- 1Shandong Provincial Key Laboratory of Animal Cells and Developmental Biology, School of Life Sciences, Shandong University, Jinan, China
- 2State Key Laboratory of Microbial Technology, Shandong University, Jinan, China
The Ras GTPase superfamily, including more than 100 members, plays a vital role in a number of cellular processes, such as cytoskeleton recombination, gene expression, and signaling pathway regulation. Some members of the superfamily participate in innate immunity in animals. However, there have been few studies of RhoA on this aspect. In the present study, we identified a RhoA GTPase in the shrimp Marsupenaeus japonicus and named it MjRhoA. Expression of MjRhoA was significantly upregulated in hemocytes and heart of shrimp challenged with Vibrio anguillarum. Overexpression of MjRhoA in shrimp caused the total bacterial number to decrease significantly and knockdown of MjRhoA increased the bacterial number obviously, with a consequent decline in shrimp survival. These results confirmed the antibacterial function of MjRhoA in shrimp. Further study showed that rate of phagocytosis of hemocytes was decreased in MjRhoA-knockdown shrimp. Interestingly, we observed that MjRhoA was translocated onto the hemocyte membrane at 1 h post V. anguillarum challenge. The expression levels of the β-integrin-mediated phagocytosis markers ROCK2 and Arp2/3 declined significantly after knockdown of MjRhoA. These results suggested that the antibacterial function of MjRhoA was related to β-integrin-mediated phagocytosis in shrimp. Our previous study identified that a C-type lectin, hFcLec4, initiated β-integrin mediated phagocytosis after bacterial infection. Thus, knockdown of hFcLec4 and β-integrin was performed. The results showed that the translocation of MjRhoA from the cytoplasm to membrane was inhibited and the expression level of MjRhoA was decreased, suggesting that MjRhoA participated in hFcLec4-integrin mediated phagocytosis. Therefore, our study identified a new hFcLec4-integrin-RhoA dependent phagocytosis against bacterial infection in shrimp.
Introduction
Small GTPases play a crucial role in the biological functions of organisms. To date, many small GTPase members have been identified and all the members belong to the Ras GTPase superfamily (1). Commonly, the Ras superfamily is divided into five groups: Ras, Ran, Rho, Rab, and Arf/Sar families (2). Except for the Arf family, the other four groups exhibit GTPase activity. All members of these families have two states, the active state (GTP-binding state) and the inactive state (GDP-binding state). The two states can be reversed by guanine nucleotide exchange factors (GEF) and GTPase-activating proteins (GAP). Through this transformation between the active and inactive states, the small GTPases successfully regulate their downstream effectors (3).
The Rho GTPase family consists of three major members, Rho, Rac, and Cdc42 (cell division cycle 42). The main functions of Rho GTPases are regulating cytoskeleton regrouping, the cell cycle, gene expression, and signal transduction (4). In particular, Cdc42 mainly functions in cell apoptosis (5), cell polarity (6), and cell motility (7). Cdc42 and Rac also regulate filopodia formation synergistically (8). In addition, various signaling pathways are regulated by Rho GTPases; for example, Cdc42 together with Rac1 regulates the transcriptional activity of the transcription factor c-Jun in JNK signaling pathway activation (9). Particular to animal cells (10), Rac1 plays a crucial role in activating the nuclear factor kappa B (NF-κB) signaling pathway via the Toll-like receptor 2 in its activated state (11). As another key member of Rho GTPases, RhoA functions in regulating the actin cytoskeleton (12). The actin cytoskeleton is crucial for cell movement, material transport, and cell differentiation by regulating the morphological structure of cells (13). In vertebrates, the expression level of RhoA is much higher in tumor cells than in normal cells (14), indicating a close association with tumor progression. In fact, RhoA-dependent cytoskeleton regulation is a fundamental molecular mechanism in tumorigenesis (15).
In contrast to vertebrates, there have been few studies of the functions of Rho GTPases family members in invertebrates. For example, Rac1 has dual functions in antibacterial and antiviral innate immunity in Fenneropenaeus chinensis (16); Cdc42 inhibits the replication of the DNA virus white spot syndrome virus (WSSV) by interacting with the arginine kinase in kuruma shrimp Marsupenaeus japonicus (17). However, there are no reports about the function of Rho in invertebrates.
In the present study, we identified a member of the Rho GTPases, RhoA, using transcriptome sequencing of kuruma shrimp, M. japonicus (denoted hereafter as MjRhoA). We observed that MjRhoA was highly expressed in the Vibrio anguillarum-challenged shrimp hemocytes and heart. Knockdown of MjRhoA led to high bacterial numbers and high mortality of shrimp. The possible mechanism of MjRhoA function in shrimp was analyzed. The results indicated that RhoA might inhibit bacterial infection by enhancing phagocytosis in shrimp.
Materials and Methods
Gene Cloning and Bioinformatic Analysis
The sequence of MjRhoA was obtained by transcriptome sequencing of hemocytes from M. japonicus. The sequence identity of MjRhoA was analyzed by the online BLASTX algorithm (https://www.ncbi.nlm.nih.gov/). The translation and isoelectric point (pI)/molecular weight (Mw) analysis of the nucleotide sequences were performed using the online tool ExPASy (https://www.expasy.org/). GeneDoc and MEGA6, respectively, were used to perform the sequence alignment and to construct the phylogenetic tree of RhoA.
Animals, Immune Challenge, and Tissue Extraction
Healthy kuruma shrimp, M. japonicus, (9–12 g) were purchased in a seafood market in Jinan, Shandong, China. The shrimp were initially cultured in a water tank at 25°C for 2 days for acclimatization to the laboratory conditions. Thereafter, for the immune challenge, the shrimp were injected with V. anguillarum (107 colony forming units (CFUs) per shrimp) suspended in 50 μL of sterile phosphate-buffered saline (PBS, 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH = 7.4). Shrimp injected with same amount of sterile PBS were used as controls.
After the shrimp were challenged by the bacteria, different organs (heart, hepatopancreas, gills, stomach, and intestine) were collected and homogenized using manual homogenizers. To collect the hemocytes, the total hemolymph was collected using a 5 mL syringe containing 1 mL anticoagulant (0.45 M NaCl, 10 mM KCl, 10 mM EDTA, and 10 mM HEPES, pH = 7.45) and then centrifuged at 800 × g for 6 min at 4°C to collect the hemocytes. The obtained hemocytes and the other organs (heart, hepatopancreas, gills, stomach, and intestine) were used for total RNA and total protein extraction.
Total RNA Extraction and cDNA Synthesis
The total RNA was isolated using TRIpure reagent (Aidlab, Beijing, China). The obtained organs and hemocytes (10 mg) were homogenized in 1 mL of Trizol reagent for RNA extraction. Based on the previous study in our lab, the cDNA was synthesized using 5 μg total RNA with the SMART cDNA synthesis kit (Clontech) following manufacterer's instuctions. The obtained cDNA was used to detect the expression level of different genes using specific primers (Table 1).
