
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 29 November 2018
Sec. Microbial Immunology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.02769
This article is part of the Research Topic Immunity to Malaria and Vaccine Strategies View all 35 articles
The quest for a licensed effective vaccine against malaria remains a global priority. Even though classical vaccine design strategies have been successful for some viral and bacterial pathogens, little success has been achieved for Plasmodium falciparum, which causes the deadliest form of malaria due to its diversity and ability to evade host immune responses. Nevertheless, recent advances in vaccinology through high throughput discovery of immune correlates of protection, lymphocyte repertoire sequencing and structural design of immunogens, provide a comprehensive approach to identifying and designing a highly efficacious vaccine for malaria. In this review, we discuss novel vaccine approaches that can be employed in malaria vaccine design.
Malaria caused by Plasmodium parasites remains a major infectious disease of public health importance. The disease is caused by five protozoan species, namely Plasmodium falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi. The deadliest of these is P. falciparum which is predominant in sub-Saharan Africa (SSA). In 2016, approximately $2.7 billion was invested globally in control and elimination programs (1). Meanwhile, it was estimated in 2016 that nearly half of the world's population was at risk of infection, with 91% of the estimated deaths being in Africa and 70% of the mortality occurring in children under 5 years (1). Notwithstanding, preventive control and intervention measures have helped decrease the burden between 2000 and 2015. For instance, the incidence of new malaria cases was down by 37% world wide and 42% for the WHO African region. In addition, the incidence of mortality over the same period decreased by about 60% globally and 66% for the African region (2). Yet, malaria imposes huge economic losses for people in the African Region and there is a need to upscale the available interventions and introduce new ones such as a licensed cost-effective vaccine (3).
Malaria eradication faces many challenges including insecticide resistance, emerging anti-malarial drug resistance and the presence of asymptomatic and submicroscopic infections.
Indoor residual spraying (IRS) and long-lasting insecticidal nets (LLINs), have been among the most effective tools for malaria control and elimination (4). So far, pyrethroids are the only recommended class of insecticides for LLINs. However, more than 30 countries have reported resistance to pyrethroids, which has the potential to spread to new areas (5–9).
The rapid development of pyrethroid resistance suggests that alternative classes of insecticides need to be identified. As a result, WHO has cautioned against the use of pyrethroids (8), raising the need for alternative measures of control. The development of resistance to malaria drugs by P. falciparum remains a major threat to malaria elimination. The WHO-recommended first line treatment for uncomplicated malaria caused by Plasmodium falciparum is the artemisinin-based combination therapies (ACTs). Historically, P. falciparum has been able to develop resistance to almost all previous first-line antimalarial drugs (10, 11). The development of resistance to these drugs almost always begins from South-East Asia, where mutant parasites resistant to antimalarial drugs are more likely to survive due to lower levels of acquired immunity, poor adherence to administered drugs and higher parasite burdens (11–14). P. falciparum resistance to artemisinin-based drugs seems to have emerged sporadically (15), with mutations for resistance found within the kelch 13 propeller gene (15, 16). An inevitable fact is that artemisinin resistance may be imminent and other intervention avenues such as the development of highly effective vaccines need to be rapidly explored.
Also, the presence of asymptomatic and submicroscopic infections poses a major threat to malaria eradication and control. Continuous exposure to infectious mosquito bites leads to the development of anti-disease and anti-parasite immunity. The level of this immunity is determined by the transmission intensity and epidemiology of the disease (17, 18). It has been shown that the microscopic prevalence of malaria is almost half of that detected by nucleic acid amplification techniques and lower in low transmission areas (19, 20). The prevalence of submicroscopic infections has been found to be high in low transmission areas and common in children, probably as a result of a less robust immune response, leading to insufficient time for the development of protective immunity. In addition, asymptomatic infections may persist for several months and serve as a major threat to malaria eradication (21) as they sustain disease transmission (22–25).
The acquisition of partial immunity and the successful treatment of clinical symptoms of malaria in children with purified immunoglobulins from semi-immune adults (26) are positive indications of the feasibility of a vaccine against malaria. This is also supported by the induction of sterile immunity in both animal models and controlled human malaria infection (CHMI) through immunization with either live or attenuated sporozoites and merozoite-infected red cells (27–29). Attenuated sporozoites, even though they still maintain their natural hepatocyte invasion ability, do not fully mature in the liver and hence do not form merozoites that are responsible for the clinical symptoms of malaria (30).
There are three stages to target for a potential malaria vaccine candidate. The first target of vaccine development is the pre-erythrocytic stage. This is the period where sporozoites travel through blood and infect hepatocytes to undergo schizogony, the vigorous multiplication stage that precedes the invasion of red blood cells (RBCs). The main purpose of developing a vaccine against this stage is to inhibit hepatocyte infections and hepatic parasite development, thus limiting RBC invasion (27, 30). The mechanisms of protection for this stage may involve antibody responses that prevent sporozoites from invading hepatocytes or cytotoxic T cells that destroy infected liver cells. So far, the licensed RTS,S, subunit vaccine remains the most advanced malaria vaccine to be developed. Other candidate vaccines include the whole-parasite vaccine candidates such as Pf sporozoite (PfSPZ), PfSPZ vaccination with chemoprophylaxis (PfSPZ-CVac) and the genetically attenuated parasite (PfSPZ-GAP).
The second target for malaria vaccine candidate design is the blood-stage of the parasite. The motivation for developing such vaccine candidates comes from evidence that people with repeated malaria infections in endemic areas develop some level of protective immunity, a state in which there is immune-controlled RBC invasion, resulting in fewer disease symptoms or asymptomatic infections (26, 31). Accordingly, vaccine candidates have been designed to elicit immune responses that will block/limit merozoite invasion of RBCs and stop the rapid replication of merozoites by targeting parasite surface proteins such merozoite surface proteins, apical membrane antigen 1 (AMA1), and the reticulocyte homolog (Rh) proteins (32–35). Other blood-stage vaccines target parasite antigens embedded in infected RBC membranes, such as P. falciparum Erythrocyte Membrane Protein-1 (PfEMP1) (36).
Despite being highly immunogenic and showing good promise as vaccine candidates, most of these antigens are also highly polymorphic and hence elicit antigen and parasite strain-specific responses (32, 33). Conversely, antigens such as the Rh proteins that show a high level of conservation (34, 35) tend to be less immunogenic (37).
The third malaria vaccine candidate target is the sexual parasite forms or gametocytes. Malaria transmission-blocking vaccines (TBVs) are designed to interrupt parasite transmission between humans and the mosquito vector through host immunological response to parasite targeted proteins such as Pfs230, Pfs45, Pfs48 (pre-fertilization antigens) and Pfs25, Pfs28 (post-fertilization antigens). Successful malaria transmission depends on the availability of infectious gametocytes in human peripheral blood that can be taken up by mosquitoes during a blood meal. Studies have reported that the degree of infectivity of gametocytes to mosquitoes is based on the gametocyte density, drug stress, clonality of infection and immune defenses of the mosquito (38–42). However, according to Churcher et al. (38), even at very low densities, gametocytes remain infectious to mosquitoes. Also, it has been reported that in the human host, transmission can be stable at very low densities and is not directly proportional to the gametocyte density in peripheral blood (43, 44). Basically, a TBV exploits the fact that there is a functional immunological activity against the sexual stage parasite proteins which is able to reduce the infectivity of the parasite, thereby decreasing malaria transmission (45, 46). Vaccine candidates that seek to interrupt malaria transmission (VIMT) are of two main types: (1) sexual, sporogenic or mosquito stage VIMT (SSM-VIMT) candidates which are expected to interrupt human-to-mosquito transmission; and (2) the pre-erythrocytic VIMT (PE-VIMT) candidates, which are expected to interrupt mosquito-to-human transmission (47). Among the TBV candidates, only Pfs25 and Pfs230 have undergone clinical trials in human (48–51). Unfortunately, a major challenge with these candidate vaccines is the inability to elicit higher antibody titers. In regards, there are considerations to conjugate these candidate vaccines (50, 52).
After decades of extensive research, the pre-erythrocytic stage vaccine, RTS,S has been licensed and is expected to undergo further testing in malaria endemic areas before possible approval for immunization. Currently, together with RTS,S, only 20 candidate vaccines are undergoing clinical trials (Table 1). For RTS,S, a recent evaluation on the safety and immunogenicity of the vaccine co-administered with the recommended expanded programme on immunization showed the vaccine to be safe and immunogenic with no related adverse events (58). The RTS,S/AS01 consists of a recombinant protein of the P. falciparum circumsporozoite protein (CSP) conjugated to a hepatitis B virus surface antigen. During clinical trials, the efficacy of the vaccine after 4 doses was observed to be 43.9% in children aged 5–17 months and 27.8% in children 6–12 weeks old (59). However, vaccine efficacy wanes with time and fails to meet the target set by the Malaria Vaccine Technology Roadmap (60). Consequently, other vaccination regimens such as the number of doses, time of immunization, and alternative approaches for vaccination are being evaluated (61).
Also, the R21, a virus-like particle vaccine which is a biosimilar of RTS,S consists of the CSP conjugated to a single hepatitis B surface antigen. The RTS,S-like vaccine has been shown to provide sterile protection in mice at very low doses. In addition, it was observed that most of the immune responses elicited against the candidate vaccine targeted the CSP in contrast to the hepatitis B surface antigen which is often targeted in the RTS,S vaccinated individuals (53). Importantly, this candidate vaccine is designed such that more epitopes of the CSP may be exposed to host immune system to enhance the efficacy of R21.
Furthermore, the whole sporozoite vaccine has been reported to provide significant protection against falciparum malaria. The whole organism candidate vaccine design approaches include the radiation-attenuated sporozoites (PfSPZ), whole PfSPZ with chemoprophylaxis (PfCVac) and the genetically modified sporozoites (PfGAP). Even though PfCVac showed complete protection to homologous P. falciparum strain, moderate protection has been observed with heterologous strains in non-exposed vaccines (62). Clinical trials with PfSPZ in endemic areas have been shown to be safe and well tolerated, however, inducing low level of immune responses compared to naïve individuals (63, 64). These may suggest that the breadth of immune responses to PfSPZ vaccines need to be increased by considering other vaccination regimens.
