
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 29 January 2019
Sec. Comparative Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.00037
This article is part of the Research Topic Vaccines and Immunostimulants for Finfish View all 21 articles
Fish immunology research is at a pivotal point with the increasing availability of functional immunoassays and major advances in omics approaches. However, studies on fish B cells and their distinct subsets remain a challenge due to the limited availability of differentially expressed surface markers. To address this constraint, cell surface proteome of Atlantic salmon IgM+ B cells were analyzed by mass spectrometry and compared to surface proteins detected from two adherent salmon head kidney cell lines, ASK and SSP-9. Out of 21 cluster of differentiation (CD) molecules identified on salmon IgM+ B cells, CD22 and CD79A were shortlisted as potential markers based on the reported B cell-specific surface expression of their mammalian homologs. Subsequent RT-qPCR analyses of flow cytometry-sorted subpopulations from head kidney leukocytes confirmed that both cd22 and cd79a genes were highly expressed in IgM+ lymphoid cells but were observed in barely detectable levels in IgM− non-lymphoid suspension and adherent cells. Similarly, significantly high cd22 and cd79a mRNA levels were observed in IgM+ or IgT+ lymphoid cells from the spleen and peritoneal cavity, but not in their corresponding IgM− IgT− non-lymphoid fractions. This suggests that the B cell restrictive expression of CD22 and CD79A extend down to the transcription level, which was consistent across different lymphoid compartments and immunoglobulin isotypes, thus strongly supporting the potential of CD22 and CD79A as pan-B cell markers for salmon. In addition, this study provides novel information on the salmon B cell surface protein repertoire, as well as insights on B cell evolution. Further investigation of the identified salmon CD molecules, including development of immunological tools for detection, will help advance our understanding of the dynamics of salmon B cell responses such as during infection, vaccination, or immunostimulation.
The sustainability of aquaculture is constantly being challenged by the occurrence and re-occurrence of infectious diseases (1). Fish vaccination has become the main prophylactic strategy against these economically-devastating pathogens. However, unlike many bacterial vaccines that are highly protective, most of the available vaccines against viral pathogens in salmon only provide suboptimal protection (2). It is not clear why the elicited immune responses of fish virus vaccines are not efficient in providing protection against subsequent infection. Consequently, an important question is: What set of host responses constitute protective immunity in salmon? Critical to this host response are the B cells with diverse functional properties that encompass both the innate and adaptive arms of the immune system, including antigen presentation (3), phagocytosis (4), production of natural (5) and antigen-specific antibodies (Abs) (6, 7), and the generation of immunological memory [reviewed in (8)].
Different lineages and subsets of B cells exist, each exhibiting specific phenotypic characteristics that respond differentially to TLR ligands, pathogens, and/or immunogens. In mammals, four subsets that belong either to the B-1 or B-2 lineages have been clearly defined. B-2 cells consist of two subpopulations: the more conventional follicular (FO) B cells that constitute the major subset in the spleen and trigger the formation of germinal centers and the subsequent production of plasmablasts, plasma cells, and memory B cells with high affinity Ag-binding capacities upon activation of T cell dependent (TD) antigens [reviewed in (9)]; and the marginal zone (MZ) B cells that integrate classical innate and adaptive signaling pathways to mount rapid antibody responses, particularly to blood-borne pathogens [reviewed in (10)]. Similar to MZ B cells, B-1 cells, which can be subdivided further into B1a and B1b based on their CD5 surface expression, have important innate functions such as phagocytosis and production of polyreactive natural Abs in a T cell independent (TI) manner (11, 12). The B-1 cells are predominantly located in the peritoneal cavity (13), but are also present in the spleen and other lymphoid organs at very low levels. While initially thought to lack memory B cell generation, recent data have shown that these innate-like B cells also generate memory B cells during TI immune responses [reviewed in (14–16)].
As the first vertebrate group that possesses all elements of adaptive immunity, teleost fish are able to execute immune functions comparable to that of mammals. Although clear differences exist between the structure and organization of the teleost and mammalian immune systems, functional equivalent lymphoid compartments have been reported [reviewed in (17, 18)]. Analogous to the mammalian bone marrow (BM), the teleost head kidney (HK) serves as both the major hematopoietic tissue and reservoir for long-lived plasma cells (6). In the absence of lymph nodes, teleost spleen constitutes as the main secondary lymphoid organ, where the majority of naïve B cells mature and circulate for continuous immune surveillance. In addition to the systemic lymphoid compartments, teleost peritoneal cavity also houses B cells whose development and migration pathways remain largely unexplored (19). In contrast to mammals, however, teleosts lack follicular structures and do not form distinguishable germinal centers (20). Little is still also known about the characteristics of teleost memory B cells but it appears to have a relatively low proliferation potential (6). Moreover, their systemic Ab responses rely on unswitched low-affinity IgM responses (21).
Three classes of immunoglobulins (Igs) have been identified in teleosts: IgM, IgD, and IgT (or IgZ in some species), with IgM+ being the predominant surface Ig isotype (22). IgD is usually co-expressed on the surface of teleost IgM+ B cells, although single positive IgM+ or IgD+ B cells (23) also exist. IgT+ only B cells comprise a separate lineage of fish B cells that appears to have a main role in mucosal immunity (24). Morphological and functional studies suggest that teleost B cells resemble mammalian B1 cells more than B2 cells (25, 26). In fact, it is hypothesized that mammalian innate-like B cells, characterized by high surface IgM expression (27), evolved from fish IgM+ B cells (18), with the B2 lineage emerging later as a more efficient subset that gradually acquired a dominant role in the mammalian adaptive immunity. Fish B cells exhibit both innate and adaptive immune functions (6), but whether these functions are performed by distinct B cell subsets or not is unknown. Specifically, which B cell subpopulation/s and/or lineages play an important role in protection against infection and/or immunity following vaccination are still open questions in fish immunology.
Cluster of differentiation (CD) is a system used for identifying cell surface markers for various leukocyte subpopulations, including B cells. At present, at least 371 CD proteins have been reported in mammals (28)—making immunophenotyping a rather trivial task. In contrast to the mammalian system where different lineages and subtypes of B cells can be identified and sorted with greater clarity through commercially available marker Abs, studying the dynamics of fish B cell responses has been challenging due to lack of pan and subset-specific markers. For Atlantic salmon, in particular, B cells are currently sorted from the total leukocyte population using surface Igs as sole markers, typically via the predominant IgM isotype (22, 29). While this approach has been extremely useful, the binding of Abs to surface Igs could trigger unwanted activation of the BCR, which may interfere with downstream assays. In addition, since the status of surface Ig expression of salmon B cells at various stages of differentiation (i.e., putative naïve B cells, plasmablasts, plasma cells, or memory B cells) is largely unknown, some of these subsets may not be detected during sorting and hence, will be excluded from further analysis.
To address this current limitation, we aimed to identify CD molecules than can be potentially used as pan- or subset-specific B cell markers and, in turn, facilitate molecular, and functional investigations of the heterogeneous salmon B cell population. Additionally, we aimed to profile the salmon B cell surface proteome in order to have a better understanding of the phenotypic characteristics of teleost IgM+ B cells.
Healthy Atlantic salmon (Salmo salar L.) QTL fish strain Aquagen standard (Aquagen, Kyrksæterøra, Norway) were obtained from the Tromsø Aquaculture Research Station (Tromsø, Norway). Fish were kept at 10°C in tanks supplied with running filtered water, natural light and fed on commercial dry feeds (Skretting, Stavanger, Norway). Estimated weight of fish used for isolation of peripheral blood leukocytes (PBL) and subsequent sorting of IgM+ B cells for proteomics analyses was 700–900 g. Head kidney leukocytes (HKL) were collected separately from the same batch of fish. Peritoneal cavity leukocytes (PeL) and splenocytes (SpL) were collected simultaneously from another batch of smaller fish (estimated mean weight: ~60 g).
Atlantic Salmon Kidney (ASK) cells (30) and Salmo salar pronephros 9 (SSP-9) cells (31), derived from the major hematopietic tissue of Atlantic salmon, were grown as monolayers at 20°C in Leibovitz (L-15) medium (Gibco, Life Technologies). ASK cell culture medium was supplemented with P/S (100 units/mL penicillin, 100 μg/mL streptomycin) and 12% fetal bovine serum (FBS), while SSP-9 cell culture medium was supplemented with 50 μg/mL gentamycin and 8% FBS. Five T-75 flasks were seeded with ASK or SSP-9 cells at a density of ~2 × 106 cells per flask and collected after 72 h at 90% confluence for subsequent cell surface protein isolation.
Blood was extracted from the caudal vein of Atlantic salmon using a vacutainer with 68 I.U. sodium heparin (Becton Dickinson) and immediately transferred into transport medium (L-15 with P/S, 2% FBS, and 20 IE/mL heparin). Spleen and HK were aseptically collected into transport medium after ensuring that all blood was drained from fish tissues. Cells from salmon peritoneal cavity were obtained by lavage and immediately stored in transport medium.
Leukocyte isolations (PBL, HKL, SpL, or PeL) were performed on Percoll gradients as described previously (32). Blood suspension was placed directly onto 54% Percoll (GE Healthcare) and centrifuged at 400 × g for 40 min at 4°C. Spleen and HK were homogenized on 100-μm cell strainers (Falcon), loaded onto 25/54% discontinuous Percoll gradients, and centrifuged as above. Similarly, peritoneal cavity cells were loaded onto 25/54% discontinuous Percoll gradient for PeL isolation. Leukocytes at the interface were collected and washed twice in L-15 with P/S before further use.
For stimulation with lipopolysaccharide (LPS), freshly isolated PBLs were seeded in two T25 flasks (Nunclon Delta Surface ThermoFisher Scientific, 6.25 × 106 cells/flask). One flask was treated with 50 μg/mL LPS (purified by Phenol extraction from Escherichia coli O111:B4, Sigma-Aldrich) diluted in Dulbecco's Phosphate Buffered Saline (DPBS; Sigma-Aldrich), while control group received only DPBS. Cells were incubated at 14°C for 72 h before staining, sorting, and surface protein isolation as detailed below.
Total leukocytes were centrifuged at 500 × g, resuspended in PBS+ (Dulbecco PBS with 1% BSA, filter-sterilized), and stained with anti-salmon IgM (IgF1-18) (1:200 dilution) and/or anti-trout IgT (2 μg/mL) monoclonal antibodies (mAbs) for 30 min. These mAbs were generously provided by Dr. Karsten Skjødt and Prof. Oriol Sunyer, respectively. Salmon anti-IgM have been shown to recognize both IgM-A and -B isotypes of Atlantic salmon (29), while trout α-IgT has been previously validated for cross-specificity with Atlantic salmon IgT (22). After two washing steps, leukocytes were incubated with isotype specific secondary Abs: IgG1-RPE (1:400 dilution) and IgG2a-APC (1:400 dilution), respectively, and viability dye FVD780 (1 μL/mL; eBioscience) in PBS+ for 20 min. All staining and centrifugation steps were done at 4°C.
Stained leukocytes were resuspended in PBS+ at 5.0 × 107 cells/mL for sorting using the BD FACS Aria III flow cytometer (BD Biosciences). Dead cells (FVD780+) and doublets (SSC-A vs. SSC-H) were excluded from the population. Remaining cells were sorted on the basis of their forward scatter (FSC) and side scatter (SSC) profiles, and then on their IgM+ (RPE fluorescence emission) and/or IgT+ (APC fluorescence emission) surface expression. FSClow SSClow subpopulation that excludes granulocytes and myeloid cells was designated lymphoid gate. Cells outside this gate were considered “non-lymphoid” (nL). PBLs with surface IgM expression within the lymphoid gate (PBL L IgM+, Figure 1A) were collected in cell culture media and used as samples for surface protein isolation by biotinylation enrichment method.
