
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 18 December 2018
Sec. Molecular Innate Immunity
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.02977
This article is part of the Research Topic Long Non-Coding RNAs and Immunity View all 6 articles
Circular ribonucleic acid (RNA) molecules (circRNAs) are covalently closed loop RNA molecules with no 5′ end caps or 3′ poly (A) tails, which are generated by back-splicing. Originally, circRNAs were considered to be byproducts of aberrant splicing. However, in recent years, development of high-throughput sequencing has led to gradual recognition of functional circRNAs, and increasing numbers of studies have elucidated their roles in cancer, neurologic diseases, and cardiovascular disorders. Nevertheless, studies of the functions of circRNAs in the immune system are relatively scarce. In this review, we detail relevant research on the biogenesis and classification of circRNAs, describe their functional mechanisms and approaches to their investigation, and summarize recent studies of circRNA function in the immune system.
Circular RNA (circRNA) is a covalently closed loop molecular form of RNA, and was discovered more than two decades ago (1, 2). Initially, circRNA was considered to be an aberrant byproduct of splicing (3–6). Recently, numerous circRNAs have been identified as a consequence of rapid developments in bioinformatics and high-throughput sequencing. Jeck et al. detected >25,000 circRNAs in fibroblasts using a genome-wide RNase R enrichment strategy (7). Memczak et al. identified 1950 circRNAs in humans, 1903 in mice, and 724 in Caenorhabditis elegans using RNA-sequencing data combined with analyses of the human leukocyte database (8). circRNAs are also expressed in fungi, plants, and protists (9–12).
Current research on circRNAs focuses on their role in cancer, neurologic diseases, and cardiovascular disorders. In this review, we cover relevant research on the biogenesis and classification of circRNAs and their functional mechanisms and methodological approaches to their study, along with summarizing recent investigations of the roles of circRNAs in the immune system.
Rather than canonical splicing, circRNAs are generated through back-splicing (13, 14) (Figure 1). Back-splicing can be accompanied by transcription (15) or may occur after transcription has been completed (16). Three main mechanisms have been reported to produce circRNAs: exon skipping, intron pairing-driven circularization, and RNA binding protein (RBP)-driven circularization.
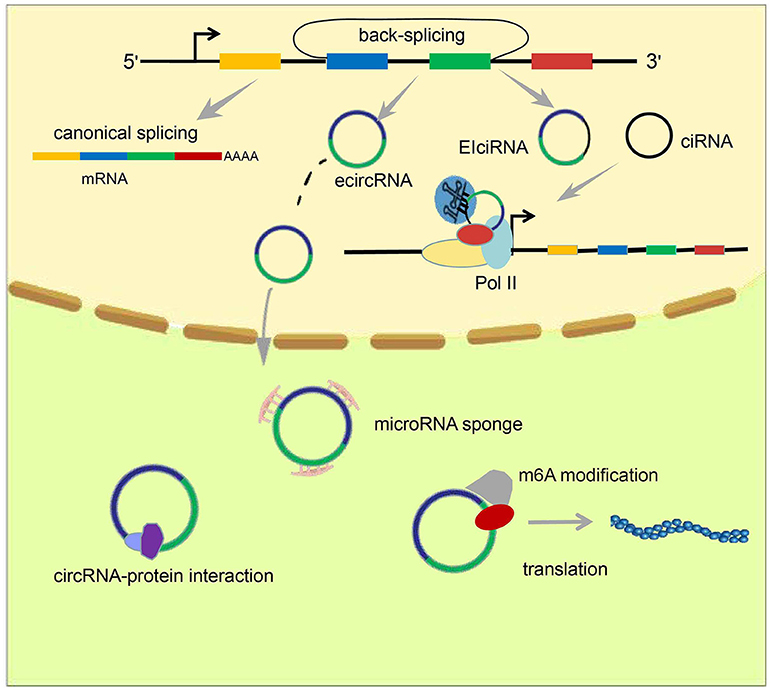
Figure 1. Formation and classification of circRNAs and models of circRNA function. circRNAs are formed by back-splicing. There are three major types of circRNA: ecircRNA, ciRNA, and EIciRNA. EcircRNAs function mainly in the cytoplasm through the “microRNA sponge” mechanism, whereas EIciRNA and ciRNA accumulate in the nucleus and facilitate transcription of their parent genes via cis-regulatory effects. In addition, some circRNAs have been reported to act by interacting with or encoding proteins.
Skipping of exons leads to the formation of a lariat structure containing the skipped exon. If introns are spliced before the lariat is unraveled by debranching enzymes, a stable exon-containing circRNA will be produced (7), along with a linear transcript excluding the skipped exon(s). This mechanism was an early understanding of how circRNAs are formed because the linear transcripts that are produced alongside circRNAs were described some years ago (17, 18).
Intron pairing-driven circularization is based on reverse complementary matches (RCMs) within flanking introns, and was established independently of the exon-skipping mechanism of circularization. RCMs can induce base-pairing between flanking introns, promoting a hairpin formation, bringing the 5′ and 3′ termini of an exon into spatial proximity, and inducing “head-to-tail” splicing. Intriguingly, the protein adenosine deaminases acting on RNA (ADAR) also participate in this process, together with RCMs. ADAR can unzip double-stranded RNA molecules by converting adenosine residues within them into creatinine molecules, thereby reducing the formation of circRNAs (19). Furthermore, Jeck et al. determined that, unlike sequence regions involved in generation of non-circular transcripts, the 200 bp upstream or downstream of a back-splice site contain canonical complementary ALU repeats, suggesting that intronic pairing may drive the circularization of exonic circRNAs (ecircRNAs). Jeck et al. also found that the length of introns flanking ecircRNAs are greater than those flanking linear RNA exons (7).
