- 1Department of Biological Sciences, School of Dental Medicine, Case Western Reserve University, Cleveland, OH, United States
- 2Experimental Immunology, Helmholtz Centre for Infection Research, Hamburg, Germany
- 3Cluster of Excellence RESIST (EXC 2155), Hannover Medical School, Hannover, Germany
Mammals co-exist with resident microbial ecosystem that is composed of an incredible number and diversity of bacteria, viruses and fungi. Owing to direct contact between resident microbes and mucosal surfaces, both parties are in continuous and complex interactions resulting in important functional consequences. These interactions govern immune homeostasis, host response to infection, vaccination and cancer, as well as predisposition to metabolic, inflammatory and neurological disorders. Here, we discuss recent studies on direct and indirect effects of resident microbiota on regulatory T cells (Tregs) and Th17 cells at the cellular and molecular level. We review mechanisms by which commensal microbes influence mucosa in the context of bioactive molecules derived from resident bacteria, immune senescence, chronic inflammation and cancer. Lastly, we discuss potential therapeutic applications of microbiota alterations and microbial derivatives, for improving resilience of mucosal immunity and combating immunopathology.
Introduction
Mammals harbor a highly diverse microbiome of at least 1000 species, and an astounding number of 10–100 trillion microbial cells, co-existing in a remarkable balance with the host immune system. Healthy human microbiome is mostly bacteria although other microbial domains such as archaea, viruses, and eukaryotes (principally fungi and protists) are also present (1). While these microbes are distributed in skin, and mucosa of ocular, nasal, oral, eye, and reproductive organs, gastrointestinal (GI) tract mucosa is the major reservoir of resident microbes in terms of abundance and species diversity (2, 3). The human colon harbors approximately 3.8 × 1013 microorganisms, followed by skin in the range of ~1011(4). Since the resurgence of microbiome research in recent years, there has been a sharp increase in understanding of how resident microbiome shapes immunity, health and disease of humans. Only a perennial holiday on a lonely island could excuse an immunologist's incognizance on intimate interrelationships between intestinal microbiota and immune balance. Direct crosstalk between resident microbes and host immune cells in mucosa emerges as a pivotal determinant of such an immune balance. Dysbiosis of resident microbes has strong association with a number of immunological disorders, including opportunistic and pathogenic infections (5–13).
Mucosal immune system has not only evolved to protect the mucosal barrier surface against external insults, it has also co-evolved with resident microbes in an interdependent harmonious relationship with them (14–21). The resulting immune balance is crucial to drive optimal immune responses without causing an over-exuberant inflammation (22–25). Past few decades have seen that an increase in hyper-hygiene mentality, mindless use of antibiotics and diet changes, have led to reduced diversity and impaired resilience in resident microbiota (26). Consequently, a disruption in aforementioned immune balance leads to rise in autoimmune and inflammatory disorders. Therefore, understanding the mechanisms of these mutualistic relationships between resident microbiota and different components of innate and adaptive immunity is vital to our understanding of immune diseases. Although gut microbiota in laboratory mice and humans differ significantly, murine models have provided a powerful tool to explore host-microbiota-pathogen interactions in mucosa (27, 28). Here we review the effects of resident microbiota on Tregs and Th17 cells, important players in determining immune balance, mucosal barrier integrity and host protective functions in mucosa. These cells mucosa can develop in mucosa independent of commensal microbiota. For example, there is evidence in germ free mice that Treg cells can be induced by dietary antigens from solid food (29). These Treg cells are of limited life span, but are distinguishable from microbiota- induced Treg cells and capable of repressing inadvertent immune responses to ingested protein antigens. Similarly, in oral mucosa, mechanical damage from mastication of food induces barrier protective Th17 cells, independent of oral commensal microbiota under homeostatic conditions (30). However, dysbiosis can lead excessive Th17 cells and lead to periodontal inflammation (31). Thus, while it is known that these cells can develop independent of microbiota, resident bacterial dysbiosis is strongly associated with alterations in these cells, causing mucosal inflammation seen in many diseases including HIV immunopathogenesis (32–41). Although other cells also play important roles in mucosal tolerance and immunity, we will not review them here.
TREGS and Th17 Cells in Mucosa Under Steady State-Conditions
Majority of the studies on mucosa-microbiota interactions discuss GI tract. Indeed, GI mucosa harbors by far the largest and most diverse microbiota, as well as abundant and dynamic population of Tregs and Th17 cells. Tregs are defined by the expression of CD25 and Foxp3, and are predominantly known for their immunosuppressive properties. These cells also express other molecules such as Cytotoxic T Lymphocyte Antigen-4 (CTLA-4), PD-1, interleukin 10 (IL-10), transforming growth factor beta 1(TGF-β1), and amphiregulin. Each of the aforementioned proteins has been shown to be either important, or dispensable for different mechanisms of Treg-mediated immunosuppression. Divergent conclusions derived from various Treg mechanism investigations have been strikingly similar to those in the popular parable of the “Blind men and an elephant.” It is now increasingly clear that suppressive and non-suppressive functions of Foxp3+ cells are largely variable, depending on local tissues, disease phenotypes, responding effector cells, and cytokine milieu (42–49).
While CD4+ effector T cell responses contribute to overt intestinal inflammation, Tregs are associated with controlling immunopathology (42, 43, 50). It is well known that Tregs are also pivotal for commensal tolerance (51–53). There have been contentions regarding the Tregs found in colon mucosa (colon Tregs; cTregs); whether they develop in thymus (thymic Tregs; tTregs), or periphery (peripheral Tregs; pTregs). The usage of Nrp-1 and Helios as markers of tTregs, and the extent to which the TCR repertoire of cTreg overlaps with that of tTregs have been debated (54, 55). Nevertheless, it is well established that cTregs require the presence of microbiota for their development, sustenance and function (56–58). There is also evidence that mucosal sites are the primary sites of development and maintenance of pTregs (59–61). First formal proof for the requirement of microbiota for the induction and maintenance of intestinal Tregs was provided by studies using germ-free (GF) animal models. GF mice show a several-fold reduction in the frequency of Helios− Tregs, when compared with conventionally housed specific pathogen free (SPF) mice. Association of individual bacterial isolates or defined consortia in GF mice is sufficient to induce intestinal Tregs (56, 57). Even antibiotic treated mice, which show depletion in resident microbiota correlating with a drastic reduction in the frequency of Tregs, lend further credence to the positive role of microbiota in sustenance of Tregs (53, 55, 62). In addition to commensal tolerance, mucosal Tregs have been shown to regulate excessive immune responses during infections (43, 63–65). Recently, they are also shown to accumulate in other tissues and provide functions such as non-suppressive tissue repair functions in muscle (66). While Tregs play diverse and often opposite roles in mucosal infections (Table 1), effects of microbiome on Tregs during these infections are largely ignored in many studies.
Th17 cells are RORγt+, CCR6+, IL-17A+, IL-17F+, with some cells expressing IL-21 and IL-22, and have been implicated both in mucosal barrier functions. Th17 cells are an important subset of effector T cells that are protective during extracellular bacterial and fungal invasion (83, 88–91). However, excessive Th17 responses are also associated with a variety of pathogenic conditions, depending on the pro-inflammatory cytokines they co-produce (30, 91–95). Littman and colleagues showed for the first time that commensal microbiota play important roles in the development of intestinal Th17 cells (22, 53, 96–100). Th17 development and differentiation is controlled by cytokine and epigenetic regulation (91, 92, 101, 102), but the mechanistic details of microbiome dependent control of Th17 development during mucosal infection is largely unclear.
Impact of Microbiome on TREGS and Th17 Cells During GI Infection and Inflammation
“Healthy” GI microbiota is mainly composed of the phyla Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, and Verrucomicrobia. Small intestine is dominated by Enterobacteriaceae and Lactobacillaceae, whereas colon contains the members of Bacteroidaceae, Lachnospiraceae, Prevotellaceae, Rikenellaceae, and Ruminococcaceae respectively (3). A number of factors including diverse environmental conditions, intake of diet and medication, as well as host genetic factors determine the dynamic composition of gut microbiota in individuals (103–107). Gut microbiota are capable of restraining the mucosal colonization by enteric pathogens, a process defined as colonization resistance (108). Thus, administration of antibiotics, and altering the resident microbiota during a mucosal infection is known to lead to post-antibiotic expansion of the pathogens. Loss of overall diversity, or even deficit in single group of bacteria can alter the susceptibility to gastrointestinal infections. For example, Clostridium difficile (C. difficile) infection, the most common cause of nosocomial diarrhea is often preceded by antibiotic usage. Colonization of C. difficile in healthy mice in fact requires a pre-exposure to a cocktail of antibiotics to alter the microbiota composition (109). However, mono-colonization of GF mice with a murine isolate from the family Lachnospiraceae could limit the colonization of C. difficile, suggesting that individual bacterial species are sufficient to confer colonization resistance to C. difficile (110). Enhanced susceptibility toward other infections after antibiotic-mediated disruption of the intestinal microbiota composition has also been reported for vancomycin-resistant Enterococcus Spp and Salmonella enterica serovar typhimurium (S. typhimurium) (108, 111). Mechanistically, mucosal carbohydrates such as fucose and sialic acid liberated by resident microbiota have been shown to control the growth of enteric pathogens. Antibiotics cause spikes in sugars that can worsen S. typhimurium and C. difficile infections (112). Microbiota alterations reduce the numbers of germinal centers in IL21-receptor knockout mice, resulting in diminished IgA+ B cells and reduced activation-induced cytidine deaminase in Peyer's patches. These events lead to the expansion of Tregs and Th17 cells, and higher bacterial burdens, but dampening of Citrobacter rodentium-induced immunopathology (113). Resident microbiota at mucosal interfaces can govern transmission and progress of parasitic protozoan infections such as Toxoplasmosis and Amoebiasis (114). In the case of Toxoplama gondii infection in mice, reduction of microbiota in the gut by prolonged antibiotic treatment leads to impaired Toll like receptor (TLR)-11 and Myeloid differentiation response 88 (MyD88) signaling and subsequent deficit in Th1 immunity, substantiating that gut commensals serve as natural molecular adjuvants during T. gondii infection (115). In a mouse model of Giardia duodenalis infection, antibiotic induced alteration of the microbiome prevents CD8 T cell activation by G. duodenalis. Conversely, GI infection can also modulate microbiota specific adaptive immunity (116). For example, a pathogenic GI infection, in parallel to specific immune reactions against the pathogen, induces immune responses to commensals and generates long-lived commensal-specific T cells. Thus an adaptive response against commensals is an integral component of mucosal immunity. However, such a commensal specific-adaptive response in a dysbiosis setting can also contribute to excessive inadvertent inflammation. In the context of HIV-1 infection, damages in GI tract and gut microbial translocation (Proteobacterial species) are associated with reduction of systemic and gut/rectal mucosal Th17 cells and Tregs (despite increased Treg/Th17 ratio) (36, 71, 72, 117, 118). A large body of evidence suggests that increased Tregs in circulation correlate to reduced immune activation in HIV+ patients, underlining the anti-inflammatory protective roles of Tregs in patients (71–73, 118–125). While combined anti-retroviral (cART) therapy in HIV+ patients generally ensures immune reconstitution in the peripheral blood, dysbiosis and Treg/Th17 abnormalities persist in gut and other mucosae (41, 126–132). This can present residual inflammation and heightened morbidities in cART treated HIV+ patients. However, in cART-treated HIV+ patients with elevated levels of immune activation, it is not clear whether altered levels and function of mucosal Tregs/Th17 cells are associated with local microbial dysbiosis (131), and if these alterations contribute to residual inflammation in HIV disease. Collectively, these findings highlight the role of microbiota in restraining pathogens and inflammation by having significant impact on Tregs and Th17 cells.
