- Department of Biochemistry and Molecular Biology, Faculty of Biosciences, Universitat Autònoma de Barcelona, Cerdanyola del Vallès, Spain
The ribonuclease A superfamily is a vertebrate-specific family of proteins that encompasses eight functional members in humans. The proteins are secreted by diverse innate immune cells, from blood cells to epithelial cells and their levels in our body fluids correlate with infection and inflammation processes. Recent studies ascribe a prominent role to secretory RNases in the extracellular space. Extracellular RNases endowed with immuno-modulatory and antimicrobial properties can participate in a wide variety of host defense tasks, from performing cellular housekeeping to maintaining body fluid sterility. Their expression and secretion are induced in response to a variety of injury stimuli. The secreted proteins can target damaged cells and facilitate their removal from the focus of infection or inflammation. Following tissue damage, RNases can participate in clearing RNA from cellular debris or work as signaling molecules to regulate the host response and contribute to tissue remodeling and repair. We provide here an overall perspective on the current knowledge of human RNases’ biological properties and their role in health and disease. The review also includes a brief description of other vertebrate family members and unrelated extracellular RNases that share common mechanisms of action. A better knowledge of RNase mechanism of actions and an understanding of their physiological roles should facilitate the development of novel therapeutics.
Introduction
Thirty years ago Steven Benner conjectured the existence of extracellular RNA communicators (1, 2). At that time, he was investigating bovine RNaseA activity and expressed skepticism that pancreatic RNases merely removed the large amount of bacterial RNA present in the ruminant digestive tract (3, 4). Based on the diverse biological properties displayed by some RNaseA family members, i.e., anti-tumoural action, angiogenesis, and neurotoxicity, he suggested that the catalytic activity of vertebrate secreted RNases intervened in the regulation of the development of higher organisms. The hypothesis was launched well before the discovery of extracellular vesicles as horizontal nanovehicle carriers and well before the discovery that angiogenin, a member of the vertebrate-specific RNaseA superfamily, generates RNA regulatory fragments (5, 6). RNaseA, the vertebrate secretory RNases’ reference family member, is a small and highly stable protein that served as a working model for biochemists during the twentieth century; several Nobel prizes in chemistry were awarded for work with RNaseA (7, 8). Many times, researchers have tried unsuccessfully to rescue the RNaseA superfamily from its purely academic role (1, 7). Despite extensive knowledge on the mechanism of catalysis (8) and phylogeny (9–12), the biological properties of some family members remained puzzling, and the ultimate physiological roles of these proteins remained elusive. Exhaustive sequencing of RNases within vertebrates and a comparative phylogenetic analysis suggested that the family emerged with a host defense role (11, 13–16). Another interesting hypothesis suggested that granulocyte-secreted proteins could play a primary role in local tissue repair and the removal of macromolecular debris following cell damage during inflammation (17). The authors considered the potential contribution to RNA clearance by eosinophil secreted RNases. Later studies also indicated an RNA scavenging role for pancreatic-type RNases (18, 19). In addition, the secreted proteins displayed immuno-regulatory properties that suggest they could participate in the transmission and amplification of local danger signals (17, 20). Therefore, extracellular RNases are key players that ensure tissue health and body homeostasis. Indeed, we find examples of genetic deficiencies in extracellular RNases that lead to immune-related diseases, such as amyotrophic lateral sclerosis (ALS), associated with human RNase5 mutations (21, 22) and cystic leukoencephalopathy, a neuronal disorder associated with RNaseT2 deficiency (23, 24). Potential RNA-targeted therapeutic applications for secretory RNases were envisaged as far back as two decades ago (25–27).
Recent methodological advances in the cellular biology and RNA fields have facilitated novel approaches to understanding the in vivo role of extracellular RNases. Hopefully, knowledge on RNases action and trafficking in biological fluids will give path to translational research from academia to pharmaceutical industry. Indeed, recent experimental trials with animal models, such as those on hematopoiesis regulation by human RNase5 (28) or attenuation of extracellular RNA (exRNA) pro-inflammatory activity by RNaseA (29), are already offering promising therapeutic results.
The review summarizes the current knowledge on the mechanism of action of the RNaseA superfamily members and their contribution to innate immunity, as sentinel proteins at the extracellular space. We also briefly compared RNaseA proteins with other extracellular RNases, such as RNaseT2 family members, which are ancient RNases that are highly conserved through taxa from viruses to humans (30, 31), and bacterial RNases (19, 32) that work as defense weapons in inter-strain warfare.
The RNase a Superfamily
The RNaseA superfamily is a vertebrate-specific gene family that has shown great divergence in a short period of time, a characteristic trait of immune-related proteins (11, 16, 33). Despite the low sequence identity between some family members (~30%), they all share a common three-dimensional structural fold and conserved motif signature (CKXXNTF). They are small secretory proteins (13–15 kDa) expressed with a short 25–27 amino acid signal peptide. The mature protein adopts an α + β kidney-shaped fold crosslinked by three to four disulfide bonds. A conserved catalytic triad formed by two His and a Lys participates in the endoribonuclease acid–base catalytic mechanism of action (8). A marked preference for cleavage of single-stranded RNA (ssRNA) is observed, with specificity for pyrimidines at the main base and a preference for purines at the secondary base site (7, 34, 35). In addition, other nucleotide-binding sites contribute to RNase-substrate specificities (36–38) and might determine the selectivity of RNases for cellular RNA. Recently, novel methodologies to identify the selective cleavage site for non-coding RNA for some family members, i.e., tRNA, have indicated their direct involvement in main cellular machinery tasks (39–41).
RNaseA superfamily members are mainly expressed in innate cells and display a variety of antimicrobial and immune modulation activities. They can participate in host immune responses, working as alarmins and safeguard molecules against infection and inflammation (16, 42–45). Table 1 summarizes most of their reported activities and suggested physiological roles. Below, we describe the eight canonical family members in humans. Table 2 indicates their reported source cell types and summarizes their constitutive and induced expression patterns. Human RNase expression and response processes activated by diverse stimuli are illustrated in Figure 1.
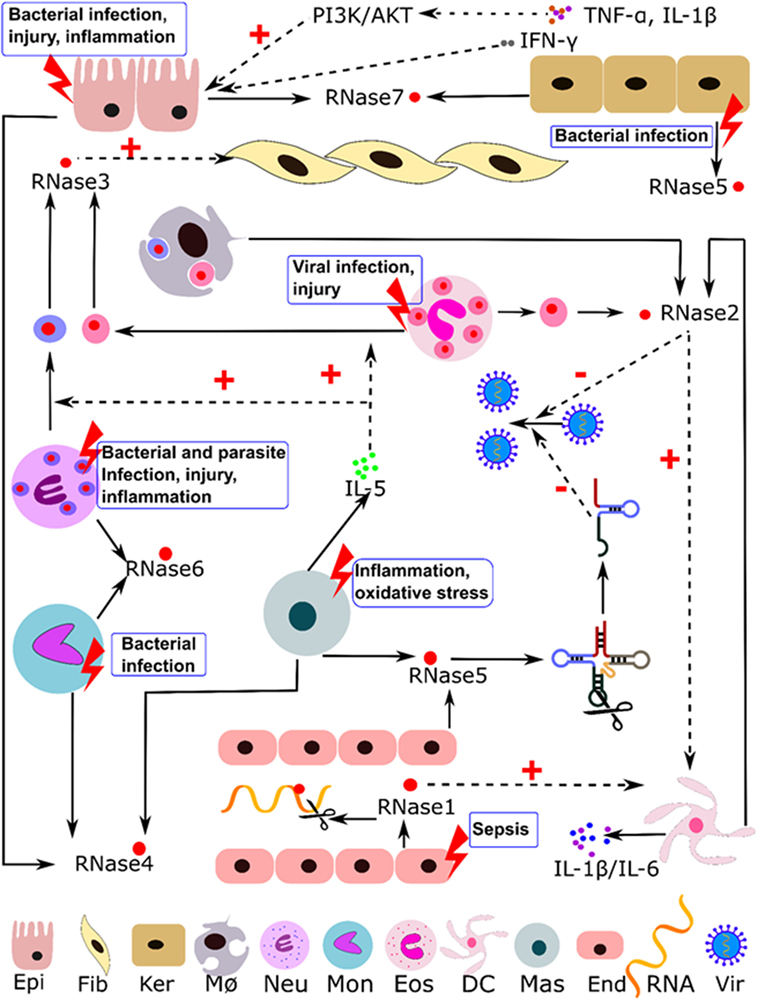
Figure 1. Illustration of the immuno-modulatory properties reported for human RNaseA family members. Induction stimuli, expression cell type, and regulation pathways are indicated. Abbreviations: Epi, epithelial; Fib, fibroblast; Ker, keratinocytes; MØ, macrophages; Neu, neutrophils; Mon, monocytes; Eos, eosinophils; DC, dendritic cells; Mas, mast cells; End, endothelial cells; Vir, virus; dotted line indicates regulation paths and solid line indicates expression and secretion processes.
hRNase1
hRNase1 is the human homolog of the family reference protein, bovine pancreatic RNaseA. A comparative evolutionary analysis indicates a divergent role for non-ruminant RNase1 family members that are unrelated to digestion (18, 142). Expression of human RNase1 was detected in almost all tissues (143). In particular, human RNase1 is abundantly expressed by endothelial cells (144, 145), and its presence in blood can modulate the content of exRNA (18, 77). An interesting hypothesis attributes an RNA scavenger role to hRNase1 (77). Supporting this proposal, Raines and co-workers (18) highlight the different optimal conditions for catalysis between ruminant and non-ruminant RNases. Whereas bovine pancreatic RNase shows a slightly acidic optimum pH for catalysis, adapted for working in the digestive tract (4), the observed optimum neutral pH for non-digestive RNases allows their enhanced activity in biological fluids such as blood (18). Likewise, the enhanced capacity to cleave double-stranded RNA (dsRNA) by non-digestive RNases (10, 18) might facilitate the removal of heterogeneous RNAs circulating in blood (18). Interestingly, clearance of exRNA would mitigate its pro-inflammatory activity (76, 77). An excess of exRNA is released in hypoxic conditions, and hRNase1 administration was reported to provide cardiac protection in a mouse model (76). Administration of the protein in vivo reduces the release of pro-inflammatory cytokines and provides a multiple-organ protection in mice (29).
hRNase2
hRNase2, together with hRNase3, is one of the main secretory proteins stored within the eosinophil secondary granules (107, 146). This eosinophil protein is partly responsible for eosinophil-induced neurotoxicity, and it is referred to as the eosinophil-derived neurotoxin (EDN), hereafter. In addition, hRNase2/EDN is expressed in other blood cell types (Table 2) such as monocytes and dendritic cells (DCs) (107). Expression of the protein is induced by viral infection (100, 107, 147), and it displays a broad antiviral activity, in particular against ssRNA viruses (104, 107). The protein inhibits the replication of respiratory syncytial (RSV), human immunodeficiency virus (HIV)-1, and hepatitis B DNA virus (100, 102, 103). RNase2 has high catalytic activity, which together with its unique protein structural features (106), is essential for its antiviral action (104). Eosinophil recruitment and degranulation is activated during RSV infection (147), and hRNase2 is a clinical marker for RSV bronchiolitis (148). Interestingly, Weller and co-workers observed the induced release of eosinophil RNases by cytokines such as eosinophil chemotactic proteins (eotaxins CCL11 and CCL24) and through the P13K/MAPK pathway in humans and mice (149). Eosinophil activation by virus infection can be mediated by the toll-like receptor (TLR)7–MyD88 signaling pathway. Likewise, specific expression of the eosinophil RNases following RSV infection in mice is reported to be mediated by the TLR7–MyD88 pathway (55). The TLR7 receptor can be activated by ssRNA, causing the eosinophil RNase to act on viral RNA to enhance the host response against the infection. In addition, viral infection of the respiratory tract is frequently accompanied by lung inflammation and hRNase2 expression. Secondarily, local eosinophil degranulation correlates with tissue damage with eosinophil degranulation activated by tissue injury (Figure 1) and the released RNases contributing to the tissue remodeling process (149). Indeed, hRNase2 immuno-regulatory properties can promote leukocyte activation, maturation, and chemotaxis (107). In particular, hRNase2 can contribute to host immunity through interactions with DCs (46, 47). The activation of DCs can be mediated by the TLR2–MyD88 pathway, and hRNase2 is classified as an alarmin (60). Direct protein binding to TLR may occur (107) taking into account the striking shape similarity between the TLR receptor and the RNase inhibitor (RI), both of which have leucine-rich repeat (LRR) domains (150). Human RNase2 and mouse RNases2 are also both expressed in macrophages (Table 2) (48, 107). Of note, the mouse eosinophil-associated RNase Ear11 works as a macrophage chemoattractant, although, in this case, the process is not directly mediated by TLR2 activation (48, 107). Because TLRs can interact with ssRNA, we can contemplate a scenario in which RNA pieces bound to RNase2, or, alternatively, the direct action of RNase cleavage products, trigger the TLR signaling cascade. As TLRs can discriminate between self and non-self molecules, cleavage of viral RNA could contribute to the host defense response. Thereby, immune regulation and antimicrobial functions would work cooperatively (151). Eosinophil degranulation during infection and inflammation can contribute locally to both eradicate the infection focus and palliate tissue injury.
hRNase3
hRNase3 is the other eosinophil RNase abundant in the secondary secretory granule, where both RNase2 and 3 together account for about one-third of the total protein content (152). The two human eosinophil RNases share 70% amino acid identity. RNases 2 and 3 emerged from a gene duplication event about 50 million years ago and underwent a divergence process at an extremely rapid rate of evolution (12). During the drift of RNase3 from a common RNase2/3 ancestor, the protein acquired much higher cationicity (pI > 10); therefore, it is called eosinophil cationic protein (ECP). Abundant surface-exposed Arg residues facilitate binding of the protein to bacterial cell membranes and subsequent destabilization through a carpet-like mechanism characteristic of many host defense antimicrobial proteins and peptides (AMPs) (153). RNase3 shares neurotoxic and antiviral activities with RNase2 (105) but it has unique bactericidal properties (86). In particular, hRNase3, together with its high cationicity, has an aggregation-prone region that promotes the protein self-aggregation and mediates the agglutination of bacterial cells (154). Bacterial cell agglutination is further enhanced by high binding affinity of the protein to anionic lipopolysaccharides in the Gram-negative bacterial wall (155). RNase3 release is induced both by infection and inflammation, and several immuno-modulatory activities have been described. The levels of protein circulating in biological fluids (Table 2) correlate with eosinophil degranulation, and it is currently used as a routine clinical marker for the diagnosis and monitoring of inflammatory disorders, such as asthma (51, 52). Airway inflammation is closely associated with eosinophil degranulation and local tissue damage follows the deposition of eosinophil secondary granule protein (156, 157). The levels of this protein are also associated with damaged airway epithelia (52). Further, skin ulceration follows eosinophil infiltration, and local protein deposits harm epithelial cells (121, 158, 159). Fortunately, the detrimental side effects of the protein tissue deposits are followed by remodeling processes. RNase3 remodeling activity is partly mediated by the upregulation of the insulin growth factor-1 receptor on epithelial cells. In addition, hRNase3 activation and chemotaxis of fibroblasts can contribute to tissue repair (49, 62, 160). Nevertheless, fibroblast activation can also lead to airway fibrosis, as observed during chronic eosinophil inflammation in asthma (49). Interestingly, a population study identified a natural genotype variant of hRNase3 (ECP97Arg) with enhanced cytotoxicity that was linked to a higher frequency of fibrosis (161). Despite the higher antimicrobial activity of the Arg97 variant, genetic selection toward a less toxic protein must have taken place in a chronic parasite infection in endemic areas of Asia to reduce the incidence of liver fibrosis (162). The Arg to Thr substitution at position 97 of hRNase3 results in a new potential N-glycosylation site at a nearby Asn residue (163). Additional glycosylation at this site blocks the cationic domain that participates in the protein antimicrobial activity (164). Similarly, a correlation between hRNase3 polymorphisms and cerebral malaria susceptibility was observed (165, 166).
Together with direct hRNase3 action on pathogens and host tissues, a series of immune-modulating activities are observed (51, 52). Eotaxin attracts eosinophils to the area of inflammation, i.e., lung in asthma, nasal mucosa in allergic rhinitis, skin in dermatitis, or gut epithelia in intestinal bowel diseases. Eosinophil degranulation is activated by IL-5, leukotriene B4 (LTB4), platelet activating factor (PAF) (52), or the P13K/MAPK pathway (149). Early experimental assays also showed mast cell activation by hRNase3 and the induction of histamine release (50). In turn, mast cells produce and secrete IL-5, PAF, and LTB4, which enhance hRNase3 release from eosinophils. Moreover, hRNase3 induces the synthesis of prostaglandin D2 by mast cells, which then acts as an eosinophil chemoattractant. This process suggests cross talk between mast cells and eosinophils, with RNase able to promote positive feedback (52). Other leukocyte cells such as neutrophils can also express hRNase3 (Table 2). Interestingly, free granules released by both eosinophils and neutrophils maintain their autonomy and functionality (167) and selectively secrete RNases upon cytokine induction (168). Moreover, the extracellular granules can also be engulfed by macrophages (Figure 1) and contribute to the immune response.
hRNase4
hRNase4 is one of the oldest representative RNase A family members within mammals, showing a static evolution history in comparison with the other family counterparts (33). RNase 4, together with RNase 5, shares some conserved structural features with non-mammalian vertebrates RNases, such as the first pyroglutamic N-terminal residue (169). Moreover, there are no reported glycosylation forms and no recognition sites for N-glycosylation (169). RNase4 retains the highest inter-species homology, close to 90%, within the family members (33, 170). Nearly ubiquitous distribution suggests a housekeeping role for this protein. The presence of hRNase4 transcripts was detected in most human tissues (143, 171) and was found particularly abundant in the liver (143) and lungs (169). Interestingly, cytoplasmic granules of monocytes also express this RNase (172). Divergence and diversification events within the oldest mammals suggest a strong evolutionary pressure that may respond to host adaptation to an ever-changing pathogen environment. In this scenario, duplication of the RNase4 gene in ancestral mammals may have led to other RNaseA family lineages that acquired a host defense function (33). There is a controversy about the physiological role of the protein. On one side, its conservation among species suggests strong evolutionary pressure to maintain an essential role. Cleaning-up of cellular RNA was first proposed (173). However, the enzyme showed enhanced preference for uridine at the main base binding site (169, 173, 174), suggesting strong selectivity for RNA recognition (169). A structural analysis highlighted particular structural features at the main base binding pocket that determine the protein enhanced preference for uridine over cytidine in comparison to other family members (174, 175). Although considerable work has been undertaken to interpret RNase4 substrate specificity, there are few reports on the biological properties of the protein. Interestingly, RNase4 was identified, along with RNase5, among the soluble factors secreted by T cells showing anti-HIV activity (176).
hRNase5
hRNase5 is considered the most ancient RNaseA family member, and it shares many structural features with non-mammalian vertebrate RNases. It is unique within the family in presenting six paired cysteine residues instead of eight. In addition, it shows rather atypical enzymatic properties, with very low catalytic efficiency for ssRNA but selective cleavage for some non-coding RNA (41, 177, 178). Its expression was detected in many adult and embryonic somatic cells (179). It was also reported in a variety of innate cells, ranging from diverse blood cell types to intestinal and skin epithelial cells (Table 2) (28, 67, 129, 180). Further expression of this protein is increased during inflammation (129, 181). RNase5 promotes angiogenesis and is, therefore, termed angiogenin (65). The purified protein was reported to display other activities, such as antimicrobial action (67, 88) and some immuno-regulatory properties (88, 182), in addition to the induction of vascularization. In particular, the protein inhibits neutrophil degranulation, a process that might induce an anti-inflammatory effect during immune response (183, 184). Interestingly, this degranulation inhibitory action was mimicked by a short tryptic peptide (183), indicating a protein activity unrelated to its enzymatic function. On the other hand, angiogenesis relies on hRNase5 catalytic activity and is inhibited by the RI (185). Distinct immuno-regulatory activities were observed to be dependant on the action of the protein on non-coding RNA, i.e., cleavage of tRNA and upregulation of rRNA. Upon nuclear translocation hRNase5 can stimulate the proliferation of several cell types, such as endothelial cells (66), by regulating rRNA transcription (63, 186). In addition, angiogenesis and cell proliferation are mediated by the activation of cellular signaling kinases such as the ERK1/2 (179, 187). In addition, hRNase5 expression is activated in response to cellular stress and promotes the formation of stress granules. The protein generates stress-induced tRNA fragments (tRFs) (tiRNA) (6, 56, 58). In turn, the tiRNA fragments can impede the formation of the translation initiation factor complex, thereby inhibiting translation (188). Accumulation of tiRNA activates the cell response to oxidative stress (59). The release of tRFs associated with hRNase5 activity is also a characteristic feature linked to endoplasmic reticulum (ER) stress, a condition that can be triggered by the accumulation of unfolded/misfolded proteins in the ER lumen (57). Likewise, hRNase5 induces the release of tiRNA by stem cells and activates hematopoietic cell regeneration (28). Generation of tRFs is also observed upon viral infection (189). Using deep sequencing methodologies, the enzyme cleavage target sites are being identified (5, 6, 39). Two tRNA halves can be produced by a unique enzymatic cut at the anticodon loop (5, 6, 58, 190). In addition, Li and co-workers observed a unique cleavage by hRNase5 at the tRNA TΨC loop (39); the resulting 3′-tRNA fragment was complementary to an endogenous human genome sequence, and the fragment was found to downregulate retroviral expression by RNA interference (39). However, potential therapeutic applications for hRNase5, such as hematopoiesis regeneration or antiviral activity (28), should be viewed with caution, considering its protein pro-tumoural properties (65). In particular, upregulation of rRNA transcription has been related to cancer cell proliferation (63, 64). A recent work correlated elevated hRNase5 expression in some tumor cell lines with the promotion of cell proliferation and development of malignant cancer (64). In addition, elevated levels of tiRNA were also observed in some cancers (190) with tRFs inducing tumorigenesis (188).
hRNase6
hRNase6, also named hRNasek6, was identified for the first time by Helen Rosenberg and Kimberly Dyer as a human ortholog of bovine kidney RNasek2 (191). The first tissue screening study to define hRNase6 expression patterns revealed a nearly ubiquitous distribution, including monocytes and neutrophils (191). The expression of this protein is upregulated following genitourinary tract bacterial infections (83) and high antimicrobial activity against Gram-positive and Gram-negative species was observed (83, 90). Very recently, hRNase6 antiviral activity was reported. Interestingly, the authors observed the protein downregulation in Th17 polarized cells upon HIV infection (192).
hRNase7
hRNase7 is probably the best studied example of an RNase that can work as a tissue safeguard sentinel (Table 1). It is also one of the most abundant antimicrobial proteins purified from skin (44, 79). RNase7 is secreted by a variety of epithelial cells (Table 2) and mostly contributes to urinary tract sterility and epidermis protection (43, 80–82). Together with high antimicrobial activity against a variety of infective microorganisms (82, 87, 89, 193), some immuno-modulatory properties were reported. RNase7 expression is upregulated during kidney infection (84). Expression is also selectively induced by inflammation signaling molecules, such as IL-1β and IFN-γ (89) or the PI3K/AKT pathway (194). The PI3K/AKT signaling pathway can modulate the innate immune response during inflammation and prevent sepsis (195, 196). Interestingly, expression of this protein in the urinary tract can be upregulated by insulin through the PI3K/AKT pathway (197). Spencer and co-workers correlated expression with increased susceptibility to infection of diabetic patients (197). Indeed, insulin induces the secretion of granulocyte content and impairment of the expression of other AMPs, which are also associated with diabetes (198). Of note, the regulation of protein expression by the P13K/AKT pathway is also observed with eosinophil-associated RNases (EARs) (149). On the other hand, RNase7 is abundantly secreted by keratinocytes (43, 44, 79) and can contribute to wound healing and tissue repair (67, 68). Protein overexpression in skin can be induced by inflammation and infection diseases (69, 81). Interestingly, protease degradation of the RI at the stratum corneum can activate hRNase7 for skin barrier protection (67). Finally, expression of the protein was recently reported to be induced in basal cells of damaged airway epithelia, reinforcing the idea of a protective role for this protein following tissue injury (Table 1) (70). A very recent study showed that this protein directly stimulated plasmocytoid DCs following tissue damage and infection, and the authors of the report proposed to classify hRNase7 as an alarmin (54). Interestingly, the immuno-modulatory activity of the RNase correlates with binding to self-DNA and activation of TLR9 receptors. The authors suggest that following tissue damage, hRNase7 detects the host DNA released by dying cells and activates the host response (54).
hRNase8
hRNase8 is the last identified and least well-characterized canonical member of the RNaseA superfamily. It was first uniquely identified in the placenta (199). Wide spectrum antimicrobial activity was observed for this protein (91), suggesting a role in amniotic fluid protection against infection. Indeed, hRNase7, the closest homolog to hRNase8 in the RNaseA family, was found recently among AMPs expressed in prenatal skin, suggesting it may contribute to amniotic cavity sterility (200). However, despite sharing a high sequence identity with hRNase7, hRNase8 shows highly reduced catalytic activity (199). A particular cysteine location within its primary sequence indicates a unique disulfide bonding among the family members. An evolutionary analysis in primates revealed a sequential cysteine gain-and-loss process, representing an unusual example of disulfide bond reshuffling (201). In addition, the protein shows an elevated rate of incorporation of non-silent mutations in its primary structure (202) suggesting functional divergence toward a distinct physiological role (141). Moreover, a unique extension at the hRNase8 N-terminus may indicate that the protein is not undergoing the secretion process shared by all other canonical members of the family (141). Recent evidence of hRNase8 gene expression in other additional tissues, such as the lung, liver, and testes (141), together with controversial reports on its recombinant protein antimicrobial activity (91, 199), urges a reconsideration of the function of this protein.
hRNases 9–13
The RNaseA superfamily was lately expanded with the discovery of several novel mammalian members (11, 203–205). The newcomers share just 15–30% identity with the eight “canonical RNases,” and they are associated mainly with male reproductive functions. Some of them, like hRNase9, are endowed with bactericidal activity and are expressed in the epididymis, with evidence of an association with sperm maturation (33, 206). The primary structure of these proteins resembles ancestral RNases, sharing the three most conserved disulfide bonds and a secretion peptide, but not the N-terminus region of mature proteins. In addition, the non-canonical RNases do not include the family signature (CKXXNTF) or the catalytic triad and their biological properties do not seem to require enzymatic activity (207).
Other Vertebrate RNaseA Family Members
The RNaseA superfamily is one of the most extensively studied gene family, since the pioneering studies on molecular evolution (208). As a vertebrate-specific family, it is an excellent working model and a deep analysis has been carried out to understand the main driving force toward a defined function in mammals (11, 33, 209, 210). Further, we will briefly comment on the non-human family members present in the extracellular compartment and displaying immune modulation properties. In particular, bovine proteins are the best characterized, being the RNaseA the bovine counterpart of human RNase1. RNaseA, the bovine pancreatic RNase1, is expressed in large amounts in the pancreas and participates in RNA degradation in the digestive tract (3).
Bovine Seminal RNase (BS-RNase)
Bovine seminal RNase is a close homolog to bovine pancreatic RNase that is solely present in seminal fluid (211), where it plays an immune-protective role (182). BS-RNase, despite sharing an 80% amino acid identity with the pancreatic RNaseA, is the only family member present in vivo as a homodimer. Owing to its natural dimeric form, BS-RNase can elude the blockage of the cytosolic RI in case of cellular internalization following endocytosis. The high cytotoxicity of the protein (212) is attributed to the degradation of cellular RNA (213). In addition, the dimerization of BS-RNase and RNaseA constructs correlates with an enhanced catalytic activity and the ability to cleave dsRNA (214–216). Degradation of dsRNA by BS-RNase is induced by IFN (217, 218) and the protein can inhibit HIV-1 replication in leukemia cells (219). Interestingly, engineered quaternary structures of RNaseA can not only mimic BS-RNase enzymatic cleavage of dsRNA, but also some of its biological properties (220, 221). The seminal RNase also differs from its pancreatic homolog by its ability to inhibit the proliferation of cancer cells mediated by autophagy induction (72). BS-RNase also inhibits the proliferation of T-lymphocytes (222, 223) and can downregulate the T cell IL-2 receptor expression (222). However, the immune-protective mechanism of BS-RNase in the seminal fluid remains unknown (182).
Bovine Milk RNases
Bovine milk RNases are another group of secretory RNases that mediate an extracellular protective role. Two proteins with RNase activity homologous to human RNases 4 and 5 were identified in bovine milk (224, 225). Both RNases were quantified in bovine milk at μM concentrations and reported to display some antimicrobial activity (225). Bovine milk RNases can participate in the host response against infection both by direct antimicrobial action and immune response activation (182). A pro-inflammatory activity is observed in epithelial cells, which is mediated by nucleic acids (53, 226). Both RNases can bind nucleic acids, and milk RNase5 induces cytokine release in leukocytes (53). Recognition of foreign pathogen nucleic acids may facilitate the activation of pattern recognition receptors and promote a pro-inflammatory response (226). Interestingly, the RNase immuno-stimulatory activity is also dependent on the protein catalytic activity (226).
Rodent RNases
Rodent RNases are another well characterized group that can help us to outline the RNaseA family involvement in the host immune response (11). Lineages of RNases 1, 2, 3, and 5 are identified in rat and mouse genomes, presenting an unusual expansion rate (11, 207, 227–229). However, no orthologs of RNases 7 and 8 have been found (11, 33). Particularly, a striking diversity of RNases 2 and 3 counterparts is observed in mice and rats. Two orthologs of the eosinophil RNases lineage were first discovered in 1996 and named EARs (230). Subsequently, up to 13 new eosinophil murine members were identified (231). The phylogenetic analysis of the distinct EAR rodent gene clusters revealed a rapid gene duplication and selection process that resulted in high diversification, a characteristic pattern of host defense protein lineages (232). Many of these EAR proteins, despite their nomenclature, are not solely secreted by eosinophil granules but can also be expressed by other cell types. For example, mEAR11 is expressed in somatic tissues, such as lungs, liver, or spleen, along with macrophages. The protein expression is induced in response to Th2 cytokines and it acts as a potent leukocyte chemoattractant (48). The immune regulation of eosinophil release of granule proteins in mice has been thoroughly studied by Weller and colleagues (149, 168). EARs similar to their human counterparts actively contribute to the host defense and tissue repair and remodeling. However, significant differences are observed in the regulation mechanism of eosinophil degranulation, limiting the use of laboratory animal models in the study of human eosinophil-associated diseases (233–235). Nonetheless, experimental studies in mice corroborate the autonomy of eosinophil cell-free granules and their activation by a common CCR3-mediated signaling pathway (149, 168). EARs can also provide immune protection against virus infection in vivo in a mice model, where the eosinophil activation and virus clearance is mediated by a TLR7-signaling pathway (55).
In addition, several homologs to human RNase5 endowed with antimicrobial properties were identified in mouse intestinal epithelium (88). In particular, the upregulation of the mouse RNase Ang4 by commensal bacteria suggests a role for this protein in the gut and systemic innate immunity, where it can establish a host defense barrier against infection (88).
Rosenberg and colleagues went further down the evolution scale and characterized avian and reptilian RNases to deepen the understanding of the role of vertebrate RNaseA family in host immunity (13, 16, 236). As mentioned previously, non-mammalian RNases are evolutionarily closely related to mammalian RNase5 members (11). Two leukocyte-associated homologs were identified in chicken, RNases A1 and A2, the last one displaying both angiogenic and bactericidal properties (13). On the other hand, the Iguana RNase is catalytically active but devoid of antimicrobial activity (236).
Frog RNases
Frog RNases are secreted by oocytes and early embryos and might protect the eggs against infection (237–241). The observed anti-proliferative properties of the RNases from early vertebrates have attracted the interest of pharmaceutical companies since their discovery (242, 243). Rana pipiens RNase, named Onconase® (ONC) after its anti-tumoural activity, is currently on phase III clinical trial. The anti-proliferative action of ONC on cancer cells is mediated by induction of the autophagy pathway (71), as reported for BS-RNase (72). Recently, its cytotoxicity was enhanced by promoting its dimerization (244). The anti-tumoural activity of ONC has also been attributed to its action on microRNA (miRNA) precursors (40). On the other hand, the frog RNase inhibits the replication of HIV-1 through directly targeting the viral RNA and host cellular tRNA (245, 246). A specific excision on host Lys-tRNA inhibits the virion replication (101). A common tRNA targeting mechanism might be shared with the oldest mammalian RNases. Moreover, a similarity between the structural fold of tRNA and miRNAs that are targeted by the frog RNase suggests that the RNA recognition and cleavage requires specific primary and secondary structures (40).
Finally, identification and characterization of fish RNases completes the overall picture of the vertebrate RNaseA family (15, 247, 248). RNases identified in zebrafish (ZF-RNases) shared the bactericidal, angiogenic and reduced catalytic properties of hRNase5 (247). Catalytic activity for ZF-RNase5 was required for angiogenesis but not for antimicrobial action (247). Interestingly, ZF-RNases 1 to 3 can activate the ERK1/2 kinase pathway similar to hRNase5 (249). Recently, the expression of several RNase2 and RNase3 paralogs in the pond-cultured blunt snout bream fish induced by bacterial infection was reported (250).
Other Extracellular RNases
RNases T2: A Family of Ancient Extracellular RNases
RNaseT2 family, in contrast with the RNaseA family, comprises a group of proteins conserved from virus to humans, suggesting a shared preserved function (31). The biological properties of human RNaseT2 have been extensively studied. The RNase that works as a signaling molecule and is secreted by damaged tissues has being classified as an alarmin (61). RNaseT2 is stored in the lysosomal compartment and contributes to the clearance of cellular macromolecular debris. Its secretion can be induced by oxidative stress and it participates in the regulation of immune response. RNaseT2 is proposed to work as an RNA scavenger in the extracellular compartment (31). Moreover, the human RNaseT2 shows macrophage chemotaxis (30) and tissue remodeling activities in vitro. Similarly, the RNaseT2 secreted by the eggs of the parasite Schistosoma mansoni, also named Omega-1, can induce the release of pro-inflammatory cytokines by macrophages during infection (251). Omega-1 can be internalized into DCs and regulates their programming pathway by the RNase-mediated cleavage of rRNAs and mRNAs and subsequent impairment of protein synthesis. Another RNaseT2 family member that has been well characterized is the yeast RNaseT2, named Rny1, which is stored in cell vacuoles, similar to the storage of other RNaseT2 members in lysosomes, and shows a selective tRNA cleavage under oxidative stress equivalent to the activity reported for hRNase5 (56). In addition, yeast RNaseT2 combines its enzymatic action with other non-catalytic properties such as binding to regulatory proteins and the destabilization of lysosomal membrane, a mechanism that can trigger the programmed cell death (56, 252). Overall, we observed common properties between the RNaseA and the RNaseT2 family members, e.g., release of stress-induced tiRNA, leukocyte activation, or exRNA scavenging (30, 31, 56, 250).
Plant Self-Incompatibility RNases (S-RNases)
Plant S-RNases prevent self-fertilization and avoid inbreeding. S-RNases exert cytotoxicity against the growing pollen tube by targeting rRNA (253, 254). Each plant is endowed with specific recognition patterns that can block the RNase activity of all the S-RNases except its own, ensuring the degradation of pollen grains corresponding to its haplotype (255). Interestingly, S-RNases exhibit a specific catalytic activity on tRNA when the plants are exposed to stress (256).
Bacterial RNases as Inter-Strain Competition Toxins
Going further down the evolutionary scale, we can find a wide variety of bacterial RNases that participate in the bacterial defense against external threats, e.g., presence of a competing bacterial species, viral infection, or the defense response of the infected host cell. Bacterial RNases can work as powerful toxins selectively targeting coding and non-coding RNAs (257–259). Among the non-coding RNAs, the specific cleavage of tRNAs is a conserved regulatory mechanism shared from bacterial to mammalian cells (56). Stress-induced tRNA cleavage is reported for the Escherichia coli endoribonuclease Prrc in response to bacteriophage infection (260). Colicins are another group of E. coli cytotoxic tRNases that block the protein synthesis machinery as a defense mechanism against other microbial competitors (261). Interestingly, comparison among the bacterial RNases suggest an evolutionary convergence to acquire structural features that enable the targeting of the tRNA anticodon loop (261). In simple eukaryotes, such as the protozoa Tetrahymena, and the budding yeast Saccharomyces cerevisiae, the release of specific tRFs during starvation is also reported (256, 262). We can establish a parallelism between the release of stress-induced tiRNA by prokaryotes, primitive eukaryotes, and human RNase5 (6), as a mechanism to downregulate protein synthesis. An intriguing question arises: have the host defense mechanisms of vertebrate RNases evolved from the ancestral prokaryotic inter-strain competition processes? Although the RNaseA superfamily is vertebrate-specific, the recent report of the structure of a bacterial RNase involved in inter-strain competition highlights a shared protein scaffold shaped for RNA recognition (32). However, the lack of sequence identity between bacterial and vertebrate RNases and the absence of any putative invertebrate intermediate suggests a convergent evolution (32, 263). Thus, the origin of the RNaseA superfamily remains unknown. In contrast, the RNaseT2 family conserves its ancestral lineage from prokaryotes to humans (56).
Similarities between unicellular self-defense and mammalian innate immune mechanisms can provide novel strategies to boost our own immune response. For example, macrophage immune regulation by Bacillus RNase (binase) can trigger the host cell anti-tumor response (19) and the RNaseT2 of the Schistosoma parasite can modulate the host response (250) and prevent the outbreak of autoimmune diseases or diabetes (264, 265). Understanding the uniqueness of RNases and their specificity for cellular RNAs will lead to the development of novel therapeutics.
RNase Traffic in the Extracellular Space
Extracellular RNases are released as secretory proteins by diverse pathways into the extracellular compartment. Recent advances in histochemical and cell analytical methodologies have unveiled the structural and functional complexity of the extracellular space. A rich world of secretory storage granules, transport vesicles, and intracellular vacuoles ensures that the organism is fit to respond to external stimuli.
Compartmentalization
Compartmentalization of RNA and RNases is an important regulatory mechanism (266). RNases packed within secretory granules will be selectively released upon action by diverse stimuli (see Figure 1). In particular, eosinophil degranulation has been thoroughly investigated and several secretory mechanisms have been described (267). Intracellular granules can undergo piecemeal degranulation, whereby small packets of derived vesicles are mobilized toward the cell surface for secretion (231). Alternatively, the cell storage granules can be freed as independent entities. Free extracellular eosinophil granules can actively release their content upon cytokine activation (167). Weller and co-workers have extensively characterized the signaling pathways that mediate the release of RNases by the free extracellular eosinophil granules in humans and mice (149, 168). Free eosinophil granules can be internalized by other innate cells, such as macrophages (Figure 1) and thereby participate in the regulatory pathways of the recipient cell. Extracellular RNases can also find a way back into cells through the endosomal pathway (240, 268). Fortunately, the cytosolic compartment of cells is protected from the potential toxicity of RNaseA superfamily members by the action of the RI, which constitutes about 0.1% of the total protein content in the cytosol of mammalian cells (269). The RI is expressed in all studied human tissues (143) and binds with an extremely high affinity to mammalian RNaseA family members (in the fM range) (270, 271). Interestingly, Raines and co-workers (270) identified the avian and reptilian counterparts of mammalian RI but no equivalent protein was detected in amphibians and fish, suggesting a specific role for RI in higher order vertebrates (270). Recent RI-knockout experiments confirmed the protective action of RI toward cytosolic RNA against endocytosed RNases (272). The inhibitor structure adopts a horseshoe conformation composed of LRRs and exposed free cysteine residues. The inhibitor is functional in its fully reduced state and is extremely sensitive to cellular oxidative stress. RI inactivation by partial oxidation can work as a mechanism to switch on the RNase-mediated degradation of cellular RNA under stress conditions (56, 270). For example, RI participates in the regulation of hRNase5 subcellular localization during stress conditions. Under stress, the cytosolic hRNase5 is liberated from the RI complex, whereas the nuclear protein is bound to the inhibitor, thereby downregulating cell growth (272). Interestingly, the hRNase5 evasion of the cytosolic RI and migration to the nucleolus is also mediated by phosphorylation (273). RI can also participate in the regulation of RNases expressed at the epidermis (67). The secreted RNases in the skin provide a protective barrier against invading pathogens. Degradation of RI by proteases at the stratum corneum can liberate the RNases’ antimicrobial action during infection (67, 274). Regulation of RNase activity by RI in the urinary tract has also been proposed by Spencer and co-workers (275).
Intercellular Communication
Nowadays, novel methodologies have led to better understanding of the functions of the extracellular compartment and have proposed previously undescribed roles for secretory proteins. RNases secreted by diverse stimuli (Figure 1) can participate in intercellular communication in an organism (56). Of note, some RNaseA family members have been detected within extracellular vesicles (117, 276, 277) (Table 2) and selective RNA packaging into the vesicles has been observed (278). This brings us back to the pioneering biochemical work on the pancreatic RNaseA family (3). The identification of the angiogenic activity of hRNase5 and the suspicion that angiogenic factors might contain RNA (279) hinted that exRNA might work as an intercellular communicator (1). Novel sequencing methodologies confirmed hRNase5 selective cleavage of non-coding RNAs and the involvement of the released products in immune regulation pathways (57). Other pancreatic RNase family members may also have evolved to acquire a non-digestive role and may contribute to the regulation of the circulating exRNA content in blood (18, 280). James Lee proposed that during vertebrate evolution the mechanism of action of granulocyte proteins might have evolved from mere localized action to an organized systemic response mechanism. The increase in size and complexity of multicellular organisms is accompanied by long distance stress signaling processes. In this context, secretory RNases originally recruited at the damaged tissue site to remove cellular RNA debris from dying cells could have acquired a selective anti-pathogen activity to provide the host protection against infection. An amoeboid-type secretory blood cell initially adapted to localized response and tissue repair duties would have acquired novel properties, allowing isolated tissue cells to communicate over an extended distance and participate in the overall systemic response (17).
Role in Health and Disease
Overall, extracellular RNases display a variety of immune-related activities that ensure that the organism is fit for survival. The RNases participate in diverse tasks, from cellular housekeeping to ensuring the sterility of body fluids (Table 1). Following tissue damage by an external injury the RNases are expressed as alarm signaling molecules (54, 60, 61). Their secretion at the inflammation site contributes to tissue repair and remodeling (52, 62). To participate in the tissue healing process, the RNases can target and remove the host-damaged cells. Selective cytotoxicity can be mediated by the activation of autophagy or apoptotic pathways (72, 75). To facilitate subsequent tissue remodeling, the RNases also function as cytokines and chemokines, displaying anti-inflammatory activities and inducing chemoattraction of innate cells, such as macrophages or DCs (29, 46–48). Other complementary activities have been reported such as binding to nucleic acids, activation of TLR receptors and removal of exRNA (54, 55, 76).
RNases’ expression can also be induced during infections and the secreted RNases can directly participate in the killing of invading microorganisms (42, 84, 89) (Table 1). Overall, we observed an organized distribution of tasks among the distinct host innate cells that can ensure the coverage of wide spectra of potential pathogens. On the other hand, there is also a downregulation in RNases’ expression after extended periods of infection (Table 2). A close inspection of the RNases’ expression patterns suggests an adaptive process by the intracellular-dwelling pathogens to inhibit the host response and extend their survival lifespan.
Fortunately, extensive research on secretory RNases is currently setting the basis for applied therapies. Clinicians are already taking advantage of the selective secretion of RNases for monitoring and diagnosing inflammation. RNase3 levels are routinely used to monitor asthma processes (51). Levels of hRNase1, hRNase3, and hRNase7 are increased during sepsis and are proposed as markers for the diagnosis of organ failure (112). Another interesting proposal is the use of RI as a cancer biomarker (281).
Furthermore, successful results achieved using experimental animal models promise therapeutic applications in the near future. Removal of circulating exRNA shows beneficial anti-inflammatory properties following tissue damage (17, 29, 76). In particular, removal of blood exRNA can protect cardiac tissue in hypoxic conditions (29, 76). Treatment with hRNase1 has been observed to reduce deposits of exRNA and inflammation in a mouse model of atherosclerosis (78). RNases can also determine the fate of RNA stress granules. Under stress conditions, hRNase5 accumulates within stress granules (282). Local accumulation of RNA and alteration of RNA self-assembly is associated with neurodegenerative diseases (283). Selective cleavage of cellular RNAs mediates response to stress stimuli (56, 57). Overall, deregulation of non-coding RNA processing is a major cause of immune-malfunctioning and serious diseases (284).
Some RNaseA members can participate in biological functions such as hematopoiesis and angiogenesis, and show anti-tumoural properties mediated by selective cellular RNA targeting (6, 40, 285). The design of RNase constructs to develop specific immunotoxins that selectively target cancer cells is currently one of the most prioritize research topics. ImmunoRNases are engineered to be internalized by tumor cells, evade RI, and degrade cellular RNA (26, 269, 286–289). A recent nanocarrier delivery system using encapsulated RNaseA effectively achieves inhibition of cancer cell proliferation (290).
RNases can also maintain the sterility of biological fluids (100, 285). Eosinophils are involved in antiviral immunity and eosinophil RNases might mediate host response by TLR7 activation (55). Expression of hRNase2 is induced by HIV-1 infection and recombinant hRNase2 administration is proposed as an anti-HIV-1 therapy (100).
Conclusion and Perspectives
The overview of the immuno-regulatory properties of secretory RNases highlights the similarities between their mechanisms of action and provides novel approaches to progress toward a deeper understanding of their ultimate in vivo biological role. We are confident that any step forward in this direction can consolidate our knowledge of the innate immune system and contribute to the development of novel treatments against immunological deregulations. In particular, the biological roles of RNases, such as in hematopoiesis regulation, tissue remodeling, prevention of infection and inflammation offer promising therapeutic applications.
Author Contributions
LL, JL, MM, and EB contributed to the original draft and edited versions. LL and JL prepared the tables and graphical material. MM and EB wrote, edited, and revised the final manuscript version. All authors approved the final manuscript version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Research work was supported by the Ministerio de Economía y Competitividad (SAF2015-66007P) and by AGAUR, Generalitat de Catalunya (2016PROD00060; 2017SGR1010), co-financed by FEDER funds. LL and JL are recipients of a CSC predoctoral fellowship.
References
1. Benner SA. Extracellular “communicator RNA.”. FEBS Lett (1988) 233:225–8. doi:10.1016/0014-5793(88)80431-9
2. Benner SA, Allemann RK. The return of pancreatic ribonucleases. Trends Biochem Sci (1989) 14:396–7. doi:10.1016/0968-0004(89)90282-X
3. Barnard EA. Biological function of pancreatic ribonuclease. Nature (1969) 221:340–4. doi:10.1038/221340a0
4. Zendzian EN, Barnard EA. Distributions of pancreatic ribonuclease, chymotrypsin, and trypsin in vertebrates. Arch Biochem Biophys (1967) 122:699–713. doi:10.1016/0003-9861(67)90180-4
5. Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett (2009) 583:437–42. doi:10.1016/j.febslet.2008.12.043
6. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol (2009) 185:35–42. doi:10.1083/jcb.200811106
8. Cuchillo CM, Nogués MV, Raines RT. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry (2011) 50:7835–41. doi:10.1021/bi201075b
9. Beintema JJ, Gaastra W, Lenstra JA, Welling GW, Fitch WM. The molecular evolution of pancreatic ribonuclease. J Mol Evol (1977) 10:49–71. doi:10.1007/BF01796134
10. Jermann TM, Opitz JG, Stackhouse J, Benner SA. Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature (1995) 374:57–9. doi:10.1038/374057a0
11. Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics (2005) 85:208–20. doi:10.1016/j.ygeno.2004.10.008
12. Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A (1998) 95:3708–13. doi:10.1073/pnas.95.7.3708
13. Nitto T, Dyer KD, Czapiga M, Rosenberg HF. Evolution and function of leukocyte RNase A ribonucleases of the avian species, Gallus gallus. J Biol Chem (2006) 281:25622–34. doi:10.1074/jbc.M604313200
14. Pizzo E, D’Alessio G. The success of the RNase scaffold in the advance of biosciences and in evolution. Gene (2007) 406:8–12. doi:10.1016/j.gene.2007.05.006
15. Cho S, Zhang J. Zebrafish ribonucleases are bactericidal: implications for the origin of the vertebrate RNase a superfamily. Mol Biol Evol (2007) 24:1259–68. doi:10.1093/molbev/msm047
16. Rosenberg HF. RNase A ribonucleases and host defense: an evolving story. J Leukoc Biol (2008) 83:1079–87. doi:10.1189/jlb.1107725
17. Lee JJ, Lee NA. Eosinophil degranulation: an evolutionary vestige or a universally destructive effector function? Clin Exp Allergy (2005) 35:986–94. doi:10.1111/j.1365-2222.2005.02302.x
18. Lomax JE, Eller CH, Raines RT. Comparative functional analysis of ribonuclease 1 homologs: molecular insights into evolving vertebrate physiology. Biochem J (2017) 474:2219–33. doi:10.1042/BCJ20170173
19. Makeeva A, Rodriguez-Montesinos J, Zelenikhin P, Nesmelov A, Preissner KT, Cabrera-Fuentes HA, et al. Antitumor macrophage response to bacillus pumilus ribonuclease (Binase). Mediators Inflamm (2017) 2017:4029641. doi:10.1155/2017/4029641
20. Matzinger P. An innate sense of danger the signals that initiate immune responses. Semin Immunol (1998) 10:399–415.
21. Wu D, Yu W, Kishikawa H, Folkerth RD, Iafrate AJ, Shen Y, et al. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol (2007) 62:609–17. doi:10.1002/ana.21221
22. Thiyagarajan N, Ferguson R, Subramanian V, Acharya R. Structural and molecular insights into the mechanism of action of human angiogenin-ALS variants in neurons. Nat Commun (2012) 3:1114–21. doi:10.1038/ncomms2126
23. Henneke M, Diekmann S, Ohlenbusch A, Kaiser J, Engelbrecht V, Kohlschütter A, et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat Genet (2009) 41:773–5. doi:10.1038/ng.398
24. Thorn A, Steinfeld R, Ziegenbein M, Grapp M, Hsiao HH, Urlaub H, et al. Structure and activity of the only human RNase T2. Nucleic Acids Res (2012) 40:8733–42. doi:10.1093/nar/gks614
25. Schein CH. From housekeeper to microsurgeon: the diagnostic and therapeutic potential of ribonucleases. Nat Biotechnol (1997) 15:529–36. doi:10.1038/nbt0697-529
26. Suzuki M, Saxena SK, Boix E, Prill RJ, Vasandani VM, Ladner JE, et al. Engineering receptor-mediated cytotoxicity into human ribonucleases by steric blockade of inhibitor interaction. Nat Biotechnol (1999) 17:265–70. doi:10.1038/7010
27. Rybak SM, Newton DL. Natural and engineered cytotoxic ribonucleases: therapeutic potential. Exp Cell Res (1999) 253:325–35. doi:10.1006/excr.1999.4718
28. Goncalves KA, Silberstein L, Li S, Severe N, Hu MG, Yang H, et al. Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell (2016) 166:894–906. doi:10.1016/j.cell.2016.06.042
29. Ma G, Chen C, Jiang H, Qiu Y, Li Y, Li X, et al. Ribonuclease attenuates hepatic ischemia reperfusion induced cognitive impairment through the inhibition of inflammatory cytokines in aged mice. Biomed Pharmacother (2017) 90:62–8. doi:10.1016/j.biopha.2017.02.094
30. Acquati F, Lualdi M, Bertilaccio S, Monti L, Turconi G, Fabbri M, et al. Loss of function of Ribonuclease T2, an ancient and phylogenetically conserved RNase, plays a crucial role in ovarian tumorigenesis. Proc Natl Acad Sci U S A (2013) 110(20):8140–5. doi:10.1073/pnas.1222079110
31. Luhtala N, Parker R. T2 Family ribonucleases: ancient enzymes with diverse roles. Trends Biochem Sci (2010) 35:253–9. doi:10.1016/j.tibs.2010.02.002
32. Batot G, Michalska K, Ekberg G, Irimpan EM, Joachimiak G, Jedrzejczak R, et al. The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily. Nucleic Acids Res (2017) 45:5013–25. doi:10.1093/nar/gkx230
33. Goo SM, Cho S. The expansion and functional diversification of the mammalian ribonuclease a superfamily epitomizes the efficiency of multigene families at generating biological novelty. Genome Biol Evol (2013) 5:2124–40. doi:10.1093/gbe/evt161
34. Sorrentino S. The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett (2010) 584:2194–200. doi:10.1016/j.febslet.2010.04.018
35. Boix E, Blanco JA, Nogués MV, Moussaoui M. Nucleotide binding architecture for secreted cytotoxic endoribonucleases. Biochimie (2013) 95:1087–97. doi:10.1016/j.biochi.2012.12.015
36. Nogués MV, Moussaoui M, Boix E, Vilanova M, Ribó M, Cuchillo CM. The contribution of noncatalytic phosphate-binding subsites to the mechanism of bovine pancreatic ribonuclease A. Cell Mol Life Sci (1998) 54:766–74. doi:10.1007/s000180050205
37. Prats-Ejarque G, Arranz-Trullén J, Blanco JA, Pulido D, Nogués MV, Moussaoui M, et al. The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement. Biochem J (2016) 473:1523–36. doi:10.1042/BCJ20160245
38. Sikriwal D, Seth D, Batra JK. Role of catalytic and non-catalytic subsite residues in ribonuclease activity of human eosinophil-derived neurotoxin. Biol Chem (2009) 390:225–34. doi:10.1515/BC.2009.025
39. Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res (2012) 40:6787–99. doi:10.1093/nar/gks307
40. Qiao M, Zu L-D, He X-H, Shen R-L, Wang Q-C, Liu M-F. Onconase downregulates microRNA expression through targeting microRNA precursors. Cell Res (2012) 22:1199–202. doi:10.1038/cr.2012.67
41. Lyons SM, Fay MM, Akiyama Y, Anderson PJ, Ivanov P. RNA biology of angiogenin: current state and perspectives. RNA Biol (2017) 14:171–8. doi:10.1080/15476286.2016.1272746
42. Boix E, Nogués MV. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Mol Biosyst (2007) 3:317–35. doi:10.1039/b617527a
43. Köten B, Simanski M, Gläser R, Podschun R, Schröder JM, Harder J. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS One (2009) 4:e6424. doi:10.1371/journal.pone.0006424
44. Simanski M, Köten B, Schröder JM, Gläser R, Harder J. Antimicrobial RNases in cutaneous defense. J Innate Immun (2012) 4:241–7. doi:10.1159/000335029
45. Koczera P, Martin L, Marx G, Schuerholz T. The ribonuclease a superfamily in humans: canonical RNases as the buttress of innate immunity. Int J Mol Sci (2016) 17:E1278. doi:10.3390/ijms17081278
46. Yang D, Rosenberg HF, Chen Q, Dyer KD, Kurosaka K, Oppenheim JJ. Eosinophil-derived neurotoxin (EDN), an antimicrobial protein with chemotactic activities for dendritic cells. Blood (2003) 102:3396–403. doi:10.1182/blood-2003-01-0151
47. Yang D, Chen Q, Rosenberg HF, Rybak SM, Newton DL, Wang ZY, et al. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J Immunol (2004) 173:6134–42. doi:10.4049/jimmunol.173.10.6134
48. Yamada KJ, Barker T, Dyer KD, Rice TA, Percopo CM, Garcia-Crespo KE, et al. Eosinophil-associated ribonuclease 11 is a macrophage chemoattractant. J Biol Chem (2015) 290:8863–75. doi:10.1074/jbc.M114.626648
49. Zagai U, Dadfar E, Lundahl J, Venge P, Sköld CM. Eosinophil cationic protein stimulates TGF-β1 release by human lung fibroblasts in vitro. Inflammation (2007) 30:153–60. doi:10.1007/s10753-007-9032-4
50. Zheutlin LM, Ackerman SJ, Gleich GJ, Thomas LL. Stimulation of basophil and rat mast cell histamine release by eosinophil granule-derived cationic proteins. J Immunol (1984) 133:2180–5.
51. Venge P, Byström J, Carlson M, Håkansson L, Karawacjzyk M, Peterson C, et al. Eosinophil cationic protein (ECP): molecular and biological properties and the use of ECP as a marker of eosinophil activation in disease. Clin Exp Allergy (1999) 29:1172–86. doi:10.1046/j.1365-2222.1999.00542.x
52. Bystrom J, Amin K, Bishop-Bailey D. Analysing the eosinophil cationic protein – a clue to the function of the eosinophil granulocyte. Respir Res (2011) 12:10. doi:10.1186/1465-9921-12-10
53. Shcheglovitova ON, Maksyanina EV, Ionova II, Rustam’yan YL, Komolova GS. Cow milk angiogenin induces cytokine production in human blood leukocytes. Bull Exp Biol Med (2003) 135:158–60. doi:10.1023/A:1023871931764
54. Kopfnagel V, Wagenknecht S, Harder J, Hofmann K, Kleine M, Buch A, et al. RNase 7 strongly promotes TLR9-mediated DNA sensing by human plasmacytoid dendritic cells. J Invest Dermatol (2017) 138:872–81. doi:10.1016/j.jid.2017.09.052
55. Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, et al. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood (2007) 110:1–3. doi:10.1182/blood-2007-01-071340
56. Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell (2009) 138:215–9. doi:10.1016/j.cell.2009.07.001
57. Mesitov MV, Soldatov RA, Zaichenko DM, Malakho SG, Klementyeva TS, Sokolovskaya AA, et al. Differential processing of small RNAs during endoplasmic reticulum stress. Sci Rep (2017) 7:1–14. doi:10.1038/srep46080
58. Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem (2010) 285:10959–68. doi:10.1074/jbc.M109.077560
59. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell (2011) 43:613–23. doi:10.1016/j.molcel.2011.06.022
60. Yang D, Chen Q, Su SB, Zhang P, Kurosaka K, Caspi RR, et al. Eosinophil-derived neurotoxin acts as an alarmin to activate the TLR2-MyD88 signal pathway in dendritic cells and enhances Th2 immune responses. J Exp Med (2008) 205:79–90. doi:10.1084/jem.20062027
61. Chan JK, Roth J, Oppenheim JJ, Tracey KJ, Vogl T, Feldmann M, et al. Alarmins: awaiting a clinical response. J Clin Invest (2012) 122:2711–9. doi:10.1172/JCI62423
62. Zagai U, Sköld CM, Trulson A, Venge P, Lundahl J. The effect of eosinophils on collagen gel contraction and implications for tissue remodelling. Clin Exp Immunol (2004) 135:427–33. doi:10.1111/j.1365-2249.2004.02396.x
63. Tsuji T, Sun Y, Kishimoto K, Olson KA, Liu S, Hirukawa S, et al. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res (2005) 65:1352–60. doi:10.1158/0008-5472.CAN-04-2058
64. Xu L, Liao WL, Lu QJ, Li CG, Yuan Y, Xu ZY, et al. ANG promotes proliferation and invasion of the cell of lung squamous carcinoma by directly up-regulating HMGA2. J Cancer (2016) 7:862–71. doi:10.7150/jca.14539
65. Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, et al. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry (1985) 24:5480–6. doi:10.1021/bi00341a030
66. Kishimoto K, Liu S, Tsuji T, Olson KA, Hu G. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene (2005) 24:445–56. doi:10.1038/sj.onc.1208223
67. Abtin A, Eckhart L, Mildner M, Ghannadan M, Harder J, Schröder J-M, et al. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J Invest Dermatol (2009) 129:2193–201. doi:10.1038/jid.2009.35
68. Bernard JJ, Gallo RL. Protecting the boundary: the sentinel role of host defense peptides in the skin. Cell Mol Life Sci (2011) 68:2189–99. doi:10.1007/s00018-011-0712-8
69. Harder J, Dressel S, Wittersheim M, Cordes J, Meyer-Hoffert U, Mrowietz U, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J Invest Dermatol (2010) 130:1355–64. doi:10.1038/jid.2009.432
70. Amatngalim GD, van Wijck Y, de Mooij-Eijk Y, Verhoosel RM, Harder J, Lekkerkerker AN, et al. Basal cells contribute to innate immunity of the airway epithelium through production of the antimicrobial protein RNase 7. J Immunol (2015) 194:3340–50. doi:10.4049/jimmunol.1402169
71. Fiorini C, Cordani M, Gotte G, Picone D, Donadelli M. Onconase induces autophagy sensitizing pancreatic cancer cells to gemcitabine and activates Akt/mTOR pathway in a ROS-dependent manner. Biochim Biophys Acta (2015) 1853:549–60. doi:10.1016/j.bbamcr.2014.12.016
72. Fiorini C, Gotte G, Donnarumma F, Picone D, Donadelli M. Bovine seminal ribonuclease triggers Beclin1-mediated autophagic cell death in pancreatic cancer cells. Biochim Biophys Acta (2014) 1843:976–84. doi:10.1016/j.bbamcr.2014.01.025
73. Navarro S, Aleu J, Jiménez M, Boix E, Cuchillo CM, Nogués MV. The cytotoxicity of eosinophil cationic protein/ribonuclease 3 on eukaryotic cell lines takes place through its aggregation on the cell membrane. Cell Mol Life Sci (2008) 65:324–37. doi:10.1007/s00018-007-7499-7
74. Spalletti-Cernia D, Sorrentino R, Di Gaetano S, Arciello A, Garbi C, Piccoli R, et al. Antineoplastic ribonucleases selectively kill thyroid carcinoma cells via caspase-mediated induction of apoptosis. J Clin Endocrinol Metab (2003) 88:2900–7. doi:10.1210/jc.2002-020373
75. Chang KC, Lo CW, Fan TC, Chang MD, Shu CW, Chang CH, et al. TNF-α mediates eosinophil cationic protein-induced apoptosis in BEAS-2B cells. BMC Cell Biol (2010) 11. doi:10.1186/1471-2121-11-6
76. Cabrera-Fuentes HA, Ruiz-Meana M, Simsekyilmaz S, Kostin S, Inserte J, Saffarzadeh M, et al. RNase1 prevents the damaging interplay between extracellular RNA and tumour necrosis factor-α in cardiac ischaemia/reperfusion injury. Thromb Haemost (2014) 112:1110–9. doi:10.1160/th14-08-0703
77. Zernecke A, Preissner KT. Extracellular ribonucleic acids (RNA) enter the stage in cardiovascular disease. Circ Res (2016) 118:469–79. doi:10.1161/CIRCRESAHA.115.307961
78. Simsekyilmaz S, Cabrera-Fuentes HA, Meiler S, Kostin S, Baumer Y, Liehn EA, et al. Role of extracellular RNA in atherosclerotic plaque formation in mice. Circulation (2014) 129:598–606. doi:10.1161/CIRCULATIONAHA.113.002562
79. Harder J, Schröder JM, Gläser R. The skin surface as antimicrobial barrier: present concepts and future outlooks. Exp Dermatol (2013) 22:1–5. doi:10.1111/exd.12046
80. Zanger P, Holzer J, Schleucher R, Steffen H, Schittek B, Gabrysch S. Constitutive expression of the antimicrobial peptide RNase 7 is associated with Staphylococcus aureus infection of the skin. J Infect Dis (2009) 200:1907–15. doi:10.1086/648408
81. Rademacher F, Simanski M, Harder J. RNase 7 in cutaneous defense. Int J Mol Sci (2016) 560:1–12. doi:10.3390/ijms17040560
82. Becknell B, Spencer JD. A review of ribonuclease 7’s structure, regulation, and contributions to host defense. Int J Mol Sci (2016) 423:1–10. doi:10.3390/ijms17030423
83. Becknell B, Eichler TE, Beceiro S, Li B, Easterling RS, Carpenter AR, et al. Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int (2015) 87:151–61. doi:10.1038/ki.2014.268
84. Spencer JD, Schwaderer AL, Wang H, Bartz J, Kline J, Eichler T, et al. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int (2013) 83:615–25. doi:10.1038/ki.2012.410
85. Ganz T. Angiogenin: an antimicrobial ribonuclease. Nat Immunol (2003) 4:213–4. doi:10.1038/ni0403-394b
86. Boix E, Salazar VA, Torrent M, Pulido D, Nogués MV, Moussaoui M. Structural determinants of the eosinophil cationic protein antimicrobial activity. Biol Chem (2012) 393:801–15. doi:10.1515/hsz-2012-0160
87. Pulido D, Torrent M, Andreu D, Nogues MV, Boix E. Two human host defense ribonucleases against mycobacteria, the eosinophil cationic protein (RNase 3) and RNase 7. Antimicrob Agents Chemother (2013) 57:3797–805. doi:10.1128/AAC.00428-13
88. Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol (2003) 4:269–73. doi:10.1038/ni888
89. Harder J, Schröder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J Biol Chem (2002) 277:46779–84. doi:10.1074/jbc.M207587200
90. Pulido D, Arranz-Trullen J, Prats-Ejarque G, Velazquez D, Torrent M, Moussaoui M, et al. Insights into the antimicrobial mechanism of action of human RNase6: structural determinants for bacterial cell agglutination and membrane permeation. Int J Mol Sci (2016) 17:552. doi:10.3390/ijms17040552
91. Rudolph B, Podschun R, Sahly H, Schubert S, Schröder JM, Harder J. Identification of RNase 8 as a novel human antimicrobial protein. Antimicrob Agents Chemother (2006) 50:3194–6. doi:10.1128/AAC.00246-06
92. Ackerman SJ, Gleich GJ, Loegering DA, Richardson BA, Butterworth AE. Comparative toxicity of purified human eosinophil granule cationic proteins for schistosomula of Schistosoma mansoni. Am J Trop Med Hyg (1985) 34:735–45. doi:10.4269/ajtmh.1985.34.735
93. Ramos AL, Discipio RG, Ferreira AM. Eosinophil cationic protein damages protoscoleces in vitro and is present in the hydatid cyst. Parasite Immunol (2006) 28:347–55. doi:10.1111/j.1365-3024.2006.00842.x
94. Hamann KJ, Gleich GJ, Checkel JL, Loegering DA, McCall JW, Barker RL. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J Immunol (1990) 144:3166–73.
95. Singh A, Batra JK. Role of unique basic residues in cytotoxic, antibacterial and antiparasitic activities of human eosinophil cationic protein. Biol Chem (2011) 392:337–46. doi:10.1515/BC.2011.037
96. Attery A, Batra JK. Mouse eosinophil associated ribonucleases: mechanism of cytotoxic, antibacterial and antiparasitic activities. Int J Biol Macromol (2017) 94:445–50. doi:10.1016/j.ijbiomac.2016.10.041
97. Blom K, Elshafie AI, Jönsson UB, Rönnelid J, Håkansson LD, Venge P. The genetically determined production of the alarmin eosinophil-derived neurotoxin is reduced in visceral leishmaniasis. APMIS (2018) 126:85–91. doi:10.1111/apm.12780
98. Rajamanickam A, Munisankar S, Bhootra Y, Dolla CK, Nutman TB, Babu S. Elevated systemic levels of eosinophil, neutrophil, and mast cell granular proteins in strongyloides stercoralis infection that diminish following treatment. Front Immunol (2018) 9:207. doi:10.3389/fimmu.2018.00207
99. Waters LS, Taverne J, Tai PC, Spry CJ, Targett GA, Playfair JH. Killing of Plasmodium falciparum by eosinophil secretory products. Infect Immun (1987) 55:877–81.
100. Bedoya VI, Boasso A, Hardy AW, Rybak S, Shearer GM, Rugeles MT. Ribonucleases in HIV type 1 inhibition: effect of recombinant RNases on infection of primary T cells and immune activation-induced RNase gene and protein expression. AIDS Res Hum Retroviruses (2006) 22:897–907. doi:10.1089/aid.2006.22.897
101. Suhasini AN, Sirdeshmukh R. Onconase action on tRNALys3, the primer for HIV-1 reverse transcription. Biochem Biophys Res Commun (2007) 363:304–9. doi:10.1016/j.bbrc.2007.08.157
102. Rugeles MT, Trubey CM, Bedoya VI, Pinto LA, Oppenheim JJ, Rybak SM, et al. Ribonuclease is partly responsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. AIDS (2003) 17:481–6. doi:10.1097/00002030-200303070-00002
103. Li Y, Zhao Y, Liu J, Huang Y, Liu Z, Xue C. A promising alternative anti-HBV agent: the targeted ribonuclease. Int J Mol Med (2009) 23:521–7. doi:10.3892/ijmm
104. Domachowske JB, Dyer KD, Bonville CA, Rosenberg HF. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J Infect Dis (1998) 177:1458–64. doi:10.1086/515322
105. Domachowske JB, Dyer KD, Adams AG, Leto TL, Rosenberg HF. Eosinophil cationic protein/RNase 3 is another RNase A-family ribonuclease with direct antiviral activity. Nucleic Acids Res (1998) 26:3358–63. doi:10.1093/nar/26.14.3358
106. Sikriwal D, Seth D, Parveen S, Malik A, Broor S, Batra JK. An insertion in loop L7 of human eosinophil-derived neurotoxin is crucial for its antiviral activity. J Cell Biochem (2012) 113:3104–12. doi:10.1002/jcb.24187
107. Rosenberg H. Eosinophil-derived neurotoxin (EDN/RNase 2) and the mouse eosinophil-associated RNases (mEars): expanding roles in promoting host defense. Int J Mol Sci (2015) 16:15442–55. doi:10.3390/ijms160715442
108. Roller BL, Monibi F, Stoker AM, Bal BS, Cook JL. Identification of novel synovial fluid biomarkers associated with meniscal pathology. J Knee Surg (2016) 29:47–62. doi:10.1055/s-0034-1394165
109. Stark M, Danielsson O, Griffiths WJ, Jörnvall H, Johansson J. Peptide repertoire of human cerebrospinal fluid: novel proteolytic fragments of neuroendocrine proteins. J Chromatogr B Biomed Sci Appl (2001) 754:357–67. doi:10.1016/S0378-4347(00)00628-9
110. Sugiyama RH, Blank A, Dekker CA. Multiple ribonucleases of human urine. Biochemistry (1981) 20(8):2268–74. doi:10.1021/bi00511a031
111. Prunotto M, Farina A, Lane L, Pernin A, Schifferli J, Hochstrasser DF, et al. Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J Proteomics (2013) 82:193–229. doi:10.1016/j.jprot.2013.01.012
112. Martin L, Koczera P, Simons N, Zechendorf E, Hoeger J, Marx G, et al. The human host defense ribonucleases 1, 3 and 7 are elevated in patients with sepsis after major surgery-A pilot study. Int J Mol Sci (2016) 17:1–11. doi:10.3390/ijms17030294
113. Schieven GL, Blank A, Dekker CA. Ribonucleases of human cerebrospinal fluid: detection of altered glycosylation relative to their serum counterparts1. Biochemistry (1982) 21:5148–55. doi:10.1021/bi00264a007
114. Reimert CM, Minuva U, Kharazmi A, Bendtzen K. Eosinophil protein X/eosinophil derived neurotoxin (EPX/EDN) detection by enzyme-linked immunosorbent assay and purification from normal human urine. J Immunol Methods (1991) 141:97–104. doi:10.1016/0022-1759(91)90214-Z
115. Levy AM, Gleich GJ, Sandborn WJ, Tremaine WJ, Steiner BL, Phillips SF. Increased eosinophil granule proteins in gut lavage fluid from patients with inflammatory bowel disease. Mayo Clin Proc (1997) 72:117–23. doi:10.4065/72.2.117
116. Rosenberg HF, Tenen DG, Ackerman SJ. Molecular cloning of the human eosinophil-derived neurotoxin: a member of the ribonuclease gene family. Proc Natl Acad Sci U S A (1989) 86:4460–4. doi:10.1073/pnas.86.12.4460
117. Raimondo F, Morosi L, Corbetta S, Chinello C, Brambilla P, Della Mina P, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst (2013) 9:1220. doi:10.1039/c3mb25582d
118. Robinson DS, Assoufi B, Durham SR, Kay AB. Eosinophil cationic protein(ECP) and eosinophil protein X (EXP) concentrations in serum and bronchial lavage fluid in asthma. effect of prednisolone treatment. Clin Exp Allergy (1995) 25:1118–27. doi:10.1111/j.1365-2222.1995.tb03259.x
119. Fahy JV, Liu J, Wong H, Boushey HA. Cellular and biochemical analysis of induced sputum from asthmatic and from healthy subjects. Am Rev Respir Dis (1993) 147:1126–31. doi:10.1164/ajrccm/147.5.1132
120. Montan PG, van Hage-Hamsten M. Eosinophil cationic protein in tears in allergic conjunctivitis. Br J Ophthalmol (1996) 80:556–60. doi:10.1136/bjo.80.6.556
121. Park YJ, Oh EJ, Park JW, Kim M, Han K. Plasma eosinophil cationic protein, interleukin-5, and ECP/Eo count ratio in patients with various eosinophilic diseases. Ann Clin Lab Sci (2006) 36:262–6.
122. Tischendorf FW, Brattig NW, Lintzel M, Buttner DW, Burchard GD, Bork K, et al. Eosinophil granule proteins in serum and urine of patients with helminth infections and atopic dermatitis. Trop Med Int Health (2000) 5:898–905. doi:10.1046/j.1365-3156.2000.00649.x
123. Rasp G, Thomas PA, Bujía J. Eosinophil inflammation of the nasal mucosa in allergic and non-allergic rhinitis measured by eosinophil cationic protein levels in native nasal fluid and serum. Clin Exp Allergy (1994) 24:1151–6. doi:10.1111/j.1365-2222.1994.tb03321.x
124. Dalli J, Montero-Melendez T, Norling LV, Yin X, Hinds C, Haskard D, et al. Heterogeneity in neutrophil microparticles reveals distinct proteome and functional properties. Mol Cell Proteomics (2013) 12:2205–19. doi:10.1074/mcp.M113.028589
125. Tollin M, Bergman P, Svenberg T, Jörnvall H, Gudmundsson GH, Agerberth B. Antimicrobial peptides in the first line defence of human colon mucosa. Peptides (2003) 24:523–30. doi:10.1016/S0196-9781(03)00114-1
126. van Herwijnen MJC, Zonneveld MI, Goerdayal S, Nolte-’t Hoen ENM, Garssen J, Stahl B, et al. Comprehensive proteomic analysis of human milk-derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol Cell Proteomics (2016) 15:3412–23. doi:10.1074/mcp.M116.060426
127. Liote F, Champy R, Moenner M, Boval-Boizard B, Badet J. Elevated angiogenin levels in synovial fluid from patients with inflammatory arthritis and secretion of angiogenin by cultured synovial fibroblasts. Clin Exp Immunol (2003) 132:163–8. doi:10.1046/j.1365-2249.2003.02117.x
128. Simcock DE, Kanabar V, Clarke GW, O’Connor BJ, Lee TH, Hirst SJ. Proangiogenic activity in bronchoalveolar lavage fluid from patients with asthma. Am J Respir Crit Care Med (2007) 176:146–53. doi:10.1164/rccm.200701-042OC
129. Koutroubakis IE, Xidakis C, Karmiris K, Sfiridaki A, Kandidaki E, Kouroumalis EA. Serum angiogenin in inflammatory bowel disease. Dig Dis Sci (2004) 49:1758–62. doi:10.1007/s10620-004-9565-4
130. McLaughlin RL, Phukan J, McCormack W, Lynch DS, Greenway M, Cronin S, et al. Angiogenin levels and ANG genotypes: dysregulation in amyotrophic lateral sclerosis. PLoS One (2010) 5:e15402. doi:10.1371/journal.pone.0015402
131. Pavlov N, Hatzi E, Bassaglia Y, Frendo J-L, Brion DE, Badet J. Angiogenin distribution in human term placenta, and expression by cultured trophoblastic cells. Angiogenesis (2003) 6:317–30. doi:10.1023/B:AGEN.0000029412.95244.81
132. Shapiro R, Vallee BL. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc Natl Acad Sci U S A (1987) 84:2238–41. doi:10.1073/pnas.84.8.2238
133. Hu GF, Xu CJ, Riordan JF. Human angiogenin is rapidly translocated to the nucleus of human umbilical vein endothelial cells and binds to DNA. J Cell Biochem (2000) 76:452–62. doi:10.1002/(SICI)1097-4644(20000301)76:3<452::AID-JCB12>3.0.CO;2-Z
134. Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci U S A (1994) 91:1677–81. doi:10.1073/pnas.91.5.1677
135. Moroianu J, Riordan JF. Identification of the nucleolar targeting signal of human angiogenin. Biochem Biophys Res Commun (1994) 203:1765–72. doi:10.1006/bbrc.1994.2391
136. Reithmayer K, Meyer KC, Kleditzsch P, Tiede S, Uppalapati SK, Gläser R, et al. Human hair follicle epithelium has an antimicrobial defence system that includes the inducible antimicrobial peptide psoriasin (S100A7) and RNase 7. Br J Dermatol (2009) 161:78–89. doi:10.1111/j.1365-2133.2009.09154.x
137. Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics (2010) 9:1324–38. doi:10.1074/mcp.M000063-MCP201
138. Wang Z, Hill S, Luther JM, Hachey DL, Schey KL. Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT). Proteomics (2012) 12:329–38. doi:10.1002/pmic.201100477
139. Kiehne K, Fincke A, Brunke G, Lange T, Fölsch UR, Herzig K-H, et al. Antimicrobial peptides in chronic anal fistula epithelium. Scand J Gastroenterol (2007) 42:1063–9. doi:10.1080/00365520701320489
140. Torres-Juarez F, Touqui L, Leon-Contreras J, Rivas-Santiago C, Enciso-Moreno JA, Hernandez-Pando R, et al. RNase7 but not psoriasin nor sPLA2-IIA associates to Mycobacterium tuberculosis during airway epithelial cells infection. Pathog Dis (2018) 76:1–19. doi:10.1093/femspd/fty005
141. Chan CC, Moser JM, Dyer KD, Percopo CM, Rosenberg HF. Genetic diversity of human RNase 8. BMC Genomics (2012) 13:40. doi:10.1186/1471-2164-13-40
142. Eller CH, Lomax JE, Raines RT. Bovine brain ribonuclease is the functional homolog of human Ribonuclease 1. J Biol Chem (2014) 289:25996–6006. doi:10.1074/jbc.M114.566166
143. Futami I, Tsushima Y, Murato Y, Tad H, Sasaki I, Seno M, et al. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol (1997) 16:413–9. doi:10.1089/dna.1997.16.413
144. Landre JBP, Hewett PW, Olivot J-M, Friedl P, Ko Y, Sachinidis A, et al. Human endothelial cells selectively express large amounts of pancreatic-type ribonuclease (RNase 1). J Cell Biochem (2002) 86:540–52. doi:10.1002/jcb.10234
145. Fischer S, Nishio M, Dadkhahi S, Gansler J, Saffarzadeh M, Shibamiyama A, et al. Expression and localisation of vascular ribonucleases in endothelial cells. Thromb Haemost (2011) 105:345–55. doi:10.1160/TH10-06-0345
146. Acharya KR, Ackerman SJJ. Eosinophil granule proteins: form and function. J Biol Chem (2014) 289:17406–15. doi:10.1074/jbc.R113.546218
147. Harrison AM, Bonville CA, Rosenberg HF, Domachowske JB. Respiratory syncytical virus-induced chemokine expression in the lower airways. Am J Respir Crit Care Med (1999) 159:1918–24. doi:10.1164/ajrccm.159.6.9805083
148. Kim C-K, Seo JK, Ban SH, Fujisawa T, Kim DW, Callaway Z. Eosinophil-derived neurotoxin levels at 3 months post-respiratory syncytial virus bronchiolitis are a predictive biomarker of recurrent wheezing. Biomarkers (2013) 18:230–5. doi:10.3109/1354750X.2013.773078
149. Shamri R, Young KM, Weller PF. PI3K, ERK, p38 MAPK and integrins regulate CCR3-mediated secretion of mouse and human eosinophil-associated RNases. Allergy (2013) 68:880–9. doi:10.1111/all.12163
150. Botos I, Segal D, Davies D. The structural biology of toll-like receptors. Structure (2011) 19:447–59. doi:10.1016/j.str.2011.02.004
151. Percopo CM, Dyer KD, Ochkur SI, Luo JL, Fischer ER, Lee JJ, et al. Activated mouse eosinophils protect against lethal respiratory virus infection. Blood (2014) 123:743–52. doi:10.1182/blood-2013-05-502443
152. Olsson I, Venge P, Spitznagel JK, Lehrer RI. Arginine-rich cationic proteins of human eosinophil granules: comparison of the constituents of eosinophilic and neutrophilic leukocytes. Lab Invest (1977) 36:493–500.
153. Torrent M, Sánchez D, Buzón V, Nogués MV, Cladera J, Boix E. Comparison of the membrane interaction mechanism of two antimicrobial RNases: RNase 3/ECP and RNase 7. Biochim Biophys Acta (2009) 1788:1116–25. doi:10.1016/j.bbamem.2009.01.013
154. Torrent M, Pulido D, Nogués MV, Boix E. Exploring new biological functions of amyloids: bacteria cell agglutination mediated by host protein aggregation. PLoS Pathog (2012) 8:e1003005. doi:10.1371/journal.ppat.1003005
155. Pulido D, Moussaoui M, Andreu D, Nogués MV, Torrent M, Boix E. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob Agents Chemother (2012) 56:2378–85. doi:10.1128/AAC.06107-11
156. Trautmann A, Schmid-Grendelmeier P, Krüger K, Crameri R, Akdis M, Akkaya A, et al. T cells and eosinophils cooperate in the induction of bronchial epithelial cell apoptosis in asthma. J Allergy Clin Immunol (2002) 109:329–37. doi:10.1067/MAI.2002.121460
157. Hogan SP, Waddell A, Fulkerson PC. Eosinophils in infection and intestinal immunity. Curr Opin Gastroenterol (2013) 29:7–14. doi:10.1097/MOG.0b013e32835ab29a
158. Plager DA, Davis MDP, Andrews AG, Coenen MJ, George TJ, Gleich GJ, et al. Eosinophil ribonucleases and their cutaneous lesion-forming activity. J Immunol (2009) 183:4013–20. doi:10.4049/jimmunol.0900055
159. Amin K, Lúdvíksdóttir D, Janson C, Nettelbladt D, Björnsson E, Roomans GM, et al. Inflammation and structural changes in the airways of patients with atopic and nonatopic asthma. BHR Group. Am J Respir Crit Care Med (2000) 162:2295–301. doi:10.1164/ajrccm.162.6.9912001
160. Hernnäs J, Särnstrand B, Lindroth P, Peterson CG, Venge P, Malmström A. Eosinophil cationic protein alters proteoglycan metabolism in human lung fibroblast cultures. Eur J Cell Biol (1992) 59:352–63.
161. Rubin J, Zagai U, Blom K, Trulson A, Engström A, Venge P. The coding ECP 434(G>C) gene polymorphism determines the cytotoxicity of ECP but has minor effects on fibroblast-mediated gel contraction and no effect on RNase activity. J Immunol (2009) 183:445–51. doi:10.4049/jimmunol.0803912
162. Eriksson J, Woschnagg C, Fernvik E, Venge P. A SELDI-TOF MS study of the genetic and post-translational molecular heterogeneity of eosinophil cationic protein. J Leukoc Biol (2007) 82:1491–500. doi:10.1189/jlb.0507272
163. Trulson A, Byström J, Engström A, Larsson R, Venge P. The functional heterogeneity of eosinophil cationic protein is determined by a gene polymorphism and post-translational modifications. Clin Exp Allergy (2007) 37:208–18. doi:10.1111/j.1365-2222.2007.02644.x
164. Salazar VA, Rubin J, Moussaoui M, Pulido D, Nogués MV, Venge P, et al. Protein post-translational modification in host defense: the antimicrobial mechanism of action of human eosinophil cationic protein native forms. FEBS J (2014) 281:5432–46. doi:10.1111/febs.13082
165. Diop G, Derbois C, Loucoubar C, Mbengue B, Ndao BN, Thiam F, et al. Genetic variants of RNASE3 (ECP) and susceptibility to severe malaria in Senegalese population. Malar J (2018) 17:1–10. doi:10.1186/s12936-018-2205-9
166. Adu B, Dodoo D, Adukpo S, Gyan BA, Hedley PL, Goka B, et al. Polymorphisms in the RNASE3 gene are associated with susceptibility to cerebral malaria in Ghanaian children. PLoS One (2011) 6:e29465. doi:10.1371/journal.pone.0029465
167. Neves JS, Harvard MS, Weller P. Functional extracellular eosinophil granules: novel implications in eosinophil immunobiology. Curr Opin Immunol (2009) 21:694–9. doi:10.1016/j.coi.2009.07.011
168. Shamri R, Melo RCN, Young KM, Bivas-Benita M, Xenakis JJ, Spencer LA, et al. CCL11 elicits secretion of RNases from mouse eosinophils and their cell-free granules. FASEB J (2012) 26:2084–93. doi:10.1096/fj.11-200246
169. Hofsteenge J, Moldow C, Vicentini AM, Zelenko O, Jarai-Kote Z, Neumann U. A single amino acid substitution changes ribonuclease 4 from a uridine-specific to a cytidine-specific enzyme. Biochemistry (1998) 37:9250–7. doi:10.1021/bi9803832
170. Hofsteenge J, Vicentini A, Zelenko O. Ribonuclease 4, an evolutionarily highly conserved member of the superfamily. Cell Mol Life Sci (1998) 54:804–10. doi:10.1007/s000180050209
171. Rosenberg HF, Dyer KD. Human ribonuclease 4 (RNase 4): coding sequence, chromosomal localization and identification of two distinct transcripts in human somatic tissues. Nucleic Acids Res (1995) 23:4290–5. doi:10.1093/nar/23.21.4290
172. Egesten A, Dyer KD, Batten D, Domachowske JB, Rosenberg HF. Ribonucleases and host defense: identification, localization and gene expression in adherent monocytes in vitro. Biochim Biophys Acta (1997) 1358:255–60. doi:10.1016/S0167-4889(97)00081-5
173. Zhou H-M, Strydom DJ. The amino acid sequence of human ribonuclease 4, a highly conserved ribonuclease that cleaves specifically on the 3’ side of uridine. Eur J Biochem (1993) 217:401–10. doi:10.1111/j.1432-1033.1993.tb18259.x
174. Liang S, Acharya KR. Structural basis of substrate specificity in porcine RNase 4. FEBS J (2016) 283:912–28. doi:10.1111/febs.13646
175. Terzyan SS, Peracaula R, de Llorens R, Tsushima Y, Yamada H, Seno M, et al. The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity. J Mol Biol (1999) 285:205–14. doi:10.1006/jmbi.1998.2288
176. Cocchi F, DeVico AL, Lu W, Popovic M, Latinovic O, Sajadi MM, et al. Soluble factors from T cells inhibiting X4 strains of HIV are a mixture of chemokines and RNases. Proc Natl Acad Sci U S A (2012) 109:5411–6. doi:10.1073/pnas.1202240109
177. Shapiro R. Structural features that determine the enzymatic potency and specificity of human angiogenin: threonine-80 and residues 58–70 and 116–123. Biochemistry (1998) 37:6847–56. doi:10.1021/bi9800146
178. Saxena SK, Rybak SM, Davey RT, Youle RJ, Ackerman EJ. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase a superfamily. J Biol Chem (1992) 267:21982–6.
179. Adams SA, Subramanian V. The angiogenins: an emerging family of ribonuclease related proteins with diverse cellular functions. Angiogenesis (1999) 3:189–99. doi:10.1023/A:1009015512200
180. Kulka M, Fukuishi N, Metcalfe DD. Human mast cells synthesize and release angiogenin, a member of the ribonuclease A (RNase A) superfamily. J Leukoc Biol (2009) 86:1217–26. doi:10.1189/jlb.0908517
181. Olson KA, Verselis SJ, Fett JW. Angiogenin is regulatedin vivoas an acute phase protein. Biochem Biophys Res Commun (1998) 242:480–3. doi:10.1006/bbrc.1997.7990
182. Gupta SK, Haigh BJ, Griffin FJ, Wheeler TT. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun (2013) 19:86–97. doi:10.1177/1753425912446955
183. Tschesche H, Kopp C, Hörl WH, Hempelmann U. Inhibition of degranulation of polymorphonuclear leukocytes by angiogenin and its tryptic fragment. J Biol Chem (1994) 269:30274–80.
184. Schmaldienst S, Oberpichler A, Tschesche H, Hörl WH. Angiogenin: a novel inhibitor of neutrophil lactoferrin release during extracorporeal circulation. Kidney Blood Press Res (2003) 26:107–12. doi:10.1159/000070992
185. Shapiro R, Ruiz-Gutierrez M, Chen CZ. Analysis of the interactions of human ribonuclease inhibitor with angiogenin and ribonuclease A by mutagenesis: importance of inhibitor residues inside versus outside the C-terminal “hot spot”. J Mol Biol (2000) 302:497–519. doi:10.1006/jmbi.2000.4075
186. Xu Z, Tsuji T, Riordan JF, Hu G. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry (2003) 42:121–8. doi:10.1021/bi020465x
187. Liu S, Yu D, Xu ZP, Riordan JF, Hu GF. Angiogenin activates Erk1/2 in human umbilical vein endothelial cells. Biochem Biophys Res Commun (2001) 287:305–10. doi:10.1006/bbrc.2001.5568
188. Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A (2015) 112:E3816–25. doi:10.1073/pnas.1510077112
189. Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther (2013) 21:368–79. doi:10.1038/mt.2012.237
190. Anderson P, Ivanov P. TRNA fragments in human health and disease. FEBS Lett (2014) 588:4297–304. doi:10.1016/j.febslet.2014.09.001
191. Rosenberg HF, Dyer KD. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res (1996) 24:3507–13. doi:10.1093/nar/24.18.3507
192. Christensen-Quick A, Lafferty M, Sun L, Marchionni L, DeVico A, Garzino-Demo A. Human Th 17 cells lack HIV-inhibitory RNases and are highly permissive to productive HIV infection. J Virol (2016) 90:7833–47. doi:10.1128/JVI.02869-15
193. Salazar VA, Arranz-Trullen J, Navarro S, Blanco JA, Sanchez D, Moussaoui M, et al. Exploring the mechanisms of action of human secretory RNase 3 and RNase 7 against Candida albicans. Microbiologyopen (2016) 5(5):830–45. doi:10.1002/mbo3.373
194. Wanke I, Steffen H, Christ C, Krismer B, Götz F, Peschel A, et al. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Invest Dermatol (2011) 131328:382–90. doi:10.1038/jid.2010.328
195. Williams DL, Li C, Ha T, Ozment-Skelton T, Kalbfleisch JH, Preiszner J, et al. Modulation of the phosphoinositide 3-kinase pathway alters innate resistance to polymicrobial sepsis. J Immunol (2004) 172:449–56. doi:10.4049/jimmunol.172.1.449
196. Schabbauer G, Tencati M, Pedersen B, Pawlinski R, Mackman N. PI3K-Akt pathway suppresses coagulation and inflammation in endotoxemic mice. Arterioscler Thromb Vasc Biol (2004) 24:1963–9. doi:10.1161/01.ATV.0000143096.15099.ce
197. Eichler TE, Becknell B, Easterling RS, Ingraham SE, Cohen DM, Schwaderer AL, et al. Insulin and the phosphatidylinositol 3-kinase signaling pathway regulate ribonuclease 7 expression in the human urinary tract. Kidney Int (2016) 90:568–79. doi:10.1016/j.kint.2016.04.025
198. Kenzel S, Mergen M, von Süßkind-Schwendi J, Wennekamp J, Deshmukh SD, Haeffner M, et al. Insulin modulates the inflammatory granulocyte response to streptococci via phosphatidylinositol 3-kinase. J Immunol (2012) 189:4582–91. doi:10.4049/jimmunol.1200205
199. Zhang J, Dyer KD, Rosenberg HF. RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Res (2002) 30:1169–75. doi:10.1093/nar/30.5.1169
200. Schuster C, Gläser R, Fiala C, Eppel W, Harder J, Schröder JM, et al. Prenatal human skin expresses the antimicrobial peptide RNase 7. Arch Dermatol Res (2013) 305:545–9. doi:10.1007/s00403-013-1340-y
201. Zhang J. Disulfide-bond reshuffling in the evolution of an ape placental ribonuclease. Mol Biol Evol (2007) 24:505–12. doi:10.1093/molbev/msl177
202. Zhang J, Zhang YP, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet (2002) 30:411–5. doi:10.1038/ng852
203. Penttinen J, Pujianto DA, Sipilä P, Huhtaniemi I, Poutanen M. Discovery in Silico and characterization in Vitro of novel genes exclusively expressed in the mouse epididymis. Mol Endocrinol (2003) 17:2138–51. doi:10.1210/me.2003-0008
204. Castella S, Fouchécourt S, Teixeira-Gomes AP, Vinh J, Belghazi M, Dacheux F, et al. Identification of a member of a new RNase a family specifically secreted by epididymal caput epithelium. Biol Reprod (2004) 70:319–28. doi:10.1095/biolreprod.103.022459
205. Cho S, Zhang J. Ancient expansion of the ribonuclease A superfamily revealed by genomic analysis of placental and marsupial mammals. Gene (2006) 373:116–25. doi:10.1016/j.gene.2006.01.018
206. Cheng GZ, Li JY, Li F, Wang HY, Shi GX. Human ribonuclease 9, a member of ribonuclease A superfamily, specifically expressed in epididymis, is a novel sperm-binding protein. Asian J Androl (2009) 11:240–51. doi:10.1038/aja.2008.30
207. Singhania NA, Dyer KD, Zhang J, Deming MS, Bonville CA, Domachowske JB, et al. Rapid evolution of the ribonuclease A superfamily: adaptive expansion of independent gene clusters in rats and mice. J Mol Evol (1999) 49:721–8. doi:10.1007/PL00006594
208. Beintema JJ, Scheffer AJ, van Dijk H, Welling GW, Zwiers H. Pancreatic ribonuclease distribution and comparisons in mammals. Nat New Biol (1973) 241:76–8. doi:10.1038/newbio241076a0
209. Rosenberg HF, Dyer KD, Tiffany HL, Gonzalez M. Rapid evolution of a unique family of primate ribonuclease genes. Nat Genet (1995) 10:219–23. doi:10.1038/ng0695-219
210. Beintema JJ, Kleineidam RG. The ribonuclease A superfamily: general discussion. Cell Mol Life Sci (1998) 54:825–32. doi:10.1007/s000180050211
211. D’Alessio G, Di Donato A, Parente A, Piccoli R. Seminal RNase: a unique member of the ribonuclease superfamily. Trends Biochem Sci (1991) 16:104–6. doi:10.1016/0968-0004(91)90042-T
212. D’alessio G. Evolution of oligomeric proteins. The unusual case of a dimeric ribonuclease. Eur J Biochem (1999) 266:699–708. doi:10.1046/j.1432-1327.1999.00912.x
213. Kim JS, Soucek J, Matousek J, Raines RT. Catalytic activity of bovine seminal ribonuclease is essential for its immunosuppressive and other biological activities. Biochem J (1995) 308:547–50. doi:10.1042/bj3080547
214. Piccoli R, Di Donato A, D’Alessio G. Co-operativity in seminal ribonuclease function. Kinetic studies. Biochem J (1988) 253:329–36.
215. Tamburrini M, Piccoli R, Picone D, Di Donato A, D’Alessio G. Dissociation and reconstitution of bovine seminal RNAase: construction of a hyperactive hybrid dimer. J Protein Chem (1989) 8:719–31. doi:10.1007/BF01024897
216. Opitz JG, Ciglic MI, Haugg M, Trautwein-Fritz K, Raillard SA, Jermann TM, et al. Origin of the catalytic activity of bovine seminal ribonuclease against double-stranded RNA†. Biochemistry (1998) 37:4023–33. doi:10.1021/bi9722047
217. Schein CH, Haugg M, Benner SA. Interferon-gamma activates the cleavage of double-stranded RNA by bovine seminal ribonuclease. FEBS Lett (1990) 270:229–32. doi:10.1016/0014-5793(90)81275-S
218. Schein CH, Hauggt M. Deletions at the C-terminus of interferon y reduce RNA binding and activation of double-stranded-RNA cleavage by bovine seminal ribonuclease. Biochem J (1995) 307:123–7. doi:10.1042/bj3070123
219. Youle RJ, Wu YN, Mikulski SM, Shogen K, Hamilton RS, Newton D, et al. RNase inhibition of human immunodeficiency virus infection of H9 cells. Proc Natl Acad Sci U S A (1994) 91:6012–6. doi:10.1073/pnas.91.13.6012
220. Libonati M, Gotte G. Oligomerization of bovine ribonuclease A: structural and functional features of its multimers. Biochem J (2004) 380:311–27. doi:10.1042/bj20031922
221. Gotte G, Mahmoud Helmy A, Ercole C, Spadaccini R, Laurents DV, Donadelli M, et al. