
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 18 March 2019
Sec. Viral Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.00440
This article is part of the Research TopicRoles of Fc Receptors in Disease and TherapyView all 25 articles
While neutralizing antibody titers measured by hemagglutination inhibition have been proposed as a correlate of protection following influenza vaccination, neutralization alone is a modest predictor of protection against seasonal influenza. Instead, emerging data point to a critical role for additional extra-neutralizing functions of antibodies in protection from infection. Specifically, beyond binding and neutralization, antibodies mediate a variety of additional immune functions via their ability to recruit and deploy innate immune effector function. Along these lines, antibody-dependent cellular cytotoxicity, antibody-mediated macrophage phagocytosis and activation, antibody-driven neutrophil activation, antibody-dependent complement deposition, and non-classical Fc-receptor antibody trafficking have all been implicated in protection from influenza infection. However, the precise mechanism(s) by which the immune system actively tunes antibody functionality to drive protective immunity has been poorly characterized. Here we review the data related to Fc-effector functional protection from influenza and discuss prospects to leverage this humoral immune activity for the development of a universal influenza vaccine.
Influenza viruses are enveloped negative-strand RNA viruses with segmented genomes that can infect a variety of birds and mammals, including humans (1). Seasonal influenza affects 10–20% of the world's population per year (2), which is estimated to cost $4.6 billion yearly for hospitalizations, doctor's visits, and medications in the United States alone (3). Additionally, influenza causes U.S. employees to miss approximately 17 million workdays due to flu, at an estimated cost of $7 billion a year in sick days and lost productivity (3). Increased infection and mortality occurred during four pandemics in the 20th and 21st centuries, in 1918, 1957, 1968, and 2009 (4), and could occur again if a new strain, such as avian influenzas H5N1 or H7N9, begins to circulate in the human population.
To address this looming threat, the National Institute of Allergy and Infectious Diseases (NIAID) has named the development of a universal influenza vaccine, defined as one that provides protection against symptomatic disease from ≥75% of influenza A strains, as one of its research priorities (5). A key component of the strategic plan is the initial characterization of the correlates of immunity against influenza infection and disease (6). While neutralization has been widely considered the major protective correlate of immunity, hemagglutinination-inhibiting neutralizing antibodies alone have been only modestly linked to protection from seasonal influenza infection, suggesting the involvement of extra-neutralizing antibody functions (7–10). Moreover, currently licensed seasonal influenza vaccines provide only moderate (10–60%) protection against specific strains of seasonal influenza, and little to no protection from emerging pandemic influenza (11). This low efficacy is caused by subtype and strain variability of the two major viral antigens, hemagglutinin (HA) and neuraminidase (NA), as well as by antigenic drift (12). All the available seasonal influenza vaccines, including the inactivated influenza vaccine (IIV), adjuvanted IIV (FluAd), and live attenuated influenza vaccine (LIAV), are given yearly due to limited response durability and the need to induce de novo immunity to novel circulating strains. The development of yearly influenza vaccines relies on predictions published by the World Health Organization to determine the strain composition for a given year (13). Due to the long lead times in producing adequate quantities of the vaccine, the strains must be selected roughly 6 months in advance of vaccine administration (13), leading to population vulnerability should a new strain enter circulation. When these predictions did not match seasonal circulating strains, the effectiveness was very low (11).
Beyond efforts to match sequences to ensure seasonal immunity, the current surrogate of protection used to evaluate influenza vaccines is the hemagglutination inhibition (HAI) assay (14). HAI was identified as a predictor of protection from infection in the initial study of egg-grown inactivated vaccine efficacy conducted in 1943 by Salk et al. (15). HAI measures the ability of an antibody or serum sample to prevent HA binding to red blood cells, and is considered a proxy for neutralization by receptor blockade (14). HAI, however, does not fully explain or predict protection in humans (7–9). Indeed, individuals lacking detectable HAI titers were found to be resistant to influenza infection. Additionally, infection risk was clearly linked to age independently of HAI titer (7). While HAI is considered a classical surrogate of protection from influenza infection, it alone is not sufficient to fully explain protection (10).
Humoral immune responses to influenza are largely directed toward the hemagglutinin molecule (HA). HA, the primary viral glycoprotein, exists as a trimer made of monomers composed of two subunits, HA1, roughly corresponding to the “head” and HA2, or “stem” (16) (Figure 1). Heterosubtypic or cross-reactive, antibodies to the HA head region are relatively rare due to heavy glycosylation and low sequence conservation in this region (17, 18). Although highly variable, cross-reactive neutralizing antibodies have been discovered against the HA head. However, these antibodies bind primarily to specific conserved epitopes, including the receptor binding domain (18). In contrast, while several protective antibodies have been identified against the HA stem, as it is more conserved, this region of the HA is poorly immunogenic (17).
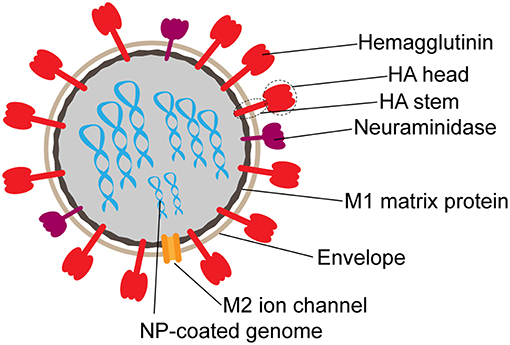
Figure 1. Schematic structure of influenza virion. Surface proteins hemagglutinin (HA) and neuraminidase (NA) are present on the surface at an approximate 3:1 ratio. The M2 ion channel also spans the envelope. M1 matrix protein forms the inner capsid, which surrounds the segmented RNA genome coated in nucleoprotein (NP).
Heterosubtypic protective antibodies against influenza primarily target either the receptor binding site on the HA head or the more conserved HA stem (19). However, the stem region is infrequently targeted compared to easily inducible strain-specific HA head responses (20). Regardless of target, antibodies against HA can mediate protection by neutralization or extra-neutralizing functions, and both modalities may be exploited by a single antibody specificity. Antibodies that target the head largely provide protection by preventing the virus from entering the target cell, and are thus referred to as neutralizing antibodies (19). Non-neutralizing functional protective influenza-specific antibodies have been documented against both the HA head and stem (21, 22); however, their mechanisms of action are more complex and varied (23). Although protective non-neutralizing antibodies have been documented across the HA molecule, these types of protective antibodies more dominantly target the stem region of HA and can exhibit wide reactivity, capturing most influenza A viruses (20). Because the neutralizing capacity of antibodies is dose-dependent (21), in this review the term non-neutralizing will be used to describe antibodies that cannot efficiently neutralize virus at the concentration currently being studied.
While the mechanism of protection mediated by neutralizing antibodies is simple to comprehend, the extra-neutralizing mechanisms of action of antibodies are less well-understood. Emerging evidence has suggested that mechanisms including the ability of antibodies to leverage the innate immune system may contribute to protection against influenza (21–31). Critically, antibodies possess two functional domains: the Fab, which recognizes the antigenic epitope, and the Fc, which interacts with Fc receptors (FcR) or complement to drive antibody-mediated effector functions (Figure 2). Passive transfer studies using both native IgG1 and FcR-binding ablated monoclonal antibodies (mAbs) clearly illustrated the importance of Fc-mediated functions in protection from infection (21, 22). Moreover, follow-up studies using FcR and complement knockout mice further clarified the critical nature of specific Fc-effector functions in protection (21–23, 25–31). Antibody mediated macrophage phagocytosis (28, 32), neutrophil production of reactive oxygen species (28), cellular cytotoxicity (29), and complement deposition (26, 27, 32, 33) have all been implicated as protective functions leveraged by antibodies to drive protection from infection and/or viral clearance. Strikingly, even broadly neutralizing HA-stem targeting and pan-strain HA-head targeting mAbs require FcRs to confer protection (22). In this review, we explore the various mechanisms beyond neutralization that are exploited by antibodies to confer protection from influenza and promote viral clearance.
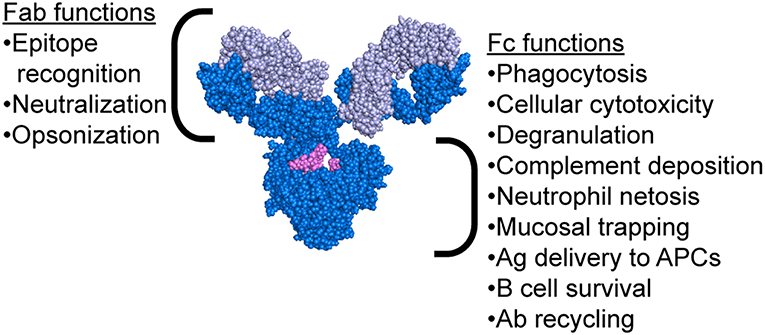
Figure 2. Antibody structure highlighting functions of both the Fab and Fc regions. Antibody image shows heavy chain in dark blue, light chain in light blue, and glycan in magenta. Antibody structure: PDB 1IGY.
Antibody-dependent cellular cytotoxicity (ADCC) is largely mediated by the interaction of pathogen or cell-surface bound antibodies with Fc gamma receptor IIIa (FcγRIIIa) on NK cells in humans (Figure 3D) and FcγRIV on monocytes, macrophages, and neutrophils in mice (34, 35). FcγRIIIa is found on the surface of human NK cells, monocytes, and macrophages (36). Engagement of FcγRIIIa by antibody causes the release of cytotoxic granules from NK cells, uptake by macrophages, apoptosis of infected targets, and secretion of antiviral cytokines and chemokines (37–40).