Alterations in resident microbiota and host immune cells, caused by host genetic makeup also play a role in the pathogenesis of inflammatory bowel diseases (IBD). One of the adaptive arms of immunity that is impacted by such changes is Tregs (133). Bacteroides fragilis for example, has been found to invade mucosa and cause excessive activation of the host intestinal immune response in genetically susceptible patients (134), while under steady-state conditions the same bacterium can enhance Treg differentiation and ensure intestinal homeostasis. Loss of autophagy protein ATG16L1 in Tregs results in aberrant type 2 responses and spontaneous intestinal inflammation (135). It is unclear whether microbiota directly induce the expression of ATG16L1 in Tregs, but it is evident that ATG16L1 and autophagic process directly promote Treg survival and metabolic adaptation in the intestine. Similarly, other genetic risk variants associated with IBD such as: NOD2, CARD9, ATG16L1, IRGM and FUT2 significantly influence the gut microbiota changes (136). For example, a decrease in Roseburia spp (known acetate to butyrate converters), Clostridiaceae family, the genera Bifidobacterium, Ruminococcus and Faecalibacterium has been observed in patients with IBD. Although many of these communities are strongly implicated in Treg maintenance, direct mechanisms of Treg regulation in the context of these genetic variants and IBD are unclear. Combined deficiency of MyD88 and JH gene, which disrupts innate interactions of immune cells with intestinal microbiota and IgA responses respectively, causes overt inflammation, highlighting the requirement of Treg-IgA mediated mechanism in tolerance (51, 137). It has also been shown that microbiota-specific Foxp3+ Treg cells can convert to interferon-γ-producing Foxp3+ T cells that have a potential to establish mucosal tolerance (138). Disruption of TLR/MyD88 signaling in Foxp3-deficient mice protect them from excessive inflammation at the environmental interfaces of skin, lungs, and intestine, showing that Tregs normally also restrain commensal dependent tonic MyD88-dependent pro-inflammatory signals (139). Mice lacking CLEC7A gene (Dectin-1), thus having dys-regulated interactions with fungal microbiome (mycobiome) show an increased susceptibility to dextran sulfate sodium (DSS) induced colitis (140). The role of Th17 cells and Tregs in this model is unknown. Certain proportion of intestinal Tregs co-expresses RORγt, the master transcription factor of the Th17 lineage, with up to 35 % in small intestine and 65 % in colon (141–143). Some of these RORγt+ Treg co-produce IL-17A (Treg17), and are substantially diminished in GF or antibiotics-treated mice. Mono-association of GF mice with a panel of 22 bacterial species from the human gastrointestinal tract shows that a number of microbes, not only Clostridiales, are capable of induce colonic RORγt+ Tregs (142). Segmented filamentous bacteria (SFB) were only mediocre inducers of RORγt+ Tregs in that study (142). These studies demonstrate that intestinal RORγt+ Tregs are highly microbiota-dependent and have functions in promoting host immunity (62). Yet, RORγt is not a perfect marker for pTregs, because recent reports show the existence of RORγt+ tTregs, particularly developing under inflammatory conditions (143–145).
While most studies have focused on in-depth characterization of mechanisms by which microbiota engage to counter-regulate their immunostimulatory properties, the reciprocal effect of Tregs on the composition and function of the intestinal microbiota was largely ignored (53, 56, 99, 146, 147). Very recently, analysis of mice harboring a reduced number of TGF-β-dependent pTregs demonstrated numerous underrepresented metabolic processes and a limited overall diversity of the microbiome, including a significant reduction of Lactobacillus johnsonii and Mucispirillum schaedleri (148). Mechanistically, it was confirmed that the impaired pTreg generation could adversely affect the microbiota niche by elevating type 2 immune responses in the host, thereby declining the microbiota abundance during the process of community assembly. In conclusion, the presence of pTregs in the intestinal immune system has a strong impact on the composition and function of the intestinal microbiota. Similarly, IL-17F deficiency induces Treg cells in the colon and modifies the composition of the intestinal microbiota and mediates protection against colitis (149). Taken together, two-way interactions between resident microbiota and host intestinal immunity confer intestinal tolerance and immunomodulation.
Impact Of Microbiota on TREGS and Th17 Cells in Oral Mucosa
Oral microbiome is vital to maintaining both oral and systemic immune homeostasis because oral mucosa is the primary gateway for the GI tract, the biggest component of the immune system (150). While a vast majority of microbiota studies has focused on intestinal mucosae and their interactions with gut microbiota, little is known about oral mucosal microenvironment colonized with a large array of resident microbes, which is structurally and functionally distinct from the GI tract (151–160). Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria are the major phyla accounting for ~96–99% of the oral microbiome, while SR1, TM7, Cyanobacteria, Spirochaetes, Synergistetes, and Tenericutes, are also found (<1% distribution). It is well established that oral-resident microbiota in poly-microbial interactions and soft-tissue biofilms avert oral diseases, but direct effect of such interactions on host oral immune cells is less clear (161–166). Oral mucosa maintains subsets of dendritic cells (DC), which produce immunomodulatory cytokines such as IL-10, TGF-β1 and Prostaglandin E2, and are predominantly tolerogenic (89, 167–169). These cells may be in intimate cross-talk with oral mucosal Tregs (58, 62, 170, 171), albeit details of such interactions between these cells are unexplored in oral mucosa. However, alterations in Tregs and Th17 functions have been implicated in human oral Candida infections and periodontitis (36, 38, 40, 69, 172–176). We and others have shown the presence of oral mucosal Foxp3+ Tregs with protective functions during local infection (89, 158, 169, 170). The interrelationship between these cells and oral commensals during an oral infection was also explored (58, 170). In the context of oropharyngeal candidiasis (OPC) infection, Treg cells play a critical role in reducing fungal burden and establishing homeostasis during post anti-fungal response (177). Tregs play rather an unconventional role of enhancing the Th17 cell response and neutrophil infiltration during early acute response, but are associated with reduced TNF-α expression in CD4 T cells at resolution phase (83, 91, 178). Candida infection in mice by itself increases the proportion of Foxp3+Tregs, in a TLR2/MyD88 dependent manner in oral mucosal tissues and draining cervical lymph nodes (58, 83, 91). A small proportion of those Foxp3+ cells co-express RORγt and IL-17A (Treg17). Antibiotic mediated depletion of resident bacteria significantly diminishes the frequency of Foxp3+Treg IL-17A− and Treg17 cells, as well as conventional Th17 cells not expressing Foxp3. Reduction of these cells is concomitant with an increase in tissue pathology and fungal burden in oral mucosa, demonstrating that resident bacteria are important for controlling Foxp3+ cells and Th17 cells, as well as mucosal immunity (Figure 1). Interestingly, Candida can also promote Th17 and Treg responses in oral mucosa (83, 179, 180). The impact of oral resident microbiome in periodontal inflammation, which is now considered a “resident microbial perturbation” rather than a disease caused by a single pathogen, is well known (181). Resident bacterium P. gingivalis, the keystone pathogen contributes to altering the abundance and composition of other normal microbiota. Shift and accumulation of gram-positive aerobes to gram-negative anaerobes such as P. gingivalis, T. denticola, F. nucleatum, and Prevotella sp. are strongly associated with damage in gingival barrier, loss of immune balance and destruction of oral tissue in periodontal disease (150). During this process, bacterial antigens from skewed microbiota can access connective tissues causing abnormal activation and expansion of inflammatory CD4+CD69+CD103− memory T cells and Th17 cells (182). Another recent study showed that periodontitis-associated expansion of Th17 cells required both IL-6 and IL-23, and was dependent on the local dysbiotic microbiome (31). Shift in resident microbiota can also include increase in C. albicans, a part of resident mycobiome in ~50–70% of healthy humans, which can rapidly transition to a pathogen and cause infections in immune-compromised and cancer patients. C. albicans is also shown to heighten P. gingivalis accumulation, worsening the series of inflammatory events associated with periodontitis severity (183, 184). It is known that Treg17 cells exist in periodontitis lesions and could be involved in inflammatory responses against periodontopathic bacteria (185). While there may be only small changes in oral microbiome in HIV+ individuals, underlying mechanisms causing dysbiosis and its association with HIV associated periodontitis during SIV/HIV infection are unclear (117, 186, 187). Precise events defining Th17 and Treg dysfunctions in the context of underlying dysbiosis and aggravating oral inflammation in HIV disease and periodontitis remain to be seen.
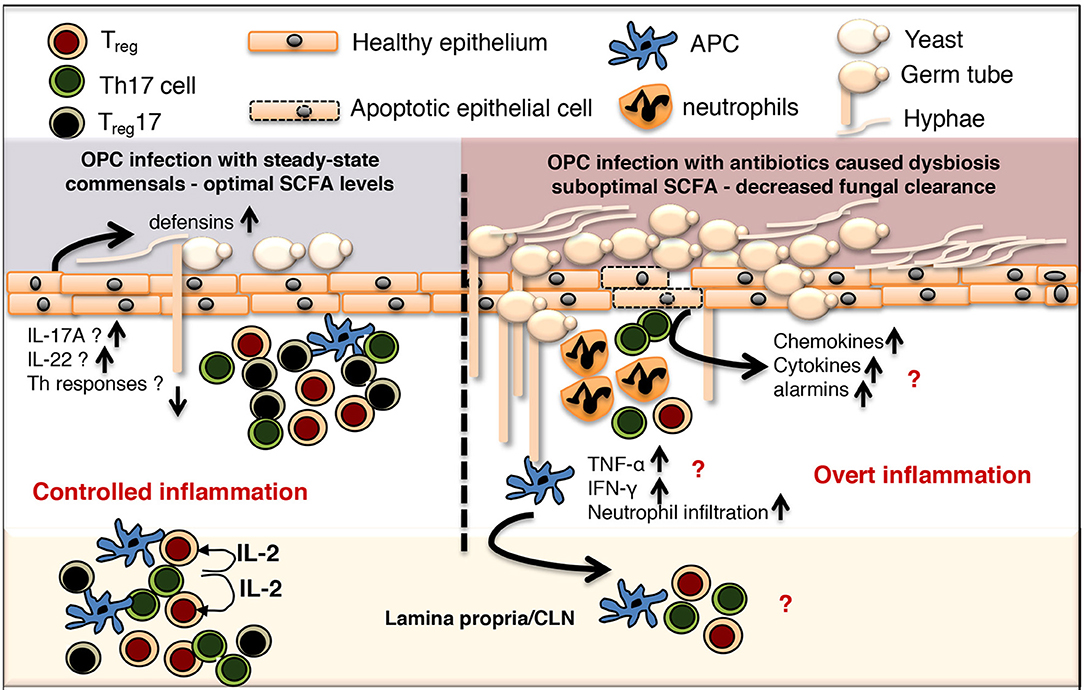
Figure 1. Controlled commensal bacteria/Treg/Treg17/Th17 cell interplay functions as a switch between protective immunity and overt inflammation in oral mucosa. OPC, Oropharyngeal candidiasis; SCFA, short chain fatty acid; CLN, cervical lymph node; APC, antigen presenting cells.
Microbiome in Mucosal Immunity and Inflammation in Other Mucosae
Lung, previously thought to be sterile, is now known to harbor a complex and dynamic microbial community of ~500 species, with a high resemblance to oral microbiome (188, 189). Lung microbiome strongly influences the development and progression of allergic responses and asthma (190). Disrupting the normal microbiome with childhood antibiotic exposure increases the risk of childhood asthma. Proteobacteria abundance in lower airway secretions correlates with pro-inflammatory Th17 cell proportions in asthmatic individuals (191, 192). Similarly, in cystic fibrosis patients, alterations of some groups in the polymicrobial community significantly affect the disease progression. Also, in chronic obstructive pulmonary disease (COPD) patients, microbial dysbiosis associated with mucus hyper-secretion and reduced airway clearance results in chronic aberrant inflammation and airway damage (193). Lung microbiota alterations are also associated with differences in pneumococcal clearance (194).
Multiple genera of microbiota exist in vaginal mucosa, often dominated by species of Lactobacillus, and a diverse array of anaerobic microorganisms, including Atopobium, Anaerococcus, Corynebacterium, Eggerthella, Gardnerella, Mobiluncus, Peptoniphilus, Prevotella, Sneathia, and Finegoldia genera (195). Lactobacilli largely impact the susceptibility to T. vaginalis infection in women. Although mechanisms are still under investigation, there is precedence that Th17 cells and Tregs can have protective and anti-inflammatory effects during T. vaginalis infection (196). During a vaginal herpes simplex virus-2 (HSV-2) infection, mice lacking Tregs fail to timely accumulate HSV-2-specific CD4 T cells and control the infection. This finding underscores the protective role of Tregs in facilitating productive mucosal immunity in vaginal mucosa (197, 198). However, mechanisms of direct control of vaginal microbiome on Tregs and Th17 cells and infection responses remain to be seen. In ocular mucosa, Corynebacterium mastiditis induces commensal specific IL-17 response γδ T cells, recruiting neutrophils and protecting the ocular mucosa from pathogenic infections (199). In nasal mucosa, on the one hand there is evidence that butyric acid-producing microorganisms associate with an impaired olfactory function (200–202). On the other, nasal microbiome is structured by IL-17 Signaling that that supports resistance to S. pneumoniae colonization in the nasal mucosa of mice (203). Collectively, while mcrobial dysbiosis and Tregs/Th17 changes are associated with many of these infections, detailed mechanisms remain to be investigated.
