- 1Laboratório de Micobacteriologia, Instituto Nacional de Pesquisas da Amazônia (INPA), Manaus, Brazil
- 2Programa de Pós-graduação em Genética, Conservação e Biologia Evolutiva (PPG-GCBEv), Instituto Nacional de Pesquisas da Amazônia (INPA), Manaus, Brazil
- 3Laboratório de Ecologia de Doenças Transmissíveis na Amazônia, Instituto Leônidas e Maria Deane, Fiocruz Amazônia, Manaus, Brazil
- 4Programa de Pós-Graduação em Biologia da Interação Patógeno-Hospedeiro, Instituto Leônidas e Maria Deane, Fiocruz Amazônia, Manaus, Brazil
- 5Programa de Pós-Graduação em Imunologia Básica e Aplicada, Universidade Federal do Amazonas, Manaus, Brazil
- 6Programa de Pós-Graduação em Biologia Celular e Molecular, Instituto Oswaldo Cruz, Rio de Janeiro, Brazil
- 7Laboratório de Genética Animal, Instituto Nacional de Pesquisas da Amazônia (INPA/CPBA), Manaus, Brazil
- 8Laboratório de Imunologia Molecular, Instituto de Ciências Biológicas, Universidade Federal do Amazonas, Manaus, Brazil
- 9Faculdade de Medicina, Universidade Nilton Lins, Manaus, Brazil
- 10Fundação de Medicina Tropical Doutor Heitor Vieira Dourado (FMT-HVD), Manaus, Brazil
- 11Laboratório de Imunoquímica, Instituto de Ciências Biológicas, Universidade Federal do Amazonas, Manaus, Brazil
Tuberculosis (TB), caused by mycobacterial species of the Mycobacterium tuberculosis complex, is a serious global health issue. Brazil is among the 22 countries with the highest number of TB cases, and the state of Amazonas has the highest incidence of TB cases in the country. Toll-like receptors (TLRs) are important pattern recognition receptors of the innate immunity and play a key role in orchestrating an effective immune response. We investigated whether the single-nucleotide polymorphisms (SNPs) 1805T/G TLR1, 2258G/A TLR2, 896A/G and 1196C/T of TLR4, 745T/C TLR6, and −1237A/G and −1486A/G of TLR9 are associated with the predisposition to TB and/or bacillary load. The SNPs genotyping was performed by nucleotide sequencing in 263 TB patients and 232 healthy controls residing in the state of Amazonas. Alleles and genotypes frequencies were similar between patients and healthy individuals for most of the investigated SNPs. Stratification of the TB patients according to their bacillary load showed that the genotype 1805TT TLR1 (rs5743618) was prevalent among paucibacillary patients [odds ratio (OR) = 0.38; 95% confidence interval (CI) = 0.19–0.76; p = 0.009] while the genotype 1805TG was common among multibacillary patients (OR = 3.72; CI = 1.65–8.4; p = 0.004). Comparison of demographic characteristics of patients to controls showed that TB is strongly associated with smoking (OR = 6.55; 95% CI = 3.2–13.6; p < 0.0001); alcohol use disorder (OR = 7.14; 95% CI = 3.7–13.9; p < 0.0001); and male gender (OR = 3.66; 95% CI = 2.52–5.3; p < 0.0001). Multivariate logistic regression demonstrated that alcoholism (OR = 2.93; 95% CI = 1.05–8.16; p = 0.03) and the 1805G allele (OR = 2.75; 95% CI = 1.33–5.7; p = 0.006) are predictive variables for multibacillary TB. Altogether, we suggest that the TLR1 1805G allele may be a relevant immunogenetic factor for the epidemiology of TB together with environmental, sociodemographic, and behavioral factors.
Introduction
Tuberculosis (TB) is a contagious disease caused by mycobacteria of the Mycobacterium tuberculosis complex. The most common clinical form is pulmonary TB. TB causes a serious public health burden worldwide and its control remains a major challenge for the scientific community and TB control programs globally. Nearly 10 million people worldwide suffered from TB each year (1). Brazil is one of the 30 countries considered a priority for TB control as more than 69 thousand new cases occur annually. The state of Amazonas has the highest incidence of TB in the country with 67.2/100 thousand inhabitants (1, 2).
Multiple socioeconomic, behavioral, medical, and genetic factors affect TB infection (3, 4). Many single-nucleotide polymorphisms (SNPs) in immune response genes are associated with TB (5–7). The most reported are HLA-DRB1, VDR, TIRAP, and NRAMP1 (6, 8). Other genes such as toll-like receptors (TLRs), TLR1, 2, 4, 6, and 9 that are involved in the initial recognition of pathogen-associated molecular patterns to trigger the activation of the effector mechanisms of innate immunity and subsequent targeting of the specific adaptive immune response to Mycobacterium tuberculosis have also been associated with susceptibility or resistance to TB (6, 9, 10).
