- 1Department of Virology and Immunology, Auckland City Hospital, Auckland, New Zealand
- 2Department of Clinical Immunology, Auckland City Hospital, Auckland, New Zealand
- 3Department of Molecular Medicine and Pathology, Faculty of Medical and Health Sciences, University of Auckland, Auckland, New Zealand
- 4School of Biological Sciences, University of Auckland, Auckland, New Zealand
Introduction
Common Variable Immunodeficiency Disorders (CVID) are a rare group of primary immunodeficiency disorders (PIDs) where late onset antibody failure leads to immune system failure (1). Onset of symptoms can occur from early childhood to the eighth decade or later (2). Current estimates suggest a prevalence between 1: 25 000 to 1: 100 000 in Caucasians (3, 4). For reasons that are unclear, CVID appears to be less frequent in Asian and African populations, although there may be ascertainment bias.
The majority of patients with CVID present with recurrent and severe infections. Untreated, patients are predisposed to chronic suppuration of the respiratory tract, often resulting in chronic sinus disease and bronchiectasis. Approximately 25% of CVID patients suffer autoimmune or inflammatory sequelae, consequent to immune dysregulation (5). There is also an increased risk of malignancy (6).
There is no single clinical feature or laboratory test, which is pathognomonic for CVID. The identification of CVID therefore relies on diagnostic criteria. There are currently three new sets of diagnostic criteria for CVID (7–9). The Ameratunga et al. (7) criteria require symptomatic primary hypogammaglobulinemia with relevant laboratory tests to establish the diagnosis. The threshold for IgG was set at 5 g/l for adults. The revised European Society for immunodeficiencies (8) are similar to the Ameratunga et al. criteria.
The most recent International Consensus Document (9) CVID criteria claim to be able to make a definite diagnosis on the basis of a single abnormal vaccine challenge result in a patient with primary hypogammaglobulinemia (9). The latter two criteria have set the threshold for IgG at 2 sd below the mean. Although immunoglobulin levels do not follow a Gaussian distribution (10), this is generally accepted as an IgG below 7 g/l. These criteria also exclude late onset combined immunodeficiency (LOCID) from the diagnosis. The immunoglobulin levels are also required to be repeated in these latter criteria.
There is ongoing debate about the utility these diagnostic criteria, the variability of IgG levels over time, the unreliability of vaccine challenge responses and flow cytometry in the diagnosis of CVID. As discussed below, this variability in protein-based assays is a strong argument for genetic testing of all patients with a CVID phenotype.
Genetics of CVID and CVID-Like Disorders
By definition the causes of CVID are unknown. In 2003, the first genetic defect was identified in Germany (11). Mutations of the Inducible T cell co-stimulator (ICOS), which plays a critical role in T and B cell communication, were discovered in patients with a CVID phenotype (11). There was a founder effect as all affected individuals in the Black Forest area shared the identical mutation. Different mutations of ICOS were subsequently identified in other parts of the world, confirming allelic heterogeneity (12).
Two years later, mutations of the T cell activator, calcium modulator and cyclophilin ligand interactor (TNFRSF13B/ TACI) were discovered (13). This molecule plays an important role in B cell signaling and immunoglobulin isotype switching. Mutations of TNFRSF13B/TACI were initially thought to cause CVID but subsequently it became apparent that identical mutations were also found in the general population at a frequency far greater than the incidence of CVID. It is now thought that mutations of TNFRSF13B/TACI predispose to CVID or have a disease modifying effect on the disorder (14).
We and others have suggested mutations associated with CVID should be categorized according to whether they cause the disorder or whether they modify or predispose to the condition (7, 15). Apart from TNFRSF13B/TACI, mutations of other genes including BAFFR (TNFRSF13C), TWEAK (TNFSF12), MSH5, and TRAIL (TNFSF10) are also thought to predispose to, or modify the disease severity of patients with CVID.
In contrast, mutations of genes such as NFKB1, NFKB2, CTLA-4, TCF3 etc. are more likely to cause the condition (16, 17). Such patients are removed from the broad category of CVID and are deemed to have a PID caused by a specific mutation. We have suggested conditions with causative mutations should be termed “CVID-like” disorders, given the close phenotypic overlap with CVID (18). None of the current diagnostic criteria for CVID allow the diagnosis if a known disorder is identified (7–9, 18). This is the basis for excluding patients with a causative mutation from the umbrella diagnosis of CVID. Since the discovery of ICOS mutations, ~30 genetic defects have been shown to modify disease severity, predispose to CVID or alternatively cause CVID-like disorders (19–21).
CVID is genetically complex. Locus heterogeneity (genocopy) is a major feature of CVID-like disorders, making it difficult to identify the affected gene purely on clinical grounds. Mutations of several genes can result in the classical phenotype of late onset antibody failure leading to recurrent and severe infections as well as autoimmunity (19).
Although clinical identification of individual CVID-like disorders is difficult, there may be subtle clues such as the presence of alopecia in combination with pituitary dysfunction, which are indicative of NFKB2 defects (19). In other cases, a careful history may reveal severe autoimmunity, which may suggest PIK3CD mutations, causing activated protein kinase 3D syndrome (APDS) or CTLA-4/LRBA mutations (22). The presence of vasculitis in the context of hypogammaglobulinemia might indicate ADA2 deficiency (19). In most cases however, such clues are absent.
