- 1Mayo Clinic Vaccine Research Group, Mayo Clinic, Rochester, MN, United States
- 2Division of Biostatistics, Mayo Clinic, Rochester, MN, United States
Background: Sex differences in immune responses to influenza vaccine may impact efficacy across populations.
Methods: In a cohort of 138 older adults (50–74 years old), we measured influenza A/H1N1 antibody titers, B-cell ELISPOT response, PBMC transcriptomics, and PBMC cell compositions at 0, 3, and 28 days post-immunization with the 2010/11 seasonal inactivated influenza vaccine.
Results: We identified higher B-cell ELISPOT responses in females than males. Potential mechanisms for sex effects were identified in four gene clusters related to T, NK, and B cells. Mediation analysis indicated that sex-dependent expression in T and NK cell genes can be partially attributed to higher CD4+ T cell and lower NK cell fractions in females. We identified strong sex effects in 135 B cell genes whose expression correlates with ELISPOT measures, and found that cell subset differences did not explain the effect of sex on these genes' expression. Post-vaccination expression of these genes, however, mediated 41% of the sex effect on ELISPOT responses.
Conclusions: These results improve our understanding of sexual dimorphism in immunity and influenza vaccine response.
Introduction
Since 2010, seasonal influenza A is believed to have killed between 12,000 and 79,000 people, has resulted in 140,000–960,000 excess hospitalizations annually in the United States (1), and incurs annual costs of nearly $90 billion (2). During the 2017–2018 influenza season, seasonal influenza caused nearly 80,000 deaths in the United States alone (3). Males and females exhibit differences in immune responses to many viral vaccines, with females generally developing significantly higher levels of humoral immunity than males, including in response to the seasonal influenza vaccine (4–16). Seasonal influenza infection in males also tends to result in worse outcomes than infection in females (17–20), an effect most pronounced in the elderly (17). As seasonal influenza vaccine efficacy is sub-optimal, and particularly variable in the older adults most likely to be seriously affected by disease (21–27), sex-based differences in vaccine response further enhance inter-individual differences in influenza protection across populations and exacerbate this major public health issue.
Sex-based differences in vaccine-induced immune responses can be observed in pre-pubertal children, throughout the reproductive years, and have been demonstrated to persist after menopause (4–13, 28–30). However, even with mounting evidence of significant differences in immune responses to vaccines based on sex, most vaccine studies do not analyze immune response outcome data by sex (31, 32). In particular, little is known about sex effects in cellular (non-antibody) immune responses to influenza vaccine, or potential mechanisms for sex-differential immune responses to vaccines. A single published study examined sex differences in gene expression after influenza vaccination, proposing the involvement of a testosterone-regulated lipid metabolism pathway differentially expressed in males and females for which normalized gene expression correlated inversely with vaccine response (adjusted odds ratio of 0.39 for males compared to 2.25 for females) (14). Additional studies are necessary to confirm and expand these results and improve our understanding of how an individual's sex affects immunity.
Methods
The study population and laboratory methods described herein are similar or identical to those published in our previous studies (33–38).
Recruitment
The original study cohort comprised 159 generally healthy older adults of Caucasian descent, ages 50–74, 62% female, who received the 2010/11 seasonal trivalent inactivated influenza vaccine (TIV; Fluarix by GlaxoSmithKline, lot AFLUA524AA; containing the A/California/7/2009 NYMC X-191 [H1N1], A/Victoria/210/2009 NYMC X-187 [H3N2; an A/Perth/16/2009-like virus], and B/Brisbane/60/2008 viral strains), which was given by standard protocol into the deltoid muscle using a 16-gauge, 1-inch needle (38, 39). Blood (100 ml each) samples were taken immediately before vaccination as well as 3 and 28 days after vaccination by professional phlebotomists in Mayo Clinic's Clinical Trials Unit (38). Recruitment was performed at Mayo Clinic, Rochester, MN. Full immune datasets (successful measurement of HAI and VNA antibody titers, B cell ELISPOT response counts, transcriptomic data, and medical records of biological sex) were successfully obtained from 138 subjects (of whom 66% were female), and these data were used for all analyses, as described previously (40). Of these 138 subjects, immune cell phenotypes were successfully obtained from 135 subjects using flow cytometry, and these data were used for plots and statistical tests incorporating immune cell subsets. A brief table with cohort characteristics is included in Supplementary Table 1. Immune outcomes and some transcriptomic data from this cohort have been previously published (36–38, 41–44); the sex-differential statistical analyses and results reported herein have not been previously published.
Ethics Statement
Written informed consent was obtained from each study participant, and the Mayo Clinic Institutional Review Board approved the study. Influenza virus was propagated in embryonated chicken eggs as described (35). Mayo Clinic's Institutional Animal Care and Use Committee (IACUC) policy does not require review of research on unhatched embryonated eggs destroyed before hatching, in accordance with the Office of Laboratory Animal Welfare and the National Institutes of Health policy.
Source of Biological Materials
Madin-Darby Canine Kidney (MDCK) cells were obtained from the American Type Culture Collection.
Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
PBMCs were isolated from subject-derived whole blood samples, as described previously using BD Vacutainer® CPT™ cell preparation tubes with sodium citrate (33). The cells were cryopreserved prior to sequencing and use in assays (33–35).
Flow Cytometric Measurement of Immune Cell Composition
Subjects' immune cell subsets were quantified using flow cytometry (45). Briefly, frozen subject PBMCs drawn at baseline were thawed and stained using fluorochrome-conjugated antibodies (Abs) from BD Biosciences (San Jose, CA). B cells, NK cells, NKT cells, monocytes, and dendritic cells were identified using the following panel of Abs: CD11c-V450; CD3V500; CD86-fluorescein isothiocyanate (FITC); CD56-phycoerythrin (PE); CD123-allophycocyanin (APC); CD20-Peridinin chlorophyll protein complex (PerCP)-Cy5.5; HLA-DR-Alexfluor 700; CD16-PE-Cy7; and CD14-APC-Cy7. CD4+, and CD8+ T cells were distinguished using the following panel of Abs: CD3-Brilliant Violet 421; CD4-AlexaFluor 700; CD25-Brilliant Violet 605; and CD127-AlexaFluor 647. Following staining, cells were fixed with 4% paraformaldehyde, and data were collected using a BD LSR II Flow Cytometer with FACSDIVA software (BD Biosciences). These antibody panels were chosen by the Human Immunology Project Consortium to allow for comparison of immunophenotyping data across studies (46). Cell population gating was performed using FLOWJO (FlowJo, LLC, Ashland, OR, United States). Debris and doublets were removed using FSC and SSC parameters. Cell subsets were identified based on the surface staining patterns indicated in Supplementary Table 2. Data represent the mean of technical quadruplicates from each subject.
Growth of Influenza Virus
Influenza A/California/7/2009/H1N1-like virus was provided by the Centers for Disease Control and Prevention (Atlanta, GA, United States). As previously described, the virus was propagated in embryonated chicken eggs and harvested from the allantoic fluid 48 h post-inoculation. Fifty-percent tissue culture infectious dose (TCID50) measurements of virus stocks were determined by infection of MDCK cells with serial dilutions of the virus, and dilution wells with successful virus replication were determined by addition of red blood cells 5 days after infection and observation of hemagglutination as per standard protocols (47–49).
Measurement of Serum Antibody Responses to Influenza Vaccine
The methods for assessing both the hemagglutination-inhibition influenza A/H1N1 antibody (HAI) titers and virus-neutralizing influenza A/H1N1antibody (VNA) titers for these subjects have been published (34–37). Ab titers were measured from sera collected in BD Vacutainer Serum tubes at each timepoint pre- and post-vaccination using standard protocols. Serum influenza A/H1N1-specific neutralizing antibody titers were measured by a cell-based microneutralization assay (assay CV = 4.7%) at each timepoint with influenza A/H1N1 virus stimulation (200 plaque-forming units per 5 μl), as previously described (37). Median values from triplicate technical replicates were used for data analysis.
Measurement of Humoral Adaptive Immune Responses to Influenza Vaccine
Influenza-specific B cells were measured by influenza A/H1N1-specific B-cell ELISPOT after in vitro stimulation of subject PBMCs with vaccine-strain A/H1N1 influenza virus, as previously described (35, 36, 40). Briefly, ELISPOT analyses using the MabTech Human IgG ELISpotPLUS Kit (Mabtech, Inc.; Cincinnati, OH) (35) were used to quantify the influenza A/H1N1-specific B cells (memory-like IgG B cells) in subjects' PBMCs. ELISPOT plates were coated with a 1:50 dilution of influenza A/H1N1 virus stock (50,000 TCID50/well). The median of four technical replicates was used for data analysis.
mRNA-seq
Methods for transcriptomic sequencing were published in our previous transcriptomics studies (43, 50). Briefly, we extracted total RNA from each cryopreserved subject PBMC sample using RNAprotect reagent and RNeasy Plus mini kits reagent (Qiagen; Valencia, CA, United States). Poly-A RNA was isolated using magnetic purification, and Mayo Clinic's Gene Sequencing Facility created cDNA libraries using the mRNA-Seq 8 Sample Prep Kit (Illumina; San Diego, CA). An Illumina HiSeq 2000 was used to perform single-end read sequencing. The human genome build 37.1 was used to align sequencing reads using TopHat (1.3.3) and Bowtie (0.12.7).
The resulting mRNA-sequencing gene-count data underwent a strict quality-control and normalization procedure, as described by Ovsyannikova et al. (43). Briefly, Conditional Quantile Normalization (51) was used to normalize gene counts; 14,197 genes were determined to have at least 32 counts at one of our three timepoints (Day 0, 3, or 28) and were used in our subsequent analyses.
Statistical Analysis of Sex Differences in Immune Cell Compositions and Single Gene Expression Levels
Wilcoxon rank-sum tests were used to test for differences between males and females in CD4+ T cell percentage and NK cells percentage and single gene-level gene expression. Spearman's correlation was used to test for correlation between NK cell percent and CD4+ T cell percent in each subject as well as for correlation between single gene expression levels and Day 28 B cell ELISPOT responses. Story and Tibshrani's method (43) for genome-wide studies was used to calculate q-values for these gene level results.
Weighted Gene Coexpression Network Analysis (WGCNA)
Because the expression of genes can be highly correlated, we chose to focus on clusters (modules) of highly correlated genes and use summary information of each cluster as a measure of gene expression of multiple correlated genes. This was accomplished by Weighted Gene Coexpression Network Analysis (WGCNA), as previously described (40). Using the Day 28 normalized gene expression data, we created data-driven gene clusters using WGCNA via creation of a co-expression similarity matrix followed by hierarchical clustering techniques (52–54). The first principal component of the gene expression levels within a gene cluster represent the cluster's eigengene and served to represent the entire cluster's gene expression activity. Pearson's correlation was used to correlate immune phenotypes with each cluster's eigengene to identify the gene clusters related to vaccine responses.
Gene Enrichment Analyses
Enrichment analysis for genes involved in known pathways or functions was previously performed on the gene clusters using the RITAN (55) package and published Blood Translation Modules (BTMs) (56), as described previously (40).