In this study, we investigated whether the SNPs rs5743618 (1805T/G) of TLR1, rs5743708 (2258G/A) of TLR2, rs4986790 (896A/G) and rs4986791 (1196C/T) of TLR4, rs5743810 (745C/T) of TLR6, and the rs5743836 (−1237A/G) and rs187084 (−1486A/G) of TLR9 are associated with TB and with the bacterial load. We identified that alcoholism and the TLR1 1805G allele may be predictive variables for multibacillary TB.
Materials and Methods
Study Population
This is a case-control study investigating the association of demographic and immunogenetics factors with susceptibility or resistance to TB. Patients with TB and healthy controls, aged 18–65 years, were recruited in Manaus, the capital city of Amazonas state, Brazil. The patients with TB were recruited at the Policlínica de Referência em Pneumologia Sanitária Cardoso Fontes, a referral center of the state for diagnosis and treatment of TB. Patients with a diagnosis of pulmonary and pleural TB were selected according to guidelines of the Brazilian Ministry of Health. Data collected included social-demographic characteristics and the presence of the following clinical symptoms: cough, expectoration, chest pain, dyspnea, hemoptysis, malaise, fever, chills, night sweats, loss of appetite, weight loss, and fatigue. The controls consisted of health professionals and healthy contacts without consanguinity to the patients with TB and without any previous history of the disease. TB patients with associated comorbidities or lack of information on the patient’s chart or subsequent diagnosis of non-tuberculous mycobacteria were excluded. TB patients were stratified into paucibacillary (including pleural TB) and multibacillary. The bacillary load was determined by bacilloscopy, as recommended by Brazilian Ministry of Health (11). This study protocol was approved by the Institutional Review Board of the National Institute of Amazonian Research (INPA) under the number CAAE: 05925212.8.0000.0006.
Identification of M. tuberculosis
Sputum samples were submitted to microscopy (12) and solid culture in according to PKO method (initials in tribute to Petroff, Kudoh, and Ogawa) (13). A smear of culture, stained by Kinyoun method, was carried out to confirm if the culture was composed of acid-fast bacilli. Later, the colony aspect (not pigmented and rough morphology) was evaluated. The DNA extraction from pure culture was made as described elsewhere (14). So, the molecular identification of M. tuberculosis complex was performed by PCR, amplification of 123 bp fragment from IS6110 insertion sequence, as described (15). If, the PCR for IS6110 was negative, the differential diagnosis of mycobacteria was performed as established (12, 16). This study also included samples (32/263) with diagnosis confirmed for TB by the molecular test, GeneXpert MTB/RIF [M. tuberculosis/rifampin (RIF) resistance] assay (Cepheid, Sunnyvale, CA, USA).
Genotyping of the Different TLR Polymorphisms
DNA from whole blood samples was extracted using a QIAamp DNA Blood kit (QIAGEN) in the automated QIAcube platform following the manufacturer’s instructions. The primers were designed to flank the different polymorphisms studied using the Primer-3 version 2.3.7 embedded in Geneious software version 6.1.7 (17) and Primer-BLAST (Table S1 in Supplementary Material). PCR conditions for the five evaluated genes were optimized separately to use only one cycling amplification program of an initial cycle of 94°C for 2 min followed by 33 cycles of 94°C for 2 min, 63°C for 30 s and 72°C for 60 s, and a final cycle of 72°C for 17 min. The PCR mix in a final volume of 25 µL contains 1× Buffer; 1.5 mM MgCl2; 0.2 mM dNTPs; 0.4 µM of sense and antisense primers; enzyme GoTaq Hot Start 1 U/μL; and approximate 100 ng of DNA. PCR products were precipitated with polyethylene glycol (PEG) (PEG 8000 at 20% P/V, 2.5 M NaCl) based on the protocol originally described by Lis and Schleif (18), with slight modifications subsequently by Lis (19) and Paithankar and Prasad (20). The purified amplicons were adjusted to the concentration range of 5–20 ng/µL, proportional to their size. The allelic discrimination was performed by direct capillary sequencing. Sequencing reaction was performed with the BigDye® Terminator Kit (3.1) (Life technologies) using the protocol suggested by Platt et al. (21). Either sense or antisense primers used in PCR of each gene was used. The sequencing product was purified with Ethanol/EDTA/Sodium Acetate, according to the recommendations of Life Technologies and submitted to capillary electrophoresis in the ABI 3130 Genetic Analyzer (Applied Biosystems) and the resulting electropherograms were edited and analyzed using the Sequencing Analysis Program (Life Technologies, version 5.3.1) and Geneious.