Molecular Mechanisms of Microbiota-Associated Alterations of TREG/Th17 Cells in Mucosae
Resident microbes have a variety of mechanisms for conferring mucosal colonization resistance (17, 204–207). They include: (1) directly competing for shared metabolites, (2) expression of inhibitory bacteriocins, (3) induction of protective mucus layer, and (4) priming of protective immune responses (208, 209). Some of the examples include commensal dependent metabolism of secondary bile acids to deoxycholate, production of organic acids, induction of antimicrobial peptides in Paneth cells, and promoting elevated antibacterial T cell responses preventing colonization and dissemination of pathogens (210–213). Although resident bacteria are known to modulate energy metabolism producing pyruvic acid, citric acid, fumaric acid and malic acid (214), how pH changes determine the mucosal immunity and T cells warrants further investigation. Resident microbiota employ multiple mechanisms that contribute to coordination of Treg/Th17 axis and safeguarding of mucosa (Figure 2). For example, microbiota dependent TLR signaling in host is one of the important mechanisms by which microbiota control inflammation and tolerance. TLR2/MyD88signaling is required for generation and expansion of Nrp1low Foxp3+ cells and Treg17 cells in oral and gut mucosa (58). In gut mucosa the capsular polysaccharide A of the Bacteroides fragilis stimulates production of IL-10 by Foxp3+ cells in a TLR2 dependent manner, thus facilitating mucosal tolerance (215). Recently it was found that this commensal also delivers immunomodulatory molecules to immune cells via secretion of outer membrane vesicles through a non-canonical autophagy pathway for inducing IL-10 expressing Foxp3+ cells. This mechanism requires the expression of host genes ATG16L1 and NOD2, whose polymorphisms are known to be associated with IBD (216). Selective deletion of Atg16l1 in T cells in mice also results in loss of Foxp3+ Treg cells and spontaneous intestinal inflammation characterized by aberrant Th2 responses. These data indicate microbiota-host interactions intimately involve the processes of autophagy and Treg differentiation. Moreover, loss of MyD88-STAT3 signaling in Tregs causes loss of mucosal Tregs and impaired T follicular regulatory cell interactions, resulting in poor IL-21 and anti-microbial IgA responses (217). Failure of this pathway results in over-growth of pathobionts, overt Th17 cell expansion and intestinal inflammation. However, the requirement of resident microbiome induced MyD88 signaling specifically in Tregs, to promote Treg sustenance and intestinal tolerance is still debated (217–219). Similar to B.fragilis, colonic Clostridium rhamnosus also potently induces IL-10+Tregs in a TGF-β1 dependent manner, which is correlated to increase in systemic IgE and resistance to colonic inflammation (56, 99). Similarly, microbiota and immune cell networks are known to control the production of IgA, which is central for mucosal barrier and intestinal tolerance. For example, Mucispirillum spp. and SFB have been directly implicated in production of intestinal IgA (137, 220, 221). Tregs are also known to promote IgA secretion, and maintenance of diversified and balanced microbiota, which in turn facilitates their expansion through a symbiotic regulatory loop, and prevent overt inflammation (222, 223). Moreover, RORγt+ Th17 cells, as well as IL-17A from other cells also promote epithelial polymeric Ig receptor and intestinal IgA expression, further contributing to intestinal homeostasis (224, 225). SFB also control commensal tolerance and anti-microbial host responses through intestinal epithelial cell fucosyl tranferase 2 expression and fucosylation, a process that is dependent on RORγt+ group 3 innate lymphoid cells (ILC3s) and IL-22 expression (226, 227). Loss of intestinal fucosylation results in increased susceptibility to infection by Salmonella typhimurium. ILC3s can also express major histocompatibility complex class II (MHCII) and mediate intestinal selection of CD4+ T cells in order to limit commensal bacteria-specific CD4 T-cell responses (228). Although IL-6, induction of Tregs, or Th17 cells were shown to be not required for ILC-mediated tolerance, alterations in Treg17 and Th17 cells in the context of fucosylation remain to be studied. Treg/Th17 cell differentiation and expansion are also independently controlled by specific members of anaerobic bacteria producing short chain fatty acids (SCFAs), such as acetate, propionate and butyrate (229, 230). Some of these bacteria include Bacteroides, Bifidobacterium, Feacalibacterium genera, and Enterobacteriaceae family, Porphyromonas gingivalis, Fusobacterium nucleatum (mouth), Clostridium cochlearium, Eubacterium multiforme (intestine), and Anaerococcus tetradius (vagina). These bacteria ferment indigestible oligosaccharides and cell surface fucosylated proteins by anaerobic glycolysis, resulting in SCFA production. SCFAs are present in the intestinal lumen at a total concentration of ~100 mM at a ratio of ~6:3:1, for acetate, propionate and butyrate respectively. Although this ratio hinges on carbohydrate availability, microbiota composition and intestinal transit time, acetate and butyrate appear to be the highest and least in abundance respectively (231). Emerging data show that SCFAs contribute to immune homeostasis in mucosa, although excessive and suboptimal levels of SCFAs are often associated with inflammation and cancer. Intestinal SCFAs have been shown to potentiate Foxp3+ cell differentiation and immunomodulatory activity in the colon (53, 99, 147, 232). Mechanistically, in addition to direct histone deacetylase (HDAC) inhibition, SCFAs can induce the expression of retinal aldehyde dehydrogenase 1 family member 1a (Aldh1a) and TGF-β1 in intestinal epithelial cells and DCs (100, 221, 233, 234). Aldh1a could further convert vitamin A into its metabolite retinoic acid in G protein–coupled receptor43 (GPCR43) and Gpr109a manner, which is capable of facilitating Treg induction. These tolerogenic DCs express CD103, sample antigens in the intestinal lamina propria, and migrate to the draining mesenteric lymph node (MLN) to induce immunomodulatory T cells (235–237). Whether SCFA mediated induction and or sustenance of mucosal Tregs require these aforementioned processes is unclear and remain to be studied. However, antibiotics precipitously decrease the oral SCFAs in saliva, showing that in the oral resident bacteria-derived-SCFA is functionally involved in controlling oral mucosal immunity and inflammation (62). Lending credence to this tenet, antibiotics treated mice show not only increased oral inflammation, but also intestinal immunopathology, when infected with oral Candida. Mechanistically, antibiotic treatment results in reduced Tregs, Th17 and Treg17 cells in oral mucosa and tissue draining cervical and axillary lymph nodes in infected mice. Intestinal inflammation in oral Candida infected mice is characterized by an increase in IFN-γ producing Th1 cells and co-producers of IFN-γ and IL-17A (Th1*) cells. Although the exact mechanism of antibiotic mediated reduction of Tregs, Th17 cells and Treg17 cells is unclear, administration of SCFA partially restored these populations and reduces oral immunopathology during the infection. SCFA administration however, only moderately ameliorates the intestinal inflammation. Therefore, the mechanism of Th1-mediated gut inflammation during oral Candida infection in the context of altered microbiota remains to be addressed. Recently, Atarashi et al. showed that oral bacterium Klebsiella spp. isolated from the salivary microbiota elicits a severe Th1 gut inflammation in the context of intestinal dysbiosis, in a genetically susceptible host (238). This finding underscores the role of oral resident microbes such as Klebsiella spp. and C. albicans in modulating T cells, possibly translocating to gut and causing overt inflammation in the gut in the context of resident microbial dysbiosis. Supporting this tenet, post oral gavage of C. albicans-infected mice pre-treated with antibiotics showed significantly altered composition of intestinal microbiota as well as CD4+ T cell mediated lung inflammation, following aerosol introduction of an allergen. However, mice without any antibiotics pre-treatment did not develop an allergic response in the airways (239, 240). Whether changes in SCFA, or Treg and Th17 cells in the lung contribute to the inflammation is unknown.
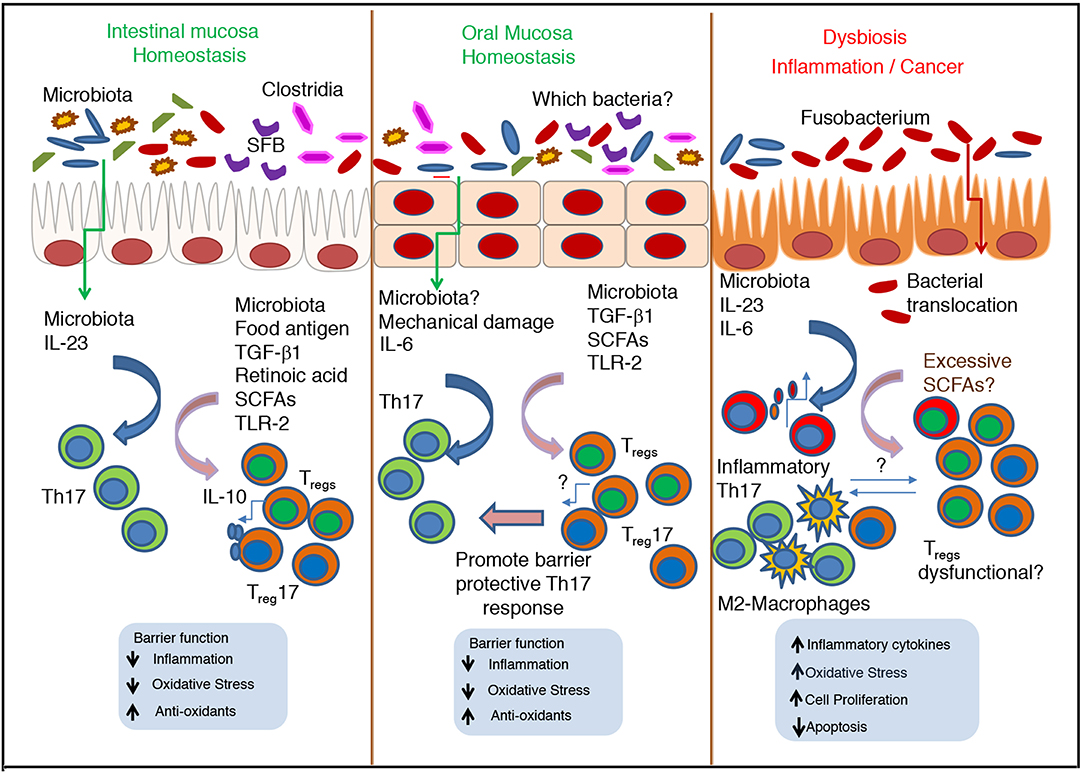
Figure 2. Cross talk between microbiota and immune cells during homeostasis and dysbiosis–Role of Th17 cells and Tregs in oral and intestinal mucosa. During homeostatic conditions, normal microbiota promote the stimulation of epithelial cells, Th17 cells and Tregs, and maintain barrier function and commensal tolerance. In oral mucosa, Th17 cells are induced by mastication induced mechanical damage, independent of commensals. However, in both mucosae SCFA mediated induction of Tregs is key for mucosal barrier function and immunomodulation. During inflammation and cancer, excessive SCFAs can increase inflammatory Th17 cells and Treg population that may be dysfunctional. The nature of their interaction with Th17 cells, tumor associated M2-type macrophages and other cells remain unclear.
Mechanistically, SCFAs also cause acetylation of p70 S6 kinase and phosphorylation rS6, promoting the mTOR activity. mTOR activity was shown to be required for generation of Th17 (T helper type 17), Th1, and IL-10+ T cells (241). Moreover phosphoinositide 3-kinase and mTOR pathways play pivotal roles in integrating growth signals in CD4+ T cell differentiation (242–249). Multiple studies support the role of mTORC1 and mTORC2 proteins in regulating Th17 and Treg fate decisions (247, 250, 251). mTORC1 signaling is constitutively active in Treg cells, and disruption of mTOR protein as well as unrestrained mTOR hyper-activation, both have been shown to cause autoimmunity by impairing Foxp3 expression and Treg functions (252–260). Another study has also shown that mTORC1 and its downstream target hypoxia-inducible factor-1α (HIF-1α) are needed for Foxp3 induction, Treg lipid and cholesterol biosynthesis from glucose, and proliferation and suppressive function in vivo (244, 254). Taken together, while direct role of SCFA in mediating mTOR activation and subsequent Treg induction in mucosa is unclear, these studies highlight the importance of how immunologically relevant microbiome can control Tregs and mucosal homeostasis through multiple mechanisms.
