
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 04 December 2018
Sec. Nutritional Immunology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.02885
This article is part of the Research Topic Human Antibodies Against The Dietary Non-Human Neu5Gc-carrying Glycans in Normal and Pathologic States View all 12 articles
Sialic acids, a group of acidic sugars abundantly expressed in the tissues of deuterostome animals but rarely found in microbes, serve as a “signature of self” for these animals. Cognate sensors for sialic acids include Siglecs, a family of transmembrane lectins of vertebrate immune systems that recognize glycans containing sialic acids. A type of sialic acid called N-glycolylneuraminic acid (Neu5Gc) is abundant in many mammalian lineages including great apes, the closest extant relatives of modern human, but was lost in the lineage leading to modern human via the pseudogenization of the CMAH gene encoding the enzyme that converts N-acetylneuraminic acid (Neu5Ac) to Neu5Gc. Loss of Neu5Gc appears to have influenced the evolution of human Siglecs, such as the adjustment of sialic acid binding preferences and the inactivation of at least one Siglec. In addition, various mechanistic studies using model systems and genetic association studies have revealed that some human Siglecs interact with pathogens and influence the outcome of infections, and these pathogens in turn likely influence the evolution of these Siglecs. By understanding the evolutionary forces affecting Siglecs, we shall achieve a better appreciation of Siglec functions, and by understanding Siglec functions, we can obtain deeper insight into the evolutionary processes driving Siglec evolution.
The role of immunity is to distinguish self vs. non-self (or what is not dangerous vs. dangerous) and to eliminate or contain the latter. Various biomolecules (nucleotides, peptides, lipids, polysaccharides, and their combinations) can be a signature of non-self (i.e., pathogen-associated molecular patterns; PAMPs), as exemplified by the diversity of ligands for Toll-like receptors, C-type lectin-like receptors, RIG-I-like receptors, and NOD-like receptors, all of which work as “pattern-recognition receptors” (1–4). Meanwhile, the signature of self (i.e., self-associated molecular patterns; SAMPs) is less well-understood, but some glycoconjugates would qualify as such (5, 6). Sialic acids are commonly synthesized by deuterostome animals and displayed on the cell surface in abundance but are rare in microbes (7), making them an ideal SAMP for distinguishing self- vs. non-self (5, 6).
For a chemical entity to be a molecular signature of self or non-self for the immune system, there must be a sensor that recognizes it. For sialic acids, Siglecs appear to be the primary pattern-recognition receptors (8–11). Siglec is a composite word from “sialic acid,” “immunoglobulin (Ig) superfamily,” and “lectins” (12). The Siglec family appears to be present only in vertebrates (13, 14). Siglecs are type 1 transmembrane proteins, with an extracellular domain consisting of multiple Ig-like domains (of which the N-terminal Ig-like domain is primarily responsible for the recognition of sialoglycans), followed by a single-pass transmembrane domain and cytoplasmic tail (Figure 1). Most of the known mammalian Siglecs are expressed on leukocytes and have an intracellular sequence motif called the immunoreceptor tyrosine-based inhibitory motif (ITIM) that recruits tyrosine phosphatase SHP-1 and thus transduces inhibitory signals. Thus, they are considered to function as sensors for sialic acids as a molecular signature of self. [However, there are some examples that imply this generalization may be somewhat too simplistic (17, 18).]. Although rodents are essential model animals for mechanistic studies in immunology, differences in primate and rodent CD33-related Siglecs (15) impose a significant challenge in the extrapolation of findings in rodents to human immunology. This situation parallels that of other immunoglobulin-like receptor families, leukocyte immunoglobulin-like receptors (LILR) and killer cell immunoglobulin-like receptors (KIR), that are encoded in a gene cluster on the same human chromosomal region as CD33-related Siglecs (chromosome 19q13.4) and are involved in self-recognition through interaction with MHC class I (19–21).
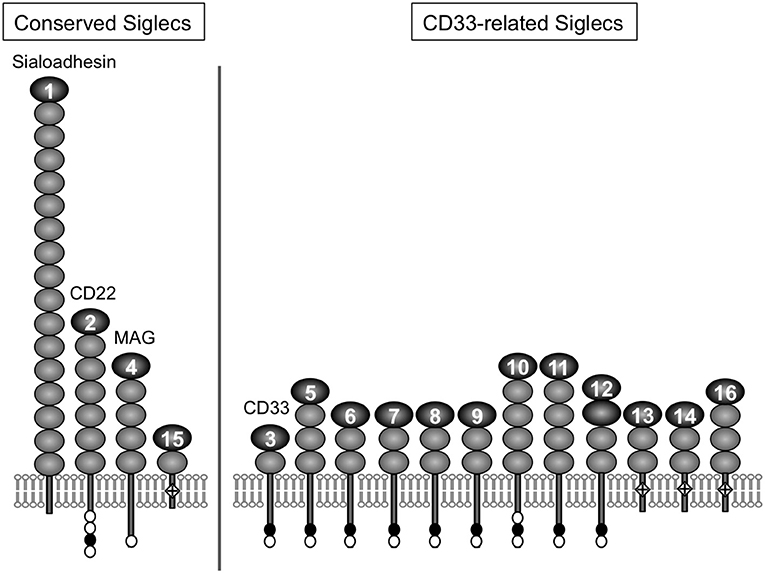
Figure 1. Illustrative representation of ape Siglecs. Mammalian Siglecs can be classified as relatively conserved members (Sialoadhesin/Siglec-1, CD22/Siglec-2, MAG/Siglec-4, and Siglec-15) and less conserved members (CD33-related Siglecs). CD33-related Siglecs are encoded in the gene cluster (15). Siglecs can be also classified based on the partner molecule involved in downstream signal transduction (i.e., those that have ITIM and interact with tyrosine phosphatase SHP-1, and those that have positively charged amino acid residue in the transmembrane domain and interact with adapter molecule DAP12, which has the “immunoreceptor tyrosine-based activating motif“ (ITAM) and recruits tyrosine kinase Syk). Siglec-13 is missing in humans but present in apes and old-world monkeys (15). Modified from (16). Closed circle, open circle, and diamond with + mark in the figure represent ITIM, ITIM-like motif, and positively charged amino acid residue in the transmembrane domain (that is required for the interaction with DAP12), respectively.
“Sialic acids” is a collective term for various naturally occurring acidic sugars with a common nine-carbon backbone (22). N-acetylneuraminic acid (Neu5Ac) is the most common type of sialic acid, and its C5-hydroxylated derivative N-glycolylneuraminic acid (Neu5Gc), along with the derivatives of Neu5Ac and Neu5Gc (mostly modified at C4 and/or C7-C9 hydroxyl groups), are generally present in mammalian tissues (22). Neu5Gc is abundant in many mammalian species, whereas humans have lost Neu5Gc, owing to the mutation (exon deletion) of the CMAH gene encoding CMP-Neu5Ac hydroxylase that is solely responsible for the de novo biosynthesis of Neu5Gc from Neu5Ac (23–26). Although some bacteria have developed ways to synthesize Neu5Ac, so far no study has demonstrated the presence of Neu5Gc on microbes (27) [A recent genomic survey (28) reported the presence of CMAH-like sequences in several microbial genomes, including those of several Helicobacter species that may express sialic acids. However, their enzymatic function has not yet been investigated]. Thus, Neu5Gc appears to be a quintessential signature of self, which is only present on deuterostome cells and missing on microbes. Indeed, some rodent Siglecs show a strong preference toward glycans containing Neu5Gc (29–32), whereas some others show a strong preference toward Neu5Ac (29, 33, 34). This imposes a conundrum: if one loses the best signature of self, the immune system may become more prone to attack its own cells (i.e., autoimmunity). How did the immune system of the human ancestor cope with the consequences of the dramatic change in the sialic acid landscape (i.e., shift of “sialome” from Neu5Gc to Neu5Ac) on the cell surface?
One possible consequence of Neu5Gc loss in human (and a possible response to the consequential autoimmune-prone state, in the evolutionary time scale) was a series of changes involving Neu5Gc-specific Siglecs, such as re-adjustment of binding specificity to Neu5Ac and “forced retirement,” as explained in the following section.
