
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 11 July 2018
Sec. Microbial Immunology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.01597
This article is part of the Research Topic The Immunology of Sepsis – Understanding Host Susceptibility, Pathogenesis of Disease, and Avenues for Future Treatment View all 27 articles
 Anina Schneider1,2
Anina Schneider1,2 Manuela Weier1,2
Manuela Weier1,2 Jacobus Herderschee2
Jacobus Herderschee2 Matthieu Perreau3
Matthieu Perreau3 Thierry Calandra2
Thierry Calandra2 Thierry Roger2
Thierry Roger2 Eric Giannoni1,2*
Eric Giannoni1,2*
Infections are a leading cause of mortality and morbidity in newborns. The high susceptibility of newborns to infection has been associated with a limited capacity to mount protective immune responses. Monocytes and macrophages are involved in the initiation, amplification, and termination of immune responses. Depending on cues received from their environment, monocytes differentiate into M1 or M2 macrophages with proinflammatory or anti-inflammatory and tissue repair properties, respectively. The purpose of this study was to characterize differences in monocyte to macrophage differentiation and polarization between newborns and adults. Monocytes from umbilical cord blood of healthy term newborns and from peripheral blood of adult healthy subjects were exposed to GM-CSF or M-CSF to induce M1 or M2 macrophages. Newborn monocytes differentiated into M1 and M2 macrophages with similar morphology and expression of differentiation/polarization markers as adult monocytes, with the exception of CD163 that was expressed at sevenfold higher levels in newborn compared to adult M1 macrophages. Upon TLR4 stimulation, newborn M1 macrophages produced threefold to sixfold lower levels of TNF than adult macrophages, while production of IL-1-β, IL-6, IL-8, IL-10, and IL-23 was at similar levels as in adults. Nuclear levels of IRF5, a transcription factor involved in M1 polarization, were markedly reduced in newborns, whereas the NF-κB and MAP kinase pathways were not altered. In line with a functional role for IRF5, adenoviral-mediated IRF5 overexpression in newborn M1 macrophages restored lipopolysaccharide-induced TNF production. Altogether, these data highlight a distinct immune response of newborn macrophages and identify IRF5 as a key regulator of macrophage TNF response in newborns.
Despite advances in perinatal care, neonatal infections remain a leading cause of mortality and morbidity worldwide (1–3). The high susceptibility to infection during the neonatal period has been linked to a developing immune system with a limited capacity to mount protective immune responses (4). Indeed, neonatal monocytes and dendritic cells (DCs) exposed to microbial products release reduced amounts of the proinflammatory and TH1-polarizing cytokines TNF, IFNγ, IL-1β, and IL-12p70 than adult cells, but similar or even higher levels of the TH17-polarizing and anti-inflammatory cytokines IL-6, IL-10, and IL-23 (5–7). Yet, uncontrolled inflammatory responses contribute to the pathogenesis of sepsis and septic shock and other conditions associated with adverse outcomes in newborns, such as necrotizing enterocolitis, bronchopulmonary dysplasia, and periventricular leucomalacia (8–11). Attempts at improving the outcome of neonatal sepsis through immune enhancing therapies including granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte transfusions, and intravenous immunoglobulins have only yielded a limited benefit (12–14). This underscores our incomplete understanding of how newborns respond to infections, and the need for new therapeutic approaches.
Tissue-resident macrophages are sentinel innate immune cells that display a spectrum of functions and produce a panel of cytokines that orchestrate innate and adaptive immune responses (15, 16). Macrophage activation and function are influenced by signals received from the local environment (17). The functional plasticity of macrophages has given rise to the notion of macrophage polarization, ranging from classically activated proinflammatory M1 macrophages to alternatively activated pro-resolving/anti-inflammatory M2 macrophages (18). The differentiation of monocytes into M1 macrophages is induced by GM-CSF, IFNγ, TNF, and bacterial lipopolysaccharide (LPS) (19–21). M1 macrophages are potent phagocytic cells that produce microbicidal molecules such as reactive oxygen and nitrogen species (ROS and NO) and TNF, IL-1β, IL-6, IL-12p70, and IL-23 (22, 23). In contrast, M-CSF, IL-4, IL-10, IL-13, adenosine, and steroid hormones induce the differentiation of monocytes into M2 macrophages (24). M2 macrophages are involved in resolving inflammation and promote tissue repair and homeostasis. M2 macrophages are characterized by the expression of scavenger receptors (CD36, CD163) and the production of high levels of IL-10 and low levels of TNF, IL-12p70, IL-23, ROS, and NO (25–27).
Monocyte to macrophage differentiation is controlled by the Janus-kinase/signal transducer and activator of transcription (JAK/STAT), MAP kinase (MAPK), and NF-κB pathways (28–31). These pathways activate suppressor of cytokine signaling (SOCS) and interferon regulatory factors (IRFs), leading to M1/M2 macrophage polarization (32, 33). In adults, IRF5, a downstream target of GM-CSF receptor (GM-CSFR), plays a critical role in driving macrophage polarization toward the M1 phenotype (23). However, the response of newborn macrophages to environmental signals driving M1 and M2 polarization and production of proinflammatory and anti-inflammatory cytokines is unknown.
Here, we report that in primary human monocytes exposed to GM-CSF, IRF5 was activated to a lower extent in newborns compared to adults during differentiation into M1 macrophages. Upon TLR4 stimulation, newborn M1 macrophages secreted lower levels of TNF compared to adult macrophages, while the production of other cytokines was not affected. Overexpression of IRF5 in newborn macrophages restored TNF production, suggesting a key role of IRF5 in shaping the distinct immune response of newborn macrophages.
Umbilical cord blood was collected after delivery of the placenta of 91 healthy term neonates. Peripheral blood was obtained from 71 healthy adult volunteers (age 18–65 years). Monocytes and macrophages from the same subjects (20 newborns and 20 adults) were used for the experiments reported in Figures 1B and 2A,C,D. Macrophages from the same subjects (10 newborns and 10 adults) were used for the experiments reported in Figures 2B and 5A–J. Different sets of newborn and adult donors were used for every other Figures 1A,C, 3A,B, 4A,B, 5A–J, and 6A–C. Blood was collected in heparinized tubes (10 U/ml). Our study was approved by the Cantonal Human Research Ethics Committee of Vaud (CER-VD, Lausanne, Switzerland).
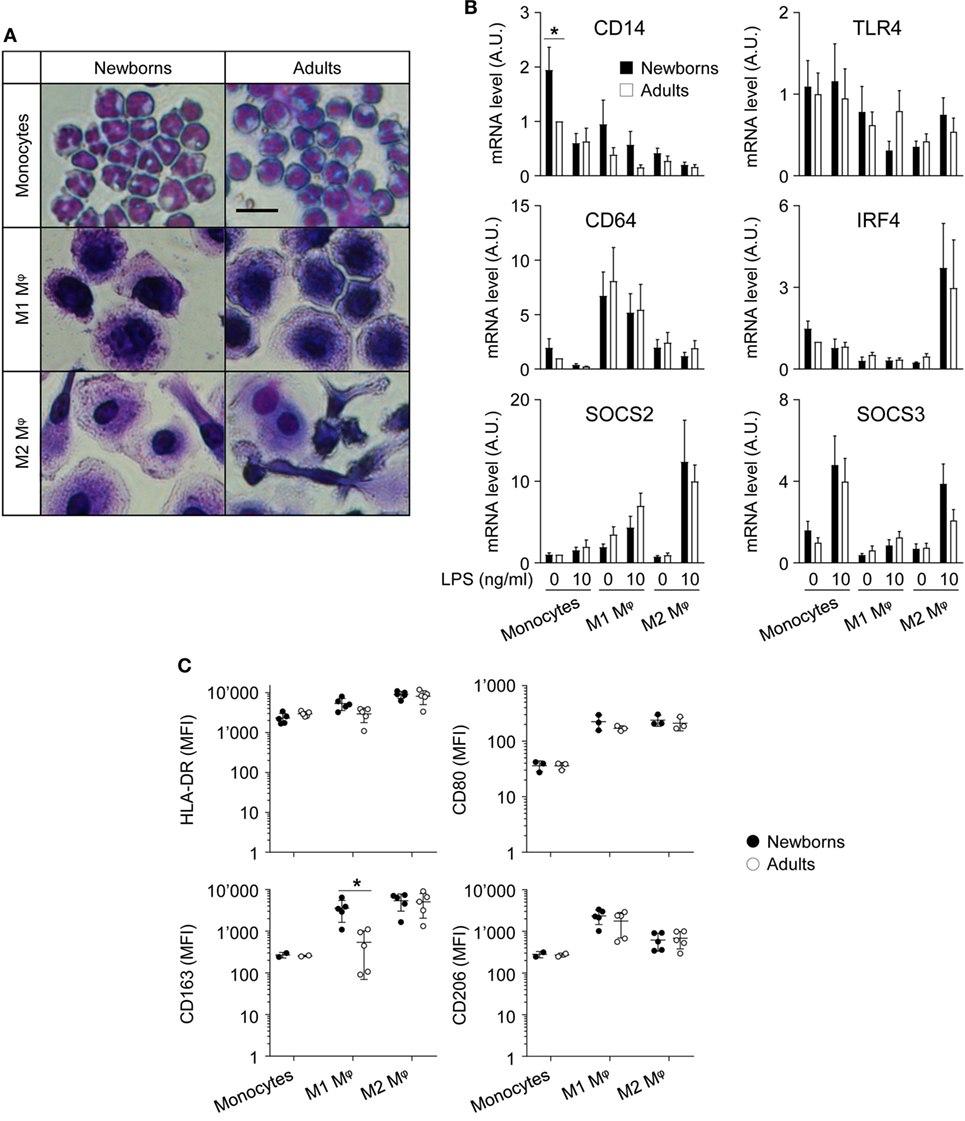
Figure 1. Adult and newborn monocytes differentiate into M1 or M2 macrophages following exposure to GM-CSF or M-CSF. Freshly isolated monocytes were cultured for 7 days with GM-CSF and M-CSF (50 ng/ml) to induce M1 and M2 macrophages. (A) Hematoxylin and eosin staining of monocytes and M1 and M2 macrophages. Scale bar = 30 µm. Data are representative of results obtained from five newborns and five adults. (B) CD14, TLR4, CD64, SOCS2, and IRF4 mRNA expression levels in newborn (black bars) and adult (white bars) monocytes and M1 and M2 macrophages were measured by RT-PCR. Data are means ± SEM from eight newborns and eight adults. (C) HLA-DR, CD80, CD163, and CD206 mean fluorescence intensity in newborn (black circles) and adult (white circles) monocytes and M1 and M2 macrophages was analyzed by flow cytometry in three to five healthy newborns and adults. Each dot represents one subject. CD163 and CD206 were not detected in monocytes from 2/5 newborns and 2/5 adults. Means ± SEM are presented. *P < 0.05.