Recent technological advances have greatly improved the prospects for designing an effective malaria vaccine through advances in high-throughput biology and computation. These alternative approaches may be focused on the parasite- or host immune system.
The technologies involved in this approach center on the identification of immunogenic antigens from the pathogen by interrogating the parasite's genome, transcriptome or proteome. It may modify the structure of the antigenic component(s) identified with the aim of targeting various strains of the pathogen. The parasite-focused approach further tests the immunogenicity and safety of the candidate antigens to design novel and improved vaccines. This approach may involve the application of reverse vaccinology, structural vaccinology, and immunoinformatics.
Reverse vaccinology, developed by Rappouli et al. is a technology first used in Meningococcus serogroup B bacteria to identify novel vaccine antigens (Figure 1). Here, the pathogen's genome is sequenced and analyzed to have access to the entire repertoire of proteins and enable comparison of conserved sequences shared among pathogens of the same species (65). Genomic data is analyzed using bioinformatics tools, taking into consideration all open reading frames. Also, with the use of computational tools, genomic sequences that are homologous to those of humans are eliminated from the vaccine candidates identified. The remaining genes are isolated and inserted into a suitable vector to obtain proteins for testing in animal models. Responses to the vaccine antigens are analyzed in immunized mice to validate their immunogenicity and efficacy levels. Importantly, molecular epidemiology studies are undertaken using various strains of the pathogen to ascertain whether the selected antigens are conserved or highly variable in a given population (66). This approach has been used to develop vaccines against serogroup B Neisseria meningitidis (67); and identify vaccine candidates for, S. agalactia and S. pyogenes (68, 69). This vaccine design approach has greatly enhanced the discovery and characterization of several pathogen antigens.
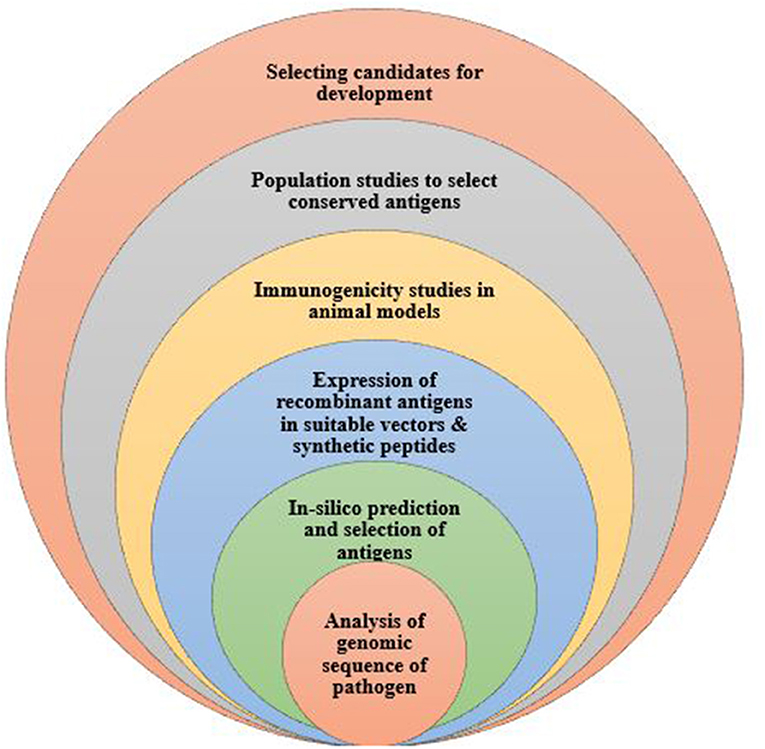
Figure 1. The process of developing a vaccine using reverse vaccinology. Reverse vaccinology starts with obtaining the genomic sequence of the pathogen and using bioinformatics tools to identify all open reading frames to predict protein antigens. The predicted antigens are subsequently expressed in suitable vectors to produce the recombinant proteins which are tested to evaluate the immunogenicity in animal models. Expressed antigens that yield high immunogenicity are selected as vaccine candidates, further tested in population studies to determine and identify conserved antigens for further vaccine development.
Reverse vaccinology has been applied in malaria to identify parasite proteins either secreted or involved in signaling for consideration as possible vaccine candidates. The genomic sequence of P. falciparum has been available since 2002 (70). In addition, the sequence of other diverse Plasmodium spp including primate (71, 72) and rodent (73) parasites have been published. Comparative analysis of these sequences has shown similar homologs between species with possibly similar functions. For instance, the conserved Pf48/45 and PfHAP2 genes, both of which are transmission blocking vaccine candidates, functions were determined based on the role of their homologs in other Plasmodium spp. The functional analysis of the P48/45 and PHAP2 genes in P. berghei established their significant role in reducing the fertility of male gametes during fertilization, promoting these genes as vaccine candidates (74, 75), which are currently in the preclinical stage (76–78).
Despite the success of reverse vaccinology, it cannot be used to identify non-peptide antigens but can identify operons that code for synthesis of such molecules (79). For pathogens with complex genomes such as malaria parasites, no successful vaccine has as yet been developed via this approach. Further progress requires, among other things, improved predictive algorithms to identify the T and B cell epitopes as well as accurate quantitative assessments before inclusion in vaccines.
An improved understanding of the native structures of biological macromolecules such as proteins and how changes in their structure affect their functions can assist the identification of suitable epitopes (80, 81). Such epitopes can be designed into accessible forms for easy uptake by immune cells. These structural considerations make it possible to improve vaccine immunogenicity and safety and mitigate the effects of sequence variability within different strains of a pathogen (82). For instance, the bacterium Meningococcus is able to evade the host's immune system with the aid of a factor H binding protein (Hbp), which inactivates the host complement pathway by blocking factor H. Structural considerations allowed immunodominant epitopes of Hbp from various meningococcal strains to be identified and grafted into a single variant molecule to form a single antigen. This antigen was used in the MenB vaccine, responses against which are able to neutralize all the targeted strains (83).
Also, in an earlier study, short conserved α-helical coiled coil structural domains were identified from the asexual blood stage of the P. falciparum by examining the Plasmodium genome (84). Upon further screening, an unstructured peptide (P27A) that unfolds in native confirmation was selected. The peptide was the target of human antibodies which were able to restrict parasite replication (85). The vaccine candidate P27A has been considered immunogenic and safe with mild adverse events after Phase1 clinical trials (86).
For pathogens like P. falciparum, structural vaccinology may also help overcome antigenic variation. For instance, the application of structural vaccinology enabled the characterization of the less polymorphic DBL4ε domain of VAR2CSA to identify novel properties in the motif that affects the functional features of the antigen (87); identification and confirmation of the three-dimensional structure of the invasion ligand Cysteine-Rich Protective Antigen (CyRPA) (88). For example, the CyRPA was identified as a protective epitope providing an additive effect with the Reticulocyte binding-like Homologous protein 5 (Pf RH5) such that antibodies against Pf RH5 and CyRPA can inhibit parasite replication in host RBCs (88). Hopefully malaria vaccines incorporating these epitopes may elicit strong protective immune responses. Combining these protective antigens to create hybrid protein vaccines with enhanced efficacy may be a viable option for malaria.
A key challenge with this approach is the identification of suitable B and T cell epitopes for incorporation into vaccine candidates.
Immunoinformatics integrates both computational approaches and experimental immunology to develop machine learning algorithms that attempt to predict the immunogenicity of antigens. These approaches can be either pattern- or theory-based and may operate at either the amino acid sequence or the protein structure level. The pattern-based approaches conceive the prediction problem as one of finding sequence/structural patterns associated with immunogenicity. In contrast, the theory-based approaches attempt to model the basis for immunogenicity, for example, by using physical principles. Examples of algorithmic tools employed by pattern-based approaches include quantitative structure-activity relationship analysis, support vector machines, and artificial neural networks (89, 90). Theory-based approaches often employ Markovian and/or Bayesian models as well as models based on statistical mechanics (91).
Immunoinformatic approaches have already been applied to P. falciparum to predict possible cytotoxic T cell epitopes coupled with HLA A/B molecules for malaria peptide vaccine design (92). For example, the PfEMP1 gene, a member of the var gene family has been associated with parasite evasion from host immune mechanisms due to its multiple variation and ability to bind to different host receptors (36). In a recent study, both in-silico and experimental approaches were used to identify antigenic epitopes from CIDR-1 and DBL-3γ conserved domains of PfEMP1. These epitopes were predicted to have good binding affinity to HLA molecules as well as the capability to induce IFN-γ, IL-4 secretion and T cell proliferation in exposed individuals (93).
Classically, HLA class I molecules optimally require peptides that are 8-10 amino acids long for presentation to CD8 T cells while HLA Class II molecules optimally require 12-25 amino acids long peptides for presentation to CD4 T cells. Of note high predictive accuracies have been achieved for bioinformatics methods for predicting peptide binding to HLA I molecules; whereas those for predicting binding to HLA II molecules require further improvement. An even greater challenge is prediction of peptide binding to B cell receptors for effective antibody responses. On-going work by us and other groups is aimed at addressing some of these challenges (91, 94). However, not all HLA binders are good epitopes for T cells and this poses a major challenge for approaches that predict HLA binders without considering the global picture of HLA-peptide-TCR interactions. Nonetheless, these computational approaches, which are quite cost-effective and are important down-selection tools in instances where there are too many peptides to evaluate experimentally, have the potential to aid in the development of effective vaccines against malaria.