To understand the consequences of a species-specific event, it is natural to compare the phenotypes between the closest relatives that have undergone the event or have not. For human, the obvious choice is great apes including chimpanzee, which is the closest extant relative of modern human. Several earlier studies have shown that at least some great ape Siglecs preferentially recognize Neu5Gc (35–37). More recent data using the sialoglycan microarray also showed that primate CD33-related Siglecs generally tend to prefer Neu5Gc (38). Reported preferences of human and chimpanzee Siglecs toward Neu5Ac and Neu5Gc are summarized in Table 1. Thus, the loss of Neu5Gc likely meant attenuation of the interactions between Siglecs and self-associated ligands in the human ancestor.
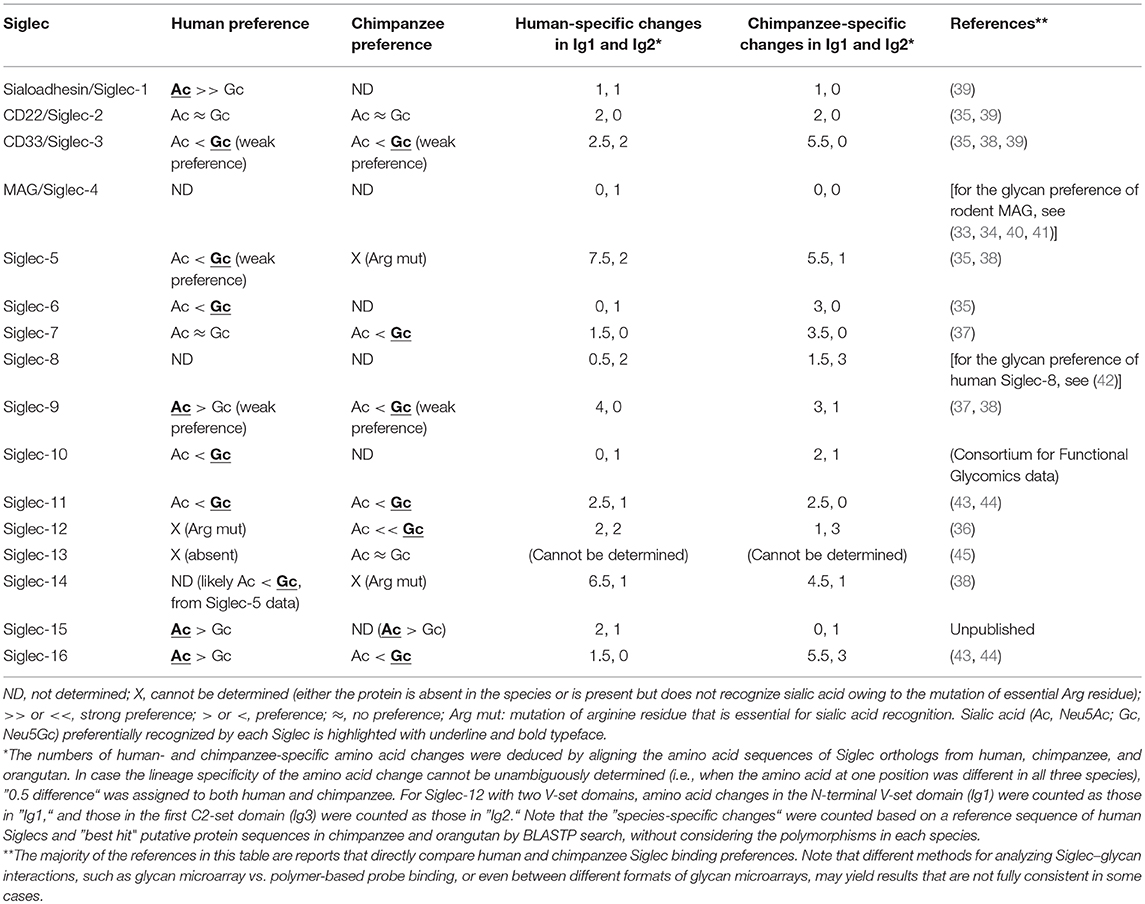
Table 1. Binding preferences (Neu5Ac vs. Neu5Gc) and lineage-specific mutations in human and chimpanzee Siglecs.
One Siglec that may have been substantially affected by the loss of Neu5Gc in the human ancestor is Siglec-12 (36). Chimpanzee Siglec-12 and human Siglec-XII are expressed on macrophages and lumenal epithelia (36, 46). Human Siglec-XII has a universal mutation (R122C) that makes the protein unable to recognize sialic acids (36). [Roman numerals are used for primate Siglecs that have a mutation at the essential arginine residue required for sialic acid recognition and thus cannot recognize sialic acid (15)]. Arginine-restored human Siglec-XII, as well as chimpanzee Siglec-12, strongly prefers Neu5Gc over Neu5Ac (36). In addition, some human SIGLEC12 alleles have acquired additional mutations (stop codon, rs16982743, and frame-shift, rs66949844) that cause premature termination of Siglec-XII protein synthesis (36, 46). These “null” mutations are common in the modern human populations (global frequency of “null” alleles: 0.19 for rs16982743, 0.59 for rs66949844). These results imply a scenario in which a Siglec that lost an endogenous ligand was forced to “retire” and then is further getting eliminated. Given that the R122C mutation is fixed in modern human populations, it is tempting to speculate that the presence of functional “Neu5Gc-recognizing” Siglec-12 may have caused a disadvantage in ancestral humans. For example, zoonotic infection of some Neu5Gc-coated envelope virus from other mammalian species may represent such selective pressure. A possible scenario for the further elimination of “signal transduction-competent but sialic acid recognition-incompetent” Siglec-XII may be that the recruitment of SHP-2 by Siglec-XII (47) on epithelial cells may assist the transformation of the epithelial cell by an oncogenic driver (e.g., receptor tyrosine kinase or RAS mutation/amplification) through activation of MAPK pathway (48–52), which may have been disadvantageous for the overall fitness of the carriers of the functional allele. However, at present there is no solid experimental evidence to support these speculations.
Primate Siglec-9 (from chimpanzee, gorilla, and baboon) also prefers Neu5Gc, whereas human Siglec-9 appears to have acquired affinity toward Neu5Ac (37, 38). Human CD33/Siglec-3 and Siglec-5 also show a similar acquired affinity to Neu5Ac compared with their counterparts in baboon, which show a strong preference for Neu5Gc (38). Given that Siglec-9 has an ortholog in rodents (Siglec-E), it may play an important role in regulating innate immunity and be indispensable (although expression patterns and functions of primate Siglec-9 and rodent Siglec-E may not completely overlap (53–55)). Human Siglec-9 may have had to undergo rapid evolution to catch up with the change in the human sialome, to resume its original functionality. It is of note that the N-terminal Ig-like domain (Ig1) of great ape Siglec-9 shows much greater inter-species sequence differences than does the adjacent C2-set Ig-like domain (Ig2) (37), which is consistent with the idea that human Siglec-9 had to evolve rapidly to respond to the loss of Neu5Gc.
In fact, the CD33-related Siglec gene cluster is among the most rapidly diversifying gene families between human and chimpanzee (56), and the N-terminal Ig-like domain of CD33-related ape Siglecs is evolving faster than the other parts of the molecule (15, 37, 57). It is of interest whether the loss of Neu5Gc contributed to the accelerated evolution of human Siglecs. Assuming this is the case, we would expect that more amino acid changes have accumulated in the first Ig-like domain (Ig1) of human Siglecs than in Ig1 of chimpanzee Siglecs. In reality, the data (Table 1) do not appear to support this prediction. Although it is true that Ig1 is undergoing faster evolution than Ig2 (total human-specific changes in Ig1 and Ig2: 33.5 and 15, respectively; total chimpanzee-specific changes in Ig1 and Ig2: 40.5 and 14, respectively; average amino acid length of Ig1 and Ig2: 126 and 96, respectively), the Ig1 of Siglecs in the lineage leading to human has accumulated less sequence changes than that leading to chimpanzee. Some of the sequence changes in human Siglecs probably represent a genuine sign of selection due to the loss of Neu5Gc, whereas the majority of them may not be. Ig1 of primate Siglecs (and likely those in other species) is evolving rapidly under selective pressure that may include, but not be limited to, the changes in the landscape of endogenous sialoglycans (38).
