
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 08 February 2019
Sec. Comparative Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.00179
This article is part of the Research Topic Vaccines and Immunostimulants for Finfish View all 21 articles
Toll-like receptors (TLRs) are pattern-recognition receptors that detect a wide variety of microbial pathogens for the initiation of host defense immunological responses. Thirteen TLRs have been identified in mammals, and teleosts contain 22 mammalian or non-mammalian TLRs. Of these, TLR9 and TLR21 are the cytosine-phosphate-guanosine-oligodeoxynucleotides (CpG-ODNs) recognition TLRs in teleosts. TLR9 is a mammalian TLR expressed in teleost but not in the avian species. TLR21 is a non-mammalian TLR expressed in both teleost and the avian species. Synthetic CpG-ODNs are potent immunostimulants that are being studied for their application against tumors, allergies, and infectious diseases, and as a vaccine adjuvant in humans. The immunostimulatory effects of CpG-ODNs as vaccine adjuvants and their antimicrobial function in domestic animals and teleosts are also being investigated. Most of our current knowledge about the molecular basis for the immunostimulatory activity of CpG-ODNs comes from earlier studies of the interaction between CpG-ODN and TLR9. More recent studies indicate that in addition to TLR9, TLR21 is another receptor for CpG-ODN recognition in teleosts to initiate immune responses. Whether these two receptors have differential functions in mediating the immunostimulatory activity of CpG-ODN in teleost has not been well-studied. Nevertheless, the existence of two recognition TLRs suggests that the molecular basis for the immunostimulatory activity of CpG-ODN in teleosts is different and more complex than in mammals. This article reviews the current knowledge of TLR9 and TLR21 activation by CpG-ODNs. The key points that need to be considered for CpG-ODNs as immunostimulants with maximum effectiveness in activation of immune responses in teleosts are discussed. This includes the structure/activity relationship of CpG-ODN activities for TLR9 and TLR21, the structure/functional relationship of these two TLRs, and differential expression levels and tissue distributions for these two TLRs.
Toll was originally identified in Drosophila as a type I transmembrane receptor involved in embryo development, and it plays an important role in innate immune responses to microbial infection in the adult fly (1–3). Thirteen toll-like receptors (TLRs), TLR1 to TLR13 were subsequently identified across all mammalian species, and humans contain ten of them, TLR1 to TLR10 (4–12). Human TLRs are well-investigated. These receptors can be divided into three subfamilies and play an essential role in innate immunity by recognizing a wide variety of pathogen-associated molecular patterns (PAMPs) from microbes (9–12). Phylogenetically, TLR1, TLR2, TLR6, and TLR10 are most closely related. TLR2 recognizes a broad range of microbial components, including lipoproteins, peptidoglycan, lipoteichoic acids, lipoarabinomannan, and zymosan (13–19). TLR2 and TLR6 form a complex that is more specific to triacyl lipopeptides; whereas, a heterodimer composed of TLR2 and TLR1 selectively recognizes triacyl lipopeptides (20–22). Ligand recognition of TLR10 has not been well-investigated; however, a recent paper showed that this TLR is a receptor for double-stranded RNA (dsRNA) (23). TLR4 is closely related to TLR5, with the former being responsible for recognizing lipopolysaccharides on the outer membrane of gram-negative bacteria and the latter recognizing flagellin, which is a component of bacterial flagella (24, 25). TLR3, TLR7, TLR8, and TLR9 comprise a TLR subfamily. These TLRs recognize nucleic acid-derived microbial PAMPs. TLR3 is activated by dsRNA generated during viral replication in infected cells (26). TLR7 and TLR8 recognize single-stranded (ss)RNA from viruses (27, 28). TLR9 is a receptor for microbial unmethylated cytosine-phosphate-guanosine (CpG) DNA (29, 30).
TLRs contain an extracellular domain (ectodomain) comprising multiple leucine-rich repeats (LLRs), a cysteine-rich motif followed by a transmembrane region, and a highly conserved cytoplasmic toll/interleukin (IL)-1 receptor (TIR) domain. The TLR ectodomain is the location of ligand binding, while the cytoplasmic TIR domain provides a key site for intracellular signaling (31, 32). Upon activation by ligand ligation, TLR monomers become dimerized. Their cytosolic domains subsequently recruit adaptor proteins from the myeloid differentiation primary response 88 (MyD88) family. These include MyD88, TIR-domain-containing adapter-inducing interferon-β (TRIF)/TIR domain-containing adapter molecule 1 (TICAM1), TIR domain-containing adapter protein (TIRAP)/MyD88 adapter-like (Mal), toll/interleukin-1 receptor protein (TIRP)/toll-like receptor adaptor molecule (TRAM), and SRAM; thereby, initiating downstream signaling pathways (31). All TLRs, except for TLR3, signal via a MyD88-dependent pathway. TLR3 and TLR4 utilize a TRIF-dependent pathway for signaling. In the MyD88-dependent pathway, a MyD88/IL-1R-associated kinase 1 (IRAK1)/IRAK4/TNFR-associated factor 6 (TRAF6) complex activates transforming growth factor beta-activated kinase 1 (TAK1), which in turn promotes the activation of several transcription factors, including factor kappa-light-chain-enhancer of activated B cells (NF-κB) and activator protein 1 (AP-1). In the TRIF-dependent pathway, the TLR recruits TRIF to activate NF-κB, AP-1, and interferon response factors (IRFs). Activation of NF-κB and AP-1 is mediated by TRAF6 and receptor-interacting protein (RIP), and IRF3/7 activation involves a TBK1-IKKε/IKKi complex (33–35). These transcription factors are key regulators of the expression of adhesion and co-stimulatory molecules and the production of various inflammatory cytokines required for triggering of innate immune responses. This subsequently leads to the activation of adaptive immune responses (36–38).
The immunostimulatory properties of microbial DNA were first discovered in a DNA fraction of bacillus Calmette–Guerin (39, 40). Additional studies have revealed that the immune stimulatory activity is present only when the DNA contains unmethylated CpG deoxynucleotides (41, 42). Synthetic phosphorothioate-modified CpG-ODNs mimic the functions of microbial CpG-deoxynucleotides containing DNA (CpG-DNA). In mammals, CpG-ODNs induce a wide variety of immune responses. Antigen presentation is promoted in dendritic cells because of the increased antigen processing and upregulated expression of costimulatory molecules. Production of inflammatory cytokines from dendritic cells, monocytes, and macrophages are increased. B-lymphocytes are activated, resulting in an increased proliferation and immunoglobulin (Ig) secretion. Natural killer (NK) cells are activated to produce IFN-γ. T-lymphocytes are also affected, resulting in initiation of T-helper (Th)1 responses. Moreover, the generation of cytotoxic T lymphocytes is increased (43–45).
By in vivo studies with gene knockout mice and in vitro studies with cell-based TLR9 activation assay, TLR9 was identified to be the cellular receptor for CpG-ODN (29, 30, 46). In mammals, TLR9 is mainly expressed in dendritic cells, monocytes/macrophages, and B cells (47–49). Activation of TLR9 by CpG-ODN results in several immunological effects, including activation of dendritic cells, monocytes, macrophages, and NK cells leading to antigen presentation and the production of cytokines. In addition, induction of TLR9 activates B cells and increases B-cell proliferation. TLR9 activation upregulates Th1 polarized cytokine productions. Cytokines including TNF-α, IL-6, IL-12, interferons, and chemokines promote T cell activation. These immunologic responses resulted by TLR9 activation replicate the in vivo function of CpG-ODNs further confirmed that TLR9 is the major cellular receptor for CpG-ODN in mammals (45, 50).
Because of these immunostimulatory activities, CpG-ODNs are being investigated for their properties against tumors, allergies, and infectious diseases for humans (50). In the last quarter of 2017, CpG-ODN was approved for the first time for application in humans. Heplisav-B, a hepatitis B vaccine containing CpG-ODN as an adjuvant, was approved by the United States Food and Drug Administration. Two doses of the new vaccine were satisfactory for immunization compared with three doses of the current hepatitis B vaccines that contain aluminum hydroxide as an adjuvant (51, 52). In addition to their application in humans, CpG-ODNs are being investigated for their adjuvant and antimicrobial activities in other species, including domestic animals and teleosts (53–56). These studies reveal the potential usages of CpG-ODNs in human health, agriculture, and aquaculture.
