- 1Laboratory of Immunology and Infectious Disease Biology, Department of Biological Sciences, Indian Institute of Science Education and Research, Bhopal, India
- 2Department of Dermatology, School of Medicine, University of California, Davis, Sacramento, CA, United States
- 3Department of Medicine, Washington University School of Medicine, Saint Louis, MO, United States
- 4Laboratory of Host Defense, WPI Immunology, Frontier Research Centre, Osaka University, Osaka, Japan
MicroRNAs are short non-coding RNAs that play a crucial role in the regulation of gene expression during cellular processes. The host-encoded miRNAs are known to modulate the antiviral defense during viral infection. In the last decade, multiple DNA and RNA viruses have been shown to produce miRNAs known as viral miRNAs (v-miRNAs) so as to evade the host immune response. In this review, we highlight the origin and biogenesis of viral miRNAs during the viral lifecycle. We also explore the role of viral miRNAs in immune evasion and hence in maintaining chronic infection and disease. Finally, we offer insights into the underexplored role of viral miRNAs as potential targets for developing therapeutics for treating complex viral diseases.
Introduction
Until the twenty first century, it was assumed that more than 95% of the eukaryotic genome is “junk” DNA; however, the advent of next-generation sequencing and high throughput functional screening has highlighted the regulatory functions of the non-coding genome. Around 50–85% of the mammalian genome is transcribed, with at least some non-coding RNA transcript, which includes microRNA (miRNA), siRNA, piwi-interacting RNA (piRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA) (1–4). In recent years, the role of miRNAs in the development of immune responses in viral infections has been a subject of immense research interest.
MicroRNAs are small RNA molecules (18–22 nt) that play a crucial role in the regulation of gene expression by binding to the 3′ untranslated region (3′UTR) of target messenger RNAs (mRNAs) (5–7). miRNA-mRNA interaction results in mRNA degradation or translation inhibition, thereby resulting in reduced gene expression, thereby modulating the biological function (8, 9). Various studies indicate that miRNA can be derived from an intronic region of coding and non-coding genes, an exonic region of non-coding genes, and intragenic regions (10, 11). In addition to the hundreds of eukaryotic cellular miRNAs, miRNAs of viral origin (also known as v-miRNAs) have been discovered that can function as post-transcriptional gene regulators to host as well as viral genes (12, 13).
In this review, we summarize the miRNA-like non-coding RNAs encoded by DNA and RNA viruses and their roles in the evasion of host immunity. Finally, we discuss the possible role of these v-miRNAs as potential targets for developing therapeutics for the treatment of viral diseases.
Discovery and Origin of Viral miRNAs
Two different approaches have been used to identify v-miRNAs. (i) The use of computational tools to predict the secondary structure of the precursor of v-miRNAs (pre-v-miRNAs). This approach usually results in a large number of false positives; however, it can lead to the identification of less abundant miRNA (14, 15). (ii) Sequencing of cloned small RNA molecules; however, less abundant miRNA may be left out (15–18). The first v-miRNAs were identified in 2004 in the Epstein-Barr virus (EBV) by cloning the small RNAs from Burkitt's lymphoma cell line latently infected with EBV. Analysis of the genomic region flanking the small RNA molecules suggested characteristic miRNA gene-like structures (19). The EBV-miRNAs miR-BHRF1-1, miR-BHRF1-2 and miR-BHRF1-3 and miR-BART1 and miR-BART2 originated from two regions in BHRF1 and BART mRNAs (19). To date, EBV is known to encode 44 v-miRNAs from 25 double-stranded RNA precursors (20). Work by Pfeffer et al. suggested that other DNA viruses might also express miRNAs to modulate host and viral gene expression (19). Notably, more than 250 v-miRNAs have been identified, and the majority of them are accounted for in the DNA viruses of the herpesvirus family (12, 21).
Studies have indicated that viruses utilize the cellular machinery to encode miRNA. Similarly to eukaryotic miRNAs, v-miRNAs are generated by the Drosha and Dicer machinery (5, 22). The viral miRNA gene is transcribed by RNA polymerase II (RNA pol II) or Pol III to generate primary miRNA (pri-miRNA) that is then processed by a complex of Drosha/DiGeorge syndrome (DGCR8) within the nucleus to generate ~70-nucleotide (nt) pre-miRNA. The pre-miRNA is exported out of the nucleus by exportin-5, where pre-miRNA is further processed by endonuclease Dicer to yield mature miRNA duplex (21–25 nt). One strand of the duplex is loaded into the RNA-induced silencing complex (RISC) containing Argonaute 2 (Ago2). The miRNA-RISC complex interacts with the target transcript and inhibits gene expression (Figure 1) (23–26).
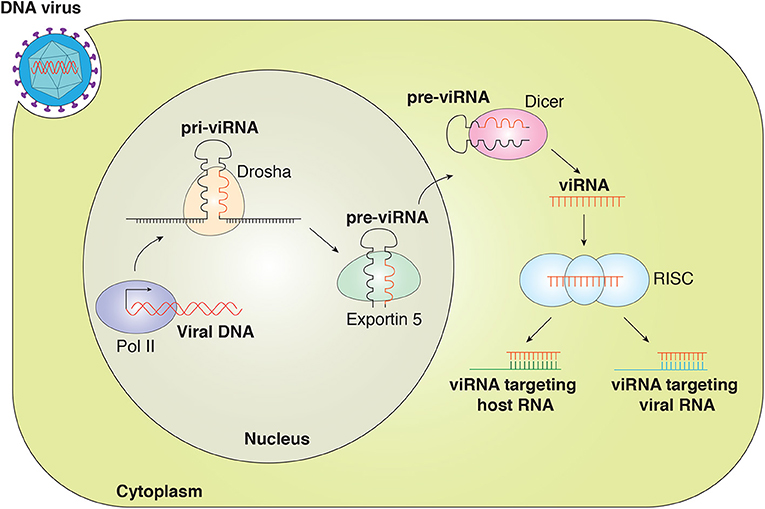
Figure 1. Canonical viral miRNAs are transcribed as pri-v-miRNAs from the genome of DNA viruses in the nucleus. Cleavage of pri-v-miRNAs by Drosha results in pre-v-miRNA that is exported to the cytoplasm via Exportin 5. In the cytoplasm, pre-v-miRNAs undergo cleavage by Dicer to generate the mature v-miRNAs that are loaded in the multiprotein RISC complex to target the host and viral mRNA transcript. v-miRNAs (viRNA).
v-miRNAs in DNA Viruses
γ-Herpesvirus-Encoded v-miRNAs
EBV is a γ-herpesvirus that infects 90% of the world population and is associated with various epithelial and lymphoid malignancies including Burkitt's lymphoma (BL), gastric carcinoma (GC), Hodgkin's lymphoma (HL), and nasopharyngeal carcinoma (NPC) (27–30). In infected cells, EBV can establish two phases of infection known as latent and lytic. EBV primarily infects the human oropharynx epithelial cells, followed by replication and spread to B cells. This results in latent infection in B cells, epithelial cells, and natural killer/T cells (31). Latent EBV infection is a substantial cause of many human malignancies (32). Under certain conditions, EBV switches from latent to lytic phase, a stage where it expresses a repertoire of more than 80 genes, accompanied by viral DNA replication and finally leading to the production of progeny virus particles (33, 34). The majority of the EBV miRNAs are transcribed from the BART and BHRF1 regions. BART and BHRF1 transcripts encode 40 and 4 mature miRNAs, respectively (34–36). BART and BHRF1 transcripts express different miRNAs during different phases of EBV latency (37). EBV-encoded BART transcripts are detected in EBV-associated NPC biopsy or EBV-positive cell lines in vitro (38, 39).