It is of note that, in contrast to the adaptation of some human Siglecs to Neu5Ac-dominant sialome, many Siglecs still appear to prefer Neu5Gc (Table 1). Possible explanations for this fact may include: (1) the adaptation of human Siglecs to Neu5Ac-dominant sialome is still incomplete, and over the time (in the scale of millions of years) most human Siglecs will eventually acquire Neu5Ac preference; (2) some Siglecs did not have strong preference toward Neu5Gc over Neu5Ac prior to the loss of Neu5Gc in human ancestor, or have already accumulated mutations to make them sufficiently suitable for Neu5Ac recognition, thus it is not necessary for them to adapt further to Neu5Ac-dominant sialome; (3) the interaction of Siglecs with exogenous ligands (e.g., bacterial nonulosonic acids) prevent complete switch from Neu5Gc to Neu5Ac preference. Although these explanations are purely speculative, some of these scenarios may be tested experimentally. For example, an independent event has eliminated Neu5Gc in the lineage leading to New World monkeys approximately 30 million years ago (58). In contrast, the timing of Neu5Gc loss in human is far more recent, which is estimated to be 3 million years ago (26). It would be interesting to see whether the Siglecs in New World monkeys prefer Neu5Ac, or some of them still prefer Neu5Gc, to test the validity of the explanation (1) above.
It is of interest to know whether there is any change in the expression patterns of Siglecs between human and chimpanzee, which might also represent a consequence of Neu5Gc loss in human. Antibody-based comparative analyses of Siglec expression patterns in human and chimpanzee (and gorilla) have revealed several examples of altered expression of Siglecs in human, as summarized in Table 2. Naturally, it is more difficult to establish the influence of the loss of Neu5Gc on the expression patterns of Siglecs than its effect on the binding preferences of Siglecs, as it is indirect. Nevertheless, it appears to be implied in some cases.
The first reported example of altered expression of Siglec in human compared with chimpanzee was the wider distribution of Sialoadhesin/Siglec-1+ macrophages in chimpanzee spleen as compared with those in human spleen (39). Although the binding specificity of chimpanzee Sialoadhesin/Siglec-1 has not been analyzed, given that both human and mouse Sialoadhesin/Siglec-1 preferentially recognize Neu5Ac (39) and the sequence differences between human and chimpanzee Sialoadhesin/Siglec-1 are small (Table 1), it is likely that chimpanzee Sialoadhesin/Siglec-1 prefers Neu5Ac. Thus, the altered distribution of human Sialoadhesin/Siglec-1+ macrophages may be a consequence of the loss of Neu5Gc in humans (39). It is possible that the altered distribution of Sialoadhesin/Siglec-1+ macrophages may be more relevant to the increased density of Neu5Ac in human tissues that may influence the migration of macrophages, rather than a change in cell types that express Sialoadhesin/Siglec-1. In this regard, it would be interesting to know whether the distribution of Sialoadhesin/Siglec-1+ macrophages in Cmah knockout mice is different from that in wild-type mice.
One of the most striking changes in Siglec expression patterns in the human immune system is the near-complete absence of Siglec-5 on human T cells, in contrast to its prominent expression on chimpanzee and gorilla T cells (61, 62). The loss of Siglec-5 from human T cells appears to be correlated with the relative hyper-activation of human T cells in response to various stimuli compared with those from other great apes. [Although Siglec-5 and Siglec-14 show extremely high sequence similarity at the extracellular domain, one study (62) used a combination of antibodies that distinguish Siglec-5 and Siglec-14 to demonstrate that Siglec-5 is expressed on chimpanzee T cells]. However, it is not clear whether the loss of Siglec-5 on human T cells has a causative relationship with the loss of Neu5Gc, as human Siglec-5 does not show strong preference for either Neu5Ac or Neu5Gc (38), and its great ape counterparts have a mutation at the essential arginine residue and lack the ability to recognize sialic acids (15, 89). It is also worth mentioning that a recent work demonstrated that Siglec-5 is inducibly expressed by the activation of human T cells (88).
Siglec-11 and Siglec-16 also have undergone unique changes in their expression patterns in humans. Whereas, human Siglec-11 and Siglec-16 are expressed on brain microglia and tissue macrophages, chimpanzee Siglec-11 and Siglec-16 appear to be absent on microglia (but present on tissue macrophages) (43, 75). The change in expression patterns appears to be a consequence of a partial gene conversion of SIGLEC11 by SIGLEC16. Of note, SIGLEC16 in humans has functional and non-functional alleles (82), and the non-functional allele appears to be the one that converted SIGLEC11 (90). SIGLEC11 and SIGLEC16 have undergone a complex series of concerted evolution through gene conversions in human lineage (90) and also in other lineages of apes (44). Both human and chimpanzee Siglec-11 and Siglec-16 appear to prefer Neu5Gc over Neu5Ac (43, 44), and thus it is tempting to speculate that the loss of Neu5Gc may have had some influence on the altered expression patterns of these Siglecs. Although it is known that the Neu5Gc level is extremely low in mammalian brains (91), Siglec-11 and Siglec-16 also preferentially recognize α2-8–linked Neu5Ac dimers, which are abundant in the brain and serve as ligands for these Siglecs on human microglia.
Siglec-6 was also reported to show different expression patterns between human and chimpanzee. Both human and chimpanzee Siglec-6 are expressed on B cells, whereas its expression on placental trophoblasts is observed only in humans (64). This altered expression is thought to be associated with the sequence change in the promoter region and transcription factor binding (64).
There are some reports of the presence of Siglec ligands in human tissues that are absent in chimpanzee tissues (64, 76). Although the exact nature of these ligands has not been identified, these findings imply that the difference in Siglec ligand expression patterns beyond the absence/presence of Neu5Gc may exist between human and chimpanzee and may also contribute to the rapid evolution of the Siglec family (particularly at Ig1) and/or their altered expression patterns.
Given that Ig1 of Siglecs (particularly that of CD33-related Siglecs) is undergoing rapid evolution (57), and not all of this may be attributed to the changing endogenous ligand landscape, there is likely an alternative driving force behind their rapid evolution. Obviously, one such force could be microbial pathogens that engage Siglecs. Indeed, recent studies have provided evidence that many Siglecs are involved in the interaction with various pathogenic microbes [for recent reviews, see (92, 93)]. These microbes include viruses, bacteria, and eukaryotic pathogens (Table 3). Many of them cover themselves with sialic acids (either by de novo biosynthesis or by “salvage” from the human body by various mechanisms), which may be considered examples of “molecular mimicry” by microbes.
The majority of the microbes reported to interact with Siglecs so far are bacteria (Table 3). This makes sense, as sialic acids (and sialic acid-like nonulosonic acids) are occasionally found in bacterial extracellular components, such as lipopolysaccharides/lipooligosaccharides (LPS/LOS), capsular polysaccharides (CPS), and flagella. For example, group B streptococcus (GBS) type III interacts with Siglec-9 through sialylated CPS and dampens inflammatory responses by neutrophils (98), whereas GBS type Ia engages Siglec-5 by β-protein and also suppresses inflammatory responses of myeloid cells (100). It should be noted that the latter case does not involve sialic acids. Similarly, non-typeable Haemophilus influenzae, an opportunistic airway pathogen, engages Siglec-5 and attenuates pro-inflammatory cytokine production by myeloid cells (102), and Escherichia coli K1 strain, a neurotropic pathogen, engages Siglec-11 and escape killing (75). Siglecs are likely under pressure to escape the exploitation by these pathogens, which may partially explain the driving force behind their rapid evolution.
It appears that Siglecs were not just escaping from these pathogens; they appear to have developed “counter-traps” against these pathogens. Some Siglecs (i.e., Siglec-5 and Siglec-14; Siglec-11 and Siglec-16) are found to be “paired receptors,” which are two Siglecs with highly homologous extracellular domains recognizing similar ligands, combined with intracellular signaling modules transducing opposing signals (i.e., one of the pair interacts with SHP-1 and transduces the inhibitory signal, whereas the other interacts with adapter protein DAP12 and tyrosine kinase Syk and transduces the activating signal). In fact, whereas the engagement of inhibitory Siglec by pathogenic bacteria suppresses anti-bacterial responses, the engagement of activating Siglec counteracts this effect (75, 102, 63). It is of note that these “paired” Siglecs appear to show more sequence differences between human and chimpanzee than other “stand-alone” Siglecs (Table 1), possibly implying that these Siglecs are under higher selective pressure to diversify than are other Siglecs. These paired Siglecs are undergoing concerted evolution through repeated gene conversions (43, 44, 89, 90), which is likely necessary to maintain the effectiveness of activating-type Siglec as “counter-traps.” It is also intriguing that, in humans, null alleles for these activating-type Siglecs (Siglec-14 and Siglec-16) are found at very high frequencies (82, 113).