Other than the mammalian TLRs, several non-mammalian TLRs have also been identified in other vertebrate lineages (57, 58). For example, ten TLRs have been identified in avian genomes. Analysis of genomic DNA of two distantly related avian species, chicken and zebra finch, have identified: TLR1La, TLR1Lb, TLR2a, TLR2b, TLR3, TLR4, TLR5, TLR7, TLR15, and TLR21. The avian TLR1La, TLR1Lb, TLR2a, TLR2b, TLR3, TLR4, TLR5, and TLR7 are orthologs to the TLR found in mammals. The TLR1La and TLR1Lb result from duplication of TLR1-like genes, and TLR2a and TLR2b result from the duplication of TLR2 genes in avian evolution (59–63). Mammalian TLR7 and TLR8 have higher homology to each other than other TLRs, which could be due to a duplication of the same gene in some evolutionary duplication event (8). The avian genome contains TLR7 but does not contain TLR8. The mammalian TLR9 and TLR10 are also missing from the avian genomes. TLR15 and TLR21 found in the avian genome do not exist in genomes of mammalian species. TLR15 is phylogenetically related to the TLR2 family and appears to be unique to the avian species. In contrast, the avian TLR21 could be an ortholog to teleost and amphibian TLR21 (57–59). Interestingly, the avian species do not contain TLR9; however, like their actions in mammalian species, CpG-ODNs also activate marked immune responses and provide protection from microbial infections in chickens (50, 55, 64–67). Further studies have revealed that chicken TLR21 is a functional homolog to mammalian TLR9 in terms of responding to CpG-ODN stimulation (68, 69). The chicken TLR21 conferred cellular responses to CpG-ODN stimulation when it was over-expressed in human embryonic kidney (HEK) 293 cells. Knockdown of this receptor by shRNA significantly reduced the CpG-ODN-induced production of IL-1, IL-6, and iNOS from chicken DH11 cells (68, 69).
In teleosts, at least 22 different TLRs have been identified, including both mammalian (TLR1–TLR4, TLR5M, TLR5S, and TLR7–TLR9) and non-mammalian TLRs (TLR13, TLR14, TLR18–TLR28). In addition, orthologs of the mammalian signaling molecules and transcription factors for TLR functions have been identified (57, 58, 70–76). These TLRs are divided into six major subfamilies: TLR1, TLR3, TLR4, TLR5, TLR7, and TLR11 (57, 58).
The structure and ligand recognition properties of fish TLR1-3, 5, and 7–9 are similar to those of their mammalian counterparts. TLR2, a member of the TLR1 family, recognizes peptidoglycan, lipoteichoic acid, and lipopeptides. TLR3 detects dsRNAs. TLR5 recognizes bacterial flagellin. Teleost TLRs 7 and 8 respond to dsRNA, as well as to ssRNA, which is also recognized by mammalian TLR7 and TLR8 (57, 58). In contrast to mammalian TLR4, fish TLR4 does not recognize lipopolysaccharides (LPSs) despite its structural conservation with the former (77). Among non-mammalian TLRs, teleost TLR19 and TLR22 recognize dsRNAs (78–83). The recognition of dsRNAs by TLR19 results in the activation of IFN and NF-κB pathways and the protection of cells from infection by the grass carp reovirus (84). TLR22 recognizes dsRNAs to induce IFN production and protect cells from birnaviruses (83). In addition, a recent study showed that in fish, TLR22 functions as an equalizer for inflammation through the selective suppression of NF-κB and the activation of the MAPK pathway (85).
The immunologic effects of CpG-ODNs have been investigated in numerous teleost species. In these teleosts, much as in mammalian and avian species, CpG-ODNs upregulate the activation of macrophages, induce the proliferation of leukocytes, stimulate cytokine expression, and protect against bacterial, viral, and parasitic infections. Thus, CpG-ODNs have been studied for their application as antimicrobial agents and vaccine adjuvants in teleosts (53, 55). There is interest in the ligand recognition and functional properties of TLR9 and TLR21 in teleosts since these two TLRs have been shown to be the cellular receptors for CpG-ODN in mammals and chickens, respectively. TLR9 and TLR21 from zebrafish (Danio rerio) were comparatively investigated (86). Direct evidence to demonstrate that these two TLRs are the functional cellular receptors for CpG-ODN came from an experiment with cell-based activation assay in which the overexpression of both zebrafish (zeb)TLR9 and zebTLR21 in HEK293 cells conferred cellular responses to CpG-ODN stimulation (86). ZebTLR9 and zebTLR21 have different recognition profiles for CpG-ODNs with different nucleotide sequences. ZebTLR9 broadly recognizes CpG-ODN sequences that have higher activity for human cells and sequences that contain higher activity for mouse cells. In contrast, zebTLR21 prefers the CpG-ODNs that have higher activity for human cells (86). The biological functions of these two TLRs were investigated further in that study. CpG-ODNs that activate both zebTLR9 and zebTLR21 are more potent than others in the activation of cytokine productions in zebrafish and are more effective in protecting teleosts from the lethal effects of bacterial infection (86). These suggest that TLR9 and TLR21 cooperatively mediate the immunostimulatory effect of CpG-ODN in zebrafish. Beside these, the functions of TLR9 and TLR21 in other teleosts have not yet comparatively investigated.
Natural CpG-DNA in microbial genomes contains a phosphodiester backbone that is quickly degraded by nucleases in vivo. Thus, the phosphorothioate backbone was developed to create synthetic CpG-ODNs by replacing oxygen with sulfur in the phosphate group of the nucleic acid to make them more resistant to nucleases (87–89). Other than this, the immunostimulatory activity of CpG-ODN is also dependent on its nucleotide sequence and structure, and it may involve different strengths of activity in different species, known as “species-specific activity.” (90–92). Most of our knowledge about the structure-dependent activity and species-specific activity of CpG-ODNs come from studies of the interaction between CpG-ODNs and mammalian TLR9 (29, 30, 46, 90–92). Because previous studies used human and mouse cells which have TLR9 only, in addition the mammalian TLR9 was identified for investigation earlier than the non-mammalian TLR21 was.
Based on their structural features, CpG-ODNs are divided mainly into four classes. Class A (also known as type D) CpG-ODNs contain a central phosphodiester palindromic region with one or more CpG-motifs in the palindrome and consist of poly (G) sequences with a phosphorothioate backbone attached to the 5′ and 3′ ends. Class B (type K) CpG-ODNs contain a phosphorothiolate backbone throughout the entire sequence with several CpG-motifs. Class C CpG-ODNs contain phosphorothioate backbone with one or two CpG-motifs and a palindromic sequence at the 3′ end. The CpG-ODNs of class P contain two palindromic sequences with phosphodiester cytosines in the palindrome (90, 93–96). Table 1 shows the structures for the four classes of CpG-ODN. Different classes of CpG-ODNs have different immunostimulatory effects. Class A CpG-ODNs stimulate the production of large amounts of IFN-α and induce the maturation of plasmacytoid dendritic cells (pDCs) but have little effect on B-cell activation. Class B CpG-ODNs strongly induce B-cell proliferation, pDC and monocyte maturation, NK cell activation, and cytokine production. They also stimulate the production of IFN-α, but to a lesser extent than class A CpG-ODNs. The extent of the capability of class C CpG-ODNs to induce B-cell proliferation and IFN-α production is between that of class A and B CpG-ODNs. The immunological activities of class P CpG-ODNs are characterized by their high capability for inducing IFN-α production and NF-κB activation. Nearly all CpG-ODNs investigated in clinical trials have been class B CpG-ODNs (90, 93–96).
Another of the major structural features of CpG-ODNs is they include one or more copies of CpG-deoxynucleotide containing hexamer (CpG-hexamer) motifs. The immunostimulatory activity of these CpG-ODNs depends on the number, position, spacing, and surrounding bases of these CpG-hexamer motifs. Their species-specific activity is determined by the nucleotide context of these CpG-hexamer motifs (90–92). For example, CpG-1826, which contains two copies of the GACGTT-hexamer motif in 20 nucleotides, is more effective in activating murine cells than CpG-2007, which contains three copies of the GTCGTT-hexamer motif in 22 nucleotides; however, CpG-2007 is more potent in activating human cells than CpG-1826 (46, 90–92, 97). In addition, the nucleotide length of CpG-ODN plays a significant role in determining its immunostimulatory activity. In rabbit cells, CpG-C46 and CpG-C4609, which each contain 12 nucleotides and have a GACGTT- and AACGTT-hexamer motif, respectively, generate stronger immune responses than CpG-1826 and CpG-2007 (98).
Several CpG-ODNs have been investigated in teleosts for their immunostimulatory activity and antimicrobial functions. There are well-written reviews for these properties of CpG-ODN in earlier works (53, 55). Table 2 summarizes the more recent work. Most of the CpG-ODNs used in these studies are class B. Like in mammals, CpG-ODN nucleotide length determines its immunostimulatory activity in teleosts. In Atlantic salmon (Salmo salar), CpG-ODNs that are 16–17 nucleotides long show less immunostimulatory effects that those that are 20–22 nucleotides long. CpG-ODNs shorter than 13 nucleotides lose their immunostimulatory properties. In addition, CpG-ODNs that are more than 30 nucleotides long have rarely been investigated for their immunologic activity in teleosts (53, 55, 120).