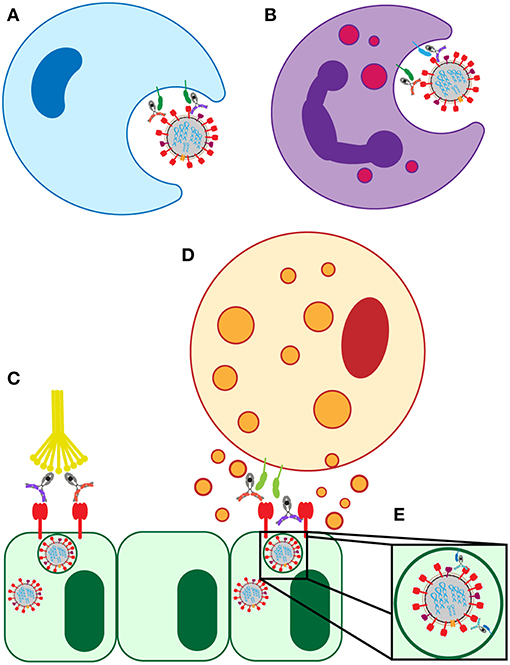
Figure 3. Known FcR-dependent innate immune effector functions acting in influenza infection. (A) Clearance of virions and infected cells by macrophage phagocytosis. (B) Clearance of virions and infected cells by neutrophil phagocytosis, and the release of cytokines and reactive oxygen species. (C) Clearance of infected lung epithelial cells and activation of the adaptive immune system by antibody interaction with C1Q. (D) Clearance of infected lung epithelial cells by ADCC. (E) Neutralization of virus by FcRn-bound HA-specific antibodies.
It was first reported in 1977 that human peripheral blood leukocytes engaged in influenza-specific ADCC, and that this effect correlated with antibody-mediated virus neutralization (41, 42). In vitro ADCC activity was linked to protection conferred by antibodies in a mouse model of influenza infection (21, 43). Mice that received an FcR-binding competent antibody capable of inducing ADCC in vitro (measured by CD107a expression) exhibited increased survival and decreased morbidity compared to mice that received either an antibody unable to bind to FcRs or a less potent ADCC-inducing antibody (21). Similarly, macaque models of repeated influenza infection confirmed the rapid development of ADCC following infection in animals previously exposed to influenza. This highlighted the presence of antibodies in the bronchoalveolar lavage (BAL) capable of inducing activation of NK cells, which were associated with increased viral clearance and decreased duration of disease (44). Analogously, in human studies, ADCC titers were associated with a reduction in disease burden in a seasonal influenza experimental infection study (45). Additionally, older adults, who had previously seen 2009 pandemic influenza-like viruses in the past, and who retained long-lived ADCC, but not neutralizing, antibody titers, were protected during the 2009 H1N1 pandemic (46). This provided further evidence that ADCC-mediating antibodies are associated with protection.
However, despite our emerging appreciation for the potential role of ADCC in protection from infection and disease, the seasonal influenza vaccine poorly induced broadly reactive ADCC-inducing antibodies in healthy children and adults (45, 47–49). Conversely, the presence of cross-reactive HA-specific antibodies that can activate NK cells in older adults suggests that these functional antibodies accumulate over the course of many years of repeated natural infection with influenza (48, 50). Despite the delay in their evolution, the data clearly suggest that these functional cross-reactive antibodies emerged naturally over time. Moreover, some healthy American adults possessed ADCC activity against avian H7N9 and H5N1 viruses that do not circulate in North America but could cause pandemic outbreaks. This indicated the natural evolution of cross-reactive functional antibodies targeting diverse HA antigens in the absence of exposure and/or other conserved viral proteins such as NP and M2 (31, 51). Furthermore, broadly cross-reactive ADCC-inducing antibodies were reported in individuals who lack broadly neutralizing influenza-specific antibodies (46, 50–52), suggesting that these functions emerge separately and may evolve under distinct stimuli. Collectively, the data clearly demonstrate that broadly protective ADCC inducing antibodies are associated with protection and evolve naturally over time.
HA head-specific mAbs induced less ADCC than stalk-specific antibodies in an in vitro NK cell activation assay (21). This difference in function has been suggested to be related to the inability of the head-specific mAbs to efficiently multimerize when bound to antigen on the cell surface and interact with low-affinity FcRs to induce functional responses (21). A recent study suggested an alternative explanation in experiments using FLAG-tagged HA to direct FLAG-specific antibody to certain regions of the HA molecule. The data from this study suggested that two points of contact were required between infected and effector cells for efficient ADCC activity (53). These direct contacts are (1) between the mAb Fc and FcR and (2) between the cell surface sialic acid and viral HA (53). However, the co-ligation of FcR and viral protein has not been borne out by studies in other infection or disease contexts, or in polyclonal pools of antibodies directed against native HA.
An additional layer of complexity in dissecting non-neutralizing antibody mediated mechanisms of protection in vivo is the comparison of polyclonal vs. monoclonal mediated antibody functions. Emerging data suggest that the level of in vitro ADCC is influenced by the ratio of ADCC-inducing to ADCC-inhibiting antibodies (22, 54). ADCC-inhibiting antibodies, which can be neutralizing, were shown to compete for binding sites on HA on the surface of viral particles and infected cells (22, 54). While the delivery of single protective ADCC-inducing mAbs demonstrated striking protection from infection in vivo (21, 22), polyclonal pools of antibodies exhibited a much more complex balance of epitopes targeted and functional competition that collectively may contribute to differential protection from infection during seasonal exposure. While it is clear that functional antibodies play a vital role in protection against influenza infection, experimental approaches able to comprehensively dissect the nature of polyclonal antibody interactions are urgently needed to further define the nature of protective antibody activity and guide vaccine design.
ADCC-inducing antibodies, as well as the direct cytopathic effects of the virus, drive infected cell apoptosis (55). These infected apoptosing cells are then cleared through phagocytosis to maintain tissue homeostasis (56). Post-infection, macrophages are rapidly recruited to the lung and are present in BAL, airway, and alveoli to support the rapid clearance of infected and/or dying cells (57). While the supernatant of influenza-infected cells can stimulate monocyte phagocytosis independently of antibody involvement (57), antibodies contribute to accelerated clearance of viral particles and infected cells through interactions with FcγRIa and FcγRIIa on immune cells (58). Antibody mediated viral phagocytosis, resulting in viral degradation, was linked to decreased spread and severity of infection (58). While this mechanism was not directly associated with prevention of infection, it was linked to reduced severity of symptoms and viral shedding, and thus attenuating disease in humans.
Antibody-dependent cellular phagocytosis (ADCP) activity (Figure 3A) in healthy human serum, mediated by monocytes/macrophages, was shown to correlate with HAI titer both for circulating and non-circulating strains of influenza (58). Interestingly, ADCP activity was still detectable in diluted serum samples, even at dilutions where neutralization was no long detectable (58). This indicated that phagocytic antibodies may mediate viral clearance even at very low levels, and thus could still provide protection or lessen the severity of disease. Along these lines, non-neutralizing protective mAbs in mice required alveolar macrophages to provide protection. This protection was partially dependent on the induction of a robust inflammatory response in the lung as shown by tissue histology and increased cytokine/chemokine production, and was partially through direct phagocytosis (30). Additionally, broadly neutralizing HA-specific mAbs also exhibited enhanced protection in the presence of alveolar macrophages (30). This macrophage-mediated protection was dependent on interactions of the antibody with FcRs on the macrophage surface, as evidenced by experiments using FcR-binding null antibodies that failed to provide protection from infection (30). Together, these studies indicate that FcR-mediated macrophage activation reduces disease burden and protects mice from lethal influenza, and that healthy human serum has influenza-specific antibodies capable of inducing this function.
Neutrophils are among the first cell populations recruited to the site of infection and/or inflammation, and have been implicated in the protective response to influenza (59). Neutrophils are involved in the phagocytic clearance of both virions and infected cells, release immunostimulatory cytokines and chemokines to recruit additional immune cells, and form neutrophil extracellular traps (NETs) to capture and inactivate the virus (60). During influenza infection, neutrophils generate the chemokine CXCL12, required for efficient recruitment of cytotoxic CD8+ T cells to the lung (61). However, beyond this indirect anti-viral role, human neutrophils express high levels of FcγRIa/b/c, FcγRIIa, and FcγRIIIb after activation, enabling them to respond rapidly and efficiently to antibody coated targets (36). In addition, neutrophils constitutively express the FcαRI, an activating receptor that binds IgA and activates cytotoxic and phagocytic responses via a shared FcR γ-chain (62).
Like macrophages, neutrophils are intimately involved in the phagocytic clearance of infected and apoptotic cells in the lung during influenza infection (Figure 3B) (57). Impaired neutrophil phagocytosis through depletion of neutrophils was linked to decreased survival in a mouse model of influenza infection (57, 63). Following phagocytosis, neutrophils form phagolysosomes containing reactive oxygen species (ROS) to eliminate the virus (60). Both HA head- and stalk-specific mAbs induced the production of ROS by neutrophils in vitro in an FcR dependent manner, shown following Fc-blockade resulting in reduced ROS production (28). Influenza-specific class-switched IgA antibodies were also implicated in neutrophil activation and ROS production (28). Despite the ability of some antibodies to recruit neutrophil activity in vitro, the critical nature of neutrophils in protection from infection remains controversial. Some animal studies using neutrophil depletions finding no significant roles for these cells in protection mediated by passively transferred non-neutralizing mAbs (30), arguing that alveolar macrophages may play a more dominant role in protection. In other studies, neutrophil recruitment and function was linked to protection from infection and reduction in disease (63, 64). Thus, additional studies will be required to ultimately define the role of neutrophils in influenza infection.