Evidence supporting the relevance of these interactions between Siglecs and bacterial pathogens in infectious diseases is emerging from genetic association studies (Table 4). Small-scale case–control studies investigating the possible correlations between the polymorphisms of SIGLEC genes and infectious disease susceptibility have revealed some correlations, such as SIGLEC14 null polymorphism and COPD exacerbation (102), pre-term delivery in the presence of GBS (63), Mycobacterium tuberculosis meningitis (138), and SIGLEC9 polymorphism and COPD exacerbation (133). In addition, large-scale genome-wide association studies (GWAS) have also revealed possible associations between SIGLEC polymorphisms and infectious diseases, such as SIGLEC5 polymorphism and leprosy (129) and severe periodontitis (128), although these GWAS did not demonstrate a direct interaction between the etiological agents and Siglec protein. Some SIGLEC genetic polymorphisms appear to influence the leukocyte counts (142); thus it is possible that the influence of SIGLEC genetic polymorphisms on antibacterial defense may be indirect. Regardless, the application of GWAS to infectious diseases may further reveal the relevance of Siglecs for immunological defense against bacterial pathogens.
With regard to viral pathogens, recent studies have revealed that Sialoadhesin/Siglec-1 (also known as CD169) may play a major role in retrovirus infection (103). For example, several groups have reported that human immunodeficiency virus (HIV) exploits Sialoadhesin/Siglec-1 to enhance infection of CD4+ T cells (the primary target cells) by trans-infection (i.e., the virus particle is captured by macrophages with Sialoadhesin/Siglec-1, which transfers the virus to CD4+ T cells and facilitates the infection) (104–107). Although a rare “null” mutation in the SIGLEC1 gene was found not to protect carriers from HIV infection (144), the low frequency of this mutation (allele frequency: ~1.3% in Europeans) may preclude us from making a definitive conclusion (145). Given that Sialoadhesin/Siglec-1 appears to be involved in retroviral infection in both mouse and human (103), one may expect that it should evolve rapidly to avoid viral infections; however, Sialoadhesin/Siglec-1 does not appear to be evolving rapidly (Table 1). This may be because enveloped viruses are coated with a host-derived membrane (a part of “self”), and thus there is no way Sialoadhesin/Siglec-1 can evolve to completely evade such an interaction (unless the virus develops a protein that binds Sialoadhesin/Siglec-1 in sialic acid-independent manner). It is worth noting that myelin-associated glycoprotein (MAG)/Siglec-4, the other Siglec known to interact with another enveloped virus (109, 110), is also highly conserved among mammals, and in both cases sialic acids are required for the interaction between the virus and Siglecs (Table 3).
Cmah null mouse is a valuable tool for the investigation of the physiological roles of Neu5Gc and the short-term consequences of its loss, although it may not be a perfect model of modern human. Using this mouse model, it was shown that the expression of Neu5Gc itself makes T cells less responsive to stimulus, without any change in Siglec expression (146). Likewise, Neu5Gc appears to have a general suppressive effect on mouse monocyte/macrophage activities, without the apparent involvement of Siglecs (147). In line with these findings, the loss of Neu5Gc has had major influences on human biology that reach far beyond Siglecs (148), explaining some of the differences between human and our close relatives (e.g., chimpanzee) in pathophysiological phenotypes (149). Although Neu5Gc from dietary sources (in the form of meat or milk from the animals that express Neu5Gc) can be incorporated into human tissue glycoproteins and glycolipids (150), the level of Neu5Gc in human tissues tends to be low, accounting for <1% of total sialic acids (151). Given that the current set of human Siglecs lack a strong preference toward Neu5Gc, and the affinity between Siglecs and sialic acids tends to be low (Kd in ~mM range), this level of Neu5Gc in human tissues may not influence human physiology by way of Siglecs. The trace amount of Neu5Gc incorporated into human tissues may be more relevant to the bacterial toxins that specifically recognize Neu5Gc (152) and xeno-autoantibodies that recognize Neu5Gc (as discussed in other articles of this series). Regardless, the loss of Neu5Gc appears to have left some footprint on the evolution of human Siglecs, as discussed above.
The evolution of human Siglecs was also likely influenced by the interaction with microbes. A recent population genetics-based study implied that some Siglecs may have been subjected to population-specific hard selective sweeps, as judged by the presence of long-range linkage disequilibrium (153). These SIGLEC genes include SIGLEC8 and SIGLEC10 among Africans, SIGLEC5, SIGLEC6, SIGLEC12, and SIGLEC14 among Europeans, and CD22 and MAG among Asians. Although it remains speculative, the population-specific difference in the signatures of selection imply that the evolution of the Siglec family in the human population is an ongoing process, and different pathogen pressures are present in different geographical locations (or through different agricultural constraints, e.g., use of different domestic animals, which may carry different kinds of bacteria/viruses).
Many questions remain with regard to the function and evolution of Siglecs. For example, do viruses really target only conserved Siglecs and are they not relevant to the rapid evolution of Siglecs? What was or is the selective force behind the spread of “null” alleles of SIGLEC14 and SIGLEC16 (and perhaps others, such as SIGLEC1) in modern human populations? Do the bacteria that express sialic acid-like nonulosonic acids (154) generally engage Siglecs to modulate immune responses and thus play a role in the evolution of Siglecs? Does the interaction between Siglecs and commensal bacteria (e.g., normal gut microbiota) play any role in the modulation of immunity and the evolution of Siglecs? Some of these questions can be addressed experimentally and will deepen our understanding of the biology of Siglecs and sialic acids.
TA analyzed the literature and wrote the manuscript.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author is grateful for supports from the Ministry of Science and Technology, Taiwan, and from Academia Sinica, Taiwan (MOST 104-2321-B-001-070, 105-2321-B-001-051, and 106-2321-B-001-032). I also thank the editors, Dr. Vered Padler-Karavani (Tel-Aviv University, Israel) for the invitation to contribute this review, and Dr. Jean-Paul Soulillou (Université de Nantes, France) for valuable comments.
1. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. (2011) 34:637–50. doi: 10.1016/j.immuni.2011.05.006
2. Geijtenbeek T, Gringhuis S. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. (2009) 9:465–79. doi: 10.1038/nri2569
3. Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. (2015) 32:48–53. doi: 10.1016/j.coi.2014.12.012
4. Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity (2007) 27:549–59. doi: 10.1016/j.immuni.2007.10.002
5. Varki A. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology (2011) 21:1121–4. doi: 10.1093/glycob/cwr087
6. Medzhitov R, Janeway CA Jr. Decoding the patterns of self and nonself by the innate immune system. Science (2002) 296:298–300. doi: 10.1126/science.1068883
7. Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. (2002) 102:439–70. doi: 10.1021/cr000407m
8. Varki A, Angata T. Siglecs - the major subfamily of I-type lectins. Glycobiology (2006) 16:1R−27R. doi: 10.1093/glycob/cwj008
9. Crocker P, Paulson J, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. (2007) 7:255–66. doi: 10.1038/nri2056
10. Pillai S, Netravali I, Cariappa A, Mattoo H. Siglecs and immune regulation. Annu Rev Immunol. (2012) 30:357–92. doi: 10.1146/annurev-immunol-020711-075018
11. Macauley M, Crocker P, Paulson J. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. (2014) 14:653–66. doi: 10.1038/nri3737
12. Crocker P, Clark E, Filbin M, Gordon S, Jones Y, Kehrl J, et al. Siglecs: a family of sialic-acid binding lectins [letter]. Glycobiology (1998) 8:v. doi: 10.1093/oxfordjournals.glycob.a018832
13. Angata T, Varki A. Cloning, characterization, and phylogenetic analysis of siglec-9, a new member of the CD33-related group of siglecs. Evidence for co-evolution with sialic acid synthesis pathways. J Biol Chem. (2000) 275:22127–35. doi: 10.1074/jbc.M002775200
14. Bornhofft KF, Goldammer T, Rebl A, Galuska SP. Siglecs: a journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev Comp Immunol. (2018) 86:219–31. doi: 10.1016/j.dci.2018.05.008
15. Angata T, Margulies E, Green E, Varki A. Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci USA. (2004) 101:13251–6. doi: 10.1073/pnas.0404833101
16. Angata T. Polymorphisms and mutations in SIGLEC genes and their associations with diseases. J Japan Biochem Soc. (2017) 89:652–9. doi: 10.14952/SEIKAGAKU.2017.890652
17. Beatson R, Tajadura-Ortega V, Achkova D, Picco G, Tsourouktsoglou TD, Klausing S, et al. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat Immunol. (2016) 17:1273–81. doi: 10.1038/ni.3552
18. Carroll DJ, O'Sullivan JA, Nix DB, Cao Y, Tiemeyer M, Bochner BS. Sialic acid-binding immunoglobulin-like lectin 8 (Siglec-8) is an activating receptor mediating beta2-integrin-dependent function in human eosinophils. J Allergy Clin Immunol. (2018) 141:2196–207. doi: 10.1016/j.jaci.2017.08.013
19. Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens (2004) 64:215–25. doi: 10.1111/j.0001-2815.2004.00290.x
20. Parham P, Guethlein LA. Genetics of natural killer cells in human health, disease, and survival. Annu Rev Immunol. (2018) 36:519–48. doi: 10.1146/annurev-immunol-042617-053149
21. Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity (2001) 15:363–74. doi: 10.1016/S1074-7613(01)00197-2
22. Varki A, Schnaar RL, Schauer R. Sialic Acids and other nonulosonic acids. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, et al., editors. Essentials of Glycobiology. 3rd ed. New York, NY: Cold Spring Harbor; Laboratory Press (2015). pp. 179–95.
