- Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, United States
Extraordinary genetic diversity is a hallmark of hepatitis C virus (HCV). Therefore, accurate measurement of the breadth of antibody neutralizing activity across diverse HCV isolates is key to defining correlates of immune protection against the virus, and essential to guide vaccine development. Panels of HCV pseudoparticle (HCVpp) or replication-competent cell culture viruses (HCVcc) can be used to measure neutralizing breadth of antibodies. These in vitro assays have been used to define neutralizing breadth of antibodies in serum, to characterize broadly neutralizing monoclonal antibodies, and to identify mechanisms of HCV resistance to antibody neutralization. Recently, larger and more diverse panels of both HCVpp and HCVcc have been described that better represent the diversity of circulating HCV strains, but further work is needed to expand and standardize these neutralization panels.
Hepatitis C Virus (HCV) Viral Diversity and the Neutralizing Antibody Response
Interferon-free therapies for HCV have revolutionized treatment of those infected with the virus, but a vaccine to prevent infection is needed (1). The extensive genetic diversity of HCV has been a major obstacle to vaccine development. HCV is genetically heterogeneous with seven genotypes and more than 80 subtypes (Figure 1). Within the envelope genes (E1 and E2), approximately 30% of amino acids differ between strains from different genotypes, while strains from different subtypes within each genotype differ at approximately 20% of their E1E2 amino acids (2–5). Even viral strains within the same subtype differ at up to 10% of their E1E2 amino acids. Within an infected individual, error-prone replication by HCV and immune selection lead to the generation of a viral swarm made up of many distinct strains, providing opportunities for expansion of antibody resistant variants (6–8). Therefore, induction of high-titer cross reactive antibodies that are capable of neutralizing diverse viruses within subtypes and across multiple genotypes may be required for an effective vaccine.
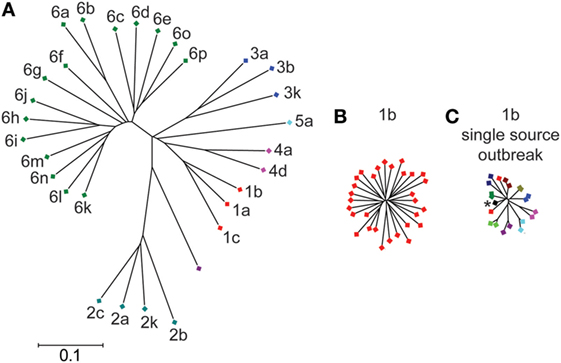
Figure 1. Genetic diversity of hepatitis C virus (HCV) E1E2 across subtypes and within a single subtype. Phylogenetic trees of E1E2 amino acid sequences. E1E2 amino acid sequences from multiple subtypes (A) or a single subtype (B) were downloaded from the LANL HCV sequence database. Not all known subtypes are shown. (C) E1E2 amino acid sequences of 10 individuals from the Irish anti-D cohort, who were all infected from the same inoculum (asterisk) between 1977 and 1978 (9). Sequences of virus from each individual are indicated with a different color. Trees were inferred using the Neighbor-joining method, with branch lengths drawn to scale. Subtypes are labeled. All three trees are on the same scale. Distances were computed using the JTT method. Analyses were performed in MEGA7 (10).
Significant effort has been devoted to development of vaccines intended to induce protective antibodies against HCV, but most vaccines tested to date in nonhuman primates or humans have induced humoral responses with relatively poor activity against heterologous HCV strains [reviewed in Ref. (11, 12)]. For example, a vaccine composed of recombinant full-length E1E2 protein from a single HCV strain has been tested in both chimpanzees and humans. Vaccination was protective against homologous HCV challenge in chimpanzees (12). However, the same vaccine reduced rates of persistence but did not prevent infection in chimpanzees after challenge with a heterologous virus (13), and neutralizing antibodies against heterologous HCV strains were induced in only three of 16 vaccines in a phase 1a human trial (14). Overall, these and other vaccine trials suggest that induction of antibodies with broad neutralizing activity across diverse HCV strains may be a critical challenge for HCV vaccine development.
Neutralizing Antibody Mediated Protection Against HCV Infection
Antibodies capable of neutralizing HCV infection (NAbs) target the viral envelope glycoproteins, E1 and E2, which are membrane-anchored proteins believed to form a heterodimer on the surface of viral particles. E2 interacts with multiple cell surface receptors, including but not limited to CD81 and scavenger receptor B1 (SR-B1), while the function of E1 remains unclear (15). Most HCV-infected individuals develop strain-specific NAbs against hypervariable region 1 (HVR1), a 27-amino acid region at the amino-terminus of E2, but viral escape mutations generally confer resistance to these antibodies (6, 16–20). In contrast, NAbs capable of neutralizing infection by multiple diverse HCV strains in vitro, commonly called broadly neutralizing antibodies (bNAbs), develop in some infected individuals. These bNAbs generally bind to relatively conserved epitopes at the CD81 receptor binding site of E2 (CD81bs) or to the E1E2 heterodimer [reviewed in Ref. (21, 22)].