Mediation Analyses and Calculation of Sex Effect in Antibody Titers, ELISPOT Result, and Gene Cluster Expression Levels
Causal mediation analysis was conducted using the “mediation” package in R version 3.4.1 (57, 58). The eigengenes from each WGCNA cluster were used as the outcome variables, and we analyzed whether the fraction of CD4 + T cells or NK cells in PBMCs mediate the relationship between sex and the gene expression eigengenes. Linear models were used for analyses, and all continuous variables were scaled to have the mean equal to zero and standard deviation equal to one. The non-parametric bootstrap method was used with 1,000 simulations to estimate variance of the model parameters and calculate the 95% confidence intervals. Results of mediation analyses include the average direct effect (ADE), average causal mediated effect (ACME), and total effect (TE). The TEs from these models were used as an estimate of the effect of sex on the gene expression clusters at each time-point (Day 0, 3, and 28). False discovery q-values were calculated using the Benjamini and Hochberg method (59). Gene clusters were reported as potentially sex-dependent if they had a sex-difference p < 0.01 and q < 0.1 at a minimum of two of the three time points measured.
We analyzed whether cluster gene expression (summarized by eigengenes) mediates the relationship between sex and B-cell ELISPOT responses and whether cell subset percentage mediates the relationship between sex and gene expression in sex-dependent gene clusters using the same framework. The TEs from these models are used to estimate the effect of sex on ELISPOT responses and antibody responses or cell subset % on gene cluster expression. The ACMEs calculated by mediation analysis are reported as estimates of the percent of the total sex effect mediated by cluster gene expression or cell subset %. False discovery q-values were calculated for the mediation analysis results using the Benjamini and Hochberg method (59).
Results
Males and Females Display Differences in Immune Outcomes After Vaccination
To assess the effect of subject sex on humoral immunity after influenza vaccination, we compared males' vs. females' influenza A/H1N1-specific B-cell ELISPOT responses in subject PBMCs harvested from each subject and cryopreserved (35) immediately prior to vaccination, 3 days after vaccination, and 28 days after vaccination (Figure 1).
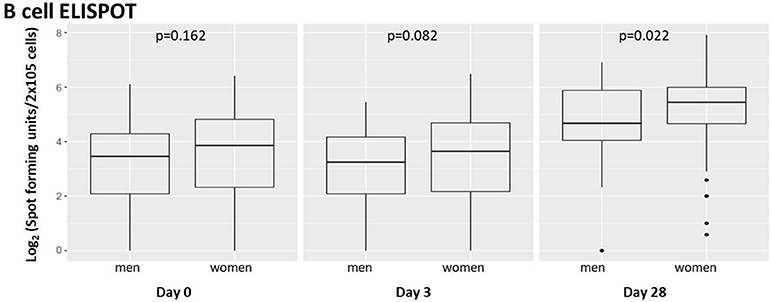
Figure 1. Memory B-cell ELISPOT responses to influenza virus differ by subject sex. Memory B cells capable of responding to vaccine strain influenza A/H1N1 were quantified using B-cell ELISPOT. Dot plots indicating individuals' responses are available in Supplementary Figure 1.
We found suggestive differences between male and female influenza-reactive B-cell populations 3 days after vaccination and significant differences (p = 0.022) 28 days post-vaccination.
We also assessed the effect of subject sex on influenza antibody titers immediately prior to vaccination, 3 days after vaccination, and 28 days after vaccination as measured by HAI and VNA assays (Figure 2).
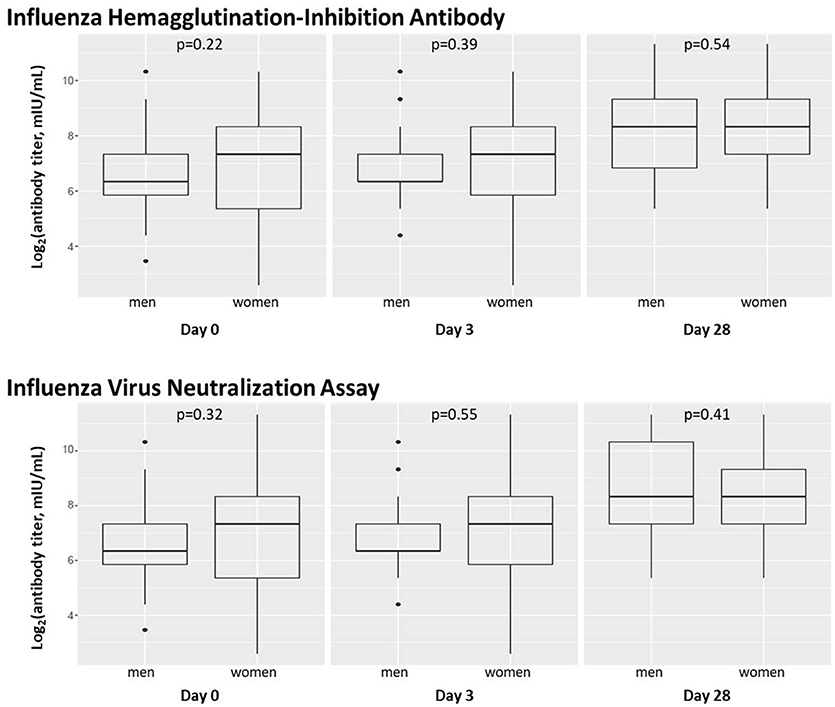
Figure 2. Subject sex does not significantly affect influenza A/H1N1 antibody titers. Box plots are shown for levels of influenza A/H1N1-reactive antibody in subject sera before and after vaccination using hemagglutination-inhibition and virus neutralization assays. Dot plots indicating individuals' responses are available in Supplementary Figure 2.
Females had higher median antibody titers at baseline and Day 3, but these differences were not statistically significant (p > 0.1). Median antibody titers in males and females were equivalent (1:320) at Day 28. HAI and VNA antibody measures were highly correlated, as previously described (36, 44, 45).
Sex Differences Are Found in Gene Expression Signatures Related to NK Cell, T cell, and B cell Activity
We also examined gene expression data to identify gene expression clusters whose expression levels are affected by subject sex. Such information may provide clues to the biological source(s) of sex differences in immune responses to influenza vaccination. In a previous study using Weighted Gene Coexpression Network Analysis (WGCNA) clustering of transcriptomic data, we identified 15 non-overlapping clusters of genes that shared similar expression patterns across the study cohort (40). In the current study, we tested the expression levels of each of these gene clusters for sex dependence; four gene clusters demonstrated significant sex dependence (p < 0.01 and FDR q < 0.1) during at least one of the three timepoints (Table 1). A full list of all quantified genes, their cluster membership, and median expression levels in males vs. females can be found in Supplementary Table 3. Genes located on the X chromosome were distributed evenly across the 15 gene clusters and comprised no more than 5% of any individual gene cluster, and thus are not responsible for observed sex dependence in overall gene cluster expression.
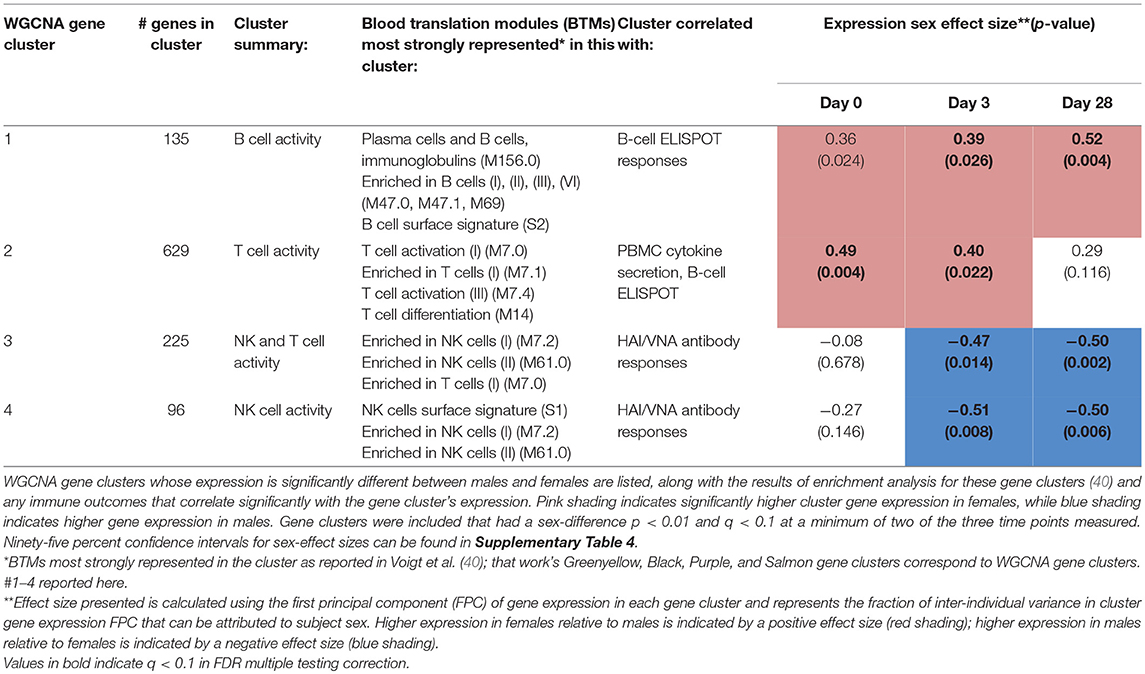
Table 1. Sex differences are found in PBMC expression of gene clusters related to immune cell activity, including CD4+ T cell, B cell, and NK cell activity.
Females demonstrated significantly higher expression of genes in a cluster of B cell-related genes (#1) than males at Day 28 (p = 0.004)—an effect that was also present during earlier timepoints (p = 0.024 at Day 0, p = 0.026 at Day 3). Females also demonstrated higher expression of T cell genes (cluster #2) than males at baseline and early in vaccine response (p = 0.004 at baseline; p = 0.022 at Day 3). In gene clusters #3 and #4 enriched for NK and cytotoxic T cell genes, we found higher gene expression in males than females during the response phase (cluster #3, p = 0.014 [Day 3] and 0.002 [Day 28]; cluster #4, p = 0.008 [Day 3], and 0.006 [Day 28]). Statistical significance of eight of these nine p-values was confirmed after FDR calculation of q-values for multiple testing correction; only the Day 1 B-cell gene cluster (#1) sex differences did not meet a q-value threshold of < 0.1.
Sex Differences in B-cell ELISPOT Levels Are Mediated by Sex-Related Differences in Expression of B-cell Related Genes
As a step toward identifying the biological source of the sex differences observed in B-cell ELISPOT results, we explored whether the sex differences observed in the expression levels of these gene clusters would statistically explain the sex differences observed in B-cell ELISPOT results. To do this, we utilized statistical mediation analysis to examine whether the gene expression of each cluster mediates the effect of subject sex on B-cell ELISPOT responses in our subjects (Figure 3).
