
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 07 November 2019
Sec. Molecular Innate Immunity
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.02613
This article is part of the Research Topic Odyssey of Surfactant Proteins SP-A and SP-D: Innate Immune Surveillance Molecules View all 15 articles
 Armen Nalian1,2
Armen Nalian1,2 Todd M. Umstead3,4
Todd M. Umstead3,4 Ching-Hui Yang2†
Ching-Hui Yang2† Patricia Silveyra3,4†
Patricia Silveyra3,4† Neal J. Thomas3,5
Neal J. Thomas3,5 Joanna Floros3,6,7
Joanna Floros3,6,7 Francis X. McCormack8
Francis X. McCormack8 Zissis C. Chroneos2,3,4,9*
Zissis C. Chroneos2,3,4,9*Surfactant protein A (SP-A) provides surfactant stability, first line host defense, and lung homeostasis by binding surfactant phospholipids, pathogens, alveolar macrophages (AMs), and epithelial cells. Non-primates express one SP-A protein whereas humans express two: SP-A1 and SP-A2 with core intra- and inter-species differences in the collagen-like domain. Here, we used macrophages and solid phase binding assays to discern structural correlates of rat (r) and human (h) SP-A function. Binding assays using recombinant rSP-A expressed in insect cells showed that lack of proline hydroxylation, truncations of amino-terminal oligomerization domains, and site-directed serine (S) or alanine (A) mutagenesis of cysteine 6 (C6S), glutamate 195 (E195A), and glutamate 171 (E171A) in the carbohydrate recognition domain (CRD) all impaired SP-A binding. Replacement of arginine 197 with alanine found in hSP-A (R197A), however, restored the binding of hydroxyproline-deficient rSP-A to the SP-A receptor SP-R210 similar to native rat and human SP-A. In silico calculation of Ca++ coordination bond length and solvent accessibility surface area revealed that the “humanized” R197A substitution alters topology and solvent accessibility of the Ca++ coordination residues of the CRD domain. Binding assays in mouse AMs that were exposed to either endogenous SP-A or hSP-A1 (6A2) and hSP-A2 (1A0) isoforms in vivo revealed that mouse SP-A is a functional hybrid of hSP-A1 and hSP-A2 in regulating SP-A receptor occupancy and binding affinity. Binding assays using neonatal and adult human AMs indicates that the interaction of SP-A1 and SP-A2 with AMs is developmentally regulated. Furthermore, our data indicate that the auxiliary ion coordination loop encompassing the conserved E171 residue may comprise a conserved site of interaction with macrophages, and SP-R210 specifically, that merits further investigation to discern conserved and divergent SP-A functions between species. In summary, our findings support the notion that complex structural adaptation of SP-A regulate conserved and species specific AM functions in vertebrates.
Surfactant protein A (SP-A) is the most abundant lipid binding and immune-surveillance component of pulmonary surfactant. SP-A belongs to the collectin family of proteins consisting of four structural domains that include an amino-terminal tail, a collagen-like domain (CDM) with Gly-X-Y repeats, an α-helical coiled-coil neck domain with heptad repeats, and a Ca++-dependent carbohydrate recognition domain (CRD) (1–3). The neck and collagen-like domains of SP-A trimerize and assemble into higher order deca-octamers via inter-chain disulfide bonds in the amino-terminal tail and non-covalent interactions between trimers. Binding to extracellular ligands depends on the neck-CRD trimer and high avidity multivalent binding of the SP-A deca-octamer. Crystallographic and molecular modeling studies of the rat SP-A CRD revealed that conformational flexibility of the Ca++ coordination site and CRD surface loops impart versatility in the ability of SP-A to bind structurally diverse endogenous and pathogen-derived ligands (2, 4–7). The interaction of SP-A with AMs and epithelial cells is receptor-mediated (8–10). Deletion, site-directed mutagenesis, and ligand competition studies showed that the amino-terminal and collagen-like domains influence receptor occupancy and functional responses in alveolar type II epithelial cells and macrophages, and interaction of the CRD domain with surfactant phospholipids (10–16). SP-A binds dipalmitoyl phosphatidylcholine, the major surfactant phospholipid, that is required for the formation of tubular myelin from secreted lamellar bodies, contributes to the adsorption of surface active phospholipids at the air-liquid interface, and facilitates turnover of spent surfactant vesicles by alveolar type II epithelial cells through the p63 receptor (7, 17–21). In host defense, SP-A binds different pathogen ligands such as the lipid A portion of lipopolysaccharide on Gram-negative bacteria (7), surface proteins and glycolipids on gram positive bacteria (22), mycobacteria (23, 24), and fungi (25, 26). These interactions facilitate pathogen clearance through agglutination, opsonization, and direct killing (22, 26–29). SP-A enhances opsonic and non-opsonic phagocytosis (22, 30–32), and shapes pathogen-dependent polarization of inflammatory responses through the SP-R210 SP-A receptor (aka Myo18A or CD245) (22, 27, 33) in macrophages.
Unlike non-primate amniotes (34), humans express two SP-A protein isoforms, SP-A1 and SP-A2 encoded by different genes SFTPA1 and SFTPA2 (35), and each gene has been identified with several variants. The hSP-A1 and hSP-A2 proteins and their respective variants differ at four core amino acids in the collagen-like domain and the variants of each gene are distinguished among themselves by additional amino acid differences present in domains other than the collagen-like domain (36–38). SP-A1 and SP-A2 differentially modulate macrophage function (39–41) and in suppressing development of idiopathic interstitial pneumonia, fibrosis, and cancer (42–45). Moreover, significant differences have been observed among SP-A1 and SP-A2 variants in survival after infection and lung function (46, 47). The presence of both proteins is required for tubular myelin formation, supra-trimeric assembly of SP-A oligomers, and optimal function of surfactant (21, 48, 49) and one SP-A gene is sufficient to exert these functions in lower vertebrates (19, 50, 51). Both human and rodent SP-As have a discrete kink peptide in the middle of the CDM that confers conformational flexibility, contributes to the quaternary organization of higher order SP-A oligomers, and spatial separation of CRD domains (52–54). The kink sequence is conserved between SP-A1 and SP-A2 but different from rodent SP-A; PCPP in human SP-As and MGLP in rodents (34, 54). The kink peptide and a unique GEC collagen triplet in SP-A1 (GER in SP-A2) result in 2 cysteine residues in the CDM of SP-A1 compared to 1 in SP-A2 and none in rodent SP-A. The GEC triplet contributes to distinct oligomeric structures in SP-A1 and the two proteins distribute differently in interfacial surfactant films (48, 49, 55). Compared to SP-A2, SP-A1 improved the biophysical activity of surfactant in lowering surface tension and resistance to inhibition by serum (56). Recombinant SP-A1 lacking the ability to form oligomers, however, retains anti-inflammatory effects on macrophages (57, 58), whereas SP-A2 variants compared to SP-A1 have been shown to exhibit higher activity in bacterial phagocytosis by AMs (59) and cytokine production in a macrophage-like cell line (39, 60).To better understand SP-A function we used binding assays and molecular modeling to define molecular and functional attributes in rodent and human SP-A.
Wild type (SP-A+/+) C57BL/6J mice were purchased from JAX labs and bred locally. Transgenic SP-A−/− and humanized mSftpa−/−,hSFTPA1(6A2) and mSftpa−/−,hSFTPA2(1A0) mice were generated as described previously (21). Sprague-Dawley rats were obtained from Harlan. All animal procedures were performed under IACUC approved protocols.
Recombinant rat SP-A proteins were synthesized in insect Sf9 cells via baculovirus expression of rat SP-A WT cDNA or mutant cDNAs generated by nested deletion or site directed mutagenesis as described in detail previously (25, 61). The present studies utilized rat SP-A with C6S, E171A, E195A, R197A, and D215A point mutations and deletion mutants lacking both amino terminal and collagen-like domains (ΔN1-G80), the collagen-like domain (ΔG8-G80), kink (ΔG8-G40) and proximal (ΔG40-G80) halves of the collagen-like domain. Recombinant rat SP-A proteins were isolated by affinity chromatography over mannose-Sepharose. Native rat SP-A was isolated from the bronchoalveolar lavage (BAL) of Sprague-Dawley rats 4 weeks after instillation of 40 mg/Kg of silica-induced alveolar proteinosis Rat BAL surfactant was separated by NaBr-density gradient centrifugation followed by delipidation, affinity and gel exclusion chromatography (12, 19, 25, 61–64). Native human SP-A was isolated from discarded alveolar proteinosis BAL by sequential isobutanol/β-octylglucoside extraction, repeated precipitation/solubilization in 20 mM CaCl2/EDTA, and extensive dialysis in 5 mM HEPES, pH 7.4 (32). Recombinant human SP-A1 (variant 6A2) and SP-A2 (variant 1A0) were expressed in CHO-K1 cells via the pEE14 expression vectors (39). SP-A1 and SP-A2 secreted in media were purified by affinity chromatography over mannose-Sepharose and gel filtration chromatography over Superose 6 (39). The purity of rat and human proteinosis SP-A was >95% and recombinant human SP-A1 and SP-A2 was >95% pure as assessed by 1 and/or 2-dimensional SDS-PAGE (39, 65), respectively. All SP-A preparations contained negligible amounts of bacterial endotoxin lipopolysaccharide below 0.1 pg/mg of protein as assessed by the Limulus Amoebocyte Lysate assay.
