
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 12 June 2018
Sec. Immunological Tolerance and Regulation
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.01338
 Johannes Baur1
Johannes Baur1 Christoph Otto2
Christoph Otto2 Ulrich Steger1
Ulrich Steger1 Stefan Klein-Hessling3
Stefan Klein-Hessling3 Khalid Muhammad3
Khalid Muhammad3 Tobias Pusch3
Tobias Pusch3 Krisna Murti3
Krisna Murti3 Rhoda Wismer3
Rhoda Wismer3 Christoph-Thomas Germer1
Christoph-Thomas Germer1 Ingo Klein4
Ingo Klein4 Nora Müller5
Nora Müller5 Edgar Serfling3*
Edgar Serfling3* Andris Avots3*
Andris Avots3*
The immune suppressants cyclosporin A (CsA) and tacrolimus (FK506) are used worldwide in transplantation medicine to suppress graft rejection. Both CsA and FK506 inhibit the phosphatase calcineurin (CN) whose activity controls the immune receptor-mediated activation of lymphocytes. Downstream targets of CN in lymphocytes are the nuclear factors of activated T cells (NFATs). We show here that the activity of NFATc1, the most prominent NFAT factor in activated lymphocytes supports the acute rejection of heterotopic heart allografts. While ablation of NFATc1 in T cells prevented graft rejection, ectopic expression of inducible NFATc1/αA isoform led to rejection of heart allografts in recipient mice. Acceptance of transplanted hearts in mice bearing NFATc1-deficient T cells was accompanied by a reduction in number and cytotoxicity of graft infiltrating cells. In CD8+ T cells, NFATc1 controls numerous intracellular signaling pathways that lead to the metabolic switch to aerobic glycolysis and the expression of numerous lymphokines, chemokines, and their receptors, including Cxcr3 that supports the rejection of allogeneic heart transplants. These findings favors NFATc1 as a molecular target for the development of new strategies to control the cytotoxicity of T cells upon organ transplantation.
The worldwide application of cyclosporin A (CsA) and tacrolimus (FK506) as immune suppressants in transplantation medicine saved the lives of thousands of patients. CsA and FK506 prevent organ rejection by inhibiting the activity of phosphatase calcineurin (CN) that orchestrates the activation of peripheral lymphocytes upon antigen contact (1). In cells, CsA binds to CypA, a low molecular weight immunophilin, and these complexes bind to CN and suppress its enzymatic activity (2). However, due to the numerous side effects caused by CsA and FK506, sustained applications of both immune suppressants not only lead to severe complications in transplant patients, in particular to nephrotoxicity, but also to neurotoxicity, hypertension, fibrosis, and cancerogenesis (3, 4). Although CsA and FK506 inhibit in lymphocytes primarily the activation of nuclear factors of activated T cell (NFAT) transcription factors (TFs), NFATs are not the only TFs whose activity is affected by CsA. Since CN supports the induction of NF-κB factors upon TCR stimulation, CsA interferes also with their TCR-mediated activation (5, 6).
Due to the many deleterious side effects of CsA and FK506 in therapy, numerous searches have been undertaken for novel CN inhibitors, or for inhibitors that suppress directly the activation of NFATs. One set of such inhibitors are cell-penetrating peptides that interfere with the interaction between CN and NFATs (7). Based on the affinity-driven selection of peptides corresponding to the main interaction site of NFATs with CN (8), several peptide versions of general structure PxIxIT (where x represents any natural amino acid) were shown to inhibit the interaction between CN and NFATs (9). They interfere with the binding of CN to an NFAT peptide that is highly conserved between all NFAT proteins and, thereby, those inhibitory peptides do not distinguish between individual NFAT members. Since, however, the individual NFAT proteins differ remarkably in their activity in vivo (10), for therapeutic interventions inhibitors have to identified that allow the specific block of individual NFAT proteins.
Among the treatments that suppressed the rejection of allogeneic transplanted hearts in mice (3) there are several therapies that affect the activation of NFATs and/or their targets in T cells. Apart from the use of CN inhibitors CsA and FK506, metabolic inhibitors and the inactivation of NF-κB or Cxcr3, a chemokine receptor gene, led to acceptance of transplanted allogeneic hearts (11–13). We showed previously that NF-κB factors support the induction of short NFATc1/αA in B lymphocytes (14). We will show here that NFATc1 ablation prolongs allograft survival, impairs the metabolic switch from oxidative phosphorylation (OXPHOS) to aerobic glycolysis in activated CD8+ T cells and the expression of chemokine receptor Cxcr3 in CTLs. These data indicate NFATc1 as a key factor in activated T cells that controls the rejection of transplanted allogeneic hearts.
Male C57BL/6J (B6, H-2b) and BALB/c (H-2d) wild-type (WT) mice were purchased from Janvier (France). Nfatc1flx/flx and Nfatc2−/− mouse lines have been described previously. Nfatc1flx/flx mice were crossed with CD4-cre mice for inactivating the Nfatc1 gene in all T cells, and with mb1-cre mice in all B cells (15–20). In dlck-cre x Nfatc1flx/flx mice, the Nfatc1 gene is inactivated in peripheral T cells. In mice of the dlck-cre x Nfatc1flx/flxx caNfatc1-STOPflx/flx line, a constitutively active version of NFATc1/αA is expressed from the Rosa26 locus upon removal of a “floxed” STOP sequence and inactivation of endogenous Nfatc1 gene in peripheral T cells (21, 22). Nfatc1P2Δ mice carry an Nfatc1 P2 promoter deletion and, due to a CMV-promoter-driven cre are deficient for P2-directed transcripts [see Figure S1 in Supplementary Material and Ref. (23)]. Bacterial artificial chromosome (BAC) transgenic (tg) mice expressing NFATc1/A-Bio protein [and BirA, the biotin-ligase from E. coli (24)] have been described previously in Ref. (25). All mice were maintained in the Central Animal Facility of the Medical Faculty (ZEMM), University of Wuerzburg, according to the institutional guidelines (acceptance AKZ 55.2-2531.01-80/10 from 22.10.2010).
For CD8+ T cell isolation, the “CD8 (Ly2) microbeads, mouse” kit (Miltenyi Biotech) was used. For αCD3/CD28 stimulation, 5 µg CD3ε (clone 145-2C11) and 2 µg CD28 (clone 37.51) (both BD Pharmingen) were used to coat multi well plates. T cells were also stimulated with 10 ng/ml tetradecanoylphorbol-13-acetate and 0.5 µM ionomycin (normally for 5 h).
Abdominal heterotopic heart transplantation into mice was performed as described previously in Ref. (26).
Heart grafts cut into small pieces were incubated in 100 U/ml collagenase at 37°C for 30 min. Cells were washed with phosphate-buffered saline (PBS), counted, and purified on a Ficoll-Hypaque gradient.
Freshly explanted heart grafts were fixed in 4% paraformaldehyde and embedded in paraffin. For hematoxylin–eosin staining, 4-µm sections were de-paraffinized with xylene, rehydrated in absolute ethanol, stained in hematoxylin solution, and counter-stained with eosin. For immune histochemical staining, de-paraffinized and rehydrated sections (1 µm) were heated for antigen unmasking in 10 mM sodium citrate buffer (pH 6.0), and stained with αCXCR3 (CD183, #bs-2209R, Bioss Antibodies, Inc. MA, USA), diluted 1:200 in antibody (Ab) dilutent (DAKO, Hamburg, Germany) at 4°C overnight. Sections were washed in PBS and incubated with 1:100 diluted horseradish-labeled goat anti-rabbit IgG (DAKO, P0448) at room temperature for 1 h. Staining was developed by adding 3,3′diaminobenzidine (DAB ready to use, DAKO) and counterstaining was done with hematoxylin.
Upon isolation, splenic CD8+ T cells were stimulated with αCD3/CD28 Abs for 24 h, attached to poly-l-Lysin-coated chamber μ-slides, fixed in 4% formaldehyde, permeabilized with 0.2% Triton-X100, and blocked with 5% BSA. Samples were incubated with primary mouse anti-NFATc1 Ab 7A6 in 1% BSA at 4° overnight.
Graft-infiltrating cells were isolated from heart grafts at day 5 after transplantation. Purification of RNA from GICs and transcriptome assays were performed at TRON GmbH (Mainz, Germany) as described previously in Ref. (27).
