
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 12 April 2019
Sec. Molecular Innate Immunity
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.00776
 Liang Jiang1,2*
Liang Jiang1,2* Weiqiang Liu1
Weiqiang Liu1 Huizhen Guo1,2
Huizhen Guo1,2 Yinghui Dang1
Yinghui Dang1 Tingcai Cheng1,2
Tingcai Cheng1,2 Wanying Yang3
Wanying Yang3 Qiang Sun1
Qiang Sun1 Bingbing Wang1
Bingbing Wang1 Yumei Wang1
Yumei Wang1 Enyu Xie1
Enyu Xie1 Qingyou Xia1,2*
Qingyou Xia1,2*Peptidoglycan recognition protein (PGRP) is an important pattern recognition receptor in innate immunity that is vital for bacterial recognition and defense in insects. Few studies report the role of PGRP in viral infection. Here we cloned two forms of PGRP from the model lepidopteran Bombyx mori: BmPGRP2-1 is a transmembrane protein, whereas BmPGRP2-2 is an intracellular protein. BmPGRP2-1 bound to diaminopimelic acid (DAP)-type peptidoglycan (PGN) to activate the canonical immune deficiency (Imd) pathway. BmPGRP2-2 knockdown reduced B. mori nucleopolyhedrovirus (BmNPV) multiplication and mortality in cell lines and in silkworm larvae, while its overexpression increased viral replication. Transcriptome and quantitative PCR (qPCR) results confirmed that BmPGRP2 negatively regulated phosphatase and tensin homolog (PTEN). BmPGRP2-2 expression was induced by BmNPV, and the protein suppressed PTEN-phosphoinositide 3-kinase (PI3K)/Akt signaling to inhibit cell apoptosis, suggesting that BmNPV modulates BmPGRP2-2-PTEN-PI3K/Akt signaling to evade host antiviral defense. These results demonstrate that the two forms of BmPGRP2 have different functions in host responses to bacteria and viruses.
Innate immunity is a self-defense mechanism against infectious non-self entities and is present in all metazoans (1, 2). The innate immune system of insects consists of humoral defenses that include the production of soluble effector molecules and cellular response like phagocytosis and encapsulation that are mediated by hemocytes (3). The innate immune response is mediated by germline-encoded pattern-recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs) that are present in pathogens but absent in the host (2, 4, 5). PAMPs include b-glucan, lipopolysaccharides (LPS) of gram-negative (G–) bacteria, and peptidoglycans (PGNs) of both gram-positive (G+) and G– bacteria, as well as bacterial and viral DNA and RNA and related molecules (2, 4, 5). LPS activates the expression of antimicrobial peptides (AMPs) in Manduca sexta (6) and malpighian tubules of Drosophila (7). PGN—an essential cell wall component in most bacteria—stimulates various immune reactions in insects (2, 5, 8). Lysine (Lys)-type PGNs are found in many G+ bacteria, whereas diaminopimelic acid (DAP)-type PGNs are found in G– and some G+ bacteria (5, 9, 10). Multiple PRRs have been identified in invertebrates, such as lectin, hemolin, G– binding proteins (GNBPs), Toll-like receptors (TLRs), and PGN recognition proteins (PGRPs); the latter two have been shown to recognize PGN (2, 5, 10).
PGRP was first purified from the hemolymph of silkworm Bombyx mori (11). Since then, nearly 100 PGRP family members have been identified from insects to mammals. These proteins are highly conserved and have a PGRP domain that is similar to the bacteriophage T7 lysozyme, an N-acetylmuramyl-alanine amidase (9, 12, 13). Insect PGRPs are categorized as short (PGRP-S), which are small extracellular proteins with signal peptides, or long (PGRP-L), which have long transcripts and are intracellular, extracellular, or transmembrane proteins (12, 14). Some PGRP-Ls have multiple splice forms; for example, the 13 Drosophila PGRP genes encode 19 proteins and the seven genes in mosquito (Anopheles gambiae) encode nine proteins (5, 12, 15, 16).
PGRPs have multiple immune-related functions. In Drosophila, PGRP-SB1 and -SB2, -SC1A/1B/2, and -LB have zinc-dependent amidase activity involving the removal of peptides from glycan chains to inhibit or scavenge the biological activity of PGN. Some of these PGRPs modulate the host immune response by eliminating PGN (8, 17–20). PGRP-SA, -SD, -LA, -LC, -LD, -LE, and -LF lack zinc-binding residues required for amidase activity while retaining the capacity to bind and recognize PGN (8, 18). Other PGRPs lacking amidase activity mediate PGN-dependent activation of the prophenoloxidase (proPO) cascade (21, 22) in which ProPO is activated to PO, leading to melanization that is toxic to microorganisms (11, 23). Transmembrane PGRP-LC and -LE recognize DAP-type PGNs and activate the immune deficiency (Imd) pathway. The former binds PGN and interacts with Imd protein via the extracellular PGRP domain and an intracellular domain, respectively (15, 24, 25). PGRP-LB has amidase activity to cleave DAP-type PGNs, limiting availability of ligand for PGRP-LC and thus inhibiting the Imd pathway (26). PGRP-SA is an extracellular protein that recognizes Lys-type PGNs and activates Toll signaling. The serine protease cascade is triggered when PGRP-SA binds to PGN, which cleaves the inactive pro Spatzle (Spz) protein to an active form, which in turn binds to and activates Toll receptor (8, 27–31).
Imd and Toll signaling activation induces AMPs against invading bacteria and fungi (5, 8, 14, 27, 32). The GATA factor Serpent is required for the onset of humoral defenses in Drosophila embryos (33). In the silkworm, the Imd pathway can be activated by Escherichia coli (E. coli) and Serratia marcescens (S. marcescens) (34), whereas the Toll pathway could be induced by Bacillus bombysepticus (35), Nosema bombycis (36), and Beauveria bassiana (34). Some studies have shown that these pathways are also involved in the antiviral immune response (37–41). In Drosophila, the replication of the RNA viruses cricket paralysis virus (CrPV) and alphavirus is increased by mutation of Imd pathway components (37, 38). Activation of Toll signaling inhibits dengue virus in Aedes aegypti (40); Toll-7 interacts with vesicular stomatitis virus and induces antiviral autophagy independently of canonical Toll signaling (39).
The model lepidopteran B. mori is an important insect because of its production of silk (42–44); as such, infection by pathogenic bacteria, fungi, and viruses can cause serious economic losses. B. mori nucleopolyhedrovirus (BmNPV) is the major threat to silkworms (45, 46). A genome analysis revealed 12 PGRP genes in B. mori (47), some of which the function has been studied. Five BmPGRP-S showed strong amidase activity toward DAP-PGN (48). BmPGRP-S4 (49) and BmPGRP-S5 (50) bound PGNs to increase proPO activation. BmPGRP-S3 could be induced by B. mori cytoplasmic polyhedrosis virus (BmCPV) (51). Recently, our study showed that BmPGRP-S2 was up-regulated upon BmCPV infection (52), overexpression of which can activate the Imd pathway and induce increased AMPs to enhance the antiviral capacity of transgenic silkworm against BmCPV (53). In this study, we cloned two forms of BmPGRP2: BmPGRP2-1 bound to DAP-type PGN to activate Imd signaling, whereas BmPGRP2-2 was induced by BmNPV and negatively regulated the phosphatase and tensin homolog (PTEN)-phosphoinositide 3-kinase (PI3K)/Akt pathway to inhibit cell apoptosis. Our results demonstrate that the two forms of BmPGRP2 have distinct functions in the host response to pathogenic bacteria and viruses. The present study confirms that PGRP was induced by viruses to escape host antiviral immunity.
B. mori strains Dazao (DZ) and 932 were maintained at the Gene Resource Library of Domesticated Silkworm (Southwest University, Chongqing, China). BmE and BmN4-SID1 cell lines (54) were cultured at 27°C. BmNPV (Guangdong strain) and BmNPV expressing green fluorescent protein (BmNPV-GFP) were collected from the haemolymph of infected silkworm larvae and the infected BmE cells, respectively (55, 56).
Based on bioinformatics analysis, primers were designed to amplify the open reading frame (ORF) of BmPGRP2 (BmPGRP2-1 ORF and BmPGRP2-2 ORF) by PCR. The 3′ untranslated region (UTR) was amplified by 3′ rapid amplification of cDNA ends. Six forward primers (5F-1,-2,-3,-4,-5, and-6) and two reverse primers (5R-1 and-2) were used to amplify the 5′ UTR. The eggs of 2-, 4-, 6-, and 8-day-old, hatched silkworm, first instar molt, second instar, second instar molt, third instar, third instar molt, fourth instar, fourth instar molt, fifth instar larvae, pupae of 2-, 4-, 6-, and 8-day-old, and moth of DZ silkworms were used for RNA extraction using Total RNA Kit (#R6834-01 and R6934-01, Omega, USA). The RNA of head, cuticle, hemocyte, midgut, fat body, silk gland, trachea, malpighian, ovary, and testis of female and male silkworms (DZ) was extracted at day-3 fifth instar. The total RNA was treated with RNase-Free DNase I (#M6101, Promega, USA) and then reverse transcribed into cDNA using M-MLV Reverse Transcriptase (#M1701, Promega, USA). The cDNA temples of different developmental stages were used for RT-PCR with primers BmPGRP2-1qRT, BmPGRP2-2qRT, and internal control TIF-4A (53, 57–59), of which the amplification cycles was 30, 30, and 25, respectively. The cDNA of each tissue was used for qPCR analysis with primers BmPGRP2-1qRT and BmPGRP2-2qRT on an ABI Prism 7500 (Applied Biosystems, USA) using an SYBR Premix Ex Taq II (#RR820A, TaKaRa, China). The thermal program of qPCR consisted of 95°C for 30 s, 40 cycles at 95°C for 5 s and 60°C for 30 s, 95°C for 15 s, 60°C for 60 s, and melt for 15 s. The control TIF-4A was used for qPCR analysis of gene expression level to standardize the variance among the different templates (53, 57–59). Each detection was performed thrice. The cycle threshold (CT) values were converted to linear values using the comparative CT method and then analyzed with statistical algorithm geNorm (57). Student's t-tests were used to analyze the statistical data. All primer sequences are shown in Table S1.