Mouse AMs were isolated by BAL using five sequential intratracheal instillation of 0.5 mL PBS/1 mM EDTA for a total of 2.5 mL. The BAL was centrifuged at 250 × g for 10 min to pellet the macrophages. The macrophages were washed once in PBS, counted, and then suspended in iodination blocking buffer. Human AMs were isolated from donated human lungs which were rejected for transplant ex vivo as described previously (66, 67). All procedures were approved by the Penn State College of Medicine Institutional Review Board. Cell purity was assessed microscopically after cytospin centrifugation and HEMA-3 differential staining (29).
The starting coordinates for molecular dynamics (MD) simulations were obtained from the recombinant rat SP-A crystal structure 1R13 (4) (http://www.rcsb.org/pdb/explore.do?structureId=1r13) consisting of the neck and CRD domain with a mutation at the glycosylation site consensus asparagine 187 (N187S) and lacking the amino-terminal and collagen-like domains (ΔN1-80) using Swiss-Pdb Viewer (68). Molecular dynamics simulations were carried out as described previously (69). We tested the impact of alanine point mutations in the primary Ca++ coordination residues of the CRD carbohydrate-lipid-LPS (CLL) binding pocket, E195A, R197A, N214A, and D215A, and E171A in the auxiliary metal ion coordination loop on Ca++- coordination bond length and solvent accessibility surface area. All residues tested are conserved between rodent and human SP-A, except for R197 which is naturally substituted by an alanine in both human SP-A1 and SPA2.
Sequence alignments were performed using ClustalW (70, 71) (Figure 1). The amino acid alignment and location of Ca++- coordination residues of rat SP-A and human SP-A2 (variant 1A0) are shown on Figure 1A and part of the CDM on Figure 1B. The numbering of amino acid residues on Figure 1A is based on the mature rat SP-A after cleavage of the respective signal peptide between residues C20-N21 or as stated otherwise (Uniprot Accession No: P08427) (72, 73). In humans, although the signal peptide cleavage has been shown to affect a few amino acids (74), here we based the numbering on one of the cleaved variants in C20-E21 of the SP-A2 variant 1A0 (Uniprot Accession No: Q8IWL1). For the CDM alignments numbering is from the start of respective signal sequences (Figure 1B) to avoid confusion with differences in amino-acid numbering for rat and human SP-A1 and SP-A2 in the literature. The numbering of all rat SP-A point and deletion mutants is based on the mature rat peptide.
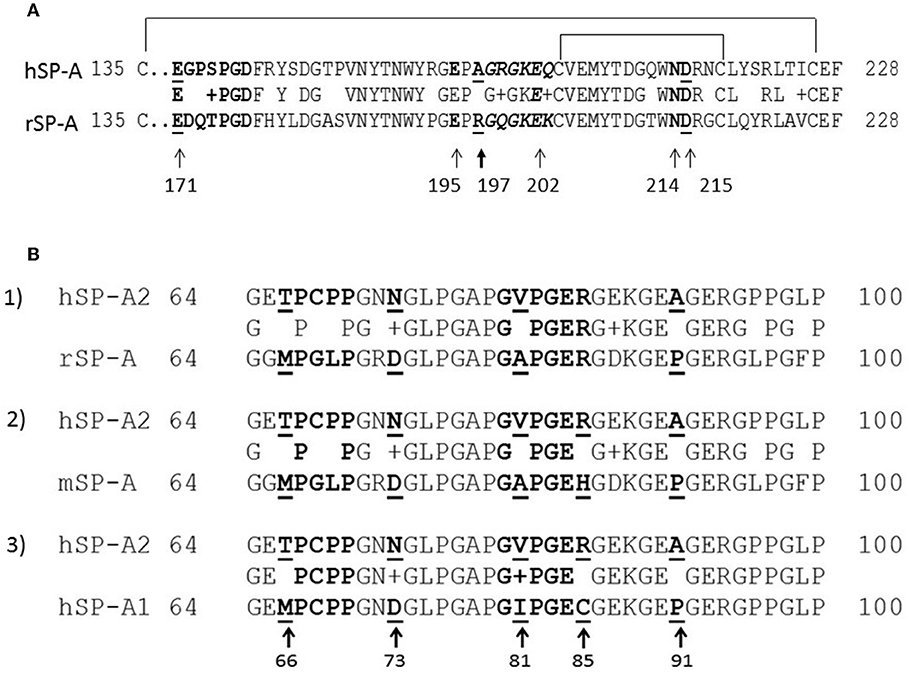
Figure 1. Partial sequence alignment of the CRD (A) and CDM (B) domains of rodent and human SP-A. (A) Alignment of the CRD domains of human SP-A2 1A0 variant (hSP-A2) and rat SP-A carbohydrate recognition domain. Amino acid numbering is based on the mature peptide of rat SP-A. Arrows point to Ca++-coordination residues. (B) Alignment of the collagen-like domains of human SP-A1 (6A2) (hSP-A1) and hSP-A2 1A0 variant, mouse SP-A (mSP-A), and rat SP-A (rSP-A). All alignments are made to hSP-A2 (1A0). The aligned sequences shown encompass the kink peptide (bolded residues 66–50) to the end of the CDM. Arrows point to core amino acids in the CDM that distinguish SP-A1 and SP-A2; the numbering is based on the precursor molecule. Residues shown in italicized red font show the sequence of embedded integrin binding motifs in the CDM. Sequence alignments were performed using ClustalW.
Macrophage binding assays were performed using mouse or human AMs, or the RAW264.7 macrophage cell line and 125I-labeled SP-A as described in detail previously (10, 32). The purified His-tagged SP-A binding neck domain of SP-R210 (32, 75) was used in solid phase binding assays with SP-A. Flat bottomed 96 well ELISA plates were treated with 0.1 M Na2CO3, pH9.3, and coated with 10 μg/mL of purified nSP-R210, and blocked in 0.1% BSA. Recombinant rat SP-A, native rat, or human SP-A1 or SP-A2 were added at increasing concentration in SP-A binding buffer (HBSS not containing phenol red, 20 mM HEPES (pH 7.4), 1 mM CaCl2, 0.2 mM MgCl2, 1% bovine serum albumin) and incubated at 37°C in a humidified chamber. Plates were washed in binding buffer, incubated with HRP-conjugated anti-rat affinity purified polyclonal rabbit anti-SP-A antibody for 1 hr. at room temperature, and then washed. Bound antibody was visualized using 3, 3′, 5, 5′-tetramethylbenzidine (TMB) HRP substrate at 450 nM.
Graphical analyses of binding data were performed with GraphPad Prism software (GraphPad Software, San Diego, CA). Binding data were analyzed by non-linear regression of saturation curves fitted to a single site equilibrium binding equation using Prism software to calculate the number of SP-A binding sites/cell (Bmax) and binding affinities (Kd). The hill equation embedded in Prizm software was used to calculate the Hill coefficient of cooperativity for human SP-A binding to murine and human alveolar macrophages.
We used deletion and point mutants of insect cell expressed rat SP-A to map SP-A binding to macrophages and recombinant SP-R210 compared to native human or rat SP-A isolated from BAL. Insect cell expressed SP-A lacks proline hydroxylation of Gly-X-Pro repeats in the CDM of SP-A (rSP-Ahyp) and also exhibits an altered pattern of glycosylation (61). Hydroxyproline imparts thermal stability that influences the degree of oligomerization of the collagen-like domain of SP-A (61). Saturation binding assays demonstrated that lack of hydroxyproline reduced SP-A binding potential to both macrophages (Figures 2A,C,E,F, Table 1) and recombinant SP-R210 (Figures 3A–D, Table 2) as demonstrated by 3–9 fold decreases in both maximal binding (Bmax) and binding affinity (Kd) compared to native SP-A.