Semiquantitative RT-PCR Analysis and Real-Time RT-PCR (qPCR) Analysis Assays
To analyze the expression level of the target gene MjRhoA at the transcriptional level, a semiquantitative method and a real-time quantitative method were performed. The semiquantitative RT-PCR was used to detect the tissue distribution of MjRhoA using a pair of specific primers (MjRhoARTF and MjRhoARTR). The RT-PCR profile was as follows: 94°C for 3 min; 35 cycles of 94°C for 20 s, 56°C for 30 s, and 72°C for 30 s; followed by 72°C for 10 min. The PCR products were analyzed using agarose gel electrophoresis (AGE) with a 1.5% agarose gel. As a control, the expression level elongation factor 1-alpha (EF1α) was determined in the same way, using a pair of primers (EF1α RTF and EF1α RTR).
To analyze the expression profiles of the target genes, qPCR was performed with primers MjRhoARTF and MjRhoARTR. EF1α RTF and EF1α RTR (Table 1) were used as the internal control. The 10 μL qPCR reaction system contained 5 μL 2 × SYBR qPCR mix, 4 μL of primers (0.5 μM), and 1 μL cDNA. The qPCR profile was as follows: 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 50 s, and reading for 2 s at 72°C; the melting curve stage was from 65 to 95°C. The 2−ΔΔCT method was used to analyze the qPCR data and the results were shown as mean ± SD. Student's t-test was used to analyze the significant difference between the PCR data of two groups.
Recombinant Expression, Purification, and Antiserum Preparation
To study the function of MjRhoA at the protein level, we first recombinantly expressed MjRhoA in the Escherichia coli. First, the cDNA fragment of MjRhoA was amplified using primers, MjRhoAexF and MjRhoAexR (Table 1). Then the cDNA fragment of MjRhoA and the empty pET30a (+) vector were digested with BamHI and XhoI and then purified using the DNA purification kit (Sangon, Shanghai, China). The fragment was ligated into the digested plasmid using T4 DNA ligase (Thermo Fisher). The constructed recombinant plasmid was then transformed into E. coli Rosetta cells for the recombinant expression of MjRhoA (induced using 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The recombinant MjRhoA was expressed in the supernatant and was purified by affinity chromatography using a Ni-resin (GenScript, Nanjing, China). The purified recombinant protein was dialyzed for 48 h at 4°C in PBS buffer containing 5% glycerinum.
The purified recombinant MjRhoA (rMjRhoA) was used to prepare antiserum in New Zealand white rabbits. Two subcutaneous injections were performed using 500 μg of protein each time. For the first injection, 500 μg of recombinant MjRhoA protein was mixed thoroughly with the same volume of Complete Freund's Adjuvant (SIGMA). After 3 weeks of sensitization, 500 μg of MjRhoA protein mixed with Incomplete Freund's Adjuvant (SIGMA) was used for the second injection. The antiserum was collected and assessed using western blotting.
DsRNA Preparation and RNA Interference Assay
To study the function of MjRhoA, RNA interference was employed. For the dsRNA preparation, the dsRNA region was analyzed and searched using Primer Premier 5, and the specific primers MjRhoA RNAi F and MjRhoA RNAi R containing a T7 promoter sequence (GCGTAATACGACTCACTATAGG) were synthesized. The primers were used to amplify the dsRNA template. The template was purified by Chloroform extraction. The dsRNA template was then used for the dsMjRhoA synthesis using T7 RNA polymerase. The 50-μL reaction system contained 2.4 μL of the four NTPs (ATP, CTP, GTP, and UTP), 4 μL of T7 RNA polymerase, 2 μL of RNase inhibitor, 1 μL of dsRNA template, 20 μL of T7 RNA polymerase buffer, and 13 μL of RNase-free water. After incubation at 37°C for 6 h, DNase I was added into the mixture and incubation was continued at 37°C for another 1.5 h to digest the DNA template. The dsRNA of MjRhoA was then extracted using Chloroform. The concentration of the extracted dsMjRhoA was detected using a micro-spectrophotometer K5500 (K.O., China). The synthesis of the control dsGFP was performed with the same way using the primers GFP RNAi F and GFP RNAi R.
For the in vivo RNA interference assay, two groups of healthy intermolting shrimp (9 g each) were prepared and each group contained more than 30 individuals. The shrimp in one group were injected with 30 μg of dsMjRhoA (diluted in 50 μL RNase-free water) and the shrimp in the control group were injected with the same amount of dsGFP (30 μg). After dsRNA injection for 48 h, at least three individuals were randomly chosen to extract the total RNA using the method detailed above for the RNAi efficiency analysis by qPCR. The remaining shrimp were used for survival rate analysis.
Survival Rate Analysis
For the survival rate assay, shrimp were divided to two groups (at least 30 individuals in each group) and injected with dsMjRhoA or dsGFP. The dsMjRhoA injection group and the control (dsGFP injection) group were simultaneously challenged with the same amount of V. anguillarum (107 CFUs/shrimp). Thereafter, the number dead shrimp were monitored every 24 h and the survival rates of the two groups were calculated.
The in vivo Bacteria Clearance Assay
For the bacteria clearance assay, the experimental groups were first injected with 30 μg dsMjRhoA (diluted in 50 μL RNase-free water) for MjRhoA RNAi or with 10 μg rMjRhoA (diluted in 50 μL PBS) for MjRhoA overexpression. The shrimp were then injected with V. anguillarum suspended in 50 μL sterile PBS (107 CFUs/shrimp) 48 h post dsMjRhoA injection or 24 h post rMjRhoA injection. Thirty minutes after injection, the hemolymph from at least three individuals in each group was collected using sterile syringes on a clean bench. To detect the total bacterial number in vivo, the Petri dishes containing 2216E medium (0.5% tryptone, 0.1% yeast extract, 0.01% FeCl3, 1.5% agar, and 2.4% NaCl) were placed on the clean bench beforehand. For each group, 50 μL shrimp of hemolymph was smeared onto the dish. After culturing at room temperature overnight, the single colonies were counted and the number of CFUs was calculated. Each independent experiment was repeated three times. The results are shown as the mean ± SD and were statistically analyzed using Student's t-test. Shrimp injected with dsGFP for RNAi and injected with a His Tag for the overexpression assay were used as controls, respectively.