CD8+ T cells from BAC tg mice expressing BirA, or from tg mice expressing BirA and Nfatc1/A-Bio, a Bio-tagged version of NFATc1/αA [(25); Figure S4 in Supplementary Material] were differentiated to CTLs in vitro. Upon re-stimulation with T + I (5 ng/ml and 0.5 µM) for 5 h, chromatin from 1 × 107 fixed cells was prepared and sonicated, followed by precipitation of chromatin on streptavidin beads (M-280, #11205D, Thermo Fisher Scientific, MA, USA) (28). After cDNA library preparation (using a NEBNext® Ultra™ DNA Library Prep Kit for Illumina®) sequencing was performed as a 50-bp single read run on an Illumina HiSeqTM2500 (Illumina, San Diego, CA, USA) using a TruSeq® Rapid SBS Kit v2 and a HiSeq Rapid Flow Cell v2. 50 bp sequence reads that passed the Illumina quality filtering were aligned to the mouse genome assembly version of July 2007 (NCB I37/mm9), using the map with Bowtie for Illumina 1.1.2. Results were visualized with the “Integrative Genomics Viewer” IGV version 2.3.81 (29).
Western blots were performed using the NFATc1-specific mAbs 7A6 and NFATc2 mAb (# 556602 and 5062574, BD Pharmingen) for NFAT detection.
Lymphocyte stainings were performed by incubation for 20 min on ice using conjugated the following mAbs (eBioscience, San Diego, CA, USA): CD8-FITC (#11-0081-82), CD4-FITC(#MCD0401), CXCR3-APC (#17-1831-82), CCR5-PE (#12-1951-81), CD62L-PE (#12-0621-81), CD44-APC (#17-0441-81), CD25-APC (#17-0251-82), B220 FITC (#11-0452-82), CD5-PE (#12-0051-82), CD23-APC (#MCD2305).
The mineral oil induced murine MOPC-315.BMP2.FUGLW55 plasmacytoma cell line (H2b) expressing eGFP and luciferase was used as target cell line. To measure luciferase during coculture the assays were performed by cultivating MOPC-luciferase target cells together with equal numbers of GICs isolated from WT B6 or Nfatc1flx/flxx CD4-cre recipient mice during the acute rejection phase (d6, effector/target ratio 4/1, followed by 20 or 36 h incubation). After centrifugation, the cell pellet was washed once with PBS and finally re-suspended in 100 µl harvesting buffer to lyse cell membranes. 50 µl of supernatant was transferred into a white, non-transparent 96-well plate. The LUMIstar Omega was primed with 1 ml of ready-to-use luciferin solution before luciferase-activity measurement was performed. 50 µl of luciferin solution per well were automatically added to the sample and measurement was performed.
The extra cellular acidification rate (ECAR) of CD8+ T cells was measured in a XF96 analyzer (Seahorse Bioscience) as described previously in Ref. (25).
Student’s t-test or the Mann–Whitney U test was used for statistical analysis with the software GraphPad Prism 6. P values above 0.05 were considered not significant.
In a previous report on the survival of heterotopic heart and skin transplants, in Nfatc2- or Nfatc3-deficient recipient mice a moderate prolongation of allogeneic transplant survival was described in Ref. (30). These observations prompted us to investigate whether NFATc1, the most prominent NFAT factor in activated peripheral T and B cells, plays a role in the rejection of allogeneic organ transplants. In a model of heterotopic heart transplantation, we used Nfatc1flx/flx mice crossed with the CD4-cre or mb1-cre mouse lines to ablate the expression of NFATc1 in peripheral T or B lymphocytes, respectively (18, 20, 22). Fully allogeneic hearts from donor BALB/c mice transplanted into WT B6 recipient mice are normally rejected within 7 days by the recipient’s immune system. However, BALB/c heart allografts transplanted into Nfatc1flx/flxxCD4-cre mice bearing both NFATc1-deficient CD4+ and CD8+ T cells survived significantly longer for more than 50 days, and more than 50% of the allografts for more than 120 days. In contrast, transplantation of allogeneic hearts into Nfatc1flx/flxx mb1-cre mice bearing Nfatc1−/− B cells had only a minor positive effect on the survival of transplanted allogeneic hearts (Figure 1A).
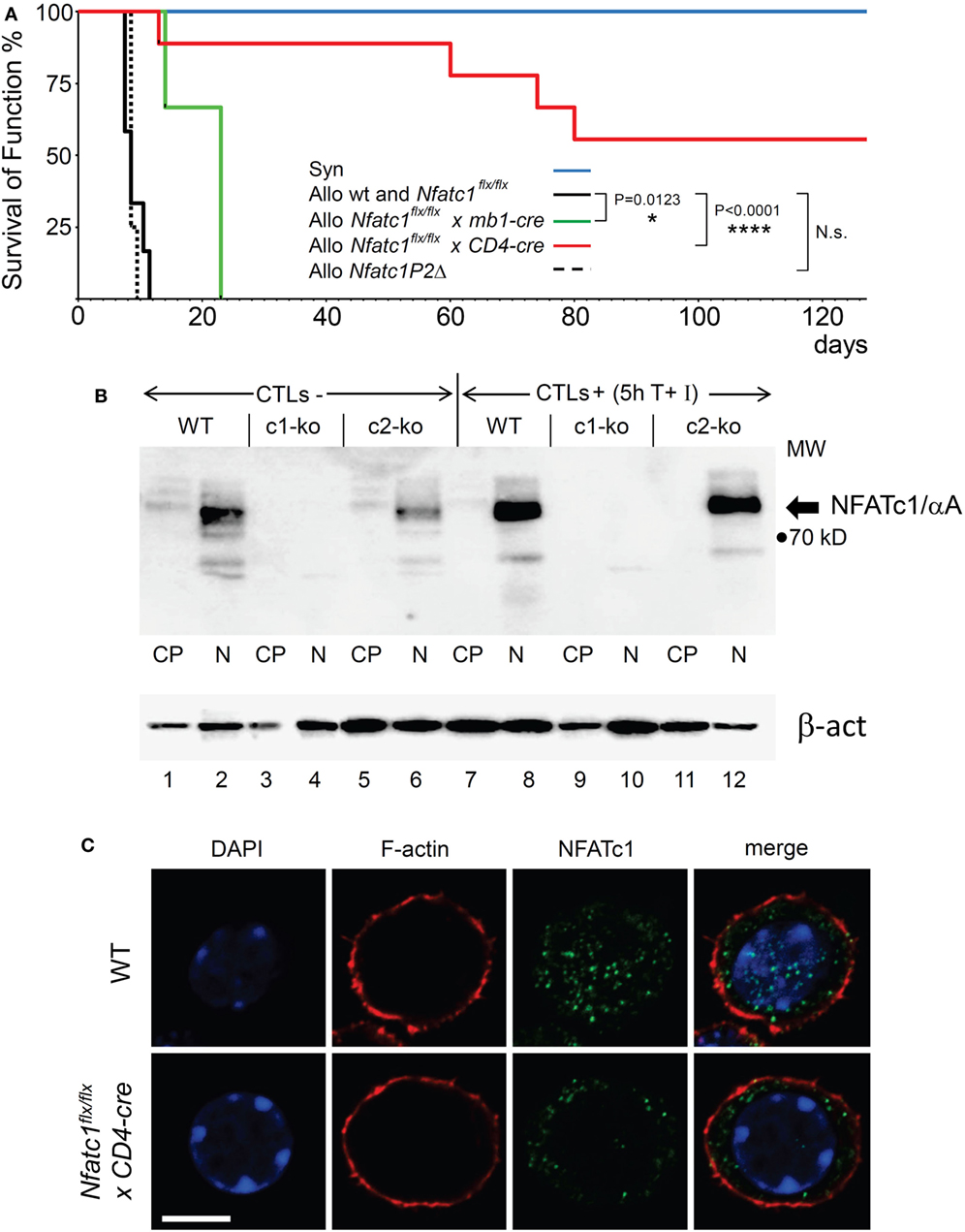
Figure 1. NFATc1 deficiency in T cells prevents acute rejection of allogeneic heterotopic heart transplants. (A) Syngeneic (C57BL/6, Syn) or fully allogenic (BALB/c, Allo) heart grafts were transplanted into wild-type (WT) C57BL6, Nfatc1flx/flx x mb1-cre, Nfatc1flx/flx x CD4-cre, or Nfatc1P2Δ recipient mice (n = 5–9 recipients per group). While Nfatc1flx/flx x mb1-cre mice bear NFATc1-deficient B cells, Nfatc1flx/flx x CD4-cre mice bear NFATc1-deficient CD4+ and CD8+ T cells. Nfatc1P2Δ mice carry an Nfatc1 P2 promoter deletion and, therefore, are deficient for P2-directed transcripts (23). The survival of heart function was monitored daily during the first 3 weeks after transplantation, then four times per week. (B) Representative western blot showing the efficient NFATc1 ablation in cytotoxic T cells (CTL) from Nfatc1flx/flx x CD4-cre (c1-ko) mice. Cytoplasmic (CP) and nuclear proteins (N) from CTLs of WT, Nfatc1flx/flx x CD4-cre (c1-ko) and Nfatc2−/− (c2-ko) mice were fractionated. For the generation of CTLs in vitro, splenic CD8+ T cells were stimulated by αCD3/CD28 antibodies (Abs) for 3 days, followed by culture in IL-2 containing medium for further 3 days. They were left un-induced (CTL-) or induced by T + I (CTL+) for 5 h (see Materials and Methods). The prominent band of NFATc1/αA, the inducible, short NFATc1 isoform, is indicated. (C) Staining of splenic CD8+ T cells from WT or Nfatc1flx/flxxCD4-cre mice activated for 24 h by αCD3/CD28 Abs in vitro. Cytospins of cells were stained with Abs raised against F-actin or NFATc1 (7A6 mAb), and counter-stained by DAPI. One typical stain of more than 50 cells analyzed is shown. Bar, 10 µm.