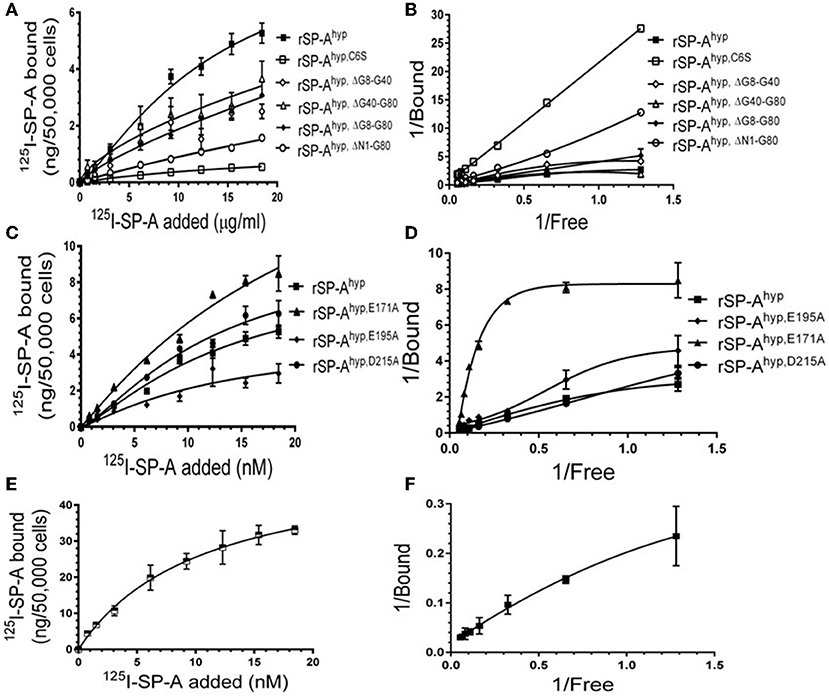
Figure 2. Impact of domain deletion and site directed mutagenesis on SP-A binding to macrophages. Binding assays used Raw264.7 macrophages in suspension. Cells were incubated with increasing concentrations of either radiolabeled recombinant rat SP-A proteins (A–D) or human SP-A (E,F) purified from alveolar proteinosis lavage. Binding data were fitted by non-linear regression to generate binding isotherms (A,C,E) or transformed into double reciprocal plots to evaluate SP-A binding behavior (B,D,F). Assays were carried out at 4°C for 1.5 h. at the 50,0000 cells/assay. Bound SP-A was separated by centrifugation over a silicon oil mixture to separate bound from free SP-A. Assays were performed in triplicate and data pooled from 2 independent experiments. Data shown are Means ± SE.
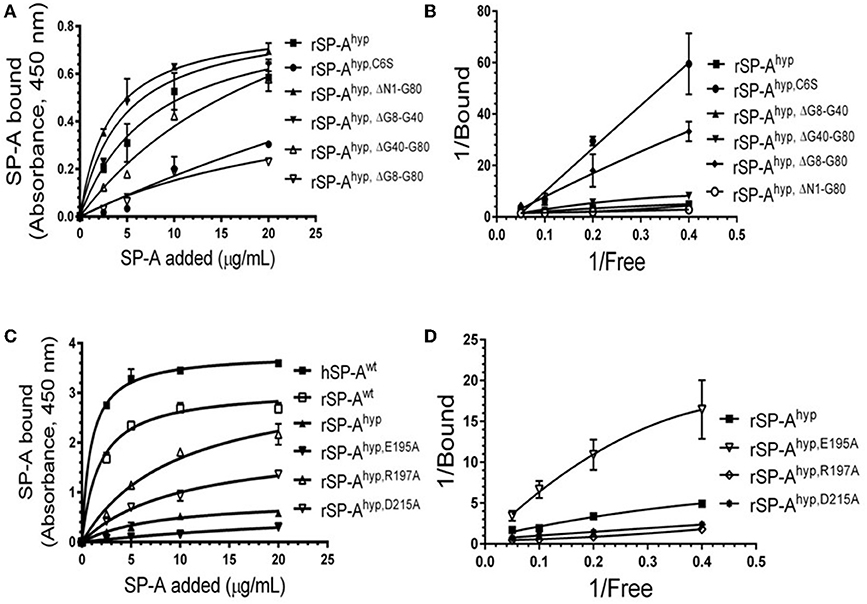
Figure 3. Impact of domain deletion and site-directed mutagenesis on SP-A binding to SP-R210. The recombinant SP-A binding domain of SP-R210 was coated onto microtiter well plates, and incubated with increasing concentration of indicated recombinant rat SP-A proteins (A–D), or native human or rat SP-A proteins (C,D). Bound protein was detected using an HRP-conjugated SP-A antibody. Binding data were fitted by non-linear regression to generate binding isotherms (A,C) or transformed into double reciprocal plots to evaluate SP-A binding behavior (B,D). Assays were performed in duplicate or triplicate and data pooled from 2 independent experiments. Data shown are Means ± SE.
Deletion and site-directed mutagenesis were then used to assess the effect of the amino-terminal and CDM oligomerization domains on the binding of rSP-Ahyp (Figures 1A,B, 2A, Tables 1, 2). Complete CDM deletion (rSP-Ahyp,ΔG8−G80) impaired binding to both macrophages and SP-R210 compared to rSP-Ahyp. Point mutation of cysteine 6 to serine in the amino terminal peptide (rSP-Ahyp,C6S) diminished Bmax with similar binding affinity to both macrophages and SP-R210 compared to rSP-Ahyp. The partial CDM deletion rSP-Ahyp,ΔG8−G40 and rSP-Ahyp,ΔG40−G80 mutants had increased high affinity binding at reduced Bmax in macrophages compared to rSP-Ahyp. The rSP-Ahyp,ΔG40−G80 mutant, however, exhibited reduced affinity to SP-R210, whereas rSP-Ahyp,ΔN1−G80 bound SP-R210 with increased affinity compared to rSP-Ahyp. The Hughes-Klotz double reciprocal plots (Figures 2B,F, 3B) indicated negative cooperativity for rSP-Ahyp, native SP-A (Figure 2F), and the partial CDM deletion mutants rSP-Ahyp,ΔG8−G40 and rSP-Ahyp,ΔG40−G80 compared to non-interacting binding sites for rSP-Ahyp,C6S, rSP-Ahyp,ΔG8−G80 and rSP-Ahyp,ΔN1−G80, indicating that oligomerization modulates the binding behavior of SP-A. Mutants to examine the role of the neck coiled-coil domain are not available. Molecular dynamics simulations with and without the coiled-coil domain showed as the CRD alone did not equilibrate in silico as indicated by the increasing root mean square deviation (RMSD) of the CRD α-carbon atoms compared to the NCRD (Figure 4).
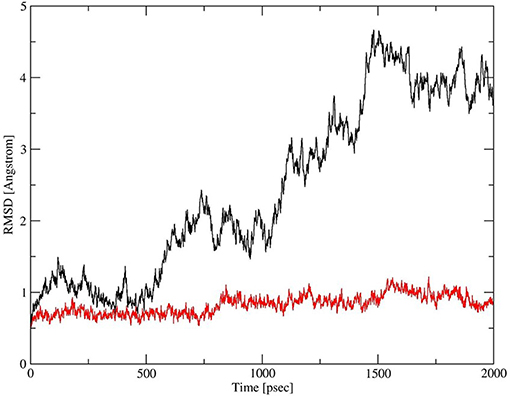
Figure 4. Dynamics of CRD and neck-CRD domains. Plot of root mean square deviation (RMSD) of α-Carbon atoms over time for CRD (red trace) and neck-CRD (black trace).