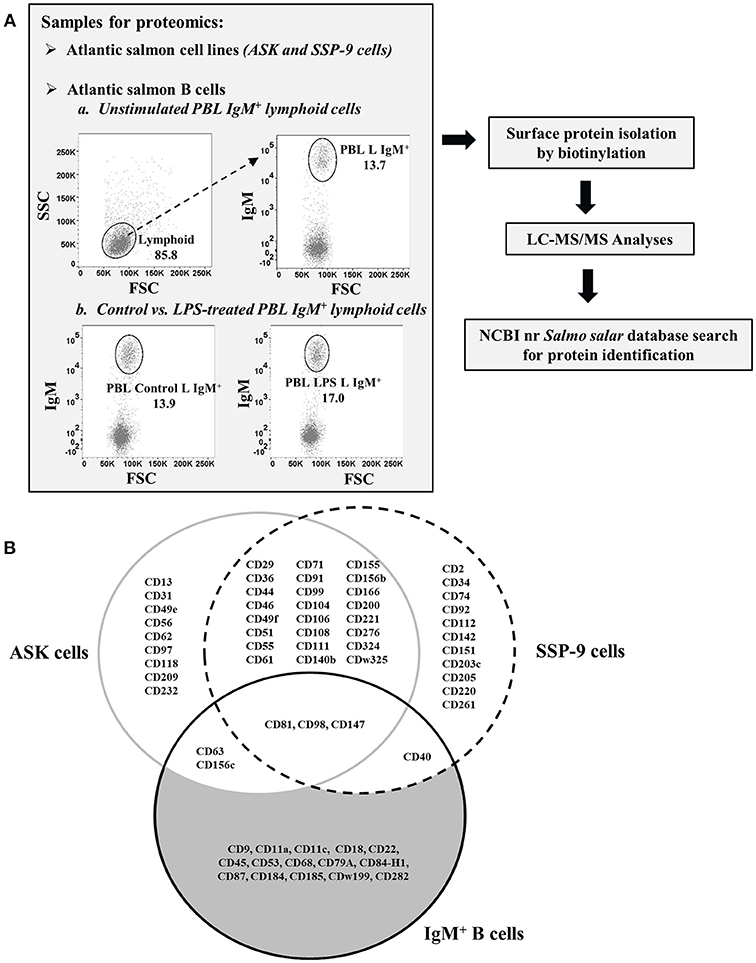
Figure 1. Isolation and identification of CD molecules on surface of Atlantic salmon B cells and head kidney cell lines. (A) Schematic diagram of proteomics workflow performed in the study. (B) Venn diagram comparing the CD molecules detected on cell surface of sorted IgM+ PBLs vs. head kidney cell lines, ASK and SSP-9, of Atlantic salmon.
For the validation of B-cell restrictive expression of candidate pan-B cell markers at the mRNA level, sorted HKL, SpL, and PeL subpopulations were used for RT-qPCR assays. Leukocytes were collected in culture either as suspension (SC) and adherent (AC) cells and stained separately as described above, and then sorted by FACS based on their FSC/SSC gating profile: lymphoid (L) vs. non-lymphoid (cells outside the lymphoid gating, nL) and their IgM and/or IgT surface expression (IgM+ vs. IgM−, IgT+ vs. IgT−). For HKLs, 4 subpopulations were obtained: SC within the lymphoid gate that was either IgM+ (HKL SC-L IgM+) or IgM− (HKL SC-L IgM−); non-lymphoid SC that was IgM− (HKL SC-nL IgM−); and non-lymphoid AC that was IgM− (HKL AC-nL IgM−) (Figure 2A). SpLs, which were mostly suspension cells, were sorted as lymphoid cells with either IgM+ (SpL L IgM+); IgT+ (SpL L IgT+), or IgM−IgT− (SpL L IgM−IgT−) surface expression; and non-lymphoid cells without IgM and IgT surface expression (SpL nL IgM−IgT−) (Figure 3A). Finally, suspension cells from PeLs were sorted as lymphoid cells expressing either IgM (PeL L IgM+) or IgT (PeL L IgT+) on their surface; and non-lymphoid cells without IgM and IgT surface expression (PeL nL IgM−IgT−) (Figure 4A). These HKL, SpL, and PeL subsets were sorted directly on RNAProtect Cell Reagent (Qiagen) and stored at −80°C until RNA extraction.
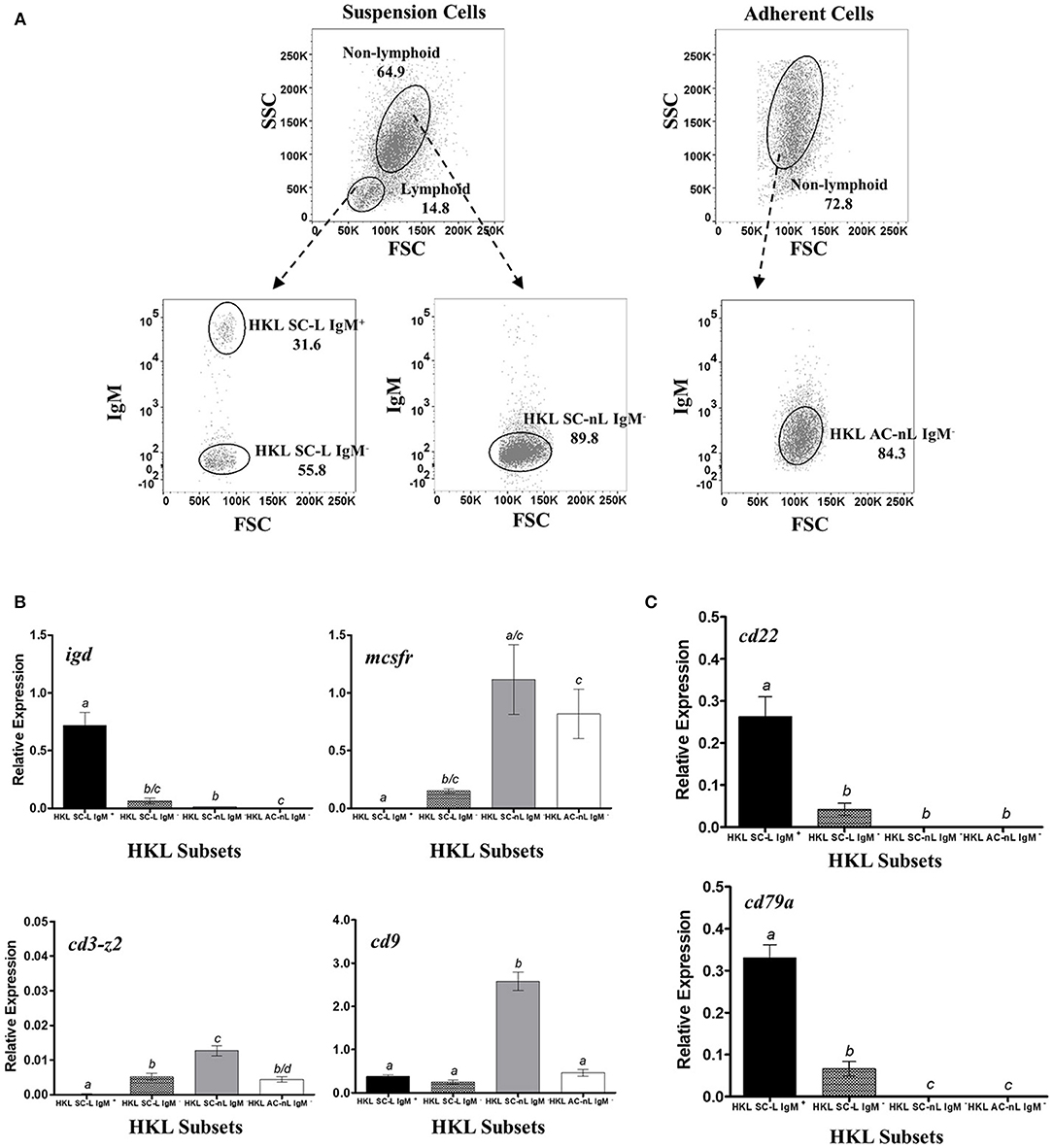
Figure 2. Restrictive gene expression of candidate pan-B cell markers in leukocytes from head kidney, a primary lymphoid organ of salmon. Suspension and adherent head kidney leukocytes (HKLs) were collected separately after 72 h in culture and subsequently sorted by flow cytometry based on cell size and granularity (FSC vs. SSC) and then surface IgM expression. Representative dot plot of the HKL subpopulations are shown in (A) with mean percentage of each fraction indicated in the graph. To ensure purity and quality of the sorted HKL subpopulations, expression of several marker genes: igd (B cell subset), mcsfr (macrophage), cd3-z2 (T cells), and cd9 (broad expression) were examined by RT-qPCR assay (B). Upon validation of sorting protocol, gene expression of cd22 and cd79a genes were subsequently determined (C). Each bar represents mean relative expression data from 3 to 4 fish ± SEM. Means with different letters are significantly different (two-tailed t-test with Welch's correction, p < 0.05).
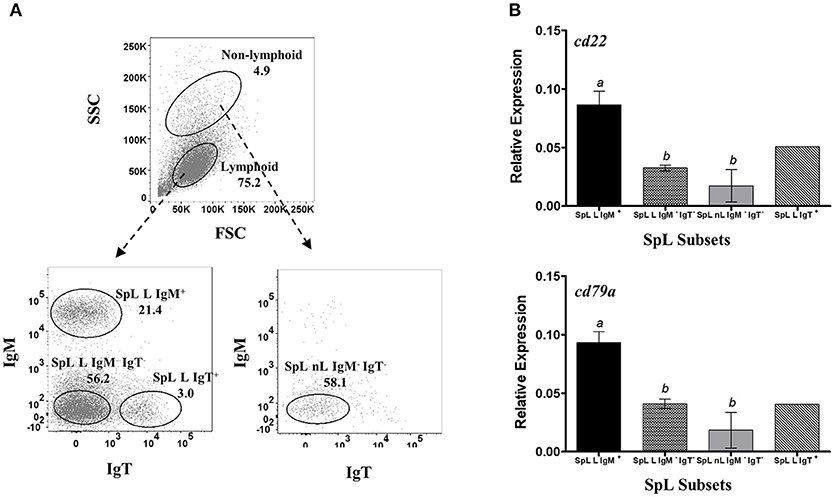
Figure 3. Restrictive gene expression of candidate pan-B cell markers in leukocytes from spleen, a secondary lymphoid organ of salmon. Freshly isolated splenocytes were sorted based on size and granularity (FSC vs. SSC) and then surface expression of IgM or IgT. Representative dot plot of the SpL subpopulations are shown in (A) with mean percentage of each fraction indicated in the graph. Cells within the lymphoid gate were sorted into IgM+, IgT+, or IgM− IgT− subsets (SpL L IgM+, SpL L IgT+, and SpL L IgM− IgT−, respectively). IgM− IgT− cells outside the lymphocyte gate (SpL nL IgM− IgT) was also collected. Expression of cd22 and cd79a genes were subsequently determined in the different splenocyte subpopulations (B). Each bar represents mean relative expression data from three pooled samples (five fish per pooled sample) ± SEM. Means with different letters are significantly different (two-tailed t-test with Welch's correction, p < 0.05). Due to very low cell frequency, sorted IgT data was obtained from a pool of 15 fish.