There is evidence from studies of HCV infection of humans and animal models that bNAbs can be protective. Notably, early-developing HCV-specific bNAb responses are associated with clearance of primary human HCV infection (23–26). Pestka et al. demonstrated that women infected during a single-source outbreak of HCV were more likely to clear the virus if they developed serum antibodies early in infection that were capable of neutralizing at least one heterologous HCV strain in vitro. Similarly, Osburn et al. demonstrated that plasma isolated immediately prior to clearance of HCV infection neutralized a median of 6 of 19 heterologous HCV strains, while acute infection plasma of control subjects with subsequent persistence of infection neutralized a median of only 1 of 19 heterologous strains. In addition, individuals who clear one infection clear subsequent infections more than 80% of the time, and clearance of reinfection is associated with rapid induction of antibodies capable of neutralizing heterologous HCV strains in vitro (27).
There is also evidence of protection in animal models of HCV infection. Infusion of immunoglobulin isolated from the serum of a chronically infected human prior to challenge with homologous virus from the same donor prevented infection of most human liver chimeric mice. Infusion of this chronic-phase human immunoglobulin prior to challenge of a chimpanzee prevented infection with homologous, but not heterologous HCV strains (28–30). In contrast, infusion of monoclonal bNAbs prior to challenge with heterologous viruses prevented infection in most human liver chimeric mice (31–33) and chimpanzees (34), and combinations of monoclonal bNAbs also abrogated established HCV infection in a human liver chimeric mice (35). Together, these studies demonstrate that induction of bNAbs may be necessary to prevent infection by diverse, heterologous HCV strains.
Experimental Systems for Quantitation of Antibody Neutralizing Activity
Assessment of antibody neutralization of HCV relies largely upon two different in vitro systems: the HCV pseudoparticle (HCVpp) system and the replication-competent HCV cell culture (HCVcc) system. HCVpp are retroviral particles with HCV envelope glycoproteins (E1 and E2) on their surface. These particles can be produced by transfection of HEK-293T cells with an E1E2-expressing plasmid and a plasmid expressing an envelope-defective HIV-1 genome with a luciferase reporter. Alternatively, cells can be transfected with an E1E2-expressing plasmid, a murine leukemia virus (MLV) Gag/Pol packaging construct, and a luciferase-encoding reporter plasmid. In either case, after transfection, enveloped particles bearing HCV E1E2 proteins on their surface are released through a retroviral budding process into culture supernatant, enabling the measurement of single rounds of viral entry into hepatoma cells or primary human hepatocytes. HCVpps were used to identify many of the cell surface receptors required for HCV entry (36–40). Unlike HCVpp, HCVcc reproduce the full replication cycle of HCV in vitro (41–45) and in animal models (46–48). HCVccs are produced through transfection of in vitro transcribed full-genomic HCV RNA into hepatoma cells. Relatively few HCV strains are capable of replication in in vitro, but recently strains from genotypes 1–3 have been adapted to allow in vitro replication (49–53). After initial transfection, culture supernatants are infectious and can be passaged serially in vitro. The HCVcc system has been used to further define the viral entry pathway (54–57).
Both HCVpp and HCVcc panels have been developed to express diverse E1E2 strains, including strains from genotypes 1–6 (HCVpp) (58, 59) and genotypes 1–7 (chimeric HCVcc) (42, 60), enabling the assessment of breadth of antibody neutralizing activity. Notably, HCVpps were used in studies described earlier in this review which demonstrated that the early development of bNAbs was associated with clearance of HCV infection (23–26). HCVccs were used in the studies described earlier which demonstrated that antibodies with broad in vitro neutralizing activity can prevent HCV infection in animal models (31–34).
The development of diverse panels of HCVpp or HCVcc has been complicated by the relatively high frequency with which primary E1E2 isolates are poorly functional or nonfunctional in vitro, producing HCVpp that do not mediate detectable entry into hepatoma cells, or chimeric HCVcc that do not replicate in in vitro (61, 62). Recently, efforts have been made to address these technical obstacles. E1E2 clones that are poorly functional when pseudotyped with HIV-1 Gag protein may produce functional HCVpp when pseudotyped with MLV Gag protein, or vice versa. The mechanisms explaining this phenomenon are unknown. In addition to pseudotyping with either HIV-1 or MLV, optimization of ratios of MLV and HCV E1E2 plasmids during transfection can also improve the function of some E1E2 strains in HCVpp (63). Along with technical improvements in HCVpp production, technology for production of HCVcc has also advanced. The development of chimeric HCVcc based on the genotype 2a JFH1 strain with Core-NS2 genes from multiple genotypes has expanded the number of diverse HCVcc available for neutralization studies. These chimeric HCVccs have an advantage relative to some full-length HCVcc strains, since adaptive mutations required for efficient replication in vitro are outside of E1E2 in most cases. Therefore, the E1E2 genes expressed accurately represent the sequences of naturally circulating viruses. Two recent studies described cloning of dozens of naturally occurring E1E2 genes into HCVcc chimeras, generating replication-competent viruses (61, 64). Wasilewski et al. used an E1E2-deleted H77/JFH-1 chimeric construct with an introduced restriction site to facilitate high-throughput insertion of diverse E1E2 genes.