In lymphocytes, the Nfatc1 gene is expressed in six individual α- and β-isoforms at the RNA and protein levels. These are generated under the control of two alternate promoters, P1 and P2, and polyA sites, pA1 and pA2, and, therefore, differ in their N- and C-terminal peptides (Figure S1 in Supplementary Material) (31). In T cells from Nfatc1flx/flxx CD4-cre mice, the expression of all isoforms should be ablated (Figure 1B and unpubl. data), but upon staining of immunoblots or cytospins prepared with NFATc1−/− CD8+ T cells using the 7A6 NFATc1 mAb various shorter protein bands or stained dots were detected. However, these NFATc1-like proteins were not imported into the nucleus upon activation of NFATc1−/− T cell cells (Figure 1C) and, therefore, should play a minor, if any role in transcriptional regulation.
To identify the role of individual NFATc1 isoforms in transplant rejection, we crossed Nfatc1-P2flx/flx mice created in our laboratory (32) with CMV-cre mice for the deletion of P2 promoter and exon 2 sequences. In such Nfatc1P2Δ mice, the generation of NFATc1/β isoforms was inhibited [see Figure S1 in Supplementary Material and Ref. (23)]. Nfatc1P2Δ mice were born at expected Mendelian ratios, were viable and developmentally indistinguishable from their Nfatc1P2flx/flx littermates, and their lymphoid compartments remained unaffected. Both T and B cells from Nfatc1P2Δ mice could be normally activated ex vivo and did not reveal any defects in proliferation or induction of Activation-Induced Cell Death (Figure S2 in Supplementary Material). However, the rejection of allogeneic hearts upon transplantation into Nfatc1P2Δ mice was detected on the same day as upon transplantation into WT B6 mice (Figure 1A). This led us to conclude that NFATc1/β proteins play a minor role in graft rejection.
In order to demonstrate that—as we assumed—the induction of the short NFATc1 isoform NFATc1/αA, the most prominent NFATc1 protein in cytotoxic T cells (Figure 1B) (25), plays an important role in the rejection of allogeneic heart transplants, we used caNfatc1-STOPflx/flx mice as recipients. Those mice express a constitutive active (ca) version of NFATc1/αA (caNFATc1) from the murine Rosa26 locus upon cre-mediated removal of a “floxed” STOP sequence (21). However, to our surprise (over-) expressing caNFATc1/αA in thymocytes of CD4-cre mice was not tolerated since all T cells expressing solely caNFATc1/αA were eliminated during positive selection and did not appear in peripheral organs of mice (unpubl. data). Therefore, we crossed caNfatc1-STOPflx/flx mice with dlck-cre x Nfatc1flx/flx mice that express cre under the control of distal lck promoter and, thereby, caNFATc1/αA in peripheral T cells (but no endogenous NFATc1). In peripheral CD8+ T cells from all dlck-cre x Nfatc1flx/flxx caNfatc1-STOPflx/flx mice that we investigated, we observed caNFATc1/αA expression. In addition, each transplantation of allogeneic BALB/c hearts into dlck-cre x Nfatc1flx/flxx caNfatc1-STOPflx/flx mice led to acute rejection around day 9 (mean survival time, MST), comparable to MST observed in C57BL/6 WT mice expressing endogenous NFATc1 (Figures 2A,B).
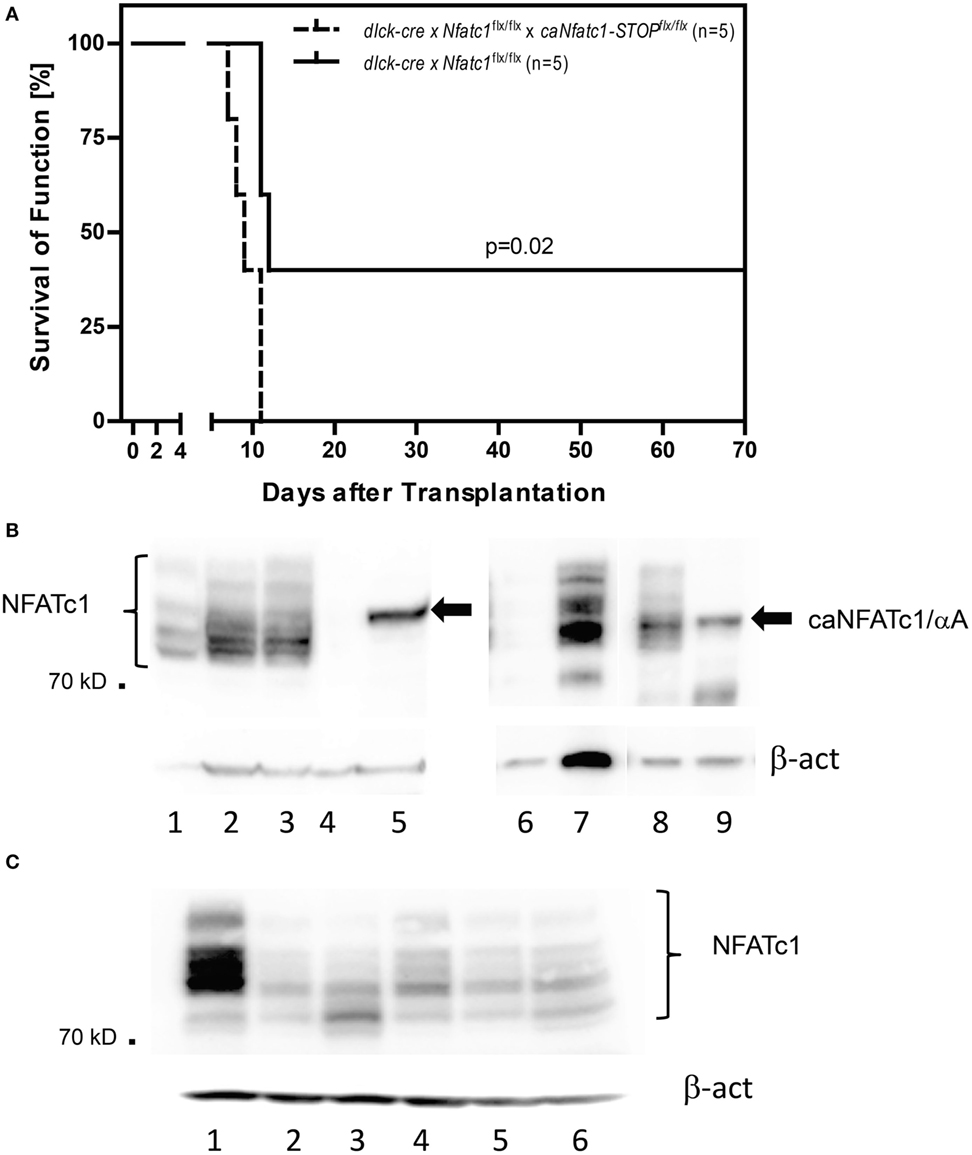
Figure 2. Expression of NFATc1/αA in peripheral T cells leads to rejection of allogeneic heterotopic heart transplants. (A) Allogenic heart grafts (of BALB/c mice) were transplanted into five dlck-cre x Nfatc1flx/flx mice, or five dlck-cre x Nfatc1flx/flx x caNfatc1-STOPflx/flx mice expressing a constitutive active (ca) version of NFATc1/αA in peripheral T cells. (B) Immunoblots showing the expression of caNFATc1/αA in CD8+ T cells from two out of five dlck-cre x Nfatc1flx/flx x caNfatc1-STOPflx/flx mice. In lanes 1 and 6, cytosolic protein, in lanes 2, 3, and 7 nuclear protein from wild-type (WT) CD8+ T cells stimulated by αCD3/CD28 antibodies (Abs) in vitro for 2 days was fractionated. In lanes 4 and 8, cytosolic protein, and in lanes 5 and 9 nuclear protein of CD8+ T cells activated for 2 days from dlck-cre x Nfatc1flx/flx x caNfatc1-STOPflx/flx mice was fractionated. (C) Immunoblot showing NFATc1 expression in CD8+ T cells from five dlck-cre x Nfatc1flxl/flx mice. In lane 1, whole cell protein of WT CD8+ T cells stimulated by αCD3/CD28 Abs in vitro for 2 days was fractionated, in lanes 2–6, whole cell protein of activated CD8+ T cells from dlck-cre x Nfatc1flx/flx mice.