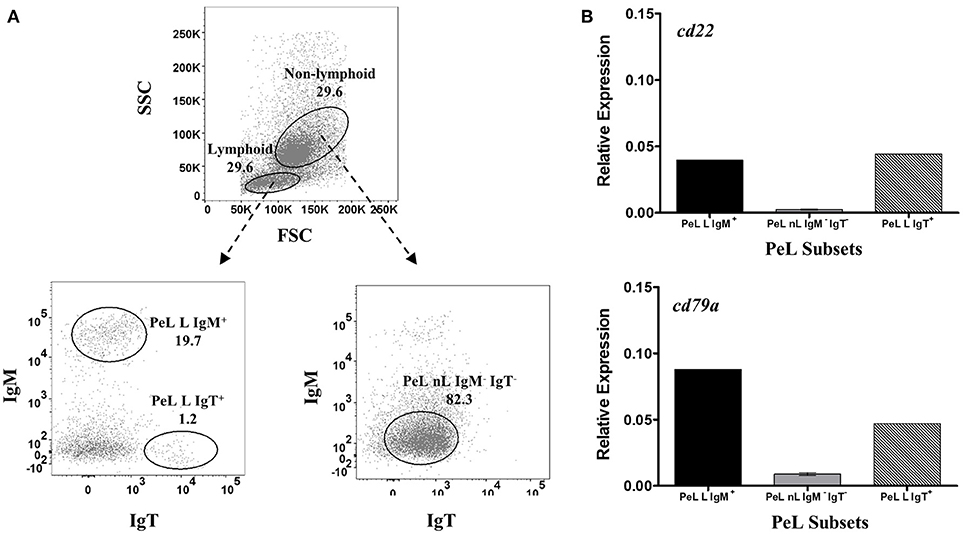
Figure 4. Restrictive gene expression of candidate pan-B cell markers in peritoneal cavity leukocytes of salmon. Freshly isolated peritoneal leukocytes were sorted based on size and granularity (FSC vs. SSC) and then surface expression of IgM or IgT. Representative dot plot of the PeL subpopulations are shown in (A) with mean percentage of each fraction indicated in the graph. PeL subsets included IgM+ or IgT+ cells within the lymphoid gate (PeL L IgM + and PeL L IgT+, respectively) and IgM− IgT− cells within the non-lymphoid gate (PeL nL IgM− IgT−). Expression of cd22 and cd79a genes were investigated in peritoneal leukocyte subpopulations (B). Due to low cell frequency, PeL L IgM+ and PeL L IgT+ bars represent the relative expression data from pooled samples of 15 fish. Relative expression data for PeL nL IgM− IgT− was obtained from the mean of 3 pooled samples (5 fish per pooled sample) ± SEM.
Cell surface proteins from Atlantic salmon cell lines and IgM+ PBLs were isolated using the Pierce® Cell Surface Protein Isolation Kit (Thermo Scientific) according to the manufacturer's protocol. ASK and SSP-9 monolayers (~4 × 107 cells) were quickly washed twice with ice-cold phosphate buffered saline (PBS) followed by incubation with 0.25 mg/mL Sulfo-NHS-SS-Biotin in ice-cold PBS (10 mL biotin solution per flask) on a rocking platform (100 rpm) for 30 min at 4°C. The biotinylation reaction was quenched by adding 500 μL of the provided Quenching Solution. Cells were harvested by gentle scraping, pooled, rinsed with Tris Buffered Saline (TBS), and lysed using the provided Lysis Buffer with Protease Inhibitor Cocktail (Halt™ ThermoFisher Scientific). Cells were sonicated on ice at low power using five 1 s bursts and then incubated for 30 min on ice with intermittent vortexing. The lysates were centrifuged at 10,000 × g for 2 min at 4°C to remove cell remnants and the resulting clarified supernatant was added to 500 μL of NeutrAvidin Agarose slurry. Biotinylated proteins were allowed to bind to the NeutrAvidin by incubating for 1 h at room temperature (RT) in the closed column with end-over-end mixing on an orbital rotator. Unbound proteins were removed by centrifugation of the column at 1,000 × g for 1 min followed by repetitive washing using the provided Wash Buffer with protease inhibitor. Finally, the captured surface proteins were eluted from the biotin-NeutrAvidin Agarose by incubation with 400 μL SDS-PAGE Sample Buffer (62.5 mM Tris-HCl pH 6.8, 1% SDS, 10% glycerol) containing 50 mM dithiothreitol (DTT) for 1 h at RT on an orbital rotator. The eluted proteins, representing the cell surface proteins, were collected by column centrifugation at 1,000 × g for 2 min.
For surface protein isolation in FACS-sorted IgM+ PBLs, cell suspension was centrifuged at 500 × g for 5 min at 4°C, washed twice with 3 mL ice-cold PBS to remove residual FBS, and then resuspended in 0.5 mL ice-cold PBS prior to biotinylation (1 × 106 IgM+ cells/mL biotin solution) as described above.
To maximize the quantity of surface proteins for subsequent mass-spectrometry analysis, eluted proteins were concentrated using the Pierce™ Protein Concentrator PES 3K MWCO (ThermoFisher Scientific) following the manufacturer's protocol.
Surface proteins isolated from the ASK and SSP-9 cell lines, naïve IgM+ PBLs, and control vs. LPS-stimulated IgM+ PBLs were subjected to proteomics analyses. Concentrated surface protein samples were directly lysed in 1 × NuPAGE LDS sample buffer (ThermoFisher Scientific), heated at 70°C for 10 min, and then fractionated by SDS-PAGE for 5 min at 200 V followed by Coomassie blue staining (SimplyBlue SafeStain; Thermo Fisher Scientific). Protein bands were cut and subjected to in-gel trypsin digestion before analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Gel slices were subjected to in-gel reduction, alkylation, and protease digestion with 6 ng/μl trypsin (V5111; Promega) (33). OMIX C18 tips (Varian, Inc., Palo Alto, CA, USA) were used for sample cleanup and concentration. Peptide mixtures containing 0.1% formic acid were loaded onto a Thermo Fisher Scientific EASY-nLC 1,000 system and EASY-spray column (C18; 2 μm; 100 Å; 50 μm; 50 cm). Peptides were fractionated using a 2–100% acetonitrile gradient in 0.1% formic acid over 50 min at a flow rate of 250 nl/min. The separated peptides were analyzed using a ThermoFisher Scientific Q-Exactive mass spectrometer.
Raw files from the QExactive were analyzed using the Proteome Discoverer 2.2 software (Thermo Fisher). The fragmentation spectra was searched against the NCBI non-redundant (nr) Salmo salar 2017_1 database using an in-house Mascot server (Matrix Sciences, UK). Peptide mass tolerances used in the search were 10 ppm, and fragment mass tolerance was 0.02 Da. Peptide ions were filtered using a false discovery rate (FDR) set to 5 % for peptide identifications.
Relative protein quantitations were done using precursor ion intensities in Proteome Discoverer 2.2. To determine the relative protein amount in each sample, Exponentially Modified Protein Abundance Index (emPAI) values were extracted from mascot search results.
Total RNA from sorted cells was extracted using either the RNeasy® Mini (≥500,000 cells) or Micro (< 500,000 cells) Kits (Qiagen), with in-column DNAse I treatment (Qiagen) according to the manufacturer's protocol. For sorted HKL, SpL, and PeL subsets, an extra centrifugation step at 5,000 × g for 10 min was performed to remove the RNAProtect Cell Reagent before proceeding to RNA extraction of the cell pellet. RNA yield and purity were determined using Nanodrop ND-1,000 Spectrophotometer (Nanodrop Tec. Wilmington, DA, USA) and stored at −80°C. Isolated RNA (75–150 ng) was reverse transcribed into cDNAs in 20 μl reaction volumes using the QuantiTect Reverse Transription Kit (Qiagen) following the manufacturer's protocol. Resulting cDNA was diluted 1:5 and stored at −20°C until further use.
Expression levels of RNA transcripts of selected genes were analyzed by RT-qPCR on an ABI Prism 7500 FAST Cycler (Applied Biosystems). cDNAs from sorted HKL subpopulations (2.5 ng total cDNA input) was used per qPCR reaction (20 μL final volume) using the Fast SYBR® Green Master Mix (Applied Biosystems). For SpL and PeL subsets, total cDNA input was 1.5 ng. Information for all primers used is listed in Table 1. The efficiency of the amplification was determined for each primer pair using serial 2-fold dilutions of pooled cDNA, and only primer pairs with efficiencies between 1.90 and 2.10 were used. Each sample was measured in duplicate under the following conditions: 95°C for 20 s followed by 40 cycles of 95°C for 3 s and 60°C for 30 s.
The expression of individual genes was normalized to that of Atlantic salmon elongation factor 1αβ (EF1αβ) and presented as relative expression using the 2−ΔCt method, where ΔCt is determined by subtracting the EF-1α value from the target Ct as described previously (35, 36). Negative controls with no template were included in all experiments. A melting curve for each PCR was determined by reading fluorescence every degree between 60 and 95°C to ensure only a single product had been amplified.
Statistical analyses of RT-qPCR data were performed in GraphPad Prism 5.04 using a two-tailed student t-test with Welch's correction when the F test indicated that the variances of both groups differed significantly. The differences between the mean values were considered significant when p ≤ 0.05.
Our mass spectrometry approach was able to identify a combined dataset of 3,140 proteins from our surface protein-enriched salmon B cell and/or HK cell line samples (full list of identified proteins is available at https://doi.org/10.18710/JU3DWE), of which 21% were deemed to be membrane proteins by GO-annotation or transmembrane predictions. This percentage, however, may have been underestimated due to incomplete annotation in some Atlantic salmon proteins. We subsequently focused on surface protein orthologs that were previously reported as part of the CD molecules in mice and/or humans.
A total of 38 and 39 CD proteins were identified from ASK and SSP-9 cells, respectively (Figure 1B). Nine CD proteins were detected only in ASK cells, while 11 CD proteins were detected only in SSP-9 cells. Twenty-four CD proteins were shared between these two cell lines.
In agreement with the established broad expression of CD81 in mammals (37), this multifunctional tetraspanin protein was detected in high quantities in all proteomics samples (Table 2). Similarly, ubiquitously expressed transmembrane glycoproteins (38–40), CD98 and CD147 were detected in salmon ASK, SSP-9, and IgM+ peripheral B cells. CD147 can bind directly to CD98, which associates with integrins, which is in turn involved in cell adhesion, fusion, proliferation, and growth (41). CD98 was more abundant in LPS-stimulated B cells than control (relative abundance > 2.00; Table 2).
The tetraspanin CD63 and transmembrane protein CD156c, also known to be expressed in many cell types (42, 43), were identified in both ASK and B cells. CD63 functions as a transport regulator implicated in intracellular protein trafficking (44). It has been shown to down-regulate CD184 (CXCR4) by serving as a molecular target of the transcriptional repressor Bcl6 (45). CD156c (ADAM10) functions as a molecular scissor that cleaves the extracellular regions of its transmembrane target proteins, which is an important mechanism for the regulation of leukocyte development and function (43).
The transmembrane protein receptor CD40 was detected in both SSP-9 and IgM+ B cells. This member of the TNFR superfamily has been initially characterized on B cells and subsequently found to be expressed on antigen-presenting cells, and many other immune and non-immune cells (46). The binding of CD40 to its CD154 ligand (CD40L) regulates a wide spectrum of cellular processes, including the activation, proliferation, and differentiation of B cells (47). CD40 exhibited the highest increase in salmon B cell surface expression upon stimulation with a TI antigen, with relative abundance value of 8.60 in LPS-treated vs. control B cells (Table 2).
Out of the 21 total CD molecules Identified from salmon IgM+ PBLs, 15 were found to be present in these B cell samples only and were not detected in the two salmon hematopoietic organ-derived cell lines (Figure 1B). The receptor-like protein tyrosine phosphatase, CD45, reported to be one of the most abundant cell surface glycoproteins expressed on mammalian leukocytes (48), was the most abundant protein detected in the salmon B cell samples with a relative quantitation value of 1.79 as estimated by emPAI (Table 2) based on protein coverage by the peptide matches in the database search result (49).
Two surface proteins (CD22 and CD79A) that previously have been used as B cell-exclusive markers in mammals were identified in the salmon B cell samples. CD22 is a transmembrane glycoprotein that belongs to the sialic acid-binding immunoglobulin-like lectins (SIGLEC) family that serves as a BCR co-receptor (50, 51). In mammals, its expression is predominantly exclusive to subsets of mature B cells, with surface expression appearing simultaneously with surface IgD and is subsequently lost on plasma cells (52–54). CD22 is a well-established regulator of innate and adaptive B cell responses in mammals [reviewed in (55)]. One of the main functions of CD22 is to down-regulate the activation threshold of BCR through its association with tyrosine phosphatases and other signaling molecules (51, 56). In addition to BCR signaling, several initial studies in mice have shown that CD22 also regulates TLR signaling and the survival of B cells (55). However, it has been suggested that CD22 likely functions differently between innate-like B-1 and conventional B-2 cells since CD22 is differentially regulated after BCR-mediated and -independent activation in these B cell lineages (57).