A synthetic sequence includes B. mori A4 promoter (A4P) and GFP was cloned into the empty vector 1180 (GenBank: U13865.1) using SalI (#R3138V, NEB, USA) and BamHI (#R3136V, NEB, USA) restriction enzymes, followed adding BmPGRP2-1 or -2 [using BamHI and NotI (#R3189V, NEB, USA)] and Simian virus (SV)40 [using NotI and HindIII (#R3104V, NEB, USA)] to construct 1180-A4P-GFP-BmPGRP2-1/2-SV40, which was transfected into BmE cells for subcellular localization analysis. PGN-EB (PGN from E. coli 0111:B4, #tlrl-pgneb), PGN-BS (PGN from Bacillus subtilis, #tlrl-pgnbs), PGN-SA (PGN from Staphylococcus aureus, #tlrl-pgnsa), and LPS-EB (LPS from E. coli 0111:B4, #tlrl-eblps) were purchased from InvivoGen (San Diego, CA, USA) and added to BmE cells. Total RNA was extracted 0, 6, 12, and 24 h after treatment to detect the presence of BmPGRP2-1, imd, and the AMP gene attacin (att)2. Fifth instar silkworms were orally infected with E. coli and S. marcescens at 109/larva, and extracted RNA was tested for the presence of BmPGRP2-1/2 and the AMP gene gloverin (glv)2. Double-stranded (ds)RNA against BmPGRP2-1 and dsRed were generated and added to BmN4-SID1 cells, whereas BmE cells were transfected with 1180 and 1180-A4P-BmPGRP2-1-SV40 (56). Total RNA was extracted to detect BmPGRP2-1, imd, spz, and att2. BmPGRP2-2 RNAi and overexpression were carried out in a similar manner, and treated cells were then infected with BmNPV-GFP; DNA was extracted at 48 h post infection (hpi) for detection of copy number of BmNPV-GFP using control gene GAPDH and virus fluorescent was observed at 72 hpi (56).
The extracellular PGRP domain of BmPGRP2-1 was cloned with the primer BmPGRP2-1pro, which was used for prokaryotic expression in E. coli with the pSKB2-MsyB vector (pSKB2 vector was added with an MsyB tag in our lab). After induction of expression with 0.1 m mol/L isopropyl β-D-thiogalactoside (IPTG, #A100487, Sangon Biotech, China) at 16°C for 16 h, the protein in the supernatant was purified and used for western blotting with a BmPGRP2-1 antibody. Purified BmPGRP2-1 was tested for binding to PGN-EB, PGN-BS, and LPS-EB by enzyme-linked immunosorbent assay.
The transgenic BmPGRP2-1 and -2 RNAi vectors was constructed using piggyBac [3 × p3 EGFP afm] vector, respectively. The non-diapausing embryos of silkworm were used for microinjection. The G1 embryos were screened for transgenic silkworm. The insertion sites of transgenic silkworms were detected using inverse PCR analysis with the transposon-specific primers pBacL and pBacR (45, 55). There was only a single band after PCR amplification using pBacL and pBacR primers (data not shown), suggesting that there was inserted as a single copy (45, 55). Sequencing of the PCR products was blasted in SilkDB (http://silkworm.swu.edu.cn/silkdb/) will reveal the inserted region in silkworm genome. Fifth instar larvae and day 2 pupae of the transgenic PGRP2-1I and non-transgenic DZ strains were used for qPCR analysis of BmPGRP2-1, imd, spz, and att2 expression. PGRP2-1I and DZ were orally infected with E. coli and S. marcescens at 109/larva at the 5th instar; 2nd day pupae were injected with S. marcescens at 10/pupa. Third instar, 3rd instar molt, 4th instar, and 4th instar molt of the transgenic PGRP2-2I and non-transgenic 932 strains were examined for BmPGRP2-2 expression by qPCR. Third instars of PGRP2-2I and 932 were orally infected with BmNPV at 3 × 105 occlusion bodies (OB)/larva, and total DNA was extracted at 48 hpi (45, 55). RNA was extracted from PGRP2-2I and 932 at 0, 3, 6, 12, and 24 hpi for analysis of BmPGRP2-2, imd, Relish, Myeloid differentiation primary response (MyD)88, Pelle, and AMP gene expression.
PGRP2-2I and 932 were orally infected with BmNPV at 2.5 × 107 OB/larva on day 3 of the 5th instar. RNA was extracted from the midgut and fat body at 3, 6, 12, and 24 hpi, and used for qPCR analysis of BmPGRP2-2 expression. The RNA libraries of the 16 samples were constructed. Raw sequencing data were generated using an Illumina HiSeq 2000 system, which has been deposited in the NCBI, the BioProject ID is PRJNA521671. The polyA tails, noncoding RNAs and low-quality reads were removed from the raw reads to generate clean reads. The qualities of raw and clean reads were analyzed using FastQC (v0.11.1). Clean reads were mapped to the silkworm genome. The gene ontology (GO) analysis and KEGG annotation of the differentially expressed genes (DEGs) were executed using WEGO online and iPathCons, respectively (52, 60). PTEN was cloned using the BmPTEN ORF primer, and its expression after BmPGRP2-2 RNAi and overexpression was evaluated. BmE cells were transfected with 1180 and 1180-A4P-PTEN-SV40; RNA was extracted and analyzed for BmPGRP2-2 expression. The 1180-A4P-GFP-PTEN-SV40 vector was constructed and transfected into BmE cells for localization analysis.
BmE cells were infected with BmNPV-GFP, and total protein was extracted for analysis of phosphorylated (p-)Akt expression at 0, 18, and 24 hpi. BmE cells were transfected with 1180 and the BmPGRP2-2 overexpression vector. BmN4-SID1 cells were incubated with dsBmPGRP2-2 and dsRed, and treated cells were infected with BmNPV-GFP (56). GAPDH, p-Akt, and total Akt levels were assessed by western blotting. The antibody of GAPDH (#CB100127, California Bioscience, USA) was used as a control. The PVDF membrane (#03010040001, Roche, Switzerland) was blocked with 5% BSA for 12 h at 4°C after transfer film, followed incubation with antibody of p-Akt (Ser505, #4054, Cell Signaling Technology, USA) or total Akt (#9272, Cell Signaling Technology, USA) for 12 h at 4 °C. And then, the PVDF membrane was washed three times for 10 min each time with TBST, incubated with secondary antibody for 30 min at 37°C, washed 3 × 10 min with TBST, and treated with SuperSignal™ West Femto (#34095, Thermo Scientific, USA). The images were analyzed following the manufacturer's instructions. Cell apoptosis was detected at 24 hpi by flow cytometry. The cells were collected from each treatment, washed with cold PBS and incubated with Annexin V-fluorescein isothiocyanate (V-FITC) and propidium iodide (#K201-100, Biovision, USA) at 25°C for 20 min in the dark on ice. These samples were then analyzed by a fluorescence-activated cell sorter and Cell Quest software (BD, USA) following the manufacturer's instructions. Each test was repeated thrice. BmE cells were treated with the 10 μM PI3K inhibitor LY294002 (#HY-10108, MedChemExpress, USA), with dimethyl sulfoxide (DMSO, #D2650, Sigma, USA) treatment serving as a control (61). The expression of p-Akt was analyzed at 24 hpi and fluorescence was observed at 72 hpi.
A bioinformatics analysis revealed that BmPGRP2 exists as two forms, BmPGRP2-1 and -2. BmPGRP2-1 consisted of exons 1, 2, and 3 with a 242-bp 3′ UTR, whereas BmPGRP2-2 consisted of exons 1, 4, 5, 6, and 7 with a 224-bp 3′ UTR (Figure 1A). To clone the 5′ UTR of BmPGRP2-1 and -2, we analyzed the 3000 bp upstream of the translation initiation site (ATG). There were four predicted transcription initiation sites (TIS) after analysis of promoter using online software (http://linux1.softberry.com/berry.phtml); forward primers (5F-1,-2,-3, and-4) were designed to target the 20–50 bp downstream of each candidate TIS. 5F-5 and 5F-6 were 300 and 600 bp downstream of 5F-4, respectively. The reverse primers 5R-1 and 5R-2 were located in exons 2 and 7, respectively (Figure 1A); 5R-1 and 5F-2,-3,-4,-5, and-6 amplified the target sequence but 5R-1 and 5F-1 did not amplify the target band; 5R-2 and 5F-6 amplified the target sequence but 5R-2 and 5F-1,-2,-3,-4, and-5 amplified none of the targets (data not shown). We confirmed by sequencing that the 1706-bp 5′ UTR of BmPGRP2-1 containing the 281-bp 5′ UTR of BmPGRP2-2 was cloned (Figure 1A). These results suggest that the two forms of BmPGRP2 are generated from different TIS.