Compared to what is known about the critical role of CpG-hexamer motif in the activity of CpG-ODN in mammalian species, a conclusion has not been reached on what type of CpG-hexamer motif is best for generating a strong immune response in teleosts. CpG-1668, which contains one copy of the GACGTT-hexamer motif in 20 nucleotides, is reported to have immunostimulatory activity, adjuvant effects, and antimicrobial properties in different teleosts, including rock bream (Oplegnathus fasciatus), olive flounder (Paralichthys olivaceus), orange-spotted grouper (Epinephelus coioides), Asian sea bass (Lates calcarifer), and Pacific red snapper (Lutjanus peru) (100–104, 115). Moreover, when fed to Atlantic salmon (Salmo salar), CpG1668 induced the expression of cytokines, such as IL-1β and IL-12β, to protect this teleost fish from infection by sea lice (Lepeophtheirus salmonis), which are the most important ectoparasites that affect the farming of Atlantic salmon (106, 107). When administered to rock bream, CpG-1668 activates stronger protective effects against viral infection than other CpG-ODNs with GTCGTT-hexamer or with the same GACGTT-hexamer motif but with different nucleotide lengths (100). CpG-2006 and CpG2007, which contain three copies of the GTCGTT-hexamer motifs in 24 and 22 nucleotides, respectively, have been shown to induce immune responses in yellowtail (Seriola quinqueradiata), olive flounder, large yellow croaker (Larimichthys crocea), grass carp (Ctenopharyngodon idella), Nile tilapia (Oreochromis niloticus), and Atlantic salmon (108, 109, 112, 113, 116, 121). In olive flounder, CpG-2007 has better protection against Edwardsiella tarda infection than CpG-1668 (112). In grass carp, CpG-1670A, which contains three copies of the AACGTT-hexamer motif in 25 nucleotides, displays a greater capacity to protect teleosts against viral infection than CpG-1668 and CpG-2006 (116).
The ability of teleost TLR9 and TLR21 to uniquely distinguish different types of CpG-hexamer motifs in teleosts may account for the different CpG-ODN sequences that have been reported to participate in the induction of immune responses in different teleost species. The zebTLR9 has been shown to broadly recognize different CpG-hexamer motifs; however, it more strongly recognizes CpG-ODN with the GACGTT- or AACGTT-hexamer motif than CpG-ODN with the GTCGTT-hexamer motif. In contrast, zebTLR21 responds more to CpG-ODN with the GTCGTT-hexamer motif. Further study suggests that CpG-ODNs with an optimized sequence for activating these two TLRs can generate the strongest immunostimulatory activity in this species (86). CpG-ODNs with the GTCGTT-hexamer motif, such as the CpG-2722 and CpG-2727, have strong effects on the TLR21 group like that required for the activation of zebTLR21; in contrast, CpG-1826 with the GACGTT-hexamer motif does not activate this TLR (99). CpG-2006, CpG-2007, and CpG-1826 are reportedly able to activate TLR21 in large yellow croakers (109). The optimized sequence for CpG-ODN to strongly activateTLR9s or TLR21s from other teleosts has not been investigated. Given the large diversity in teleost species, there is not expected to be a universal CpG-ODN sequence for strong activation of TLR9 or TLR21 from different teleost species. This means that the interaction of CpG-ODN with TLR9 or TLR21 from different teleost species must be investigated individually to generate conclusions about how to design a sequence for CpG-ODN with a strong immunostimulatory activity in the teleost species.
Along with the requirement of an optimized nucleotide sequence for CpG-ODN to strongly activate TLR9 and TLR21, whether CpG-ODN can generate a strong immune response in a teleost species is also determined by the intrinsic functional activity of TLR9 and TLR21 in that teleost. Furthermore, although both TLR9 and TLR21 in zebrafish are active in response to CpG-ODN stimulation (86), it is still unclear whether both are functional in other teleost species. Nevertheless, some suggestions can be made from the protein sequences analysis of these two TLRs from different teleost species and the study of the structure/functional activity relationship of mammalian TLR8.
In mammals, TLR7, TLR8, and TLR9 are phylogenetically closely related and are a subfamily of TLRs (5, 8). These three TLRs have an ectodomain in a horseshoe-like shape that consists of 25 copies of LRRs and a unique undefined region (also called a Z-loop) between LRR14 and LRR15 (122, 123), as shown in Figures 1A,B for the ectodomains of TLR9s. This unique undefined region plays an important role in ligand activation of members of this TLR subfamily (124–126). Previous studies have shown that TLR8s from several non-rodent species, including cat, horse, sheep, and bovine, are activated by their agonists; whereas, TLR8s from the mouse and rat, two rodent species, do not respond to ligand stimulation (127). Another study revealed that rabbit TLR8 (also a rodent TLR8) has very little activity after ligand stimulation compared to that of humans (128). Inspection of the ectodomains of these TLR8s reveals that the lengths of amino acid residues within the undefined regions varies between TLR8s from the non-rodent group and those from the rodent group. Compared to non-rodent TLR8s, the undefined regions of mouse and rat TLR8s are shorter by five amino acid residues; whereas, the undefined region of rabbit TLR8 is longer by 34 amino acid residues. Although the structural base is still unclear, it has been suggested that the lesser functional activity of these rodent TLR8s is a result of the varied lengths of their undefined regions (127, 128). Distinct from TLR8, non-functional TLR7 and TLR9 have not been reported in mammalian species. Consistently, the length of the undefined regions in mammalian TLR7s and TLR9s are more conserved than that in the TLR8s (127, 128).
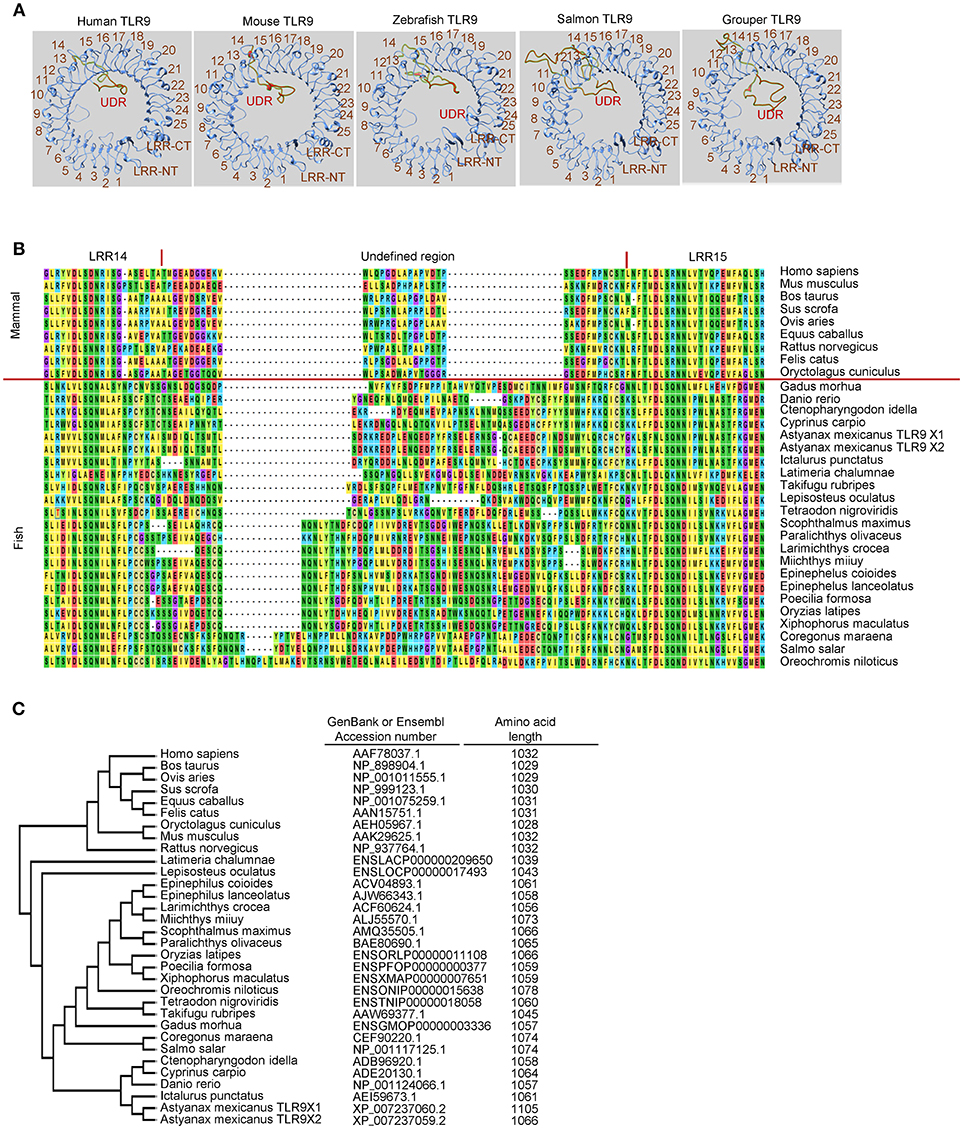
Figure 1. Undefined region of toll-like receptor 9 (TLR9) from different teleost species. (A) Computational modeling of the ectodomain protein structures of TLR9 from different species as indicated. These structural models were predicted with SWISS MODEL (www.swissmodel.expasy.org). (B) Alignment of protein sequences for the undefined regions between leucine-reach repeat (LRR)14 and LRR15 in the ectodomain of TLR9 from different species. Multiple alignments of the amino acid sequences of TLR21s were performed using ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2). (C) Phylogenetic analysis of TLR9s from different species. The GenBank accession numbers of these TLR9 protein sequences are listed in the left column. Numbers in the right column are the amino acid lengths of these TLR9s.