Following the identification of EBV-encoded miRNA, several reports identified miRNA expressed by Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8), another member of the γ-herpesvirus family. KSHV is known to cause Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), Multicentric Castleman's disease (MCD), and KSHV inflammatory cytokine syndrome (40–42). A total of 25 mature miRNA processed from 12 different pre-miRNAs have been identified in KSHV and are in the latency-associated region (43, 44). Out of the 12 pre-miRNAs, 10 pre-miRNAs are located between the kaposin and open reading frame 71 (ORF71) genes, while miR-K10 is located within the kaposin gene, and miR-K12 is mapped to the 3′-UTR of kaposin. Although all known KSHV v-miRNAs are expressed during the viral-latent phase, v-miRNAs originating from pre-miR-K10 and pre-miR-K12 are further enhanced during the viral-lytic phase (15, 16, 44, 45). Also, several of the KSHV-encoded v-miRNAs share the seed sequence with human-encoded miRNAs and therefore regulate many target genes. KSHV miRNAs help in maintaining KSHV latency and interfere with the host immune system by regulating viral and cellular gene expression, ultimately contributing to KS development (46).
β-Herpesvirus-Encoded v-miRNAs
Like γ-herpesvirus, α- and β-herpesvirus are also found to express v-miRNAs. The β-herpesvirus human cytomegalovirus (HCMV) is commonly found in the human population and has the largest genome, 230 kb of double-stranded DNA (dsDNA), among the herpesvirus family (47). HCMV causes serious life-threatening diseases in patients with a compromised immune system such as the human immunodeficiency virus (HIV) infection or patients undergoing immunosuppressive therapies (48, 49). v-miRNAs encoded by HCMV were first identified in 2005 (16). The study predicted and cloned nine pre-miRNAs, which were later validated in two independent studies by Northern blotting (50, 51). More studies on HCMV miRNAs identified additional miRNAs using deep sequencing technology (ref). A total of 26 HCMV v-miRNAs have been identified to date, dispersed throughout the genome (16, 51, 52). The HCMV miRNAs target multiple host genes involved in immune response and cell cycle control and thereby enhance HCMV virulence (53, 54).
HCMV miRs, namely, miR-UL-112-1, US25-1, US25-2, US25-2-5p, US5-1, US33-5p, and ULD148D, have been shown to inhibit HCMV DNA viral replication by targeting multiple host and viral regulatory genes (discussed in the following sections) (55–63). miR-UL112, US25-2-3p, and US4-1 modulate immune recognition by cytotoxic T lymphocyte (CTL) and natural killer (NK) cells (64–67). Similarly, the HCMV miRs UL112-3p, US5-1, UL112-1, US25-1-5p, and UL148D target multiple host inflammatory genes and result in reduced inflammatory response (53, 54, 68–71). Also, UL148D and UL36-5p are found to inhibit programmed cell death by repressing the expression of cellular genes involved in the regulation of apoptosis (72–74). Altogether, HCMV miRNAs play an important role in regulating the expression of host and viral genes to induce latent infection.
α-Herpesvirus-Encoded v-miRNAs
Herpes simplex virus (HSV) has two members, HSV-1 and HSV-2, which are known to cause oral or genital herpes lesions (75). v-miRNAs in HSV were first identified in 2006, and, to date, HSV-1 and HSV-2 are known to encode 27 and 24 functional v-miRNAs, respectively (76–78). Few of the HSV-1 and HSV-2 v-miRNAs share the same seed sequence (77, 78). Like γ-herpesvirus, HSV-1 and HSV-2 v-miRNAs are associated with latency-associated transcript and are expressed during the latent phase of infection (79).
Almost all herpesviruses encode their own v-miRNAs except varicella-zoster virus (VZV). Many small-RNA sequencing studies have been performed for VZV, but VZV v-miRNAs have not yet been identified (80).
Papillomavirus (PV)
Human papillomaviruses (HPV) preferentially infect the keratinocytes of mucous membranes or skin and cause numerous benign and malignant lesions at different anatomical locations. HPV infection is a common cause of cervical cancer (81–84). HPV infection is associated with varying proportions of other cancers of the anogenital tract, head and neck region, and skin (85).
The first report for the prediction of HPV-encoded miRNAs came in 2011. In that study, the authors predicted the HPV-encoded miRNAs in several HPV types. They predicted the pre-miRNAs using a computational algorithm and compared the conserved mature miRNAs with currently known miRNAs. Predicted HPV miRNAs related to miR-466,-467, and -669 were common and specific to the mucosal HPV types. Also, the authors observed that HPV-38 expresses a miRNA conserved to human let-7a (86). In another study, the authors generated small RNA libraries of 10 HPV-associated cervical cancer and two HPV16-harboring cell lines. From the sequencing data, nine putative HPV miRNAs were discovered. Four HPV-encoded miRNAs (two by HPV16, one by HPV38, and one by HPV68) were validated (87).
Similarly, another study developed miRNA discovery by using a forced genome expression (miDGE) tool for the identification of miRNAs. The study screened 73 different PV genomes using miDGE and observed that most of the PV genomes are unlikely to encode viral miRNAs. However, they could identify and validate five different miRNAs (hpv17-miR-H1, hpv37-miRH1, hpv41-miR-H1, fcpv1-miR-F1, and fcpv1-miR-F2 encoded by four different PVs: HPV17, 37, 41, and FcPV1). These HPV miRNAs targeted transcripts corresponding to the early region of the HPV genome (18).
Hepadnavirus
Hepatitis B virus (HBV) is the best-known member of the Hepadna virus family. Infection with HBV can cause chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC). HBV has infected 2 billion people worldwide (88). In comparison to healthy individuals, HBV-infected patients have a 100-fold higher risk of development of HCC (89).
To date, only one HBV-encoded miRNA has been identified by deep sequencing of HBV-positive HCC tissue. The corresponding study identified and confirmed the expression of HBV-miR-3. HBV-miR-3 downregulated HBV protein and HBV replication by reducing the expression of HBcAg, a positive regulator of HBV transcription and pregenomic RNA (pgRNA), which inhibits HBV replication overall (90).
Adenovirus
The adenovirus genome encodes two non-coding RNAs, namely VA-RNA I and VA-RNA II (91). VA RNAs are structurally like pre-miRNAs and bind and block Dicer (92, 93). However, 2–5% of VA RNAs can still be processed by Dicer in a manner similar to cellular miRNAs and can produce VA-RNA-derived miRNAs (called mivaRNAs) (92, 94). VA-RNA-I produces the two most abundant mivaRNAs (mivaRNAI-137 and mivaRNAI-138), and VA-RNA-II produces a single mivaRNA (mivaRNAII-138) (95). mivaRNAs have the potential to regulate cellular gene expression, which could be important in the adenovirus life cycle. Ectopic expression of mivaRNAI-138 in HeLa cells downregulated the expression of several genes controlling DNA repair, cell growth, apoptosis, and RNA metabolism (96). Recently, it was shown that mivaRNAII-138 enhanced Jun N-terminal kinase (JNK) signaling by downregulating CUL4, a negative regulator of the JNK signaling cascade (97). JNK signaling is crucial for viral replication, and several viruses have been reported to activate the JNK pathway upon infection (98–100). However, VA RNA-derived miRNA was dispensable for lytic replication of adenovirus in tissue culture cells. VA RNAI is a known suppressor of interferon-induced PKR enzyme, and, in the absence of PKR, the deletion of VA RNAI rescued viral late gene expression, suggesting that VA RNA-derived mivaRNAs might play an important role in inhibiting the PKR pathway to promote late gene expression (101).
Polyomavirus
Polyomavirus are non-enveloped viruses that infect a wide range of species including humans, primates, rodents, birds, and cattle (102). Few reports have characterized miRNAs in polyomaviruses. The betapolyomaviruses BK virus (BKV), JC virus (JCV), simian virus 40 (SV40), Merkel cell polyomavirus (MKV), and simian agent 12 (SA12) have one pre-miRNA at the 3′ end that encodes two mature miRNAs. These miRNAs control the viral replication by autoregulating the viral gene expression or inhibiting the viral T antigen expression to suppress antiviral T cell response (17, 103–105).
v-miRNAs in RNA Viruses
Most of our current understanding of v-miRNAs has been attributed to v-miRNAs that were produced by DNA viruses. The detection of v-miRNAs in RNA viruses has been controversial, with a few reports suggesting non-canonical miRNA-like small RNAs produced during RNA virus infections; however, these small RNAs lack the canonical stem-loop structure found in miRNAs, so their biogenesis and function are not well-understood (106, 107). The following reasons might explain the lack of v-miRNAs produced by RNA viruses during infection: (i) the RNA viruses consists either +/– sense or double-stranded RNA (dsRNA) and replicate in the cytoplasm of the host cell, so the RNA molecules are not accessible to the miRNA biogenesis machinery in the nuclei (108); (ii) excision of pre-miRNA from the primary transcript might result in the destruction of RNA-based viral genomes; (iii) the generated v-miRNA may target the viral genome itself, resulting in cleavage of the viral genome (109).
