
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 19 September 2019
Sec. Mucosal Immunity
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.02233
This article is part of the Research TopicThe Role of Physical and Biological Gut Barriers in Modulating Crosstalk between the Microbiota and the Immune SystemView all 16 articles
 Alba Miranda-Ribera1,2
Alba Miranda-Ribera1,2 Maria Ennamorati1
Maria Ennamorati1 Gloria Serena1,2
Gloria Serena1,2 Murat Cetinbas3,4
Murat Cetinbas3,4 Jinggang Lan1
Jinggang Lan1 Ruslan I. Sadreyev3,5
Ruslan I. Sadreyev3,5 Nitya Jain1,2
Nitya Jain1,2 Alessio Fasano1,2†
Alessio Fasano1,2† Maria Fiorentino1,2*†
Maria Fiorentino1,2*†The balanced interplay between epithelial barrier, immune system, and microbiota maintains gut homeostasis, while disruption of this interplay may lead to inflammation. Paracellular permeability is governed by intercellular tight-junctions (TJs). Zonulin is, to date, the only known physiological regulator of intestinal TJs. We used a zonulin transgenic mouse (Ztm) model characterized by increased small intestinal permeability to elucidate the role of a primary impaired gut barrier on microbiome composition and/or immune profile. Ztm exhibit an altered gene expression profile of TJs in the gut compared to wild-type mice (WT): Claudin-15, Claudin-5, Jam-3, and Myosin-1C are decreased in the male duodenum whereas Claudin-15, Claudin-7, and ZO-2 are reduced in the female colon. These results are compatible with loss of gut barrier function and are paralleled by an altered microbiota composition with reduced abundance of the genus Akkermansia, known to have positive effects on gut barrier integrity and strengthening, and an increased abundance of the Rikenella genus, associated to low-grade inflammatory conditions. Immune profile analysis shows a subtly skewed distribution of immune cell subsets toward a pro-inflammatory phenotype with more IL-17 producing adaptive and innate-like T cells in Ztm. Interestingly, microbiota “normalization” involving the transfer of WT microbiota into Ztm, did not rescue the altered immune profile. Our data suggest that a primary impaired gut barrier causing an uncontrolled trafficking of microbial products leads to a latent pro-inflammatory status, with a skewed microbiota composition and immune profile that, in the presence of an environmental trigger, as we have previously described (1), might promote the onset of overt inflammation and an increased risk of chronic disease.
The interplay among host genetics, immune response, intestinal microbiota composition and function, and exposure to environmental factors is critical in the breakdown of the normally coordinated, homeostatic mucosal immune tolerance leading to the development of chronic inflammatory disorders. In this scenario, the role of increased gut permeability has been recently appreciated and seems to be a critical factor in the chain of events leading to break of tolerance and onset of inflammation, whose sequence is still largely unknown. Increased intestinal permeability has been reported more than 30 years ago in celiac and Crohn's disease patients and first-degree relatives suggesting that barrier dysfunction may be necessary but not sufficient to disease development (2–5). Animal studies suggest that epithelial barrier dysfunction can trigger intestinal disease (6–9) as the development of spontaneous colitis in the IL-10 KO mouse is prevented by blocking increased small intestinal permeability with the zonulin inhibitor AT1001 (8). A recent in-vitro study using a human “gut-on-a-chip” model of inflammation that allows manipulations of the gut barrier and inflammatory factors, has shown that barrier dysfunction plays a critical role in the initiation of inflammation (10). Increased intestinal permeability has been proposed in the pathogenesis of several diseases not only affecting the gastrointestinal system but also metabolic disorders such as obesity and diabetes and/or diseases of the brain and the nervous system such as Parkinson's disease, multiple sclerosis, and the autism spectrum (11–16).
Intestinal hyperpermeability may precede the clinical manifestations of disease (2, 6, 17) but since inflammation can induce permeability defects (18–20), the controversy regarding the role of barrier defects as the cause or the consequence of disease has not been resolved. Growing evidence suggest mutual influence of impaired gut barrier, microbiome composition/function, and immune profile (21–23), with subsequent break of tolerance leading to chronic inflammatory diseases in genetically susceptible individuals. The currently accepted view is that besides genetic predisposition, exposure to environmental stimuli, loss of gut barrier function, inappropriate immune response, and changes in gut microbiota composition and function seem to be all at play in the pathogenesis of CID (24, 25). Also, intriguing is the evidence that these five factors can influence each other in an intricate “interactome” that remains largely undefined. Identifying the key elements that initiate break of intestinal mucosal tolerance is hence of paramount importance to guide the development of therapeutic approaches to specifically target the initiator of the entire cascade that leads to inflammation. Despite research efforts, a primary role of increased intestinal permeability in the onset of inflammation has not been established.
Pre-haptoglobin 2 (pHP2) is the archetype of the zonulin family (26) that reversibly regulates intestinal permeability by modulating intercellular TJs (27, 28) and its release is triggered by specific microbiota (29) and exposure to gliadin (30, 31). Zonulin is integrally involved in the pathogenesis of several CID (30–32) and it is augmented in chronic inflammatory conditions associated with TJ dysfunction, both at mucosal level (for example celiac disease) (28) and systemically (for example type-1 diabetes) (33). Zonulin plays a role as specific modulator of innate immune response involving antigen trafficking through both epithelial and endothelial compartments as well as M2 polarized macrophages expansion (22, 34).
Mice do not express pHP2, which is present only in humans (26). The Zonulin transgenic mouse (Ztm), genetically engineered to express murine pHP2 under its natural promoter (35), is a model of zonulin-induced increased intestinal permeability and represents a unique and extremely valuable tool to understand how loss of control of antigen trafficking may influence the gut microbiota composition and the immune system development.
In this study, we have leveraged on the Ztm as an animal model of increased gut permeability and inflammation susceptibility (1) to get insights on the role of the gut barrier in orchestrating early mucosal events that eventually lead to the onset of inflammation and development of disease.
A colony of C57Bl/6 (WT) was maintained in our facility at Massachusetts General Hospital (MGH). Breeding pairs of Ztm were created as previously described (35), and generously donated by Andrew Levy. Both colonies were maintained in our facilities under standard conditions (12 h light: 12 h dark cycle, standard humidity, and temperature), and housed in separate cages within the same facility during the study. Animals were fed standard pellet food and water ad libitum. Mice 4–8 weeks of age were used in this study. All animal studies in this paper were approved by the Institutional Animal Care and Use Committee at the MGH (2013N000013).
WT and Ztm mice were anesthetized using Forane (isoflurane, USP) from Baxter, and euthanized by cervical dislocation. Tissues (duodenum, jejunum and colon) were immediately collected and kept at −80°C until processed. Total RNA was extracted from frozen tissues using TRIzol™ reagent (Thermo Fisher) and the Direct-Zol RNA mini prep Kit (ZymoReserach) following the manufacturer's protocol. To ensure full removal of genomic DNA, samples were treated with DNA free™ Kit (Thermo Fisher,). Total mRNA was quantified and A260/280 and A260/230 measured using a Nanodrop 2000 (Thermo Fisher). cDNA was made with random hexamer primers plus Maxima H− First Strand cDNA Synthesis Kit (Thermo Fisher) according to manufacturer's instructions. Quantitative real-time PCR was performed using PerfeCTa SYBR® Green SuperMix (Quanta) and run in a Bio Rad CFX connect thermocycler. All primers were designed in house (Table 1) using NIH's Primer Blast and obtained from Integrated DNA Technologies. We analyzed for gene expression the two most proximal sections of the small intestine (duodenum and jejunum). Only the sections of the intestine where we found significant gene expression changes are shown on graphs.