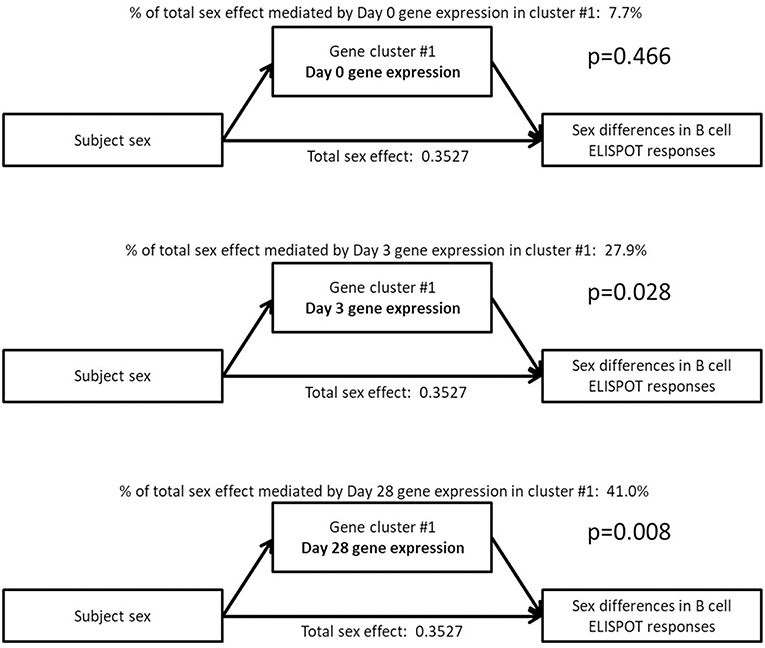
Figure 3. Mediation analysis indicates that the sex effect on B-cell ELISPOT responses is mediated by Day 3 and Day 28 gene expression in a small gene cluster of 135 genes highly enriched for B cell-related genes. The total sex effect represents the fraction of the standard deviation in ELISPOT response that can be explained by sex.
Sex differences in Day 28 B-cell ELISPOT results were determined to be mediated by sex-dependent gene expression of a single gene cluster after vaccination (cluster #1: B cell genes, mediating effect p-values of 0.028 and 0.008 for Day 3, and Day 28 cluster expression, respectively). This gene cluster represents 135 B cell genes whose postvaccination expression levels we previously showed to correlate strongly with Day 28 B-cell ELISPOT responses (Day 3 correlation 0.28, p = 0.001; Day 28 correlation 0.3, p = 3 × 10−4) (40). Sex-dependent gene expression in gene cluster #1 at 28 days postvaccination explains 41% of the effect of sex on Day 28 B-cell ELISPOT responses, according to our mediation analysis. Gene expression in other gene clusters did not significantly mediate the effect of sex on B-cell ELISPOT responses (p > 0.05).
Males and Females Have Different Immune Cell Compositions That May Affect Immune Responses
One variable that could account for these observed sex differences in gene expression is sex differences in immune cell subset composition and quantity. To explore differences in resting immune states between males and females, we examined baseline differences in immune cell composition between males and females. PBMCs from the 135 subjects with sufficient remaining cryopreserved samples were stained with a panel of fluorescent antibodies to cell markers designed to differentiate and quantify subsets of immune cells via flow cytometry. Specifically, populations of T cells (with CD4+ and CD8+ subsets quantified), B cells, NK cells, NK T cells, monocytes, and dendritic cells were quantified relative to one another (Figure 4).
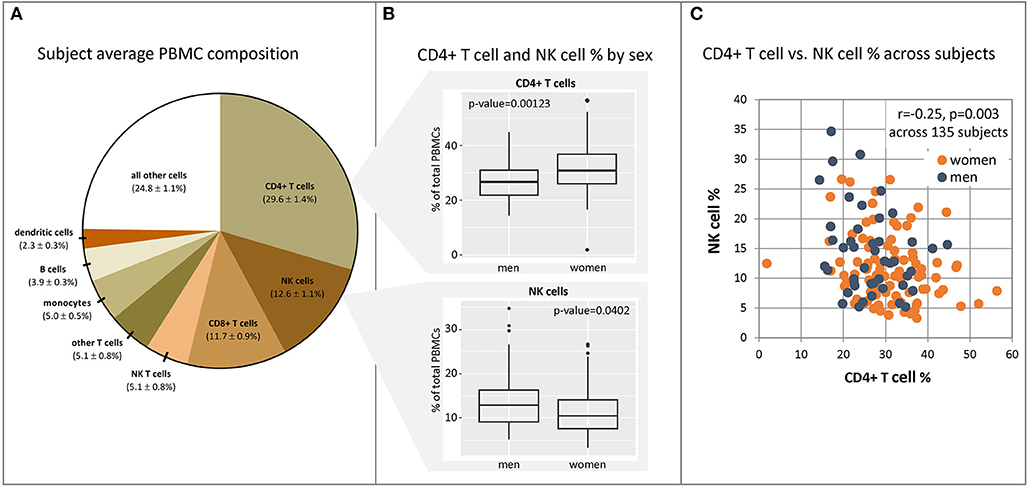
Figure 4. CD4+ and NK cell fractions in PBMCs differ between males and females. PBMC samples were harvested from our cohort subjects immediately prior to vaccination, and populations of immune cell subsets were quantified in each subject sample using fluorescent antibody staining for unique cellular markers and flow cytometry. (A) mean PBMC composition and 95% confidence intervals across 135 subjects. (B) Significantly different fractions of CD4+ T cells and NK cells were found in males (n = 45) vs. females (n = 90). Dot plots indicating individuals' responses are available in Supplementary Figure 3. (C) CD4+ T cell and NK cell fractions in each subject show weak correlation with one another. Source data may be found in Supplementary Table 5.
The average composition of subject PBMCs prior to vaccination is displayed in Figure 4A. Analysis demonstrated a significantly higher fraction of CD4+ T cells in females relative to males (Wilcoxon Rank Sum test, 31.2 vs. 27.1%, p = 0.001), and correspondingly higher fractions of NK cells in males relative to females (13.5 vs. 10.5%, p = 0.04) (panel B). Other cell populations (B cells, monocytes, dendritic cells, NK T cells, and CD8+ T cells) were not found to be significantly different between males and females. An effect of age was observed for baseline CD8+ T cell and NK cell levels; there was a modest decrease in CD8+ T cell fractions as age increased (Spearman rank correlation = −0.18, p = 0.034) as well as an increase in NK cell fractions (Spearman rank correlation = 0.21, p = 0.011). No significant effect of age was seen on any other cell type across the age range (50–74 years, p > 0.2 for all cell subsets, Spearman rank correlation). CD4+ T cell and NK cell fractions in each subject demonstrated a weak inverse correlation with one another, suggesting that these populations may or may not be regulated in part by the same factors (Panel C).
Sex Differences in Subject PBMC Immune Cell Compositions Mediate Sex Differences in NK and T cell Gene Cluster Expression Levels, but do not Affect B cell Gene Clusters or ELISPOT Measures
We observed higher levels of gene expression in females than males in gene clusters that corresponded to T helper cell activity (cluster #2), which also corresponded with the higher fractions of CD4+ T cells observed in females. Similarly, the lower levels of NK cells in females' overall PBMC compositions corresponded with lower gene expression in NK cell-related gene clusters (clusters #3 and #4). To test if the differences in cell subset composition could statistically explain the observed sex differences in gene cluster expression levels, we conducted mediation analyses to determine whether cell subset composition mediates the effect of subject sex on gene expression levels in these gene clusters (Figure 5).
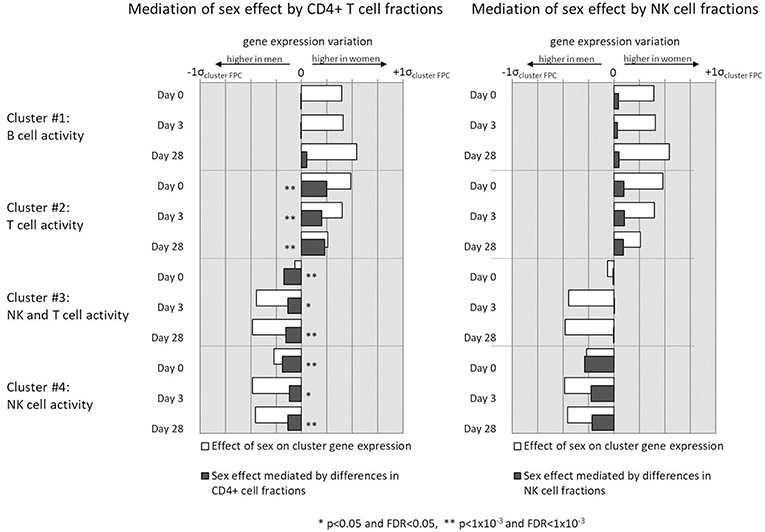
Figure 5. Mediation analysis indicates whether sex-related differences in gene expression of select gene clusters is mediated by observed sex differences in CD4+ cell (left column) and NK cell (right column) fractions of subject PBMCs.
Sex differences in CD4+ cell counts were found to partially mediate sex differences in NK and T cell gene clusters #2, #3, and #4 to a highly significant degree. NK cell differences also mediate the effect of sex on gene clusters #2 and #3, which are involved in NK and T cell activity, but to a less statistically significant degree that did not retain significance after multiple testing correction (0.01 < p < 0.05, q > 0.1). In no gene cluster did differences in immune cell quantities mediate the entirety of the observed sex effect on gene expression. Neither NK cell nor CD4+ T cell sex differences mediated the sex differences found in B cell gene activity (cluster #1), likely reflecting other biological mechanisms as primary contributors to sex effects on B cell gene expression. Finally, neither B cell, NK cell, nor CD4+ T cell fractions between subjects were found to mediate sex differences in ELISPOT response (p > 0.1, data not shown).
B cell Genes That Mediate the Effect of Sex on B-cell ELISPOT Responses Control B cell Proliferation, Differentiation, and BCR Signaling
In order to identify other potential mechanisms for the larger B-cell ELISPOT responses observed in females, we further examined genes in cluster #1 to identify specific genes in this cluster that appear to be expressed in a sex-dependent manner and may therefore be causally mediating these effects. We examined expression levels of individual genes in this cluster and tested for sex-differential expression. The 20 most significantly sex-dependent genes are shown in Table 2 (see Supplementary Table 6 for the complete list of cluster #1 genes).
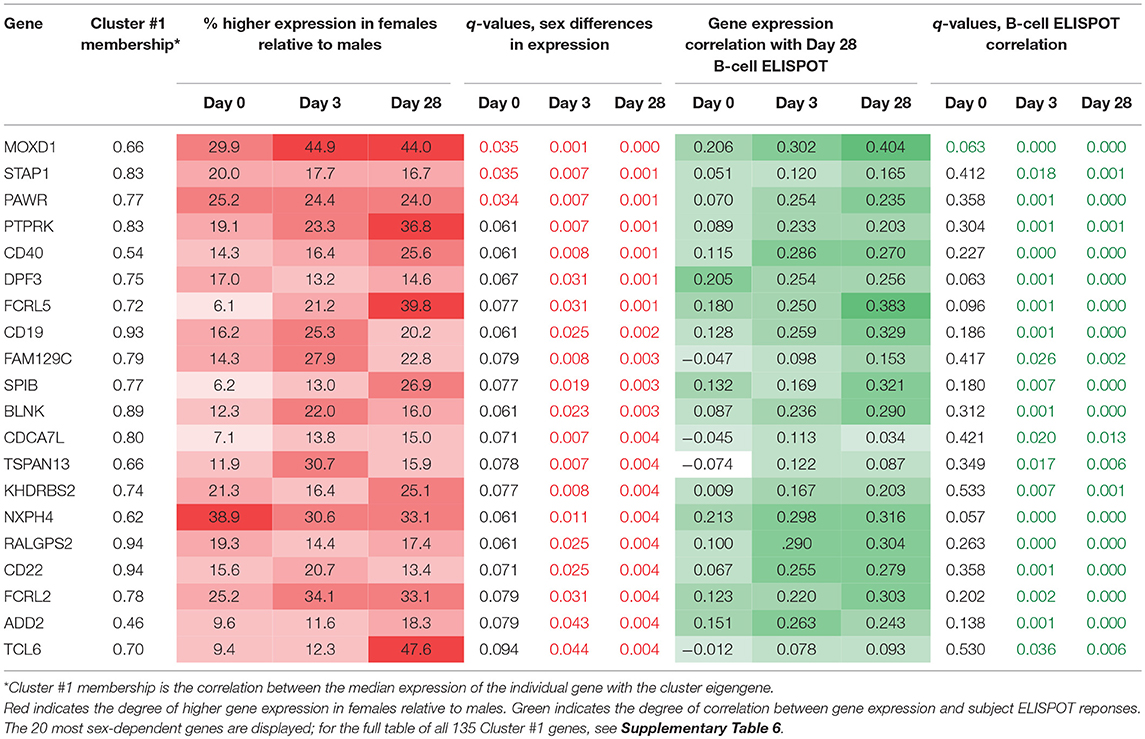
Table 2. Gene cluster #1 genes with significant differences in expression levels between males and females.