Influenza Virus
The influenza virus replicates inside the host nucleus and it therefore differs from most of the other RNA viruses. As the virus replicates inside the host nucleus, it can utilize the nuclear miRNA processing factor Drosha to express v-miRNAs (108). One of the hallmarks of H5N1 influenza virus infection is a strong and rapid production of antiviral cytokines, a process known as a cytokine storm. A study using an influenza virus engineered by incorporating a primary (pri) form of cellular miRNA-124 into the genome showed the ability to produce functional miR-124 without any deleterious effects on the viral life cycle. This study suggests that RNA viruses have the ability to exploit the host's small RNA machinery to produce v-miRNAs (110). The ability to produce functional miRNAs can be harnessed to mediate the delivery of miRNAs or siRNAs using RNA virus-based vectors (111). H5N1 encodes miRNA-like small RNA named miR-HA-3p, which accentuates the production of antiviral cytokines by targeting PCBP2, a known regulator of RIG/MAVS signaling. Suppression of PCBP2 leads to high levels of cytokine production and results in high mortality (112). In another study, it was shown that the 5′ end of all the eight segments of the influenza virus encodes small viral leader RNAs (leRNAs). LeRNAs are known to play a key role in the genomic RNA encapsidation to newly generate progeny virions, suggesting an important role in the viral life cycle (113).
Ebola Virus
Ebola virus (EBOV) is a negative-sense RNA virus that causes severe infectious disease with a high mortality rate of 83–90%. To date, few studies have identified miRNA-like small RNA encoded by EBOV (114–117). The first report was by Liang et al., who predicted that three mature miRNAs (EBOV-miR-1-5p,-3p, and miR-2-3p) are formed from two pre-miRNAs (EBOV-pre-miR-1 and EBOV-pre-miR-2) using the host cellular machinery. Further, they validated the production of mature miRNA by transfecting the pre-forms in HEK293T and showed reduced production of the mature miRNAs in Dicer-deficient cells (115). Similarly, another group performed genome-wide screening and predicted the formation of seven mature miRNAs from four pre-miRNAs (EBOV-pre-miRNA-T1, -T2, -T3, and -T4) (117). The third study identified two mature miRNAs and found that one miRNA, EBOV-miR-1-5p, serves as an analog of human miR-155 (116). Overexpression of EBOV-miR-1-5p inhibits the expression of importin-alpha5 in HEK293T cells. Reduced expression of importin-alpha5 could facilitate evasion of the host immune system (116). Although these reports identify miRNAs expressed in EBOV infection, the underlying mechanism is not so well-understood. With the development of a reverse genetics system for EBOV, characterizing the roles of these miRNAs in regulating the viral life cycle and immune response could yield insights into controlling EBOV pathogenesis. One more study showed that EBOV expresses a putative miRNA-like RNA fragment, EBOV-miR-VP-3p, which is highly conserved among other strains. EBOV-miR-VP-3p was found in the exosomes and was abundantly present in the sera of individuals infected with EBOV. Interestingly, EBOV-miR-VP-3p was detectable in the serum even before the detection of viral genomic RNA, indicating that EBOV miRNAs may serve as a biomarker for early diagnosis of EBOV infection (114).
HIV-1
Using a computational prediction tool, one study predicted that HIV-1 might encode five putative pre-miRNAs (118). Furthermore, another study showed that nef-derived miR-N367 suppresses nef expression by targeting HIV-1 nef transcript to regulate HIV-1 virulence (119). Subsequently, two independent studies found that the HIV-1 TAR element located at the 5′ end of HIV-1 encodes two microRNAs, namely miR-TAR-5p and miR-TAR-3p. TAR miRNAs target host apoptotic genes such as ERCC1 and IER3 that are involved in DNA repair to inhibit apoptosis (120–122). Another study found miR-H3 located in the active region of reverse transcriptase. miR-H3 was found to interact with HIV-1 5′LTR and enhance promoter activity, thereby increasing viral production (123). Next-generation sequencing technology has been widely used to identify and validate HIV-1 miRNAs. However, there have been some discrepancies in the miRNAs reported in different studies. Sequencing analysis of HIV-1-infected cell lines such as TZM-bl and CD8166 and primary human CD4+ PBMCs did not detect any HIV-1 miRNAs (124). More importantly, the HIV-1 miRNAs reported fail to satisfy the essential criteria for classification as authentic viral miRNAs. The majority of the HIV-1 miRNAs are derived from a few locations in the genome, their size is <20 nucleotides in length, and the pri-miRNA stem loop lacks the defining properties of canonical pri-miRNA stem loops (125). These outstanding questions on the functional interplay between HIV-1 miRNAs and cellular targets provide a significant opportunity to understand the viral pathogenesis better so as to develop anti-HIV-1 therapies.
Viral miRNAs in Immune Evasion
Targeting the Viral Gene Expression
One of the major functions of viral miRNAs involves targeting viral gene expression to control latency or as a switch from latency to activation (Figure 2A). SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells (17). HSV1-induced latency is driven by LAT (Latency Associated Transcript), which encodes for non-coding RNAs such as miR-H2-3p and miR-H6. These miRNAs target viral reactivation factors ICP0 and ICP4, which are essential in controlling viral reactivation from latency of HSV-1 (79). Similarly, for HSV-2, miR-I, miR-II, and miR-III expressed by LAT reduce the expression of ICP34.5, a key viral neurovirulence factor (126). miR-I is also expressed in human sacral dorsal root ganglia of neurons latently infected with HSV-2, suggesting the role of v-miRNAs in HSV-2 latency in human neurons (127).
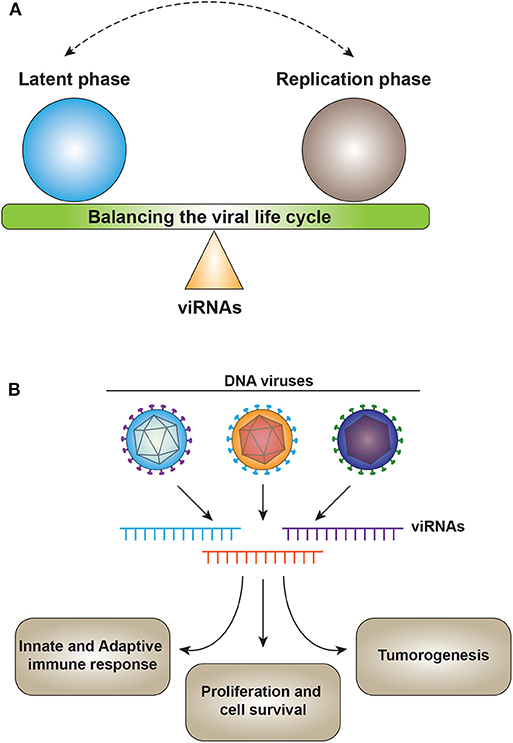
Figure 2. (A) v-miRNAs act as a fine switch between latency and the active replication phase of the viral lifecycle. (B) v-miRNAs control the host physiology by targeting multiple processes including immune response, cell survival, and tumorigenesis. v-miRNAs (viRNA).
For HCMV, miR-UL112-1 downregulated the expression of IE72 (UL123, IE1), UL112/113, and UL120/121 by translational inhibition rather than transcript degradation (55). miR-UL112 also targets the UL114 gene, a viral uracil DNA glycosylase, with minimal effects on viral growth. In addition, human fibroblast cells (HFF) ectopically expressing miR-US25-1 and miR-US25-2 reduced viral DNA synthesis for HCMV by downregulating IE72 and pp65, one of the most abundant proteins of the virion tegument of HCMV (62). Interestingly, the antiviral effects of the ectopically expressed HCMV miRNAs miR-US25-1 and miR-US25-2 were also observed for other DNA viruses such as HSV-1 and adenovirus, suggesting that these two miRNAs might target cellular genes that are essential for virus growth or to maintain latency (62).
