- 1Division of Vaccine Discovery, La Jolla Institute for Immunology, La Jolla, CA, United States
- 2Department of Medicine, University of California, San Diego, La Jolla, CA, United States
DENV is a major public health problem worldwide, thus underlining the overall significance of the proposed Program. The four dengue virus (DENV) serotypes (1–4) cause the most common mosquito-borne viral disease of humans, with 3 billion people at risk for infection and up to 100 million cases each year, most often affecting children. The protective role of T cells during viral infection is well-established. Generally, CD8 T cells can control viral infection through several mechanisms, including direct cytotoxicity, and production of pro-inflammatory cytokines such as IFN-γ and TNF-α. Similarly, CD4 T cells are thought to control viral infection through multiple mechanisms, including enhancement of B and CD8 T cell responses, production of inflammatory and anti-viral cytokines, cytotoxicity, and promotion of memory responses. To probe the phenotype of virus-specific T cells, epitopes derived from viral sequences need to be known. Here we discuss the identification of CD4 and CD8 T cell epitopes derived from DENV and how these epitopes have been used by researchers to interrogate the phenotype and function of DENV-specific T cell populations.
DENV Infection and the Complex Roles of T Cells
Dengue virus (DENV) belongs to the genus Flavivirus and is closely related to several other flaviviruses including Zika virus (ZIKV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), and West Nile virus (WNV) (1). DENV is a serious public health issue especially in tropical and subtropical areas, and it is estimated that ~390 million people are infected yearly with DENV (2). DENV infection is associated with a range of clinical manifestations, from asymptomatic to more severe presentations including dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). There is currently no specific therapy available for the treatment of dengue diseases other than supportive care. Furthermore, Dengvaxia® (Sanofi Pasteur), the first licensed DENV vaccine, is associated with efficacy and safety concerns (3–7). Sridhar et al. integratively analyzed data from three clinical trials and reported that Dengvaxia® increases the risk of severe dengue and hospitalization among vaccinees who have not been exposed to DENV before the vaccination (8). In order to develop effective DENV therapeutics and vaccines, it is important to define immunological correlates of protection against DENV infection as well as biomarkers that can be used to access their safety and efficacy.
Although T cells have important functions in combating viral pathogens, both pathological and protective effects of T cells have been reported in the context of DENV infection (9–14). According to T cell original antigenic sin, cross-reactive T cells that are specific for a primary DENV serotype become predominant during a secondary heterologous infection (9–16). Consequently, the expansion of preexisting cross-reactive and low-affinity memory T cells results in ineffective viral control and contributes to immunopathology and severe dengue disease through excessive production of inflammatory cytokines (9–16). In contrast to the implications of original antigenic sin, several lines of evidence indicate that T cells contribute to the control of DENV infection. Murine studies demonstrate that CD4 T cells and especially CD8 T cells can play a protective role against DENV challenge (17–24). Furthermore, HLA alleles associated with protection from severe dengue disease are also associated with strong and multifunctional T cell responses, supporting the notion that T cells have protective functions during DENV infection (25–28). The main characteristic of an efficient vaccine is the prophylactic effect provided by protective neutralizing antibodies. Therefore, it is possible that in Dengvaxia® vaccines, native conserved masked conformational DENV (1–4) epitopes are not unmasked and therefore not accessible for highly neutralizing and broadly protective antibodies. Nevertheless, Dengvaxia® is a yellow fever dengue chimeric vaccine and lacks DENV non-structural (NS) proteins that contain a large proportion of T cell epitopes (25, 28, 29). Therefore, the suboptimal efficacy of Dengvaxia® may partially due to its defective ability to induce T cell responses (30). Indeed, a single dose of the live attenuated tetravalent DENV vaccine TV003 provides complete protection against infection with a DENV-2 challenge virus (31), potentially highlighting the importance of harnessing the protective functions of both humoral and cellular antiviral immunity.
Metadata Analysis of DENV-Derived CD4 and CD8 T Cell Epitopes
Human antigen-specific T cell immune responses are driven by two factors that are host specific. First the capability of antigen-derived peptides to be bound and presented in the context of HLA class I and II molecules. Second, the immunogenicity of those peptides that depends on the capability of T cells to recognize through T cell receptor (TCR) the HLA-peptide complex and trigger T-cell specific immune responses. Several studies have identified the DENV epitopes able to induce CD8 and/or CD4 T cells specific-response and consecutively the immunodominance of DENV proteins for DENV-specific T cell response. In this review, we summarize previous published data of all the DENV-epitopes experimentally identified by us and others by performing an overall analysis of data available in Immune Epitope Database (www.IEDB.org).
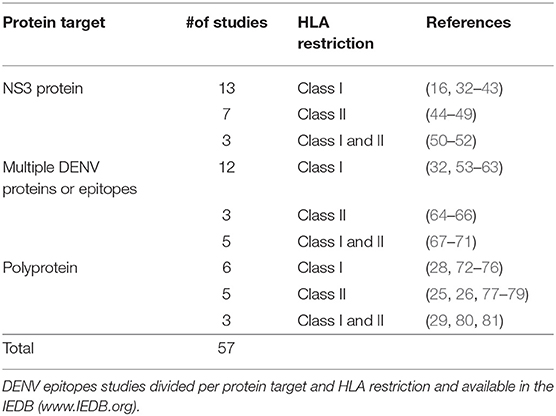
The IEDB database was queried on July 8th 2019 using the following search parameters: Positive assays only, Organism: Dengue virus (ID:12637), No B cell assays, No MHC ligand assays, Host: Homo sapiens (Human). This query retrieved a total of 57 different publications (Table 1). Most of the studies were focused on NS3 protein or multiple DENV proteins defined as immunodominant region based on previous studies (Table 1). Additional studies evaluated the full DENV polyprotein targeting either HLA-Transgenic DENV infected mice (32, 72) or human samples. Specifically, we considered human samples derived from different geographical locations and collected from either healthy DENV seropositive blood donors DENV (25, 26, 28, 73, 77, 80) or patients during acute dengue infection (29, 74, 75) or healthy donor after experimental vaccination with dengue virus (76, 78, 81) (Table 1). A total of 2191 epitopes were described in the 57 references retrieved by the query (Table 1); of those, 825 were restricted by HLA class I molecules, and 1345 epitopes were restricted by HLA class II. To define the most dominant epitopes, we considered epitopes that were reported positive in multiple donors and/or multiple studies. For example, the NS31608−1618 is the most dominant HLA class I epitope, being independently identified by several studies also incorporating different amino acid variants (16, 28, 33–35, 53, 73) (Supplementary Table 1A). Conversely, in the case of HLA class II molecules the C50−64 and C72−86 epitopes are examples of dominant epitopes, recognized in the majority of donors tested (12 positive out of 17 and 9 positive out of 11, respectively) in several independent studies (25, 77, 78). Supplementary Tables 1A,B list the most dominant class I and class II epitopes. Specifically, to identify the top ~80 epitopes we selected class I epitopes recognized in 5 donors or more (84 epitopes), and class II epitopes recognized in 8 donors of more (77 epitopes).