CD79A, on the other hand, is an integral membrane protein belonging to the Ig gene superfamily that associates with membrane Ig on B cell surface and, together with CD79B, forms the signal transduction region of the B cell antigen receptor (BCR) complex (58, 59). Due to their importance in B cell development and induction of B cell activation, CD79 proteins are expressed in virtually all subsets of B cells (60), from the very early stages of B cell development to plasma cells (61–65).
Of the four tetraspanin proteins that were identified by proteomics, detectable quantities of CD9 and CD53 were only present in salmon B cells, despite the reported surface expression in different hematopoietic cell types (66, 67) and/or endothelial cells (68) of their mammalian homologs. This protein family consists of four-span transmembrane proteins that have been described as “master organizers” of the plasma membrane (69) and “molecular facilitators” (42) in a variety of biological processes.
CD18 (ITGB2), CD11a (ITGAL), and CD11c (ITGAX) that were detected on salmon B cell surface belong to the β2 integrin family of adhesion and signaling molecules. Integrin β chain subunit CD18 can pair with one of four alpha chain subunits (CD11a, CD11b, CD11c, or CD11d) to form the CD11/CD18 complex that play important roles in the recruitment of immune cells to sites of inflammation, cell–cell contact formation, and regulation of downstream effects on cellular signaling (70). In mammals, these β2 integrin complexes are found exclusively on leukocytes, particularly myeloid cells and NK cells, and to a lower expression level, B and T lymphocytes.
Three chemokine receptors, CD184 (CXCR4), CD185 (CXCR5), and CDw199 (CCR9) were identified in Atlantic salmon B cells. These are G-protein coupled, seven-transmembrane receptors with CD184 and CD185 classified into the C-X-C (alpha) class and CDw199 into the CC (beta) class. CD184 and CD185 are known to have broad expression across many cell types, including lymphocytes, endothelial, epithelial and hematopoietic stem cells (71). Similarly, CDw199 is expressed in different leukocyte subpopulations such as macrophages, dendritic cells, T cells, and B cells (72–75). These chemokine receptors are important in the migration, maturation, and function of B cells (76–78).
In addition to members of the protein families described above, several other surface proteins were found present on salmon B cells. These included CD68, CD84-H1, CD87, and CD282.
CD68, the human homolog of macrosialin, is a highly glycosylated type I transmembrane protein belonging to the lysosomal-associated membrane protein (LAMP) family of glycoproteins (79). While this protein was initially regarded as a macrophage marker (80), CD68 expression on other hematopoietic and non-hematopoietic cell types have been subsequently reported, including B cell lines (81–83).
CD84-H1 (a.k.a. CD2-F10 and SLAM9) belongs to the signaling lymphocyte activation molecule (SLAM) family of cell surface receptors within the immunoglobulin superfamily (84, 85). This protein has been shown to be widely expressed in many immune cells of humans, including B cells (84, 85). However, the exact function of CD84-H1 in B cells is still unclear. In general, SLAM proteins are said to contribute to the generation of long-term humoral immune response (86, 87).
CD87, also called urokinase plasminogen activator receptor (uPAR), is a surface glycoprotein that mediates a wide range of biological processes beyond plasminogen activation, including cellular adhesion, migration, chemotaxis, and proliferation [reviewed in (88, 89)]. In humans, CD87 is known to be highly expressed on monocytes and granulocytes, particularly on mature cells (90). As such, it is used as a surface marker for terminal granulocytic maturation (91). Resting B and T lymphocytes and cell lines of lymphoid lineage do not seem to express CD87 (90, 92), although surface expression has been reported on activated T cells and NK cells (90). Next to CD45 and CD18, salmon CD87 had the highest emPAI obtained in naïve B cells, with relative abundance comparable between control and LPS-activated B cell samples.
Another surface protein detected on salmon B cell surface was CD282, more commonly known as toll-like receptor 2 (TLR2), a membrane-bound protein that recognizes the evolutionarily conserved bacterial lipopeptides (93). In humans, surface expression of CD282 is found to be highest on innate cells such as monocytes and granulocytes (94). Detectable level of CD282 expression has also been observed on activated but not resting T cells (95, 96), as well as on surface of different B cell subpopulations, albeit mostly at low level (94, 97, 98).
Given the known B cell-exclusive surface expression of their mammalian homologs, CD22 and CD79A were shortlisted as potential pan-B cell markers for salmon. In the absence of definitive Abs against these candidate markers, we resorted to qPCR assays for validating their B cell restrictive expression using leukocyte subsets from different lymphoid organs.
Sorted cells from HK (Figure 2A) were used as the main source of different leukocyte subpopulations for the RT-qPCR validation assays. As the major hematopoietic organ in teleost, HK consists of a heterogeneous mixture of leukocytes belonging to the lymphoid and myeloid lineages (99). HKL subsets were first examined for gene expression of known markers for B cells, macrophages, and T cells in order to confirm the nature of cells present in each subset, as well as to ensure that no cross-contamination of cell subsets occurred during FACS sorting.
As expected, only the lymphoid subsets had detectable igd expression, with transcript levels highest in the HK suspension cell IgM+ lymphoid subset (HKL SC-L IgM+, Figure 2B), in accordance with the reported dual IgM and IgD expression in majority of naïve mature peripheral B cells of mammals [reviewed in (100–102)] and trout (103). Significant albeit much lower igd expression was also observed in HK suspension cell IgM− lymphoid subset (HKL SC-L IgM−), which could suggest the presence of IgD+ only B cells in naïve HKLs. By contrast, the suspension and adherent IgM− non-lymphoid subsets (HKL SC-nL IgM− and HKL AC-nL IgM−, respectively), assumed to consist of myeloid cells (macrophages, monocytes, and/or granulocytes), had very low to non-existent igd expression.
The HK suspension cell IgM+ lymphoid subset had no detectable transcription of the macrophage marker, mcsfr, which was expressed in high levels in the putative myeloid cell subsets, HKL SC-nL IgM− and HKL AC-nL IgM−. Detectable levels of mcsfr transcripts was present in the HK suspension cell IgM− lymphoid subset, which could indicate the presence of contaminating macrophages in this leukocyte subpopulation. Similarly, no detectable cd3-z2 gene expression was observed in the HK suspension cell IgM+ lymphoid subset, while the remaining HKL subsets exhibited relatively low expression of this T cell marker.
Additionally, to ensure that any observed absence of detectable transcripts was not due to poor cDNA/RNA quality of our samples, the mRNA levels of the broadly expressed CD9 tetraspanin were also determined in the sorted HKL subpopulations. In general, relative expression of cd9 was comparably high across all HKL subsets, with the HK suspension cell IgM− non-lymphoid subset having the most expression.
The gene expression of our candidate B cell markers was subsequently investigated. HK suspension cell IgM+ lymphoid subset with a putative B cell phenotype (i.e., significant gene expression of igd and cd9, but not mcsfr and cd3-z2) exhibited the highest cd22 and cd79a expression (Figure 2C). By contrast, very low to non-existent mRNA levels were observed in both the suspension and adherent IgM− non-lymphoid subsets (HKL SnL IgM− and HKL AnLIgM−, respectively), consisting mostly of cells from the myeloid lineage and some T lymphocytes as per mcsfr and cd3z gene expression profile. Low but detectable levels of cd22 and cd79a were also observed in the HKL SL IgM− subset, likely due to the presence of IgD+ and IgT+ B cells in this leukocyte subpopulation.
To check whether the apparent B cell restrictive expression of cd22 and cd79a is consistent across different B cells from different lymphoid sources, we also performed the same gene expression assay in leukocyte subsets from spleen (Figure 3A) and peritoneal cavity (Figure 4A). IgT subpopulation was also added in the analysis to determine cd22 and cd79a expression in a non-IgM B cell isotype. Similar to what was observed in HKLs, highest and lowest cd22 and cd79a mRNA levels were obtained in the spleen IgM+ lymphoid (SpL L IgM+) and IgM− non-lymphoid subsets (SpL nL IgM−), respectively (Figure 3B). High mRNA expression for these candidate markers was also found in the spleen IgT+ lymphoid subset (SpL L IgT+) as well as in the IgM− IgT− lymphoid subset (SpL L IgM− IgT−), which could contain IgD only B cells.
For PeLs, comparable levels of cd22 transcription were observed between IgM+ (PeL L IgM+) and IgT+ (PeL L IgT+) lymphoid subsets, while very low cd22 gene expression was obtained in the PeL IgM− IgT− non-lymphoid subset (PeL nL IgM− IgT−) (Figure 4B). Gene expression of cd79a, on the other hand, was higher in the IgM+ than IgT+ peritoneal lymphoid cells. Lowest cd79a expression was observed in the IgM− IgT− non-lymphoid subset of the peritoneal leukocytes (PeL nL IgM− IgT−).
To our knowledge, this is the first report on the profiling of salmon B cell surface protein repertoire. Although proteomics has been employed in some investigations of fish immune responses to various infections [reviewed in (104)], it is not commonly used to characterize the surface protein expression of specific immune-related cells in teleosts. A previous proteomics study on Atlantic salmon focused on profiling changes in fish serum proteins following infection with salmonid alphavirus (105). Our approach focused on B cells and was useful in the identification of 21 CD molecules from salmon IgM+ B cells. The relatively limited number of CD proteins identified could be partially explained by the initial low number of B cells in the starting material (3 × 106 viable IgM+ B cells after 72 h in culture) used in the surface protein isolation. In addition, despite being one of the popular choices for cell surface proteome profiling, the biotinylation technique employed for the enrichment of plasma membrane proteins may not be the most effective method for isolating glycosylated proteins (106). Since majority of cell surface proteins are glycosylated (107), many surface glycoproteins may not have been included in our mass spectrometry samples.
Peripheral blood was used as source of salmon B cells for proteomics due to higher total leukocyte yield [i.e., 15 × and 4 × more total leukocytes than the spleen and the HK, respectively (108)] and the abundance of IgM+ B cells in this compartment (22). For comparison, surface proteins from two Atlantic salmon head kidney cell lines, ASK and SSP-9, were also identified. Although the exact cell composition of these hematopoietic-tissue derived cell lines is unknown, these seem to include cells from the myeloid lineage based on their respective proteomics profile. Given its epithelial-like morphology and previously reported gene expression profile (31), SSP-9 is likely comprised of macrophage-like cells. Specifically, majority of the identified SSP-9-exclusive surface proteins (CD2, CD34, CD74, CD151, CD205) are associated with expression on a macrophage subset with antigen-presenting capacity (109–113). Detection of CD203c, a known basophil and mast cell marker in humans (114), suggest that SSP-9 may contain granulocytes.
All the surface proteins identified from our salmon B cell samples have been previously detected on the surface of mammalian B cells, except for CD87. In teleost, only a few of these B cell-associated proteins have been studied so far. Constitutive gene expression of tetraspanins cd9 and cd63 have been reported in sorted IgM+ B cells of trout from different immune organs (115). Surface expression of CD22 has also been shown in PBLs of tongue sole (116).
In keeping with their established roles in B cell activation, CD40 (117) and CD98 (118) surface expression were significantly higher in LPS-stimulated IgM+ B cell samples. Co-expression of CD40 and IgM+ has been established on B cell surface of zebrafish, with significant up-regulation of CD40 similarly observed following LPS treatment (119). In addition, modest increase in CD40 transcripts has been reported for Atlantic salmon IgM+ B cells in the presence of another TLR agonist, CpG oligodeoxynucleotides (22).
Surprisingly, CD87, which has been reported to be absent on surface of resting B lymphocytes and B cell lines of mammals (90, 92), was detected in our naïve salmon IgM+ B cell samples. It should be noted that CD87 acts as a ligand for β2 integrins such as CD18 and CD11a/c identified on salmon B cells, and thus usually found in close association with these protein complexes on leukocytes (120). CD87 has been demonstrated to mediate cell-cell adhesion by interaction with integrins on the same as well as apposing cells (121). Hence, the CD87 proteins detected in our proteomics experiment may be an artifact from a CD87-β2 integrin complex formed by trans-interaction of salmon B cells with CD87-expressing monocytes or granulocytes. It is unlikely that the proteins were contributed directly by contaminating monocytes or granulocytes since the samples were checked for purity (>98% IgM+ PBLs) before surface protein isolation. Alternatively, it is also possible that salmon, in contrast to mammals, express CD87 on their B cell surface.
