- 1Division of Gastroenterology, Hepatology and Nutrition, Boston Children's Hospital, Boston, MA, United States
- 2Center for Motility and Functional Gastrointestinal Disorders Boston Children's Hospital, Boston, MA, United States
- 3Aerodigestive Center, Boston Children's Hospital, Boston, MA, United States
- 4Department of Pathology, Boston Children's Hospital, Boston, MA, United States
Eosinophilic esophagitis (EoE), a Th2-type allergic immune disorder characterized by an eosinophil-rich esophageal immune infiltrate, is often associated with food impaction (FI) in pediatric patients but the molecular mechanisms underlying the development of this complication are not well understood. We aim to identify molecular pathways involved in the development of FI. Due to large variations in disease presentation, our analysis was further geared to find markers capable of distinguishing EoE patients that are prone to develop food impactions and thus expand an established medical algorithm for EoE by developing a secondary analysis that allows for the identification of patients with food impactions as a distinct patient population. To this end, mRNA patterns from esophageal biopsies of pediatric EoE patients presenting with and without food impactions were compared and machine learning techniques were employed to establish a diagnostic probability score to identify patients with food impactions (EoE+FI). Our analysis showed that EoE patients with food impaction were indistinguishable from other EoE patients based on their tissue eosinophil count, serum IgE levels, or the mRNA transcriptome-based p(EoE). Irrespectively, an additional analysis loop of the medical algorithm was able to separate EoE+FI patients and a composite FI-score was established that identified such patients with a sensitivity of 93% and a specificity of 100%. The esophageal mRNA pattern of EoE+FI patients was typified by lower expression levels of mast cell markers and Th2 associated transcripts, such as FCERIB, CPA3, CCL2, IL4, and IL5. Furthermore, lower expression levels of regulators of esophageal motility (NOS2 and HIF1A) were detected in EoE+FI. The EoE+FI -specific mRNA pattern indicates that impaired motility may be one underlying factor for the development of food impactions in pediatric patients. The availability of improved diagnostic tools such as a medical algorithm for EoE subpopulations will have a direct impact on clinical practice because such strategies can identify molecular inflammatory characteristics of individual EoE patients, which, in turn, will facilitate the development of individualized therapeutic approaches that target the relevant pathways affected in each patient.
Introduction
Eosinophilic esophagitis (EoE) is an allergic disorder that is characterized by an eosinophil-rich immune infiltrate of the esophagus. Recent epidemiological reports estimate the incidence of EoE at 1/10,000 new cases per year with the current prevalence of EoE as 25.9–56.7/100,000 in the United States and suggest a rapid increase in incidence in developed countries (1–4).
EoE remains a challenge to diagnose and manage due to the high clinical variations of the disease (5–7). The diagnosis is made clinicopathologically by demonstrating at least 15 eosinophils per high-power field in at least one esophageal biopsy, along with the presence of typical EoE symptoms (8). Evaluation of 2–4 biopsies from both proximal and distal esophagus is recommended because of the patchy nature of the immune infiltrate in EoE (9). Symptomology is age and gender dependent with younger children commonly presenting with nonspecific symptoms such as failure to thrive, feeding difficulties, and choking on solid foods. Older children on the other hand, more capable of communicating their symptoms, frequently complain of abdominal pain, dysphagia, and vomiting (10–13). In contrast to children, adolescents and adults typically present with symptoms more specific to esophageal dysfunction and ongoing fibrosis, such as dysphagia, food impaction, and esophageal strictures (10, 14). In addition to the wide array of presenting symptoms the biological mechanisms underlying disease onset and progression are varied, which has led to the definition of many EoE subpopulations (15). This increasing prevalence and the heterogeneous nature of the disease underlie the pressing need to improve disease management, treatment, and diagnosis.
Currently, the use of machine learning diagnostic algorithms using mRNA pattern stamps is at the forefront of emerging diagnostic strategies for EoE. These approaches rely on an automatized evaluation of the transcriptional profile of the inflamed esophagus to calculate probability scores (16, 17). The use of a machine learning approach allows for an unbiased assessment of patient biopsies independent of human error and is designed to be self-improving with the addition of data from new patients as they become available.
The automatization of data analysis in EoE also opens the possibility of an individualized approach to EoE diagnostics and therapy. This strategy will allow clinicians to evaluate the phenotype of EoE presentation of each patient and potentially assign patients into subpopulations of EoE. Currently, multiple subpopulations of EoE patients are defined by traits such as clinical symptoms, responses to therapies (PPI-REE), and/or underlying gene expression patterns (LTC4S-EoE, IL23-EoE, iNKT-EoE, IGHEhi-EoE) (15–20). In a recently published medical algorithm, a diagnostic score for patients with increased local IgE production has been developed as a secondary diagnosis loop to identify patients that potentially suffer from local esophageal allergies (17). Further expansion of this algorithm to define additional subpopulations of EoE has the potential to improve the understanding of the disease, and to assist in the prediction of symptom onset, response to therapy, and stratification of the underlying EoE etiology.
A significant portion of patients with EoE suffer from food impactions, making EoE the leading cause of food impaction and dysphagia in the pediatric population (21). Nevertheless, the pathologic mechanisms that lead to food impactions in EoE are not well understood (22, 23). Among patients with EoE, food impaction can result from both obstructive anatomical features of the esophagus, such as esophageal stenosis, narrow-caliber esophagus, and strictures, as well as motility dysregulations such as achalasia, and diffuse esophageal spasms (23–27). The majority of food impactions in EoE patients are thought to be the result of a natural progression from an inflammatory phenotype of EoE to a fibrotic one (28–30). However, particularly in the pediatric population, there are EoE patients who present with food impactions without showing any endoscopic features of fibrosis (27).
Hence, the goal of the current study was to examine a large dataset of mRNA pattern stamps of EoE patients to define a transcriptional signature that could identify EoE patients with food impactions and yield insights about possible pathological causes for this condition in the context of EoE. We further aimed to modify our medical algorithm that calculates a composite probability score for EoE (pEoE) to establish an additional diagnostic score for the identification of EoE patients with food impaction.