Immunodominant DENV Proteins Recognized by CD4 and CD8 T Cell
Dissecting the immunodominance of T cell response has considerable implications in vaccine design as the first licensed Dengvaxia® vaccine (Sanofi Pasteur) is composed of DENV structural proteins (prM, E) in a yellow fever backbone while both TV003 (NIH) and TDV (Takeda) vaccines contains both DENV structural and non-structural proteins (31, 82, 83). To assess DENV immunodominance without the bias of adding multiple studies that take in consideration only a single or few DENV proteins, we focused on the 14 studies performed on the full DENV polyprotein and summarized in Table 1.
Epitopes derived from those 14 studies have been extracted from IEDB, redundant epitopes independently defined in donors with different HLA restriction have been included to give prominence to promiscuous peptides able to be presented and recognized by different HLA molecules. Finally, epitopes have been divided based on class I or class II restriction and plotted on a DENV reference sequence (ID:12637) by using the Immunobrowser tool (http://tools.iedb.org/immunomebrowser/) (84) freely available on the IEDB. Figure 1 shows the immunodominance of CD8 (Figure 1A, HLA class I restriction) and CD4 T cells (Figure 1B, HLA class II restriction), respectively.
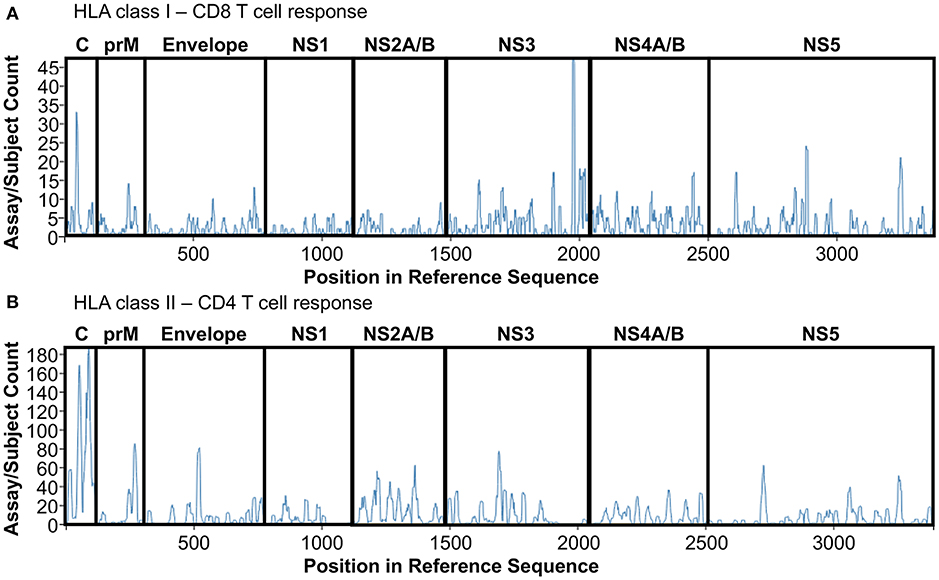
Figure 1. DENV T cell immunodominance. List of epitopes derived from DENV polyprotein studies summarized in Table 1 have been extracted and plotted on a reference DENV sequence (ID:12637) by using Immunobrowser. (A) HLA class I restriction. (B) HLA class II restriction.
CD8 T cell responses targets mainly NS3 protein, followed by Capsid, NS5 and NS4A/B proteins (Figure 1A). NS3 immunodominance is confirmed throughout all the different studies used for the analysis, representing the most frequent target of CD8 T cell response, disregarding geographical location, and HLA restriction.
Conversely, CD4 T cell responses targets mainly Capisd followed by Envelope, NS3, NS2A/B, and NS5 proteins (Figure 1B). Capsid immunodominance is consistently identified in all the different studies analyzed, representing the most frequent target of CD4 T cell response. Finally, protein immunodominance for both CD4 and CD8 T cells is also function of the multiple exposure of DENV infection, that tends to skew protein immunodominance toward epitopes highly conserved across the different DENV serotypes as previously reported (85). Overall, T cell protein immunodominance is quite complex and widely focused on different protein targets, suggesting that in order to trigger an efficient DENV-specific T cell response both Structural (C, prM, Envelope) and Non-Structural (NS1-5) proteins are required.
Characterization of Human DENV-Specific T cell Responses
Megapool Approach to Detect DENV-Specific T Cell Responses
Epitope identification studies have provided the basis for phenotyping DENV-specific T cell responses directly ex vivo without the need for in vitro stimulation that could potentially alter T cell phenotypes. Utilizing the knowledge of the epitopes recognized we developed the megapool approach, which allows for combining a large number of peptides into one peptide pool based on sequential lyophilization. This enables detection of DENV-specific T cell responses irrespective of HLA types and DENV serotypes in various immunological contexts where only small amounts of blood are available (78). DENV megapools have been generated for both CD4 and CD8 T cells, which consist of 180 and 268 peptides, respectively (25, 27, 28, 73, 77). These peptides are pooled, lyophilized, and resuspended to form a master mix, which is then used to stimulate T cell ex vivo (86). DENV CD4 and CD8 megapools account for 62 and 90% of the IFN-γ response in Sri Lankan and Nicaraguan cohorts, respectively, and have been validated in different geographical locations supporting their global applicability (25, 27, 28, 73, 77).
