- 1Shandong Provincial Key Laboratory of Animal Biotechnology and Disease Control and Prevention, College of Animal Science and Technology, Shandong Agricultural University, Tai'an, China
- 2College of Life Sciences, Shandong Agricultural University, Tai'an, China
Circular RNAs (circRNAs) are a class of endogenous noncoding RNA, which is different from linear RNA. CircRNA is an RNA molecule with a closed loop structure formed by reverse splicing. CircRNAs have been studied in several organisms, however, the circRNAs associated with the response to Salmonella enterica serovar Enteritidis (SE) inoculation in chickens are still unclear. In the current study, Jining Bairi chickens were inoculated with SE. CircRNAs involved in the response to SE inoculation were identified through next-generation sequencing. Our results showed that there were 5,118 circRNAs identified in the control and treated groups. There were 62 circRNAs significantly differentially expressed following SE inoculation. Functional classification revealed that those significantly differentially expressed circRNAs were associated with immune system process, rhythmic process and signaling following SE inoculation. CircRNAs NC_006091.4: 65510578|65515090, NC_006099.4: 16132825|16236906, and NC_006099.4: 15993284|16006290 play important roles in the response to SE inoculation. The findings in the current study provide evidence that circRNA alterations are involved in the response to SE inoculation in the chicken.
Introduction
Salmonella enterica serovar Enteritidis (SE) is one of the most common serotypes of the Salmonella bacteria reported worldwide and is the primary source of human intestinal infection (1). From the total confirmed Salmonella infections, 18% were caused by SE, and the incidence was 2.83 per 100,000 people in the United States (2). Egg-related salmonellosis costs $44 million per year in Australia (3). SE has a close relationship with the chicken, as poultry meat and egg are regarded as the main source of human foodborne infection (4). SE infection is mainly caused by oral intake of contaminated feed or water. SE can enter the bloodstream and colonize the internal organs. The cecum is the main site of Salmonella colonization (5). Many studies showed that genetic selection is an efficient way to control Salmonella infection in the chicken (6).
Circular RNAs (CircRNAs), a novel type of noncoding RNAs, compose a class of RNA developing covalently closed loop structures without 5′-3′ polarities (7) but with widespread, abundant, and tissue-specific expression across animals (8). To date, circRNA can be divided into three types, namely, intronic circRNA, exonic circRNA and exon-intron circRNA (9–11). Many circRNAs comprise one to several exons of protein-coding genes (12). CircRNA production may occur posttranscriptionally (13). Several functions have been identified for circRNAs, including transcription regulation, RNA transport (14), protein binding (15), and regulation of translation (9). CircRNAs were identified as efficient microRNA (miRNA) sponges (16, 17). Moreover, circRNAs have been found to be associated with diseases, including Alzheimer's disease, colorectal and ovarian cancer, idiopathic lung fibrosis and hepatocellular carcinoma in humans (12, 18, 19). Recent studies indicated that circular RNA expression alterations were enriched and stable in exosomes and could be a promising biomarker for cancer diagnosis (20, 21). It has been reported that circRNA alterations are involved in resistance to ALV-J-induced tumor formation in the chicken (22).
The technological breakthroughs in high-throughput deep sequencing and functional genome promote the study of circRNAs (23). A large number of circRNAs has been identified in Archaea, mice, and humans (10, 24). However, the identification of circRNAs related to certain traits in chickens is limited. In the current study, next-generation sequencing was used to detect circRNAs involved in the response to SE inoculation in the chicken. The study will lay the foundation in which circRNAs may be used as molecular markers related to the host response to SE inoculation in chickens.
Materials and Methods
Animals and SE Inoculation
The Jining Bairi Chicken, a China local chicken breed, used in the current study was provided by Shandong Bairi Chicken Breeding Co., Ltd (Jining, Shandong, China). The SE strain (CVCC3377) used for the inoculation was purchased from China Veterinary Culture Collection Center (http://cvcc.ivdc.org.cn/). To make the inoculant, SE were enriched in LB broth at 37°C for 16 h, pelleted at 4,000 rpm for 5 min, and diluted with sterilized PBS to an adjusted OD value of 1. The concentration of SE in the inoculant was measured by the plating method. The experimental design of animal inoculation was described in detail previously (25). In brief, 168 2-day-old SE negative Jining Bairi chickens were randomly divided into 2 groups, 84 chickens in each group. Chickens in one group were orally inoculated with 0.3 mL inoculants of 109 cfu/ml as the treated (T) group, and chickens in another group were mock inoculated with the same amount of sterile PBS as the control (C) group. Twelve chickens from each of the T group and C group were euthanized by cervical dislocation for sample collection at 1, 3, 7, 14, 21, 28, and 35 days postinoculation (dpi). The cecum samples were frozen in liquid nitrogen and stored at −80°C until further RNA isolation. Samples collected at 7 dpi were selected for the current study based on the bacterial number in cecal content in the treated group. All animal procedures were approved by the Shandong Agricultural University Animal Care and Use Committee.
RNA Extraction and circRNA Sequencing
Four samples from each T and C groups at 7 dpi were randomly selected for RNA isolation. Total RNA was isolated from each sample using the TRIzol reagent (Invitrogen, Grand Island, NY) according to the manufacturer's instructions. The concentration of RNA sample was measured using a DS-11 Spectrophotometer (DeNovix, Wilmington, DE, USA). The integrity of the RNA sample was assessed by agarose gel electrophoresis. Three qualified RNA samples were selected in each group and encoded as C1, C2, C3 and T1, T2, T3 to construct the library. In total, six libraries were constructed. Subsequently, the RNA libraries were sequenced by Illumina HiSeq2500 platform according to the manufacturer's instructions at BioMarker Technologies (Beijing, China).
Data Analysis
Raw data were first processed using a custom Perl script. Clean reads were obtained after removing the adaptors, poly-N reads, and low quality reads and used for the downstream analysis. The clean reads were mapped to the chicken genome sequence (galGal 5.0) using the TopHat2 version 2.0.10 (26) and bowtie2 software (27). CircRNAs were predicted using CIRI (CircRNA Identifier) (28). Annotation of the circRNA was performed based on the following databases: Nr (NCBI nonredundant protein sequences), Pfam (protein family), KOG/COG (Cluster of Orthologous Groups of proteins), and Swiss-Prot (http://www.ebi.ac.uk/swissprot/). All the data have been deposited into the SRA database with an accession number of SRP158084.
Function Analysis and Identification of Differentially Expressed circRNAs
The raw junction reads for all the samples were normalized to the number of total mapped reads and log2 transformed. The significantly differentially expressed (SDE) circRNAs between the T and C groups were identified using the DESeq 2 (29). The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. Fold change > 2.0 and P < 0.05 were considered significant.
Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) biological pathway enrichment analysis for SDE circRNAs were performed using the KOBAS 3.0 software (http://kobas.cbi.pku.edu.cn/index.php) (30).
Correlation Between circRNAs, miRNAs, and Genes
miRNAs mediated by SDE circRNAs were predicted using miRanda (31). The number of miRNAs interacting with each circRNA and the number of circRNAs interacting with each miRNA were counted. The genes targeted by circRNA-mediated miRNAs were mapped to the chicken functional interaction network in the Reactome database using the Reactome FI network plugin in the Cytoscape 3.5.1 software (32).