23. Chou H, Takematsu H, Diaz S, Iber J, Nickerson E, Wright K, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci USA. (1998) 95:11751–6. doi: 10.1073/pnas.95.20.11751
24. Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem. (1998) 273:15866–71. doi: 10.1074/jbc.273.25.15866
25. Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N. Alu-mediated inactivation of the human CMP- N-acetylneuraminic acid hydroxylase gene. Proc Natl Acad Sci USA. (2001) 98:11399–404. doi: 10.1073/pnas.191268198
26. Chou H, Hayakawa T, Diaz S, Krings M, Indriati E, Leakey M, et al. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci USA. (2002) 99:11736–41. doi: 10.1073/pnas.182257399
27. Varki A. Multiple changes in sialic acid biology during human evolution. Glycoconj J. (2009) 26:231–45. doi: 10.1007/s10719-008-9183-z
28. Peri S, Kulkarni A, Feyertag F, Berninsone PM, Alvarez-Ponce D. Phylogenetic distribution of CMP-Neu5Ac hydroxylase (CMAH), the enzyme synthetizing the proinflammatory human xenoantigen Neu5Gc. Genome Biol Evol. (2018) 10:207–19. doi: 10.1093/gbe/evx251
29. Kelm S, Schauer R, Manuguerra JC, Gross HJ, Crocker PR. Modifications of cell surface sialic acids modulate cell adhesion mediated by sialoadhesin and CD22. Glycoconj J. (1994) 11:576–85. doi: 10.1007/BF00731309
30. Naito Y, Takematsu H, Koyama S, Miyake S, Yamamoto H, Fujinawa R, et al. Germinal center marker GL7 probes activation-dependent repression of N-glycolylneuraminic acid, a sialic acid species involved in the negative modulation of B-cell activation. Mol Cell Biol. (2007) 27:3008–22. doi: 10.1128/MCB.02047-06
31. Macauley MS, Kawasaki N, Peng W, Wang SH, He Y, Arlian BM, et al. Unmasking of CD22 co-receptor on germinal center b-cells occurs by alternative mechanisms in mouse and man. J Biol Chem. (2015) 290:30066–77. doi: 10.1074/jbc.M115.691337
32. Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, et al. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J Exp Med. (2010) 207:173–87. doi: 10.1084/jem.20091873
33. Collins B, Kiso M, Hasegawa A, Tropak M, Roder J, Crocker P, et al. Binding specificities of the sialoadhesin family of I-type lectins - Sialic acid linkage and substructure requirements for binding of myelin-associated glycoprotein, Schwann cell myelin protein, and sialoadhesin. J Biol Chem. (1997) 272:16889–95. doi: 10.1074/jbc.272.27.16889
34. Collins B, Yang L, Mukhopadhyay G, Filbin M, Kiso M, Hasegawa A, et al. Sialic acid specificity of myelin-associated glycoprotein binding. J Biol Chem. (1997) 272:1248–55. doi: 10.1074/jbc.272.2.1248
35. Brinkman-Van der Linden E, Varki A. New aspects of siglec binding specificities, including the significance of fucosylation and of the sialyl-Tn epitope. J Biol Chem. (2000) 275:8625–32. doi: 10.1074/jbc.275.12.8625
36. Angata T, Varki N, Varki A. A second uniquely human mutation affecting sialic acid biology. J Biol Chem. (2001) 276:40282–7. doi: 10.1074/jbc.M105926200
37. Sonnenburg J, Altheide T, Varki A. A uniquely human consequence of domain-specific functional adaptation in a sialic acid-binding receptor. Glycobiology (2004) 14:339–46. doi: 10.1093/glycob/cwh039
38. Padler-Karavani V, Hurtado-Ziola N, Chang Y, Sonnenburg J, Ronaghy A, Yu H, et al. Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J. (2014) 28:1280–93. doi: 10.1096/fj.13-241497
39. Brinkman-Van der Linden E, Sjoberg E, Juneja L, Crocker P, Varki N, Varki A. Loss of N-glycolylneuraminic acid in human evolution - Implications for sialic acid recognition by siglecs. J Biol Chem. (2000) 275:8633–40. doi: 10.1074/jbc.275.12.8633
40. Blixt O, Collins B, Van Den Nieuwenhof I, Crocker P, Paulson J. Sialoside specificity of the Siglec family assessed using novel multivalent probes: Identification of potent inhibitors of myelin associated glycoprotein. J Biol Chem. (2003) 278 31007–19. doi: 10.1074/jbc.M304331200
41. Collins B, Ito H, Sawada N, Ishida H, Kiso M, Schnaar R. Enhanced binding of the neural siglecs, myelin-associated glycoprotein and Schwann cell myelin protein, to Chol-1 (alpha-series) gangliosides and novel sulfated Chol-1 analogs. J Biol Chem. (1999) 274:37637–43. doi: 10.1074/jbc.274.53.37637
42. Bochner B, Alvarez R, Mehta P, Bovin N, Blixt O, White J, et al. Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem. (2005) 280:4307–12. doi: 10.1074/jbc.M412378200
43. Hayakawa T, Angata T, Lewis A, Mikkelsen T, Varki N, Varki A. A human-specific gene in microglia. Science. (2005) 309:1693. doi: 10.1126/science.1114321
44. Hayakawa T, Khedri Z, Schwarz F, Landig C, Liang SY, Yu H, et al. Coevolution of Siglec−11 and Siglec−16 via gene conversion in primates. BMC Evol Biol. (2017) 17:228. doi: 10.1186/s12862-017-1075-z
45. Wang X, Mitra N, Secundino I, Banda K, Cruz P, Padler-Karavani V, et al. Specific inactivation of two immunomodulatory SIGLEC genes during human evolution. Proc Natl Acad Sci USA. (2012) 109:9935–40. doi: 10.1073/pnas.1119459109
46. Mitra N, Banda K, Altheide T, Schaffer L, Johnson-Pais T, Beuten J, et al. SIGLEC12, a human-specific segregating (pseudo)gene, encodes a signaling molecule expressed in prostate carcinomas. J Biol Chem. (2011) 286:23003–11. doi: 10.1074/jbc.M111.244152
47. Yu Z, Lai C, Maoui M, Banville D, Shen S. Identification and characterization of S2V, a novel putative siglec that contains two V set Ig-like domains and recruits protein-tyrosine phosphatases SHPs. J Biol Chem. (2001) 276:23816–24. doi: 10.1074/jbc.M102394200
48. Dardaei L, Wang HQ, Singh M, Fordjour P, Shaw KX, Yoda S, et al. SHP2 inhibition restores sensitivity in ALK-rearranged non-small-cell lung cancer resistant to ALK inhibitors. Nat Med. (2018) 24:512–7. doi: 10.1038/nm.4497
49. Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med. (2018) 24:961–7. doi: 10.1038/s41591-018-0023-9
50. Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med. (2018) 24:954–60. doi: 10.1038/s41591-018-0024-8
51. Wong GS, Zhou J, Liu JB, Wu Z, Xu X, Li T, et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med. (2018) 24:968–77. doi: 10.1038/s41591-018-0022-x
52. Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol. (2018) 20:1064–73. doi: 10.1038/s41556-018-0169-1
53. Zhang J, Biedermann B, Nitschke L, Crocker P. The murine inhibitory receptor mSiglec-E is expressed broadly on cells of the innate immune system whereas mSiglec-F is restricted to eosinophils. Eur J Immunol. (2004) 34:1175–84. doi: 10.1002/eji.200324723
54. von Gunten S, Yousefi S, Seitz M, Jakob S, Schaffner T, Seger R, et al. Siglec-9 transduces apoptotic and nonapoptotic death signals into neutrophils depending on the proinflammatory cytokine environment. Blood (2005) 106:1423–31. doi: 10.1182/blood-2004-10-4112