Compared to the sustained survival of allogeneic transplanted hearts in the majority of Nfatc1flx/flxx CD4-cre recipient mice for 60 days and longer (Figure 1A), we detected a long-term survival (>60 days) of heart allografts in two out of five dlck-cre x Nfatc1flx/flx recipient mice (Figure 2A). In immunoblots using protein of activated CD8+ T cells from five dlck-cre x Nfatc1flx/flx mice, we detected numerous, albeit weak bands with the same or a similar size of the endogenous NFATc1 (Figure 2C). This might be due to an incomplete ablation of endogenous NFATc1 in splenic CD8+ T cells of dlck-cre x Nfatc1flx/flx mice. A deletion efficiency of approximately 80% has been reported for the dlck-cre line in CD8+ T cells, whereas a markedly higher efficiency rate was detected for the murine CD4-cre line (33).
In allogeneic BALB/c hearts, we observed a threefold to fivefold increase of GICs at days 5 and 6 during the acute rejection phase, compared to the number of GICs in transplanted syngeneic hearts (Figures 3A,B). However, approximately the same number of GICs we detected in allogeneic hearts transplanted into Nfatc1flx/flxx CD4-cre mice (Figure 3B), and in long-term accepted heart transplants (>100 days after transplantation) a further decrease in number of GICs was detected (Figure 3B). Compared to GICs isolated from rejected hearts, the cytotoxicity of GICs from accepted hearts of Nfatc1flx/flxx CD4-cre mice was significantly impaired, reaching approximately one-third of cytotoxicity of GICs from rejected hearts (Figure 3C).
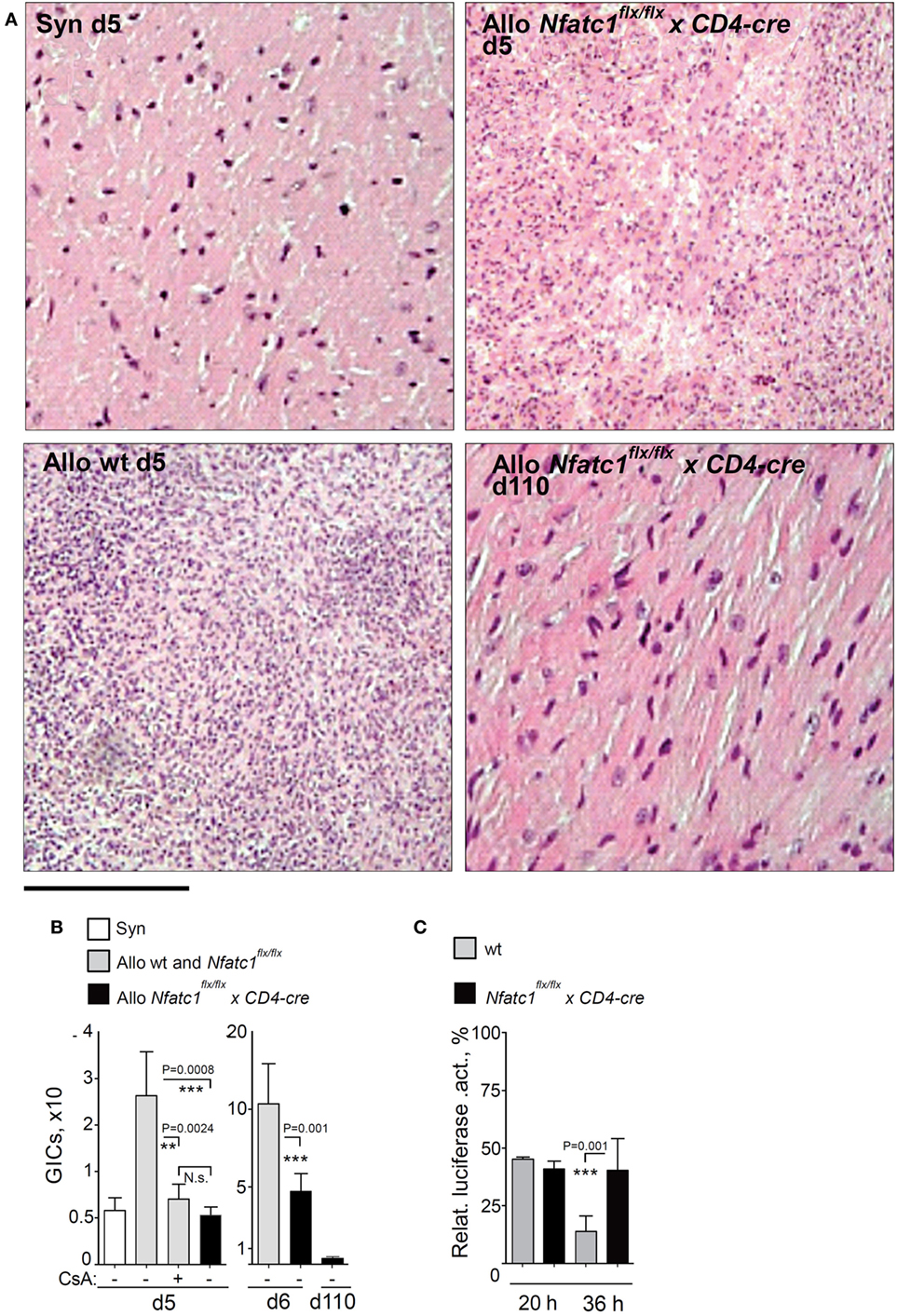
Figure 3. Graft-Infiltrating Cells (GICs) in transplanted hearts. (A) Representative hematoxylin–eosin stains of grafts from syngeneic and allogeneic wild-type (WT) recipients, and from Nfatc1flx/flx x CD4-cre recipients during the acute rejection phase (d5), and from Nfatc1flx/flx x CD4-cre recipients at d110. Bar, 300 µm. (B) Quantification of GICs in transplanted allogeneic hearts with (+) and without (−) cyclosporin A (CsA) treatment of animals. (C) Cytotoxicity of GICs. Luciferase assays were performed after cultivation of MOPC-luciferase target cells together with equal numbers of GICs isolated from WT or Nfatc1flx/flx xCD4-cre recipient mice during the acute rejection phase (d6, effector/target ratio 4/1, followed by 20 or 36 h incubation).
To define molecular targets of NFATc1 in GICs of transplanted hearts, we determined by next-generation sequencing (NGS) the transcriptomes of GICs isolated from syngeneic and allogeneic grafts on day 5 after transplantation, i.e., during the acute rejection phase of allogeneic grafts, and from allogeneic grafts of Nfatc1flx/flxx CD4-cre mice. Some of the B6 WT recipients of allogeneic grafts were treated with CsA (n = 5). In GICs from untreated allografts, the transcript levels of T cell signature genes (e.g., of Cd3g,d,e, Cd4, Cd8a,b, and Il2ra,rb genes) and of genes coding for signal molecules in T cells (of Lck, Zap70, and Fyn genes) were found to be increased, compared to GICs from syngeneic mice. In GICs from Nfatc1fl/flx CD4-cre and CsA-treated WT recipients the transcript levels were lower, compared to GICs from untreated allogeneic recipients (Figure 4A). This is also true for the Gzmb and Prf1 genes encoding the most prominent granzymes and perforins in CTLs.