Similarly, phenotypic heterogeneity makes diagnosis difficult as the clinical manifestations can vary widely, even within the same family carrying the identical mutation. We have recently described the pleomorphic clinical presentation of a family with NFKB1 deficiency (23). One heterozygous brother carrying the mutation was asymptomatic with normal immunoglobulins, while his heterozygous sister had severe disease with features of late onset combined immunodeficiency (LOCID) (23). We have used our CVID disease severity score (CDSS) to quantify the phenotypic severity of individual family members (24). The phenotypic heterogeneity may be the result of variable penetrance and expressivity, epigenetic influences or epistasis caused by gene-gene interactions.
As noted in the case of ICOS deficiency, CVID-like disorders also manifest allelic heterogeneity where different mutations of the same gene can result in a similar phenotype. Because of genetic and phenotypic heterogeneity, there has been understandable reluctance to routinely sequence CVID patients because of the low yield (25). Serial Sanger sequencing of an ever-increasing list of individual genes was not an efficient use of valuable resources (25).
Given the rapid progress in the understanding of these conditions in recent years, we believe there is now a strong case for routine diagnostic genetic testing of patients with a CVID phenotype (Table 1). This change in approach is both the result of identifying increasing numbers of genetic defects as well as advances in technology, particularly NGS. We have previously discussed diagnosing CVID in the era of genome sequencing (19). In this current viewpoint article, we have incorporated new information, mostly from our recent studies, to strengthen the arguments for routine diagnostic sequencing of patients with a CVID phenotype (26, 27). This article will serve as the evidence base for what is becoming routine practice in the care of CVID patients. It will assist clinical services in implementing such a strategy.
NGS Allows Efficient Sequencing of Genes from Disorders With Locus Heterogeneity
Over the last decade, NGS has revolutionized the approach to molecular diagnosis. Multiple genes can now be sequenced simultaneously, with either gene panels or by Whole Exome Sequencing (WES) and targeted analysis (19). In non-consanguineous populations, the causative mutation may be identified in ~25% of CVID patients (28). The diagnostic yield is much higher in kindreds with more than one affected individual, founder populations or those with high rates of consanguinity (29–31). In some cohorts, the presence of parental consanguinity was associated with more severe disease (32).
The advent of NGS is the principal reason for the feasibility of routine genetic sequencing of patients with CVID-like disorders, who have locus heterogeneity (33). A similar approach is now being undertaken for other disorders with locus heterogeneity such at atypical hemolytic uremic syndrome (aHUS) and hemophagocytic lymphohistiocytosis (HLH) (34).
Although there was initial concern about copy number variants contributing to CVID (35) a more recent paper did not support these initial observations (36). Therefore WES is a reasonable option for investigating these patients.
We next discuss specific advantages as well as potential disadvantages undertaking routine diagnostic sequencing of patients with a CVID phenotype.
Specific Advantages of Identifying the Causative Mutation in Patients with CVID-Like Disorders (Table 1)
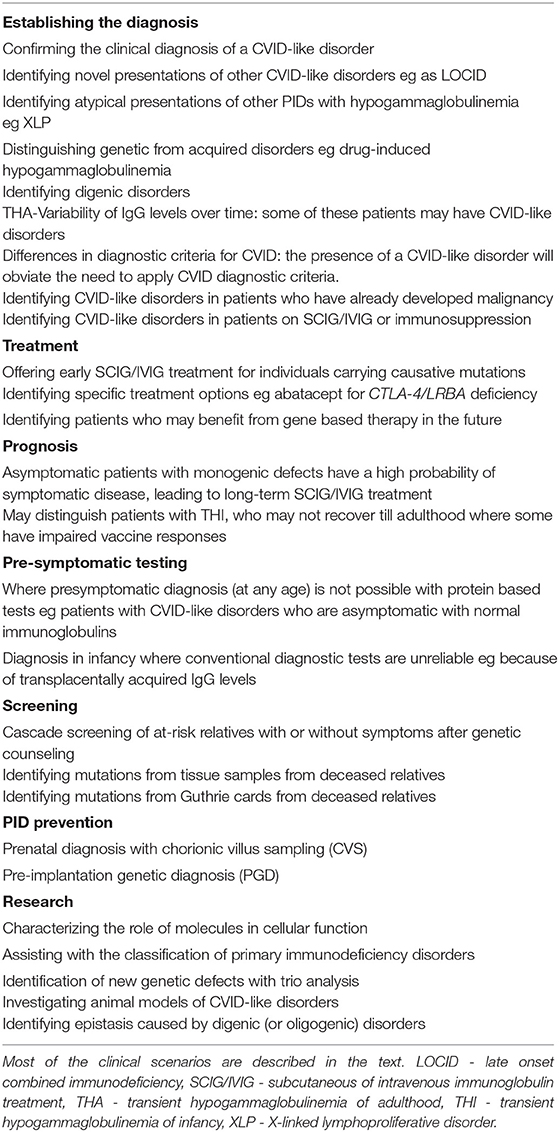
Identifying the causative genetic defect is now the standard of care of PID patients (25). We have outlined the many overlapping advantages (and some disadvantages) in identifying the underlying genetic defect in PIDs (37–39). We have listed in detail the specific advantages of genetic diagnosis in CVID-like disorders in Table 1.