In addition to the involvement of introns and exons, some RNA-binding proteins also contribute to the formation of circRNAs. For example, in Drosophila, the splicing factor muscleblind (MBL) drives the circularization of the second exon of MBL to produce circMBL, by binding specifically to MBL binding sites within introns flanking circMBL sequences (15). More importantly, the interaction between MBL and circMBL contributes to regulation of the levels of MBL protein. When MBL is present in excess, it reduces the production of MBL mRNA by promoting circMBL formation. A protein called “quaking” (QKI) was identified by Conn et al. and shown to promote the production of circRNAs during human epithelial–mesenchymal transition by binding to a motif within circRNA flanking introns (20). RBM20 is also an RNA-binding protein that regulates numerous cardiac-specific gene-editing processes. Mutation of RMB20 is involved in dilated cardiomyopathy through its effects on the generation of circRNAs from Titin (21). The RNA-binding protein FUS can mediate circRNA formation by mediating RNA back-splicing in neurons (22). Moreover, HNRNPL, an RNA splicing factor, participates in regulation of circRNA formation in prostate cancer (23). In conclusion, the formation of circRNAs is dependent upon the regulation of cis elements and trans-factors (24).
Studies have identified three main types of circRNA: ecircRNA, circular intronic RNA (ciRNA), and exon–intron circRNA (EIciRNA) (Figure 1). Published data suggest that ecircRNAs function mainly through the microRNA “sponge” mechanism, proposed first by Memczak et al. who found there were 63 microRNA-7 binding sites on CDR1as, and hence designated CDR1as a “microRNA sponge.” EcircRNA can enhance levels of microRNA target genes through adsorption of microRNA molecules. Unlike ecircRNAs, intron-containing circRNAs (ciRNAs or EIciRNAs), in general, reside in the nucleus and regulate gene transcription (25–27). Chen et al. found that the function of ci-ankrd52, which is derived from the second intron of ANKRD52, may depend on a consensus motif containing a 7-nucleotide (nt) GU-rich element near its 5′ splice site and an 11-nt C-rich element close to the branchpoint to avoid being debranched. Ci-ankrd52 accumulates mainly in the nucleus and promotes transcription of ANKRD52 through a cis-regulatory effect of RNAPol II (25).
circRNAs are not degraded readily by RNase R, and are more stable than their cognate linear mRNAs, with half-lives exceeding 48 h (7). circRNAs also exhibit a degree of conservation between species. Jeck et al. identified ecircRNAs from 14.4% of actively transcribed genes in human fibroblasts. Furthermore, they identified 69 ecircRNAs from murine testes that mapped to precisely orthologous chromosome locations compared with human circRNAs (7).
Numerous circRNAs exhibit specificity to tissue, developmental period, or cell type. Memczak et al. found that different cells express specific circRNAs through identification of circRNAs in four human cell types: cluster of differentiation (CD)19+ leukocytes, CD34+ leukocytes, neutrophils, and HEK293 cells. Analogously, they noted that several nematode circRNAs were expressed in oocytes but absent in 1- or 2-cell embryos (8).
Although most circRNAs do not have the potential for translation (28), several recent studies have confirmed that circRNAs can be translated into proteins. Legnini et al. screened differentially expressed circRNAs in Duchenne muscular dystrophy by RNA sequencing. They determined that circ-ZNF609 can affect muscle formation through regulation of myoblast proliferation. Surprisingly, they also found that circ-ZNF609 can encode a protein (29), but whether this protein has a role in myoblast proliferation is not known. Yang et al. demonstrated that modification of N6-methyladenosine (m6A), which is induced by the methyltransferase METTL3/14 and suppressed by the demethylase FTO, promotes protein translation through recruiting an initiation factor, eIF4G2, and a m6A reader, YTHDF3. Furthermore, they found that m6A-containing circRNAs, with the potential to be translated into proteins, are common in the human transcriptome (30). Yang et al. also demonstrated that circ-FBXW7 can be translated into the protein FBXW7-185aa, thereby co-regulating the stability of c-Myc, along with its parent gene-encoded protein, FBXW7, to inhibit the progress of malignant glioma (31). Pamudurti et al. described the discovery of numerous circRNA-translated proteins or peptides based on ribosome “footprinting” experiments in Drosophila brains. They demonstrated that ribosomes can bind at the stop codon of circMbl and that a protein encoded by circMbl can be identified by protein spectroscopy (32).
Several reports have detailed many chemical modifications present on DNA and RNA. Surprisingly, recent breakthrough investigations of chemical modification of circRNAs have demonstrated that a m6A modification occurs in circRNA. This was discovered first by Yang et al. who further demonstrated that this modification promotes protein translation through recruitment of the initiation factor eIF4G2, and the m6A reader YTHDF3 (30) (Figure 1). Zhou et al. further confirmed the existence of the m6A modification in circRNAs, and also proposed some characteristics of m6A-modified circRNAs (33).