Materials and Methods
Study Population
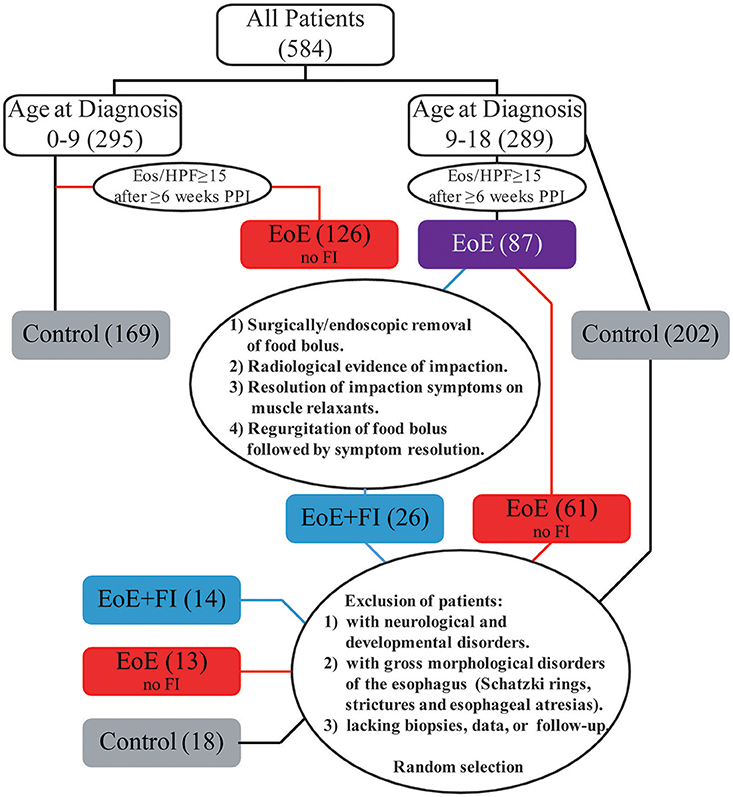
The patients included in this study were enrolled in an observational longitudinal cohort study, performed at Boston Children's Hospital that concentrates on the understanding of the pathophysiology and diagnosis of EoE (17–19, 31, 32). Children between 1 and 18 years of age, who were scheduled for an elective upper gastrointestinal endoscopy at the Division of Gastroenterology at Boston Children's Hospital, due to a clinical suspicion for EoE (such as presenting with dysphagia, regurgitation, feeding intolerance or failure to thrive), were invited to participate. Following written informed consent by the patients and/or their legal guardians, caregivers filled out a questionnaire regarding the subject's medical history, current and past symptomatology, allergic comorbidity, and dietary habits. In addition to clinical biopsies, two study biopsies, one from the proximal and one from the distal esophagus, were obtained during endoscopy from each patient. Additional information on each subject's medical history was obtained by retrospective chart review. All patients were treated independently of this study. Patients were approached to provide follow-up information and additional esophageal biopsies at follow-up hospital visits and endoscopies. Approval for this study was obtained by the institutional Investigational Review Board of Boston Children's Hospital (Harvard Medical School, Boston MA, approval number: 07-11-0460).
Clinicopathological Diagnosis by Reference Standards
Patients were classified as having EoE according to existent clinicopathological diagnostic guidelines at the time of sample collection, using the gold standard criteria: esophagitis that is histologically characterized by ≥15 eosinophil per high power field in at least one biopsy obtained after ≥4 weeks of treatment with a PPI and exclusion of other causes of esophageal eosinophilia. Patients that have normal esophageal histology with no clinical evidence of an underlying esophageal disease were classified as controls. Gastroesophageal reflux disease was diagnosed when there was evidence of esophagitis (< 15 eosinophils per hpf after >8 weeks PPI treatment) and symptoms associated with reflux. These patients were excluded from analysis along with those with an unknown or ambiguous diagnosis, received steroid therapy or other immunomodulatory medications at the time of inclusion and/or showed evidence of a narrow esophagus or esophageal strictures.
Food impaction in EoE patients (EoE+FI) was defined as following: (1) Experiencing an episode of food impaction that requires removal of the food bolus via endoscopy or surgery, (2) experiencing an episode in which the patient presents to an emergency department and has radiological evidence of impaction and/or relief of symptoms following administration of esophageal relaxants, (3) experiencing an episode of self-reported food impaction that was resolved by bolus regurgitation. EoE patients that do not meet any of these criteria were designated as EoE without food impaction (EoE no FI). Patients who have been diagnosed with esophageal motility disorders such as achalasia, or who have anatomical abnormalities in the esophagus such as esophageal atresias, or tracheoesophageal fistulas were excluded from the study.
Biopsy Processing and Digital mRNA Profiling
Study biopsies from the esophagus were collected in RNAlater (Qiagen, Valencia, CA) and stored at −80°C. For mRNA profiling, the biopsies were thawed and homogenized in RTL buffer (Qiagen). Further processing of the samples was done with the nCounter® Prep Station and Digital Analyzer, following the manufacturer's instructions (nCounter® system; NanoString Technologies, Seattle, WA; www.nanostring.com) using a previously published panel as established based on the published EoE transcriptome (33).
Statistical Analysis
Transcript data was normalized by performing background subtraction and normalizing to the geometric mean of the internal positive controls and to the geometric mean of 5 housekeeping genes. The normalized data was analyzed using Kruskal-Wallis test with Dunn's multiple comparisons test. Disease probability scores were calculated using an established algorithm (17). Statistical analysis of clinical characteristics of patients was performed with Fisher's exact test. All statistical tests were performed using python 3.5.2 with the following modules: scipy 0.18.1, numpy 1.11.1, pandas 0.18.1, scikit-learn 0.18, and matplotlib 1.5.3, or IBM SPSS Statistics for Windows, Version 23.0 (Armonk, NY), or GraphPad Prism 7.00 for Windows (La Jolla, CA).
Results
Basic Characteristics of EoE Patients With Food Impactions
The incidence of FI in the EoE population was 12.1% (26/215) referred to as EoE+FI for the rest of the manuscript). While the median age of onset in the pediatric EoE cohort was 10.38 (1.23–18.90), the median age of EoE+FI patients was 14.79 (10.70–16.99), implying that FI is found in a subpopulation of patients with onset and/or diagnosis later in childhood. When restricting the analysis to EoE patients that were diagnosed after the age of nine, the incidence of FI increased to 29.2% (26/89). To control for age as a confounding factor for the mRNA pattern comparison, we randomly selected 13 age-matched EoE patients who did not present with a food impaction event prior or at least 2 years after diagnosis and 18 age-matched control patients with no esophageal eosinophilia or esophageal inflammation (Figure 1). This patient population was used for comparative mRNA pattern analysis and machine learning strategies presented in the rest of the manuscript.