We found 116 genes that demonstrated sex-differential expression at a minimum of one timepoint after multiple testing correction (q < 0.05). The expression of most of these genes also correlated significantly with Day 28 B-cell ELISPOT measures across the subjects and represent potential links between sex-dependent gene expression and B-cell ELISPOT measures.
These genes fall into two distinct categories. Approximately half of these genes have known direct functions in B cell signaling, proliferation, and differentiation (e.g., CD40, FCRL5, CD19, BLNK, EBF1). That these genes are more highly expressed in females relative to males indicates the heightened activity of females' B cells after vaccination relative to those of males. The remaining sex-dependent genes are largely transcription factors and are also likely to be active in B cell responses to vaccination, with links to cell fate decision making (e.g., SOX5, DPF3), cell proliferation/differentiation (e.g., SPIB), and cell signaling (e.g., MOXD1, PTPRK, SPIB, KHDRBS2). The sex-differential expression of these genes after vaccination reinforces that while B cell fractions are similar in males and females, intrinsic B cell signaling, proliferation, and differentiation activities are significantly higher in females. Our evidence points to this collection of genes as likely to be critically involved in the sex-differential immune outcomes observed after influenza vaccination.
Discussion
Summary of Findings
We found higher levels of influenza A-specific memory B cells in females relative to males after seasonal influenza vaccination. Previous studies investigating sex differences in seasonal influenza vaccine responses have largely examined antibody titers, reaching no clear consensus (8, 9, 44, 60, 61). Additionally, the ability to identify subtle sex differences in antibody responses is limited by the variability and log-based nature of antibody titer assays. Our study is the first to identify sex differences using the sensitive influenza-specific memory B cell ELISPOT assay as a key measure of long-term vaccine-induced immune memory.
Transcriptional sex differences in vaccine responses were observed in four gene clusters highly enriched for NK cell, T cell, or B cell genes. The strongest sex effect was noted for a small gene cluster of 135 B cell genes (p = 0.004). The post-vaccination expression of genes in this cluster alone was found to statistically mediate a substantial fraction of the effect of subject sex on B-cell ELISPOT responses (mediation p-value 0.008). No other gene clusters were found to mediate the sex effects on ELISPOT, regardless of the sex dependence of their expression levels.
Immune cell composition has previously been shown to affect responses to vaccination (36, 62). Here, we demonstrate that males and females have different PBMC fractions of CD4+ T cells and NK cells, and that these differences mediate a significant part of the sex-differential expression levels in T cell and NK cell gene clusters after influenza vaccination (Figure 6). However, the sex-differential gene expression of B cells that ultimately leads to higher B-cell ELISPOT responses in females appears to be independent of CD4+ and NK cell sex differences.
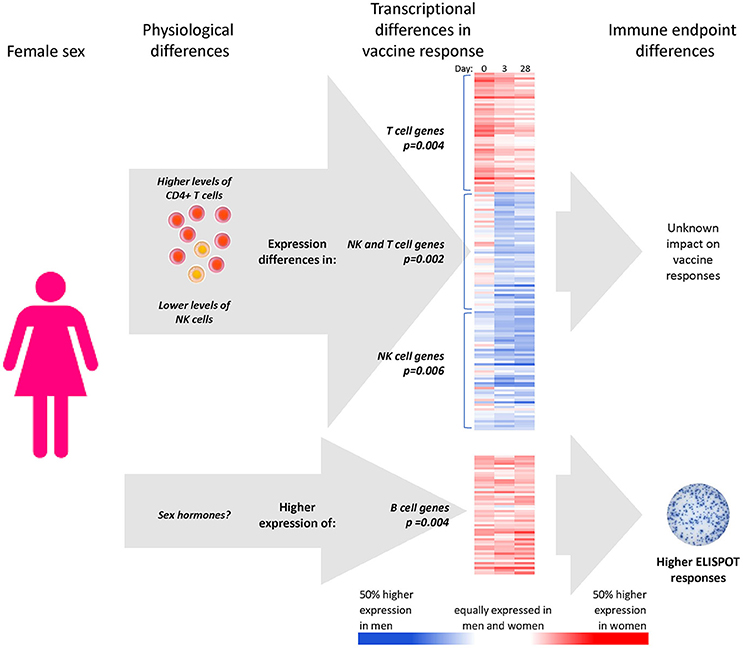
Figure 6. Apparent mechanisms of sex effects on influenza vaccine responses. CD4+ T cell and NK cell numbers mediate part of the effect of subject sex on the expression levels of gene clusters involved in NK cell and T cell genes, with unknown impact on immune outcomes. Higher expression of 135 B cell genes in females was not found to relate to sex differences in immune cells; however, it was determined that these B cell genes were, however, statistical mediators of the higher B-cell ELISPOT responses to seasonal influenza vaccine found in females relative to males. *Gene expression heatmaps present the sex-differential expression of the 50 most representative genes of each sex-dependent gene cluster (i.e., most highly correlated with the cluster eigengene).
Sex Differences in CD4+ T cells
Influenza-specific memory T cells play an important role in the protection of individuals against influenza disease (63–65). While serum antibody measures are often used as a correlate of protection, influenza antibody titers are limited as measures of vaccine efficacy, particularly in older individuals (63–65). Populations of influenza-reactive memory CD4+ T cells, on the other hand, have been repeatedly demonstrated to correlate well with protection in both younger and elderly adults (64, 65). These CD4+ T cell responses are induced in the weeks after influenza vaccination and may be better predictors of long-term immunity in healthy adults than other measures of immunity (66). Higher net levels of CD4+ T cells in women relative to men may, therefore, significantly affect an individual's ability to respond effectively to influenza vaccines. Indeed, female mice have been shown to have greater levels of CD4+ T cell responses than males when exposed to the 2009 H1N1 strain of influenza virus (15).
Ours is not the first study to have identified larger fractions of total CD4+ T cells in females' PBMCs relative to males'. In large studies of healthy individuals spanning a broad age range, Amadori et al. and Wikby et al. also found no effect of sex on CD8+ T cell populations but higher CD4+ T cell counts and higher CD4/CD8 ratios in females (67, 68). Interestingly, while CD4/CD8 ratios have been previously found to remain relatively constant in individuals over multiple-year time frames (67), inverted CD4/CD8 ratios have also been shown to increase with age over decades-long time scales and have been associated with a higher mortality risk in individuals over the age of 60 (68, 69).
These sex-differential CD4/CD8 cell ratios appear to be at least partly sex hormone-mediated, as indicated in studies of androgen-deficient men, and women after surgical menopause (70–74). Indeed, hormone therapy was demonstrated to reverse the hormone-deficiency effects on CD4/CD8 ratios in both these situations; however, the timeframe necessary for hormone treatment to achieve such effects is unclear. Natural monthly menstrual changes in sex hormones were found to have no apparent effect on CD4/CD8 T cell ratios (67); thus, hormone differences may need to exist for extended periods of time—from many months to years—for effects on CD4+ T cell populations to be observed (75).
In addition to higher numbers of CD4+ T cells in females relative to males, the ability of these cells to respond to vaccination appears to differ substantially between sexes. For example, CD4+ T cells express estrogen receptors (76), and estrogen has been demonstrated to impact T cell development, mature T-cell function, and the establishment of T cell memory (77–81). The methylation and transcription patterns of males' and females' T cells are, as a result, quite distinct (82, 83).
In older individuals, this sex effect on T cell activity may exacerbate concomitant age effects on immunity. Studies have previously demonstrated that naïve CD4 functions decrease dramatically with age and contribute to reduced humoral responses to vaccination (63, 78, 84–87). Our analysis showed higher age-corrected gene expression of many genes relative to naïve T cell functions in female subjects relative to males. Examples include baseline expression of CD28 (p = 0.004) and the genome-organizer SATB1 (p = 0.003), which play critical roles in activating T cell genes and shaping the T cell pool (88, 89). We hypothesize that higher levels of CD4+ T cells, combined with potentially higher activity as suggested by others (90), may help older females retain net Th cell function over time and maintain the ability to successfully respond to vaccination.
Sex Differences in NK Cells and Inflammation
Others have also observed higher levels of NK cells in males relative to females. NK-cell numbers also appear to be regulated by sex hormones (75). The role of NK cells in vaccine response has particular implications in older populations. Proinflammatory cytokines, such as IL-6, IL-1β, and TNFα, are produced at elevated levels in elderly persons (91). This phenomenon, termed “inflammaging,” may be due to chronic stimulation of innate immunity, which has been demonstrated to negatively impact immune responses to vaccination (92–94), perhaps by masking the stimuli induced by vaccines (95). Larger fractions of NK cells in male PBMCs and greater activity of proinflammatory NK cell-related genes suggest that older males may experience a higher degree of inflammaging than older females. This may add to the difficulty in developing protective vaccine-induced immune responses in older males.
Sex Differences in B cell Responses to Vaccination
We found that total B cell fractions did not differ significantly between men and women; however, the numbers of influenza-specific B cells after vaccination as measured by ELISPOT were significantly higher in women than men. These results suggest that sex differences do not originate in differences in total numbers of B cells, but in a heightened ability of women to respond to vaccination and generate long-lasting influenza-reactive memory B cells. Supporting this hypothesis is the fact that we observed clear sex-related differences in B cell gene expression after vaccination. For example, CD40 gene expression in PBMCs was significantly higher in females than males across the timepoints in our study (p < 0.01) (Supplementary Table 6), implying that female antigen-presenting cells may be inherently better poised to proliferate and differentiate in response to vaccine-activated CD4+ T cells (96, 97).