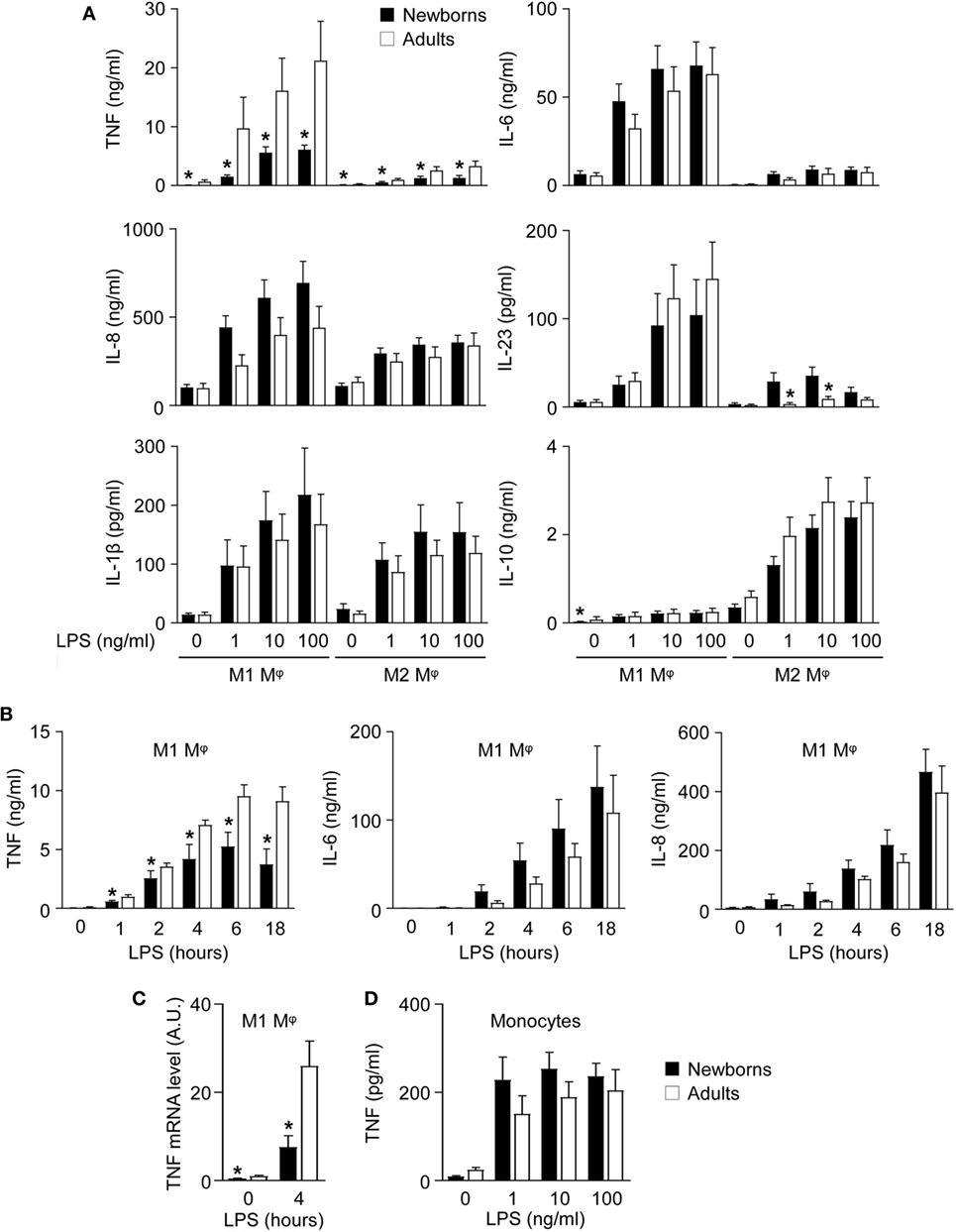
Figure 2. Newborn M1 macrophages secrete reduced amounts of TNF after lipopolysaccharide (LPS) stimulation. Newborn (black bars) and adult (white bars) monocytes were cultured for 7 days with GM-CSF (50 ng/ml) and with M-CSF (50 ng/ml) to induce M1 (A–C) and M2 macrophages (A). Macrophages (A–C) and monocytes (D) were stimulated with 0–100 ng/ml LPS. (A) Cytokine levels were measured in cell culture supernatants collected after 20 h. Data are means ± SEM from 20 (TNF, IL-6, IL-8, IL-1β, and IL-10) or 10 (IL-23) newborn and adult subjects. (B) TNF, IL-6, and IL-8 levels were measured in cell culture supernatants collected after 0–18 h. Data are means ± SEM from eight newborns and six adults. (C) TNF mRNA expression levels in M1 macrophages exposed for 0 and 4 h to 100 ng/ml LPS were measured by RT-PCR. Data are means ± SEM from 10 newborns and 9 adults. (D) TNF concentrations in cell culture supernatants of monocytes exposed for 20 h to 0–100 ng/ml LPS. Data are means ± SEM from 20 newborns and 10 adults. *P < 0.05.
Mononuclear cells were isolated by Ficoll Hypaque (GE Healthcare) gradient density centrifugation. Monocytes were extracted from blood mononuclear cells by positive selection using magnetic microbeads coupled to anti-CD14 antibodies (Miltenyi Biotec) (34–36). Purity assessed by flow cytometry was >95%. Viability determined by trypan blue exclusion was >95%. Monocytes were cultured in RPMI medium 1640 supplemented with 10% (vol/vol) FCS (GE Healthcare) and GM-CSF (50 ng/ml) (Peprotech) or M-CSF (50 ng/ml) (Peprotech) for 1 week to induce M1 or M2 macrophages, respectively. Ultrapure E. coli O111:B4 LPS was purchased from List Biological Laboratories. Polyclonal and monoclonal antibodies (pAbs and mAbs) used for flow cytometry, Western blotting and cytometry by time of flight (CyTOF) are described in Table S1 in Supplementary Material. Unless specified otherwise, all other reagents were obtained from Sigma-Aldrich.
RNA was extracted, reverse transcribed, and used in real-time PCR as described (37). The primers (5′–3′ sequences, sense and antisense) used for amplification were: HPRT, GAACGTCTTGCTCGAGATGTG and CCAGCAGGTCAGCAAAGAATT; CD14, CGCCCTGAACTCCCTCAAT and CTTGGCTGGCAGTCCTTTAGG; TLR4, AGTTTCCTGCAATGGATCAAGG and CTGCTTATCTGAAGGTGTTGCAC; CD64, TGCCACAGAGGATGGAAATG and CTGGAGGCCAAGCACTTGA; IRF4, AATCCTCGTGAAGGAGCTGA and GTAGATCGTGCTCTGGCACA; SOCS2, GGATGGTACTGGGGAAGTATGACTG and AGTCGATCAGATGAACCACACTGTC; SOCS3, GCTCCAAGAGCGAGTACCAG and CTGTCGCGGATCAGAAAGGT; TNF, CAGAGGGCCTGTACCTCATC and GGAAGACCCCTCCCAGATAG. Gene-specific expression was normalized to the expression of HPRT and was expressed in arbitrary units (A.U.).
Mononuclear cells and macrophages were stained using mAbs (Table S1 in Supplementary Material) as described (38). Thirty thousand events were acquired with a LSR-II flow cytometer (BD Biosciences). Data were analyzed using the BD FACSDiva™ software (BD Biosciences).
Cytokine concentrations in cell-culture supernatants were measured by ELISA (BD Biosciences, for TNF, IFNγ, IL-1β, IL-6, IL-8, and IL-10) or by the Luminex technology (Affymetrix eBioscience, for IL-12p70, IL-20, IL-23, and IL-27).