55. Jandus C, Boligan K, Chijioke O, Liu H, Dahlhaus M, Demoulins T, et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest. (2014) 124:1810–20. doi: 10.1172/JCI65899
56. The Chimpanzee Sequencing and Analysis Consortium. Initial Sequence of the Chimpanzee Genome and Comparison with the Human Genome. Nature. (2005) 437:69–87. doi: 10.1038/nature04072
57. Altheide TK, Hayakawa T, Mikkelsen TS, Diaz S, Varki N, Varki A. System-wide genomic and biochemical comparisons of sialic acid biology among primates and rodents: Evidence for two modes of rapid evolution. J Biol Chem. (2006) 281:25689–702. doi: 10.1074/jbc.M604221200
58. Springer SA, Diaz SL, Gagneux P. Parallel evolution of a self-signal: humans and new world monkeys independently lost the cell surface sugar Neu5Gc. Immunogenetics (2014) 66:671–4. doi: 10.1007/s00251-014-0795-0
59. Moller JR. Rapid conversion of myelin-associated glycoprotein to a soluble derivative in primates. Brain Res. (1996) 741:27–31. doi: 10.1016/S0006-8993(96)00882-7
60. Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, et al. Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci USA. (2012) 109:16480–5. doi: 10.1073/pnas.1117943109
61. Nguyen D, Hurtado-Ziola N, Gagneux P, Varki A. Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci USA. (2006) 103:7765–70. doi: 10.1073/pnas.0510484103
62. Soto PC, Stein LL, Hurtado-Ziola N, Hedrick SM, Varki A. Relative over-reactivity of human versus chimpanzee lymphocytes: implications for the human diseases associated with immune activation. J Immunol. (2010) 184:4185–95. doi: 10.4049/jimmunol.0903420
63. Ali S, Fong J, Carlin A, Busch T, Linden R, Angata T, et al. Siglec-5 and Siglec−14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med. (2014) 211:1231–42. doi: 10.1084/jem.20131853
64. Brinkman-Van der Linden EC, Hurtado-Ziola N, Hayakawa T, Wiggleton L, Benirschke K, Varki A, et al. Human-specific expression of Siglec-6 in the placenta. Glycobiology (2007) 17:922–31. doi: 10.1093/glycob/cwm065
65. Falco M, Biassoni R, Bottino C, Vitale M, Sivori S, Augugliaro R, et al. Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J Exp Med. (1999) 190:793–801. doi: 10.1084/jem.190.6.793
66. Nicoll G, Ni J, Liu D, Klenerman P, Munday J, Dubock S, et al. Identification and characterization of a novel siglec, siglec-7, expressed by human natural killer cells and monocytes. J Biol Chem. (1999) 274:34089–95. doi: 10.1074/jbc.274.48.34089
67. Vitale C, Romagnani C, Falco M, Ponte M, Vitale M, Moretta A, et al. Engagement of p75/AIRM1 or CD33 inhibits the proliferation of normal or leukemic myeloid cells. Proc Natl Acad Sci USA. (1999) 96:15091–6. doi: 10.1073/pnas.96.26.15091
68. Ikehara Y, Ikehara S, Paulson J. Negative regulation of T cell receptor signaling by Siglec-7 (p70/AIRM) and Siglec-9. J Biol Chem. (2004) 279:43117–25. doi: 10.1074/jbc.M403538200
69. Mizrahi S, Gibbs B, Karra L, Ben-Zimra M, Levi-Schaffer F. Siglec-7 is an inhibitory receptor on human mast cells and basophils. J Allergy Clin Immunol. (2014) 134:230–3. doi: 10.1016/j.jaci.2014.03.031
70. Nguyen K, Hamzeh-Cognasse H, Palle S, Anselme-Bertrand I, Arthaud C, Chavarin P, et al. Role of Siglec-7 in apoptosis in human platelets. PLoS One. (2014) 9:e106239. doi: 10.1371/journal.pone.0106239
71. Floyd H, Ni J, Cornish A, Zeng Z, Liu D, Carter K, et al. Siglec-8. A novel eosinophil-specific member of the immunoglobulin superfamily. J Biol Chem. (2000) 275:861–6. doi: 10.1074/jbc.275.2.861
72. Kikly K, Bochner B, Freeman S, Tan K, Gallagher K, D'alessio K, et al. Identification of SAF-2, a novel siglec expressed on eosinophils, mast cells, and basophils. J Allergy Clin Immunol. (2000) 105(6 Pt 1):1093–100. doi: 10.1067/mai.2000.107127
73. Munday J, Kerr S, Ni J, Cornish AL, Zhang JQ, Nicoll G, et al. Identification, characterization and leucocyte expression of Siglec−10, a novel human sialic acid-binding receptor. Biochem J. (2001) 355(Pt 2):489–97. doi: 10.1042/bj3550489
74. Stephenson HN, Mills DC, Jones H, Milioris E, Copland A, Dorrell N, et al. Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL−10 expression via Siglec−10 receptor: a novel flagellin-host interaction. J Infect Dis. (2014) 210:1487–98. doi: 10.1093/infdis/jiu287
75. Schwarz F, Landig CS, Siddiqui S, Secundino I, Olson J, Varki N, et al. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J. (2017) 36:751–60. doi: 10.15252/embj.201695581
76. Wang X, Chow R, Deng L, Anderson D, Weidner N, Godwin A, et al. Expression of Siglec−11 by human and chimpanzee ovarian stromal cells, with uniquely human ligands: implications for human ovarian physiology and pathology. Glycobiology (2011) 21:1038–48. doi: 10.1093/glycob/cwr039
77. Angata T, Kerr S, Greaves D, Varki N, Crocker P, Varki A. Cloning and characterization of human Siglec−11. A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem. (2002) 277:24466–74. doi: 10.1074/jbc.M202833200
78. Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec−15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology (2007) 17:838–46. doi: 10.1093/glycob/cwm049
79. Hiruma Y, Hirai T, Tsuda E. Siglec−15, a member of the sialic acid-binding lectin, is a novel regulator for osteoclast differentiation. Biochem Biophys Res Commun. (2011) 409:424–9. doi: 10.1016/j.bbrc.2011.05.015
80. Kameda Y, Takahata M, Komatsu M, Mikuni S, Hatakeyama S, Shimizu T, et al. Siglec−15 regulates osteoclast differentiation by modulating RANKL-induced phosphatidylinositol 3-kinase/Akt and Erk pathways in association with signaling Adaptor DAP12. J Bone Miner Res. (2013) 28:2463–75. doi: 10.1002/jbmr.1989
81. Takamiya R, Ohtsubo K, Takamatsu S, Taniguchi N, Angata T. The interaction between Siglec−15 and tumor-associated sialyl-Tn antigen enhances TGF-beta secretion from monocytes/macrophages through the DAP12-Syk pathway. Glycobiology (2013) 23:178–87. doi: 10.1093/glycob/cws139
82. Cao H, Lakner U, de Bono B, Traherne J, Trowsdale J, Barrow A. SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol. (2008) 38:2303–15. doi: 10.1002/eji.200738078
83. Han K, Kim Y, Lee J, Lim J, Lee KY, Kang CS, et al. Human basophils express CD22 without expression of CD19. Cytometry (1999) 37:178–83. doi: 10.1002/(SICI)1097-0320(19991101)37:3<178::AID-CYTO3>3.0.CO;2-Z
84. Valent P, Ashman LK, Hinterberger W, Eckersberger F, Majdic O, Lechner K, et al. Mast cell typing: demonstration of a distinct hematopoietic cell type and evidence for immunophenotypic relationship to mononuclear phagocytes. Blood (1989) 73:1778–85.
85. Stain C, Stockinger H, Scharf M, Jager U, Gossinger H, Lechner K, et al. Human blood basophils display a unique phenotype including activation linked membrane structures. Blood. (1987) 70:1872–9.