As more E1E2-matched HCVpp and HCVcc are produced, some phenotypic differences between E1E2 expressed using the two systems have been identified. For reasons that are still unknown, some E1E2 strains that produce replication-competent HCVcc chimeras produce poorly functional HCVpp, and, conversely, some E1E2 strains that are functional in HCVpp do not produce replication-competent HCVcc chimeras (62). In addition, HCVpp tend to be generally more neutralization sensitive than HCVcc (61, 62), perhaps due to structural differences between HCVcc and HCVpp, including the association of apolipoproteins with HCVcc but not HCVpp (65, 66). Fortunately, despite these differences, multiple studies have demonstrated concordance between the neutralization results of identical E1E2 clones expressed in either HCVpp or HCVcc, including concordance in the rank order of neutralization sensitivity of different E1E2 strains and concordance in resistance phenotypes of specific mutations (61, 62, 64, 67). These results suggest that, for most experiments, either HCVpp or HCVcc can be used to measure antibody neutralizing activity, expanding the diversity of E1E2 isolates available for neutralization assays.
HCV Panels Used to Quantitate Neutralizing Breadth
Defining the neutralizing breadth of anti-HCV monoclonal antibodies and immune sera is critical to understand the antibody response required to protect against circulating HCV strains. Due to the previously limited availability of diverse E1E2 isolates in HCVcc and HCVpp, the neutralizing breadth of anti-HCV antibodies has traditionally been measured using relatively small panels of HCVcc or HCVpp (31, 36, 42, 59, 68, 69). As described earlier in this review, these smaller panels have greatly advanced understanding of virus neutralization, but they do not express many of the polymorphisms common in natural HCV isolates, and, therefore, they may overestimate neutralizing breadth of antibodies or sera. In addition, many of these studies have measured neutralization of historically important reference strains of HCV (31, 36, 42, 59, 68–70), such as the genotype 1a isolate, H77, which is highly sensitive to neutralization (69).
Recently, panels of HCVpp and HCVcc have been expanded to encompass more of the diversity of circulating HCV strains (Table 1). A panel of HCVpp from the University of Nottingham includes 78 HCVpp predominantly from genotype 1, but also including some isolates from genotypes 2–6 (61). A second panel developed at Johns Hopkins University comprises natural genotype 1a and 1b E1E2 isolates from cohorts in the US and Ireland. This panel includes 113 HCVpp and expresses 97% of amino acid polymorphisms present at greater than 5% frequency in a reference set of 643 genotype 1 HCV isolates submitted to GenBank from around the world (71). Sensitivity of HCVpp in this panel to neutralization by two bNAbs varied by more than 100-fold (67). A 19-HCVpp subset of this panel expresses 94% of amino acid polymorphisms present at greater than 5% frequency in worldwide genotype 1 sequence (72). This 19 HCVpp genotype 1 panel has been used extensively to define neutralizing breadth of mAbs and sera (9, 23, 27, 62, 64, 72, 73). While significantly fewer replication-competent HCVcc strains are available, progress has been made in expanding the number of isolates (61–63, 74). Most recently, Carlsen et al. applied a panel of chimeric HCVcc constructs expressing 16 E1E2 genes from genotypes 1–3. Like the diverse HCVpp in the Nottingham and Hopkins panels, these HCVcc also varied widely in sensitivity to neutralization by monoclonal antibodies (74).
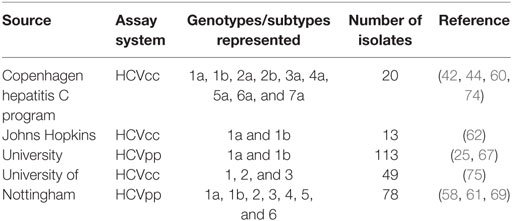
Table 1. HCV cell culture (HCVcc) and HCV pseudoparticle (HCVpp) panels for neutralization breadth testing.
Neutralization Sensitivity Varies by HCV Strain, not by Genotype
Heterologous neutralizing activity of HCV-infected human sera is not primarily dictated by the infecting viral genotype. Some studies demonstrated that sera from genotype 1 or 4-infected individuals neutralized genotype 1, 4, 5, and 6 viruses more efficiently than viruses from genotypes 2 and 3 (59, 76). However, these findings may be attributable to strain-specific, rather than genotype-specific effects, since IgG isolated in a different study from serum of subjects infected with either genotype 1, 2, or 3 viruses showed similar neutralization of genotype 1, 2, and 3 HCVpp. There was no relationship between infecting genotype and the genotype of HCVpp that was neutralized by each sample (69). Similarly, in a third study, neutralizing breadth of plasma samples measured using a diverse panel of genotype 1 HCVpp did not differ between individuals infected with genotype 1, 2, or 3 viruses (25). Together, these studies suggest that genetic subtypes of HCV are not neutralization serotypes.