The complement system can recognize and eliminate viruses directly or can contribute to viral clearance via antibody mediated activation (Figure 3C) (33). The requirement for complement in protection from lethal influenza infection in mice was established in 1978 and has been more recently replicated on novel influenza strains (26, 65). Influenza virions were shown to be susceptible to both classical and alternative complement mediated lysis in vitro only when opsonized by antibodies (27). However, the level of susceptibility varied by strain. Further supporting the involvement of antibody-mediated complement elimination in the influenza immune response, synergy between the classical and alternative complement pathways was shown to provide protection against pandemic H1N1 strains in mice and the cooperativity of both pathways is associated with enhanced viral clearance (27). In these experiments, C3 knockout mice (deficient in all complement pathways because C3 is the central point of the cascade), C4 knockout mice (deficient in classical and lectin pathways), and complement factor B (FB) knockout mice (deficient in alternative pathway) were infected with influenza and disease progression was compared. While both C4- and FB-deficient mice showed increased mortality, neither pathway alone nor the additive mortality approached the level of mortality in C3 knockout mice, who have both pathways of complement ablated. This indicated that the two complement pathways work synergistically to clear infection (27).
Beyond antibody driven virion elimination, the complement protein C3 was also shown to promote higher titers of influenza-specific IgG antibodies. C3 also improved CD4+ and CD8+ T cell responses in the mouse models of influenza infection (33). Vaccination was administered to C3 knockout mice, resulting in dampened antibody titers leading to increased mortality when compared to wild type mice. The role of complement in driving immunity was proposed to be effectuated by the formation of pro-inflammatory complement degradation products C5a and C3a, which can serve a dual role of directly recruiting T cells and enhancing T cell priming by recruiting and stimulating antigen-presenting cells to the site of infection (33).
In human serum, neutralization and complement-dependent lysis activities by mAbs have not always correlated, although neutralizing antibodies can induce complement-dependent lysis (66). Both IgG1 and IgM antibodies have been implicated in the activation of the complement system in influenza infection (27). Complement-stimulating antibodies correlated with protection from infection in children in a serosurveillance study of seasonal influenza (50), which was potentially attributable to their generally higher cross-reactivity compared to neutralizing antibodies (31). Importantly, if complement-inducing antibodies do in fact generally possess higher cross-reactivity when compared to neutralizing antibodies, complement lysis of virus is an attractive strategy for limiting initial infection with influenza by otherwise non-protective non-neutralizing antibodies, broadening the epitopes that can be targeted by vaccination.
The neonatal Fc receptor, FcRn, is involved in transcytosing IgG across the placenta during fetal development, across the vascular endothelium to increase extravascular antibody levels, and across the mucosal epithelium to provide humoral defense within the mucosa (67). Additionally, FcRn has a non-canonical role in antiviral immunity against influenza. FcRn was implicated in facilitating antibody-mediated neutralization of influenza virions by HA head-specific antibodies that bind to the virus at acid pH (Figure 3E) (68). These unusual head-specific antibodies were then shown to neutralize the virus by preventing trafficking of the viral ribonucleoproteins into the nucleus for replication (68).
Systems level analyses aimed at defining biomarkers of productive immunity to flu vaccination identified pre-existing antibody titers as a negative predictor of response to vaccination (69), thought to act by capturing, destroying, and preventing response to vaccine antigens (historically called original antigenic sin) (70). However, recent studies suggested that pre-existing antibodies shape the immune response to influenza vaccination in ways that could be utilized to improve protection. Individuals with the most influenza-specific antibody affinity maturation had significant changes in antibody glycosylation, namely increased sialic acid (71). A mechanism was proposed in which pre-existing cross-reactive influenza antibodies, which opsonize incoming vaccine antigen, drove the delivery of immune complexes to germinal centers of lymph nodes by subcapsular sinus macrophages or non-cognate B cells, preferentially when antibodies were sialylated (72, 73). This delivery relied on interaction of immune complexes with the non-canonical IgG Fc-receptor CD23 to capture antigen and move it to germinal center (71). This delivery of antigen to lymph nodes was speculated to increase and extend the contact between B cells and antigens of interest to drive affinity maturation, which can increase both the affinity and potential evolution of neutralization (74, 75). Identified broadly cross-reactive neutralizing mAbs specific to influenza HA were shown to be highly affinity matured, indicating that this pathway may be essential for the development of broad humoral immunity (76).
In addition to trapping and delivery, these sialylated Fcs were also shown to increase B cell inhibitory FcR, FcγRIIb, expression, resulting in elevated thresholds required to activate B cells during development in the germinal center (77). With these elevated activation thresholds, B cells that require higher affinity interactions or an ability to capture more antigen to become fully activated within the germinal center may then experience more aggressive somatic hypermutation and consequent affinity maturation. Sialylated immune complexes bind to non-cognate B cells at high levels in the presence of CD23 (77) to increase affinity maturation. This may offer a novel approach for the design of next generation vaccines able to leverage the potent immunomodulatory activity of the Fc-domain of antibodies.
Together, these data suggest that the quality of the antibody response may not only influence direct antiviral activity, but may also be critical in influencing the response to vaccination. Seasonal influenza infection in humans has been shown to induce moderate antibody cross-reactivity (78), and these pre-existing cross-reactive antibodies may be molding the affinity maturation of new antibodies following vaccination through mechanisms that increase somatic hypermutation, including increased antigen retention within germinal centers (71). Harnessing, increasing, and improving this pathway presents a novel method of improving the breadth and binding affinity of antibodies following influenza vaccination.
The functional potency of an antibody is significantly affected by the antibody's subclass, which determines the binding affinity of the antibody for FcRs (79). Antibody function is determined at the time of B cell programming via class switch recombination (IgA, IgM, IgG, IgD, or IgE; Figure 4). Because each isotype can interact with a distinct family of FcRs present on innate immune cells within disparate compartments, each isotype has the capacity to drive unique antibody effector functions. Beyond the isotypes, there is additional capacity to select for subclasses of particular antibody isotypes. In humans, four IgG subclasses can be additionally selected during an immune response, each of which have further differential affinities for individual FcRs (36). IgG1 antibodies are the most prevalent at approximately 65% of total serum IgG, with the other three subclasses in decreasing fractions in numerical order (80). Because individual subclasses have different affinities for FcRs (36), subclasses drive different antibody effector functions. IgG1 and IgG3 are considered to be the most functional subclasses due to their enhanced ability to bind to FcRs, while IgG2 and IgG4 have lower affinities for FcRs (81, 82). However, IgG3 has a shorter half-life, related to decreased binding affinity to FcRn and due to a proteolytically vulnerable hinge, although multiple allotypes of IgG3 with longer half-lives have been reported among non-Caucasian populations (83).
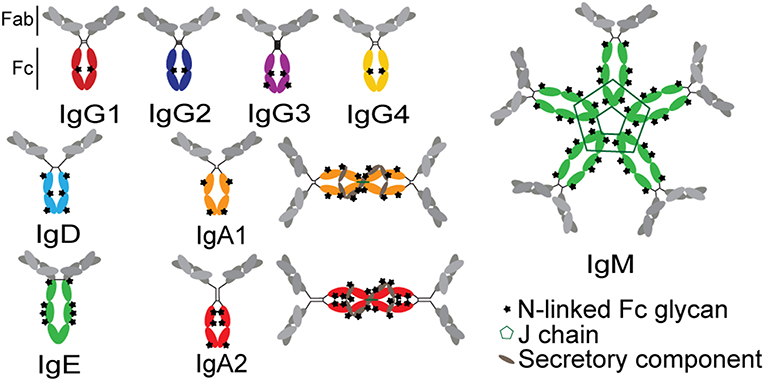
Figure 4. Structures of antibody isotypes and subclasses. Fc domains are in color while Fab domains are in gray. Stars indicate N-linked Fc glycans. IgA isotypes are shown as both monomers (predominant in serum) and dimers (predominant at mucosal surfaces).
The relative magnitude and distribution of IgG subclass responses vary between acute influenza infection and vaccination. Vaccination increased IgG3 production when compared to acute infection in adults and children who were previously exposed to natural influenza but not previously vaccinated (84). IgG3 levels following seasonal influenza vaccination correlated with cytokine production by peripheral blood mononuclear cells (PBMCs) stimulated ex vivo with infectious influenza virus, suggesting that enhanced IgG3 responses were a marker of a more effective response to vaccination (80). While IgG3 is widely considered to be the most functional subclass due to its affinity for FcRs, the specific effects of IgG3 in protection from influenza remain largely unclear.
While IgG is present at higher levels in the blood, IgA antibodies are produced at considerable levels in mucosal tissues (85). Secreted IgA represents ~70% of the body's total Ig production and, in mucosa, is primarily dimeric, with only small fractions of monomer, trimer, and tetrameric IgA in the mucosa. In serum, IgA is primarily monomeric (86). Mucosal IgA can prevent influenza infection in the nasal and upper respiratory mucosa with higher heterologous neutralization than IgG with the same Fab (85, 87–89). The protective activity of IgAs was linked to both direct viral neutralization as well as viral capture and cross-linking to the mucosal surface, preventing cell entry in the absence of classical neutralization (85). However, beyond its direct antiviral effects, IgA may also recruit the indirect activity of the innate immune system, via the Fc-receptor for IgA, FcαRI, which is constitutively expressed on neutrophils and increases in expression as neutrophils mature (62). Stimulation of this receptor by influenza-specific IgA was linked to increased ROS production (28), although the precise effect of this activation during influenza infection is unclear.