We then examined the effect of alanine (A) mutagenesis of residues that influence geometry and Ca++ coordination in the CRD domain (4, 5) using binding assays and MD simulations (Table 1). Compared to the crystal structure, the MD simulations indicate that the side chain carbonyl for asparagine (N) 214 and backbone carbonyl of aspartate (D) 215 occupy spaces further away from Ca++ in the WT CRD. The side chain hydroxyl of D215 and both oxygens in the E202 side chain are oriented closer to the Ca++ ion (Table 3). The invariant glutamate 171 (E171) influences Ca++ coordination of the E202 residue side chain, conformation of the double-loop structure surrounding the CRD carbohydrate binding pocket via a side-chain salt bridge with lysine K203, and coordination of an auxiliary ion (4, 5). The E171A substitution mutant (rSP-Ahyp,E171A) exhibited increased binding at high concentrations to macrophages compared to rSP-Ahyp (Figure 2C). The double-reciprocal Hughes-Klotz plot (76) of rSP-Ahyp,E171A showed strongly increased negative cooperativity compared to rSP-Ahyp (Figure 2D), suggesting abortive interaction with macrophages at low concentration. Binding to SP-R210 was not be determined. The E202 side chain of rSP-Ahyp,E171A did not coordinate Ca++, consistent with crystallographic data (4). The average surface area solvent accessibility (SASA) of the Ca++ ion and Ca++ coordination amino acids, however, was not altered by the E171A substitution compared to WT (Tables 4, 5). In previous studies, mutation of residues E195 (rSP-Ahyp,E195A) and D215 (rSP-Ahyp,D215A), two invariant residues that coordinate Ca++, demonstrated critical roles in Ca++-dependent binding and aggregation of DPPC, and binding to alveolar type II epithelial cells (13). E195 coordinates Ca++ via its side chain oxygens. Here, the binding affinity of the rSP-Ahyp,E195A mutant could not be accurately determined from the binding curves (Table 1). The double-reciprocal plot of rSP-Ahyp,E195A binding data, however, revealed atypical sigmoidal behavior suggesting interaction with low and high affinity binding sites in macrophages (Figure 2D), whereas this mutant exhibits negative cooperativity with recombinant SP-R210 (Figure 3D). In the E195A substitution, the R197 backbone carbonyl moved 4.63 Å away from Ca++, and also altered the orientation of asparagine (N) 214, D215, and E202 side chains relative to the Ca++ ion (Table 3 and Figures 5A,B). These changes were accompanied by increased H2O occupancy and average SASA of the Ca++ coordination site residues (Tables 4, 5), suggesting that H2O modifies the ligand binding properties of this mutant. The rSP-Ahyp,D215A mutant, on the other hand, displayed increased Bmax and low affinity non-cooperative binding compared to rSP-Ahyp (Figures 2C, 3C and Tables 1, 2). The D215 residue contributes both its side chain and peptide bond carbonyl to Ca++-coordination. The in silico calculations showed that the D215 backbone carbonyl moves closer to Ca++ by 1.33 Å when the side chain is replaced by Alanine (Table 3). The D215A substitution also influenced the orientation of E195 and E202 side chains, Ca++ proximity of the R197 carbonyl, and increased Ca++-H2O occupancy (Tables 3–5). The arginine (R) 197 residue, which coordinates Ca++-H2O occupancy via its peptide bond carbonyl, is naturally switched with alanine in human SP-A. Interestingly, the R197A substitution increased the binding of rSP-Ahyp,R197A to SP-R210 similar to the level of native rat and human SP-A, neutralizing the negative impact of proline hydroxylation deficiency in the CDM (Figure 3C and Table 2). The computational analysis revealed that the R197A switch brings the hydroxyl oxygens of E195 and N214 side chains and peptide chain carbonyl oxygen of D215 closer to Ca++ by 1-2.5 Å while the hydroxyl oxygen of D215 and both side chain oxygens of E202 orient away from Ca++ (Table 3). The hydroxyl oxygen of the D215 side chain rotated the farthest, 4.15 Å away from Ca++ compared to WT. These adjustments in Ca++ coordination are accompanied by >50% decrease in hydrophilic SASA of the Ca++ coordination shell from 227.38 ± 23.94 to 106 ± 11.50 Å2 of WT and rSP-Ahyp,R197A, respectively, suggesting a more compact structure of the “humanized” CRD (Tables 4, 5).
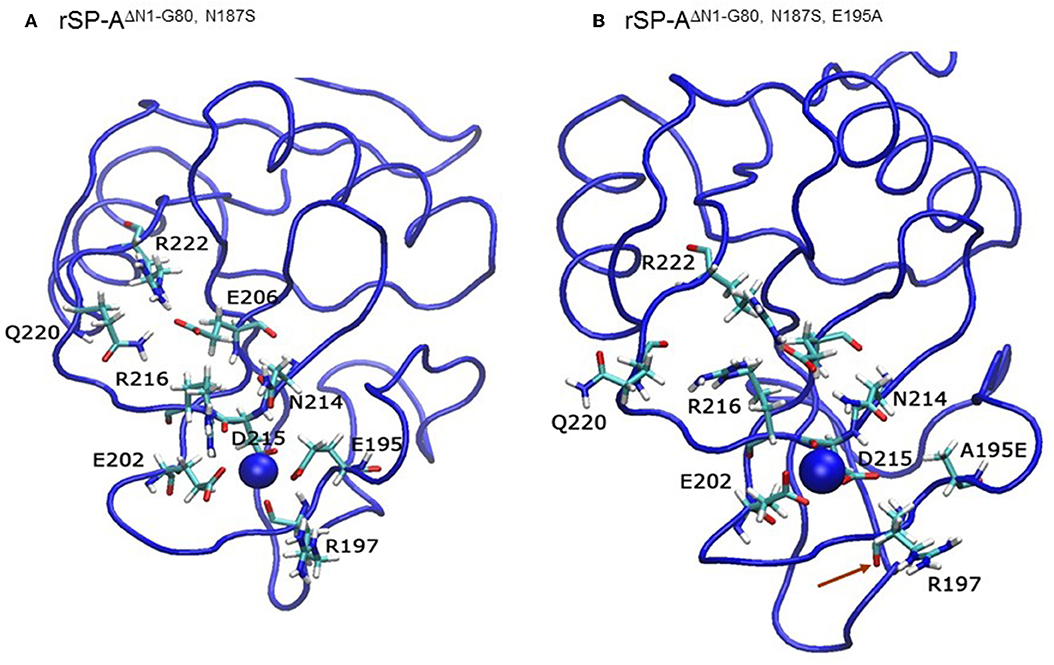
Figure 5. The E195A substitution alters the Ca++-coordination mode in the CLL binding pocket. Ribbon diagrams depicting the geometry of Ca++-coordination site of the native (A) and mutant E195A NCRD (B). Red arrows point to the carbonyl oxygen in the R197 residue.
We took advantage of previously developed “humanized” SP-A transgenic mice to determine the impact of SP-A isoforms on AM binding phenotype. The hSP-A1 and hSP-A2 isoforms were expressed in the lungs of SP-A−/− mice. Binding assays were carried out in AMs from WT (SP-A+/+), SP-A-deficient (SP-A−/−), or SP-A−/− mice carrying either expression of human SP-A genes SFTPA1 (6A2) or SFTPA2 (1A0) via the alveolar epithelial Sftpc promoter (21). The coding sequences of 6A2 and 1A0 hSP-A1 and hSP-A2 variants, respectively, differ in four core amino acids in their collagen-like domain plus amino acids 19 and 91 (77), based on the numbering of the precursor molecule. The binding assays in Figures 6A–D and Table 6 show that in vivo expression of hSP-A2 resulted in ligand-induced upregulation of hSPA2 binding to AMs compared to SP-A−/− and hSP-A1 exposed AMs as demonstrated by the 2-2.5 increase in Bmax (Table 6). hSP-A2 also improved the binding potential of hSP-A1 by increasing binding affinity. hSPA1 did not bind SP-A−/− AMs and its expression alone induced only low affinity binding of hSPA1 compared to high affinity binding for hSP-A2, although at lower Bmax compared to the hSPA2 expression. High affinity binding of hSP-A1 was observed only in AMs from hSP-A2 or WT mice. The endogenous SP-A, however, had a hybrid effect enhancing both binding affinity and Bmax with positive cooperativity. Compared to one another, the Bmax of hSP-A1 binding was 2–3-fold lower than that of hSP-A2 binding to murine AMs, indicating that the CDM is responsible for differences in receptor binding capacity of hSP-A isoforms with identical CRD.