Microbiota and TREG/Th17 Cell Regulation of Immune Senescence and Chronic Inflammation
While resident microbes have aforementioned protective functions in mucosa, they can also trigger and sustain inflammation during aging and other chronic inflammatory conditions. Some studies demonstrate direct relationship between aging and changes in microbiota, albeit the mechanisms remain largely unstudied. Aging causes increased accumulation of gut Enterobacteria, Streptococci, and yeasts but declining levels of Akkermansia muciniphila, Bifidobacteria and Bacteroides (261–266). Reduced Akkermansia muciniphila is associated with reduced butyrate and impaired intestinal barrier. Consequently, aged mice display endotoxin leakage, and triggering of 4-1BB receptor signaling and insulin resistance. In oral mucosa, aging causes higher levels of RANKL+ cells, and increased inflammatory Th17 cell accumulation, with concomitant loss of alveolar bone, which are dependent on the presence of commensal microbiota (30, 267, 268). In contrast, these events do not occur in in germfree mice periodontium, showing potentially pathogenic roles of commensal microbiota in aging associated dysbiosis setting. Similarly, resident microbiota have been implicated in the onset and progression of experimental autoimmune encephalomyelitis (EAE) (269). GF mice exhibit lower levels of the pro-inflammatory cytokine IFN-γ and IL-17A producing cells, and a reciprocal increase in Tregs in the intestine and spinal cord. These changes in GF mice correlate with a significantly attenuated EAE, compared with conventionally raised mice. Remarkably, intestinal colonization with SFB alone can promote Th17 cells in the gut and in the central nervous system (CNS), enhancing disease progression (270). Furthermore, partial elimination of intestinal microbiota ameliorates established collagen-induced arthritis by dampening Th17 responses in mice (271). Some bacteria also provide inflammatory signals resulting in chronic inflammation and tumorigenesis, likely by inducing genetic and epigenetic changes in host cells. For example, Fusobabacteria spp. has been implicated in increased risk of IBD and colorectal cancer (272–275). Also, in oral mucosa, the abundance of Fusobacterium increases, while the number of Streptococcus, Haemophilus, Porphyromonas, and Actinomyces decreases with cancer progression in oral squamous cell carcinoma (276). Interestingly, Fusobabacteria, and several other bacteria of oral mucosal origin, including genera of Streptococcus, Staphylococcus, Peptostreptococcus may translocate to intestine in the context of gut inflammation and carcinogenesis (277–279), similar to Klebsiella spp and C. albicans in susceptible host (62, 238). It is tempting to speculate that loss of Treg functions in the context of dybiosis, excessive SCFA and oral microbial translocation may have contributed to exuberant intestinal inflammation and predisposition to carcinogenesis in these studies (Figure 2). However, whether the mouth- to -gut translocation is a cause, or consequence of dysbiosis and intestinal inflammation, and the underlying mechanisms still remain to be understood and warrant further investigation.
Therapeutic Applications of Microbiota Alterations and Microbiota Derived Metabolites
As we discussed above, studies on patient cohorts, mechanistic studies on mice and epidemiological studies have led to a better understanding of how microbiota changes impact mucosal immunity, and vice versa. Mechanistic “proof-of principle” studies using disease models have opened ways to manipulate these processes, providing therapeutic approaches. Some of the widely used approaches include administration of sodium butyrate and pre- and pro-biotics, and transplantation of fecal microbiota (280–283). However, there are hurdles in pro-biotic and microbiota transplantation approaches. Existing microbiota, whether it is healthy or dysbiotic is largely stable over time in an individual. Without profound perturbation of the existing microbiota, it is challenging to introduce microbiota exogenously. The effects of exogenous bacteria introduced by probiotic and transplant approaches are greatly influenced by existing microbiota in a competitive niche, and are inconsistent. Therefore, approaches to target these niches in favor of exogenous bacteria are being studied (283, 284). Direct administration of microbial derivatives appears to be a promising venue. Butyrate has been shown to alleviate high-fat-diet induced non-alcoholic fatty liver disease. It potently down modulates peroxisome proliferator-activated receptor α-mediated activation of β oxidation, causing reduced inflammation (285). For cART treated HIV+ individuals, aside from cART treatment, probiotics have been studied to combat persistent systemic inflammation. This approach in the context of cART may lead to improved and holistic management of inflammatory events and higher cancer susceptibility in HIV+ patients. Application of probiotics has also shown positive effect on the course of pneumonia, acute exacerbation of bronchial asthma and COPD in mice models, but warrants further studies in humans (286). SCFA has been shown to have therapeutic potential in microbiome-targeted interventions in anti-aging medicine. Butyrate and dietary fibers have been shown to promote anti-inflammatory effects in the context of aging associated neuro-inflammation in mice (287). Adult and aged mice fed with 5% inulin (high fiber) diet for 4 weeks show an altered gut microbiome and increased butyrate, acetate, and total SCFA production, coinciding with a reduction in neuro-inflammation. High fiber supplementation in aging is a non-invasive strategy to increase butyrate levels, and these data suggest that an increase in butyrate through added soluble fiber such as inulin could counterbalance the age-related microbiota dysbiosis, potentially leading to neurological benefits (287, 288). Similarly, dietary fiber also suppresses colon carcinogenesis in polyposis mice (289). Mechanistically it has been shown to inhibit colorectal cancer cell migration through micro-RNA regulation (290). In summary, alterations of mechanisms of microbiota-host interactions are proving to hold promise for treating a variety of disorders in humans.
Conclusion
It is now well established that resident microbes provide enormous advantages to the host, while dysbiosis can trigger acute and chronic inflammatory conditions. One of the mechanisms by which these microbes regulate immunity id through controlling Tregs and Th17 cells. These cells present in various mucosal locations and share various signaling pathways for their development and sustenance, as stated above. However, signals modulating these subsets unique to each mucosal environment in different epithelial cell contexts are unclear. Most mechanistic studies showing Treg/Th17 developmental regulation were performed using the in vitro cultures using cells isolated from blood (human), spleen and lymph nodes (mice). While there is enough evidence to show that these cells could be regulated by overlapping signaling mechanisms, cells from these mucosae were not directly compared for similarities and differences in their development and functions. Such studies are warranted to get further insights in to homeostatic and dysbiotic conditions in different mucosae. Such studies in the context of microbial manipulation approach will offer new avenues to manipulate their interactions with the host for treating immune-mediated and metabolic disorders. While mono-association of certain genera in GF mice have proven to alter mucosal Tregs and Th17 cells and offer some beneficial effects in some experimental settings (98), from a therapeutic perspective, the field is still at its infancy and warrants intense mechanistic investigations. Taken together, further research in microbiota targeted approaches will enable the field to take the center stage in the management of health and disease in humans.
Author Contributions
PP and JH wrote the manuscript. NB, ES, MZ, and SJ contributed to the discussion.
Funding
PP was supported by startup funding by SODM, CWRU and NIH 1RO1DE026923 grant to PP. JH was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy—EXC 2155 RESIST—Project ID 39087428, and the Priority Program Intestinal Microbiota (SPP1656).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. (2016) 8:51. doi: 10.1186/s13073-016-0307-y
2. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. (1977) 31:107–33. doi: 10.1146/annurev.mi.31.100177.000543
3. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. (2016) 14:20–32. doi: 10.1038/nrmicro3552
4. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
5. Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol. (2011) 131:1974–80. doi: 10.1038/jid.2011.182
6. Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. (2012) 37:158–70. doi: 10.1016/j.immuni.2012.04.011
7. Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. (2012) 9:577–89. doi: 10.1038/nrgastro.2012.156
8. Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. (2012) 18:538–46. doi: 10.1038/nm.2657
9. Abt MC, Artis D. The dynamic influence of commensal bacteria on the immune response to pathogens. Curr Opin Microbiol. (2013) 16:4–9. doi: 10.1016/j.mib.2012.12.002
10. Hill DA, Artis D. The influence of commensal bacteria-derived signals on basophil-associated allergic inflammation. Gut Microb. (2013) 4:76–83. doi: 10.4161/gmic.22759
11. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. (2013) 14:685–90. doi: 10.1038/ni.2608
12. Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. (2013) 13:321–35. doi: 10.1038/nri3430
13. Spasova DS, Surh CD. Blowing on embers: commensal microbiota and our immune system. Front Immunol. (2014) 5:318. doi: 10.3389/fimmu.2014.00318
14. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. (2004) 4:478–85. doi: 10.1038/nri1373
15. Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. (2006) 124:823–35. doi: 10.1016/j.cell.2006.02.016
16. Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. (2010) 330:1768–73. doi: 10.1126/science.1195568
17. Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. (2013) 14:646–53. doi: 10.1038/ni.2604
18. Brito F, Zaltman C, Carvalho AT, Fischer RG, Persson R, Gustafsson A, et al. Subgingival microflora in inflammatory bowel disease patients with untreated periodontitis. Eur J Gastroenterol Hepatol. (2013) 25:239–45. doi: 10.1097/MEG.0b013e32835a2b70
19. Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol. (2013) 21:334–41. doi: 10.1016/j.tim.2013.04.002
20. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. (2014) 157:121–41. doi: 10.1016/j.cell.2014.03.011
21. Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol. (2017) 18:851–60. doi: 10.1038/ni.3780
22. Bilate A. M., Bousbaine, D., Mesin, L., Agudelo, M., Leube, J., Kratzert, A., et al. (2016). Tissue-specific emergence of regulatory and intraepithelial T cells from a clonal T cell precursor. Sci Immunol 1:eaaf7471. doi: 10.1126/sciimmunol.aaf7471
23. Calderon-Gomez E, Bassolas-Molina H, Mora-Buch R, Dotti I, Planell N, Esteller M, et al. Commensal-Specific CD4(+) Cells From Patients With Crohn's Disease Have a T-Helper 17 Inflammatory Profile. Gastroenterology. (2016) 151:489–500 e483. doi: 10.1053/j.gastro.2016.05.050
24. Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, et al. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. (2017) 153:1320–37 e1316. doi: 10.1053/j.gastro.2017.07.047
25. Navabi N, Whitt J, Wu SE, Woo V, Moncivaiz J, Jordan MB, et al. Epithelial histone deacetylase 3 instructs intestinal immunity by coordinating local lymphocyte activation. Cell Rep. (2017) 19:1165–75. doi: 10.1016/j.celrep.2017.04.046
26. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. (2016) 529:212–5. doi: 10.1038/nature16504
27. Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. (2016) 534:191–9. doi: 10.1038/nature18285
28. Willyard C. Squeaky clean mice could be ruining research. Nature. (2018) 556:16–8. doi: 10.1038/d41586-018-03916-9
29. Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. (2016) 351:858–63. doi: 10.1126/science.aac5560
30. Dutzan N, Abusleme L, Bridgeman H, Greenwell-Wild T, Zangerle-Murray T, Fife ME, et al. On-going mechanical damage from mastication drives homeostatic Th17 cell responses at the oral barrier. Immunity. (2017) 46:133–47. doi: 10.1016/j.immuni.2016.12.010
31. Dutzan N, Kajikawa T, Abusleme L, Greenwell-Wild T, Zuazo CE, Ikeuchi T, et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med. (2018) 10:eaat0797. doi: 10.1126/scitranslmed.aat0797
32. Klein RS, Harris CA, Small CB, Moll B, Lesser M, Friedland GH. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N Engl J Med. (1984) 311:354–8. doi: 10.1056/NEJM198408093110602
33. Patton LL. Sensitivity, specificity, and positive predictive value of oral opportunistic infections in adults with HIV/AIDS as markers of immune suppression and viral burden. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. (2000) 90:182–8. doi: 10.1067/moe.2000.108799
34. Patton LL, McKaig R, Strauss R, Rogers D, Eron JJ Jr. Changing prevalence of oral manifestations of human immuno-deficiency virus in the era of protease inhibitor therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. (2000) 89:299–304. doi: 10.1016/S1079-2104(00)70092-8
35. Gaitan Cepeda LA, Ceballos Salobrena A, Lopez Ortega K, Arzate Mora N, Jimenez Soriano Y. Oral lesions and immune reconstitution syndrome in HIV+/AIDS patients receiving highly active antiretroviral therapy. Epidemiological evidence. Med Oral Patol Oral Cir Bucal. (2008) 13:E85–93.