DENV-Specific CD8 T Cells: Activated, Skin-Homing, and Functional
Using tetramers incorporating three variants of the HLA-A*1101-restricted DENV NS3133−142 epitope, Friberg et al. reported that cross-reactive CD8 T cells develop following both primary and secondary DENV infections and that the magnitude of tetramer+ CD8 T cell response does not correlated with disease severity (35). Although tetramer+ CD8 T cells upregulate the activation marker CD38 during the acute phase of infection (35), the phenotype of these DENV-specific CD8 T cells was not further assessed in this study. More recent studies using DENV peptide pools shows that higher magnitude and more polyfunctional CD8 T cell responses correlate with HLA alleles that are associated with reduced risk of severe dengue disease (27, 28), which is consistent with the report that the frequency of DENV-specific cytokine-producing CD8 T cells is higher among children who subsequently developed subclinical secondary infection than those who developed symptomatic secondary infection (87). Using a pool containing 268 CD8 T cell epitopes derived from DENV (termed megapool), de Alwis et al. demonstrated that the majority of DENV-specific IFN-γ+ CD8 T cells have a CD45RA−CCR7− effector memory (Tem) or CD45RA+CCR7− effector memory re-expressing CD45RA (Temra) phenotype (27). Notably, DENV-specific CD8 T cells are also associated with increased PD-1 expression in donors expressing the immunodominant allele HLA-B*35:01. In contrast to classical exhausted CD8 T cells, these DENV-specific PD-1+ CD8 T cells do not co-express other exhaustion makers and are apparently proliferative and functional (27), suggesting that PD-1 may serve as a marker of activated and highly functional antigen-specific CD8 T cells in the context of DENV infection.
Since DENV infection initiates at the site of the mosquito bite in the host skin, it is possible that CD8 T cells may migrate to the site of infection and mediate localized responses. Indeed, DENV NS3 27-specific CD8 T cells in the periphery blood upregulate the expression of several chemokine receptors including CCR5, CXCR3, and CXCR6 as well as the skin-homing molecule cutaneous lymphocyte-associated antigen (CLA) during acute DENV infection (67). Moreover, DENV-specific CD8 T cells are readily detectable in the skin of DENV-infected individuals at the acute stage (67), suggesting that these cells may exert effector functions at the site of infection. Tissue-resident memory T (Trm) cells reside in non-lymphoid tissues including the skin and can serve as a front line of defense against invading pathogens such as vaccinia virus (88). It would be interesting to investigate whether DENV-specific CD8 T cells could differentiate into Trm cells in the skin that may mount rapid and localized protective immunity upon reinfection.
Comprehensive transcriptomic profiling of DENV-specific CD8 T cells has also been carried out. Chandele et al. performed microarray analysis on HLA-DR+CD38+ activated CD8 T cells isolated from the PBMCs of DENV-infected patients and found that these cells upregulate genes involved in T cell proliferation, activation, migration and cytotoxicity (89). Interestingly, HLA-DR+CD38+ CD8 T cells also display increased expression of multiple inhibitory receptors and downregulate several genes that are involved in TCR signaling (89). A more recent study from our group characterized the transcriptomic profiles of DENV-specific CD8 Tem and Temra subsets identified by their production of IFN-γ following simulation with the megapool of DENV-derived epitopes (90). DENV-specific Tem and especially Temra cells display specialized gene expression profiles and upregulated genes that are associated with activation, co-stimulation, and effector functions (90), which is consistent with the previous study from Chandele et al. Since these DENV-specific Tem and Temra cells were isolated from healthy donors with secondary DENV infection, these studies suggest that DENV-specific CD8 T cell populations may maintain an activated phenotype in donors that have been infected multiple times with DENV. Interestingly, DENV-specific Temra cells may have higher expression of a few killer cell immunoglobulin-like receptor (KIR) genes including KIR2DL3 by comparison with DENV-specific Tem cells (90). In addition, DENV-specific CD8 T cells may show preferential usage of TCR beta-chain variable (TRBV) genes (90), which is in line with the report that DENV NS3133-specific CD8 T cells targeting HLA-A*11:01-restricted epitope variants derived from DENV1, DENV3, and DENV4 but not DENV2 preferentially use a few TRBV segments including TRBV9, TRBV12-3/4, and especially TRBV11-2 (33).
DENV-Specific CD4 T Cells: A Tale of Cytotoxicity
The majority of antigen-specific CD4 T cells differentiate into T helper type 1 (Th1) and follicular helper T (Tfh) cells following viral infections and provide help to CD8 T cells and B cells (91–93). Indeed, DENV-specific CD4 T cells produce Th1 cell-associated cytokines including IFN-γ, TNF-α, and IL-2 following both infection and vaccination (87, 94, 95). In addition, DENV-specific CD4 T cells with cytotoxic activity have been reported by numerous studies (12) and their frequency may be lower in patients with more severe dengue disease (96). Interestingly, a subset human CD4 T cells, which is CD45RA+CCR7− and termed effector memory re-expressing CD45RA T (Temra) cells, expands in individuals that have been infected with DENV multiple times, and the frequency of DENV-specific CD4 Temra cells is higher in donors expressing an HLA allele associated with protection from severe dengue disease (26). Despite the production of IFN-γ, these cells may not represent classical Th1 cells as they lack the expression of CXCR3 (26). CD4 Temra cells have increased expression of several cytotoxic molecules including CD107a, perforin, granzyme B as well as the CX3CL1 (fractalkine) receptor, CX3CR1 (26). Notably, CX3CR1 has recently been reported to be a member of a 20-gene set that can predict severe dengue disease (97). Subsequent transcriptomic profiling studies further revealed the gene expression patterns and heterogeneity of CD4 Temra cells and identified additional phenotypic markers such as GPR56 and CD244 that are specifically expressed by cytotoxic CD4 Temra cells and confirmed their expression on DENV-specific Temra cells (98, 99). Additionally, cytotoxic CD4 Temra cells may have undergone extensive clonal expansions based upon TCR analysis (98, 99), supporting the notion that these cells are induced by repeated DENV infections.
Both Foxp3+ regulatory T (Treg) cells and Foxp3− type 1 regulatory T (Tr1) cells can suppress inflammation and exert immunoregulatory effects (100, 101). However, their functional significance in the context of DENV infection is less well-defined (14). It has been reported that the frequency of Treg cells and the ratio of Treg cells to effector T cells are significantly higher during acute DENV infection than after recovery in patients with mild disease but not in those with severe disease (102). However, subsequent studies indicate that the frequency of Treg cells is not associated with viral load or disease severity (103). Therefore, whether and how Treg and Tr1 cells influence antiviral immune response and disease progression during DENV infection warrants further investigation.
