
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 31 May 2019
Sec. Microbial Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.01203
This article is part of the Research TopicUnderstanding and Exploiting Host-Commensal Interactions to Combat PathogensView all 17 articles
A diverse community of trillions of commensal bacteria inhabits mucosal and epidermal surfaces in humans and plays an important role in defense against pathogens, including respiratory pathogens. Commensal bacteria act on the host's immune system to induce protective responses that prevent colonization and invasion by pathogens. On the other hand, these bacteria can directly inhibit the growth of respiratory pathogens by producing antimicrobial products/signals and competing for nutrients and adhesion sites. Such mechanisms preserve the niche for commensal bacteria and support the host in containing respiratory infections. Herein, we discuss current evidence on the role of commensal bacteria in conferring protection against respiratory pathogens and the underlying mechanisms by which these bacteria do so. A deeper knowledge of how commensal bacteria interact with the host and pathogens might provide new insights that are poised to aid in the development of vaccines and therapeutics that target infectious diseases.
Since the inception of the Human Microbiome Project in 2007, a plethora of knowledge has accumulated that throws light on diverse and crucial roles played by commensal bacteria in homeostasis and disease (1, 2). With the help of advances in omic and systems biology technologies, our knowledge of the composition, genetics, and functional capacity of commensal bacteria is growing at a fast pace. It is becoming clear that commensal bacteria, which reside in various parts of the human body, such as the gut and airways, correspond approximately to the total number of human cells (about 1:1 ratio), and exert a profound impact on regulation of immunophysiological functions, including but not limited to, metabolism, ontogeny, and pathogen defense (3, 4). Several recent studies have shown that commensals promote resistance to gut pathogens that is mutually beneficial to the host and the commensal microbiota (5–7). However, imbalances in the microbial communities can occur, and are linked to many diseases, such as inflammatory bowel disease, allergies, asthma, diabetes, and obesity (8). It remains scantily understood how these bacteria execute their functional activities against respiratory pathogens. Respiratory infections are of utmost importance, as they inflict substantial social and economic burden on people across the world in general and in low and lower-middle income countries in particular (9–11). Additionally, current therapeutic and prophylactic interventions against respiratory diseases have major constraints, such as rapid emergence of anti-microbial resistance and disruption of the normal microbiota by use of antibiotics. Unraveling the interactions between commensals and pathogens may allow the exploitation of inhibitory properties of commensals to combat pathogens causing respiratory diseases. In this review article, we provide an overview of the current state of knowledge about the role of commensal bacteria in protective immunity to respiratory pathogens and the mechanisms involved in commensal bacteria mediated defenses. Understanding the relationship between commensal bacteria, host, and pathogen is a way forward to develop safe and effective prophylactics and therapeutics against pathogens.
Much of what is known about the direct role of commensal bacteria in protection against respiratory pathogens stems from studies using various mouse models, including germ-free and antibiotic-treated mice (12–23). Following lung infection with Streptococcus pneumoniae, numerous studies have shown that antibiotic-treated mice display significantly higher pathogen loads and increased pathologies in the lungs compared with sham-treated mice (15, 21). Likewise, germ-free mice showed enhanced levels of bacterial burden when subjected to S. pneumoniae and Klebsiella pneumoniae lung infections (15). Similar to the protection conferred by fecal microbiota transplant (FMT) against intestinal pathogens, FMT in gut microbiota-depleted mice restored pulmonary bacterial clearance early after S. pneumoniae infection (21). In case of mouse models of Mycobacterium tuberculosis infection, gut microbiota disruption after pre- and post-antibiotic treatment showed decreased resistance to infection in the lungs, which was associated with severe histopathological changes, such as pulmonary granulomas (24). Furthermore, antibiotic-induced dysbiosis changed the microbiota diversity in the gut and promoted lung colonization by M. tuberculosis (25). Similar protective effect was conferred after mice having antibiotic-induced disrupted microbiota received an intranasal infectious dose of influenza A virus (12). In a different study, FMT into germ-free mice led to reconstitution of the gut microbiota that facilitated increased survival against lethal influenza A virus infection (26). Overall, these studies employing multiple experimental approaches provide in vivo evidence that underscores a profound contribution of commensal bacteria in defense against diverse respiratory pathogens.
Even though antibiotic-treated and germ-free mice have proven to be a crucial tool in understanding the role of the microbiota in pathogen defense, there are potential pitfalls that need consideration while interpreting results from studies involving these animals. Germ-free animals lack all detectable microbes in different organs and have an impaired immune system, whereas antibiotics are used to deplete specific microbiota (27). Although these two approaches provide crucial information on the function of the microbiota in general, the specific contribution of the microbiota found in different body compartments, such as the lung microbiota, in immunity to respiratory infections is unclear. This is important because the lung microbiota, which in healthy adults seems to mainly consist of a small number of bacteria originating from the oral cavity, plays a significant role in respiratory health, and disease (28–30). Additionally, there is a need for models that can answer a more direct question about protection in the presence of a fully developed immune system. To address this issue, researchers have attempted to deplete the lung microbiota in mice by a combination of aerosolized vancomycin and neomycin via nasal route, which resulted in a significant reduction in the lung commensal microbiota, with the advantage of minimally affecting the gut microbiota (31, 32). But the possibility of antibiotic spread to the nearby tissues/organs harboring different microbiota remains, requiring future studies to focus on developing better models to fill in this pitfall.