Evidence in the literature corroborates the existence of sex-based differences in B cell immunity. For example, a large microarray study (n = 5,241) of normal adults in the absence of any immune stimulation found strong female-biased expression of genes involved in immune system processes, including regulation of immunity, response to cytokines, and regulation and activation of leukocyte differentiation (98). In mice exposed to the 2009 strain of H1N1 influenza, antibody from females demonstrated both greater antibody responses and increased antibody avidity and specificity than in males (15). While the mechanism behind differences in B cell gene expression is not well-defined, evidence exists for both sex hormone-mediated and sex hormone-independent components. The existence of sex differences in infants' responses to vaccination indicates that non-hormonal sex differences in immunity exist. However, estrogens and other sex hormones also play clear roles in immunity; in the aforementioned microarray study (98), the gene transcription differences between males and females appear to be at least partially linked to estrogen levels—largest in females using hormonal contraceptives and smaller, though still present, in postmenopausal females. Similarly, in vivo estrogen levels were found to relate to lymphocyte homeostasis in another study (99). These results confirm that sex differences in B cell responses to immunization are likely a combination of hormone-mediated and hormone-independent effects.
Sex Hormone Effects on Immunity
Estrogen, testosterone, and other sex hormones have been observed to clearly impact B cell activity; these effects may be direct, indirect, or both. B cells express, for example, ERα/β estrogen receptors, allowing estrogen-related compounds to directly modulate lymphocyte function. In vitro treatment of both male and female human PBMCs with 17-beta estradiol was demonstrated decades ago to significantly enhance B cell differentiation (100, 101). Estrogen-related compounds have been demonstrated to directly increase B cells' expression of anti-apoptotic molecules critical for B cell activity and development, while androgen treatment decreases expression of such molecules (102–104). Such hormone-modulated gene expression appears to affect diverse B cell functions from lymphopoesis to BCR signaling, B cell expansion, and B cell maturation (103–106), as reviewed by Sakiani et al. (107). Direct binding of estrogen receptors has several effects on activated B cells, including increased antibody production, somatic hypermutation, class-switch recombination, and on the development and persistence of B cell memory (108–113).
Testosterone, while not demonstrated to directly influence B cells in vitro to the extent of estrogen (100, 101, 114), has also been implicated in sex-based differential vaccine responses. In a systems biology analysis of data from 91 young and old individuals by Furman et al. (14), high testosterone levels in men were associated with poor virus-neutralizing response. This study further identified a cluster of 35 lipid metabolism genes known to be testosterone-regulated whose expression related to poor virus-neutralizing activity in men. We successfully quantified 22 of these 35 lipid metabolism-related genes in our current study, yet we did not find any consistent sex differences in these genes' expression to replicate this result despite a significantly larger sample size. As Furman et al. noted, the effect of these 35 genes' expression was most notable in males with high testosterone levels. As our cohort comprises solely older individuals with likely lower testosterone levels due to age, the effects of high testosterone males may simply no longer be observable; however, we did not measure testosterone levels in our study.
The presence of estrogen and testosterone may also impact B cells indirectly via their effects on other immune cells. Though at levels lower than in B cells, estrogen receptors are present on a large array of immune cells, including T cells, NK cells, dendritic cells, and monocytes (76, 115). It is well-established that sex hormones regulate innate immune functions of monocytes, dendritic cells, and macrophages, which affect NFκB and TLR signaling, interferon secretion, and proinflammatory cytokine production (98, 116–122). Evidence exists for the involvement of monocytes and/or T cells in estrogen-based augmentation of antibody responses in PBMCs both in vitro and in vivo (123, 124).
While the precise cellular mechanisms behind the effects of sex hormones on immunity are still being fully defined, the impact of estrogen and other sex hormones on humoral and cellular immune responses is not a new concept. It has been suggested for decades that estrogen enhances humoral and possibly cellular immune responses and is involved in the pathogenesis of various autoimmune diseases found predominantly in females, such as systemic lupus erythematosus (114, 125, 126).
Hormone-Independent Sex Effects on Immunity
While sex-differential immune responses to vaccines have been noted in infants and pre-pubertal children (4, 127–130) in whom large hormonal differences do not exist, non-hormonal sex differences have not been well-studied or understood. The existence of different immune-regulating genes and miRNAs on the X and Y chromosomes has been suggested as a mechanism for genetic and epigenetic, rather than hormonal, sex-related differences in immune responses (131–133). While our study demonstrated that X-chromosome genes comprised < 5% of any gene cluster, transcriptional differences in a few key regulatory genes could significantly affect the immune response system and contribute to the observed sex differences. Our study provides a list of 135 B cell genes that may be important mediators of this effect. In a follow-up study, it would be interesting to examine the influence of X-linked genes on the expression of these mediators of sex-differential ELISPOT responses. This information may lead to further refinement of our understanding of how biological sex influences immunity.
Some have additionally proposed that sex differences in immunity may be mediated by the microbiome, as differences appear to exist between male and female microbiomes, and microbiome compositions in turn affect immune responses (134–140). However, the source of many of these sex differences in microbiome composition is proposed to be sex-hormone mediated (135, 136, 141); thus, it is questionable whether such a mechanism would in fact be hormone-independent or explain infant sex-based differences in immunity. Additional study should be done of sex effects on microbiome compositions and any effects this may have on vaccine responses.
Study Strengths and Limitations
The strengths of this study include the comprehensive datasets collected from the 138-subject cohort. These data allowed us to examine the sources and potential mechanisms of sex effects on immunity at a level previously impossible in most studies. Additionally, the WGCNA hierarchical gene clustering algorithm, which clustered genes effectively based on expression levels across our 138 subjects (40) may, in fact, be identifying and incorporating sex differences in gene expression into the creation of the gene clusters, despite WGCNA being blind to subject sex. For example, the WGCNA algorithm created multiple gene clusters enriched for T cell genes, one of which (cluster #2 described here) showed strong sex-dependent gene expression while others demonstrated no sex dependence in gene expression and are, as a result, not discussed in detail here. Similarly, the introduction of mediation analysis to this study adds a novel and valuable tool to the field of systems vaccinology by allowing us to statistically connect sex differences in cell subset compositions to sex differences in immune gene expression and immune outcomes.
Our study does, nonetheless, have several limitations. The cohort size of 138 subjects remains relatively small for detecting subtle differences between populations. While our Caucasian cohort allows for excellent male/female comparisons, generalizability to other populations would require further work. Pre-existing immunity is also a potential confounding factor in all influenza vaccine studies. Invariably, some individuals have high levels of pre-existing cellular and/or antibody responses and paradoxically appear not to respond well to vaccination. This may reflect high levels of pre-existing immunity rather than an inability to respond to the vaccine (37). While others have created elaborate algorithms to deal with pre-existing immunity in influenza vaccine transcriptomic studies (142), we previously demonstrated that the WGCNA gene clustering algorithm does not require normalization to baseline expression levels, thus avoiding the need to correct for pre-existing immunity using complex methods (50). Finally, this study was conducted using a seasonal influenza vaccine containing influenza A/H1N1, influenza A/H3N2, and influenza B. While the presented PBMC gene expression data reflect responses to vaccination with all three influenza strains, only influenza A/H1N1-specific neutralizing antibody and memory B-cell ELISPOT assays were conducted due to costs and practical limitations. With additional funding, further studies examining sex differences in ELISPOT and antibody measures of specific responses to the A/H3N2 and influenza B vaccine strains may be performed in the future.
Summary
We demonstrate in this study that sexually dimorphic components of the immune responses to seasonal influenza vaccination include differences in immune cell populations; expression levels of gene clusters related to T cell, B cell, and NK cell activity; and memory B cell activity as measured by B-cell ELISPOT. Higher memory B cell activity after vaccination was identified in females relative to males—a result correlated to, and statistically mediated by, the expression levels of a small cluster of 135 B cell-related genes. The biological source of these genes' sex-differential expression is unknown, but may be related to human sex hormones and their myriad effects on B cells. This study identifies key genes mediating these effects for future study of the underlying mechanisms. Sex differential responses to seasonal influenza vaccine may impact vaccine efficacy across populations, particularly for older adults with weaker immune responses. Knowledge gained by this and future studies may lead to the development of better vaccines to optimally induce protective immunity in individuals of both sexes, and to the possibility of individualized vaccine practice (143, 144) utilizing adjuvants and other mechanisms.
Author Contributions
All authors were involved in study conceptualization and reviewed, edited the manuscript. EV and DG prepared the original draft of the manuscript. RK and IO supervised the laboratory work. EV, DG, KG, and DS designed the methodology used for analysis, and KG, DG, and EV performed the formal analysis. GP and IO supervised the research efforts. KG and EV prepared the figures.
Conflict of Interest Statement
GP is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories. GP offers consultative advice on vaccine development to Merck & Co. Inc., Avianax, Adjuvance, Valneva, Medicago, Sanofi Pasteur, GlaxoSmithKline, and Emergent Biosolutions. RK has received funding from Merck Research Laboratories to study waning immunity to mumps vaccine. GP and IO hold patents related to vaccinia and measles peptide research. RK holds a patent related to vaccinia peptide vaccines. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Caroline L. Vitse for her editorial assistance with this manuscript. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number U01AI089859. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research was also supported in part by funding from the Mayo Clinic Women's Health Research Center through their Early Career Investigator Award to EV.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00180/full#supplementary-material
References
1. Centers for Disease Control and Prevention. Estimated Influenza Illnesses, Medical visits, Hospitalizations, and Deaths in the United States–2017–2018 Influenza Season (2018). Available online at: https://www.cdc.gov/flu/about/burden/estimates.htm
2. Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine (2007) 25:5086–96. doi: 10.1016/j.vaccine.2007.03.046
3. Flannery B, Chung JR, Belongia EA, Mclean HQ, Gaglani M, Murthy K, et al. Interim estimates of 2017-18 seasonal influenza vaccine effectiveness - United States, february 2018. MMWR (2018) 67:180–5. doi: 10.15585/mmwr.mm6706a2
4. Fang JWS, Lai CL, Chung HT, Wu PC, Lau JYN. Female children respond to recombinant hepatitis B vaccine with a higher titre than male. J Tropical Pediatr. (1994) 40:104–7. doi: 10.1093/tropej/40.2.104
5. Stanberry LR, Spruance SL, Cunningham AL, Bernstein DI, Mindel A, Sacks S, et al. Glycoprotein-D-adjuvant vaccine to prevent genital herpes. New Engl J Med. (2002) 347:1652–61. doi: 10.1056/NEJMoa011915
6. Kanesa-Thasan N, Sun W, Ludwig GV, Rossi C, Putnak JR, Mangiafico JA, et al. Atypical antibody responses in dengue vaccine recipients. Am J Trop Med Hygiene (2003) 69:32–8. doi: 10.4269/ajtmh.2003.69.32
7. Cook IF, Barr I, Hartel G, Pond D, Hampson AW. Reactogenicity and immunogenicity of an inactivated influenza vaccine administered by intramuscular or subcutaneous injection in elderly adults. Vaccine (2006) 24:2395–402. doi: 10.1016/j.vaccine.2005.11.057
8. Couch RB, Winokur P, Brady R, Belshe R, Chen WH, Cate TR, et al. Safety and immunogenicity of a high dosage trivalent influenza vaccine among elderly subjects. Vaccine (2007) 25:7656–63. doi: 10.1016/j.vaccine.2007.08.042
9. Engler RJ, Nelson MR, Klote MM, Vanraden MJ, Huang CY, Cox NJ, et al. Half- vs full-dose trivalent inactivated influenza vaccine (2004-2005): age, dose, and sex effects on immune responses. Arch Intern Med. (2008) 168:2405–14. doi: 10.1001/archinternmed.2008.513
10. Kennedy RB, Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Ryan MA, et al. Gender effects on humoral immune responses to smallpox vaccine. Vaccine (2009) 27:3319–23. doi: 10.1016/j.vaccine.2009.01.086
11. Veit O, Niedrig M, Chapuis-Taillard C, Cavassini M, Mossdorf E, Schmid P, et al. Immunogenicity and safety of yellow fever vaccination for 102 HIV-infected patients. Clin Infect Dis. (2009) 48:659–66. doi: 10.1086/597006
12. Lorenzo ME, Hodgson A, Robinson DP, Kaplan JB, Pekosz A, Klein SL. Antibody responses and cross protection against lethal influenza A viruses differ between the sexes in C57BL/6 mice. Vaccine (2011) 29:9246–55. doi: 10.1016/j.vaccine.2011.09.110
13. Klein SL, Hodgson A, Robinson DP. Mechanisms of sex disparities in influenza pathogenesis. J Leuk Biol. (2012) 92:67–73. doi: 10.1189/jlb.0811427
14. Furman D, Hejblum BP, Simon N, Jojic V, Dekker CL, Thiebaut R, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc Natl Acad Sci USA. (2014) 111:869–74. doi: 10.1073/pnas.1321060111
15. Fink AL, Engle K, Ursin RL, Tang WY, Klein SL. Biological sex affects vaccine efficacy and protection against influenza in mice. Proc Natl Acad Sciences USA. (2018) 115:12477–82. doi: 10.1073/pnas.1805268115
16. Fink AL, Klein SL. The evolution of greater humoral immunity in females than males: implications for vaccine efficacy. Curr Opin Physiol. (2018) 6:16–20. doi: 10.1016/j.cophys.2018.03.010
17. Crighton EJ, Moineddin R, Mamdani M, Upshur RE. Influenza and pneumonia hospitalizations in Ontario: a time-series analysis. Epidemiol Infect. (2004) 132:1167–74. doi: 10.1017/S0950268804002924
18. Crighton EJ, Elliott SJ, Moineddin R, Kanaroglou P, Upshur RE. An exploratory spatial analysis of pneumonia and influenza hospitalizations in Ontario by age and gender. Epidemiol Infect. (2007) 135:253–61. doi: 10.1017/S095026880600690X
19. Vom Steeg LG, Vermillion MS, Hall OJ, Alam O, Mcfarland R, Chen H, et al. Age and testosterone mediate influenza pathogenesis in male mice. Am J Physiol Lung Cell Mol Physiol. (2016) 311:L1234–44. doi: 10.1152/ajplung.00352.2016
20. Vom Steeg LG, Klein SL. Sex and sex steroids impact influenza pathogenesis across the life course. Semin Immunopathol. (2018). doi: 10.1007/s00281-018-0718-5 [Epub ahead of print].