Immunocytochemistry and Phagocytosis Assay
For the immunocytochemistry assay, hemocytes were washed with PBS twice and then dropped on a prepared glass slide (coated with polylysine) for 1 h to sediment the hemocytes. The glass slides were then treated with 0.2% Triton X-100 (in PBS) for 5 min to enhance the permeability of the cell membrane. Afterwards, the glass slides were washed with PBS for 5 min six times. The hemocytes were then blocked in 3% bovine serum albumin (BSA) at 37°C for 30 min. The hemocytes were incubated with MjRhoA antiserum as the primary antibody diluted with 3% BSA (1:500) at 37°C for 3 h. Thereafter, the glass slides were washed with PBS for 5 min six times. After blocking in 3% BSA for 30 min for a second time, the hemocytes were incubated with the secondary antibody (fluorescein isothiocyanate (FITC)-goat anti rabbit, diluted with 3% BSA, 1:10,000) at 37°C for 1 h. The slides were then washed with PBS six times, and the cell nuclei were stained with 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) (diluted with PBS, 1:1,000) for 10 min. After washing with PBS six times, the hemocytes were observed under a fluorescence microscope (Olympus BX51, Japan).
For the phagocytosis assay, untreated shrimp were first injected with FITC-labeled V. anguillarum. To prepare the FITC-labeled V. anguillarum, the overnight cultured bacteria were first killed at 72°C for 20 min, and then suspended in 0.1 M NaHCO3 (pH = 9.0); the bacteria were labeled with 1 mg/mL FITC at 28°C for 1 h. After washing with sterile PBS six times (5 min each), the bacteria were injected into shrimp. Thirty minutes after injection, the shrimp hemocytes from different groups were collected and washed with PBS twice. The hemocytes were dropped onto a glass slide (coated with polylysine) for 1 h. After treatment with 0.2% Triton X-100 (in PBS) for 5 min, the cells were washed with PBS five times. The nuclei was stained with DAPI for 10 min. After washing with PBS, the hemocytes were observed under a fluorescence microscope and the phagocytosis rate (the proportion of phagocytic cells among the total cells) and the phagocytic index (the proportion of bacteria phagocytosed by the shrimp hemocytes among the total cells) were calculated.
Results
Sequence Alignment and Phylogenetic Tree of RhoA
The mRNA sequence of RhoA was obtained by transcriptome sequencing in kuruma shrimp M. japonicus and was named MjRhoA. The full-length cDNA sequence of MjRhoA consists of 1,640 bp, with a 579 bp open reading frame (Figure S1). MjRhoA encodes a polypeptide of 192 amino acids, with theoretical pI and Mw of polypeptide 6.0 and 21.60 KDa.
To analyze the sequence similarity with RhoAs from different species, sequence alignment was performed and a phylogenetic tree was built. The analysis was showed that RhoAs from different species are relatively conserved (Figure S2A). Notably, Rho1 proteins from D. melanogaster and other species were quite similar to RhoA, suggesting that Rho1s belonging to the RhoA family. In the phylogenetic analysis, the RhoAs proteins were mainly divided into two branches: RhoAs of invertebrates and RhoAs of most vertebrates. Though MjRhoA is phylogenetically close to those RhoAs of different invertebrates, it belongs to one specific branch (Figure S2B). It is worth mentioning that the RhoA from the freshwater fish Poeciliopsis prolifica is quite different from the other RhoAs (Figure S2B).
MjRhoA Was Upregulated in Shrimp Challenged by V. anguillarum
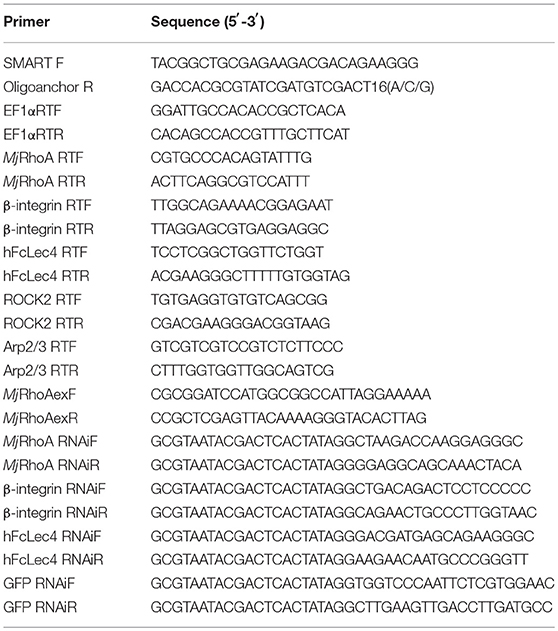
First, polyclonal antibodies against MjRhoA was prepared using rMjRhoA with a His tag (Figure 1A). The anti-MjRhoA polyclonal antibodies could recognize native MjRhoA in shrimp with appropriate molecular mass although there were also a few non-specific bands recognized by the antibodies (Figure 1B), this might be the impurity of the protein. The tissue distribution of MjRhoA in shrimp was then analyzed by qPCR and western blotting. The results showed that MjRhoA was expressed in different tested tissues and was highly expressed in hemocytes and heart, with quite low expression in the hepatopancreas (Figure 1C). The expression patterns of MjRhoA in hemocytes and heart were detected using qPCR. The results indicated that the expression of MjRhoA was upregulated in hemocytes at 12 h post V. anguillarum challenge and remained at a high level from 12 to 48 h (Figure 1D). The expression level of MjRhoA was also upregulated in the heart at 12 h post V. anguillarum infection, and the expression level at 24 h post infection was relatively lower than 12 h and 48 h; however, it was still higher than that in the normal (unchallenged) control samples (Figure 1D). These results indicated that MjRhoA might play a role in resisting infection by V. anguillarum in shrimp.