Statistical Analysis
Parametric Student’s t-test was used for means age comparison. Two-tailed Fisher’s exact test was applied for association of TB with BCG vaccine, sex, smoking, and alcoholism. The frequency of the alleles and genotypes between groups was compared with two-tailed Fisher’s exact test. Variables with a p value were considered significant. Genetic models also were applied to elucidate the association of the SNPs with susceptibility to TB and bacillary load. Akaike information criterion (AIC) was considered in the evaluation of applied genetic models. The free software R, version 3.1.2 and the embedded function to study SNP association—SNPassoc1 were used. The multivariate logistic regression analysis with stepwise approach was applied in the choice of variables predictive of multibacillary TB. The analyzed variables were age, sex, BCG, alcoholism, and the presence of the 1805G allele. Variables with a p value <0.05 were considered and the variables p >0.1 were excluded from the regression analysis. For each regression model proposed, the discrimination capacity of each model was analyzed as described previously by Boechat et al. (22, 23). The model with the largest area under the curve (AUC) was considered. Statistical analyses were performed using MedCalc Statistical Software version for Windows, version 15.2 (MedCalc Software, Ostend, Belgium2).
Results
Baseline Characteristics of All the Participants of the Study
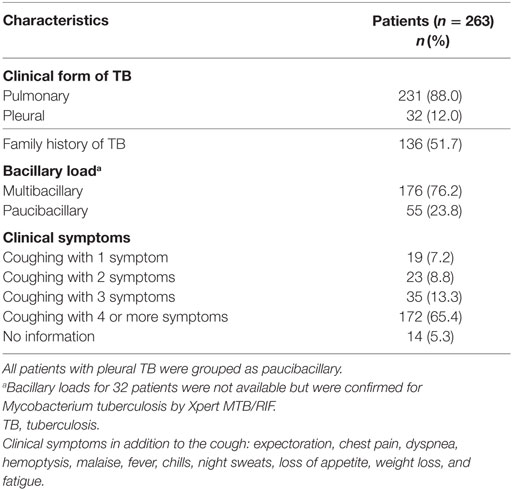
This study included 263 patients with TB and 232 healthy controls. The baseline characteristics of the study population are shown in Table 1. The age distribution between TB patients and controls was similar. TB patients had a mean of 33.4 ± 11.9 years of age and were predominantly male (65.7%). By contrast, the control group was predominantly female (65.9%). BCG vaccine adherence was higher among the controls, 92.0% compared to 78.7% among the TB patients. Smoking and alcoholism were significantly associated with a greater predisposition to TB. The clinical characteristics of the patients with TB are shown in Table 2. Pulmonary TB (88%) was the most prevalent and the majority of the patients reported more than three clinical symptoms.
Comparison of the Genotypes and Alleles Frequencies of the Polymorphisms of the Different TLRs Between TB Patients and Healthy Controls
The genotypes and alleles frequencies of the different SNPs studied are shown in Table 3. Differences in the number of samples for each SNP are due to the rigorous adoption of only high-quality sequences analyzed. The Hardy–Weinberg equilibrium (HWE) was observed for the different SNPs in both cases and healthy controls except for both SNPs of TLR4. Further analysis of the SNPs of TLR4 was abandoned due the deviations from the HWE. The frequencies of the genotypes and alleles of the other SNPs were similar between patients and controls except for the SNPs of TLR9. Notably, TLR2 SNP 2258G/A was not observed in our population of study. TLR9 −1237A/G and −1486A/G SNPs showed minor allele frequency (MAF) for patient populations and controls of 15.3 and 17.4% for the first SNP and 38.5 and 33.3% for the second, respectively. The genotype −1237AG was associated with TB protection [odds ratio (OR): 0.62; confidence interval (CI): 0.39–0.97; p = 0.04].
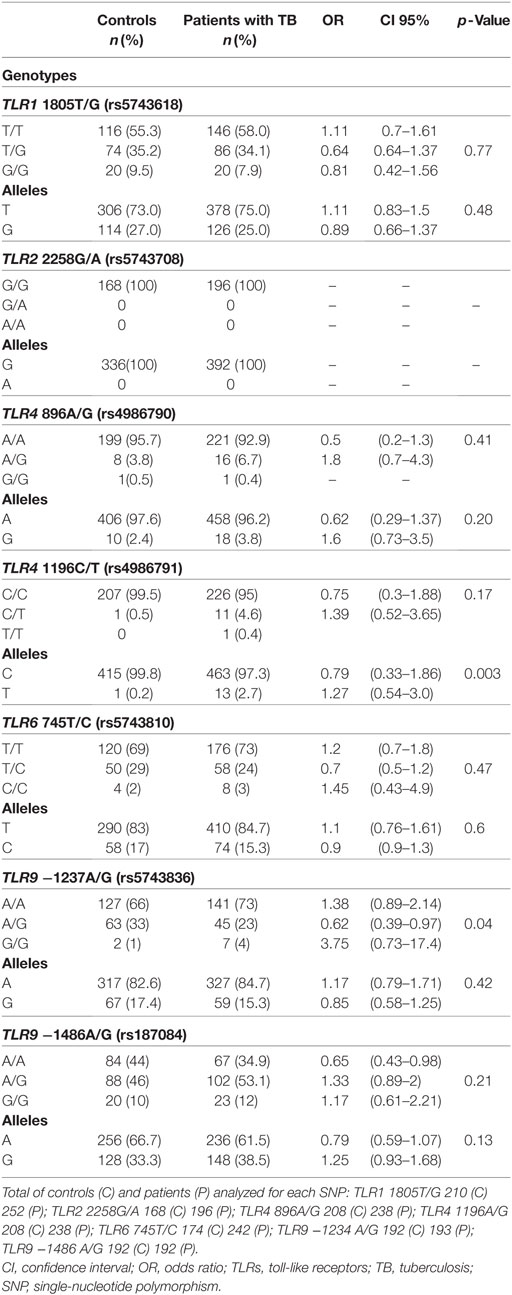
Table 3. Genotypes and allele frequencies of the TLRs polymorphisms in healthy controls and patients with TB.