T cells help to B cells is provided by a CD4 T cells subset termed follicular T helper cells (Tfh) (104). Tfh cells have been associated with protective roles in human infectious disease (105–107) and vaccinees (108–110). They provide several forms of T cell help to B cells such as signals that promote survival, proliferation, plasma cell differentiation, hypermutation, class-switch recombination, adhesion and chemoattraction (cell migration) (111). Tfh cells are essential for the generation of most isotype switched and affinity matured antibodies, and therefore they have an obvious role in protective immunity against pathogens. A recent breakthrough has been the ability to detect Tfh cells in peripheral blood (105, 112) thus allowing their assessment in PBMC samples based on surface markers. A central marker of Tfh cells is the CXC-chemokine receptor 5 (CXCR5) which is required for T and B cells to enter into follicles. OX40, and PD-L1 have further been identified as TCR activation-dependent markers of human Tfh cells (113, 114). Recent studies have reported an expansion of peripheral Tfh cells in DENV-infected children during the acute phase (115). Furthermore, Tfh cells are more abundant in patients with secondary DENV infection and in those who developed a more severe dengue disease (115). Although Tfh cells has been shown to promote DENV-specific antibody responses in mice (116), the differentiation and functional significance of DENV-specific Tfh cells in humans warrants further investigation.
Conclusion and Perspective
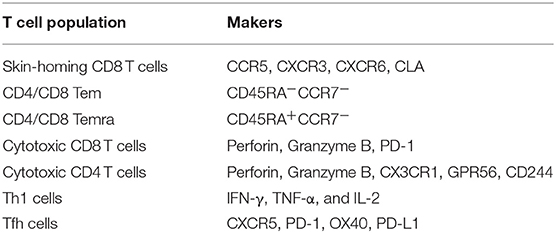
All DENV-specific T cell phenotypes discussed in this review as well as the markers they express are summarized in Figure 2 and Table 2, respectively. Comprehensive epitope identification over the last few years has provided the tools that allow one to probe for DENV specific T cell responses in donors exposed to natural infection and vaccination. Global assessment of dengue virus-specific CD4 and CD8 T cell responses in dengue-endemic areas led to the development of new tools (megapools) that allow analysis of small samples typically available from pediatric and hospital cohorts. It has also been demonstrated that DENV-specific CD4 and CD8 responses are more complex than previously thought, with different subsets revealed by in depth phenotypic and transcriptomic analyses. The hypothesis that these different subsets have unique roles and dictate and shape clinical outcomes and vaccine efficacy will have to be explored in the near future.
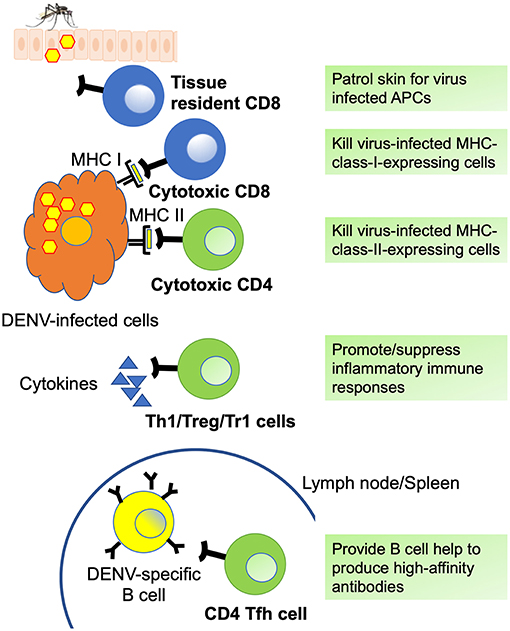
Figure 2. DENV specific T cell phenotypes. Summary of DENV-specific CD8 (blue) and CD4 (green) T cell phenotypes and functions. Tissue resident CD8 T cells patrol skin for infected antigen presenting cells (APCs) and can generate immediate effector functions. Cytotoxic CD8 and CD4 T cells express cytotoxic molecules such as granzyme B and perforin and can kill virus-infected cells via MHC I- and MHC II-dependent mechanisms. Th1 cells mediate and promote antiviral immune responses via the production of inflammatory cytokines such as IFN-γ and TNF-α, whereas regulatory CD4 T cells including Treg and Tr1 cells suppress inflammatory immune responses by producing cytokines such as IL-10 and TGF-β. Tfh cells provide help to DENV-specific germinal center B cells (yellow) and are essential for optimal germinal center reactions, thus promoting the generation of high-affinity antibodies, memory B cells, and long-lived plasma cells.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases grants U19 AI118626 and P01 AI106695 and IEDB contract 75N93019C00001 as well as National Institutes of Health contracts HHSN272200900042C and HHSN27220140045C. YT was supported through The American Association of Immunologists Intersect Fellowship Program for Computational Scientists and Immunologists.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02125/full#supplementary-material
References
1. Burke D, Monath T. Flaviviruses. In: Knipe D, Howley P, editors. Field's Virology. Philadelphia: Lippincott, Williams and Wilkins (2001). p. 1043–126.
2. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. (2013) 496:50a4–7. doi: 10.1038/nature12060
3. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. (2012) 380:1559–67. doi: 10.1016/S0140-6736(12)61428-7
4. Villar L, Dayan GH, Arredondo-Garcia JL, Rivera DM, Cunha R, Deseda C, et al. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med. (2015) 372:113–23. doi: 10.1056/NEJMoa1411037
5. Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, et al. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med. (2015) 373:1195–206. doi: 10.1056/NEJMoa1506223
6. Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. (2014) 384:1358–65. doi: 10.1016/S0140-6736(14)61060-6
7. Halstead SB. Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine. (2017) 35:6355–8. doi: 10.1016/j.vaccine.2017.09.089
8. Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, et al. Effect of dengue serostatus on dengue vaccine safety and efficacy. N Engl J Med. (2018) 379:327–40. doi: 10.1056/NEJMoa1800820
9. Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. (2011) 11:532–43. doi: 10.1038/nri3014
10. Weiskopf D, Sette A. T-cell immunity to infection with dengue virus in humans. Front Immunol. (2014) 5:93. doi: 10.3389/fimmu.2014.00093
11. Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol. (2015) 15:745–59. doi: 10.1038/nri3916
12. Tian Y, Sette A, Weiskopf D. Cytotoxic CD4 T cells: differentiation, function, and application to dengue virus infection. Front. Immunol. (2016) 7:531. doi: 10.3389/fimmu.2016.00531
13. Ngono AE, Shresta S. Immune response to dengue and Zika. Annu Rev Immunol. (2018) 36:279–308. doi: 10.1146/annurev-immunol-042617-053142
14. St. John AL, Rathore APS. Adaptive immune responses to primary and secondary dengue virus infections. Nat Rev Immunol. (2019) 19:218–30. doi: 10.1038/s41577-019-0123-x
15. Halstead SB, Rojanasuphot S, Sangkawibha N. Original antigenic sin in dengue. Am J Trop Med Hyg. (1983) 32:154–6. doi: 10.4269/ajtmh.1983.32.154
16. Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. (2003) 9:921–7. doi: 10.1038/nm887
17. Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, Yauch LE, et al. Gamma interferon. (IFN-gamma) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-alpha/beta receptor-deficient mice. J Virol. (2012) 86:12561–70. doi: 10.1128/JVI.06743-11
18. Zompi S, Santich BH, Beatty PR, Harris E. Protection from secondary dengue virus infection in a mouse model reveals the role of serotype cross-reactive B and T cells. J Immunol. (2012) 188:404–16. doi: 10.4049/jimmunol.1102124
19. Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, et al. A protective role for dengue virus-specific CD8+ T cells. J Immunol. (2009) 182:4865–73. doi: 10.4049/jimmunol.0801974
20. Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, et al. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol. (2010) 185:5405–16. doi: 10.4049/jimmunol.1001709
21. Zellweger RM, Tang WW, Eddy WE, King K, Sanchez MC, Shresta S. CD8+ T cells can mediate short-term protection against heterotypic dengue virus reinfection in mice. J Virol. (2015) 89:6494–505. doi: 10.1128/JVI.00036-15
22. Elong Ngono A, Chen HW, Tang WW, Joo Y, King K, Weiskopf D, et al. Protective role of cross-reactive CD8 T cells against dengue virus infection. EBioMed. (2016) 13:284–93. doi: 10.1016/j.ebiom.2016.10.006
23. Zellweger RM, Miller R, Eddy WE, White LJ, Johnston RE, Shresta S. Role of humoral versus cellular responses induced by a protective dengue vaccine candidate. PLoS Pathog. (2013) 9:e1003723. doi: 10.1371/journal.ppat.1003723
24. Zellweger RM, Eddy WE, Tang WW, Miller R, Shresta S. CD8+ T cells prevent antigen-induced antibody-dependent enhancement of dengue disease in mice. J Immunol. (2014) 193:4117–24. doi: 10.4049/jimmunol.1401597
25. Grifoni A, Angelo MA, Lopez B, O'Rourke PH, Sidney J, Cerpas C, et al. Global assessment of dengue virus-specific CD4. (+) T cell responses in dengue-endemic areas. Front Immunol. (2017) 8:1309. doi: 10.3389/fimmu.2017.01309
26. Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci USA. (2015) 112:E4256–63. doi: 10.1073/pnas.1505956112
27. de Alwis R, Bangs DJ, Angelo MA, Cerpas C, Fernando A, Sidney J, et al. Immunodominant dengue virus-specific CD8+ T cell responses are associated with a memory PD-1+ phenotype. J Virol. (2016) 90:4771–9. doi: 10.1128/JVI.02892-15
28. Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci USA. (2013) 110:E2046–53. doi: 10.1073/pnas.1305227110
29. Rivino L, Kumaran EA, Jovanovic V, Nadua K, Teo EW, Pang SW, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol. (2013) 87:2693–706. doi: 10.1128/JVI.02675-12
30. Halstead SB. Which dengue vaccine approach is the most promising, and should we be concerned about enhanced disease after vaccination? There is only one true winner. Cold Spring Harb Perspect Biol. (2018) 10:a030700. doi: 10.1101/cshperspect.a030700
31. Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med. (2016) 8:330ra36. doi: 10.1126/scitranslmed.aaf1517
32. Jaiswal S, Pazoles P, Woda M, Shultz LD, Greiner DL, Brehm MA, et al. Enhanced humoral and HLA-A2-restricted dengue virus-specific T-cell responses in humanized BLT NSG mice. Immunology. (2012) 136:334–43. doi: 10.1111/j.1365-2567.2012.03585.x
33. Culshaw A, Ladell K, Gras S, McLaren JE, Miners KL, Farenc C, et al. Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8. (+) T cell response. Nat Immunol. (2017) 18:1228–37. doi: 10.1038/ni.3850
34. Friberg H, Burns L, Woda M, Kalayanarooj S, Endy TP, Stephens HA, et al. Memory CD8+ T cells from naturally acquired primary dengue virus infection are highly cross-reactive. Immunol Cell Biol. (2011) 89:122–9. doi: 10.1038/icb.2010.61
35. Friberg H, Bashyam H, Toyosaki-Maeda T, Potts JA, Greenough T, Kalayanarooj S, et al. Cross-reactivity and expansion of dengue-specific T cells during acute primary and secondary infections in humans. Sci Rep. (2011) 1:51. doi: 10.1038/srep00051
36. Zivny J, Kurane I, Leporati AM, Ibe M, Takiguchi M, Zeng LL, et al. A single nine-amino acid peptide induces virus-specific, CD8+ human cytotoxic T lymphocyte clones of heterogeneous serotype specificities. J Exp Med. (1995) 182:853–63. doi: 10.1084/jem.182.3.853
37. Livingston PG, Kurane I, Dai LC, Okamoto Y, Lai CJ, Men R, et al. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte. (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J Immunol. (1995) 154:1287–95.
38. Zivny J, DeFronzo M, Jarry W, Jameson J, Cruz J, Ennis FA, et al. Partial agonist effect influences the CTL response to a heterologous dengue virus serotype. J Immunol. (1999) 163:2754–60.
39. Zivna I, Green S, Vaughn DW, Kalayanarooj S, Stephens HA, Chandanayingyong D, et al. T cell responses to an HLA-B*07-restricted epitope on the dengue NS3 protein correlate with disease severity. J Immunol. (2002) 168:5959–65. doi: 10.4049/jimmunol.168.11.5959
40. Sanchez V, Gimenez S, Tomlinson B, Chan PK, Thomas GN, Forrat R, et al. Innate and adaptive cellular immunity in flavivirus-naive human recipients of a live-attenuated dengue serotype 3 vaccine produced in Vero cells. (VDV3). Vaccine. (2006) 24:4914–26. doi: 10.1016/j.vaccine.2006.03.066
41. Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. (2006) 176:3821–9. doi: 10.4049/jimmunol.176.6.3821
42. Chang CX, Tan AT, Or MY, Toh KY, Lim PY, Chia AS, et al. Conditional ligands for Asian HLA variants facilitate the definition of CD8+ T-cell responses in acute and chronic viral diseases. Eur J Immunol. (2013) 43:1109–20. doi: 10.1002/eji.201243088
43. Piazza P, Campbell D, Marques E, Hildebrand WH, Buchli R, Mailliard R, et al. Dengue virus-infected human dendritic cells reveal hierarchies of naturally expressed novel NS3 CD8 T cell epitopes. Clin Exp Immunol. (2014) 177:696–702. doi: 10.1111/cei.12373
44. Kurane I, Dai LC, Livingston PG, Reed E, Ennis FA. Definition of an HLA-DPw2-restricted epitope on NS3, recognized by a dengue virus serotype-cross-reactive human CD4+ CD8- cytotoxic T-cell clone. J Virol. (1993) 67:6285–8.