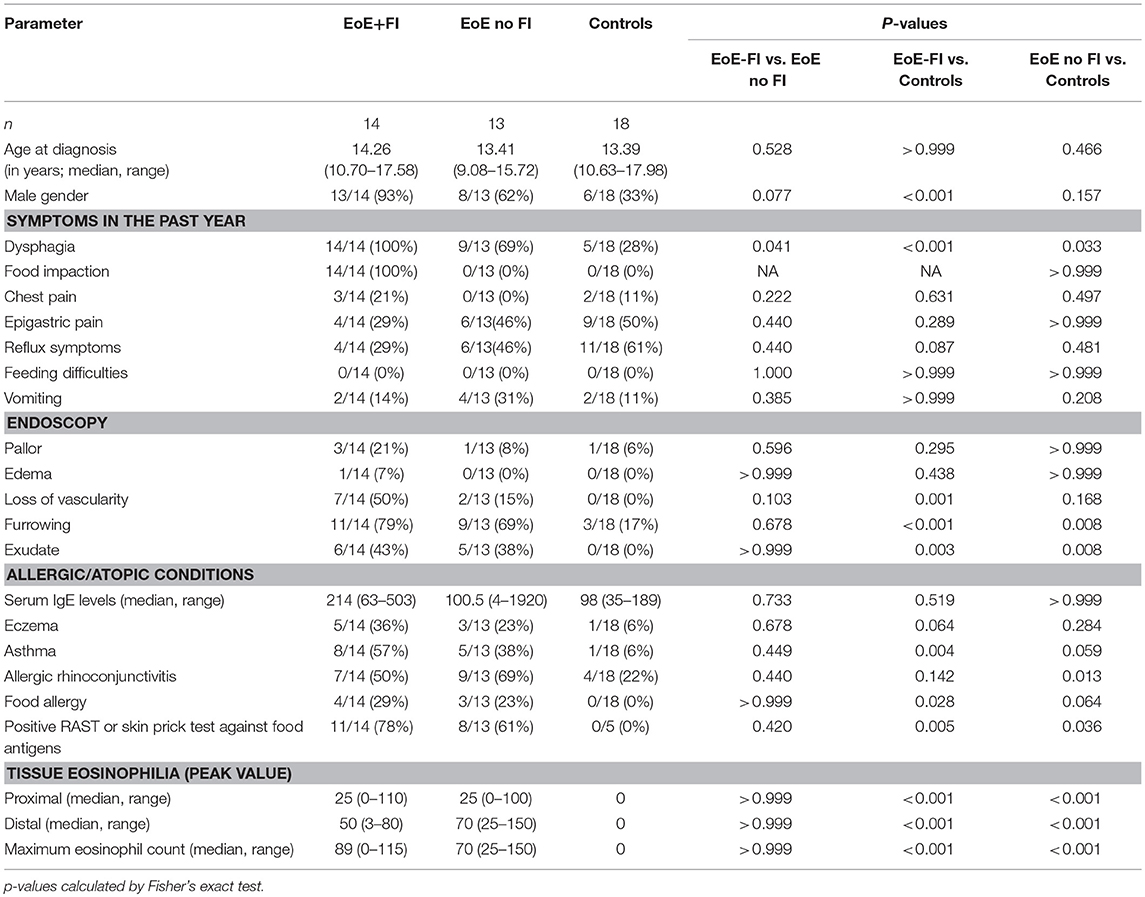
EoE is predominantly found in males (34). In our entire cohort, 67.0% (144/215) of patients were male. In a sub-analysis of the age–matched study population an even higher male predominance of cases presenting with FI was observed with 93% (13/14) of EoE+FI being male compared to 62% (8/13) in the age-matched EoE group without FI.
The incidences of common gastrointestinal symptoms such as epigastric pain, reflux symptoms, and vomiting were comparable between EoE patients who did or did not present with FI. In contrast, dysphagia was significantly more common in the EoE+FI group. The gross endoscopic findings, such as esophageal pallor, edemas, furrowing, loss of vascularity, and the presence of exudate, were comparable between EoE patients with and without FI (Table 1).
EoE Patients With Food Impactions Cannot be Differentiated From Other EoE Patients Based on Standard Diagnostic Measures
Histological and transcriptional measures of eosinophil infiltration were analyzed to determine whether common markers of disease severity could be used to distinguish EoE from EoE+FI patients. No differences in the degree of eosinophilia in either proximal or distal biopsies (Figures 2A–C), or in the maximum count throughout the esophagus was noted (Figure 2D) between the two patient groups. Additionally, the esophageal expression of the eosinotrophic chemokine CCL26 was not significantly different between EoE no FI and EoE+FI patients (Figure 2E). A recently published diagnostic algorithm uses machine learning approaches to calculate a probability score for EoE diagnosis (p(EoE)) based on the transcriptional profile of the esophageal tissue in EoE patients (17). Based on this algorithm, an EoE diagnosis could be made when p(EoE) values are >25. When calculating the three probability values for EoE, GERD and control patients, EoE no FI, and EoE+FI patients cluster together and separate from control patients (Figure 2F). The p(EoE) of patients with and without FI was not significantly different (Figure 2G). This data suggests that the presentation of food impactions in EoE does not result from a quantitative difference in tissue eosinophilia, which indicates that an additional analysis loop is needed to expand the medical algorithm to better define the EoE+FI subpopulation.
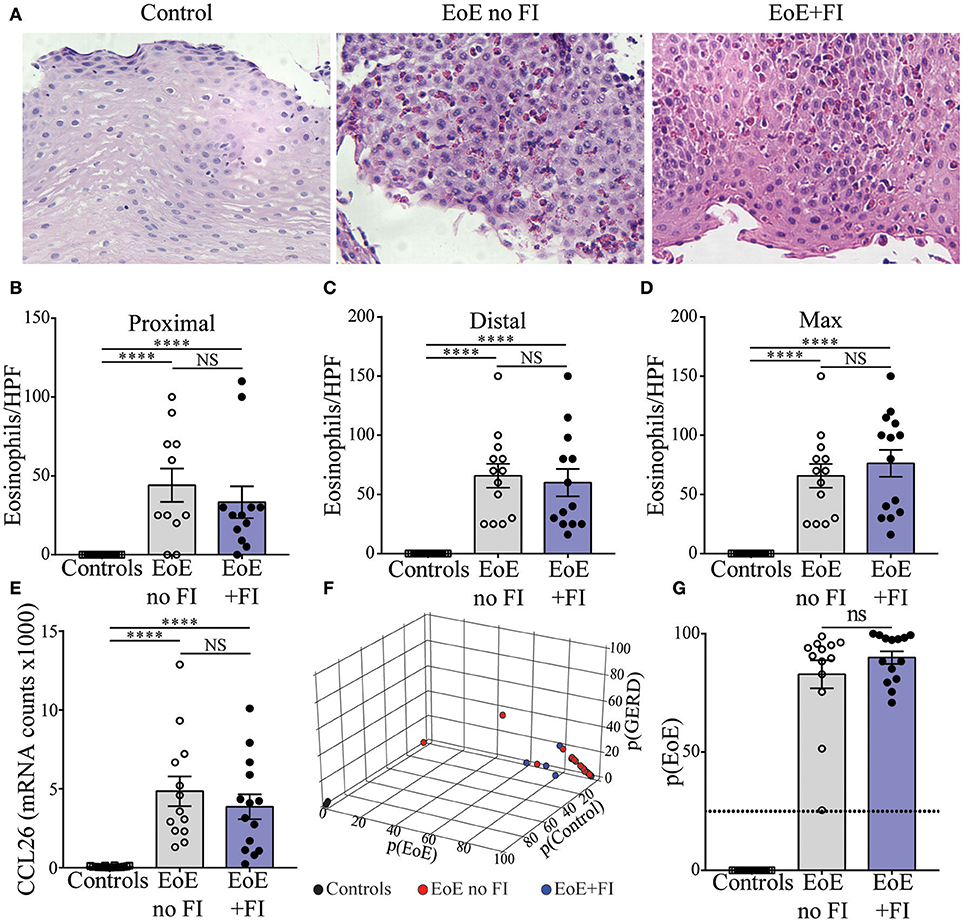
Figure 2. Measures of EoE severity.(A) Representative hematoxylin and eosin staining of distal esophageal biopsies of Control, EoE no FI, and EoE+FI patients. Comparison of eosinophil counts in (B) proximal and (C) distal esophageal biopsies. (D) Maximum eosinophil infiltration. (E) CCL2 mRNA transcript levels in the esophagus. (F,G) disease probability scores (p(EoE), p(Control), p(GERD)) in Control, EoE no FI, and EoE+FI patients. ****p < 0.0001 as calculated by Dunn's multiple comparison test after Kruskal-Wallis test.