36. Kanwar B, Favre D, McCune JM. Th17 and regulatory T cells: implications for AIDS pathogenesis. Curr Opin HIV AIDS. (2010) 5:151–7. doi: 10.1097/COH.0b013e328335c0c1
37. Fidel P. L., and Jr. (2011). Candida-host interactions in HIV disease: implications for oropharyngeal candidiasis. Adv Dent Res. 23, 45–49. doi: 10.1177/0022034511399284
38. Li D, Chen J, Jia M, Hong K, Ruan Y, Liang H, et al. Loss of balance between T helper type 17 and regulatory T cells in chronic human immunodeficiency virus infection. Clin Exp Immunol. (2011) 165:363–71. doi: 10.1111/j.1365-2249.2011.04435.x
39. Cassone A, Cauda R. Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. AIDS. (2012) 26:1457–72. doi: 10.1097/QAD.0b013e3283536ba8
40. Huppler AR, Bishu S, Gaffen SL. Mucocutaneous candidiasis: the IL-17 pathway and implications for targeted immunotherapy. Arthr Res Ther. (2012) 14:217. doi: 10.1186/ar3893
41. Pandiyan P, Younes S, Ribeiro S, Talla A, Bhaskaran N, McDonald D, et al. Mucosal regulatory T cells and T helper 17 cells in HIV associated immune activation. Front immunol. (2016) 7:228. doi: 10.3389/fimmu.2016.00228
42. Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4(+)CD25(+)Foxp3(+) regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4(+) T cells. Nat Immunol. (2007) 8:1353–62. doi: 10.1038/ni1536
43. Pandiyan P, Zheng L, Lenardo MJ. The molecular mechanisms of regulatory T cell immunosuppression. Front Immunol. (2011) 2:60. doi: 10.3389/fimmu.2011.00060
44. Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, et al. A special population of regulatory T cells potentiates muscle repair. Cell. (2013) 155:1282–95. doi: 10.1016/j.cell.2013.10.054
45. Edwards JP, Fujii H, Zhou AX, Creemers J, Unutmaz D, Shevach EM. Regulation of the expression of GARP/latent TGF-beta1 complexes on mouse T cells and their role in regulatory T cell and Th17 differentiation. J Immunol. (2013) 190:5506–15. doi: 10.4049/jimmunol.1300199
46. Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, et al. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell. (2015) 162:1078–89. doi: 10.1016/j.cell.2015.08.021
47. Worthington JJ, Kelly A, Smedley C, Bauche D, Campbell S, Marie JC, et al. Integrin alphavbeta8-Mediated TGF-beta Activation by Effector Regulatory T Cells Is Essential for Suppression of T-Cell-Mediated Inflammation. Immunity. (2015) 42:903–15. doi: 10.1016/j.immuni.2015.04.012
48. Jin RM, Warunek J, Wohlfert EA. Therapeutic administration of IL-10 and amphiregulin alleviates chronic skeletal muscle inflammation and damage induced by infection. Immunohorizons. (2018) 2:142–54. doi: 10.4049/immunohorizons.1800024
49. Povoleri GAM, Nova-Lamperti E, Scotta C, Fanelli G, Chen YC, Becker PD, et al. Human retinoic acid-regulated CD161(+) regulatory T cells support wound repair in intestinal mucosa. Nat Immunol. (2018) 19:1403–14. doi: 10.1038/s41590-018-0230-z
50. Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. (2009) 27:313–38. doi: 10.1146/annurev.immunol.021908.132657
51. Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. (2009) 106:19256–61. doi: 10.1073/pnas.0812681106
52. Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. (2013) 497:258–62. doi: 10.1038/nature12079
53. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
54. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. (2011) 478:250–4. doi: 10.1038/nature10434
55. Nutsch K, Chai JN, Ai TL, Russler-Germain E, Feehley T, Nagler CR, et al. Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Rep. (2016) 17:206–20. doi: 10.1016/j.celrep.2016.08.092
56. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
57. Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. (2011) 34:794–806. doi: 10.1016/j.immuni.2011.03.021
58. Bhaskaran N, Cohen S, Zhang Y, Weinberg A, Pandiyan P. TLR-2 signaling promotes IL-17A production in CD4+CD25+Foxp3+ regulatory cells during oropharyngeal candidiasis. Pathogens. (2015) 4:90–110. doi: 10.3390/pathogens4010090
59. Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. (2011) 34:237–46. doi: 10.1016/j.immuni.2011.01.016
60. Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. (2012) 209:1723–42, S1721. doi: 10.1084/jem.20120914
61. Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. (2012) 209:1713–22, S1711-9. doi: 10.1084/jem.20120822
62. Bhaskaran N, Quigley C, Paw C, Butala S, Schneider E, Pandiyan P. Role of short chain fatty acids in controlling tregs and immunopathology during mucosal infection. Front Microbiol. (2018) 9:1995. doi: 10.3389/fmicb.2018.01995
63. Sakaguchi S. The origin of FOXP3-expressing CD4+ regulatory T cells: thymus or periphery. J Clin Invest. (2003) 112:1310–2. doi: 10.1172/JCI200320274
64. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. (2008) 133:775–87. doi: 10.1016/j.cell.2008.05.009
65. Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. (2009) 30:636–45. doi: 10.1016/j.immuni.2009.04.010
66. Panduro M, Benoist C, Mathis D. Tissue Tregs. Annu Rev Immunol. (2016) 34:609–33. doi: 10.1146/annurev-immunol-032712-095948
67. Rowe JH, Ertelt JM, Aguilera MN, Farrar MA, Way SS. Foxp3(+) regulatory T cell expansion required for sustaining pregnancy compromises host defense against prenatal bacterial pathogens. Cell Host Microbe. (2011) 10:54–64. doi: 10.1016/j.chom.2011.06.005
68. Johanns TM, Ertelt JM, Rowe JH, Way SS. Regulatory T cell suppressive potency dictates the balance between bacterial proliferation and clearance during persistent Salmonella infection. PLoS Pathog. (2010) 6:e1001043. doi: 10.1371/journal.ppat.1001043
69. Garlet GP, Cardoso CR, Mariano FS, Claudino M, de Assis GF, Campanelli AP, et al. Regulatory T cells attenuate experimental periodontitis progression in mice. J Clin Periodontol. (2010) 37:591–600. doi: 10.1111/j.1600-051X.2010.01586.x
70. Zhong Y, Cantwell A, Dube PH. Transforming growth factor beta and CD25 are important for controlling systemic dissemination following Yersinia enterocolitica infection of the gut. Infect Immun. (2010) 78:3716–25. doi: 10.1128/IAI.00203-10
71. Eggena MP, Barugahare B, Jones N, Okello M, Mutalya S, Kityo C, et al. Depletion of regulatory T cells in HIV infection is associated with immune activation. J Immunol. (2005) 174:4407–14.
72. Chase AJ, Sedaghat AR, German JR, Gama L, Zink MC, Clements JE, et al. Severe depletion of CD4+ CD25+ regulatory T cells from the intestinal lamina propria but not peripheral blood or lymph nodes during acute simian immunodeficiency virus infection. J Virol. (2007) 81:12748–57. doi: 10.1128/JVI.00841-07
73. Chase AJ, Yang HC, Zhang H, Blankson JN, Siliciano RF. Preservation of FoxP3+ regulatory T cells in the peripheral blood of human immunodeficiency virus type 1-infected elite suppressors correlates with low CD4+ T-cell activation. J Virol. (2008) 82:8307–15. doi: 10.1128/JVI.00520-08
74. Elahi S, Dinges WL, Lejarcegui N, Laing KJ, Collier AC, Koelle DM, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med. (2011) 17:989–95. doi: 10.1038/nm.2422
75. Lanteri MC, O'Brien KM, Purtha WE, Cameron MJ, Lund JM, Owen RE, et al. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest. (2009) 119:3266–77. doi: 10.1172/JCI39387
76. Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. (2009) 31:772–86. doi: 10.1016/j.immuni.2009.10.001
77. Morampudi V, De Craeye S, Le Moine A, Detienne S, Braun MY, D'Souza S. Partial depletion of CD4(+)CD25(+)Foxp3(+) T regulatory cells significantly increases morbidity during acute phase Toxoplasma gondii infection in resistant BALB/c mice. Microbes Infect. (2011) 13:394–404. doi: 10.1016/j.micinf.2011.01.006
78. Tenorio EP, Olguin JE, Fernandez J, Vieyra P, Saavedra R. Reduction of Foxp3+ cells by depletion with the PC61 mAb induces mortality in resistant BALB/c mice infected with Toxoplasma gondii. J Biomed Biotechnol. (2010) 2010:786078. doi: 10.1155/2010/786078
79. Rausch S, Huehn J, Loddenkemper C, Hepworth MR, Klotz C, Sparwasser T, et al. Establishment of nematode infection despite increased Th2 responses and immunopathology after selective depletion of Foxp3+ cells. Eur J Immunol. (2009) 39:3066–77. doi: 10.1002/eji.200939644
80. Mendez S, Reckling SK, Piccirillo CA, Sacks D, Belkaid Y. Role for CD4(+) CD25(+) regulatory T cells in reactivation of persistent leishmaniasis and control of concomitant immunity. J Exp Med. (2004) 200:201–10. doi: 10.1084/jem.20040298
81. Baumgart M, Tompkins F, Leng J, Hesse M. Naturally occurring CD4+Foxp3+ regulatory T cells are an essential, IL-10-independent part of the immunoregulatory network in Schistosoma mansoni egg-induced inflammation. J Immunol. (2006) 176:5374–87. doi: 10.4049/jimmunol.176.9.5374
82. Montagnoli C, Bacci A, Bozza S, Gaziano R, Mosci P, Sharpe AH, et al. B7/CD28-dependent CD4+CD25+ regulatory T cells are essential components of the memory-protective immunity to Candida albicans. J Immunol. (2002) 169:6298–308. doi: 10.4049/jimmunol.169.11.6298
83. Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, et al. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. (2011) 34:422–34. doi: 10.1016/j.immuni.2011.03.002
84. Montagnoli C, Fallarino F, Gaziano R, Bozza S, Bellocchio S, Zelante T, et al. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J Immunol. (2006) 176:1712–23. doi: 10.4049/jimmunol.176.3.1712
85. Hori S, Carvalho TL, Demengeot J. CD25+CD4+ regulatory T cells suppress CD4+ T cell-mediated pulmonary hyperinflammation driven by Pneumocystis carinii in immunodeficient mice. Eur J Immunol. (2002) 32:1282–91. doi: 10.1002/1521-4141(200205)32:5<1282::AID-IMMU1282>3.0.CO;2-#
86. Scott-Browne JP, Shafiani S, Tucker-Heard G, Ishida-Tsubota K, Fontenot JD, Rudensky AY, et al. Expansion and function of Foxp3-expressing T regulatory cells during tuberculosis. J Exp Med. (2007) 204:2159–69. doi: 10.1084/jem.20062105
87. Chen CY, Huang D, Yao S, Halliday L, Zeng G, Wang RC, et al. IL-2 simultaneously expands Foxp3+ T regulatory and T effector cells and confers resistance to severe tuberculosis (TB): implicative Treg-T effector cooperation in immunity to TB. J Immunol. (2012) 188:4278–88. doi: 10.4049/jimmunol.1101291
88. Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. (2009) 206:299–311. doi: 10.1084/jem.20081463
89. Allam JP, Duan Y, Winter J, Stojanovski G, Fronhoffs F, Wenghoefer M, et al. Tolerogenic T cells, Th1/Th17 cytokines and TLR2/TLR4 expressing dendritic cells predominate the microenvironment within distinct oral mucosal sites. Allergy. (2011) 66:532–9. doi: 10.1111/j.1398-9995.2010.02510.x
90. Cheng SC, van de Veerdonk FL, Lenardon M, Stoffels M, Plantinga T, Smeekens S, et al. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J Leukoc Biol. (2011) 90:357–66. doi: 10.1189/jlb.1210702
91. Bhaskaran N, Liu Z, Saravanamuthu SS, Yan C, Hu Y, Dong L, et al. Identification of Casz1 as a regulatory protein controlling T helper cell differentiation, inflammation, and immunity. Front Immunol. (2018) 9:184. doi: 10.3389/fimmu.2018.00184
92. Martinez GJ, Nurieva RI, Yang XO, Dong C. Regulation and function of proinflammatory TH17 cells. Ann NY Acad Sci. (2008) 1143:188–211. doi: 10.1196/annals.1443.021
93. Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. (2012) 11:763–76. doi: 10.1038/nrd3794
94. Pandiyan P, Yang XP, Saravanamuthu SS, Zheng L, Ishihara S, O'Shea JJ, et al. The role of IL-15 in activating STAT5 and fine-tuning IL-17A production in CD4 T lymphocytes. J Immunol. (2012) 189:4237–46. doi: 10.4049/jimmunol.1201476
95. Duhen R, Glatigny S, Arbelaez CA, Blair TC, Oukka M, Bettelli E. Cutting edge: the pathogenicity of IFN-gamma-producing Th17 cells is independent of T-bet. J Immunol. (2013) 190:4478–82. doi: 10.4049/jimmunol.1203172
96. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
97. Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. (2011) 10:311–23. doi: 10.1016/j.chom.2011.10.004
98. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. (2013) 500:232–6. doi: 10.1038/nature12331
99. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
100. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. (2014) 40:128–39. doi: 10.1016/j.immuni.2013.12.007
101. Bettelli E, Korn T, Kuchroo VK. Th17: the third member of the effector T cell trilogy. Curr Opin Immunol. (2007) 19:652–7. doi: 10.1016/j.coi.2007.07.020
102. Jiang Y, Liu Y, Lu H, Sun SC, Jin W, Wang X, et al. Epigenetic activation during T helper 17 cell differentiation is mediated by Tripartite motif containing 28. Nat Commun. (2018) 9:1424. doi: 10.1038/s41467-018-03852-2
103. Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. (2010) 120:4332–41. doi: 10.1172/JCI43918
104. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. (2011) 145:745–57. doi: 10.1016/j.cell.2011.04.022
105. Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. (2011) 60:1354–62. doi: 10.1136/gut.2010.216259
106. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
107. Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM. Host adaptive immunity alters gut microbiota. ISME J. (2015) 9:770–81. doi: 10.1038/ismej.2014.165