The microbiota consists of a large number of bacterial species, and therefore, it is of great interest to specifically identify commensal species that protect from respiratory pathogens. Recent studies have evaluated the protective efficacy of commensal bacterial species in respiratory infections. Oral administration of Bifidobacterium longum (BB536), but not saline, in mice significantly reduced viral loads, pulmonary pathology, and body weight loss following intranasal challenge with influenza virus, suggesting a protective role for this commensal bacterium in influenza infection (33). Similarly, oral or nasal inoculation of mice with different strains of Lactobacillus, e.g., L. gasseri (TMC0356), L. rhamnosus (CRL 1505), and L. brevis (KB290), conferred protective immunity to influenza virus infections (34–36). Furthermore, L. rhamnosus (CRL 1505) exerted a protective effect in mice subjected to an intranasal challenge with respiratory syncytial virus infection (37, 38). These data indicate a prophylactic role for commensal bacteria against viral pathogens. In order to assess therapeutic significance, B. longum (MM2) was orally administered in mice infected with influenza virus. Mice that received B. longum (MM2) ameliorated infection, as determined by decreased body weight loss, viral titers, and inflammation, compared with control mice (39). Protective effect induced by these probiotic bacteria is not confined to respiratory infections with viruses, but can be applicable to bacterial pathogens (37–40). Intake of B. longum (51A) in mice not only demonstrated protective effect against infection with K. pneumoniae, but also suppressed inflammatory changes in the lung (40). Very recently, we have demonstrated that intranasal immunization of mice with the commensal Streptococcus mitis conferred protection against lung infection with S. pneumoniae strains D39 (serotype 2) and TIGR4 (serotype 4), which illuminates the unique ability of S. mitis to offer resistance to different pneumococcal serotypes (41). Two recent studies performed by independent groups further show that the gut colonizer bacterium Helicobacter hepaticus influences the composition of the gut microbiota and the outcome of M. tuberculosis infection in mouse models (42, 43). Mice subjected to intestinal colonization with H. hepaticus, when challenged by intranasal route with M. tuberculosis, reflected higher mycobacterial burden in the lungs compared with the controls (42). This increased mycobacterial burden in the H. hepaticus-colonized mice coincided with severe M. tuberculosis-mediated pulmonary pathologies, mainly characterized by granulomas and tissue damage, and accumulation/production of inflammatory leukocytes/cytokines (42). Similar to these results, mice colonized with H. hepaticus eliminated subunit-vaccine-induced protective immunity to lung infection with M. tuberculosis (43). Taken together, these findings indicate that commensal bacteria can be harnessed for prophylactic and therapeutic purposes, provided utmost precaution on the possible negative effects of enriching for specific colonizers of the microbiota.
Relatively little information is available on whether commensal bacteria can prevent respiratory infections in humans. Oral commensals, such as Streptococcus oralis and Streptococcus salivarius, can induce protection against middle ear inflammation, referred to as otitis media, which is primarily caused by respiratory pathogens, such as S. pneumoniae and Haemophilus influenzae (44–46). Upon intranasal administration of S. salivarius and S. oralis, children susceptible to acute otitis media displayed reduced recurrences of disease with no side effects (44). Contrarily, a nasal spray containing oral commensals, e.g., S. mitis and S. oralis, in susceptible children under 4 years of age showed no significant effect regarding the number of episodes of recurrent otitis media compared to the placebo group (46). The discrepancy in these studies might be due to differences in bacterial doses, inoculation regimens, and combinations, which need to be analyzed in light of new technologies (e.g., metagenomics and next generation sequencing) and concepts like dysbiosis. The fact that antibiotics were used together with the streptococcal nasal spray in the first study, but not in the second, is also an important factor to consider. In controlled infection studies in humans, nasopharyngeal colonization by the commensal Neisseria lactamica provided protection against the respiratory pathogen Neisseria meningitidis (47, 48). Furthermore, in a block-randomized challenge trial, 310 healthy individuals (18–25 years) were intranasally inoculated with live N. lactamica or sham and the bacterial carriage was monitored for 26 weeks (48). All those who developed nasopharyngeal colonization by N. lactamica revealed a significant reduction in the N. meningitidis carriage compared with sham-treated ones (48). These studies show that commensal bacteria not only show inhibitory/displacing effects on the carriage of respiratory pathogens but also highlight the ease and safety with which these bacteria can be used to contain infections in humans. It is however notable that most bacteria with high pathogenic potential, such as those in the above examples, are also part of the healthy microbiome (49). The reason as to why these pathogens cause diseases is attributed to various host and microbial factors, including viral infections (49). Dysbiosis in particular, such as a result of antibiotic use has been associated with a reduction in the prevalence of respiratory commensal bacteria like Corynebacterium spp. and Dolosigranulum spp. in the nasopharynx of healthy infants. These are considered to reduce the colonization by S. pneumoniae, H. influenzae, and S. aureus in the respiratory tract (50). It is further shown that respiratory syncytial virus infection in children below 2 years of age was positively correlated with nasopharyngeal H. influenzae and Streptococcus microbiota clusters and inversely correlated with Staphylococcus aureus (51). Transcriptomic analysis of the children infected with H. influenzae and Streptococcus clusters presented greater expression of immune components, suggesting that nasopharyngeal microbiota can influence host immunity (51). In line with this, prolonged antibiotic treatment in early life has also been annexed with an increased risk for respiratory infections in infants (52, 53). Thus, these studies shine light on the effect of the microbiota perturbations caused by antibiotics on host susceptibility to respiratory infections, particularly during the critical life period of immune maturation.
A pertinent question however remains as to what are the underlying mechanisms by which commensal bacteria perform their protective function against respiratory pathogens. Emerging data thus far indicate that commensal bacteria confer protection in two ways: host-mediated immunity (acting on the host's immune system) and direct action (directly inhibiting/killing pathogens and competing for colonization).