Identification of a causative mutation will confirm the presence of a CVID-like disorder and will enable diagnosis of patients with atypical presentations. This is particularly important given the genetic and phenotypic heterogeneity in CVID-like disorders, outlined above.
Some patients with well-characterized PIDs such as X-linked lymphoproliferative (XLP) disorder or STAT3 mutations can present rarely with predominant hypogammaglobulinemia. If other characteristic features of these disorders are not obvious, such atypical presentations may cause confusion with CVID. Given there may be specific treatments for these conditions, early identification is of paramount importance. Pre-emptive bone marrow transplantation prior to EBV infection in pre-symptomatic male relatives, carrying the mutation, can be life-saving in XLP (40). NGS will rapidly identify the majority of “non-CVID” patients presenting with hypogammaglobulinemia. We distinguish PIDs such as XLP and STAT3 mutations, which do not typically present with antibody deficiency (41–43) from disorders such as NFKB1 mutations which, most often present with hypogammaglobulinemia, that are more appropriately termed CVID-like disorders (19).
Identification of the mutation will offer prognostic information. We have recently shown that many children with transient hypogammaglobulinemia of infancy (THI) do not recover until early adulthood (27). CVID/CVID-like disorders are thus the principal differential diagnosis until patients with THI recover. Identification of a causative mutation in a child with persistent hypogammaglobulinemia will exclude THI and will indicate the patient is likely to require long-term subcutaneous or intravenous immunoglobulin (SCIG/IVIG) therapy. As noted above, genetic sequencing of children with severe symptomatic immunodeficiencies is now the standard of care.
Hypogammaglobulinemia can be caused by a wide range of non-immunological disorders and it can sometimes be difficult to exclude these secondary causes. If a causative genetic defect is identified, this will exclude secondary causes, such as anticonvulsant drugs, gut disease or other rare conditions (44–48).
As with other genetic disorders, identification of a mutation has profound implications for family members. The presence of a genetic defect may allow early diagnosis and prompt commencement of SCIG/IVIG treatment of affected family members, when they develop symptoms. We suggest patients with CVID-like disorders are offered SCIG/IVIG on the basis of clinical symptoms and vaccine challenge responses may not be necessary. Such pre-symptomatic individuals, carrying the family mutation, could be made aware of potential risks and complications. This may either prevent catastrophic infections and mitigate ongoing target organ damage leading to bronchiectasis and other disabling complications (24).
Detection of a causative mutation may allow a future reduction in the numbers of PIDs by preimplantation genetic diagnosis (PGD). The specific mutation allows prenatal diagnosis with chorionic villus sampling (CVS) and/or PGD. An individualized approach is required. As we have previously stated, it is not appropriate to consider CVS or PGD for families carrying only mutations predisposing to CVID such as TNFRSF13B/TACI (49). Even within mutations causing CVID-like disorders, penetrance and expressivity vary widely. Some variants such as the TCF3 mutation we have described appear to be fully penetrant, while one member of our family with NFKB1 haploinsufficiency is phenotypically normal. There will need to be careful counseling of such families.
While current technologies will not prevent disease caused by new mutations, PGD could lead to a substantial decrease in the prevalence of disease within a generation. This will result in a major reduction in the burden of suffering as well as healthcare costs. The NZ government offers free in vitro fertilization and PGD for families carrying severe genetic defects. Delay in access to this innovative but under-resourced program in NZ is however a significant barrier.
Identification of the specific mutation may lead to new therapeutic options. Patients with mutations of CTLA-4 or LRBA may be candidates for abatacept. Those with gain of function mutations of GOF-PIK3CD or loss of function LOF-PIK3R1 (APDS 1 and 2) may improve with mTOR inhibitors such as rapamycin or newer agents such as Idelalisib. Patients with a severe CVID-like disorder caused by mutations of ADA2 may benefit from early bone marrow transplantation.
Discovery of the mutation may in the future lead to gene-based therapies including retroviral gene transfer or gene editing with CRISPR-Cas9. CRISPR-Cas9 has been used to repair CYBB gene mutations in X-linked chronic granulomatous disease cells (50). As discussed previously, off-target effects of the CRISPR-Cas9 system may limit its in vivo use (51), although there has been progress to mitigate these risks (52). We are unaware of any current in vivo trials of retroviral gene therapy or CRISPR-Cas9 gene editing studies in patients with CVID-like disorders. Given the variable penetrance an expressivity, such gene-based therapies should only be considered for severely symptomatic individuals in the future.
The use of NGS has resulted in new discoveries including novel mechanisms of disease (32). We have recently shown the existence of quantitative epistasis in a patient with digenic inheritance leading to a CVID-like disorder (49, 53). Epistasis is the synergistic, non-linear interaction of two or more genetic loci leading either to a much more severe disorder or to a completely different phenotype. We have suggested the synergistic interaction of genes is termed quantitative epistasis, while those leading to a different phenotype are termed qualitative epistasis (49).
