- 1InnatOss Laboratories B.V., Oss, Netherlands
- 2EpiVax, Inc., Providence, RI, United States
- 3Department of Cell and Molecular Biology, Institute for Immunology and Informatics, University of Rhode Island, Providence, RI, United States
- 4Department of Biomedical Sciences, Colorado State University, Fort Collins, CO, United States
- 5Vaccine and Immunotherapy Center, Massachusetts General Hospital, Boston, MA, United States
- 6Department of Medicine, Yale University School of Medicine, New Haven, CT, United States
Coxiella burnetii, the causative agent of Q fever, is a Gram-negative intracellular bacterium transmitted via aerosol. Regulatory approval of the Australian whole-cell vaccine Q-VAX® in the US and Europe is hindered by reactogenicity in previously exposed individuals. The aim of this study was to identify and rationally select C. burnetii epitopes for design of a safe, effective, and less reactogenic T-cell targeted human Q fever vaccine. Immunoinformatic methods were used to predict 65 HLA class I epitopes and 50 promiscuous HLA class II C. burnetii epitope clusters, which are conserved across strains of C. burnetii. HLA binding assays confirmed 89% of class I and 75% of class II predictions, and 11 HLA class II epitopes elicited IFNγ responses following heterologous DNA/DNA/peptide/peptide prime-boost immunizations of HLA-DR3 transgenic mice. Human immune responses to the predicted epitopes were characterized in individuals naturally exposed to C. burnetii during the 2007–2010 Dutch Q fever outbreak. Subjects were divided into three groups: controls with no immunological evidence of previous infection and individuals with responses to heat-killed C. burnetii in a whole blood IFNγ release assay (IGRA) who remained asymptomatic or who experienced clinical Q fever during the outbreak. Recall responses to C. burnetii epitopes were assessed by cultured IFNγ ELISpot. While HLA class I epitope responses were sparse in this cohort, we identified 21 HLA class II epitopes that recalled T-cell IFNγ responses in 10–28% of IGRA+ subjects. IGRA+ individuals with past asymptomatic and symptomatic C. burnetii infection showed a comparable response pattern and cumulative peptide response which correlated with IGRA responses. None of the peptides elicited reactogenicity in a C. burnetii exposure-primed guinea pig model. These data demonstrate that a substantial proportion of immunoinformatically identified HLA class II epitopes show long-lived immunoreactivity in naturally infected individuals, making them desirable candidates for a novel human multi-epitope Q fever vaccine.
Introduction
Q fever is a zoonotic disease that is transmitted to humans predominantly by aerosol from infected ruminants such as goats, sheep, and cattle and has a global public health impact (1). Its causative agent, the small Gram-negative coccobacillus Coxiella burnetii, infects a wide range of vertebrate and invertebrate hosts, is very stable in the environment and highly contagious; it is estimated that a single inhaled organism can result in infection (2). As a result, C. burnetii is also considered to be a potential biothreat agent (3). Q fever is endemic in many countries worldwide, with outbreaks occurring mainly in occupational settings, including the livestock industry and deployed military personnel (1). The largest reported outbreak occurred in the Netherlands from 2007 to 2010 with an estimated 40,000 infections at the center of the epidemic area alone (4). Infection remains asymptomatic in an estimated 50–60% of individuals (1). Acute C. burnetii infection, when identified clinically and serologically, can be treated with antibiotics such as doxycycline. However, long-term complications of infection are common; 10–20% of patients with acute Q fever later develop Q fever fatigue syndrome, and 1–5% of (often asymptomatically) infected individuals progress to persistent infection known as chronic Q fever, manifesting as endocarditis, aneurysms or vascular infections in individuals with specific risk factors (1, 5). Therefore, a preventive Q fever vaccine is considered critical in occupational and biodefense settings (6).
The two currently available Q fever vaccine formulations, Q-VAX® for humans (licensed for use in Australia only) and COXEVAC® for ruminant animals such as goats (licensed in the European Union), are inactivated whole cell vaccines based on phase I C. burnetii. Although Q-VAX® is strongly protective (7, 8), reported side effects in previously exposed individuals, in particular at the site of injection, necessitate pre-vaccination screening and are a daunting hurdle to regulatory approval in the US and Europe (6, 9). Hence, there is a clear need for a less reactogenic and at least equally efficacious vaccine for occupational and biodefense purposes.
Subunit vaccines have the advantage over traditional whole cell vaccines that they are better tolerated and offer the opportunity for rational design, both in terms of the target antigens and the appropriate delivery or adjuvant system selected to elicit the desired immune response (10). While both humoral and cellular immune responses are elicited upon C. burnetii infection and administration of whole cell vaccines (11), antibodies alone are insufficient to resolve infection (12, 13). Results from studies in murine infection models suggest that T-cell responses, particularly Th1 responses, are critical for clearance of the bacteria (13–15). The Th1 cytokine IFNγ has been shown to restore phagosome maturation and facilitate intracellular killing of C. burnetii (16, 17). Accordingly, a proof of concept study showed that partial protection in C57BL/6 mice can be elicited by a vaccine comprising seven C. burnetii CD4 epitopes (18).
In this context, the objective of the Q-VaxCelerate consortium is to develop a non-reactogenic T-cell-targeted vaccine to prevent Q fever disease in humans (19). To rationally select epitopes for inclusion in such a vaccine, we set out to identify HLA class I and class II C. burnetii epitopes using a combination of immunoinformatic and experimental methods. A library of computationally predicted human T-cell epitopes derived from C. burnetii was assessed for human HLA binding in vitro, immunogenicity and reactogenicity were determined in animal models, and antigenicity was investigated in a cohort of individuals naturally exposed to C. burnetii during the 2007–2010 Dutch Q fever outbreak. Using this systematic approach, we successfully identified a set of epitopes that recalls long-term memory IFNγ T-cell responses in humans and thus represents a promising first step in the development of a T-cell based human multi-epitope Q fever vaccine.
Materials and Methods
Ethics Statement
Animal research protocols for studies with HLA-DR3 transgenic mice performed by EpiVax were reviewed and approved by TGA Sciences Incorporated Institutional Animal Care and Use Committee (P07-10R20-EV69, P07-10R20-EV71). Animal research protocols for guinea pig experiments were reviewed and approved by the Colorado State University Institutional Animal Care and Use Committee (14-5305A, 16-6844A). All animal experimental activities were conducted in full compliance with university, federal and international regulations and the standards of the DoD Animal Care and Use Review Office. Methods of euthanasia as described below were consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association (AVMA).
The human study was carried out in accordance with the recommendations of the Medical Ethical Committee Brabant (Tilburg, Netherlands). All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was reviewed and approved by the Medical Ethical Committee Brabant (Tilburg, Netherlands, NL51305.028.15).
Immunoinformatics
Sequence Collection
C. burnetii antigens used in epitope predictions were immune-dominant antigens that were recognized by sera from C. burnetii-exposed humans and demonstrated to stimulate CD4+ T-cell and/or antibody responses in mice, as reported in the literature (18, 20–29), and type IV secretion system (T4SS) effectors (21, 23, 30–37). Of 94 T4SS protein sequences retrieved from the UniProt database (38), 53 were selected for immunoinformatic analysis; 20 putative effectors lacking experimental support for secretion, eight hypothetical proteins from pseudogenes and 13 proteins with discontinued database records were rejected. An additional 40 sequences were retrieved from UniProt for the immune-dominant antigen set. All sequences were retrieved from the reference RSA 493/Nine Mile phase I C. burnetii strain. Six additional complete C. burnetii genomes available in 2015 were obtained from UniProt for homology analyses: (i) RSA 331/Henzerling II, (ii) CbuK_Q154, (iii) Namibia, (iv) MSU Goat Q177, (v) CbuG_Q212, and (vi) Dugway 5J108-111. Of note, genome sequences were completed for two C. burnetii strains associated with the Dutch Q fever outbreak (39) during the course of this work, and all but one of the selected epitopes (p69) were 100% conserved in the two Dutch strains.
Epitope Prediction
Sequences for all antigens from the reference Nine Mile strain were scored for binding potential against a panel of HLA class II and class I alleles using the EpiMatrix algorithm (40). This algorithm as well as the ClustiMer and JanusMatrix algorithms discussed below are part of the proprietary iVAX toolkit developed by EpiVax, which is available for use under license or by collaboration (http://epivax.com/immunogenicity-screening/ivax-web-based-vaccine-design). Epitopes which were 100% identical in the corresponding antigens from at least six of the seven genomes (HLA class I epitopes) or all seven genomes (HLA class II epitopes) were prioritized. Class II epitopes were identified for eight supertype HLA-DR alleles: DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*0801, DRB1*1101, DRB1*1301, and DRB1*1501. For class I epitopes, selections were made based on predictions for six HLA-A and HLA-B supertype alleles: A*0101, A*0201, A*0301, A*2402, B*0702, B*4403. Each set of supertype alleles covers >95% of the human population worldwide (41, 42). EpiMatrix assigns a score for each peptide/allele pair on a normalized Z-scale. Peptides with Z-Scores ≥1.64 are considered hits and have a significant likelihood of binding to HLA molecules. These sequences represent the top 5% of any randomly generated set of 9-mer peptides (43).
Regions of high class II epitope density per antigen were identified using the ClustiMer algorithm for selection of vaccine candidates with increased breadth of reactivity (40). ClustiMer searches for contiguous segments of 15–30 amino acids with elevated class II HLA binding promiscuity. Epitope density within a cluster is estimated by EpiMatrix Cluster Scores, where scores of 10 and above are considered elevated. Peptide sequences with elevated EpiMatrix Cluster Scores usually contain class II HLA binding motifs to most, if not all, HLA-DR supertype alleles. MHC Class II epitope clusters were further filtered to remove sequences derived from signal sequences and transmembrane domains to avoid peptide synthesis and solubility complications. For class I peptides, epitopes with Z-Scores in the top 1% of a normal distribution (Z-Score ≥2.32) were prioritized. A greater weight was given to the top 1% EpiMatrix scoring peptides per HLA class I supertype that were also predicted to bind to at least one additional HLA class I supertype.
Homology Analysis
To eliminate peptide candidates unlikely to stimulate effector T-cell responses due to potential cross-reactivity with T-cells previously trained on host or commensal antigens, predicted HLA class I and class II Coxiella sequences were screened for T-cell receptor-face homology against host and commensal proteins using the JanusMatrix algorithm (44). Databases comprising the UniProt reviewed human proteome (http://www.uniprot.org/proteomes/UP000005640) and Human Microbiome Project gut commensal proteomes were used. Epitopes with low human or human microbiome JanusMatrix homology scores were prioritized. The JanusMatrix human homology score cutoff was 2, for both HLA class I and II, and the microbiome cutoff was 25 for HLA class I and 37 for HLA class II. JanusMatrix cutoffs are defined such that only 5% of randomly generated HLA ligands exceed the thresholds.
Peptide Synthesis
Five milligrams of peptide for each epitope selected as defined above were prepared by Fmoc solid phase peptide synthesis (Twentyfirst Century Biochemicals, Marlborough, MA, United States) and HPLC purified to >90% purity (peak area). The peptide mass was verified by ESI-MS (QSTAR XL Pro, Qo-TOF) and the sequence of each peptide was verified by tandem mass spectrometry (CID MSMS).
HLA Binding Assays
HLA Class II Binding Assay
A competition-based assay based on Steere et al. was employed (45). In 96-well plates, non-biotinylated test peptides over a three-log concentration range competed for binding to 50 nM soluble class II molecules (Benaroya Institute, Seattle, WA) against a biotinylated standard peptide at a fixed concentration (25 nM) at 37°C for 24 h to reach equilibrium. Class II molecules were then captured on ELISA plates using pan anti-class II antibodies (clone L243, anti-HLA-DR; Biolegend). Plates were washed and incubated with DELFIA Europium-labeled streptavidin (Perkin Elmer) for 1 h at room temperature. DELFIA Europium activation buffer (Perkin Elmer) was added to develop the plates for 15–20 min at room temperature before data acquisition on a Time Resolved Fluorescence (TRF) plate reader. All assays were performed in triplicate. Binding assays were performed for 8 alleles: DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, DRB1*0801, DRB1*1101, DRB1*1301, and DRB1*1501. Dose-dependent binding data were fitted by non-linear regression to calculate IC50 values for each peptide-HLA allele pair (GraphPad Prism Version 7). Peptide binding affinity was classified according to the following: very high (IC50 < 0.1 μM), high (IC50 0.1–1 μM), moderate (IC50 1–10 μM) and low affinity (IC50 10–100 μM). Epitopes with IC50 values too high to accurately measure under binding conditions tested (>100 μM) or with no dose-dependent responses were considered non-binders.