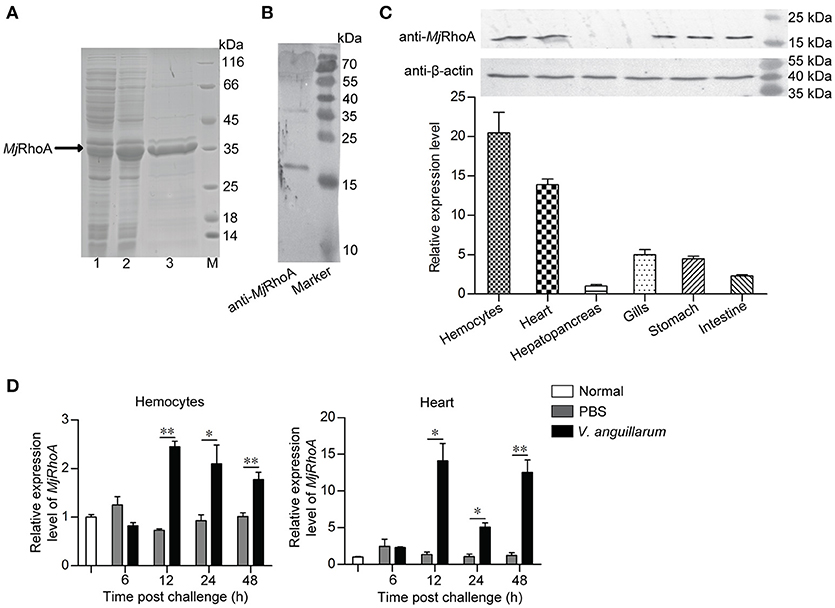
Figure 1. MjRhoA was upregulated in shrimp challenged by V. anguillarum. (A) Recombinant expression of MjRhoA in E. coli cells detected by SDS-PAGE. Lane 1, total proteins in the E. coli Rosetta cells transformed with pET30a-MjRhoA before induction with IPTG; Lane 2, total proteins in the E. coli Rosetta cells containing pET30a-MjRhoA after induction with 0.5 mM IPTG; Lane 3, purified recombinant MjRhoA using affinity chromatography with a His-binding resin; Lane M, protein standard. (B) The specificity of the anti-MjRhoA prepared using the recombinant MjRhoA was detected by western blotting. (C) Expression level of MjRhoA in different tissues of normal shrimp. The expression level of MjRhoA in hemocytes and other five tissues (heart, hepatopancreas, gills, stomach, and intestine) were detected by western blot (upper) and qPCR (bottom). In the qPCR results, the relative expression levels in different tissues were normalized by the lowest expression level of MjRhoA in the hepatopancreas. (D) The expression patterns of MjRhoA at different time points post V. anguillarum challenge. Untreated shrimp and the shrimp injected with PBS were used as controls. All the results were from at least three repeated experiments and are shown as the mean ± SD. The significance of the differences was analyzed using Student's t-test. *p < 0.05, **p < 0.01.
MjRhoA Suppresses Bacterial Infection in vivo
To investigate the function of MjRhoA in bacterial infection, overexpression and knockdown of MjRhoA were performed, and the bacterial number in the shrimp was analyzed. For the overexpression, we injected 10 μg rMjRhoA and the His tag into two shrimp groups, separately, before V. anguillarum infection and then the total bacterial number was analyzed. After overexpression of MjRhoA, the number of bacteria in vivo was significantly decreased compared with that in the control (Figure 2A). To confirm the function of MjRhoA, we then performed an RNAi assay. After knockdown of MjRhoA (Figure 2B), we assessed the bacterial number in the hemolymph of the shrimp. The result showed the number of total bacteria in vivo was significantly increased after knockdown of MjRhoA (Figure 2C) compared with that in the control. To analyze the survival rate, shrimp were divided into two groups (shrimp injected with dsMjRhoA as the experimental group and shrimp injected with dsGFP as the control), followed by infection with V. anguillarum. As shown in Figure 2D, after knockdown of MjRhoA expression, the survival rate was markedly decreased compared with that in the dsGFP group (Figure 2D). These results indicated that the small GTPase MjRhoA contributes to resistance to bacterial infection in vivo.
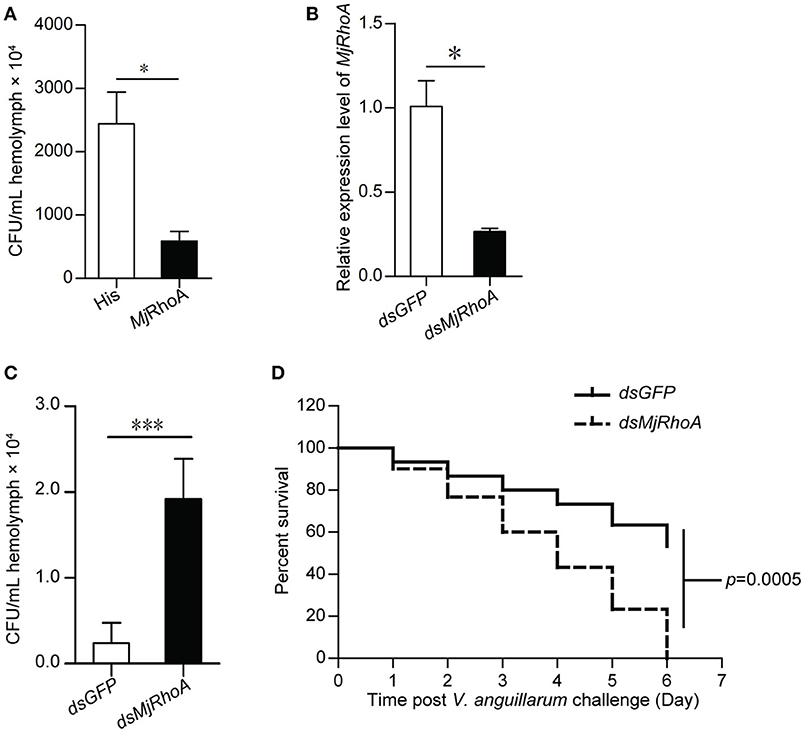
Figure 2. The total bacteria number and survival rate analysis after overexpression and knockdown of MjRhoA. (A) Total bacteria detected after treatment with MjRhoA (“overexpression”) in the shrimp infected with V. anguillarum. After overexpression of MjRhoA, the total bacteria from the whole hemolymph were counted. The bacteria number in the His-tag injection group was used as a control. (B) The efficiency of MjRhoA RNAi. Shrimp were injected with 30 μg of dsMjRhoA and the same amount of dsGFP, respectively. After 48 h, the shrimp hemocytes were collected and the total RNA was extracted for qPCR analysis. Shrimp injected with dsGFP were used as a control. (C) Total bacteria in vivo detecting after knockdown of MjRhoA. The whole hemolymph of shrimp were collected and spread onto the LB solid medium. After growing at room temperature, the total bacteria were counted. The total bacteria number in the dsGFP injected shrimp was used as a control. (D) Survival rate of shrimp infected with V. anguillarum after knockdown of MjRhoA. The survival rate of shrimp injected with dsGFP was used as a control. The data were statistically analyzed using Student's t-test. *p < 0.05, ***p < 0.001.
The Bacterial Clearance Ability of MjRhoA Was Related to Phagocytosis
To analyze the cellular mechanism of MjRhoA's bacterial clearance function, we performed immunocytochemistry in hemocytes from MjRhoA-knockdown shrimp injected with FITC labeled V. anguillarum. The results showed that the amount of the FITC-labeled bacteria in the hemocytes was reduced in MjRhoA-knockdown shrimp compared with that in the control (Figure 3A). The phagocytosis rate and phagocytic index were then analyzed. The results showed that both of the phagocytosis rate and the phagocytic index decreased after knockdown of MjRhoA (Figure 3B). These results suggested that the bacterial clearance ability of MjRhoA was dependent on hemocyte phagocytosis.
Figure 3. MjRhoA bacterial clearance was dependent on phagocytosis. (A) Bacterial phagocytosis of hemocytes was analyzed by immunocytochemistry. The phagocytosis detection with hemocytes from dsGFP-injected shrimp was used as a control. (B) The phagocytosis rate and the phagocytic index calculated after counting hemocytes injected with FITC- V. anguillarum. At least 100 cells were counted and three different views were chosen for statistical analysis. *p < 0.05, **p < 0.01.