MicroRNAs produced by Dicer processing enzymes from single-stranded RNA precursors with a hairpin structure of ~70–90 nt are single-stranded small RNAs of ~21–23 nt (34, 35). Through initiation of RNA-induced silencing complex (RISC), mRNAs are degraded or their translation is hindered by base pairing with target gene transcripts (36, 37). Memczak et al. was the first to propose the microRNA sponge model. They found that there were 63 microRNA-7 binding sites within the circular transcript CDR1as (ciRS-7). Hansen et al. determined that a testes-specific circRNA called Sry can also serve as a sponge for microRNA-138 (38). Recent studies have shown that various circRNAs can function to adsorb microRNAs, thereby releasing target mRNAs (Figure 1). This process is associated with several diseases. In 2017, Han et al. demonstrated that circMTO1 upregulates p21 by competitively binding to microRNA-9, thereby inhibiting the proliferation of hepatocellular carcinoma (39). Zhong et al. clarified that the circRNA MYLK acts as a competing endogenous RNA (ceRNA) to bind directly to microRNA-29a, thereby promoting the development of bladder cancer by activating VEGFA/VEGFR2 signaling pathway and the Ras-Erk pathway (40). The circRNA circEPSTI1 was discovered to be an endogenous competitive RNA and sponge for microRNAs. Through binding to microRNA-4753 and microRNA-6809, circEPSTI1 upregulates expression of B cell CLL/lymphoma 11A (BCL11A), promotes the proliferation of triple-negative breast cancer cells, and inhibits their apoptosis (41).
Rather than acting as microRNA sponges in the cytoplasm, some circRNAs act by interacting with corresponding proteins (Figure 1). In 2016, circ-foxo3 was found to impede cell-cycle progression by forming a ternary complex with p21 and Cdk2 proteins (42). In addition, another circRNA, circ-Amotl1, which is highly expressed in cancerous cell lines, can increase nuclear retention of the oncogenic protein c-myc to promote its stability, and increase its affinity for binding to several promoters, thereby upregulating c-myc targets such as HIF1α, Cdc25a, ELK-1, and JUN (43). This observation reveals a novel function of circRNAs in tumorigenesis. Conceivably, there are circRNAs other than circ-Amotl1 that act by similar mechanisms. In 2017, Abdelmohsen et al. was the first to report competition between a circRNA and its cognate mRNA for the RNA-binding protein HuR, which has been studied extensively and can regulate protein expression through interaction with a wide range of RNAs. They proposed that high levels of circPABPN1, a circRNA derived from PABPN1, repress HuR binding to PABPN1 mRNA through binding to HuR itself, causing a reduction in PABPN1 mRNA translation (44). This mechanism, based on the interaction between a circRNA and its cognate mRNA, provided new insights and spurred further investigation of the roles of circRNAs in the nucleus.
Multiple non-coding RNAs, including HOTAIR and MALAT1, are known to regulate gene transcription in trans; that is, they influence the transcription of genes other than their parent genes. However, there have been studies that have shown circRNAs, mainly intron-containing circRNAs (ciRNAs or EIciRNAs), can have cis-regulatory effects on gene expression. In 2013, Chen et al. reported that ci-ankrd52, which is generated from the second intron of ANKRD52, accumulates primarily in the nucleus and promotes transcription of ANKRD52 via the cis-regulatory effects of RNAPol II (25). In 2014, Li et al. proposed a model for the cis-regulatory effects of EIciRNAs based on their findings from investigation of two EIciRNAs: circEIF3J and circPAIP2. They proposed that EIciRNAs may interact with proteins such as U1 snRNP via RNA–RNA interplay between U1 snRNA and EIciRNA. Then, EIciRNA–U1 snRNP complexes could interact with Pol II at the promoter regions of parental genes, thereby enhancing their transcription (Figure 1). This phenomenon produces a positive feedback loop because, once transcription has been initiated, EIciRNA generation will increase, further promoting gene transcription (26).
In addition to being detected inside cells, circRNA has also been reported to be present in extracellular fluids. Li et al. enriched exosomes in serum samples from patients suffering from colon cancer to examine circRNA expression in exosomes. Compared with healthy controls, hundreds of circRNAs were expressed differentially in serum exosomes from patients with colon cancer (45). In 2016, Guarnerio et al. was the first to report that chromosomal translocations can produce fusion circRNAs in acute promyelocytic leukemia. Furthermore, using an MLL/AF9-AML model, they found that f-circM9 could contribute to progression of acute myeloid leukemia (46). Based on those findings, Tan et al. confirmed that the common fusion gene in non-small-cell lung cancer (NSCLC), EML4-ALK, can produce a fused circRNA called F-circEA, and demonstrated that F-circEA promotes the proliferation and migration of cells. F-circEA was also detected in the serum of EML4-ALK-positive patients, indicating that this circRNA is highly likely to be useful as a diagnostic marker for EML4-ALK-positive NSCLC (47). With further research it is likely that disease-specific circRNAs will be developed as disease biomarkers.
Identification of circRNA after sequencing is the first step in circRNA research. Various identification tools have been developed. Whether the identification of circRNAs is accurate and comprehensive is dependent upon the rigor and reliability of the algorithm. The Find_circ algorithm uses bowtie2 to map the original reads to the reference genome, discards all mapped sequences, takes 20 nt of each unmapped read as an anchor, and then determines the location of the anchor in the genome again to identify whether the splicing of circRNA is present (8). The CIRCexplorer algorithm uses the TopHat algorithm to map the RNA-sequencing reads to the human hg19 reference genome, and then maps the unmapped reads with the TopHat-Fusion algorithm. Such reads, unmapped with TopHat but mapped with TopHat-Fusion on the same chromosome in a back-spliced order, are extracted as candidate back-spliced junctionreads (48). The CIRI algorithm proposes paired chiastic clipping (PCC) signals to identify circRNAs. The PCC signal is detected by collecting and comparing the alignment information of all the segments of a read. PCC signals do not rely on existing annotation information, so back-splicing can be identified from zero to predict various types of circRNA, including intronic and intergenic circRNAs (49). In 2015, Hansen et al. found that short circRNAs (especially circRNAs < 200 nt in length) are degraded readily by RNase R, and that a circRNA predicted by a single algorithm specifically has a higher false-positive rate. Conclusively, they suggested that identification of circRNAs can be done using a combination of different tools or RNA libraries with linear RNA being removed (50). We have summarized the four commonly used circRNA algorithms in Table 1.