HLA Class I Binding Assay
A competition-based assay based on fluorescence polarization, where reference fluorescent-labeled peptide is incubated with soluble HLA with or without competitor test peptides, was performed according to Buchli et al. (46). Peptide binding over a three-log concentration range was assessed for the A*0101, A*0201, A*0301, A*2402, B*0702, and B*4402 alleles. The latter was used to test B*4403 predictions. A dose response curve fit was obtained by non-linear regression to calculate IC50 values for each peptide-HLA allele pair. Binding strength was classified according to IC50 value: high (<5 μM), moderate (5–50 μM), low (50–1,000 μM) or no affinity (>1,000 μM or no dose-dependent response).
Assessment of Class II Epitope Immunogenicity in tgHLA-DR3 Mice
Multi-Epitope DNA Vaccine Engineering
Multi-epitope DNA vaccines were engineered by concatenating epitope sequences to form five multi-epitope genes, each containing 10 HLA class II epitopes. To avoid production of neo-epitopes at epitope junctions, the VaxCAD algorithm was used to arrange epitopes in an order that diminishes potential junctional immunogenicity (40). Where re-ordering by VaxCAD did not sufficiently reduce potential junctional immunogenicity, Gly-Pro-Gly-Pro-Gly spacer sequences were engineered between epitopes to further remove junctional epitopes. Genes were synthesized by GeneArt and subcloned at pre-determined flanking restriction sites downstream of the tissue plasminogen activator leader sequence in pNTC8682-eRNA41H (Nature Technology Corporation), a DNA vaccine vector that accommodates FDA recommendations for construction of plasmid DNA vaccines.
Plasmid DNA Vaccine Production
High purity plasmids for immunization were prepared by Nature Technology Corporation, Inc. at research grade. Each plasmid underwent quality control testing including spectrophotometric concentration and A260/A280 ratio determination (1.97), restriction digest analysis to assure the presence of the multi-epitope genes, agarose gel electrophoresis determination of residual host RNA and DNA (none detected), and quantitative endotoxin testing (<2.0 EU/mg).
Peptide Vaccine Preparation
Peptides produced by twentyfirst Century Biochemicals corresponding to epitopes in the DNA vaccines were formulated in incomplete Freund's adjuvant (IFA) with 10 μg each of immunostimulatory CpG oligodeoxynucleotide 1826 (5′-TCCATGACGTTCCTGACGTT-3′), muramyl dipeptide (MDP) and CL097 (InvivoGen).
Mice
HLA-DR3 transgenic mice were obtained from Dr. Chella David (Mayo Medical School) under commercial license to EpiVax. The mice express the HLA-DRA and DRB1*0301 genes on a B.10-Ab0 mouse class II-negative background (47).
Vaccinations
Vaccine and placebo-treated mice (n = 3/group) were all female and 6–8 weeks old at the start of heterologous DNA/DNA/peptide/peptide prime-boost immunizations. Epitopes were arranged into five groups of 10 epitopes in generally descending order of EpiMatrix epitope cluster score as follows: Group 1 (p9, p12, p14, p22, p27, p28, p31, p32, p45, p49), group 2 (p3, p7, p8, p15, p16, p38, p39, p42, p44, p46), group 3 (p4, p10, p19, p20, p21, p23, p25, p34, p40, p48), group 4 (p1, p2, p5, p6, p11, p13, p30, p36, p41, p47), group 5 (p17, p18, p24, p26, p29, p33, p35, p37, p43, p50). DR3 epitope scores correlated with cluster scores across the groups. DNA-prime vaccine was administered to mice intramuscularly by electroporation using the Ichor Medical Systems TriGrid platform with 20 μL of 10 μg naked plasmid DNA in sterile PBS injected into the quadriceps muscle. Mice received the DNA vaccine twice spaced by a 2-week interval. Two weeks later, they were boosted twice with peptide vaccine at a 2-week interval. For peptide immunizations, each mouse was administered 100 μl IFA emulsion (50 μg peptide) subcutaneously by needle stick injection. Four weeks after the final vaccination, tgHLA-DR3 mice were euthanized by administration of ketamine/xylazine intraperitoneally at 4–5 times the anesthetic dose (ketamine 80–100 mg/kg; xylazine 8–10 mg/kg).
Ex vivo ELISpot Assay in Mouse Splenocytes
The frequency of epitope-specific splenocytes was determined by IFNγ ELISpot assay using the colorimetric Mabtech IFNγ ELISpot Kit with pre-coated plates according to the manufacturer's protocol. Washed splenocytes in RPMI 1640 (Gibco) supplemented with 10% fetal calf serum (FCS, Atlanta Biologicals) were added at 2.5 × 105 cells per well. Individual peptides were added at 10 μg/ml in triplicate wells. Peptide pools were added at 10 μg/ml, equating to 1 μg/ml per peptide. Triplicate wells were stimulated with 2 μg/ml Concanavalin A (ConA; Sigma Aldrich) as a positive control, and six replicate wells with medium containing 0.02% DMSO were used for background determination. Raw spot counts were recorded by ZellNet Consulting, Inc. using a Zeiss high-resolution automated ELISpot reader system and companion KS ELISpot software. Results were calculated as the average number of spots in the peptide wells, adjusted to spots per one million cells. A response in immunized mice was considered positive if the number of averaged spots was (i) at least twice as high as background (stimulation index ≥2), (ii) >50 spot forming cells above background per million splenocytes (1 response per 20,000 cells), and (iii) statistically different (p < 0.05) from that of mock immunized mice by Student's t-test.
Human Study Cohort
Q fever exposed individuals were recruited from a cohort characterized in a previous large Q fever study conducted in the village of Herpen, the Netherlands (48), which experienced a high incidence of C. burnetii infection during the 2007–2010 Q fever outbreak (49). All subjects had been tested using a Q fever interferon-γ release assay (IGRA, Q-detect™) assay of cellular immunity during a previous study in which 80% of the adult population of Herpen were screened for Q fever (48). Individuals were invited to participate in the current study following preselection based on clinical history (disease, comorbidities, and medication) as well as IGRA and serological data generated during the previous study (48). To maximize the potential to detect C. burnetii epitope-specific T-cells, preference was given to donors with strong responses to whole heat-killed C. burnetii in the IGRA and without potentially confounding immune disorders. In addition, five individuals with known past symptomatic Q fever consented to participate. In total, 143 participants provided written informed consent. IGRA responses were re-assessed upon inclusion in October 2015. Volunteers who had no history of Q fever disease (48) and scored negative by immunofluorescence assay (50) as well as by IGRA in spring 2014 and upon inclusion into the present study in autumn 2015 were allocated to control group A (n = 26). Seven volunteers that were IGRA positive in 2014 but did not meet positive scoring thresholds anymore 1.5 years later upon inclusion into the present study were excluded from further analysis. The remaining 110 volunteers that were positive by IGRA in both 2014 and 2015 were subdivided based on past Q fever disease [either registered (notified) in the national surveillance system, or self-reported] into groups B (asymptomatic, n = 73) and C (symptomatic, n = 37).
Whole Blood IFNγ Release Assay (Q-Detect™)
Whole lithium-heparin anti-coagulated blood was stimulated with C. burnetii antigen (heat killed Cb02629, Wageningen Bioveterinay Research, lot 14VRIM014) in 96-well polypropylene plates (Greiner BioOne) by adding 180 μl blood to 20 μl C. burnetii antigen diluted in phenol red-free RPMI supplemented with Glutamax (2 mM), Gentamycin (5 μg/ml) and sodium pyruvate (1 mM, all ThermoFisher Scientific). A 1.5 % (v/v, final concentration) solution of PHA-M (ThermoFisher Scientific), was added to separate wells as a positive control. Medium only was added to the negative control wells. All stimulations were performed in duplicate. After 22 ± 1 h whole blood cultures were re-suspended and IFNγ concentrations were assessed in whole blood by ELISA, using the IFNγ Pelipair protocol (Sanquin) with minor modifications. The upper detection limit of IGRA under these conditions is 1,050 pg/ml. A subject was scored positive by IGRA if the C. burnetii-induced IFNγ production was ≥16 pg/ml above background and the ratio of the logarithmic value of background-subtracted C. burnetii and PHA responses {(log[C. burnetii]–log[neg control])/(log[PHA]-log[neg control])} was ≥0.4.
HLA Typing of Human Subjects
HLA typing was performed at the HLA laboratory at the Laboratory of Translational Immunology at the UMC Utrecht, the Netherlands. Genomic DNA was isolated from EDTA anti-coagulated blood within 48 h upon collection using the MagNA Pure Compact system (Roche Diagnostics). The DNA samples were typed for HLA-A, -B (5′-UTR-3′UTR), and -DRB1 (exon 2–3′-UTR) by Next Generation Sequencing (NGS). Firstly, HLA target sequences were generated by long-range PCR using the Qiagen LongRange PCR kit and HLA-A, -B, and -DRB1 primers as described previously (51). Library preparation was performed using the GenDX NGSgo®-LibrX and NGSgo®-IndX kits following the manufacturers' recommendations (GenDX). Pooled samples were sequenced on an Illumina MiSeq by 2 × 250 paired end reading using the MiSeq reagent kit v2 (500 cycles). Sequences were analyzed with the NGSEngine software (GenDX). The resulting HLA-A, HLA-B, and HLA-DRB1 alleles were assigned to supertype families as defined by (42, 52) and/or based on homology of the HLA binding pockets. For determining allelic frequencies, donors homozygous for a given HLA allele were counted once.
Cultured ELISpot Assay Screening of Peptides in Human PBMCs
A combination of antigen-specific T-cell expansion culture and enzyme-linked immune spot assay (cultured ELISpot) was chosen as the primary assay for peptide screening, to achieve high sensitivity for detecting low frequency antigen-specific T-cell responses to C. burnetii peptides and facilitate detection of central memory T-cells (53). The protocol was adapted from Subbramanian et al. (54) and optimized at Innatoss using two reference peptide pools of HLA class I and class II peptides of CEF (Cytomegalovirus, Epstein-Barr virus, and influenza; JPT Peptide Technologies). A comparison of three media showed that RPMI medium supplemented with 10% FCS (HyClone) as a blocking and culture medium gave the best signal to background ratio in this ELISpot assay.
Antigen-Specific Expansion
PBMCs were isolated from lithium-heparin anti-coagulated blood using Leukosep tubes prefilled with Ficoll (Greiner BioOne) according to the manufacturer's recommendations. Isolated PBMCs were used for antigen-specific expansion cultures at 5 × 106 cells per well in 48-well flat bottom plates (Corning) in 150 μl complete RPMI (phenol red-free RPMI supplemented with Glutamax (2 mM), Gentamycin (5 μg/ml), sodium pyruvate (1 mM, all Thermo Fisher Scientific) and 10% fetal bovine serum (HyClone). Antigen-specific expansion was performed using stimulation with pools of 10 peptides each (final concentration 2 μg/ml per peptide, 0.2% DMSO). On day 3 and 6 of culture, medium was refreshed by addition of an equal volume of complete RPMI with recombinant human IL-2 (Immunotools, final concentration 20 units/ml). On day 8, cells were harvested, counted and individual peptide responses assessed by ELISpot.
ELISpot Assay
MultiScreen IP filter plates (Merck Millipore) were pre-wetted with 35% ethanol, coated overnight with IFNγ capture antibody at the recommended concentration (Diaclone) in DPBS (Thermo Fisher Scientific) and blocked for at least 60 min with complete RPMI prior to addition of cells for re-stimulation. For each expansion culture, recovered cells were evenly distributed for peptide re-stimulation, negative and positive controls. Based on cell availability, a median of 38,000 cells per expansion pool (interquartile range (IQR) 27,000–52,000) were plated per replicate well. Assay wells were re-stimulated with each of the 10 respective individual peptides in quadruplicate (final concentration 2 μg/ml per peptide, 0.02% DMSO). Control wells were stimulated with either medium only (containing 0.02% DMSO; eight replicates) or with Staphylococcus enterotoxin B (SEB, final concentration 1 μg/ml, Sigma Aldrich; quadruplicate assays). After 20 h incubation at 37°C, plates were washed, and sequentially incubated with a biotinylated anti-human IFNγ detection antibody and streptavidin-alkaline phosphatase conjugate in DPBS/0.5% FCS and developed using BCIP-NBT (all Diaclone) according to the manufacturer's recommendation. ELISpot plates were dried overnight, scanned on an AID Classic reader system and analyzed using the AID ELISpot software v7.0 (both AID Diagnostika GmbH). Spot forming units were counted using identical settings for all plates and all counts were reviewed and adjusted manually only where necessary to remove artifacts.
Data Analysis
To account for variation in background responses between expansion cultures and donors following cytokine-assisted T-cell expansion and to decrease the likelihood of detecting false positive responses in plates with either high background (when only considering an absolute difference to background) or low background (when only considering a fold difference above background), three combined threshold criteria were applied. Peptide re-stimulation responses were defined as positive when they were (i) significantly higher than spot counts in matched negative control wells from the same expansion culture by one-way ANOVA with Holm-Šídák multiple comparison correction post-hoc test, reached (ii) a stimulation index of at least 2 above the matched negative control wells, and (iii) an absolute cut-off of >10 SFU/well.