V. anguillarum Challenge Affects the Subcellular Location of MjRhoA in Hemocytes
Although the above results indicated that the anti-bacterial function of MjRhoA was related to phagocytosis, the molecular mechanism was still unclear. Rho GTPases family members are recruited to the phagocytic site and converted from their inactive GDP-bound forms to their active GTP-bound forms in the process of phagocytosis (18). We first analyzed the subcellular distribution of MjRhoA in shrimp after bacterial challenge. We found that MjRhoA was located in the cytoplasm under normal conditions, and showed no obvious change in hemocytes at the early stage of V. anguillarum infection (Figures 4A,B). MjRhoA translocated onto the membrane of the hemocytes at 1 h post V. anguillarum challenge, whereas it was still in the cytoplasm in the PBS-treated group (Figures 4A,B). We then analyzed the expression level of genes related to phagocytosis (ROCK2 and Arp2/3). The expression levels of these phagocytosis-related genes were downregulated (Figure 4D) after knockdown of MjRhoA (Figure 4C). The results confirmed that MjRhoA responds to bacterial infection and exerts its anti-bacterial function via phagocytosis.
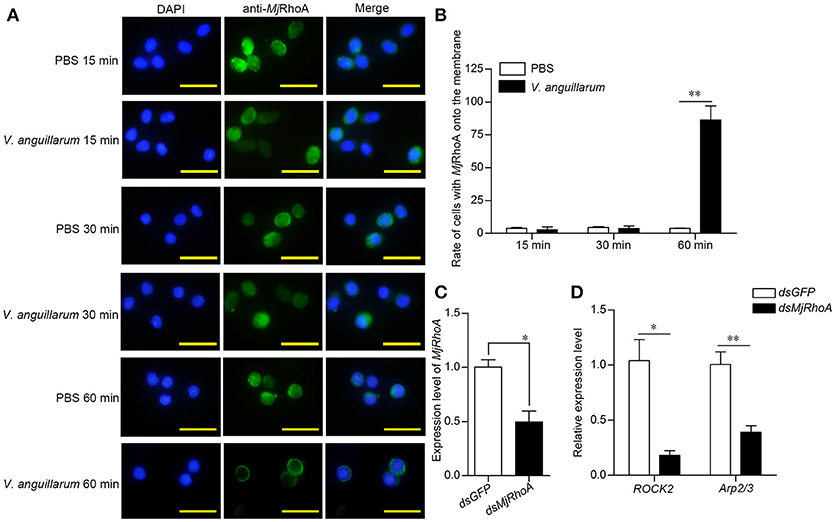
Figure 4. MjRhoA transfers to the cell membrane and regulates phagocytosis related genes after bacterial challenge. (A) The location of native MjRhoA in hemocytes, detected using immunocytochemistry. Normal shrimp were injected with 50 μL V. anguillarum (107 cells per shrimp) and the same volume of PBS for 1 h, separately. The MjRhoA location in vivo was detected using anti-MjRhoA antiserum. The nuclei were stained with DAPI. The location of MjRhoA in PBS-injected shrimp hemocytes was used as a control. Scale bar = 20 μm. (B) The proportion of hemocytes with MjRhoA on the cell membrane among the total cells among shrimp challenged by V. anguillarum at different time points. Shrimp injected with PBS was used as a control. (C) The RNAi efficiency of MjRhoA after knockdown for 48 h. Shrimp injected with dsGFP were used as a control. (D) The expression level of phagocytosis-related genes was detected using qPCR after knockdown of MjRhoA. Two genes, ROCK2 and Arp2/3, were chosen to detect phagocytosis. DsGFP injection was injected as a control. *p < 0.05, **p < 0.01.
MjRhoA Is Required for β-Integrin-Mediated Phagocytosis
ROCK2 and Arp2/3 were reported as two crucial members of the integrin-mediated phagocytosis pathway (19, 20). In addition, RhoA is related to integrin-mediated phagocytosis (21, 22). Thus, we detected the expression level and subcellular location of MjRhoA after knockdown of β-integrin followed by V. anguillarum infection for 60 min. After knockdown of β-integrin (Figure 5A), MjRhoA expression was also decreased (Figure 5B), indicating that β-integrin could regulate the downstream MjRhoA. MjRhoA was located on the cell membrane in the dsGFP-injection shrimp 1 h post V. anguillarum challenge (Figures 5C,c). However, the translocation of MjRhoA to the cell membrane in hemocytes was inhibited after knockdown of β-integrin in shrimp challenged by the bacteria (Figure 5C). These results suggested that MjRhoA participates in β-integrin-dependent phagocytosis of hemocytes against V. anguillarum infection.
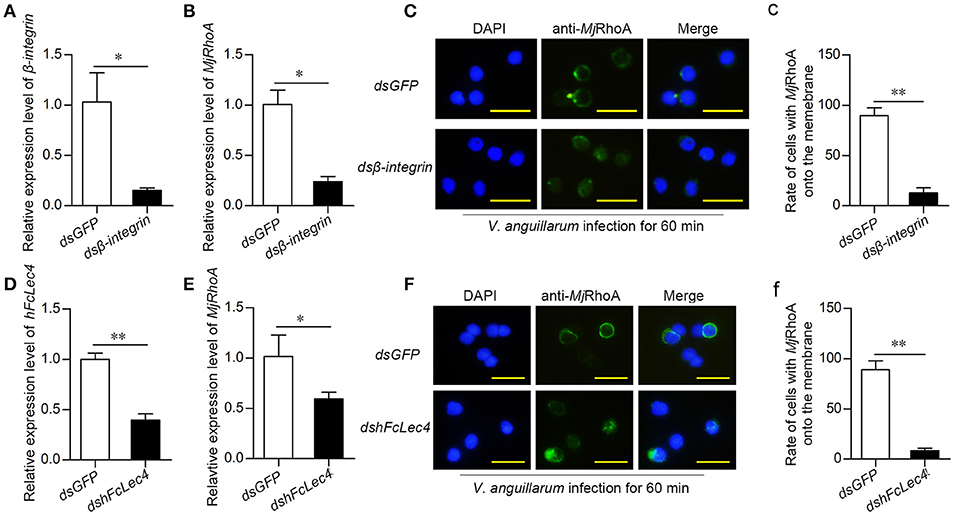
Figure 5. MjRhoA participates in hFcLec4-β-integrin-mediated phagocytosis. (A) The knockdown efficiency after injection of β-integrin dsRNA. Shrimp injected with the same amount of dsGFP (30 μg per shrimp) were used as a control. (B) MjRhoA expression detection after knockdown of β-integrin; dsGFP was used as a control. (C) The subcellular location of MjRhoA analysis by immunocytochemistry assay after knockdown of dsβ-integrin. Normal shrimp were first injected with 30 μg β-integrin dsRNA for 48 h and then injected with V. anguillarum (107 cells per shrimp) for 1 h. The location of MjRhoA in dsGFP-injected shrimp hemocytes was used as a control. Scale bar = 20 μm. (c) The statistical analysis of the cells with MjRhoA located on the membrane after injection of dsβ-integrin; at least three different views were chosen for cell counting; dsGFP injection was used as a control. (D) The knockdown efficiency of hFcLec4. Injection of dsGFP was used as a control. (E) The MjRhoA expression level analyzed by qPCR after knockdown of hFcLec4; dsGFP was used as a control. (F) The subcellular location of MjRhoA after knockdown of hFcLec4; dsGFP was used as a control. (f) Rate of cells with MjRhoA transferred to the cell membrane after knockdown of hFcLec4. *p < 0.05, **p < 0.01.