RNase R is a member of the Escherichia coli RNR superfamily. It can cleave RNA in the 3′-5′ direction and digest almost all linear RNAs, but it cannot digest circRNAs readily (52). High-throughput sequencing for genome-wide identification of circRNAs requires enrichment of circRNA through treatment of samples with RNase R to remove linear RNAs, followed by enhancement of the concentration of circRNAs to facilitate their identification. Two approaches are used for the identification of circRNAs: reverse transcription-polymerase chain reaction (RT-PCR) and northern blotting. In RT-PCR, after digestion by RNase R, cDNA samples are amplified with divergent primers and convergent primers. Subsequent agarose-gel electrophoresis reveals that amplification with divergent primers generates a band in the RNase R(+) group, whereas the convergent primers produce no band. Divergent and convergent primers generate bands in the RNase R(–) group, indicating that circRNAs are present and resistant to digestion by RNase R (31, 53). Northern blotting is conducted using probes specific for circRNA and mRNA. The results show that linear mRNA cannot be detected in the RNase R(+) group, whereas its corresponding circRNA is visible, indicating that mRNAs are digested, whereas circRNAs are not (54). Notably, some circRNAs will also decrease in abundance after long-term digestion by RNase R, probably because of susceptibility to RNase R.
Technology based on RNA interference is used widely to knockdown circRNA expression. To eliminate non-specific knockdown effects on cognate linear RNA, a specific small interfering RNA (siRNA) or short-hairpin RNA (shRNA) targeting circRNA must be directed to the back-splicing site. This strategy limits the design of siRNA sequences, and siRNA or shRNA targeting back-splicing sites will be partially complementary to the cognate linear RNA, raising the possibility of unwanted effects on expression of linear RNA. To solve this problem, Li et al. proposed that the control sequence should be partially replaced (~10 nt) by a siRNA sequence targeting the back-splicing site (24). Similarly, knockout of circRNAs in animals risks simultaneously influencing expression of the cognate linear RNA and completely knocking out a gene is highly likely to affect expression of neighboring genes (55, 56). Based on the mechanism of circRNA production, Zhang et al. achieved knockout of circGCN1LI in human PA1 cells by removing an intron complementary sequence using CRISPR/Cas9 (16).
Plasmids used commonly to overexpress circRNAs are universal loop-forming framework vectors and gene-specific flanking sequence vectors. For example, Liang et al. cloned the exon2/3 and flanking sequences of ZKSCAN1 to construct a vector, and then overexpressed circ-ZKSCAN1. Then, the flanking sequence was modified, and exons2/3 of ZKSCAN1 was replaced with a polyclonal restriction site to construct an empty vector: pcDNA3.1(+) CircRNA Mini Vector (57). Overexpression using a gene-specific flanking vector sequence is consistent with generation of natural circRNAs in vivo. Based on the characteristics of circRNA flanking sequences, length-appropriate flanking repeats (Alu elements) and circRNA sequences are cloned to generate a eukaryotic expression vector construct. In 2015, Lu et al. found that tRNA introns can form circular RNAs (tricRNAs) (58) and, in 2016, Schmidt et al. created a new circRNA expression vector based on tRNA splicing and transformation. First, they designed restriction enzyme sites in the two bulge–helix–bulge (BHB) regions of the tRNA intron and inserted the circRNA sequence for expression (59). In brief, the vector produced a circRNA by joining BHB motifs via an RtcB enzyme, which was not dependent upon a reverse-complementary sequence.
In general, different circRNAs with different loop-forming characteristics exhibit variation in looping efficiency. Moreover, circRNA production by overexpression vectors is accompanied by the production of linear isoforms, which is an additional challenge for strategies employing circRNA overexpression.
Few reports have elucidated the role of circRNA in immune responses. Nevertheless, Chen et al. attempted to construct circRNAs to transfect cells based on in vitro transcription followed by auto-splicing circulation. Surprisingly, they found that in vitro circRNAs induced activation of cellular immune response pathways and inhibited RNA virus infection, which is mediated by retinoic acid-inducible gene-I (RIG-I); however, endogenous circRNAs did not induce this pathway because of their binding to specific RNA-binding proteins (60) (Figure 2). These findings suggest that exogenous introduction of circRNAs could be used to activate antiviral immune responses for therapeutic purposes. Nevertheless, some interesting questions must be solved: How do RBPs recognize self and non-self circRNAs, and then induce activation of non-self circRNAs on the RIG-1 pathway?