Association of TLR1 SNP 1805T/G and Multibacillary TB
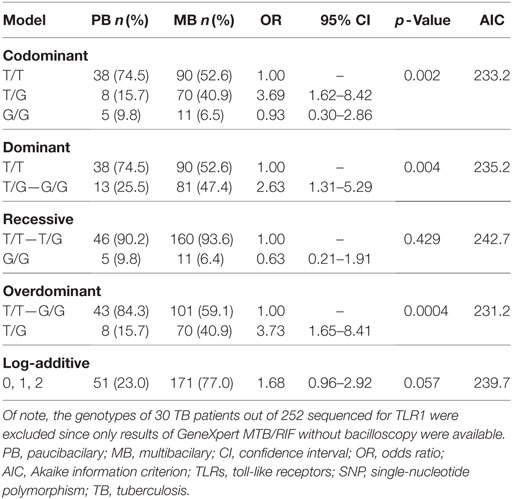
Patients with TB were stratified into paucibacillary and multibacillary TB. Comparison of genotypes and allele frequencies of all the SNPs between multibacillary and paucibacillary TB revealed that the genotype 1805TT TLR1 (rs5743618) was prevalent among paucibacillary patients (OR = 0.38; 95% CI = 0.19–0.76; p = 0.009) while the genotype 1805TG was common among multibacillary patients (OR = 3.72; CI = 1.65–8.4; p = 0.004). The 1805G allele (OR = 1.71; CI = 0.97–3.01; p = 0.05) showed statistical trend for association. Further analysis applying four genetic models as shown in Table 4 showed that the codominant model with the genotype (1805TG) was associated with the multibacillary TB form (OR = 3.69; CI = 1.62–8.42; p = 0.002). The same genotype was associated with the multibacillary form in the overdominant model (OR = 3.73; CI = 1.65–8.41; p = 0.0004). In the dominant model, the genotypes (1805TG + 1805GG) also showed association with the multibacillary TB (OR = 2.63; CI = 1.31–5.29; p = 0.004). The overdominant model better explains the association to multibacillary TB according to AICs results.
Alcoholism and the TLR1 SNP Guide Predisposition Multibacillary TB
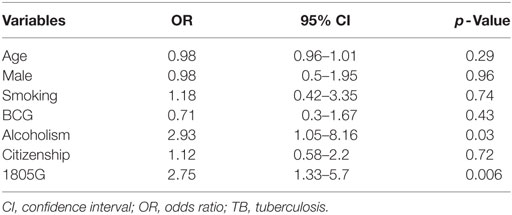
Multivariate logistic regression analysis was applied to adjust baseline characteristics, using a logistic regression model for multibacillary TB shown in Table 5, including gender, age, smoking status, alcoholism, and citizenship. There was no influence for gender in the analysis (p = 0.96). Logistic regression showed the variables alcoholism (OR = 2.93; CI = 1.05–8.16; p = 0.03) and allele 1805G (OR = 2.75; CI = 1.33–5.7; p = 0.006) were associated with the multibacillary TB (Table 5).
Logistic regression with a stepwise approach only showed alcoholism (OR = 3.04; CI = 1.21–7.67; p = 0.01) and allele 1805G (OR = 2.59; CI = 1.28–5.26; p = 0.008) were associated with the multibacillary TB. The two variables were subsequently challenged in two regression models. Model 1 that included only the variable alcoholism, obtained an AUC of 0.58. In the model 2 that included both the variables alcoholism and SNP 1805T/G, the AUC increased to 0.64 suggesting that both alcoholism and SNP 1805T/G act together in the establishment of multibacillary TB.
Discussion
Tuberculosis continues to be a health problem worldwide and with the emergence of HIV there is a resurgence of TB cases among AIDS patients and becomes barriers to the roll back of TB. Enormous headway has been achieved in the understanding of the immunopathology of TB. Understanding the puzzle of the host genetics contribution to the resistance or susceptibility to MTB infection may help in the strategy of designing new vaccines to TB or in the combination of chemotherapy with immunotherapy.