21. Govaert TM, Thijs CT, Masurel N, Sprenger MJ, Dinant GJ, Knottnerus JA. The efficacy of influenza vaccination in elderly individuals. A randomized double-blind placebo-controlled trial. J Amer Med Assoc. (1994) 272:1661–5. doi: 10.1001/jama.1994.03520210045030
22. Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA (2003) 289:179–86. doi: 10.1001/jama.289.2.179
23. Jefferson T, Di PC, Al-Ansary LA, Ferroni E, Thorning S, Thomas RE. Vaccines for preventing influenza in the elderly. Cochrane Database Syst Rev. (2010) 2:CD004876. doi: 10.1002/14651858.CD004876.pub3
24. Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. (2011) 12:36–44. doi: 10.1016/S1473-3099(11)70295-X
25. Lambert ND, Ovsyannikova IG, Pankratz VS, Jacobson RM, Poland GA. Understanding the immune response to seasonal influenza vaccination in older adults: a systems biology approach. Exp Rev Vaccines (2012) 11:985–94. doi: 10.1586/erv.12.61
26. D'mello T, Brammer L, Blanton L, Kniss K, Smith S, Mustaquim D, et al. Update: influenza activity–United States, September 28, 2014-February 21, 2015. MMWR (2015) 64:206–12.
27. Centers for Disease Control and Prevention. 2014-2015 Influenza Season Week 15 Ending April 18, 2015 (2015). Available online at: http://www.cdc.gov/flu/weekly/
28. Klein SL, Jedlicka A, Pekosz A. The Xs and Y of immune responses to viral vaccines. Lancet Infect Dis. (2010) 10:338–49. doi: 10.1016/S1473-3099(10)70049-9
29. Klein SL, Pekosz A. Sex-based biology and the rational design of influenza vaccination strategies. J Infect Dis. (2014) 209(Suppl. 3):S114–9. doi: 10.1093/infdis/jiu066
30. Flanagan KL, Fink AL, Plebanski M, Klein SL. Sex and gender differences in the outcomes of vaccination over the life course. Ann Rev Cell Dev Biol. (2017) 33:577–99. doi: 10.1146/annurev-cellbio-100616-060718
31. Beery AK, Zucker I. Sex bias in neuroscience and biomedical research. Neurosci Biobehavioral Rev. (2011) 35:565–72. doi: 10.1016/j.neubiorev.2010.07.002
32. Klein SL. Immune cells have sex and so should journal articles. Endocrinology (2012) 153:2544–50. doi: 10.1210/en.2011-2120
33. Umlauf BJ, Haralambieva IH, Ovsyannikova IG, Kennedy RB, Pankratz VS, Jacobson RM, et al. Associations between demographic variables and multiple measles-specific innate and cell-mediated immune responses after measles vaccination. Viral Immunol. (2012) 25:29–36. doi: 10.1089/vim.2011.0051
34. Salk HM, Haralambieva IH, Ovsyannikova IG, Goergen KM, Poland GA. Granzyme B ELISPOT assay to measure influenza-specific cellular immunity. J Immunol Methods (2013) 398–399:44–50. doi: 10.1016/j.jim.2013.09.007
35. Painter SD, Haralambieva IH, Ovsyannikova IG, Grill DE, Poland GA. Detection of Influenza A/H1N1-specific human igg-secreting B cells in older adults by ELISPOT assay. Viral Immunology (2014) 27:32–8. doi: 10.1089/vim.2013.0099
36. Haralambieva IH, Painter SD, Kennedy RB, Ovsyannikova IG, Lambert ND, Goergen KM, et al. The impact of immunosenescence on humoral immune response variation after influenza A/H1N1 vaccination in older subjects. PLoS ONE (2015) 10:e0122282. doi: 10.1371/journal.pone.0122282
37. Jacobson RM, Grill DE, Oberg AL, Tosh PK, Ovsyannikova IG, Poland GA. Profiles of influenza A/H1N1 vaccine response using hemagglutination-inhibition titers. Hum Vaccines Immunother. (2015) 11:961–9. doi: 10.1080/21645515.2015.1011990
38. Kennedy RB, Tosh PK, Goergen KM, Grill DE, Oberg AL, Poland GA. Statistical modeling using early markers of innate immunity to explain variation in humoral responses to influenza vaccine in older adults. Vaccine (2015) 33:3682–8. doi: 10.1016/j.vaccine.2015.06.031
39. Fluarix (2010). Highlights of Prescribing Information. Package insert. Available online at: http://id-ea.org/wp-content/uploads/2012/05/Fluarix-Package-Insert.pdf
40. Voigt EA, Grill DE, Zimmermann MT, Simon WL, Ovsyannikova IG, Kennedy RB, et al. Transcriptomic signatures of cellular and humoral immune responses in older adults after seasonal influenza vaccination identified by data-driven clustering. Sci Rep. (2018) 8:739. doi: 10.1038/s41598-017-17735-x
41. Ovsyannikova IG, White SJ, Larrabee BR, Grill DE, Jacobson RM, Poland GA. Leptin and leptin-related gene polymorphisms, obesity, and influenza A/H1N1 vaccine-induced immune responses in older individuals. Vaccine (2014) 32:881–7. doi: 10.1016/j.vaccine.2013.12.009
42. Haralambieva IH, Ovsyannikova IG, Kennedy RB, Zimmermann MT, Grill DE, Oberg AL, et al. Transcriptional signatures of influenza A/H1N1-specific IgG memory-like B cell response in older individuals. Vaccine (2016) 34:3993–4002. doi: 10.1016/j.vaccine.2016.06.034
43. Ovsyannikova IG, Oberg AL, Kennedy RB, Zimmermann MT, Haralambieva IH, Goergen KM, et al. Gene signatures related to HAI response following influenza A/H1N1 vaccine in older individuals. Heliyon (2016) 2:e00098. doi: 10.1016/j.heliyon.2016.e00098
44. Ovsyannikova IG, Salk HM, Kennedy RB, Haralambieva IH, Zimmermann MT, Grill DE, et al. Gene signatures associated with adaptive humoral immunity following seasonal influenza A/H1N1 vaccination. Genes Immun. (2016) 17:371–9. doi: 10.1038/gene.2016.34
45. Kennedy RB, Simon WL, Gibson MJ, Goergen KM, Grill DE, Oberg AL, et al. The composition of immune cells serves as a predictor of adaptive immunity in a cohort of 50- to 74-year-old adults. Immunology (2016) 148:266–75. doi: 10.1111/imm.12599
46. HIPC-CHI Project Team HIPC-I Consortium. Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci Immunol. (2017). 14:eaal4656. doi: 10.1126/sciimmunol.aal4656
47. Webster R, Cox N, Stohr K. WHO Manual on Animal Influenza Diagnosis and Surveillance Geneva: World Health Organization. (2002).
48. Wang S, Taaffe J, Parker C, Solorzano A, Cao H, Garcia-Sastre A, et al. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codon-optimized HA DNA vaccines. J Virol. (2006) 80:11628–37. doi: 10.1128/JVI.01065-06
49. World Health Organization. WHO Global Influenza Surveillance Network: Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza (2011). Available online at: http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf
50. Voigt EA, Haralambieva IH, Larrabee BL, Kennedy RB, Ovsyannikova IG, Schaid DJ, et al. Polymorphisms in the wilms tumor gene are associated with interindividual variations in rubella virus-specific cellular immunity after measles-mumps-rubella II vaccination. J Infect Dis. (2018) 217:560–6. doi: 10.1093/infdis/jix538
51. Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics (2012) 13:204–16. doi: 10.1093/biostatistics/kxr054
52. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics (2008) 9:559. doi: 10.1186/1471-2105-9-559