The proband had mutations of both TCF3 and TNFRSF13B/ TACI genes, which caused a severe defect in antibody production leading to a CVID-like disorder. These two genetic loci lie in tandem, along the immunoglobulin production and isotype switching pathways. The synergistic interaction of these two mutations caused quantitative epistasis, leading to a severe CVID-like disorder. Our study confirms genes such as TNFRSF13B/TACI have disease modifying effects on the severity of CVID-like disorders and supports the separation these two groups of mutations. Such digenic patients can only be identified by NGS (16, 54) and are a strong argument for sequencing both groups of genes, either causing CVID-like disorders (NFKB1, NFKB2, etc.) or those modifying the severity of CVID, such as TNFRSF13B/ TACI (19, 55).
Diagnostic Uncertainty Caused by Variability of Protein Based Laboratory Tests
As discussed above, the diagnosis of CVID relies on diagnostic criteria. In our long-term prospective NZ hypogammaglobulinemia study (NZHS), we have shown marked fluctuations in IgG levels in patients with hypogammaglobulinemia. Of concern was that 41.6% (20/48) of symptomatic patients were able to normalize their IgG on at least one occasion, when measured over time. Seven of twelve hypogammaglobulinemic patients with bronchiectasis were also able to normalize their IgG on at least one occasion. We have termed this phenomenon transient hypogammaglobulinemia of adulthood (THA) (26).
Some of these patients with hypogammaglobulinemia may have a CVID-like disorder and in time will experience progressive clinical deterioration (23). Identifying a causative genetic defect will establish the diagnosis and assist with monitoring and therapeutic decisions. Patients with profound hypogammaglobulinemia (<3 g/l) and those who are symptomatic with persistent hypogammaglobulinemia should be considered for genetic testing. Patients with asymptomatic THA with subsequent sustained normal IgG levels do not need testing, with the possible exception of those with a family history of an immunological disorder. We have shown that some family members carrying mutations of CVID-like disorders can be asymptomatic with normal IgG levels (23). We acknowledge patients with THA will need to be carefully assessed.
Most patients with CVID have reduced memory B cells and these constitute a diagnostic criterion in Category C of our criteria (56). It is however important for memory B cell subsets to be measured on at least two occasions, as we have shown the numbers can vary on repeat testing (57). We assessed memory B cell subsets on a monthly basis in a cohort of CVID patients being treated with IVIG. Our results showed there was considerable variability leading to changes in diagnostic categories on a monthly basis, particularly for the Freiburg and Paris criteria. The variability was less marked for the EUROclass trial guideline. This again illustrates the variability of protein based assays for CVID assessment and is an argument for genetic testing.
Unreliability of Vaccine Responses in Current Diagnostic Criteria for CVID
We have discussed the difficulties with the previous ESID/PAGID (1999) criteria for CVID (58). They lacked precision and asymptomatic patients with trivial hypogammaglobulinemia of IgG and IgA, with mildly impaired responses to the diphtheria vaccine could be designated as having CVID and offered life-long SCIG/IVIG.
We have recently reviewed diagnostic criteria for CVID in the NZHS (26). We showed while there was general congruence of diagnostic criteria, there were important differences. In our study, many asymptomatic individuals with mild hypogammaglobulinemia qualified as having definite CVID by ICON (2016) criteria, because of impaired vaccine responses to Pneumovax 23® or the diphtheria vaccine (26). Given their excellent health over a mean follow-up of 106 months (to date), it is unlikely these asymptomatic patients have definite CVID or any other immunological disorder.
In our study, both symptomatic and asymptomatic patients with hypogammaglobulinemia had excellent responses to H. influenzae type B (HIB) and tetanus vaccines (59). Vaccine responses were thus non-discriminatory in the NZHS. Similarly, we also recently showed some patients with THI, who subsequently recovered, had impaired vaccine responses, which could potentially lead to misdiagnosis of definite CVID if ICON (2016) criteria are applied (27). IgM and IgA levels in particular, can be difficult to interpret in young children.
Identifying the causative mutation would obviate the need to apply CVID diagnostic criteria, as the patient would then be reclassified as having a CVID-like disorder (60). Thus, the primary aim of genetic sequencing is to remove these patients from the umbrella diagnosis of CVID so they can be more accurately classified as having a specific PID.
Caveats
While we advocate routine diagnostic WES or WGS for all patients with a CVID phenotype, there are important caveats. We have discussed the technical limitations of NGS including lack of uniform coverage with WES leading to errors (19, 61). These errors are less likely with WGS but currently this technology is more expensive than WES. NGS is not available in all parts of the world. However, several commercial companies are now offering these tests, some using gene panels, while others offer WES with targeted analysis. One company offering WES with targeted analysis releases raw data for an additional fee, which allows future analysis of gene mutations, which have yet to be discovered. With the appropriate consents and ethics approvals, this data can also be converted to parents:child trio analysis for gene-discovery research studies.
It is important to counsel patients before offering these studies as there is a risk of identifying variants of unknown significance (VUS) (19). This can be frustrating for both patient and physician (62). In some cases the pathogenicity of a VUS can be resolved by collateral techniques such as functional studies (63). Another important caveat is the risk of assigning disease causality to ethnic specific variants. What may fit all decision criteria for a mutation causing a rare disease may simply be a common benign variant (polymorphism) in an under-surveyed ethnic group. Current databases comprise predominantly Caucasian individuals, while other ethnicities such as Maori are poorly represented.