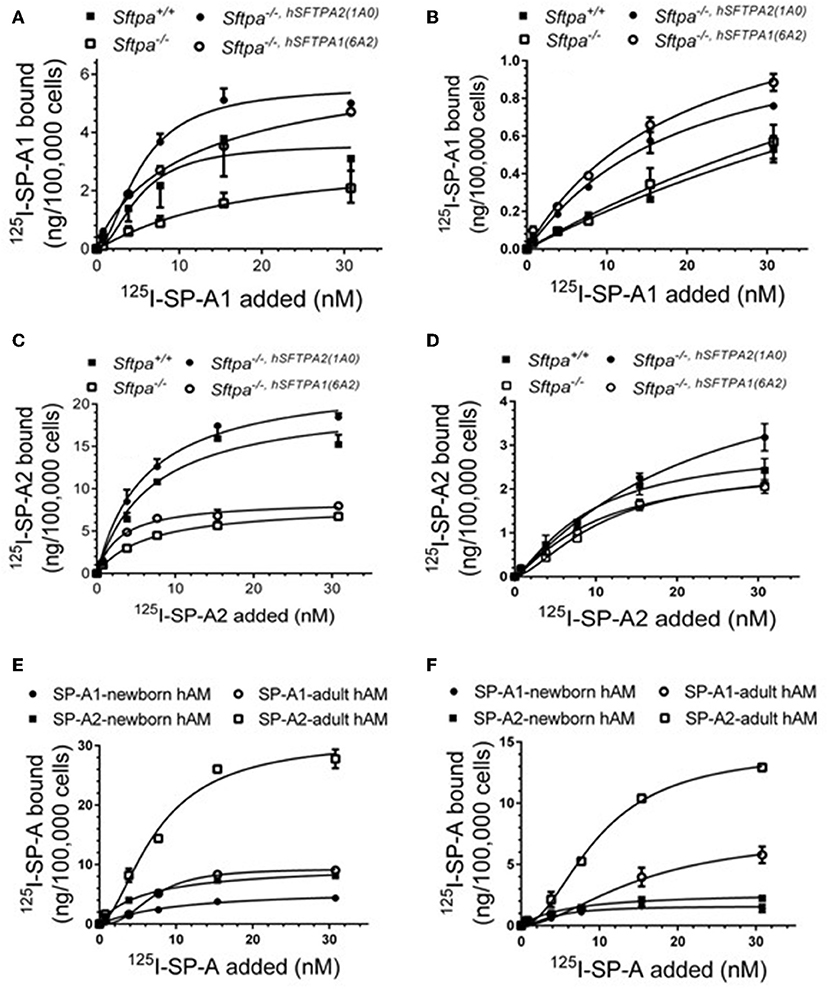
Figure 6. Murine and human SP-A variants are paracrine regulators of SP-A binding to alveolar macrophages in postnatal lung. Binding assays were carried out with alveolar macrophages obtained from either murine lung (A–D) or human lung (E,F) using radiolabeled human SP-A1 or SP-A2 in the presence (A,C,E) or absence of calcium (B,D,F). Alveolar macrophages from murine lungs (A–D) were obtained from WT (SP-A+/+), SP-A-deficient (SP-A−/−), or humanized mice expressing either SP-A1 variant 1A0 (SP-A−/−,hSFTPA2(1A0) ), or SP-A2 variant 6A2 (SP-A−/−,hSFTPA2(6A2) ) in the absence of endogenous murine SP-A (Sftpa). Human alveolar macrophages were obtained from 6 month (E) and 20 years old (F) rejected transplant lungs. Assays were carried out at 4°C for 1.5 h at 50,0000 cells/assay. Bound SP-A was separated by centrifugation over a silicon oil mixture. Assays were performed in duplicate and data pooled from 2 independent experiments. Alveolar macrophages from mouse lungs were pooled from 10 mice per genotype to obtain sufficient number of cells per experiment. Binding isotherms were generated using Prizm software by non-linear regression analysis using the Hill equation. Data shown are Means ± SE.
To address differences in receptor binding properties between mouse and human AMs, we performed binding assays with human AMs obtained from donated non-transplanted lung from a 6 month old newborn and a 20 year old adult individual. Figure 6E and Table 7 demonstrate that the Bmax for hSP-A2 was 3-fold higher in adult AMs than newborn AMs compared to a 1.6-fold increase for hSP-A1. The binding affinities were similar. Non-linear regression analysis of saturation data (Figure 6E, Table 7) revealed a change from non-cooperative to cooperative binding in newborn vs. adult AMs, respectively. The binding assays in mouse AMs exposed to endogenous or human SP-As in vivo indicate that hSP-A2 and endogenous mouse SP-A shaped the positive cooperativity binding phenotype of AMs (Figures 6A,B, and Table 6), whereas hSP-A1 did not. Additional assays in the presence of EDTA revealed that 30–50% of Bmax for hSP-A1 and hSP-A2 represent Ca++-independent binding sites (Figure 6F). The cooperative binding behavior in the absence of Ca++ was not altered. The binding affinities for both hSP-A isoforms were higher in newborn AMs compared to adult AMs, suggesting developmental regulation of SP-A binding in postnatal life.
The present studies refine our understanding of structural features that modulate rodent and human SP-A binding properties to macrophages. To our knowledge, the present study is the first direct comparison of rodent and human SP-A variants. Nested deletion mutagenesis, point mutations, and in silico simulations of rat SP-A indicated that high affinity binding to macrophages or SP-R210 depends on inter-domain interactions and binding sites in the CDM and CRD domains. Previous studies reported that the CDM of rat and human SP-A mediates binding to macrophages (10, 14). Here, deletion of the entire CDM abolished high affinity binding to both macrophages and SP-R210, whereas the rSP-Ahyp,ΔN1−80 mutant lacking both domains displayed low affinity binding relative to SP-Ahyp, suggesting that the amino-terminal peptide plays a conformational role in modulating binding distally. Partial CDM deletion, however, indicated that the amino-terminal half of the CDM that follows the kink peptide binds macrophages and SP-R210. On the other hand, site directed mutagenesis in the CRD domain showed that the E171A, D215A, and E195A point mutants altered binding behavior. All three mutations altered the conformation of Ca++ coordination and, furthermore, D215A and E195A increased accessibility to solvent. The E171 residue stabilizes the conformation of the CRD surface loops, while D215 and E195 are critical for binding to carbohydrates, lipids, and LPS (4–7). The present findings support the notion that the SP-R210 binding site on the CRD domain lies outside the CLL binding pocket.
Species adaptation in the CDM domain produced two alternate SP-A isoforms, SP-A1 and SP-A2, and changes in the CRD produced four variants, two for SP-A1 and two for SP-A2. These demarcate differences in ligand-induced modulation of receptor occupancy on AMs in vivo. In the present study we studied two of the CRD variants, one for SP-A1 (6A2) and one for SP-A2 (1A0). The amino-terminal half of the CDM of rat SP-A (Figure 1B) contains contiguous 85RGDKGE90 integrin binding motifs that are partially conserved in mouse SP-A with a corresponding 85HGDKGE90 but diverged in human SP-A with 85CGEKGE90 in SP-A1 and 85RGEKGE90 in SP-A2 (Figure 1B). Although RGD, HGD, and KGE motifs are well-known for binding to integrins (78, 79), other studies have reported interactions with the C2 membrane binding domain of phospholipases and other proteins (80–82). The interaction of this motif with SP-R210 remains to be determined. Evolutionary adaptation in the CRD domain of both isoforms introduced a natural R197A substitution between rodent and human SP-A2 (1A0) (Figure 1A). This substitution appears to isolate the impact of post-translational hydroxylation of proline on receptor occupancy by altering the interaction of the CRD domain with solvent and topology of Ca++ coordination residues in the CLL. These CDM and CRD adaptations may compensate for differences in dietary requirements for ascorbic acid and proline hydroxylation between rodents and humans (83). The present findings, however, indicate that single amino acid changes can profoundly affect CRD conformation, domain-domain interaction, and binding to macrophage receptors via CDM and CRD domains. In this regard, the CRD domain of human SP-A1 and SP-A2 is polymorphic resulting in diverse allelic combinations of SP-A1 and SP-A2 variants. Both coding and non-coding polymorphisms have been associated with increased or decreased risk to lung disease in numerous studies (77, 84). Understanding the structure-function relationship on human SP-A isoforms will require better understanding of copy number variation and biochemical characterization of polymorphic chain composition of SP-A1 and SP-A2 isoforms.
SP-A plays versatile roles in host defense through concomitant interactions with pathogens, surfactant lipids, and immune cells (1). SP-A controls the direction of macrophage activation, suppressing or inducing macrophage inflammatory responses as well as polarization toward either M1 or M2 types of differentiation and intrinsic regulation of innate immune receptor expression and function through the coordination of two SP-R210 isoforms (14, 27, 29, 33, 85, 86). Given present evidence that the SP-A CLL interacts with pattern recognition receptors (87), a distinct receptor binding site could be used to facilitate transfer of diverse SP-A cargo to innate immune receptors on macrophages. The SP-A structural determinants that differentially modulate receptor occupancy and signaling in AMs in the face of diverse immune challenges in the local lung microenvironment are subject to extensive investigation at the molecular level.
Human SP-A isolated from alveolar proteinosis fluid has been utilized extensively to discern function utilizing rodent alveolar macrophages without knowledge of the donor SP-A genotype. These studies generated invaluable information but also produced controversial as well as disparate results on SP-A immune functions and identity of SP-A receptors. The present findings in mouse AMs exposed to human SP-A isoforms not only support cross-species conservation and validity of ex vivo studies but also indicate genetic origin of SP-A preparations as an important variable in understanding versatility of SP-A function in humans. In this regard, the presence of SP-A1 may attenuate receptor occupancy, which likely regulates functional interactions with AMs at physiologically high levels of SP-A at homeostasis. Furthermore, although the present studies are limited by the small numbers of samples, the findings in newborn and adult AMs suggest increased receptor complexity that is developmentally regulated in humans.