Beyond isotype and subclass selection, the humoral immune response additionally modifies antibodies via post-translational changes in Fc-glycosylation to further tune antibody affinity for FcRs, and thus to modulate antibody effector function (90). Each IgG molecule contains two N-glycosylation sites, at asparagine 297 (N297) on each heavy chain (Figure 4). The core Fc glycan structure is biantennary, with a structure consisting of two-branched linked N-acetylglucosamine (GlcNAc), a mannose, followed by 2 branched mannoses, each followed by an additional GlcNAc on each mannose (Figure 5). Three additional sugars can then be added at variable levels, including a core fucose on the first GlcNAc, galactoses that can be added to each terminal GlcNAc, sialic acids that can subsequently be added to the each galactose, and finally the addition of a bisecting GlcNAc to the core mannose (Figure 5) (91). Given the variable addition of each of the 4 additional sugars, a total of 36 distinct glycan structures can be added to any given IgG (92). Importantly, while glycans do not interact directly with FcRs, they influence the flexibility and structure of the antibody Fc, thereby changing interactions with FcRs (93). Complete removal of the Fc glycan ablates low affinity FcR binding, with only high affinity FcγRI retaining measurable binding ability (94). Additionally, IgA and IgM antibodies are also Fc-glycosylated (Figure 4), although it is unclear how this glycosylation changes affinity for FcRs.
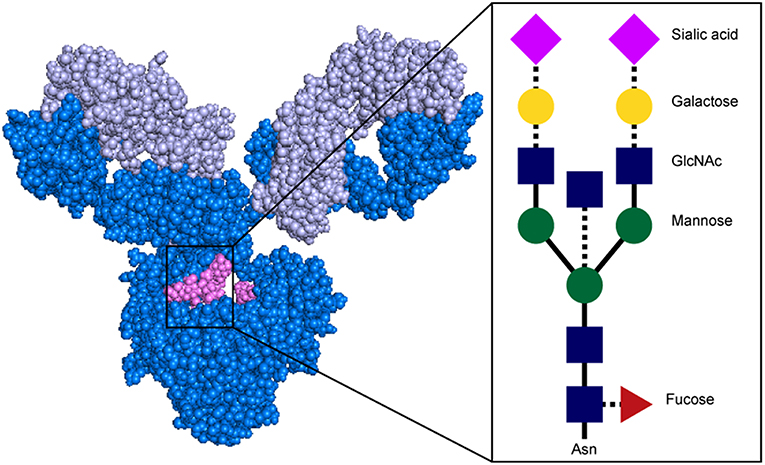
Figure 5. Structure of antibody glycan. Antibody image shows heavy chain in dark blue, light chain in light blue, and glycan in magenta. In glycan schematic, solid lines indicate core glycan consisting of two-branched linked N-acetylglucosamine (GlcNAc; blue rectangle), a mannose (green circle), followed by 2 branched mannoses, each followed by an additional GlcNAc on each mannose. Dotted lines indicate additional sugars that can be added at variable levels, including a core fucose on the first GlcNAc (red triangle), galactoses (yellow circle) to each terminal GlcNAc, sialic acids (pink diamond) to the each galactose, and a bisecting GlcNAc to the core mannose. Antibody structure: PDB 1IGY.
Across diseases, dramatic shifts have been identified in IgG glycosylation, such as a significant increase in agalactosylated antibodies in chronic inflammatory diseases such as autoimmune flares and HIV infection (95, 96). In the monoclonal therapeutics community, the systematic removal of specific components of the Fc N-glycan have highlighted the critical role of individual sugars in shaping antibody effector function (97). Specifically, the presence of sialylation drives anti-inflammatory activity in vivo (98, 99). The removal of fucose either directly or indirectly, through the upregulation of the bisecting GlcNAc, results in increased antibody affinity for FcγRIIIa and consequently enhanced ADCC (90, 100). Conversely, agalactosylated antibodies decrease ADCC and drive pro-inflammatory responses (90).
Beyond our emerging appreciation for a role of sialylated antibodies in vaccine induced affinity maturation (71, 101) described above, influenza vaccination is known to alter Fc glycosylation (102, 103). Soon after immunization, influenza-specific antibodies rose rapidly, and had increased galactosylation, increased sialylation, and reduced bisection compared to pre-existing influenza-specific antibodies (102, 103). However, these glycan shifts normalized after a month, suggesting that these transient changes in influenza-specific antibody glycan profiles may reflect differences in glycosylation by plasmablasts, not plasma cells. Even in the absence of vaccination, HA-specific antibodies exhibited unique glycan profiles compared to HIV-specific antibodies from the same individuals. Specifically, HA-specific antibodies were more highly galactosylated and sialylated and contained reduced b-GlcNAc (102), highlighting the unique glycan profiles that are naturally selected on influenza-specific antibodies. Whether individuals who control the virus more effectively tune antibody glycosylation in a specific or selective manner is unclear, but represents a simple strategy to modulate antibody function.
One of the greatest hurdles of influenza vaccination is overcoming response anergy caused by previous exposures to influenza in the aging immune system. An attractive strategy to overcome this anergy and generate protective humoral immunity, particularly for novel strains or universal vaccine formulations, is the use of adjuvants to enhance and tune the response to vaccination. There are currently four adjuvants licensed for use in influenza vaccines in the United States and/or in Europe: aluminum salts (alum), MF59, AS03, and virosomes (104). In addition to adsorbing antigens and creating multivalent lattices of antigens, alum activates the inflammasome promoting more effective responses upon antigen-presenting cell delivery (105). Alum induces primarily a Th2-driven response (104). However, Th1 responses are likely to be more critical in the clearance of intracellular pathogens, including influenza (105). Oil-in-water emulsions, such as MF59 and AS03, are predicted to work through a more balanced Th1/Th2 response, enhancing both T cell and antibody responses via delivery to antigen presenting cells as well as through the recruitment of specific innate immune cells to the site of injection (106). Specifically, in an adjuvanted HIV vaccine trial in macaques, MF59 increased recruitment of neutrophils, monocytes, and MDCs to the site of injection and recruitment of neutrophils to the draining lymph node (107). In the context of influenza vaccination, MF59 increased the heterosubtypic, or broadly reactive, antibody response and increased neutralizing antibody responses to influenza (108, 109). Unfortunately, MF59 also shifted the response even further toward the immunodominant HA head and away from the HA stem (110). Yet, MF59 increased the affinity of antibodies developed following both seasonal and novel pandemic influenza vaccines, suggesting that if skewed selectively to particular target antigenic sites, this adjuvant could drive enhanced affinity maturation to the correct sites of vulnerability (110). Virosomes or phospholipid vesicles, have also been studied in the context of influenza HA and NA vaccines, showing similar profiles to MF59 (104). The effects of these adjuvants on FcR-mediated antibody functionality are only beginning to be studied (107).
Other adjuvants are currently under investigation to specifically and selectively enhance influenza specific immunity. For example, liposomes provide unique scaffolds for antigen delivery (105), and were shown to increase the humoral and Th1 response, boosting neutralization, in mice following influenza vaccination (104). Additionally, virus-like particles, or nanoparticles, which deliver antigens in a multivalent manner, similar to their native conformation, increased heterosubtypic IgG2a neutralizing antibody titers in mice, the mouse analog of IgG3, the most functional antibody subclass in humans (111). Presentation of antigens in the form of a viral particle may play an essential role in driving functional antibody responses (112–114). Another type of adjuvant, ISCOMS (antigen, cholesterol, phospholipid and saponin-defined immunomodulatory complexes), created a balanced, protective immune response based on strong MHC class I presentation in trials with a pandemic influenza antigen (104). However, tests of ISCOMS with cancer antigens showed that this adjuvant shifted the response away from antibodies, toward CD4+ and CD8+ T cells, with limited changes seen to antibody responses (115). Finally, Toll-like receptor (TLR) agonists, involved in early pathogen sensing, are known to tune the inflammatory response to tailor immunity in a pathogen specific manner (104). Several TLR agonists were shown to increase influenza-specific antibody titers following vaccination, however their effects on antibody-mediated functions beyond neutralization are unexplored (104, 116). Thus, while previous studies with these adjuvants have primarily focused on neutralizing antibody responses, additional insights on the specific effects of adjuvants on shaping protective FcR activity will provide additional avenues to tune and direct protective immunity against influenza infection.
In addition to efforts to promote more effective immune stimulation through adjuvants, intense investigation has focused on the development and design of unique antigens able to selectively direct the immune response away from strain-specific immunodominant sites to those that are more conserved (17). These include the design of computationally enhanced globally relevant HA sequences, the design of mini-antigens and chimeric antigens, and glycan-enhanced antigens.
Computationally optimized broadly reactive antigens (COBRAs), designed based on computational modeling of influenza strains to create mosaic antigens aimed at focusing the immune response on the evolution of heterosubtypic responses, had some success in eliciting broadly reactive HAI titers that target both seasonal and pandemic strains of influenza (117–120). In a recent study, COBRA H3s did not increase the breadth of HAI reactivity by vaccine-induced antibodies across a panel of strains. However, these COBRA H3s did increase the phylogenetic diversity of neutralized strains (120), meaning that the COBRA-induced responses altered which strains were recognized and neutralized without increasing the total number of strains recognized (120). These data suggest that COBRA antigens can increase heterosubtypic responses to conserved epitopes on the immunodominant head, but are unable to create broadly reactive responses to the conserved HA stem.