A wealth of emerging evidence indicates that both the lung and gut microbiota are involved in the regulation of immune responses during lung infections (28, 54). However, it is difficult to assess the specific contributions of the lung and gut microbiota to protective immunity to respiratory pathogens, mainly due to three reasons: (1) the gut microbiota is the largest and most diverse community of commensals that significantly influences the outcome of immunity in the lungs as well as gut; (2) the gut is the largest lymphoid organ in the body because of which it occupies a central position in host-microbiota studies; and (3) we do not have optimal models to ascertain their specific roles in immunity. Despite an important role for these commensal bacteria in promoting resistance against respiratory pathogens, the mechanistic basis for this resistance remains unclear. Several studies have shown a potential defect in innate immunity and subsequent adaptive immunity in the lung, when signals from commensal bacteria are abrogated (12, 14, 25, 55). Key innate immune cells that are recruited to the lungs and are involved in protective immunity include macrophages, natural killer (NK) cells, and mucosal-associated invariant T (MAIT) cells (12, 14, 25, 55). The pulmonary macrophages in mice depleted of the microbiota by antibiotics reflected reduced expression of the macrophage-associated antiviral genes, such as Irf 7, Ifnb, Mx1, Tnfa, Il6, and Il1b following influenza virus infection (14). This corresponded with reduced response to IFN-γ, IFN-α, or influenza infection in macrophages from the mice treated with antibiotics. In vivo experiments in mice also indicated that the alveolar macrophage response was impaired during viral infection, which was characterized by down-regulation of most of the antiviral genes activated ex vivo (14). Wang et al. demonstrated a new mechanism in mice colonized with S. aureus where CD11b+ M2 alveolar macrophages, stimulated with Toll-like receptor (TLR) 2, play a protective role in influenza infection (56). Another innate immune cell type is MAIT cell that is shown to play an important role in microbiota mediated mycobacterial immunity (25). Flow cytometric analysis reflected that mice depleted of the microbiota had reduced number of lung MAIT cells, characterized by MR1-5-OP-RU tetramer+TCRβ+ phenotype, which expressed significantly lower IL-17A compared with control mice, suggesting that lung MAIT cells may function to contain early pulmonary M. tuberculosis infection (25). Furthermore, NK cells from germ-free mice did not induce anti-influenza immunity because macrophages and dendritic cells failed to produce type 1 IFN in response to infection (55). Cumulatively, these data suggest that microbiota-derived signals provide a stimulus that maintains the potency of the lung innate immune system needed for invoking effective immunity (Figure 1).
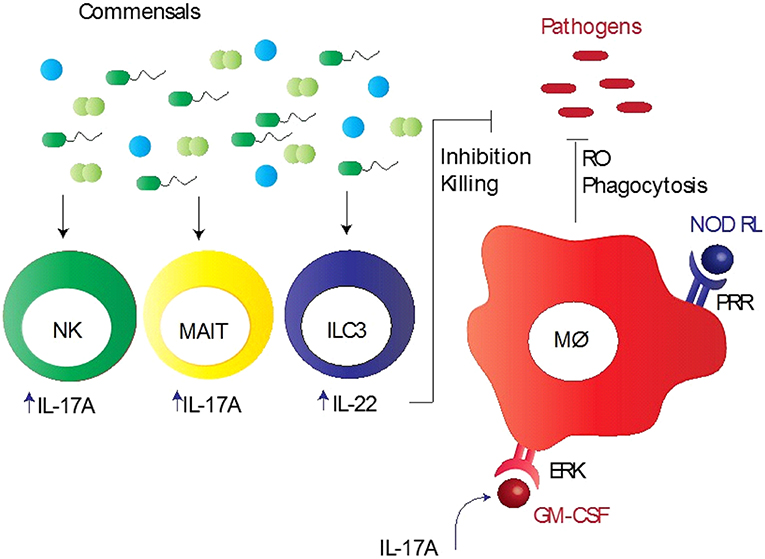
Figure 1. Commensal bacteria mediated innate immunity to respiratory pathogens. Commensal bacteria stimulate various innate immune cells, particularly alveolar macrophages (Mϕ), mucosa-associated invariant T (MAIT) cells, group 3 innate lymphoid cells (ILC3), and natural killer (NK) cells, to induce early protection. These bacteria promote pathogen killing via granulocyte–macrophage colony-stimulating factor (GM-CSF), which stimulates pathogen killing and clearance by alveolar macrophages (Mϕ) through phagocytosis, reduced reactive oxygen species (RO), and extracellular signal regulated kinase (ERK) signaling. Intrapulmonary GM-CSF production in response to infection is regulated by the microbiota via interleukin-17A (IL-17A). Pattern recognition receptor (PRR) expressed by Mϕ recognizes PRR ligands, such as nod-like receptor ligands (NOD RL), leading to the activation of Mϕ. NK and MAIT cells when activated by commensal bacteria produce large quantities of IL-17A, whereas ILC3 cells secrete IL-22, aiding in inhibition/killing of various respiratory pathogens.