Guinea Pig Reactogenicity Against Class I and II Peptides
Female Dunkin-Hartley guinea pigs were sensitized by intranasal inoculation with 106 genome equivalents of C. burnetii Nine Mile strain or saline in 100 μL volume, as described previously (55). Peptides were tested by intradermal challenge, delivered at day 42 post sensitization. HLA class II and I peptides were administered in pools of five, with each peptide being tested twice in two different pool preparations. Peptide pools were created with 2 μg of each peptide (10 μg peptide per pool in total) in 100 μL saline with 1% DMSO. Challenge with 2 μg COXEVAC® whole cell vaccine (Ceva Sante Animale, Libourne, France) was used as a positive control; negative controls consisted of saline or 1% DMSO injections. On day 7, animals were anesthetized (ketamine 40 mg/kg and xylazine 5 mg/kg, i.p.) and euthanized with beuthanasia (i.p.). Gross reactions were monitored daily and skin biopsies obtained at day 7 post-challenge were fixed, sectioned, and stained with hematoxylin and eosin. Histological reactions were scored by an experimenter blinded to the treatment group, using the criteria previously described (55). Briefly, a score of 0 indicates no inflammation, 1 indicates localized macrophage dominated inflammation, 2 macrophage dominated inflammation with limited tissue infiltrations, 3 lymphocytic inflammatory infiltrates extending into the deep dermis, 4 edema and increased pyogranulomatous inflammation extending deep into the subcutis, and 5 widespread pyogranulomatous inflammation including necrosis.
Results
In silico Identification of Predicted C. burnetii T-Cell Epitopes
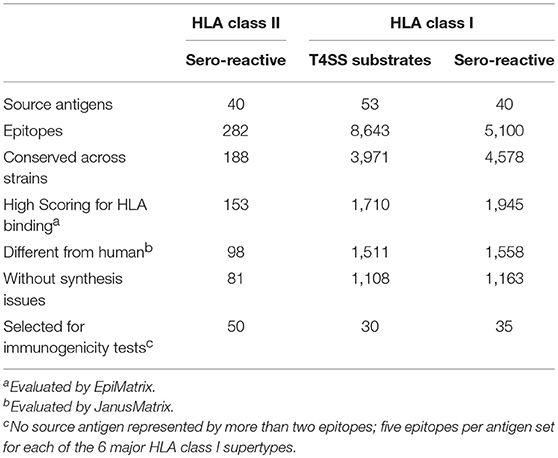
Two C. burnetii antigen sets were selected as the basis for immunoinformatic identification of HLA class I and II T-cell epitopes. The first set (for HLA class I epitope prediction) was comprised of 53 published substrates of the type IV secretion system (T4SS), which are translocated from C. burnetii to the host cytoplasm where they are expected to enter the class I antigen processing pathway and trigger CD8+ T cell responses (21, 23, 30–37). The second set (for both HLA class I and II epitope prediction) covered 40 sero-reactive C. burnetii antigens based on antibody responses in humans and mice, as well as evidence of processing and presentation to stimulate murine CD4+ T-cells (18, 26, 28, 29, 56). Using the EpiMatrix algorithm, 8,643 putative 9- and 10-mer T-cell epitopes predicted to bind to at least one of six HLA class I supertype alleles and 282 promiscuous epitope clusters, spanning 14–25 amino acids, predicted to bind HLA class II alleles covering >90% of the world-wide human population, were identified from the reference C. burnetii Nine Mile strain (Table 1).
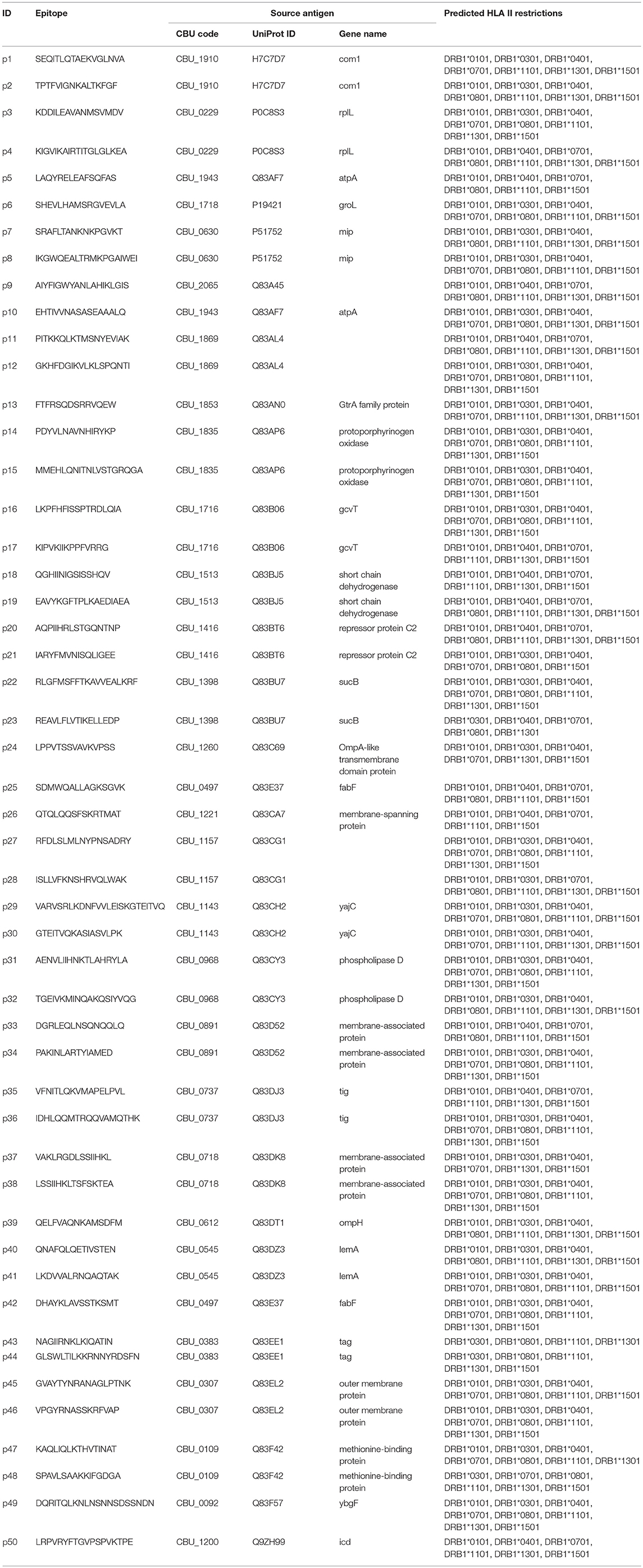
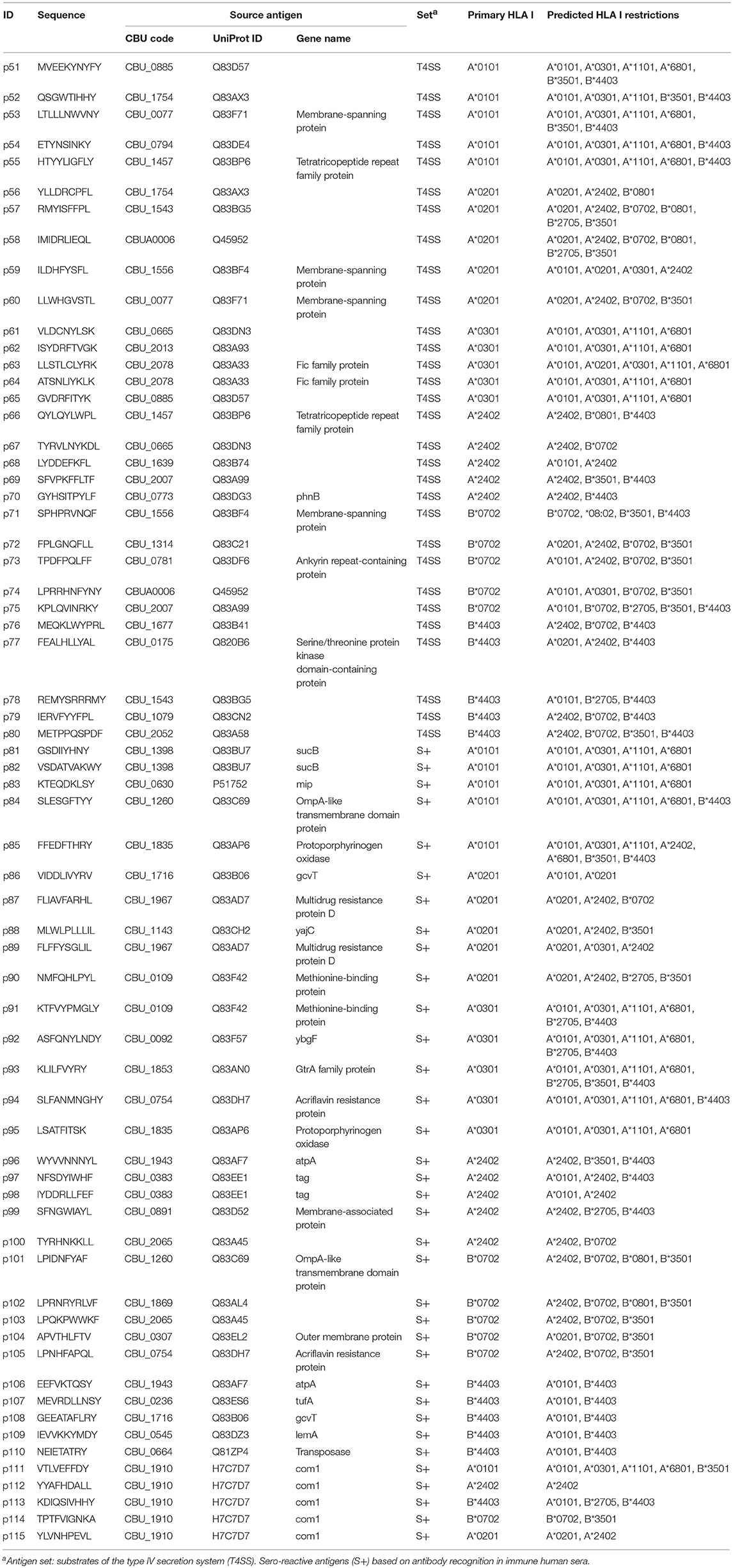
The derived HLA class I epitopes and HLA class II promiscuous epitope clusters were then filtered to focus on sequences that (i) are conserved with other C. burnetii strains; (ii) have very high likelihood of binding human HLA alleles; (iii) exhibit low potential for cross-reactivity with peptides derived from the human proteome or microbiome based on the JanusMatrix algorithm (44); and (iv) do not present obvious issues for peptide synthesis or stability. Finally, 50 HLA class II epitope clusters (Table 2) and 65 HLA class I epitopes (Table 3) were selected for immune reactivity screening such that no source antigen was represented more than twice. Five epitopes were selected for each of the six HLA class I supertypes and for each antigen set (T4SS substrates and sero-reactive antigens), if possible giving preference to HLA class I ligands predicted to bind to at least two class I supertype alleles. An additional five HLA class I epitopes were specifically selected from the immunodominant C. burnetii antigen com1 (28, 56).
HLA Binding of Predicted C. burnetii Epitopes
Epitope predictions were validated using HLA binding assays. Peptides representing the 50 promiscuous C. burnetii HLA class II epitope clusters were assayed in competition binding assays against each of the eight class II HLA supertype alleles (Figure 1). Each of the peptides bound as predicted to at least two HLA-DR alleles: 6% bound to two HLA-DR alleles, 2% to three alleles, 10% to four alleles, 4% to five alleles, 28% to six alleles, 24% to seven alleles, and 26% to all eight alleles. Independent of original binding predictions, amongst all 400 peptide-allele pairs tested, 6.5% of peptides bound with very high affinity (IC50 < 0.1 μM), 16% of peptides bound with high affinity (IC50 0.1–1 μM), 30% bound with moderate affinity (IC50 1–10 μM), 25.5% bound with low affinity (IC50 10–100 μM), and 22% exhibited no detectable affinity for the HLA-DR tested (IC50 >100 μM or no dose-dependent response). Overall, 81% of predicted binding events and 41% of predicted non-binding events were verified in vitro. Collectively, the agreement of computational predictions with binding assay results was 75% (χ2, p < 0.001), ranging from 60 to 88% for individual alleles (Table S1), consistent with published studies using the same algorithms and assay conditions (57, 58).