Due to the sophisticated immune-evasion mechanisms of P. falciparum that allow it to coexist with the host, vaccinologists require new paradigms in vaccine development. One such new paradigm that has been developed to target these pathogens is the immune-focused approach (Figure 2). In contrast to the parasite-focused approach, which centers on the pathogen of interest, this new approach seeks to harness the host immune system to more rapidly design effective vaccines. It focuses on studying the host immune system to discover protective immune signatures. It is expected that these protective signatures can be induced de novo in susceptible hosts to protect them against infection and/or disease. Compared to the pathogen-focused approach, the immune-focused approach has, in principle, a greater potential for success against pathogens like malaria parasites, which have highly variable genomes. In particular, it may be able to identify and design immune cells with broadly neutralizing antibodies (95) and enhanced cellular immune responses, which has proved difficult to accomplish by using conventional approaches. To provide context for the discussion of opportunities for vaccine development, we begin with a brief overview of human immunity to malaria.
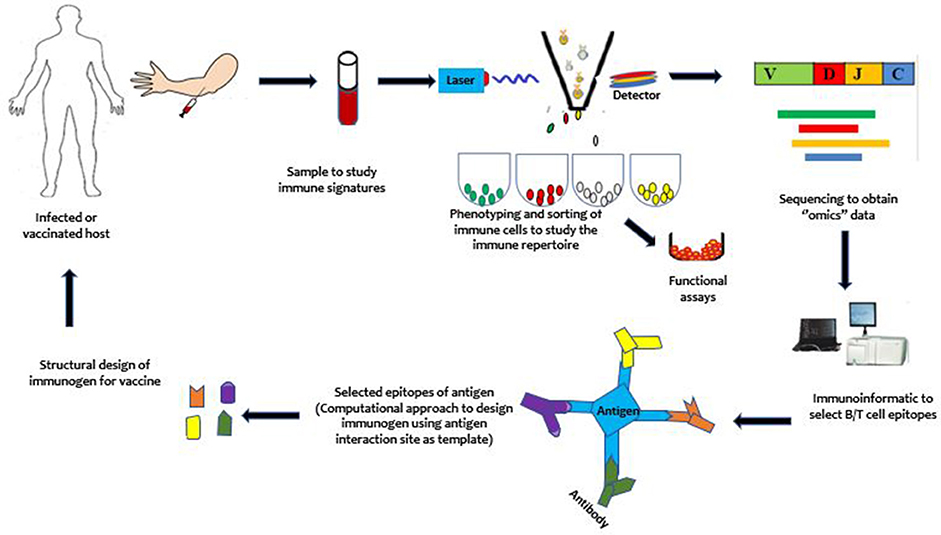
Figure 2. An illustration of the immune-focused approach to vaccine development. Briefly, from top left, samples are obtained from an infected but protected or a vaccinated host. Immune cells are phenotyped and sorted using single cell sorting technologies such as the flow cytometry. The sorted cells may be sequenced directly or proliferated for functional studies before sequencing to study the immune repertoires (B/T cell receptor repertoire). With the use of computational approaches, the immunogen is designed and taken through further stages of development to yield a vaccine. After vaccination, mechanisms underlying vaccine efficacy and safety can be studied to further enhance the developed vaccine to achieve maximum protection against the targeted pathogen.
In contrast to many pathogens against which highly potent, long-lived immunity is achieved, human immunity to malarial parasites is less potent and relatively short-lived (17). In malaria, acquired immunity to infection is rare; rather, what develops naturally, generally over a long period, is acquired immunity to disease (96). Such clinical immunity generally targets the disease-causing asexual blood stage of malarial parasites. It tends to be acquired faster in moderate-to-high transmission settings compared to low transmission areas, and with a higher number of clinical episodes (97–99). As with other infectious diseases, the development of clinical immunity to malaria is dependent on the adaptive arm of the immune system, and the principal mediators consist of specific subsets of B and T cells. Some progress has been made to elucidate the underlying mechanisms, although the key immune determinants remain unclear.
In addition, the ability to predict the beginning and end of transmission seasons have made it possible to study host responses to infection and some immune dynamics that occur before, during and after infection as well as, drug interventions (100) and how they may affect the immunity acquired (101, 102).
In natural infections, acquisition of immunity to sporozoite stage infections is limited, probably due to the low number of sporozoites that are inoculated as well as the limited time that sporozoites have extracellular, prior to hepatocyte invasion. In addition, it has been reported that Plasmodium sporozoites are able to modulate the cytokine environment by downregulating Th1 responses and antigen presentation to T cells (103). Recently, it was reported that continuous exposure to P. falciparum leads to the induction and expression of immunoregulatory cytokines such as IL-10 and affects the function of dendritic cells (104). These, coupled with frequent infection and immune activation, may profoundly impact on the tolerogenic environment leading to the escape of sporozoites from immune cells. Nevertheless, functional properties of antibodies to sporozoite-stage infections have been associated with natural protection from clinical disease. It has been reported that these antibodies kill sporozoites through complement fixation and inhibit hepatocyte invasion. However, the response to sporozoite antigens was age-dependent and acquired slowly compared to blood-stage antigens (105).
The blood-stage parasite is associated with the clinical symptoms of the disease as it causes an upregulation of pro-inflammatory cytokines, regulatory T cells and parasite sequestration in small blood vessels in host organs. Antibodies have been reported to play functional roles in preventing parasite invasion of red blood cells (106, 107). Antibodies to parasite antigens are associated with clinical immunity in endemic areas (108–110). The mechanisms of antibody activity may include blocking invasion of erythrocytes (111); opsonizing parasites to facilitate their clearance (110, 112) enhancing the killing of infected cells by monocytes (113); complement-mediated lysis of infected cells (114); and inhibiting adherence of infected erythrocytes to vascular endothelium (115). However, the generation of atypical memory B cells which have reduced effector functions has been observed under chronic conditions (116, 117).
T cells have also been shown to play protective roles during blood-stage infection. For instance, protection from the disease has been associated with FOXP3− Th1 cells which are self-regulatory and produce IFNγ, TNF and IL-10 (101, 118, 119). These cells are believed to prevent the production of pyrogenic factors that may lead to the manifestation of clinical disease. However, these immune responses are not long-lasting and easily decay after infection has waned. T cell responses are hampered by the upregulation of negative immune regulatory receptors which may blunt or cause anergic responses (116, 120, 121). Our recent study found higher levels of T regulatory cells to be associated with higher blood levels of P. falciparum in children (118), suggesting less effective control of the parasite. Indeed, trying to understand these various aspects of the immune responses is a quite complex task (122).
Compared to natural infections, inducing sterile immunity in naïve individuals has been achieved through whole sporozoite immunization (29, 123) although similar outcomes have not been seen in individuals from malaria endemic areas (63, 64). Vaccination of volunteers with radiation-attenuated sporozoites has shown that both T cells and antibody responses play a significant role in protecting vaccinated cohorts against clinical challenge. It was observed that T cells from the periphery of these individuals, when stimulated with P. falciparum sporozoites in vitro, produced effector cytokines in a dose-dependent manner whereas antibody levels increased and prevented hepatocyte invasion (124–127).
The challenges of inducing immunity to malaria by natural or artificial means are compounded by the sophisticated immune-evasion strategies of the parasite. The parasite has a large genome consisting of about 5,300–5,500 possible antigenic targets (70). This extensive gene repertoire coupled with the parasite's high mutation rate allows for extensive variation of antigens that can be potential vaccine targets. Moreover, the epitopes targeted by the immune system exhibit a hierarchy of immunogenicity, with immunodominant epitopes that induce large amounts of antibodies, not all of which are neutralizing and may mask sub-dominant epitopes bound by neutralizing antibodies (73). In addition, the parasite switches off antigenic phenotypes, associated with the variant antigens resulting in functional diversity. Consequently, infections are mostly characterized by successive parasitemia waves caused by different parasite variants, making the development of long-lived immunity to the parasite very challenging (36, 128). Furthermore, key antigens such as CSP contain tandem repeats that have been implicated in immune evasion by suppressing antibody responses against adjacent antigens (129).
The age and genetics of a person may modulate the immune responses elicited during infections and vaccinations (130, 131). Nonetheless, these responses that modulate infection may help to systematically define factors associated with protection from disease. Conventional approaches to understanding immune correlates of protection against P. falciparum includes, but is not limited to, ELISA, Elispot and Western blots. However, recent advances in high throughput assays have allowed in-depth analysis of immune correlates of protection to multiple falciparum antigens. Individuals in malaria-endemic areas generate antibodies to different P. falciparum proteins which may be protective or serve as a serological marker for exposure.
High throughput assays that probe the genomic, proteomic and transcriptomic data of immune responses are useful means of determining correlates of protection in exposed and vaccine trial cohorts. Independent studies using library expression and protein microarray has characterized host immune reactivity to different P. falciparum antigens. Using these approaches, Doolan et al. (132) were able to identify stage-specific P. falciparum antigens associated with protection in naturally exposed individuals, vaccine protected and non-protected individuals using a protein microarray chip with 250 proteins. They observed distinctive antibody profiles in the various groups to these antigens. Also, in an independent study, involving a large cohort of children naturally exposed to malaria (≤10 years old) in Kenya, it was observed that responses to fewer proteins from the 39 P. falciparum antigens analyzed were significantly associated with protection, and these included AMA1 and MSP2. Also, antibodies to the top 10 proteins provided an additive effect whereas most antibody responses to the other antigens were markers of malaria exposure (133).
A similar study conducted in Mali probed sera from malaria-exposed children and adults against 1204 proteins. Among these proteins, 91 were associated with sexual stage-specific immunity with specific-IgG responses culminating during the transmission season. It was further observed that immunity to these sexual stage vaccine candidates (Pfs48/45 and Pfs230 but not Pfs25) can be boosted in natural infections (134). These studies showed evidence that the breadth and magnitude of the antibody response is a better correlate of immune protection. Furthermore, in analyzing PBMCs for non-humoral immune responses associated with protection using DNA microarrays, qRT-PCR and flow cytometry, it was observed that repeated exposure to malaria in children was associated with the upregulation of genes involved in immune regulation (such as IL-10 secretion from CD4+Foxp3-), phagocytosis and activation of adaptive immune system. In contrast, gene expression levels of chemokines and cytokines associated with fever and inflammation (such as IL-1β, TNF, CXCL2 and IL-8) were downregulated (101).