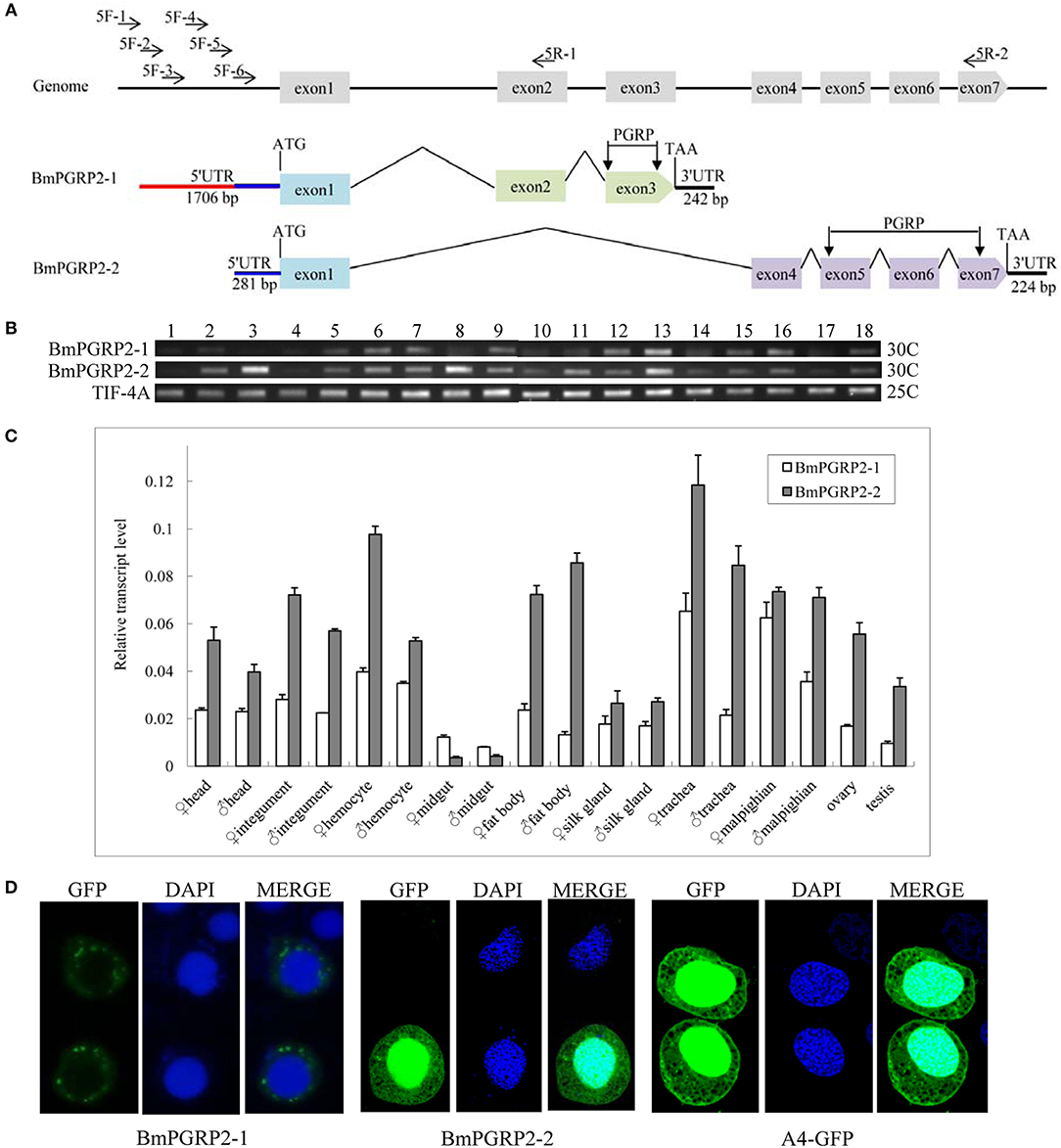
Figure 1. BmPGRP2 expression patterns. (A) Schematic representation of BmPGRP2 gene structure from DZ silkworm. Two forms of BmPGRP2, BmPGRP2-1, and -2, were cloned. The 5′ UTR was amplified with primers 5F-1,-2,-3,-4,-5, and-6 and 5R-1 and-2. The 5′ UTR of BmPGRP2-1 was longer and contained that of BmPGRP2-2. (B) Analysis of BmPGRP2 expression at whole individual of different developmental stages by RT-PCR. Points 1 to 4 represent 2-, 4-, 6-, and 8-day-old eggs; 5 to 13 represent hatched silkworm, first instar molt, second instar, second instar molt, third instar, third instar molt, fourth instar, fourth instar molt, and fifth instar larvae, respectively; 14 to 17 represent 2-, 4-, 6-, and 8-day-old pupae, respectively; and 18 represents moth. TIF-4A (GenBank: DQ443290.1) was the internal control. 25C and 30C represented the PCR amplification cycles was 25 and 30, respectively. (C) qPCR analysis of BmPGRP2 expression in the head, cuticle, hemocyte, midgut, fat body, silk gland, trachea, malpighian, ovary, and testis of female and male silkworms. TIF-4A was used as the internal control. Bars represent standard deviation. (D) Subcellular localization of BmPGRP2. The vector 1180-A4P-GFP-BmPGRP2-1/2-SV40 and 1180-A4P-GFP-SV40 (A4-GFP) was transfected into BmE cells, respectively. The nucleus was dye blue by DAPI, and green fluorescence represented the location of target protein.
A sequence analysis suggested that BmPGRP2-1 and-2 contained a PGRP domain, while a phylogenetic analysis revealed that BmPGRP2-1 and-2 clustered into distinct classes (Figure S1). The RT-PCR results showed that BmPGRP2-1 was more highly expressed in the larva than that in the egg (Figure 1B). The qPCR analysis showed that BmPGRP2-1 and -2 levels were lowest in the midgut, and that the expression of BmPGRP2-2 was higher than that of BmPGRP2-1 in all tissues of male and female larvae except for the midgut (Figure 1C). The predicted results from WoLF PSORT program on website showed that BmPGRP2-1 and-2 have no nucleotide localization sequences (NLS). A subcellular localization analysis indicated that BmPGRP2-1 was a transmembrane protein whereas BmPGRP2-2 was an intracellular protein (Figure 1D). The differences in phylogenetic position (Figure S1), temporal and spatial expression patterns (Figures 1B,C), and subcellular localization (Figure 1D) between the BmPGRP2-1 and-2 suggest that the two isoforms play distinct roles in host immune response to pathogens.
Multiple sequence alignment revealed that the zinc-binding cysteine residue of T7 lysozyme was replaced by a serine residue in BmPGRP2-1; the other seven PGRP proteins showed no amidase activity (Figure S2), suggesting that BmPGRP2-1 also lacks this activity. We tested whether BmPGRP2-1 can be induced by PGN. BmPGRP2-1 and glv2 but not BmPGRP2-2 levels were induced in silkworm larvae by the G– bacteria E. coli and S. marcescens (Figure S3). A qPCR analysis showed that BmPGRP2-1 expression was induced by DAP-type PGN-EB and PGN-BS but not LPS-EB or Lys-type PGN-SA (Figure 2A); imd and att2 levels were also increased in BmE cells treated with PGN-EB and -BS (Figure 2B). However, Toll pathway activation in cells and individuals was unaltered by the treatment (data not shown). In addition, BmPGRP2-1 overexpression in BmE cells increased the levels of imd and att2 (Figure 2C), whereas silence of BmPGRP2-1 had the opposite effect (Figure 2D) and spz level was unaltered. Thus, a DAP-type PGN induces BmPGRP2-1 to activate Imd signaling and AMP.
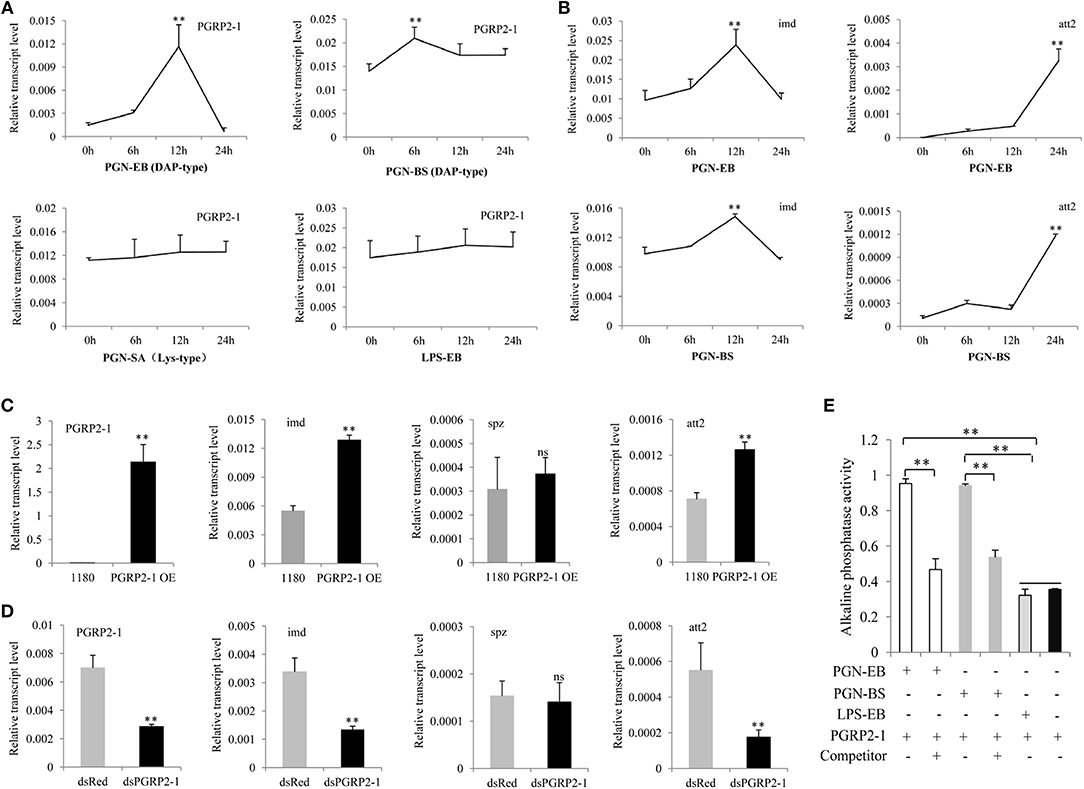
Figure 2. BmPGRP2-1 binds to DAP-type PGN and activates the Imd pathway. (A) qPCR analysis of BmPGRP2-1 expression after PGN-EB (PGN from E. coli 0111:B4), -BS (PGN from Bacillus subtilis), and -SA (PGN from Staphylococcus aureus) and LPS-EB (LPS from E. coli 0111:B4) treatment. (B) Detection of imd and att2 by qPCR in cells treated with PGN-EB and -BS. (C) BmPGRP2-1 overexpression. The 1180-A4P-BmPGRP2-1-SV40 (PGRP2-1 OE) or empty 1180 vector was transfected into BmE cells. (D) RNAi of BmPGRP2-1. DsRNA targeting BmPGRP2-1 (dsPGRP2-1) and control dsRed were added to BmN4-SID1 cells. BmPGRP2-1, imd, spz, and att2 expressions were evaluated by qPCR. TIF-4A was used as the internal control of qPCR analysis, and each detection was performed thrice. (E) Interaction between BmPGRP2-1 and PGN-EB or -BS as determined by enzyme-linked immunosorbent assay. PGN-EB and -BS were set to each other's competitors; BmPGRP2-1 was pretreated with competitor (or left untreated) and the binding of BmPGRP2-1 with PGN-EB or -BS was evaluated based on alkaline phosphatase activity. LPS-EB was used as a control. Each assay was performed in triplicate. Student's t-tests were used to analyze the statistical data. ns: not significant. Bars represent standard deviation. **P < 0.01.