Like mammalian TLR9s, teleost TLR9s also contain an undefined region in their ectodomain, which results in an extruded loop in the horseshoe-shaped ectodomain of these TLRs (Figures 1A,B). Interestingly, there are large variations in the length of undefined regions in teleost TLR9s. The regions in teleost TLR9s are longer than in mammalian TLR9s (Figure 1B). Moreover, the length of these undefined regions is more consistent in the more phylogenetically-related teleost TLR9s than in the more distantly-related teleost TLR9s. For example, TLR9s of zebrafish, grass carp, common carp (Cyprinus carpio), Mexican tetra (Astyanax mexicanus), and channel catfish (Ictalurus punctatus) are more closely phylogenetically related, and the lengths of their undefined regions are more consistent than in the TLR9s of the Atlantic salmon and orange-spotted grouper, which are more distantly related (Figures 1B,C). Given that the undefined regions play a role in the functional activity of TLR8, this structural analysis of undefined regions within TLR9s from different teleosts suggests that there is a large difference in the intrinsic functional activities of TLR9s from different teleost species. Furthermore, because the more phylogenetically-related teleost TLR9s contain more conserved undefined regions, it also suggests that there are more similar functional activities for the more closely related teleost TLR9s.
In contrast, although TLR21 is functionally related to TLR9 in response to CpG-ODN stimulation, TLR21 is more phylogenetically related to members of the TLR11 subfamily and is an ortholog closer to the TLR13 subfamily (129, 130). Analysis of chicken TLR21 revealed that it does not have an undefined region, as in TLR9. In addition, a study of TLR21 proteins from different species shows that these TLR21s are highly homologous (130). The same is true for teleost TLR21s. As Figure 2 illustrates, undefined regions are not found in teleost TLR21s whether the TLR21s are closely related or distantly related to each other; therefore, the highly-diversified ectodomains of TLR9s from different teleost are not observed in teleost TLR21s. In general, the teleost TLR9s contain more than 1,000 amino acid residues, and the teleost TLR21s have < 1,000. A lack of the undefined region in these teleost TLR21s is the main reason why teleost TLR9s contain more amino acid residues than TLR21s (Figures 1C, 2C). The more conserved ectodomains of teleost TLR21s suggest a more stable functional activity of TLR21s within different teleost species. Nevertheless, these suggestions made by structural analyses of the teleost TLR9s and TLR21s are waiting for confirmation by experimental investigation.
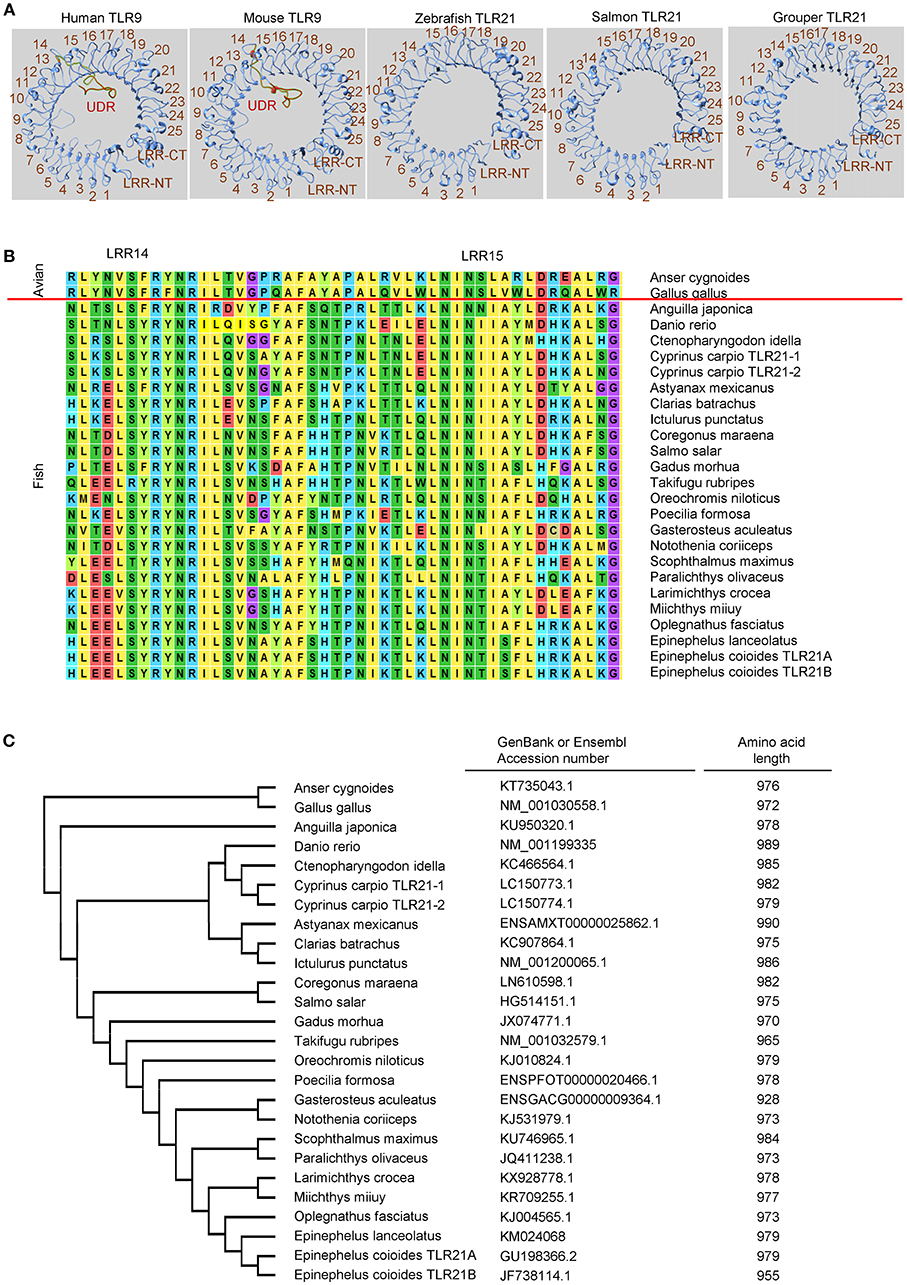
Figure 2. Toll-like receptor 21 (TLR21) from different teleost species does not contain an undefined region. (A) Computational modeling of the ectodomain protein structures of TLR21 from different species as indicated. These structural models were predicted with SWISS MODEL (www.swissmodel.expasy.org). (B) Alignment of protein sequences for the regions from leucine-reach repeat (LRR)14 to LRR15 in the ectodomain of TLR21 from different species. ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2) was used to perform multiple alignments of the amino acid sequences of TLR21s. (C) Phylogenetic analysis of TLR21 from different species. The GenBank accession numbers of these TLR21 protein sequences are listed in the left column. Numbers in the right column are the amino acid lengths of these TLR21s.
In addition to their functional activity, the differential expression levels and tissue distributions of TLR9 and TLR21 are likely to be another level of determinant of CpG-ODN efficacy in teleosts. The expression profile of TLR9 has been investigated in several different species of teleost and has been shown to be broadly expressed in different tissue types and development stages (131–135). In gilthead sea bream (Sparus aurata), the expression levels of TLR9 transcripts are detected in the gill, head kidney, and spleen (119). In channel catfish, TLR9 is expressed in the skin, gill, head kidney, and spleen (135). In addition, TLR9 expression is inducible by responding to different stimuli and microbial infections (105, 135). For example, TLR9 is broadly expressed in larval, juvenile, and adult stages of cobia (Rachycentron canadum) in all analyzed tissues, including the gill, intestine, head kidney, liver, skin, and spleen. Cobia challenged with Photobacterium damselae subsp. piscicida results in increased TLR9 expression in these tissues with different dynamic profiles (105). TLR9 expression in the skin and gills of channel catfish is induced by infection with Ichthyophthirius multifiliis (135).