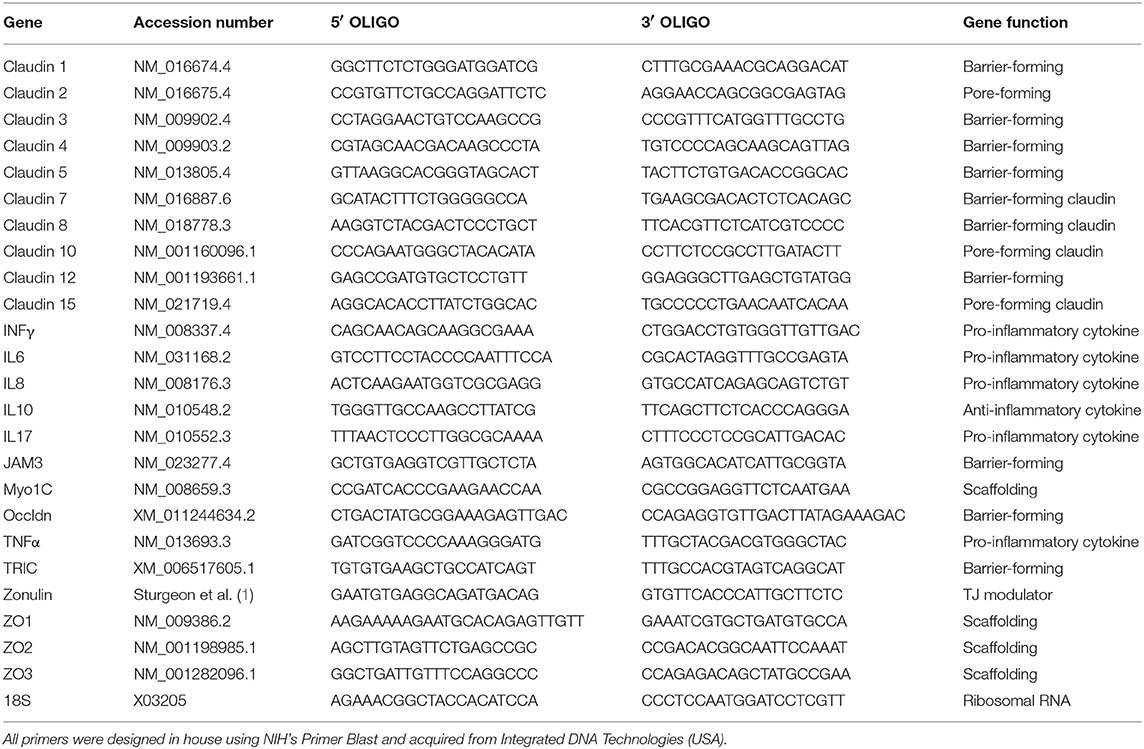
Table 1. List of all primers and FASTA accession numbers of the genes analyzed by real time qPCR analysis.
Stools were collected from a total of 71 mice divided into 3 groups: WT (n = 38), Ztm (n = 28), and ZtmbWT (n = 5) (Ztm receiving bedding from WT). To minimize cage effects (36), mice within each genotype were cohoused for 2 weeks before sample collection. Fecal samples were collected from each animal, flash frozen, and kept at −80°C until further processing. Genomic DNA, for each sample, was extracted using the DNeasy powersoil extraction kit (Qiagen) following Qiagen's instructions. DNA was quantified using Nanodrop. In order to carry out the phylogenetic profiling, the hypervariable V4 region of the 16S rRNA gene was amplified by PCR using 5X prime master mix (Prime), reverse 806 primers were barcoded and a unique forward 515 primer (Integrated DNA Technologies) was used. To confirm correct amplification of the V4 regions, a regular gel electrophoresis was run. PCR products were purified using the QIAquick PCR purification kit (Qiagen) and their concentration was measured by Quant-iT Picogreen dsDNA kit following manufacturer's instructions. Sequencing of the samples was done at the MGH NextGen Sequencing Core facility (Boston, US), on the Illumina system using the MiSeq v2 500 cycles reagent kit as per manufacturer's instructions. To allow maximum coverage of the amplicon the system sequenced a total of 250 paired-end cycles. The following primers were used for the sequencing (37):
read 1 (TATGGTAATT GT GTGYCAGCMGCCGCGGTAA)
read 2 (AGTCAGCCAGCCGGACTACNVGGGTWTCTAAT)
index (AATGATACGGCGACCACCGAGATCTACACGCT).
Timed mating of C57BL/6 (WT) and Ztm mice were set up and monitored for live births. On the day of birth, half the soiled bedding from a WT cage was mixed with autoclaved sterile bedding in a new cage. Ztm breeders with their newborn pups were transferred to this new cage with WT bedding (ZtmbWT mice). This process was repeated twice a week for 4 weeks until the pups were weaned. Colonic fecal content was collected 28 days after birth and analyzed for microbial composition.
Spleens were mechanically disrupted, and single cell suspensions were depleted of RBCs with ACK lysis buffer (Invitrogen). Lymphocytes from intestines were prepared as described (38). Small and large intestines from 28 to 35 days old mice were collected in ice-cold HBSS, cleaned and chopped into 1 mm pieces. Samples were pooled from 2 mice and intestinal epithelial cells were removed by shaking in HBSS containing 5 mM EDTA, 1 mM DTT, 5% FCS, and 15 mM HEPES for 30 min at 37°C. Tissues were digested in enzyme solution prepared in EDTA-free HBSS buffer containing 0.15 mg/mL Liberase TL (Roche) and 0.1 mg/mL DNase1 (Roche) for 45 min at 37°C. The LPLs liberated into the supernatant were filtered in a 40 μm strainer and stained for flow cytometry.
Cells were first stained for surface markers and then fixed in eBioscience Fix/Perm buffer. Cells were subsequently permeabilized in eBioscience permeabilization buffer and stained with antibodies against transcription factors. All data were acquired on custom-built BD FACS Aria or Fortessa X20 and data analyzed using FlowJo software (Treestar, CA). mCD1d-PBS57 tetramers were from NIH tetramer core. All batches of reagent were titrated for optimal staining of C57BL/6 splenocytes, with unloaded CD1d tetramers serving as negative controls. Changes in antibody staining panels (e.g., antibody clones, reagent batches) across experiments were minimized. Strong discrepancies in staining from different experiments were noted and the data in question were excluded from final analyses.
Sequencing data were processed and analyzed with QIIME2 software package v. 2018.2.0 (39). The sequencing reads with low quality score (average Q < 25) were truncated to 240 bp followed by filtering using deblur algorithm with default settings (40). The remaining high quality reads were aligned to the reference library using mafft (41). Next, the aligned reads were masked to remove highly variable positions, and a phylogenetic tree was generated from the masked alignment using the FastTree method (42). Alpha and beta diversity metrics and Principal Component Analysis plots based on Jaccard distance were generated using default QIIME2 plugins (39). Taxonomy assignment was performed using feature-classifier method and naïve Bayes classifier trained on the Greengenes 13_8 99% operational taxonomic units (OTUs). Differential abundance analysis of OTUs was performed using ANCOM (43). The Kruskal-Wallis test was used to assess the statistical significance of abundance differences, with the multiple testing corrections using Benjamini-Hochberg false discovery rate (FDR). The FDR cutoff was set at 0.05.
Statistical analysis was performed in Graph Pad Prism 8, all data are expressed as means ± standard error of the mean (SEM) using the non-parametric Mann-Whitney test for comparisons between 2 groups for the qPCR analysis and unpaired parametric t-test for comparisons between 2 groups for the flow cytometry data. P < 0.05 was considered statistically significant in both tests. Figures 1, 2 represent the expression levels of the genes analyzed. Multiple genes are shown in each graph for convenience. Please note that each gene has been compared to its WT counterpart by the method described above and represented as fold change. In Table 2 is reported the fold change of all genes analyzed. Zonulin gene expression in the Ztm intestine has been expressed as dCt (Table 2) since no zonulin is present in the WT.
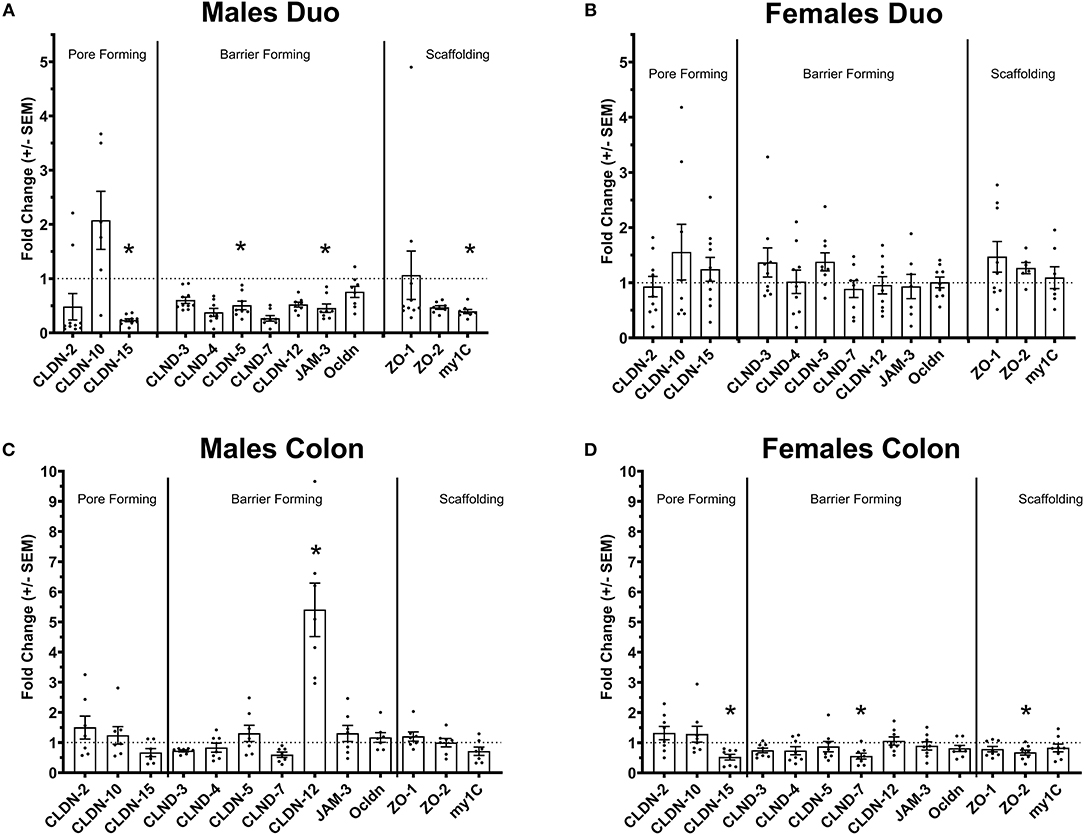
Figure 1. Tight-junctions gene expression profile in the intestine of Ztm. (A) Down regulated tight-junction genes in the duodenum Ztm males (n = 6–10). (B) Unaltered tight-junction profile in the duodenum of Ztm females (n = 7–12). (C) Increased CLDN-12 in the colon of Ztm males (n = 7). (D) Downregulation of tight junction genes in the colon of Ztm females (n = 8). mRNA expression was analyzed by qPCR, normalized to WT, and expressed as fold regulation. Statistical analysis was performed by the non-parametric Mann-Whitney test comparing each gene to its WT counterpart. *p < 0.05.