Finally, our simulation and binding data indicate that the auxiliary ion coordination loop encompassing the conserved E171 residue may comprise a conserved site of interaction with macrophages, and SP-R210 specifically, that merits further investigation to discern conserved and divergent SP-A functions between species. In this context, we recognize that there are limitations inherent to MD simulations that impact the behavior of ions. MD simulations, however, have the potential to unveil how structural differences alter dynamic properties that are crucial for understanding the interplay of structure and function as well as providing guidelines for experiments. The purpose of the simulations, however, was not to reproduce the exact theoretical or experimental ion interactions but instead, to carry out a comparative study of wild type and mutated structures and specifically investigate the distances and their stability. This comparative in silico analysis of the R197A mutant in combination with our binding results showing that this species adapted residue modulates binding to the receptor opens exciting questions on future structure-function studies to understand the basis of molecular adaptation of the human SP-A molecules.
The raw datasets supporting the conclusions of this manuscript will be made available by the authors upon request.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee, Pennsylvania State University College of Medicine. All procedures with isolation of human alveolar macrophages were approved by the Penn State College of Medicine Institutional Review Board.
AN acquired and generated figures and tables for all the molecular dynamics simulation data. TU performed all binding assays with hSP-A1 and hSP-A2, isolated macrophages from mouse and lung BAL, and purified human SP-A proteins. C-HY performed all binding assays with recombinant rat SP-A, purified recombinant SP-R210, and cultured cells. PS and NT coordinated the procurement and logistics of donated human lung and contributed to the isolation of human alveolar macrophages. JF provided humanized SP-A1 and SP-A2 mice for the isolation of alveolar macrophages, and edited the manuscript critically. FM produced all recombinant rat SP-A proteins. ZC conceptualized, designed, led the study, and wrote the manuscript.
This work was supported by the National Institutes of Health [grant numbers HL068127 and EHS009882]; The Department of Pediatrics, Penn State College of Medicine, and The Children's Miracle Network.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AMs, alveolar macrophages; BAL, bronchoalveolar lavage; CDM, collagen-like domain; CLL, carbohydrate, lipids, LPS binding site; CRD, carbohydrate recognition domain; GEC, Glycine Glutamate Cysteine; GER, Glycine Glutamate Arginine; MDLG, Methionine Aspartate Leucine Glycine; NCRD, coiled-coil neck and CRD domain; PCPP, Proline Cysteine Proline Proline; SP-A, surfactant protein A; rSP-A, native rat SP-A; hSP-A, native human SP-A; hSP-A1 6A2, human SP-A isoform 1 variant 6A2; hSP-A2 1A0, human SP-A isoform 2 variant 1A0; rSP-Ahyp, hydroxyproline-deficient recombinant rat SP-A; SP-R210, SP-A receptor 210; Myo18A, Myosin 18A.
1. Chroneos ZC, Sever-Chroneos Z, Shepherd VL. Pulmonary surfactant: an immunological perspective. Cell Physiol Biochem. (2010) 25:13–26. doi: 10.1159/000272047
2. Seaton BA, Crouch EC, McCormack FX, Head JF, Hartshorn KL, Mendelsohn R. Review: structural determinants of pattern recognition by lung collectins. Innate Immun. (2010) 16:143–50. doi: 10.1177/1753425910368716
3. Nathan N, Taytard J, Duquesnoy P, Thouvenin G, Corvol H, Amselem S, et al. Surfactant protein A: a key player in lung homeostasis. Int J Biochem Cell Biol. (2016) 81:151–5. doi: 10.1016/j.biocel.2016.11.003
4. Head JF, Mealy TR, McCormack FX, Seaton BA. Crystal structure of trimeric carbohydrate recognition and neck domains of surfactant protein A. J Biol Chem. (2003) 278:43254–60. doi: 10.1074/jbc.M305628200
5. Shang F, Rynkiewicz MJ, McCormack FX, Wu H, Cafarella TM, Head JF, et al. Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change. J Biol Chem. (2011) 286:757–65. doi: 10.1074/jbc.M110.175265
6. Goh BC, Wu H, Rynkiewicz MJ, Schulten K, Seaton BA, McCormack FX. Elucidation of Lipid binding sites on lung surfactant protein A using X-ray crystallography, mutagenesis, and molecular dynamics simulations. Biochemistry. (2016) 55:3692–701. doi: 10.1021/acs.biochem.6b00048
7. Rynkiewicz MJ, Wu H, Cafarella TR, Nikolaidis NM, Head JF, Seaton BA, et al. Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop. Biochemistry. (2017) 56:4095–105. doi: 10.1021/acs.biochem.6b01313
8. Kuroki Y, Mason RJ, Voelker DR. Alveolar type II cells express a high-affinity receptor for pulmonary surfactant protein A. Proc Natl Acad Sci USA. (1988) 85:5566–70. doi: 10.1073/pnas.85.15.5566
9. Wright JR, Borchelt JD, Hawgood S. Lung surfactant apoprotein SP-A (26-36 kDa) binds with high affinity to isolated alveolar type II cells. Proc Natl Acad Sci USA. (1989) 86:5410–4. doi: 10.1073/pnas.86.14.5410
10. Chroneos ZC, Abdolrasulnia R, Whitsett JA, Rice WR, Shepherd VL. Purification of a cell-surface receptor for surfactant protein A. J Biol Chem. (1996) 271:16375–83. doi: 10.1074/jbc.271.27.16375
11. Pison U, Wright JR, Hawgood S. Specific binding of surfactant apoprotein SP-A to rat alveolar macrophages. Am J Physiol. (1992) 262:L412–L417. doi: 10.1152/ajplung.1992.262.4.L412
12. McCormack FX, Kuroki Y, Stewart JJ, Mason RJ, Voelker DR. Surfactant protein A amino acids Glu195 and Arg197 are essential for receptor binding, phospholipid aggregation, regulation of secretion, and the facilitated uptake of phospholipid by type II cells. J Biol Chem. (1994) 269:29801–7.
13. McCormack FX, Stewart J, Voelker DR, Damodarasamy M. Alanine mutagenesis of surfactant protein A reveals that lipid binding and pH-dependent liposome aggregation are mediated by the carbohydrate recognition domain. Biochemistry. (1997) 36:13963–71. doi: 10.1021/bi970745q
14. Borron P, McCormack FX, Elhalwagi BM, Chroneos ZC, Lewis JF, Zhu S, et al. Surfactant protein A inhibits T cell proliferation via its collagen-like tail and a 210-kDa receptor. Am J Physiol. (1998) 275:L679–686. doi: 10.1152/ajplung.1998.275.4.L679
15. Chiba H, Sano H, Saitoh M, Sohma H, Voelker DR, Akino T, et al. Introduction of mannose binding protein-type phosphatidylinositol recognition into pulmonary surfactant protein A. Biochemistry. (1999) 38:7321–31. doi: 10.1021/bi990353e
16. Yu SH, McCormack FX, Voelker DR, Possmayer F. Interactions of pulmonary surfactant protein SP-A with monolayers of dipalmitoylphosphatidylcholine and cholesterol: roles of SP-A domains. J Lipid Res. (1999) 40:920–9.