Interestingly, the application of next-generation sequencing techniques such as lymphocyte immune repertoire sequencing, including T cell receptor (TCR), membrane-bound B cell receptor (BCR) or secreted BCR can allow an in-depth analysis of host factors associated with pathogen recognition, identification and protection from disease. The TCR structure is heterodimeric with two protein subunits; an alpha and beta chain or gamma and delta chain with both a constant and variable region. Similarly, the BCR consists of two heavy and light chains which are joined together by disulphide bonds to form a Y shaped immunoglobulin together with a variable and a constant region. The lymphocyte receptors (TCR/BCR) have similar structures including a variable, diversity and joining regions that enable diversification in identifying different host pathogens. In the generation of receptor diversity, there is a recombination of a V, D, and J segment of a beta or heavy chain, and a V and J segment for the alpha or light chain. For the BCR, this process helps expose very potent neutralizing antibodies that may be public in protecting against clinical disease. The generation of the variable regions may help guarantee higher levels of somatic mutation at the antigen binding site which may be shared or unique to an individual(s).
Despite the documented importance of lymphocyte receptors for antigen recognition and, hence, for the initiation of adaptive immune responses, the specific TCRs/BCRs that determine immunity to particular pathogens remain poorly understood. To our knowledge, no previous study has comprehensively mapped these receptors and analyzed how their expression profiles may correlate with individual variations in immune protection against malaria. In addition, the application of machine learning algorithms such as random forests, support vector machines may allow the identification of patterns on immune correlates that may predict protection against disease (118, 135).
Moreover, these approaches generate huge amounts of data that can be computationally analyzed to generate new, experimentally testable hypotheses. These may yield novel insights into the mechanisms underlying vaccine safety and efficacy. Importantly, data from such studies will inform pathways to which vaccine strategies should focus.
Effective vaccines are supposed to elicit and provide long-term protection as well as require both B and T cells to produce effective antibodies to neutralize surface-expressed antigens. B cell lineage vaccine design is an immune-focused approach that combines human immunology, structural biology, and computational protein design to develop a vaccine. The aim is to identify in both naïve and memory B cell receptors, paratopes (antigen binding sites) that interact with immunogens of interest. For a vaccine to be designed through this approach, memory B cell clones from the same lineage (or clone) are first identified and isolated from patients that produce broadly neutralizing antibodies or protective antibodies. These clones are then sequenced to obtain the V(D)J and VJ gene pairs that make up the B cell receptors in order to identify the paratope. Computational approaches are used to design an immunogen that interacts with the identified paratope (95). For P. falciparum, neutralizing antibodies produced by activated B cells are required to prevent the infection of new RBCs. By isolating such protective B cells from malaria patients and sequencing and analyzing their antigen receptors, it might be possible to identify immunogens able to induce protective immunity in susceptible individuals.
In malaria, the identification of broadly neutralizing antibodies remained elusive partly due to the high polymorphic nature of P. falciparum antigens. In addition, malaria vaccine candidates tend to induce antibodies with weak neutralizing ability, low breadth and strain-specific. However, Tan et al. (136) have recently identified monoclonal antibodies that can recognize P. falciparum-infected RBCs (iRBCs) from different strains of parasites. These antibodies recognize and bind to iRBCs through the RIFIN proteins, a group of variant antigens that are extracellularly expressed on the surface of iRBCs and have been associated with immune evasion (128) to initialize opsonization.
Another remarkable example is the identification of the novel antigenic target NPDP (part of the sequence in the N-terminal junction peptide) that is found between genes for the CSP and NANP and NVDP tandem repeats (137, 138). Independent studies in 2018 by Tan et al. (137) and Kisalu et al. (138) identified and isolated neutralizing antibodies from memory B cells and plasmablasts that could inhibit hepatocyte infection by PfSPZ. Through structural information, they were able to identify that these antibodies bind to conserved epitopes in the N-terminus of the CSP that is not found within the RTS,S vaccine.
Furthermore, mAbs that can inhibit parasite replication to about 97% have been isolated from CHMI donors immunized with RTS,S. Deciphering the structure and functionality of these antibodies have provided an informed overview on the structure of the CSP in vivo. Thus provides positive implications in the design of CSP immunogens against P. falciparum (139). However, there are still unsolved questions on the antibody responses to the PfCSP which have been described to be protective (139) and non-protective (129) as well as more structural information is needed to induce such potent neutralizing antibodies during vaccination Nonetheless, it is very interesting since they have implications in designing immunogens that can target specific immune responses and probably improve the efficacy of the RTS,S vaccine.
Protective immunity to malaria liver-stage infection has been attributed to T cells in both human and rodent models. In studying immune responses to malaria such as cerebral malaria, murine models have provided significant understanding of various immunological properties that have impacted our understanding of the immune activity in humans.
For instance, Lau et al. (140) characterize MHC-restricted TCR that have potential in enhancing antigen presentation to T cells to enhance T cell immunity. They developed a novel CD8+ T cell receptor to P. berghei termed PbT-I from transgenic mouse with immune specificity for liver-stage and blood-stage infections. Isolated TCR genes from Vα8.3 and Vβ10 were isolated from a restricted hybridoma T cell line generated from Plasmodium berghei ANKA (PbA) blood-stage infection. Despite been developed for PbA, this transgenic MHC-I restricted T cell line was cross-reactive to P. chabaudi and P. yoelli. This implies that they may recognize conserved regions in rodent Plasmodium spp. Functional analysis revealed that the PbT-I cells produced effector cytokines (IFNγ, TNFα) and was positive for the degranulation marker (CD107a) showing their involvement in immune activity during the PbA infection. Using PbT-I CD8+ T cells, the peptides responsible for their activation were elucidated.
In a subsequent research, they identified and developed PbT-II CD4+ T cells from mouse transgenic line using the TCRα (Vα2.7, Jα12, Cα) and TCRβ gene (Vβ12, Dβ2, Jβ2.4) segments to blood-stage PbA infection (141). These cells were cross-reactive to rodent parasites (P. berghei, yoelli and chabaudi) and to P. falciparum. These MHC-II restricted PbT CD4+ T cells enhanced both humoral activity of B cells and cytotoxic activity of CD8+ T cells. In addition, the study confirmed that immunity to antigens in both blood stage and liver-stage development can restrict parasite replication in the hepatic stage and characterize the impact of blood stage antigen presentation to T cells that can enhance such T-cell immunity during infection. The uses of these target antigens may delineate protective immune responses and possibly circumvent pathologic outcomes. More importantly, further work should be focused on identifying and understanding such broadly reactive Plasmodium-specific T cells in host infections.
The structure-based vaccine approach can be employed in both the parasite and immune-focused approach. However, in the immune-focused approach, the principle is based on understanding the structural properties of the immune cell providing the desired response. Here, the properties of the antigenic binding site on the immune cell is studied at the atomic level (80, 81). By understanding these properties, the approach seeks to design and develop immunogens to target the protective response or develop these immune cells for use as interventions.
Structural-based vaccine design has aided in unmasking immunodominant epitopes in the haemagglutinin-stem of the influenza virus (142), the fusion protein in the respiratory syncytial virus (143) and CD4 binding site in HIV-1 virus. For example, identifying conserved immunogenic epitopes in HIV has been quite challenging. However, elucidating the structure of broadly neutralizing antibodies (bNAbs) has been very useful. Using NAb, subdominant epitopes in the CD4 binding site by the gp120 viral protein were identified. Probing the structure of the antigenic binding site on CD4, the structural properties helped in the development of a recombinant protein (RSC3) with specificity to the NAb. The RSC3 was further used to identify and isolate B cells that expressed broadly neutralizing antibodies with increased breadth. VRC01 and 3BNC117, highly potent monoclonal bnMAb with reactivity to about 91% to HIV-1 isolates were developed (144). Phase I clinical trials of the VRC01 were reported as safe with no allergenicity (145, 146). It is currently being evaluated in a Phase IIB trials with a projected overall efficacy of 53 and 82% (147). These observations indicate that using structural properties, subdominant epitopes can be uncovered to design immunogens to target a specific immune response.
Currently, there are few examples of the successful use of these approaches in malaria vaccine design. For instance, using invasion-inhibitory monoclonal antibodies, the novel structure of PfRh5 in complex with basigin was characterized, together with novel protective epitopes found in the complex (148). Similarly, for P. vivax infections, bNAbs that confer strain-specific immune responses (149) were isolated. These bNAbs, enabled the characterization of protective epitopes in the duffy binding protein that can be included in the design of a potent P. vivax vaccine (150).
The development of a highly efficacious malaria vaccine faces many challenges, both technical and biological. Partly because the parasite is equipped with a variety of evasion mechanisms allow it to co-exist with the host. With the recent advent of high throughput approaches such as lymphocyte repertoire sequencing and structural design of immunogens, the breadth of protection of previous and current vaccine candidates may be enhanced as well as the identification of new candidate vaccines. In addition, vaccinologist may be able to design vaccines that drive the immune system through unusual yet protective pathways. Likewise, the application of mathematical modeling and computational approaches to the data thus obtained will open new pathways toward designing highly effective vaccines against malaria and aid in achieving the targets set by the malaria vaccine technology roadmap for 2030.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Augustina Frimpong is supported by a Ph.D. fellowship from a World Bank African Centers of Excellence Grant (ACE02-WACCBIP: Awandare) and the International Development Research Center grant from the African Institute for Mathematical Sciences, Ghana, the Carnegie Corporation of New York and the University of Ghana BanGA Ph.D. Research Grant. WN is supported by the AIMS Global Secretariat.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACTs, artemisinin-based combination therapies; PfEMP1, P. falciparum Erythrocyte Membrane Protein-1; MSP1, Merozoite Surface Protein; AMA1, Apical Membrane Antigen 1; Rh, Reticulocyte homolog (Rh); MHC, Major histocompatibility complex; HLA, Human leukocyte antigen; PfSPZ, Pf sporozoite; PfCVac, Pf chemoprophylaxis vaccination; PfGAP, Pf genetically attenuated parasite; TBV, transmission-blocking vaccines; BCR, B cell receptor; bnMAb, broadly neutralizing monoclonal antibody; TCR, T cell receptor; VIMT, Vaccine that interrupt malaria transmission; SSM-VIMT, Sexual, sporogenic or mosquito stage VIMT; PE-VIMT, pre-erythrocytic VIMT.