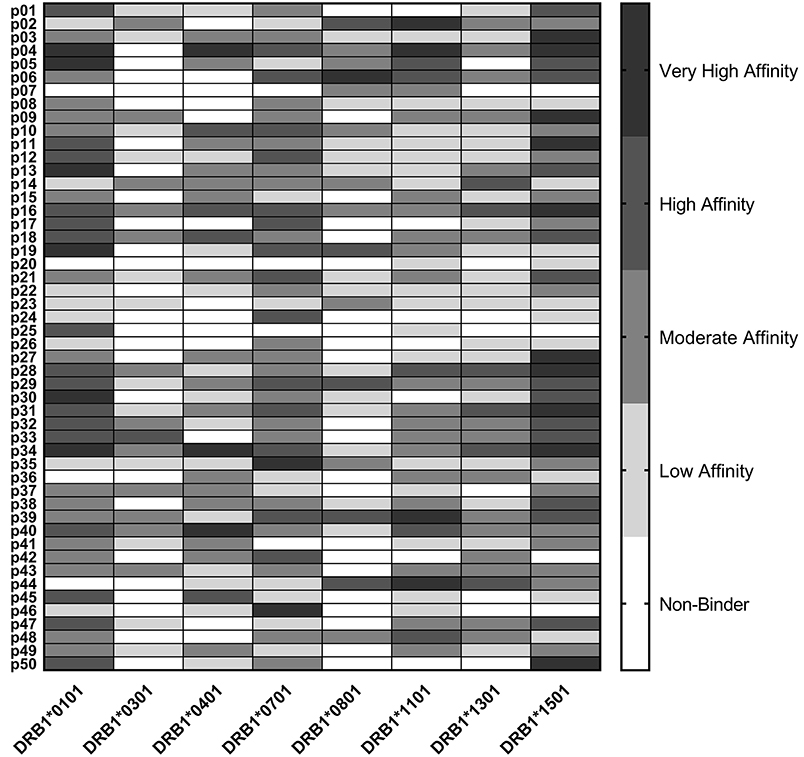
Figure 1. HLA Class II Binding Assay Results. C. burnetii HLA class II epitope affinities for eight common DR alleles were assessed in competition binding assays. Peptide binding strength was classified according to IC50 value in the following affinity categories: very high (<0.1 μM), high (0.1–1 μM), moderate (1–10 μM), and low (10–100 μM). Epitopes with IC50 values >100 μM or with no dose-dependent response were considered non-binders.
HLA class I binding affinities were determined for the primary predicted HLA-A/B supertype allele for each peptide (Figure 2). Among the 65 peptide-allele pairs tested, 56% bound with high affinity (IC50 < 5 μM), 21.5% with moderate affinity (IC50 5–50 μM), 7.5% with low affinity (IC50 50–1,000 μM), and 14% exhibited no detectable affinity for the HLA class I allele tested (IC50 >1,000 μM or no dose-dependent response). All predicted A*0201, A*0301, and A*2402 peptides, and 8/11 A*0101 and B*0702 peptides bound HLA as predicted. An assay using B*4403 molecules was not available, but an assay using the related B*4402 allele showed eight out of 11 B*4403 peptides bound HLA as predicted. Based on these results, epitope prediction accuracy was 100% for A*0201, A*0301, and A*2402 and 73% for A*0101, B*0702, and B*4403. Overall, predictive accuracy for this set of HLA class I peptides was 89% (Table S1). Taken together, the binding assay results suggest that the predicted HLA class II epitope clusters and HLA class I epitopes represent a set of sequences with meaningful potential for stimulating C. burnetii-specific immune responses across a broad range of HLA types.
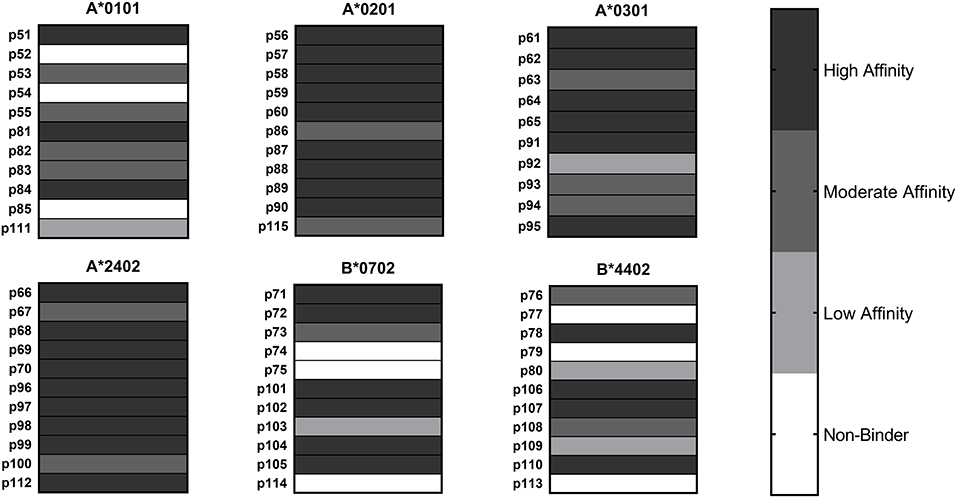
Figure 2. HLA Class I Binding Assay Results. C. burnetii HLA class I epitope affinities for six supertype HLA-A and HLA-B alleles were assessed in competition binding assays. Peptide binding strength was classified according to IC50 value in the following categories: high affinity (<5 μM), moderate affinity (5–50 μM), and low affinity (50–1,000 μM). Epitopes with IC50 values >1,000 μM or with no dose-dependent response were considered non-binders.
Immunogenicity of Class II Epitopes in tgHLA-DR3 Mice
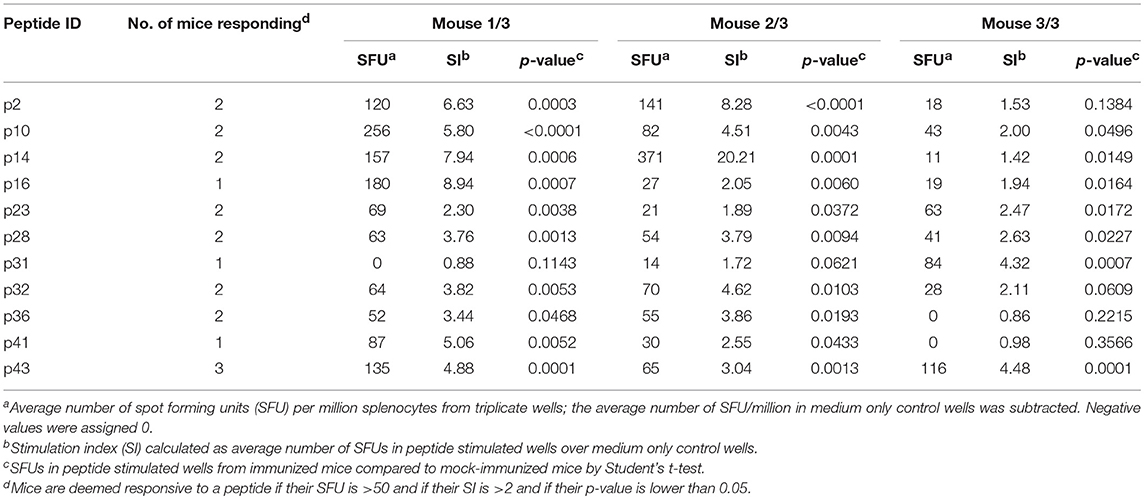
While peptide binding to HLA is necessary to induce a T-cell response, it is not sufficient. To determine whether the predicted C. burnetii HLA class II epitope clusters are capable of eliciting a de novo immune response via a cognate human HLA in vivo, the class II peptides were screened for immunogenicity in transgenic mice expressing human HLA-DR3 (tgHLA-DR3). The 50 HLA class II C. burnetii peptides were arranged into five groups of 10 peptides each based on predicted HLA-DR3 immunogenicity and three mice per group were subjected to heterologous DNA/DNA/peptide/peptide prime-boost immunizations. Peptide-specific responses of splenocytes from tgHLA-DR3 mice were assessed by ex vivo IFNγ ELISpot assay. Positive peptide-specific IFNγ responses in vivo were observed for 11 peptides, eight of which evoked responses in at least 2/3 mice (Table 4). All 11 of these immunogenic peptides were predicted to bind HLA-DR3, and all except one (p36) also showed binding to DRB1*0301 in vitro. Therefore, altogether 10/21 peptides both predicted and confirmed to bind HLA-DR3 were immunogenic in vivo. None of the 13 peptides that were predicted not to bind HLA-DR3 were immunogenic. An odds ratio, calculated to determine the association of epitope prediction and immunogenicity, showed a statistically significant association for immunogenicity given an HLA-DR3 prediction (Fisher's exact test, p = 0.0229). No peptide-specific responses were detected in mock-immunized mice that received vehicle vaccine controls.
Selection of Coxiella-Exposed Human Subjects for Epitope Antigenicity Screening
The transgenic mouse study provided a snapshot of peptide-specific immunogenicity for a single HLA class II allele. To assess antigenicity across multiple HLA types and determine whether the selected class I and II peptides are capable of recalling long-lasting IFNγ memory responses, we next turned to a unique cohort of individuals naturally exposed to C. burnetii. Subjects for this study were recruited from a well-characterized population in the village of Herpen in the Netherlands (48), which experienced a high incidence of C. burnetii infection during the 2007–2010 Q fever outbreak. Recruited subjects were categorized based on clinical Q fever history and recall responses to heat-killed C. burnetii in a whole blood IFNγ release assay (IGRA) (Table 5). Group A and B subjects had no clinical history of Q fever, while group C individuals had recovered from an acute episode of diagnosed Q fever. Group A controls had no IFNγ recall responses to C. burnetii. Group B (past asymptomatic infection) and group C (past symptomatic infection) subjects showed varying degrees of IFNγ recall responses to C. burnetii (both p < 0.0001 compared to group A controls by one-way ANOVA with Holm-Šídák multiple comparisons test), with no significant difference between the two groups (p = 0.9). HLA types were sufficiently diverse within the total cohort to have all desired HLA types represented in all three clinical groups (Tables S2, S3), with the exception of HLA-DR8, which is underrepresented particularly in group C, in accordance with this allele's expected low frequency in the Dutch population (42, 59, 60). Assessment of the binding potential of all peptides with respect to the HLA types of the 136 subjects in groups A-C showed no significant differences across groups, with 75–82% of all class I peptides and between 93 and 96% of all class II peptides predicted to be recognized by each group.
Two partially overlapping subgroups of the total cohort were selected based on a broad representation of HLA-A/HLA-B and HLA-DR supertypes and allocated to HLA class I and II peptide screening, respectively. The screening cohort for the 50 HLA class II peptides consisted of 21 group A control donors (IGRA-, no clinical disease), and 56 IGRA+ donors from groups B (asymptomatic, n = 33) and C (symptomatic, n = 23) (Table S2). The screening cohort for the 65 HLA class I peptides included 20 group A control donors (IGRA-, no clinical disease), and 57 IGRA+ donors from groups B (asymptomatic, n = 32) and C (symptomatic, n = 25) (Table S3). Care was taken to maintain a distribution of IGRA responses comparable to the total cohort (Tables S2, S3).
Human Antigenic Responses to Predicted HLA Class II Epitopes
Human antigenic T-cell IFNγ recall responses against the predicted epitope clusters from C. burnetii were evaluated ex vivo using freshly isolated peripheral blood mononuclear cells (PBMCs) in a cultured ELISpot assay for detection of central memory T-cells. A cultured ELISpot assay was used to achieve high sensitivity for detecting low frequency antigen-specific T-cell responses. Amongst the 3,843 assessments made for the HLA class II peptides (50 peptides screened against 77 donors with some exceptions due to technical error or insufficient cell numbers), 307 yielded positive responses [stimulation index (SI) ≥2, Figures S1, S2]. These antigenic responses covered 44/50 HLA class II peptides. Only six peptides were not recognized by any subject (p9, p11, p34, p35, p40, and p49). For four of these peptides, a second peptide from the same respective source antigen induced recall responses by at least one IGRA+ subject. Therefore, responses were detected to all but two (CBU_2065 and CBU_0092/YbgF) of the 29 source antigens from which HLA class II epitope clusters were predicted and screened.
The cumulative HLA class II peptide response per subject was more frequent in IGRA+ compared to IGRA- subjects (Figure 3A). The majority of IGRA- individuals recognized none (10/21 donors) or only one or two peptides (8/21 donors). While only 14% (3/21) of IGRA- group A individuals recognized three or more peptides, this was true for 46% (26/56) IGRA+ group B and C donors (Figure 3B). Amongst IGRA+ individuals, there was a positive correlation between the cumulative number of recognized peptides per donor, and the IFNγ responses to whole heat-killed C. burnetii as assessed 12–17 months prior to peptide screening (Figure 3C). Of note, all but one individual that showed responses to three or more peptides had an IGRA response of at least 250 pg/ml IFNγ in this standardized assay (Figure 3D).