None of the SNPs investigated in this study were associated with a greater predisposition to TB. Only the TLR9 −1237AG genotype was associated with TB protection. Stratification of the patients into multibacillary and paucibacillary TB did not show any association with the bacillary load. Few studies evaluated the importance of SNPs −1237A/G and −1486A/G of TLR9 in TB, and results are conflicting. Two cited no association with TB (24, 25) while two reported association with higher risk for TB (26, 27). The predisposition to TB is known to be complex and is determined by many genetic variations in a multiple of genes contributing to smaller inputs in the development of the disease (5, 10).
We observed the 1805TG genotype is associated for the first time to the best of our knowledge with multibacillary TB. Interestingly, it has been shown that TLR1 with this variation is hyporesponsive to its agonists and impedes its migration to the surface of cells expressing it (28–30). Among the demographic and behavioral factors investigated, only alcoholism contributed to the multibacillary form.
A previous study reported that TLR1 regulates polypeptide-induced signaling and responds to extracts of M. tuberculosis, suggesting that TLR1 may substantially regulate the immune response against the bacillus and impact on the outcome of TB. Authors also demonstrated that subjects with TLR1 SNP1805G/G genotype produce low concentrations of IL-6, an important cytokine in the inflammatory response. Heterozygous individuals produce intermediate levels suggesting that the G and T allele are codominant. The association of heterozygous individuals to multibacillary TB in this study may in part be explained by the influence of the G allele that correlates to low level of IL-6 to inefficiently control the multiplication of the bacillus (29). However, the effect of the hypo-responsiveness of this variation on the risk of TB and the bacillary load is not clearly understood due to the complex immune response to TB and the immune escape developed by M. tuberculosis (31, 32).
In this study, we observed that males, smokers, and alcoholism were more common among the TB patients. Furthermore, family history of TB was reported in more than 50% of the patients reinforcing the importance of intra-domiciliary TB transmission or the component of genetic factors in the predisposition to TB. Previous studies have also observed similar features and reinforced that smoking, alcohol, and other drug abuse, along with socioeconomic factors, increase the risk for TB development (33).
There is biological evidence to explain the association of smoking with TB. Chronic exposure to cigarette components alters the normal functioning of mucociliary clearance. This facilitates the access of the bacilli to the pulmonary alveoli, giving rise to an infectious focus. Macrophages resident in the alveola of smokers also present reduced phagocytic action and decreased level of pro-inflammatory cytokines. Nicotine, one of many toxic constituents found in cigarettes, acts directly on acetylcholine receptors in macrophages and leads to a lower secretion of TNF-alpha, generating a favorable environment for bacillary survival in the macrophages and contributes to immune evasion (34, 35).
Variations in the frequency of SNPs suggest distinct evolutionary histories among populations in recent human evolution due to selective pressure exerted by diseases, such as TB and Malaria (36). This study observed MAF for SNP 1805T/G of TLR1 of 25.0 and 27.0% in TB patients and controls, respectively. Previous studies of other infectious diseases in Brazil showed frequencies varying between 11.0 and 40.0% (37–40). Frequency of the MAF in other populations such as Spain, Turkey, and Nepal are 30.0, 43.0, and 6.0%, respectively (28, 41, 42). The 1805G mutant allele is rare in Vietnamese population (28).
TLR2 SNP 2258G/A was not observed in our study. This SNP is also rare in four populations investigated by the HapMap project and in a population from Colombia (43). The TLR4 896A/G and 1196C/T SNPs are co-segregated and the frequency of haplotype of the mutant alleles ranges from 0 to 5.0% among different populations worldwide. However, this frequency is 10–20 times higher among African populations and it is suggested that may be a result of selective pressure due to malaria infection (44, 45). Our study observed an MAF of 3.8 and 2.4% for 896A/G, and 2.7 and 0.2% for the 1196C/T SNP among patients and controls, respectively. Other studies in Brazil showed higher MAF varying from 4.6 to 7.0% (37, 38, 46). However, one study of case-controls of malaria from the Amazonas observed similar MAF to ours indicating the frequency is a reflex of the local population (40).
The MAF of TLR6 745C/T varies from 0 to 46.0% among 11 populations investigated by the HapMap. In this study, the MAF is 15.3 and 17% in TB patients and controls, respectively. Another study in Brazil reported an MAF of 18.0 and 10.0% in individuals of symptomatic and asymptomatic malaria, respectively (37). The MAF of TLR9 −1237A/G and −1486A/G SNPs are 15.3 and 17.4% for the first SNP and 38.5 and 33.3% for the second in patients and controls, respectively. Similar high MAF for these SNPs were observed in other studies in the Amazonas (37, 40).
Our study has few limitations. Our sample size is small, and male is prevalent among the TB cases. However, this should not have a great influence on the study as the frequencies of the alleles of males and females are similar among the controls. Moreover, new study with a higher sample size is needed to confirm the results observed in this study.