3. Alonso PL, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, Collins F, et al. A Research Agenda to Underpin Malaria Eradication. PLoS Med. (2011) 8:e1000406. doi: 10.1371/journal.pmed.1000406
4. Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature (2015) 526:207. doi: 10.1038/nature15535
5. WHO. Global Plan for Insecticide Resistance Management in Malaria Vectors (GPIRM). Geneva: World Health Organization (2012).
6. Müller P, Donnelly MJ, Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics (2007) 8:36. doi: 10.1186/1471-2164-8-36
7. Toé KH, N'Falé S, Dabiré RK, Ranson H, Jones CM. The recent escalation in strength of pyrethroid resistance in Anopheles coluzzi in West Africa is linked to increased expression of multiple gene families. BMC Genomics (2015) 16:146. doi: 10.1186/s12864-015-1342-6
8. Ranson H, Lissenden N. Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. (2016) 32:187–96. doi: 10.1016/j.pt.2015.11.010
9. Choi KS, Christian R, Nardini L, Wood OR, Agubuzo E, Muleba M, et al. Insecticide resistance and role in malaria transmission of Anopheles funestus populations from Zambia and Zimbabwe. Parasit Vectors (2014) 7:464. doi: 10.1186/s13071-014-0464-z
10. Moore DV, Lanier JE. Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am J Trop Med Hyg. (1961) 10:5–9. doi: 10.4269/ajtmh.1961.10.5
11. Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science (2004) 305:1124. doi: 10.1126/science.1098876
12. Hanboonkunupakarn B, White NJ. The threat of antimalarial drug resistance. Trop Dis Travel Med Vaccin. (2016) 2:10. doi: 10.1186/s40794-016-0027-8
13. Ataíde R, Powell R, Moore K, McLean A, Phyo AP, Nair S, et al. Declining transmission and immunity to malaria and emerging artemisinin resistance in Thailand: a longitudinal study. J Infect Dis. (2017) 216:723–31. doi: 10.1093/infdis/jix371
14. Ataide R, Ashley EA, Powell R, Chan J-A, Malloy MJ, O'Flaherty K, et al. Host immunity to Plasmodium falciparum and the assessment of emerging artemisinin resistance in a multinational cohort. Proc Natl. Acad. Sci. (2017) 114:3515–20. doi: 10.1073/pnas.1615875114
15. Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. (2014) 371:411–23. doi: 10.1056/NEJMoa1314981
16. Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis. (2017) 17:491–7. doi: 10.1016/S1473-3099(17)30048-8
17. Doolan DL, Dobaño C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. (2009) 22:13–36. doi: 10.1128/CMR.00025-08
18. Tran TM, Li S, Doumbo S, Doumtabe D, Huang C-Y, Dia S, et al. An intensive longitudinal cohort study of malian children and adults reveals no evidence of acquired immunity to Plasmodium falciparum infection. Clin Infect Dis (2013) 57:40-7. doi: 10.1093/cid/cit17
19. Okell LC, Ghani AC, Lyons E, Drakeley CJ. Submicroscopic infection in Plasmodium falciparum-endemic populations: a systematic review and meta-analysis. J Infect Dis. (2009) 200:1509–17. doi: 10.1086/644781
20. Idris ZM, Chan CW, Kongere J, Gitaka J, Logedi J, Omar A, et al. High and heterogeneous prevalence of asymptomatic and sub-microscopic malaria infections on islands in Lake Victoria, Kenya. Sci Reports (2016) 6:36958. doi: 10.1038/srep36958
21. Roucher C, Rogier C, Dieye-Ba F, Sokhna C, Tall A, Trape J-F. Changing Malaria Epidemiology and Diagnostic Criteria for Plasmodium falciparum Clinical Malaria. PLoS ONE (2012) 7:e46188. doi: 10.1371/journal.pone.0046188
22. Baum E, Sattabongkot J, Sirichaisinthop J, Kiattibutr K, Jain A, Taghavian O, et al. Common asymptomatic and submicroscopic malaria infections in Western Thailand revealed in longitudinal molecular and serological studies: a challenge to malaria elimination. Malaria J. (2016) 15:333. doi: 10.1186/s12936-016-1393-4
23. Lindblade KA, Steinhardt L, Samuels A, Kachur SP, Slutsker L. The silent threat: asymptomatic parasitemia and malaria transmission. Expert Rev Anti Infect Ther. (2013) 11:623–39. doi: 10.1586/eri.13.45
24. Bousema JT, Gouagna LC, Drakeley CJ, Meutstege AM, Okech BA, Akim IN, et al. Plasmodium falciparum gametocyte carriage in asymptomatic children in western Kenya. Mal J. (2004) 3:18. doi: 10.1186/1475-2875-3-18
25. Coleman RE, Kumpitak C, Ponlawat A, Maneechai N, Phunkitchar V, Rachapaew N, et al. Infectivity of asymptomatic Plasmodium-infected human populations to Anopheles dirus mosquitoes in western Thailand. J Med Entomol. (2004) 41:201–8. doi: 10.1603/0022-2585-41.2.201
26. Cohen S, McGregor I, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature 1961:733–7. doi: 10.1038/192733a0
27. Roestenberg M, McCall M, Hopman J, Wiersma J, Luty AJ, van Gemert GJ, et al. Protection against a malaria challenge by sporozoite inoculation. N Engl J Med. (2009) 361:468–77. doi: 10.1056/NEJMoa0805832
28. Pombo DJ, Lawrence G, Hirunpetcharat C, Rzepczyk C, Bryden M, Cloonan N, et al. Immunity to malaria after administration of ultra-low doses of red cells infected with Plasmodium falciparum. Lancet (2002) 360:610–7. doi: 10.1016/S0140-6736(02)09784-2
29. Hoffman SL, Goh LM, Luke TC, Schneider I, Le TP, Doolan DL, et al. Protection of humans against malaria by immunization with radiation-attenuated Plasmodium falciparum sporozoites. J Infect Dis (2002) 185:1155–64. doi: 10.1086/339409
30. Nussenzweig V, Nussenzweig RS. Rationale for the development of an engineered sporozoite malaria vaccine. Adv Immunol. (1989) 45:283–334. doi: 10.1016/S0065-2776(08)60695-1
31. Baruch D, Gormely J, Ma C, Howard R, Pasloske B. Plasmodium falciparum erythrocyte membrane protein 1 is a parasitized erythrocyte receptor for adherence to CD36, thrombospondin, and intercellular adhesion molecule 1. Proc Natl Acad Sci USA. (1996) 93:3497–502. doi: 10.1073/pnas.93.8.3497
32. Hill DL, Wilson DW, Sampaio NG, Eriksson EM, Ryg-Cornejo V, Harrison GLA, et al. Merozoite Antigens of Plasmodium falciparum elicit strain-transcending opsonizing immunity. Infect Immun (2016) 84:2175–84. doi: 10.1128/IAI.00145-16
33. Kusi KA, Dodoo D, Bosomprah S, van der Eijk M, Faber BW, Kocken CHM, et al. Measurement of the plasma levels of antibodies against the polymorphic vaccine candidate apical membrane antigen 1 in a malaria-exposed population. BMC Infect Dis. (2012) 12:32. doi: 10.1186/1471-2334-12-32
34. Baum J, Chen L, Healer J, Lopaticki S, Boyle M, Triglia T, et al. Reticulocyte-binding protein homologue 5–an essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. Int J Parasitol. (2009) 39:371–80. doi: 10.1016/j.ijpara.2008.10.006
35. Rodriguez M, Lustigman S, Montero E, Oksov Y, Lobo CA. PfRH5: a novel reticulocyte-binding family homolog of Plasmodium falciparum that binds to the erythrocyte, and an investigation of its receptor. PLoS ONE (2008) 3:e3300. doi: 10.1371/journal.pone.0003300
36. Mkumbaye SI, Wang CW, Lyimo E, Jespersen JS, Manjurano A, Mosha J, et al. The severity of Plasmodium falciparum infection is associated with transcript levels of var genes encoding EPCR-binding PfEMP1. Infect Immun. (2017) 85:e00841-16. doi: 10.1128/IAI.00841-16
37. Partey FD, Castberg FC, Sarbah EW, Silk SE, Awandare GA, Draper SJ, et al. Kinetics of antibody responses to PfRH5-complex antigens in Ghanaian children with Plasmodium falciparum malaria. PLoS ONE (2018) 13:e0198371. doi: 10.1371/journal.pone.0198371
38. Churcher TS, Bousema T, Walker M, Drakeley C, Schneider P, Ouédraogo AL, et al. Predicting mosquito infection from Plasmodium falciparum gametocyte density and estimating the reservoir of infection. Elife (2013) 2:e00626. doi: 10.7554/eLife.00626
39. Bousema T, Okell L, Shekalaghe S, Griffin JT, Omar S, Sawa P, et al. Revisiting the circulation time of Plasmodium falciparum gametocytes: molecular detection methods to estimate the duration of gametocyte carriage and the effect of gametocytocidal drugs. Mal J. (2010) 9:136. doi: 10.1186/1475-2875-9-136