Some studies have reported that PGRP binds to PGN through the extracellular PGRP domain to activate Imd signaling (15, 24, 25). To determine whether BmPGRP2-1 can bind to a DAP-type PGN, we purified a prokaryotic protein with a PGRP domain (Figure S4A). Western blotting detected the target band in the purified protein (Figure S4B). Alkaline phosphatase activity was higher in BmPGRP2-1+PGN-EB and BmPGRP2-1+PGN-BS than in BmPGRP2-1+LPS-EB and BmPGRP2-1. In a competition assay using PGN-EB and PGN-BS, pretreatment of BmPGRP2-1 with a competitor decreased alkaline phosphatase activity (Figure 2E), demonstrating that BmPGRP2-1 directly binds to DAP-type PGN.
To further clarify the function of BmPGRP2-1 in immune defense of silkworm against bacteria, we injected the transgenic RNAi vector pb-PGRP2-1I (Figure S5A) into embryos to generate a transgenic strain of PGRP2-1I (Figure S5B). The results of inverse PCR showed that a single copy of the transgene was detected in an intergenic region of the genome (Figure S5C). A qPCR analysis showed that BmPGRP2-1 and imd levels were lower in PGRP2-1I as compared to DZ larvae (Figure 3Aa). However, the viability of PGRP2-1I and DZ strains was unaffected by oral infection with E. coli and S. marcescens. The expression of BmPGRP2-1, imd, and att2 was also reduced in PGRP2-1I as compared to DZ pupae (Figure 3Ab), and the survival rate after S. marcescens injection was lower in PGRP2-1I than in DZ (Figure 3Ac). These results suggest that downregulation of BmPGRP2-1 inhibits Imd signaling and reduces the resistance of silkworm to G– bacteria.
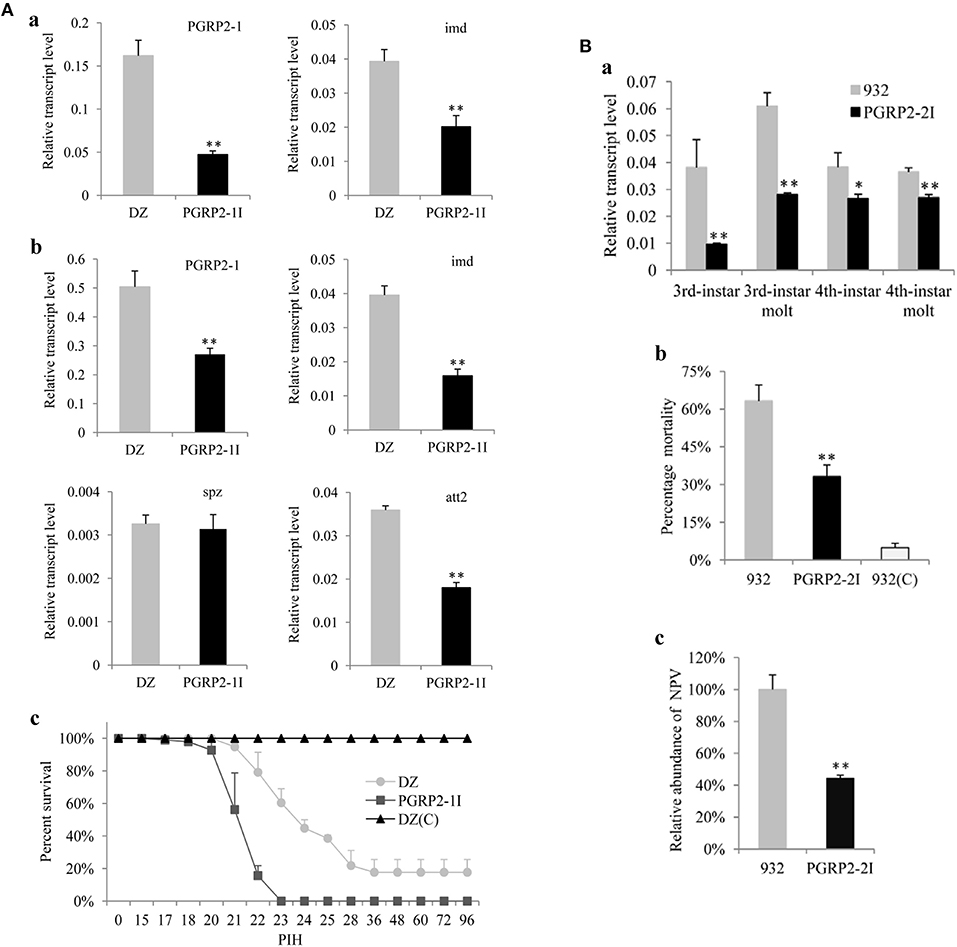
Figure 3. Analysis of transgenic RNAi silkworms. (A) Analysis of the transgenic line PGRP2-1I. Five larvae (a) and five pupae (b) were used to extract RNA for analysis of BmPGRP2-1, imd, spz, and att2 expression by qPCR. TIF-4A was used as a control, and each assay was performed thrice. (c) Resistance analysis. Pupae of PGRP2-1I and DZ were injected with S. marcescens at 10/pupa. DZ(C) was the uninfected control. Each treatment had four repeats, and each repeat contained 25 pupae. PIH, post infection hour. (B) Analysis of PGRP2-2I. (a) BmPGRP2-2 expression, as determined by qPCR. Each RNA sample was extracted from five larvae. TIF-4A was used as a control of qPCR, and each assay was performed thrice. (b) Mortality of B. mori. PGRP2-2I and 932 were orally infected at the 3rd instar stage with BmNPV using 3 × 105 occlusion bodies (OB)/larva; 932(C) was the uninfected control. Each line contained triplicate replicates and each repeat had 70 larvae. (c) Analysis of viral DNA content by qPCR 48 h after BmNPV infection. Each DNA sample was extracted from 10 treated larvae. GAPDH (GenBank: AB262581.1) was used as the internal control, and each assay was performed thrice. Student's t-tests were used to analyze the statistical data. Bars represent standard deviation. *P < 0.05; **P < 0.01.
BmPGRP2-2 but not BmPGRP2-1 expression was induced in BmE cells by BmNPV (Figure S6). To determine the role of BmPGRP2-2 in BmNPV infection, BmE cells were transfected with a BmPGRP2-2 overexpression vector containing a flag epitope. RT-PCR and western blot analyses confirmed that BmPGRP2-2 was upregulated in the cells (Figure 4Aa). Correspondingly, the fluorescence of the virus was enhanced (Figure 4Ab) and viral load was increased in cells overexpressing BmPGRP2-2 relative to controls (Figure 4Ac). These results suggest that BmPGRP2-2 overexpression promotes BmNPV multiplication. Furthermore, when BmPGRP2-2 dsRNA was added to BmN4-SID1 cells, BmPGRP2-2 expression was suppressed (Figure 4Ba), and viral fluorescence (Figure 4Bb) and viral DNA level (Figure 4Bc) were reduced, implying that BmPGRP2-2 knockdown inhibits BmNPV replication. Together, these data demonstrated that BmNPV induces BmPGRP2-2 to promote viral replication.