The predisposition to TB and its bacillary load involves numerous factors that are determined mainly by the human intrinsic factors and the virulence of the mycobacterial strain. Further studies are necessary to evaluate the combined effect of these factors in TB. In addition, more studies are needed to understand the functional effect of this TLR1 variant to elucidate its importance in the regulation of the immune response and in the immunopathogenesis of TB.
Ethics Statement
This study protocol was approved by the Institutional Review Board of the Instituto Nacional de Pesquisas da Amazônia (INPA) under the number CAAE: 05925212.8.0000.0006. All patients gave written informed consent.
Author Contributions
RB-N, FN, AS, AB, RR, and JP conceived and designed the study. RB-N, VA, MO, and AS were responsible for acquisition and quality of the clinical and laboratory data. RB-N, VA, AS, and MO performed the identification of Mycobacterium tuberculosis. RB-N, FN, and GS performed the genotyping of the different TR polymorphisms. RB-N, FN, and AB conducted data mining and statistical analysis. RB-N, FN, MO, AS, AB, and RR wrote the first version of the paper. All authors were involved in the interpretation of the data and participated in the final writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MS and handling Editor declared their shared affiliation.
Acknowledgments
We are thankful to the staff of Policlínica de Referência em Pneumologia Sanitária Cardoso Fontes, Manaus, Amazonas, Brazil. We are also grateful to the Programa de Apoio à Pós-Graduação-Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (PROAP/CAPES), Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM), and the Programa de Desenvolvimento Tecnológico em Insumos para a Saúde—PDTIS-FIOCRUZ for the use of the nucleotide sequencing facilities at ILMD—Fiocruz Amazônia. We are grateful to Aileen Chang, from George Washington University, for English reviewing of the manuscript. RB-N received a CAPES fellowship.
Funding
This work was supported by FAPEAM, SUSAM, DECIT, CNPq (Call: Programa de Pesquisa para o SUS: Gestão compartilhada em Saúde—PPSUS 2009), and FAPEAM Call: UNIVERSAL AMAZONAS (grant number: 472/2012).
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.01455/full#supplementary-material.
Footnotes
- ^https://cran.r-project.org/web/packages/SNPassoc/index.html (Accessed: March 09, 2016).
- ^https://www.medcalc.org/ (Accessed: January 18, 2016).
References
1. World Health Organization. Global Tuberculosis Report 2017. Geneva: World Health Organization (2017).
2. Brasil. Secretaria de Vigilância em Saúde − Ministério da Saúde. Indicadores prioritários para o monitoramento do Plano Nacional pelo Fim da Tuberculose como Problema de Saúde Pública no Brasil. (2017). Available from: http://portalarquivos2.saude.gov.br/images/pdf/2017/marco/23/2017-V-48-N-8-Indicadores-priorit--rios-para-o-monitoramento-do-Plano-Nacional-pelo-Fim-da-Tuberculose-como-Problema-de-Sa--de-P--blica-no-Brasil.pdf (Accessed: December 19, 2017).
3. Casanova JL, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol (2011) 29:447–91. doi:10.1146/annurev-immunol-030409-101335
4. Shaler CR, Horvath C, Lai R, Xing Z. Understanding delayed T-cell priming, lung recruitment, and airway luminal T-cell responses in host defense against pulmonary tuberculosis. Clin Dev Immunol (2012) 2012:628293. doi:10.1155/2012/628293
5. van Tong H, Velavan TP, Thye T, Meyer CG. Human genetic factors in tuberculosis: an update. Trop Med Int Health (2017) 22:1063–71. doi:10.1111/tmi.12923
6. Stein CM, Sausville L, Wejse C, Sobota RS, Zetola NM, Hill PC, et al. Genomics of human pulmonary tuberculosis: from genes to pathways. Curr Genet Med Rep (2017) 5:149–66. doi:10.1007/s40142-017-0130-9
7. Wang Y, Zhang MM, Huang WW, Wu SQ, Wang MG, Tang XY, et al. Polymorphisms in toll-like receptor 10 and tuberculosis susceptibility: evidence from three independent series. Front Immunol (2018) 9:309. doi:10.3389/fimmu.2018.00309
8. Souza de Lima D, Morishi Ogusku M, Pôrto Dos Santos M, de Melo Silva CM, Alves de Almeida V, Assumpção Antunes I, et al. Alleles of HLA-DRB1*04 associated with pulmonary tuberculosis in Amazon Brazilian population. PLoS One (2016) 11:e0147543. doi:10.1371/journal.pone.0147543
9. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34:637–50. doi:10.1016/j.immuni.2011.05.006
10. Schurz H, Daya M, Möller M, Hoal EG, Salie M. TLR1, 2, 4, 6 and 9 variants associated with tuberculosis susceptibility: a systematic review and meta-analysis. PLoS One (2015) 10:e0139711. doi:10.1371/journal.pone.0139711
11. BRASIL. Ministério da Saúde. Manual de recomendações para o controle da tuberculose no Brasil. (2011). Available from: http://bvsms.saude.gov.br/bvs/publicacoes/manual_recomendacoes_controle_tuberculose_brasil.pdf (Accessed: May 20, 2018).