Validation of circRNAs Expression Through qRT-PCR (Quantitative Real-Time PCR)
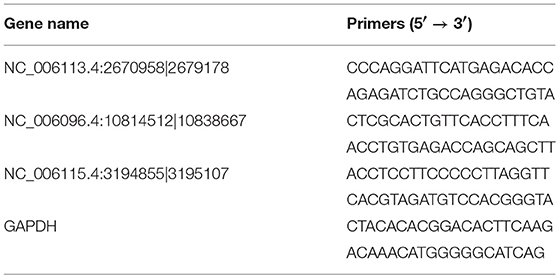
The RNA samples used for quantitative real-time PCR (qRT-PCR) validation were the same as those used for sequencing. A total amount of 10 μg RNA per sample were digested with 20 U RNase R (Epicenter, Chicago, IL) at 37°C for 1 h and purified with phenol/chloroform/isoamyl alcohol. One microgram of digested RNA was reverse transcribed into cDNA using a PrimeScriptTM RT reagent Kit with genomic DNA Eraser (Takara, Dalian, China) according to the manufacturer's instructions. The qRT-PCR was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) with a PCR mixture (20 μL) containing 10 μL SYBR Green qPCR Mix (2x), 0.5 μL (0.2 μM) forward primer, 0.5 μL (0.2 μM) reverse primer, 2 μL cDNA, and 7 μL ddH2O. The amplification conditions were as follows: 1 cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 5 s, 54°C for 15 s. GAPDH was used as the internal standard. All reactions were performed in triplicate. The sequences of specific outward-facing primers used in the qRT-PCR are listed in Table 1 and were synthesized by Sangon Biotech (Shanghai, China). The relative expression of the validated circRNA was analyzed using the 2−ΔΔCT method (33). The data were represented as the mean ± standard deviation. The student's t-test was used to assess the difference in expression of each validated circRNA between the two groups. P < 0.05 was considered significant.
Results
Data Quality and circRNAs Identification
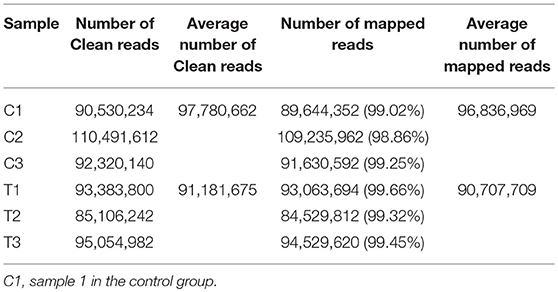
In total, 83.37 Gb clean data were obtained from the 6 samples. The percentage of bases with Q30 was 90.95%, 90.42%, 90.49% and 90.05%, 90.39%, 90.12% in C1, C2, C3 and T1, T2, T3 groups, respectively. The average number of clean reads in the C group and T group was 97,780,662 and 91,181,675, respectively. The clean reads from each sample were aligned with the chicken reference genome (galGal5.0); the rate of mapped reads was 98.86–99.66% (Table 2).
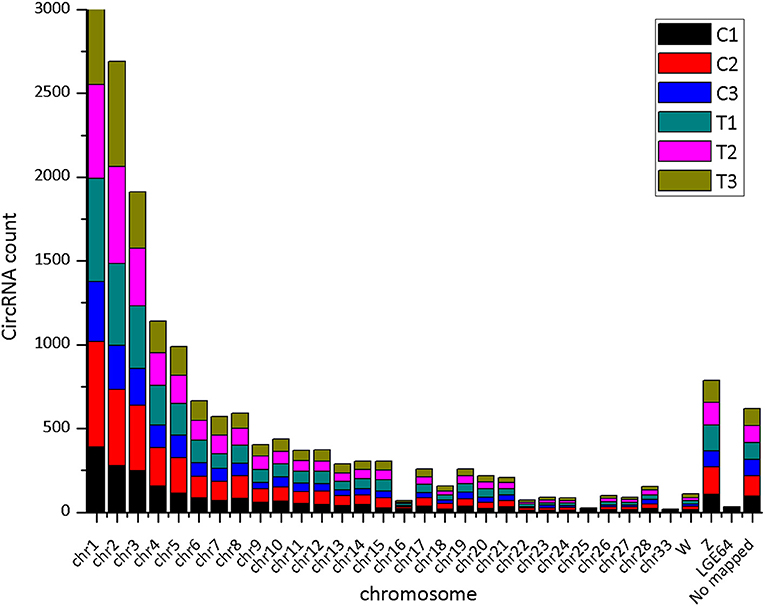
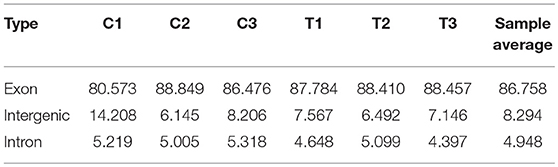
The CIRI software was used to predict circRNAs. There were 5,118 circRNAs identified across the six samples. The number of circRNAs in C1, C2, C3, T1, T2, and T3 was 2,239, 3,418, 2,091, 3,264, 2,989, and 2,893, respectively. The distribution of identified circRNAs on the chromosomes was not even. There were 96.7% circRNAs mapped on Chr1-28, Chr33, ChrW, ChrZ, and LGF64. The accumulative number of circRNAs across the six samples was more than 1,000 on the Chr1-4, 500-1,000 in Chr5-8 and ChrZ, 100-500 in Chr9-15, Chr17-21, Chr26, Chr28, and ChrW, 50-100 in Chr16, Chr22-24, and Chr27, and less than 50 in Chr25, Chr33, and ChrLGE64 (Figure 1). CircRNAs were mainly located in the exon, intergenic, and intron regions. There were 86.8% circRNAs mapped to the exon region (Table 3). The coverage of circRNAs (average length of the circRNA × number of circRNAs/the length of the chromosome) on each chromosome ranged from 0.003 to 1.479. The coverage of circRNAs on Chr16 was the largest (1.479). The coverage of circRNAs was >0.1 on Chr17 and ChrLEG64, < 0.01 on Chr25 and Chr33 (Supplementary File 1).
Differentially Expressed circRNAs Responding to SE Inoculation
The junction reads in each sample were counted as the expression level of circRNAs. The expression of circRNAs varied across different regions within each chromosome and was different between the T and C groups (Figure 2).
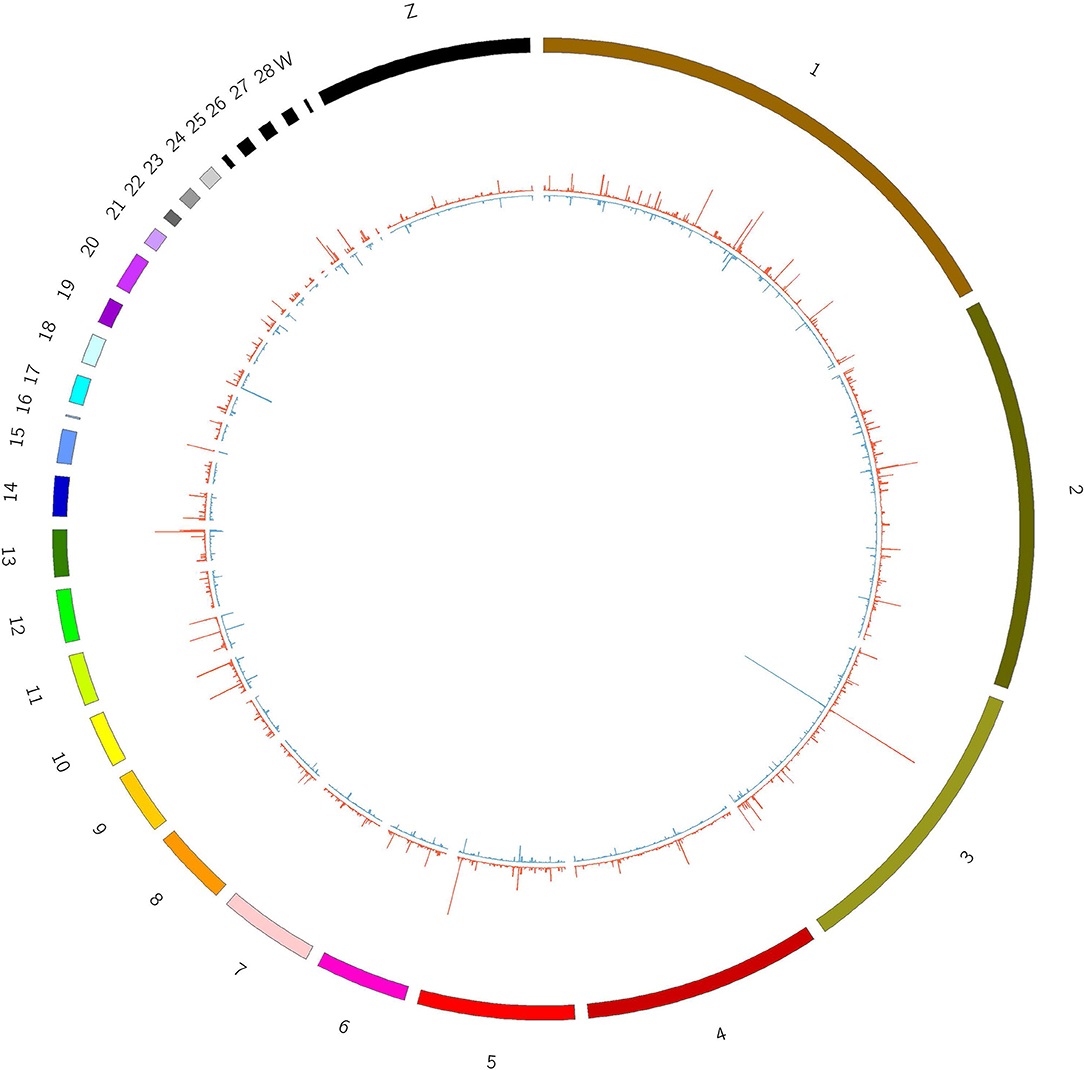
Figure 2. Expression of circRNAs in T and C groups. Outline corresponds to the reference genome. The inside corresponds to the average chromosome coverage across the C and T samples. Blue = C group, red = T group.