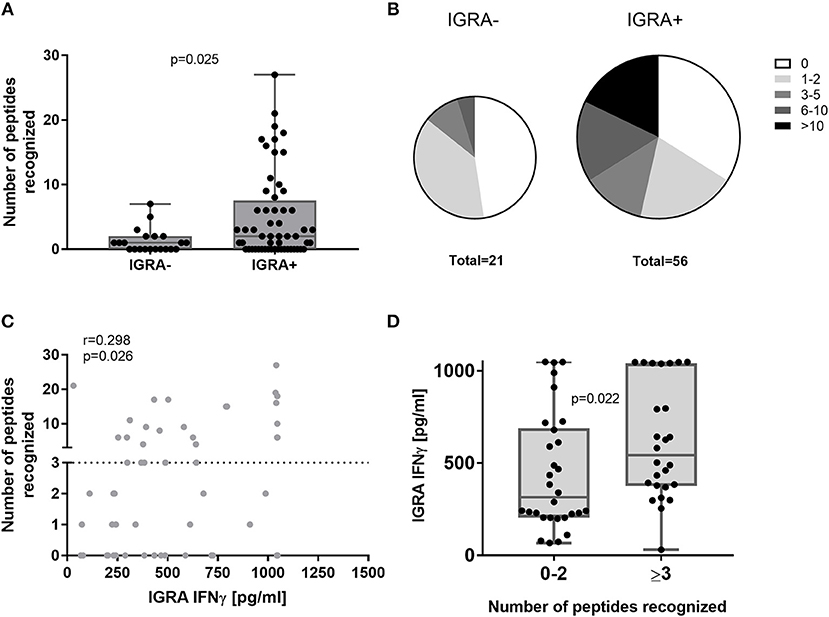
Figure 3. Cumulative HLA class II peptide responses across clinical groups and in relation to IGRA. Data are shown for IGRA- and IGRA+ as the cumulative peptide response (SI ≥2) per donor (A) or as the proportion of subjects recognizing 0, 1–2, 3–5, 6–10, or >10 peptides (B). (C) The cumulative number of peptides recognized by individual IGRA+ subjects is plotted against their IGRA response to whole heat-killed C. burnetii and analyzed by Spearman correlation. (D) IGRA responses are shown for IGRA+ individuals recognizing 0–2 or ≥3 peptides (cut-off SI≥2). Groups in (A,D) were compared by Mann-Whitney-test. Whisker-dot-plots show the median and interquartile range (25 and 75th percentile) with whiskers extending from min to max values.
Of the 44 antigenic HLA class II peptides, 21 peptides were recognized by >10% of all IGRA+ individuals (at least 6/56, Figure 4A) and were hence considered highly antigenic. These 21 highly antigenic peptides accounted for 78.3% of all positive peptide responses of IGRA+ individuals that reached a SI≥2 (220/281). A large proportion of these responses was well above the cut-off of SI ≥2: Amongst responding IGRA+ individuals, responses to 13 of these 21 peptides showed a median SI between 3.0 and 4.8 (Figure 4B), and for 11 of these peptides >10% of all IGRA+ donors reached at least SI ≥3. Amongst IGRA- control subjects group A, these peptides elicited either no recall responses (11/21), or responses from only one or two individuals. Only for a single peptide (p21) did the recall response frequency reach a similar proportion in IGRA- (3/21; 14.3%) as in IGRA+ donors (8/56; 14.3%). Amongst the 21 highly antigenic peptides, there were five source proteins that were represented by two epitopes each (p14+p15, CBU_1835/protoporphyrinogen oxidase; p18+p19, CBU_1513/short chain dehydrogenase; p22+p23, CBU_1398/SucB, p37+p38, CBU_0718, p45+p46, CBU_0307/outer membrane protein). Notably, not only were all 11 of the tgHLA-DR3 mice immunogenic peptides also antigenic in at least one IGRA+ individual, but five of the tgHLA-DR3 immunogenic peptides were amongst the 21 highly reactive peptides in the human cohort (p2, p14, p16, p22, p31).
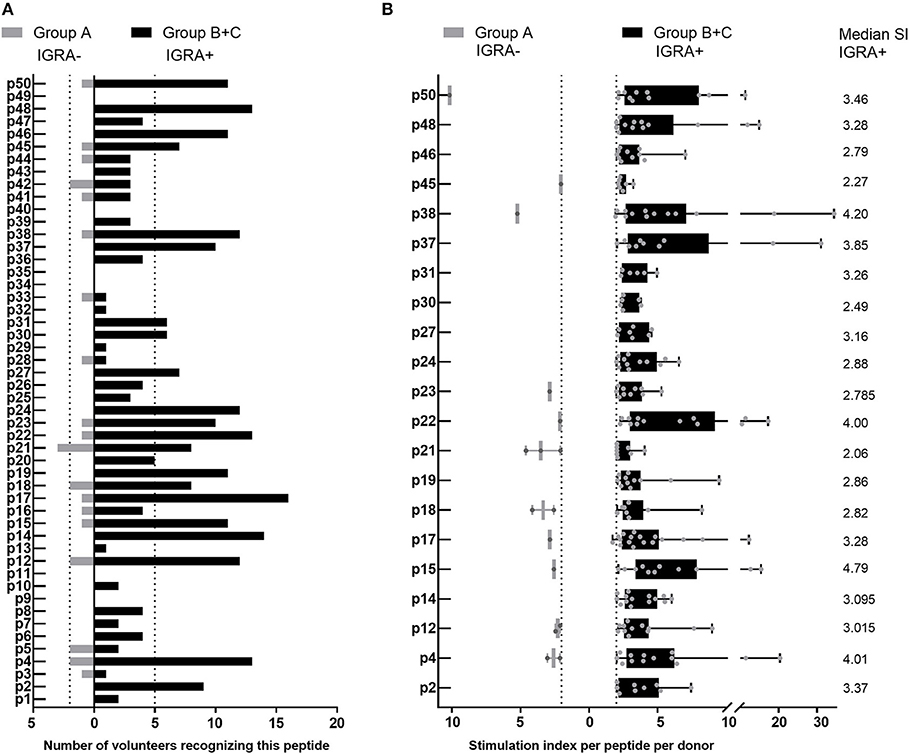
Figure 4. Highly antigenic HLA class II peptides identified by cultured IFNγ ELISpot assay. Data are shown as the number of individuals recognizing the 50 individual peptides, depending on whether subjects are IGRA- (Group A, no clinical disease, n = 21; gray bars) or IGRA+ (Group B, asymptomatic; Group C, symptomatic; n = 56; black bars). (A) Shows peptide responses reaching a SI ≥2. Bars extending over dotted lines indicate those peptides that were recognized by more than 10% of IGRA- subjects (>2/21) or IGRA+ subjects (>5/56). (B) Shows the stimulation index for each positive response per subject for the 21 highly antigenic peptides identified in (A). Whisker-dot-plots show the interquartile range (25 and 75th percentile) with whiskers extending from min to max values. Numbers indicate the median SI per peptide for IGRA+ subjects.
Amongst IGRA+ donors, group C of past symptomatic individuals had a higher proportion of high responders (recognizing >10 peptides) and a smaller proportion of non-responders to HLA class II epitope clusters than past asymptomatic subjects (Group B, Figure 5A). However, the two groups did not differ significantly in their cumulative peptide response per subject (Figure 5B). When comparing responses to individual peptides between group B and C, there was again a trend that responses particularly to the most antigenic peptides were more frequent amongst past symptomatic donors and a number of peptides were recognized only within this group by either a single (p3, p33) or multiple donors (p1, p6, p10) (Figure 5C). There were some peptides, however, that were recognized solely by past asymptomatic subjects (p25, p42, and p43, 3/33 each; p13, p28, p29, and p33, 1/33 each).
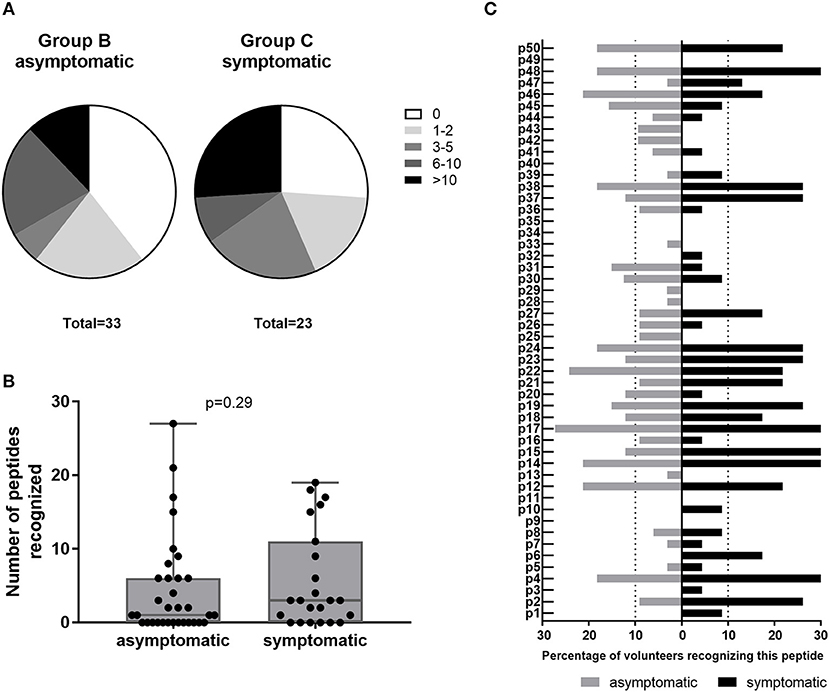
Figure 5. Class II peptide antigenicity patterns in past asymptomatic and symptomatic, IGRA+ individuals. Data are shown as (A) the proportion of subjects recognizing 0, 1–2, 3–5, 6–10, or >10 peptides for IGRA+ donors with past asymptomatic or symptomatic Q fever or (B) the cumulative peptide response (SI ≥2) per donor. Groups are compared by Mann-Whitney-test. Whisker-dot-plots show the median, 25 and 75th percentile with whiskers extending from min to max values. (C) Data are shown as the proportion of subjects recognizing the 50 individual peptides reaching a SI ≥2, depending on whether IGRA+ individuals were asymptomatic (Group B, n = 32; gray bars) or symptomatic (Group C, n = 23; black bars) during the Q fever outbreak. Bars extending over dotted lines indicate those peptides that were recognized by more than 10% of group B (>3/32) or group C subjects (>2/23).
Human Antigenic Responses to Predicted HLA Class I Epitopes
Recall responses to the 65 HLA class I peptides were also assessed using the cultured IFNγ ELISpot approach. In 4,794 assessments (65 peptides screened in 77 donors with some exceptions due to technical error or insufficient cell numbers), only 23 positive responses were detected (Figure S3). Four of these positive responses came from IGRA- group A control subjects. The remaining 19 responses were detected in IGRA+ subjects, with one individual recognizing 8 different peptides (Figures S3, S4). Of the 15 HLA class I peptides that were found to be antigenic in humans, ten peptides were recognized by only a single donor each and only five HLA class I peptides were recognized more than once: p53, p91, and p92 were each recognized by 2/57 donors, and two epitopes (p111 and p113) from com1 were recognized by 3/57 and 4/57 donors, respectively. Only two of the reactive peptides came from the T4SS data set (p53, p54), the remaining 13 were derived from sero-reactive source antigens. Albeit rare, HLA class I peptide responses were clearly detectable, with nearly half (11/23) of the positive responses reaching a stimulation index (SI) of ≥3 (Figures S3, S4), and all responses were consistent with predictions for primary or secondary HLA-A/B allele binding.
Evaluation of Class I and Class II Epitopes in Guinea Pig Model of Vaccine Reactogenicity
Due to reactogenicity issues observed with the only Q fever vaccine licensed for humans, all C. burnetii HLA class I and class II peptides examined in this study were screened for potential reactogenicity in a guinea pig model of exposure-primed delayed-type hypersensitivity (55, 61), with the goal of eliminating reactogenic peptides from further consideration for inclusion in a vaccine. In contrast to reactogenicity mediated by the whole cell phase I vaccine COXEVAC®, no gross reactions were noted at any of the negative control or peptide pool injection sites (Figure S5). Mild lymphocytic inflammation (score = 1) was noted in two HLA class II peptide pools; no overlap in peptides was noted between the two pools and no other pools with the same peptides showed histological changes. Therefore, these reactions were not likely to be considered a reactogenic response to C. burnetii peptides.
Discussion
In the present study, we aimed to identify HLA class I and class II epitopes for inclusion in a non-reactogenic T-cell-targeted vaccine to control C. burnetii infection and hence prevent Q fever disease in humans. HLA binding competition assays demonstrated a high accuracy of the immunoinformatic predictions for HLA class I and class II recognition, and a murine immunogenicity study demonstrated that half of the peptides both predicted and shown to bind HLA-DR3 were also capable of eliciting de novo responses in tgHLA-DR3 mice. Notably, while responses to the 65 selected HLA class I epitopes were barely detectable in PBMCs from individuals naturally exposed to C. burnetii during the Dutch Q fever outbreak 7–10 years prior to this study, 21 out of the 50 predicted HLA class II epitope clusters recalled responses in 10–28% of those individuals that also showed an ex vivo response to whole heat-killed C. burnetii. We thereby demonstrate for the first time that natural exposure of humans to C. burnetii induces long-lived responses to immunodominant, promiscuous, and conserved HLA class II T-cell epitope clusters. These peptide sequences are desirable candidates for inclusion in a vaccine aiming to elicit sustained T-cell memory in a broad and immunogenetically diverse human population that can be boosted and recalled by natural exposure.