86. Yokoi H, Myers A, Matsumoto K, Crocker P, Saito H, Bochner B. Alteration and acquisition of Siglecs during in vitro maturation of CD34+ progenitors into human mast cells. Allergy (2006) 61:769–76. doi: 10.1111/j.1398-9995.2006.01133.x
87. Kardava L, Moir S, Wang W, Ho J, Buckner CM, Posada JG, et al. Attenuation of HIV-associated human B cell exhaustion by siRNA downregulation of inhibitory receptors. J Clin Invest. (2011) 121:2614–24. doi: 10.1172/JCI45685
88. Stanczak MA, Siddiqui SS, Trefny MP, Thommen DS, Boligan KF, von Gunten S, et al. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J Clin Invest. (2018) 128:4912–23. doi: 10.1172/JCI120612
89. Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec−14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. (2006) 20:1964–73. doi: 10.1096/fj.06-5800com
90. Wang X, Mitra N, Cruz P, Deng L, Varki N, Angata T, et al. Evolution of siglec−11 and siglec−16 genes in hominins. Mol Biol Evol. (2012) 29:2073–86. doi: 10.1093/molbev/mss077
91. Davies LR, Varki A. Why is N-glycolylneuraminic acid rare in the vertebrate brain? Top Curr Chem. (2015) 366:31–54. doi: 10.1007/128_2013_419
92. Chang YC, Nizet V. The interplay between Siglecs and sialylated pathogens. Glycobiology (2014) 24:818–25. doi: 10.1093/glycob/cwu067
93. Angata T, Varki A. Siglec interactions with pathogens. In: Taniguchi N, Endo T, Hart GW, Seeberger PH, Wong C-H, editors. Glycoscience: Biology and Medicine. (Tokyo: Springer) (2015). pp. 633–42.
94. Jones C, Virji M, Crocker P. Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol Microbiol. (2003) 49:1213–25. doi: 10.1046/j.1365-2958.2003.03634.x
95. Avril T, Wagner E, Willison H, Crocker P. Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect Immun. (2006) 74:4133–41. doi: 10.1128/IAI.02094-05
96. Heikema A, Bergman M, Richards H, Crocker P, Gilbert M, Samsom J, et al. Characterization of the specific interaction between sialoadhesin and sialylated Campylobacter jejuni lipooligosaccharides. Infect Immun. (2010) 78:3237–46. doi: 10.1128/IAI.01273-09
97. Bax M, Kuijf M, Heikema A, van Rijs W, Bruijns S, Garcia-Vallejo J, et al. Campylobacter jejuni lipooligosaccharides modulate dendritic cell-mediated T cell polarization in a sialic acid linkage-dependent manner. Infect Immun. (2011) 79:2681–9. doi: 10.1128/IAI.00009-11
98. Carlin A, Uchiyama S, Chang Y, Lewis A, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood (2009) 113:3333–6. doi: 10.1182/blood-2008-11-187302
99. Carlin A, Lewis A, Varki A, Nizet V. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol. (2007) 189:1231–7. doi: 10.1128/JB.01155-06
100. Carlin A, Chang Y, Areschoug T, Lindahl G, Hurtado-Ziola N, King C, et al. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. (2009) 206:1691–9. doi: 10.1084/jem.20090691
101. Khatua B, Bhattacharya K, Mandal C. Sialoglycoproteins adsorbed by Pseudomonas aeruginosa facilitate their survival by impeding neutrophil extracellular trap through siglec-9. J Leukoc Biol. (2012) 91:641–55. doi: 10.1189/jlb.0511260
102. Angata T, Ishii T, Motegi T, Oka R, Taylor R, Soto P, et al. Loss of Siglec−14 reduces the risk of chronic obstructive pulmonary disease exacerbation. Cell Mol Life Sci. (2013) 70:3199–210. doi: 10.1007/s00018-013-1311-7
103. Sewald X, Ladinsky MS, Uchil PD, Beloor J, Pi R, Herrmann C, et al. Retroviruses use CD169-mediated trans-infection of permissive lymphocytes to establish infection. Science (2015) 350:563–7. doi: 10.1126/science.aab2749
104. Rempel H, Calosing C, Sun B, Pulliam L. Sialoadhesin expressed on IFN-induced monocytes binds HIV-1 and enhances infectivity. PLoS ONE (2008) 3:e1967. doi: 10.1371/journal.pone.0001967
105. Zou Z, Chastain A, Moir S, Ford J, Trandem K, Martinelli E, et al. Siglecs facilitate HIV-1 infection of macrophages through adhesion with viral sialic acids. PLoS ONE (2011) 6:e24559. doi: 10.1371/journal.pone.0024559
106. Izquierdo-Useros N, Lorizate M, Puertas M, Rodriguez-Plata M, Zangger N, Erikson E, et al. Siglec-1 is a novel dendritic cell receptor that mediates HIV-1 trans-infection through recognition of viral membrane gangliosides. PLoS Biol. (2012) 10:e1001448. doi: 10.1371/journal.pbio.1001448
107. Puryear W, Akiyama H, Geer S, Ramirez N, Yu X, Reinhard B, et al. Interferon-inducible mechanism of dendritic cell-mediated HIV-1 dissemination is dependent on Siglec-1/CD169. PLoS Pathog. (2013) 9:e1003291. doi: 10.1371/journal.ppat.1003291
108. Varchetta S, Lusso P, Hudspeth K, Mikulak J, Mele D, Paolucci S, et al. Sialic acid-binding Ig-like lectin-7 interacts with HIV-1 gp120 and facilitates infection of CD4pos T cells and macrophages. Retrovirology (2013) 10:154. doi: 10.1186/1742-4690-10-154
109. Suenaga T, Satoh T, Somboonthum P, Kawaguchi Y, Mori Y, Arase H. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc Natl Acad Sci USA. (2010) 107:866–71. doi: 10.1073/pnas.0913351107
110. Suenaga T, Matsumoto M, Arisawa F, Kohyama M, Hirayasu K, Mori Y, et al. Sialic acids on varicella-zoster virus glycoprotein b are required for cell-cell fusion. J Biol Chem. (2015) 290:19833–43. doi: 10.1074/jbc.M114.635508
111. Varchetta S, Brunetta E, Roberto A, Mikulak J, Hudspeth K, Mondelli M, et al. Engagement of siglec-7 receptor induces a pro-inflammatory response selectively in monocytes. PLoS ONE (2012) 7:e45821. doi: 10.1371/journal.pone.0045821
112. Roy S, Mandal C. Leishmania donovani utilize sialic acids for binding and phagocytosis in the macrophages through selective utilization of siglecs and impair the innate immune arm. PLoS Negl Trop Dis. (2016) 10:e0004904. doi: 10.1371/journal.pntd.0004904
113. Yamanaka M, Kato Y, Angata T, Narimatsu H. Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology (2009) 19:841–6. doi: 10.1093/glycob/cwp052
114. Bukvic BK, Blekic M, Simpson A, Marinho S, Curtin JA, Hankinson J, et al. Asthma severity, polymorphisms in 20p13 and their interaction with tobacco smoke exposure. Pediatr Allergy Immunol. (2013) 24:10–8. doi: 10.1111/pai.12019
115. Jonsson S, Sveinbjornsson G, de Lapuente Portilla AL, Swaminathan B, Plomp R, Dekkers G, et al. Identification of sequence variants influencing immunoglobulin levels. Nat Genet. (2017) 49:1182–91. doi: 10.1038/ng.3897
116. Hitomi Y, Tsuchiya N, Hasegawa M, Fujimoto M, Takehara K, Tokunaga K, et al. Association of CD22 gene polymorphism with susceptibility to limited cutaneous systemic sclerosis. Tissue Antigens (2007) 69:242–9. doi: 10.1111/j.1399-0039.2007.00801.x
117. Ma H, Qazi S, Ozer Z, Gaynon P, Reaman G, Uckun F. CD22 Exon 12 deletion is a characteristic genetic defect of therapy-refractory clones in paediatric acute lymphoblastic leukaemia. Br J Haematol. (2012) 156:89–98. doi: 10.1111/j.1365-2141.2011.08901.x
118. Uckun F, Goodman P, Ma H, Dibirdik I, Qazi S. CD22 EXON 12 deletion as a pathogenic mechanism of human B-precursor leukemia. Proc Natl Acad Sci USA. (2010) 107:16852–7. doi: 10.1073/pnas.1007896107
119. Naj A, Jun G, Beecham G, Wang L, Vardarajan B, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. (2011) 43:436–41. doi: 10.1038/ng.801
120. Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J, Carrasquillo M, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. (2011) 43:429–35. doi: 10.1038/ng.803