The SDE circRNAs between the treated and control groups were identified through the DESeq2 software. There were 62 SDE circRNAs between the two groups including 30 upregulated and 32 downregulated circRNAs (P < 0.05, fold change >2). There were more than 5 SDE circRNAs located on Chr1, 2, 3, and 4. There was one SDE circRNA located on Chr8, 14, 16, 17, 20, 24, 28 and W (Supplementary File 2).
The heatmap based on the expression of SDE circRNAs across the six samples showed that all the SDE circRNAs were clustered into 4 groups. Group 1 composed of upregulated circRNAs in the treated group including subgroups B and C, Group 2 composed of downregulated circRNAs in the treated group (subgroup D). Group 3 composed of circRNAs with low expression in both the treated and control groups (subgroup E). Group 4 composed of circRNAs with high expression in both the treated and control groups (subgroup A) (Figure 3).
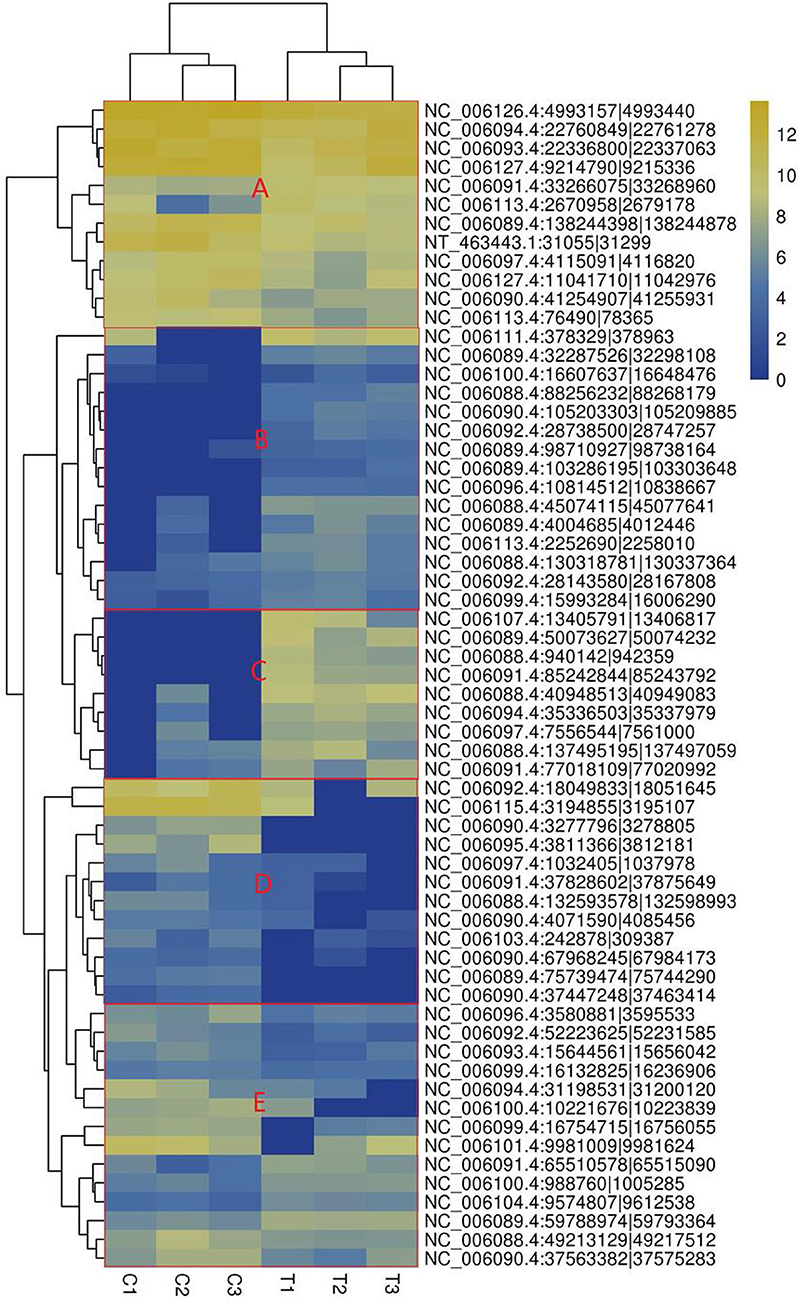
Figure 3. Heatmap of significantly differentially expressed circRNAs. (A) circRNAs with high expression in both the treated and control groups, (B) circRNAs highly expressed in the treated group with small difference, (C) circRNAs highly expressed in the treated group with great difference, (D) circRNAs with lower expression in the treated group. The columns represent different samples. The rows represent different circRNAs. The colors represent the level of expression of the circRNA in the sample (log2TPM). Yellow indicates higher circRNA expression level and blue shows lower circRNA expression level. T1, T2, and T3 are the samples in the T group; C1, C2, and C3 are the samples in the C group.
COG Function Classification of Parental Genes
The COG (Clusters of Orthologous Groups) function classification showed that the SDE circRNAs were associated with five categories: general function prediction only (R), transcription (K), replication, recombination and repair (L), signal transduction mechanisms (T), and posttranslational modification, protein turnover, chaperoned (O) with proportions of 26.32, 15.79, 15.79, 13.16, and 10.53%, respectively (Figure 4).
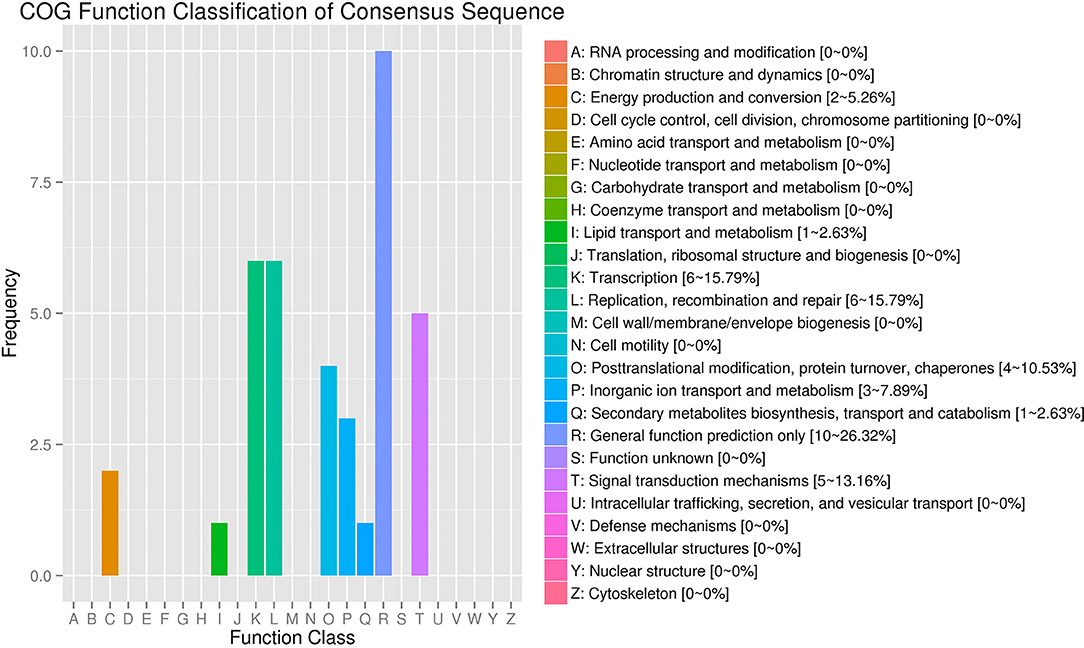
Figure 4. COG annotation of circRNA parental genes. The X-axis shows the COG function classification of the consensus sequence. The Y-axis shows the percentage of genes in each functional class.