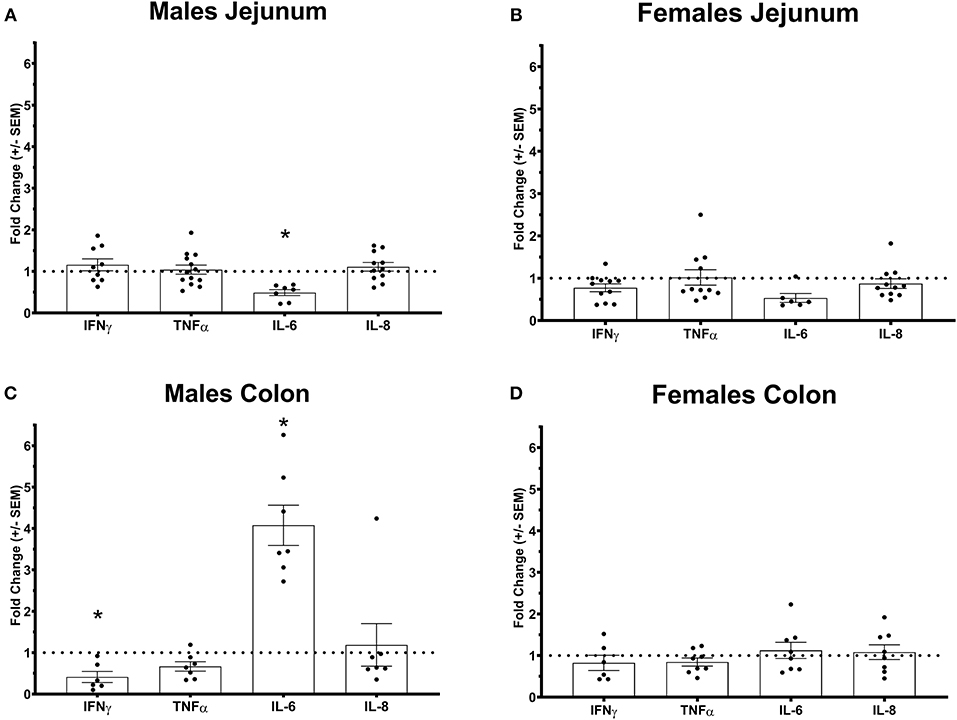
Figure 2. Pro-inflammatory cytokines gene expression profile in Ztm intestine: A) Down-regulated IL-6 in the jejunum of Ztm males (n = 13). B) Pro-inflammatory cytokines profile in and g the jejunum of Ztm females (n = 10). C) Down-regulated IFN up-regulated IL-6 in the colon Ztm males (n = 7). D) Unaltered Pro-inflammatory cytokines profile in the colon of Ztm females (n = 8). mRNA expression was analyzed by qPCR, normalized to WT, and expressed as fold regulation. Statistical analysis was performed by the non-parametric Mann-Whitney test comparing each gene to its WT counterpart. *p < 0.05.
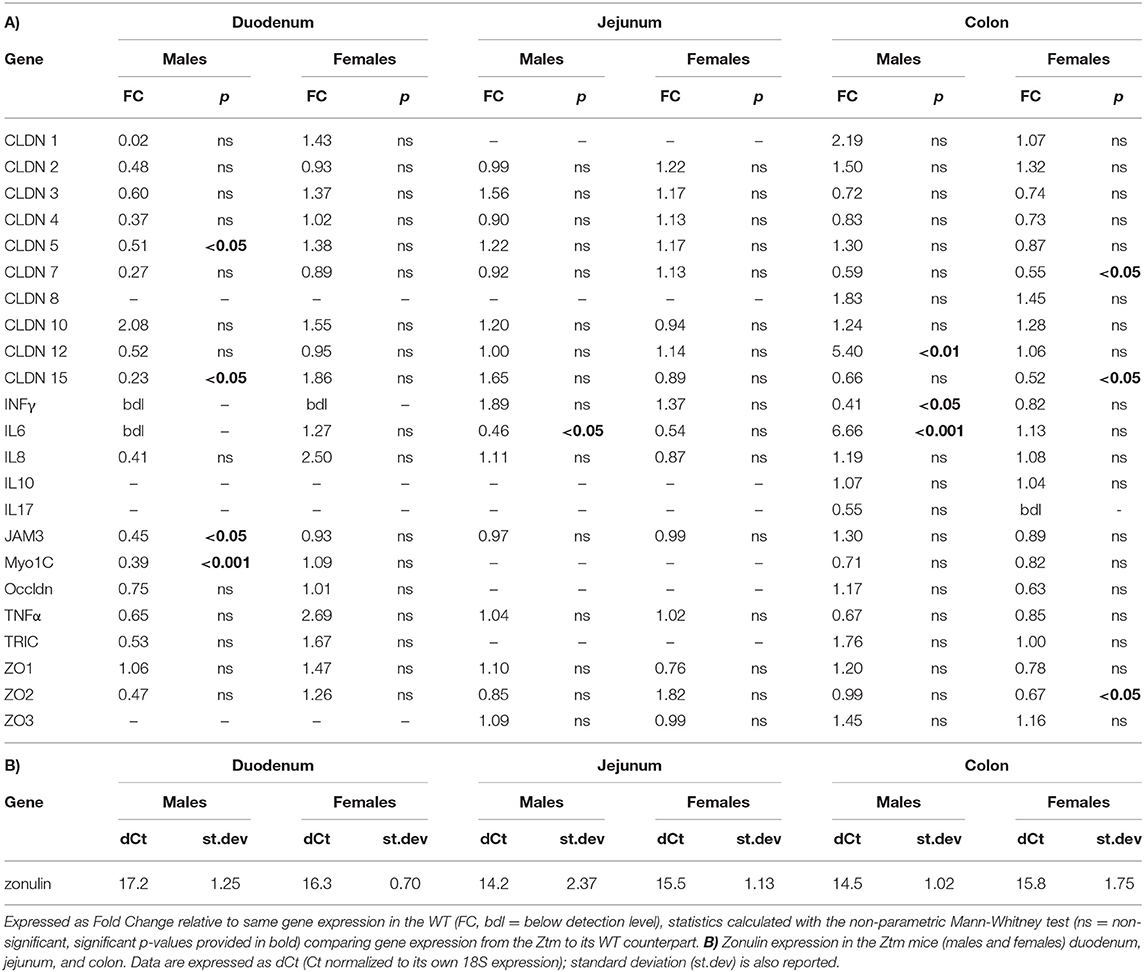
Table 2. (A) List of all genes analyzed by real time PCR in the duodenum, jejunum, and colon of adult mice (n = 6–12).