Peripheral blood provides the means for cells to move systemically within the animal body; hence, one would expect a mixture of circulating B cell subsets in this compartment. Surface detection of chemokine receptors CXCR4, CXCR5, and CCR9 important in trafficking naïve B lymphocytes (76) seem to suggest that our peripheral IgM B cell population is comprised of a migrating population of B cells from different lymphoid compartments. In humans, the majority of peripheral B cells are of the conventional B-2 cell type bearing both membrane IgM and membrane IgD, which accounts for about 10% of total PBLs (122). Given the current technical constraints, it was not possible to further sort the largely undefined subsets of the salmon IgM+ peripheral B cell subpopulation.
More recently, it has been shown that, contrary to the dogma that all plasma cells have permanently switched off expression of membrane-bound Ig molecules to produce their secreted version (antibodies), some mature plasma cells in mice retain their expression of surface IgM and functional BCR (123, 124). Thus, it is possible that our LPS-treated peripheral B cells contained some differentiating IgM+ plasmablasts, plasma cells, or memory B cells. However, none of the known markers for these differentiated B cells [i.e., CD27 for memory B cells (125), CD138 for plasma cells (126)] was identified in the stimulated B cell samples. Based on previous studies in trout, this can be partially explained by the relatively short stimulation period (72 h incubation in culture) used in our study. In naïve trout PBLs, significant antibody secreting cell response is observed only after 4 days of LPS stimulation using 4 times higher dose, with peak responses occurring by Day 7 in culture (127). In addition, memory B cell responses to TI and TD antigens are observed only after 4–7 days of in vitro re-stimulation of PBLs from previously immunized trout (108).
It should be emphasized that several other mammalian pan-B cell and subset markers known to be encoded in the Atlantic salmon genome were not detected in our salmon IgM+ B cell samples. Failure to detect a particular surface protein should be interpreted with caution due to previously indicated technical limitations of our experimental approach or their presence at low frequency in our heterogeneous pool of IgM+ B cell subpopulations. Moreover, it should be noted that our shortlist of salmon CD proteins was based on their mammalian counterparts. Other surface proteins that were detected exclusively in the IgM+ samples but without homologs or are not classified as CD molecules in mammals were not included in this list. Further investigation of the “non-CD” surface proteins is therefore needed in order to determine whether or not these can serve as unique B cell markers in salmon.
Due to lack of access to antibodies against salmon IgD and myeloid markers, our analysis was constrained to leukocyte subpopulations sorted by cell size and granularity and subsequent positive selection for IgM+ or IgT+ cells, representing independent salmon B cell lineages. Despite this clear limitation, our current sorting protocol is still a valid useful tool in studying the different salmon leukocyte populations as evidenced by the results of our qPCR validation assays using different leukocyte markers.
In general, cd22 vs. cd79a gene expression patterns of the different leukocyte subsets were comparable across different lymphoid organs. Highest relative expression were observed in all IgM+ B cell subsets, while very low to below detectable level of expression were found in IgM−/IgT− non-lymphoid subset (likely consisting of myeloid cells). Relatively lower but significant level of expression was also found in non-IgM subsets within the lymphoid gate, which likely consist of either a mixed population of IgT+ or IgD+ only B cells (i.e., HKL SC-L IgM−) or IgD+ only B cells (i.e., SpL L IgM− IgT−). While generally regarded as B cell-exclusive proteins (52, 128), it is important to note that mammalian CD22 and CD79A have also been reported to be expressed on the surface of some non-B cell populations, including T cells (129) and various myeloid cells (130–133). However, this is likely not the case for Atlantic salmon given the negligible transcript levels of cd22 and cd79a in all the myeloid-containing leukocyte subsets tested.
Although mRNA levels do not necessarily correlate directly with surface protein expression, gene expression profiles of salmon cd22 and cd79a clearly showed that the B cell exclusivity of our candidate CD molecules extend down to the level of transcription. This B cell-restrictive expression was consistent across different lymphoid compartments (peripheral blood, head kidney, spleen, peritoneal cavity) and different Ig isotypes (IgM and IgT), thus providing strong support to the potential of CD22 and CD79A as pan-B cell markers for salmon.
Extensive gene duplications in the Atlantic salmon genome (134, 135) should be taken into account in finding appropriate B cell markers. Specifically, Atlantic salmon encodes an unusually high number of CD22-like paralogs and isoforms (Supplementary Table 1) that could have different regulatory expression and conformation on B cells. Interestingly, peptide sequences specific to only one CD22 paralog was detected on the salmon B cell surface in our mass spectrometry analysis. Based on our qPCR assay, mRNA expression of this particular variant was also specific in salmon B cells. In humans, a CD22 molecule with a different conformation to that found on B cells has been detected on basophils (130). Whether other CD22 variants/isoforms are also expressed in other subpopulations of salmon leukocytes, or in specific subsets of salmon B cells, is still unknown, but is beyond the scope of this study. For simplicity, the particular CD22 paralog (XP_014021065.1) identified in this study was referred as CD22.
CD45, the most abundant surface protein identified in our salmon B cell samples, exists in different alternative splicing isoforms, which are expressed in cell-type specific patterns on functional subsets of mammalian lymphocytes (48). In mice, a long isoform of CD45 called B220 (136) has been used as a pan B-cell marker due to its specific expression throughout stages of B cell development, including entry into the memory B cell pool (137). A similar long CD45 isoform (CD45RABC) has been identified in human B cells (96), which has been used as marker for certain human B cell subsets (138). This B-cell specific CD45 isoform in mammals consists of all 33 exons of the CD45 gene (139). Atlantic salmon genome encodes for seven CD45 transcript variants in Chromosome 10 (Supplementary Table 1), with alternate splicing of three exons (4, 5, 6) producing six CD45 isoforms [GenBank Assembly Accession No. GCA_000233375.4 (135)]. Given that all exons are present in the B cell-specific isoform, it was not possible to determine whether the long CD45 isoform was the only isoform detected on salmon B cell surface. Hence, further investigation for the presence of other CD45 isoforms in sorted salmon B cell subpopulations (i.e., qPCR assays using primers specific for the other splice variants) is still needed before its potential as another salmon pan-B cell marker can be fully appreciated.
Several other CD molecules identified on salmon B cell samples can be explored for their potential in discriminating specific B cell subsets despite their non-exclusive B cell surface expression. Murine CD9 is expressed on the surface of innate-like B cells and plasma cells, but not on naïve conventional B2 cells (140, 141). In our proteomics data, CD9 was detected in high abundance in all samples of salmon IgM+ peripheral B cells but not in either of the two salmon head kidney cell lines. This implies that the salmon IgM+ peripheral B cell subpopulation are comprised of cells with significantly high expression of this protein. In rainbow trout, cd9 mRNA levels are similar in IgM+ and IgM− B cells from blood and HK (115). In agreement with this, comparable levels of cd9 expression was also observed in the IgM+ and IgM− fractions of our sorted HKL lymphoid cells, but highest constitutive gene expression was still found in the myeloid fractions. Thus, while CD9 cannot be used as the sole and primary marker for salmon B cells, its potential for identifying specific B cell subsets should be explored further.
Other potential B cell subset markers are the CD11 β2 integrins. In contrast to mammals wherein four β2 integrins have been identified, Atlantic salmon genome encodes only two: CD11a and CD11c. Both of these proteins were detected in our salmon IgM+ B cell samples, which could suggest their potential use as markers to functionally-equivalent B cell subsets in mammals. High surface CD11a expression has been associated with a new subpopulation of IFN-γ-secreting innate B cells (142). Expression of CD11c has also been recently used as marker for distinct B cell subsets in mice and humans (143, 144), including CD11clow-expressing plasmablasts that predominantly secrete Ag-specific IgM antibodies in a T cell-independent manner (145) and CD11c+ ‘atypical memory B cells’ (144).
Interestingly, CD11b, which is used as a marker to differentiate B1 cells that reside in the peritoneal cavity and those that recently migrated into the spleen (146), has not been detected until present. Among salmonids, only coho and Chinook salmon have CD11b-like protein recorded in the sequence database. However, the salmonid sequences of CD11c (ITGAX) is very similar to that of CD11b (70). In fact, Atlantic salmon CD11c (XP_014014772.1) has 70 and 90% homology with CD11c-like proteins of coho (XP_020308950.1) and Chinook (XP_024269535.1) salmon, respectively. Similarly, CD11d, which is only found in coho (XP_020334852.1), has 95% homology with Atlantic salmon CD11a (XP_014032228.1). At this point, it is unknown whether Atlantic salmon CD11c-like and CD11a-like proteins have overlapping functions with the mammalian CD11b and CD11d proteins, and whether expression patterns of these mammalian B cell subset markers are similar to their ancestral counterpart in teleosts.
It is worthy to note that several of the surface proteins (CD9, CD11a, CD11c) identified on salmon IgM+ B cells in high abundance are associated with innate-like B cells in mice and humans. CD22, one of the identified potential salmon B cell marker in this study, is also known to be expressed the highest in MZ B cell precursors out of any mammalian B cell subset (147). Additionally, detection of CD282 (TLR2), which has been previously shown to be expressed higher in naïve innate-like B cells than conventional B2 cells (97), implies a possible function of salmon IgM+ peripheral B cells in TI-responses. Altogether, these are consistent with a salmon IgM+ B cell phenotype closer to innate-like B cells (B1 and MZ B2) than conventional B2 cells in mammals. This supports the previously proposed B cell evolution hypothesis (18) which suggests that teleost IgM B cells are the ancestors of the innate-like B cells of mammals (25, 148). Whether these innate-like B cell-associated proteins detected from our B cell samples are evenly expressed on the surface of all salmon IgM+ B cells or only expressed at very high levels on certain sub-populations, is a question that warrants further investigations. This can be determined upon availability of the necessary immunological tools and antibodies to analyze the frequency and level of their surface expression.
Speculations regarding the possible characteristics of salmon IgM B cell populations based solely on pre-conceived notions of the surface phenotype, function, and anatomical distribution of their mammalian counterparts should come with a caveat. Indeed, the conservation of protein functions between mammals and teleosts for many of the B cell-associated CD molecules identified in this study has to be established. Therefore, subsets comprising the salmon IgM+ B cells need to be better defined, in order to facilitate further functional studies of these individual surface proteins.
In summary, our salmon B cell proteomics approach provide novel information on the surface phenotype of salmon IgM B cells as well as some evolutionary insights in reference to their mammalian counterparts. While possible exclusion of several glycosylated and/or less abundant proteins cannot be discounted due to technical limitations of the protocols used, our detection of 21 CD molecules remains a significant advancement in profiling the salmon B cell surface proteome. In addition, identification of CD22 and CD79A as potential pan-B cell markers represents a considerable positive step toward salmon B cell marker development. Further investigation and evaluation of the salmon CD molecules identified in this study would improve our understanding of B cell dynamics in salmon in the presence of TI and TD antigens during immunostimulation, pathogen infection, or vaccination.
The authors confirm that the experimental protocols used for the live fish experiments were based on the Animal Welfare Act (https://www.regjeringen.no/en/dokumenter/animal-welfare-act/id571188/) and performed in accordance with relevant guidelines and regulations given by the Norwegian Animal Research Authority.
MP performed most of the experimental work (leukocyte isolation, FACS sorting, surface protein isolation, qPCR analyses) and wrote the manuscript. J-AB was responsible for the mass spectrometry analyses. LT performed RNA extraction and cDNA syntheses. IJ and JJ collaborated in obtaining funding, helped designed the experiments, and reviewed the manuscript.