EoE Patients With Food Impaction Present With Comparable Measures of Systemic Allergy but Decreased Levels of Esophageal Allergy Based on the IGHE Score
Since EoE is classified as an allergic disorder, allergic comorbidities were analyzed as a potential factor in distinguishing the clinically-defined EoE+FI patient subpopulation. No significant difference in the occurrence of food allergies, asthma, eczema, or seasonal allergies was observed in EoE+FI patients compared to EoE patients without FI (Figures 3A,B). The frequency of sensitized patients as determined by RAST or skin prick tests was also comparable between both patient groups (Figure 3C). Furthermore, there was no significant difference in total serum IgE titers (Figure 3D). These data suggest that there is no relationship between IgE mediated allergic comorbidities and food impactions in EoE patients. It is important to note here, however, that the correlation between serum IgE titers and EoE is low (35, 36). We recently established esophageal IgE production as an additional readout for tissue allergy in EoE patients (17). To test if esophageal tissue allergy can be used to identify EoE+FI patients, we analyzed the composite IGHE score. This score was defined as a secondary analysis loop of the published EoE diagnostic algorithm as a correlative measure of increased esophageal allergic Th2-type inflammation (17). Using the published cutoff score of 37.5, we found that 6/13 patients without FI presented with an elevated IGHE-score while none of the patients with FI did (Figure 3E). Based on this analysis, the systemic allergic phenotype is unlikely an eliciting and/or contributing factor for the development of food impaction in EoE patients, however the extent of the local esophageal allergic phenotype may negatively correlate with FI symptoms.
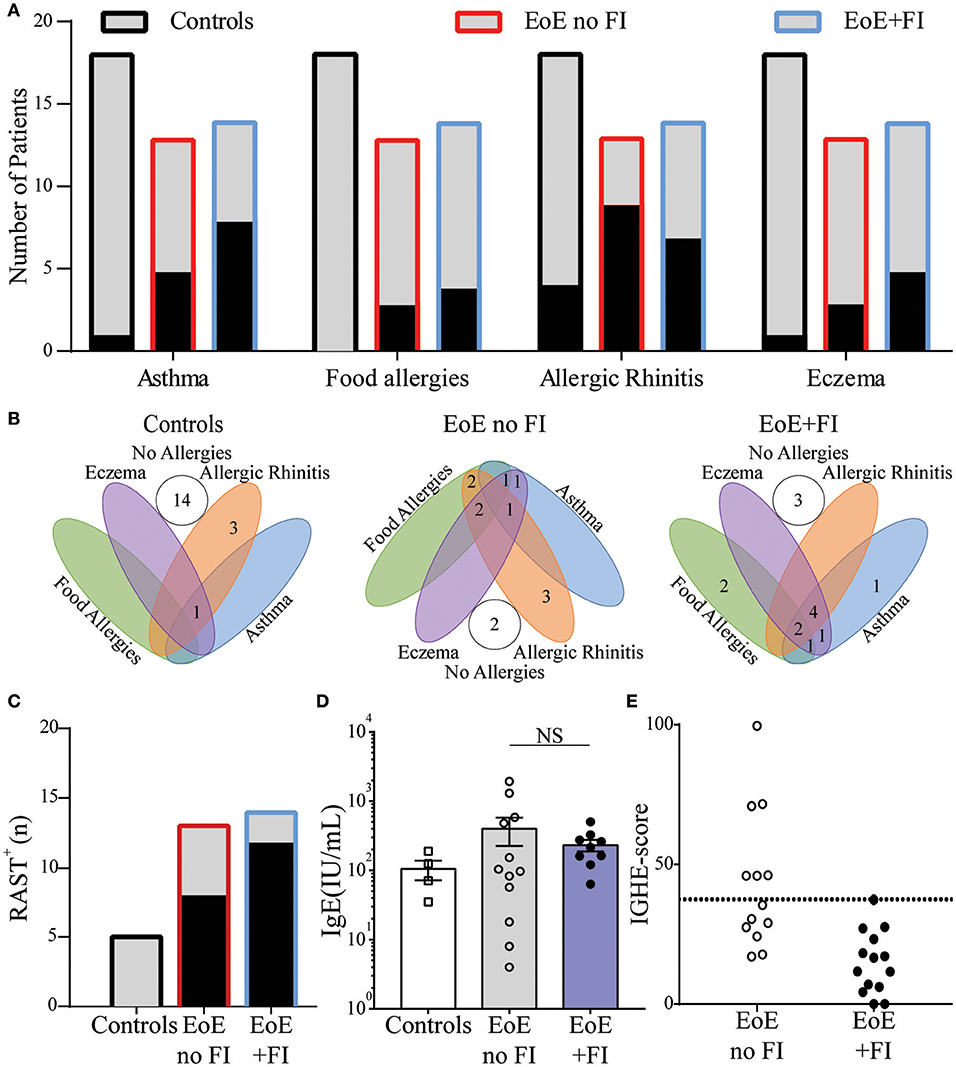
Figure 3. Comparison of clinical allergies and measurements of allergic sensitization in EoE no FI and EoE+FI patients. (A) Frequency of individual allergic comorbidities, and (B) distribution throughout the patient population. (C) Patients with a positive RAST to a food allergen. (D) Serum concentrations of IgE. (E) Esophageal allergy scores (IGHE score).