FcLec4 Homolog Is the Pattern Recognition Receptor in β-Integrin-RhoA-Mediated Phagocytosis
A previous study showed that the C-type lectin, hFcLec4, interacts with β-integrin to promote hemocyte phagocytosis (23). To confirm that MjRhoA participates in hFcLec4-integrin-mediated phagocytosis, we detected the effects of hFcLec4 on the expression level and subcellular location of MjRhoA. The results showed that MjRhoA expression was downregulated after knockdown of hFcLec4 (Figures 5D,E). As expected, the transfer of MjRhoA onto the membrane was also suppressed after knockdown of hFcLec4 (Figures 5F,f). Taken together, our results identified a new hFcLec4-β-integrin-RhoA-mediated phagocytic pathway against bacterial infection. The intracellular hFcLec4 works as a pattern recognition receptor to sense bacterial infection. After binding to infecting bacteria, hFcLec4 interacts with β-integrin and initiates MjRhoA-mediated phagocytosis.
Discussion
The present study found that a member of the Rho GTPases, MjRhoA, participates in hFcLec4-β-integrin-mediated hemocyte phagocytosis to protect the host from infection by V. anguillarum. β-Integrin has been identified to play a role in anti-microbial immunity in Drosophila (24). However, the Drosophila integrin that is involved in bacterial uptake is still unknown. An ortholog of the integrin (BINT2) that has been identified in the mosquito Anopheles gambia is related to phagocytosis because its gene knockout resulted in reduced phagocytosis of E. coli by 70% (25). In the previous study, we identified a C-type Lectin, hFcLec4, which interacts with β-integrin and initiates phagocytosis against bacteria (23). We hypothesized that MjRhoA might be involved in hFcLec4-integrin-mediated phagocytosis. Therefore, we performed RNAi to knock down hFcLec4 and β-integrin expression, and found that the expression level of MjRhoA decreased and subcellular location change of MjRhoA in response to bacterial challenge was inhibited. These results indicated that MjRhoA was involved in the phagocytotic pathway and located in the downstream section.
RNA interference is now a common method of choice for researchers doing for loss of function studies for its ease, speed, and specificity. It is worth mentioning that the knockdown efficiency is sometimes related to different genes: some genes are easily knockdown at low dosage of dsRNA, but some other genes are not even using high dosage of dsRNA. Thus, the injection dosage of dsRNA depends on different situations. For example, 120 μg of dsPl-TEP was injected into crayfish (20 ± 2 g) for the RNAi of Pl-TEP (26); in Penaeus monodon, the dosage of PmTBC1D20 dsRNA was 10 μg/g shrimp used in the RNAi assay of PmTBC1D20 (27). Here in our study, we injected 30 μg dsRNA/shrimp to get the optimal RNAi efficiency.
There are many reports in vertebrates about RhoA functions in cell morphology. In multiciliated epithelial cells (MCCs), RhoA regulates actin assembly and functions in the apical emergence of MCCs (28). In endothelial cells, the activation of local RhoA regulates the F-actin structure formation (29). In addition, there is a kind of synergistic effect between RhoA and actomyosin in regulating the cell-cell junction and functions (30). In vertebrates, the major functions of RhoA are regulating the actin cytoskeleton (31), stress fiber formation (32), and cell polarization (33). In the present study, we found that knockdown of MjRhoA inhibited the phagocytosis rate of shrimp hemocytes after injection of V. anguillarum. At the same time, the expression levels of phagocytosis-related genes (ROCK2 and Arp2/3) were also downregulated. These results indicated that MjRhoA suppresses bacterial infection in a phagocytosis-related manner. Coincidently, many small GTPases participate in innate immunity in the same manner. Multiple members of the Rab family function in the phagocytosis (34); Rab6 rearranges actin to regulate phagocytosis and protect kuruma shrimp M. japonicus from WSSV infection in Ye et al. (35). As a feedback regulation gene (36), Ran interacts with myosin and regulates hemocytic phagocytosis (37). Ran GTPases function as a kind of immunostimulant target that affects phagocytosis against WSSV infection (38). However, all above studies have not referred to the initial step of the phagocytosis, how infected pathogens are recognized? In this study, we provided data for connection of pathogen recognition and phagocytosis in immune response against bacterial infection.
A previous study reported two types of phagocytosis regulated by Rho GTPases (39): Fc gamma receptor (Fcγ)-mediated phagocytosis and integrin-mediated phagocytosis. The first type is regulated by Cdc42 and Rac (40), and another small GTPase, ARF6, is necessary for this type of phagocytosis (41). For integrin-mediated phagocytosis, RhoA plays a crucial role in the pathway and functions in actin polymerization (42). Electron microscopy observation showed that integrin-mediated phagocytosis is not associated with the formation of membrane protrusions (22). Phagocytosis is regulated by RhoA activity, in which activated RhoA is recruited to the site of phagocytosis, and particles that are bound to integrin sink into the cells without generating major protrusions (21). Our results showed that MjRhoA was transferred onto the cytomembrane after V. anguillarum challenge, suggesting that a GTP-bound active form of MjRhoA should take part in this type of phagocytosis. After being activated by the upstream integrin, RhoA regulates actin remodeling and then initiates phagosome formation via the downstream effectors, ROCK and Arp2/3 (22). ROCK activity mediates the local recruitment of the actin-branching protein Arp2/3 and the activation of myosin IIA underneath the bound particles (43). As two markers of the β-integrin-mediated phagocytosis pathway (19, 20, 44), the expression levels of ROCK2 and Arp2/3 were affected by changes in MjRhoA levels in the present study (Figure 4D).
In summary, our study found that RhoA participates in hFcLec4-β-integrin-mediated phagocytosis, which protects the host against bacterial infection (Figure 6). Our study identified a new hFcLec4-β-integrin-RhoA mediated phagocytic pathway against bacterial infection in shrimp innate immunity.