We have previously shown that Ztm mice exhibit a significantly increased small intestinal permeability both in-vivo and in-vitro at baseline associated to a higher susceptibility to DSS colitis, particularly in male mice, compared to WT (1). To elucidate the molecular basis of this altered permeability, we performed real time PCR of TJ and pro-inflammatory genes in different sections of the intestine (i.e., small intestine and colon) of WT and Ztm. Our results show an altered regulation of several TJs genes in Ztm compared to WT (Figure 1). In the small intestine of male Ztm we observed a significantly reduced expression of Claudin (CLDN)-15, CLDN-5, JAM-3, and Myo1C (p < 0.05; Figure 1A). We did not detect alterations in the expression of any of the analyzed genes in the small intestine of female Ztm (Figure 1B), in line with our previous finding of higher susceptibility to DSS-induced increased morbidity and mortality in male mice (1). Conversely, while in the colon of male Ztm only CLDN-12 gene expression appeared significantly (p < 0.05) increased (Figure 1C), in the female colon CLDN-15, CLDN-7, and ZO-2 were significantly (p < 0.05) reduced (Figure 1D). When looking at the expression of pro-inflammatory genes in the small intestine and colon, we detected a significant downregulation of IL-6 expression in the jejunum (p < 0.05) and its upregulation in the colon (p < 0.05) in Ztm males compared to WT males (Figures 2A,C). IFNγ was also significantly downregulated in the Ztm male colon (p < 0.05). Females exhibited a slightly decreased IL-6 expression in the jejunum (Figure 2B), while no changes were observed in the colon (Figure 2D) in any of the inflammatory cytokines analyzed. Please see Table 2 for the fold change of all genes analyzed.
To investigate whether constitutively increased gut permeability affects the host gut microbial composition, we analyzed the microbiome of 4–8 weeks old Ztm and WT mice. The gut microbiota composition in 4–8 week mice is established (44) but still susceptible to fluctuations by external factors (i.e., diet). Hence, this timeframe represents a window of opportunity to influence changes in the immune system and microbiome composition, and as such to investigate the impact of gut permeability. Principle component analysis (PCA) showed that Ztm microbiota clustered distinct from WT gut microbiota (Figure 3A). Analysis at phylum level showed that microbiota in Ztm was characterized by a marked reduced abundance of Verrucomicrobia (0.2%) compared to WT (5.2%), and the almost complete absence of Cyanobacteria (Ztm = 0.1% WT = 1.3%) (Figure 3B). At the class level, Clostridiacea were also significantly reduced in Ztm compared to WT. A deeper analysis confirmed a significantly reduced abundance of the orders RF32 (Alphaproteobacteria) and YS2 (Cyanobacteria) and the genus Akkermansia in Ztm compared to WT mice. Conversely, Betaproteobacteria and the genus Rikenella were significantly more abundant in the gut of Ztm compared to WT (Figure 3C), pointing to a pro-inflammatory microbiota composition in Ztm.
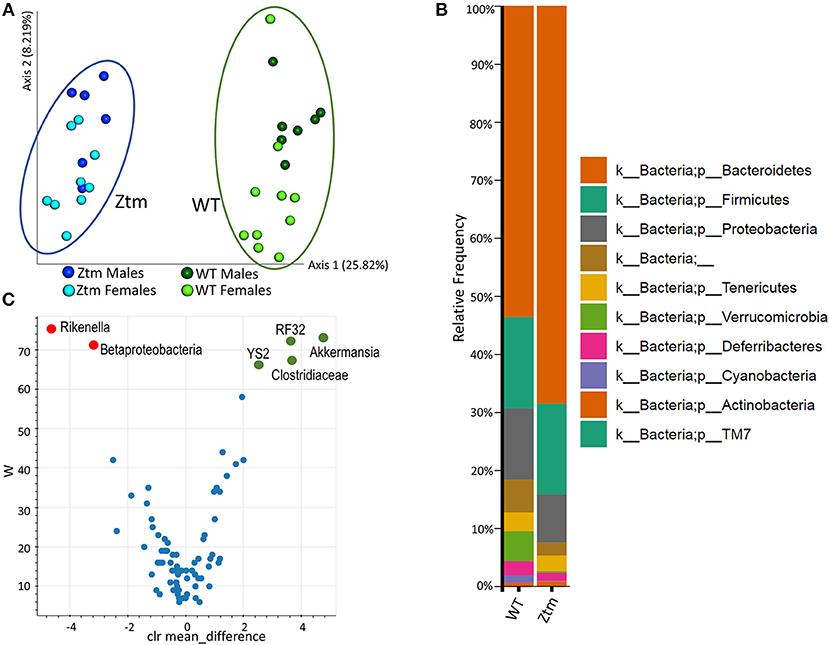
Figure 3. Baseline dysbiosis in Ztm gut microbiota. Stools from 15 WT mice (WT males n = 6, WT females n = 9) and 17 Ztm (Ztm males n = 7, Ztm females n = 10) were analyzed. (A) Principal Component Analysis (PCA) of Jaccard distances for Ztm (n = 17) and WT (n = 15) mice show distinct clustering between Ztm and WT. No significant differences between males and females within each group were detected. (B) Microbiota composition at phylum level. WT (n = 15), Ztm (n = 17). Males and females within each group were analyzed together. Absence of Verrucomicrobia and Cyanobacteria in the Ztm is evident. (C) Volcano plot showing the statistical differences between Ztm and WT at genus level. In green overrepresented genus in WT mice, in red overrepresented genus in Ztm. Differential abundance analysis of OTUs was performed using ANCOM. Statistical significance was tested by the Kruskal-Wallis of abundance differences, with the multiple testing corrections using Benjamini-Hochberg false discovery rate (FDR) set at 0.05.
To understand whether changes in the Ztm microbiota affected the development of the immune system (described below), we also analyzed the microbiota in Ztm mice that had received at birth bedding from a cage housing WT mouse (ZtmbWT, Ztm receiving bedding from WT) with the goal of normalizing microbiota between WT and Ztm mice. The Shannon Diversity Index (indication of the microbiota's complexity within the group) showed a significant difference between WT and Ztm (q = 0.007) but not between WT and the bedding-transferred ZtmbWT (Figure 4A). Consistently with the Shannon Diversity Index, the PCA plot showed that Ztm microbial composition segregated from the WT cluster, while ZtmbWT microbiota clustered with the WT microbiota (Figure 4B), indicating the successful engraftment of WT microbiota in the ZtmbWT. Verrucomicrobia and Cyanobacteria, that were severely reduced in Ztm, were engrafted in ZtmbWT where we observed the successful colonization of Akkermansia (Figure 4C) and, normalization of Rikenella abundance (data not shown).
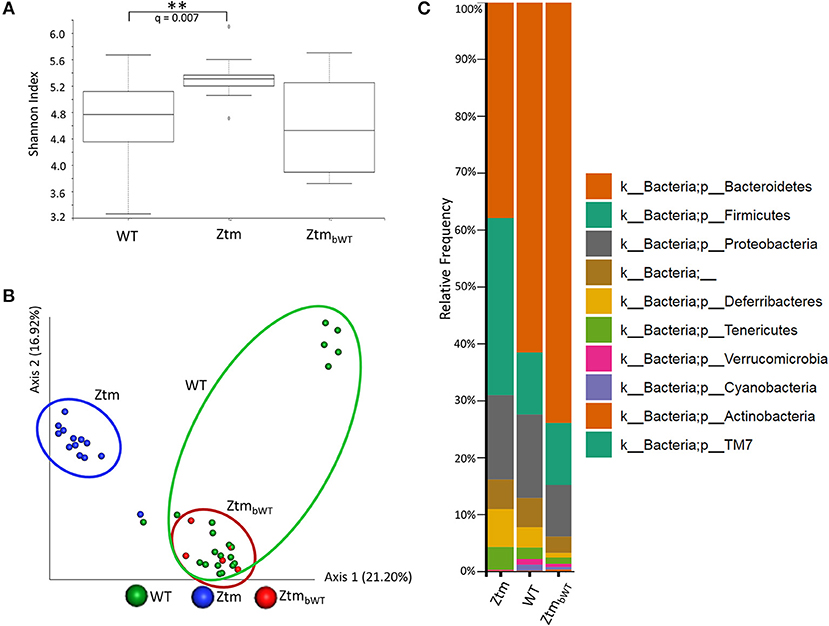
Figure 4. Microbiota analysis of Ztm mice with and without bedding transfer. Stools recovered from the colon of both male and female pups were analyzed as described in the material and methods. Stools were collected at weaning. ZtmbWT represent pups that were subjected to bedding transfer from WT. Bedding transfer was carried out twice a week since birth. (A) Shannon Index, expression of alpha diversity (variation/complexity of the microbiome within the group). WT n = 21, Ztm n = 13, ZtmbWT n = 5. WT and Ztm have a significantly different (q = 0.007) microbiota diversity, whereas no statistical difference is observed between WT and ZtmbWT. (B) Microbiota composition at phylum level. Successful engraftment of Verrucomicrobia and Cyanobacteria from WT to ZtmbWT following bedding transfer. (C) Principal component Analysis (PCA) of Jaccard distances. ZtmbWT (red) cluster with WT (green) and separately from Ztm (in blue). Differential abundance analysis of OTUs was performed using ANCOM. Statistical significance was tested by the Kruskal-Wallis of abundance differences, with the multiple testing corrections using Benjamini-Hochberg false discovery rate (FDR) set at 0.05. **q < 0.01.