Monocytes were exposed for 0, 15, 30, 60, or 120 min to GM-CSF and fixed with formaldehyde at a final concentration of 1.5%. Cells were stained using an anti-CD14 mAb conjugated with the Fluidigm MaxPar conjugation kit (Fludigm). Cells were washed with Cell Staining Media and PBS, fixed with 2% formaldehyde, and bar-coded using Scn-Bn-EDTA-palladium barcode reagents (39). After barcoding, cells were pooled, permeabilized for 30 min at −20°C using 100% methanol, washed twice with 6 ml Cell Staining Media containing 0.3% saponin, and incubated for 30 min with mAbs directed against intracellular targets. Finally, cells were incubated overnight at 4°C in intercalation solution (PBS, 0.3% saponin, 1% formaldehyde, 125 nM Cell-ID Intercalator-Ir, Fluidigm) before acquisition on a CyTOF 1 upgraded to a CyTOF 2. Individual data files were concatenated, normalized, and deconvoluted as described (40) and were analyzed using Cytobank (Cytobank Inc.).
Whole cellular extracts and cytoplasmic and nuclear extracts were prepared as described previously (34). Equal amounts of protein extracts were electrophoresed through SDS/PAGE. Proteins were transferred onto nitrocellulose membranes (Schleicher and Schuell). Membranes were incubated with Abs (listed in Table S1 in Supplementary Material) directed against NF-κBp65, IκBα, total and phosphorylated p38, ERK1/2, and JNK MAPKs, MAP kinase phosphatase-1 (MKP-1), IRF5, IRF8, total and phosphorylated Akt, GAPDH, β-actin, and TATA-binding protein. After washing, membranes were incubated with horseradish peroxidase-conjugated secondary Abs (Pierce Biotechnology Inc.). Signals were revealed using enhanced chemiluminescence detection (GE Healthcare). Images were recorded using a Fusion Fx system (Viber Lourmat).
IRF5-encoding and control empty adenoviral vectors (Applied Biochemical Materials Inc.) were amplified in HEK-293 cells (ATCC CRL-1573) and stored at −80°C in 10% glycerol. Macrophages were transduced with the adenoviral preparations (50 µl for 105 cells, 1 ml for 2.5 × 106), and used 24 h later for functional studies.
Chromatin immunoprecipitation analyses were performed according to the manufacturer’s recommendations (MAGnifiy Chromatin Immunoprecipitation System, Thermo Fisher). Briefly, 1 × 106 M1 macrophages were fixed with 1% formaldehyde. Chromatin was sheared by 16 cycles of 30-s pulse/30-s rest with an amplitude of 14% using an Ultrasonic Liquid Processor (Branson). Chromatin was incubated overnight at 4°C with 5 µg of antibodies directed against IRF5 (Cell Signaling Technology), or RNA polymerase II (Pol II, Table S1 in Supplementary Material), or with control IgGs (provided in the kit). Real-time PCR was performed with a 7500 Fast Real-Time PCR System using the SYBR Kapa Fast Mix (Sigma-Aldrich). The following sense and antisense primers (5′–3′ sequences) were used for amplification: TNF, TGCTTGTTCCTCAGCCTCTT, and TCACCCATCCCATCTCTCTC.
Statistical analyses were performed using PRISM (Graphpad Software Inc.). Data are expressed as means ± SEMs. Comparisons between the different groups were performed by two-two-tailed t tests. Findings were considered statistically significant when P < 0.05.
The differentiation and polarization of freshly isolated newborn and adult monocytes into M1 and M2 macrophages following 7 days of culture with recombinant GM-CSF and M-CSF were analyzed by hematoxylin and eosin staining (Figure 1A) and by measuring the expression of maturation/differentiation markers by RT-PCR (CD14, CD64, SOCS2, SOCS3, and IRF4; Figure 1B) and flow cytometry (HLA-DR, CD80, CD163, CD206; Figure 1C). Monocytes and M1 macrophages from healthy term newborns and adult volunteers showed a round shape, while M2 macrophages displayed a more elongated shape, consistent with the expected phenotype (41, 42). No difference in morphology and viability (91 ± 1 versus 95 ± 1% and 91 ± 1 versus 94 ± 1% for newborn and adult M1 and M2 macrophages, respectively) was noticed between newborn and adult cells.
CD14 mRNA levels were higher (2.1- to 4.7-fold) in monocytes than in macrophages (Figure 1B), as anticipated (43). Unexpectedly, CD14 was more expressed (1.9-fold) in newborn than in adult monocytes. When compared to monocytes, TLR4 was expressed at lower levels in M2 macrophages (2.4- to 3.2-fold), while the M1 marker CD64 was enriched (5.2- to 8.1-fold) in M1 macrophages, and the M2 markers SOCS2 and IRF4 were enriched (5.1- to 8.1 and 3.6- to 4.8-fold) in LPS-stimulated M2 macrophages (44–47). SOCS3, a gene implicated in the repression of the M1 phenotype (48), was expressed at lower levels in M1 macrophages than in monocytes. Newborns and adult cells expressed similar levels of TLR4, CD64, SOCS2, SOCS3, and IRF4.
HLA-DR, CD80, CD163, and CD206 were expressed at similar levels by newborn and adult monocytes (Figure 1C). The mean fluorescence intensity (MFI) of each of the molecules increased, albeit to different extents, in M1 and M2 macrophages (MFI fold increase versus monocytes: HLA-DR: 2.3–3.9; CD80: 6.2–6.6; CD163: 33–50; CD206: 5.5–16.5). HLA-DR and CD80 MFI were similar in newborn and adult M1 and M2 macrophages. CD163 was previously reported as an M2 marker in adults (41, 49, 50). However, in newborns, CD163 MFI strongly increased in both M1 and M2 macrophages compared to monocytes (33- and 50-fold). CD206, an M2 polarization marker at the transcript level (27), was more expressed in GM-CSF than M-CSF-derived macrophages by flow cytometry (41, 49). Accordingly, CD206 MFI was higher in newborn and adult M1 macrophages than in M2 macrophages, without noticeable difference of expression between newborns and adults. Overall, following exposure to GM-CSF and M-CSF, newborn monocytes differentiated into cells adopting morphological features and expressing markers of M1 and M2 macrophages similar to adult cells, with the exception of CD163 that was expressed at higher levels in newborn than adult M1 macrophages.
Functional studies were performed to compare the capacity of newborn and adult M1 and M2 macrophages to secrete proinflammatory and anti-inflammatory cytokines in response to TLR4 stimulation. In response to LPS, newborn and adult M1 macrophages secreted higher levels of TNF, IL-6, IL-8, and IL-23 and lower levels of IL-10 than M2 macrophages (Figure 2A). IL-1β secretion was similar between M1 and M2 macrophages. IFNγ, IL-12p70, IL-20, and IL-27 were undetectable.
Interestingly, newborn M1 macrophages secreted 3- to 6-fold less TNF (6.1 versus 21.2 ng/ml using 100 ng/ml LPS), while they produced IL-1β, IL-6, IL-8, IL-10, and IL-23 in the same range as adult M1 macrophages. Reduced TNF secretion was detected as early as 1 h following LPS stimulation (Figure 2B) and was associated with lower TNF mRNA expression in newborn M1 macrophages (Figure 2C).
We then evaluated whether the diminished TNF secretion by newborn M1 macrophages was also present in monocytes (Figure 2D). LPS-induced TNF secretion was much lower in monocytes than in macrophages and was similar in newborn and adult monocytes. These findings suggested that GM-CSF triggered a different response in newborn and adult cells resulting in a specific reduction of TNF production by newborn M1 macrophages. This was unlikely to be due to gender differences, as macrophages from males released similar amounts of TNF compared to macrophages from females, both in newborns and adults.
As a first approach to decipher the impact of GM-CSF on macrophage differentiation, we analyzed the expression CSF receptors and the activation of downstream signaling pathways in monocytes. Classical (CD14++CD16−), intermediate (CD14++CD16+SLAN−), and non-classical (CD14+CD16++SLAN+) monocyte subsets (51, 52) were equally distributed in newborns and adults and expressed similar levels of GM-CSFR, M-CSFR, and HLA-DR (Figure 3A; Figure S1 in Supplementary Material).