Several recent studies have also demonstrated that neutralization sensitivity varies by HCV strain, rather than by genotype. In a study by Carlsen et al., which measured neutralization of 16 HCVcc from genotypes 1–3 by 10 mAbs, 50% inhibitory concentrations (IC50) of the same mAb across different HCVcc isolates varied by more than 1,000-fold, with many of the mAbs failing to neutralize all strains in the panel. Notably, this variation in neutralization sensitivity was discernable between isolates from the same subtype as well as between isolates from different genotypes (74). Another study by Urbanowicz et al. measured neutralization of more than 70 HCVpp from genotypes 1–6 by 5 monoclonal antibodies, and also observed wide variation in neutralization sensitivity (61). Again, this neutralization sensitivity varied widely across isolates within each of the subtypes tested. Notably, neutralizing breadth of 7 mAbs was similar when measured in 2 independent studies using either 16 genotype 1–3 HCVcc from the Copenhagen Hepatitis C Program panel (74) or the Johns Hopkins University panel of 19 genotype 1 HCVpp (72), further supporting the concept that neutralization sensitivity to many bNAbs varies by HCV strain, rather than by genotype (Figure 2). Together, these studies suggest that it may be most important for neutralization panels to include many diverse E1E2 strains, even if they represent a single genotype, rather than fewer strains from multiple genotypes.
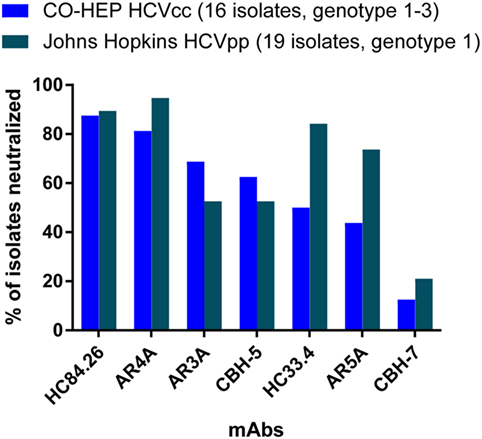
Figure 2. Neutralizing breadth of seven monoclonal antibodies measured using HCV cell culture (HCVcc) and HCV pseudoparticle (HCVpp) panels. Values shown are the percentage of 16 genotype 1–3 HCVcc isolates from the Copenhagen Hepatitis C Program (CO-HEP) neutralized with an IC50 less than 10 µg/mL (74), or the percentage of 19 genotype 1 HCVpp isolates from Johns Hopkins University neutralized more than 50% by 10 µg/mL of mAb (72).
Resistance is Conferred by Polymorphisms within and Distant from BNAB Epitopes
Studies using genetically diverse HCVpp and HCVcc have identified multiple mechanisms of HCV resistance to bNAbs. Polymorphisms have been identified in primary HCV isolates that confer resistance to many individual bNAbs and groups of bNAbs. Some bNAb resistance mutations fall within identified bNAb epitopes. When given monotherapy with the monoclonal bNAb designated HCV1, HCV-infected liver transplant recipients developed neutralization resistance mutations at polyprotein positions 415 and 417, within the HCV1 binding epitope (77). Similarly, a naturally occurring F442I polymorphism within the binding epitope of another bNAb, designated HC84.26, confers resistance to neutralization by that bNAb (72). Notably, however, polymorphisms distant from bNAb epitopes can also confer resistance to neutralization. Several studies have demonstrated no association between the level of bNAb resistance of natural HCV strains and mutations within the epitopes of those bNAbs (25, 72). Other studies have identified few mutations in bNAb epitopes in HCV-infected individuals (78), even though bNAbs commonly develop during chronic infection. This observation may be explained in part by the discovery that mutations outside of bNAb epitopes are capable of conferring bNAb resistance (67, 72, 79). Mutations in the central beta sheet of E2, distant from known binding epitopes, can confer resistance to antibodies binding at the CD81 binding site of E2 (72, 79), and a mutation in HVR1 that modulates E2-scavenger receptor-B1 interaction confers resistance to bNAbs AR4A and HC33.4, even if their binding epitopes are fully conserved (67). Prentoe et al. also demonstrate the role of HVR1 in modulating sensitivity of different HCV strains to bNAbs targeting conserved epitopes (80). Although bNAbs in the study did not bind to HVR1, deletion of HVR1 reduced differences in bNAb sensitivity between many HCV strains. Together, these studies suggest that neutralizing breadth of an antibody cannot be predicted solely from the level of conservation of the binding epitope of that antibody.