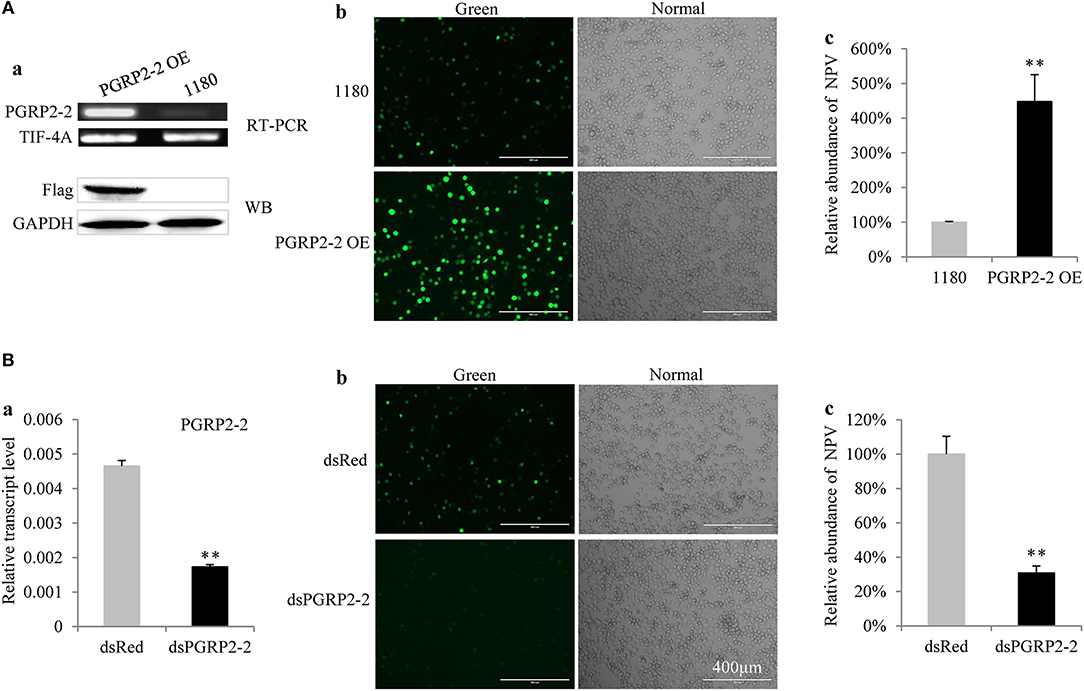
Figure 4. BmPGRP2-2 regulates BmNPV multiplication. (A) BmPGRP2-2 overexpression promoted BmNPV replication. A BmPGRP2-2 overexpression vector with a flag tag (PGRP2-2 OE) was transfected into BmE cells; BmPGRP2-2 transcript and protein were detected by RT-PCR (using control TIF-4A) and western blotting (using an anti-Flag antibody and control GAPDH antibody), respectively (a). The treated cells were infected with BmNPV-GFP. The green fluorescence of the virus was visualized at 72 hpi (b) and accumulated viral DNA content was detected by qPCR (using control GAPDH) at 48 hpi (c). (B) BmPGRP2-2 knockdown inhibited BmNPV. The dsRNA of BmPGRP2-2 (dsPGRP2-2) was added to BmN4-SID1 cells, and BmPGRP2-2 expression was determined by qPCR (using control TIF-4A) (a); viral green fluorescence (b) and BmNPV DNA content (using control GAPDH) were detected (c) after infection with BmNPV-GFP. Bars represent standard deviation. **P < 0.01.
To clarify the role of BmPGRP2-2 in the immune response of silkworm larvae to BmNPV, we used the transgenic RNAi vector pb-PGRP2-2I (Figure S7A) and generated the transgenic 932 strains PGRP2-2I and PGRP2-2Ia (Figure S7B) containing the transgene insertion in an intergenic region and in the intron of a predicted gene, respectively (Figures S7C). PGRP2-2I was selected for subsequent analysis. BmPGRP2-2 expression was decreased in PGRP2-2I as compared to the control 932 strain (Figure 3Ba). Moreover, mortality after BmNPV infection (Figure 3Bb) and accumulated BmNPV DNA content were significantly lower in PGRP2-2I than in 932 (Figure 3Bc). These results indicate that silencing BmPGRP2-2 enhances the antiviral capacity of silkworm.
Since Imd and Toll signaling pathways are involved in the antiviral immune response (37–40), we investigated whether BmPGRP2-2 activates these pathways. BmPGRP2-2 was induced by BmNPV in larvae, with lower expression in PGRP2-2I than in 932 (Figure S8A). However, there was no difference in the levels of imd, Relish, MyD88, Pelle, glv3, and glv4 of PGRP2-2I and 932 upon BmNPV infection (Figure S8B). These results suggest that BmPGRP2-2 mediates its antiviral effects through a pathway other than canonical Imd and Toll signaling.
To identify the signaling pathway downstream of BmPGRP2-2, RNA was extracted from the midgut and fat body of PGRP2-2I and 932 at 3, 6, 12, and 24 hpi (Figure S9A). We confirmed that BmPGRP2-2 expression in both tissues was significantly lower in PGRP2-2I than in 932 by qPCR (Figure S9B). And then, the 16 RNA samples were used for RNA-seq (Supplementary File 1). The FPKM values of silkworm immune-related genes were shown in Table S2 after information analysis. The transcriptome analysis revealed that BmPTEN expression was higher in the midgut and fat body of PGRP2-2I than in those of 932 (Figure S10); this was confirmed by qPCR (Figure 5A). We also found that BmPGRP2-2 knockdown and overexpression increased and decreased the expression level of BmPTEN, respectively (Figure 5B), indicating a negative regulatory relationship. We also examined whether BmPGRP2-2 is regulated by BmPTEN; however, BmPTEN overexpression did not affect BmPGRP2-2 expression (Figure 5C). BmPTEN was an intracellular protein (Figure 5D). These results indicate that BmPTEN is downstream of and negatively regulated by BmPGRP2-2.
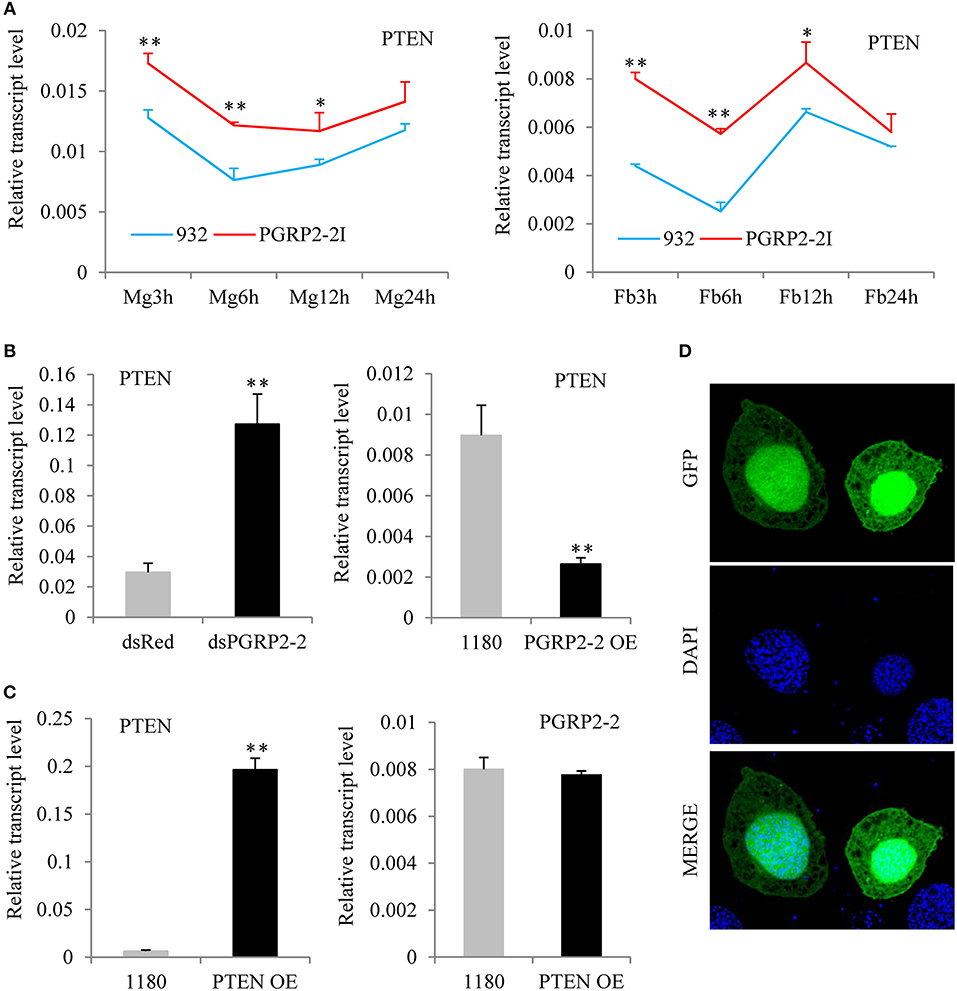
Figure 5. BmPGRP2-2 negatively regulates PTEN. (A) qPCR analysis of PTEN expression (using control TIF-4A) in PGRP2-2I. PGRP2-2I and 932 were orally infected with BmNPV on day 3 of the 5th instar. RNA was extracted from the midgut (MG) and fat body (Fb) at 3, 6, 12, and 24 hpi. (B) Detection of PTEN (using control TIF-4A) after BmPGRP2-2 RNAi and overexpression. (C) qPCR analysis of BmPGRP2-2 (using control TIF-4A) after PTEN overexpression (D) Subcellular localization of PTEN. The vector 1180-A4P-GFP-PTEN-SV40 was transfected into BmE cells. The green fluorescence represented the location of target protein and blue indicated nucleus. Bars, standard deviations. Significant differences, **P < 0.01, *P < 0.05.
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) infection increases Akt phosphorylation in Sf9 cells (61). Akt is a downstream effector of PI3K-dependent cell survival, whereas PTEN is an inhibitor of PI3K/Akt signaling (62, 63). We therefore investigated whether p-Akt is upregulated in BmE cells after BmNPV infection. BmNPV induced Akt phosphorylation, with the maximum level detected at 24 hpi (Figure S11A). We also examined found that BmPGRP2-2 overexpression and knockdown increased and decreased Akt phosphorylation, respectively, at 24 hpi (Figure 6A). Moreover, pharmacological inhibition of p-Akt by treatment with the PI3K inhibitor LY294002 suppressed BmNPV replication (Figure 6B).