45. Zeng L, Kurane I, Okamoto Y, Ennis FA, Brinton MA. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J Virol. (1996) 70:3108–17.
46. Okamoto Y, Kurane I, Leporati AM, Ennis FA. Definition of the region on NS3 which contains multiple epitopes recognized by dengue virus serotype-cross-reactive and flavivirus-cross-reactive, HLA-DPw2-restricted CD4+ T cell clones. J General Virol. (1998) 79(Pt 4):697–704. doi: 10.1099/0022-1317-79-4-697
47. Kurane I, Zeng L, Brinton MA, Ennis FA. Definition of an epitope on NS3 recognized by human CD4+ cytotoxic T lymphocyte clones cross-reactive for dengue virus types 2, 3, and 4. Virology. (1998) 240:169–74. doi: 10.1006/viro.1997.8925
48. Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. (2005) 175:2676–83. doi: 10.4049/jimmunol.175.4.2676
49. Moran E, Simmons C, Chau NV, Luhn K, Wills B, Phuong Dung N, et al. Preservation of a critical epitope core region is associated with the high degree of flaviviral cross-reactivity exhibited by a dengue-specific CD4+ T cell clone. Eur J Immunol. (2008) 38:1050–7. doi: 10.1002/eji.200737699
50. Kurane I, Okamoto Y, Dai LC, Zeng LL, Brinton MA, Ennis FA. Flavivirus-cross-reactive, HLA-DR15-restricted epitope on NS3 recognized by human CD4+ CD8- cytotoxic T lymphocyte clones. J General Virol. (1995) 76(Pt 9):2243–9. doi: 10.1099/0022-1317-76-9-2243
51. Guy B, Nougarede N, Begue S, Sanchez V, Souag N, Carre M, et al. Cell-mediated immunity induced by chimeric tetravalent dengue vaccine in naive or flavivirus-primed subjects. Vaccine. (2008) 26:5712–21. doi: 10.1016/j.vaccine.2008.08.019
52. Lim MQ, Kumaran EAP, Tan HC, Lye DC, Leo YS, Ooi EE, et al. Cross-reactivity and anti-viral function of dengue capsid and NS3-specific memory T cells toward zika virus. Front Immunol. (2018) 9:2225. doi: 10.3389/fimmu.2018.02225
53. Townsley E, Woda M, Thomas SJ, Kalayanarooj S, Gibbons RV, Nisalak A, et al. Distinct activation phenotype of a highly conserved novel HLA-B57-restricted epitope during dengue virus infection. Immunology. (2014) 141:27–38. doi: 10.1111/imm.12161
54. Wen J, Duan Z, Jiang L. Identification of a dengue virus-specific HLA-A*0201-restricted CD8+ T cell epitope. J Med Virol. (2010) 82:642–8. doi: 10.1002/jmv.21736
55. Testa JS, Shetty V, Sinnathamby G, Nickens Z, Hafner J, Kamal S, et al. Conserved MHC class I-presented dengue virus epitopes identified by immunoproteomics analysis are targets for cross-serotype reactive T-cell response. J Infect Dis. (2012) 205:647–55. doi: 10.1093/infdis/jir814
56. Simmons CP, Dong T, Chau NV, Dung NT, Chau TN, Thao le TT, et al. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J Virol. (2005) 79:5665–75. doi: 10.1128/JVI.79.9.5665-5675.2005
57. Duan Z, Guo J, Huang X, Liu H, Chen X, Jiang M, et al. Identification of cytotoxic T lymphocyte epitopes in dengue virus serotype 1. J Med Virol. (2015) 87:1077–89. doi: 10.1002/jmv.24167
58. Imrie A, Meeks J, Gurary A, Sukhbataar M, Kitsutani P, Effler P, et al. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J Virol. (2007) 81:10081–91. doi: 10.1128/JVI.00330-07
59. Bashyam HS, Green S, Rothman AL. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotypes. J Immunol. (2006) 176:2817–24. doi: 10.4049/jimmunol.176.5.2817
60. Nascimento EJ, Mailliard RB, Khan AM, Sidney J, Sette A, Guzman N, et al. Identification of conserved and HLA promiscuous DENV3 T-cell epitopes. PLoS Negl Trop Dis. (2013) 7:e2497. doi: 10.1371/journal.pntd.0002497
61. Townsley E, O'Connor G, Cosgrove C, Woda M, Co M, Thomas SJ, et al. Interaction of a dengue virus NS1-derived peptide with the inhibitory receptor KIR3DL1 on natural killer cells. Clin Exp Immunol. (2016) 183:419–30. doi: 10.1111/cei.12722
62. Mathew A, Kurane I, Green S, Stephens HA, Vaughn DW, Kalayanarooj S, et al. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J Virol. (1998) 72:3999–4004.
63. Loke H, Bethell DB, Phuong CX, Dung M, Schneider J, White NJ, et al. Strong HLA class I–restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J Infect Dis. (2001) 184:1369–73. doi: 10.1086/324320
64. Gagnon SJ, Zeng W, Kurane I, Ennis FA. Identification of two epitopes on the dengue 4 virus capsid protein recognized by a serotype-specific and a panel of serotype-cross-reactive human CD4+ cytotoxic T-lymphocyte clones. J Virol. (1996) 70:141–7.