53. Team RDC. R: A Language for Statistical Computing. Vienna: R Foundation for Statistical Computing (2009).
54. Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Statist Softw. (2012) 46:i11. doi: 10.18637/jss.v046.i11
55. Zimmermann MT. Rapid Integration of Term Annotation and Network Resources (2017). Available online at: https://github.com/MTZimmer/RITAN
56. Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. (2014) 15:195–204. doi: 10.1038/ni.2789
57. Imai K, Keele L, Tingley D. A general approach to causal mediation analysis. Psychol Methods (2010) 15:309–34. doi: 10.1037/a0020761
58. Tingley D, Yamamoto T, Hirose K, Keele L, Imai K. Mediation: Causal Mediation Analysis (2017). Available online at: https://cran.r-project.org/web/packages/mediation/index.html
59. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B (1995) 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
60. Falsey AR, Treanor JJ, Tornieporth N, Capellan J, Gorse GJ. Randomized, double-blind controlled phase 3 trial comparing the immunogenicity of high-dose and standard-dose influenza vaccine in adults 65 years of age and older. J Infect Dis. (2009) 200:172–80. doi: 10.1086/599790
61. Talbot HK, Coleman LA, Zhu Y, Spencer S, Thompson M, Cheng PY, et al. Factors associated with maintenance of antibody responses to influenza vaccine in older, community-dwelling adults. BMC Infect Dis. (2015) 15:195. doi: 10.1186/s12879-015-0926-8
62. Zimmermann MT, Kennedy RB, Grill DE, Oberg AL, Goergen KM, Ovsyannikova IG, et al. Integration of immune cell populations, mRNA-Seq, and CpG methylation to better predict humoral immunity to influenza vaccination: dependence of mRNA-Seq/CpG methylation on immune cell populations. Front Immunol. (2017) 8:445. doi: 10.3389/fimmu.2017.00445
63. Murasko DM, Bernstein ED, Gardner EM, Gross P, Munk G, Dran S, et al. Role of humoral and cell-mediated immunity in protection from influenza disease after immunization of healthy elderly. Exper Gerontol (2002) 37:427–39. doi: 10.1016/S0531-5565(01)00210-8
64. Mcelhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, et al. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. (2006) 176:6333–9. doi: 10.4049/jimmunol.176.10.6333
65. Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. (2012) 18:274–80. doi: 10.1038/nm.2612
66. Galli G, Medini D, Borgogni E, Zedda L, Bardelli M, Malzone C, et al. Adjuvanted H5N1 vaccine induces early CD4+ T cell response that predicts long-term persistence of protective antibody levels. Proc Natl Acad Sci USA. (2009) 106:3877–82. doi: 10.1073/pnas.0813390106
67. Amadori A, Zamarchi R, De Silvestro G, Forza G, Cavatton G, Danieli GA, et al. Genetic control of the CD4/CD8 T-cell ratio in humans. Nat Med. (1995) 1:1279–83. doi: 10.1038/nm1295-1279
68. Wikby A, Mansson IA, Johansson B, Strindhall J, Nilsson SE. The immune risk profile is associated with age and gender: findings from three Swedish population studies of individuals 20-100 years of age. Biogerontology (2008) 9:299–308. doi: 10.1007/s10522-008-9138-6
69. Prabhu SB, Rathore DK, Nair D, Chaudhary A, Raza S, Kanodia P, et al. Comparison of human neonatal and adult blood leukocyte subset composition phenotypes. PLoS ONE (2016) 11:e0162242. doi: 10.1371/journal.pone.0162242
70. Bizzarro A, Valentini G, Di Martino G, Daponte A, De Bellis A, Iacono G. Influence of testosterone therapy on clinical and immunological features of autoimmune diseases associated with Klinefelter's syndrome. J Clin Endocrinol Metab. (1987) 64:32–6. doi: 10.1210/jcem-64-1-32
71. Giglio T, Imro MA, Filaci G, Scudeletti M, Puppo F, De Cecco L, et al. Immune cell circulating subsets are affected by gonadal function. Life Sci. (1994) 54:1305–12. doi: 10.1016/0024-3205(94)00508-7
72. Kocar IH, Yesilova Z, Ozata M, Turan M, Sengul A, Ozdemir I. The effect of testosterone replacement treatment on immunological features of patients with Klinefelter's syndrome. Clin Exper Immunol. (2000) 121:448–52. doi: 10.1046/j.1365-2249.2000.01329.x
73. Porter VR, Greendale GA, Schocken M, Zhu X, Effros RB. Immune effects of hormone replacement therapy in post-menopausal women. Exper Gerontol. (2001) 36:311–26. doi: 10.1016/S0531-5565(00)00195-9
74. Kumru S, Godekmerdan A, Yilmaz B. Immune effects of surgical menopause and estrogen replacement therapy in peri-menopausal women. J Reprod Immunol. (2004) 63:31–8. doi: 10.1016/j.jri.2004.02.001
75. Giltay EJ, Fonk JC, Von Blomberg BM, Drexhage HA, Schalkwijk C, Gooren LJ. In vivo effects of sex steroids on lymphocyte responsiveness and immunoglobulin levels in humans. J Clin Endocrinol Metabol. (2000) 85:1648–57. doi: 10.1210/jcem.85.4.6562
76. Phiel KL, Henderson RA, Adelman SJ, Elloso MM. Differential estrogen receptor gene expression in human peripheral blood mononuclear cell populations. Immunol Lett. (2005) 97:107–13. doi: 10.1016/j.imlet.2004.10.007
77. Michalek RD, Gerriets VA, Nichols AG, Inoue M, Kazmin D, Chang CY, et al. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proc Natl Acad Sci USA. (2011) 108:18348–53. doi: 10.1073/pnas.1108856108
78. Nacka-Aleksic M, Pilipovic I, Stojic-Vukanic Z, Kosec D, Bufan B, Vujnovic I, et al. Sexual dimorphism in the aged rat CD4+ T lymphocyte-mediated immune response elicited by inoculation with spinal cord homogenate. Mech Ageing Dev. (2015) 152:15–31. doi: 10.1016/j.mad.2015.09.004
79. Aggelakopoulou M, Kourepini E, Paschalidis N, Simoes DC, Kalavrizioti D, Dimisianos N, et al. ERbeta-dependent direct suppression of human and murine Th17 cells and treatment of established central nervous system autoimmunity by a neurosteroid. J Immunol. (2016) 197:2598–609. doi: 10.4049/jimmunol.1601038
80. Anipindi VC, Bagri P, Roth K, Dizzell SE, Nguyen PV, Shaler CR, et al. Estradiol enhances CD4+ T-cell anti-viral immunity by priming vaginal DCs to induce Th17 responses via an IL-1-dependent pathway. PLoS Pathog. (2016) 12:e1005589. doi: 10.1371/journal.ppat.1005589
81. Stojic-Vukanic Z, Bufan B, Pilipovic I, Vujnovic I, Nacka-Aleksic M, Petrovic R, et al. Estradiol enhances capacity of TLR-matured splenic dendritic cells to polarize CD4+ lymphocytes into IL-17/GM-CSF-producing cells in vitro. Int Immunopharmacol. (2016) 40:244–53. doi: 10.1016/j.intimp.2016.09.001
82. Mo R, Chen J, Grolleau-Julius A, Murphy HS, Richardson BC, Yung RL. Estrogen regulates CCR gene expression and function in T lymphocytes. J Immunol. (2005) 174:6023–9. doi: 10.4049/jimmunol.174.10.6023
83. Mamrut S, Avidan N, Staun-Ram E, Ginzburg E, Truffault F, Berrih-Aknin S, et al. Integrative analysis of methylome and transcriptome in human blood identifies extensive sex- and immune cell-specific differentially methylated regions. Epigenetics (2015) 10:943–57. doi: 10.1080/15592294.2015.1084462
84. Haynes L, Linton PJ, Eaton SM, Tonkonogy SL, Swain SL. Interleukin 2, but not other common gamma chain-binding cytokines, can reverse the defect in generation of CD4 effector T cells from naive T cells of aged mice. J Exper Med. (1999) 190:1013–24. doi: 10.1084/jem.190.7.1013
85. Gomez I, Hainz U, Jenewein B, Schwaiger S, Wolf AM, Grubeck-Loebenstein B. Changes in the expression of CD31 and CXCR3 in CD4+ naive T cells in elderly persons. Mech Ageing Dev. (2003) 124:395–402. doi: 10.1016/S0047-6374(03)00014-9
86. Eaton SM, Burns EM, Kusser K, Randall TD, Haynes L. Age-related defects in CD4 T cell cognate helper function lead to reductions in humoral responses. J Exper Med. (2004) 200:1613–22. doi: 10.1084/jem.20041395
87. Haynes L, Eaton SM, Burns EM, Rincon M, Swain SL. Inflammatory cytokines overcome age-related defects in CD4 T cell responses in vivo. J Immunol. (2004) 172:5194–9. doi: 10.4049/jimmunol.172.9.5194
88. Kakugawa K, Kojo S, Tanaka H, Seo W, Endo TA, Kitagawa Y, et al. Essential roles of SATB1 in specifying T lymphocyte subsets. Cell Rep. (2017) 19:1176–88. doi: 10.1016/j.celrep.2017.04.038
89. Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nature Immunol. (2017) 18:173–83. doi: 10.1038/ni.3646
90. Hirokawa K, Utsuyama M, Hayashi Y, Kitagawa M, Makinodan T, Fulop T. Slower immune system aging in women versus men in the Japanese population. Immun Ageing (2013) 10:19. doi: 10.1186/1742-4933-10-19
91. Franceschi C, Bonafe M, Valensin S, Olivieri F, De LM, Ottaviani E, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann NY Acad Sci. (2000) 908:244–54. doi: 10.1111/j.1749-6632.2000.tb06651.x
92. Haq K, Mcelhaney JE. Immunosenescence: influenza vaccination and the elderly. Curr Opin Immunol. (2014) 29C:38–42. doi: 10.1016/j.coi.2014.03.008
93. Pawelec G, Goldeck D, Derhovanessian E. Inflammation, ageing and chronic disease. Curr Opin Immunol. (2014) 29:23–8. doi: 10.1016/j.coi.2014.03.007
94. Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity (2015) 43:1186–98. doi: 10.1016/j.immuni.2015.11.012
95. Weinberger B, Herndler-Brandstetter D, Schwanninger A, Weiskopf D, Grubeck-Loebenstein B. Biology of immune responses to vaccines in elderly persons. Clin Infect Dis. (2008) 46:1078–84. doi: 10.1086/529197
96. Schwartz MA, Kolhatkar NS, Thouvenel C, Khim S, Rawlings DJ. CD4+ T cells and CD40 participate in selection and homeostasis of peripheral B cells. J Immunol. (2014) 193:3492–502. doi: 10.4049/jimmunol.1400798
97. Upadhyay M, Priya GK, Ramesh P, Madhavi MB, Rath S, Bal V, et al. CD40 signaling drives B lymphocytes into an intermediate memory-like state, poised between naive and plasma cells. J Cellular Physiol. (2014) 229:1387–96. doi: 10.1002/jcp.24572
98. Jansen R, Batista S, Brooks AI, Tischfield JA, Willemsen G, Van Grootheest G, et al. Sex differences in the human peripheral blood transcriptome. BMC Genomics (2014) 15:33. doi: 10.1186/1471-2164-15-33
99. Engelmann F, Rivera A, Park B, Messerle-Forbes M, Jensen JT, Messaoudi I. Impact of estrogen therapy on lymphocyte homeostasis and the response to seasonal influenza vaccine in post-menopausal women. PLoS ONE (2016) 11:e0149045. doi: 10.1371/journal.pone.0149045
100. Paavonen T, Andersson LC, Adlercreutz H. Sex hormone regulation of in vitro immune response. Estradiol enhances human B cell maturation via inhibition of suppressor T cells in pokeweed mitogen-stimulated cultures. J Exper Med. (1981) 154:1935–45. doi: 10.1084/jem.154.6.1935
101. Sthoeger ZM, Chiorazzi N, Lahita RG. Regulation of the immune response by sex hormones In vitro effects of estradiol and testosterone on pokeweed mitogen-induced human B cell differentiation. J Immunol. (1988) 141:91–8.