GO and KEGG Pathway Analysis of Parental Genes
The parental genes of the SDE circRNAs were predicted. The GO and KEGG pathway analyses were performed for those parental genes. The results of BP (biological processes), MF (molecular functions), and CC (cellular components) were shown in Figure 5. For the GO-BP, the SDE circRNAs were associated with localization, biological adhesion, immune system process, reproductive process, growth, signaling, multi-organism process, rhythmic process, biological phase, and cell aggregation. In terms of the GO-CC, the SDE circRNAs were mainly located in the synapse, organelle and the macromolecular complex. For the GO-MF, the SDE circRNAs were associated with nucleic acid-binding transcription factor activity and protein-binding transcription factor activity (Figure 5). The CLOCK gene was the parental gene of circRNA NC_006091.4: 65510578|65515090 and was involved in the rhythmic process.
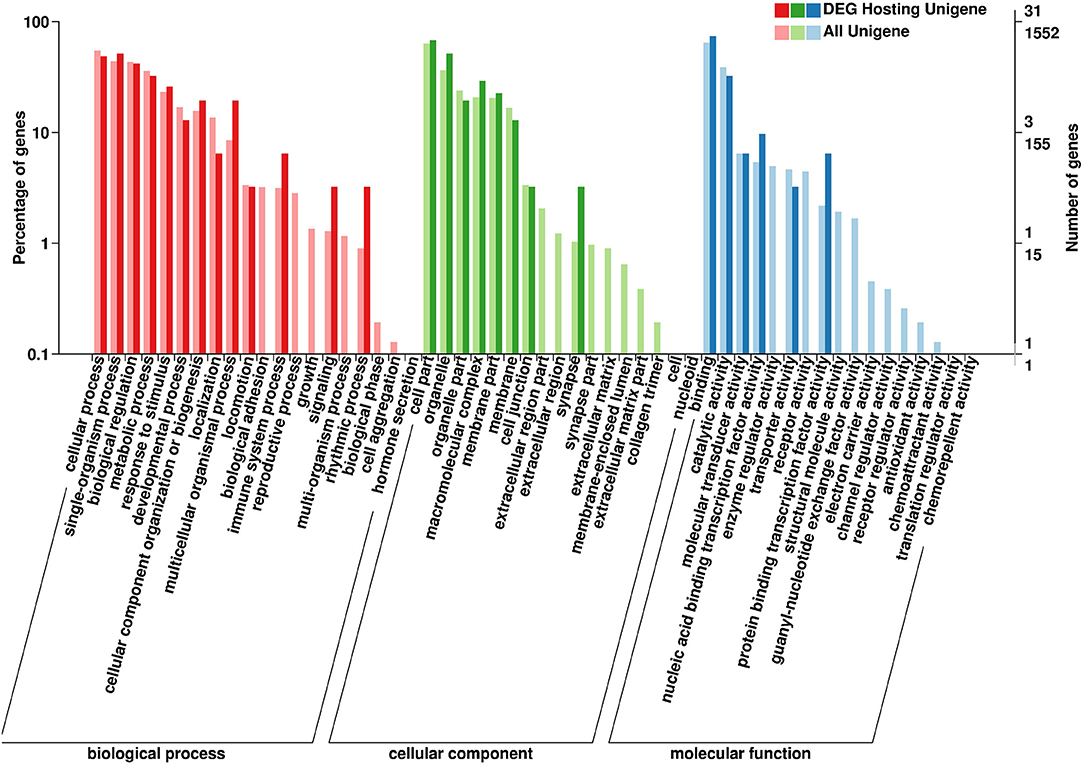
Figure 5. Gene Ontology (GO) annotation of circRNA parental genes. The X-axis shows the GO function classification. The Y-axis shows the percentage of genes (Left) and the number of genes (Right).
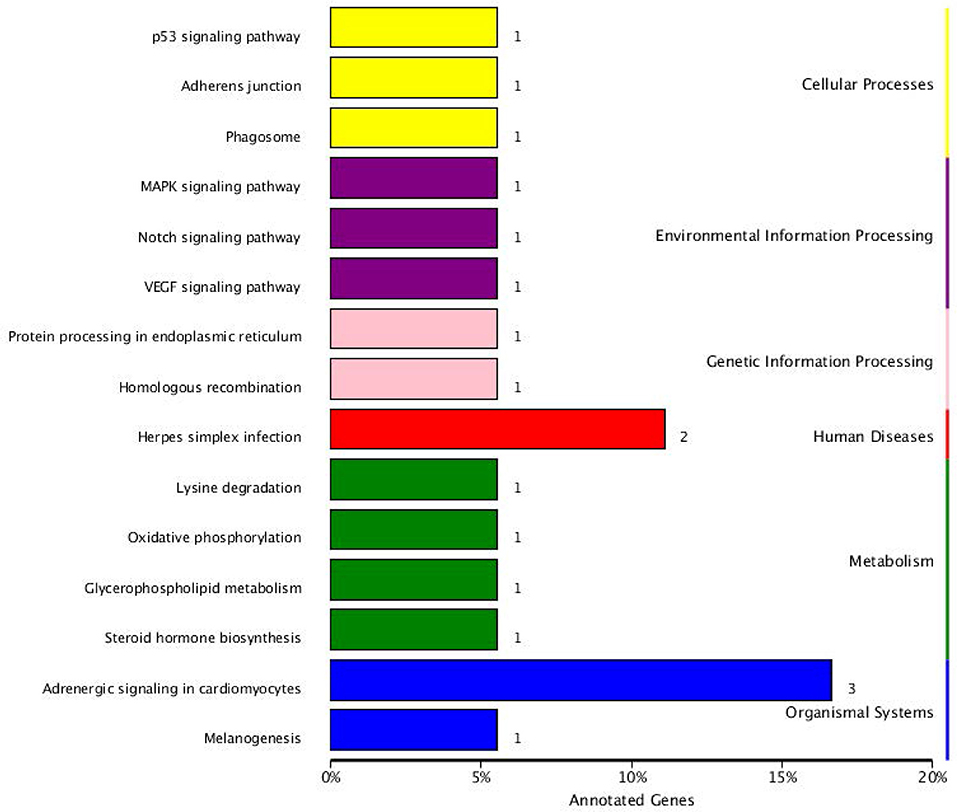
The KEGG pathway analysis results showed that the parental genes of those SDE circRNAs were enriched in 15 pathways. Those enriched pathways were divided into three categories. (1) The metabolism-related including oxidative phosphorylation pathway, lysine degradation, glycerophospholipid metabolism, and steroid hormone biosynthesis. (2) The immune-related including p53 signaling pathway, MAPK signaling pathway, Notch signaling pathway, VEGF signaling pathway, Herpes simplex infection, and Adrenergic signaling in cardiomyocytes. (3) Other pathways including Adherens junction, Phagosome, Protein processing in the endoplasmic reticulum, Homologous recombination and Melanogenesis. The RYR2, TPM1, and TPM2 genes were included in the Adrenergic signaling in cardiomyocytes pathway. The CLOCK and USP7 genes were included in the Herpes simplex infection pathway (Figure 6).