This work was supported by the Aquaculture program of The Research Council of Norway (Grant No. 254892): Multiple routes to B cell memory in Atlantic salmon and UiT The Arctic University of Norway. The publication charges for this article have been funded by a grant from the publication fund of UiT The Arctic University of Norway.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Dr. Karsten Skjødt and Prof. Oriol Sunyer for generously providing the mAbs for sorting IgM and IgT B cells. Toril Anne Grønset, Shiferaw Jenberie, Guro Strandskog, Mikael Fjeld Wold, and Morten Bay Styrvold are also acknowledged for their excellent technical assistance (processing of mass spectrometry samples, cell isolation).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00037/full#supplementary-material
1. Thorarinsson R, Powell DB. Effects of disease risk, vaccine efficacy, and market price on the economics of fish vaccination. Aquaculture (2006) 256:42–9. doi: 10.1016/j.aquaculture.2006.01.033
2. Holvold LB, Myhr AI, Dalmo RA. Strategies and hurdles using DNA vaccines to fish. Vet Res, (2014) 45:21. doi: 10.1186/1297-9716-45-21
3. Zhu LY, Lin AF, Shao T, Nie L, Dong WR, Xiang LX, et al. B cells in teleost fish act as pivotal initiating APCs in priming adaptive immunity: an evolutionary perspective on the origin of the B-1 cell subset and B7 molecules. J Immunol. (2014) 192:2699–714. doi: 10.4049/jimmunol.1301312
4. Li J, Barreda DR, Zhang YA, Boshra H, Gelman AE, Lapatra S, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol. (2006) 7:1116–24. doi: 10.1038/ni1389
5. Sinyakov MS, Dror M, Zhevelev HM, Margel S, Avtalion RR. Natural antibodies and their significance in active immunization and protection against a defined pathogen in fish. Vaccine (2002). 20:3668–3674. doi: 10.1016/S0264-410X(02)00379-1
6. Ye J, Kaattari IM, Ma C, Kaattari S. The teleost humoral immune response. Fish Shellfish Immunol. (2013) 35:1719–28. doi: 10.1016/j.fsi.2013.10.015
7. Zwollo P. The humoral immune system of anadromous fish. Dev Comp Immunol. (2018) 80:24–33. doi: 10.1016/j.dci.2016.12.008
8. Yamaguchi T, Quillet E, Boudinot P, Fischer U. What could be the mechanisms of immunological memory in fish? Fish Shellfish Immunol. (2019) 85:3–8. doi: 10.1016/j.fsi.2018.01.035
9. Vale AM, Kearney JF, Nobrega A, Schroeder HW. Chapter 7–Development and function of B cell subsets. In: Alt FW, Honjo T, Radbruch A, Reth M, editors. Molecular Biology of B Cells, 2nd ed. London: Academic Press (2015). p. 99–119. doi: 10.1016/B978-0-12-397933-9.00007-2
10. Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol. (2013) 13:118–32. doi: 10.1038/nri3383
11. Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity (2005) 23:7–18. doi: 10.1016/j.immuni.2005.04.011
12. Baumgarth N. B-1 Cell heterogeneity and the regulation of natural and antigen-induced IgM production. Front Immunol. (2016) 7:324. doi: 10.3389/fimmu.2016.00324
13. Baumgarth N. Innate-like B cells and their rules of engagement. Adv Exp Med Biol. (2013) 785:57–66. doi: 10.1007/978-1-4614-6217-0_7
14. Yang Y, Ghosn EE, Cole LE, Obukhanych TV, Sadate-Ngatchou P, Vogel SN, et al. Antigen-specific memory in B-1a and its relationship to natural immunity. Proc Natl Acad Sci USA. (2012) 109:5388–93. doi: 10.1073/pnas.1121627109
15. Yang Y, Ghosn EE, Cole LE, Obukhanych TV, Sadate-Ngatchou P, Vogel SN, et al. Antigen-specific antibody responses in B-1a and their relationship to natural immunity. Proc Natl Acad Sci USA. (2012) 109:5382–7. doi: 10.1073/pnas.1121631109
16. Kurosaki T, Kometani K, Ise W. Memory B cells. Nat Rev Immunol. (2015) 15:149. doi: 10.1038/nri3802
17. Sunyer JO. Fishing for mammalian paradigms in the teleost immune system. Nat Immunol. (2013) 14:320–6. doi: 10.1038/ni.2549
18. Sunyer JO. Evolutionary and functional relationships of B cells from fish and mammals: insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect Disord Drug Targets (2012) 12:200–12. doi: 10.2174/187152612800564419
19. Korytár T, Jaros J, Verleih M, Rebl A, Kotterba G, Kühn C, et al. Novel insights into the peritoneal inflammation of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. (2013) 35:1192–9. doi: 10.1016/j.fsi.2013.07.032
20. Zapata AG, Torroba M, Vicente A, Varas A, Sacedón R, Jiménez E. The relevance of cell microenvironments for the appearance of lympho-haemopoietic tissues in primitive vertebrates. Histol Histopathol. (1995) 10:761–78.
21. Cain KD, Jones DR, Raison RL. Antibody-antigen kinetics following immunization of rainbow trout (Oncorhynchus mykiss) with a T-cell dependent antigen. Dev Comp Immunol. (2002) 26:181–90. doi: 10.1016/S0145-305X(01)00063-5
22. Jenberie S, Thim HL, Sunyer JO, Skjødt K, Jensen I, Jørgensen JB, et al. Profiling Atlantic salmon B cell populations: CpG-mediated TLR-ligation enhances IgM secretion and modulates immune gene expression. Sci Rep. (2018) 8:3565. doi: 10.1038/s41598-018-21895-9
23. Edholm ES, Bengtén E, Stafford JL, Sahoo M, Taylor EB, Miller NW, et al. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J Immunol. (2010) 185:4082–94. doi: 10.4049/jimmunol.1000631
24. Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. (2010) 11:827–35. doi: 10.1038/ni.1913
25. Abós B, Bird S, Granja AG, Morel E, More Bayona JA, Barreda DR, et al. Identification of the first teleost CD5 molecule: additional evidence on phenotypical and functional similarities between Fish IgM(+) B cells and mammalian B1 cells. J Immunol. (2018) 201:465–480. doi: 10.4049/jimmunol.1701546
26. Gonzalez R, Matsiota P, Torchy C, De Kinkelin P, Avrameas S. Natural anti-TNP antibodies from rainbow trout interfere with viral infection in vitro. Res Immunol. (1989) 140:675–84. doi: 10.1016/0923-2494(89)90021-7
27. Chumley MJ, Dal Porto JM, Cambier JC. The unique antigen receptor signaling phenotype of B-1 cells is influenced by locale but induced by antigen. J Immunol. (2002) 169:1735–43. doi: 10.4049/jimmunol.169.4.1735
28. Clark G, Stockinger H, Balderas R, van Zelm MC, Zola H, Hart D, et al. Nomenclature of CD molecules from the tenth human leucocyte differentiation antigen workshop. Clin Transl Immunol. (2016) 5:e57. doi: 10.1038/cti.2015.38
29. Hedfors IA, Bakke H, Skjødt K, Grimholt U. Antibodies recognizing both IgM isotypes in Atlantic salmon. Fish Shellfish Immunol. (2012) 33:1199–206. doi: 10.1016/j.fsi.2012.09.009
30. Devold M, Krossøy B, Aspehaug V, Nylund A. Use of RT-PCR for diagnosis of infectious salmon anaemia virus (ISAV) in carrier sea trout Salmo trutta after experimental infection. Dis Aquat Organ. (2000) 40:9–18. doi: 10.3354/dao040009
31. Rodriguez Saint-Jean S, González C, Monrás M, Romero A, Ballesteros N, Enríquez R, et al. Establishment and characterization of a new cell line (SSP-9) derived from Atlantic salmon Salmo salar that expresses type I ifn. J Fish Biol. (2014) 85:1526–45. doi: 10.1111/jfb.12503
32. Jørgensen JB, Johansen A, Stenersen B, Sommer AI. CpG oligodeoxynucleotides and plasmid DNA stimulate Atlantic salmon (Salmo salar L.) leucocytes to produce supernatants with antiviral activity. Dev Comp Immunol. (2001) 25:313–21. doi: 10.1016/S0145-305X(00)00068-9
33. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. (1996) 68:850–8. doi: 10.1021/ac950914h
34. Tadiso Lie KK, Hordvik I. Molecular cloning of IgT from Atlantic salmon, and analysis of the relative expression of τ, μ and δ in different tissues. Vet Immunol Immunopathol. (2011) 139:17–26. doi: 10.1016/j.vetimm.2010.07.024
35. Cuesta A, Meseguer J, Esteban MA. The antimicrobial peptide hepcidin exerts an important role in the innate immunity against bacteria in the bony fish gilthead seabream. Mol Immunol. (2008) 45:2333–42. doi: 10.1016/j.molimm.2007.11.007
36. Wang T, Bird S, Koussounadis A, Holland JW, Carrington A, Zou J, et al. Identification of a novel IL-1 cytokine family member in teleost fish. J Immunol. (2009) 183:962–74. doi: 10.4049/jimmunol.0802953
37. Levy S, Todd SC, Maecker HT. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. (1998) 16:89–109. doi: 10.1146/annurev.immunol.16.1.89
38. Haynes BF, Hemler ME, Mann DL, Eisenbarth GS, Shelhamer J, Mostowski HS, et al. Characterization of a monoclonal antibody (4F2) that binds to human monocytes and to a subset of activated lymphocytes. J Immunol. (1981) 126:1409–14.
39. Moretta A, Mingari MC, Haynes BF, Sekaly RP, Moretta L, Fauci AS. Phenotypic characterization of human cytolytic T lymphocytes in mixed lymphocyte culture. J Exp Med. (1981) 153:213–8. doi: 10.1084/jem.153.1.213
40. Attia M, Huet E, Delbé J, Ledoux D, Menashi S, Martelly I. Extracellular matrix metalloproteinase inducer (EMMPRIN/CD147) as a novel regulator of myogenic cell differentiation. J Cell Physiol. (2011) 226:141–9. doi: 10.1002/jcp.22315
41. Nguyen HT, Merlin D. Homeostatic and innate immune responses: role of the transmembrane glycoprotein CD98. Cell Mol Life Sci. (2012) 69:3015–26. doi: 10.1007/s00018-012-0963-z
42. Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. Faseb J. (1997) 11:428–42. doi: 10.1096/fasebj.11.6.9194523
43. Matthews AL, Koo CZ, Szyroka J, Harrison N, Kanhere A, Tomlinson MG. Regulation of leukocytes by tspanC8 tetraspanins and the “Molecular Scissor” ADAM10. Front Immunol. (2018) 9:1451. doi: 10.3389/fimmu.2018.01451
44. Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. (2009) 315:1584–92. doi: 10.1016/j.yexcr.2008.09.020
45. Yoshida N, Kitayama D, Arima M, Sakamoto A, Inamine A, Watanabe-Takano H, et al. CXCR4 expression on activated B cells is downregulated by CD63 and IL-21. J Immunol. (2011) 186:2800–8. doi: 10.4049/jimmunol.1003401
46. van Kooten C, Banchereau J. Functions of CD40 on B cells, dendritic cells and other cells. Curr Opin Immunol. (1997) 9:330–7.
47. Durie FH, Foy TM, Masters SR, Laman JD, Noelle RJ. The role of CD40 in the regulation of humoral and cell-mediated immunity. Immunol Today. (1994) 15:406–11. doi: 10.1016/0167-5699(94)90269-0
48. Trowbridge IS, Thomas ML. CD45: an emerging role as a protein tyrosine phosphatase required for lymphocyte activation and development. Annu Rev Immunol. (1994) 12:85–116.
49. Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomic (2005) 4:1265–72. doi: 10.1074/mcp.M500061-MCP200
50. Nitschke L. CD22 and Siglec-G: B-cell inhibitory receptors with distinct functions. Immunol Rev. (2009) 230:128–43. doi: 10.1111/j.1600-065X.2009.00801.x
51. Nitschke L. CD22 and Siglec-G regulate inhibition of B-cell signaling by sialic acid ligand binding and control B-cell tolerance. Glycobiology (2014) 24:807–17. doi: 10.1093/glycob/cwu066
52. Clark EA. CD22, a B cell-specific receptor, mediates adhesion and signal transduction. J Immunol. (1993) 150:4715–8.
53. Erickson LD1 TL, Bhatia SK, Grabstein KH, Waldschmidt TJ. Differential expression of CD22 (Lyb8) on murine B cells. Int Immunol. (1996) 8:1121–9. doi: 10.1093/intimm/8.7.1121
54. Moyron-Quiroz JE, Partida-Sánchez S, Donís-Hernández R, Sandoval-Montes C, Santos-Argumedo L. Expression and function of CD22, a B-cell restricted molecule. Scand J Immunol. (2002) 55:343–51. doi: 10.1046/j.1365-3083.2002.01063.x
55. Clark EA, Giltiay NV. CD22: a regulator of innate and adaptive b cell responses and autoimmunity. Front Immunol. (2018) 9:2235 doi: 10.3389/fimmu.2018.02235
56. Nitschke L, Carsetti R, Ocker B, Köhler G, Lamers MC. CD22 is a negative regulator of B-cell receptor signalling. Curr Biol. (1997) 7:133–43.
57. Lajaunias F, Nitschke L, Moll T, Martinez-Soria E, Semac I, Chicheportiche Y, et al. Differentially regulated expression and function of CD22 in activated B-1 and B-2 lymphocytes. J Immunol. (2002) 168:6078–83. doi: 10.4049/jimmunol.168.12.6078
58. Wienands J. The B-cell antigen receptor: formation of signaling complexes and the function of adaptor proteins. Curr Top Microbiol Immunol. (2000) 245:53–76. doi: 10.1007/978-3-642-57066-7_2
59. van Noesel CJ, van Lier RA, Cordell JL, Tse AG, van Schijndel GM, de Vries EF, et al. The membrane IgM-associated heterodimer on human B cells is a newly defined B cell antigen that contains the protein product of the mb-1 gene. J Immunol. (1991) 146:3881–8.
61. Patterson HC, Kraus M, Wang D, Shahsafaei A, Henderson JM, Seagal J, et al. Cytoplasmic Ig alpha serine/threonines fine-tune Ig alpha tyrosine phosphorylation and limit bone marrow plasma cell formation. J Immunol. (2011) 187:2853–8. doi: 10.4049/jimmunol.1101143
62. Leduc I, Preud'homme JL, Cogne M. Structure and expression of the mb-1 transcript in human lymphoid cells. Clin Exp Immunol. (1992) 90:141–6. doi: 10.1111/j.1365-2249.1992.tb05846.x
63. Mason D, Cordell JL, Tse AG, van Dongen JJ, van Noesel CJ, Micklem K, et al. The IgM-associated protein mb-1 as a marker of normal and neoplastic B cells. J Immunol. (1991) 147:2474–2482.
64. Dworzak MN, Fritsch G, Fröschl G, Printz D, Gadner H. Four-color flow cytometric investigation of terminal deoxynucleotidyl transferase-positive lymphoid precursors in pediatric bone marrow: CD79a expression precedes CD19 in early B-cell ontogeny. Blood (1998) 92:3203–9.
65. Tanaka T, Ichimura K, Sato Y, Takata K, Morito T, Tamura M, et al. Frequent downregulation or loss of CD79a expression in plasma cell myelomas: potential clue for diagnosis. Pathol Int. (2009) 59:804–8. doi: 10.1111/j.1440-1827.2009.02448.x
66. Horejsi V, Vlcek C. Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett. (1991) 288:1–4.
67. Tohami T, Drucker L, Radnay J, Shapira H, Lishner M. Expression of tetraspanins in peripheral blood leukocytes: a comparison between normal and infectious conditions. Tissue Antigens. (2004) 64:235–42. doi: 10.1111/j.1399-0039.2004.00271.x
68. Gutierrez-Lopez MD, Ovalle S, Yanez-Mo M, Sanchez-Sanchez N, Rubinstein E, Olmo N, et al. A functionally relevant conformational epitope on the CD9 tetraspanin depends on the association with activated beta1 integrin. J Biol Chem. (2003) 278:208–18. doi: 10.1074/jbc.M207805200
69. Florin L, Lang T. Tetraspanin assemblies in virus infection. Front Immunol. (2018) 9:1140. doi: 10.3389/fimmu.2018.01140
70. Schittenhelm L, Hilkens CM, Morrison VL. Beta2 integrins as regulators of dendritic cell, monocyte, and macrophage function. Front Immunol. (2017) 8:1866. doi: 10.3389/fimmu.2017.01866
71. Rossi D, Zlotnik A. The biology of chemokines and their receptors. Ann Rev Immunol. (2000) 18:217–42. doi: 10.1146/annurev.immunol.18.1.217
72. Demberg T, Mohanram V, Venzon D, Robert-Guroff M. Phenotypes and distribution of mucosal memory B-cell populations in the SIV/SHIV rhesus macaque model. Clin Immunol. (2014) 153:264–76. doi: 10.1016/j.clim.2014.04.017
73. Lee HS, Kim HR, Lee EH, Jang MH, Kim SB, Park JW, et al. Characterization of CCR9 expression and thymus-expressed chemokine responsiveness of the murine thymus, spleen and mesenteric lymph node. Immunobiology (2012) 217:402–11. doi: 10.1016/j.imbio.2011.10.014
74. Amiya T, Nakamoto N, Chu PS, Teratani T, Nakajima H, Fukuchi Y, et al. Bone marrow-derived macrophages distinct from tissue-resident macrophages play a pivotal role in concanavalin a-induced murine liver injury via CCR9 axis. Sci Rep. (2016) 6:35146. doi: 10.1038/srep35146
75. Mizuno S, Kanai T, Mikami Y, Sujino T, Ono Y, Hayashi A, et al. CCR9+ plasmacytoid dendritic cells in the small intestine suppress development of intestinal inflammation in mice. Immunol Lett. (2012) 146:64–9. doi: 10.1016/j.imlet.2012.05.001
76. Stein JV, Nombela-Arrieta C. Chemokine control of lymphocyte trafficking: a general overview. Immunology (2005) 116:1–12. doi: 10.1111/j.1365-2567.2005.02183.x
77. Wurbel MA, Malissen M, Guy-Grand D, Meffre E, Nussenzweig MC, Richelme M, et al. Mice lacking the CCR9 CC-chemokine receptor show a mild impairment of early T- and B-cell development and a reduction in T-cell receptor gammadelta(+) gut intraepithelial lymphocytes. Blood (2001) 98:2626–32. doi: 10.1182/blood.V98.9.2626
78. Mora JR, von Andrian UH. Differentiation and homing of IgA-secreting cells. Mucosal Immunol. (2008) 1:96–109. doi: 10.1038/mi.2007.14
79. Holness CL, da Silva RP, Fawcett J, Gordon S, Simmons DL. Macrosialin, a mouse macrophage-restricted glycoprotein, is a member of the lamp/lgp family. J Biol Chem. (1993) 268:9661–6.
80. Gordon S. Macrophage-restricted molecules: role in differentiation and activation. Immunol Lett. (1999) 65:5–8. doi: 10.1016/S0165-2478(98)00116-3
81. Gottfried E, Kunz-Schughart LA, Weber A, Rehli M, Peuker A, Müller A, et al. Expression of CD68 in non-myeloid cell types. Scand J Immunol. (2008) 67:453–63. doi: 10.1111/j.1365-3083.2008.02091.x
82. Kunisch E, Fuhrmann R, Roth A, Winter R, Lungershausen W, Kinne RW. Macrophage specificity of three anti-CD68 monoclonal antibodies (KP1, EBM11, and PGM1) widely used for immunohistochemistry and flow cytometry. Ann Rheum Dis. (2004) 63:774–84. doi: 10.1136/ard.2003.013029
83. O'Reilly D, Greaves DR. Cell-type-specific expression of the human CD68 gene is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping. Genomics (2007) 90:407–15. doi: 10.1016/j.ygeno.2007.04.010
84. Fennelly JA, Tiwari B, Davis SJ, Evans EJ. CD2F-10: a new member of the CD2 subset of the immunoglobulin superfamily. Immunogenetics (2001) 53:599–602. doi: 10.1007/s002510100364
85. Zhang W, Wan T, Li N, Yuan Z, He L, Zhu X, et al. Genetic approach to insight into the immunobiology of human dendritic cells and identification of CD84-H1, a novel CD84 homologue. Clin Cancer Res. (2001) 7(Suppl. 3):822–9s.
86. De Salort J, Sintes J, Llinàs L, Matesanz-Isabel J, Engel P. Expression of SLAM (CD150) cell-surface receptors on human B-cell subsets: from pro-B to plasma cells. Immunol Lett. (2011) 134:129–36. doi: 10.1016/j.imlet.2010.09.021
87. Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. (2009) 9:39. doi: 10.1038/nri2456
88. Reuning U, Magdolen V, Hapke S, Schmitt M. Molecular and functional interdependence of the urokinase-type plasminogen activator system with integrins. Biol Chem. (2003) 384:1119–31. doi: 10.1515/BC.2003.125
89. Ge Y, Elghetany MT. Urokinase plasminogen activator receptor (CD87): something old, something new. Lab Hematol. (2003) 9:67–71.
90. Plesner T, Behrendt N, Ploug M. Structure, function and expression on blood and bone marrow cells of the urokinase-type plasminogen activator receptor. uPAStem Cells R (1997) 15:398–408. doi: 10.1002/stem.150398
91. Elghetany MT, Patel J, Martinez J, Schwab H. CD87 as a marker for terminal granulocytic maturation: assessment of its expression during granulopoiesis. Cytometry B Clin Cytom. (2003) 51:9–13. doi: 10.1002/cyto.b.10008
92. Jardí M, Inglés-Esteve J, Burgal M, Azqueta C, Velasco F, López-Pedrera C, et al. Distinct patterns of urokinase receptor (uPAR) expression by leukemic cells and peripheral blood cells. Thromb Haemost. (1996) 76:1009–19.
93. Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. (1999) 274:17406–9. doi: 10.1074/jbc.274.25.17406
94. Flo TH, Halaas O, Torp S, Ryan L, Lien E, Dybdahl B, et al. Differential expression of Toll-like receptor 2 in human cells. J Leukoc Biol. (2001) 69:474–81. doi: 10.1189/jlb.69.3.474
95. Lee SM, Joo YD, Seo SK. Expression and Function of TLR2 on CD4 Versus CD8 T Cells. Immune Netw. (2009) 9:127–32. doi: 10.4110/in.2009.9.4.127
96. Bleesing JJ, Fleisher TA. Human B cells express a CD45 isoform that is similar to murine B220 and is downregulated with acquisition of the memory B-cell marker CD27. Cytometry B Clin Cytom. (2003) 51:1–8. doi: 10.1002/cyto.b.10007
97. Dasari P, Nicholson IC, Hodge G, Dandie GW, Zola H. Expression of toll-like receptors on B lymphocytes. Cell Immunol. (2005) 236:140–5. doi: 10.1016/j.cellimm.2005.08.020
98. Booth J, Wilson H, Jimbo S, Mutwiri G. Modulation of B cell responses by Toll-like receptors. Cell Tissue Res. (2011) 343:131–40. doi: 10.1007/s00441-010-1031-3
99. Kobayashi I, Katakura F, Moritomo T. Isolation and characterization of hematopoietic stem cells in teleost fish. Dev Comp Immunol. (2016) 58:86–94. doi: 10.1016/j.dci.2016.01.003
100. Geisberger R, Lamers M, Achatz G. The riddle of the dual expression of IgM and IgD. Immunology (2006) 118:429–37. doi: 10.1111/j.1365-2567.2006.02386.x
101. Edholm ES, Bengten E, Wilson M. Insights into the function of IgD. Dev Comp Immunol. (2011) 35:1309–16. doi: 10.1016/j.dci.2011.03.002
102. Gutzeit C, Chen K, Cerutti A. The enigmatic function of IgD: some answers at last. Eur J Immunol. (2018) 48:1101–13. doi: 10.1002/eji.201646547
103. Ramirez-Gomez F, Greene W, Rego K, Hansen JD, Costa G, Kataria P, et al. Discovery and characterization of secretory IgD in rainbow trout: secretory IgD is produced through a novel splicing mechanism. J Immunol. (2012) 188:1341–9. doi: 10.4049/jimmunol.1101938
104. Ye H, Lin Q, Luo H. Applications of transcriptomics and proteomics in understanding fish immunity. Fish Shellfish Immunol. (2018) 77:319–27. doi: 10.1016/j.fsi.2018.03.046
105. Braceland M, Bickerdike R, Tinsley J, Cockerill D, Mcloughlin MF, Graham DA, et al. The serum proteome of Atlantic salmon, Salmo salar, during pancreas disease (PD) following infection with salmonid alphavirus subtype 3 (SAV3). J Proteomics (2013) 94:423–36. doi: 10.1016/j.jprot.2013.10.016
106. Elschenbroich S, Kim Y, Medin JA, Kislinger T. Isolation of cell surface proteins for mass spectrometry-based proteomics. Expert Rev Proteomics (2010) 7:141–54. doi: 10.1586/epr.09.97
107. Schiess R, Mueller LN, Schmidt A, Mueller M, Wollscheid B, Aebersold R. Analysis of cell surface proteome changes via label-free, quantitative mass spectrometry. Mol Cell Proteomics (2009) 8:624–38. doi: 10.1074/mcp.M800172-MCP200
108. Ma C, Ye J, Kaattari SL. Differential compartmentalization of memory B cells versus plasma cells in salmonid fish. Eur J Immunol. (2013) 43:360–70. doi: 10.1002/eji.201242570
109. Crawford K, Gabuzda D, Pantazopoulos V, Xu J, Clement C, Reinherz E, et al. Circulating CD2+ monocytes are dendritic cells. J Immunol. (1999) 163:5920–8.