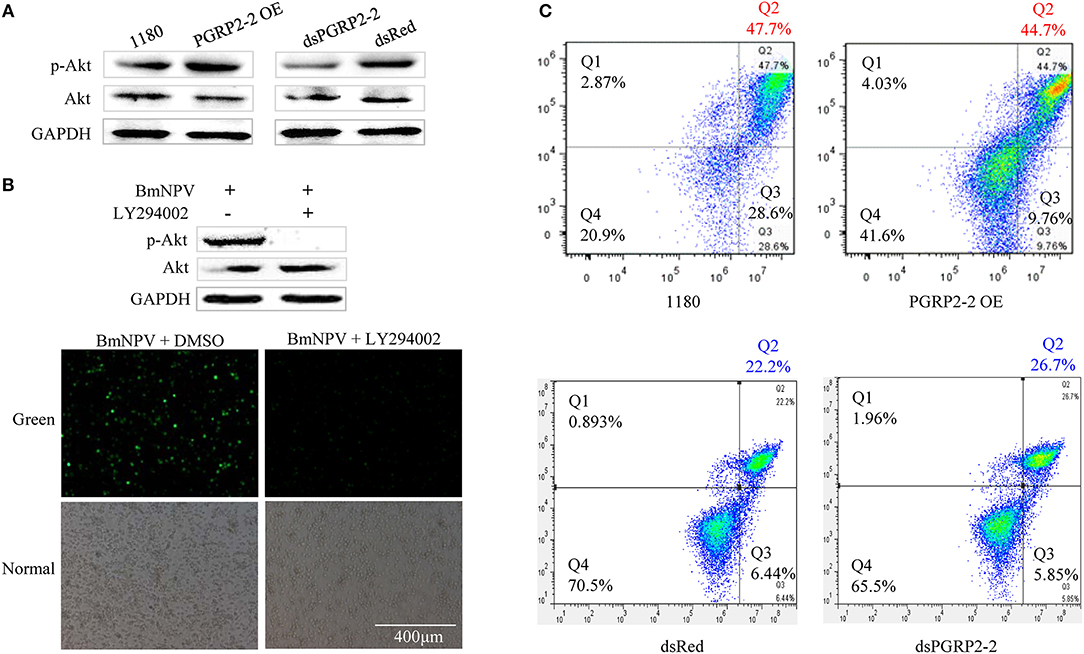
Figure 6. BmPGRP2-2 regulates Akt phosphorylation and cell apoptosis. (A) Detection of p-Akt by western blotting. Following BmPGRP2-2 RNAi and overexpression, cells were infected with BmNPV-GFP; protein was extracted at 24 hpi. (B) Treatment with the PI3K inhibitor LY294002 (10 μM) inhibits Akt phosphorylation and BmNPV replication. BmE cells were treated with BmNPV-GFP+LY294002; BmNPV-GFP+dimethyl sulfoxide (DMSO) was used as a control. Akt phosphorylation was evaluated at 24 hpi, and fluorescence associated with the virus was detected. (C) Analysis of cell apoptosis. Following BmPGRP2-2 RNAi and overexpression, apoptotic cells were detected by flow cytometry at 24 hpi. X and Y axis represented Annexin V-fluorescein isothiocyanate (V-FITC) and propidium iodide (PI), respectively. Late apoptotic, early apoptotic, and non-apoptotic cells were assigned to Q2, Q3, and Q4 section, respectively.
Previous studied have shown that Akt plays an important role in promoting cell survival by suppressing apoptosis (63, 64). We examined whether BmPGRP2-2 regulates apoptosis via modulation of Akt activation. BmNPV infection of BmE cells resulted in the formation of apoptotic bodies (Figure S11B). A flow cytometry analysis showed that BmPGRP2-2 overexpression and knockdown reduced and increased the apoptotic fraction, respectively (Figure 6C). These results indicate that BmPGRP2-2 induced by BmNPV inhibits apoptosis via Akt activation.
PGRP is well-known to play an important role in immune defense of insects against bacteria. In this study, we demonstrated that BmPGRP2-1 binds to DAP-type PGN to activate Imd signaling and inhibit bacteria, whereas BmNPV-induced BmPGRP2-2 suppresses host cell apoptosis to enable viral replication.
Multiple splice forms of PGRP-L have been identified in Drosophila and A. gambiae, although it is unclear how these are generated (5, 12, 15, 16). Our results indicate that the two forms of BmPGRP2 are transcribed from different TIS. BmPGRP2-1 was generated from the distal TIS and contained a long 5′ UTR, whereas BmPGRP2-2 with a short 5′ UTR was transcribed from the proximal TIS. BmNPV is the primary pathogen to silkworm (46); we speculate that BmPGRP2-1 is the evolutionarily more ancient form, and that BmPGRP2-2 arose through interactions between the silkworm and BmNPV.
The two forms of BmPGRP2 showed distinct expression patterns, subcellular localization, and roles in the immune response to different pathogens. BmPGRP2-1 is a transmembrane protein that binds to DAP-type PGN via the extracellular PGRP domain, thereby activating the Imd pathway and AMP, similar to PGRP-LC in Drosophila. We therefore speculate that BmPGRP2-1 interacts with Imd to activate downstream signaling molecules through an PGRP-LC-like intracellular domain (15, 24, 25). Previous studies have demonstrated that DAP-type PGNs activates Imd signaling, whereas Lys-type PGNs activate the Toll pathway (8, 65). We found here that BmPGRP2-1 was induced by G– bacteria and that silencing BmPGRP2-1 expression suppressed Imd signaling and AMP and reduced the resistance of transgenic silkworm to G– bacteria, confirming that BmPGRP2-1 plays a key role in the immune defense against G– bacteria and possibly against some G+ bacteria with DAP-type PGNs, although the latter requires confirmation.
BmPGRP2-2 was induced by BmNPV to promote viral replication; on the other hand, BmPGRP2-2 knockdown inhibited BmNPV. Some studies have reported that the Imd and Toll pathways are involved in antiviral immune response; inhibition of Imd signaling increased CrPV and alphavirus replication in Drosophila (37, 38) whereas Toll pathway activation suppressed dengue virus replication in A. aegypti (40). The expression of PGRP-LA and PGRP-SC1A was up-regulated in Drosophila after infection of Nora virus (66). Sigma virus infection induced the expression of PGRP-SB1, PGRP-SD, and some AMP genes without altering Toll and Imd signaling in Drosophila (67). Imd pathway activation inhibited BmCPV in silkworm (53). The results of the present study demonstrate that antiviral signaling of BmPGRP2-2 against BmNPV is independent of the two pathways. The results of our previous study (53) and present study revealed the different responses of Imd pathway to different viruses in silkworm. JAK/STAT pathway could be activated by BmNPV but not BmCPV (34). This was unexpected, given that BmPGRP2-2 was found to negatively regulate PTEN and activate Akt to inhibit apoptosis. PGRP-LB has been shown to function as an negative regulator of Imd pathways in Drosophila (26). It was previously reported that PI3K-Akt signaling is required for efficient Baculovirus replication (61). The Hepatitis B virus HBx protein suppressed PTEN expression and activated Akt phosphorylation (68), while the hepatitis C virus NS5A protein-induced suppression of PTEN abrogated its inhibitory effect on PI3K/Akt signaling, triggering Akt activation to promote cell survival (69). This study demonstrated that virus-induced PRRs in the host negatively regulate the PTEN-PI3K/Akt pathway. Thus, the mechanism of BmNPV infection is similar to those of some human viruses, suggesting that B. mori and BmNPV can serve as a model of human–virus interaction, for instance to identify new host genes that are involved in viral infection and to screen antiviral drugs targeting the PTEN-PI3K/Akt pathway.
We found that BmPGRP2-2 negatively regulates PTEN-PI3K/Akt signaling to inhibit cell apoptosis, in contrast to the classic Imd and Toll pathways that are activated by PGRP. Apoptosis plays a key role in host defense against viral infection; however, viruses use various strategies to support cell survival, which is in turn beneficial for viral propagation (69). BmPGRP2-1 was induced by G– bacteria to activate Imd signaling and inhibit bacterial proliferation, whereas BmPGRP2-2 was induced by BmNPV to promote cell survival and consequently, viral replication. Thus, PGRP not only mediates host immune defense against bacteria but is also used by viruses to evade host antiviral defense systems (Figure 7). Some open questions are whether BmPGRP2-2 and other PRRs are used by other viruses; which viral components are recognized by BmPGRP2-2; the mechanism underlying negative regulation of PTEN by BmPGRP2-2; and whether there are other host genes or immune components involved in BmPGRP2-PTEN-PI3K/Akt signaling.
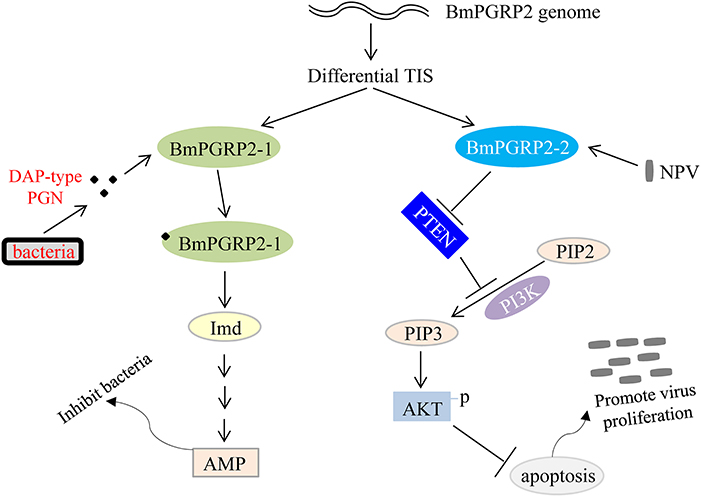
Figure 7. Model of BmPGRP2 functional differentiation in host immune response to pathogens. The two isoforms of BmPGRP2 were generated from different transcription initiation sites (TIS). BmPGRP2-1 recognizes and binds to DAP-type PGN through the extracellular PGRP domain and activates the Imd pathway and AMP to inhibit bacteria; BmPGRP2-2 is induced by BmNPV to suppress PTEN, thereby relieving its inhibition of PI3K/Akt signaling and triggering an increase in Akt phosphorylation and activation to inhibit cell apoptosis. The increased cell survival is beneficial for viral replication. (Part of the evidence for protein levels is lacking).