Esophageal Inflammation in EoE Patients With Food Impaction Differs From Those Without Food Impaction
EoE is a complex multifactorial disease, so in order to understand the immunological pathways contributing to the differences in disease presentation the transcriptional profile of 74 genes was analyzed (Figure 4A). EoE+FI patients present with significantly lower transcript counts of CPA3, a mast cell specific transcript that encodes for carboxypeptidase a3, as well as lower FCER1B, the mast cell and basophil specific beta chain of the high-affinity IgE receptor (Figures 4B,C). In agreement with the IGHE score, this data set suggests that EoE+FI patients present with a lesser contribution of the esophageal mast cell compartment to the local inflammation. CCL2 is the primary chemokine responsible for the recruitment of mast cell precursors. In line with the probable lower frequency of mast cells, patients presenting with food impactions express lower transcript levels of CCL2 in their esophageal biopsies indicating that the recruitment of this cell type might be altered (Figure 4D). Finally, it is important to note that the Th2-type cytokines IL4 and IL5 are significantly less expressed in patients with food impaction, suggesting a less pronounced Th2-type inflammatory tissue environment in patients with FI (Figures 4A,E). Combined these data demonstrate that patients with food impaction present with similar levels of tissue eosinophilia but the mast cell infiltration and Th2 inflammation are reduced.
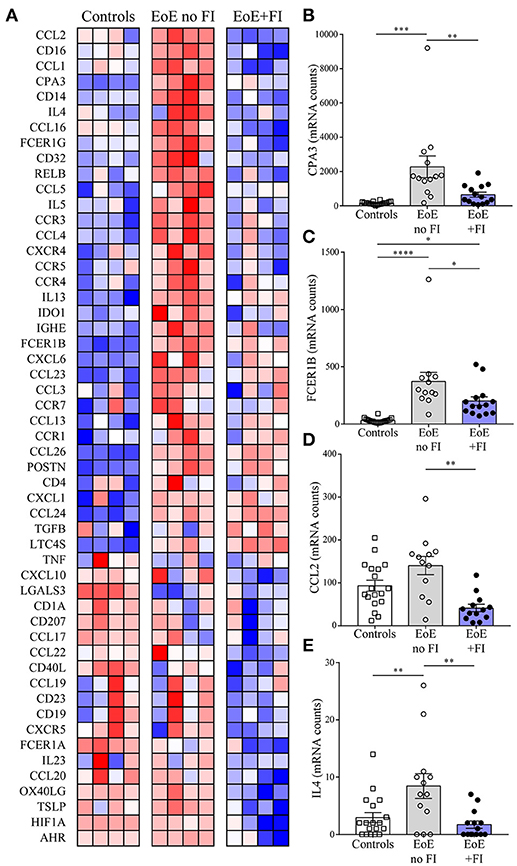
Figure 4. EoE+FI specific mRNA pattern. (A) Heat map comparison of esophageal mRNA patterns in control, EoE no FI, and EoE+FI patients. Relative expression of (B) CPA3, (C) FCER1B, (D) CCL2, (E) IL4 in control, EoE no FI, and EoE+FI patients. *p < 0.05 **p < 0.01, ***p < 0.001,****p < 0.0001 as calculated by Dunn's multiple comparison test after Kruskal-Wallis test.
Establishing an Analysis Loop That Allows for the Differentiation of EoE Patients With Food Impactions
Given the unique characteristics in immunological mRNA transcript pattern of EoE patients with FI, we hypothesized that an algorithm-based score could be developed to differentiate EoE patients with and without FI. By weighing transcripts based on the fold difference in their expression and their adjusted significance, transcript weights were calculated (Figures 5A,B). These weights were used to calculate a raw composite FI-score for each EoE patient using weighted factor analysis. This raw score was then standardized and a cutoff determined by ROC analysis. With the calculated cutoff of 0.03 the FI-score distinguished patients with and without FI with a sensitivity of 0.93 and a specificity of 1.00 (Figures 5C,D). The AUC for the ROC was 0.99 with a standard error of 0.02 and a positive predictive value of 100.00% and a negative predictive value of 92.86%. This secondary analysis can be used to expand the previously published diagnostic algorithm and may serve to predict the risk of food impactions prospectively.
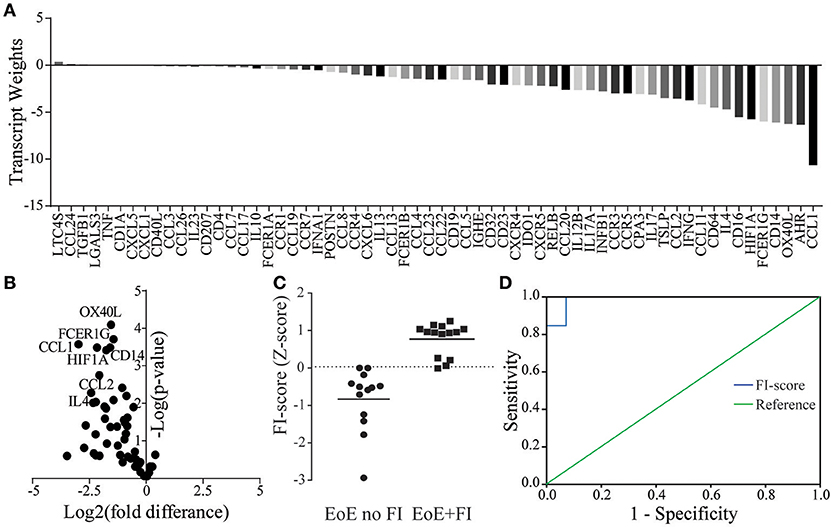
Figure 5. FI-score in EoE no FI and EoE+FI patients. (A) Transcript weights of the factors differentiating EoE and EoE+FI. (B) Volcano plots of normalized mRNA transcripts displayed as fold difference (x-axis) and significance (y-axis) used for the calculation of the factor weights. (C) Calculated standardized FI-score. (D) ROC analysis for differentiating EoE no FI and EoE+FI patients based on FI-score (AUC = 0.99, Sensitivity = 0.93 Specificity = 1).
The Inflammatory mRNA Pattern of EoE+FI Patients Indicates Underlying Esophageal Dysmotility
When analyzing the mRNA pattern for transcripts with potential influence on motility, we found that the inducible nitric oxide (NO) synthase NOS2 was significantly less expressed in EoE patients with FI (Figure 6A). Therefore these patients may not produce sufficient esophageal NO, a key signal for relaxation of smooth muscles (37, 38). Additionally, the expression levels of the transcription factor hypoxia induced factor 1a (HIF1A, Figure 6B) were significantly lower in EoE+FI patients. Loss of HIF1A expression has previously been described to result in an increase in smooth muscle contractility (39, 40). Combined, these results show altered expression levels of key regulators of motility, which implies that esophageal dysmotility may underlie the development of FI in EoE.