Accurate Assessment of Neutralizing Breadth Informs Vaccine Development
Given the extraordinary genetic and phenotypic diversity of HCV, accurate measurement of antibody neutralizing breadth is critical to guide vaccine development. Some studies have suggested that very high titers of bNAbs, as measured using in vitro neutralization assays, may be necessary for protection in in vivo. Bukh et al. demonstrated that IgG with high cross-neutralizing titers in vitro protected chimpanzees against homologous, but not heterologous challenge with HCVcc expressing E1E2 strains identical to those used for in vitro testing (28). In addition, measurement of neutralizing breadth of bNAbs using diverse HCVpp and HCVcc panels has shown that no bNAb or serum sample identified to date potently neutralizing all HCV strains (25, 61, 72, 74). These studies suggest that vaccine induction of bNAbs targeting a single HCV epitope may be insufficient for protection. Importantly, neutralizing breadth of bNAb combinations can exceed that of individual bNAbs. In one study, multiple bNAbs, including AR3A and HC84.26, displayed enhanced neutralization against multiple HCV strains when combined with a second bNAb, designated AR4A (74). In another study, bNAbs targeting distinct epitopes displayed enhanced neutralizing breadth when used in combination (73). Two NAbs described in the Mankowski et al. study, designated HEPC74 and HEPC98, displayed both enhanced neutralizing breadth and enhanced potency (synergy) when used in combination. HEPC74 binds at the CD81 binding site of E2 and acts primarily by blocking E2 binding to the CD81 cell surface receptor. In contrast, HEPC98 binds to HVR1 of E2 and acts primarily by blocking E2 binding to the co-receptor SR-B1. Together, these studies suggest that vaccine induction of multiple bNAbs targeting distinct epitopes may be desirable.
Future Directions
Diverse panels of HCVpp and HCVcc have been used to identify bNAbs, to define neutralizing breadth of antibodies in serum, and to identify mechanisms of HCV resistance to antibody neutralization. However, methods used to define neutralizing breadth can be improved. The vast genetic diversity of HCV is well-described, but the phenotypic diversity of the virus as it relates to neutralization sensitivity remains incompletely understood. Recently, larger and more diverse panels of both HCVpp and HCVcc have been described to better represent the diversity of circulating HCV strains. Currently, available genotype 1 panels encompass most commonly occurring polymorphisms of genotype 1 that isolates worldwide, but fewer isolates are available from genotypes 2 through 7. The expansion of HCVpp and HCVcc panels has identified natural E1E2 strains with resistance to even the most broadly neutralizing mAbs. It is not known whether further expansion of HCVpp and HCVcc panels will identify additional resistant clones, or if strains currently in use accurately represent worldwide phenotypic diversity. Given this limited understanding of the relationship between genetic and phenotypic differences between E1E2 strains, comparison of antibody responses induced by different vaccines has been hampered by the use of different neutralization panels in different laboratories. Therefore, future goals for the field should include optimization and standardization of a genotype 1 neutralization panel representing the many diverse E1E2 isolates already available, along with expansion and standardization of neutralization panels encompassing other genotypes. These standardized panels could be used to accurately evaluate and compare neutralizing antibodies and post-vaccination immune sera tested in different laboratories, accelerating development of an HCV vaccine.
Author Contributions
VK wrote the manuscript. JB edited the manuscript and prepared figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a past co-authorship with the authors.
Acknowledgments
The authors would like to thank Dr. Stuart Ray, Dr. Andrea Cox, and Dr. Steven Foung for useful discussions. This work was supported by NIH grants R01AI127469 and U19AI088791.
References
1. Hill AM, Nath S, Simmons B. The road to elimination of hepatitis C: analysis of cures versus new infections in 91 countries. J Virus Erad (2017) 3:117–23.
2. Bukh J. The history of hepatitis C virus (HCV): basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol (2016) 65:S2–21. doi:10.1016/j.jhep.2016.07.035
3. Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, et al. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol (2017) 98:2–3. doi:10.1099/jgv.0.000672
4. Smith DB, Becher P, Bukh J, Gould EA, Meyers G, Monath T, et al. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol (2016) 97:2894–907. doi:10.1099/jgv.0.000612
5. Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology (2014) 59:318–27. doi:10.1002/hep.26744
6. Farci P, Bukh J, Purcell RH. The quasispecies of hepatitis C virus and the host immune response. Springer Semin Immunopathol (1997) 19:5–26. doi:10.1007/BF00945022
7. Forns X, Purcell RH, Bukh J. Quasispecies in viral persistence and pathogenesis of hepatitis C virus. Trends Microbiol (1999) 7:402–10. doi:10.1016/S0966-842X(99)01590-5
8. Martell M, Esteban JI, Quer J, Genesca J, Weiner A, Esteban R, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol (1992) 66:3225–9.