108. Bohnhoff M., M. C. (1962). Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J Infect Dis 111:11.
109. Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, et al. A mouse model of Clostridium difficile-associated dieases. Gastroenterology. (2008) 135:1984–92. doi: 10.1053/j.gastro.2008.09.002
110. Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun. (2012) 80:3786–94. doi: 10.1128/IAI.00647-12
111. Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. (2008) 76:4726–36. doi: 10.1128/IAI.00319-08
112. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. (2013) 502:96–9. doi: 10.1038/nature12503
113. Cho H, Jaime H, de Oliveira RP, Kang B, Spolski R, Vaziri T, et al. Defective IgA response to atypical intestinal commensals in IL-21 receptor deficiency reshapes immune cell homeostasis and mucosal immunity. Mucosal Immunol. (2018). doi: 10.1038/s41385-018-0056-x
114. Bar AK, Phukan N, Pinheiro J, Simoes-Barbosa A. The interplay of host microbiota and parasitic protozoans at mucosal interfaces: implications for the outcomes of infections and diseases. PLoS Negl Trop Dis. (2015) 9:e0004176. doi: 10.1371/journal.pntd.0004176
115. Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe. (2009) 6:187–96. doi: 10.1016/j.chom.2009.06.005
116. Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. (2012) 337:1553–6. doi: 10.1126/science.1220961
117. Nilsson J, Boasso A, Velilla PA, Zhang R, Vaccari M, Franchini G, et al. HIV-1-driven regulatory T-cell accumulation in lymphoid tissues is associated with disease progression in HIV/AIDS. Blood. (2006) 108:3808–17. doi: 10.1182/blood-2006-05-021576
118. Baker CA, Clark R, Ventura F, Jones NG, Guzman D, Bangsberg DR, et al. Peripheral CD4 loss of regulatory T cells is associated with persistent viraemia in chronic HIV infection. Clin Exp Immunol. (2007) 147:533–9. doi: 10.1111/j.1365-2249.2006.03319.x
119. Kinter AL, Hennessey M, Bell A, Kern S, Lin Y, Daucher M, et al. CD25(+)CD4(+) regulatory T cells from the peripheral blood of asymptomatic HIV-infected individuals regulate CD4(+) and CD8(+) HIV-specific T cell immune responses in vitro and are associated with favorable clinical markers of disease status. J Exp Med. (2004) 200:331–43. doi: 10.1084/jem.20032069
120. Kornfeld C, Ploquin MJ, Pandrea I, Faye A, Onanga R, Apetrei C, et al. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J Clin Invest. (2005) 115:1082–91. doi: 10.1172/JCI23006
121. Mozos A, Garrido M, Carreras J, Plana M, Diaz A, Alos L, et al. Redistribution of FOXP3-positive regulatory T cells from lymphoid tissues to peripheral blood in HIV-infected patients. J Acquir Immune Defic Syndr. (2007) 46:529–37. doi: 10.1097/QAI.0b013e31815b69ae
122. Tenorio AR, Martinson J, Pollard D, Baum L, Landay A. The relationship of T-regulatory cell subsets to disease stage, immune activation, and pathogen-specific immunity in HIV infection. J Acquir Immune Defic Syndr. (2008) 48:577–80. doi: 10.1097/QAI.0b013e31817bbea5
123. Jiao Y, Fu J, Xing S, Fu B, Zhang Z, Shi M, et al. The decrease of regulatory T cells correlates with excessive activation and apoptosis of CD8+ T cells in HIV-1-infected typical progressors, but not in long-term non-progressors. Immunology. (2009) 128:e366–375. doi: 10.1111/j.1365-2567.2008.02978.x
124. Owen RE, Heitman JW, Hirschkorn DF, Lanteri MC, Biswas HH, Martin JN, et al. HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. AIDS. (2010) 24:1095–105. doi: 10.1097/QAD.0b013e3283377a1e
125. Angin M, Kwon DS, Streeck H, Wen F, King M, Rezai A, et al. Preserved function of regulatory T cells in chronic HIV-1 infection despite decreased numbers in blood and tissue. J Infect Dis. (2012) 205:1495–500. doi: 10.1093/infdis/jis236
126. Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. (2010) 2:32ra36. doi: 10.1126/scitranslmed.3000632
127. Mavigner M, Cazabat M, Dubois M, L'Faqihi F. E., Requena M, Pasquier C, et al. (2012). Altered CD4+ T cell homing to the gut impairs mucosal immune reconstitution in treated HIV-infected individuals. J Clin Invest. 122, 62–69. doi: 10.1172/JCI59011
128. Brenchley JM. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal Immunol. (2013) 6:657–65. doi: 10.1038/mi.2013.15
129. Kim CJ, McKinnon LR, Kovacs C, Kandel G, Huibner S, Chege D, et al. Mucosal Th17 cell function is altered during HIV infection and is an independent predictor of systemic immune activation. J Immunol. (2013) 191:2164–73. doi: 10.4049/jimmunol.1300829
130. Pallikkuth S, Micci L, Ende ZS, Iriele RI, Cervasi B, Lawson B, et al. Maintenance of intestinal Th17 cells and reduced microbial translocation in SIV-infected rhesus macaques treated with interleukin (IL)-21. PLoS Pathog. (2013) 9:e1003471. doi: 10.1371/journal.ppat.1003471
131. Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. (2015) 8:1009–20. doi: 10.1038/mi.2014.128
132. Hensley-McBain T, Berard AR, Manuzak JA, Miller CJ, Zevin AS, Polacino P, et al. Intestinal damage precedes mucosal immune dysfunction in SIV infection. Mucosal Immunol. (2018) 11:1429–40. doi: 10.1038/s41385-018-0032-5
133. Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. (2011) 474:298–306. doi: 10.1038/nature10208
134. Zhang M, Sun K, Wu Y, Yang Y, Tso P, Wu Z. Interactions between intestinal microbiota and host immune response in inflammatory bowel disease. Front Immunol. (2017) 8:942. doi: 10.3389/fimmu.2017.00942
135. Kabat AM, Pott J, Maloy KJ. The mucosal immune system and its regulation by autophagy. Front Immunol. (2016) 7:240. doi: 10.3389/fimmu.2016.00240
136. Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. (2018) 67:108–19. doi: 10.1136/gutjnl-2016-312135
137. Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. (2009) 325:617–20. doi: 10.1126/science.1172747
138. Feng T, Cao AT, Weaver CT, Elson CO, Cong Y. Interleukin-12 converts Foxp3+ regulatory T cells to interferon-gamma-producing Foxp3+ T cells that inhibit colitis. Gastroenterology. (2011) 140:2031–43. doi: 10.1053/j.gastro.2011.03.009
139. Rivas MN, Koh YT, Chen A, Nguyen A, Lee YH, Lawson G, et al. MyD88 is critically involved in immune tolerance breakdown at environmental interfaces of Foxp3-deficient mice. J Clin Invest. (2012) 122:1933–47. doi: 10.1172/JCI40591
140. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. (2012) 336:1314–7. doi: 10.1126/science.1221789
141. Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat+ T cells. Science. (2015) 349:989–93. doi: 10.1126/science.aac4263
142. Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. (2015) 349:993–7. doi: 10.1126/science.aaa9420
143. Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, et al. Foxp3+ T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. (2016) 9:444–57. doi: 10.1038/mi.2015.74
144. Kim BS, Lu H, Ichiyama K, Chen X, Zhang YB, Mistry NA, et al. Generation of RORgammat(+) Antigen-Specific T Regulatory 17 Cells from Foxp3(+) Precursors in Autoimmunity. Cell Rep. (2017) 21:195–207. doi: 10.1016/j.celrep.2017.09.021
145. Yang J, Zou M, Pezoldt J, Zhou X, Huehn J. Thymus-derived Foxp3(+) regulatory T cells upregulate RORgammat expression under inflammatory conditions. J Mol Med. (2018) 96:1387–94. doi: 10.1007/s00109-018-1706-x
146. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. (2008) 453:620–5. doi: 10.1038/nature07008
147. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
148. Campbell C, Dikiy S, Bhattarai SK, Chinen T, Matheis F, Calafiore M, et al. Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity. (2018) 48:1245-1257 e1249. doi: 10.1016/j.immuni.2018.04.013
149. Tang C, Kakuta S, Shimizu K, Kadoki M, Kamiya T, Shimazu T, et al. Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing Treg cells through modification of the intestinal microbiota. Nat Immunol. (2018) 19:755–65. doi: 10.1038/s41590-018-0134-y
150. Sultan AS, Kong EF, Rizk AM, Jabra-Rizk MA. The oral microbiome: a Lesson in coexistence. PLoS Pathog. (2018) 14:e1006719. doi: 10.1371/journal.ppat.1006719
151. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. (2005) 43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005
152. Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. (2009) 28:405–11. doi: 10.1089/dna.2009.0874
153. Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. (2009) 9:259. doi: 10.1186/1471-2180-9-259
154. Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. (2010) 6:e1000713. doi: 10.1371/journal.ppat.1000713
155. Ahn J, Yang L, Paster BJ, Ganly I, Morris L, Pei Z, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS ONE. (2011) 6:e22788. doi: 10.1371/journal.pone.0022788
156. Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simon-Soro A, Pignatelli M, et al. The oral metagenome in health and disease. ISME J. (2012) 6:46–56. doi: 10.1038/ismej.2011.85
157. Dang AT, Cotton S, Sankaran-Walters S, Li CS, Lee CY, Dandekar S, et al. Evidence of an increased pathogenic footprint in the lingual microbiome of untreated HIV infected patients. BMC Microbiol. (2012) 12:153. doi: 10.1186/1471-2180-12-153
158. Yamazaki S, Maruyama A, Okada K, Matsumoto M, Morita A, Seya T. Dendritic cells from oral cavity induce Foxp3(+) regulatory T cells upon antigen stimulation. PLoS ONE. (2012) 7:e51665. doi: 10.1371/journal.pone.0051665
159. Bashan A, Gibson TE, Friedman J, Carey VJ, Weiss ST, Hohmann EL, et al. Universality of human microbial dynamics. Nature. (2016) 534:259–62. doi: 10.1038/nature18301
160. Teng F, Darveekaran Nair SS, Zhu P, Li S, Huang S, Li X, et al. Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Sci Rep. (2018) 8:16321. doi: 10.1038/s41598-018-34294-x
161. Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. (2009) 299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x
162. Peters BM, Jabra-Rizk MA, O'May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. (2012) 25:193–213. doi: 10.1128/CMR.00013-11
163. Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, et al. Microbial interactions in building of communities. Mol Oral Microbiol. (2013) 28:83–101. doi: 10.1111/omi.12012
164. Guo L, He X, Shi W. Intercellular communications in multispecies oral microbial communities. Front Microbiol. (2014) 5:328. doi: 10.3389/fmicb.2014.00328
165. Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol. (2014) 52:188–99. doi: 10.1007/s12275-014-4067-3
166. Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. (2014) 16:214–31. doi: 10.1111/cmi.12216
167. Cutler CW, Jotwani R. Dendritic cells at the oral mucosal interface. J Dent Res. (2006) 85:678–89. doi: 10.1177/154405910608500801
168. Novak N, Haberstok J, Bieber T, Allam JP. The immune privilege of the oral mucosa. Trends Mol Med. (2008) 14:191–8. doi: 10.1016/j.molmed.2008.03.001
169. Hovav AH. Dendritic cells of the oral mucosa. Mucosal Immunol. (2013) 7:27–37. doi: 10.1038/mi.2013.42
170. Pandiyan P, Bhaskaran N, Zhang Y, Weinberg A. Isolation of T cells from mouse oral tissues. Biol Proced Online. (2014) 16:4. doi: 10.1186/1480-9222-16-4
171. Park JY, Chung H, DiPalma DT, Tai X, Park JH. Immune quiescence in the oral mucosa is maintained by a uniquely large population of highly activated Foxp3(+) regulatory T cells. Mucosal Immunol. (2018). doi: 10.1038/s41385-018-0027-2
172. Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. (2008) 452:773–6. doi: 10.1038/nature06764
173. Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. (2010) 30:196–203. doi: 10.1007/s10875-010-9368-7
174. Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. (2010) 8:481–90. doi: 10.1038/nrmicro2337