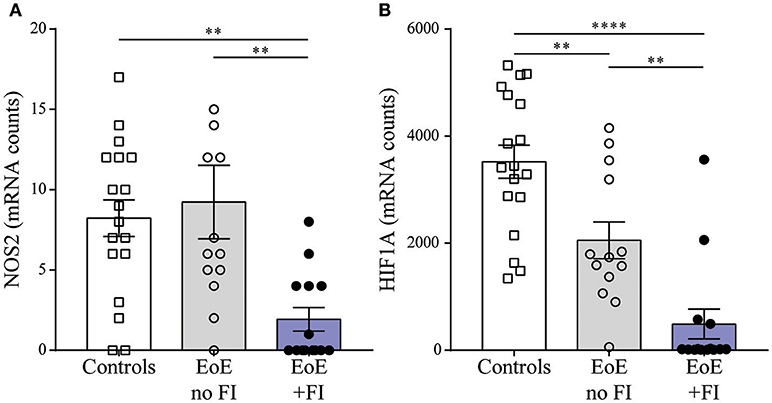
Figure 6. Motility related transcript levels. Comparison of (A) NOS2 and (B) HIF1A expression between patient groups. **p < 0.01, ***p < 0.001 as calculated by Dunn's multiple comparison test after Kruskal-Wallis test.
Discussion
Our work confirmed that EoE patients who suffer from food impactions cannot be identified as a distinct subgroup of EoE using gold standard diagnostic markers, such as eosinophil infiltration, histological features, or comorbidities. Yet, we were able to identify a unique esophageal mRNA profile in EoE patients with food impactions who did not show any evidence of anatomical narrowing of the esophagus by endoscopy. A modification of a published medical algorithm used the FI-specific mRNA pattern stamp to create a diagnostic score for this patient subpopulation (EoE+FI), which is characterized by lower expression levels of mast cell specific transcripts, Th2 cytokines, and decreased expression of key regulators of smooth muscle contractility and relaxation.
Food impaction is a common complication in EoE. In adult onset EoE, FI occurs with high frequency (55%) and is one of the main indicators of the disease (41). However, in our pediatric cohort, it occurred in only 12% of our cohort, which probably reflects the fact that in children FI start to occur in older children. As in adults we also observed a male predominance (35). Currently, no diagnostic tool for the identification of patients prone to develop this comorbidity exists. Generally, EoE is a chronic inflammatory disorder that leads to fibrosis and strictures in the esophagus and it has been thought that food impactions occur due to the fibrostenotic complications. Indeed strictures are found and identified as the cause of food impaction in some of the patients. However, in many patients, food impactions occur in the absence of such anatomical problems or any other identifiable cause. These observations imply that impaired esophageal motility contributes to food impactions among pediatric EoE patients (27). The hypothesis that food impactions are not necessarily connected to fibrosis is strongly supported by the findings from our study, which indicate that markers of mast cell expansion and Th2 cytokines, both of which are key regulators of fibrosis, are downregulated in EoE patients with food impactions. The observed inflammatory mRNA pattern implies that FI in pediatric EoE patients may develop independent of fibrosis and rather, manifests as a result of esophageal dysmotility.
Transcripts for inducible nitric oxide synthase (NOS2) were significantly downregulated in EoE+FI patients. Lower expression of NOS2 suggests diminished production and lower bioavailability of NO. This may lead to an increase in smooth muscle cell contractility because lack of NO prevents smooth muscle relaxation, which in turn results in poor peristalsis. Additionally, HIF1A was downregulated in EoE+FI patients in our cohort. HIF1A expression levels are acknowledged for contributing to the regulation of vascular smooth muscle tone and low levels of HIF1A expression have been shown to be associated with hypertension due to hypercontractile vascular smooth musculature (39, 40). It is currently unknown whether the role of HIF1A in smooth muscle contractility applies to esophageal smooth muscle cells. However, it is tempting to speculate that the reduction in esophageal HIF1A expression levels contributes to motility dysfunction by exacerbating the difficulty of esophageal smooth muscles to relax during swallowing. The combination of low NOS2 expression to regulate relaxation and low HIF1A expression to regulate smooth muscle tone and contractility can potentially result in smooth muscle cramping and spasm, and subsequently to food getting stuck in the esophagus in a fibrosis-independent manner.
Using both the immunological and non-immunological transcriptional differences in the expression patterns of EoE patients with food impactions, we modified our existing algorithm for diagnosing EoE patients to include a secondary analysis for assessing the risk for food impaction. The predictive power of the current study is limited by the small number of pediatric EoE patients with food impactions. It will, therefore, be important to confirm the accuracy of our algorithm by recruiting additional patients who can be integrated into the current analysis in a forward-feeding way to expand the training set and to generate an additional test set. Ideally, such an analysis would be performed as a multicenter study. Another limitation of the current study is its retrospective nature which allows for the identification of a gene signature that identifies patients with food impactions as a subpopulation but cannot predict the development of this complication. For the latter purpose, a longitudinal cohort study needs to be designed to predictively analyze the esophageal mRNA signature changes in EoE patients as they age and as they develop food impaction, or phenotypes specific to other subpopulations of EoE. Such analysis will determine if the differences in gene expression, which we have identified as specific for EoE+FI can be observed before the first food impaction event. Furthermore, at the inception of our cohort, the definition of EoE still included a lack of response to 8 weeks of PPI, so all patients included in the present cohort are EoE patients that did not respond to PPI. We do not have any patient with PPI responsive eosinophilia, in the study cohort so it is not clear if the mRNA pattern of those patients will be the same.
Currently, the unpredictability of EoE therapy response and disease progression necessitates physicians to find the best treatment strategy for each patient using a trial and error approach monitored by gold standard diagnosis which significantly affects the quality of life of the EoE patients (42). For this purpose, EoE algorithms and their future expansions with secondary diagnostic loops appear as attractive strategies because they will permit the prediction of the development of symptoms and treatment responsiveness and can, thus, help inform the diagnosis and treatment of EoE reducing the time between initial presentation and effective treatment. Improved diagnostic strategies will assist physicians in educating EoE patients and their families if their score implies the risk of developing food impactions, thus helping the child and family anticipate how the disease will impact them particularly. Additionally, the FI-score will assist physicians in assessing the risk for food impactions in each patient objectively, making it a highly useful diagnostic tool when dealing with patients that are unable to accurately express their symptoms.
Author Contributions
BS, UA, CK, RR, SB, and EF designed experiments, performed research, and analyzed data. KH, RR, JV, JG, and SN were involved in patient recruitment, evaluation of clinical patient status, evaluation of biopsies, and analysis of human serum data. BS, SN, and EF wrote the first draft of the manuscript. All co-authors contributed to writing the final manuscript and approved its last version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with the authors and the reviewer HO.