EBV-encoded BART miRNAs play a key role in maintaining latency and controlling the viral life cycle. BART miRNAs such as miR-BART16, miR-BART17-5p, and miR-BART1-5p target the 3′ UTR of the LMP1 gene and negatively regulate LMP1 protein expression. LMP1 is capable of inducing cell growth and proliferation, but overexpression of LMP1 can result in inhibition of growth and apoptosis. Therefore, the downregulation of LMP1 expression may favor EBV-associated cancer development by exerting tight control on LMP1 expression (128). The EBV-encoded miRNA miR-BART2 inhibits the viral transition from the latent to the lytic part of the life cycle by suppressing the viral DNA polymerase BALF5 (129). Similarly, BART20-derived v-miRNA maintains the latency phase in EBV-associated tumors by targeting two EBV immediate-early genes, BZLF1, and BRLF1 (130).
Few reports have identified v-miRNAs involved in the maintenance of KSHV latency. The KSHV-encoded miRNA miRK9 targets the viral protein RTA, a major lytic switch protein. RTA plays an important role in controlling viral reactivation from latency. v-miRNA-mediated regulation of RTA fine-tunes viral reactivation in the KSHV life cycle (119). In addition to miR-K9, miR-K12-5 can inhibit RTA expression. However, unlike miR-K9, which targets a sequence in the 3′ UTR of RTA, the 3′ UTR of RTA does not contain a favorable seed sequence for miR-K12-5, suggesting an indirect effect on RTA expression (131).
As discussed in the previous sections, the expression of v-miRNAs by RNA viruses is highly controversial. Viral miRNAs are not detected in the majority of RNA virus families, mostly due to the inaccessibility of the host machinery required for miRNA biogenesis. Small viral RNAs (svRNAs) have been identified in influenza virus infections that play a role in switching the viral polymerase from transcription toward genome replication by interacting with the polymerase machinery. However, whether svRNAs target viral or host transcripts is unknown (132). Using in silico tools, the HIV-1 genome was putatively shown to encode five pre-miRNAs. Based on the mature miRNA sequence deduced from the pre-miRNAs, these miRNAs were computationally predicted to target a large set of host cellular genes to establish a favorable cellular milieu for viral replication (118).
Targeting the Host Cellular Genes
Evidence has been accumulating for v-miRNAs modulating the host immune response to enable a favorable intracellular milieu (Figure 2B). To date, studies of v-miRNAs targeting host cellular genes have been mainly focused on KSHV and EBV infections (Table 1). Host target cellular genes have been identified with the help of gene expression profiling in HEK293 cells, ectopically expressing the KSHV miRNA cluster. KSHV miRNA suppressed the expression of thrombospondin 1 (THBS1), strong tumor suppressor, and anti-angiogenic factor (159). KSHV miR-K12-1 controls cell survival and proliferation by targeting p21, a key tumor suppressor and inducer of cell cycle arrest (160). miR-K5, along with K12-9 and miR-K12-10b, targets Bcl-2-associated factor (BCLAF1), a known apoptotic factor (123). In addition, KSHV v-miRNAs reduce expression of C/EBPβ p20 (LIP), a known negative regulator of IL6 and IL10 cytokines, to regulate the cytokine signaling in infected cells (165). KSHV miR-K1 regulates the NF-κB pathway by directly targeting IκB. Suppressing IκB enhances NF-κB activity and inhibits viral lytic replication (161). KSHV miR-K12-10a suppresses the expression of TWEAKR (TNF-like weak inducer of apoptosis receptor) (169), whereas miR-K12-9 and miR-K12-5 target the TLR/Interleukin-1R signaling pathway by targeting IRAK1 and MYD88, thereby controlling inflammation (168). KSHV viral miRNAs also modulate the host gene expression to control pathogenesis. miR-K12-6 and miR-K12-11 direct the transcriptional reprogramming in latently infected cells by targeting the cellular transcription factor MAF (167). Several KSHV viral miRNAs target Retinoblastoma (Rb)-like protein 2 (Rbl-2), a negative regulator of DNA methyltransferases, to maintain latency (131). Using Ago2-based RIP-Chip in multiple B cell lines latently infected with KSHV or stably transduced to express 10 KSHV miRNAs identified genes involved in lymphocyte activation and pre-mRNA splicing such as LRRC8D and NHP2L1, respectively (139).
The same study identified Epstein-Barr virus (EBV) miRNAs targeting host cellular genes in EBV-positive B cell lines. EBV miRNAs were shown to regulate cellular transport processes by targeting key genes such as TOMM22 and IPO7 (139). EBV miR-BART5 controlled proliferation and established latent infection by targeting PUMA. PUMA is known to modulate apoptosis by p53, so by suppressing PUMA, EBV miRNAs alter the susceptibility to apoptotic agents and improve host cell survival (143). In EBV-associated non-Hodgkin's lymphomas, miR-BHRF1-3 targets T cell-attracting chemokine CXCL11. Sequestering miR-BHRF1-3 by antisense oligos reversed the suppression of CXCL11 in primary cultures derived from patients with EBV-positive Burkitt's lymphoma (153).
HCMV miRNAs have been shown to target host genes involved in the antiviral immune response. miR-UL112 blocks the natural killer (NK) cell-mediated recognition of virus-infected cells by inhibiting the expression of MICB, a stress-induced ligand essential for NK-cell activity. Suppression of MICB results in decreased binding to the NKG2D receptor, thereby leading to decreased killing of virally infected cells by NK cells (67). Interestingly, MICB expression is suppressed by KSHV miR-K12-7 and EBV miR-BART2 by binding to different sites in the 3′ UTR, highlighting a common strategy for immune evasion commonly used by multiple DNA viruses (140). Additionally, HCMV miR-US4-1 targets the endoplasmic reticulum-resident aminopeptidase ERAP1, which is required for MHC class I antigen presentation on CD8 T cells, resulting in less clearance of infected cells by HCMV-specific cytotoxic T cells (66).
Therapeutic Potential of Viral miRNAs
Combating viral diseases and virus-associated cancers is an ongoing health-related challenge present globally. Recent studies have focused on identifying miRNAs as targets for treating several diseases, including viral diseases. Antagomirs designed to sequester host miR-122 involved in HCV infection entered the phase II of human clinical trial and show promising effects against the infection (180, 181). Using similar approaches, v-miRNAs could also be targeted by using inhibitor or sponge-based antagomirs in DNA virus infections. A few reports support the notion of developing antagomirs against v-miRNAs rather than cellular miRNAs, as this could reduce the possibility of side-effects or off-target effects in human subjects and might solve the problem of non-toxic/site-specific, targeted delivery (182). In mouse cytomegalovirus (MCMV) infection, mice receiving antagomirs against MCMV v-miRNAs had reduced occurrence of MCMV upon challenge. In another study, gold nanoparticles containing anti-EBV-miR-BART7-3p were shown to therapeutically deliver anti-miRNAs against EBV-miR-BART7-3p, inhibiting the tumorigenicity of EBV-positive cells in mice (20, 183).
Similarly, using EBV promoters such as EBER2 promoter was reported to effectively express miRNA sponge to silence specific genes in EBV-infected cells and might be useful in the targeting of EBV-positive NPC cells (184). In addition, v-miRNAs were also considered as biomarkers in many virally infected diseases. miR-VP-3p was found to be present in the sera of EBOV-infected patients but not in healthy controls, and this v-miRNA was detectable in the serum prior to the detection of viral genomic RNA, indicating that miR-VP-3p may serve as a biomarker for early diagnosis of EBOV (114). HCMV-encoded miR-US4-1 serves as a biomarker for IFNα treatment potency in the serum of hepatitis B patients (135). Moreover, certain v-miRNA adapters (HSUR2) were discovered that recruit miRNA to target transcripts through alternative base-pairing. Inhibitors against these supportive v-miRNAs adaptors could be considered as an alternative therapeutic candidate (185). Finally, critical design and validation of miRNA-based studies in cell lines and animal models can help identify novel therapeutic candidates for treatments in the future.