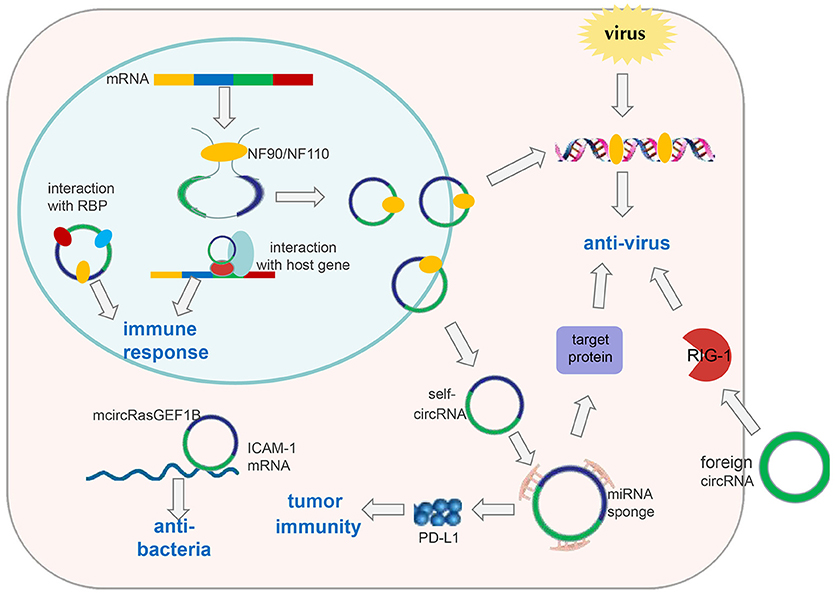
Figure 2. CircRNAs in immune responses. Exogenous circRNAs can activate the RIG-1 cellular immune response pathway to suppress viral replication. The immune factor NF90/NF110 can promote circRNA formation by stabilizing the intron complementary sequence. Under viral infection, NF90/NF110 is exported from the nucleus to the cytoplasm to inhibit virus replication. Meanwhile, circRNA expression in the nucleus is decreased. Thus, endogenous circRNAs can be used as “molecular indicators” of antiviral proteins to prompt antiviral immune responses. circRNAs can also function as “microRNA sponges” to increase expression of target proteins involved in antiviral responses or tumor immunity, such as PD-L1. mcircRasGEF1B can help to protect cells from bacterial infection by enhancing the stability of mature ICAM-1 mRNA. In addition, circRNAs can be involved in immune responses by interacting with proteins or their host genes.
In addition, Li et al. found that circRNAs are involved in viral infection via the immune response factor NF90/NF110 (61). First, they applied a genome-wide siRNA screening strategy targeting all unique human genes with a Dox-inducible circmCherry expression vector to profile proteins involved in circRNA biogenesis. Consequently, they determined that NF90/NF110, encoded by the interleukin enhancer binding factor 3 (ILF3) gene, promoted circRNA formation by stabilizing the intron complementary sequence. Upon viral infection, NF90/NF110 was exported from the nucleus to the cytoplasm to inhibit virus replication. Meanwhile, circRNA expression in the nucleus was decreased (Figure 2). In this way, circRNAs may be used as “molecular indicators” of NF90/NF110 to prompt antiviral immune responses. However, the detailed mechanisms by which antiviral proteins promote circRNA biogenesis and key elements within circRNAs to interact with antiviral proteins have yet to be explored.
The two studies mentioned above provide evidence to support circRNA involvement in antiviral immunity through interactions with specific antiviral proteins (62). However, one of the studies elucidated the function of exogenous circRNAs in antiviral immunity, and the other study suggested endogenous circRNAs may be used as molecular indicators of antiviral proteins to prompt antiviral immune responses. In addition to interacting with antiviral proteins, circRNAs may have other roles during antiviral immune responses. Through sequencing of the whole transcriptome, Shi et al. found that expression of 536 circRNAs was dysregulated significantly in herpes simplex virus 1 (HSV-1) -infected cells in contrast to uninfected human fibroblasts. Similarly, they screened differentially expressed genes and microRNAs in HSV-1-infected cells. Furthermore, they undertook analyses of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Their results suggested that these differentially expressed genes were very enriched in the pathways of immune responses, such as the NOD-like receptor signaling pathway and JAK-STAT signaling pathway (63). An integrated analysis of the circRNA–microRNA–gene axis revealed circRNAs can regulate the genes associated with host immune responses, which was mediated by microRNAs. These data suggest that circRNAs can regulate host antiviral immune responses through interactions with the corresponding microRNAs (Figure 2). However, these results based on large-scale bioinformation analyses should be validated experimentally.
Interestingly, several studies have demonstrated that some viruses can encode their own microRNAs, which facilitates the entry, replication, and virulence of viruses by targeting host transcripts, including some antiviral signaling molecules (64–67). For example, microRNA-BHRF1-3 encoded by the Epstein–Barr virus diminishes the levels of CXC-chemokine ligand 11, a chemoattractant in immune responses (68). In recent years, ceRNAs, including long non-coding RNA and circRNA (which can combine with microRNAs completely with a protein-coding target) have been studied widely. Ghosal et al. established a database named “HumanViCe” in which circRNAs that can sponge virus microRNAs can be predicted, and these circRNAs are enriched in pathways associated with the entry and replication of viruses and host immune responses (69). In summary, HumanViCe can aid exploration of the roles of circRNAs in viral infection but also circRNAs may act as potential antiviral targets.