110. Eto H, Ishimine H, Kinoshita K, Watanabe-Susaki K, Kato H, Doi K, et al. Characterization of human adipose tissue-resident hematopoietic cell populations reveals a novel macrophage subpopulation with CD34 expression and mesenchymal multipotency. Stem Cells Dev. (2013) 22:985–97. doi: 10.1089/scd.2012.0442
111. Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochim Biophys Acta (2002) 1542:1–13. doi: 10.1016/S0167-4889(01)00166-5
112. Champion TC, Partridge LJ, Ong SM, Malleret B, Wong SC, Monk PN. Monocyte subsets have distinct patterns of tetraspanin expression and different capacities to form multinucleate giant cells. Front Immunol. (2018) 9:1247. doi: 10.3389/fimmu.2018.01247
113. Bonifaz L, Bonnyay D, Mahnke K, Rivera M, Nussenzweig MC, Steinman RM. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J Exp Med. (2002) 196:1627–38. doi: 10.1084/jem.20021598
114. Bühring HJ, Simmons PJ, Pudney M, Müller R, Jarrossay D, van Agthoven A, et al. The monoclonal antibody 97A6 defines a novel surface antigen expressed on human basophils and their multipotent and unipotent progenitors. Blood (1999) 94:2343–56.
115. Castro R, Abós B, González L, Aquilino C, Pignatelli J, Tafalla C. Molecular characterization of CD9 and CD63, two tetraspanin family members expressed in trout B lymphocytes. Dev Comp Immunol. (2015) 51:116–25. doi: 10.1016/j.dci.2015.03.002
116. Li Y-Q, Zhang J, Li J, Sun L. First characterization of fish CD22: An inhibitory role in the activation of peripheral blood leukocytes. Vet Immunol Immunopathol. (2017) 190:39–44. doi: 10.1016/j.vetimm.2017.07.006
117. Xu H, Liew LN, Kuo IC, Huang CH, Goh DL, Chua KY. The modulatory effects of lipopolysaccharide-stimulated B cells on differential T-cell polarization. Immunology (2008) 125:218–28. doi: 10.1111/j.1365-2567.2008.02832.x
118. Kehrl JH, Fauci AS. Identification, purification, and characterization of antigen-activated and antigen-specific human B lymphocytes. Trans Assoc Am Physicians (1983) 96:182–7.
119. Gong YF, Xiang LX, Shao JZ. CD154-CD40 interactions are essential for thymus-dependent antibody production in zebrafish: insights into the origin of costimulatory pathway in helper T cell-regulated adaptive immunity in early vertebrates. J Immunol. (2009) 182:7749–62. doi: 10.4049/jimmunol.0804370
120. Chapman HA, Wei Y, Simon DI, Waltz DA. Role of urokinase receptor and caveolin in regulation of integrin signaling. Thromb Haemost. (1999) 82:291–7.
121. Tarui T, Mazar AP, Cines DB, Takada Y. Urokinase-type plasminogen activator receptor (CD87) is a ligand for integrins and mediates cell-cell interaction. J Biol Chem. (2001) 276:3983–90. doi: 10.1074/jbc.M008220200
122. Vuillier F, Scott-Algara D, Dighiero G. Extensive analysis of lymphocyte subsets in normal subjects by three-color immunofluorescence. Nouv Rev Fr Hematol. (1991) 33:31–8.
123. Blanc P, Moro-Sibilot L, Barthly L, Jagot F, This S, de Bernard S, et al. Mature IgM-expressing plasma cells sense antigen and develop competence for cytokine production upon antigenic challenge. Nat Commun. (2016) 7:13600. doi: 10.1038/ncomms13600
124. Racine R, McLaughlin M, Jones DD, Wittmer ST, MacNamara KC, Woodland DL, et al. IgM production by bone marrow plasmablasts contributes to long-term protection against intracellular bacterial infection. J Immunol. (2011) 186:1011–21. doi: 10.4049/jimmunol.1002836
125. Agematsu K. Memory B cells and CD27. Histol Histopathol. (2000) 15:573–6. doi: 10.14670/HH-15.573
126. Chilosi M, Adami F, Lestani M, Montagna L, Cimarosto L, Semenzato G, et al. CD138/syndecan-1: a useful immunohistochemical marker of normal and neoplastic plasma cells on routine trephine bone marrow biopsies. Mod Pathol. (1999) 12:1101–6.
127. Bromage ES, Kaattari IM, Zwollo P, Kaattari SL. Plasmablast and plasma cell production and distribution in trout immune tissues. J Immunol. (2004) 173:7317–23. doi: 10.4049/jimmunol.173.12.7317
128. Herren B, Burrows PD. B cell-restricted human mb-1 gene: expression, function, and lineage infidelity. Immunol Res. (2002) 26:35–43. doi: 10.1385/IR:26:1-3:035
129. Sathish JG, Walters J, Luo JC, Johnson KG, Leroy FG, Brennan P, et al. CD22 is a functional ligand for SH2 domain-containing protein-tyrosine phosphatase-1 in primary T cells. J Biol Chem. (2004) 279:47783–91. doi: 10.1074/jbc.M402354200
130. Toba K, Hanawa H, Fuse I, Sakaue M, Watanabe K, Uesugi Y, et al. Difference in CD22 molecules in human B cells and basophils. Exp Hematol. (2002) 30:205–11. doi: 10.1016/S0301-472X(01)00791-3
131. Wen T, Mingler MK, Blanchard C, Wahl B, Pabst O, Rothenberg ME. The pan-B cell marker CD22 is expressed on gastrointestinal eosinophils and negatively regulates tissue eosinophilia. J Immunol. (2012) 188:1075–82. doi: 10.4049/jimmunol.1102222
132. Luger D, Yang YA, Raviv A, Weinberg D, Banerjee S, Lee MJ, et al. Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS ONE (2013) 8:e76115. doi: 10.1371/journal.pone.0076115
133. Bhargava P, Kallakury BV, Ross JS, Azumi N, Bagg A. CD79a is heterogeneously expressed in neoplastic and normal myeloid precursors and megakaryocytes in an antibody clone-dependent manner. Am J Clin Pathol. (2007) 128:306–13. doi: 10.1309/UXCDG9PWN7G89Y54
134. Warren IA, Ciborowski KL, Casadei E, Hazlerigg DG, Martin S, Jordan WC, et al. Extensive local gene duplication and functional divergence among paralogs in Atlantic salmon. Genome Biol Evol. (2014) 6:1790–1805. doi: 10.1093/gbe/evu131
135. Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic salmon genome provides insights into rediploidization. Nature (2016) 533:200. doi: 10.1038/nature17164
136. Coffman RL, Weissman IL. B220: a B cell-specific member of the T200 glycoprotein family. Nature (1981) 289:681. doi: 10.1038/289681a0
137. Hardy RR, Hayakawa K. B Cell Development Pathways. Ann Rev Immunol. (2001) 19:595–621. doi: 10.1146/annurev.immunol.19.1.595
138. Rodig SJ, Shahsafaei A, Li B, Dorfman DM. The CD45 isoform B220 identifies select subsets of human B cells and B-cell lymphoproliferative disorders. Hum Pathol. (2005) 36:51–7. doi: 10.1016/j.humpath.2004.10.016
139. Zikherman J, Weiss A. Alternative splicing of CD45: the tip of the iceberg. Immunity (2008) 29:839–41. doi: 10.1016/j.immuni.2008.12.005
140. Yoon SO, Zhang X, Lee IY, Spencer N, Vo P, Choi YS. CD9 is a novel marker for plasma cell precursors in human germinal centers. Biochem Biophys Res Commun. (2013) 431:41–6. doi: 10.1016/j.bbrc.2012.12.102
141. Won WJ, Kearney JF. CD9 is a unique marker for marginal zone B cells, B1 cells, and plasma cells in mice. J Immunol. (2002) 168:5605–11. doi: 10.4049/jimmunol.168.11.5605
142. Bao Y, Liu X, Han C, Xu S, Xie B, Zhang Q, et al. Identification of IFN-gamma-producing innate B cells. Cell Res. (2014) 24:161–76. doi: 10.1038/cr.2013.155
143. Winslow GM, Papillion Am, Kenderes KJ, Levack RC. CD11c+ T-bet+ memory B cells: Immune maintenance during chronic infection and inflammation? Cell Immunol. (2017) 321:8–17. doi: 10.1016/j.cellimm.2017.07.006
144. Karnell JL, Kumar V, Wang J, Wang S, Voynova E, Ettinger R. Role of CD11c(+) T-bet(+) B cells in human health and disease. Cell Immunol. (2017) 321:40–45. doi: 10.1016/j.cellimm.2017.05.008
145. Racine R, Chatterjee M, Winslow GM. CD11c expression identifies a population of extrafollicular antigen-specific splenic plasmablasts responsible for CD4 T-independent antibody responses during intracellular bacterial infection. J Immunol. (2008) 181:1375–85. doi: 10.4049/jimmunol.181.2.1375
146. Ghosn EE, Yang Y, Tung J, Herzenberg LA, Herzenberg LA. CD11b expression distinguishes sequential stages of peritoneal B-1 development. Proc Natl Acad Sci USA. (2008) 105:5195–200. doi: 10.1073/pnas.0712350105
147. Santos L, Draves KE, Boton M, Grewal PK, Marth JD, Clark EA. Dendritic cell-dependent inhibition of B cell proliferation requires CD22. J Immunol. (2008) 180:4561–9. doi: 10.4049/jimmunol.180.7.4561
Keywords: B cells, cell surface markers, teleost fish, salmon, CD22, CD79A, IgM, proteomics
Citation: Peñaranda MMD, Jensen I, Tollersrud LG, Bruun J-A and Jørgensen JB (2019) Profiling the Atlantic Salmon IgM+ B Cell Surface Proteome: Novel Information on Teleost Fish B Cell Protein Repertoire and Identification of Potential B Cell Markers. Front. Immunol. 10:37. doi: 10.3389/fimmu.2019.00037
Received: 31 October 2018; Accepted: 08 January 2019;
Published: 29 January 2019.
Edited by:
Hetron Mweemba Munang'andu, Norwegian University of Life Sciences, NorwayCopyright © 2019 Peñaranda, Jensen, Tollersrud, Bruun and Jørgensen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ma. Michelle D. Peñaranda, bWljaGVsbGUucGVuYXJhbmRhQHVpdC5ubw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.