Changes in intestinal barrier function may cause bacterial products including endotoxins and other molecules to leak through the gut barrier and disseminate in both the lamina propria and systemically, potentially instigating changes in the immune phenotype. To determine if increased intestinal permeability and/or an altered microbiota influence lymphocyte distribution in the gut and in secondary lymphoid organs, we compared WT mice with Ztm and ZtmbWT, as described in the above paragraph (Figures 4B,C).
Overall, Ztm and ZtmbWT mice had similar distribution of major immune cell subsets compared to WT mice (Figures 5, 6). Flow cytometry analysis of lymphocytes in the colon and small intestine revealed no significant differences in the frequency of IL7R+ conventional γδ-TCR+ cells, CD4+CD25+ cells, conventional CD4+CD25neg and CD8+ T cells, and CD19+ B cells (Figures 5A,C). However, we noted an overall statistically significant increase in the frequency of IL7R+RORγt+ cells in the small intestine but not in the colon of both Ztm and ZtmbWT mice (Figure 5A, red slice; Figures 5B,C). This increase was most likely due to elevated IL7R+CD3negRORγt+ cells that may be innate lymphoid cells (ILCs) (Figure 5B, bottom row; Figure 5C). Splenic cellularity was also comparable across all groups (Figure 6A) as was the distribution of CD4+ and CD8+ T cells (Figure 6B). There was no significant difference in the frequency of CD4+CD25+ cells in the spleen of Ztm and ZtmbWT mice compared to WT mice (Figure 6C). However, the frequency and numbers of CD4+CD25+ cells expressing RORγt+ was significantly increased in ZtmbWT mice (Figure 6C). We also noted an increased frequency and numbers of CD4+RORγt+ Th17 cells in both Ztm and ZtmbWT mice (Figure 6C). Further analyses revealed an interesting distribution of transcription factor RORγt-expressing innate-like T cells. While the frequency and numbers of mCD1d-PBS57 tetramer+ invariant NKT (iNKT) cells was decreased, the frequency of RORγt expressing subset of iNKT cells (NKT17 cells) increased in both Ztm and ZtmbWT compared to WT mice (Figure 6D). Similarly, there was a modest increase in the frequency of γδ T cells (γδ TCR+ innate-like T cells) that expressed RORγt (Figure 6E). NKT17 cells and γδ-17 T cells are pro-inflammatory innate-like T cell subsets that produce IL-17 and have been implicated in the pathogenesis of various autoimmune diseases including type 1 diabetes and celiac disease (45, 46). Finally, we determined the frequency and phenotype of conventional CD11chi dendritic cells (CDC) and CD11cloSiglecH+ plasmacytoid dendritic cells (PDC) and noted a small but significant increase in the frequency of splenic PDCs but not CDCs in both Ztm and ZtmbWT compared to WT mice (Figure 6F). These data suggest that altered gut permeability subtly increased the baseline frequency of IL-17 producing T cells in mucosal tissue and in secondary lymphoid organs of Ztm mice in the absence of overt inflammation and disease. The fact that the engraftment of WT microbiota did not affect the immune phenotype in ZtmbWT (no significant differences were detected between Ztm and ZtmbWT), suggests that the increased trafficking of microbial products through an impaired gut barrier rather than the function of an imbalanced microbiota primarily imprints the development of the immune system in the Ztm.
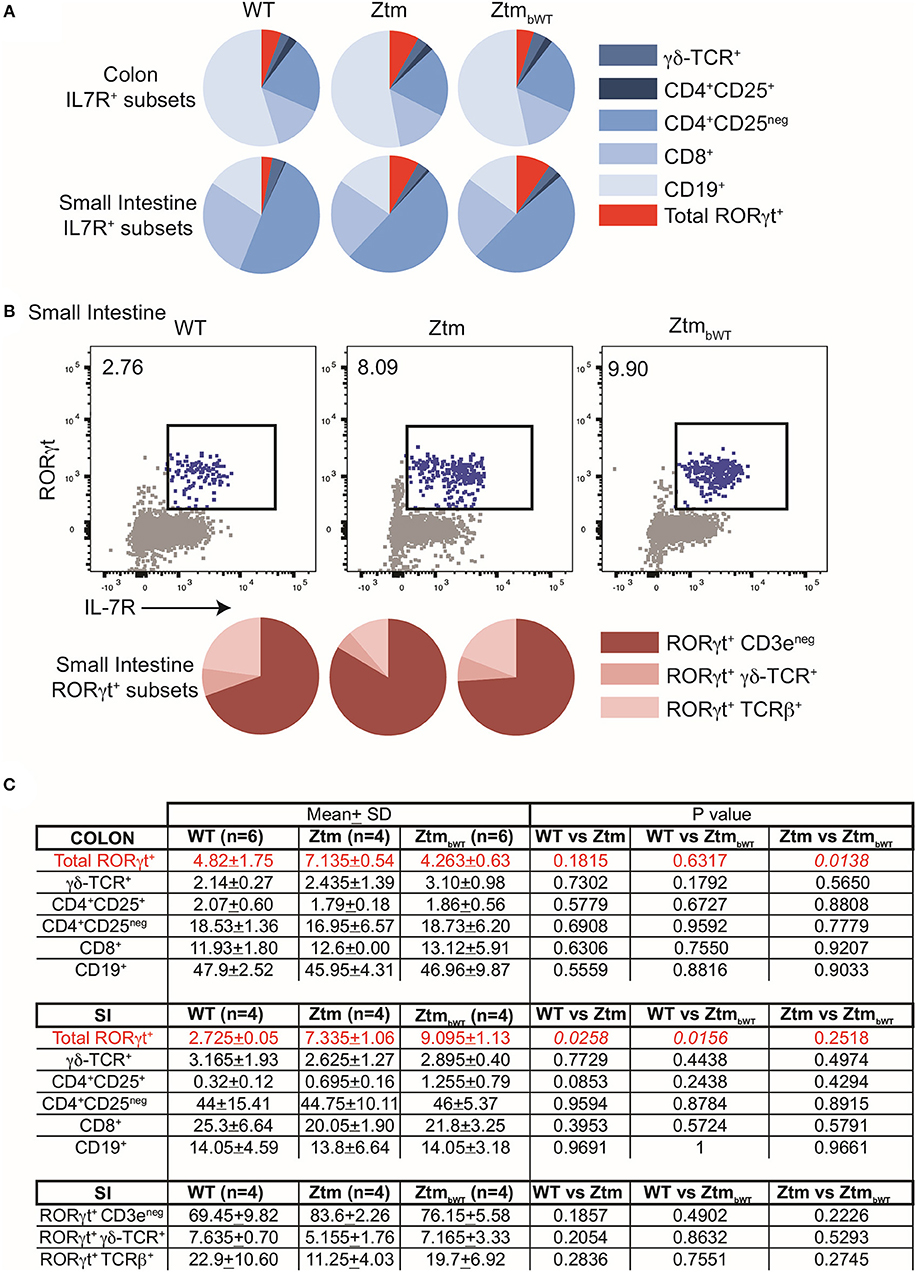
Figure 5. Distribution of immune cell subsets in colon and SI of WT and Ztm/ZtmbWT mice. Lamina propria and epithelial compartment lymphocytes from the colon (WT n = 6; Ztm n = 4; ZtmbWT n = 6) and small intestine (SI) (WT n = 4; Ztm n = 4; ZtmbWT n = 4) of 35 days old mice were analyzed by flow cytometry. (A) Pie chart representation of distribution of indicated IL7R+ subsets in colon (Top row) and small intestine (SI) (Bottom Row). (B) (Top row) Representative flow cytometry dot plots show expression of IL-7R (x-axis) and RORγt (y-axis) in lymphocytes from the SI. (Bottom Row) Pie chart representation of distribution of indicated IL7R+RORγt+ subsets in SI. (C) Summary of distribution of immune cell subsets in colon and SI. Mean, SD, and p-values are indicated. Unpaired parametric t-test was used for comparisons between the 2 groups with significance set at p < 0.05.