65. Wen JS, Jiang LF, Zhou JM, Yan HJ, Fang DY. Computational prediction and identification of dengue virus-specific CD4. (+) T-cell epitopes. Virus Res. (2008) 132:42–8. doi: 10.1016/j.virusres.2007.10.010
66. Bowerman NA, Falta MT, Mack DG, Kappler JW, Fontenot AP. Mutagenesis of beryllium-specific TCRs suggests an unusual binding topology for antigen recognition. J Immunol. (2011) 187:3694–703. doi: 10.4049/jimmunol.1101872
67. Rivino L, Kumaran EA, Thein TL, Too CT, Gan VC, Hanson BJ, et al. Virus-specific T lymphocytes home to the skin during natural dengue infection. Sci Transl Med. (2015) 7:278ra35. doi: 10.1126/scitranslmed.aaa0526
68. Malavige GN, Jeewandara C, Alles KM, Salimi M, Gomes L, Kamaladasa A, et al. Suppression of virus specific immune responses by IL-10 in acute dengue infection. PLoS Negl Trop Dis. (2013) 7:e2409. doi: 10.1371/journal.pntd.0002409
69. Grifoni A, Pham J, Sidney J, O'Rourke PH, Paul S, Peters B, et al. Prior Dengue virus exposure shapes T cell immunity to Zika virus in humans. J Virol. (2017) 91:e01469–17. doi: 10.1128/JVI.01469-17
70. Turtle L, Bali T, Buxton G, Chib S, Chan S, Soni M, et al. Human T cell responses to Japanese encephalitis virus in health and disease. J Exp Med. (2016) 213:1331–52. doi: 10.1084/jem.20151517
71. Turtle L, Tatullo F, Bali T, Ravi V, Soni M, Chan S, et al. Cellular immune responses to live attenuated japanese encephalitis. (JE) Vaccine SA14–14-2 in adults in a je/dengue co-endemic area. PLoS Negl Trop Dis. (2017) 11:e0005263. doi: 10.1371/journal.pntd.0005263
72. Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, et al. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J Immunol. (2011) 187:4268–79. doi: 10.4049/jimmunol.1101970
73. Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, et al. Human CD8+ T-cell responses against the 4 dengue virus serotypes are associated with distinct patterns of protein targets. J Infect Dis. (2015) 212:1743–51. doi: 10.1093/infdis/jiv289
74. Rivino L, Tan AT, Chia A, Kumaran EA, Grotenbreg GM, MacAry PA, et al. Defining CD8+ T cell determinants during human viral infection in populations of Asian ethnicity. J Immunol. (2013) 191:4010–9. doi: 10.4049/jimmunol.1301507
75. Appanna R, Huat TL, See LL, Tan PL, Vadivelu J, Devi S. Cross-reactive T-cell responses to the nonstructural regions of dengue viruses among dengue fever and dengue hemorrhagic fever patients in Malaysia. Clin Vaccine Immunol. (2007) 14:969–77. doi: 10.1128/CVI.00069-07
76. Weiskopf D, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, et al. The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes. J Virol. (2015) 89:120–8. doi: 10.1128/JVI.02129-14
77. Weiskopf D, Angelo MA, Grifoni A, O'Rourke PH, Sidney J, Paul S, et al. HLA-DRB1 alleles are associated with different magnitudes of dengue virus-specific CD4+ T-cell responses. J Infect Dis. (2016) 214:1117–24. doi: 10.1093/infdis/jiw309
78. Angelo MA, Grifoni A, O'Rourke PH, Sidney J, Paul S, Peters B, et al. Human CD4+ T cell responses to an attenuated tetravalent dengue vaccine parallel those induced by natural infection in magnitude, HLA restriction, and antigen specificity. J Virol. (2017) 91:e02147–16. doi: 10.1128/JVI.02147-16
79. Grifoni A, Moore E, Voic H, Sidney J, Phillips E, Jadi R, et al. Characterization of magnitude and antigen specificity of HLA-DP, DQ, and DRB3/4/5 Restricted DENV-Specific CD4+ T Cell Responses. Front Immunol. (2019). doi: 10.3389/fimmu.2019.01568
80. Malavige GN, McGowan S, Atukorale V, Salimi M, Peelawatta M, Fernando N, et al. Identification of serotype-specific T cell responses to highly conserved regions of the dengue viruses. Clin Exp Immunol. (2012) 168:215–23. doi: 10.1111/j.1365-2249.2012.04566.x
81. Grifoni A, Angelo M, Sidney J, Paul S, Peters B, de Silva AD, et al. Patterns of cellular immunity associated with experimental infection with rDEN2Delta30. (Tonga/74) support its suitability as a human dengue virus challenge strain. J Virol. (2017) 91:e02133–16. doi: 10.1128/JVI.02133-16
82. Osorio JE, Wallace D, Stinchcomb DT. A recombinant, chimeric tetravalent dengue vaccine candidate based on a dengue virus serotype 2 backbone. Exp Rev Vaccines. (2016) 15:497–508. doi: 10.1586/14760584.2016.1128328
83. Kirkpatrick BD, Durbin AP, Pierce KK, Carmolli MP, Tibery CM, Grier PL, et al. Robust and balanced immune responses to all 4 dengue virus serotypes following administration of a single dose of a live attenuated tetravalent dengue vaccine to healthy, flavivirus-naive adults. J Infect Dis. (2015) 212:702–10. doi: 10.1093/infdis/jiv082
84. Dhanda SK, Vita R, Ha B, Grifoni A, Peters B, Sette A. ImmunomeBrowser: a tool to aggregate and visualize complex and heterogeneous epitopes in reference protein. Bioinformatics. (2018) 34:3931–3. doi: 10.1093/bioinformatics/bty463
85. Weiskopf D, Angelo MA, Sidney J, Peters B, Shresta S, Sette A. Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J Virol. (2014) 88:11383–94. doi: 10.1128/JVI.01108-14
86. Carrasco Pro S, Sidney J, Paul S, Lindestam Arlehamn C, Weiskopf D, Peters B, et al. Automatic generation of validated specific epitope sets. J Immunol Res. (2015) 2015:763461. doi: 10.1155/2015/763461
87. Hatch S, Endy TP, Thomas S, Mathew A, Potts J, Pazoles P, et al. Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J Infect Dis. (2011) 203:1282–91. doi: 10.1093/infdis/jir012
88. Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. (2014) 41:886–97. doi: 10.1016/j.immuni.2014.12.007
89. Chandele A, Sewatanon J, Gunisetty S, Singla M, Onlamoon N, Akondy RS, et al. Characterization of human CD8 T cell responses in dengue virus-infected patients from India. J Virol. (2016) 90:11259–78. doi: 10.1128/JVI.01424-16