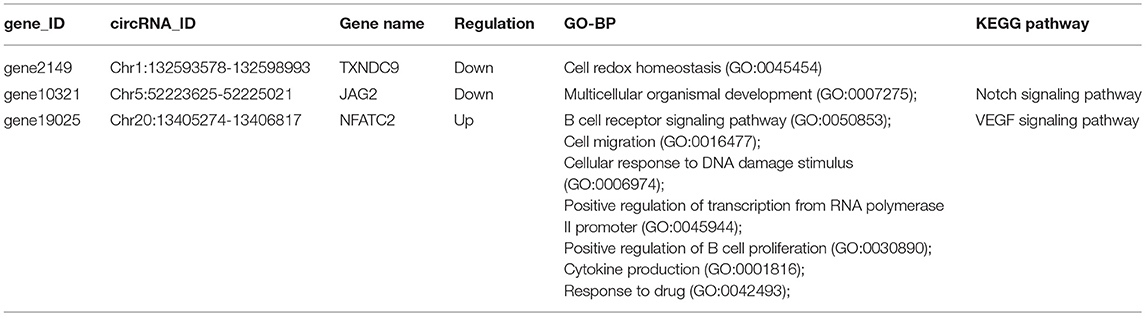
The genes included in the GO terms of the immune system were retrieved from BioMart (http://asia.ensembl.org/biomart). Three immune-related genes, TXNDC9, JAG2, and NFATC2, were parental genes of SDE circRNAs NC_006088.4:132593578|132598993, NC_006092.4:52223625|52225021, NC_006092.4:52223625|52231585, NC_006107.4:13405274|13406817, NC_006107.4:13405274|13430298, NC_006107.4:13405277|13406817, NC_006107.4:13405791|13406817, NC_006107.4:13405791|13430298, NC_006107.4:13405791|13448229, NC_006107.4:13419173|13420895, and NC_006107.4:13444867|13448229 and related to cell redox homeostasis, multicellular organismal development, the B cell receptor signaling pathway, cellular response to DNA damage stimulus, positive regulation of transcription from RNA polymerase II promoter, positive regulation of B cell proliferation, cytokine production, and response to drug biological process (Table 4).
Interaction Between circRNAs, miRNAs, and Genes
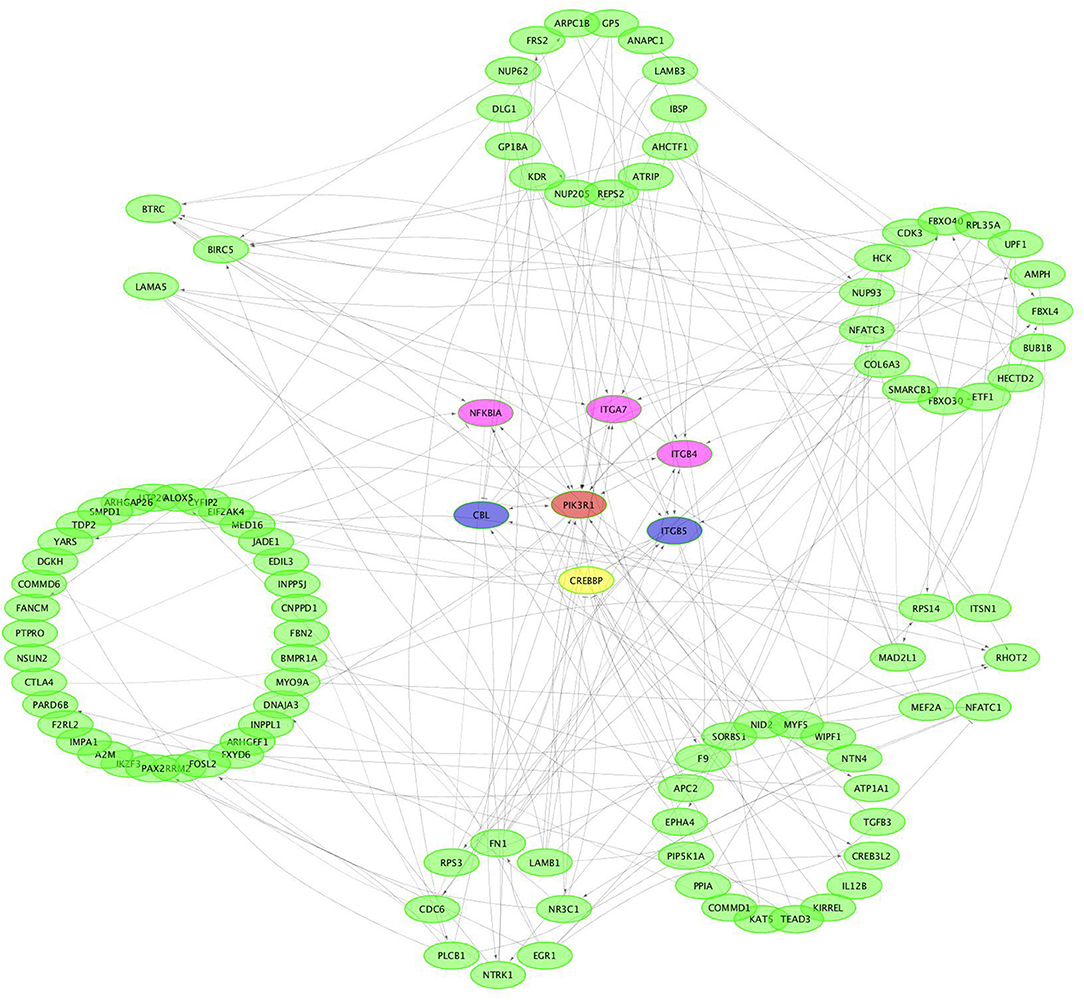
CircRNAs can act as miRNA sponges. The miRNAs interacting with the SDE circRNAs were predicted using miRanda. There were 1,787 interaction incidents identified between the 60 SDE circRNAs and 624 miRNAs (Supplementary File 3). The number of miRNAs interacting with different circRNAs was not even. CircRNA NC_006103.4:242878|309387, NC_006099.4:16132825|16236906, and NC_006100.4:16607637|16648476 interacted with more than 100 miRNAs. Ten circRNAs interacted with 50–100 miRNAs. Twenty-seven circRNAs interacted with 10–50 miRNAs. Twenty circRNAs interacted with < 10 miRNAs. CircRNA NC_006101.4:9981009|9981624 interacted with only one miRNA of gga-miR-1756a (Figure 7). One miRNA interacted with different circRNAs. Gga-miR-6545-3p, gga-miR-1696, gga-miR-1768, gga-miR-6553-5p, gga-miR-6573-5p, gga-miR-34a-5p, gga-miR-449c-5p, and gga-miR-6549-5p interacted with more than 10 circRNAs. One hundred and nineteen miRNAs interacted with 5–9 circRNAs. More than one third (212/624) miRNAs interacted with one circRNA. The immune function related gga-mir-34a-5p, located on Chr16, regulated 629 target genes. The interaction network of proteins encoded by those target genes was constructed through the Reactome FI network plug-in using Cytoscape 3.5.1. Seven proteins connected with more than 7 other proteins. PIK3R1 connected with 20 proteins, CBL and ITGB5 connected with 10 proteins, CREBBP connected with 9 proteins. NFKBIA, ITGAT, and ITGB4 connected with 8 proteins. Thirty-seven proteins connected with 4–7 other proteins, and 94 proteins connected with < 4 other proteins (Figure 8).