Ng et al. identified a lipopolysaccharide (LPS)-inducible circRNA generated from linear RasGEF1B, named mcircRasGEF1B, the expression of which is dependent upon the LPS-Toll-like receptor-4-nuclear factor-kappa B (LPS-TLR4-NF-κB) pathway. mcircRasGEF1B is cell type-specific, exhibits evolutionary conservation between mice and humans, and localizes preferentially to the cytoplasm (70). Also, its knockdown using the corresponding antisense oligodeoxynucleotide (ASOs) reduces intercellular adhesion molecule (ICAM-1) expression in the LPS/TLR4 signaling pathway by affecting the stability of the mature ICAM-1 mRNA, but not mRNA splicing (70) (Figure 2). In the immune system, ICAM-1 recruits leukocytes to sites of tissue inflammation, as well as enhancing adhesion between antigen-presenting cells and T cells (71, 72). In addition, ICAM-1 has been reported to inhibit M2 polarization of macrophages in tumors (73). Therefore, we speculate that mcircRasGEF1B may contribute to suppression of polarization of M2 macrophages during immune responses. These discoveries broaden our understanding and suggest that circRNAs may be important for the “fine tuning” of immune responses, and may help to protect cells from microbial infection.
The relationship between microRNAs and immunity has been well-studied, leading to the hypothesis that circRNA may contribute to immune regulation through interactions with microRNAs. Zhang et al. reported that hsa_circ_0020397 can upregulate expression of PD-L1 (the target gene of microRNA-138) by binding to microRNA-138 in colorectal cancer cells. The consequent increase in PD-L1 levels contributes to tumor escape from immune responses (74, 75) (Figure 2). This information provides new insights for “checkpoint therapy” in cancer patients. Zheng et al. demonstrated that circHIPK3 rescues the downregulation of microRNA-124 on expression of interleukin (IL)-6R (76), implying that circHIPK3 may function in tumor immunity response. There are several bioinformatics databases that can be used to predict circRNAs that could bind to microRNAs, including circbase, starBasev2.0, and circinteractome (77).
In addition to microRNAs, circRNAs can interact directly with proteins that function in immune responses. Similarly, we can predict such circRNAs using bioinformatics databases, then verify the predictions using in vitro and in vivo experiments. Using the circinteractome database, we found that the host gene of hsa_circ_0032139 is HIF1A, which plays an important part in inflammation via NF-κB and mitogen-activated protein kinase (MAPK) pathways (78). Under hypoxia, degradation of HIF1A protein is prevented, leading to its accumulation, and association with HIF1B to exert transcription regulation on target genes, including pro-inflammatory cytokines, most glycolytic enzymes, and glucose transporters, among others (79–83). Presumably, hsa_circ_0032139 can regulate inflammation through its association with HIF1A. Another circRNA, hsa_circ_0038481, has been predicted to associate with TLR4, a classical pattern-recognition receptor. This implies that hsa_circ_0038481 may be involved in LPS-stimulated signaling pathways, such as the NF-κB and MAPK.
Rheumatoid arthritis (RA) is a chronic, inflammatory synovitis-based systemic disease of unknown etiology (84). Zheng et al. screened the top-ten upregulated, and downregulated circRNAs in RA patients based on analyses of peripheral-blood mononuclear cell chips, and selected the top-five corresponding microRNAs for each circRNA (85). We can speculate that these differentially expressed circRNAs may function in RA by acting as sponges of the corresponding microRNAs, which have been reported to be associated with RA. More importantly, the study by Zheng et al. provides clues that circRNAs may have the same effects as their host genes in RA. For example, hsa_circ_0038644, one of the dysregulated circRNAs in RA, is spliced from PRKCB, which is associated with LPS-induced activation of the NF-κB signaling pathway (86). Therefore, we hypothesized that hsa_circ_0038644 can aggravate inflammation in RA patients.
Type-2 diabetes mellitus (T2DM) is characterized by hyperglycemia, insulin resistance, and chronic inflammation (87). Fang et al. found circANKRD36 to be upregulated markedly in the peripheral blood cells of T2DM patients. Furthermore, an association was noted between circANKRD36 expression and inflammatory factors (88). Therefore, circANKRD36 may serve as a potential biomarker and be involved in inflammation in T2DM.
Similarly, Li et al. measured circRNA expression in the T cells of patients with systemic lupus erythematosus (SLE). They found hsa_circ_0045272 to be downregulated significantly. Furthermore, they demonstrated that hsa_circ_0045272 regulated apoptosis and IL-2 production negatively (89). However, the mechanisms underlying the involvement of hsa_circ_0045272 in SLE pathogenesis merits further exploration.
Numerous studies have shown that a high proportion of tumor-infiltrating lymphocytes (TILs) in the tumor microenvironment can improve clinical outcomes. Weng et al. showed that high expression of hsa_circ_0064428 is associated with a low proportion of TILs, poor survival, large tumor volume, and tumor metastasis in patients with hepatocellular carcinoma (90). These observations suggest that hsa_circ_0064428 can act as a potential immune-associated prognosis biomarker for hepatocellular carcinoma.
Overall, the roles of circRNAs in immune diseases have been studied based on large-scale microarray and RNA sequencing analyses, by which differentially expressed circRNAs were screened to further verify their functions experimentally. However, the mechanisms by which circRNAs regulate disease development merit further exploration.
Although circRNAs have become a hot topic in RNA research in recent years, several important areas merit investigation. For example, Dong et al. showed that circRNA can be reverse-transcribed in vivo and fused into the genome to generate a pseudogene (91). Further research is needed to elucidate the molecular mechanisms underlying circRNA trans-transcriptional translocation and the effects of pseudogenes derived from circRNAs. In addition, unlike linear mRNA, construction of overexpression vectors for circRNA is challenging because it requires splicing of the two termini of amplified fragments to generate loop structures. Moreover, data in the mouse circRNA database is incomplete, which poses specific obstacles for in vivo experiments. Finally, circRNAs in the cytoplasm, which act as microRNA sponges, have been studied widely, but circRNAs that can function through this mechanism are in the minority. Most circRNAs rarely contain so many miRNA binding sites and have low expression (24, 28), so future research should explore other mechanisms of circRNAs.