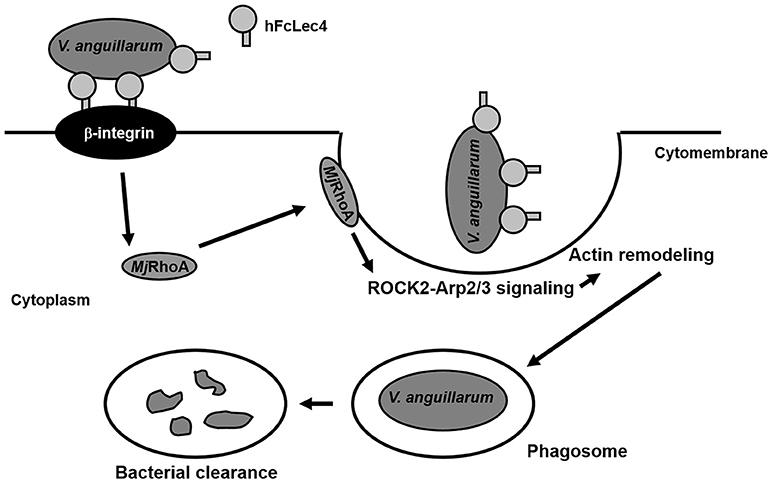
Figure 6. MjRhoA participates in hFcLec4-β-integrin-mediated phagocytosis in shrimp hemocytes during bacterial infection. FcLec4 homolog serves as a pattern recognition receptor (PRR) and recognizes the infecting bacteria. After binding to the bacterial surface, hFcLec4 activates β-integrin signaling and regulates the expression and subcellular location of downstream MjRhoA. Activated MjRhoA is recruited onto the cytomembrane and regulates actin remodeling through the downstream effectors, ROCK2 and Arp2/3. Actin remodeling triggers cell morphological changes and phagosome formation to destroy the bacteria.
Author Contributions
J-XW, X-FZ, and J-DX conceived and designed the experiments. J-DX and J-XW wrote the manuscript. J-DX performed the majority of the experiments. J-DX, J-XW, X-FZ, X-WW, G-JN, and M-QD contributed experimental suggestions and revised the manuscript.
Funding
This work was finically supported by the National Natural Science Foundation of China (grants 31630084 and 31472303).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.01928/full#supplementary-material
Figure S1. The full-length nucleotide sequence of MjRhoA cDNA and predicted amino acid sequence of MjRhoA.
Figure S2. Multiple sequence alignment and phylogenetic tree of MjRhoA. (A) Multiple sequence alignment of MjRhoA with RhoAs of 19 other species: Acanthaster planci, Callorhinchus milii, Ciona intestinalis, Danaus plexippus, Danio rerio, Drosophila melanogaste, Felis catus, Gallus gallus, Homo sapiens, Meleagris gallopavo, Mus musculus, Myotis lucifugus, Ovis aries, Papilio xuthus, Poeciliopsis prolifica, Strongylocentrotus intermedius, Sus scrofa, Tigriopus japonicus, Xenopus laevis. (B) Phylogenetic tree of MjRhoA with RhoA amino acid sequences of 19 other species.
References
1. Johnson DS, Chen YH. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol. (2012) 12:458–63. doi: 10.1016/j.coph.2012.02.003
2. Kahn RA, Der CJ, Bokoch GM. The ras superfamily of GTP-binding proteins: guidelines on nomenclature. FASEB J (1992) 6:2512–3.
3. Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature (2002) 420:629–35. doi: 10.1038/nature01148
5. Chu JY, Dransfield I, Rossi AG, Vermeren S. Non-canonical PI3K-Cdc42-Pak-Mek-Erk signaling promotes immune-complex-induced apoptosis in human neutrophils. Cell Rep. (2016) 17:374–86. doi: 10.1016/j.celrep.2016.09.006
6. Rincon SA, Estravis M, Perez P. Cdc42 regulates polarized growth and cell integrity in fission yeast. Biochem Soc Trans. (2014) 42:201–5. doi: 10.1042/BST20130155
7. Burbage M, Keppler SJ, Montaner B, Mattila PK, Batista FD. The small Rho GTPase TC10 modulates B cell immune responses. J Immunol. (2017) 199:1682–95. doi: 10.4049/jimmunol.1602167
8. Gallo G.Letourneau PC. Regulation of growth cone actin filaments by guidance cues. J Neurobiol. (2004) 58:92–102. doi: 10.1002/neu.10282
9. Teramoto H, Malek RL, Behbahani B, Castellone MD, Lee NH, Gutkind JS. Identification of H-Ras, RhoA, Rac1 and Cdc42 responsive genes. Oncogene (2003) 22:2689–97 doi: 10.1038/sj.onc.1206364
10. Mathur J, Hulskamp M. Signal transduction: Rho-like proteins in plants. Curr Biol. (2002) 12:R526–8. doi: 10.1016/S0960-9822(02)01029-1
11. Arbibe L, Mira JP, Teusch N, Kline L, Guha M, Mackman N, et al. Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat Immunol. (2000) 1:533–40. doi: 10.1038/82797
12. Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, et al: Local translation of RhoA regulates growth cone collapse. Nature (2005) 436:1020–24. doi: 10.1038/nature03885
13. Singh V, Davidson AC, Hume PJ, Humphreys D, Koronakis V. Arf GTPase interplay with Rho GTPases in regulation of the actin cytoskeleton. Small GTPases (2017) 19:1−8. doi: 10.1080/21541248.2017.1329691
14. Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer (2002) 87:635–44. doi: 10.1038/sj.bjc.6600510
15. O'Connor K, Chen M. Dynamic functions of RhoA in tumor cell migration and invasion. Small GTPases (2013) 4:141–7. doi: 10.4161/sgtp.25131
16. Chi Y, Li F, Sun Y, Wen R, Li S. Expression and function analysis of Rac1 homolog in Chinese shrimp Fenneropenaeus chinensis. Fish Shellfish Immunol. (2013) 35:927–32. doi: 10.1016/j.fsi.2013.07.006
17. Xu JD, Jiang HS, Wei TD, Zhang KY, Wang XW, Zhao XF, et al. Interaction of the Small GTPase Cdc42 with arginine kinase restricts white spot syndrome virus in shrimp. J Virol. (2017) 91. doi: 10.1128/JVI.01916-16
18. Chimini G, Chavrier P. Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nat Cell Biol. (2000) 2:E191–6. doi: 10.1038/35036454
19. Marek I, Becker R, Fahlbusch FB, Menendez-Castro C, Rascher W, Daniel C, et al. Expression of the Alpha8 integrin chain facilitates phagocytosis by renal mesangial cells. Cell Physiol Biochem. (2018) 45:2161–173. doi: 10.1159/000488160
20. Rotty JD, Brighton HE, Craig SL, Asokan SB, Cheng N, Ting JP, et al. Arp2/3 complex is required for macrophage integrin functions but is dispensable for FcR phagocytosis and in vivo motility. Dev Cell (2017) 42:498–513. doi: 10.1016/j.devcel.2017.08.003
21. Wiedemann A, Patel JC, Lim J, Tsun A, van Kooyk Y, Caron E. Two distinct cytoplasmic regions of the beta2 integrin chain regulate RhoA function during phagocytosis. J Cell Biol. (2006) 172:1069–79. doi: 10.1083/jcb.200508075
22. Dupuy AG, Caron E. Integrin-dependent phagocytosis: spreading from microadhesion to new concepts. J Cell Sci. (2008) 121:1773–83. doi: 10.1242/jcs.018036
23. Wang XW, Zhao XF, Wang JX. C-type lectin binds to beta-integrin to promote hemocytic phagocytosis in an invertebrate. J Biol Chem. (2014) 289:2405–14. doi: 10.1074/jbc.M113.528885
24. Foukas LC, Katsoulas HL, Paraskevopoulou N, Metheniti A, Lambropoulou M, Marmaras VJ. Phagocytosis of Escherichia coli by insect hemocytes requires both activation of the Ras/mitogen-activated protein kinase signal transduction pathway for attachment and beta3 integrin for internalization. J Biol Chem. (1998) 273:14813–8.