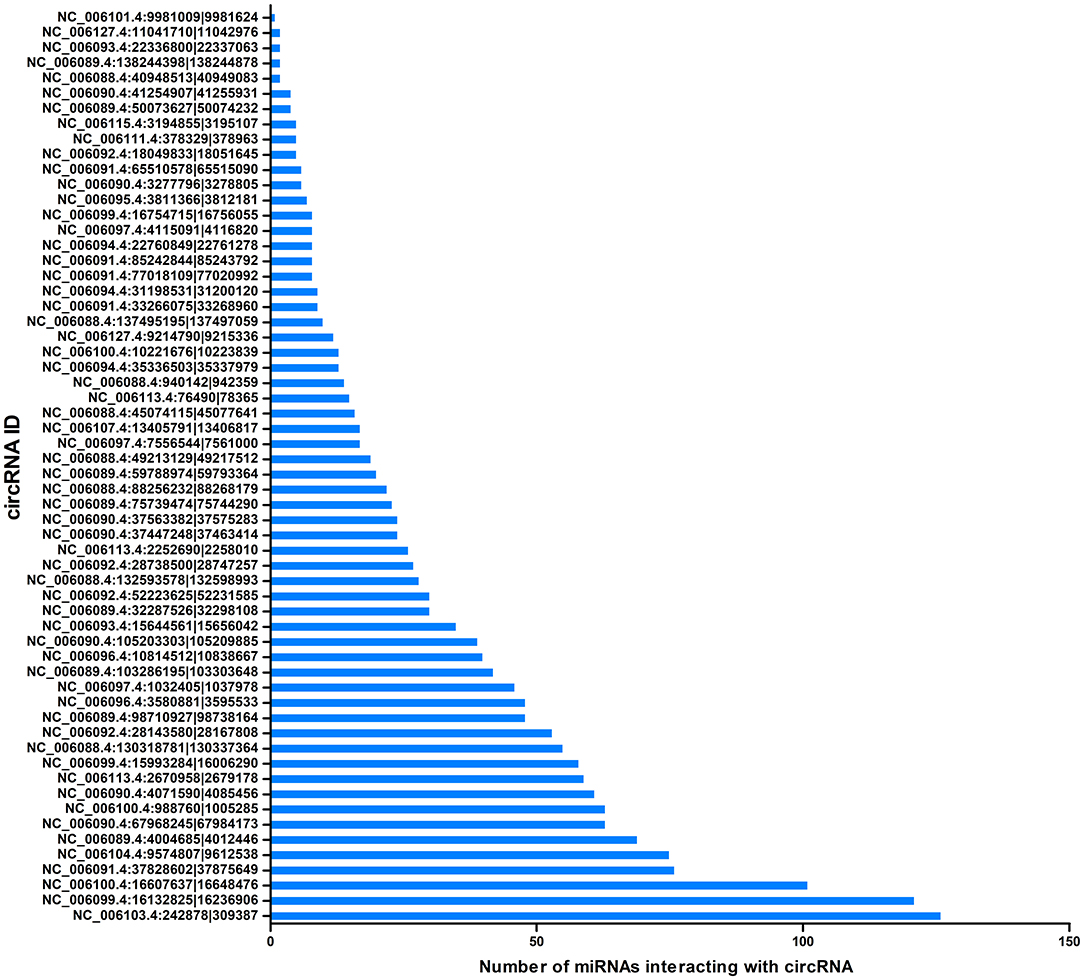
Figure 7. Correlation between miRNAs and circRNAs. The X-axis represents the number of miRNAs interacting with circRNA. The Y-axis represents the circRNA name.
Validation of circRNA Expression by qRT-PCR
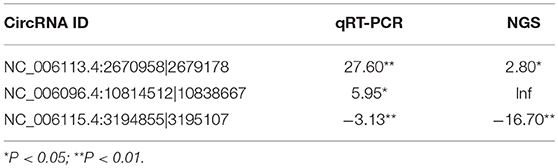
Three circRNAs including two upregulated (NC_006113.4:2670958|2679178, NC_006096.4:10814512|10838667) and one downregulated (NC_006115.4:3194855|3195107) were randomly selected for validation by qRT-PCR. The expression of each of all the validated circRNAs detected by qRT-PCR was consistent with that detected by the sequencing in terms of the regulatory direction and significance. CircRNAs NC_006113.4:2670958|2679178 and NC_006096.4:10814512|10838667 were upregulated with a fold change of 27.60 and 5.95 by qRT-PCR, respectively. CircRNA NC_006115.4:3194855|3195107 was downregulated with a fold change of 3.13 (Table 5).
Discussion
CircRNAs are recently discovered noncoding RNAs and have attracted significant attention (34). In the current study, circRNAs related to SE inoculation in chickens have been revealed through next-generation sequencing. A close number of upregulated and downregulated circRNAs were identified in several circRNAs profiling studies (18, 34). Similar results were found in the current study since the number of significantly upregulated and downregulated circRNAs was very close (30 vs. 32) in the chicken cecum following SE inoculation (Supplementary File 2). It has been reported that the number of upregulated and downregulated genes (115 vs. 37) was significantly different; however, the number of upregulated and downregulated miRNAs (22 vs. 15) following SE inoculation was similar (35, 36). When a persistent immunity to SE inoculation is established, the host immune signaling pathways can be modulated (37). Many immune-related functional terms and pathways have been found following Salmonella inoculation in the chicken (38–40). In the current study, only two immune-related functional terms (response to stimulus and immune system process) were enriched (Figure 5). The regulatory roles played by the genes, miRNAs, and circRNAs in the response to SE inoculation in the chicken were different. It has been reported that circadian rhythms can influence mammal immune response through regulating the blood circulation during diurnal sleeping/waking cycles (41, 42). Domestic pigs exhibit diurnal rhythms in peripheral blood immune cell numbers (43). It has been reported that the circadian rhythm was significantly enriched in the chicken cecum following Campylobacter jejuni (C. jejuni) infection (44). In the current study, the circadian rhythm-associated circRNAs were significantly triggered by SE inoculation. A different circadian rhythm regulation mechanism could exist in response to C. jejuni compared to that of SE inoculation.
Metabolism is important to facilitate the requirements for energy and biosynthesis and directly regulate immune cell functions. A bacterial infection competes for nutrients with immune cells (45). The differentially expressed circRNAs were enriched in metabolism-related KEGG pathways including the oxidative phosphorylation pathway, lysine degradation, glycerophospholipid metabolism, and steroid hormone biosynthesis following SE inoculation (Figure 6). Upon encountering an antigen, lymphocytes switch into the specific effector state with metabolism changes (46). Those metabolic changes are required not only for lymphocyte plasticity but also for T cell fate (47). Salmonella infection induces rapid and robust T-cell activation in mammalians (48). Changes in metabolic activity have been shown to intimately support T cell differentiation and effector functions (49). The oxidative phosphorylation pathway is the significant energy generating system in animals and it is highly conserved in insects and vertebrates (50). Naïve T cells rely on oxidative phosphorylation to maintain the energy demand; in contrast, activated T cells engage in aerobic glycolysis consuming massive amounts of glucose (51). Oxygen and reactive oxygen species metabolism were enriched following SE infection in chickens (35). The oxidative phosphorylation pathway is a prime candidate for cytonuclear genomic incompatibilities, and ATPases are composed of subunits from both the nuclear and mitochondrial genomes (52, 53). It has been reported that supplementation with lysine-yielding Bacillus subtilis in the diet increased intestinal immune response in Linwu ducks (54). It has been reported the balance between metabolism and the immune system contributes to the response to SE inoculation in chickens (36, 55).
CircRNAs were identified as efficient microRNA (miRNA) sponges (16, 17). Many studies have indicated that circRNAs regulate the function of miRNAs acting as competing endogenous RNAs (ceRNAs) (16, 56, 57). miR-143 and miR-26 are differentially expressed in whole blood after Salmonella inoculation in pigs (58). Gga-miR-101-3p and gga-miR-155 were identified as candidates potentially associated with SE infection in the chicken (59). It has been reported that gga-miR-125b-5p, gga-miR-34a-5p, gga-miR-1416-5p, and gga-miR-1662 play an important role in SE infection (55). miRNAs buffer and alter the variance of relatively low expressed genes in response to Salmonella infection in pigs (60). In the current study, gga-miR-125b-5p and gga-miR-34a-5p interacted with 3 and 10 circRNAs, respectively (Supplementary File 3). Proteins encoded by gga-miR-34a-5p-mediated genes had close interaction (Figure 8). The results showed that circRNA may have interacted with miRNA in the response to SE inoculation in chickens.
In poultry, circadian rhythms are generated from the transcription/translation-based oscillatory loop including Per2, Per3, CLOCK, and Bmal1 (61–63). Circadian disruptions have been well-documented in adverse effects on human health through influencing lipid and glucose homeostasis, inflammation, and cardiovascular functions (64). Studies have shown that a set of cytokines, IL-6, IL- 1β, IL-18, IL-2, TGF-β4, K60, and IL-8, and circadian clock genes (cry1/2, per2/3, Bmal1/2, and CLOCK) have a 24-h periodic expression pattern in response to bacterial colonization (65, 66). Furthermore, the CLOCK gene was involved in the herpes simplex infection pathway and related to human disease. The CLOCK gene was significantly changed post C. jejuni inoculation (67). In the current study, the SDE circRNA NC_006091.4:65510578|65515090 originated from the CLOCK gene was significantly upregulated (Supplementary File 2) and could play an important role in response to SE inoculation.