175. Pion M, Jaramillo-Ruiz D, Martinez A, Munoz-Fernandez MA, Correa-Rocha R. HIV infection of human regulatory T cells downregulates Foxp3 expression by increasing DNMT3b levels and DNA methylation in the FOXP3 gene. AIDS. (2013) 27:2019–29. doi: 10.1097/QAD.0b013e32836253fd
176. Cheng WC, Hughes FJ, Taams LS. The presence, function and regulation of IL-17 and Th17 cells in periodontitis. J Clin Periodontol. (2014) 41:541–9. doi: 10.1111/jcpe.12238
177. Bhaskaran N, Quigley C, Weinberg A, Huang A, Popkin D, Pandiyan P. Transforming growth factor-beta1 sustains the survival of Foxp3 regulatory cells during late phase of oropharyngeal candidiasis infection. Mucosal Immunol. (2016) 9:1015–26. doi: 10.1038/mi.2015.115
178. Bhaskaran N, Weinberg A, Pandiyan P. Th17 inflammation model of oropharyngeal candidiasis in immunodeficient mice. J Vis Exp. (2015). doi: 10.3791/52538
179. Hernandez-Santos N, Huppler AR, Peterson AC, Khader SA, McKenna KC, Gaffen SL. Th17 cells confer long-term adaptive immunity to oral mucosal Candida albicans infections. Mucosal Immunol. (2013) 6:900–10. doi: 10.1038/mi.2012.128
180. Whibley N, Gaffen SL. Brothers in arms: Th17 and Treg responses in Candida albicans immunity. PLoS Pathog. (2014) 10:e1004456. doi: 10.1371/journal.ppat.1004456
181. Khan SA, Kong EF, Meiller TF, Jabra-Rizk MA. Periodontal diseases: bug induced, host promoted. PLoS Pathog. (2015) 11:e1004952. doi: 10.1371/journal.ppat.1004952
182. Mahanonda R, Champaiboon C, Subbalekha K, Sa-Ard-Iam N, Yongyuth A, Isaraphithakkul B, et al. Memory T cell subsets in healthy gingiva and periodontitis tissues. J Periodontol. (2018) 89:1121–30. doi: 10.1002/JPER.17-0674
183. Tamai R, Sugamata M, Kiyoura Y. Candida albicans enhances invasion of human gingival epithelial cells and gingival fibroblasts by Porphyromonas gingivalis. Microb Pathog. (2011) 51:250–4. doi: 10.1016/j.micpath.2011.06.009
184. Canabarro A, Valle C, Farias MR, Santos FB, Lazera M, Wanke B. Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J Periodontal Res. (2013) 48:428–32. doi: 10.1111/jre.12022
185. Okui T, Aoki Y, Ito H, Honda T, Yamazaki K. The presence of IL-17+/FOXP3+ double-positive cells in periodontitis. J Dent Res. (2012) 91:574–9. doi: 10.1177/0022034512446341
186. Andersson J, Fehniger TE, Patterson BK, Pottage J, Agnoli M, Jones P, et al. Early reduction of immune activation in lymphoid tissue following highly active HIV therapy. AIDS. (1998) 12:F123–129.
187. Starr JR, Huang Y, Lee KH, Murphy CM, Moscicki AB, Shiboski CH, et al. Oral microbiota in youth with perinatally acquired HIV infection. Microbiome. (2018) 6:100. doi: 10.1186/s40168-018-0484-6
188. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. (2015) 6:e00037. doi: 10.1128/mBio.00037-15
189. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. (2015) 12:821–30. doi: 10.1513/AnnalsATS.201501-029OC
190. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. (2015) 17:704–15. doi: 10.1016/j.chom.2015.03.008
191. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. (2010) 5:e8578. doi: 10.1371/journal.pone.0008578
192. Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. (2015) 136:874–84. doi: 10.1016/j.jaci.2015.05.044
193. Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio. (2012) 3:e00251–12. doi: 10.1128/mBio.00251-12
194. Krone CL, Biesbroek G, Trzcinski K, Sanders EA, Bogaert D. Respiratory microbiota dynamics following Streptococcus pneumoniae acquisition in young and elderly mice. Infect Immun. (2014) 82:1725–31. doi: 10.1128/IAI.01290-13
195. Ma B, Forney LJ, Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol. (2012) 66:371–89. doi: 10.1146/annurev-micro-092611-150157
196. Nemati M, Malla N, Yadav M, Khorramdelazad H, Jafarzadeh A. Humoral and T cell-mediated immune response against trichomoniasis. Parasite Immunol. (2018) 40:e12510. doi: 10.1111/pim.12510
197. Lund JM, Hsing L, Pham TT, Rudensky AY. Coordination of early protective immunity to viral infection by regulatory T cells. Science. (2008) 320:1220–4. doi: 10.1126/science.1155209
198. Soerens AG, Da Costa A, Lund JM. Regulatory T cells are essential to promote proper CD4 T-cell priming upon mucosal infection. Mucosal Immunol. (2016) 9:1395–406. doi: 10.1038/mi.2016.19
199. St Leger AJ, Desai JV, Drummond RA, Kugadas A, Almaghrabi F, Silver P, et al. An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal gammadelta T Cells. Immunity. (2017) 47:148–58 e145. doi: 10.1016/j.immuni.2017.06.014
200. Mahdavinia M, Keshavarzian A, Tobin MC, Landay AL, Schleimer RP. A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin Exp Aller. (2016) 46:21–41. doi: 10.1111/cea.12666
201. Koskinen K, Reichert JL, Hoier S, Schachenreiter J, Duller S, Moissl-Eichinger C, et al. The nasal microbiome mirrors and potentially shapes olfactory function. Sci Rep. (2018) 8:1296. doi: 10.1038/s41598-018-19438-3
202. Yang HJ, LoSavio PS, Engen PA, Naqib A, Mehta A, Kota R, et al. Association of nasal microbiome and asthma control in patients with chronic rhinosinusitis. Clin Exp Aller. (2018) 48:1744–7. doi: 10.1111/cea.13255
203. Ritchie ND, Ijaz UZ, Evans TJ. IL-17 signalling restructures the nasal microbiome and drives dynamic changes following Streptococcus pneumoniae colonization. BMC Genomics. (2017) 18:807. doi: 10.1186/s12864-017-4215-3
204. Oliveira MR, Tafuri WL, Afonso LC, Oliveira MA, Nicoli JR, Vieira EC, et al. Germ-free mice produce high levels of interferon-gamma in response to infection with Leishmania major but fail to heal lesions. Parasitology. (2005) 131(Pt 4), 477–488. doi: 10.1017/S0031182005008073
205. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. (2006) 124:837–48. doi: 10.1016/j.cell.2006.02.017
206. Kelly D, King T, Aminov R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res. (2007) 622:58–69. doi: 10.1016/j.mrfmmm.2007.03.011
207. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336:489–93. doi: 10.1126/science.1219328
208. Caballero S, Pamer EG. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol. (2015) 33:227–56. doi: 10.1146/annurev-immunol-032713-120238
209. Thiemann S, Smit N, Strowig T. Antibiotics and the intestinal microbiome: individual responses, resilience of the ecosystem, and the susceptibility to infections. Curr Topics Microbiol Immunol. (2016) 398:124–38. doi: 10.1007/82_2016_504
210. Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. (2008) 190:2505–12. doi: 10.1128/JB.01765-07
211. Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. (2008) 105:20858–63. doi: 10.1073/pnas.0808723105
212. Sun Y, O'Riordan MX. Regulation of bacterial pathogenesis by intestinal short-chain Fatty acids. Adv Appl Microbiol. (2013) 85:93–118. doi: 10.1016/B978-0-12-407672-3.00003-4
213. Thiemann S, Smit N, Roy U, Lesker TR, Galvez EJC, Helmecke J, et al. Enhancement of IFNgamma production by distinct commensals ameliorates salmonella-induced disease. Cell Host Microbe. (2017) 21:682–94 e685. doi: 10.1016/j.chom.2017.05.005
214. Shukla SD, Budden KF, Neal R, Hansbro PM. Microbiome effects on immunity, health and disease in the lung. Clin Transl Immunol. (2017) 6:e133. doi: 10.1038/cti.2017.6
215. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. (2011) 332:974–7. doi: 10.1126/science.1206095
216. Kabat AM, Harrison OJ, Riffelmacher T, Moghaddam AE, Pearson CF, Laing A, et al. The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. Elife. (2016) 5:e12444. doi: 10.7554/eLife.12444
217. Wang S, Charbonnier LM, Noval Rivas M, Georgiev P, Li N, Gerber G, et al. MyD88 Adaptor-dependent microbial sensing by regulatory t cells promotes mucosal tolerance and enforces commensalism. Immunity. (2015) 43:289–303. doi: 10.1016/j.immuni.2015.06.014
218. Schenten D, Nish SA, Yu S, Yan X, Lee HK, Brodsky I, et al. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity. (2014) 40:78–90. doi: 10.1016/j.immuni.2013.10.023
219. Cording S, Fleissner D, Heimesaat MM, Bereswill S, Loddenkemper C, Uematsu S, et al. Commensal microbiota drive proliferation of conventional and Foxp3(+) regulatory CD4(+) T cells in mesenteric lymph nodes and Peyer's patches. Eur J Microbiol Immunol. (2013) 3:1–10. doi: 10.1556/EuJMI.3.2013.1.1
220. Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. (2017) 544:498–502. doi: 10.1038/nature22058
221. Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao Y, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. (2017) 10:946–56. doi: 10.1038/mi.2016.114
222. Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. (2009) 323:1488–92. doi: 10.1126/science.1169152
223. Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. (2014) 41:152–65. doi: 10.1016/j.immuni.2014.05.016
224. Cao AT, Yao S, Gong B, Elson CO, Cong Y. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. (2012) 189:4666–73. doi: 10.4049/jimmunol.1200955
225. Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, et al. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. (2013) 14:372–9. doi: 10.1038/ni.2552
226. Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. (2014) 345:1254009. doi: 10.1126/science.1254009
227. Goto Y, Uematsu S, Kiyono H. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol. (2016) 17:1244–51. doi: 10.1038/ni.3587
228. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. (2013) 498:113–7. doi: 10.1038/nature12240
229. Victoriano AF, Imai K, Okamoto T. Interaction between endogenous bacterial flora and latent HIV infection. Clin Vaccine Immunol. (2013) 20:773–9. doi: 10.1128/CVI.00766-12
230. Yu X, Shahir AM, Sha J, Feng Z, Eapen B, Nithianantham S, et al. Short-chain fatty acids from periodontal pathogens suppress histone deacetylases, EZH2, and SUV39H1 to promote Kaposi's sarcoma-associated herpesvirus replication. J Virol. (2014) 88:4466–79. doi: 10.1128/JVI.03326-13
231. Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. (2003) 62:67–72. doi: 10.1079/PNS2002207
232. Zeng H, Chi H. Metabolic control of regulatory T cell development and function. Trends Immunol. (2015) 36:3–12. doi: 10.1016/j.it.2014.08.003
233. Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. (2007) 204:1757–64. doi: 10.1084/jem.20070590
234. Iliev ID, Mileti E, Matteoli G, Chieppa M, Rescigno M. Intestinal epithelial cells promote colitis-protective regulatory T-cell differentiation through dendritic cell conditioning. Mucosal Immunol. (2009) 2:340–50. doi: 10.1038/mi.2009.13
235. Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. (2007) 8:1086–94. doi: 10.1038/ni1511
236. Molenaar R, Knippenberg M, Goverse G, Olivier BJ, de Vos AF, O'Toole T, et al. Expression of retinaldehyde dehydrogenase enzymes in mucosal dendritic cells and gut-draining lymph node stromal cells is controlled by dietary vitamin A. J Immunol. (2011) 186:1934–42. doi: 10.4049/jimmunol.1001672
237. Goverse G, Molenaar R, Macia L, Tan J, Erkelens MN, Konijn T, et al. Diet-derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J Immunol. (2017) 198:2172–81. doi: 10.4049/jimmunol.1600165
238. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. (2017) 358:359–65. doi: 10.1126/science.aan4526
239. Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. (2004) 72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004
240. Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol. (2015) 6:1085. doi: 10.3389/fmicb.2015.01085
241. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. (2015) 8:80–93. doi: 10.1038/mi.2014.44
242. Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. (1999) 13:2905–27.