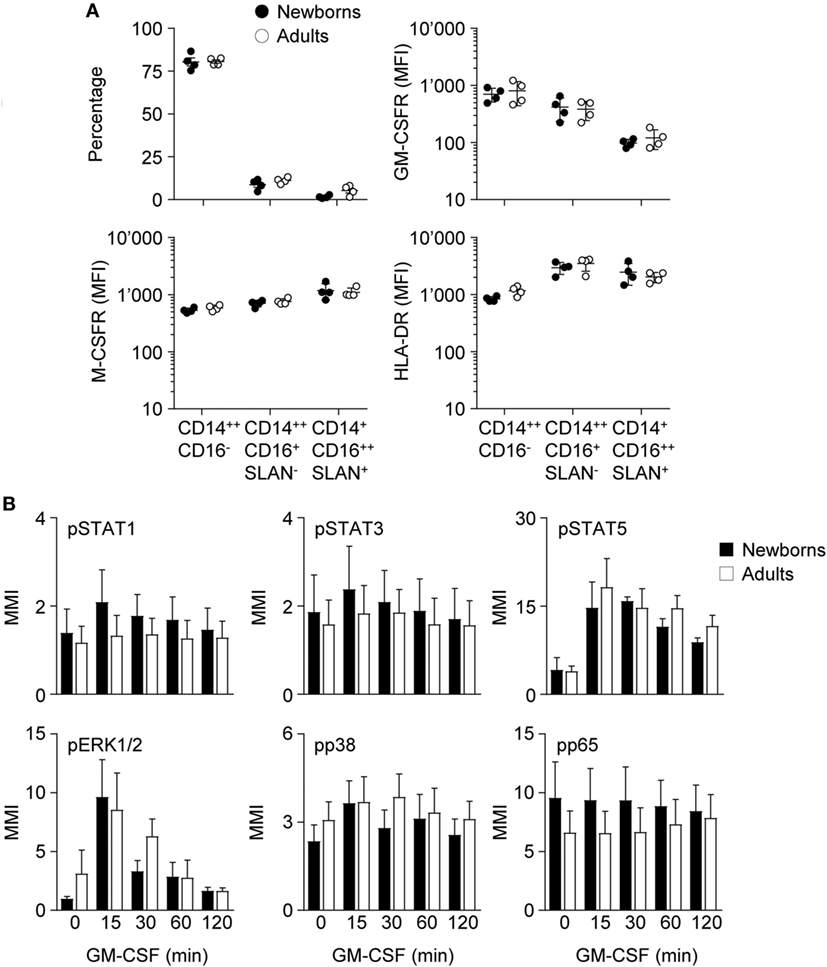
Figure 3. Proportions of monocyte subpopulations, expression of GM-CSF receptor (GM-CSFR), M-CSFR and HLA-DR and activation of intracellular signaling pathways by GM-CSF in newborn and adult monocytes. (A) Percentages of classical CD14++CD16−, intermediate CD14++CD16+SLAN−, and non-classical CD14+CD16++SLAN+ subpopulations and mean fluorescence intensity of GM-CSFR, M-CSFR, and HLA-DR in newborn (black circles) and adult (white circles) monocytes were determined by flow cytometry. Each dot represents one healthy subject. Means ± SEM are depicted. (B) Nuclear levels pSTAT1, pSTAT3, pSTAT5, pp38, pERK, and pNF-κBp65 in newborn (black bars) and adult (white bars) monocytes exposed for 15–120 min to 50 ng/ml GM-CSF were analyzed by CyTOF. Mean magnetic intensities were determined. Data are means ± SEM from six newborns and six adults.
Binding of GM-CSF to the GM-CSFR initiates the JAK/STAT, MAPK, and NF-ĸB intracellular signaling pathways and activates IRF5 in DCs (53). The pathways activated by GM-CSF in newborn and adult monocytes were investigated by mass cytometry. Exposure of newborn and adult monocytes to GM-CSF increased the phosphorylation of STAT5 (3.5- to 4.6-fold) and ERK1/2 (2.8- to 9.9-fold), but not that of STAT1, STAT3, p38, and NFĸBp65 (Figure 3B). No difference was detected between newborns and adults.
IRF5 is a downstream target of GM-CSFR signaling and plays a key role in M1 polarization (23). IRF5 is activated by phosphorylation, leading to its dimerization and nuclear translocation to promote the expression of immune response genes (54). Intracellular levels of IRF5 were similar in newborn and adult monocytes (Figure 4A). We then quantified IRF5 in cytoplasmic and nuclear fractions obtained from monocytes exposed for 0–7 days to GM-CSF (Figure 4B). Cytoplasmic levels of IRF5 started to rise at day 1, peaked at day 3–6, and declined at day 7, while nuclear levels of IRF5 increased from day 2 to day 7. Cytoplasmic levels of IRF5 were 1.4- to 2.3-fold higher in adults than in newborns at days 1–7, while nuclear levels were 2.0- to 4.5-fold higher in adults from day 0 to day 7. Of note, IRF5 was detected at 15–20 lower levels in the nucleus than in the cytoplasm at day 7. Thus, during GM-CSF-induced monocyte to M1 macrophage differentiation, IRF5 was expressed at lower levels and translocated to the nucleus to a lower extent in newborn than in adult cells, a difference that might well explain the reduced expression of TNF in newborn M1 macrophages.
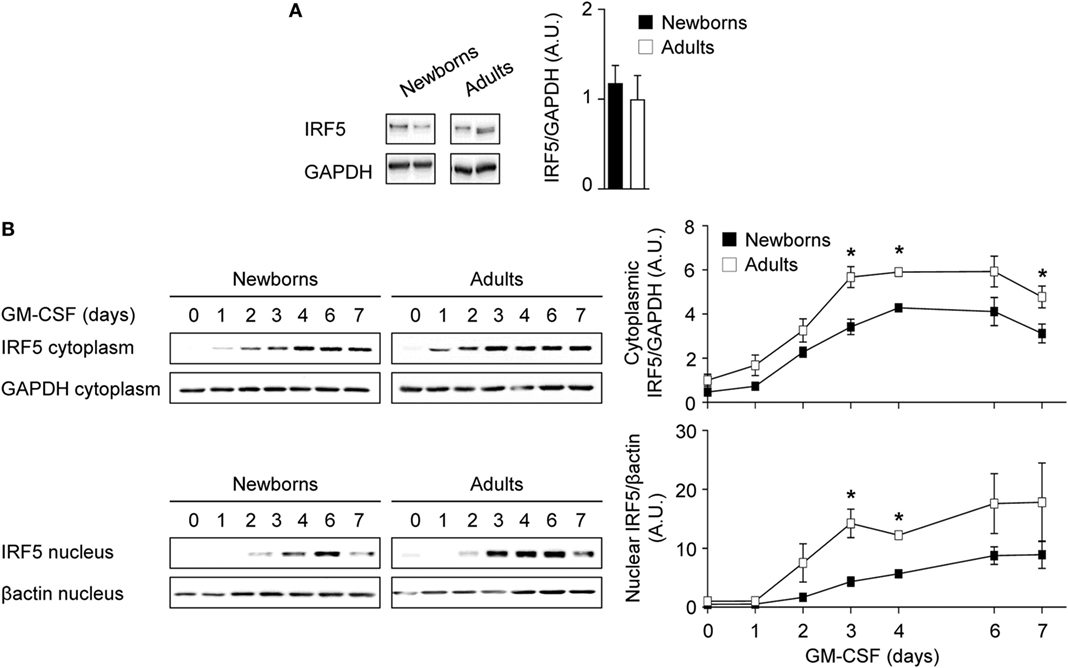
Figure 4. Reduced cytoplasmic and nuclear IRF5 levels during GM-CSF-induced monocyte to macrophage differentiation in newborns. (A) Intracellular IRF5 levels in freshly isolated newborn (black bars) and adult (white bars) monocytes. Data are means ± SEM from 10 newborns and 10 adults. (B) Cytoplasmic and nuclear IRF5 levels in newborn (black) and adults (white) monocytes cultured for 0–7 days with 50 ng/ml GM-CSF. IRF5 levels were analyzed by Western blotting (left panels) and quantified by imaging (right panels). Data are means ± SEM from nine newborns and nine adults. *P < 0.05.
To further characterize the mechanisms underlying M1 macrophage polarization, the expression of IRF5 and IRF8, another transcription factor implicated in M1 polarization (55), and the activation of NF-κB, MAPK, and Akt signaling pathways were analyzed in GM-CSF-induced M1 macrophages exposed to LPS for 0, 15, 30, and 60 min. Cytosolic IRF5 levels were lower in newborn than in adult M1 macrophages before and following LPS exposure (Figure 5A). Nuclear IRF5 levels decreased following LPS stimulation and were lower in newborns than in adults at all time points, although differences were not statistically significant (Figure 5B). Newborn M1 macrophages expressed lower levels of cytosolic IκBα and higher levels of nuclear NF-ĸBp65 before and 15 min after LPS stimulation (Figures 5C,D). Phosphorylation of ERK1/2 and p38, but not of JNK, was higher in newborn M1 macrophages at baseline and upon LPS stimulation (Figures 5E–G). In line with these findings, expression of MKP-1/dual specificity phosphatase (DUSP1), a DUSP that inactivates ERK1/2 and p38, was reduced in newborn M1 macrophages (1.4- to 1.9-fold less at baseline and 15 min after LPS stimulation; Figure 5H). No difference in IRF8 expression was noticed between newborns and adults (Figure 5I). Phospho-Akt levels were not affected by LPS stimulation in newborn and adult M1 macrophages (Figure 5J). Combined altogether, and considering that NF-κB and MAPK signaling pathways were not impaired in newborns, our data pointed toward IRF5 as a possible regulator, which decreased expression in newborn M1 macrophages could be involved in a selectively reduced TNF production.