TLR21 has an expression profile like that of TLR9. In yellow catfish, the TLR21 gene is detected in fertilized eggs and in the young up to 30 days after hatching. In adult fish, this gene is detected in the muscles, stomach, skin, swim bladder, midgut, brain, spleen, trunk kidney, skin mucus, head kidney, liver, heart, gill, and blood, with the highest expression in the spleen. TLR21 mRNA expression levels in the spleen, head kidney, trunk kidney, liver, and blood of yellow catfish are upregulated after challenging the fish with killed Aeromonas hydrophila (136). In turbot (Scophthalmus maximus), TLR21 transcripts are broadly expressed in different tissues, with the highest expression in the spleen followed by the head kidney and liver. In addition, after infection with turbot reddish body iridovirus or stimulation with polyinosinic:polycytidylic acid and CpG-2395, which contain a GTCGTT-hexamer motif within 22 nucleotides, the expression of the turbot TLR21 transcript is upregulated in the gills, head kidney, spleen, and muscle (114). In large yellow croakers, TLR21 is expressed in all tested tissues, with higher levels in immune-related tissues such as the spleen, head kidney, and gills (109). In rock bream, TLR21 transcripts are ubiquitously expressed in different tissues, with higher expression in the spleen followed by the liver and blood. In contrast, the kidney, heart, gill, head kidney, and skin have lower expression levels of these transcripts. In addition, mRNA of the rock bream TLR21 is significantly upregulated in the spleen after stimulation with Streptococcus iniae, rock bream iridovirus, and Edwardsiella tarda (137).
Interestingly, the induction of gene expression in different tissues of cobia by CpG-ODNs is reported to be CpG-ODN-sequence dependent. CpG-1668 and CpG-2006 induce high expression levels of TLR9 in the spleen; whereas, CpG-1668 is more potent in the induction of TLR9 expression in the liver. In the liver and spleen, CpG-1668 and CpG-2006 induce higher expressions of IL-1β and CC chemokines than CpG-2395 and the control CpG-2137; however, in these tissues, CpG-2006 induces high levels of immunoglobulin M (IgM), and CpG-2395 induces high expression levels of Mx (105). The underlying reason for this CpG-ODN sequence- and tissue type-dependent induction of gene expressions is unclear. However, it may reflect that the different expression levels of TLR9 and TLR21 in a tissue type and the ability of TLR9 and TLR21 to differentially recognize different type of CpG-ODN are the main causes for the different activity levels of a CpG-ODN in different tissues.
Most of the knowledge on how to design a nucleotide sequence for CpG-ODN to achieve strong in vivo immunostimulatory activity has come from early studies on mammalian species that express TLR9 only and not TLR21. The discovery of TLR21 as another CpG-ODN receptor in teleosts may explain why previous experience on the activities of various CpG-ODNs in mammals cannot be replicated in teleosts. This also suggests that more understanding of both TLR9 and TLR21 is required for design of CpG-ODN sequence to have strong activity in teleosts. Given that the functional activity of TLR9 and TLR21 may vary among different teleosts, further investigations with cell-based TLR9 and TLR21 activation assays are required to determine whether both TLRs in a teleost are functional or if only one of the two TLRs has the dominant functional activity.
Aquaculture is one of the fastest growing areas of agriculture. The production of farmed teleosts has exceeded that of captured teleosts. Farmed teleosts are susceptible to viral, bacterial, and parasitic infections. Thus, effective immune modulators, including vaccines, and vaccine adjuvants, are required for the aquaculture of farmed teleosts (138–140). CpG-ODN has proven to be an effective adjuvant and antimicrobial agent in teleost (53, 55). The approval of its usage as a vaccine adjuvant in humans (51, 52) further supports its effectiveness and safety as an immunostimulant in agricultural areas, including aquaculture, for food production.
C-YL, G-YY, YL, RX, and T-HC the review results from the opinions and concepts of all authors listed. The review was written by C-YL and T-HC with the help of G-YY, YL, and RX.
This work was supported by grants from the National Health Research Institutes, Taiwan (IM-107-PP-02, NHRI-EX107-10630SI), and Ministry of Science and Technology of Taiwan (MOST 105-2314-B-400-006, and MOST 105-2320-B-400-013-MY3).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell (1996) 86:973–83. doi: 10.1016/S0092-8674(00)80172-5
2. Imler JL, Hoffmann JA. Toll receptors in Drosophila: a family of molecules regulating development and immunity. Curr Top Microbiol Immunol. (2002) 270:63–79. doi: 10.1007/978-3-642-59430-4_4
3. Valanne S, Wang JH, Ramet M. The Drosophila Toll signaling pathway. J Immunol. (2011) 186:649–56. doi: 10.4049/jimmunol.1002302
4. Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA. (1998) 95:588–93. doi: 10.1073/pnas.95.2.588
5. Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. (2000) 11:372–8.
6. Takeuchi O, Kawai T, Sanjo H, Copeland NG, Gilbert DJ, Jenkins NA, et al. TLR6: a novel member of an expanding toll-like receptor family. Gene (1999) 231:59–65. doi: 10.1016/S0378-1119(99)00098-0
7. Du X, Poltorak A, Wei Y, Beutler B. Three novel mammalian toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw. (2000) 11:362–71.
8. Chuang TH, Ulevitch RJ. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta (2001) 1518:157–61. doi: 10.1016/S0167-4781(00)00289-X
9. Medzhitov R, Preston-Hurlburt P, Janeway CA Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature (1997) 388:394–7. doi: 10.1038/41131
10. Beutler B. Innate immune responses to microbial poisons: discovery and function of the Toll-like receptors. Annu Rev Pharmacol Toxicol. (2003) 43:609–28. doi: 10.1146/annurev.pharmtox.43.100901.135729
11. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
12. O'Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. (2013) 13:453–60. doi: 10.1038/nri3446
13. Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science (1999) 285:736–9. doi: 10.1126/science.285.5428.736
14. Coelho PS, Klein A, Talvani A, Coutinho SF, Takeuchi O, Akira S, et al. Glycosylphosphatidylinositol-anchored mucin-like glycoproteins isolated from Trypanosoma cruzi trypomastigotes induce in vivo leukocyte recruitment dependent on MCP-1 production by IFN-gamma-primed-macrophages. J Leukoc Biol. (2002) 71:837–44. doi: 10.1189/jlb.71.5.837
15. Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
16. Massari P, Henneke P, Ho Y, Latz E, Golenbock DT, Wetzler LM. Cutting edge: immune stimulation by neisserial porins is toll-like receptor 2 and MyD88 dependent. J Immunol. (2002) 168:1533–7. doi: 10.4049/jimmunol.168.4.1533
17. Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J Immunol. (1999) 163:3920–7.
18. Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. (1999) 274:17406–9. doi: 10.1074/jbc.274.25.17406
19. Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, et al. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature (1999) 401:811–5. doi: 10.1038/44605
20. Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci USA. (2000) 97:13766–71. doi: 10.1073/pnas.250476497
21. Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. (2001) 13:933–40. doi: 10.1093/intimm/13.7.933
22. Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. (2002) 169:10–4. doi: 10.4049/jimmunol.169.1.10
23. Lee SM, Yip TF, Yan S, Jin DY, Wei HL, Guo RT, et al. Recognition of double-stranded RNA and regulation of interferon pathway by toll-like receptor 10. Front Immunol. (2018) 9:516. doi: 10.3389/fimmu.2018.00516
24. Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science (1998) 282:2085–8. doi: 10.1126/science.282.5396.2085
25. Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature (2001) 410:1099–103. doi: 10.1038/35074106
26. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature (2001) 413:732–8. doi: 10.1038/35099560
27. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (2004) 303:1529–31. doi: 10.1126/science.1093616
28. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science (2004) 303:1526–9. doi: 10.1126/science.1093620
29. Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, et al. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci USA. (2001) 98:9237–42. doi: 10.1073/pnas.161293498
30. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature (2000) 408:740–5. doi: 10.1038/35047123
31. O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. (2007) 7:353–64. doi: 10.1038/nri2079
32. Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity (2008) 29:182–91. doi: 10.1016/j.immuni.2008.07.007
33. Moynagh PN. TLR signalling and activation of IRFs: revisiting old friends from the NF-kappaB pathway. Trends Immunol. (2005) 26:469–76. doi: 10.1016/j.it.2005.06.009
34. Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, et al. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. (2003) 198:1043–55. doi: 10.1084/jem.20031023
35. Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. (2004) 5:503–7. doi: 10.1038/ni1061
36. West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. (2006) 22:409–37. doi: 10.1146/annurev.cellbio.21.122303.115827
37. Kawai T, Akira S. TLR signaling. Semin Immunol. (2007) 19:24–32. doi: 10.1016/j.smim.2006.12.004
38. Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. (2007) 76:447–80. doi: 10.1146/annurev.biochem.76.060605.122847
39. Tokunaga T, Yamamoto H, Shimada S, Abe H, Fukuda T, Fujisawa Y, et al. Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG. I. Isolation, physicochemical characterization, and antitumor activity. J Natl Cancer Inst. (1984) 72:955–62.