Conclusion
Although the biogenesis, mechanism and function of virally encoded miRNAs are not well-characterized, substantial progress has been made in the last few years. With the emergence of high-throughput sequencing technologies and computational analysis tools, the number of newly discovered v-miRNAs is increasing. Multiple lines of evidence have strengthened the “classical” hypothesis of v-miRNAs solely originating from DNA viruses; however, some non-canonical miRNA-like RNA fragments have been detected during RNA virus infections. While the major functions of v-miRNAs across divergent virus families have been broadly attributed to immune evasion, autoregulation of the viral life cycle and tumorigenesis, there is still a broad gap in annotating the exact molecular determinants underlying these functions. The holy grail of the functional importance of v-miRNAs warrants more investigation to provide therapeutically amenable leads for targeting infectious diseases in the future.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. (2011) 147:1537–50. doi: 10.1016/j.cell.2011.11.055
2. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
3. Czech B, Munafo M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, et al. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet. (2018) 52:131–57. doi: 10.1146/annurev-genet-120417-031441
4. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
6. Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. (2008) 9:831–42. doi: 10.1038/nrg2455
7. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. (2004) 5:522–31. doi: 10.1038/nrg1379
8. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. (2004) 305:1437–41. doi: 10.1126/science.1102513
9. Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. (2004) 15:185–97. doi: 10.1016/j.molcel.2004.07.007
10. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. (2004) 23:4051–60. doi: 10.1038/sj.emboj.7600385
11. Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. (2006) 13:1097–101. doi: 10.1038/nsmb1167
12. Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. (2011) 411:325–43. doi: 10.1016/j.virol.2011.01.002
13. Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. (2012) 8:e1003018. doi: 10.1371/journal.ppat.1003018
14. Grundhoff A. Computational prediction of viral miRNAs. Methods Mol Biol. (2011) 721:143–52. doi: 10.1007/978-1-61779-037-9_8
15. Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. (2006) 12:733–50. doi: 10.1261/rna.2326106
16. Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. (2005) 2:269–76. doi: 10.1038/nmeth746
17. Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. (2005) 435:682–6. doi: 10.1038/nature03576
18. Chirayil R, Kincaid RP, Dahlke C, Kuny CV, Dalken N, Spohn M, et al. Identification of virus-encoded microRNAs in divergent Papillomaviruses. PLoS Pathog. (2018) 14:e1007156. doi: 10.1371/journal.ppat.1007156
19. Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science. (2004) 304:734–36. doi: 10.1126/science.1096781
20. Wang M, Gu B, Chen X, Wang Y, Li P, Wang K. The function and therapeutic potential of epstein-barr virus-encoded microRNAs in cancer. Mol Ther Nucleic Acids. (2019) 17:657–68. doi: 10.1016/j.omtn.2019.07.002
21. Kim H, Iizasa H, Kanehiro Y, Fekadu S, Yoshiyama H. Herpesviral microRNAs in cellular metabolism and immune responses. Front Microbiol. (2017) 8:1318. doi: 10.3389/fmicb.2017.01318
22. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. (2009) 136:642–55. doi: 10.1016/j.cell.2009.01.035
23. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. (2003) 425:415–9. doi: 10.1038/nature01957
24. Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. (2005) 24:138–48. doi: 10.1038/sj.emboj.7600491
25. Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. (2002) 297:2056–60. doi: 10.1126/science.1073827
26. Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. (2002) 110:563–74. doi: 10.1016/S0092-8674(02)00908-X
27. Cohen JI, Fauci AS, Varmus H, Nabel GJ. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med. (2011) 3:107fs7. doi: 10.1126/scitranslmed.3002878
28. Jha HC, Pei Y, Robertson ES. Epstein-Barr Virus: diseases linked to infection and transformation. Front Microbiol. (2016) 7:1602. doi: 10.3389/fmicb.2016.01602
29. Javier RT, Butel JS. The history of tumor virology. Cancer Res. (2008) 68:7693–706. doi: 10.1158/0008-5472.CAN-08-3301
31. Fields BN, Knipe DM, Howley PM. Fields virology. Philadelphia, PA: Wolters Kluwer Health/Lippincott Williams & Wilkins (2013).
32. Niedobitek G, Agathanggelou A, Herbst H, Whitehead L, Wright DH, Young LS. Epstein-Barr virus (EBV) infection in infectious mononucleosis: virus latency, replication and phenotype of EBV-infected cells. J Pathol. (1997) 182:151–9. doi: 10.1002/(SICI)1096-9896(199706)182:2<151::AID-PATH824>3.0.CO;2-3
33. Ressing ME, van Gent M, Gram AM, Hooykaas MJ, Piersma SJ, Wiertz EJ. Immune evasion by Epstein-Barr virus. Curr Top Microbiol Immunol. (2015) 391:355–81. doi: 10.1007/978-3-319-22834-1_12
34. Kenney SC, Mertz JE. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin Cancer Biol. (2014) 26:60–8. doi: 10.1016/j.semcancer.2014.01.002
35. Klinke O, Feederle R, Delecluse HJ. Genetics of Epstein-Barr virus microRNAs. Semin Cancer Biol. (2014) 26:52–9. doi: 10.1016/j.semcancer.2014.02.002
36. Barth S, Meister G, Grasser FA. EBV-encoded miRNAs. Biochim Biophys Acta. (2011) 1809:631–40. doi: 10.1016/j.bbagrm.2011.05.010
37. Kang MS, Kieff E. Epstein-Barr virus latent genes. Exp Mol Med. (2015) 47:e131. doi: 10.1038/emm.2014.84
38. Amoroso R, Fitzsimmons L, Thomas WA, Kelly GL, Rowe M, Bell AI. Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J Virol. (2011) 85:996–1010. doi: 10.1128/JVI.01528-10
39. Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, et al. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol. (2009) 83:2357–67. doi: 10.1128/JVI.02104-08
40. Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. (1995) 86:1276–80. doi: 10.1182/blood.V86.4.1276.bloodjournal8641276
41. Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. (1995) 332:1186–91. doi: 10.1056/NEJM199505043321802
42. Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. (1994) 266:1865–9. doi: 10.1126/science.7997879
43. Cai X, Cullen BR. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J Virol. (2006) 80:2234–42. doi: 10.1128/JVI.80.5.2234-2242.2006
44. Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. (2005) 102:5570–5. doi: 10.1073/pnas.0408192102
45. Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. (2005) 79:9301–5. doi: 10.1128/JVI.79.14.9301-9305.2005
46. Lei X, Zhu Y, Jones T, Bai Z, Huang Y, Gao SJ. A Kaposi's sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor β pathway to promote cell survival. J Virol. (2012) 86:11698–711. doi: 10.1128/JVI.06855-11
47. Gandhi MK, Khanna R. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect Dis. (2004) 4:725–38. doi: 10.1016/S1473-3099(04)01202-2
48. Ljungman P, Hakki M, Boeckh M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol Oncol Clin North Am. (2011) 25:151–69. doi: 10.1016/j.hoc.2010.11.011
49. Ramanan P, Razonable RR. Cytomegalovirus infections in solid organ transplantation: a review. Infect Chemother. (2013) 45:260–71. doi: 10.3947/ic.2013.45.3.260
50. Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. (2005) 7:1684–95. doi: 10.1111/j.1462-5822.2005.00598.x
51. Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, et al. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. (2005) 79:12095–9. doi: 10.1128/JVI.79.18.12095-12099.2005
52. Stark TJ, Arnold JD, Spector DH, Yeo GW. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol. (2012) 86:226–35. doi: 10.1128/JVI.05903-11
53. Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA. Human cytomegalovirus miR-UL112–3p targets TLR2 and modulates the TLR2/IRAK1/NFκB signaling pathway. PLoS Pathog. (2015) 11:e1004881. doi: 10.1371/journal.ppat.1004881
54. Hancock MH, Hook LM, Mitchell J, Nelson JA. Human cytomegalovirus MicroRNAs miR-US5–1 and miR-UL112–3p block proinflammatory cytokine production in response to NF-κB-activating factors through direct downregulation of IKKα and IKKβ. mBio. (2017) 8:e00109–17. doi: 10.1128/mBio.00109-17
55. Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. (2007) 3:e163. doi: 10.1371/journal.ppat.0030163
56. Jiang S, Huang Y, Qi Y, He R, Liu Z, Ma Y, et al. Human cytomegalovirus miR-US5–1 inhibits viral replication by targeting Geminin mRNA. Virol Sin. (2017) 32:431–9. doi: 10.1007/s12250-017-4064-x
57. Pan C, Zhu D, Wang Y, Li L, Li D, Liu F, et al. Human cytomegalovirus miR-UL148D facilitates latent viral infection by targeting host cell immediate early response gene 5. PLoS Pathog. (2016) 12:e1006007. doi: 10.1371/journal.ppat.1006007
58. Jiang S, Qi Y, He R, Huang Y, Liu Z, Ma Y, et al. Human cytomegalovirus microRNA miR-US25–1-5p inhibits viral replication by targeting multiple cellular genes during infection. Gene. (2015) 570:108–14. doi: 10.1016/j.gene.2015.06.009
59. Guo X, Qi Y, Huang Y, Liu Z, Ma Y, Shao Y, et al. Human cytomegalovirus miR-US33–5p inhibits viral DNA synthesis and viral replication by down-regulating expression of the host Syntaxin3. FEBS Lett. (2015) 589:440–6. doi: 10.1016/j.febslet.2014.12.030
60. Pavelin J, Reynolds N, Chiweshe S, Wu G, Tiribassi R, Grey F. Systematic microRNA analysis identifies ATP6V0C as an essential host factor for human cytomegalovirus replication. PLoS Pathog. (2013) 9:e1003820. doi: 10.1371/journal.ppat.1003820
61. Qi M, Qi Y, Ma Y, He R, Ji Y, Sun Z, et al. Over-expression of human cytomegalovirus miR-US25–2-3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Lett. (2013) 587:2266–71. doi: 10.1016/j.febslet.2013.05.057
62. Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol. (2009) 83:10684–93. doi: 10.1128/JVI.01292-09
63. Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci USA. (2008) 105:5453–8. doi: 10.1073/pnas.0711910105
64. Huang Y, Chen D, He J, Cai J, Shen K, Liu X, et al. Hcmv-miR-UL112 attenuates NK cell activity by inhibition type I interferon secretion. Immunol Lett. (2015) 163:151–6. doi: 10.1016/j.imlet.2014.12.003
65. Esteso G, Luzon E, Sarmiento E, Gomez-Caro R, Steinle A, Murphy G, et al. Altered microRNA expression after infection with human cytomegalovirus leads to TIMP3 downregulation and increased shedding of metalloprotease substrates, including MICA. J Immunol. (2014) 193:1344–52. doi: 10.4049/jimmunol.1303441
66. Kim S, Lee S, Shin J, Kim Y, Evnouchidou I, Kim D, et al. Human cytomegalovirus microRNA miR-US4–1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat Immunol. (2011) 12:984–91. doi: 10.1038/ni.2097
67. Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, et al. Host immune system gene targeting by a viral miRNA. Science. (2007) 317:376–81. doi: 10.1126/science.1140956
68. Chen J, Xia S, Yang X, Chen H, Li F, Liu F, et al. Human cytomegalovirus encoded miR-US25–1-5p attenuates CD147/EMMPRIN-mediated early antiviral response. Viruses. (2017) 9:365. doi: 10.3390/v9120365
69. Lau B, Poole E, Krishna B, Sellart I, Wills MR, Murphy E, et al. The expression of human cytomegalovirus MicroRNA MiR-UL148D during latent infection in primary myeloid cells inhibits activin A-triggered secretion of IL-6. Sci Rep. (2016) 6:31205. doi: 10.1038/srep33771
70. Huang Y, Qi Y, Ma Y, He R, Ji Y, Sun Z, et al. The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by hcmv-miR-UL112–1. Virol J. (2013) 10:51. doi: 10.1186/1743-422X-10-51
71. Kim Y, Lee S, Kim S, Kim D, Ahn JH, Ahn K. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. (2012) 8:e1002577. doi: 10.1371/journal.ppat.1002577
72. Guo X, Huang Y, Qi Y, Liu Z, Ma Y, Shao Y, et al. Human cytomegalovirus miR-UL36–5p inhibits apoptosis via downregulation of adenine nucleotide translocator 3 in cultured cells. Arch Virol. (2015) 160:2483–90. doi: 10.1007/s00705-015-2498-8
73. Babu SG, Pandeya A, Verma N, Shukla N, Kumar RV, Saxena S. Role of HCMV miR-UL70–3p and miR-UL148D in overcoming the cellular apoptosis. Mol Cell Biochem. (2014) 393:89–98. doi: 10.1007/s11010-014-2049-8
74. Wang YP, Qi Y, Huang YJ, Qi ML, Ma YP, He R, et al. Identification of immediate early gene X-1 as a cellular target gene of hcmv-mir-UL148D. Int J Mol Med. (2013) 31:959–66. doi: 10.3892/ijmm.2013.1271
75. Wagner EK, Devi-Rao G, Feldman LT, Dobson AT, Zhang YF, Flanagan WM, et al. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. (1988) 62:1194–202. doi: 10.1128/JVI.62.4.1194-1202.1988
76. Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. (2006) 80:5499–508. doi: 10.1128/JVI.00200-06
77. Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, et al. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. (2010) 84:4659–72. doi: 10.1128/JVI.02725-09
78. Umbach JL, Wang K, Tang S, Krause PR, Mont EK, Cohen JI, et al. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J Virol. (2010) 84:1189–92. doi: 10.1128/JVI.01712-09
79. Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. (2008) 454:780–3. doi: 10.1038/nature07103
80. Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. (2009) 83:10677–83. doi: 10.1128/JVI.01185-09
81. Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. (2003) 348:518–27. doi: 10.1056/NEJMoa021641
82. Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. (2013) 382:889–99. doi: 10.1016/S0140-6736(13)60022-7
83. Munoz N, Bosch FX, de Sanjose S, Tafur L, Izarzugaza I, Gili M, et al. The causal link between human papillomavirus and invasive cervical cancer: a population-based case-control study in Colombia and Spain. Int J Cancer. (1992) 52:743–9. doi: 10.1002/ijc.2910520513
84. Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. (1999) 189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F
85. Parkin DM. The global health burden of infection-associated cancers in the year (2002). Int J Cancer. (2006) 118:3030–44. doi: 10.1002/ijc.21731
86. Gu W, An J, Ye P, Zhao KN, Antonsson A. Prediction of conserved microRNAs from skin and mucosal human papillomaviruses. Arch Virol. (2011) 156:1161–71. doi: 10.1007/s00705-011-0974-3
87. Qian K, Pietila T, Ronty M, Michon F, Frilander MJ, Ritari J, et al. Identification and validation of human papillomavirus encoded microRNAs. PLoS ONE. (2013) 8:e70202. doi: 10.1371/journal.pone.0070202
88. Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. (2014) 384:2053–63. doi: 10.1016/S0140-6736(14)60220-8
89. Xu C, Zhou W, Wang Y, Qiao L. Hepatitis B virus-induced hepatocellular carcinoma. Cancer Lett. (2014) 345:216–22. doi: 10.1016/j.canlet.2013.08.035
90. Yang X, Li H, Sun H, Fan H, Hu Y, Liu M, et al. Hepatitis B virus-encoded MicroRNA controls viral replication. J Virol. (2017) 91:e01919–16. doi: 10.1128/JVI.01919-16
91. Ma Y, Mathews MB. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J Virol. (1996) 70:5083–99. doi: 10.1128/JVI.70.8.5083-5099.1996
92. Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. (2005) 79:9556–65. doi: 10.1128/JVI.79.15.9556-9565.2005
93. Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. (2004) 78:12868–76. doi: 10.1128/JVI.78.23.12868-12876.2004
94. Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol. (2006) 80:1376–84. doi: 10.1128/JVI.80.3.1376-1384.2006
95. Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol. (2007) 81:10540–9. doi: 10.1128/JVI.00885-07
96. Aparicio O, Carnero E, Abad X, Razquin N, Guruceaga E, Segura V, et al. Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res. (2010) 38:750–63. doi: 10.1093/nar/gkp1028