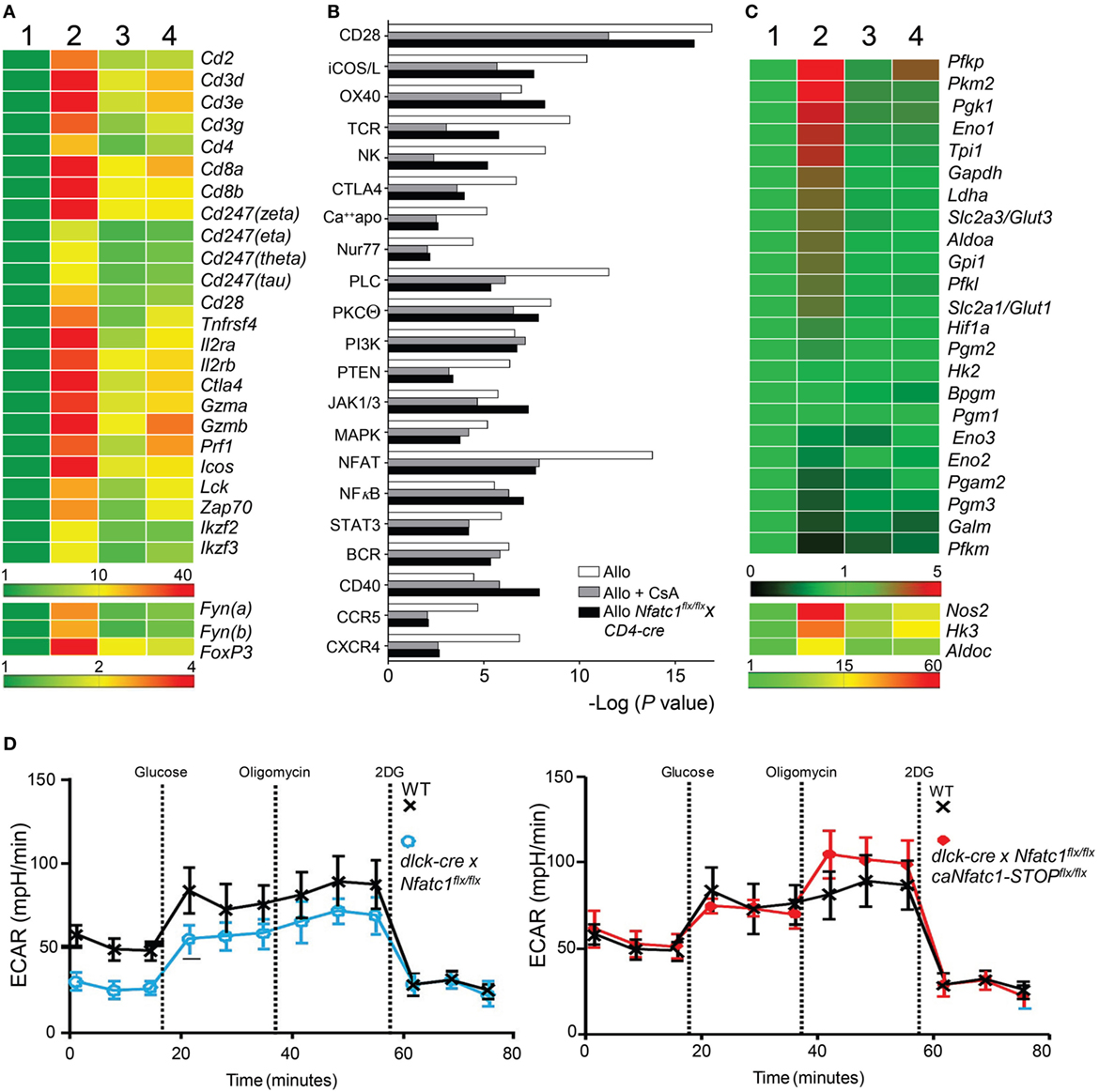
Figure 4. Cyclosporin A (CsA)-sensitive nuclear factors of activated T cell (NFAT) signaling is a major transcriptional pathway in Graft-Infiltrating Cells (GICs) from allogeneic heterotopic heart transplants. Syngeneic (C57BL/6, Syn) or fully allogeneic (BALB/c, Allo) heart grafts were transplanted into wild-type (WT) or NFATc1-deficient (Nfatc1flx/flxxCD4-cre) C57BL/6 recipients. A subgroup of WT recipients was treated daily with CsA (15 mg/kg, Allo CsA). GICs were purified during acute rejection phase (d5) and subjected to next-generation sequencing (NGS) assays. (A) RNA expression profiles (“heat map”) of T cell signature genes. GICs were purified from syngeneic recipients (lane 1), allogeneic recipients in the absence or presence of CsA (lanes 2 and 3, respectively), and from recipients bearing Nfatc1−/− T cells (lane 4). Their RNAs were isolated and subjected to NGS. (B) Ingenious Pathway Analyses. Expression data were normalized to the transcriptome of GICs in syngeneic transplantations. (C) RNA expression of genes encoding glycolytic enzymes. (D) Extracellular flux analyses of activated CD8+ Ts from WT and dlck-cre x Nfatc1flx/flx mice (left), and from WT and dlck-cre x Nfatc1flx/flx x caNfatc1-STOPflx/flx mice (right). Data of three independent assays are shown.
The rejection of heart allografts is executed by various subsets of cells of adaptive and innate immune system. The population of GICs has been characterized by flow cytometry (34) but not at the transcriptome level. We hypothesized that the transcription of numerous genes that control signaling pathways for the activation, survival, and migration of GICs become activated in the majority of GICs, and suppressed by CsA. Analyses of signaling pathway activity (Figure 4B) revealed the expected activation and CsA-sensitivity of several T cell activation pathways, including the signatures of NK cell activity and activation of CD28 co-stimulatory pathway (35). Among them are the NF-κB and NFAT signatures that were found to be activated during the acute rejection phase. However, while NF-κB signaling remained unaffected, NFAT signaling in GICs was markedly suppressed by CsA treatment and in Nfatc1flx/flxx CD4-cre mice (Figure 4B). From these data, one may conclude that NFAT signaling is a major CsA-sensitive transcriptional pathway activated in GICs during acute rejection phase of fully allogeneic heart transplants.
Among the genes whose transcript levels increased upon transplantation and decreased by CsA treatment or NFATc1 ablation, there were numerous genes that encode enzymes of aerobic glycolysis (Figure 4C). This metabolic switch from OXPHOS to aerobic glycolysis is a typical sign of activated lymphocytes which have a high demand for energy and cellular building blocks, such as nucleotides and amino acids (36). To elucidate whether NFATc1 indeed plays a role in the control of aerobic glycolysis, we studied glycolysis by extracellular flux analysis in WT and Nfatc1−/− CD8+ T cells from dlck-cre x Nfatc1flx/flx mice that we used in transplantation experiments (Figure 2A). As seen in Figure 4D, NFATc1 ablation in peripheral T cells by dlck-cre led to a marked decrease in ECAR which reflects the metabolic switch to glycolysis upon T cell activation. However, when—on the background of deficiency of endogenous NFATc1—the ca version of NFATc1/αA was expressed (Figure 4B), ectopic expression of NFATc1/αA exerted an overall increase in ECAR (Figure 4D), reflecting a stimulation of glycolysis and glycolytic capacity of CD8+ T cells by NFATc1/αA. These data suggest that NFATc1 supports the switch from OXPHOS to aerobic glycolysis during activation of peripheral CD8+ T cells. They are in line with the observation that inhibiting aerobic glycolysis in T cells suppressed the rejection of allogeneic heart transplants (37).
We were also interested in elucidating whether the transplantation of an allogeneic heart at d6, i.e., during the acute rejection phase, affected the peripheral T cell compartment in WT mice, in WT mice treated with CsA and in Nfatc1flx/flxx CD4-cre mice bearing NFATc1-deficient T cells. However, upon transplantation we detected only subtle differences in the composition of splenic T cells from these three types of mice (Figure S3 in Supplementary Material).
The expression of chemokines and chemokine receptors plays a crucial role in the migration of peripheral T cells to inflamed organs. Cxcr3 expression was shown to determine the balance between effector and memory T cells (38) and to be required for acute allogeneic heart rejection (12). Histochemical stains of Cxcr3 expression revealed high levels of Cxcr3 in cells of allogeneic heart transplants whereas a strong decrease in Cxcr3 levels was observed in heart allografts of CsA-treated B6 WT or in Nfatc1flx/flxx CD4-cre mice (Figure 5A). In addition, the data of transcriptome assays showed a strong increase in Cxcr3 transcripts in GICs isolated from allogeneic heart transplants and much less Cxcr3 transcripts in GICs isolated from hearts transplanted into CsA-treated or Nfatc1flx/flxx CD4-cre mice (Figure 5B). These findings are supported by quantitative real-time polymerase chain reaction assays of RNAs from murine CTLs that show a marked decrease of Cxcr3 transcripts in Nfatc1−/− CTLs (Figure 5C).