Adaptive immunity follows innate immunity and is crucial for specific immunity against respiratory pathogens (57–59). Rabbit antisera raised against S. mitis show cross-reactivity with S. pneumoniae (59). Similar to IgG mediated cross-reactivity, IgA antibodies from the sera, nasal wash, and bronchoalveolar lavage of mice vaccinated with S. mitis cross-reacted with S. pneumoniae serotypes 2 and 4 (41). On the other hand, human CD4+ T cells expressing IL-17A, which are reactive to S. mitis, show cross-reactivity toward S. pneumoniae in an in vitro culture system (60). Intranasal vaccination of mice with S. mitis led to an increased production of IL-17A by CD4+ T cells in the lungs compared to PBS-treated control mice (41). These results are in line with the evidence that the gut commensal segmented filamentous bacteria (SFB) regulate pulmonary Th17 immunity to the fungal pathogen Aspergillus fumigatus (19). In a mouse model of influenza viral infection, it is shown that commensal bacteria regulate virus-specific CD4+ and CD8+ T cells and antibodies following lung infection with virus (12). Overall, commensal bacteria mediated adaptive immunity to respiratory pathogens include both humoral (IgG and IgA) and T cell-mediated responses.
Involvement of the gut microbiota in protective immunity to pulmonary pathogens illustrates a pathogenic nexus between the microbiota and the “gut-lung axis,” underscoring a profound protective influence of the gut commensals over several pathogens residing at distant anatomical compartments of the body (61). The gut microbiota mediated control of the lung immunopathology is also evident from studies demonstrating the susceptibility of animals with the altered gut microbiota to allergic lung diseases (20). On the other hand, dysbiosis in the lung microbiota can change the composition of the gut microbiota. For example, mice subjected to lung infection with influenza virus infection or intranasal instillation of lipopolysaccharide disturbed the gut microbiota homeostasis, which supports the fact that the gut and lungs are closely linked in a way that they affect each other's microbiology and physiology (62, 63). Moreover, how the gut microbiota controls the lung immunity has recently been explored by few key studies in mouse models of pulmonary bacterial infections. Brown et al. performed a well-designed and comprehensive study that sheds light on a major innate immune mechanism used by the microbiota to clear lung infections in mouse models (15). In antibiotic-treated mice, there was an increased growth of S. pneumoniae and K. pneumoniae in the lungs after bacterial inoculation compared to sham-treated mice, which was associated with reduced production of innate immune factors, such granulocyte–macrophage colony-stimulating factor (GM-CSF) (15). In vivo neutralization of GM-CSF into antibiotic-treated mice, which received the microbiota from the sham-treated mice and had restored pulmonary bacterial clearance, resulted in making these mice prone to infections (15). These findings suggest that GM-CSF is essential for the microbiota to execute their functional activities against both Gram-positive (S. pneumoniae) and Gram-negative (K. pneumoniae) pathogens. It was further demonstrated that GM-CSF programs alveolar macrophage function via an extracellular signal-regulated kinase (ERK)-specific signaling pathway leading to increased pathogen killing via reactive oxygen species (ROS) (15). Several studies have implicated pattern recognition receptor (PRR) ligands produced by the gut microbiota in controlling immune responses outside the intestinal tract (16, 64). Following antibiotic-mediated depletion of the microbiota in mice, early clearance of K. pneumoniae was impaired and this could be rescued by injection of bacterial Nod-like receptor (NLR) ligands (the NOD1 ligand MurNAcTri(DAP) and NOD2 ligand muramyl dipeptide [MDP]), but not bacterial TLR ligands (16). Defects in early innate immunity were found to be due to reduced ROS-mediated killing of bacteria by alveolar macrophages (16). Interestingly, upon treatment of mice with antibiotics and NLR ligands orally prior to S. pneumoniae lung infection, neutralization of GM-CSF abrogated the rescue of respiratory clearance (15). Taking account of all these data, it is clear that the microbiota and NLR ligands regulate lung innate immunity to respiratory pathogens via GM-CSF, highlighting crucial mechanisms of the gut-lung axis of communication. In addition, the gut commensal SFB has been reported to provide protection in immunocompromised (Rag−/−) mice by partially enhancing neutrophil resolution during pneumococcal lung infection, which corresponded with reduced expression of the anti-phagocytic molecule CD47 (65). Like NK cells, another lymphoid cell population referred to as group 3 lymphoid cells that produce IL-22 (IL-22+ILC3), a cytokine involved in pathogen defense, has been implicated in gut commensal bacteria-induced protection against S. pneumoniae (66). Disruption of commensal bacteria by antibiotics decreased the influx of IL-22+ILC3 cells into the lungs of new born mice, which made them more prone to pneumococcal infection compared with control mice. This effect was reversed when ILC3 cells were adoptively transferred or exogenous IL-22 administered in mice (66). Thus, these immune mechanisms furnish crucial information on how the gut microbiota controls protective immunity to lung infections (Figure 1).
Commensal bacteria resist colonization of pathogens by using wide range of direct mechanisms for niche competition, such as secretion of inhibitory substances and nutrient competition, enlisting the exploitative, and interference modes of competition (67). Recent studies dissected novel mechanisms used by resident commensals to inhibit and contain respiratory pathogens, such as disruption of biofilms, exploitation of host resources to generate antimicrobial products, and down-regulation of virulence genes. This highlights the complexity and diversity of mechanisms involved in direct inhibition (68–70). The well-documented mechanism by which commensal bacteria can directly inhibit the pathogen growth and compete with them is the production of ribosomally produced antimicrobials called bacteriocins (71, 72). For example, S. salivarius produces a wide range of bacteriocins, which is a major mechanism that antagonizes S. pneumoniae (71, 73–75). More recently, it is also demonstrated that S. salivarius reduces the S. pneumoniae colonization by blocking the adhesion sites, suggesting multiple mechanisms used by this commensal to inhibit pathogens (76). Apart from ribosomally encoded bacteriocins, commensal bacteria encode non-ribosomally produced bioactive antimicrobials to compete with pathogens (77). Zipperer et al. showed that the nasal commensal Staphylococcus lugdunensis directly inhibits the growth of S. aureus through a novel cyclic antimicrobial peptide named “Lugdunin.” Lugdunin possessed bactericidal activity against all tested strains of S. aureus in vitro. Moreover, in animal model, the co-colonization of S. lugdunensis and S. aureus resulted in competitive exclusion of S. aureus (77). The use of purified antimicrobials or bacterial strains encoding antimicrobials may serve as a source of new generation of antibiotics to deal with multidrug resistant strains, such as methicillin resistant S. aureus. One mechanism, which contributes to competitive advantage for colonization of commensal bacteria to preserve their niche and to suppress the growth of pathogens, includes the production of hydrogen peroxide. Epidemiological data show a negative correlation between S. pneumoniae and S. aureus and presumably, the reason for increased S. aureus related otitis media after use of pneumococcal vaccine (78, 79). One possible mechanism implicated to define this negative association is hydrogen peroxide mediated inhibition of S. aureus by pneumococcal hydrogen peroxide (80).