25. Moita LF, Wang-Sattler R, Michel K, Zimmermann T, Blandin S, Levashina EA, et al. Kafatos: in vivo identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity (2005) 23:65–73. doi: 10.1016/j.immuni.2005.05.006
26. Wu C, Noonin C, Jiravanichpaisal P, Soderhall I, Soderhall K. An insect TEP in a crustacean is specific for cuticular tissues and involved in intestinal defense. Insect Biochem Mol Biol. (2012) 42:71–80. doi: 10.1016/j.ibmb.2011.10.006
27. Yingvilasprasert W, Supungul P, Tassanakajon A. PmTBC1D20, a Rab GTPase-activating protein from the black tiger shrimp, Penaeus monodon, is involved in white spot syndrome virus infection. Dev Comp Immunol. (2014) 42:302–10. doi: 10.1016/j.dci.2013.09.008
28. Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB. RhoA regulates actin network dynamics during apical surface emergence in multiciliated epithelial cells. J Cell Sci. (2017) 130:420–28. doi: 10.1242/jcs.194704
29. Heemskerk N, Schimmel L, Oort C, van Rijssel J, Yin T, Ma B, et al. F-actin-rich contractile endothelial pores prevent vascular leakage during leukocyte diapedesis through local RhoA signalling. Nat Commun. (2016) 7:10493. doi: 10.1038/ncomms10493
30. Arnold TR, Stephenson RE, Miller AL. Rho GTPases and actomyosin: partners in regulating epithelial cell-cell junction structure and function. Exp Cell Res. (2017) 358:20–30. doi: 10.1016/j.yexcr.2017.03.053
31. Girouard MP, Pool M, Alchini R, Rambaldi I, Fournier AE. RhoA proteolysis regulates the actin cytoskeleton in response to oxidative stress. PLoS ONE (2016) 11:e0168641. doi: 10.1371/journal.pone.0168641
32. Oakes PW, Wagner E, Brand CA, Probst D, Linke M, Schwarz US, et al. Gardel: optogenetic control of RhoA reveals zyxin-mediated elasticity of stress fibres. Nat Commun. (2017) 8:15817. doi: 10.1038/ncomms15817
33. Datta A, Sandilands E, Mostov KE, Bryant DM. Fibroblast-derived HGF drives acinar lung cancer cell polarization through integrin-dependent RhoA-ROCK1 inhibition. Cell Signal. (2017) 40:91–8. doi: 10.1016/j.cellsig.2017.09.001
34. Yeo JC, Wall AA, Luo L, Stow JL. Sequential recruitment of Rab GTPases during early stages of phagocytosis. Cell Logist. (2016) 6:e1140615. doi: 10.1080/21592799.2016.1140615
35. Ye T, Tang W, Zhang X. Involvement of Rab6 in the regulation of phagocytosis against virus infection in invertebrates. J Proteome Res. (2012) 11:4834–46. doi: 10.1021/pr300274k
36. Zhao Z, Wang J, Zhang X. Feedback regulation of Ran gene expression by Ran protein. Gene (2011) 485:85–90. doi: 10.1016/j.gene.2011.06.008
37. Liu W, Han F, Zhang X. Ran GTPase regulates hemocytic phagocytosis of shrimp by interaction with myosin. J Proteome Res. (2009) 8:1198–206. doi: 10.1021/pr800840x
38. Zhao Z, Jiang C, Zhang X. Effects of immunostimulants targeting Ran GTPase on phagocytosis against virus infection in shrimp. Fish Shellfish Immunol. (2011) 31:1013–8. doi: 10.1016/j.fsi.2011.08.022
39. Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science (1998) 282:1717–21.
40. Cougoule C, Hoshino S, Dart A, Lim J, Caron E. Dissociation of recruitment and activation of the small G-protein Rac during Fcgamma receptor-mediated phagocytosis. J Biol Chem. (2006) 281:8756–64. doi: 10.1074/jbc.M513731200
42. Werner E. Integrin clustering drives phagocytosis coupled to collagenase 1 induction through RhoA GTPase and superoxide production. Antioxid Redox Signal. (2005) 7:318–26. doi: 10.1089/ars.2005.7.318
43. Olazabal IM, Caron E, May RC, Schilling K, Knecht DA, Machesky LM. Rho-kinase and myosin-II control phagocytic cup formation during CR, but not FcgammaR, phagocytosis. Curr Biol. (2002) 12:1413–18. doi: 10.1016/S0960-9822(02)01069-2
Keywords: RhoA, innate immunity, integrin dependent phagocytosis, antibacterial cellular immunity, Marsupenaeus japonicus
Citation: Xu J-D, Diao M-Q, Niu G-J, Wang X-W, Zhao X-F and Wang J-X (2018) A Small GTPase, RhoA, Inhibits Bacterial Infection Through Integrin Mediated Phagocytosis in Invertebrates. Front. Immunol. 9:1928. doi: 10.3389/fimmu.2018.01928
Received: 16 May 2018; Accepted: 06 August 2018;
Published: 30 August 2018.
Edited by:
Chu-Fang Lo, National Cheng Kung University, TaiwanReviewed by:
Anchalee Tassanakajon, Chulalongkorn University, ThailandArun K. Dhar, University of Arizona, United States
Copyright © 2018 Xu, Diao, Niu, Wang, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Xing Wang, anh3YW5nQHNkdS5lZHUuY24=
 Ji-Dong Xu1
Ji-Dong Xu1 Xian-Wei Wang
Xian-Wei Wang Xiao-Fan Zhao
Xiao-Fan Zhao Jin-Xing Wang
Jin-Xing Wang