12. BRASIL. Ministério da Saúde. Secretaria de Vigilância em Saúde. Departamento de Vigilância. Manual Nacional de Vigilância Laboratorial da Tuberculose e outras Micobactérias. Brasília: Ministério da Saúde (2008). 434 p.
13. Salem JI, Carvalho CM, Ogusku MM, Maia R, Ruffino-Netto A. PKO: alternative method for isolating mycobacteria from sputum. Acta Amaz (2007) 37:419–24. doi:10.1590/S0044-59672007000300013
14. Leão SC, Martin A, Mejia GI, Palomino JC, Robledo J, da Silva Telles MA, et al. Practical Handbook for the Phenotypic and Genotypic Identification of Mycobacteria. Brussels: European Commission International Co-operation (INCO) for Developing Countries (DEV) Concerted Action (CA) (2004). 165 p.
15. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol (1993) 31:406–9.
16. Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol (1993) 31:175–8.
17. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (2012) 28:1647–9. doi:10.1093/bioinformatics/bts199
18. Lis JT, Schleif R. Size fractionation of double-stranded DNA by precipitation with polyethylene glycol. Nucleic Acids Res (1975) 2:383–9. doi:10.1093/nar/2.3.383
19. Lis JT. Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods Enzymol (1980) 65:347–53. doi:10.1016/S0076-6879(80)65044-7
20. Paithankar KR, Prasad KSN. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res (1991) 19:1346–1346. doi:10.1093/nar/19.6.1346
21. Platt AR, Woodhall RW, George AL. Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. Biotechniques (2007) 43:60–2. doi:10.2144/000112499
22. Boechat N de O, Ogusku MM, Boechat AL, Sadahiro A. Interaction between smoking and HLA-DRB1*04 gene is associated with a high cardiovascular risk in Brazilian Amazon patients with rheumatoid arthritis. PLoS One (2012) 7:e41588. doi:10.1371/journal.pone.0041588
23. Boechat AL, Ogusku MM, Sadahiro A, Dos-Santos MC. Association between the PTPN22 1858C/T gene polymorphism and tuberculosis resistance. Infect Genet Evol (2013) 16:310–3. doi:10.1016/j.meegid.2013.02.019
24. Selvaraj P, Harishankar M, Singh B, Jawahar MS, Banurekha VV. Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis (Edinb) (2010) 90:306–10. doi:10.1016/j.tube.2010.08.001
25. Jahantigh D, Salimi S, Alavi-Naini R, Emamdadi A, Owaysee Osquee H, Farajian Mashhadi F. Association between TLR4 and TLR9 gene polymorphisms with development of pulmonary tuberculosis in Zahedan, southeastern Iran. ScientificWorldJournal (2013) 2013:534053–7. doi:10.1155/2013/534053
26. Velez DR, Wejse C, Stryjewski ME, Abbate E, Hulme WF, Myers JL, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet (2010) 127:65–73. doi:10.1007/s00439-009-0741-7
27. Torres-García D, Cruz-Lagunas A, García-Sancho Figueroa MC, Fernández-Plata R, Baez-Saldaña R, Mendoza-Milla C, et al. Variants in toll-like receptor 9 gene influence susceptibility to tuberculosis in a Mexican population. J Transl Med (2013) 11:220. doi:10.1186/1479-5876-11-220
28. Johnson CM, Lyle EA, Omueti KO, Stepensky VA, Yegin O, Alpsoy E, et al. Cutting edge: a common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol (2007) 178:7520–4. doi:10.4049/jimmunol.178.12.7520
29. Hawn TR, Misch EA, Dunstan SJ, Thwaites GE, Lan NTN, Quy HT, et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur J Immunol (2007) 37:2280–9. doi:10.1002/eji.200737034
30. Qi H, Sun L, Wu X, Jin Y, Xiao J, Wang S, et al. Toll-like receptor 1(TLR1) Gene SNP rs5743618 is associated with increased risk for tuberculosis in Han Chinese children. Tuberculosis (Edinb) (2015) 95:197–203. doi:10.1016/j.tube.2014.12.001
31. Hart BE, Tapping RI. Cell surface trafficking of TLR1 is differentially regulated by the chaperones PRAT4A and PRAT4B. J Biol Chem (2012) 287:16550–62. doi:10.1074/jbc.M112.342717
32. Hart BE, Tapping RI. Differential trafficking of TLR1 I602S underlies host protection against pathogenic mycobacteria. J Immunol (2012) 189:5347–55. doi:10.4049/jimmunol.1201545