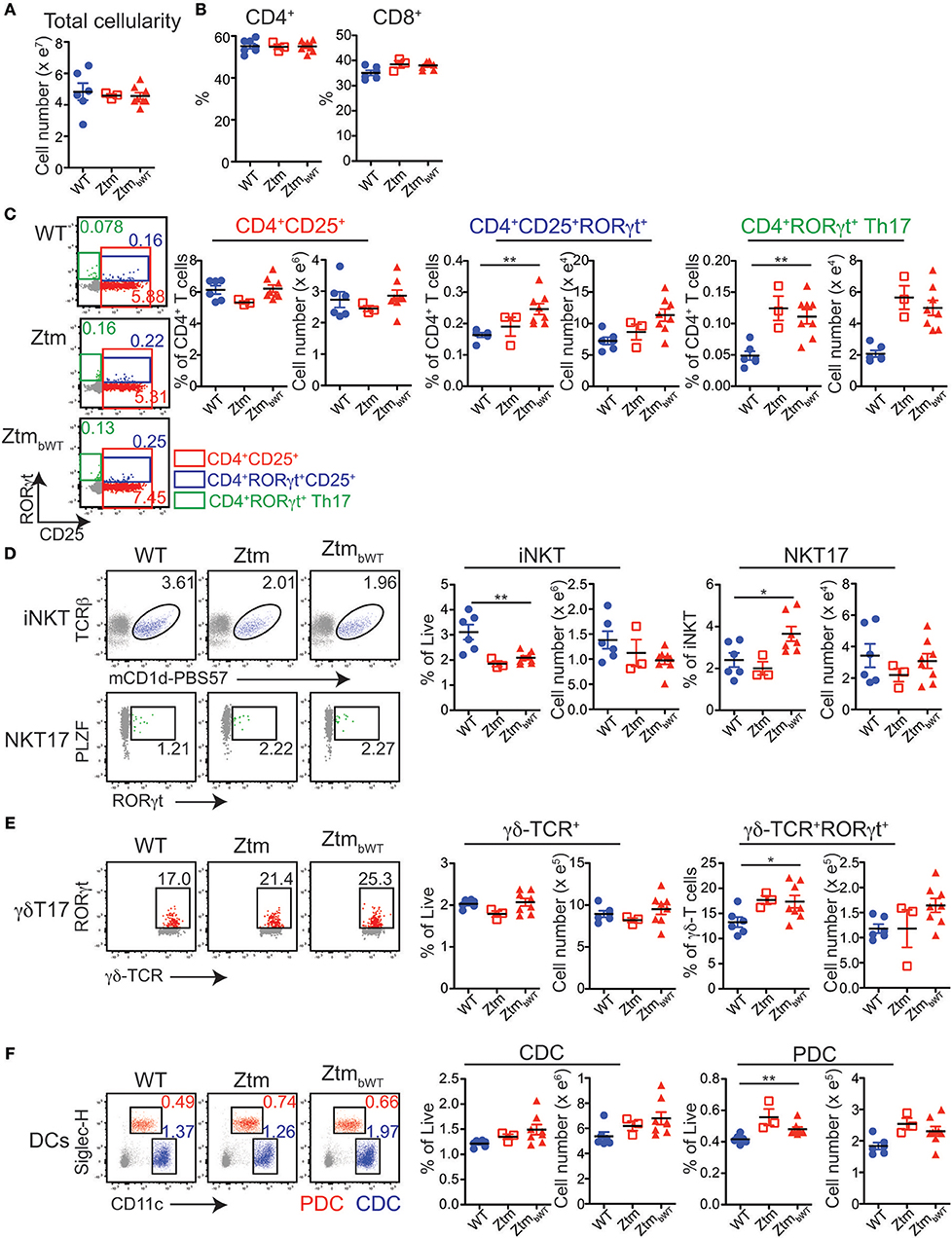
Figure 6. Distribution of immune cell subsets in spleen of WT and Ztm/ZtmbWT mice: single cell preparations from the spleen of 28–35 days old WT (n = 6), Ztm (n = 3), and ZtmbWT (n = 8) mice analyzed by flow cytometry. (A) Total splenic cellularity. (B) Vertical scatter plots show frequency of IL7R+TCRβ+ CD4+ and CD8+ T cells. (C) (Left) Representative flow cytometry dot plots show expression of CD25 (x-axis) and RORγt (y-axis) on CD4+ T cells. (Right) Vertical scatter plots show frequency and cell numbers of CD4+CD25+ cells, CD4+CD25+RORγt+ cells, and CD4+RORγt+ Th17 cells. (D) (Left) Representative flow cytometry dot plots show expression of (Top row) mCD1d-PBS57 tetramer (x-axis) and TCRβ (y-axis) on IL7R+ cells and (Bottom row) RORγt (x-axis) and PLZF (y-axis) on mCD1d-PBS57 tetramer+ TCRβ+ iNKT cells. (Right) Vertical scatter plots show frequency and cell numbers of iNKT cells and RORγt+ NKT17 cells. (E) (Left) Representative flow cytometry dot plots show expression of δ-TCR (x-axis) and RORγt (y-axis) on IL7R+ cells. (Right) Vertical scatter plots show frequency and cell numbers of γδ T cells and RORγt+ γδ-T17 cells. (F) (Left) Representative flow cytometry dot plots show expression of CD11c (x-axis) and Siglec-H (y-axis) on live cells (Red cells: CD11cloSiglec-H+ plasmacytoid dendritic cells or PDCs; Blue cells: CD11chiSiglec-Hneg conventional dendritic cells or CDCs). (Right) Vertical scatter plots show frequency and cell numbers of CDCs and PDCs. Data in (A–F) are representative of 3 independent experiments. Error bars are SEMs. Unpaired parametric t-test was used for comparisons between 2 groups with significance set at *p < 0.05 and **p < 0.01.
We have previously shown that Ztm express zonulin, exhibit constitutive increased gut permeability in-vivo and are more susceptible to DSS induced colitis (1). The increased morbidity and mortality affected predominantly Ztm males and is supported by our molecular data showing an altered TJ gene expression profile in the duodenum of male Ztm, with the significant decreased expression levels of several genes suggesting an impairment of small intestinal barrier function. Although, its function of gut barrier- or pore-forming claudin still has not been well-characterized, CLDN-12 significant increase in the colon of Ztm males could represent a compensatory mechanism to re-establish barrier function and/or electrolytes absorption (47). Of interest was the significantly reduced expression of Myosin-1C gene, because of its proven involvement in the zonulin pathway and its disassociation from ZO-1, leading to TJ disassembly and increased intestinal permeability (48). It's important to point out that although changes in gene expression not necessarily correspond to changes in protein expression, the fact that multiple TJ genes are affected, strongly suggests an impaired gut barrier. Also, intriguing are our findings of IL6 gene expression upregulation in the colon of Ztm males (and not females), suggesting that the increased permeability and trafficking of microbial products in the duodenum generates a pre-inflammatory imprinting in the immune system predisposing Ztm to develop colitis when exposed to an external inflammatory stimulus (1). Increased colonic expression of IL-6 might also be the consequence of a constitutive increased passage of microbiota species into the colonic lamina propria.
Gut permeability defects in the Ztm seem to have a critical effect on both the gut microbiota composition and the immune system. The analysis of stool microbiota showed dysbiosis in the Ztm gut. Striking was the marked reduced presence of Akkermansia (sp. muciniphila) vs. a significantly more abundant Rikenella genus in the Ztm gut. A. muciniphila contribute to enterocyte monolayer integrity and the overall strengthening of the gut epithelial barrier (49–51) and low levels have been associated to many human diseases (52–54) and have been reported as intestinal microbiota feature of both genetic and diet-induced obese and diabetic mice (55, 56).
While A. muciniphila are indicators of a gut healthy status, on the other end high abundance of Rikenella has been found in obesity and diabetes (57, 58) and hence associated with a status of low chronic non-infective inflammation. Overall, the low abundance of A. muciniphila and the enriched Rikenella presence in the gut of Ztm, suggest that the Ztm gut microbiota is skewed vs. a more maladaptive and pathogenic profile.
It is well-known that commensal bacteria are involved in the development of the immune system and the establishment of oral tolerance (59–61). The balance between pro-inflammatory IL-17+ helper Th17 cells and the anti-inflammatory Foxp3+ regulatory T cells (Treg) in the mucosa (62, 63) plays a key role in the development of chronic inflammatory diseases (64, 65). Microbial products have been shown to regulate the balance between Treg cells and IL-17 producing RORγt+ Th17 cells (59, 66). It has been shown that specific components of the commensal microbiota induce Th17 cells in the lamina propria of the small intestine and that antibiotic treatment prevented such differentiation (67).
In the Ztm, despite an altered baseline trafficking of microbial products, major immune cell subset distribution appears unaffected. However, the modest increase in pro-inflammatory IL-17 producing innate and innate-like cells in the intestine as well as in gut distal secondary lymphoid sites indicate that the threshold of immune reactivity in these mice might be altered, rendering them more susceptible to lose tolerance to non-self-antigens with subsequent onset of inflammation. In fact, when challenged with DSS, Ztm develop a more severe colitis and exhibit a significantly increased morbidity and mortality that is associated to elevated zonulin gene expression (1).
Immune profile studies following bedding transfer with the successful colonization of WT microbiota into Ztm mice, show no changes indicating that the Ztm immunophenotype is largely unaffected by the microbial composition and suggest that increased intestinal trafficking of microbial products primarily drives changes in the host immune system. In other words, the reduced barrier function in Ztm supersedes an imbalanced microbiota for the enhanced differentiation of RORγt+ and IL-17 producing subsets.