Some drugs targeting the PTEN-PI3K/Akt pathway can inhibit viral infection. Blocking virus-induced PI3K/Akt signaling inhibited the replication of influenza virus (70). Emodin suppressed p-Akt to induce hepatocellular carcinoma cell apoptosis (71), whereas geridonin combined with paclitaxel induced apoptosis and inhibited the proliferation of gastric cancer cells via upregulation of PTEN and suppression of Akt phosphorylation (72). The PI3K inhibitor LY294002 was shown to block Akt activation and thereby inhibit AcMNPV (61) and BmNPV (this study). These drugs may be used in sericulture to control BmNPV infection if the production process and purity can be optimized. We have generated transgenic antiviral silkworms that block BmNPV infection via overexpression of Bmlipase-1 (55); by suppressing BmNPV mRNA by viral gene RNAi (45); and by inhibiting BmNPV protein synthesis via hycu-ep32 overexpression (73). In the present study, we confirmed that modulating host immune defense can inhibit BmNPV; overexpressing antiviral genes and dsRNA targeting viral genes can further enhance host resistance (74). Hence, combining four antiviral strategies (Bmlipase-1 and hycu-ep32 overexpression and silencing of BmPGRP2-2 and various viral genes) can potentially yield a transgenic silkworm with high resistance to BmNPV.
In conclusion, we cloned two forms of BmPGRP2 generated from different TIS and characterized their distinct functions in the host immune response to pathogens (Figure 7). Our findings indicate that BmPGRP2-2 does not function in canonical immune signaling pathways and PGRP2 is not only involved in host immune defense against invading pathogens, but is also used by viruses to evade host antiviral mechanisms.
LJ and QX designed research. LJ, HG, TC, WY, and QX analyzed data. LJ, WL, YD, QS, BW, YW, and EX performed experiments. LJ and QX wrote the manuscript.
This work was funded by the National Natural Science Foundation of China (No. 31501875, No. 31802014), the Natural Science Foundation of Chongqing, China (cstc2018jcyjAX0211, cstc2018jcyjAX0487, cstc2016jcyjA0524), and Chongqing Postdoctoral Science Foundation (XmT2018006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Xiaofeng Wu for gifting the BmNPV-GFP virus, Takahiro Kusakabe for gifting the BmN4-SID1 cells, Dr. Pingzhen Xu, Dr. Qiong Yang, and Mrs. Yang Xiao for the help in experiment, Dr. Jun Duan and Dr. Yuqian Wu for the help in RNA-seq data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00776/full#supplementary-material
PRRs, pattern-recognition receptors; PAMPs, pathogen-associated molecular patterns; LPS, lipopolysaccharides; PGRP, peptidoglycan recognition protein; PGRP-S, short PGRPs; PGRP-L, long PGRPs; PGNs, peptidoglycans; DAP-type, diaminopimelic acid type; Lys-typ, Lysine typ; Imd, immune deficiency; G–, gram-negative; G+, gram-positive; AMPs, antimicrobial peptides; GNBPs, G– binding proteins; TLRs, Toll-like receptors; proPO, prophenoloxidase; Spz, Spatzle; E. coli, Escherichia coli; S. marcescens, Serratia marcescens; CrPV, cricket paralysis virus; B. mori, Bombyx mori; BmNPV, B. mori nucleopolyhedrovirus; BmCPV, B. mori cytoplasmic polyhedrosis virus; AcMNPV, Autographa californica multiple nucleopolyhedrovirus; PTEN, phosphatase and tensin homolog; PI3K, phosphoinositide 3-kinase; DZ, Dazao; BmNPV-GFP, BmNPV expressing green fluorescent protein; RT-PCR, reverse transcription PCR; qPCR, quantitative PCR; ORF, open reading frame; UTR, untranslated region; CT, cycle threshold; RNAi, RNA interference; A4P, A4 promoter; SV40, Simian virus 40; PGN-EB, PGN from E. coli 0111:B4; PGN-BS, PGN from Bacillus subtilis; PGN-SA, PGN from Staphylococcus aureus; LPS-EB, LPS from E. coli 0111:B4; att2, attacin 2; glv2, gloverin 2; dsRNA, double-stranded RNA; hpi, h post infection; pSKB2-MsyB, pSKB2 vector with an MsyB tag; MyD88, Myeloid differentiation primary response 88; OB, occlusion bodies; GO, gene ontology; DEGs, differentially expressed genes; p-Akt, phosphorylated Akt; V-FITC, V-fluorescein isothiocyanate; IPTG, isopropyl β-D-thiogalactoside; DMSO, dimethyl sulfoxide; ATG, translation initiation site; TIS, transcription initiation sites; NLS, nucleotide localization sequences.
1. Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. (2002) 3:121–6. doi: 10.1038/ni0202-121
2. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
3. Strand MR. The insect cellular immune response. Insect Science. (2008) 15:1–14. doi: 10.1111/j.1744-7917.2008.00183.x
4. Medzhitov R, Janeway CA Jr. Decoding the patterns of self and nonself by the innate immune system. Science. (2002) 296:298–300. doi: 10.1126/science.1068883
5. Kurata S. Peptidoglycan recognition proteins in Drosophila immunity. Dev Comp Immunol. (2014) 42:36–41. doi: 10.1016/j.dci.2013.06.006
6. Rao XJ, Yu XQ. Lipoteichoic acid and lipopolysaccharide can activate antimicrobial peptide expression in the tobacco hornworm Manduca sexta. Dev. Comp. Immunol. (2010) 34:1119–28. doi: 10.1016/j.dci.2010.06.007
7. Tapadia MG, Verma P. Immune response and anti-microbial peptides expression in malpighian tubules of drosophila melanogaster is under developmental regulation. PLoS ONE. (2012) 7:e040714. doi: 10.1371/journal.pone.0040714
8. Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol. (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
9. Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev. (2004) 198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x
10. Ni DJ, Song LS, Wu LT, Chang YQ, Yu YD, Qiu LM, et al. Molecular cloning and mRNA expression of peptidoglycan recognition protein (PGRP) gene in bay scallop (Argopecten irradians, Lamarck 1819). Dev Comp Immunol. (2007) 31:548–58. doi: 10.1016/j.dci.2006.09.001
11. Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem. (1996) 271:13854–60. doi: 10.1074/jbc.271.23.13854
12. Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. (2000) 97:13772–7. doi: 10.1073/pnas.97.25.13772
13. Royet J, Reichhart JM, Hoffmann JA. Sensing and signaling during infection in Drosophila. Curr Opin Immunol. (2005) 17:11–7. doi: 10.1016/j.coi.2004.12.002
14. Dziarski R. Peptidoglycan recognition proteins (PGRPs). Mol Immunol. (2004) 40:877–86. doi: 10.1016/j.molimm.2003.10.011
15. Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in Drosophila. Science. (2002) 296:359–62. doi: 10.1126/science.1070216
16. Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem. (2003) 278:26319–22. doi: 10.1074/jbc.C300184200
17. Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. (2003) 278:7059–64. doi: 10.1074/jbc.M208900200
18. Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. (2006) 24:463–73. doi: 10.1016/j.immuni.2006.02.012
19. Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem Biophys Res Commun. (2006) 350:994–99. doi: 10.1016/j.bbrc.2006.09.139
20. Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathogens. (2006) 2:139–47. doi: 10.1371/journal.ppat.0020014
21. Schmidt RL, Trejo TR, Plummer TB, Platt JL, Tang AH. Infection-induced proteolvsis of PGRP-LC controls the IMD activation and melanization cascades in. FASEB J. (2008) 22:918–29. doi: 10.1096/fj.06-7907com
22. Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA. (2002) 99:13705–10. doi: 10.1073/pnas.212301199
23. Soderhall K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol. (1998) 10:23–8. doi: 10.1016/S0952-7915(98)80026-5
24. Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. (2002) 416:640–4. doi: 10.1038/nature734
25. Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RAB. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E-coli. Nature. (2002) 416:644–8. doi: 10.1038/nature735
26. Kounatidis I, Ligoxygakis P. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. (2012) 2:120075. doi: 10.1098/rsob.120075
27. Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. (2007) 7:862–74. doi: 10.1038/nri2194
28. Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, et al. A spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. (2006) 10:45–55. doi: 10.1016/j.devcel.2005.11.013
29. Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. (2002) 297:114–6. doi: 10.1126/science.1072391
30. Weber ANR, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, et al. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat Immunol. (2003) 4:794–800. doi: 10.1038/ni955
31. Hu XD, Yagi Y, Tanji T, Zhou SL, Ip YT. Multimerization and interaction of Toll and Spatzle in Drosophila. Proc Natl Acad Sci USA. (2004) 101:9369–74. doi: 10.1073/pnas.0307062101
32. Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity (Reprinted from Nature, vol 292, pg 246–248, 1981). J Immunol. (2009) 182:6635–37. Available online at: http://www.jimmunol.org/content/182/11/6635
33. Tingvall TO, Roos E, Engstrom Y. The GATA factor Serpent is required for the onset of the humoral immune response in Drosophila embryos. Proc Natl Acad Sci USA. (2001) 98:3884–8. doi: 10.1073/pnas.061230198
34. Liu W, Liu J, Lu Y, Gong Y, Zhu M, Chen F, Liang Z, Zhu L, Kuang S, Hu X, et al Immune signaling pathways activated in response to different pathogenic micro-organisms in Bombyx mori. Mol Immunol. (2015) 65:391–7. doi: 10.1016/j.molimm.2015.02.018
35. Huang L, Cheng T, Xu P, Cheng D, Fang T, Xia Q. A genome-wide survey for host response of silkworm, Bombyx mori during pathogen Bacillus bombyseptieus infection. PLoS ONE. (2009) 4:e8098. doi: 10.1371/journal.pone.0008098
36. Ma Z, Li C, Pan G, Li Z, Han B, Xu J, et al. Genome-wide transcriptional response of silkworm (Bombyx mori) to infection by the microsporidian Nosema bombycis. PLoS ONE. (2013) 8:e84137. doi: 10.1371/journal.pone.0084137
37. Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the imd pathway in arthropod antiviral response. PLoS Pathogens. (2009) 5:1000582. doi: 10.1371/journal.ppat.1000582
38. Costa A, Jan E, Sarnow P, Schneider D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE. (2009) 4:e7436. doi: 10.1371/journal.pone.0007436
39. Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. (2012) 36:658–67. doi: 10.1016/j.immuni.2012.03.003
40. Xi ZY, Ramirez JL, Dimopoulos G. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathogens. (2008) 4:e1000098. doi: 10.1371/journal.ppat.1000098
41. Kingsolver MB, Huang Z, Hardy RW. Insect antiviral innate immunity: pathways, effectors, and connections. J Mol Biol. (2013) 425:4921–36. doi: 10.1016/j.jmb.2013.10.006
42. Xia QY, Guo YR, Zhang Z, Li D, Xuan ZL, Li Z, et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science. (2009) 326:433–6. doi: 10.1126/science.1176620
43. Xia QY, Wang J, Zhou ZY, Li RQ, Fan W, Cheng DJ, et al. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. (2008) 38:1036–45. doi: 10.1016/j.ibmb.2008.11.004
44. Xia QY, Zhou ZY, Lu C, Cheng DJ, Dai FY, Li B, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. (2004) 306:1937–40. doi: 10.1126/science.1102210
45. Jiang L, Zhao P, Wang GH, Cheng TC, Yang Q, Jin SK, et al. Comparison of factors that may affect the inhibitory efficacy of transgenic RNAi targeting of baculoviral genes in silkworm, Bombyx mori. Antiviral Res. (2013) 97:255–63. doi: 10.1016/j.antiviral.2012.12.020
46. Jiang L, Xia QY. The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect Biochem Mol Biol. (2014) 48:1–7. doi: 10.1016/j.ibmb.2014.02.003
47. Cheng TC, Xia QY, Xu PZ, Tan X, Fang T, Xiang ZH. Identification and comparative analysis of immune-related genes and signaling pathways in the silkworm, Bombyx mori. Acta Entomol Sinica. (2009) 52:235–45.