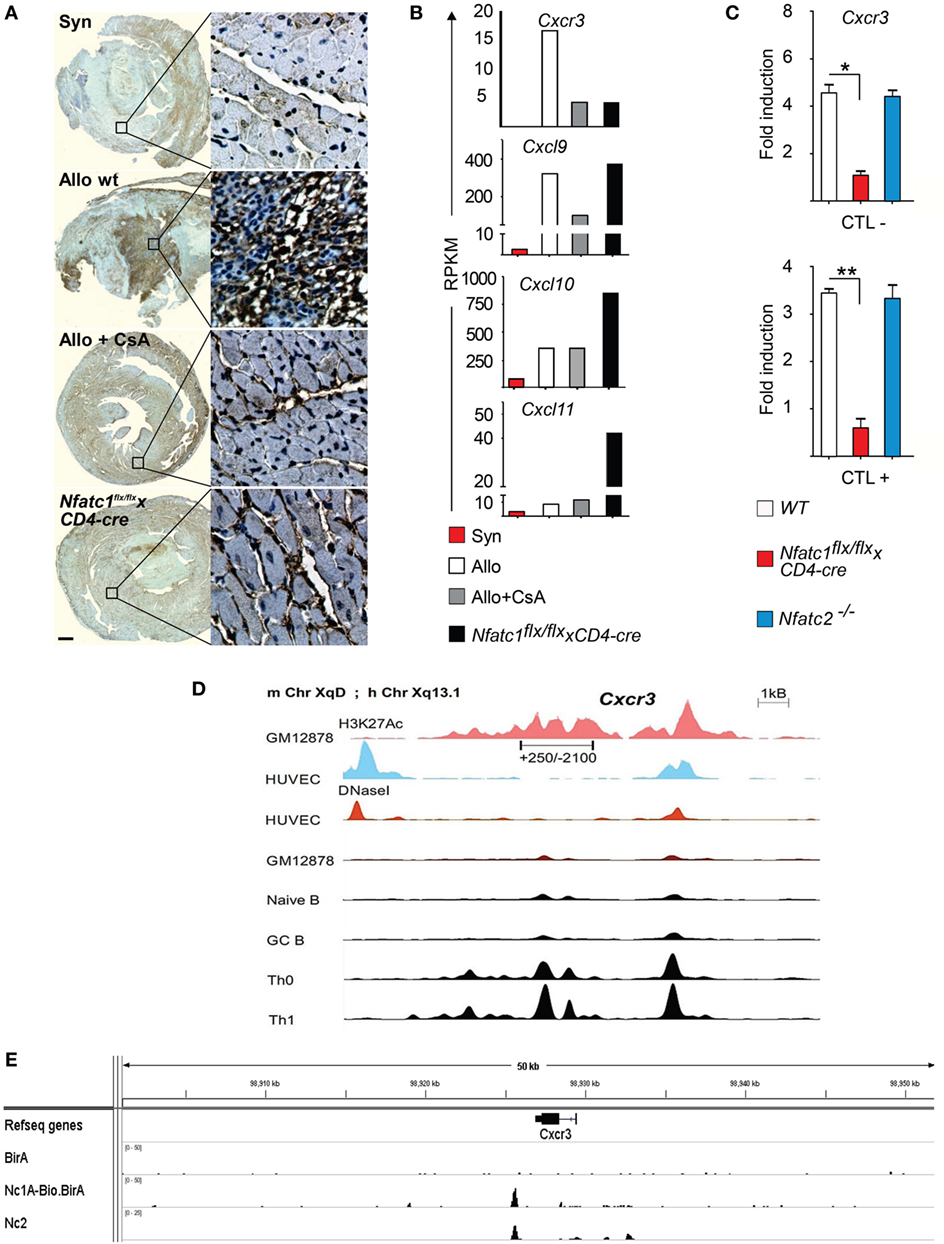
Figure 5. The Cxcr3 gene is a direct NFATc1 target. (A) Representative immune histochemical stains of Cxcr3. Syngeneic (Syn) or allogeneic (Allo) heart grafts were transplanted into wild-type (WT) or cyclosporin A (CsA)-treated WT mice, or in recipient mice bearing NFATc1-deficient T cells (Nfatc1flx/flx x CD4-cre). Grafts and graft-infiltrating cells (GICs) were analyzed during the acute rejection phase on day 5. Bar, 500 µm. (B) Next-generation sequencing assays. Normalized RNA expression of Cxcr3 gene and of relevant ligand genes—Cxcl9, Cxcl10, and Cxcl11—in the transcriptome of GICs. RPKM, reads per kilobase per million reads. (C) Induction of Cxcr3 transcripts in CTL cells derived from WT, Nfatc1flx/flx x CD4-cre or Nfatc2−/− mice. Real-time PCR assays of RNAs were normalized to Actb expression. (D) Genomic organization of the Cxcr3 locus. The distribution of epigenetic modification histone mark H3K27Ac and the presence of DNaseI hypersensitive sites in non-expressing and cells expressing (as Th1 cells) is indicated. (E) Binding of NFATc1/A-Bio and NFATc2 (39) to the Cxcr3 locus in CTLs. ChIP seq assays using a bacterial artificial chromosome transgenic mice expressing NFATc1/A-Bio protein (see Figure S4 in Supplementary Material).
In murine T cells, the Cxcr3 locus is characterized by numerous DNase I hypersensitive chromatin sites in front of the Cxcr3 gene, and extended H3K27 acetylation both in front and within the gene (Figure 5D). However, we were unable to trans-activate the immediate upstream promoter region (spanning the nucleotides from +250 to −2100) cloned in front of a luciferase reporter gene by NFATc1 (over-) expression in Jurkat T cells (data not shown). ChIP seq assays of CTLs that were generated in vitro from splenic CD8+ T cells of a novel tg mouse line expressing NFATc1/A-Bio protein (Figure S4 in Supplementary Material) (25) revealed strong NFATc1 binding downstream of the Cxcr3 gene, approximately 1 kb 3′ from its poly A addition site (Figure 5E, below). These data suggest that NFATc1 plays a critical role in the expression of Cxcr3 gene in T cells (Figure 5A). Moreover, the data suggest that in part the effect of NFATc1 on heart allograft rejection is mediated by Cxcr3 expression.
The findings of our study presented here showed that ablation of Nfatc1 induction in T cells prevented the rejection of heterotropic heart transplants. In contrast, the expression of a ca version NFATc1/αA in T cells supported heart graft rejection indicating an important function for NFATc1/αA in the T cell-mediated immune reaction against allogeneic heart grafts.
The deficiency of NFATc1 in recipient mice resulted in an impaired infiltration of graft and decreased cytotoxicity of GICs. While during acute heart rejection GICs correspond to lymphocytes, monocytes, and neutrophils (40), organ graft rejection is a T cell-dependent process (41, 42) and a massive CD8+ T cell infiltration and a high ratio of CD8+/CD4+ cells was reported for heart allograft rejection in mice (43). The fate of organ transplants is determined to a large part by the number of effector T cells. Regulatory T cells (Tregs) that are known to control the activation and expansion of effector T cells can also affect the fate of a transplanted organ. In a MHC class II-mismatched cardiac allograft model, acute rejection of the donor heart grafts was inhibited by CD4+CD25+ Tregs that restricted the clonal expansion of alloreactive T cells (44). While we did not observe an effect of NFATs on the generation of thymus-derived naturally occurring regulatory T cells (nTreg), the generation of peripherally induced Treg by TGF-β was highly dependent on NFAT expression (15). However, neither inactivation of NFATs did impair the suppressive capacity of Tregs (15, 45), nor did activation of Tregs lead to any NFATc1/αA induction (46). These findings prompted us to conclude that Tregs play only a minor, if any role during induction of transplant tolerance in mice bearing NFATc1-deficient T cells.
One may hypothesize that due to the MHC I and II mismatches of allogeneic graft, NFATc1 expression is rapidly induced in T lymphocytes of peripheral lymphoid organs, in particular in spleen (47). This might lead to the fast re-stimulation of effector CTLs and their migration into the allograft in which CTLs destroy the graft (48, 49). Among the “best” target genes of NFATc1 that we determined by NGS and ChIP seq assays of murine CTLs generated in vitro we detected numerous lymphokine, chemokine, and chemokine receptor genes (25). We showed here that the expression of Cxcr3 gene, which was shown previously to play an important role in acute allograft rejection (12) is controlled by NFATc1 in T cells. However, Cxcr3 is not the only molecule of chemokine network that affects heart transplants (50). Ablation of Ccr4, a further chemokine receptor, led to prolonged survival of allogeneic murine hearts in a chronic transplant model (51), and priming of T cells through the receptor of hepatocyte growth factor, c-Met, in vitro resulted in the generation of “T cell cardiotropism,” including Cxcr4 and Ccr4 expression (52). The priming through c-Met affected also the expression of Ccl3 and Ccl4, two ligands of chemokine receptor Ccr5 that are strongly induced by NFATc1 in murine CTLs (25). Among the direct targets immediately downstream of NFATc1 in T cells is the Irf4 gene (25). The TF IRF4 has been shown recently to control the rejection of allogeneic murine hearts upon heterotopic transplantation (53).