These long-lived epitope-specific T-cell responses are consistent with previous studies in Q fever convalescent individuals and recipients of the whole cell formalin-inactivated phase I vaccine Q-VAX®, which have consistently reported a long-lived T-cell response to whole cell C. burnetii antigen. The T-cell response, in this context, develops within 2 weeks post-vaccination (62), is detectable for at least 8–10 years following natural exposure or vaccination (63, 64) and in some studies was found to be considerably more durable than the mounted antibody response (62, 63). However, studies investigating the specific antigen targets of the induced immune response in humans have so far solely focused on antibodies (20, 21, 23, 24, 26, 27, 29, 33, 65–71). T-cell antigen targets have only been assessed in a limited number of murine studies (18, 21, 22, 72), including one study that investigated T-cell responses to recombinant C. burnetii proteins by ELISpot in HLA-DR4 transgenic mice (21). T-cell antigen targets of murine CD4 T-cells in these studies were all chosen based on antigen recognition by human sera of Q fever exposed individuals (18, 21, 22). While this B-cell antigen-guided screening approach for T-cell targets precludes identification of T-cell epitopes for source antigens that do not elicit a dominant antibody response, it has the advantage of significantly reducing and focusing the number of antigens to be evaluated. In the present study, 44/50 HLA class II peptides (88%) from 27/29 selected sero-reactive source antigens (93%) elicited T-cell recall responses from at least one donor with a history of C. burnetii exposure during the Dutch Q fever outbreak. This large hit rate for HLA class II epitope clusters illustrated the feasibility of this approach to focus on sero-reactive antigens for T-cell epitope prediction.
One major challenge when focusing a vaccine on individual epitopes is to choose epitopes that are capable of eliciting responses from a large part of the human population. Despite the expected HLA diversity in this human cohort, 21/50 (42%) of the screened HLA class II peptides, representing 16/29 source antigens, were recognized by more than 10% of the subjects exposed 7–10 years prior. In comparison, IFNγ recall responses to H2 I-Ab-restricted C. burnetii epitopes were only detectable for 17–26 and 5.3–6.8% of the predicted epitopes in inbred C57BL/6 mice immediately following immunization with the selected source antigens or whole cell antigen, respectively (18, 22). Of note, all but one of the antigens in the murine CD4 epitope vaccine study (18) were also amongst the HLA class II source antigens in the present study, and one of the seven vaccine epitope sequences (OmpA146–160), covered by p45 in our peptide screening library, was one of the highly antigenic peptides recognized by 7/56 (12.5%) of IGRA+ subjects. These results validate the ability of the immunoinformatic algorithms employed to predict promiscuous HLA class II epitopes for humans. Moreover, the highly antigenic HLA class II epitopes identified herein are not only promiscuous across HLA-DR supertypes (76% predicted binding to 6 or more alleles) but are also conserved across all C. burnetii genomes publicly available in 2015, as well as in the Dutch outbreak strain (39). Such conserved immunodominant T-cell epitopes have also been found for Mycobacterium tuberculosis (73–75), hepatitis C virus (76, 77), and HIV (78, 79) and are interesting vaccine targets, since they are likely under positive selection pressure and hence unlikely to be subject to vaccine-induced immune evasion (73).
A somewhat unexpected finding of the present study was the scarcity of detectable responses to the predicted HLA class I epitopes in human subjects. Earlier studies indicated that human CD8 T-cells contribute to C. burnetii-specific IFNγ production (80), and in murine models CD8 T-cells appear to be more critical than CD4 T-cells for resolving infection (15, 81). Using a similar approach to the present study, a recent report identified 29 class I epitopes from T4SS substrates that conferred partial protection in a murine vaccine-challenge study (72). One of these 29 was amongst the few HLA class I epitopes recognized by two human donors (p92 from the tol-pal system protein YbgF). The proportion of peptides recognized at least once in the human cohort (15/65, 23%) was comparable to the recognition frequency observed in the murine study (29/157, 18%), suggesting that human HLA class I epitopes were predicted effectively in the present study.
The much smaller frequency of HLA class I compared to HLA class II responses could be due to a number of reasons. Firstly, while HLA class II peptides are promiscuous epitope clusters each containing 5–20 9-mers with various HLA-DR binding motifs, HLA class I epitopes do not display this promiscuity. Each of the 65 HLA class I peptides have, on average, 0.4 binding motifs across the class I supertype alleles modeled by EpiMatrix and represented in over 95% of the global human population. This is in stark contrast to 1.36 binding motifs found, on average, for the 50 HLA class II peptides across class II supertype alleles, representing a 3.4-fold increase over the number of motifs for the HLA class I peptides. The larger number of binding opportunities for HLA class II peptides may thus contribute to the greater number of responses they recalled.
Secondly, reports have shown that the TCR repertoire of CD4 T-cells is estimated to be about five times greater than the TCR repertoire of CD8 T-cells (82), suggesting that there are more opportunities for CD4 T-cell reactivity. Finally, it is also possible that circulating C. burnetii-specific CD8 T-cells are less well detected than CD4 T-cells at such a late time point (7–10 years) post-exposure, which would not be unprecedented: following smallpox infection or vaccination, antigen-specific CD8 T-cells show a much faster contraction to undetectable levels than do CD4 T-cells (83, 84). Similarly, a significant contraction in Mycobacterium tuberculosis-specific CD8 but not CD4 effector memory responses was reported (85) as well as a significant decrease in total and antigen-specific central memory CD8 T-cells (86). Of note, C. burnetii epitope-specific murine CD8 T-cell responses were assessed only 10 days post-infection (72), and challenge experiments were carried out no later than 28 days following vaccination or transfer of infection-induced CD8 T-cells (15, 72). The most obvious starting point for evaluating whether responses to a common set of human HLA class I epitopes are induced by natural exposure to C. burnetii would therefore be to screen in much more recently exposed donors. In the absence of another outbreak, inclusion of a sufficiently large cohort in a reasonable time frame is, however, unlikely. Alternatively, one could assess HLA class I epitope responses in individuals that recently received the whole cell vaccine Q-VAX®. This would provide additional insights into the contribution of the predicted epitopes to protection, which would also be valuable for the identified highly reactive HLA class II epitope clusters. Given that the vaccine is only licensed for use in Australia, however, pursuit of such a study would be restricted to this location. Finally, we cannot strictly conclude that the observed HLA class II recall responses arise solely from CD4 T-cells and not CD8 T-cells. Although rare, HLA class II-restricted CD8 T-cells have been identified in both mice (87) and humans (88). Definitively addressing the relative responses would require further experimentation with purified T-cell populations rather than total PBMCs.
In the present study, we further compared epitope-specific responses between C. burnetii exposed individuals with or without a history of an acute clinical episode of Q fever. Although responses tended to be more frequent in past symptomatic donors and some peptides were recognized only by either past symptomatic or asymptomatic individuals, these observations are based on a relatively small number of responding subjects. Moreover, none of the observed differences were striking enough to suggest a significant difference in the quality of the induced T-cell response between these two groups that would explain why some individuals developed symptoms and others did not. What would be of interest in future studies is to compare this group of convalescent individuals with patients that developed persistent infection with C. burnetii (chronic Q fever) (1, 89), to identify those epitopes for which recognition may be crucial to control and clear infection and hence might be lacking in individuals with persistent infection. This would be analogous to observations from herpes simplex virus vaccine research, where distinct epitope-specific T-cell repertoires have been identified for infected individuals that either successfully control infection (asymptomatic) or not, leading to a proposed emphasis on so-called “asymptomatic epitopes” (90).
The ultimate aim of the Q-VaxCelerate consortium is to develop a non-reactogenic Q fever vaccine that could be administered without screening for pre-exposure, especially in an outbreak setting or when occupational hazard warrants immediate broad-scale vaccination. Since none of the peptides as such elicited a reactogenic response in the guinea pig model, no peptides were eliminated from further consideration by this screen. To minimize chances for reactogenicity of the epitope vaccine in its final formulation, one delivery strategy would be to use adenoviral vectors delivered intramuscularly. In this way one could avoid the use of adjuvants that cause localized inflammation at the injection site; such vectors have had a good safety profile in human clinical trials and have been shown to induce potent CD4 and CD8 T-cell responses (91–93). Reactogenicity of the candidate vaccine in its final formulation should then also include a parallel evaluation of antigenicity in the same animal model as an additional control.
In this study we focused on the identification of antigenic and immunogenic T-cell epitopes and hence the cellular component of the envisaged non-reactogenic multi-epitope vaccine. However, we do not preclude the additional involvement of a significant component of antibody-mediated protection in vaccine design. While studies in murine models using formalin-inactivated C. burnetii phase I vaccines (94) and of the human whole cell vaccine Q-VAX® (9) suggest a strong contribution of antibodies to protection, how antibodies could provide efficient protection against this intracellular pathogen is not known. Specificity of the antibody response to Q-VAX® has only been analyzed by immunofluorescence assay against whole cell C. burnetii, with the exception of a single study assessing the targets of anti-sera from two Q-VAX® vaccinees by protein microarray (20). As an additional caveat, anti-sera from patients with chronic Q fever recognizing phase I antigens were shown to enhance C. burnetii infectivity and replication in macrophages in vitro (95). Nevertheless, the presence of C. burnetii-specific antibodies after both natural infection and immunization (1, 9, 64), and the protective role of these in animal models (12, 13, 94) warrant the consideration of humoral responses in human vaccine design. In this context, elicitation of epitope-specific antibody responses has been described for a HLA class II multi-epitope DNA vaccine (96). Further evaluation of a candidate multi-epitope vaccine should thus also include assessment of humoral responses. Additionally, vaccine optimization could involve the inclusion of known immunogenic whole proteins (24) or specifically selected sequences for (conformational) B-cell epitopes, including a recently described lipopolysaccharide-targeted peptide mimic (97).
In conclusion, we herein identify for the first time a set of C. burnetii-specific HLA class II T-cell epitope clusters that was computationally predicted and shown to bind a broad range of HLA-DR types, elicit immunogenicity in tgHLA-DR3 mice and recalled long-lived memory responses in naturally exposed individuals. Further research will focus on evaluating the immunogenicity and protective efficacy of these desirable candidates for a novel promiscuous multi-epitope-based Q fever vaccine in animal models in an appropriate and non-reactogenic formulation that can be transferred directly into clinical studies in humans.
Data Availability Statement
All relevant data generated for this study are included in the manuscript and the supplementary files. The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation within the scope allowed by patient privacy regulations, to any qualified researcher.
Author Contributions
AS, LM, LB, PR, WM, TB, RB, RAB, AG, ADG, AES, and MP conceptualized and designed the study and experiments. Experiments were performed by AS, GR, LM, LB, CB, and AG. Data were analyzed and interpreted by AS, GR, LM, LB, and AES. GR, LM, WM, RAB, AG, ADG, and MP contributed critical reagents, materials, and analytic tools. TB, RB, RAB, AG, ADG, AES, and MP acquired funding and supervised research activities. AS, GR, LM, LB, and AES wrote the manuscript and PR, WM, TB, CB, SR, RB, RAB, AG, ADG, and MP critically reviewed and approved the manuscript.
Funding
This research was supported by contract HDTRA1-15-C-0020 from the US Defense Threat Reduction Agency (www.dtra.mil), awarded to Massachusetts General Hospital (MP Lead Principal Investigator); work by authors at other institutions was supported by subcontracts under the prime contract award to MGH.