Forkhead box proteins (FOXP) are part of a large transcription factor family with diverse functions in development, metabolism, organogenesis, and cancer (68). FOXP1, a member of the “FOXP” subfamily, is an essential transcriptional regulator for B lymphopoiesis (69, 70) and the generation of quiescent naïve T cells during thymocyte development (71). Disruption of FOXP1 leads to cognitive dysfunction including intellectual disability and autism spectrum disorder together with language impairment (72). A gene could be spliced into one or more circRNAs (73). circ-SHKBP1 regulated the angiogenesis of glioma-exposed endothelial cells through the miR-544a/FOXP1 and miR-379/FOXP2 Pathways (74). The level of miR-152 and FOXP1 was inversely correlated in grade 3 and 4 ovarian tumor tissues (75). Two circRNAs NC_006099.4:16132825|16236906 and NC_006099.4:15993284|16006290 originated from FOXP1 were significantly expressed with reverse regulatory direction (Supplementary File 2). Those two circRNAs could regulate the response to SE inoculation through FOXP1 and miRNAs in the chicken. The mechanism of interaction between circRNAs and FOXP1 in the response to SE inoculation in the chicken needs to be further warranted.
Conclusions
In conclusion, circRNAs were involved in the response to SE inoculation in the chicken. CircRNAs associated with the immune system process, the rhythmic process and metabolic process contribute to the response to SE inoculation. CircRNAs NC_006091.4:65510578|65515090, NC_006099.4:16132825|16236906, and NC_006099.4:15993284|16006290 play critical roles in the response to SE inoculation. The findings herein will provide fundamental information on the mechanism of circRNAs regulating the response to SE inoculation in the chicken.
Ethics Statement
All animal procedures were approved by Shandong Agricultural University Animal Care and Use Committee.
Author Contributions
LZ and LLu performed the experiments, analyzed the data, and drafted the manuscript. LLn performed the experiments and collected samples. HT and XF provided chickens and helped to analyze the data. HL reviewed the manuscript. XL designed the experiment and reviewed the manuscript.
Funding
This project was supported by The National Natural Science Foundation of China (31601980, 31872343), the Shandong Modern Agricultural Industry & Technology System (SDAIT-11-02), the Funds of Shandong Double Tops Program (SYL2017YSTD12), the Shandong Provincial Natural Science Foundation, China (ZR2018MC026), the Shandong Province Agricultural Seed Project (2017LZN007), the Taishan Scholar and China Scholarship Council (201508370050).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01186/full#supplementary-material
Supplementary File 1. The coverage of circRNAs in each chromosome.
Supplementary File 2. Information of the significantly differentially expressed circRNAs.
Supplementary File 3. Correlation between circRNAs and miRNAs.
References
1. Karaffová V, Bobíková K, Husáková E, Levkut M, Herich R, Revajová V, et al. Interaction of TGF-β4 and IL-17 with IgA secretion in the intestine of chickens fed with E. faecium AL41 and challenged with S. enteritidis. Res Vet Sci. (2015) 100:75–9. doi: 10.1016/j.rvsc.2015.04.005
2. Hu L, Zhang G, Allard MW, Yao K, Stones R, Hoffmann M, et al. Complete Genome Sequences of Two Salmonella enterica subsp. enterica serovar enteritidis strains isolated from egg products in the United States. Genome Announce. (2017) 5:e00614-17. doi: 10.1128/genomeA.00614-17
3. Samiullah, Chousalkar K, Roberts JR, Sexton M, May D, Kiermeier A. Effects of egg shell quality and washing on Salmonella Infantis penetration. Int J Food Microbiol. (2013) 165:77–83. doi: 10.1016/j.ijfoodmicro.2013.05.002
4. Barrow PA, Jones MA, Smith AL, Wigley P. The long view: Salmonella–the last forty years. Avian Pathol. (2012) 41:413–20. doi: 10.1080/03079457.2012.718071
5. Chappell L, Kaiser P, Barrow P, Jones MA, Johnston C, Wigley P, et al. The immunobiology of avian systemic salmonellosis. Vet Immunol Immunopathol. (2009) 128:53–9. doi: 10.1016/j.vetimm.2008.10.295
6. Kaiser MG, Lamont SJ. Genetic line differences in survival and pathogen load in young layer chicks after Salmonella enterica serovar enteritidis exposure. Poultry Sci. (2001) 80:1105–8. doi: 10.1093/ps/80.8.1105
7. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
8. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdisc Rev RNA. (2015) 6:563–79. doi: 10.1002/wrna.1294
9. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
10. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. (2012) 40:3131–42. doi: 10.1093/nar/gkr1009
11. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. (2012) 7:e30733. doi: 10.1371/journal.pone.0030733
12. Bachmayrheyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitnerhofmann T, et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. (2015) 5:8057. doi: 10.1038/srep08057
13. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. (2014) 28:2233–47. doi: 10.1101/gad.251926.114
14. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
15. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
16. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
17. Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. (2007) 4:721–6. doi: 10.1038/nmeth1079
18. Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet. (2013) 4:307. doi: 10.3389/fgene.2013.00307
19. Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. (2016) 95:e3811. doi: 10.1097/MD.0000000000003811
20. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. (2015) 25:981–4. doi: 10.1038/cr.2015.82
21. Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen MT, et al. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. (2016) 471:52–6. doi: 10.1016/j.bbrc.2016.01.183
22. Zhang X, Yan Y, Lei X, Li A, Zhang H, Dai Z, et al. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget. (2017) 8:34961–70. doi: 10.18632/oncotarget.16442
23. Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. (2015) 208:88–95. doi: 10.1111/nph.13585
24. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. (2014) 15:409. doi: 10.1186/s13059-014-0409-z
25. Liu L, Lin L, Zheng L, Hui T, Fan X, Xue N, et al. Cecal microbiome profile altered by Salmonella enterica, serovar Enteritidis inoculation in chicken. Gut Pathogens. (2018) 10:34. doi: 10.1186/s13099-018-0261-x
26. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. (2013) 14:R36. doi: 10.1186/gb-2013-14-4-r36
27. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. (2012) 9:357. doi: 10.1038/nmeth.1923
28. Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. (2015) 16:4. doi: 10.1186/s13059-014-0571-3
29. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
30. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. (2011) 39:316–22. doi: 10.1093/nar/gkr483
31. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. (2004) 2:e363. doi: 10.1371/journal.pbio.0020363
32. Bachman BJ, Venner E, Lua RC, Erdin S, Lichtarge O. ETAscape: analyzing protein networks to predict enzymatic function and substrates in Cytoscape. Bioinformatics. (2012) 28:2186–8. doi: 10.1093/bioinformatics/bts331
33. Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, et al. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE. (2008) 3:e2945. doi: 10.1371/journal.pone.0002945
34. Sand M, Bechara FG, Gambichler T, Sand D, Bromba M, Hahn SA, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. (2016) 83:210–8. doi: 10.1016/j.jdermsci.2016.05.012
35. Chiang HI, Swaggerty CL, Kogut MH, Dowd SE, Li X, Pevzner IY, et al. Gene expression profiling in chicken heterophils with Salmonella enteritidis stimulation using a chicken 44 K Agilent microarray. BMC Genomics. (2008) 9:526. doi: 10.1186/1471-2164-9-526
36. Wu G, Liu L, Qi Y, Sun Y, Yang N, Xu G, et al. Splenic gene expression profiling in White Leghorn layer inoculated with the Salmonella enterica serovar Enteritidis. Anim Genet. (2016) 46:617–26. doi: 10.1111/age.12341
37. Kogut MH, Swaggerty CL, Byrd JA, Selvaraj R, Arsenault RJ. Chicken-specific kinome array reveals that Salmonella enterica serovar enteritidis modulates host immune signaling pathways in the cecum to establish a persistence infection. Int J Mol Sci. (2016) 17:1207. doi: 10.3390/ijms17081207
38. Szmolka A, Wiener Z, Matulova ME, Varmuzova K, Rychlik I. Gene expression profiles of chicken embryo fibroblasts in response to salmonella enteritidis infection. PLoS ONE. (2015) 10:e0127708. doi: 10.1371/journal.pone.0127708
39. He H, Arsenault RJ, Genovese KJ, Johnson C, Kogut MH. Chicken macrophages infected with Salmonella (S.) Enteritidis or S. Heidelberg produce differential responses in immune and metabolic signaling pathways. Vet Immunol Immunopathol. (2018) 195:46–55. doi: 10.1016/j.vetimm.2017.11.002
40. Swaggerty CL, Kogut MH, He H, Genovese KJ, Johnson C, Arsenault RJ. Differential levels of cecal colonization by salmonella enteritidis in chickens triggers distinct immune kinome profiles. Front Vet Sci. (2017) 4:214. doi: 10.3389/fvets.2017.00214
41. Born J, Lange T, Hansen K, Mölle M, Fehm HL, Born J, et al. Effects of sleep and circadian rhythm on human circulating immune cells. J Immunol. (1997) 158:4454–64.