There is also the problem of de novo pathogenic mutations in databases, where the disease is yet to manifest. These will need careful analysis. In silico analysis and the frequency of such alleles may indicate their true significance. If the frequency of homozygous healthy individuals is lower than expected (given the variant allele frequency), it may suggest that the homozygous state is disease causing.
In some cases, WES and WGS may identify potentially important mutations in unrelated genes such as those associated with cancer, cardiovascular disease, severe neurological disorders etc. The American College of Medical Genetics (ACMG) has published guidelines for the analysis and disclosure of these “medically actionable” incidental findings in patients undergoing NGS (64). We have discussed the difficulties with these ACMG guidelines (19). Studies have shown low yield from these guidelines (65, 66).
There is also a possibility of identifying VUS in these genes. A single expert may not be able to resolve the significance of all of these variants in different organ systems. This is likely to cause great anxiety and expense in societies without a socialized health system, if there is no insurance coverage. At the time of consent, we encourage our patients to opt out of disclosing these incidental findings in unrelated organ systems.
We have also discussed other social and financial disadvantages of identifying the mutation, such as genetic discrimination in the domains of insurance or employment (39). The Americans with Disabilities Act 1990 (ADA) and the Genetic Information Non-discrimination Act of 2008 (GINA) protects Americans from such discrimination. Such enabling legislation is however not universally enacted in all jurisdictions. In spite of robust legislation protecting individual rights, NZ does not currently have laws forbidding genetic discrimination.
Securing funding for these tests is a common problem. Clinical services and insurance providers have been slow to recognize the value of such technology and the far-reaching benefits of testing (25). Prevention of a single case could lead to lifetime savings of over $2M, which would fund NGS for a large cohort of CVID patients. If funding is not immediately available from clinical services, in many cases NGS can be undertaken as part of research studies, with the appropriate consents. In many cases, our patients have self-funded these tests.
In spite of these limitations, we believe all patients with a CVID phenotype should now be routinely offered diagnostic NGS sequencing if resources permit. If a causal mutation is not found, such patients can be enrolled in gene discovery research studies with the appropriate consents and ethics approvals (19).
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank our patients for participating in our studies for the benefit of others. We hope our studies will be of direct benefit to patients and their families. We hope this monograph will assist colleagues in making the case to insurance companies and funders of clinical services to routinely offer these tests to patients with CVID. We thank the A+ Trust, AMRF, IDFNZ, and ASCIA for grant support. The references cited are mainly from our own studies in this personal viewpoint article.
References
1. Abbott JK, Gelfand EW. Common variable immunodeficiency : diagnosis, management, and treatment. Immunol Allergy Clin North Am. (2015) 35:637–58. doi: 10.1016/j.iac.2015.07.009
2. Gathmann B, Mahlaoui N, Gérard L, Oksenhendler E, Warnatz K, et al. Clinical picture and treatment of 2212 patients with common variable immunodeficiency. J Allergy Clin Immunol. (2014) 134:116–26. doi: 10.1016/j.jaci.2013.12.1077
3. Selenius JS, Martelius T, Pikkarainen S, Siitonen S, Mattila E, Pietikäinen R, et al. Unexpectedly high prevalence of common variable immunodeficiency in Finland. Front Immunol. (2017) 8:1190. doi: 10.3389/fimmu.2017.01190
4. Westh L, Mogensen TH, Dalgaard LS, Bernth Jensen JM, Katzenstein T, Hansen AE, et al. Identification and characterization of a nationwide danish adult common variable immunodeficiency Cohort. Scand J Immunol. (2017) 85:450–61. doi: 10.1111/sji.12551
5. Ameratunga R, Becroft DM, Hunter W. The simultaneous presentation of sarcoidosis and common variable immune deficiency. Pathology. (2000) 32:280–2. doi: 10.1080/pat.32.4.280.282
6. Kralickova P, Milota T, Litzman J, Malkusova I, Jilek D, Petanova J, et al. CVID-Associated Tumors: Czech Nationwide Study Focused on Epidemiology, Immunology, and Genetic Background in a Cohort of Patients With CVID. Front Immunol. (2018) 9:3135. doi: 10.3389/fimmu.2018.03135
7. Ameratunga R, Woon ST, Gillis D, Koopmans W, Steele R. New diagnostic criteria for common variable immune deficiency (CVID), which may assist with decisions to treat with intravenous or subcutaneous immunoglobulin. Clin Exp Immunol. (2013) 174:203–11. doi: 10.1111/cei.12178
8. Seidel MG, Kindle G, Gathmann B, Quinti I, Buckland M, van Montfrans J, et al. The European Society for Immunodeficiencies (ESID) registry working definitions for the clinical diagnosis of inborn errors of immunity. J Allergy Clin Immunol Pract. (2019). 7:1763–70. doi: 10.1016/j.jaip.2019.02.004
9. Bonilla FA, Barlan I, Chapel H, Costa-Carvalho BT, Cunningham-Rundles C, de la Morena MT, et al. International Consensus Document (ICON): Common Variable Immunodeficiency Disorders. J Allergy Clin Immunol Pract. (2016) 4:38–59. doi: 10.1016/j.jaip.2015.07.025
10. Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O, Ledue TB, Craig WY. Reference distributions for immunoglobulins A, G, and M: a practical, simple, and clinically relevant approach in a large cohort. J Clin Lab Anal. (1998) 12:363–70.