33. Lin H-H, Ezzati M, Murray M. Tobacco smoke, indoor air pollution and tuberculosis: a systematic review and meta-analysis. PLoS Med (2007) 4:e20. doi:10.1371/journal.pmed.0040020
34. Sopori M. Effects of cigarette smoke on the immune system. Nat Rev Immunol (2002) 2:372–7. doi:10.1038/nri803
35. Wang H, Yu M, Ochani M, Amella CA, Tanovic M, Susarla S, et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature (2003) 421:384–8. doi:10.1038/nature01339
36. Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet (2001) 2:967–77. doi:10.1038/35103577
37. Leoratti FMS, Farias L, Alves FP, Suarez-Mútis MC, Coura JR, Kalil J, et al. Variants in the toll-like receptor signaling pathway and clinical outcomes of malaria. J Infect Dis (2008) 198:772–80. doi:10.1086/590440
38. Ramasawmy R, Cunha-Neto E, Fae KC, Borba SCP, Teixeira PC, Ferreira SCP, et al. Heterozygosity for the S180L variant of MAL/TIRAP, a gene expressing an adaptor protein in the toll-like receptor pathway, is associated with lower risk of developing chronic Chagas cardiomyopathy. J Infect Dis (2009) 199:1838–45. doi:10.1086/599212
39. Marques C de S, Brito-de-Souza VN, Guerreiro LTA, Martins JH, Amaral EP, Cardoso CC. Toll-like receptor 1 N248S single-nucleotide polymorphism is associated with leprosy risk and regulates immune activation during mycobacterial infection. J Infect Dis (2013) 208:120–9. doi:10.1093/infdis/jit133
40. Costa AG, Ramasawmy R, Ibiapina HNS, Sampaio VS, Xábregas LA, Brasil LW, et al. Association of TLR variants with susceptibility to Plasmodium vivax malaria and parasitemia in the Amazon region of Brazil. PLoS One (2017) 12:e0183840. doi:10.1371/journal.pone.0183840
41. Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One (2007) 2:e1318. doi:10.1371/journal.pone.0001318
42. Misch EA, Hawn TR. Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci (2008) 114:347–60. doi:10.1042/CS20070214
43. Sánchez D, Lefebvre C, Rioux J, García LF, Barrera LF. Evaluation of Toll-like receptor and adaptor molecule polymorphisms for susceptibility to tuberculosis in a Colombian population. Int J Immunogenet (2012) 39:216–23. doi:10.1111/j.1744-313X.2011.01077.x
44. Ferwerda B, McCall MB, Verheijen K, Kullberg B-J, van der Ven AJ, Van der Meer JW, et al. Functional consequences of toll-like receptor 4 polymorphisms. Mol Med (2008) 14:346–52. doi:10.2119/2007-00135.Ferwerda
45. Ferwerda B, Kibiki GS, Netea MG, Dolmans WMV, van der Ven AJ. The toll-like receptor 4 Asp299Gly variant and tuberculosis susceptibility in HIV-infected patients in Tanzania. AIDS (2007) 21:1375–7. doi:10.1097/QAD.0b013e32814e6b2d
Keywords: tuberculosis, immunogenetics, single-nucleotide polymorphism, toll-like receptors, Brazil
Citation: Barletta-Naveca RH, Naveca FG, Almeida VAd, Porto JIR, Silva GAVd, Ogusku MM, Sadahiro A, Ramasawmy R and Boechat AL (2018) Toll-Like Receptor-1 Single-Nucleotide Polymorphism 1805T/G Is Associated With Predisposition to Multibacillary Tuberculosis. Front. Immunol. 9:1455. doi: 10.3389/fimmu.2018.01455
Received: 28 February 2018; Accepted: 12 June 2018;
Published: 25 June 2018
Edited by:
Antonio C. R. Vallinoto, Institute of Biological Sciences (ICB) of Federal University of Pará, BrazilReviewed by:
Shashank Gupta, Brown University, United StatesMaísa Silva De Sousa, Universidade Federal do Pará, Brazil
Gaddam Suman Latha, Osmania University, India
Copyright: © 2018 Barletta-Naveca, Naveca, Almeida, Porto, Silva, Ogusku, Sadahiro, Ramasawmy and Boechat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raphaela Honorato Barletta-Naveca, cmFwaGFlbGFuYXZlY2FAZ21haWwuY29t;
Felipe Gomes Naveca, ZmVsaXBlLm5hdmVjYUBmaW9jcnV6LmJy
 Raphaela Honorato Barletta-Naveca
Raphaela Honorato Barletta-Naveca Felipe Gomes Naveca
Felipe Gomes Naveca Vanessa Alves de Almeida
Vanessa Alves de Almeida Jorge Ivan Rebelo Porto
Jorge Ivan Rebelo Porto George Allan Villarouco da Silva
George Allan Villarouco da Silva Mauricio Morishi Ogusku
Mauricio Morishi Ogusku Aya Sadahiro
Aya Sadahiro Rajendranath Ramasawmy
Rajendranath Ramasawmy Antonio Luiz Boechat
Antonio Luiz Boechat