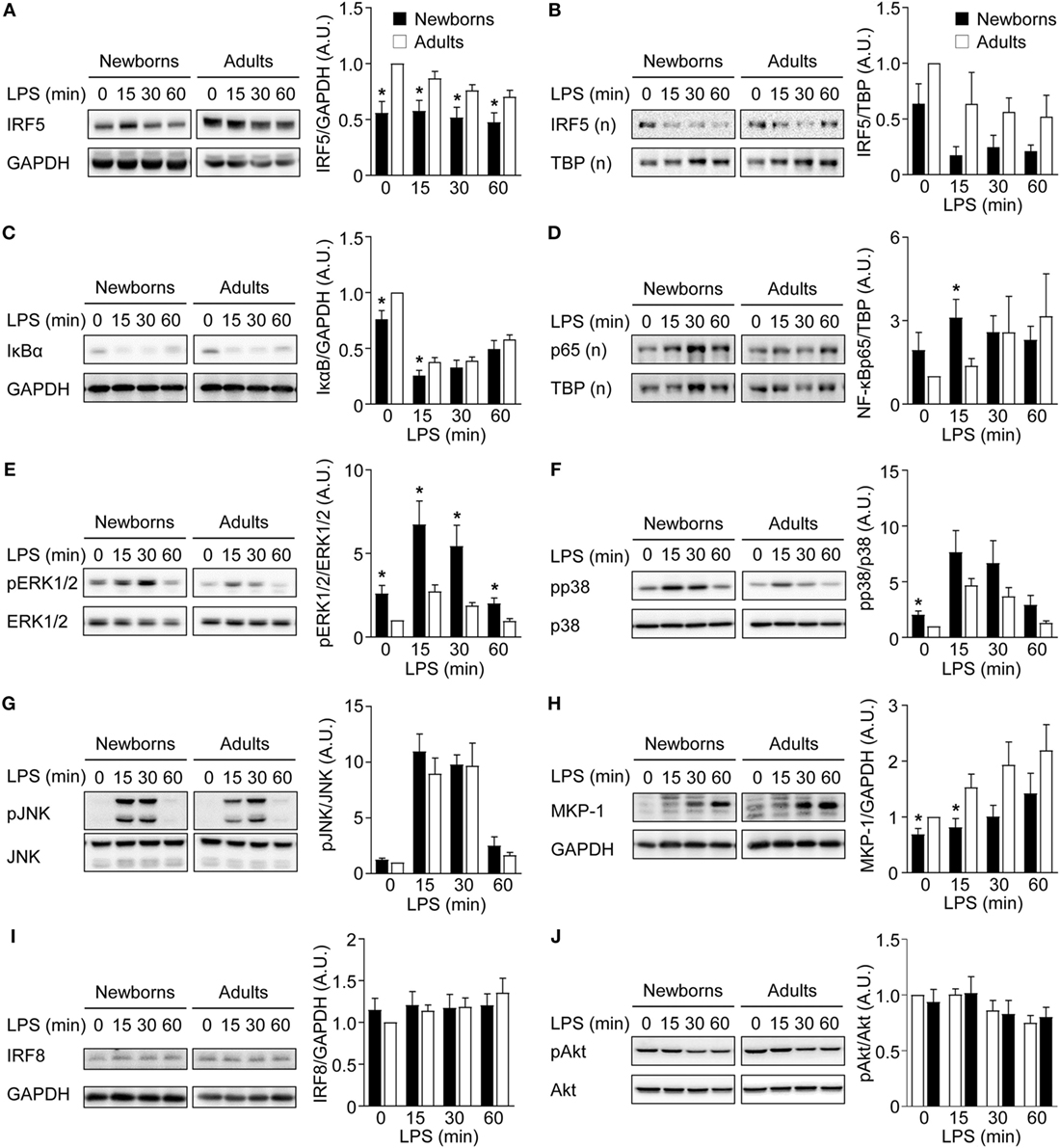
Figure 5. Reduced IRF5 expression levels in newborn M1 macrophages. Newborn (black bars) and adult (white bars) M1 macrophages were stimulated with 100 ng/ml lipopolysaccharide for 0–60 min. IRF5 (A,B), IκBα (C), NF-κBp65 (D), phosphorylated and total ERK1/2 (E), p38 (F) and JNK (G), MAP kinase phosphatase-1 (H), IRF8 (I), and phosphorylated and total Akt (J) were analyzed by Western blotting using total (A,C,E–J) or nuclear (B,D) cellular extracts and quantified by imaging. Values were normalized to those obtained from resting adult M1 macrophages set at 1. Data are means ± SEM from 8 to 10 newborns and 8 to 10 adults. *P < 0.05.
To investigate the relationship between lower levels of IRF5 and reduced TNF secretion in newborn M1 macrophages, we transduced newborn M1 macrophages with an IRF5 expressing adenoviral vector. Transduction increased IRF5 expression 1.6-fold (Figure 6A) and markedly increased (2.1- to 4.0-fold) TNF secretion, while it did not affect IL-6, IL-8, and IL-10 secretion (Figure 6B). Next, we examined the recruitment of IRF5 and RNA Pol II to the TNF promoter by ChIP. IRF5 binding was detected in unstimulated M1 macrophages and strongly decreased 1 h after exposure to LPS in both newborns and adults (Figure 6C). LPS stimulation for 1 h led to the recruitment of RNA Pol II to the TNF promoter in newborn and adult M1 macrophages. In summary, a selective increase in LPS-induced TNF production following IRF5 overexpression in newborn M1 macrophages strongly suggests an important role for IRF5 in shaping the TNF response in newborns. Yet, the mechanism of action of IRF5 might be independent of its recruitment to the TNF promoter following LPS stimulation.
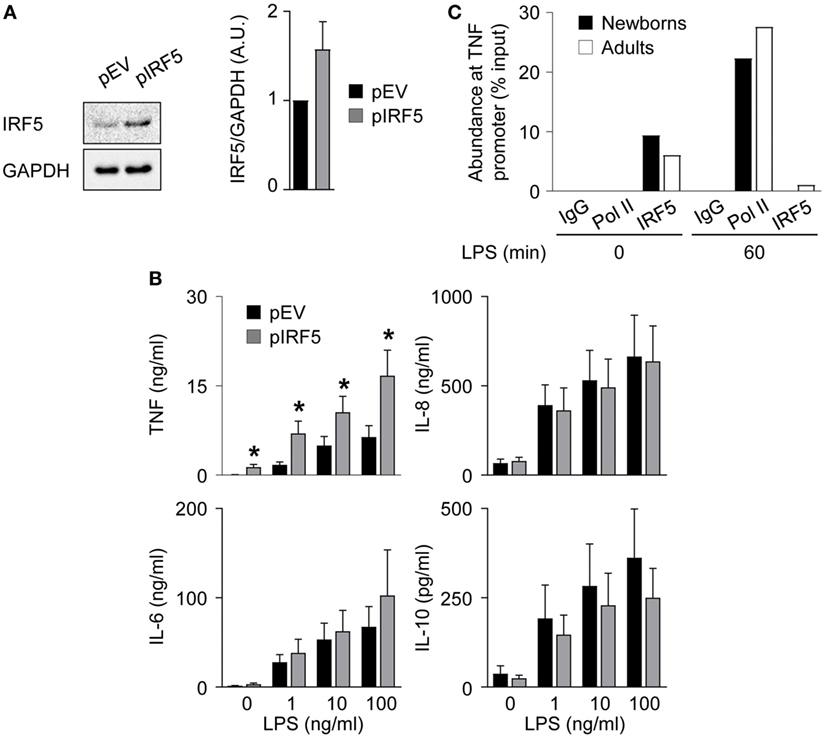
Figure 6. IRF5 overexpression restores TNF secretion by newborn M1 macrophages. Newborn M1 macrophages were transduced with empty control (pEV) (black bars) and IRF5-expressing (gray bars) adenoviral vectors (A,B). (A) IRF5 levels were analyzed by Western blotting (left panel) and quantified by imaging (right panel). (B) Newborn M1 macrophages were stimulated with 0–100 ng/ml lipopolysaccharide (LPS). TNF, IL-6, IL-8, and IL-10 concentrations were measured in cell culture supernatants collected after 20 h. Data are means ± SEM from 20 (TNF) or 10 (IL-6, IL-10, and IL-8) newborns. *P < 0.05. (C) The recruitment of RNA polymerase II (Pol II) and IRF5 to the TNF promoter in M1 macrophages before and 1 h after stimulation with 100 ng/ml LPS was assessed by chromatin immunoprecipitation. Data from one experiment representative of two experiments are presented as the percentage input relative to genomic DNA set at 100%.
We report that monocyte-derived M1 macrophages from newborns exhibit a strongly reduced capability to release TNF upon TLR4 stimulation, while the production of other cytokines is at similar levels as in adults. IRF5 is a key factor shaping this important functional characteristic of newborn macrophages (Figure 7).