Nutrient competition is also a strategy used by commensal bacteria to reduce the fitness of pathogens by competing for the same pool of resources (81). Stubbendieck et al. recently showed that isolates of Corynebacterium spp. inhibited Staphylococcus spp. in vitro. This inhibition was due to reduced iron bioavailability, mediated by siderophore–induced sequestration of iron by Corynebacterium spp. (82). Another novel mechanism of commensal mediated inhibition is through the production of secreted enzymes. Iwase et al. first demonstrated the negative correlation between the commensal Staphylococcus epidermidis and pathogenic S. aureus in human nasal samples. To gain further insight to explain this negative association, they identified the inhibitory factor produced by S. epidermidis as serine protease, which inhibits the biofilm formation and human nasal colonization by S. aureus (68). Follow-up study from the same group showed that intranasal colonization of mice with serine protease producing S. epidermidis inhibited colonization with methicillin resistant S. aureus (83). Commensal bacteria also exploit the host resources to generate metabolic compounds with antimicrobial properties to suppress the growth of respiratory pathogens. An elegant study by Bomar et al. investigated the mechanistic explanation for correlation between increased abundance of Corynebacterium species and reduced S. pneumoniae colonization (69). Interestingly, they found that Corynebacterium accolens encodes lipase, which catalyzes the hydrolysis of host triacylglycerolsto to produce free fatty acids with antibacterial properties that suppress the growth of S. pneumoniae (69). Taken together, the above examples evidently suggest that antagonistic interactions exist in the polymicrobial community utilizing wide range of mechanisms by which commensal bacteria inhibit respiratory pathogens. Advanced understanding of existing mechanisms using both in vitro and in vivo models and further elucidation of novel mechanisms may enable us to exploit commensals to inhibit respiratory pathogens. Mechanisms used by commensal bacteria to directly inhibit/kill respiratory pathogens are exemplified in Table 1.
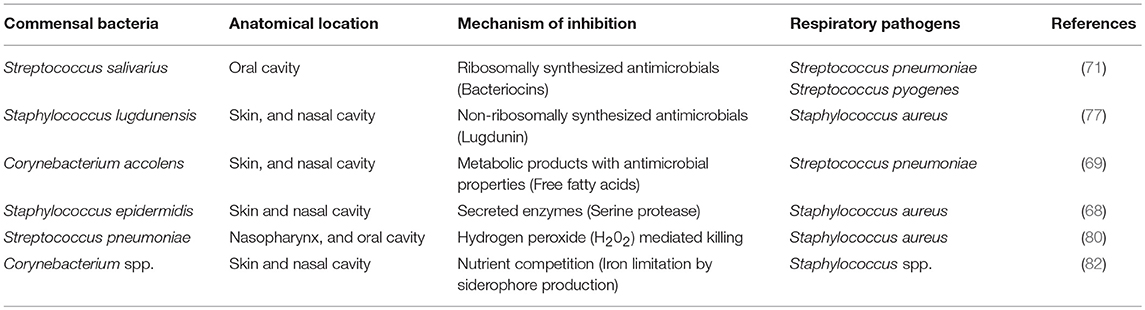
Table 1. Examples of direct mechanisms of colonization resistance used by commensal bacteria against respiratory pathogens.
Advanced research technologies have been applied to evaluate the contribution of commensal bacteria to respiratory infections. Accumulating evidence indicates an important role for commensal bacteria in defense against respiratory pathogens, which paves the way to target these bacteria for the development of vaccines and therapeutics that provide optimal protection with safety and low cost. Moreover, the use of modern experimental tools to decipher the novel mechanisms used by commensals to inhibit pathogens may assist in designing novel therapeutics with targeted approach focusing exclusively on the pathogen inhibition without disrupting the homeostatic microbial community. Future studies are required to address the following questions: (1) What are the underlying mechanisms by which the trio of commensals, pathogens, and host interact with each other? (2) What could be the long-term consequences of using commensal bacteria-based vaccines/therapeutics on the host, pathogens, and the microbiota? (3) What are the effects of medical manipulations, such as antibiotics and probiotics, on the biology of commensal bacteria? (4) How can we use commensal bacteria-expressed bacteriocins for protection against respiratory pathogens? (5) Which specific commensal bacterial species of the microbiota are directly involved in protection immunity to different pathogens? (6) How can we use commensal microbiota/bacteria to correct dysbiosis? A sincere exploration of these questions may have implications for the clinical use of commensal bacteria with inhibitory properties against pathogens. This may be important to bypass the drawbacks associated with currently available options, such as antimicrobial resistance.