40. Shimada S, Yano O, Inoue H, Kuramoto E, Fukuda T, Yamamoto H, et al. Antitumor activity of the DNA fraction from Mycobacterium bovis BCG. II. Effects on various syngeneic mouse tumors. J Natl Cancer Inst. (1985) 74:681–8.
41. Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature (1995) 374:546–9. doi: 10.1038/374546a0
42. Roman M, Martin-Orozco E, Goodman JS, Nguyen MD, Sato Y, Ronaghy A, et al. Immunostimulatory DNA sequences function as T helper-1-promoting adjuvants. Nat Med. (1997) 3:849–54. doi: 10.1038/nm0897-849
43. Ballas ZK, Rasmussen WL, Krieg AM. Induction of NK activity in murine and human cells by CpG motifs in oligodeoxynucleotides and bacterial DNA. J Immunol. (1996) 157:1840–5.
44. Chu RS, Targoni OS, Krieg AM, Lehmann PV, Harding CV. CpG oligodeoxynucleotides act as adjuvants that switch on T helper 1 (Th1) immunity. J Exp Med. (1997) 186:1623–31. doi: 10.1084/jem.186.10.1623
45. Klinman DM, Yi AK, Beaucage SL, Conover J, Krieg AM. CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc Natl Acad Sci USA. (1996) 93:2879–83. doi: 10.1073/pnas.93.7.2879
46. Chuang TH, Lee J, Kline L, Mathison JC, Ulevitch RJ. Toll-like receptor 9 mediates CpG-DNA signaling. J Leukoc Biol. (2002) 71:538–44. doi: 10.1189/jlb.71.3.538
47. Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T, et al. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. (2002) 168:4531–7. doi: 10.4049/jimmunol.168.9.4531
48. Jarrossay D, Napolitani G, Colonna M, Sallusto F, Lanzavecchia A. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur J Immunol. (2001) 31:3388–93. doi: 10.1002/1521-4141(200111)31:11 < 3388::AID-IMMU3388>3.0.CO;2-Q
49. Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, et al. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J Exp Med. (2001) 194:863–9. doi: 10.1084/jem.194.6.863
50. Vollmer J, Krieg AM. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv Drug Deliv Rev. (2009) 61:195–204. doi: 10.1016/j.addr.2008.12.008
51. A Two-Dose Hepatitis B Vaccine for Adults (Heplisav-B). JAMA (2018) 319:822–823. doi: 10.1001/jama.2018.1097
52. Jackson S, Lentino J, Kopp J, Murray L, Ellison W, Rhee M, et al. Immunogenicity of a two-dose investigational hepatitis B vaccine, HBsAg-1018, using a toll-like receptor 9 agonist adjuvant compared with a licensed hepatitis B vaccine in adults. Vaccine (2018) 36:668–674. doi: 10.1016/j.vaccine.2017.12.038
53. Carrington AC, Secombes CJ. A review of CpGs and their relevance to aquaculture. Vet Immunol Immunopathol. (2006) 112:87–101. doi: 10.1016/j.vetimm.2006.03.015
54. Bode C, Zhao G, Steinhagen F, Kinjo T, Klinman DM. CpG DNA as a vaccine adjuvant. Expert Rev Vaccines (2011) 10:499–511. doi: 10.1586/erv.10.174
55. Chaung HC. CpG oligodeoxynucleotides as DNA adjuvants in vertebrates and their applications in immunotherapy. Int Immunopharmacol. (2006) 6:1586–96. doi: 10.1016/j.intimp.2006.06.001
56. Mutwiri G, Pontarollo R, Babiuk S, Griebel P, van Drunen Littel-van den Hurk S, Mena A, et al. Biological activity of immunostimulatory CpG DNA motifs in domestic animals. Vet Immunol Immunopathol. (2003) 91:89–103. doi: 10.1016/S0165-2427(02)00246-5
57. Nie L, Cai SY, Shao JZ, Chen J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol. (2018) 9:1523. doi: 10.3389/fimmu.2018.01523
58. Zhang J, Kong X, Zhou C, Li L, Nie G, Li X. Toll-like receptor recognition of bacteria in fish: ligand specificity and signal pathways. Fish Shellfish Immunol. (2014) 41:380–8. doi: 10.1016/j.fsi.2014.09.022
59. Brownlie R, Allan B. Avian toll-like receptors. Cell Tissue Res. (2011) 343:121–30. doi: 10.1007/s00441-010-1026-0
60. Smith J, Speed D, Law AS, Glass EJ, Burt DW. In-silico identification of chicken immune-related genes. Immunogenetics (2004) 56:122–33. doi: 10.1007/s00251-004-0669-y
61. Yilmaz A, Shen S, Adelson DL, Xavier S, Zhu JJ. Identification and sequence analysis of chicken Toll-like receptors. Immunogenetics (2005) 56:743–53. doi: 10.1007/s00251-004-0740-8
62. Boyd A, Philbin VJ, Smith AL. Conserved and distinct aspects of the avian Toll-like receptor (TLR) system: implications for transmission and control of bird-borne zoonoses. Biochem Soc Trans. (2007) 35:1504–7. doi: 10.1042/BST0351504
63. Temperley ND, Berlin S, Paton IR, Griffin DK, Burt DW. Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics (2008) 9:62. doi: 10.1186/1471-2164-9-62
64. Gupta SK, Deb R, Dey S, Chellappa MM. Toll-like receptor-based adjuvants: enhancing the immune response to vaccines against infectious diseases of chicken. Expert Rev Vaccines (2014) 13:909–25. doi: 10.1586/14760584.2014.920236
65. Gomis S, Babiuk L, Allan B, Willson P, Waters E, Ambrose N, et al. Protection of neonatal chicks against a lethal challenge of Escherichia coli using DNA containing cytosine-phosphodiester-guanine motifs. Avian Dis. (2004) 48:813–22. doi: 10.1637/7194-041204R
66. Patel BA, Gomis S, Dar A, Willson PJ, Babiuk LA, Potter A, et al. Oligodeoxynucleotides containing CpG motifs (CpG-ODN) predominantly induce Th1-type immune response in neonatal chicks. Dev Comp Immunol. (2008) 32:1041–9. doi: 10.1016/j.dci.2008.02.007
67. Haygreen L, Davison F, Kaiser P. DNA vaccines for poultry: the jump from theory to practice. Expert Rev Vaccines (2005) 4:51–62. doi: 10.1586/14760584.4.1.51
68. Brownlie R, Zhu J, Allan B, Mutwiri GK, Babiuk LA, Potter A, et al. Chicken TLR21 acts as a functional homologue to mammalian TLR9 in the recognition of CpG oligodeoxynucleotides. Mol Immunol. (2009) 46:3163–70. doi: 10.1016/j.molimm.2009.06.002
69. Keestra AM, de Zoete MR, Bouwman LI, van Putten JP. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J Immunol. (2010) 185:460–7. doi: 10.4049/jimmunol.0901921
70. Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. (2008) 25:341–50. doi: 10.1016/j.fsi.2008.05.005
71. Meijer AH, Spaink HP. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets (2011) 12:1000–17. doi: 10.2174/138945011795677809
72. Meijer AH, Gabby Krens SF, Medina Rodriguez IA, He S, Bitter W, Ewa Snaar-Jagalska B, et al. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. (2004) 40:773–83. doi: 10.1016/j.molimm.2003.10.003
73. Phelan PE, Mellon MT, Kim CH. Functional characterization of full-length TLR3, IRAK-4, and TRAF6 in zebrafish (Danio rerio). Mol Immunol. (2005) 42:1057–71. doi: 10.1016/j.molimm.2004.11.005
74. Wang J, Zhang Z, Liu J, Li F, Chang F, Fu H, et al. Structural characterization and evolutionary analysis of fish-specific TLR27. Fish Shellfish Immunol. (2015) 45:940–5. doi: 10.1016/j.fsi.2015.06.017
75. Wang Y, Li J, Han J, Shu C, Xu T. Identification and characteristic analysis of TLR28: a novel member of the TLR1 family in teleost. Dev Comp Immunol. (2016) 62:102–7. doi: 10.1016/j.dci.2016.05.001
76. van der Vaart M, Spaink HP, Meijer AH. Pathogen recognition and activation of the innate immune response in zebrafish. Adv Hematol. (2012) 2012:159807. doi: 10.1155/2012/159807
77. Sepulcre MP, Alcaraz-Perez F, Lopez-Munoz A, Roca FJ, Meseguer J, Cayuela ML, et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol. (2009) 182:1836–45. doi: 10.4049/jimmunol.0801755
78. Quiniou SM, Boudinot P, Bengten E. Comprehensive survey and genomic characterization of Toll-like receptors (TLRs) in channel catfish, Ictalurus punctatus: identification of novel fish TLRs. Immunogenetics (2013) 65:511–30. doi: 10.1007/s00251-013-0694-9
79. Munoz I, Sepulcre MP, Meseguer J, Mulero V. Toll-like receptor 22 of gilthead seabream, Sparus aurata: molecular cloning, expression profiles and post-transcriptional regulation. Dev Comp Immunol. (2014) 44:173–9. doi: 10.1016/j.dci.2013.12.003
80. Ding X, Lu DQ, Hou QH, Li SS, Liu XC, Zhang Y, et al. Orange-spotted grouper (Epinephelus coioides) toll-like receptor 22: molecular characterization, expression pattern and pertinent signaling pathways. Fish Shellfish Immunol. (2012) 33:494–503. doi: 10.1016/j.fsi.2012.05.034
81. Lv J, Huang R, Li H, Luo D, Liao L, Zhu Z, et al. Cloning and characterization of the grass carp (Ctenopharyngodon idella) Toll-like receptor 22 gene, a fish-specific gene. Fish Shellfish Immunol. (2012) 32:1022–31. doi: 10.1016/j.fsi.2012.02.024
82. Sundaram AY, Consuegra S, Kiron V, Fernandes JM. Positive selection pressure within teleost Toll-like receptors tlr21 and tlr22 subfamilies and their response to temperature stress and microbial components in zebrafish. Mol Biol Rep. (2012) 39:8965–75. doi: 10.1007/s11033-012-1765-y
83. Matsuo A, Oshiumi H, Tsujita T, Mitani H, Kasai H, Yoshimizu M, et al. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J Immunol. (2008) 181:3474–85. doi: 10.4049/jimmunol.181.5.3474
84. Ji J, Rao Y, Wan Q, Liao Z, Su J. Teleost-Specific TLR19 Localizes to endosome, recognizes dsRNA, recruits TRIF, triggers both IFN and NF-kappaB pathways, and protects cells from grass carp reovirus infection. J Immunol. (2018) 200:573–85. doi: 10.4049/jimmunol.1701149
85. Ding X, Liang Y, Peng W, Li R, Lin H, Zhang Y, et al. Intracellular TLR22 acts as an inflammation equalizer via suppression of NF-kappaB and selective activation of MAPK pathway in fish. Fish Shellfish Immunol. (2018) 72:646–57. doi: 10.1016/j.fsi.2017.11.042
86. Yeh DW, Liu YL, Lo YC, Yuh CH, Yu GY, Lo JF, et al. Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc Natl Acad Sci USA. (2013) 110:20711–6. doi: 10.1073/pnas.1305273110
87. Shaw JP, Kent K, Bird J, Fishback J, Froehler B. Modified deoxyoligonucleotides stable To exonuclease degradation in serum. Nucleic Acids Res. (1991) 19:747–50.