Acknowledgments
We thank all members of the Fiebiger, the Nurko, and the Rosen groups for discussions and technical assistance. EF was supported by a Bridge Grant from the Research Council of Boston Children's Hospital, an Emerging Investigator Award from FARE, a Senior Research Award from the Crohn's and Colitis Foundation, and an unrestricted gift from the Mead Johnson Nutrition Company. This work was further supported by an NIH grant of the Harvard Digestive Diseases Center (P30DK034854, Core C). RR was supported by NIH (R01 DK097112).
Abbreviations
FI, Food impaction; AUC, area under the curve; EoE, eosinophilic esophagitis; EoE+FI, eosinophilic esophagitis presenting with food impaction; GERD, gastroesophageal reflux disease; PPI-REE, proton pump responsive eosinophilic esophagitis; NO, nitric oxide; HIF1A, hypoxia induced factor 1a; CCL26, C-C motif chemokine ligand 26; NOS2, Nitric oxide synthase 2, inducible nitric oxide synthase IL4, Interleukin 4; IL5, Interleukin 5; FCER1B, Fc fragment of IgE, high affinity I, receptor for; beta polypeptide, High affinity IgE receptor beta polypeptide; CPA3, carboxypeptidase a3.
References
1. Mansoor E, Cooper GS. The 2010-2015 prevalence of eosinophilic esophagitis in the USA: a population-based study. Dig Dis Sci. (2016) 61:2928–34. doi: 10.1007/s10620-016-4204-4
2. Heine RG. Insights into the emerging epidemic of eosinophilic oesophagitis. best practice and research. Clin Gastroenterol. (2015) 29:731–7. doi: 10.1016/j.bpg.2015.09.004
3. Dellon ES, Jensen ET, Martin CF, Shaheen NJ, Kappelman MD. Prevalence of eosinophilic esophagitis in the United States. Clin Gastroenterol Hepatol. (2014) 12:589–96 e1. doi: 10.1016/j.cgh.2013.09.008
4. Dellon ES. Epidemiology of eosinophilic esophagitis. Gastroenterol Clin North Am. (2014) 43:201–18. doi: 10.1016/j.gtc.2014.02.002
5. Moawad FJ, Cheng E, Schoepfer A, Al-Haddad S, Bellizzi AM, Dawson H, et al. Eosinophilic esophagitis: current perspectives from diagnosis to management. Ann N Y Acad Sci. (2016) 1380:204–17. doi: 10.1111/nyas.13164
6. Cianferoni A, Spergel J. Eosinophilic esophagitis: a comprehensive review. Clin Rev Allergy Immunol. (2016) 50:159–74. doi: 10.1007/s12016-015-8501-z
7. Furuta GT, Katzka DA. Eosinophilic esophagitis. N Engl J Med. (2015) 373:1640–8. doi: 10.1056/NEJMra1502863
8. Clayton F, Peterson K. Eosinophilic esophagitis: pathophysiology and definition. Gastrointest Endosc Clin N Am. (2018) 28:1–14. doi: 10.1016/j.giec.2017.07.011
9. Simon D, Straumann A, Schoepfer AM, Simon HU. Current concepts in eosinophilic esophagitis. Allergo J Int. (2017) 26:258–66. doi: 10.1007/s40629-017-0037-8
10. Markowitz JE, Clayton SB. Eosinophilic esophagitis in children and adults. Gastrointest Endosc Clin N Am. (2018) 28:59–75. doi: 10.1016/j.giec.2017.07.004
11. Lynch KL, Dhalla S, Chedid V, Ravich WJ, Stein EM, Montgomery EA, et al. Gender is a determinative factor in the initial clinical presentation of eosinophilic esophagitis. Dis Esophagus (2016) 29:174–8. doi: 10.1111/dote.12307
12. Gonsalves N. Distinct features in the clinical presentations of eosinophilic esophagitis in children and adults: is this the same disease? Dig Dis. (2014) 32:89–92. doi: 10.1159/000357078
13. Lucendo AJ, Sanchez-Cazalilla M. Adult versus pediatric eosinophilic esophagitis: important differences and similarities for the clinician to understand. Expert Rev Clin Immunol. (2012) 8:733–45. doi: 10.1586/eci.12.68
14. Miehlke S. Clinical features of Eosinophilic esophagitis in children and adults. best practice & research. Clin Gastroenterol. (2015) 29:739–48. doi: 10.1016/j.bpg.2015.09.005
15. Mudde AC, Lexmond WS, Blumberg RS, Nurko S, Fiebiger E. Eosinophilic esophagitis: published evidences for disease subtypes, indications for patient subpopulations, and how to translate patient observations to murine experimental models. World Allergy Organ J. (2016) 9:23. doi: 10.1186/s40413-016-0114-3
16. Dellon ES, Veerappan R, Selitsky SR, Parker JS, Higgins LL, Beitia R, et al. A gene expression panel is accurate for diagnosis and monitoring treatment of eosinophilic esophagitis in adults. Clin Trans Gastroenterol. (2017) 8:e74. doi: 10.1038/ctg.2017.2
17. Sallis BF, Erkert L, Monino-Romero S, Acar U, Wu R, Konnikova L, et al. An algorithm for the classification of mRNA patterns in eosinophilic esophagitis: Integration of machine learning. J Allergy Clin Immunol. (2017) 141:1354–64.e9. doi: 10.1016/j.jaci.2017.11.027
18. Lexmond WS, Neves JF, Nurko S, Olszak T, Exley MA, Blumberg RS, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am J Gastroenterol. (2014) 109:646–57. doi: 10.1038/ajg.2014.12
19. Lexmond WS, Pardo M, Rooney K, Goettel JA, Snapper SB, Yen EH, et al. Elevated levels of leukotriene C4 synthase mRNA distinguish a subpopulation of eosinophilic oesophagitis patients. Clin Exp Allergy (2013) 43:902–13. doi: 10.1111/cea.12146
20. Shoda T, Wen T, Aceves SS, Abonia JP, Atkins D, Bonis PA, Caldwell JM, et al. Consortium of eosinophilic gastrointestinal disease, eosinophilic oesophagitis endotype classification by molecular, clinical, and histopathological analyses: a cross-sectional study. Lancet Gastroenterol Hepatol. (2018) 3:477–88. doi: 10.1016/S2468-1253(18)30096-7
21. Munoz-Persy M, Lucendo AJ. Treatment of eosinophilic esophagitis in the pediatric patient: an evidence-based approach. Eur J Pediatr. (2018) 177:649–63. doi: 10.1007/s00431-018-3129-7
22. Hurtado CW, Furuta GT, Kramer RE. Etiology of esophageal food impactions in children. J Pediatr Gastroenterol Nutr. (2011) 52:43–6. doi: 10.1097/MPG.0b013e3181e67072