102. Verthelyi DI, Ahmed SA. Estrogen increases the number of plasma cells and enhances their autoantibody production in non-autoimmune C57BL/6 mice. Cell Immunol. (1998) 189:125–34. doi: 10.1006/cimm.1998.1372
103. Grimaldi CM, Cleary J, Dagtas AS, Moussai D, Diamond B. Estrogen alters thresholds for B cell apoptosis and activation. J Clin Invest. (2002) 109:1625–33. doi: 10.1172/JCI0214873
104. Altuwaijri S, Chuang KH, Lai KP, Lai JJ, Lin HY, Young FM, et al. Susceptibility to autoimmunity and B cell resistance to apoptosis in mice lacking androgen receptor in B cells. Molec Endocrinol. (2009) 23:444–53. doi: 10.1210/me.2008-0106
105. Hill L, Jeganathan V, Chinnasamy P, Grimaldi C, Diamond B. Differential roles of estrogen receptors alpha and beta in control of B-cell maturation and selection. Molec Med. (2011) 17:211–20. doi: 10.2119/molmed.2010.00172
106. Asaba J, Bandyopadhyay M, Kindy M, Dasgupta S. Estrogen receptor signal in regulation of B cell activation during diverse immune responses. Intl J Biochem Cell Biol. (2015) 68:42–7. doi: 10.1016/j.biocel.2015.08.012
107. Sakiani S, Olsen NJ, Kovacs WJ. Gonadal steroids and humoral immunity. Nat Rev Endocrinol. (2013) 9:56–62. doi: 10.1038/nrendo.2012.206
108. Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, et al. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat Immunol. (2009) 10:540–50. doi: 10.1038/ni.1725
109. Pauklin S, Petersen-Mahrt SK. Progesterone inhibits activation-induced deaminase by binding to the promoter. J Immunol. (2009) 183:1238–44. doi: 10.4049/jimmunol.0803915
110. Pauklin S, Sernandez IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK. Estrogen directly activates AID transcription and function. J Exper Med. (2009) 206:99–111. doi: 10.1084/jem.20080521
111. Mai T, Zan H, Zhang J, Hawkins JS, Xu Z, Casali P. Estrogen receptors bind to and activate the HOXC4/HoxC4 promoter to potentiate HoxC4-mediated activation-induced cytosine deaminase induction, immunoglobulin class switch DNA recombination, and somatic hypermutation. J Biol Chem. (2010) 285:37797–810. doi: 10.1074/jbc.M110.169086
112. Gitlin AD, Von Boehmer L, Gazumyan A, Shulman Z, Oliveira TY, Nussenzweig MC. Independent roles of switching and hypermutation in the development and persistence of b lymphocyte memory. Immunity (2016) 44:769–81. doi: 10.1016/j.immuni.2016.01.011
113. Jones BG, Penkert RR, Xu B, Fan Y, Neale G, Gearhart PJ, et al. Binding of estrogen receptors to switch sites and regulatory elements in the immunoglobulin heavy chain locus of activated B cells suggests a direct influence of estrogen on antibody expression. Molec Immunol. (2016) 77:97–102. doi: 10.1016/j.molimm.2016.07.015
114. Mori H, Sawairi M, Itoh N, Hanabayashi T, Tamaya T. Effects of sex steroids on immunoglobulin M production by Epstein-Barr virus-transformed B-cell line SKW6-CL4. J In Vitro Fert Embryo Transf. (1991) 8:329–35. doi: 10.1007/BF01133023
115. Laffont S, Rouquie N, Azar P, Seillet C, Plumas J, Aspord C, et al. X-Chromosome complement and estrogen receptor signaling independently contribute to the enhanced TLR7-mediated IFN-alpha production of plasmacytoid dendritic cells from women. J Immunol. (2014) 193:5444–52. doi: 10.4049/jimmunol.1303400
116. Berghofer B, Frommer T, Haley G, Fink L, Bein G, Hackstein H. TLR7 ligands induce higher IFN-alpha production in females. J Immunol. (2006) 177:2088–96. doi: 10.4049/jimmunol.177.4.2088
117. Meier A, Chang JJ, Chan ES, Pollard RB, Sidhu HK, Kulkarni S, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. (2009) 15:955–9. doi: 10.1038/nm.2004
118. Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF-kappa B activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J Immunol. (2010) 184:5029–37. doi: 10.4049/jimmunol.0903463
119. Panchanathan R, Shen H, Zhang X, Ho SM, Choubey D. Mutually positive regulatory feedback loop between interferons and estrogen receptor-alpha in mice: implications for sex bias in autoimmunity. PLoS ONE (2010) 5:e10868. doi: 10.1371/journal.pone.0010868
120. Torcia MG, Nencioni L, Clemente AM, Civitelli L, Celestino I, Limongi D, et al. Sex differences in the response to viral infections: TLR8 and TLR9 ligand stimulation induce higher IL10 production in males. PLoS ONE (2012) 7:e39853. doi: 10.1371/journal.pone.0039853
121. Young NA, Wu LC, Burd CJ, Friedman AK, Kaffenberger BH, Rajaram MV, et al. Estrogen modulation of endosome-associated toll-like receptor 8, an IFNalpha-independent mechanism of sex-bias in systemic lupus erythematosus. Clin Immunol. (2014) 151:66–77. doi: 10.1016/j.clim.2014.01.006
122. Jitprasertwong P, Charadram N, Kumphune S, Pongcharoen S, Sirisinha S. Female sex hormones modulate Porphyromonas gingivalis lipopolysaccharide-induced Toll-like receptor signaling in primary human monocytes. J Periodont Res. (2016) 51:395–406. doi: 10.1111/jre.12320
123. Kanda N, Tamaki K. Estrogen enhances immunoglobulin production by human PBMCs. J Allergy Clin Immunol. (1999) 103:282–8. doi: 10.1016/S0091-6749(99)70503-8
124. Adori M, Kiss E, Barad Z, Barabas K, Kiszely E, Schneider A, et al. Estrogen augments the T cell-dependent but not the T-independent immune response. Cellular Molec Life Sci. (2010) 67:1661–74. doi: 10.1007/s00018-010-0270-5
125. Carlsten H, Tarkowski A, Holmdahl R, Nilsson LA. Oestrogen is a potent disease accelerator in SLE-prone MRL lpr/lpr mice. Clin Exper Immunol. (1990) 80:467–73. doi: 10.1111/j.1365-2249.1990.tb03311.x
126. Ansar Ahmed S, Talal N. Importance of sex hormones in systemic lupus erythematosus. In: Wallace DJ, Han BH, editors. Duboi's Lupus Erythematosus. Philadelphia: Lea and Febiger (1993). p. 148–56.
127. Mossong J, O'callaghan CJ, Ratnam S. Modelling antibody response to measles vaccine and subsequent waning of immunity in a low exposure population. Vaccine (2000) 19:523–9. doi: 10.1016/S0264-410X(00)00175-4
128. Stensballe LG, Nante E, Jensen IP, Kofoed PE, Poulsen A, Jensen H, et al. Acute lower respiratory tract infections and respiratory syncytial virus in infants in Guinea-Bissau: a beneficial effect of BCG vaccination for girls community based case-control study. Vaccine (2005) 23:1251–7. doi: 10.1016/j.vaccine.2004.09.006
129. Cook IF. Sexual dimorphism of humoral immunity with human vaccines. Vaccine (2008) 26:3551–5. doi: 10.1016/j.vaccine.2008.04.054
130. Biering-Sorensen S, Aaby P, Napirna BM, Roth A, Ravn H, Rodrigues A, et al. Small randomized trial among low-birth-weight children receiving bacillus Calmette-Guerin vaccination at first health center contact. Pediat Infect Dis J. (2012) 31:306–8. doi: 10.1097/INF.0b013e3182458289
131. Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol. (2010) 10:594–604. doi: 10.1038/nri2815
132. Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays (2011) 33:791–802. doi: 10.1002/bies.201100047
133. Case LK, Wall EH, Dragon JA, Saligrama N, Krementsov DN, Moussawi M, et al. The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res. (2013) 23:1474–85. doi: 10.1101/gr.156703.113
134. Flak MB, Neves JF, Blumberg RS. Immunology. Welcome to the microgenderome. Science (2013) 339:1044–5. doi: 10.1126/science.1236226
135. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science (2013) 339:1084–8. doi: 10.1126/science.1233521
136. Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity (2013) 3:400–12. doi: 10.1016/j.immuni.2013.08.013
137. Brotman RM, Ravel J, Bavoil PM, Gravitt PE, Ghanem KG. Microbiome, sex hormones, and immune responses in the reproductive tract: challenges for vaccine development against sexually transmitted infections. Vaccine (2014) 32:1543–52. doi: 10.1016/j.vaccine.2013.10.010
138. Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol. (2014) 35:507–17. doi: 10.1016/j.it.2014.07.010
139. Shapiro H, Thaiss CA, Levy M, Elinav E. The cross talk between microbiota and the immune system: metabolites take center stage. Curr Opin Immunol. (2014) 30:54–62. doi: 10.1016/j.coi.2014.07.003
140. Vemuri R, Sylvia KE, Klein SL, Forster SC, Plebanski M, Eri R, et al. The microgenderome revealed: sex differences in bidirectional interactions between the microbiota, hormones, immunity and disease susceptibility. Semin Immunopathol. (2018). doi: 10.1007/s00281-018-0716-7 [Epub ahead of print].
141. Vom Steeg LG, Klein SL. sex steroids mediate bidirectional interactions between hosts and microbes. Horm Behav. (2017) 88:45–51. doi: 10.1016/j.yhbeh.2016.10.016
142. Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell (2014) 157:499–513. doi: 10.1016/j.cell.2014.03.031
143. Klein SL, Poland GA. Personalized vaccinology: one size and dose might not fit both sexes. Vaccine (2013) 31:2599–600. doi: 10.1016/j.vaccine.2013.02.070
Keywords: sexual dimorphism, influenza vaccine, influenza, elderly, vaccinomics, systems biology, sex differences, immunity
Citation: Voigt EA, Ovsyannikova IG, Kennedy RB, Grill DE, Goergen KM, Schaid DJ and Poland GA (2019) Sex Differences in Older Adults' Immune Responses to Seasonal Influenza Vaccination. Front. Immunol. 10:180. doi: 10.3389/fimmu.2019.00180
Received: 06 November 2018; Accepted: 21 January 2019;
Published: 27 February 2019.
Edited by:
Rashika El Ridi, Cairo University, EgyptReviewed by:
Randy A. Albrecht, Icahn School of Medicine at Mount Sinai, United StatesGeert Leroux-Roels, Ghent University, Belgium
Raffael Nachbagauer, Icahn School of Medicine at Mount Sinai, United States
Copyright © 2019 Voigt, Ovsyannikova, Kennedy, Grill, Goergen, Schaid and Poland. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory A. Poland, cG9sYW5kLmdyZWdvcnlAbWF5by5lZHU=
 Emily A. Voigt
Emily A. Voigt Inna G. Ovsyannikova
Inna G. Ovsyannikova Richard B. Kennedy
Richard B. Kennedy Diane E. Grill
Diane E. Grill Krista M. Goergen
Krista M. Goergen Daniel J. Schaid
Daniel J. Schaid Gregory A. Poland
Gregory A. Poland