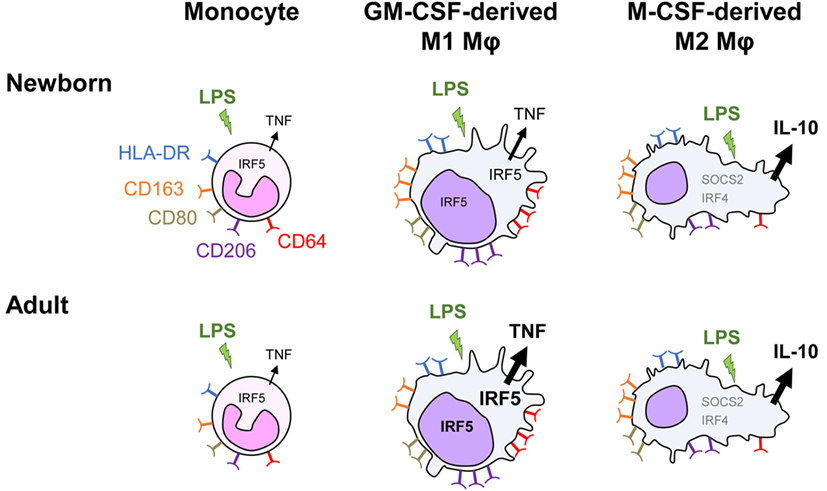
Figure 7. Distinct TLR4 response in neonatal monocyte-derived macrophages. Exposure of newborn and adult monocytes to GM-CSF and M-CSF drives their differentiation into macrophages having an M1 (CD64high and CD206high round cells) and an M2 (SOCS2high, IRF4high and CD163high elongated cells) phenotype. Macrophages express high levels of membrane-bound HLA-DR, CD80, CD163, and CD206, with newborn M1 macrophages expressing higher levels of the hemoglobin scavenger receptor CD163 than adult M1 macrophages. Following TLR4 stimulation, newborn monocytes secrete TNF to a similar level as adult monocytes. M1 macrophages secrete higher levels of TNF but low levels of IL-10, whereas M2 macrophages secrete low levels of TNF but high levels of IL-10. TNF secretion is threefold to sixfold lower in newborn than in adult M1 macrophages, while the production of other cytokines (IL-1β, IL-6, IL-8, and IL-23) is at similar levels. IRF5, a transcription factor implicated in M1 polarization, increases in the cytoplasm and in the nucleus during monocyte to M1 macrophage differentiation. Both cytoplasmic and nuclear levels of IRF5 are lower in newborn than adult M1 macrophages. IRF5 overexpression in newborn M1 macrophages restores LPS-induced TNF production in newborn M1 macrophages.
Studies in mice, rats, and monkeys have described organ, tissue, and species-specific phenotypic and functional differences between newborn and adult macrophages. Globally, newborn macrophages display reduced capacities to kill bacteria (56–59) and to produce proinflammatory cytokines (59–62) while they release anti-inflammatory cytokines at the same levels as adult macrophages (61–63). Previous studies in humans have investigated mixed populations of umbilical cord blood mononuclear cells or monocyte-derived cells, without a phenotypic characterization of differentiated cells (64–67). Cord blood-derived macrophage-like cells have a reduced capacity to kill group B Streptococcus and Candida, and release lower amounts of TNF, IL-1β, IL-6, and IL-12 in response to LPS (64, 68).
M-CSF is constitutively expressed by several cell types including fibroblasts, endothelial cells, stromal cells, and osteoblasts (69). Besides promoting survival, proliferation, and differentiation of bone marrow progenitors and monocytes, steady-state expression of M-CSF contributes to polarize macrophages toward an M2 phenotype (70). GM-CSF is expressed at low levels in the circulation and in tissues at homeostasis and plays a critical role in the terminal differentiation and functions of alveolar macrophages (71). Inflammation and infections trigger the production of GM-CSF by endothelial cells, fibroblasts macrophages, T cells, mast cells, and natural killer cells. GM-CSF drives M1 polarization, which is essential to mount efficient antimicrobial responses. Morphological and phenotypical analyses confirmed that newborn monocytes differentiate into cells adopting features of M1 and M2 macrophages, similar to adult cells. Uniquely, CD163 was strongly upregulated by both M1 and M2 macrophages in newborns, while this molecule is commonly used as an M2 marker in adults [(41, 49, 50) and our data]. Reduced activation of IRF5 in newborns might be implicated as IRF5 downregulates CD163 expression in adult macrophages (23). CD163 is a scavenger receptor involved in the clearance of free hemoglobin (72). During the neonatal period, high expression of CD163 in both M1 and M2 macrophages could be relevant, since newborn infants have an elevated turnover of erythrocytes under physiologic conditions and are prone to hemolysis during infection (8, 73, 74).
The lower capacity of newborn M1 macrophages to release TNF is most likely acquired during the process of monocyte to macrophage differentiation. Indeed, newborn monocytes released similar levels of TNF as adult monocytes under the experimental conditions used in the present study. Clearly, newborn M1 macrophages are not globally defective in TLR4 signaling, considering that TLR4 expression, NF-κBp65 nuclear translocation, ERK1/2 phosphorylation and MKP-1 expression, and production of IL-1β, IL-6, IL-8, and IL-23 are not diminished in newborn M1 macrophages.
IRF5 regulatory axis shapes the phenotype of newborn macrophages and plays an important role in systemic inflammation (54), as IRF5-deficient mice are protected from LPS-induced systemic inflammation and autoimmune diseases (75). Freshly isolated newborn monocytes expressed IRF5 to a similar extent as adult monocytes but had reduced expression and nuclear translocation of IRF5 when cultured with GM-CSF. Moreover, adenoviral-mediated IRF5 overexpression in newborn M1 macrophages restored TLR4-mediated TNF secretion, while it did not impact IL-6, IL-8, and IL-10 production, indicating that IRF5 might play a key role in the selective reduction of TNF secretion observed in newborn macrophages. In contrast, germline deletion of IRF5 impairs LPS-induced production of Th1/Th17 cytokines in mice (75), and IRF5 overexpression in adult human macrophages increases expression of TNF, IL-1β, IL-12p70, and IL-23 and reduces secretion of IL-10 (23). These data suggest that IRF5 has a broader impact on cytokine production in adult than in newborn cells. Further studies will be required to define whether IRF5 differential expression impacts on immune functions besides TNF production in newborns.
Following exposure of macrophages to LPS, IRF5 is recruited to regulatory elements of the TNF gene and stimulates transcription (75, 76). In adult M1 macrophages, NOD2 stimulation triggers an IRF5-dependent activation of MAPKs, NFκB, and Akt2, increasing TNF, IL-1β, and IL-12 production (77). However, in our study, LPS stimulation did not increase IRF5 expression, nuclear translocation, and recruitment to the TNF promoter in M1 macrophages. Moreover, NF-κB and MAPKs signaling pathways were not impaired in newborn M1 macrophages, and Akt was not activated following LPS stimulation. Chromatin remodeling is implicated in monocyte to macrophage differentiation and macrophage polarization (78, 79), and histone acetylation and methylation are regulators of TNF gene expression (80). Further studies will be required to address whether GM-CSF induced a specific epigenetic reprogramming in newborn monocytes making newborn M1 macrophages less prone to transcribe TNF in response to TLR4 stimulation. It will be also important to define whether posttranscriptional modifications of IRF5 required for optimal TNF transcription are reduced in newborn macrophages.
Previous studies have identified reduced activation of IRF family members as mechanisms underlying the limited capacity of neonatal DCs to mount proinflammatory responses. Lower IRF3 activity in newborn monocyte-derived DCs in response to TLR4 stimulation is associated with reduced expression of IFN-β, IL-12p70, and the IFN-inducible chemokines CXCL9, CXCL10, and CXCL11 (5). Moreover, the limited production of typeI/III IFNs by newborn plasmocytoid DCs exposed to herpes simplex virus-1 is linked to a reduced nuclear translocation of IRF7 (81). Combined altogether, these studies put forward a major role of IRFs in shaping the unique characteristics of newborn myeloid cells.
Our findings recognize characteristics of newborn macrophages that could be relevant to the vulnerability to infections observed during the neonatal period. Indeed, TNF is an early response cytokine that plays a crucial role in recruiting innate immune cells to sites of infection and promoting microbicidal activities. However, during established infections, excessive levels of TNF participate to the dysregulated immune responses that contribute to the pathogenesis of sepsis (74). Moreover, inflammation can cause considerable damage to developing organs, resulting in death or long-term disability (10). Thus, lower production of TNF by newborn macrophages exposed to microbial products could be advantageous to limit inflammatory responses during postnatal colonization of the skin and gastrointestinal tract and to reduce organ dysfunction and damage during systemic infection. The observation of a selective reduction in TNF secretion by newborn macrophages, while activation of major signaling pathways and production of other cytokines is maintained, supports the concept that immune responses are highly regulated to meet the specific requirements of early life. While we focused on differences between the developing neonatal immune system and the fully developed adult immune system, the absence of data from children is a limitation.
In summary, we identified distinct characteristics of the monocytic lineage in newborns that show limited IRF5 activation during monocyte to macrophage differentiation, and a specific reduction of TNF production upon TLR4 stimulation in M1 macrophages. These observations are relevant in the context of neonatal inflammation and infection and may provide a new potential target for immune modulating therapies during the neonatal period.
This study was carried out in accordance with the recommendations of Swiss Ethics Comittees on research involving human subjects. The protocol was approved by the Cantonal Human research Ethics Committee of Vaud (CER-VD, Lausanne, Switzerland). All subjects gave written informed consent in accordance with the Declaration of Helsinki.
EG and AS designed the study and wrote the first draft of the manuscript. AS and MW performed experiments. JH performed CyTOF studies. AS, JH, MP, TC, TR, and EG analyzed and interpreted the data. All the authors revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Société Académique Vaudoise to AS, the Lucien Picard Foundation, the ProTechno Foundation, the WEGH Foundation, and the Leenaards Foundation to EG and by grants from the Swiss National Science Foundation (146838 to EG and 173123 to TR). We thank Craig Fenwick and Giuseppe Pantaleo for their assistance in mass cytometry studies.