97. Wakabayashi K, Machitani M, Tachibana M, Sakurai F, Mizuguchi H. A MicroRNA derived from adenovirus virus-associated RNAII promotes virus infection via posttranscriptional gene silencing. J Virol. (2019) 93:e01265–18. doi: 10.1128/JVI.01265-18
98. Ceballos-Olvera I, Chavez-Salinas S, Medina F, Ludert JE, del Angel RM. JNK phosphorylation, induced during dengue virus infection, is important for viral infection and requires the presence of cholesterol. Virology. (2010) 396:30–6. doi: 10.1016/j.virol.2009.10.019
99. McLean TI, Bachenheimer SL. Activation of cJUN N-terminal kinase by herpes simplex virus type 1 enhances viral replication. J Virol. (1999) 73:8415–26. doi: 10.1128/JVI.73.10.8415-8426.1999
100. Ludwig S, Ehrhardt C, Neumeier ER, Kracht M, Rapp UR, Pleschka S. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J Biol Chem. (2001) 276:10990–8. doi: 10.1074/jbc.M009902200
101. Kamel W, Segerman B, Oberg D, Punga T, Akusjarvi G. The adenovirus VA RNA-derived miRNAs are not essential for lytic virus growth in tissue culture cells. Nucleic Acids Res. (2013) 41:4802–12. doi: 10.1093/nar/gkt172
102. Moens U, Calvignac-Spencer S, Lauber C, Ramqvist T, Feltkamp MCW, Daugherty MD, et al. ICTV Virus taxonomy profile: polyomaviridae. J Gen Virol. (2017) 98:1159–60. doi: 10.1099/jgv.0.000839
103. Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. (2009) 383:183–7. doi: 10.1016/j.virol.2008.11.001
104. Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. (2008) 82:9823–8. doi: 10.1128/JVI.01144-08
105. Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, et al. Complete nucleotide sequence of polyomavirus SA12. J Virol. (2005) 79:13094–104. doi: 10.1128/JVI.79.20.13094-13104.2005
106. Varble A, ten Oever BR. Implications of RNA virus-produced miRNAs. RNA Biol. (2011) 8:190–4. doi: 10.4161/rna.8.2.13983
107. Aguado LC, tenOever B. RNA virus building blocks-miRNAs not included. PLoS Pathog. (2018) 14:e1006963. doi: 10.1371/journal.ppat.1006963
108. Neumann G, Brownlee GG, Fodor E, Kawaoka Y. Orthomyxovirus replication, transcription, and polyadenylation. Curr Top Microbiol Immunol. (2004) 283:121–43. doi: 10.1007/978-3-662-06099-5_4
109. Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. (2010) 6:e1000787. doi: 10.1371/journal.ppat.1000787
110. Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci USA. (2010) 107:11519–24. doi: 10.1073/pnas.1003115107
111. tenOever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. (2013) 11:169–80. doi: 10.1038/nrmicro2971
112. Li X, Fu Z, Liang H, Wang Y, Qi X, Ding M, et al. H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly(rC)-binding protein 2. Cell Res. (2018) 28:157–71. doi: 10.1038/cr.2018.3
113. Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio. (2010) 1:e00204–10. doi: 10.1128/mBio.00204-10
114. Chen Z, Liang H, Chen X, Ke Y, Zhou Z, Yang M, et al. An Ebola virus-encoded microRNA-like fragment serves as a biomarker for early diagnosis of Ebola virus disease. Cell Res. (2016) 26:380–3. doi: 10.1038/cr.2016.21
115. Liang H, Zhou Z, Zhang S, Zen K, Chen X, Zhang C. Identification of Ebola virus microRNAs and their putative pathological function. Sci China Life Sci. (2014) 57:973–81. doi: 10.1007/s11427-014-4759-2
116. Liu Y, Sun J, Zhang H, Wang M, Gao GF, Li X. Ebola virus encodes a miR-155 analog to regulate importin-α5 expression. Cell Mol Life Sci. (2016) 73:3733–44. doi: 10.1007/s00018-016-2215-0
117. Teng Y, Wang Y, Zhang X, Liu W, Fan H, Yao H, et al. Systematic genome-wide screening and prediction of microRNAs in EBOV during the 2014 ebolavirus outbreak. Sci Rep. (2015) 5:9912. doi: 10.1038/srep09912
118. Bennasser Y, Le SY, Yeung ML, Jeang KT. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. (2004) 1:43. doi: 10.1186/1742-4690-1-43
119. Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. (2004) 1:44. doi: 10.1186/1742-4690-1-44
120. Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. (2008) 36:2353–65. doi: 10.1093/nar/gkn076
121. Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. (2007) 8:63. doi: 10.1186/1471-2199-8-63
122. Klase Z, Winograd R, Davis J, Carpio L, Hildreth R, Heydarian M, et al. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology. (2009) 6:18. doi: 10.1186/1742-4690-6-18
123. Zhang Y, Fan M, Geng G, Liu B, Huang Z, Luo H, et al. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology. (2014) 11:23. doi: 10.1186/1742-4690-11-23
124. Whisnant AW, Kehl T, Bao Q, Materniak M, Kuzmak J, Lochelt M, et al. Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J Virol. (2014) 88:4679–86. doi: 10.1128/JVI.03587-13
125. Balasubramaniam M, Pandhare J, Dash C. Are microRNAs important players in HIV-1 infection? An update. Viruses. (2018) 10:E110. doi: 10.3390/v10030110
126. Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. (2009) 83:1433–42. doi: 10.1128/JVI.01723-08
127. Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci USA. (2008) 105:10931–6. doi: 10.1073/pnas.0801845105
128. Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. (2007) 104:16164–9. doi: 10.1073/pnas.0702896104
129. Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. (2008) 36:666–75. doi: 10.1093/nar/gkm1080
130. Jung YJ, Choi H, Kim H, Lee SK. MicroRNA miR-BART20–5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J Virol. (2014) 88:9027–37. doi: 10.1128/JVI.00721-14
131. Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J Virol. (2010) 84:2697–706. doi: 10.1128/JVI.01997-09
132. Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci USA. (2010) 107:11525–30. doi: 10.1073/pnas.1001984107
133. Enk J, Levi A, Weisblum Y, Yamin R, Charpak-Amikam Y, Wolf DG, et al. HSV1 MicroRNA modulation of GPI anchoring and downstream immune evasion. Cell Rep. (2016) 17:949–56. doi: 10.1016/j.celrep.2016.09.077
134. Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, et al. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe. (2014) 15:363–73. doi: 10.1016/j.chom.2014.02.004
135. Pan Y, Wang N, Zhou Z, Liang H, Pan C, Zhu D, et al. Circulating human cytomegalovirus-encoded HCMV-miR-US4–1 as an indicator for predicting the efficacy of IFNα treatment in chronic hepatitis B patients. Sci Rep. (2016) 6:23007. doi: 10.1038/srep23007
136. Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL, et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. (2009) 11:1174–84. doi: 10.1593/neo.09888
137. Kanda T, Miyata M, Kano M, Kondo S, Yoshizaki T, Iizasa H. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J Virol. (2015) 89:2684–97. doi: 10.1128/JVI.03189-14
138. Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J, et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc Natl Acad Sci USA. (2016) 113:E6467–75. doi: 10.1073/pnas.1605884113
139. Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. (2010) 7:324–34. doi: 10.1016/j.chom.2010.03.008
140. Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. (2009) 5:376–85. doi: 10.1016/j.chom.2009.03.003
141. Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. (2000) 1:119–26. doi: 10.1038/77793
142. Lisnic VJ, Krmpotic A, Jonjic S. Modulation of natural killer cell activity by viruses. Curr Opin Microbiol. (2010) 13:530–9. doi: 10.1016/j.mib.2010.05.011
143. Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. (2008) 205:2551–60. doi: 10.1084/jem.20072581
144. Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y, et al. Epstein-Barr virus MicroRNA miR-BART5–3p Inhibits p53 expression. J Virol. (2018) 92:e01022–18. doi: 10.1128/JVI.01022-18
145. Huang WT, Lin CW. EBV-encoded miR-BART20–5p and miR-BART8 inhibit the IFN- γ-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am J Pathol. (2014) 184:1185–97. doi: 10.1016/j.ajpath.2013.12.024
146. Ross N, Gandhi MK, Nourse JP. The Epstein-Barr virus microRNA BART11–5p targets the early B-cell transcription factor EBF1. Am J Blood Res. (2013) 3:210–24.