RK, FP, and SS wrote and revised the manuscript. All authors assisted in the conception of this review and acquisition of relevant literature. All authors gave approval of the last version to be published.
This work was supported by a grant from the Norwegian Research Council (Grant number - 241011).
The authors declare that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
2. Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, et al. The human microbiome project: a community resource for the healthy human microbiome. PLoS Biol. (2012) 10:e1001377. doi: 10.1371/journal.pbio.1001377
3. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
4. Wang BH, Yao MF, Lv LX, Ling ZX, Li LJ. The human microbiota in health and disease. Eng Prc. (2017) 3:71–82. doi: 10.1016/J.ENG.2017.01.008
5. Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. (2013) 13:321–35. doi: 10.1038/nri3430
6. Abt MC, Pamer EG. Commensal bacteria mediated defenses against pathogens. Curr Opin Immunol. (2014) 29:16–22. doi: 10.1016/j.coi.2014.03.003
7. Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. (2013) 13:790–801. doi: 10.1038/nri3535
8. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. (2015) 26:26191. doi: 10.3402/mehd.v26.26191
9. Epaud R. An update on paediatric respiratory diseases. Eur Respir Rev. (2018) 27:180013. doi: 10.1183/16000617.0013-2018
10. Alimi Y, Lim WS, Lansbury L, Leonardi-Bee J, Nguyen-Van-Tam JS. Systematic review of respiratory viral pathogens identified in adults with community-acquired pneumonia in Europe. J Clin Virol. (2017) 95:26–35. doi: 10.1016/j.jcv.2017.07.019
11. Fogel N Tuberculosis: A disease without boundaries. Tuberculosis. (2015) 95:527–531. doi: 10.1016/j.tube.2015.05.017
12. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. (2011) 108:5354–9. doi: 10.1073/pnas.1019378108
13. Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, et al. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol. (2012) 188:1411–20. doi: 10.4049/jimmunol.1101682
14. Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. (2012) 37:158–70. doi: 10.1016/j.immuni.2012.04.011
15. Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun. (2017) 8:1512. doi: 10.1038/s41467-017-01803-x
16. Clarke TB Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun. (2014) 82:4596–606. doi: 10.1128/IAI.02212-14
17. Gauguet S, D'Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, et al. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun. (2015) 83:4003–14. doi: 10.1128/IAI.00037-15
18. Lankelma JM, Birnie E, Weehuizen AF, Scicluna BP, Belzer C, Houtkooper RH, et al. The gut microbiota as a modulator of innate immunity during melioidosis. PLoS Neglect Trop D. (2017) 11:e0005548. doi: 10.1371/journal.pntd.0005548
19. McAleer JP, Nguyen LH, Chen K, Kumar P, Ricks DM, Binnie M, et al. Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J Immunol. (2016) 197:97–107. doi: 10.4049/jimmunol.1502566
20. Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol. (2015) 6:1085. doi: 10.3389/fmicb.2015.01085
21. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–83. doi: 10.1136/gutjnl-2015-309728
22. Wu S, Jiang ZY, Sun YF, Yu B, Chen J, Dai CQ, et al. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza A virus infection. Curr Microbiol. (2013) 67:414–22. doi: 10.1007/s00284-013-0380-z
23. Chen LW, Chen PH, Hsu CM. Commensal microflora contribute to host defense against escherichia coli pneumonia through Toll-Like Receptors. Shock. (2011) 36:67–75. doi: 10.1097/SHK.0b013e3182184ee7
24. Khan N, Vidyarthi A, Nadeem S, Negi S, Nair G, Agrewala JN. Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front Immunol. (2016) 7:529. doi: 10.3389/fimmu.2016.00529
25. Dumas A, Corral D, Colom A, Levillain F, Peixoto A, Hudrisier D, et al., The host microbiota contributes to early protection against lung colonization by Mycobacterium tuberculosis. Front Immunol. (2018) 9:2656. doi: 10.3389/fimmu.2018.02656
26. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. (2017) 171:1015–28.e13. doi: 10.1016/j.cell.2017.09.016
27. Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. (2018) 9:1534. doi: 10.3389/fphys.2018.01534
28. Mathieu E, Escribano-Vazquez U, Descamps D, Cherbuy C, Langella P, Riffault S, et al. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front Physiol. (2018) 9:1168. doi: 10.3389/fphys.2018.01168
29. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. (2010) 5:e8578. doi: 10.1371/journal.pone.0008578
30. Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, et al. Application of a neutral community model to assess structuring of the human lung microbiome. MBio. (2015) 6:e02284–14. doi: 10.1128/mBio.02284-14
31. Le Noci V, Guglielmetti S, Arioli S, Camisaschi C, Bianchi F, Sommariva M, et al. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: a strategy to promote immunosurveillance against lung metastases. Cell Rep. (2018) 24:3528–38. doi: 10.1016/j.celrep.2018.08.090
32. Zarogoulidis P, Kioumis I, Porpodis K, Spyratos D, Tsakiridis K, Huang H, et al. Clinical experimentation with aerosol antibiotics: current and future methods of administration. Drug Des Devel Ther. (2013) 7:1115–34. doi: 10.2147/DDDT.S51303