Given the complexity of altering immunodominance using whole HA molecules, additional efforts have aimed to direct immunity against minimal antigenic regions associated with broadly protective responses including the stem (121, 122) and the receptor binding site (123). Although broadly protective non-neutralizing responses can target the stem region of HA, these responses are typically subdominant (124). HA stem-only antigens or antigens with conserved stem domains but altered HA heads have been developed (17). For example, a “headless” HA vaccine tested in mice created a broadly protective non-neutralizing immune response (125). Furthermore, chimeric HA vaccines, which were used to immunize animals with “exotic” chimeric molecules that coupled unusual heads to a single stem region, have shown promise. Specifically, using heads that have not circulated in the population, coupled to conserved stems, this vaccine strategy drove robust focused stem-specific protective immune responses and higher cross-reactive HA-specific antibody titers, and are now in clinical trials (126, 127).
Seasonal influenza vaccines have been produced in eggs since the introduction of yearly vaccination, and the manufacturing techniques have remained largely unchanged for decades (128). Emerging data and technical advances are increasing the attractiveness of cell culture-based production strategies, rather than egg-based production. Beyond issues related to speed and cost of vaccine production across these platforms, qualitative differences in antigens from egg-based vaccines compared to circulating viruses may necessitate this shift. HA is highly N-glycosylated in a host-cell dependent manner (129–131). The glycosylation of egg-grown vaccine virus is different than that of naturally infecting virus (132). Emerging data points to the importance of glycosylation not only in shaping antigen-exposure on the surface of the HA molecule, such as masking of specific epitopes (131, 133), but also in contributing to the antigenicity of mAb binding epitopes (134–136). Differential HA glycosylation between egg- and cell culture-grown virus impacted innate immune interactions with the virus in the lung, including neutralization by surfactant protein D (SP-D) (137) and binding to mannose-specific lectins (138). Moreover, altered glycosylation was shown to change both cellular and humoral response kinetics in vitro and in vivo (131, 139). Vaccination of mice with de-glycosylated HA led to decreased CD4+ T cell activation and cytokine production, resulting in reduced HA-specific antibody titers and HAI titers (131, 139). Studies of the antibody response using differentially glycosylated (not de-glycosylated) HAs showed that glycosylation alters the binding and neutralization of monoclonal antibodies, but lacked further detail about the effects of glycosylation on polyclonal antibody pools or on Fc-mediated function (131, 139, 140).
Epidemiological studies in recent years investigating poor vaccine protective efficacy have shown that antigen glycosylation had a direct impact. In the 2016–2017 flu season, the circulating H3N2 virus had a new glycosylation site compared to previous seasons. However, the egg-adapted version of the viral strain used to produce the vaccine lacked this site through an amino acid mismatch in an antigenic site, resulting in decreased vaccine effectiveness (134). Given that glycosylation can strongly impact epitope antigenicity, a vital mismatch at a site of neutralization sensitivity resulted in the induction of non-protective immunity and rendered the circulating virus invisible to vaccine induced immune responses. Shaping glycosylation to produce representative antigens is critical to achieving protective immune responses to vaccination.
While the majority of the humoral immune response is directed toward the immunodominant HA molecule, antibodies also emerge against other targets including neuraminidase (NA), nucleoprotein (NP), and Matrix-2 (M2) (Figure 1). Antibodies targeting NA, while not neutralizing, can prevent viral exit from infected cells to block subsequent rounds of infection (12), and have been associated with seasonal protection (12, 141–143). However, NA-specific antibodies have also been shown to drive ADCC (51, 144), suggesting that this less immunodominant target is vulnerable to multiple modes of antibody mediated targeting.
The highly conserved internal viral proteins NP and M2 have been shown to induce an immune response that is also broadly reactive (145). NP-specific antibodies, which are always non-neutralizing (146), mediated viral clearance through FcRs and protection in mouse models of influenza infection (143, 147). Similarly, non-neutralizing M2-specific antibodies mediated ADCC and ADCP to clear infected cells and promoted rapid viral clearance (141, 142). Thus, while current vaccination strategies largely focus on the development of broadly reactive immunity against HA, additional largely non-neutralizing conserved antigens exist within influenza that may represent next generation targets for protective universal immunity.
More than 4 decades of research has clearly illustrated the importance of both direct neutralization and non-neutralizing functional antibodies in protection against influenza infection and disease. Because neutralizing and non-neutralizing antibody activities are not induced in a mutually exclusive manner, vaccine strategies able to leverage both functions of antibodies are likely to confer the greatest level of protection. However, the precise innate immune effector functions to precise sites of viral vulnerability on HA or other target antigens remain to be determined. With emerging novel vaccine design strategies, coupled to emerging immune modulatory adjuvants, opportunities to drive universal protection are on the horizon.
CB and GA wrote the review article, contributed to revision, and approved the submitted version.
This work was supported by the following: the Ragon Institute, the Samana Cay MGH scholar program, and NIH grants AI080289 (5R37AI080289-09), AI102660-01 (5R01AI102660-04), and AI129797-01 (1R01A1129797-01) as well as the NIH T32 AI007245 (CB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would also like to acknowledge Harvard CFAR for ongoing support P30 AI060354-02. We thank J. Burke and M. Slein for comments, and M. Davis for administrative support.
1. Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol Mech Dis. (2008) 3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316
2. WHO. Influenza (2008) Available online at: http://www.who.int/immunization/topics/influenza/en/
3. Director NI for OS and HO of the. Influenza in the Workplace (2018) Available online at: https://www.cdc.gov/niosh/topics/flu/activities.html
4. WHO. Influenza Virus Infections in Humans. (2014) Available online at: http://www.who.int/influenza/human_animal_interface/virology_laboratories_and_vaccines/influenza_virus_infections_humans_feb14.pdf
5. Paules CI, Marston HD, Eisinger RW, Baltimore D, Fauci AS. The pathway to a universal influenza vaccine. Immunity. (2017) 47:599–603. doi: 10.1016/j.immuni.2017.09.007
6. Erbelding EJ, Post DJ, Stemmy EJ, Roberts PC, Augustine AD, Ferguson S, et al. A universal influenza vaccine: the strategic plan for the national institute of allergy and infectious diseases. J Infect Dis. (2018) 218:347–54. doi: 10.1093/infdis/jiy103
7. Fox A, Mai LQ, Thanh LT, Wolbers M, Le Khanh Hang N, Thai PQ, et al. Hemagglutination inhibiting antibodies and protection against seasonal and pandemic influenza infection. J Infect. (2015) 70:187–96. doi: 10.1016/j.jinf.2014.09.003
8. Zhao X, Fang VJ, Ohmit SE, Monto AS, Cook AR, Cowling BJ. Quantifying protection against influenza virus infection measured by hemagglutination-inhibition assays in vaccine trials. Epidemiology. (2016) 27:143–51. doi: 10.1097/EDE.0000000000000402
9. Ohmit SE, Petrie JG, Cross RT, Johnson E, Monto AS. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J Infect Dis. (2011) 204:1879–85. doi: 10.1093/infdis/jir661
10. Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, et al. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio. (2016) 7:e00417–16. doi: 10.1128/mBio.00417-16
11. CDC. Seasonal Influenza Vaccine Effectiveness, 2005-2017 (2017) Available online at: https://www.cdc.gov/flu/professionals/vaccination/effectiveness-studies.htm
12. Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. (2010) 285:28403–9. doi: 10.1074/jbc.R110.129809
13. WHO. WHO Recommendations on the Composition of Influenza Virus Vaccines (2017) Available online at: http://www.who.int/influenza/vaccines/virus/recommendations/en/ (Accessed November 15, 2017).
14. CDC. Influenza (Flu) (2018) Available online at: https://www.cdc.gov/flu/index.htm
15. Salk JE, Menke WJ, Francis TJ. A clinical, epidemiological and immunological evaluation op vaccination against epidemic influenza. Am J Epidemiol. (1945) 42:57–93. doi: 10.1093/oxfordjournals.aje.a119025
16. Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol. (2015) 386:323–41. doi: 10.1007/82_2014_413
17. Angeletti D, Yewdell JW. Is it possible to develop a “universal” influenza virus vaccine? Outflanking antibody immunodominance on the road to universal influenza vaccination. Cold Spring Harb Perspect Biol. (2017) 10:a028852. doi: 10.1101/cshperspect.a028852
18. Ellebedy AH, Ahmed R. Re-engaging cross-reactive memory B cells: the influenza puzzle. Front Immunol. (2012) 3:53. doi: 10.3389/fimmu.2012.00053
19. Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr Opin Virol. (2012) 2:134–41. doi: 10.1016/j.coviro.2012.02.005
20. Tan GS, Krammer F, Eggink D, Kongchanagul A, Moran TM, Palese P. A Pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol. (2012) 86:6179–88. doi: 10.1128/JVI.00469-12
21. Dilillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk – specific antibodies require Fc γ R interactions for protection against influenza virus in vivo. Nat Med. (2014) 20:143–51. doi: 10.1038/nm.3443
22. DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. (2016) 126:605–10. doi: 10.1172/JCI84428
23. Jegaskanda S, Vanderven HA, Wheatley AK, Kent SJ. Fc or not Fc; that is the question: antibody Fc-receptor interactions are key to universal influenza vaccine design. Hum Vaccines Immunother. (2017) 13:1288–96. doi: 10.1080/21645515.2017.1290018
24. Arunkumar GA, Ioannou A, Wohlbold TJ, Meade P, Aslam S, Amanat F, et al. Broadly cross-reactive, non-neutralizing antibodies against the influenza B virus hemagglutinin demonstrate effector function dependent protection against lethal viral challenge in mice. J Virol. (2019). doi: 10.1128/JVI.01696-18 [Epub ahead of print].