Few studies have focused on circRNAs in the immune response. Reminiscent of studies in cancer, circRNA research in immunity also requires measuring circRNA expression, and then elucidating the function of differentially expressed circRNAs and the mechanisms underlying it. Notably, circRNA localization could aid exploration of the mechanisms by which circRNAs regulate immune responses. circRNAs in the cytoplasm are likely to act as microRNA sponges, whereas circRNAs in the nucleus are likely to interact with proteins, or elicit effects by promoting/suppressing the role of their host genes (Figure 2). Excitingly, circRNAs are promising markers or drug targets for some immune diseases, which could facilitate rapid diagnosis and treatment.
LY the first author, contributed to collection of references and manuscript preparation. JF and YZ contributed to manuscript modifications.
This work was supported by grants from the National Key R&D Program of China (2016YFC1305102), The National Natural Science Foundation of China (NSFC, 81671561), 1000 Young Talents Plan Program of China, Initial Funding for New PI, Fudan Children's Hospital and Fudan University, and Shanghai Municipal Planning Commission of Science and Research Fund (201740065) (to YZ). Shanghai Pujiang Program 16PJ1401600 (to JF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell (1993) 73:1019–30. doi: 10.1016/0092-8674(93)90279-Y
2. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science (1995) 268:415–7. doi: 10.1126/science.7536344
3. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. (1993) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
4. Pasman Z, Been MD. Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA (1996) 2:603–10.
5. Cocquerelle C, Daubersies P, Majérus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. (1992) 11:1095–8. doi: 10.1002/j.1460-2075.1992.tb05148.x
6. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell (1991) 64:607–13. doi: 10.1016/0092-8674(91)90244-S
7. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (2013) 19:141–57. doi: 10.1261/rna.035667.112
8. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495:333–8. doi: 10.1038/nature11928
9. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife (2015) 4:e07540. doi: 10.7554/eLife.07540
10. Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, Sabeti PC. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics (2015) 16:454. doi: 10.1186/s12864-015-1603-4
11. Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA (2015) 21:2076–87. doi: 10.1261/rna.052282.115
12. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE (2014) 9:e90859. doi: 10.1371/journal.pone.0090859
13. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA (2014) 20:1829−42. doi: 10.1261/rna.047126.114
14. Wilusz JE. A 360 degrees view of circular RNAs: From biogenesis to functions. Wiley Interdisc Rev RNA (2018) 9:e1478. doi: 10.1002/wrna.1478
15. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
16. Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, et al. The biogenesis of nascent circular RNAs. Cell Rep. (2016) 15:611–24. doi: 10.1016/j.celrep.2016.03.058
17. Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. (1997) 17:2985–93. doi: 10.1128/MCB.17.6.2985
18. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA. (1996) 93:6536–41. doi: 10.1073/pnas.93.13.6536
19. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
20. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
21. Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. (2016) 119:996–1003. doi: 10.1161/CIRCRESAHA.116.30956810.1161/CIRCRESAHA.116.309568
22. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. (2017) 8:14741. doi: 10.1038/ncomms14741
23. Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci USA. (2017) 114:E5207–15. doi: 10.1073/pnas.1617467114
24. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell (2018) 71:428–42. doi: 10.1016/j.molcel.2018.06.034
25. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
26. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
27. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Corrigendum: exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2017) 24:194. doi: 10.1038/nsmb0217-194a
28. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. (2014) 15:409. doi: 10.1186/s13059-014-0409-z
29. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell (2017) 66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017
30. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N-methyladenosine. Cell Res. (2017) 27:626–41. doi: 10.1038/cr.2017.31
31. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. (2018) 110, 304–15. doi: 10.1093/jnci/djx166
32. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021
33. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. (2017) 20:2262–76. doi: 10.1016/j.celrep.2017.08.027
34. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
35. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
36. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer (2006) 6:259–69. doi: 10.1038/nrc1840
37. Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. (2012) 6:590–610. doi: 10.1016/j.molonc.2012.09.006
38. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495:384–8. doi: 10.1038/nature11993
39. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (2017) 66:1151–64. doi: 10.1002/hep.29270
40. Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. (2017) 403:305–17. doi: 10.1016/j.canlet.2017.06.027
41. Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics (2018) 8:4003–15. doi: 10.7150/thno.24106
42. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. (2016) 44:2846–58. doi: 10.1093/nar/gkw027
43. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. (2017) 24:1609–20. doi: 10.1038/cdd.2017.86
44. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. (2017) 14:361–9. doi: 10.1080/15476286.2017.1279788
45. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. (2015) 25:981–4. doi: 10.1038/cr.2015.82
46. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell (2016) 166:1055–6. doi: 10.1016/j.cell.2016.07.035
47. Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. (2018) 28:693–5. doi: 10.1038/s41422-018-0033-7
48. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
49. Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. (2015) 16:4. doi: 10.1186/s13059-014-0571-3
50. Hansen TB, Venø MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. (2016) 44:e58. doi: 10.1093/nar/gkv1458
51. Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. (2010) 38:e178. doi: 10.1093/nar/gkq622
52. Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. (2002) 277:21624–9. doi: 10.1074/jbc.M202942200
53. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. (2006) 34:e63. doi: 10.1093/nar/gkl151
54. Rybak-Wolf A, Stottmeister C, GlaŽar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell (2015) 58:870–85. doi: 10.1016/j.molcel.2015.03.027
55. Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature (2017) 548:343–6. doi: 10.1038/nature23451
56. Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. CRISPRi-based genome-scale identification of functional long non-coding RNA loci in human cells. Science (2017) 355:aah7111. doi: 10.1126/science.aah7111
57. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. (2014) 28:2233–47. doi: 10.1101/gad.251926.114
58. Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA (2015) 21:1554–65. doi: 10.1261/rna.052944.115
59. Schmidt CA, Noto JJ, Filonov GS, Matera AG. A method for expressing and imaging abundant, stable, circular RNAs in vivo using tRNA splicing. Methods Enzymol. (2016) 572:215–36. doi: 10.1016/bs.mie.2016.02.018
60. Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell (2017) 67:228–38 e5. doi: 10.1016/j.molcel.2017.05.022
61. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell (2017) 67:214–27 e7. doi: 10.1016/j.molcel.2017.05.023
62. Cadena C, Hur S. Antiviral immunity and circular RNA: no end in sight. Mol Cell (2017) 67:163–4. doi: 10.1016/j.molcel.2017.07.005
63. Shi J, Hu N, Mo L, Zeng Z, Sun J, Hu Y. Deep RNA Sequencing reveals a repertoire of human fibroblast circular RNAs associated with cellular responses to herpes simplex virus 1 infection. Cell Physiol Biochem. (2018) 47:2031–45. doi: 10.1159/000491471
64. Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. (2005) 7:1684–95. doi: 10.1111/j.1462-5822.2005.00598.x
65. Nair V, Zavolan M. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. (2006) 14:169–75. doi: 10.1016/j.tim.2006.02.007
66. Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science (2004) 304:734–6. doi: 10.1126/science.1096781
67. Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature (2008) 454:780–3. doi: 10.1038/nature07103
68. Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, et al. EBV MicroRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. (2008) 68:1436–42. doi: 10.1158/0008-5472.CAN-07-5126
69. Ghosal S, Das S, Sen R, Chakrabarti J. HumanViCe: host ceRNA network in virus infected cells in human. Front Genet. (2014) 5:249. doi: 10.3389/fgene.2014.00249
70. Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. (2016) 13:861–71. doi: 10.1080/15476286.2016.1207036
71. Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell (1989) 56:849–53. doi: 10.1016/0092-8674(89)90689-2
72. Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med. (2000) 28:1379–86. doi: 10.1016/S0891-5849(00)00223-9
73. Yang M, Liu J, Piao C, Shao J, Du J. ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis. (2015) 6:e1780. doi: 10.1038/cddis.2015.144
74. Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. (2017) 41:1056–64. doi: 10.1002/cbin.10826
75. Xu Z, Li P, Fan L, Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol. (2018) 9:9. doi: 10.3389/fimmu.2018.00009
76. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. (2016) 7:11215. doi: 10.1038/ncomms11215
77. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer (2017) 16:58. doi: 10.1186/s12943-017-0630-y
78. Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-kappaB. Biochem J. (2006) 396:517–27. doi: 10.1042/BJ20051839
79. Dengler VL, Galbraith M, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. (2014) 49:1–15. doi: 10.3109/10409238.2013.838205
80. Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell (2003) 112:645–57. doi: 10.1016/S0092-8674(03)00154-5
81. Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. (2009) 9:609–17. doi: 10.1038/nri2607
82. Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. (2006) 3:187–97. doi: 10.1016/j.cmet.2006.01.012
83. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. (2008) 7:11–20. doi: 10.1016/j.cmet.2007.10.002
84. Firestein GS. Evolving concepts of rheumatoid arthritis. Nature (2003) 423:356–61. doi: 10.1038/nature01661
85. Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep. (2017) 16:8029–36. doi: 10.3892/mmr.2017.7638
86. Sheng YJ, Gao JP, Li J, Han JW, Xu Q, Hu WL, et al. Follow-up study identifies two novel susceptibility loci PRKCB and 8p11.21 for systemic lupus erythematosus. Rheumatology (Oxford) (2011) 50:682–8. doi: 10.1093/rheumatology/keq313
87. Seidell JC. Obesity, insulin resistance and diabetes–a worldwide epidemic. Br J Nutr. (2000) 83 (Suppl. 1):S5–8. doi: 10.1017/S000711450000088X
88. Fang Y, Wang X, Li W, Han J, Jin J, Su F, et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. (2018) 42:1865–74. doi: 10.3892/ijmm.2018.3783
89. Li LJ, Zhu ZW, Zhao W, Tao SS, Li BZ, Xu SZ, et al. Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology (2018) 155:137–49. doi: 10.1111/imm.12940
90. Weng Q, Chen M, Li M, Zheng YF, Shao G, Fan W, et al. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. (2018) doi: 10.1136/jmedgenet-2018-105440. [Epub ahead of print].
Keywords: circRNA, biogenesis, function, research approaches, immune regulation
Citation: Yang L, Fu J and Zhou Y (2018) Circular RNAs and Their Emerging Roles in Immune Regulation. Front. Immunol. 9:2977. doi: 10.3389/fimmu.2018.02977
Received: 18 September 2018; Accepted: 04 December 2018;
Published: 18 December 2018.
Edited by:
Adam Williams, Jackson Laboratory, United StatesCopyright © 2018 Yang, Fu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yufeng Zhou, eWZ6aG91MUBmdWRhbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.