40. Nsango SE, Abate L, Thoma M, Pompon J, Fraiture M, Rademacher A, et al. Genetic clonality of Plasmodium falciparum affects the outcome of infection in Anopheles gambiae. Int J Parasitol. (2012) 42:589–95. doi: 10.1016/j.ijpara.2012.03.008
41. Lefèvre T, Vantaux A, Dabiré KR, Mouline K, Cohuet A. Non-Genetic Determinants of Mosquito Competence for Malaria Parasites. PLoS Pathogens (2013) 9:e1003365. doi: 10.1371/journal.ppat.1003365
42. Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathogens (2009) 5:e1000423. doi: 10.1371/journal.ppat.1000423
43. Schneider P, Bousema JT, Gouagna LC, Otieno S, Van de Vegte-Bolmer M, Omar SA, et al. Submicroscopic Plasmodium falciparum gametocyte densities frequently result in mosquito infection. Am J Tropic Med Hygiene (2007) 76:470–4. doi: 10.4269/ajtmh.2007.76.470
44. Hallett RL, Dunyo S, Ord R, Jawara M, Pinder M, Randall A, et al. Chloroquine/sulphadoxine-pyrimethamine for gambian children with malaria: transmission to mosquitoes of multidrug-resistant Plasmodium falciparum. PLoS Clin Trials (2006) 1:e15. doi: 10.1371/journal.pctr.0010015
45. Saul A. Efficacy model for mosquito stage transmission blocking vaccines for malaria. Parasitology (2008) 135:1497–506. doi: 10.1017/S0031182008000280
46. Carter R, Mendis KN, Miller LH, Molineaux L, Saul A. Malaria transmission-blocking vaccines—how can their development be supported? (2000) 6:241–4. doi: 10.1038/73062
47. Jones S, Grignard L, Nebie I, Chilongola J, Dodoo D, Sauerwein R, Theisen M, Roeffen W, Singh SK, Singh RK: Naturally acquired antibody responses to recombinant Pfs230 and Pfs48/45 transmission blocking vaccine candidates. J Infect (2015) 71:117–27. doi: 10.1016/j.jinf.2015.03.007
48. Doumbo OK, Niaré K, Healy SA, Sagara I, Duffy PE. Malaria transmission-blocking vaccines: present status and future perspectives. In: Towards Malaria Elimination-A Leap Forward. IntechOpen. (2018). doi: 10.5772/intechopen.77241
49. MacDonald NJ, Nguyen V, Shimp R, Reiter K, Herrera R, Burkhardt M, et al. Structural and immunological characterization of recombinant 6-cysteine domains of the Plasmodium falciparum sexual stage protein Pfs230. J Bioll Chem. (2016) 291:19913–22. doi: 10.1074/jbc.M116.732305
50. Talaat KR, Ellis RD, Hurd J, Hentrich A, Gabriel E, Hynes NA, et al. Safety and immunogenicity of Pfs25-EPA/Alhydrogel®, a transmission blocking vaccine against Plasmodium falciparum: an open label study in malaria naïve adults. PLoS ONE (2016) 11:e0163144. doi: 10.1371/journal.pone.0163144
51. Wu Y, Ellis RD, Shaffer D, Fontes E, Malkin EM, Mahanty S, et al. Phase 1 trial of malaria transmission blocking vaccine candidates Pfs25 and Pvs25 formulated with montanide ISA 51. PLoS ONE (2008) 3:e2636. doi: 10.1371/journal.pone.0002636
52. Sagara I, Healy SA, Assadou MH, Gabriel EE, Kone M, Sissoko K, et al. Safety and immunogenicity of Pfs25H-EPA/Alhydrogel, a transmission-blocking vaccine against Plasmodium falciparum: a randomised, double-blind, comparator-controlled, dose-escalation study in healthy Malian adults. Lancet Infect Dis (2018) 18:969–82. doi: 10.1016/S1473-3099(18)30344-X
53. Collins KA, Snaith R, Cottingham MG, Gilbert SC, Hill AVS. Enhancing protective immunity to malaria with a highly immunogenic virus-like particle vaccine. Sci Reports (2017) 7:46621. doi: 10.1038/srep46621
54. Ogwang C, Kimani D, Edwards NJ, Roberts R, Mwacharo J, Bowyer G, et al. Prime-boost vaccination with chimpanzee adenovirus and modified vaccinia Ankara encoding TRAP provides partial protection against Plasmodium falciparum infection in Kenyan adults. Sci Transl Med. (2015) 7:286re285. doi: 10.1126/scitranslmed.aaa2373
55. Bliss CM, Drammeh A, Bowyer G, Sanou GS, Jagne YJ, Ouedraogo O, et al. Viral vector malaria vaccines induce high-level T cell and antibody responses in West African children and infants. Mol Ther. (2017) 25:547–59. doi: 10.1016/j.ymthe.2016.11.003
56. Mensah VA, Roetynck S, Kanteh EK, Bowyer G, Ndaw A, Oko F, et al. Safety and immunogenicity of malaria vectored vaccines given with routine expanded program on immunization vaccines in gambian infants and neonates: a randomized controlled trial. Front Immunol. (2017) 8:1551. doi: 10.3389/fimmu.2017.01551
57. Sirima S, Durier C, Kara L, Houard S, Gansane A, Loulergue P, et al. Safety and immunogenicity of a recombinant Plasmodium falciparum AMA1-DiCo malaria vaccine adjuvanted with GLA-SE or Alhydrogel® in European and African adults: a phase 1a/1b, randomized, double-blind multi-centre trial. Vaccine (2017) 35:6218–27. doi: 10.1016/j.vaccine.2017.09.027
58. Valéa I, Adjei S, Usuf E, Traore O, Ansong D, Tinto H, et al. Immune response to the hepatitis B antigen in the RTS, S/AS01 malaria vaccine, and co-administration with pneumococcal conjugate and rotavirus vaccines in African children: a randomized controlled trial. Hum Vaccin Immunotherapeut. (2018) 14: 1489–1500. doi: 10.1080/21645515.2018.1442996
59. Rts S. Efficacy and safety of RTS, S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet (2015) 386:31–45. doi: 10.1016/S0140-6736(15)60721-8
60. Gosling R, von Seidlein L. The future of the RTS, S/AS01 malaria vaccine: an alternative development plan. PLoS Med. (2016) 13:e1001994. doi: 10.1371/journal.pmed.1001994
61. Chaudhury S, Regules JA, Darko CA, Dutta S, Wallqvist A, Waters NC, et al. Delayed fractional dose regimen of the RTS, S/AS01 malaria vaccine candidate enhances an IgG4 response that inhibits serum opsonophagocytosis. Sci Reports (2017) 7:7998. doi: 10.1038/s41598-017-08526-5
62. Walk J, Reuling IJ, Behet MC, Meerstein-Kessel L, Graumans W, van Gemert G-J, et al. Modest heterologous protection after Plasmodium falciparum sporozoite immunization: a double-blind randomized controlled clinical trial. BMC Med. (2017) 15:168. doi: 10.1186/s12916-017-0923-4
63. Sissoko MS, Healy SA, Katile A, Omaswa F, Zaidi I, Gabriel EE, et al. Safety and efficacy of PfSPZ Vaccine against Plasmodium falciparum via direct venous inoculation in healthy malaria-exposed adults in Mali: a randomised, double-blind phase 1 trial. Lancet Infect Dis. (2017) 17:498–509. doi: 10.1016/S1473-3099(17)30104-4
64. Olotu A, Urbano V, Hamad A, Eka M, Chemba M, Nyakarungu E, et al. Advancing global health through development and clinical trials partnerships: a randomized, placebo-controlled, double-blind assessment of safety, tolerability, and immunogenicity of PfSPZ vaccine for malaria in healthy equatoguinean men. Am J Tropic Med Hyg. (2018) 98:308–18. doi: 10.4269/ajtmh.17-0449
65. Seib KL, Zhao X, Rappuoli R. Developing vaccines in the era of genomics: a decade of reverse vaccinology. Clin. Microbiol. Infect (2012) 18:109–116. doi: 10.1111/j.1469-0691.2012.03939.x
66. Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. (2000) 3:445–50. doi: 10.1016/S1369-5274(00)00119-3
67. Watson PS, Turner DP. Clinical experience with the meningococcal B vaccine, Bexsero®: Prospects for reducing the burden of meningococcal serogroup B disease. Vaccine (2016) 34:875–80. doi: 10.1016/j.vaccine.2015.11.057
68. Vesikari T, Esposito S, Prymula R, Ypma E, Kohl I, Toneatto D, et al. Immunogenicity and safety of an investigational multicomponent, recombinant, meningococcal serogroup B vaccine (4CMenB) administered concomitantly with routine infant and child vaccinations: results of two randomised trials. Lancet (2013) 381:825–35. doi: 10.1016/S0140-6736(12)61961-8
69. Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, et al. Identification of a Universal Group B Streptococcus Vaccine by Multiple Genome Screen. Science (2005) 309:148–50. doi: 10.1126/science.1109869
70. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature (2002) 419:498. doi: 10.1038/nature01097
71. Sundararaman SA, Plenderleith LJ, Liu W, Loy DE, Learn GH, Li Y, et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun. (2016) 7:11078. doi: 10.1038/ncomms11078
72. Otto TD, Rayner JC, Böhme U, Pain A, Spottiswoode N, Sanders M, et al. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nature Commun. (2014) 5:4754. doi: 10.1038/ncomms5754
73. Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science (2005) 307:82–6. doi: 10.1126/science.1103717
74. van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JA, Dodemont HJ, et al. A central role for P48/45 in malaria parasite male gamete fertility. Cell (2001) 104:153–64. doi: 10.1016/S0092-8674(01)00199-4
75. Blagborough AM, Sinden RE. Plasmodium berghei HAP2 induces strong malaria transmission-blocking immunity in vivo and in vitro. Vaccine (2009) 27:5187–94. doi: 10.1016/j.vaccine.2009.06.069
76. Pritsch M, Ben-Khaled N, Chaloupka M, Kobold S, Berens-Riha N, Peter A, et al. Comparison of intranasal outer membrane vesicles with cholera toxin and injected MF59C. 1 as adjuvants for malaria transmission blocking antigens AnAPN1 and Pfs48/45. J Immunol Res. (2016) 2016:3576028. doi: 10.1155/2016/3576028
77. Merino KM, Bansal GP, Kumar N. Reduced immunogenicity of Plasmodium falciparum gamete surface antigen (Pfs48/45) in mice after disruption of disulphide bonds–evaluating effect of interferon-γ-inducible lysosomal thiol reductase. Immunology (2016) 148:433–47. doi: 10.1111/imm.12621
78. Miura K, Takashima E, Deng B, Tullo G, Diouf A, Moretz SE, et al. Functional comparison of Plasmodium falciparum transmission-blocking vaccine candidates by the standard membrane-feeding assay. Infect Immun. (2013) 81:4377–82. doi: 10.1128/IAI.01056-13
79. Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity (2010) 33:530–41. doi: 10.1016/j.immuni.2010.09.017
80. Delany I, Rappuoli R, De Gregorio E. Vaccines for the 21st century. EMBO Molr Med. (2014) 6:708–20. doi: 10.1002/emmm.201403876
81. Thomas S, Luxon BA. Vaccines based on structure-based design provide protection against infectious diseases. Expert Rev Vaccin. (2013) 12:1301–11. doi: 10.1586/14760584.2013.840092
82. Nuccitelli A, Cozzi R, Gourlay LJ, Donnarumma D, Necchi F, Norais N, et al. Structure-based approach to rationally design a chimeric protein for an effective vaccine against Group B Streptococcus infections. Proc Natl Acad Sci. USA. (2011) 108:10278–83. doi: 10.1073/pnas.1106590108
83. Scarselli M, Aricò B, Brunelli B, Savino S, Di Marcello F, Palumbo E, et al. Rational design of a meningococcal antigen inducing broad protective immunity. Sci Transl Med. (2011) 3:91ra62. doi: 10.1126/scitranslmed.3002234
84. Villard V, Agak GW, Frank G, Jafarshad A, Servis C, Nébié I, et al. Rapid identification of malaria vaccine candidates based on α-helical coiled coil protein motif. PLoS ONE (2007) 2:e645. doi: 10.1371/journal.pone.0000645
85. Olugbile S, Kulangara C, Bang G, Bertholet S, Suzarte E, Villard V, et al. Vaccine potentials of an intrinsically unstructured fragment derived from the blood stage-associated Plasmodium falciparum protein PFF0165c. Infect immun. (2009) 77:5701–9. doi: 10.1128/IAI.00652-09
86. Steiner-Monard V, Kamaka K, Karoui O, Roethlisberger S, Audran R, Daubenberger C, et al. The candidate blood stage malaria vaccine P27A induces a robust humoral response in a fast track to the field phase I trial in exposed and non exposed volunteers. Clin Infect Dis. (2018) ciy514. doi: 10.1093/cid/ciy514
87. Gangnard S, Lewit-Bentley A, Dechavanne S, Srivastava A, Amirat F, Bentley GA, et al. Structure of the DBL3X-DBL4ε region of the VAR2CSA placental malaria vaccine candidate: insight into DBL domain interactions. Sci Reports (2015) 5:14868. doi: 10.1038/srep14868
88. Favuzza P, Guffart E, Tamborrini M, Scherer B, Dreyer AM, Rufer AC, et al. Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody. Elife (2017) 6:e20383. doi: 10.7554/eLife.20383
89. Wan J, Liu W, Xu Q, Ren Y, Flower DR, Li T. SVRMHC prediction server for MHC-binding peptides. BMC Bioinformat. (2006) 7:463. doi: 10.1186/1471-2105-7-463
90. Liu W, Meng X, Xu Q, Flower DR, Li T. Quantitative prediction of mouse class I MHC peptide binding affinity using support vector machine regression (SVR) models. BMC Bioinformat. (2006) 7:182. doi: 10.1186/1471-2105-7-182
91. Degoot AM, Chirove F, Ndifon W. Trans-Allelic Model for Prediction of Peptide:MHC-II Interactions. Front Immunol. (2018) 9:1410. doi: 10.3389/fimmu.2018.01410
92. Doolan DL. Plasmodium immunomics. Int J Parasitol. (2011) 41:3–20. doi: 10.1016/j.ijpara.2010.08.002
93. Khan N, Kumar R, Chauhan S, Farooq U. An immunoinformatics approach to promiscuous peptide design for the Plasmodium falciparum erythrocyte membrane protein-1. Mol BioSyst. (2017) 13:2160–7. doi: 10.1039/C7MB00332C
94. Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology (2018) 154:394–406. doi: 10.1111/imm.12889
95. Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature Biotechnol. (2012) 30:423–33. doi: 10.1038/nbt.2197
96. Stanisic DI, Good MF. Examining cellular immune responses to inform development of a blood-stage malaria vaccine. Parasitology (2016) 143:208–23. doi: 10.1017/S0031182015001092
97. Rodriguez-Barraquer I, Arinaitwe E, Jagannathan P, Kamya MR, Rosenthal PJ, Rek J, et al. Quantification of anti-parasite and anti-disease immunity to malaria as a function of age and exposure. eLife (2018) 7:e35832. doi: 10.7554/eLife.35832
98. Rodriguez-Barraquer I, Arinaitwe E, Jagannathan P, Boyle MJ, Tappero J, Muhindo M, et al. Quantifying heterogeneous malaria exposure and clinical protection in a cohort of Ugandan children. J Infect Dis. (2016) 214:1072–80. doi: 10.1093/infdis/jiw301
99. Baird JK, Basri H, Bangs MJ, Andersen EM, Jones TR, Masbar S, et al. Age-specific prevalence of Plasmodium falciparum among six populations with limited histories of exposure to endemic malaria. Am J Tropic Med Hyg. (1993) 49:707–19. doi: 10.4269/ajtmh.1993.49.707
100. Portugal S, Tran TM, Ongoiba A, Bathily A, Li S, Doumbo S, et al. Treatment of chronic asymptomatic Plasmodium falciparum infection does not increase the risk of clinical malaria upon reinfection. Clin Infect Dis. (2016) 64:645–53. doi: 10.1093/cid/ciw849
101. Portugal S, Moebius J, Skinner J, Doumbo S, Doumtabe D, Kone Y, et al. Exposure-dependent control of malaria-induced inflammation in children. PLoS Pathog. (2014) 10:e1004079. doi: 10.1371/journal.ppat.1004079
102. Boyle MJ, Jagannathan P, Farrington LA, Eccles-James I, Wamala S, McIntyre TI, et al. Decline of FoxP3+ Regulatory CD4 T cells in peripheral blood of children heavily exposed to malaria. PLoS Pathog. (2015) 11:e1005041. doi: 10.1371/journal.ppat.1005041
103. Guilbride DL, Gawlinski P, Guilbride PD. Why functional pre-erythrocytic and bloodstage malaria vaccines fail: a meta-analysis of fully protective immunizations and novel immunological model. PLoS ONE (2010) 5:e10685. doi: 10.1371/journal.pone.0010685
104. Loughland JR, Woodberry T, Boyle MJ, Tipping PE, Piera KA, Amante FH, et al. Plasmodium falciparum activates CD16+ dendritic cells to produce TNF and IL-10 in subpatent malaria. J Infect Dis. (2018) jiy555. doi: 10.1093/infdis/jiy555. [Epub ahead of print].
105. Kurtovic L, Behet MC, Feng G, Reiling L, Chelimo K, Dent AE, et al. Human antibodies activate complement against Plasmodium falciparum sporozoites, and are associated with protection against malaria in children. BMC Med. (2018) 16:61. doi: 10.1186/s12916-018-1054-2
106. Teo A, Feng G, Brown GV, Beeson JG, Rogerson SJ. Functional antibodies and protection against blood-stage malaria. Trends Parasitol. (2016) 32:887–898. doi: 10.1016/j.pt.2016.07.003
107. Hart GT, Tran T, Theorell J, Arora G, Crompton P, Bryceson Y, et al. A new malaria killer: Fc receptor gamma chain and PLZF identify NK cell subsets that correlate with reduced Plasmodium falciparum parasitemia and increased antibody dependent cellular cytotoxicity against opsonized infected RBCs. Am Assoc Immnol. (2018).
108. Jäschke A, Coulibaly B, Remarque EJ, Bujard H, Epp C. Merozoite surface protein-1 from P. falciparum is a major target of opsonizing antibodies in individuals with acquired immunity against malaria. Clin Vaccine Immunol. (2017).