42. Liu J, Malkani G, Mankani G, Shi X, Meyer M, Cunninghamrunddles S, et al. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. (2006) 74:4750–6. doi: 10.1128/IAI.00287-06
43. Engert LC, Weiler U, Pfaffinger B, Stefanski V, Schmucker SS. Diurnal rhythms in peripheral blood immune cell numbers of domestic pigs. Dev Compar Immunol. (2017) 79:11–20. doi: 10.1016/j.dci.2017.10.003
44. Li X, Swaggerty CL, Kogut MH, Chiang H, Ying W, Genovese KJ, et al. Gene expression profiling of the local cecal response of genetic chicken lines that differ in their susceptibility to Campylobacter jejuni colonization. PLoS ONE. (2010) 5:e11827. doi: 10.1371/journal.pone.0011827
45. Loftus RM, Finlay DK. Immunometabolism; cellular metabolism turns immune regulator. J Biol Chem. (2015) 291:1–10. doi: 10.1074/jbc.R115.693903
46. Walker MA, Volpi S, Sims KB, Walter JE, Traggiai E. Powering the immune system: mitochondria in immune function and deficiency. J Immunol Res. (2014) 2014:164309. doi: 10.1155/2014/164309
47. Windt GJWVD, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. (2012) 249:27–42. doi: 10.1111/j.1600-065X.2012.01150.x
48. Ravindran R, Mcsorley SJ. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology. (2005) 114:450–8. doi: 10.1111/j.1365-2567.2005.02140.x
49. Maciver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. (2013) 31:259–83. doi: 10.1146/annurev-immunol-032712-095956
50. Porcelli D, Barsanti P, Pesole G, Caggese C. The nuclear OXPHOS genes in insecta: a common evolutionary origin, a common cis-regulatory motif, a common destiny for gene duplicates. BMC Evol Biol. (2007) 7:215. doi: 10.1186/1471-2148-7-215
51. Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. (2014) 20:61–72. doi: 10.1016/j.cmet.2014.05.004
52. Blier PU, Dufresne F, Burton RS. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet Tig. (2001) 17:400–6. doi: 10.1016/S0168-9525(01)02338-1
53. Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. (2004) 19:645–53. doi: 10.1016/j.tree.2004.10.003
54. Xing Y, Wang S, Fan J, Oso AO, Kim SW, Xiao D, et al. Effects of dietary supplementation with lysine-yielding on gut morphology, cecal microflora, and intestinal immune response of Linwu ducks. J Anim Sci. (2015) 93:3449–57. doi: 10.2527/jas.2014-8090
55. Wu G, Qi Y, Liu X, Yang N, Xu G, Liu L, et al. Cecal MicroRNAome response to Salmonella enterica serovar Enteritidis infection in White Leghorn Layer. BMC Genomics. (2017) 18:77. doi: 10.1186/s12864-016-3413-8
56. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
57. Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. (2015) 6:6001–13. doi: 10.18632/oncotarget.3469
58. Yao M, Gao W, Tao H, Yang J, Liu G, Huang T. Regulation signature of miR-143 and miR-26 in porcine Salmonella infection identified by binding site enrichment analysis. Mol Genet Genomics. (2016) 291:789–99. doi: 10.1007/s00438-015-1146-z
59. Li P, Fan W, Li Q, Wang J, Liu R, Everaert N, et al. Splenic microRNA Expression profiles and integration analyses involved in host responses to Salmonella enteritidis infection in chickens. Front Cell Infect Microbiol. (2017) 7:377. doi: 10.3389/fcimb.2017.00377
60. Bao H, Kommadath A, Plastow GS, Tuggle CK, Guan LL, Stothard P. MicroRNA buffering and altered variance of gene expression in response to salmonella infection. PLoS ONE. (2014) 9:e94352. doi: 10.1371/journal.pone.0094352
61. Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. (2001) 2:702. doi: 10.1038/35088576
62. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. (2002) 418:935–41. doi: 10.1038/nature00965
63. Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. (2005) 37:187–92. doi: 10.1038/ng1504
64. Maury E, Hong HK, Bass J. Circadian disruption in the pathogenesis of metabolic syndrome. Diabetes Metab. (2014) 40:338–46. doi: 10.1016/j.diabet.2013.12.005
65. Li YP, Ingmer H, Madsen M, Bang DD. Cytokine responses in primary chicken embryo intestinal cells infected with Campylobacter jejuni strains of human and chicken origin and the expression of bacterial virulence-associated genes. BMC Microbiol. (2008) 8: 107. doi: 10.1186/1471-2180-8-107
66. Naidu KS, Morgan LW, Bailey MJ. Inflammation in the avian spleen: timing is everything. BMC Mol Biol. (2010) 11:104. doi: 10.1186/1471-2199-11-104
67. Liu X, Liu L, Zhang M, Yang N, Qi Y, Sun Y, et al. Messenger RNA expression of chicken CLOCK gene in the response to Campylobacter jejuni inoculation. Poult Sci. (2015) 94:2124–30. doi: 10.3382/ps/pev203
68. Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. (2002) 250:1–23. doi: 10.1006/dbio.2002.0780
69. Li C, Tucker PW. DNA-binding properties and secondary structural model of the hepatocyte nuclear factor 3/fork head domain. Proc Natl Acad Sci USA. (1993) 90:11583. doi: 10.1073/pnas.90.24.11583
70. Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, et al. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. (2006) 7:819. doi: 10.1038/ni1358
71. Feng X, Ippolito GC, Tian L, Wiehagen K, Oh S, Sambandam A, et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. (2015) 115:510–8. doi: 10.1182/blood-2009-07-232694
72. Bacon C, Rappold GA. The distinct and overlapping phenotypic spectra of FOXP1 and FOXP2 in cognitive disorders. Hum Genet. (2012) 131:1687–98. doi: 10.1007/s00439-012-1193-z
73. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary Sequence-Mediated Exon Circularization. Cell. (2014) 159:134. doi: 10.1016/j.cell.2014.09.001
74. He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H, et al. circ-SHKBP1 Regulates the Angiogenesis of U87 Glioma-Exposed Endothelial Cells through miR-544a/FOXP1 and miR-379/FOXP2 Pathways. Mol Therapy Nucleic Acids. (2018) 10:331–48. doi: 10.1016/j.omtn.2017.12.014
Keywords: chicken, circular RNA, Salmonella enterica serovar Enteritidis, expression profile, metabolism, immunology
Citation: Zheng L, Liu L, Lin L, Tang H, Fan X, Lin H and Li X (2019) Cecal CircRNAs Are Associated With the Response to Salmonella Enterica Serovar Enteritidis Inoculation in the Chicken. Front. Immunol. 10:1186. doi: 10.3389/fimmu.2019.01186
Received: 25 April 2018; Accepted: 09 May 2019;
Published: 31 May 2019.
Edited by:
Janice C. Telfer, University of Massachusetts Amherst, United StatesReviewed by:
Harald Osmundsen, University of Oslo, NorwayQingmei Xie, South China Agricultural University, China
Copyright © 2019 Zheng, Liu, Lin, Tang, Fan, Lin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Lin, aGFpbGluQHNkYXUuZWR1LmNu; Xianyao Li, eHlsaUBzZGF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Linna Zheng
Linna Zheng Liying Liu
Liying Liu Lili Lin1
Lili Lin1 Hui Tang
Hui Tang Xinzhong Fan
Xinzhong Fan Xianyao Li
Xianyao Li