In summary, our data show that ablation of Nfatc1 induction in T cells is sufficient to prevent the rejection of allogeneic heart transplants, whereas (over-) expressing NFATc1/αA led to fulminant rejection. These findings favor NFATc1/αA as molecular target to prevent graft rejection upon heart transplantation.
Animal experiments were performed according to project licenses (acceptance AKZ 55.2-2531.01-80/10 from 22.10.2010 and AKZ 55.2-2531.01-63/11), which are approved and controlled by the Regierung von Unterfranken, Würzburg.
JB, CO, US, SK-H, KhM, TP, KrM, RW, C-TG, IK, and NM performed experiments, ES and AA designed the study, performed experiments, analyzed data, and wrote the manuscript, along with CO.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are very much indebted to Dr. Irina Chodnewskaja, Bettina Mühling, Erwin Schmitt, Manuela Hofmann, and Doris Michel for their skillful assistance. The authors are thankful to Dr. Meinrad Busslinger (Vienna) for advice regarding the Bio-tagged technology, to Drs. Valesca Bukur and Ugur Sahin (Mainz) as well as Drs. Matthias Klein and Tobias Bopp (Mainz) for support in DNA sequencing, Dr. Almut Schulze (Würzburg) in extracellular flux analysis, and to Drs. Volker Ellenrieder (Göttingen) and Anjana Rao (San Diego) for providing us with mouse lines. This work was supported by the Deutsche Forschungsgemeinschaft, grant TRR52 (to ES and AA), the Wilhelm-Sander foundation (to ES and SK-H), the Scheel foundation for Cancer Research (to ES and AA) and the IZKF Würzburg (D-235, to JB and AA).
This work was supported by the Deutsche Forschungsgemeinschaft, grant TRR52 (to ES and AA), the Wilhelm-Sander foundation (to ES and SK-H), the Scheel foundation for Cancer Research (to ES and AA) and the IZKF Würzburg (D-235, to JB and AA).
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.01338/full#supplementary-material.
Ab, antibody; AICD, activation-induced cell death; BAC, bacterial artificial chromosome; CsA, cyclosporin A; CN, Calcineurin; ECAR, extra cellular acidification rate; FK506, tacrolimus; GICs, graft-infiltrating cells; NFATs, nuclear factors of activated T cells; NGS, next-generation sequencing; OXPHOS, oxidative phosphorylation; qRT-PCR, quantitative real-time polymerase chain reaction; T + I, 12-O-tetradecanoylphorbol-13-acetate (TPA) + Ionomycin; TF, transcription factor; tg, transgenic; WT, wild type.
1. Azzi JR, Sayegh MH, Mallat SG. Calcineurin inhibitors: 40 years later, can’t live without. J Immunol (2013) 191(12):5785–91. doi:10.4049/jimmunol.1390055
2. Liu J, Farmer JD Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell (1991) 66(4):807–15. doi:10.1016/0092-8674(91)90124-H
3. Aliabadi A, Cochrane AB, Zuckermann AO. Current strategies and future trends in immunosuppression after heart transplantation. Curr Opin Organ Transplant (2012) 17(5):540–5. doi:10.1097/MOT.0b013e328358000c
4. Sieber M, Baumgrass R. Novel inhibitors of the calcineurin/NFATc hub – alternatives to CsA and FK506? Cell Commun Signal (2009) 7:25. doi:10.1186/1478-811X-7-25
5. Palkowitsch L, Marienfeld U, Brunner C, Eitelhuber A, Krappmann D, Marienfeld RB. The Ca2+-dependent phosphatase calcineurin controls the formation of the Carma1-Bcl10-Malt1 complex during T cell receptor-induced NF-kappaB activation. J Biol Chem (2011) 286(9):7522–34. doi:10.1074/jbc.M110.155895
6. Frischbutter S, Gabriel C, Bendfeldt H, Radbruch A, Baumgrass R. Dephosphorylation of Bcl-10 by calcineurin is essential for canonical NF-kappaB activation in Th cells. Eur J Immunol (2011) 41(8):2349–57. doi:10.1002/eji.201041052
7. Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol (2011) 21(2):91–103. doi:10.1016/j.tcb.2010.09.011
8. Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science (1999) 285(5436):2129–33. doi:10.1126/science.285.5436.2129
9. Choi JM, Sohn JH, Park TY, Park JW, Lee SK. Cell permeable NFAT inhibitory peptide Sim-2-VIVIT inhibits T-cell activation and alleviates allergic airway inflammation and hyper-responsiveness. Immunol Lett (2012) 143(2):170–6. doi:10.1016/j.imlet.2012.01.016
10. Serfling E, Avots A, Klein-Hessling S, Rudolf R, Vaeth M, Berberich-Siebelt F. NFATc1/alphaA: the other face of NFAT factors in lymphocytes. Cell Commun Signal (2012) 10(1):16. doi:10.1186/1478-811X-10-16
11. Orosz CG, Wakely E, Bergese SD, VanBuskirk AM, Ferguson RM, Mullet D, et al. Prevention of murine cardiac allograft rejection with gallium nitrate. Comparison with anti-CD4 monoclonal antibody. Transplantation (1996) 61(5):783–91. doi:10.1097/00007890-199603150-00019
12. Hancock WW, Lu B, Gao W, Csizmadia V, Faia K, King JA, et al. Requirement of the chemokine receptor CXCR3 for acute allograft rejection. J Exp Med (2000) 192(10):1515–20. doi:10.1084/jem.192.10.1515
13. Zhou P, Hwang KW, Palucki DA, Guo Z, Boothby M, Newell KA, et al. Impaired NF-kappaB activation in T cells permits tolerance to primary heart allografts and to secondary donor skin grafts. Am J Transplant (2003) 3(2):139–47. doi:10.1034/j.1600-6143.2003.00033.x
14. Muhammad K, Alrefai H, Marienfeld R, Pham DA, Murti K, Patra AK, et al. NF-kappaB factors control the induction of NFATc1 in B lymphocytes. Eur J Immunol (2014) 44(11):3392–402. doi:10.1002/eji.201444756
15. Vaeth M, Schliesser U, Muller G, Reissig S, Satoh K, Tuettenberg A, et al. Dependence on nuclear factor of activated T-cells (NFAT) levels discriminates conventional T cells from Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A (2012) 109(40):16258–63. doi:10.1073/pnas.1203870109
16. Oh-Hora M, Komatsu N, Pishyareh M, Feske S, Hori S, Taniguchi M, et al. Agonist-selected T cell development requires strong T cell receptor signaling and store-operated calcium entry. Immunity (2013) 38(5):881–95. doi:10.1016/j.immuni.2013.02.008
17. Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, et al. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest (2008) 118(11):3775–89. doi:10.1172/JCI35711
18. Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A (2006) 103(37):13789–94. doi:10.1073/pnas.0605944103
19. Hodge MR, Ranger AM, Charles de la Brousse F, Hoey T, Grusby MJ, Glimcher LH. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity (1996) 4(4):397–405. doi:10.1016/S1074-7613(00)80253-8
20. Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity (2001) 15(5):763–74. doi:10.1016/S1074-7613(01)00227-8
21. Baumgart S, Chen NM, Siveke JT, Konig A, Zhang JS, Singh SK, et al. Inflammation-induced NFATc1-STAT3 transcription complex promotes pancreatic cancer initiation by KrasG12D. Cancer Discov (2014) 4(6):688–701. doi:10.1158/2159-8290.CD-13-0593
22. Zhang DJ, Wang Q, Wei J, Baimukanova G, Buchholz F, Stewart AF, et al. Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol (2005) 174(11):6725–31. doi:10.4049/jimmunol.174.11.6725
23. Busch R, Murti K, Liu J, Patra AK, Muhammad K, Knobeloch KP, et al. NFATc1 releases BCL6-dependent repression of CCR2 agonist expression in peritoneal macrophages from Saccharomyces cerevisiae infected mice. Eur J Immunol (2016) 46(3):634–46. doi:10.1002/eji.201545925
24. de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol (2003) 33(2):314–25. doi:10.1002/immu.200310005