11. Grimbacher B, Hutloff A, Schlesier M, Glocker E, Warnatz K, Dräger R, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. (2003) 4:261–8. doi: 10.1038/ni902
12. Schepp J, Chou J, Skrabl-Baumgartner A, Arkwright PD, Engelhardt KR, Hambleton S, et al. 14 years after discovery: clinical follow-up on 15 patients with inducible co-stimulator deficiency. Front Immunol. (2017) 8:964. doi: 10.3389/fimmu.2017.00964
13. Salzer U, Chapel HM, Webster AD, Pan-Hammarström Q, Schmitt-Graeff A, Schlesier M, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. (2005) 37:820–8. doi: 10.1038/ng1600
14. Pan-Hammarstrom Q, Salzer U, Du L, Rachid R, Bonilla F, Schneider L, et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat Genet. (2007) 39:429–30. doi: 10.1038/ng0407-429
15. Ameratunga R, Woon ST, Gillis D, Koopmans W, Steele R. New diagnostic criteria for CVID. Expert Rev Clin Immunol. (2014) 10:183–6. doi: 10.1586/1744666X.2014.875274
16. de Valles-Ibáñez G, Esteve-Solé A, Piquer M, González-Navarro EA, Hernandez-Rodriguez J, Laayouni H, et al. Evaluating the Genetics of common variable immunodeficiency: monogenetic model and beyond. Front Immunol. (2018) 9:636. doi: 10.3389/fimmu.2018.00636
17. Fliegauf M, L., Bryant V, Frede N, Slade C, Woon ST, Lehnert K, et al. Haploinsufficiency of the NF-κB1 subunit p50 in common variable immunodeficiency. Am Jo Hum Genet. (2015) 97:389–403. doi: 10.1016/j.ajhg.2015.07.008
18. Ameratunga R, Brewerton M, Slade C, Jordan A, Gillis D, Steele R, et al. Comparison of diagnostic criteria for common variable immunodeficiency disorder. Front Immunol. (2014) 5:415. doi: 10.3389/fimmu.2014.00415
19. Ameratunga R, Lehnert K, Woon ST, Gillis D, Bryant VL, Slade CA, et al. Review: diagnosing common variable immunodeficiency disorder in the Era of genome sequencing. Clin Rev Allergy Immunol. (2018) 54:261–8. doi: 10.1007/s12016-017-8645-0
20. Bogaert DJ, Dullaers M, Lambrecht BN, Vermaelen KY, De Baere E, Haerynck F. Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J Med Genet. (2016) 53:575–90. doi: 10.1136/jmedgenet-2015-103690
21. Picard C, Bobby Gaspar H, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, et al. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J Clin Immunol. (2018) 38:96–128. doi: 10.1007/s10875-017-0464-9
22. Cagdas D, Halaçli SO, Tan Ç, Lo B, Çetinkaya PG, Esenboga S, et al. A spectrum of clinical findings from ALPS to CVID: several novel LRBA defects. J Clin Immunol. (2019) 39:726–38. doi: 10.1007/s10875-019-00677-6
23. Ameratunga R, Ahn Y, Jordan A, Lehnert K, Brothers S, Woon ST. Keeping it in the family: the case for considering late onset combined immunodeficiency a subset of common variable immunodeficiency disorders. Expert Rev Clin Immunol. (2018) 14:549–56. doi: 10.1080/1744666X.2018.1481750
24. Ameratunga R. Assessing Disease Severity in Common Variable Immunodeficiency Disorders (CVID) and CVID-Like Disorders. Front Immunol. (2018) 9:2130. doi: 10.3389/fimmu.2018.02130
25. Ameratunga R, Steele R, Jordan A, Preece K, Barker R, Brewerton M, et al. The case for a national service for primary immune deficiency disorders in New Zealand. N Z Med J. (2016) 129:75–90.