88. Stein CA, Subasinghe C, Shinozuka K, Cohen JS. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. (1988) 16:3209–21. doi: 10.1093/nar/16.8.3209
89. Mutwiri GK, Nichani AK, Babiuk S, Babiuk LA. Strategies for enhancing the immunostimulatory effects of CpG oligodeoxynucleotides. J Control Release (2004) 97:1–17. doi: 10.1016/j.jconrel.2004.02.022
90. Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. (2002) 20:709–60. doi: 10.1146/annurev.immunol.20.100301.064842
91. Pisetsky DS. Mechanisms of immune stimulation by bacterial DNA. Springer Semin Immunopathol. (2000) 22:21–33. doi: 10.1007/s002810000021
92. Yamamoto S, Yamamoto T, Tokunaga T. The discovery of immunostimulatory DNA sequence. Springer Semin Immunopathol. (2000) 22:11–9. doi: 10.1007/s002810000019
93. Gursel M, Verthelyi D, Gursel I, Ishii KJ, Klinman DM. Differential and competitive activation of human immune cells by distinct classes of CpG oligodeoxynucleotide. J Leukoc Biol. (2002) 71:813–20. doi: 10.1189/jlb.71.5.813
94. Vollmer J, Weeratna R, Payette P, Jurk M, Schetter C, Laucht M, et al. Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur J Immunol. (2004) 34:251–62. doi: 10.1002/eji.200324032
95. Samulowitz U, Weber M, Weeratna R, Uhlmann E, Noll B, Krieg AM, et al. A novel class of immune-stimulatory CpG oligodeoxynucleotides unifies high potency in type I interferon induction with preferred structural properties. Oligonucleotides (2010) 20:93–101. doi: 10.1089/oli.2009.0210
96. Hanagata N. Structure-dependent immunostimulatory effect of CpG-oligodeoxynucleotides and their delivery system. Int J Nanomed. (2012) 7:2181–95. doi: 10.2147/IJN.S30197
97. Rankin R, Pontarollo R, Ioannou X, Krieg AM, Hecker R, Babiuk LA, et al. CpG motif identification for veterinary and laboratory species demonstrates that sequence recognition is highly conserved. Antisense Nucleic Acid Drug Dev. (2001) 11:333–40. doi: 10.1089/108729001753231713
98. Chuang TH, Lai CY, Tseng PH, Yuan CJ, Hsu LC. Development of CpG-oligodeoxynucleotides for effective activation of rabbit TLR9 mediated immune responses. PLoS ONE (2014) 9:e108808. doi: 10.1371/journal.pone.0108808
99. Yeh DW, Lai CY, Liu YL, Lu CH, Tseng PH, Yuh CH, et al. CpG-oligodeoxynucleotides developed for grouper toll-like receptor (TLR) 21s effectively activate mouse and human TLR9s mediated immune responses. Sci Rep. (2017) 7:17297. doi: 10.1038/s41598-017-17609-2
100. Jung MH, Jung SJ. CpG ODN 1668 induce innate and adaptive immune responses in rock bream. Fish Shellfish Immunol. (2017) 69:247–57. doi: 10.1016/j.fsi.2017.08.030
101. Jung MH, Lee J, Ortega-Villaizan M, Perez L, Jung SJ. Protective immunity against Megalocytivirus infection in rock bream (Oplegnathus fasciatus) following CpG ODN administration. Vaccine (2017) 35:3691–9. doi: 10.1016/j.vaccine.2017.05.073
102. Nguyen HT, Nguyen T TT, Wang YT, Wang PC, Chen SC. Effectiveness of formalin-killed vaccines containing CpG oligodeoxynucleotide 1668 adjuvants against Vibrio harveyi in orange-spotted grouper. Fish Shellfish Immunol. (2017) 68:124–31. doi: 10.1016/j.fsi.2017.07.018
103. Cardenas-Reyna T, Angulo C, Hori-Oshima S, Velazquez-Lizarraga E, Reyes-Becerril M. B-cell activating CpG ODN 1668 enhance the immune response of Pacific red snapper (Lutjanus peru) exposed to Vibrio parahaemolitycus. Dev Comp Immunol. (2016) 62:72–81. doi: 10.1016/j.dci.2016.04.022
104. Lee EH, Kim KH. CpG-ODN increases resistance of olive flounder (Paralichthys olivaceus) against Philasterides dicentrarchi (Ciliophora: Scuticociliatia) infection. Fish Shellfish Immunol. (2009) 26:29–32. doi: 10.1016/j.fsi.2008.10.001
105. Byadgi O, Puteri D, Lee YH, Lee JW, Cheng TC. Identification and expression analysis of cobia (Rachycentron canadum) Toll-like receptor 9 gene. Fish Shellfish Immunol. (2014) 36:417–27. doi: 10.1016/j.fsi.2013.12.017
106. Purcell SL, Friend SE, Covello J M, Donkin A, Groman DB, Poley J, et al. CpG inclusion in feed reduces sea lice, Lepeophtheirus salmonis, numbers following re-infection. J Fish Dis. (2013) 36:229–40. doi: 10.1111/jfd.12024
107. Covello JM, Friend SE, Purcell SL, Burka JF, Markham R JF, Donkin AW, et al. Effects of orally administered immunostimulants on inflammatory gene expression and sea lice (Lepeophtheirus salmonis) burdens on Atlantic salmon (Salmo salar) Aquaculture (2012) 366–367. doi: 10.1016/j.aquaculture.2012.08.051
108. Angulo C, Alamillo E, Hirono I, Kondo H, Jirapongpairoj W, Perez-Urbiola JC, et al. Class B CpG-ODN2006 is highly associated with IgM and antimicrobial peptide gene expression through TLR9 pathway in yellowtail Seriola lalandi. Fish Shellfish Immunol. (2018) 77:71–82. doi: 10.1016/j.fsi.2018.03.025
109. Sun M, Mu Y, Ding Y, Ao J, Chen X. Molecular and functional characterization of Toll-like receptor 21 in large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. (2016) 59:179–88. doi: 10.1016/j.fsi.2016.10.024
110. Jenberie S, Thim HL, Sunyer JO, Skjodt K, Jensen I, Jorgensen JB. Profiling Atlantic salmon B cell populations: CpG-mediated TLR-ligation enhances IgM secretion and modulates immune gene expression. Sci Rep. (2018) 8:3565. doi: 10.1038/s41598-018-21895-9
111. Iliev DB, Hansen T, Jorgensen SM, Krasnov A, Jorgensen JB. CpG- and LPS-activated MAPK signaling in in vitro cultured salmon (Salmo salar) mononuclear phagocytes. Fish Shellfish Immunol. (2013) 35:1079–85. doi: 10.1016/j.fsi.2013.07.014
112. Cha YJ, Lee CR, Kwon JY, Kang YJ. Protective effects of CpG-ODN 2007 administration against Edwardsiella tarda infection in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. (2017) 68:327–31. doi: 10.1016/j.fsi.2017.07.037
113. Pridgeon JW, Klesius PH, Mu X, Yancey RJ, Kievit MS, Dominowski PJ. Efficacy of QCDCR formulated CpG ODN 2007 in Nile tilapia against Streptococcus iniae and identification of upregulated genes. Vet Immunol Immunopathol. (2012) 145:179–90. doi: 10.1016/j.vetimm.2011.11.001
114. Li S, Wang G, Liu D, Liu Q, Hu G. Cloning and expression analysis of a Toll-like receptor 21 (TLR21) gene from turbot, Scophthalmus maximus. Dev Comp Immunol. (2017) 73:163–8. doi: 10.1016/j.dci.2017.03.021
