† Donghua Xu
Donghua Xu