90. Tian Y, Babor M, Lane J, Seumois G, Liang S, Goonawardhana NDS, et al. Dengue-specific CD8+ T cell subsets display specialized transcriptomic and TCR profiles. J Clin Invest. (2019) 130:1727–41. doi: 10.1172/JCI123726
91. Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. (2012) 12:749–61. doi: 10.1038/nri3307
92. Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4. (+) T cells in immunity to viruses. Nat Rev Immunol. (2012) 12:136–48. doi: 10.1038/nri3152
93. Tian Y, Zajac AJ. IL-21 and T cell differentiation: consider the context. Trends Immunol. (2016) 37:557–68. doi: 10.1016/j.it.2016.06.001
94. Lindow JC, Borochoff-Porte N, Durbin AP, Whitehead SS, Fimlaid KA, Bunn JY, et al. Primary vaccination with low dose live dengue 1 virus generates a proinflammatory, multifunctional T cell response in humans. PLoS Negl Trop Dis. (2012) 6:e1742. doi: 10.1371/journal.pntd.0001742
95. Gwinn W, Sun W, Innis BL, Caudill J, King AD. Serotype-specific T. (H)1 responses in recipients of two doses of candidate live-attenuated dengue virus vaccines. Am J Trop Med Hyg. (2003) 69:39–47. doi: 10.4269/ajtmh.2003.69.39
96. Duangchinda T, Dejnirattisai W, Vasanawathana S, Limpitikul W, Tangthawornchaikul N, Malasit P, et al. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc Natl Acad Sci USA. (2010) 107:16922–7. doi: 10.1073/pnas.1010867107
97. Robinson M, Sweeney TE, Barouch-Bentov R, Sahoo MK, Kalesinskas L, Vallania F, et al. A 20-Gene set predictive of progression to severe dengue. Cell Rep. (2019) 26:1104–11.e4. doi: 10.1016/j.celrep.2019.01.033
98. Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O'Rourke P, de Silva AD, et al. Precursors of human CD4. (+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol. (2018) 3:eaan8664. doi: 10.1126/sciimmunol.aan8664
99. Tian Y, Babor M, Lane J, Schulten V, Patil VS, Seumois G, et al. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat Commun. (2017) 8:1473. doi: 10.1038/s41467-017-01728-5
100. Plitas G, Rudensky AY. Regulatory T cells: differentiation and function. Cancer Immunol Res. (2016) 4:721–5. doi: 10.1158/2326-6066.CIR-16-0193
101. Zeng H, Zhang R, Jin B, Chen L. Type 1 regulatory T cells: a new mechanism of peripheral immune tolerance. Cell Mol Immunol. (2015) 12:566–71. doi: 10.1038/cmi.2015.44
102. Luhn K, Simmons CP, Moran E, Dung NT, Chau TN, Quyen NT, et al. Increased frequencies of CD4+ CD25. (high) regulatory T cells in acute dengue infection. J Exp Med. (2007) 204:979–85. doi: 10.1084/jem.20061381
103. Jayaratne HE, Wijeratne D, Fernando S, Kamaladasa A, Gomes L, Wijewickrama A, et al. Regulatory T-cells in acute dengue viral infection. Immunology. (2018) 154:89–97. doi: 10.1111/imm.12863
104. Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. (2014) 41:529–42. doi: 10.1016/j.immuni.2014.10.004
105. Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. (2013) 39:758–69. doi: 10.1016/j.immuni.2013.08.031
106. Obeng-Adjei N, Portugal S, Tran TM, Yazew TB, Skinner J, Li S, et al. Circulating Th1-Cell-type Tfh cells that exhibit impaired B cell help are preferentially activated during acute malaria in children. Cell Rep. (2015) 13:425–39. doi: 10.1016/j.celrep.2015.09.004
107. Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, et al. BACH2 regulates CD8. (+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol. (2016) 17:851–60. doi: 10.1038/ni.3441
108. Havenar-Daughton C, Carnathan DG, Torrents de la Pena A, Pauthner M, Briney B, Reiss SM, et al. Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. (2016) 17:2195–209. doi: 10.1016/j.celrep.2016.10.085
109. Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. (2013) 5:176ra32. doi: 10.1126/scitranslmed.3005191
110. Onabajo OO, George J, Lewis MG, Mattapallil JJ. Rhesus macaque lymph node PD-1. (hi)CD4+ T cells express high levels of CXCR5 and IL-21 and display a CCR7. (lo)ICOS+Bcl6+ T-follicular helper. (Tfh) cell phenotype. PLoS ONE. (2013) 8:e59758. doi: 10.1371/journal.pone.0059758
111. Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. (2015) 15:185–9. doi: 10.1038/nri3803
112. Schmitt N, Ueno H. Blood Tfh cells come with colors. Immunity. (2013) 39:629–30. doi: 10.1016/j.immuni.2013.09.011
113. Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar-Daughton C, Reiss SM, et al. A cytokine-independent approach to identify antigen-specific human germinal center t follicular helper cells and rare antigen-specific CD4+ T cells in blood. J Immunol. (2016) 197:983–93. doi: 10.4049/jimmunol.1600318
114. Havenar-Daughton C, Reiss SM, Carnathan DG, Wu JE, Kendric K, Torrents de la Pena A, et al. Cytokine-independent detection of antigen-specific germinal center T follicular helper cells in immunized nonhuman primates using a live cell activation-induced marker technique. J Immunol. (2016) 197:994–1002. doi: 10.4049/jimmunol.1600320
115. Haltaufderhyde K, Srikiatkhachorn A, Green S, Macareo L, Park S, Kalayanarooj S, et al. Activation of peripheral T follicular helper cells during acute dengue virus infection. J Infect Dis. (2018) 218:1675–85. doi: 10.1093/infdis/jiy360
Keywords: dengue, CD4 T cell, CD8 T cell, T cell epitope, vaccine
Citation: Tian Y, Grifoni A, Sette A and Weiskopf D (2019) Human T Cell Response to Dengue Virus Infection. Front. Immunol. 10:2125. doi: 10.3389/fimmu.2019.02125
Received: 01 March 2019; Accepted: 23 August 2019;
Published: 04 September 2019.
Edited by:
Mario Mago Clerici, University of Milan, ItalyReviewed by:
Jianzhong Zhu, Yangzhou University, ChinaFrancisco Veas, Institut de Recherche pour le Développement (IRD), France
Copyright © 2019 Tian, Grifoni, Sette and Weiskopf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Weiskopf, ZGFuaWVsYUBsamkub3Jn
 Yuan Tian
Yuan Tian Alba Grifoni
Alba Grifoni Alessandro Sette
Alessandro Sette Daniela Weiskopf
Daniela Weiskopf