Our data indicate that zonulin-dependent increased gut permeability and microbial products trafficking orchestrate the development of the immune system and microbiota composition toward a pro-inflammatory status in the Ztm, with increased IL-17 producing cells and the lack of gut protective microbial species (i.e., Akkermansia) vs. an over-representation of pro-inflammatory strains (i.e., Rikenella). This low-grade pro-inflammatory-skewed status renders Ztm critically susceptible to develop overt inflammation leading to disease in the presence of exogenous stimuli (i.e., DSS) (1). Taken together, these data suggest that increased trafficking of microbial products in the small intestine might cause break of tolerance and onset of inflammation either locally or at distance. Indeed, the DSS experiments in Ztm (1) support our hypothesis of a key role of small intestinal microbial products trafficking in triggering inflammation at a distal site (colon). The observation that the onset of colitis with loss of the stem cell niche and its regenerative capacity can be rescued by blocking the zonulin pathway suggests a pivotal role of the mutually-influenced increased small intestinal trafficking of microbial products, a pro-inflammatory immune system, and gut dysbiosis shown in this paper, rather than merely a local effect of DSS once it reaches the colon. This is also in line with data generated in the IL-10 KO mouse, another model of colitis in which increased small intestinal permeability was shown to have a pathogenetic role in leading to colitis (8).
Although, further investigations are needed, these observations support our hypothesis and a new paradigm in which the presence in Ztm of an increased gut permeability, an altered immune system and dysbiosis of the gut microbiota are not sufficient to initiate the chain of events leading to overt inflammation. In the absence of an exogenous stimulus, Ztm remain healthy. It is the action of an external trigger to drive Ztm over the threshold where they develop disease. The same chain of events can be hypothesized in CID, in which in genetically predisposed individuals, exogenous factors that trigger epithelial barrier dysfunction and activation of the immune system could selectively promote the growth of microbial species in the gut that in turn worsen changes in the intestinal tissue resulting in breaking mucosal tolerance and onset of chronic inflammation, locally, and/or systemically and clinical symptoms.
In conclusion, this study provides important insights on the understanding of the role of gut permeability in the initiation of inflammatory diseases, offering the basis for innovative and yet unexplored therapeutic approaches for the management of these debilitating chronic diseases aimed at re-establishing the intestinal barrier function by downregulating the zonulin pathway.
All datasets generated for this study are included in the manuscript/supplementary files.
This study was carried out in accordance with the recommendations and guidelines of the Institutional Animal Care and Use Committee at the MGH. The protocol was approved by the Institutional Animal Care and Use Committee at the MGH (2013N000013).
MF, AM-R, NJ, RS, and AF conceived and designed the experiments. AM-R, ME, GS, JL, and MC carried out the experiments. MF, AM-R, NJ, RS, MC, and AF contributed to the interpretation of the results. AM-R, MF, NJ, and AF wrote the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
This work was supported partially by The National Institutes of Health (NIH) grants DK048373 and DK104344 to AF, NIH/NIAID Career Transition award 1K22AI116661 to NJ and P30 DK40561 to RS.
AF is a stock holder at Alba Therapeutics, serves as a consultant for Inova Diagnostics and Innovate Biopharmaceuticals, is an advisory board member for Axial Biotherapeutics and Ubiome, and has a speaker agreement with Mead Johnson Nutrition.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Sturgeon C, Lan J, Fasano A. Zonulin transgenic mice show altered gut permeability and increased morbidity/mortality in the DSS colitis model. Ann N Y Acad Sci. (2017) 1397:130–42. doi: 10.1111/nyas.13343
2. Katz KD, Hollander D, Vadheim CM, McElree C, Delahunty T, Dadufalza VD, et al. Intestinal permeability in patients with Crohn's disease and their healthy relatives. Gastroenterology. (1989) 97:927–31. doi: 10.1016/0016-5085(89)91499-6
3. van Elburg RM, Uil JJ, Mulder CJ, Heymans HS. Intestinal permeability in patients with coeliac disease and relatives of patients with coeliac disease. Gut. (1993) 34:354–7. doi: 10.1136/gut.34.3.354
4. Clayburgh DR, Shen L, Turner JR. A porous defense: the leaky epithelial barrier in intestinal disease. Lab Invest. (2004) 84:282–91. doi: 10.1038/labinvest.3700050
5. Pearson AD, Eastham EJ, Laker MF, Craft AW, Nelson R. Intestinal permeability in children with Crohn's disease and coeliac disease. Br Med J. (1982) 285:20–1. doi: 10.1136/bmj.285.6334.20
6. Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, Fedorak RN. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis. (1999) 5:262–70. doi: 10.1097/00054725-199911000-00004
7. Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, et al. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. (2009) 136:551–63. doi: 10.1053/j.gastro.2008.10.081
8. Arrieta MC, Madsen K, Doyle J, Meddings J. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut. (2009) 58:41–8. doi: 10.1136/gut.2008.150888
9. Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science. (1995) 270:1203–7. doi: 10.1126/science.270.5239.1203
10. Shin W, Kim HJ. Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc Natl Acad Sci USA. (2018) 115:E10539–47. doi: 10.1073/pnas.1810819115
11. Antalis TM, Shea-Donohue T, Vogel SN, Sears C, Fasano A. Mechanisms of disease: protease functions in intestinal mucosal pathobiology. Nat Clin Pract Gastroenterol Hepatol. (2007) 4:393–402. doi: 10.1038/ncpgasthep0846
12. Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. (2008) 57:463–7. doi: 10.1136/gut.2007.133132
13. Anderson G, Seo M, Berk M, Carvalho AF, Maes M. Gut permeability and microbiota in Parkinson's disease: role of depression, tryptophan catabolites, oxidative and nitrosative stress and melatonergic pathways. Curr Pharm Des. (2016) 22:6142–51. doi: 10.2174/1381612822666160906161513
14. Buscarinu MC, Cerasoli B, Annibali V, Policano C, Lionetto L, Capi M, et al. Altered intestinal permeability in patients with relapsing-remitting multiple sclerosis: a pilot study. Mult Scler. (2017) 23:442–6. doi: 10.1177/1352458516652498
15. de Magistris L, Familiari V, Pascotto A, Sapone A, Frolli A, Iardino P, et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J Pediatr Gastroenterol Nutr. (2010) 51:418–24. doi: 10.1097/MPG.0b013e3181dcc4a5
16. Fiorentino M, Sapone A, Senger S, Camhi SS, Kadzielski SM, Buie TM, et al. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol Autism. (2016) 7:49. doi: 10.1186/s13229-016-0110-z
17. Olaison G, Sjödahl R, Tagesson C. Abnormal intestinal permeability in Crohn's disease. A possible pathogenic factor. Scand J Gastroenterol. (1990) 25:321–8. doi: 10.3109/00365529009095493
18. Alican I, Kubes P. A critical role for nitric oxide in intestinal barrier function and dysfunction. Am J Physiol. (1996) 270(2 Pt 1):G225–37. doi: 10.1152/ajpgi.1996.270.2.G225
19. Rao RK. Acetaldehyde-induced barrier disruption and paracellular permeability in Caco-2 cell monolayer. Methods Mol Biol. (2008) 447:171–83. doi: 10.1007/978-1-59745-242-7_13
20. Al-Sadi R, Guo S, Ye D, Ma TY. TNF-alpha modulation of intestinal epithelial tight junction barrier is regulated by ERK1/2 activation of Elk-1. Am J Pathol. (2013) 183:1871–84. doi: 10.1016/j.ajpath.2013.09.001
21. Stevens BR, Goel R, Seungbum K, Richards EM, Holbert RC, Pepine CJ, et al. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut. (2017) 67:1555–7. doi: 10.1136/gutjnl-2017-314759
22. Ciccia F, Guggino G, Rizzo A, Alessandro R, Luchetti MM, Milling S, et al. Dysbiosis and zonulin upregulation alter gut epithelial and vascular barriers in patients with ankylosing spondylitis. Ann Rheum Dis. (2017) 76:1123–32. doi: 10.1136/annrheumdis-2016-210000
23. Damms-Machado A, Louis S, Schnitzer A, Volynets V, Rings A, Basrai M, et al. Gut permeability is related to body weight, fatty liver disease, and insulin resistance in obese individuals undergoing weight reduction. Am J Clin Nutr. (2017) 105:127–35. doi: 10.3945/ajcn.116.131110
24. Bernstein CN, Forbes JD. Gut microbiome in inflammatory bowel disease and other chronic immune-mediated inflammatory diseases. Inflamm Intest Dis. (2017) 2:116–23. doi: 10.1159/000481401