25. Wong SS, Duan S, DeBeauchamp J, Zanin M, Kercher L, Sonnberg S, et al. The immune correlates of protection for an avian influenza H5N1 vaccine in the ferret model using oil-in-water adjuvants. Sci Rep. (2017) 7:44727. doi: 10.1038/srep44727
26. O'Brien KB, Morrison TE, Dundore DY, Heise MT, Schultz-Cherry S. A protective role for complement C3 protein during pandemic 2009 H1N1 and H5N1 influenza a virus infection. PLoS ONE. (2011) 6:e17377. doi: 10.1371/journal.pone.0017377
27. Rattan A, Pawar SD, Nawadkar R, Kulkarni N, Lal G, Mullick J, et al. Synergy between the classical and alternative pathways of complement is essential for conferring effective protection against the pandemic influenza A(H1N1) 2009 virus infection. PLoS Pathog. (2017) 13:e1006248. doi: 10.1371/journal.ppat.1006248
28. Mullarkey CE, Bailey MJ, Golubeva DA, Tan GS, Nachbagauer R, He W, et al. Broadly neutralizing hemagglutinin stalk-specific antibodies induce potent phagocytosis of immune complexes by neutrophils in an Fc-dependent manner. MBio. (2016) 7:1–12. doi: 10.1128/mBio.01624-16
29. Huber VC, Lynch JM, Bucher DJ, Le J, Metzger DW. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J Immunol. (2001) 166:7381–8. doi: 10.4049/jimmunol.166.12.7381
30. He W, Chen CJ, Mullarkey CE, Hamilton JR, Wong CK, Leon PE, et al. Alveolar macrophages are critical for broadly-reactive antibody-mediated protection against influenza A virus in mice. Nat Commun. (2017) 8:846. doi: 10.1038/s41467-017-00928-3
31. Terajima M, Co MDT, Cruz J, Ennis FA. High antibody-dependent cellular cytotoxicity antibody titers to H5N1 and H7N9 avian influenza A viruses in healthy US adults and older children. J Infect Dis. (2015) 212:1052–60. doi: 10.1093/infdis/jiv181
32. Jegaskanda S. The potential role of fc-receptor functions in the development of a universal influenza vaccine. Vaccines. (2018) 6:E27. doi: 10.3390/vaccines6020027
33. Kopf M, Abel B, Gallimore A, Carroll M, Bachmann MF. Complement component C3 promotes T-cell priming and lung migration to control acute influenza virus infection. Nat Med. (2002) 8:373–8. doi: 10.1038/nm0402-373
34. Perussia B, Acuto O, Terhorst C, Faust J, Lazarus R, Perussia B, et al. Human natural killer cells analyzed by B73.1, a monoclonal antibody blocking Fc receptor functions. II. Studies of B73.1 antibody-antigen interaction on the lymphocyte membrane. J Immunol. (1983) 130:2142–8.
35. Ritz J, Schmidt RE, Michon J, Hercend T, Schlossman SF. Characterization of functional surface structures on human natural killer cells. Adv Immunol. (1988) 42:181–211. doi: 10.1016/S0065-2776(08)60845-7
36. Bournazos S, Woof JM, Hart SP, Dransfield I. Functional and clinical consequences of Fc receptor polymorphic and copy number variants. Clin Exp Immunol. (2009) 157:244–54. doi: 10.1111/j.1365-2249.2009.03980.x
37. Seidel UJE, Schlegel P, Lang P. Natural killer cell mediated antibody-dependent cellular cytotoxicity in tumor immunotherapy with therapeutic antibodies. Front Immunol. (2013) 4:76. doi: 10.3389/fimmu.2013.00076
38. Werfel T, Uciechowski P, Tetteroo PA, Kurrle R, Deicher H, Werfel T, et al. Activation of cloned human natural killer cells via Fc gamma RIII. J Immunol. (1989) 142:1102–6.
39. De Maria A, Bozzano F, Cantoni C, Moretta L. Revisiting human natural killer cell subset function revealed cytolytic CD56dimCD16+ NK cells as rapid producers of abundant IFN- on activation. Proc Natl Acad Sci USA. (2011) 108:728–32. doi: 10.1073/pnas.1012356108
40. Oliva A, Kinter AL, Vaccarezza M, Rubbert A, Catanzaro A, Moir S, Monaco J, et al. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J Clin Invest. (1998) 102:223–31.
41. Greenberg SB, Criswell BS, Six HR, Couch RB. Lymphocyte cytotoxicity to influenza virus-infected cells. II. Requirement for antibody and non-T lymphocytes. J Immunol. (1977) 119:2100–6.
42. Greenberg SB, Criswell BSUE, Six HR, Couch RB. Lymphocyte cytotoxicity to influenza virus-infected cells : response to vaccination and virus infection. Infect Immun. (1978) 20:640–5.
43. Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RMB, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. (2015) 349:1301–6. doi: 10.1126/science.aac7263
44. Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol. (2013) 87:5512–22. doi: 10.1128/JVI.03030-12
45. Jegaskanda S, Luke C, Hickman HD, Sangster MY, Wieland-Alter WF, McBride JM, et al. Generation and protective ability of influenza virus-specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J Infect Dis. (2016) 214:945–52. doi: 10.1093/infdis/jiw262
46. Jegaskanda S, Laurie KL, Amarasena TH, Winnall WR, Kramski M, De Rose R, et al. Age-associated cross-reactive antibody-dependent cellular cytotoxicity toward 2009 pandemic influenza A virus subtype H1N1. J Infect Dis. (2013) 208:1051–61. doi: 10.1093/infdis/jit294
47. Jacobsen H, Rajendran M, Choi A, Sjursen H, Brokstad KA, Cox RJ, et al. Influenza virus hemagglutinin stalk-specific antibodies in human serum are a surrogate marker for in vivo protection in a serum transfer mouse challenge model. MBio. (2017) 8:e01463–17. doi: 10.1128/mBio.01463-17
48. Vanderven HA, Jegaskanda S, Wines BD, Hogarth PM, Carmuglia S, Rockman S, et al. Antibody-dependent cellular cytotoxicity responses to seasonal influenza vaccination in older adults. J Infect Dis. (2017) 217:12–23. doi: 10.1093/infdis/jix554
49. Hashimoto G, Wright PF, David T. Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells. J Infect Dis. (1983) 148:785–94. doi: 10.1093/infdis/148.5.785
50. Co MDT, Terajima M, Thomas SJ, Jarman RG, Rungrojcharoenkit K, Fernandez S, et al. Relationship of preexisting influenza hemagglutination inhibition, complement-dependent lytic, and antibody-dependent cellular cytotoxicity antibodies to the development of clinical illness in a prospective study of A(H1N1)pdm09 influenza in children. Viral Immunol. (2014) 27:375–82. doi: 10.1089/vim.2014.0061
51. Jegaskanda S, Co MDT, Cruz J, Subbarao K, Ennis FA, Terajima M. Induction of H7N9-cross-reactive antibody-dependent cellular cytotoxicity antibodies by human seasonal influenza a viruses that are directed toward the nucleoprotein. J Infect Dis. (2017) 215:818–23. doi: 10.1093/infdis/jiw629
52. Mesman AW, Westerhuis BM, Ten Hulscher HI, Jacobi RH, De Bruin E, Van Beek J, et al. Influenza virus A(H1N1)2009 antibody-dependent cellular cytotoxicity in young children prior to the H1N1 pandemic. J Gen Virol. (2016) 97:2157–65. doi: 10.1099/jgv.0.000552
53. Leon PE, He W, Mullarkey CE, Bailey MJ, Miller MS, Krammer F, et al. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc Natl Acad Sci USA. (2016) 113:E5944–51. doi: 10.1073/pnas.1613225113
54. He W, Tan GS, Mullarkey CE, Lee AJ, Lam MMW, Krammer F, et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci USA. (2016) 113:11931–6. doi: 10.1073/pnas.1609316113
55. Barber GN. Host defense, viruses and apoptosis. Cell Death Differ. (2001) 8:113–26. doi: 10.1038/sj.cdd.4400823
56. Ren Y, Savill J. Apoptosis: the importance of being eaten. Cell Death Differ. (1998) 5:563–8. doi: 10.1038/sj.cdd.4400407
57. Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol. (2007) 178:2448–57. doi: 10.4049/jimmunol.178.4.2448
58. Ana-Sosa-Batiz F, Vanderven H, Jegaskanda S, Johnston A, Rockman S, Laurie K, et al. Influenza-specific antibody-dependent phagocytosis. PLoS ONE. (2016) 11:e0154461. doi: 10.1371/journal.pone.0154461
59. Mócsai A. Diverse novel functions of neutrophils in immunity, inflammation, and beyond. J Exp Med. (2013) 210:1283–99. doi: 10.1084/jem.20122220
60. Brinkmann V, Zychlinsky A. Beneficial suicide: why neutrophils die to make NETs. Nat Rev Microbiol. (2007) 5:577–82. doi: 10.1038/nrmicro1710
61. Lim K, Hyun YM, Lambert-Emo K, Capece T, Bae S, Miller R, et al. Neutrophil trails guide influenza specific CD8+ T cells in the airways. Science. (2015) 349:aaa4352. doi: 10.1126/science.aaa4352
62. Otten MA, Leusen JHW, Rudolph E, van der Linden JA, Beelen RHJ, Jan GJ, et al. FcR gamma-chain dependent signaling in immature neutrophils is mediated by Fc alpha RI, but not by Fc gamma RI 1. J Immunol. (2007) 179:2918–24. doi: 10.4049/jimmunol.179.5.2918
63. Fujisawa H. Neutrophils play an essential role in cooperation with antibody in both protection against and recovery from pulmonary infection with influenza virus in mice. J Virol. (2008) 82:2772–83. doi: 10.1128/JVI.01210-07
64. Fujisawa H, Tsuru S, Taniguchi M, Zinnaka Y, Nomoto K. Protective mechanisms against pulmonary infection with influenza virus. I. Relative contribution of polymorphonuclear leukocytes and of alveolar macrophages to protection during the early phase of intranasal infection. J Gen Virol. (1987) 68:425–32. doi: 10.1099/0022-1317-68-2-425
65. Hicks JT, Ennis FA, Kim E, Verbonitz M. The importance of an intact complement pathway in recovery from a primary viral infection: influenza in decomplemented and in C5-deficient mice. J Immunol. (1978) 121:1437–45.