48. Zhang RN, Li CT, Ren FF, Ye MQ, Deng XJ, Yi HY, et al. Functional characterization of short-type peptidoglycan recognition proteins (PGRPs) from silkworm Bombyx mori in innate immunity. Dev Comp Immunol. (2019) 95:59–67. doi: 10.1016/j.dci.2019.01.015
49. Yang PJ, Zhan MY, Ye C, Yu XQ, Rao XJ. Molecular cloning and characterization of a short peptidoglycan recognition protein from silkworm Bombyx mori. Insect Mol Biol. (2017) 26:665–76. doi: 10.1111/imb.12330
50. Chen K, Liu C, He Y, Jiang H, Lu Z. A short-type peptidoglycan recognition protein from the silkworm: expression, characterization and involvement in the prophenoloxidase activation pathway. Dev Comp Immunol. (2014) 45:1–9. doi: 10.1016/j.dci.2014.01.017
51. Gao K, Deng XY, Qian HY, Qin GX, Hou CX, Guo XJ. Cloning and expression analysis of a peptidoglycan recognition protein in silkworm related to virus infection. Gene. (2014) 552:24–31. doi: 10.1016/j.gene.2014.09.008
52. Jiang L, Peng ZW, Guo YB, Cheng TC, Guo HZ, Sun Q, et al. Transcriptome analysis of interactions between silkworm and cytoplasmic polyhedrosis virus. Sci Rep. (2016) 6:24894. doi: 10.1038/srep24894
53. Zhao P, Xia F, Jiang L, Guo HZ, Xu GW, Sun Q, et al. Enhanced antiviral immunity against Bombyx mori cytoplasmic polyhedrosis virus via overexpression of peptidoglycan recognition protein S2 in transgenic silkworms. Dev Comp Immunol. (2018) 87:84–9. doi: 10.1016/j.dci.2018.05.021
54. Mon H, Kobayashi I, Ohkubo S, Tomita S, Lee J, Sezutsu H, et al. Effective RNA interference in cultured silkworm cells mediated by overexpression of Caenorhabditis elegans SID-1. RNA Biol. (2012) 9:40–6. doi: 10.4161/rna.9.1.18084
55. Jiang L, Wang G, Cheng T, Yang Q, Jin S, Lu G, et al. Resistance to Bombyx mori nucleopolyhedrovirus via overexpression of an endogenous antiviral gene in transgenic silkworms. Arch Virol. (2012) 157:1323–8. doi: 10.1007/s00705-012-1309-8
56. Jin S, Cheng T, Jiang L, Lin P, Yang Q, Xiao Y, et al. Identification of a new Sprouty protein responsible for the inhibition of the Bombyx mori nucleopolyhedrovirus reproduction. PLoS ONE. (2014) 9:e99200. doi: 10.1371/journal.pone.0099200
57. Wang G, Chen Y, Zhang X, Bai B, Yan H, Qin D, et al. Selection of reference genes for tissue/organ samples on day 3 fifth-instar larvae in silkworm, Bombyx mori. Arch Insect Biochem Physiol. (2018) 98:e21458. doi: 10.1002/arch.21458
58. Guo HZ, Jiang L, Xia QY. Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol Gene Genomics. (2016) 291:999–1004. doi: 10.1007/s00438-015-1125-4
59. Guo HZ, Xu GW, Wang BB, Xia F, Sun Q, Wang YM, et al. Phosphoenolpyruvate carboxykinase is involved in antiviral immunity against Bombyx mori nucleopolyhedrovirus. Dev Comp Immunol. (2019) 92:193–8. doi: 10.1016/j.dci.2018.11.015
60. Guo HZ, Huang CL, Jiang L, Cheng TC, Feng TS, Xia QY. Transcriptome analysis of the response of silkworm to drastic changes in ambient temperature. Appl Microbiol Biotechnol. (2018) 102:10161–70. doi: 10.1007/s00253-018-9387-5
61. Xiao W, Yang Y, Weng QB, Lin TH, Yuan MJ, Yang K, et al. The role of the PI3K-Akt signal transduction pathway in Autographa californica multiple nucleopolyhedrovirus infection of Spodoptera frugiperda cells. Virology. (2009) 391:83–9. doi: 10.1016/j.virol.2009.06.007
62. Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase AKT pathway. Proc Natl Acad Sci USA. (1999) 96:4240–5. doi: 10.1073/pnas.96.8.4240
63. Blanco-Aparicio C, Renner O, Leal JFM, Carnero A. PTEN, more than the AKT pathway. Carcinogenesis. (2007) 28:1379–86. doi: 10.1093/carcin/bgm052
64. Downward J. PI 3-kinase, Akt and cell survival. Semi Cell Dev Biol. (2004) 15:177–82. doi: 10.1016/j.semcdb.2004.01.002
65. Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. (2003) 4:478–84. doi: 10.1038/ni922
66. Lopez W, Page AM, Carlson DJ, Ericson BL, Cserhati MF, Guda C, Carlson KA. Analysis of immune-related genes during Nora virus infection of Drosophila melanogaster using next generation sequencing. Aims Microbiol. (2018) 4:123–39. doi: 10.3934/microbiol.2018.1.123
67. Tsai CW, McGraw EA, Ammar ED, Dietzgen RG, Hogenhout SA. Drosophila melanogaster mounts a unique immune response to the rhabdovirus Sigma virus. Appl Environ Microbiol. (2008) 74:3251–6. doi: 10.1128/AEM.02248-07
68. Chung TW, Lee YC, Ko JH, Kim CH. Hepatitis B virus X protein modulates the expression of PTEN by inhibiting the function of p53, a transcriptional activator in liver cells. Cancer Res. (2003) 63:3453–8.
69. Cheng D, Zhang LL, Yang GB, Zhao L, Peng F, Tian Y, et al. Hepatitis C virus NS5A drives a PTEN-PI3K/Akt feedback loop to support cell survival. Liver Int. (2015) 35:1682–91. doi: 10.1111/liv.12733
70. Wu MS, Yen HR, Chang CW, Peng TY, Hsieh CF, Chen CJ, et al. Mechanism of action of the suppression of influenza virus replication by Ko-Ken Tang through inhibition of the phosphatidylinositol 3-kinase/Akt signaling pathway and viral RNP nuclear export. J Ethnopharmacol. (2011) 134:614–23. doi: 10.1016/j.jep.2011.01.005
71. Lin WF, Zhong MF, Yin HX, Chen YG, Cao QX, Wang C, et al. Emodin induces hepatocellular carcinoma cell apoptosis through MAPK and PI3K/AKT signaling pathways in vitro and in vivo. Oncol Rep. (2016) 36:961–7. doi: 10.3892/or.2016.4861
72. Wang SQ, Wang C, Chang LM, Zhou KR, Wang JW, Ke Y, et al. Geridonin and paclitaxel act synergistically to inhibit the proliferation of gastric cancer cells through ROS-mediated regulation of the PTEN/PI3K/Akt pathway. Oncotarget. (2016) 7:72990–3002. doi: 10.18632/oncotarget.12166
73. Jiang L, Cheng TC, Zhao P, Yang Q, Wang GH, Jin SK, et al. Resistance to BmNPV via overexpression of an exogenous gene controlled by an inducible promoter and enhancer in transgenic silkworm, Bombyx mori. PLoS ONE. (2012) 7:e41838. doi: 10.1371/journal.pone.0041838
Keywords: DAP-type PGN, PGRP, Imd, silkworm, BmNPV, PTEN, Akt, apoptosis
Citation: Jiang L, Liu W, Guo H, Dang Y, Cheng T, Yang W, Sun Q, Wang B, Wang Y, Xie E and Xia Q (2019) Distinct Functions of Bombyx mori Peptidoglycan Recognition Protein 2 in Immune Responses to Bacteria and Viruses. Front. Immunol. 10:776. doi: 10.3389/fimmu.2019.00776
Received: 11 December 2018; Accepted: 25 March 2019;
Published: 12 April 2019.
Edited by:
Alexandre Corthay, Oslo University Hospital, NorwayReviewed by:
Ilias Kounatidis, Diamond Light Source, United KingdomCopyright © 2019 Jiang, Liu, Guo, Dang, Cheng, Yang, Sun, Wang, Wang, Xie and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Jiang, amlhbmdsaWFuZ0Bzd3UuZWR1LmNu
Qingyou Xia, eGlhcXlAc3d1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.