33. Iwabuchi N, Xiao JZ, Yaeshima T, Iwatsuki K. Oral administration of bifidobacterium longum ameliorates influenza virus infection in mice. Biol Pharm Bull. (2011) 34:1352–5. doi: 10.1248/bpb.34.1352
34. Kawase M, He F, Kubota A, Yoda K, Miyazawa K, Hiramatsu M. Heat-killed Lactobacillus gasseri TMC0356 protects mice against influenza virus infection by stimulating gut and respiratory immune responses. FEMS Immunol Med Microbiol. (2012) 64:280–8. doi: 10.1111/j.1574-695X.2011.00903.x
35. Zelaya H, Tada A, Vizoso-Pinto MG, Salva S, Kanmani P, Aguero G, et al. Nasal priming with immunobiotic Lactobacillus rhamnosus modulates inflammation-coagulation interactions and reduces influenza virus-associated pulmonary damage. Inflamm Res. (2015) 64:589–602. doi: 10.1007/s00011-015-0837-6
36. Waki N, Yajima N, Suganuma H, Buddle BM, Luo D, Heiser A, et al. Oral administration of Lactobacillus brevis KB290 to mice alleviates clinical symptoms following influenza virus infection. Lett Appl Microbiol. (2014) 58:87–93. doi: 10.1111/lam.12160
37. Chiba E, Tomosada Y, Vizoso-Pinto MG, Salva S, Takahashi T, Tsukida K, et al. Immunobiotic Lactobacillus rhamnosus improves resistance of infant mice against respiratory syncytial virus infection. Int Immunopharmacol. (2013) 17:373–82. doi: 10.1016/j.intimp.2013.06.024
38. Tomosada Y, Chiba E, Zelaya H, Takahashi T, Tsukida K, Kitazawa H, et al. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. (2013) 14:40. doi: 10.1186/1471-2172-14-40
39. Kawahara T, Takahashi T, Oishi K, Tanaka H, Masuda M, Takahashi S, et al. Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol Immunol. (2015) 59:1–12. doi: 10.1111/1348-0421.12210
40. Vieira AT, Rocha VM, Tavares L, Garcia CC, Teixeira MM, Oliveira SC, et al. Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 5(1A). Microbes Infect. (2016) 18:180–9. doi: 10.1016/j.micinf.2015.10.008
41. Shekhar S, Khan R, Schenck K, Petersen FC. Intranasal immunization with the commensal Streptococcus mitis confers protective immunity against pneumococcal lung infection. Appl Environ Microbiol. (2019). 85:e02235–18. doi: 10.1128/AEM.02235-18
42. Majlessi L, Sayes F, Bureau JF, Pawlik A, Michel V, Jouvion G, et al. Colonization with Helicobacter is concomitant with modified gut microbiota and drastic failure of the immune control of Mycobacterium tuberculosis. Mucosal Immunol. (2017) 10:1178–89. doi: 10.1038/mi.2016.140
43. Arnold IC, Hutchings C, Kondova I, Hey A, Powrie F, Beverley P, et al. Helicobacter hepaticus infection in BALB/c mice abolishes subunit-vaccine-induced protection against M. tuberculosis. Vaccine. (2015) 33:1808–14. doi: 10.1016/j.vaccine.2015.02.041
44. La Mantia I, Varricchio A, Ciprandi G. Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for preventing recurrent acute otitis media in children: a real-life clinical experience. Int J Gen Med. (2017) 10:171–5. doi: 10.2147/IJGM.S137614
45. Marchisio P, Santagati M, Scillato M, Baggi E, Fattizzo M, Rosazza C, et al. Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis-prone children. Eur J Clin Microbiol Infect Dis. (2015) 34:2377–83. doi: 10.1007/s10096-015-2491-x
46. Tano K, Hakansson EG, Holm SE, Hellstrom S. A nasal spray with alpha-haemolytic streptococci as long term prophylaxis against recurrent otitis media. Int J Pediatr Otorhi. (2002) 62:17–23. doi: 10.1016/S0165-5876(01)00581-X
47. Evans CM, Pratt CB, Matheson M, Vaughan TE, Findlow J, Borrow R, et al. Nasopharyngeal colonization by Neisseria lactamica and induction of protective immunity against Neisseria meningitidis. Clin Infect Dis. (2011) 52:70–7. doi: 10.1093/cid/ciq065
48. Deasy AM, Guccione E, Dale AP, Andrews N, Evans CM, Bennett JS, et al. Nasal inoculation of the commensal neisseria lactamica inhibits carriage of neisseria meningitidis by young adults: a controlled human infection study. Clin Infect Dis. (2015) 60:1512–20. doi: 10.1093/cid/civ098
49. Henriques-Normark B, Normark S. Commensal pathogens, with a focus on Streptococcus pneumoniae, and interactions with the human host. Exp Cell Res. (2010) 316:1408–14. doi: 10.1016/j.yexcr.2010.03.003
50. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. (2015) 17:704–15. doi: 10.1016/j.chom.2015.03.008
51. de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. (2016) 194:1104–15. doi: 10.1164/rccm.201602-0220OC
52. Arboleya S, Sanchez B, Milani C, Duranti S, Solis G, Fernandez N, et al. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr. (2015) 166:538–44. doi: 10.1016/j.jpeds.2014.09.041
53. Kuppala VS, Meinzen-Derr J, Morrow AL, Schibler KR. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J Pediatr. (2011) 159:720–5. doi: 10.1016/j.jpeds.2011.05.033
54. Man WH, de Steenhuijsen Piters WA, Bogaert D The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. (2017) 15:259–70. doi: 10.1038/nrmicro.2017.14
55. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. (2012) 37:171–86. doi: 10.1016/j.immuni.2012.05.020
56. Wang J, Li F, Sun R, Gao X, Wei H, Li LJ, et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat Commun. (2013) 4:2106. doi: 10.1038/ncomms3106