25. Klein-Hessling S, Muhammad K, Klein M, Pusch T, Rudolf R, Floter J, et al. NFATc1 controls the cytotoxicity of CD8(+) T cells. Nat Commun (2017) 8(1):511. doi:10.1038/s41467-017-00612-6
26. Sitaru AG, Timmermann W, Ulrichs K, Otto C. Hierarchical immunogenicity of donor MHC class I peptides in allotransplantation. Hum Immunol (2002) 63(10):871–9. doi:10.1016/S0198-8859(02)00452-4
27. Alrefai H, Muhammad K, Rudolf R, Pham DA, Klein-Hessling S, Patra AK, et al. NFATc1 supports imiquimod-induced skin inflammation by suppressing IL-10 synthesis in B cells. Nat Commun (2016) 7:11724. doi:10.1038/ncomms11724
28. Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, et al. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity (2011) 34(2):175–87. doi:10.1016/j.immuni.2011.02.005
29. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol (2011) 29(1):24–6. doi:10.1038/nbt.1754
30. Ueno T, Yamada A, Ito T, Yeung MY, Gorbatov R, Shimizu T, et al. Role of nuclear factor of activated T cell (NFAT) transcription factors in skin and vascularized cardiac allograft rejection. Transplantation (2011) 92(5):e26–7. doi:10.1097/TP.0b013e318228061c
31. Serfling E, Klein-Hessling S, Palmetshofer A, Bopp T, Stassen M, Schmitt E. NFAT transcription factors in control of peripheral T cell tolerance. Eur J Immunol (2006) 36(11):2837–43. doi:10.1002/eji.200536618
32. Klein-Hessling S, Rudolf R, Muhammad K, Knobeloch KP, Maqbool MA, Cauchy P, et al. A threshold level of NFATc1 activity facilitates thymocyte differentiation and opposes notch-driven leukaemia development. Nat Commun (2016) 7:11841. doi:10.1038/ncomms11841
33. Sharma S, Zhu J. Immunologic applications of conditional gene modification technology in the mouse. Curr Protoc Immunol (2014) 105:10.34.1–13. doi:10.1002/0471142735.im1034s105
34. Stinn JL, Taylor MK, Becker G, Nagano H, Hasegawa S, Furakawa Y, et al. Interferon-gamma-secreting T-cell populations in rejecting murine cardiac allografts: assessment by flow cytometry. Am J Pathol (1998) 153(5):1383–92. doi:10.1016/S0002-9440(10)65725-2
35. Maier S, Tertilt C, Chambron N, Gerauer K, Huser N, Heidecke CD, et al. Inhibition of natural killer cells results in acceptance of cardiac allografts in CD28-/- mice. Nat Med (2001) 7(5):557–62. doi:10.1038/87880
36. Finlay D, Cantrell DA. Metabolism, migration and memory in cytotoxic T cells. Nat Rev Immunol (2011) 11(2):109–17. doi:10.1038/nri2888
37. Lee CF, Lo YC, Cheng CH, Furtmuller GJ, Oh B, Andrade-Oliveira V, et al. Preventing allograft rejection by targeting immune metabolism. Cell Rep (2015) 13(4):760–70. doi:10.1016/j.celrep.2015.09.036
38. Hu JK, Kagari T, Clingan JM, Matloubian M. Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc Natl Acad Sci U S A (2011) 108(21):E118–27. doi:10.1073/pnas.1101881108
39. Martinez GJ, Pereira RM, Aijo T, Kim EY, Marangoni F, Pipkin ME, et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity (2015) 42(2):265–78. doi:10.1016/j.immuni.2015.01.006
40. Reichel CA, Khandoga A, Anders HJ, Schlondorff D, Luckow B, Krombach F. Chemokine receptors Ccr1, Ccr2, and Ccr5 mediate neutrophil migration to postischemic tissue. J Leukoc Biol (2006) 79(1):114–22. doi:10.1189/jlb.0605337
41. Whitby EH, Sparshott SM, Bell EB. Allograft rejection in athymic nude rats by transferred T-cell subsets. I. The response of naive CD4+ and CD8+ thoracic duct lymphocytes to complete allogeneic incompatibilities. Immunology (1990) 69(1):78–84.
42. Jones ND, Van Maurik A, Hara M, Gilot BJ, Morris PJ, Wood KJ. T-cell activation, proliferation, and memory after cardiac transplantation in vivo. Ann Surg (1999) 229(4):570–8. doi:10.1097/00000658-199904000-00018
43. Szot GL, Zhou P, Rulifson I, Wang J, Guo Z, Kim O, et al. Different mechanisms of cardiac allograft rejection in wildtype and CD28-deficient mice. Am J Transplant (2001) 1(1):38–46. doi:10.1034/j.1600-6143.2001.010108.x
44. Schenk S, Kish DD, He C, El-Sawy T, Chiffoleau E, Chen C, et al. Alloreactive T cell responses and acute rejection of single class II MHC-disparate heart allografts are under strict regulation by CD4+ CD25+ T cells. J Immunol (2005) 174(6):3741–8. doi:10.4049/jimmunol.174.8.5135-a
45. Bopp T, Palmetshofer A, Serfling E, Heib V, Schmitt S, Richter C, et al. NFATc2 and NFATc3 transcription factors play a crucial role in suppression of CD4+ T lymphocytes by CD4+ CD25+ regulatory T cells. J Exp Med (2005) 201(2):181–7. doi:10.1084/jem.20041538
46. Vaeth M, Gogishvili T, Bopp T, Klein M, Berberich-Siebelt F, Gattenloehner S, et al. Regulatory T cells facilitate the nuclear accumulation of inducible cAMP early repressor (ICER) and suppress nuclear factor of activated T cell c1 (NFATc1). Proc Natl Acad Sci U S A (2011) 108(6):2480–5. doi:10.1073/pnas.1009463108
47. Larsen CP, Morris PJ, Austyn JM. Migration of dendritic leukocytes from cardiac allografts into host spleens. A novel pathway for initiation of rejection. J Exp Med (1990) 171(1):307–14. doi:10.1084/jem.171.1.307
48. Gilot BJ, Hara M, Jones ND, van Maurik A, Niimi M, Hadjianastassiou V, et al. Visualization of the in vivo generation of donor antigen-specific effector CD8+ T cells during mouse cardiac allograft rejection: in vivo effector CD8+ T cell generation during allograft rejection. Transplantation (2000) 69(4):639–48. doi:10.1097/00007890-200002270-00028
49. Delfs MW, Furukawa Y, Mitchell RN, Lichtman AH. CD8+ T cell subsets TC1 and TC2 cause different histopathologic forms of murine cardiac allograft rejection. Transplantation (2001) 71(5):606–10. doi:10.1097/00007890-200103150-00005
50. O’Boyle G, Ali S, Kirby JA. Chemokines in transplantation: what can atypical receptors teach us about anti-inflammatory therapy? Transplant Rev (Orlando) (2011) 25(4):136–44. doi:10.1016/j.trre.2010.10.005
51. Huser N, Tertilt C, Gerauer K, Maier S, Traeger T, Assfalg V, et al. CCR4-deficient mice show prolonged graft survival in a chronic cardiac transplant rejection model. Eur J Immunol (2005) 35(1):128–38. doi:10.1002/eji.200324745
52. Komarowska I, Coe D, Wang G, Haas R, Mauro C, Kishore M, et al. Hepatocyte growth factor receptor c-Met instructs T cell cardiotropism and promotes T cell migration to the heart via autocrine chemokine release. Immunity (2015) 42(6):1087–99. doi:10.1016/j.immuni.2015.05.014
Keywords: NFATc1, transplantation, heterologous, CD8+ T cells, ChIPseq, metabolism
Citation: Baur J, Otto C, Steger U, Klein-Hessling S, Muhammad K, Pusch T, Murti K, Wismer R, Germer C-T, Klein I, Müller N, Serfling E and Avots A (2018) The Transcription Factor NFATc1 Supports the Rejection of Heterotopic Heart Allografts. Front. Immunol. 9:1338. doi: 10.3389/fimmu.2018.01338
Received: 13 February 2018; Accepted: 29 May 2018;
Published: 12 June 2018
Edited by:
Yong Zhao, Institute of Zoology (CAS), ChinaReviewed by:
Bruce Milne Hall, University of New South Wales, AustraliaCopyright: © 2018 Baur, Otto, Steger, Klein-Hessling, Muhammad, Pusch, Murti, Wismer, Germer, Klein, Müller, Serfling and Avots. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edgar Serfling, c2VyZmxpbmcuZUBtYWlsLnVuaS13dWVyemJ1cmcuZGU=;
Andris Avots, YW5keV9hdm90c0Bob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.