9. Bailey JR, Laskey S, Wasilewski LN, Munshaw S, Fanning LJ, Kenny-Walsh E, et al. Constraints on viral evolution during chronic hepatitis C virus infection arising from a common-source exposure. J Virol (2012) 86:12582–90. doi:10.1128/JVI.01440-12
10. Kumar S, Stecher G, Tamura K. MEGA7: molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol (2016) 33:1870–4. doi:10.1093/molbev/msw054
11. Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med (2013) 19:869–78. doi:10.1038/nm.3183
12. Choo QL, Kuo G, Ralston R, Weiner A, Chien D, van Nest G, et al. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci U S A (1994) 91:1294–8. doi:10.1073/pnas.91.4.1294
13. Houghton M. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol Rev (2011) 239:99–108. doi:10.1111/j.1600-065X.2010.00977.x
14. Law JL, Chen C, Wong J, Hockman D, Santer DM, Frey SE, et al. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One (2013) 8:e59776. doi:10.1371/journal.pone.0059776
15. Colpitts CC, Baumert TF. Hepatitis C virus cell entry: a target for novel antiviral strategies to address limitations of direct acting antivirals. Hepatol Int (2016) 10:741–8. doi:10.1007/s12072-016-9724-7
16. Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology (2009) 136:2377–86. doi:10.1053/j.gastro.2009.02.080
17. Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, et al. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A (1994) 91:7792–6. doi:10.1073/pnas.91.16.7792
18. Gal-Tanamy M, Keck ZY, Yi M, McKeating JA, Patel AH, Foung SK, et al. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proc Natl Acad Sci U S A (2008) 105:19450–5. doi:10.1073/pnas.0809879105
19. Shimizu YK, Hijikata M, Iwamoto A, Alter HJ, Purcell RH, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol (1994) 68:1494–500.
20. von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology (2007) 132:667–78. doi:10.1053/j.gastro.2006.12.008
21. Fuerst TR, Pierce BG, Keck ZY, Foung SKH. Designing a B cell-based vaccine against a highly variable hepatitis C virus. Front Microbiol (2017) 8:2692. doi:10.3389/fmicb.2017.02692
22. Kinchen VJ, Cox AL, Bailey JR. Can broadly neutralizing monoclonal antibodies lead to a hepatitis C virus vaccine? Trends Microbiol (2018). doi:10.1016/j.tim.2018.04.002
23. Bailey JR, Flyak AI, Cohen VJ, Li H, Wasilewski LN, Snider AE, et al. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight (2017) 2:e92872. doi:10.1172/jci.insight.92872
24. Logvinoff C, Major ME, Oldach D, Heyward S, Talal A, Balfe P, et al. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci U S A (2004) 101:10149–54. doi:10.1073/pnas.0403519101
25. Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology (2014) 59:2140–51. doi:10.1002/hep.27013
26. Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A (2007) 104:6025–30. doi:10.1073/pnas.0607026104
27. Osburn WO, Fisher BE, Dowd KA, Urban G, Liu L, Ray SC, et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology (2010) 138:315–24. doi:10.1053/j.gastro.2009.09.017
28. Bukh J, Engle RE, Faulk K, Wang RY, Farci P, Alter HJ, et al. Immunoglobulin with high-titer in vitro cross-neutralizing hepatitis C virus antibodies passively protects chimpanzees from homologous, but not heterologous, challenge. J Virol (2015) 89:9128–32. doi:10.1128/JVI.01194-15
29. Meuleman P, Bukh J, Verhoye L, Farhoudi A, Vanwolleghem T, Wang RY, et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology (2011) 53:755–62. doi:10.1002/hep.24171
30. Vanwolleghem T, Bukh J, Meuleman P, Desombere I, Meunier JC, Alter H, et al. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology (2008) 47:1846–55. doi:10.1002/hep.22244
31. Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A (2012) 109:6205–10. doi:10.1073/pnas.1114927109
32. Keck ZY, Wang Y, Lau P, Lund G, Rangarajan S, Fauvelle C, et al. Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute HCV infection in mice. Hepatology (2016) 64:1922–33. doi:10.1002/hep.28850
33. Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med (2008) 14:25–7. doi:10.1038/nm1698
34. Morin TJ, Broering TJ, Leav BA, Blair BM, Rowley KJ, Boucher EN, et al. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog (2012) 8:e1002895. doi:10.1371/journal.ppat.1002895
35. de Jong YP, Dorner M, Mommersteeg MC, Xiao JW, Balazs AB, Robbins JB, et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med (2014) 6:254ra129. doi:10.1126/scitranslmed.3009512
36. Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med (2003) 197:633–42. doi:10.1084/jem.20021756
37. Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, et al. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem (2003) 278:41624–30. doi:10.1074/jbc.M305289200
38. Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc Natl Acad Sci U S A (2004) 101:7270–4. doi:10.1073/pnas.0402253101
39. McKeating JA, Zhang LQ, Logvinoff C, Flint M, Zhang J, Yu J, et al. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J Virol (2004) 78:8496–505. doi:10.1128/JVI.78.16.8496-8505.2004
40. Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol (2004) 78:1448–55. doi:10.1128/JVI.78.3.1448-1455.2004
41. Gottwein JM, Scheel TK, Hoegh AM, Lademann JB, Eugen-Olsen J, Lisby G, et al. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology (2007) 133:1614–26. doi:10.1053/j.gastro.2007.08.005
42. Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, et al. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology (2009) 49:364–77. doi:10.1002/hep.22673
43. Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science (2005) 309:623–6. doi:10.1126/science.1114016
44. Scheel TK, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. Recombinant HCV variants with NS5A from genotypes 1-7 have different sensitivities to an NS5A inhibitor but not interferon-alpha. Gastroenterology (2011) 140:1032–42. doi:10.1053/j.gastro.2010.11.036
45. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med (2005) 11:791–6. doi:10.1038/nm0805-905b
46. Bukh J, Purcell RH. A milestone for hepatitis C virus research: a virus generated in cell culture is fully viable in vivo. Proc Natl Acad Sci U S A (2006) 103:3500–1. doi:10.1073/pnas.0600551103
47. Kato T, Choi Y, Elmowalid G, Sapp RK, Barth H, Furusaka A, et al. Hepatitis C virus JFH-1 strain infection in chimpanzees is associated with low pathogenicity and emergence of an adaptive mutation. Hepatology (2008) 48:732–40. doi:10.1002/hep.22422
48. Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A (2006) 103:3805–9. doi:10.1073/pnas.0511218103
49. Li YP, Ramirez S, Gottwein JM, Scheel TK, Mikkelsen L, Purcell RH, et al. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc Natl Acad Sci U S A (2012) 109:E1101–10. doi:10.1073/pnas.1203829109
50. Li YP, Ramirez S, Jensen SB, Purcell RH, Gottwein JM, Bukh J. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc Natl Acad Sci U S A (2012) 109:19757–62. doi:10.1073/pnas.1218260109
51. Li YP, Ramirez S, Mikkelsen L, Bukh J. Efficient infectious cell culture systems of the hepatitis C virus (HCV) prototype strains HCV-1 and H77. J Virol (2015) 89:811–23. doi:10.1128/JVI.02877-14
52. Ramirez S, Li YP, Jensen SB, Pedersen J, Gottwein JM, Bukh J. Highly efficient infectious cell culture of three hepatitis C virus genotype 2b strains and sensitivity to lead protease, nonstructural protein 5A, and polymerase inhibitors. Hepatology (2014) 59:395–407. doi:10.1002/hep.26660
53. Ramirez S, Mikkelsen LS, Gottwein JM, Bukh J. Robust HCV genotype 3a infectious cell culture system permits identification of escape variants with resistance to sofosbuvir. Gastroenterology (2016) 151(973–985):e972. doi:10.1053/j.gastro.2016.07.013
54. Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, et al. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol (2011) 85:596–605. doi:10.1128/JVI.01592-10
55. Carlsen TH, Scheel TK, Ramirez S, Foung SK, Bukh J. Characterization of hepatitis C virus recombinants with chimeric E1/E2 envelope proteins and identification of single amino acids in the E2 stem region important for entry. J Virol (2013) 87:1385–99. doi:10.1128/JVI.00684-12
56. Mathiesen CK, Prentoe J, Meredith LW, Jensen TB, Krarup H, McKeating JA, et al. Adaptive mutations enhance assembly and cell-to-cell transmission of a high-titer hepatitis C virus genotype 5a core-NS2 JFH1-based recombinant. J Virol (2015) 89:7758–75. doi:10.1128/JVI.00039-15
57. Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology (2008) 47:17–24. doi:10.1002/hep.21959
58. Lavillette D, Tarr AW, Voisset C, Donot P, Bartosch B, Bain C, et al. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology (2005) 41:265–74. doi:10.1002/hep.20542
59. Meunier JC, Engle RE, Faulk K, Zhao M, Bartosch B, Alter H, et al. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci U S A (2005) 102:4560–5. doi:10.1073/pnas.0501275102
60. Pedersen J, Carlsen TH, Prentoe J, Ramirez S, Jensen TB, Forns X, et al. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology (2013) 58:1587–97. doi:10.1002/hep.26524
61. Urbanowicz RA, McClure CP, Brown RJ, Tsoleridis T, Persson MA, Krey T, et al. A diverse panel of hepatitis C virus glycoproteins for use in vaccine research reveals extremes of monoclonal antibody neutralization resistance. J Virol (2016) 90:3288–301. doi:10.1128/JVI.02700-15