Conflict of Interest Statement
AG is a senior officer and shareholder and AS is an employee of Innatoss Laboratories B.V., which provides diagnostic screening for Q fever. ADG and WM are senior officers and shareholders, and LM, GR, and CB are employees of EpiVax, Inc., a company specializing in immunoinformatic analysis. Innatoss Laboratories B.V. and EpiVax, Inc., own patents to technologies utilized by associated authors in the research reported here.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the volunteers from the village of Herpen, NL, for their participation in this study. This study utilized the results of the Q-Herpen-II database [described in (50)] established by the GGD Hart voor Brabant (s'Hertogenbosch, NL) in collaboration with the Jeroen Bosch Hospital (s'Hertogenbosch, NL) and Innatoss Laboratories (Oss, NL). We thank the GGD Hart voor Brabant for making parts of the Herpen-II database available for analysis as the basis for subject selection and for help with the recruitment of volunteers. Dr. Thomas Luijkx is acknowledged for his contribution to volunteer inclusion at the start of the project, Dr. Hendrik-Jan Roest and Dr. Alex Bossers for providing pre-publication access to the Dutch strain genome sequence, Katinka Mulder for expert technical assistance with human epitope screening, Frances Terry for contributing to the immunoinformatics analysis, Pooja Hindocha for assistance in data analysis and figure preparation, Skylar Korek for assistance in manuscript formatting, and Nicole Marlenee and Paul Gordy for their technical assistance with the guinea pig studies. We thank Ceva Biomune for providing the COXEVAC® vaccine.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00207/full#supplementary-material
References
1. Eldin C, Melenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, et al. From Q fever to Coxiella burnetii infection: a paradigm change. Clin Microbiol Rev. (2017) 30:115–90. doi: 10.1128/CMR.00045-16
3. Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. Q fever: a biological weapon in your backyard. Lancet Infect Dis. (2003) 3:709–21. doi: 10.1016/S1473-3099(03)00804-1
4. Kampschreur LM, Hagenaars JC, Wielders CC, Elsman P, Lestrade PJ, Koning OH, et al. Screening for Coxiella burnetii seroprevalence in chronic Q fever high-risk groups reveals the magnitude of the Dutch Q fever outbreak. Epidemiol Infect. (2013) 141:847–51. doi: 10.1017/S0950268812001203
5. Kampschreur LM, Oosterheert JJ, Hoepelman AI, Lestrade PJ, Renders NH, Elsman P, et al. Prevalence of chronic Q fever in patients with a history of cardiac valve surgery in an area where Coxiella burnetii is epidemic. Clin Vaccine Immunol. (2012) 19:1165–9. doi: 10.1128/CVI.00185-12
6. Ruiz S, Wolfe DN. Vaccination against Q fever for biodefense and public health indications. Front Microbiol. (2014) 5:726. doi: 10.3389/fmicb.2014.00726
7. Chiu CK, Durrheim DN. A review of the efficacy of human Q fever vaccine registered in Australia. N S W Public Health Bull. (2007) 18:133–6. doi: 10.1071/NB07057
8. Gefenaite G, Munster JM, van Houdt R, Hak E. Effectiveness of the Q fever vaccine: a meta-analysis. Vaccine (2011) 29:395–8. doi: 10.1016/j.vaccine.2010.11.008
9. Marmion BP, Ormsbee RA, Kyrkou M, Wright J, Worswick DA, Izzo AA, et al. Vaccine prophylaxis of abattoir-associated Q fever: eight years' experience in Australian abattoirs. Epidemiol Infect. (1990) 104:275–87. doi: 10.1017/S0950268800059458
10. Karch CP, Burkhard P. Vaccine technologies: from whole organisms to rationally designed protein assemblies. Biochem Pharmacol. (2016) 120:1–14. doi: 10.1016/j.bcp.2016.05.001
11. Zhang G, Zhang Y, Samuel JE. Components of protective immunity. Adv Exp Med Biol. (2012) 984:91–104. doi: 10.1007/978-94-007-4315-1_5
12. Humphres RC, Hinrichs DJ. Role of antibody in Coxiella burnetii infection. Infect Immun. (1981) 31:641–5.
13. Zhang G, Russell-Lodrigue KE, Andoh M, Zhang Y, Hendrix LR, Samuel JE. Mechanisms of vaccine-induced protective immunity against Coxiella burnetii infection in BALB/c mice. J Immunol. (2007) 179:8372–80. doi: 10.4049/jimmunol.179.12.8372
14. Andoh M, Zhang G, Russell-Lodrigue KE, Shive HR, Weeks BR, Samuel JE. T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infect Immun. (2007) 75:3245–55. doi: 10.1128/IAI.01767-06
15. Read AJ, Erickson S, Harmsen AG. Role of CD4+ and CD8+ T cells in clearance of primary pulmonary infection with Coxiella burnetii. Infect Immun. (2010) 78:3019–26. doi: 10.1128/IAI.00101-10
16. Dellacasagrande J, Capo C, Raoult D, Mege JL. IFN-gamma-mediated control of Coxiella burnetii survival in monocytes: the role of cell apoptosis and TNF. J Immunol. (1999) 162:2259–65.
17. Ghigo E, Capo C, Tung CH, Raoult D, Gorvel JP, Mege JL. Coxiella burnetii survival in THP-1 monocytes involves the impairment of phagosome maturation: IFN-gamma mediates its restoration and bacterial killing. J Immunol. (2002) 169:4488–95. doi: 10.4049/jimmunol.169.8.4488
18. Xiong X, Qi Y, Jiao J, Gong W, Duan C, Wen B. Exploratory study on Th1 epitope-induced protective immunity against Coxiella burnetii infection. PLoS ONE (2014) 9:e87206. doi: 10.1371/journal.pone.0087206
19. Reeves PM, Paul SR, Sluder AE, Brauns TA, Poznansky MC. Q-vaxcelerate: a distributed development approach for a new Coxiella burnetii vaccine. Hum Vaccine Immunother. (2017) 13:2977–81. doi: 10.1080/21645515.2017.1371377
20. Beare PA, Chen C, Bouman T, Pablo J, Unal B, Cockrell DC, et al. Candidate antigens for Q fever serodiagnosis revealed by immunoscreening of a Coxiella burnetii protein microarray. Clin Vaccine Immunol. (2008) 15:1771–9. doi: 10.1128/CVI.00300-08
21. Chen C, Bouman TJ, Beare PA, Mertens K, Zhang GQ, Russell-Lodrigue KE, et al. A systematic approach to evaluate humoral and cellular immune responses to Coxiella burnetii immunoreactive antigens. Clin Microbiol Infect. (2009) 15 (Suppl. 2):156–7. doi: 10.1111/j.1469-0691.2008.02206.x
22. Chen C, Dow C, Wang P, Sidney J, Read A, Harmsen A, et al. Identification of CD4+ T cell epitopes in C. burnetii antigens targeted by antibody responses. PLoS ONE (2011) 6:e17712. doi: 10.1371/journal.pone.0017712
23. Coleman SA, Fischer ER, Cockrell DC, Voth DE, Howe D, Mead DJ, et al. Proteome and antigen profiling of Coxiella burnetii developmental forms. Infect Immun. (2007) 75:290–8. doi: 10.1128/IAI.00883-06
24. Kowalczewska M, Sekeyova Z, Raoult D. Proteomics paves the way for Q fever diagnostics. Genome Med. (2011) 3:50. doi: 10.1186/gm266
25. Li Q, Niu D, Wen B, Chen M, Qiu L, Zhang J. Protective immunity against Q fever induced with a recombinant P1 antigen fused with HspB of Coxiella burnetii. Ann N Y Acad Sci. (2005) 1063:130–42. doi: 10.1196/annals.1355.021
26. Vigil A, Ortega R, Nakajima-Sasaki R, Pablo J, Molina DM, Chao CC, et al. Genome-wide profiling of humoral immune response to Coxiella burnetii infection by protein microarray. Proteomics (2010) 10:2259–69. doi: 10.1002/pmic.201000064
27. Wang X, Xiong X, Graves S, Stenos J, Wen B. Protein array of Coxiella burnetii probed with Q fever sera. Sci China Life Sci. (2013) 56:453–9. doi: 10.1007/s11427-013-4472-6
28. Wei Y, Wang X, Xiong X, Wen B. Coxiella burnetii antigen-stimulated dendritic cells mediated protection against Coxiella burnetii in BALB/c mice. J Infect Dis. (2011) 203:283–91. doi: 10.1093/infdis/jiq037
29. Xiong X, Wang X, Wen B, Graves S, Stenos J. Potential serodiagnostic markers for Q fever identified in Coxiella burnetii by immunoproteomic and protein microarray approaches. BMC Microbiol. (2012) 12:35. doi: 10.1186/1471-2180-12-35
30. Carey KL, Newton HJ, Luhrmann A, Roy CR. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog. (2011) 7:e1002056. doi: 10.1371/journal.ppat.1002056
31. Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, et al. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci USA. (2010) 107:21755–60. doi: 10.1073/pnas.1010485107
32. Luhrmann A, Nogueira CV, Carey KL, Roy CR. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci USA. (2010) 107:18997–9001. doi: 10.1073/pnas.1004380107
33. Sekeyova Z, Kowalczewska M, Vincentelli R, Decloquement P, Flores-Ramirez G, Skultety L, et al. Characterization of antigens for Q fever serodiagnostics. Acta Virol. (2010) 54:173–80. doi: 10.4149/av_2010_03_173
34. van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol. (2013) 11:561–73. doi: 10.1038/nrmicro3049
35. Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, et al. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol. (2011) 193:1493–503. doi: 10.1128/JB.01359-10
36. Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, et al. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. (2009) 191:4232–42. doi: 10.1128/JB.01656-08
37. Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, et al. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol. (2013) 195:3914–24. doi: 10.1128/JB.00071-13
38. UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. (2018) 46:2699. doi: 10.1093/nar/gky092
39. Kuley R, Smith HE, Janse I, Harders FL, Baas F, Schijlen E, et al. First complete genome sequence of the Dutch veterinary Coxiella burnetii strain NL3262, originating from the largest global Q fever outbreak, and draft genome sequence of its epidemiologically linked chronic human isolate NLhu3345937. Genome Announc. (2016) 4:e00245–16. doi: 10.1128/genomeA.00245-16
40. Moise L, Gutierrez A, Kibria F, Martin R, Tassone R, Liu R, et al. iVAX: an integrated toolkit for the selection and optimization of antigens and the design of epitope-driven vaccines. Hum Vaccine Immunother. (2015) 11:2312–21. doi: 10.1080/21645515.2015.1061159
41. Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics (1999) 50:201–12. doi: 10.1007/s002510050594
42. Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. (1998) 160:3363–73.
43. De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol. (2009) 131:189–201. doi: 10.1016/j.clim.2009.01.009
44. Moise L, Gutierrez AH, Bailey-Kellogg C, Terry F, Leng Q, Abdel Hady KM, et al. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix. Hum Vaccine Immunother. (2013) 9:1577–86. doi: 10.4161/hv.24615
45. Steere AC, Klitz W, Drouin EE, Falk BA, Kwok WW, Nepom GT, et al. Antibiotic-refractory Lyme arthritis is associated with HLA-DR molecules that bind a Borrelia burgdorferi peptide. J Exp Med. (2006) 203:961–71. doi: 10.1084/jem.20052471
46. Buchli R, VanGundy RS, Hickman-Miller HD, Giberson CF, Bardet W, Hildebrand WH. Real-time measurement of in vitro peptide binding to soluble HLA-A*0201 by fluorescence polarization. Biochemistry (2004) 43:14852–63. doi: 10.1021/bi048580q
47. Mangalam AK, Khare M, Krco C, Rodriguez M, David C. Identification of T cell epitopes on human proteolipid protein and induction of experimental autoimmune encephalomyelitis in HLA class II-transgenic mice. Eur J Immunol. (2004) 34:280–90. doi: 10.1002/eji.200324597
48. Morroy G, Van Der Hoek W, Nanver ZD, Schneeberger PM, Bleeker-Rovers CP, Van Der Velden J, et al. The health status of a village population, 7 years after a major Q fever outbreak. Epidemiol Infect. (2016) 144:1153–62. doi: 10.1017/S0950268815002472
49. Karagiannis I, Schimmer B, Van Lier A, Timen A, Schneeberger P, Van Rotterdam B, et al. Investigation of a Q fever outbreak in a rural area of The Netherlands. Epidemiol Infect. (2009) 137:1283–94. doi: 10.1017/S0950268808001908
50. Morroy G, van der Hoek W, Albers J, Coutinho RA, Bleeker-Rovers CP, Schneeberger PM. Population screening for chronic Q-fever seven years after a major outbreak. PLoS ONE (2015) 10:e0131777. doi: 10.1371/journal.pone.0131777
51. Shiina T, Suzuki S, Ozaki Y, Taira H, Kikkawa E, Shigenari A, et al. Super high resolution for single molecule-sequence-based typing of classical HLA loci at the 8-digit level using next generation sequencers. Tissue Antigens (2012) 80:305–16. doi: 10.1111/j.1399-0039.2012.01941.x
52. Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. (2008) 9:1. doi: 10.1186/1471-2172-9-1
53. Calarota SA, Baldanti F. Enumeration and characterization of human memory T cells by enzyme-linked immunospot assays. Clin Dev Immunol. (2013) 2013:637649. doi: 10.1155/2013/637649
54. Subbramanian RA, Basha S, Brady RC, Hazenfeld S, Shata MT, Bernstein DI. Age-related changes in magnitude and diversity of cross-reactive CD4+ T-cell responses to the novel pandemic H1N1 influenza hemagglutinin. Hum Immunol. (2010) 71:957–63. doi: 10.1016/j.humimm.2010.07.005
55. Baeten LA, Podell BK, Sluder AE, Garritsen A, Bowen RA, Poznansky MC. Standardized guinea pig model for Q fever vaccine reactogenicity. PLoS ONE (2018) 13:e0205882. doi: 10.1371/journal.pone.0205882
56. Xiong X, Meng Y, Wang X, Qi Y, Li J, Duan C, et al. Mice immunized with bone marrow-derived dendritic cells stimulated with recombinant Coxiella burnetii Com1 and Mip demonstrate enhanced bacterial clearance in association with a Th1 immune response. Vaccine (2012) 30:6809–15. doi: 10.1016/j.vaccine.2012.09.017
57. Moise L, Tassone R, Latimer H, Terry F, Levitz L, Haran JP, et al. Immunization with cross-conserved H1N1 influenza CD4+ T-cell epitopes lowers viral burden in HLA DR3 transgenic mice. Hum Vaccine Immunother. (2013) 9:2060–8. doi: 10.4161/hv.26511
58. Moise L, Terry F, Ardito M, Tassone R, Latimer H, Boyle C, et al. Universal H1N1 influenza vaccine development: identification of consensus class II hemagglutinin and neuraminidase epitopes derived from strains circulating between 1980 and 2011. Hum Vaccine Immunother. (2013) 9:1598–607. doi: 10.4161/hv.25598
59. Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, daSilva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. (2015) 43:D784–8. doi: 10.1093/nar/gku1166
60. Schipper RF, Schreuder GM, D'Amaro J, Oudshoorn M. HLA gene and haplotype frequencies in Dutch blood donors. Tissue Antigens (1996) 48:562–74. doi: 10.1111/j.1399-0039.1996.tb02670.x
61. Wilhelmsen CL, Waag DM. Guinea pig abscess/hypersensitivity model for study of adverse vaccination reactions induced by use of Q fever vaccines. Comp Med. (2000) 50:374–8.