23. Weiss AH, Iorio N, Schey R. Esophageal motility in eosinophilic esophagitis. Rev Gastroenterol Mex. (2015) 80:205–13. doi: 10.1016/j.rgmxen.2015.05.002
24. Davis BP. Pathophysiology of eosinophilic esophagitis. Clin Rev Allergy Immunol. (2018) 154:333–45. doi: 10.1053/j.gastro.2017.06.065
25. Eluri S, Runge TM, Cotton CC, Burk CM, Wolf WA, Woosley JT, et al. The extremely narrow-caliber esophagus is a treatment-resistant subphenotype of eosinophilic esophagitis. Gastrointest Endosc. (2016) 83:1142–8. doi: 10.1016/j.gie.2015.11.019
26. Nhu QM, Aceves SS. Tissue remodeling in chronic eosinophilic esophageal inflammation: parallels in asthma and therapeutic perspectives. Front Med. (2017) 4:128. doi: 10.3389/fmed.2017.00128
27. Nurko S, Rosen R, Furuta GT. Esophageal dysmotility in children with eosinophilic esophagitis: a study using prolonged esophageal manometry. Am J Gastroenterol. (2009) 104:3050–7. doi: 10.1038/ajg.2009.543
28. DeBrosse CW, Franciosi JP, King EC, Butz BK, Greenberg AB, Collins MH, et al. Long-term outcomes in pediatric-onset esophageal eosinophilia. J Allergy Clin Immunol. (2011) 128:132–8. doi: 10.1016/j.jaci.2011.05.006
29. Menard-Katcher C, Benitez AJ, Pan Z, Ahmed FN, Wilkins BJ, Capocelli KE, et al. Influence of age and eosinophilic esophagitis on esophageal distensibility in a pediatric cohort. Am J Gastroenterol. (2017) 112:1466–73. doi: 10.1038/ajg.2017.131
30. Warners MJ, R.Oude Nijhuis AB, L.de Wijkerslooth RH, Smout A, Bredenoord AJ. The natural course of eosinophilic esophagitis and long-term consequences of undiagnosed disease in a large cohort. Am J Gastroenterol. (2018) 113:836–44. doi: 10.1038/s41395-018-0052-5
31. Lexmond WS, Hu L, Pardo M, Heinz N, Rooney K, LaRosa J, et al. Accuracy of digital mRNA profiling of oesophageal biopsies as a novel diagnostic approach to eosinophilic oesophagitis. Clin Exp Allergy (2015) 45:1317–27. doi: 10.1111/cea.12523
32. Yen EH, Hornick JL, Dehlink E, Dokter M, Baker A, Fiebiger E, et al. Comparative analysis of FcepsilonRI expression patterns in patients with eosinophilic and reflux esophagitis. J Pediatr Gastroenterol Nutr. (2010) 51:584–92. doi: 10.1097/MPG.0b013e3181de7685
33. Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. (2006) 116:536–47. doi: 10.1172/JCI26679
34. Lipowska AM, Kavitt RT. Demographic features of eosinophilic esophagitis. Gastrointest Endosc Clin N Am. (2018) 28:27–33. doi: 10.1016/j.giec.2017.07.002
35. Clayton F, Fang JC, Gleich GJ, Lucendo AJ, Olalla JM, Vinson LA, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology (2014) 147:602–9. doi: 10.1053/j.gastro.2014.05.036
36. Rodriguez-Sanchez J, Gomez Torrijos E, Lopez Viedma B, de la Santa Belda E, Martin Davila F, Garcia Rodriguez C, et al. Efficacy of IgE-targeted vs empiric six-food elimination diets for adult eosinophilic oesophagitis. Allergy (2014) 69:936–42. doi: 10.1111/all.12420
37. Mori D, Watanabe N, Kaminuma O, Murata T, Hiroi T, Ozaki H, et al. IL-17A induces hypo-contraction of intestinal smooth muscle via induction of iNOS in muscularis macrophages. J Pharmacol Sci. (2014) 125:394–405. doi: 10.1254/jphs.14060FP
38. Van Geldre LA, Lefebvre RA. Interaction of NO and VIP in gastrointestinal smooth muscle relaxation. Curr Pharm Des. (2004) 10:2483–97. doi: 10.2174/1381612043383890
39. Barnes EA, Chen CH, Sedan O, Cornfield DN. Loss of smooth muscle cell hypoxia inducible factor-1alpha underlies increased vascular contractility in pulmonary hypertension. FASEB J. (2017) 31:650–62. doi: 10.1096/fj.201600557R
40. Kim YM, Barnes EA, Alvira CM, Ying L, Reddy S, Cornfield DN. Hypoxia-inducible factor-1alpha in pulmonary artery smooth muscle cells lowers vascular tone by decreasing myosin light chain phosphorylation. Circ Res. (2013) 112:1230–3. doi: 10.1161/CIRCRESAHA.112.300646
41. Pasha SF, DiBaise JK, Kim HJ, De Petris G, Crowell MD, Fleischer DE, et al. Patient characteristics, clinical, endoscopic, and histologic findings in adult eosinophilic esophagitis: a case series and systematic review of the medical literature. Dis Esophagus (2007) 20:311–9. doi: 10.1111/j.1442-2050.2007.00721.x
42. Hiremath G, Kodroff E, Strobel MJ, Scott M, Book W, Reidy C, et al. Individuals affected by eosinophilic gastrointestinal disorders have complex unmet needs and frequently experience unique barriers to care. Clin Res Hepatol Gastroenterol. (2018) S2210-7401(18)30042-1. doi: 10.1016/j.clinre.2018.03.003
Keywords: eosinophilic esophagitis, food impaction, machine learning classification, medical algorithm, esophageal motility, eosinophils
Citation: Sallis BF, Acar U, Hawthorne K, Babcock SJ, Kanagaratham C, Goldsmith JD, Rosen R, Vanderhoof JA, Nurko S and Fiebiger E (2018) A Distinct Esophageal mRNA Pattern Identifies Eosinophilic Esophagitis Patients With Food Impactions. Front. Immunol. 9:2059. doi: 10.3389/fimmu.2018.02059
Received: 22 May 2018; Accepted: 21 August 2018;
Published: 05 November 2018.
Edited by:
Michiko Oyoshi, Boston Children's Hospital, Harvard University, United StatesReviewed by:
Hans Oettgen, Harvard University, United StatesJulio Villena, Centro de Referencia para Lactobacilos, Argentina
Copyright © 2018 Sallis, Acar, Hawthorne, Babcock, Kanagaratham, Goldsmith, Rosen, Vanderhoof, Nurko and Fiebiger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samuel Nurko, c2FtdWVsLm51cmtvQGNoaWxkcmVucy5oYXJ2YXJkLmVkdQ==
†Co-senior authors
 Benjamin F. Sallis
Benjamin F. Sallis Utkucan Acar
Utkucan Acar Kelsey Hawthorne
Kelsey Hawthorne Stephen J. Babcock1
Stephen J. Babcock1 Rachel Rosen
Rachel Rosen Edda Fiebiger
Edda Fiebiger