115. K UT, Chirapongsatonkul N, Itami T, Tantikitti C. CpG ODN mimicking CpG rich region of myxosporean Myxobolus supamattayai stimulates innate immunity in Asian sea bass (Lates calcarifer) and defense against Streptococcus iniae. Fish Shellfish Immunol. (2016) 58:116–24. doi: 10.1016/j.fsi.2016.09.005
116. Su H, Yuan G, Su J. A specific CpG oligodeoxynucleotide induces protective antiviral responses against grass carp reovirus in grass carp Ctenopharyngodon idella. Dev Comp Immunol. (2016) 60:218–27. doi: 10.1016/j.dci.2016.03.007
117. Zhou ZX, Zhang J, Sun L. C7: a CpG oligodeoxynucleotide that induces protective immune response against megalocytivirus in Japanese flounder (Paralichthys olivaceus) via Toll-like receptor 9-mediated signaling pathway. Dev Comp Immunol. (2014) 44:124–32. doi: 10.1016/j.dci.2013.12.002
118. Liu CS, Sun Y, Hu YH, Sun L. Identification and analysis of a CpG motif that protects turbot (Scophthalmus maximus) against bacterial challenge and enhances vaccine-induced specific immunity. Vaccine (2010) 28:4153–61. doi: 10.1016/j.vaccine.2010.04.016
119. Cuesta A, Esteban MA, Meseguer J. The expression profile of TLR9 mRNA and CpG ODNs immunostimulatory actions in the teleost gilthead seabream points to a major role of lymphocytes. Cell Mol Life Sci. (2008) 65:2091–104. doi: 10.1007/s00018-008-8146-7
120. Jorgensen JB, Johansen LH, Steiro K, Johansen A. CpG DNA induces protective antiviral ;immune responses in Atlantic salmon (Salmo salar L.). J Virol. (2003) 77:11471–9. doi: 10.1128/JVI.77.21.11471-11479.2003
121. Strandskog G, Ellingsen T, Jorgensen JB. Characterization of three distinct CpG oligonucleotide classes which differ in ability to induce IFN alpha/beta activity and cell proliferation in Atlantic salmon (Salmo salar L.) leukocytes. Dev Comp Immunol. (2007) 31:39–51. doi: 10.1016/j.dci.2006.05.004
122. Bell JK, Mullen GE, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. (2003) 24:528–33. doi: 10.1016/S1471-4906(03)00242-4
123. Werling D, Jann OC, Offord V, Glass EJ, Coffey TJ. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. (2009) 30:124–30. doi: 10.1016/j.it.2008.12.001
124. Zhang Z, Ohto U, Shimizu T. Toward a structural understanding of nucleic acid-sensing Toll-like receptors in the innate immune system. FEBS Lett. (2017) 591:3167–81. doi: 10.1002/1873-3468.12749
125. Shimizu T. Structural insights into ligand recognition and regulation of nucleic acid-sensing Toll-like receptors. Curr Opin Struct Biol. (2017) 47:52–9. doi: 10.1016/j.sbi.2017.05.010
126. Maeda K, Akira S. TLR7 Structure: Cut in Z-Loop. Immunity (2016) 45:705–7. doi: 10.1016/j.immuni.2016.10.003
127. Liu J, Xu C, Hsu LC, Luo Y, Xiang R, Chuang TH. A five-amino-acid motif in the undefined region of the TLR8 ectodomain is required for species-specific ligand recognition. Mol Immunol. (2010) 47:1083–90. doi: 10.1016/j.molimm.2009.11.003
128. Lai CY, Liu YL, Yu GY, Maa MC, Leu TH, Xu C, et al. TLR7/8 agonists activate a mild immune response in rabbits through TLR8 but not TLR7. Vaccine (2014) 32:5593–9. doi: 10.1016/j.vaccine.2014.07.104
129. Matsushima N, Tanaka T, Enkhbayar P, Mikami T, Taga M, Yamada K, et al. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics (2007) 8:124. doi: 10.1186/1471-2164-8-124
130. Wu H, Wang H, Jiang W, Lian Z. The evolutionary characteristics and structural biology of Gallus toll-like receptor 21. J Mol Recognit. (2018) 6:e2696. doi: 10.1002/jmr.2696
131. Jault C, Pichon L, Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol Immunol. (2004) 40:759–71. doi: 10.1016/j.molimm.2003.10.001
132. Skjaeveland I, Iliev DB, Zou J, Jorgensen T, Jorgensen JB. A TLR9 homolog that is up-regulated by IFN-gamma in Atlantic salmon (Salmo salar). Dev Comp Immunol. (2008) 32:603–7. doi: 10.1016/j.dci.2007.10.011
133. Yu Y, Zhong Q, Li C, Jiang L, Yan F, Wang Z, et al. Isolation and characterization of Toll-like receptor 9 in half-smooth tongue sole Cynoglossus semilaevis. Fish Shellfish Immunol. (2009) 26:492–9. doi: 10.1016/j.fsi.2009.02.005
134. Ortega-Villaizan M, Chico V, Falco A, Perez L, Coll JM, Estepa A. The rainbow trout TLR9 gene and its role in the immune responses elicited by a plasmid encoding the glycoprotein G of the viral haemorrhagic septicaemia rhabdovirus (VHSV). Mol Immunol. (2009) 46:1710–7. doi: 10.1016/j.molimm.2009.02.006
135. Zhao F, Li YW, Pan HJ, Shi CB, Luo XC, Li AX, et al. Expression profiles of toll-like receptors in channel catfish (Ictalurus punctatus) after infection with Ichthyophthirius multifiliis. Fish Shellfish Immunol. (2013) 35:993–7. doi: 10.1016/j.fsi.2013.05.023
136. Wang KL, Ji W, Zhang GR, Wei KJ, Shi ZC, Zhang XT, et al. Molecular characterization and expression analysis of three TLR genes in yellow catfish (Pelteobagrus fulvidraco): responses to stimulation of Aeromonas hydrophila and TLR ligands. Fish Shellfish Immunol. (2017) 66:466–79. doi: 10.1016/j.fsi.2017.05.056
137. Priyathilaka TT, Elvitigala DA, Whang I, Lim BS, Jeong HB, Yeo SY, et al. Molecular characterization and transcriptional analysis of non-mammalian type Toll like receptor (TLR21) from rock bream (Oplegnathus fasciatus). Gene (2014) 553:105–16. doi: 10.1016/j.gene.2014.10.008
138. Tafalla C, Bøgwald J, Dalmo RA. Adjuvants and immunostimulants in fish vaccines: current knowledge and future perspectives. Fish Shellfish Immunol. (2013) 35:1740–50. doi: 10.1016/j.fsi.2013.02.029
139. Dhar AK, Manna SK, Thomas Allnutt FC. Viral vaccines for farmed finfish. Virus Dis. (2014) 25:1–17. doi: 10.1007/s13337-013-0186-4
Keywords: adjuvant, CpG-ODN, immune modulator, innate immunity, toll-like receptor
Citation: Lai C-Y, Yu G-Y, Luo Y, Xiang R and Chuang T-H (2019) Immunostimulatory Activities of CpG-Oligodeoxynucleotides in Teleosts: Toll-Like Receptors 9 and 21. Front. Immunol. 10:179. doi: 10.3389/fimmu.2019.00179
Received: 30 October 2018; Accepted: 21 January 2019;
Published: 08 February 2019.
Edited by:
Irene Salinas, University of New Mexico, United StatesReviewed by:
Kevin R. Maisey, Universidad de Santiago de Chile, ChileCopyright © 2019 Lai, Yu, Luo, Xiang and Chuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsung-Hsien Chuang, dGhjaHVhbmdAbmhyaS5vcmcudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.