66. Terajima M, Cruz J, Co MDT, Lee J-H, Kaur K, Wilson PC, et al. Complement-dependent lysis of influenza A virus-infected cells by broadly cross-reactive human monoclonal antibodies. J Virol. (2011) 85:13463–7. doi: 10.1128/JVI.05193-11
67. Baker K, Qiao SW, Kuo T, Kobayashi K, Yoshida M, Lencer WI, et al. Immune and non-immune functions of the (not so) neonatal Fc receptor, FcRn. Semin Immunopathol. (2009) 31:223–36. doi: 10.1007/s00281-009-0160-9
68. Bai Y, Ye L, Tesar DB, Song H, Zhao D, Bjorkman PJ, Roopenian DC. Intracellular neutralization of viral infection in polarized epithelial cells by neonatal Fc receptor (FcRn)-mediated IgG transport. Proc Natl Acad Sci USA. (2011) 108, 18406–18411. doi: 10.1073/pnas.1115348108/DC
69. Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J Immunol. (2009) 183:3294–301. doi: 10.4049/jimmunol.0900398
71. Wang TT, Bournazos S, Ravetch JV. Immunological responses to influenza vaccination: lessons for improving vaccine efficacy. Curr Opin Immunol. (2018) 53:124–9. doi: 10.1016/j.coi.2018.04.026
72. Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. (2012) 30:429–57. doi: 10.1146/annurev-immunol-020711-075032
73. Phan TG, Green JA, Xu Y, Cyster JG. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat Immunol. (2009) 10:786–96. doi: 10.1038/ni.1745
74. Heesters BA, Chatterjee P, Kim YA, Gonzalez SF, Kuligowski MP, Kirchhausen T, et al. Endocytosis and recycling of immune complexes by follicular dendritic cells enhances B cell antigen binding and activation. Immunity. (2013) 38:1164–75. doi: 10.1016/j.immuni.2013.02.023
75. Tew JG, Mandel TE. Prolonged antigen half-life in the lymphoid follicles of specifically immunized mice. Immunology. (1979) 37:69–76.
76. Fu Y, Zhang Z, Sheehan J, Avnir Y, Ridenour C, Sachnik T, et al. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat Commun. (2016) 7:12780. doi: 10.1038/ncomms12780
77. Wang TT, Maamary J, Tan GS, Bournazos S, Davis CW, Krammer F, et al. Anti-HA glycoforms drive B cell affinity selection and determine influenza vaccine efficacy. Cell. (2015) 162:160–9. doi: 10.1016/j.cell.2015.06.026
78. Nachbagauer R, Choi A, Hirsh A, Margine I, Iida S, Barrera A, et al. Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nat Immunol. (2017) 18:464–73. doi: 10.1038/ni.3684
79. Nimmerjahn F, Ravetch JV. Immunology: divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science. (2005) 310:1510–12. doi: 10.1126/science.1118948
80. Frasca D, Diaz A, Romero M, Mendez NV, Landin AM, Blomberg BB. Effects of age on H1N1-specific serum IgG1 and IgG3 levels evaluated during the 2011-2012 influenza vaccine season. Immun Ageing. (2013) 10:14. doi: 10.1186/1742-4933-10-14
81. Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S. Specificity and affinity of human Fc gamma receptors and their polymorphic variants for human IgG subclasses. Receptor. (2009) 113:3716–25. doi: 10.1182/blood-2008-09-179754
82. Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. (2014) 5:520. doi: 10.3389/fimmu.2014.00520
83. Einarsdottir H, Ji Y, Visser R, Mo C, Luo G, Scherjon S, et al. H435-containing immunoglobulin G3 allotypes are transported efficiently across the human placenta: implications for alloantibody-mediated diseases of the newborn. Transfusion. (2014) 54:665–71. doi: 10.1111/trf.12334
84. El-Madhun AS, Cox RJ, Haaheim LR. The effect of age and natural priming on the IgG and IgA subclass responses after parenteral influenza vaccination. J Infect Dis. (1999) 180:1356–60. doi: 10.1086/315003
85. Muramatsu M, Yoshida R, Yokoyama A, Miyamoto H, Kajihara M, Maruyama J, et al. Comparison of antiviral activity between IgA and IgG specific to influenza virus hemagglutinin: increased potential of IgA for heterosubtypic immunity. PLoS ONE. (2014) 9:e85582. doi: 10.1371/journal.pone.0085582
86. Snoeck V, Peters IR, Cox E. The IgA system: a comparison of structure and function in different species. Vet Res. (2006) 37:455–67. doi: 10.1051/vetres:2006010
87. Renegar KB, Small PA, Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol. (2004) 173:1978–86. doi: 10.4049/jimmunol.173.3.1978
88. van Riet E, Ainai A, Suzuki T, Hasegawa H. Mucosal IgA responses in influenza virus infections; thoughts for vaccine design. Vaccine. (2012) 30:5893–900. doi: 10.1016/j.vaccine.2012.04.109
89. Seibert CW, Rahmat S, Krause JC, Eggink D, Albrecht RA, Goff PH, et al. Recombinant IgA is sufficient to prevent influenza virus transmission in guinea pigs. J Virol. (2013) 87:7793–804. doi: 10.1128/JVI.00979-13
90. Jennewein MF, Alter G. The immunoregulatory roles of antibody glycosylation. Trends Immunol. (2017) 38:358–72. doi: 10.1016/j.it.2017.02.004
91. Stanley P, Taniguchi N, Aebi M. Chapter 9: N-Glycans. In: Varki A, Cummings R, Esko J, et al. editors. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press (2015–2017). doi: 10.1101/glycobiology.3e.009
92. Pučić M, KneŽević A, Vidič J, Adamczyk B, Novokmet M, Polašek O, et al. High throughput isolation and glycosylation analysis of IgG–variability and heritability of the IgG glycome in three isolated human populations. Mol Cell Proteomics. (2011) 10:M111.010090. doi: 10.1074/mcp.M111.010090
93. Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol. (2003) 325:979–89. doi: 10.1016/S0022-2836(02)01250-0
94. Subedi GP, Barb AW. The structural role of antibody N-glycosylation in receptor interactions. Structure. (2015) 23:1573–83. doi: 10.1016/j.str.2015.06.015
95. Ackerman ME, Crispin M, Yu X, Baruah K, Boesch AW, Harvey DJ, et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J Clin Invest. (2013) 123:2183–92. doi: 10.1172/JCI65708
96. Parekh R, Dwek R, Sutton B, Fernandes D, Leung A, Stanworth D, et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature. (1985) 316:452–7. doi: 10.1038/316452a0
97. Li T, Dilillo DJ, Bournazos S, Giddens JP, Ravetch JV, Wang L. Modulating IgG effector function by Fc glycan engineering. Proc Natl Acad Sci USA. (2017) 114:3485–90. doi: 10.1073/pnas.1702173114
98. Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. (2008) 320:373–6. doi: 10.1126/science.1154315
99. Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-Inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. (2006) 313:670–3. doi: 10.1126/science.1129594
100. Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Gloria Meng Y, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J Biol Chem. (2002) 277:26733–40. doi: 10.1074/jbc.M202069200
101. Lofano G, Gorman MJ, Yousif AS, Yu W-H, Fox JM, Dugast A-S, et al. Antigen-specific antibody Fc glycosylation enhances humoral immunity via the recruitment of complement. Sci Immunol. (2018) 3:eaat7796. doi: 10.1126/sciimmunol.aat7796
102. Mahan AE, Jennewein MF, Suscovich T, Dionne K, Tedesco J, Chung AW, et al. Antigen-specific antibody glycosylation is regulated via vaccination. PLoS Pathog. (2016) 12:e1005456. doi: 10.1371/journal.ppat.1005456
103. Selman MHJ, de Jong SE, Soonawala D, Kroon FP, Adegnika AA, Deelder AM, et al. Changes in antigen-specific IgG1 Fc N -glycosylation upon influenza and tetanus vaccination. Mol Cell Proteomics. (2012) 11:M111.014563. doi: 10.1074/mcp.M111.014563
104. Tregoning JS, Russell RF, Kinnear E. Adjuvanted influenza vaccines. Hum Vaccines Immunother. (2018) 14:550–64. doi: 10.1080/21645515.2017.1415684
105. Leroux-Roels G. Unmet needs in modern vaccinology. Adjuvants to improve the immune response. Vaccine. (2010) 28(Suppl. 3):C25–36. doi: 10.1016/j.vaccine.2010.07.021
106. O'Hagan DT, Ott GS, De Gregorio E, Seubert A. The mechanism of action of MF59 - an innately attractive adjuvant formulation. Vaccine. (2012) 30:4341–8. doi: 10.1016/j.vaccine.2011.09.061
107. Liang F, Lindgren G, Sandgren KJ, Thompson EA, Francica JR, Seubert A, et al. Vaccine priming is restricted to draining lymph nodes & controlled by adjuvant-mediated antigen uptake. Sci Transl Med. (2017) 9:eaal2094. doi: 10.1126/scitranslmed.aal2094
108. O'Hagan DT, Rappuoli R, De Gregorio E, Tsai T, Del Giudice G. MF59 adjuvant: the best insurance against influenza strain diversity. Expert Rev Vaccines. (2011) 10:447–62. doi: 10.1586/erv.11.23
109. Bernstein DI, Edwards KM, Dekker CL, Belshe R, Talbot HKB, Graham IL, et al. Effects of adjuvants on the safety and immunogenicity of an avian influenza H5N1 vaccine in adults. J Infect Dis. (2008) 197:667–75. doi: 10.1086/527489
110. Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, et al. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. (2011) 3:85ra48. doi: 10.1126/scitranslmed.3002336
111. Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. (2012) 119:5640–50. doi: 10.1182/blood-2012-01-380121
112. Hocart MJ, Mackenzie JS, Stewart GA. The immunoglobulin G subclass responses of mice to influenza A virus: the effect of mouse strain, and the neutralizing abilities of individual protein A-purified subclass antibodies. J Gen Virol. (1989) 70:2439–48. doi: 10.1099/0022-1317-70-9-2439
113. Coutelier J, Van Der Logt JTM, Heessen FWA, Warnier G, Snick JVAN. IgG2a restriction of murine antibodies elicited by viral infections. J Exp Med. (1987) 165:64–9. doi: 10.1084/jem.165.1.64
114. Savard C, Guérin A, Drouin K, Bolduc M, Laliberté-Gagné ME, Dumas MC, et al. Improvement of the trivalent inactivated flu vaccine using papmv nanoparticles. PLoS ONE. (2011) 6:e21522. doi: 10.1371/journal.pone.0021522
115. Davis ID, Chen W, Jackson H, Parente P, Shackleton M, Hopkins W, et al. Recombinant NY-ESO-1 protein with ISCOMATRIX adjuvant induces broad integrated antibody and CD4+ and CD8+ T cell responses in humans. Proc Natl Acad Sci USA. (2004) 101:10697–702. doi: 10.1073/pnas.0403572101
116. Borriello F, Pietrasanta C, Lai JCY, Walsh LM, Sharma P, O'Driscoll DN, et al. Identification and characterization of stimulator of interferon genes as a robust adjuvant target for early life immunization. Front Immunol. (2017) 8:1772. doi: 10.3389/fimmu.2017.01772
117. Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, et al. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis. (2012) 205:1562–70. doi: 10.1093/infdis/jis232
118. Giles BM, Bissel SJ, DeAlmeida DR, Wiley CA, Ross TM. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol. (2012) 19:128–39. doi: 10.1128/CVI.05533-11
119. Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, et al. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol. (2016) 90:4720–34. doi: 10.1128/JVI.03152-15
120. Wong TM, Allen JD, Bebin-Blackwell A-G, Carter DM, Alefantis T, DiNapoli J, et al. Computationally optimized broadly reactive hemagglutinin elicits hemagglutination inhibition antibodies against a panel of H3N2 influenza virus cocirculating variants. J Virol. (2017) 91:e01581–17. doi: 10.1128/JVI.01581-17
121. Van Der Lubbe JEM, Verspuij JWA, Huizingh J, Schmit-Tillemans SPR, Tolboom JTBM, Dekking LEHA, et al. Mini-HA is superior to full length hemagglutinin immunization in inducing stem-specific antibodies and protection against group 1 influenza virus challenges in mice. Front Immunol. (2018) 9:2350. doi: 10.3389/fimmu.2018.02350
122. Liu W-C, Jan J-T, Huang Y-J, Chen T-H, Wu S-C. Unmasking stem-specific neutralizing epitopes by abolishing N-linked glycosylation sites of influenza virus hemagglutinin proteins for vaccine design. J Virol. (2016) 90:8496–508. doi: 10.1128/JVI.00880-16
123. Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, et al. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci USA. (2018) 115:168–73. doi: 10.1073/pnas.1715471115
124. Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, et al. Defining B cell immunodominance to viruses. Nat Immunol. (2017) 18:456–63. doi: 10.1038/ni.3680
125. Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. (2010) 1:e00018–10. doi: 10.1128/mBio.00018-10
126. Nachbagauer R, Liu WC, Choi A, Wohlbold TJ, Atlas T, Rajendran M, et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. npj Vaccines. (2017) 2:26. doi: 10.1038/s41541-017-0026-4
127. Sunwoo S-Y, Schotsaert M, Morozov I, Davis A, Li Y, Lee J, et al. A universal influenza virus vaccine candidate tested in a pig vaccination-infection model in the presence of maternal antibodies. Vaccines. (2018) 6:E64. doi: 10.3390/vaccines6030064
128. Harding AT, Heaton NS. Efforts to improve the seasonal influenza vaccine. Vaccines. (2018) 6:19. doi: 10.3390/vaccines6020019
129. Roedig JV, Rapp E, Höper D, Genzel Y, Reichl U. Impact of host cell line adaptation on quasispecies composition and glycosylation of influenza a virus hemagglutinin. PLoS ONE. (2011) 6:e27989. doi: 10.1371/journal.pone.0027989
130. Schwarzer J, Rapp E, Hennig R, Genzel Y, Jordan I, Sandig V, et al. Glycan analysis in cell culture-based influenza vaccine production: influence of host cell line and virus strain on the glycosylation pattern of viral hemagglutinin. Vaccine. (2009) 27:4325–36. doi: 10.1016/j.vaccine.2009.04.076
131. Hutter J, Rodig JV, Hoper D, Seeberger PH, Reichl U, Rapp E, et al. Toward animal cell culture-based influenza vaccine design: viral hemagglutinin N-glycosylation markedly impacts immunogenicity. J Immunol. (2013) 190:220–30. doi: 10.4049/jimmunol.1201060
132. Gambaryan AS, Marinina VP, Tuzikov AB, Bovin NV, Rudneva IA, Sinitsyn BV, et al. Effects of host-dependent glycosylation of hemagglutinin on receptor- binding properties of H1N1 human influenza A virus grown in MDCK cells and in embryonated eggs. Virology. (1998) 247:170–7.
133. Ekiert DC, Bhabha G, Elsliger M, Friesen RHE, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. (2009) 324:246–51. doi: 10.1126/science.1171491
134. Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci USA. (2017) 114:12578–83. doi: 10.1073/pnas.1712377114
135. Abe Y, Takashita E, Sugawara K, Matsuzaki Y, Muraki Y. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol. (2004) 78:9605–11. doi: 10.1128/JVI.78.18.9605-9611.2004
136. Wang W, Lu B, Zhou H, Suguitan AL, Cheng X, Subbarao K, et al. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol. (2010) 84:6570–7. doi: 10.1128/JVI.00221-10
137. Tate MD, Job ER, Brooks AG, Reading PC. Glycosylation of the hemagglutinin modulates the sensitivity of H3N2 influenza viruses to innate proteins in airway secretions and virulence in mice. Virology. (2011) 413:84–92. doi: 10.1016/j.virol.2011.01.036
138. Tate MD, Brooks AG, Reading PC. Specific sites of N-linked glycosylation on the hemagglutinin of H1N1 subtype influenza A virus determine sensitivity to inhibitors of the innate immune system and virulence in mice. J Immunol. (2011) 187:1884–94. doi: 10.4049/jimmunol.1100295
139. Wu C-Y, Lin C-W, Tsai T-I, Lee C-CD, Chuang H-Y, Chen J-B, et al. Influenza A surface glycosylation and vaccine design. Proc Natl Acad Sci USA. (2017) 114:280–5. doi: 10.1073/pnas.1617174114
140. Sun X, Jayaraman A, Maniprasad P, Raman R, Houser KV, Pappas C, et al. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J Virol. (2013) 87:8756–66. doi: 10.1128/JVI.00593-13
141. El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, et al. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol. (2011) 186:1022–31. doi: 10.4049/jimmunol.0902147
142. Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. (2004) 172:5598–605. doi: 10.4049/jimmunol.172.9.5598
143. LaMere MW, Lam H-T, Moquin A, Haynes L, Lund FE, Randall TD, et al. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J Immunol. (2011) 186:4331–9. doi: 10.4049/jimmunol.1003057
144. Marcelin G, Sandbulte MR, Webby RJ. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol. (2012) 22:267–79. doi: 10.1002/rmv.1713
145. Zheng M, Luo J, Chen Z. Development of universal influenza vaccines based on influenza virus M and NP genes. Infection. (2014) 42:251–62. doi: 10.1007/s15010-013-0546-4
146. Jegaskanda S, Reading PC, Kent SJ. Influenza-specific antibody-dependent cellular cytotoxicity: toward a universal influenza vaccine. J Immunol. (2014) 193:469–75. doi: 10.4049/jimmunol.1400432
Keywords: influenza, antibody, Fc receptor, vaccine, ADCC, glycosylation, adjuvant
Citation: Boudreau CM and Alter G (2019) Extra-Neutralizing FcR-Mediated Antibody Functions for a Universal Influenza Vaccine. Front. Immunol. 10:440. doi: 10.3389/fimmu.2019.00440
Received: 13 December 2018; Accepted: 19 February 2019;
Published: 18 March 2019.
Edited by:
Gestur Vidarsson, Sanquin Research, NetherlandsReviewed by:
Hillary Anne Vanderven, James Cook University, AustraliaCopyright © 2019 Boudreau and Alter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Galit Alter, Z2F0bGVyQG1naC5oYXJ2YXJkLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.