243. Pandiyan P, Gartner D, Soezeri O, Radbruch A, Schulze-Osthoff K, Brunner-Weinzierl MC. CD152 (CTLA-4) determines the unequal resistance of Th1 and Th2 cells against activation-induced cell death by a mechanism requiring PI3 kinase function. J Exp Med. (2004) 199:831–42. doi: 10.1084/jem.20031058
244. Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. (2009) 30:832–44. doi: 10.1016/j.immuni.2009.04.014
245. Esposito M, Ruffini F, Bellone M, Gagliani N, Battaglia M, Martino G, et al. Rapamycin inhibits relapsing experimental autoimmune encephalomyelitis by both effector and regulatory T cells modulation. J Neuroimmunol. (2010) 220:52–63. doi: 10.1016/j.jneuroim.2010.01.001
246. Procaccini C, De Rosa V, Galgani M, Abanni L, Cali G, Porcellini A, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. (2010) 33:929–41. doi: 10.1016/j.immuni.2010.11.024
247. Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. (2011) 12:295–303. doi: 10.1038/ni.2005
248. So T, Croft M. Regulation of PI-3-Kinase and Akt Signaling in T lymphocytes and other cells by TNFR family molecules. Front Immunol. (2013) 4:139. doi: 10.3389/fimmu.2013.00139
249. Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature. (2016) 539:437–42. doi: 10.1038/nature19834
250. Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. (2012) 12:325–38. doi: 10.1038/nri3198
251. Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. (2012) 30:39–68. doi: 10.1146/annurev-immunol-020711-075024
252. Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA. (2008) 105:7797–802. doi: 10.1073/pnas.0800928105
253. Clambey ET, McNamee EN, Westrich JA, Glover LE, Campbell EL, Jedlicka P, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci USA. (2012) 109:E2784–2793. doi: 10.1073/pnas.1202366109
254. Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. (2013) 499:485–90. doi: 10.1038/nature12297
255. Carbone F, De Rosa V, Carrieri PB, Montella S, Bruzzese D, Porcellini A, et al. Regulatory T cell proliferative potential is impaired in human autoimmune disease. Nat Med. (2014) 20:69–74. doi: 10.1038/nm.3411
256. Hurez V, Dao V, Liu A, Pandeswara S, Gelfond J, Sun L, et al. Chronic mTOR inhibition in mice with rapamycin alters T, B, myeloid, and innate lymphoid cells and gut flora and prolongs life of immune-deficient mice. Aging Cell. (2015) 14:945–56. doi: 10.1111/acel.12380
257. Shrestha S, Yang K, Guy C, Vogel P, Neale G, Chi H. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol. (2015) 16:178–87. doi: 10.1038/ni.3076
258. Apostolidis SA, Rodriguez-Rodriguez N, Suarez-Fueyo A, Dioufa N, Ozcan E, Crispin JC, et al. Phosphatase PP2A is requisite for the function of regulatory T cells. Nat Immunol. (2016) 17:556–64. doi: 10.1038/ni.3390
259. Gabriel SS, Kallies A. Sugars and fat - A healthy way to generate functional regulatory T cells. Eur J Immunol. (2016) 46:2705–9. doi: 10.1002/eji.201646663
260. Kasper IR, Apostolidis SA, Sharabi A, Tsokos GC. Empowering Regulatory T Cells in Autoimmunity. Trends Mol Med. (2016) 22:784–97. doi: 10.1016/j.molmed.2016.07.003
261. Merchant HA, Liu F, Orlu Gul M, Basit AW. Age-mediated changes in the gastrointestinal tract. Int J Pharm. (2016) 512:382–95. doi: 10.1016/j.ijpharm.2016.04.024
262. Shibagaki N, Suda W, Clavaud C, Bastien P, Takayasu L, Iioka E, et al. Aging-related changes in the diversity of women's skin microbiomes associated with oral bacteria. Sci Rep. (2017) 7:10567. doi: 10.1038/s41598-017-10834-9
263. Zhang L, Wang Y, Xiayu X, Shi C, Chen W, Song N, et al. Altered Gut Microbiota in a Mouse Model of Alzheimer's Disease. J Alzheimers Dis. (2017) 60:1241–57. doi: 10.3233/JAD-170020
264. Zhang T, Xiang J, Cui B, He Z, Li P, Chen H, et al. Cost-effectiveness analysis of fecal microbiota transplantation for inflammatory bowel disease. Oncotarget. (2017) 8:88894–903. doi: 10.18632/oncotarget.21491
265. Zhang W, Zhu YH, Zhou D, Wu Q, Song D, Dicksved J, et al. Oral administration of a select mixture of bacillus probiotics affects the gut microbiota and goblet cell function following Escherichia coli challenge in newly weaned pigs of genotype MUC4 that are supposed to be enterotoxigenic E. coli F4ab/ac receptor negative. Appl Environ Microbiol. (2017) 83: e02747–16. doi: 10.1128/AEM.02747-16
266. Bodogai M, O'Connell J, Kim K, Kim Y, Moritoh K, Chen C, et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci Transl Med. (2018) 10:eaat4271. doi: 10.1126/scitranslmed.aat4271
267. Irie K, Novince CM, Darveau RP. Impact of the oral commensal flora on alveolar bone homeostasis. J Dent Res. (2014) 93:801–6. doi: 10.1177/0022034514540173
268. Irie K, Tomofuji T, Ekuni D, Fukuhara D, Uchida Y, Kataoka K, et al. Age-related changes of CD4(+) T cell migration and cytokine expression in germ-free and SPF mice periodontium. Arch Oral Biol. (2018) 87:72–8. doi: 10.1016/j.archoralbio.2017.12.007
269. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. (2011) 108(Suppl. 1):4615–22. doi: 10.1073/pnas.1000082107
270. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. (2010) 32:815–27. doi: 10.1016/j.immuni.2010.06.001
271. Rogier R, Evans-Marin H, Manasson J, van der Kraan PM, Walgreen B, Helsen MM, et al. Alteration of the intestinal microbiome characterizes preclinical inflammatory arthritis in mice and its modulation attenuates established arthritis. Sci Rep. (2017) 7:15613. doi: 10.1038/s41598-017-15802-x
272. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. (2013) 14:195–206. doi: 10.1016/j.chom.2013.07.012
273. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. (2015) 1:653–61. doi: 10.1001/jamaoncol.2015.1377
274. Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol Res. (2018) 6:1327–36. doi: 10.1158/2326-6066.CIR-18-0174
275. Liu L, Tabung FK, Zhang X, Nowak JA, Qian ZR, Hamada T, et al. Diets that promote colon inflammation associate with risk of colorectal carcinomas that contain Fusobacterium nucleatum. Clin Gastroenterol Hepatol. (2018) 16:1622–1631 e1623. doi: 10.1016/j.cgh.2018.04.030
276. Yang CY, Yeh YM, Yu HY, Chin CY, Hsu CW, Liu H, et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol. (2018) 9:862. doi: 10.3389/fmicb.2018.00862
277. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. (2019) 17:156–166. doi: 10.1038/s41579-018-0129-6
278. Dinakaran V, Mandape SN, Shuba K, Pratap S, Sakhare SS, Tabatabai MA, et al. Identification of specific oral and gut pathogens in full thickness colon of colitis patients: implications for colon motility. Front Microbiol. (2018) 9:3220. doi: 10.3389/fmicb.2018.03220
279. Sears CL. The who, where and how of fusobacteria and colon cancer. Elife. (2018) 7:e28434. doi: 10.7554/eLife.28434
280. Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. (2013) 501:426–9. doi: 10.1038/nature12447
281. D'Souza B, Slack T, Wong SW, Lam F, Muhlmann M, Koestenbauer J, et al. Randomized controlled trial of probiotics after colonoscopy. ANZ J Surg. (2017) 87:E65–E69. doi: 10.1111/ans.13225
282. Bagga D, Reichert JL, Koschutnig K, Aigner CS, Holzer P, Koskinen K, et al. Probiotics drive gut microbiome triggering emotional brain signatures. Gut Microbes. (2018) 9:486–96. doi: 10.1080/19490976.2018.1460015
283. Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. (2018) 557:434–8. doi: 10.1038/s41586-018-0092-4
284. Hoffmann D, Palumbo F, Ravel J, Roghmann MC, Rowthorn V, von Rosenvinge E. Improving regulation of microbiota transplants. Science. (2017) 358:1390–1. doi: 10.1126/science.aaq0034
285. Sun B, Jia Y, Hong J, Sun Q, Gao S, Hu Y, et al. Sodium butyrate ameliorates high-fat-diet-induced non-alcoholic fatty liver disease through peroxisome proliferator-activated receptor alpha-mediated activation of beta oxidation and suppression of inflammation. J Agric Food Chem. (2018) 66:7633–42. doi: 10.1021/acs.jafc.8b01189
286. Mortaz E, Adcock IM, Folkerts G, Barnes PJ, Paul Vos A, Garssen J. Probiotics in the management of lung diseases. Mediat Inflamm. (2013) 2013:751068. doi: 10.1155/2013/751068
287. Matt SM, Allen JM, Lawson MA, Mailing LJ, Woods JA, Johnson RW. Butyrate and dietary soluble fiber improve neuroinflammation associated with aging in mice. Front Immunol. (2018) 9:1832. doi: 10.3389/fimmu.2018.01832
288. Lu M, Wang Z. Microbiota and Aging. Adv Exp Med Biol. (2018) 1086:141–56. doi: 10.1007/978-981-13-1117-8_9
289. Bishehsari F, Engen PA, Preite NZ, Tuncil YE, Naqib A, Shaikh M, et al. Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes. (2018) 9:E102. doi: 10.3390/genes9020102
Keywords: microbiome, mucosa, Treg, mucosal immunity, inflammation, Th17, antibiotics, resident microbes
Citation: Pandiyan P, Bhaskaran N, Zou M, Schneider E, Jayaraman S and Huehn J (2019) Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front. Immunol. 10:426. doi: 10.3389/fimmu.2019.00426
Received: 02 December 2018; Accepted: 18 February 2019;
Published: 08 March 2019.
Edited by:
Sudhanshu Shekhar, University of Oslo, NorwayReviewed by:
Nobuhiko Kamada, University of Michigan Health System, United StatesShimpei Kawamoto, Osaka University, Japan
Copyright © 2019 Pandiyan, Bhaskaran, Zou, Schneider, Jayaraman and Huehn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pushpa Pandiyan, cHhwMjI2QGNhc2UuZWR1
 Pushpa Pandiyan
Pushpa Pandiyan Natarajan Bhaskaran
Natarajan Bhaskaran Mangge Zou2,3
Mangge Zou2,3 Elizabeth Schneider
Elizabeth Schneider Sangeetha Jayaraman
Sangeetha Jayaraman Jochen Huehn
Jochen Huehn