147. Choi H, Lee H, Kim SR, Gho YS, Lee SK. Epstein-Barr virus-encoded microRNA BART15–3p promotes cell apoptosis partially by targeting BRUCE. J Virol. (2013) 87:8135–44. doi: 10.1128/JVI.03159-12
148. Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J Immunol. (2012) 189:3795–9. doi: 10.4049/jimmunol.1200312
149. Hooykaas MJG, van Gent M, Soppe JA, Kruse E, Boer IGJ, van Leenen D, et al. EBV MicroRNA BART16 suppresses type I IFN signaling. J Immunol. (2017) 198:4062–73. doi: 10.4049/jimmunol.1501605
150. Qiu J, Thorley-Lawson DA. EBV microRNA BART 18–5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci USA. (2014) 111:11157–62. doi: 10.1073/pnas.1406136111
151. Kim H, Choi H, Lee SK. Epstein-Barr virus miR-BART20–5p regulates cell proliferation and apoptosis by targeting BAD. Cancer Lett. (2015) 356:733–42. doi: 10.1016/j.canlet.2014.10.023
152. Ma J, Nie K, Redmond D, Liu Y, Elemento O, Knowles DM, et al. EBV-miR-BHRF1–2 targets PRDM1/Blimp1: potential role in EBV lymphomagenesis. Leukemia. (2016) 30:594–604. doi: 10.1038/leu.2015.285
153. Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1–3. Cancer Res. (2008) 68:1436–42. doi: 10.1158/0008-5472.CAN-07-5126
154. Wahl A, Linnstaedt SD, Esoda C, Krisko JF, Martinez-Torres F, Delecluse HJ, et al. A cluster of virus-encoded microRNAs accelerates acute systemic Epstein-Barr virus infection but does not significantly enhance virus-induced oncogenesis in vivo. J Virol. (2013) 87:5437–46. doi: 10.1128/JVI.00281-13
155. Feederle R, Linnstaedt SD, Bannert H, Lips H, Bencun M, Cullen BR, et al. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. (2011) 7:e1001294. doi: 10.1371/journal.ppat.1001294
156. Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. (2010) 6:e1001063. doi: 10.1371/journal.ppat.1001063
157. Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K. miR-K12–7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS ONE. (2011) 6:e16224. doi: 10.1371/journal.pone.0016224
158. Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. (2009) 6:570–5. doi: 10.1016/j.chom.2009.11.008
159. Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. (2007) 3:e65. doi: 10.1371/journal.ppat.0030065
160. Gottwein E, Cullen BR. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol. (2010) 84:5229–37. doi: 10.1128/JVI.00202-10
161. Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, et al. Regulation of NF-κB inhibitor IκBα and viral replication by a KSHV microRNA. Nat Cell Biol. (2010) 12:193–9. doi: 10.1038/ncb2019
162. Moody R, Zhu Y, Huang Y, Cui X, Jones T, Bedolla R, et al. KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog. (2013) 9:e1003857. doi: 10.1371/journal.ppat.1003857
163. Suffert G, Malterer G, Hausser J, Viiliainen J, Fender A, Contrant M, et al. Kaposi's sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog. (2011) 7:e1002405. doi: 10.1371/journal.ppat.1002405
164. Lu CC, Li Z, Chu CY, Feng J, Feng J, Sun R, et al. MicroRNAs encoded by Kaposi's sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. (2010) 11:784–90. doi: 10.1038/embor.2010.132
165. Qin Z, Kearney P, Plaisance K, Parsons CH. Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol. (2010) 87:25–34. doi: 10.1189/jlb.0409251
166. Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. (2009) 41:130–4. doi: 10.1038/ng.266
167. Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, et al. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. (2010) 24:195–205. doi: 10.1101/gad.553410
168. Abend JR, Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM. Kaposi's sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J Virol. (2012) 86:11663–74. doi: 10.1128/JVI.01147-12
169. Abend JR, Uldrick T, Ziegelbauer JM. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol. (2010) 84:12139–51. doi: 10.1128/JVI.00884-10
170. Sin SH, Kim YB, Dittmer DP. Latency locus complements MicroRNA 155 deficiency in vivo. J Virol. (2013) 87:11908–11. doi: 10.1128/JVI.01620-13
171. Boss IW, Nadeau PE, Abbott JR, Yang Y, Mergia A, Renne R. A Kaposi's sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rγnull mice. J Virol. (2011) 85:9877–86. doi: 10.1128/JVI.05558-11
172. Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. (2007) 450:1096–9. doi: 10.1038/nature05992
173. Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. (2007) 81:12836–45. doi: 10.1128/JVI.01804-07
174. Duy J, Honko AN, Altamura LA, Bixler SL, Wollen-Roberts S, Wauquier N, et al. Virus-encoded miRNAs in Ebola virus disease. Sci Rep. (2018) 8:6480. doi: 10.1038/s41598-018-23916-z
175. Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, et al. West Nile virus encodes a microRNA-like small RNA in the 3' untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. (2012) 40:2210–23. doi: 10.1093/nar/gkr848
176. Hussain M, Asgari S. MicroRNA-like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc Natl Acad Sci USA. (2014) 111:2746–51. doi: 10.1073/pnas.1320123111
177. Islam MS, Khan MA, Murad MW, Karim M, Islam A. In silico analysis revealed Zika virus miRNAs associated with viral pathogenesis through alteration of host genes involved in immune response and neurological functions. J Med Virol. (2019) 91:1584–94. doi: 10.1002/jmv.25505
178. Shi J, Duan Z, Sun J, Wu M, Wang B, Zhang J, et al. Identification and validation of a novel microRNA-like molecule derived from a cytoplasmic RNA virus antigenome by bioinformatics and experimental approaches. Virol J. (2014) 11:121. doi: 10.1186/1743-422X-11-121
179. Ouellet DL, Vigneault-Edwards J, Letourneau K, Gobeil LA, Plante I, Burnett JC, et al. Regulation of host gene expression by HIV-1 TAR microRNAs. Retrovirology. (2013) 10:86. doi: 10.1186/1742-4690-10-86
180. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. (2013) 368:1685–94. doi: 10.1056/NEJMoa1209026
181. van der Ree MH, de Vree JM, Stelma F, Willemse S, van der Valk M, Rietdijk S, et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: a phase 1B, double-blind, randomised controlled trial. Lancet. (2017) 389:709–17. doi: 10.1016/S0140-6736(16)31715-9
182. Tahamtan A, Inchley CS, Marzban M, Tavakoli-Yaraki M, Teymoori-Rad M, Nakstad B, et al. The role of microRNAs in respiratory viral infection: friend or foe? Rev Med Virol. (2016) 26:389–407. doi: 10.1002/rmv.1894
183. Cai L, Li J, Zhang X, Lu Y, Wang J, Lyu X, et al. Gold nano-particles (AuNPs) carrying anti-EBV-miR-BART7–3p inhibit growth of EBV-positive nasopharyngeal carcinoma. Oncotarget. (2015) 6:7838–50. doi: 10.18632/oncotarget.3046
184. Choy EY, Kok KH, Tsao SW, Jin DY. Utility of Epstein-Barr virus-encoded small RNA promoters for driving the expression of fusion transcripts harboring short hairpin RNAs. Gene Ther. (2008) 15:191–202. doi: 10.1038/sj.gt.3303055
Keywords: host immunity, viruses, MicroRNAs, infection, pathogenesis
Citation: Mishra R, Kumar A, Ingle H and Kumar H (2020) The Interplay Between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 10:3079. doi: 10.3389/fimmu.2019.03079
Received: 31 October 2019; Accepted: 17 December 2019;
Published: 23 January 2020.
Edited by:
Afsar Raza Naqvi, University of Illinois at Chicago, United StatesReviewed by:
Niels A. W. Lemmermann, Johannes Gutenberg-University of Mainz, GermanyJulio Martin-Garcia, Drexel University, United States
Copyright © 2020 Mishra, Kumar, Ingle and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Himanshu Kumar, aGt1bWFyQGlpc2VyYi5hYy5pbg==
†These authors have contributed equally to this work
 Richa Mishra1†
Richa Mishra1† Ashish Kumar
Ashish Kumar Harshad Ingle
Harshad Ingle Himanshu Kumar
Himanshu Kumar