62. Izzo AA, Marmion BP, Worswick DA. Markers of cell-mediated immunity after vaccination with an inactivated, whole-cell Q fever vaccine. J Infect Dis. (1988) 157:781–9. doi: 10.1093/infdis/157.4.781
63. Jerrells TR, Mallavia LP, Hinrichs DJ. Detection of long-term cellular immunity to Coxiella burnetii as assayed by lymphocyte transformation. Infect Immun. (1975) 11:280–6.
64. Kersh GJ, Fitzpatrick KA, Self JS, Biggerstaff BJ, Massung RF. Long-Term immune responses to Coxiella burnetii after vaccination. Clin Vaccine Immunol. (2013) 20:129–33. doi: 10.1128/CVI.00613-12
65. Chao CC, Chen HW, Li X, Xu WB, Hanson B, Ching WM. Identification, cloning, and expression of potential diagnostic markers for Q fever. Ann N Y Acad Sci. (2005) 1063:76–8. doi: 10.1196/annals.1355.010
66. Flores-Ramirez G, Danchenko M, Quevedo-Diaz M, Skultety L. Reliable tool for detection of novel Coxiella burnetii antigens, using immobilized human polyclonal antibodies. J Chromatogr B Analyt Technol Biomed Life Sci. (2017) 1047:84–91. doi: 10.1016/j.jchromb.2016.08.044
67. Gerlach C, Skultety L, Henning K, Neubauer H, Mertens K. Coxiella burnetii immunogenic proteins as a basis for new Q fever diagnostic and vaccine development. Acta Virol. (2017) 61:377–90. doi: 10.4149/av_2017_320
68. Jiao J, Xiong X, Qi Y, Gong W, Duan C, Yang X, et al. Serological characterization of surface-exposed proteins of Coxiella burnetii. Microbiology (2014) 160 (Pt 12):2718–31. doi: 10.1099/mic.0.082131-0
69. Papadioti A, Markoutsa S, Vranakis I, Tselentis Y, Karas M, Psaroulaki A, et al. A proteomic approach to investigate the differential antigenic profile of two Coxiella burnetii strains. J Proteomics (2011) 74:1150–9. doi: 10.1016/j.jprot.2011.04.016
70. Sekeyova Z, Kowalczewska M, Decloquement P, Pelletier N, Spitalska E, Raoult D. Identification of protein candidates for the serodiagnosis of Q fever endocarditis by an immunoproteomic approach. Eur J Clin Microbiol Infect Dis. (2009) 28:287–95. doi: 10.1007/s10096-008-0621-4
71. Vigil A, Chen C, Jain A, Nakajima-Sasaki R, Jasinskas A, Pablo J, et al. Profiling the humoral immune response of acute and chronic Q fever by protein microarray. Mol Cell Proteomics (2011) 10:M110.006304. doi: 10.1074/mcp.m110.006304
72. Xiong X, Jiao J, Gregory AE, Wang P, Bi Y, Wang X, et al. Identification of Coxiella burnetii CD8+ T-cell epitopes and delivery by attenuated listeria monocytogenes as a vaccine vector in a C57BL/6 mouse model. J Infect Dis. (2017) 215:1580–9. doi: 10.1093/infdis/jiw470
73. Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. (2010) 42:498–503. doi: 10.1038/ng.590
74. Ernst JD. Antigenic variation and immune escape in the MTBC. Adv Exp Med Biol. (2017) 1019:171–90. doi: 10.1007/978-3-319-64371-7_9
75. Tientcheu LD, Koch A, Ndengane M, Andoseh G, Kampmann B, Wilkinson RJ. Immunological consequences of strain variation within the Mycobacterium tuberculosis complex. Eur J Immunol. (2017) 47:432–45. doi: 10.1002/eji.201646562
76. Lamonaca V, Missale G, Urbani S, Pilli M, Boni C, Mori C, et al. Conserved hepatitis C virus sequences are highly immunogenic for CD4+ T cells: implications for vaccine development. Hepatology (1999) 30:1088–98. doi: 10.1002/hep.510300435
77. Penna A, Missale G, Lamonaca V, Pilli M, Mori C, Zanelli P, et al. Intrahepatic and circulating HLA class II-restricted, hepatitis C virus-specific T cells: functional characterization in patients with chronic hepatitis C. Hepatology (2002) 35:1225–36. doi: 10.1053/jhep.2002.33153
78. Almeida RR, Rosa DS, Ribeiro SP, Santana VC, Kallas EG, Sidney J, et al. Broad and cross-clade CD4+ T-cell responses elicited by a DNA vaccine encoding highly conserved and promiscuous HIV-1 M-group consensus peptides. PLoS ONE (2012) 7:e45267. doi: 10.1371/journal.pone.0045267
79. Fonseca SG, Coutinho-Silva A, Fonseca LA, Segurado AC, Moraes SL, Rodrigues H, et al. Identification of novel consensus CD4 T-cell epitopes from clade B HIV-1 whole genome that are frequently recognized by HIV-1 infected patients. AIDS (2006) 20:2263–73. doi: 10.1097/01.aids.0000253353.48331.5f
80. Izzo AA, Marmion BP. Variation in interferon-gamma responses to Coxiella burnetii antigens with lymphocytes from vaccinated or naturally infected subjects. Clin Exp Immunol. (1993) 94:507–15. doi: 10.1111/j.1365-2249.1993.tb08226.x
81. Buttrum L, Ledbetter L, Cherla R, Zhang Y, Mitchell WJ, Zhang G. Both major histocompatibility complex class I (MHC-I) and MHC-II molecules are required, while MHC-I appears to play a critical role in host defense against primary Coxiella burnetii infection. Infect Immun. (2018) 86:e00602–17. doi: 10.1128/IAI.00602-17
82. Li HM, Hiroi T, Zhang Y, Shi A, Chen G, De S, et al. TCRbeta repertoire of CD4+ and CD8+ T cells is distinct in richness, distribution, and CDR3 amino acid composition. J Leukoc Biol. (2016) 99:505–13. doi: 10.1189/jlb.6A0215-071RR
83. Amara RR, Nigam P, Sharma S, Liu J, Bostik V. Long-lived poxvirus immunity, robust CD4 help, and better persistence of CD4 than CD8 T cells. J Virol. (2004) 78:3811–6. doi: 10.1128/JVI.78.8.3811-3816.2004
84. Hammarlund E, Lewis MW, Hanifin JM, Mori M, Koudelka CW, Slifka MK. Antiviral immunity following smallpox virus infection: a case-control study. J Virol. (2010) 84:12754–60. doi: 10.1128/JVI.01763-10
85. Nyendak MR, Park B, Null MD, Baseke J, Swarbrick G, Mayanja-Kizza H, et al. Mycobacterium tuberculosis specific CD8+ T cells rapidly decline with antituberculosis treatment. PLoS ONE (2013) 8:e81564. doi: 10.1371/journal.pone.0081564
86. Axelsson-Robertson R, Rao M, Loxton AG, Walzl G, Bates M, Zumla A, et al. Frequency of Mycobacterium tuberculosis-specific CD8+ T-cells in the course of anti-tuberculosis treatment. Int J Infect Dis. (2015) 32:23–9. doi: 10.1016/j.ijid.2015.01.017
87. Pearce EL, Shedlock DJ, Shen H. Functional characterization of MHC class II-restricted CD8+CD4− and CD8−CD4− T cell responses to infection in CD4−/− mice. J Immunol. (2004) 173:2494–9. doi: 10.4049/jimmunol.173.4.2494
88. Ranasinghe S, Lamothe PA, Soghoian DZ, Kazer SW, Cole MB, Shalek AK, et al. Antiviral CD8+ T cells restricted by human leukocyte antigen class II exist during natural HIV infection and exhibit clonal expansion. Immunity (2016) 45:917–30. doi: 10.1016/j.immuni.2016.09.015
89. Kampschreur LM, Wegdam-Blans MC, Wever PC, Renders NH, Delsing CE, Sprong T, et al. Chronic Q fever diagnosis- consensus guideline versus expert opinion. Emerg Infect Dis. (2015) 21:1183–8. doi: 10.3201/eid2107.130955
90. Chentoufi AA, Kritzer E, Yu DM, Nesburn AB, Benmohamed L. Towards a rational design of an asymptomatic clinical herpes vaccine: the old, the new, and the unknown. Clin Dev Immunol. (2012) 2012:187585. doi: 10.1155/2012/187585
91. Gilbert SC. T-cell-inducing vaccines—what's the future. Immunology (2012) 135:19–26. doi: 10.1111/j.1365-2567.2011.03517.x
92. Humphreys IR, Sebastian S. Novel viral vectors in infectious diseases. Immunology (2018) 153:1–9. doi: 10.1111/imm.12829
93. Zhang C, Zhou D. Adenoviral vector-based strategies against infectious disease and cancer. Hum Vaccine Immunother. (2016) 12:2064–74. doi: 10.1080/21645515.2016.1165908
94. Zhang G, Peng Y, Schoenlaub L, Elliott A, Mitchell W, Zhang Y. Formalin-inactivated Coxiella burnetii phase I vaccine-induced protection depends on B cells to produce protective IgM and IgG. Infect Immun. (2013) 81:2112–22. doi: 10.1128/IAI.00297-13
95. Desnues B, Imbert G, Raoult D, Mege JL, Ghigo E. Role of specific antibodies in Coxiella burnetii infection of macrophages. Clin Microbiol Infect. (2009) 15 (Suppl. 2):161–2. doi: 10.1111/j.1469-0691.2008.02208.x
96. Bounds CE, Terry FE, Moise L, Hannaman D, Martin WD, De Groot AS, et al. An immunoinformatics-derived DNA vaccine encoding human class II T cell epitopes of Ebola virus, Sudan virus, and Venezuelan equine encephalitis virus is immunogenic in HLA transgenic mice. Hum Vaccine Immunother. (2017) 13:2824–36. doi: 10.1080/21645515.2017.1329788
Keywords: Coxiella burnetii, Q fever, multi-epitope vaccine, immunoinformatics, IFNγ, T cell
Citation: Scholzen A, Richard G, Moise L, Baeten LA, Reeves PM, Martin WD, Brauns TA, Boyle CM, Raju Paul S, Bucala R, Bowen RA, Garritsen A, De Groot AS, Sluder AE and Poznansky MC (2019) Promiscuous Coxiella burnetii CD4 Epitope Clusters Associated With Human Recall Responses Are Candidates for a Novel T-Cell Targeted Multi-Epitope Q Fever Vaccine. Front. Immunol. 10:207. doi: 10.3389/fimmu.2019.00207
Received: 04 October 2018; Accepted: 23 January 2019;
Published: 15 February 2019.
Edited by:
Fabio Bagnoli, GlaxoSmithKline, ItalyReviewed by:
Said Dermime, National Center for Cancer Care and Research, QatarAllen Jay Rosenspire, Wayne State University, United States
Copyright © 2019 Scholzen, Richard, Moise, Baeten, Reeves, Martin, Brauns, Boyle, Raju Paul, Bucala, Bowen, Garritsen, De Groot, Sluder and Poznansky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ann E. Sluder, YXNsdWRlckBtZ2guaGFydmFyZC5lZHU=
†These authors have contributed equally to this work
‡Present Address: Timothy A. Brauns, SmartPharm Therapeutics, Inc., Cambridge, MA, United States
 Anja Scholzen
Anja Scholzen Guilhem Richard
Guilhem Richard Leonard Moise
Leonard Moise Laurie A. Baeten4
Laurie A. Baeten4 William D. Martin
William D. Martin Susan Raju Paul
Susan Raju Paul Richard A. Bowen
Richard A. Bowen Anne S. De Groot
Anne S. De Groot Ann E. Sluder
Ann E. Sluder