121. Malik M, Simpson J, Parikh I, Wilfred B, Fardo D, Nelson P, et al. CD33 Alzheimer's risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. (2013) 33:13320–5. doi: 10.1523/JNEUROSCI.1224-13.2013
122. Raj T, Ryan K, Replogle J, Chibnik L, Rosenkrantz L, Tang A, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer's disease susceptibility. Hum Mol Genet. (2014) 23:2729-36. doi: 10.1093/hmg/ddt666
123. Lamba JK, Pounds S, Cao X, Downing JR, Campana D, Ribeiro RC, et al. Coding polymorphisms in CD33 and response to gemtuzumab ozogamicin in pediatric patients with AML: a pilot study. Leukemia (2009) 23:402–4. doi: 10.1038/leu.2008.185
124. Mortland L, Alonzo T, Walter R, Gerbing R, Mitra A, Pollard J, et al. Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin-containing chemotherapy. Clin Cancer Res. (2013) 19:1620–7. doi: 10.1158/1078-0432.CCR-12-3115
125. Wan C, Yang Y, Feng G, Gu N, Liu H, Zhu S, et al. Polymorphisms of myelin-associated glycoprotein gene are associated with schizophrenia in the Chinese Han population. Neurosci Lett. (2005) 388:126–31. doi: 10.1016/j.neulet.2005.06.051
126. Yang Y, Qin W, Shugart Y, He G, Liu X, Zhou J, et al. Possible association of the MAG locus with schizophrenia in a Chinese Han cohort of family trios. Schizophr Res. (2005) 75:11–9. doi: 10.1016/j.schres.2004.11.013
127. Jitoku D, Hattori E, Iwayama Y, Yamada K, Toyota T, Kikuchi M, et al. Association study of Nogo-related genes with schizophrenia in a Japanese case-control sample. Am J Med Genet B Neuropsychiatr Genet. (2011) 156B:581–92. doi: 10.1002/ajmg.b.31199
128. Munz M, Willenborg C, Richter GM, Jockel-Schneider Y, Graetz C, Staufenbiel I, et al. A genome-wide association study identifies nucleotide variants at SIGLEC5 and DEFA1A3 as risk loci for periodontitis. Hum Mol Genet. (2017) 26:2577–88. doi: 10.1093/hmg/ddx151
129. Liu H, Irwanto A, Fu X, Yu G, Yu Y, Sun Y, et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat Genet. (2015) 47:267–71. doi: 10.1038/ng.3212
130. Sun C, Molineros JE, Looger LL, Zhou XJ, Kim K, Okada Y, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet. (2016) 48:323–30. doi: 10.1038/ng.3496
131. Gao P, Shimizu K, Grant A, Rafaels N, Zhou L, Hudson S, et al. Polymorphisms in the sialic acid-binding immunoglobulin-like lectin-8 (Siglec-8) gene are associated with susceptibility to asthma. Eur J Hum Genet. (2010) 18:713–9. doi: 10.1038/ejhg.2009.239
132. Laubli H, Pearce O, Schwarz F, Siddiqui S, Deng L, Stanczak M, et al. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc Natl Acad Sci USA. (2014) 111:14211–6. doi: 10.1073/pnas.1409580111
133. Ishii T, Angata T, Wan ES, Cho MH, Motegi T, Gao C, et al. Influence of SIGLEC9 polymorphisms on COPD phenotypes including exacerbation frequency. Respirology (2017) 22:684–90. doi: 10.1111/resp.12952
134. McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. (2017) 49:1126–32. doi: 10.1038/ng.3892
135. McDonough C, Gong Y, Padmanabhan S, Burkley B, Langaee T, Melander O, et al. Pharmacogenomic association of nonsynonymous SNPs in SIGLEC12, A1BG, and the selectin region and cardiovascular outcomes. Hypertension (2013) 62:48–54. doi: 10.1161/HYPERTENSIONAHA.111.00823
136. Friedman E, Moran DS, Ben-Avraham D, Yanovich R, Atzmon G. Novel candidate genes putatively involved in stress fracture predisposition detected by whole-exome sequencing. Genet Res (Camb). (2014) 96:e004. doi: 10.1017/S001667231400007X
137. Ma Q, Ozel AB, Ramdas S, McGee B, Khoriaty R, Siemieniak D, et al. Genetic variants in PLG, LPA, and SIGLEC 14 as well as smoking contribute to plasma plasminogen levels. Blood (2014) 124:3155–64. doi: 10.1182/blood-2014-03-560086
138. Graustein AD, Horne DJ, Fong JJ, Schwarz F, Mefford HC, Peterson GJ, et al. The SIGLEC14 null allele is associated with Mycobacterium tuberculosis- and BCG-induced clinical and immunologic outcomes. Tuberculosis (Edinb). (2017) 104:38–45. doi: 10.1016/j.tube.2017.02.005
139. Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. (2017) 8:14357. doi: 10.1038/ncomms14357
140. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature. (2018) 558:73–9. doi: 10.1038/s41586-018-0175-2
141. Sasayama D, Hattori K, Ogawa S, Yokota Y, Matsumura R, Teraishi T, et al. Genome-wide quantitative trait loci mapping of the human cerebrospinal fluid proteome. Hum Mol Genet. (2017) 26:44–51. doi: 10.1093/hmg/ddw366
142. Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. (2016) 167:1415–29 e19. doi: 10.1016/j.cell.2016.10.042
143. Angata T. Associations of genetic polymorphisms of Siglecs with human diseases. Glycobiology (2014) 24:785–93. doi: 10.1093/glycob/cwu043
144. Martinez-Picado J, McLaren PJ, Erkizia I, Martin MP, Benet S, Rotger M, et al. Identification of Siglec-1 null individuals infected with HIV-1x. Nat Commun. (2016) 7:12412. doi: 10.1038/ncomms12412
145. Martinez-Picado J, McLaren PJ, Telenti A, Izquierdo-Useros N. Retroviruses As Myeloid Cell Riders: What Natural Human Siglec-1 “Knockouts” Tell Us About Pathogenesis. Front Immunol. (2017) 8:1593. doi: 10.3389/fimmu.2017.01593
146. Buchlis G, Odorizzi P, Soto PC, Pearce OM, Hui DJ, Jordan MS, et al. Enhanced T cell function in a mouse model of human glycosylation. J Immunol. (2013) 191:228–37. doi: 10.4049/jimmunol.1202905
147. Okerblom JJ, Schwarz F, Olson J, Fletes W, Ali SR, Martin PT, et al. Loss of CMAH during human evolution primed the monocyte-macrophage lineage toward a more inflammatory and phagocytic state. J Immunol. (2017) 198:2366–73. doi: 10.4049/jimmunol.1601471
148. Okerblom J, Varki A. Biochemical, cellular, physiological, and pathological consequences of human loss of N-glycolylneuraminic acid. Chembiochem (2017) 18:1155–71. doi: 10.1002/cbic.201700077
149. Varki A, Altheide TK. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. (2005) 15:1746–58. doi: 10.1101/gr.3737405
150. Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, et al. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc Natl Acad Sci USA. (2003) 100:12045–50. doi: 10.1073/pnas.2131556100
151. Malykh YN, Schauer R, Shaw L. N-Glycolylneuraminic acid in human tumours. Biochimie (2001) 83:623–34. doi: 10.1016/S0300-9084(01)01303-7
152. Byres E, Paton AW, Paton JC, Lofling JC, Smith DF, Wilce MC, et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature (2008) 456:648–52. doi: 10.1038/nature07428
153. Moon JM, Aronoff DM, Capra JA, Abbot P, Rokas A. Examination of signatures of recent positive selection on genes involved in human sialic acid biology. G3 (Bethesda) (2018) 8:1315–25. doi: 10.1534/g3.118.200035
Keywords: Siglec, sialic acid, Neu5Ac, Neu5Gc, immunity, microbes
Citation: Angata T (2018) Possible Influences of Endogenous and Exogenous Ligands on the Evolution of Human Siglecs. Front. Immunol. 9:2885. doi: 10.3389/fimmu.2018.02885
Received: 31 August 2018; Accepted: 23 November 2018;
Published: 04 December 2018.
Edited by:
Vered Padler-Karavani, Tel Aviv University, IsraelReviewed by:
Stephan von Gunten, University of Bern, SwitzerlandCopyright © 2018 Angata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Angata, YW5nYXRhQGdhdGUuc2luaWNhLmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.