17. Korfhagen TR, Levine AM, Whitsett JA. Surfactant protein A (SP-A) gene targeted mice. Biochim Biophys Acta. (1998) 1408:296–302. doi: 10.1016/S0925-4439(98)00075-1
18. Ikegami M, Elhalwagi BM, Palaniyar N, Dienger K, Korfhagen T, Whitsett JA, et al. The collagen-like region of surfactant protein A (SP-A) is required for correction of surfactant structural and functional defects in the SP-A null mouse. J Biol Chem. (2001) 276:38542–8. doi: 10.1074/jbc.M102054200
19. Palaniyar N, Ikegami M, Korfhagen T, Whitsett J, McCormack FX. Domains of surfactant protein A that affect protein oligomerization, lipid structure and surface tension. Comp Biochem Physiol A Mol Integr Physiol. (2001) 129:109–27. doi: 10.1016/S1095-6433(01)00309-9
20. Bates SR. P63 (CKAP4) as an SP-A receptor: implications for surfactant turnover. Cell Physiol Biochem. (2010) 25:41–54. doi: 10.1159/000272062
21. Wang G, Guo X, DiAngelo S, Thomas NJ, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: formation of tubular myelin in vivo requires both gene products. J Biol Chem. (2010) 285:11998–2010. doi: 10.1074/jbc.M109.046243
22. Sever-Chroneos Z, Krupa A, Davis J, Hasan M, Yang CH, Szeliga J, et al. Surfactant protein A (SP-A)-mediated clearance of Staphylococcus aureus involves binding of SP-A to the staphylococcal adhesin eap and the macrophage receptors SP-A receptor 210 and scavenger receptor class A. J Biol Chem. (2011) 286:4854–70. doi: 10.1074/jbc.M110.125567
23. Sidobre S, Nigou J, Puzo G, Riviere M. Lipoglycans are putative ligands for the human pulmonary surfactant protein A attachment to mycobacteria. Critical role of the lipids for lectin-carbohydrate recognition. J Biol Chem. (2000) 275:2415–22. doi: 10.1074/jbc.275.4.2415
24. Ragas A, Roussel L, Puzo G, Riviere M. The Mycobacterium tuberculosis cell-surface glycoprotein apa as a potential adhesin to colonize target cells via the innate immune system pulmonary C-type lectin surfactant protein A. J Biol Chem. (2007) 282:5133–42. doi: 10.1074/jbc.M610183200
25. McCormack FX, Festa AL, Andrews RP, Linke M, Walzer PD. The carbohydrate recognition domain of surfactant protein A mediates binding to the major surface glycoprotein of Pneumocystis carinii. Biochemistry. (1997) 36:8092–9. doi: 10.1021/bi970313f
26. McCormack FX, Gibbons R, Ward SR, Kuzmenko A, Wu H, Deepe GS Jr. Macrophage-independent fungicidal action of the pulmonary collectins. J Biol Chem. (2003) 278:36250–6. doi: 10.1074/jbc.M303086200
27. Weikert LF, Lopez JP, Abdolrasulnia R, Chroneos ZC, Shepherd VL. Surfactant protein A enhances mycobacterial killing by rat macrophages through a nitric oxide-dependent pathway. Am J Physiol Lung Cell Mol Physiol. (2000) 279:L216–223. doi: 10.1152/ajplung.2000.279.2.L216
28. Wu H, Kuzmenko A, Wan S, Schaffer L, Weiss A, Fisher JH, et al. Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J Clin Invest. (2003) 111:1589–602. doi: 10.1172/JCI16889
29. Yang L, Carrillo M, Wu YM, DiAngelo SL, Silveyra P, Umstead TM, et al. SP-R210 (Myo18A) isoforms as intrinsic modulators of macrophage priming and activation. PLoS ONE. (2015) 10:e0126576. doi: 10.1371/journal.pone.0126576
30. Tenner AJ, Robinson SL, Borchelt J, Wright JR. Human pulmonary surfactant protein (SP-A), a protein structurally homologous to C1q, can enhance FcR- and CR1-mediated phagocytosis. J Biol Chem. (1989) 264:13923–8.
31. Crowther JE, Kutala VK, Kuppusamy P, Ferguson JS, Beharka AA, Zweier JL, et al. Pulmonary surfactant protein a inhibits macrophage reactive oxygen intermediate production in response to stimuli by reducing NADPH oxidase activity. J Immunol. (2004) 172:6866–74. doi: 10.4049/jimmunol.172.11.6866
32. Yang CH, Szeliga J, Jordan J, Faske S, Sever-Chroneos Z, Dorsett B, et al. Identification of the surfactant protein A receptor 210 as the unconventional myosin 18A. J Biol Chem. (2005) 280:34447–57. doi: 10.1074/jbc.M505229200
33. Minutti CM, Jackson-Jones LH, Garcia-Fojeda B, Knipper JA, Sutherland TE, Logan N, et al. Local amplifiers of IL-4Ralpha-mediated macrophage activation promote repair in lung and liver. Science. (2017) 356:1076–80. doi: 10.1126/science.aaj2067
34. Floros J, Wang G, Mikerov AN. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2–impact on function. Crit Rev Eukaryot Gene Expr. (2009) 19:125–37. doi: 10.1615/CritRevEukarGeneExpr.v19.i2.30
35. Floros J, Steinbrink R, Jacobs K, Phelps D, Kriz R, Recny M, et al. Isolation and characterization of cDNA clones for the 35-kDa pulmonary surfactant-associated protein. J Biol Chem. (1986) 261:9029–33.
36. Karinch AM, Floros J. 5' splicing and allelic variants of the human pulmonary surfactant protein A genes. Am J Respir Cell Mol Biol. (1995) 12:77–88. doi: 10.1165/ajrcmb.12.1.7811473
37. Floros J, Hoover RR. Genetics of the hydrophilic surfactant proteins A and D. Biochim Biophys Acta. (1998) 1408:312–22. doi: 10.1016/S0925-4439(98)00077-5
38. DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, et al. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers. (1999) 15:269–81. doi: 10.1155/1999/961430
39. Wang G, Umstead TM, Phelps DS, Al-Mondhiry H, Floros J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ Health Perspect. (2002) 110:79–84. doi: 10.1289/ehp.0211079
40. Mikerov AN, Wang G, Umstead TM, Zacharatos M, Thomas NJ, Phelps DS, et al. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. (2007) 75:1403–12. doi: 10.1128/IAI.01341-06
41. Phelps DS, Umstead TM, Floros J. Sex differences in the acute in vivo effects of different human SP-A variants on the mouse alveolar macrophage proteome. J Proteomics. (2014) 108:427–44. doi: 10.1016/j.jprot.2014.06.007
42. Wang Y, Kuan PJ, Xing C, Cronkhite JT, Torres F, Rosenblatt RL, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. (2009) 84:52–9. doi: 10.1016/j.ajhg.2008.11.010
43. Mitsuhashi A, Goto H, Kuramoto T, Tabata S, Yukishige S, Abe S, et al. Surfactant protein A suppresses lung cancer progression by regulating the polarization of tumor-associated macrophages. Am J Pathol. (2013) 182:1843–53. doi: 10.1016/j.ajpath.2013.01.030
44. Phelps DS, Umstead TM, Silveyra P, Hu S, Wang G, Floros J. Differences in the alveolar macrophage proteome in transgenic mice expressing human SP-A1 and SP-A2. J Proteom Genom Res. (2013) 1:2–26. doi: 10.14302/issn.2326-0793.jpgr-12-207
45. Nathan N, Giraud V, Picard C, Nunes H, Dastot-Le Moal F, Copin B, et al. Germline SFTPA1 mutation in familial idiopathic interstitial pneumonia and lung cancer. Hum Mol Genet. (2016) 25:1457–67. doi: 10.1093/hmg/ddw014
46. Thorenoor N, Umstead TM, Zhang X, Phelps DS, Floros J. Survival of surfactant protein-A1 and SP-A2 transgenic mice after Klebsiella pneumoniae infection, exhibits sex-, gene-, and variant specific differences; treatment with surfactant protein improves survival. Front Immunol. (2018) 9:2404. doi: 10.3389/fimmu.2018.02404
47. Thorenoor N, Zhang X, Umstead TM, Scott Halstead E, Phelps DS, Floros J. Differential effects of innate immune variants of surfactant protein-A1 (SFTPA1) and SP-A2 (SFTPA2) in airway function after Klebsiella pneumoniae infection and sex differences. Respir Res. (2018) 19:23. doi: 10.1186/s12931-018-0723-1
48. Sanchez-Barbero F, Rivas G, Steinhilber W, Casals C. Structural and functional differences among human surfactant proteins SP-A1, SP-A2 and co-expressed SP-A1/SP-A2: role of supratrimeric oligomerization. Biochem J. (2007) 406:479–89. doi: 10.1042/BJ20070275
49. Wang G, Myers C, Mikerov A, Floros J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry. (2007) 46:8425–35. doi: 10.1021/bi7004569
50. Korfhagen TR, Bruno MD, Ross GF, Huelsman KM, Ikegami M, Jobe AH, et al. Altered surfactant function and structure in SP-A gene targeted mice. Proc Natl Acad Sci USA. (1996) 93:9594–9. doi: 10.1073/pnas.93.18.9594
51. Veldhuizen RA, Yao LJ, Hearn SA, Possmayer F, Lewis JF. Surfactant-associated protein A is important for maintaining surfactant large-aggregate forms during surface-area cycling. Biochem J. (1996) 313 (Pt 3):835–40. doi: 10.1042/bj3130835
52. Voss T, Schafer KP, Nielsen PF, Schafer A, Maier C, Hannappel E, et al. Primary structure differences of human surfactant-associated proteins isolated from normal and proteinosis lung. Biochim Biophys Acta. (1992) 1138:261–7. doi: 10.1016/0925-4439(92)90002-5
53. Ridsdale RA, Palaniyar N, Holterman CE, Inchley K, Possmayer F, Harauz G. Cation-mediated conformational variants of surfactant protein A. Biochim Biophys Acta. (1999) 1453:23–34. doi: 10.1016/S0925-4439(98)00057-X
54. Uemura T, Sano H, Katoh T, Nishitani C, Mitsuzawa H, Shimizu T, et al. Surfactant protein A without the interruption of Gly-X-Y repeats loses a kink of oligomeric structure and exhibits impaired phospholipid liposome aggregation ability. Biochemistry. (2006) 45:14543–51. doi: 10.1021/bi061338u
55. Wang G, Taneva S, Keough KM, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta. (2007) 1768:2060–9. doi: 10.1016/j.bbamem.2007.06.025
56. Lopez-Rodriguez E, Pascual A, Arroyo R, Floros J, Perez-Gil J. Human pulmonary surfactant protein SP-A1 provides maximal efficiency of lung interfacial films. Biophys J. (2016) 111:524–36. doi: 10.1016/j.bpj.2016.06.025
57. Sanchez-Barbero F, Strassner J, Garcia-Canero R, Steinhilber W, Casals C. Role of the degree of oligomerization in the structure and function of human surfactant protein A. J Biol Chem. (2005) 280:7659–70. doi: 10.1074/jbc.M410266200
58. Watson A, Kronqvist N, Spalluto CM, Griffiths M, Staples KJ, Wilkinson T, et al. Novel expression of a functional trimeric fragment of human SP-A with efficacy in neutralisation of RSV. Immunobiology. (2017) 222:111–8. doi: 10.1016/j.imbio.2016.10.015
59. Mikerov AN, Umstead TM, Gan X, Huang W, Guo X, Wang G, et al. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. (2008) 294:L121–L130. doi: 10.1152/ajplung.00288.2007
60. Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol. (2004) 286:L546–553. doi: 10.1152/ajplung.00267.2003
61. McCormack FX, Calvert HM, Watson PA, Smith DL, Mason RJ, Voelker DR. The structure and function of surfactant protein A. Hydroxyproline- and carbohydrate-deficient mutant proteins J Biol Chem. (1994) 269:5833–41.