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.01597/full#supplementary-material.
1. Agyeman P, Schlapbach LJ, Giannoni E, Stocker M, Posfay-Barbe KM, Heininger U, et al. Epidemiology of blood culture-proven bacterial sepsis in children in Switzerland: a population-based cohort study. Lancet Child Adolesc Health (2017) 10(1):124–33. doi:10.1016/S2352-4642(17)30010-X
2. Liu L, Oza S, Hogan D, Chu Y, Perin J, Zhu J, et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet (2017) 388(10063):3027–35. doi:10.1016/S0140-6736(16)31593-8
3. Giannoni E, Agyeman P, Stocker M, Posfay-Barbe KM, Heininger U, Spycher BD, et al. Early-onset, hospital-acquired and community-acquired neonatal sepsis: a prospective population-based cohort study. J Pediatr (2018).
4. Zhang X, Zhivaki D, Lo-Man R. Unique aspects of the perinatal immune system. Nat Rev Immunol (2017) 17(8):495–507. doi:10.1038/nri.2017.54
5. Aksoy E, Albarani V, Nguyen M, Laes JF, Ruelle JL, De Wit D, et al. Interferon regulatory factor 3-dependent responses to lipopolysaccharide are selectively blunted in cord blood cells. Blood (2007) 109(7):2887–93. doi:10.1182/blood-2006-06-027862
6. Sharma AA, Jen R, Kan B, Sharma A, Marchant E, Tang A, et al. Impaired NLRP3 inflammasome activity during fetal development regulates IL-1beta production in human monocytes. Eur J Immunol (2015) 45(1):238–49. doi:10.1002/eji.201444707
7. Kollmann TR, Crabtree J, Rein-Weston A, Blimkie D, Thommai F, Wang XY, et al. Neonatal innate TLR-mediated responses are distinct from those of adults. J Immunol (2009) 183(11):7150–60. doi:10.4049/jimmunol.0901481
8. Shane AL, Sanchez PJ, Stoll BJ. Neonatal sepsis. Lancet (2017) 390(10104):1770–80. doi:10.1016/S0140-6736(17)31002-4
9. Balany J, Bhandari V. Understanding the impact of infection, inflammation, and their persistence in the pathogenesis of bronchopulmonary dysplasia. Front Med (2015) 2:90. doi:10.3389/fmed.2015.00090
10. Hagberg H, Mallard C, Ferriero DM, Vannucci SJ, Levison SW, Vexler ZS, et al. The role of inflammation in perinatal brain injury. Nat Rev Neurol (2015) 11(4):192–208. doi:10.1038/nrneurol.2015.13
11. Hodzic Z, Bolock AM, Good M. The role of mucosal immunity in the pathogenesis of necrotizing enterocolitis. Front Pediatr (2017) 5:40. doi:10.3389/fped.2017.00040
12. Carr R, Brocklehurst P, Dore CJ, Modi N. Granulocyte-macrophage colony stimulating factor administered as prophylaxis for reduction of sepsis in extremely preterm, small for gestational age neonates (the PROGRAMS trial): a single-blind, multicentre, randomised controlled trial. Lancet (2009) 373(9659):226–33. doi:10.1016/S0140-6736(09)60071-4
13. Carr R, Modi N, Dore C. G-CSF and GM-CSF for treating or preventing neonatal infections. Cochrane Database Syst Rev (2003) (3):CD003066. doi:10.1002/14651858.CD003066
14. Group IC, Brocklehurst P, Farrell B, King A, Juszczak E, Darlow B, et al. Treatment of neonatal sepsis with intravenous immune globulin. N Engl J Med (2011) 365(13):1201–11. doi:10.1056/NEJMoa1100441
15. Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol (2013) 14(10):986–95. doi:10.1038/ni.2705
16. Borges da Silva H, Fonseca R, Pereira RM, Cassado Ados A, Alvarez JM, D’Imperio Lima MR. Splenic macrophage subsets and their function during blood-borne infections. Front Immunol (2015) 6:480. doi:10.3389/fimmu.2015.00480
17. Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol (2015) 17(1):34–40. doi:10.1038/ni.3324
18. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity (2014) 41(1):14–20. doi:10.1016/j.immuni.2014.06.008
19. Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol (2016) 17(1):26–33. doi:10.1038/ni.3306
20. Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, et al. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol (2012) 188(11):5752–65. doi:10.4049/jimmunol.1103426
21. Hamilton TA, Zhao C, Pavicic PG Jr, Datta S. Myeloid colony-stimulating factors as regulators of macrophage polarization. Front Immunol (2014) 5:554. doi:10.3389/fimmu.2014.00554
22. Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci U S A (2004) 101(13):4560–5. doi:10.1073/pnas.0400983101
23. Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol (2011) 12(3):231–8. doi:10.1038/ni.1990
24. Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol (2009) 27:451–83. doi:10.1146/annurev.immunol.021908.132532
25. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol (2011) 11(11):723–37. doi:10.1038/nri3073
26. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity (2010) 32(5):593–604. doi:10.1016/j.immuni.2010.05.007
27. Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol (2006) 177(10):7303–11. doi:10.4049/jimmunol.177.10.7303
28. Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol (2014) 14(6):392–404. doi:10.1038/nri3671
29. Richardson ET, Shukla S, Nagy N, Boom WH, Beck RC, Zhou L, et al. ERK signaling is essential for macrophage development. PLoS One (2015) 10(10):e0140064. doi:10.1371/journal.pone.0140064
30. Yun MR, Seo JM, Park HY. Visfatin contributes to the differentiation of monocytes into macrophages through the differential regulation of inflammatory cytokines in THP-1 cells. Cell Signal (2014) 26(4):705–15. doi:10.1016/j.cellsig.2013.12.010
31. Coccia EM, Del Russo N, Stellacci E, Testa U, Marziali G, Battistini A. STAT1 activation during monocyte to macrophage maturation: role of adhesion molecules. Int Immunol (1999) 11(7):1075–83. doi:10.1093/intimm/11.7.1075
32. Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol (2014) 5:614. doi:10.3389/fimmu.2014.00614
33. Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol (2011) 11(11):750–61. doi:10.1038/nri3088
34. Giannoni E, Guignard L, Knaup Reymond M, Perreau M, Roth-Kleiner M, Calandra T, et al. Estradiol and progesterone strongly inhibit the innate immune response of mononuclear cells in newborns. Infect Immun (2011) 79(7):2690–8. doi:10.1128/IAI.00076-11
35. Roger T, Schneider A, Weier M, Sweep FC, Le Roy D, Bernhagen J, et al. High expression levels of macrophage migration inhibitory factor sustain the innate immune responses of neonates. Proc Natl Acad Sci U S A (2016) 113(8):E997–1005. doi:10.1073/pnas.1514018113
36. Roger T, Schlapbach LJ, Schneider A, Weier M, Wellmann S, Marquis P, et al. Plasma levels of macrophage migration inhibitory factor and d-dopachrome tautomerase show a highly specific profile in early life. Front Immunol (2017) 8:26. doi:10.3389/fimmu.2017.00026
37. Mombelli M, Lugrin J, Rubino I, Chanson AL, Giddey M, Calandra T, et al. Histone deacetylase inhibitors impair antibacterial defenses of macrophages. J Infect Dis (2011) 204(9):1367–74. doi:10.1093/infdis/jir553
38. Perreau M, Vigano S, Bellanger F, Pellaton C, Buss G, Comte D, et al. Exhaustion of bacteria-specific CD4 T cells and microbial translocation in common variable immunodeficiency disorders. J Exp Med (2014) 211(10):2033–45. doi:10.1084/jem.20140039
39. Zunder ER, Finck R, Behbehani GK, Amir el AD, Krishnaswamy S, Gonzalez VD, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc (2015) 10(2):316–33. doi:10.1038/nprot.2015.020
40. Ciarlo E, Heinonen T, Herderschee J, Fenwick C, Mombelli M, Le Roy D, et al. Impact of the microbial derived short chain fatty acid propionate on host susceptibility to bacterial and fungal infections in vivo. Sci Rep (2016) 6:37944. doi:10.1038/srep37944
41. Kittan NA, Allen RM, Dhaliwal A, Cavassani KA, Schaller M, Gallagher KA, et al. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One (2013) 8(10):e78045. doi:10.1371/journal.pone.0078045
42. McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF. Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci U S A (2013) 110(43):17253–8. doi:10.1073/pnas.1308887110
43. Gantner F, Kupferschmidt R, Schudt C, Wendel A, Hatzelmann A. In vitro differentiation of human monocytes to macrophages: change of PDE profile and its relationship to suppression of tumour necrosis factor-alpha release by PDE inhibitors. Br J Pharmacol (1997) 121(2):221–31. doi:10.1038/sj.bjp.0701124
44. Ambarus CA, Krausz S, van Eijk M, Hamann J, Radstake TR, Reedquist KA, et al. Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. J Immunol Methods (2012) 375(1–2):196–206. doi:10.1016/j.jim.2011.10.013