109. Laurens MB, Kouriba B, Bergmann-Leitner E, Angov E, Coulibaly D, Diarra I, et al. Strain-specific Plasmodium falciparum growth inhibition among Malian children immunized with a blood-stage malaria vaccine. PLoS One (2017) 12:e0173294. doi: 10.1371/journal.pone.0173294
110. Chiu CYH, Hodder AN, Lin CS, Hill DL, Li Wai Suen CSN, Schofield L, et al. Antibodies to the Plasmodium falciparum Proteins MSPDBL1 and MSPDBL2 Opsonize Merozoites, Inhibit Parasite Growth, and Predict Protection From Clinical Malaria. J Infect Dis. (2015) 212:406–15. doi: 10.1093/infdis/jiv057
111. Persson KEM, Fowkes FJI, McCallum FJ, Gicheru N, Reiling L, Richards JS, et al. Erythrocyte-binding antigens of Plasmodium falciparum are targets of human inhibitory antibodies and function to evade naturally acquired immunity. J Immunol. (2013) 191:785–94. doi: 10.4049/jimmunol.1300444
112. Osier FH, Feng G, Boyle MJ, Langer C, Zhou J, Richards JS, et al. Opsonic phagocytosis of Plasmodium falciparum merozoites: mechanism in human immunity and a correlate of protection against malaria. BMC Med. (2014) 12:108. doi: 10.1186/1741-7015-12-108
113. Hill DL, Schofield L, Wilson DW. IgG opsonization of merozoites: multiple immune mechanisms for malaria vaccine development. Int J Parasitol. (2017) 47:585–95. doi: 10.1016/j.ijpara.2017.05.004
114. Boyle Michelle J, Reiling L, Feng G, Langer C, Osier Faith H, Aspeling-Jones H, et al. Human antibodies fix complement to inhibit Plasmodium falciparum invasion of erythrocytes and are associated with protection against malaria. Immunity (2015) 42:580–90. doi: 10.1016/j.immuni.2015.02.012
115. Olsen RW, Ecklu-Mensah G, Bengtsson A, Ofori MF, Lusingu JPA, Castberg FC, et al. Natural and Vaccine-Induced Acquisition of Cross-Reactive IgG-Inhibiting ICAM-1-Specific Binding of a Plasmodium falciparum PfEMP1 Subtype Associated Specifically with Cerebral Malaria. Infect Immun. (2018) 86:e00622-17. doi: 10.1128/IAI.00622-17
116. Illingworth J, Butler NS, Roetynck S, Mwacharo J, Pierce SK, Bejon P, et al. Chronic exposure to Plasmodium falciparum is associated with phenotypic evidence of B and T cell exhaustion. J Immunol (2013) 190:1038–47. doi: 10.4049/jimmunol.1202438
117. Portugal S, Tipton CM, Sohn H, Kone Y, Wang J, Li S, et al. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. Elife (2015) 4:e07218. doi: 10.7554/eLife.07218
118. Frimpong A, Kusi KA, Tornyigah B, Ofori MF, Ndifon W. Characterization of T cell activation and regulation in children with asymptomatic Plasmodium falciparum infection. Mal J. (2018) 17:263. doi: 10.1186/s12936-018-2410-6
119. Walther M, Jeffries D, Finney OC, Njie M, Ebonyi A, Deininger S, et al. Distinct roles for FOXP3+ and FOXP3– CD4+ T cells in regulating cellular immunity to uncomplicated and severe Plasmodium falciparum malaria. PLoS Pathog. (2009) 5:e1000364. doi: 10.1371/journal.ppat.1000364
120. Kurup SP, Obeng-Adjei N, Anthony SM, Traore B, Doumbo OK, Butler NS, et al. Regulatory T cells impede acute and long-term immunity to blood-stage malaria through CTLA-4. Nature Med. (2017) 23:1220. doi: 10.1038/nm.4395
121. Abel A, Steeg C, Aminkiah F, Addai-Mensah O, Addo M, Gagliani N, et al. Differential expression pattern of co-inhibitory molecules on CD4+ T cells in uncomplicated versus complicated malaria. Sci Reports (2018) 8:4789. doi: 10.1038/s41598-018-22659-1
122. Valletta JJ, Recker M. Identification of immune signatures predictive of clinical protection from malaria. PLoS Comput Biol. (2017) 13:e1005812. doi: 10.1371/journal.pcbi.1005812
123. Mordmüller B, Surat G, Lagler H, Chakravarty S, Ishizuka AS, Lalremruata A, et al. Sterile protection against human malaria by chemoattenuated PfSPZ vaccine. Nature (2017) 542:445. doi: 10.1038/nature21060
124. Lyke KE, Ishizuka AS, Berry AA, Chakravarty S, DeZure A, Enama ME, et al. Attenuated PfSPZ Vaccine induces strain-transcending T cells and durable protection against heterologous controlled human malaria infection. Proc Natl Acad Sci USA. (2017) 114:2711–16. doi: 10.1073/pnas.1615324114
125. Ishizuka AS, Lyke KE, DeZure A, Berry AA, Richie TL, Mendoza FH, et al. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat Med. (2016) 22:614. doi: 10.1038/nm.4110
126. Seder RA, Chang L-J, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science (2013) 341:1359–65. doi: 10.1126/science.1241800
127. Epstein JE, Tewari K, Lyke K, Sim B, Billingsley P, Laurens M, et al. Live attenuated malaria vaccine designed to protect through hepatic CD8+ T cell immunity. Science 2011:1211548. doi: 10.1126/science.1211548
128. Saito F, Hirayasu K, Satoh T, Wang CW, Lusingu J, Arimori T, et al. Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors. Nature (2017) 552:101. doi: 10.1038/nature24994
129. Scally SW, Murugan R, Bosch A, Triller G, Costa G, Mordmüller B, et al. Rare PfCSP C-terminal antibodies induced by live sporozoite vaccination are ineffective against malaria infection. J Exp Med. (2018) 215:63–75. doi: 10.1084/jem.20170869
130. Pulendran B. Systems vaccinology: probing humanity's diverse immune systems with vaccines. Proc Natl Acad Sci USA. (2014) 111:12300–6. doi: 10.1073/pnas.1400476111
131. Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity (2010) 33:516–29. doi: 10.1016/j.immuni.2010.10.006
132. Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics (2008) 8:4680–94. doi: 10.1002/pmic.200800194
133. Osier FH, Mackinnon MJ, Crosnier C, Fegan G, Kamuyu G, Wanaguru M, et al. New antigens for a multicomponent blood-stage malaria vaccine. Sci Transl Med. (2014) 6:247ra102. doi: 10.1126/scitranslmed.3008705
134. Skinner J, Huang C-Y, Waisberg M, Felgner PL, Doumbo OK, Ongoiba A, et al. Plasmodium falciparum gametocyte-specific antibody profiling reveals boosting through natural infection and identifies potential markers of gametocyte exposure. Infect Immun. (2015) 83:4229–36. doi: 10.1128/IAI.00644-15
135. Arevalillo JM, Sztein MB, Kotloff KL, Levine MM, Simon JK. Identification of immune correlates of protection in Shigella infection by application of machine learning. J Biomed Informat. (2017) 74:1–9. doi: 10.1016/j.jbi.2017.08.005
136. Tan J, Pieper K, Piccoli L, Abdi A, Foglierini M, Geiger R, et al. A LAIR1 insertion generates broadly reactive antibodies against malaria variant antigens. Nature (2016) 529:105. doi: 10.1038/nature16450
137. Tan J, Sack BK, Oyen D, Zenklusen I, Piccoli L, Barbieri S, et al. A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein. Nature Med. (2018) 24:401. doi: 10.1038/nm.4513
138. Kisalu NK, Idris AH, Weidle C, Flores-Garcia Y, Flynn BJ, Sack BK, et al. A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite. Nature Med. (2018) 24:408. doi: 10.1038/nm.4512
139. Oyen D, Torres JL, Wille-Reece U, Ockenhouse CF, Emerling D, Glanville J, et al. Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein. Proc Natl Acad Sci USA. (2017) 114:e10438–445. doi: 10.1073/pnas.1715812114
140. Lau LS, Fernandez-Ruiz D, Mollard V, Sturm A, Neller MA, Cozijnsen A, et al. CD8+ T Cells from a Novel T cell receptor transgenic mouse induce liver-stage immunity that can be boosted by blood-stage infection in rodent malaria. PLoS Pathog. (2014) 10:e1004135. doi: 10.1371/journal.ppat.1004135
141. Fernandez-Ruiz D, Lau LS, Ghazanfari N, Jones CM, Ng WY, Davey GM, et al. Development of a Novel CD4+ TCR transgenic line that reveals a dominant role for CD8+ dendritic cells and CD40 signaling in the generation of helper and CTL responses to blood-stage malaria. J Immunol. (2017) 199:4165–79. doi: 10.4049/jimmunol.1700186
142. Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science (2015) 349:1301–6. doi: 10.1126/science.aac7263
143. Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, et al. Prefusion F–specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Science Transl Med. (2015) 7:309ra162. doi: 10.1126/scitranslmed.aac4241
144. Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (2010) 329:856–61. doi: 10.1126/science.1187659
145. Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Science Transl Med. (2015) 7:319ra206. doi: 10.1126/scitranslmed.aad5752
146. Ledgerwood J, Coates E, Yamshchikov G, Saunders J, Holman L, Enama M, et al. Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin Exper Immunol. (2015) 182:289–301. doi: 10.1111/cei.12692
147. Huang Y, Karuna S, Carpp LN, Reeves D, Pegu A, Seaton K, et al. Modeling cumulative overall prevention efficacy for the VRC01 phase 2b efficacy trials. Hum Vaccin Immunother. (2018) 14:2116–27. doi: 10.1080/21645515.2018.1462640
148. Wright KE, Hjerrild KA, Bartlett J, Douglas AD, Jin J, Brown RE, et al. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature (2014) 515:427. doi: 10.1038/nature13715
149. Payne RO, Silk SE, Elias SC, Milne KH, Rawlinson TA, Llewellyn D, et al. Human vaccination against Plasmodium vivax Duffy-binding protein induces strain-transcending antibodies. JCI Insight (2017) 2:e93683. doi: 10.1172/jci.insight.93683
Keywords: Plasmodium falciparum, malaria, vaccine, immunoinformatics, structure-based, lymphocyte repertoire sequencing
Citation: Frimpong A, Kusi KA, Ofori MF and Ndifon W (2018) Novel Strategies for Malaria Vaccine Design. Front. Immunol. 9:2769. doi: 10.3389/fimmu.2018.02769
Received: 27 August 2018; Accepted: 12 November 2018;
Published: 29 November 2018.
Edited by:
Kevin Couper, University of Manchester, United KingdomReviewed by:
Katie Louise Flanagan, RMIT University, AustraliaCopyright © 2018 Frimpong, Kusi, Ofori and Ndifon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Augustina Frimpong, dGluYWZyaW1wQGdtYWlsLmNvbQ==
Wilfred Ndifon, d25kaWZvbkBhaW1zLmFjLnph
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.