26. Ameratunga R, Ahn Y, Steele R, Woon ST. The natural history of untreated primary hypogammaglobulinemia in adults: Implications for the diagnosis and treatment of Common Variable Immunodeficiency Disorders (CVID). Frontiers Immunology. (2019) 17;10:1541. doi: 10.3389/fimmu.2019.01541
27. Ameratunga R, Ahn Y, Steele R, Woon ST. Transient hypogammaglobulinemia of infancy: many patients recover in adolescence and adulthood. Clin Exp Immunol. (2019) 198:224–32. doi: 10.1111/cei.13345
28. Maffucci P, Filion CA, Boisson B, Itan Y, Shang L, Casanova JL, et al. Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front Immunol. (2016) 7:220. doi: 10.3389/fimmu.2016.00220
29. Abolhassani H, Aghamohammadi A, Fang M, Rezaei N, Jiang C, Liu X, et al. Clinical implications of systematic phenotyping and exome sequencing in patients with primary antibody deficiency. Genetics in medicine : official journal of the American College of Med Genet. (2019) 21:243–51. doi: 10.1038/s41436-018-0012-x
30. Al-Herz W, Chou J, Delmonte OM, Massaad MJ, Bainter W, Castagnoli R, et al. Comprehensive genetic results for primary immunodeficiency disorders in a highly consanguineous population. Front Immunol. (2018) 9:3146. doi: 10.3389/fimmu.2018.03146
31. Al-Herz W, Al-Ahmad M, Al-Khabaz A, Husain A, Sadek A, Othman Y. The Kuwait National Primary Immunodeficiency Registry 2004–2018. Front Immunol. (2019) 10:1754. doi: 10.3389/fimmu.2019.01754
32. Rivoisy C, Gérard L, Boutboul D, Malphettes M, Fieschi C, Durieu I, et al. Parental consanguinity is associated with a severe phenotype in common variable immunodeficiency. J Clin Immunol. (2012) 32:98–105. doi: 10.1007/s10875-011-9604-9
33. Woon ST, Ameratunga R. Comprehensive genetic testing for primary immunodeficiency disorders in a tertiary hospital: 10-year experience in Auckland, New Zealand. Allergy Asthma Clin Immunol. (2016) 12:65. doi: 10.1186/s13223-016-0169-2
34. Schulert GS, Zhang M, Fall N, Husami A, Kissell D, Hanosh A, et al. Whole-Exome Sequencing Reveals Mutations in Genes Linked to Hemophagocytic Lymphohistiocytosis and Macrophage Activation Syndrome in Fatal Cases of H1N1 Influenza. J Infect Dis. (2016) 213:1180–8. doi: 10.1093/infdis/jiv550
35. Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J Allergy Clin Immunol. (2011) 127:1360–7 e6. doi: 10.1016/j.jaci.2011.02.039
36. Keller M, Glessner J, Resnick E, Perez E, Chapel H, Lucas M, et al. Burden of copy number variation in common variable immunodeficiency. Clin Exp Immunol. (2014) 177:269–71. doi: 10.1111/cei.12255
37. Ameratunga R, Woon ST. Customised molecular diagnosis of primary immune deficiency disorders in New Zealand: an efficient strategy for a small developed country. N Z Med J. (2009) 122:46–53.
38. Ameratunga R, Woon ST, Brewerton M, Koopmans W, Jordan A, Brothers S, et al. Primary immune deficiency disorders in the South Pacific: the clinical utility of a customized genetic testing program in New Zealand. Ann N Y Acad Sci. (2011) 1238:53–64. doi: 10.1111/j.1749-6632.2011.06238.x
39. Ameratunga R, Woon ST, Neas K, Love DR. The clinical utility of molecular diagnostic testing for primary immune deficiency disorders: a case based review. Allergy Asthma Clin Immunol. (2010) 6:12. doi: 10.1186/1710-1492-6-12
40. Woon ST, Ameratunga R, Croxson M, Taylor G, Neas K, Edkins E, et al. Follicular lymphoma in a X-linked lymphoproliferative syndrome carrier female. Scand J Immunol. (2008) 68:153–8. doi: 10.1111/j.1365-3083.2008.02128.x
41. Lee WI, Huang JL, Kuo ML, Lin SJ, Chen LC, Chen MT, et al. Analysis of genetic defects in patients with the common variable immunodeficiency phenotype in a single Taiwanese tertiary care hospital. Ann Allergy Asthma Immunol. (2007) 99:433–42. doi: 10.1016/S1081-1206(10)60569-8
42. Vince N, Mouillot G, Malphettes M, Limou S, Boutboul D, Guignet A, et al. Genetic screening of male patients with primary hypogammaglobulinemia can guide diagnosis and clinical management. Hum Immunol. (2018) 79:571–7. doi: 10.1016/j.humimm.2018.04.014
43. Buchbinder D, Baker R, Lee YN, Ravell J, Zhang Y, McElwee J, et al. Identification of patients with RAG mutations previously diagnosed with common variable immunodeficiency disorders. J Clin Immunol. (2015) 35:119–24. doi: 10.1007/s10875-014-0121-5
44. Ameratunga R, Lindsay K, Woon S-T, Jordan A, Anderson NE, Koopmans W. New diagnostic criteria could distinguish common variable immunodeficiency disorder from anticonvulsant-induced hypogammaglobulinemia. Clin Exp Neuroimmunol. (2015) 6:83–8. doi: 10.1111/cen3.12135
45. Smith J, Fernando T, McGrath N, Ameratunga R. Lamotrigine-induced common variable immune deficiency. Neurology. (2004) 62:833–4. doi: 10.1212/01.WNL.0000113754.29225.5D
46. Ameratunga R, Casey P, Parry S, Kendi C. Hypogammaglobulinemia factitia. Munchausen syndrome presenting as Common Variable Immune Deficiency. Allergy Asthma Clin Immunol. (2013) 9:36. doi: 10.1186/1710-1492-9-36
47. Ameratunga R, Barker RW, Steele RH, Deo M, Woon ST, Yeong ML, et al. Profound reversible hypogammaglobulinemia caused by celiac disease in the absence of protein losing enteropathy. J Clin Immunol. (2015) 35:589–94. doi: 10.1007/s10875-015-0189-6
48. Duraisingham SS, Buckland MS, Grigoriadou S, Longhurst HJ. Secondary antibody deficiency. Expert Rev Clin Immunol. (2014) 10:583–91. doi: 10.1586/1744666X.2014.902314
49. Ameratunga R, Woon ST, Bryant VL, Steele R, Slade C, Leung EY, et al. Clinical Implications of Digenic inheritiance and epistasis in primary immunodeficiency disorders. Front Immunol. (2018) 8:1965 doi: 10.3389/fimmu.2017.01965
50. De Ravin SS, Li L, Wu X, Choi U, Allen C, Koontz S, et al. CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci Transl Med. (2017) 9:eaah3480. doi: 10.1126/scitranslmed.aah3480
51. Ameratunga R, Bartlett A, McCall J, Steele R, Woon ST, Katelaris CH. Hereditary angioedema as a metabolic liver disorder: novel therapeutic options and prospects for cure. Front Immunol. (2016) 7:547. doi: 10.3389/fimmu.2016.00547
52. Bak RO, Dever DP, Porteus MH. CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nature Protoc. (2018) 13:358–76. doi: 10.1038/nprot.2017.143
53. Ameratunga R, Koopmans W, Woon ST, Leung E, Lehnert K, Slade CA, et al. Epistatic interactions between mutations of TACI (TNFRSF13B) and TCF3 result in a severe primary immunodeficiency disorder and systemic lupus erythematosus. Clin Transl Immunol. (2017) 6:e159. doi: 10.1038/cti.2017.41
54. Chiriaco M, Di Matteo G, Conti F, Petricone D, De Luca M, Di Cesare S, et al. First Case of Patient With Two Homozygous Mutations in MYD88 and CARD9 Genes Presenting With Pyogenic Bacterial Infections, Elevated IgE, and Persistent EBV Viremia. Front Immunol. (2019) 10:130. doi: 10.3389/fimmu.2019.00130
55. Li R, Zheng Y, Li Y, Zhang R, Wang F, Yang D, et al. Common Variable Immunodeficiency with Genetic Defects Identified by Whole Exome Sequencing. BioMed Res Int. (2018) 2018:3724630. doi: 10.1155/2018/3724630
56. Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. (2008) 111:77–85. doi: 10.1182/blood-2007-06-091744
57. Koopmans W, Woon ST, Zeng IS, Koopmans W, Jordan A, Brothers S, et al. Variability of memory B cell markers in a cohort of Common Variable Immune Deficiency patients over six months. Scand J Immunol. (2013) 77:470–5. doi: 10.1111/sji.12028
58. Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies). Clin Immunol. (1999) 93:190–7. doi: 10.1006/clim.1999.4799
59. Ameratunga SN, Lennon DR, Entwistle B, Robinson E, Ameratunga RV. The immunogenicity of Haemophilus influenzae: meningococcal protein conjugate vaccine in Polynesian and non-Polynesian New Zealand infants. J Paediatr Child Health. (1997) 33:138–41. doi: 10.1111/j.1440-1754.1997.tb01016.x
60. Ameratunga R, Storey P, Barker R, Jordan A, Koopmans W, Woon ST. Application of diagnostic and treatment criteria for common variable immunodeficiency disorder. Expert Rev Clin Immunol. (2015) 12:257–66. doi: 10.1586/1744666X.2016.1126509
61. Rudilla F, Franco-Jarava C, Martínez-Gallo M, et al. Expanding the Clinical and Genetic Spectra of Primary Immunodeficiency-Related Disorders With Clinical Exome Sequencing: Expected and Unexpected Findings. Frontiers in immunology. (2019) 10:2325. doi: 10.3389/fimmu.2019.02325
62. Vears DF, Sénécal K, Clarke AJ, Jackson L, Laberge AM, Lovrecic L, et al. Points to consider for laboratories reporting results from diagnostic genomic sequencing. Eur J Hum Genet. (2018) 26:36–43. doi: 10.1038/s41431-017-0043-9
63. Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. (2017) 9:26. doi: 10.1186/s13073-017-0412-6
64. Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. (2013) 15:565–74. doi: 10.1038/gim.2013.135
65. Ormond KE, O'Daniel JM, Kalia SS. Secondary findings: How did we get here, and where are we going? J Genet Counseling. (2019) 28:326–33. doi: 10.1002/jgc4.1098
Keywords: CVID - common variable immunodeficiency disorders, hypogammaglobinaemia, genetic sequence analysis, transient hypogammaglobulinemia of adulthood (THA), transient hypogammaglobulinemia of infancy
Citation: Ameratunga R, Lehnert K and Woon S-T (2019) All Patients With Common Variable Immunodeficiency Disorders (CVID) Should Be Routinely Offered Diagnostic Genetic Testing. Front. Immunol. 10:2678. doi: 10.3389/fimmu.2019.02678
Received: 22 August 2019; Accepted: 30 October 2019;
Published: 22 November 2019.
Edited by:
Stuart G. Tangye, Garvan Institute of Medical Research, AustraliaReviewed by:
Bertrand Boisson, The Rockefeller University, United StatesLennart Hammarström, Karolinska Institute (KI), Sweden
Copyright © 2019 Ameratunga, Lehnert and Woon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rohan Ameratunga, RXJvaGFuYUBhZGhiLmdvdnQubno=
 Rohan Ameratunga
Rohan Ameratunga Klaus Lehnert4
Klaus Lehnert4 See-Tarn Woon
See-Tarn Woon