45. Beyer M, Mallmann MR, Xue J, Staratschek-Jox A, Vorholt D, Krebs W, et al. High-resolution transcriptome of human macrophages. PLoS One (2012) 7(9):e45466. doi:10.1371/journal.pone.0045466
46. Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood (2013) 121(9):e57–69. doi:10.1182/blood-2012-06-436212
47. Spence S, Fitzsimons A, Boyd CR, Kessler J, Fitzgerald D, Elliott J, et al. Suppressors of cytokine signaling 2 and 3 diametrically control macrophage polarization. Immunity (2013) 38(1):66–78. doi:10.1016/j.immuni.2012.09.013
48. Qin H, Holdbrooks AT, Liu Y, Reynolds SL, Yanagisawa LL, Benveniste EN. SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J Immunol (2012) 189(7):3439–48. doi:10.4049/jimmunol.1201168
49. Svensson J, Jenmalm MC, Matussek A, Geffers R, Berg G, Ernerudh J. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J Immunol (2011) 187(7):3671–82. doi:10.4049/jimmunol.1100130
50. Vogel DY, Glim JE, Stavenuiter AW, Breur M, Heijnen P, Amor S, et al. Human macrophage polarization in vitro: maturation and activation methods compared. Immunobiology (2014) 219(9):695–703. doi:10.1016/j.imbio.2014.05.002
51. Hofer TP, Zawada AM, Frankenberger M, Skokann K, Satzl AA, Gesierich W, et al. slan-defined subsets of CD16-positive monocytes: impact of granulomatous inflammation and M-CSF receptor mutation. Blood (2015) 126(24):2601–10. doi:10.1182/blood-2015-06-651331
52. Ancuta P. A slan-based nomenclature for monocytes? Blood (2015) 126(24):2536–8. doi:10.1182/blood-2015-10-675470
53. van de Laar L, Coffer PJ, Woltman AM. Regulation of dendritic cell development by GM-CSF: molecular control and implications for immune homeostasis and therapy. Blood (2012) 119(15):3383–93. doi:10.1182/blood-2011-11-370130
54. Ryzhakov G, Eames HL, Udalova IA. Activation and function of interferon regulatory factor 5. J Interferon Cytokine Res (2015) 35(2):71–8. doi:10.1089/jir.2014.0023
55. Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol (2012) 13(7):642–50. doi:10.1038/ni.2304
56. Zhang Q, Coveney AP, Yu S, Liu JH, Li Y, Blankson S, et al. Inefficient antimicrobial functions of innate phagocytes render infant mice more susceptible to bacterial infection. Eur J Immunol (2013) 43(5):1322–32. doi:10.1002/eji.201243077
57. Saito F, Kuwata H, Oiki E, Koike M, Uchiyama Y, Honda K, et al. Inefficient phagosome maturation in infant macrophages. Biochem Biophys Res Commun (2008) 375(1):113–8. doi:10.1016/j.bbrc.2008.07.141
58. Kurland G, Cheung AT, Miller ME, Ayin SA, Cho MM, Ford EW. The ontogeny of pulmonary defenses: alveolar macrophage function in neonatal and juvenile rhesus monkeys. Pediatr Res (1988) 23(3):293–7. doi:10.1203/00006450-198803000-00013
59. Bakker JM, Broug-Holub E, Kroes H, van Rees EP, Kraal G, van Iwaarden JF. Functional immaturity of rat alveolar macrophages during postnatal development. Immunology (1998) 94(3):304–9. doi:10.1046/j.1365-2567.1998.00518.x
60. Ballinger MN, Peters-Golden M, Moore BB. Impaired neonatal macrophage phagocytosis is not explained by overproduction of prostaglandin E2. Respir Res (2011) 12:155. doi:10.1186/1465-9921-12-155
61. Chelvarajan RL, Collins SM, Doubinskaia IE, Goes S, Van Willigen J, Flanagan D, et al. Defective macrophage function in neonates and its impact on unresponsiveness of neonates to polysaccharide antigens. J Leukoc Biol (2004) 75(6):982–94. doi:10.1189/jlb.0403179
62. Chelvarajan L, Popa D, Liu Y, Getchell TV, Stromberg AJ, Bondada S. Molecular mechanisms underlying anti-inflammatory phenotype of neonatal splenic macrophages. J Leukoc Biol (2007) 82(2):403–16. doi:10.1189/jlb.0107071
63. Lee PT, Holt PG, McWilliam AS. Ontogeny of rat pulmonary alveolar macrophage function: evidence for a selective deficiency in il-10 and nitric oxide production by newborn alveolar macrophages. Cytokine (2001) 15(1):53–7. doi:10.1006/cyto.2001.0894
64. Marodi L, Kaposzta R, Nemes E. Survival of group B streptococcus type III in mononuclear phagocytes: differential regulation of bacterial killing in cord macrophages by human recombinant gamma interferon and granulocyte-macrophage colony-stimulating factor. Infect Immun (2000) 68(4):2167–70. doi:10.1128/IAI.68.4.2167-2170.2000
65. Marodi L, Goda K, Palicz A, Szabo G. Cytokine receptor signalling in neonatal macrophages: defective STAT-1 phosphorylation in response to stimulation with IFN-gamma. Clin Exp Immunol (2001) 126(3):456–60. doi:10.1046/j.1365-2249.2001.01693.x
66. Kondo Y, Yasui K, Yashiro M, Tsuge M, Kotani N, Morishima T. Multi-nucleated giant cell formation from human cord blood monocytes in vitro, in comparison with adult peripheral blood monocytes. Clin Exp Immunol (2009) 158(1):84–90. doi:10.1111/j.1365-2249.2009.03990.x
67. Ulas T, Pirr S, Fehlhaber B, Bickes MS, Loof TG, Vogl T, et al. S100-alarmin-induced innate immune programming protects newborn infants from sepsis. Nat Immunol (2017) 18(6):622–32. doi:10.1038/ni.3745
68. Marodi L. Innate cellular immune responses in newborns. Clin Immunol (2006) 118(2–3):137–44. doi:10.1016/j.clim.2005.10.012
69. Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol (2008) 8(7):533–44. doi:10.1038/nri2356
70. Fleetwood AJ, Dinh H, Cook AD, Hertzog PJ, Hamilton JA. GM-CSF- and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. J Leukoc Biol (2009) 86(2):411–21. doi:10.1189/jlb.1108702
71. Trapnell BC, Whitsett JA. Gm-CSF regulates pulmonary surfactant homeostasis and alveolar macrophage-mediated innate host defense. Annu Rev Physiol (2002) 64:775–802. doi:10.1146/annurev.physiol.64.090601.113847
72. Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, et al. Identification of the haemoglobin scavenger receptor. Nature (2001) 409(6817):198–201. doi:10.1038/35051594
73. Dennery PA, Seidman DS, Stevenson DK. Neonatal hyperbilirubinemia. N Engl J Med (2001) 344(8):581–90. doi:10.1056/NEJM200102223440807
74. Wynn JL, Wong HR. Pathophysiology and treatment of septic shock in neonates. Clin Perinatol (2010) 37(2):439–79. doi:10.1016/j.clp.2010.04.002
75. Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, et al. Integral role of IRF-5 in the gene induction programme activated by toll-like receptors. Nature (2005) 434(7030):243–9. doi:10.1038/nature03308
76. Saliba DG, Heger A, Eames HL, Oikonomopoulos S, Teixeira A, Blazek K, et al. IRF5:RelA interaction targets inflammatory genes in macrophages. Cell Rep (2014) 8(5):1308–17. doi:10.1016/j.celrep.2014.07.034
77. Hedl M, Yan J, Abraham C. IRF5 and IRF5 disease-risk variants increase glycolysis and human M1 macrophage polarization by regulating proximal signaling and Akt2 activation. Cell Rep (2016) 16(9):2442–55. doi:10.1016/j.celrep.2016.07.060
78. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science (2014) 345(6204):1251086. doi:10.1126/science.1251086
79. Schmidt SV, Krebs W, Ulas T, Xue J, Bassler K, Gunther P, et al. The transcriptional regulator network of human inflammatory macrophages is defined by open chromatin. Cell Res (2016) 26(2):151–70. doi:10.1038/cr.2016.1
80. Sullivan KE, Reddy AB, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, et al. Epigenetic regulation of tumor necrosis factor alpha. Mol Cell Biol (2007) 27(14):5147–60. doi:10.1128/MCB.02429-06
Keywords: M1/M2 macrophages, newborns, innate immunity, interferon regulatory factor 5, monocytes, GM-CSF, LPS, tumor necrosis factor
Citation: Schneider A, Weier M, Herderschee J, Perreau M, Calandra T, Roger T and Giannoni E (2018) IRF5 Is a Key Regulator of Macrophage Response to Lipopolysaccharide in Newborns. Front. Immunol. 9:1597. doi: 10.3389/fimmu.2018.01597
Received: 06 April 2018; Accepted: 27 June 2018;
Published: 11 July 2018
Edited by:
Laurel L. Lenz, University of Colorado, United StatesReviewed by:
Evangelos Giamarellos-Bourboulis, National and Kapodistrian University of Athens, GreeceCopyright: © 2018 Schneider, Weier, Herderschee, Perreau, Calandra, Roger and Giannoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eric Giannoni, ZXJpYy5naWFubm9uaUBjaHV2LmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.