62. Pattanajitvilai S, Kuroki Y, Tsunezawa W, McCormack FX, Voelker DR. Mutational analysis of Arg197 of rat surfactant protein A. His197 creates specific lipid uptake defects. J Biol Chem. (1998) 273:5702–7. doi: 10.1074/jbc.273.10.5702
63. Tsunezawa W, Sano H, Sohma H, McCormack FX, Voelker DR, Kuroki Y. Site-directed mutagenesis of surfactant protein A reveals dissociation of lipid aggregation and lipid uptake by alveolar type II cells. Biochim Biophys Acta. (1998) 1387:433–46. doi: 10.1016/S0167-4838(98)00159-9
64. McCormack FX, Damodarasamy M, Elhalwagi BM. Deletion mapping of N-terminal domains of surfactant protein A. The N-terminal segment is required for phospholipid aggregation and specific inhibition of surfactant secretion. J Biol Chem. (1999) 274:3173–81. doi: 10.1074/jbc.274.5.3173
65. Wang G, Phelps DS, Umstead TM, Floros J. Human SP-A protein variants derived from one or both genes stimulate TNF-alpha production in the THP-1 cell line. Am J Physiol Lung Cell Mol Physiol. (2000) 278:L946–L954. doi: 10.1152/ajplung.2000.278.5.L946
66. Yu WC, Chan RW, Wang J, Travanty EA, Nicholls JM, Peiris JS, et al. Viral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 viruses. J Virol. (2011) 85:6844–55. doi: 10.1128/JVI.02200-10
67. Silveyra P, Chroneos ZC, DiAngelo SL, Thomas NJ, Noutsios GT, Tsotakos N, et al. Knockdown of Drosha in human alveolar type II cells alters expression of SP-A in culture: a pilot study. Exp Lung Res. (2014) 40:354–66. doi: 10.3109/01902148.2014.929757
68. Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. (1997) 18:2714–23. doi: 10.1002/elps.1150181505
69. Iakhiaev AV, Nalian A, Koenig K, Idell S. Thrombin-thrombomodulin inhibits prourokinase-mediated pleural mesothelial cell-dependent fibrinolysis. Thromb Res. (2007) 120:715–25. doi: 10.1016/j.thromres.2006.12.001
70. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
71. Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, et al. Clustal W and clustal X version 2.0. Bioinformatics. (2007) 23:2947–8. doi: 10.1093/bioinformatics/btm404
72. McCormack FX. Structure, processing and properties of surfactant protein A. Biochim Biophys Acta. (1998) 1408:109–31. doi: 10.1016/S0925-4439(98)00062-3
73. Lin Z, Wang Y, Zhu K, Floros J. Differential allele expression of host defense genes, pulmonary surfactant protein-A and osteopontin, in rat. Mol Immunol. (2004) 41:1155–65. doi: 10.1016/j.molimm.2004.06.006
74. Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry. (2004) 43:4227–39. doi: 10.1021/bi036023i
75. Szeliga J, Jordan J, Yang CH, Sever-Chroneos Z, Chroneos ZC. Bacterial expression of recombinant MyoXVIIIA domains. Anal Biochem. (2005) 346:179–81. doi: 10.1016/j.ab.2005.07.021
76. Yoshida M, Ikegami M, Reed JA, Chroneos ZC, Whitsett JA. GM-CSF regulates protein and lipid catabolism by alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. (2001) 280:L379–386. doi: 10.1152/ajplung.2001.280.3.L379
77. Silveyra P, Floros J. Genetic complexity of the human surfactant-associated proteins SP-A1 and SP-A2. Gene. (2013) 531:126–32. doi: 10.1016/j.gene.2012.09.111
78. Xu J, Rodriguez D, Petitclerc E, Kim JJ, Hangai M, Moon YS, et al. Proteolytic exposure of a cryptic site within collagen type IV is required for angiogenesis and tumor growth in vivo. J Cell Biol. (2001) 154:1069–79. doi: 10.1083/jcb.200103111
79. Lu X, Lu D, Scully MF, Kakkar VV. Integrins in drug targeting-RGD templates in toxins. Curr Pharm Des. (2006) 12:2749–69. doi: 10.2174/138161206777947713
80. Andersen MH, Graversen H, Fedosov SN, Petersen TE, Rasmussen JT. Functional analyses of two cellular binding domains of bovine lactadherin. Biochemistry. (2000) 39:6200–6. doi: 10.1021/bi992221r
81. Simoes I, Mueller EC, Otto A, Bur D, Cheung AY, Faro C, et al. Molecular analysis of the interaction between cardosin A and phospholipase D(alpha). Identification of RGD/KGE sequences as binding motifs for C2 domains. FEBS J. (2005) 272:5786–98. doi: 10.1111/j.1742-4658.2005.04967.x
82. Rothwangl KB, Rong L. Analysis of a conserved RGE/RGD motif in HCV E2 in mediating entry. Virol J. (2009) 6:12. doi: 10.1186/1743-422X-6-12
83. Parsons KK, Maeda N, Yamauchi M, Banes AJ, Koller BH. Ascorbic acid-independent synthesis of collagen in mice. Am J Physiol Endocrinol Metab. (2006) 290:E1131–E1139. doi: 10.1152/ajpendo.00339.2005
84. Noutsios GT, Floros J. Childhood asthma: causes, risks, and protective factors; a role of innate immunity. Swiss Med Wkly. (2014) 144:w14036. doi: 10.4414/smw.2014.14036
85. Weikert LF, Edwards K, Chroneos ZC, Hager C, Hoffman L, Shepherd VL. SP-A enhances uptake of bacillus Calmette-Guerin by macrophages through a specific SP-A receptor. Am J Physiol. (1997) 272:L989–995. doi: 10.1152/ajplung.1997.272.5.L989
86. Samten B, Townsend JC, Sever-Chroneos Z, Pasquinelli V, Barnes PF, Chroneos ZC. An antibody against the surfactant protein A (SP-A)-binding domain of the SP-A receptor inhibits T cell-mediated immune responses to Mycobacterium tuberculosis. J Leukoc Biol. (2008) 84:115–23. doi: 10.1189/jlb.1207835
Keywords: alveolar macrophages, binding, lung, surfactant protein A, receptor
Citation: Nalian A, Umstead TM, Yang C-H, Silveyra P, Thomas NJ, Floros J, McCormack FX and Chroneos ZC (2019) Structural and Functional Determinants of Rodent and Human Surfactant Protein A: A Synthesis of Binding and Computational Data. Front. Immunol. 10:2613. doi: 10.3389/fimmu.2019.02613
Received: 17 July 2019; Accepted: 21 October 2019;
Published: 07 November 2019.
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Thomas Vorup-Jensen, Aarhus University, DenmarkCopyright © 2019 Nalian, Umstead, Yang, Silveyra, Thomas, Floros, McCormack and Chroneos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zissis C. Chroneos, emNocm9uZW9zQHBlbm5zdGF0ZWhlYWx0aC5wc3UuZWR1
†Present address: Ching-Hui Yang, Corteva AgriScience, Johnston, IA, United States
Patricia Silveyra, Behavioral Laboratory, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.