25. Slyepchenko A, Maes M, Machado-Vieira R, Anderson G, Solmi M, Sanz Y, et al. Intestinal dysbiosis, gut hyperpermeability and bacterial translocation: missing links between depression, obesity and type 2 diabetes. Curr Pharm Des. (2016) 22:6087–106. doi: 10.2174/1381612822666160922165706
26. Tripathi A, Lammers KM, Goldblum S, Shea-Donohue T, Netzel-Arnett S, Buzza MS, et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc Natl Acad Sci USA. (2009) 106:16799–804. doi: 10.1073/pnas.0906773106
27. Fasano A. Regulation of intercellular tight junctions by zonula occludens toxin and its eukaryotic analogue zonulin. Ann N Y Acad Sci. (2000) 915:214–22. doi: 10.1111/j.1749-6632.2000.tb05244.x
28. Fasano A, Not T, Wang W, Uzzau S, Berti I, Tommasini A, et al. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet. (2000) 355:1518–9. doi: 10.1016/S0140-6736(00)02169-3
29. Soderborg TK, Friedman JE. Imbalance in gut microbes from babies born to obese mothers increases gut permeability and myeloid cell adaptations that provoke obesity and NAFLD. Microb Cell. (2018) 6:102–4. doi: 10.15698/mic2019.01.666
30. Watts T, Berti I, Sapone A, Gerarduzzi T, Not T, Zielke R, et al. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc Natl Acad Sci USA. (2005) 102:2916–21. doi: 10.1073/pnas.0500178102
31. Paterson BM, Lammers KM, Arrieta MC, Fasano A, Meddings JB. The safety, tolerance, pharmacokinetic and pharmacodynamic effects of single doses of AT-1001 in coeliac disease subjects: a proof of concept study. Aliment Pharmacol Ther. (2007) 26:757–66. doi: 10.1111/j.1365-2036.2007.03413.x
32. Sturgeon C, Fasano A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers. (2016) 4:e1251384. doi: 10.1080/21688370.2016.1251384
33. Sapone A, de Magistris L, Pietzak M, Clemente MG, Tripathi A, Cucca F, et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes. (2006) 55:1443–9. doi: 10.2337/db05-1593
34. Thomas KE, Sapone A, Fasano A, Vogel SN. Gliadin stimulation of murine macrophage inflammatory gene expression and intestinal permeability are MyD88-dependent: role of the innate immune response in Celiac disease. J Immunol. (2006) 176:2512–21. doi: 10.4049/jimmunol.176.4.2512
35. Levy AP, Levy JE, Kalet-Litman S, Miller-Lotan R, Levy NS, Asaf R, et al. Haptoglobin genotype is a determinant of iron, lipid peroxidation, and macrophage accumulation in the atherosclerotic plaque. Arterioscler Thromb Vasc Biol. (2007) 27:134–40. doi: 10.1161/01.ATV.0000251020.24399.a2
36. McCoy KD, Geuking MB, Ronchi F. Gut microbiome standardization in control and experimental mice. Curr Protoc Immunol. (2017) 117:23.1.1–23.1.13. doi: 10.1002/cpim.25
37. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. (2011) 108 (Suppl 1):4516–22. doi: 10.1073/pnas.1000080107
38. Porter C, Ennamorati M, Jain N. In vivo photolabeling of cells in the colon to assess migratory potential of hematopoietic cells in neonatal mice. J Vis Exp. (2018) e57929. doi: 10.3791/57929
39. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. doi: 10.1038/nmeth.f.303
40. Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. (2017) 2:e00191–16. doi: 10.1128/mSystems.00191-16
41. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
42. Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. (2010) 5:e9490. doi: 10.1371/journal.pone.0009490
43. Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. (2015) 26:27663. doi: 10.3402/mehd.v26.27663
44. Schloss PD, Schubert AM, Zackular JP, Iverson KD, Young VB, Petrosino JF. Stabilization of the murine gut microbiome following weaning. Gut Microbes. (2012) 3:383–93. doi: 10.4161/gmic.21008
45. De Giorgi L, Sorini C, Cosorich I, Ferrarese R, Canducci F, Falcone M. Increased iNKT17 cell frequency in the intestine of non-obese diabetic mice correlates with high bacterioidales and low clostridiales abundance. Front Immunol. (2018) 9:1752. doi: 10.3389/fimmu.2018.01752
46. Dunne MR, Elliott L, Hussey S, Mahmud N, Kelly J, Doherty DG, et al. Persistent changes in circulating and intestinal gammadelta T cell subsets, invariant natural killer T cells and mucosal-associated invariant T cells in children and adults with coeliac disease. PLoS ONE. (2013) 8:e76008. doi: 10.1371/journal.pone.0076008
47. Fujita H, Sugimoto K, Inatomi S, Maeda T, Osanai M, Uchiyama Y, et al. Tight junction proteins claudin-2 and−12 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Mol Biol Cell. (2008) 19:1912–21. doi: 10.1091/mbc.e07-09-0973
48. Goldblum SE, Rai U, Tripathi A, Thakar M, De Leo L, Di Toro N, et al. The active Zot domain (aa 288-293) increases ZO-1 and myosin 1C serine/threonine phosphorylation, alters interaction between ZO-1 and its binding partners, and induces tight junction disassembly through proteinase activated receptor 2 activation. FASEB J. (2011) 25:144–58. doi: 10.1096/fj.10-158972
49. Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol. (2015) 81:3655–62. doi: 10.1128/AEM.04050-14
50. Belzer C, de Vos WM. Microbes inside–from diversity to function: the case of Akkermansia. ISME J. (2012) 6:1449–58. doi: 10.1038/ismej.2012.6
51. Earley H, Lennon G, Balfe A, Kilcoyne M, Clyne M, Joshi L, et al. A preliminary study examining the binding capacity of Akkermansia muciniphila and Desulfovibrio spp. to colonic mucin in health and ulcerative colitis. PLoS ONE. (2015) 10:e0135280. doi: 10.1371/journal.pone.0135280
52. Grander C, Adolph TE, Wieser V, Lowe P, Wrzosek L, Gyongyosi B, et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. (2018) 67:891–901. doi: 10.1136/gutjnl-2016-313432
53. Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol. (2011) 77:6718–21. doi: 10.1128/AEM.05212-11
54. Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. (2010) 105:2420–8. doi: 10.1038/ajg.2010.281
55. Schneeberger M, Everard A, Gómez-Valadés AG, Matamoros S, Ramírez S, Delzenne NM, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. (2015) 5:16643. doi: 10.1038/srep16643
56. Song H, Chu Q, Yan F, Yang Y, Han W, Zheng X. Red pitaya betacyanins protects from diet-induced obesity, liver steatosis and insulin resistance in association with modulation of gut microbiota in mice. J Gastroenterol Hepatol. (2016) 31:1462–9. doi: 10.1111/jgh.13278
57. Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, et al. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front Microbiol. (2011) 2:149. doi: 10.3389/fmicb.2011.00149
58. Leiva-Gea I, Sánchez-Alcoholado L, Martín-Tejedor B, Castellano-Castillo D, Moreno-Indias I, Urda-Cardona A, et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care. (2018) 41:2385–95. doi: 10.2337/dc18-0253
59. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
60. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. (2004) 4:478–85. doi: 10.1038/nri1373
61. Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Bérard M, Kleinschek M, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. (2011) 12:320–6. doi: 10.1038/ni.2002
62. Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. (2006) 441:235–8. doi: 10.1038/nature04753
63. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. (2006) 126:1121–33. doi: 10.1016/j.cell.2006.07.035
64. Shen X, Du J, Guan W, Zhao Y. The balance of intestinal Foxp3+ regulatory T cells and Th17 cells and its biological significance. Expert Rev Clin Immunol. (2014) 10:353–62. doi: 10.1586/1744666X.2014.882232
65. Ueno A, Jeffery L, Kobayashi T, Hibi T, Ghosh S, Jijon H. Th17 plasticity and its relevance to inflammatory bowel disease. J Autoimmun. (2018) 87:38–49. doi: 10.1016/j.jaut.2017.12.004
66. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
Keywords: gut permeability, microbial products trafficking, tight-junctions, zonulin transgenic mouse, microbiota, dysbiosis, immunity, chronic inflammatory diseases
Citation: Miranda-Ribera A, Ennamorati M, Serena G, Cetinbas M, Lan J, Sadreyev RI, Jain N, Fasano A and Fiorentino M (2019) Exploiting the Zonulin Mouse Model to Establish the Role of Primary Impaired Gut Barrier Function on Microbiota Composition and Immune Profiles. Front. Immunol. 10:2233. doi: 10.3389/fimmu.2019.02233
Received: 20 March 2019; Accepted: 03 September 2019;
Published: 19 September 2019.
Edited by:
Marika Falcone, San Raffaele Hospital (IRCCS), ItalyReviewed by:
Claudio Nicoletti, University of Florence, ItalyCopyright © 2019 Miranda-Ribera, Ennamorati, Serena, Cetinbas, Lan, Sadreyev, Jain, Fasano and Fiorentino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Fiorentino, bXJmaW9yZW50aW5vQG1naC5oYXJ2YXJkLmVkdQ==
†These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.