57. Shekhar S, Khan R, Ferreira DM, Mitsi E, German E, Rorvik GH, et al. Antibodies reactive to commensal Streptococcus mitis show cross-reactivity with virulent Streptococcus pneumoniae serotypes. Front Immunol. (2018) 9:747. doi: 10.3389/fimmu.2018.00747
58. Shekhar S, Schenck K, Petersen FC. Exploring host-commensal interactions in the respiratory tract. Front Immunol. (2017) 8:1971. doi: 10.3389/fimmu.2017.01971
59. Skov Sorensen UB, Yao K, Yang Y, Tettelin H, Kilian M. Capsular polysaccharide expression in commensal streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae. MBio. (2016) 7: e01844–16. doi: 10.1128/mBio.01844-16
60. Engen SA, Rukke HV, Becattini S, Jarrossay D, Blix IJ, Petersen FC, et al. The oral commensal streptococcus mitis shows a mixed memory Th cell signature that is similar to and cross-reactive with Streptococcus pneumoniae. PLoS ONE. (2014) 9:e104306. doi: 10.1371/journal.pone.0104306
61. Dumas A, Bernard L, Poquet Y, Lugo-Villarino G, Neyrolles O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. (2018) 20:e12966. doi: 10.1111/cmi.12966
62. Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. (2012) 3:463–7. doi: 10.4161/gmic.21288
63. Sze MA, Tsuruta M, Yang SW, Oh Y, Man SF, Hogg JC, et al. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS ONE. (2014) 9:e111228. doi: 10.1371/journal.pone.0111228
64. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. (2013) 14:668–75. doi: 10.1038/ni.2635
65. Felix KM, Jaimez IA, Nguyen TV, Ma H, Raslan WA, Klinger CN, et al. Gut microbiota contributes to resistance against pneumococcal pneumonia in immunodeficient Rag(-/-) mice. Front Cell Infect Microbiol. (2018) 8:118. doi: 10.3389/fcimb.2018.00118
66. Gray J, Oehrle K, Worthen G, Alenghat T, Whitsett J, Deshmukh H. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med. (2017) 9:eaaf9412. doi: 10.1126/scitranslmed.aaf9412
67. Stubbendieck RM, Straight PD. Multifaceted interfaces of bacterial competition. J Bacteriol. (2016) 198:2145–55. doi: 10.1128/JB.00275-16
68. Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. (2010) 465:346–U100. doi: 10.1038/nature09074
69. Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. Mbio. (2016) 7:e01725–15. doi: 10.1128/mBio.01725-15
70. Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus shifts toward commensalism in response to corynebacterium species. Front Microbiol. (2016) 7:1230. doi: 10.3389/fmicb.2016.01230
71. Santagati M, Scillato M, Patane F, Aiello C, Stefani S. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol Med Microbiol. (2012) 65:23–31. doi: 10.1111/j.1574-695X.2012.00928.x
72. Janek D, Zipperer A, Kulik A, Krismer B, Peschel A. High frequency and diversity of antimicrobial activities produced by nasal staphylococcus strains against bacterial competitors. PLoS Pathog. (2016) 12:e1005812. doi: 10.1371/journal.ppat.1005812
73. Tagg JR Streptococcal bacteriocin-like inhibitory substances: some personal insights into the bacteriocin-like activities produced by streptococci good and bad. Probiot Antimicro. (2009) 1:60–6. doi: 10.1007/s12602-008-9002-7
74. Walls T, Power D, Tagg J. Bacteriocin-like inhibitory substance (BLIS) production by the normal flora of the nasopharynx: potential to protect against otitis media? J Med Microbiol. (2003) 52:829–33. doi: 10.1099/jmm.0.05259-0
75. Wescombe PA, Heng CK, Burton JP, Chilcott CN, Tagg JR. Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future Microbiol. (2009) 4:819–35. doi: 10.2217/fmb.09.61
76. Manning J, Dunne EM, Wescombe PA, Hale JD, Mulholland EK, Tagg JR, et al. Investigation of Streptococcus salivarius-mediated inhibition of pneumococcal adherence to pharyngeal epithelial cells. BMC Microbiol. (2016) 16:225. doi: 10.1186/s12866-016-0843-z
77. Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. (2016) 535:511–6. doi: 10.1038/nature18634
78. Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in children. Jama-J Am Med Assoc. (2004) 292:716–20. doi: 10.1001/jama.292.6.716
79. Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rumke HC, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. (2004) 363:1871–2. doi: 10.1016/S0140-6736(04)16357-5
80. Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol. (2006) 188:4996–5001. doi: 10.1128/JB.00317-06
81. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. (2013) 14:685–90. doi: 10.1038/ni.2608
82. Stubbendieck RM, May DS, Chevrette MG, Temkin MI, Wendt-Pienkowski E, Cagnazzo J, et al. Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl Environ Microbiol. (2018) AEM.02406–18. doi: 10.1128/AEM.02406-18
Keywords: host, commensal, pathogen, lungs, vaccine
Citation: Khan R, Petersen FC and Shekhar S (2019) Commensal Bacteria: An Emerging Player in Defense Against Respiratory Pathogens. Front. Immunol. 10:1203. doi: 10.3389/fimmu.2019.01203
Received: 01 February 2019; Accepted: 13 May 2019;
Published: 31 May 2019.
Edited by:
Laurel L. Lenz, University of Colorado Denver, United StatesReviewed by:
Theresa Alenghat, Cincinnati Children's Hospital Medical Center, United StatesCopyright © 2019 Khan, Petersen and Shekhar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rabia Khan, cmFiaWFfbmFlZW1raGFuQGhvdG1haWwuY29t; Sudhanshu Shekhar, c3VkaGFuc2h1LnNoZWtoYXJAb2RvbnQudWlvLm5v
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.