62. Wasilewski LN, Ray SC, Bailey JR. Hepatitis C virus resistance to broadly neutralizing antibodies measured using replication-competent virus and pseudoparticles. J Gen Virol (2016) 97:2883–93. doi:10.1099/jgv.0.000608
63. Urbanowicz RA, McClure CP, King B, Mason CP, Ball JK, Tarr AW. Novel functional hepatitis C virus glycoprotein isolates identified using an optimized viral pseudotype entry assay. J Gen Virol (2016) 97:2265–79. doi:10.1099/jgv.0.000537
64. Wasilewski LN, El-Diwany R, Munshaw S, Snider AE, Brady JK, Osburn WO, et al. A hepatitis C virus envelope polymorphism confers resistance to neutralization by polyclonal sera and broadly neutralizing monoclonal antibodies. J Virol (2016) 90:3773–82. doi:10.1128/JVI.02837-15
65. Fauvelle C, Felmlee DJ, Crouchet E, Lee J, Heydmann L, Lefevre M, et al. Apolipoprotein E mediates evasion from hepatitis C virus neutralizing antibodies. Gastroenterology (2016) 150:206–217. doi:10.1053/j.gastro.2015.09.014
66. Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M Jr, et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A (2007) 104:5848–53. doi:10.1073/pnas.0700760104
67. El-Diwany R, Cohen VJ, Mankowski MC, Wasilewski LN, Brady JK, Snider AE, et al. Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1. PLoS Pathog (2017) 13:e1006235. doi:10.1371/journal.ppat.1006235
68. Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV e2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog (2012) 8:e1002653. doi:10.1371/journal.ppat.1002653
69. Tarr AW, Urbanowicz RA, Hamed MR, Albecka A, McClure CP, Brown RJ, et al. Hepatitis C patient-derived glycoproteins exhibit marked differences in susceptibility to serum neutralizing antibodies: genetic subtype defines antigenic but not neutralization serotype. J Virol (2011) 85:4246–57. doi:10.1128/JVI.01332-10
70. Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, et al. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A (2003) 100:14199–204. doi:10.1073/pnas.2335981100
71. Munshaw S, Bailey JR, Liu L, Osburn WO, Burke KP, Cox AL, et al. Computational reconstruction of bole1a, a representative synthetic hepatitis C virus subtype 1a genome. J Virol (2012) 86:5915–21. doi:10.1128/JVI.05959-11
72. Bailey JR, Wasilewski LN, Snider AE, El-Diwany R, Osburn WO, Keck Z, et al. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest (2015) 125:437–47. doi:10.1172/JCI78794
73. Mankowski MC, Kinchen VJ, Wasilewski LN, Flyak AI, Ray SC, Crowe JE, et al. Synergistic anti-HCV broadly neutralizing human monoclonal antibodies with independent mechanisms. Proc Natl Acad Sci U S A (2018) 115:E82–91. doi:10.1073/pnas.1718441115
74. Carlsen TH, Pedersen J, Prentoe JC, Giang E, Keck ZY, Mikkelsen LS, et al. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology (2014) 60:1551–62. doi:10.1002/hep.27298
75. McClure CP, Urbanowicz RA, King BJ, Cano-Crespo S, Tarr AW, Ball JK. Flexible and rapid construction of viral chimeras applied to hepatitis C virus. J Gen Virol (2016) 97:2187–93. doi:10.1099/jgv.0.000530
76. Scheel TK, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter HJ, et al. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A (2008) 105:997–1002. doi:10.1073/pnas.0711044105
77. Chung RT, Gordon FD, Curry MP, Schiano TD, Emre S, Corey K, et al. Human monoclonal antibody MBL-HCV1 delays HCV viral rebound following liver transplantation: a randomized controlled study. Am J Transplant (2013) 13:1047–54. doi:10.1111/ajt.12083
78. Rodrigo C, Walker MR, Leung P, Eltahla AA, Grebely J, Dore GJ, et al. Limited naturally occurring escape in broadly neutralizing antibody epitopes in hepatitis C glycoprotein E2 and constrained sequence usage in acute infection. Infect Genet Evol (2017) 49:88–96. doi:10.1016/j.meegid.2017.01.006
79. Keck ZY, Li SH, Xia J, von Hahn T, Balfe P, McKeating JA, et al. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J Virol (2009) 83:6149–60. doi:10.1128/JVI.00248-09
Keywords: hepatitis C virus, Flaviviridae, antibodies, neutralizing breadth, viral diversity
Citation: Kinchen VJ and Bailey JR (2018) Defining Breadth of Hepatitis C Virus Neutralization. Front. Immunol. 9:1703. doi: 10.3389/fimmu.2018.01703
Received: 03 April 2018; Accepted: 10 July 2018;
Published: 02 August 2018
Edited by:
Steven Foung, Stanford University, United StatesReviewed by:
Joseph Torresi, University of Melbourne, AustraliaJonathan Ball, University of Nottingham, United Kingdom
Copyright: © 2018 Kinchen and Bailey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justin R. Bailey, amJhaWxleTdAamhtaS5lZHU=
 Valerie J. Kinchen
Valerie J. Kinchen Justin R. Bailey
Justin R. Bailey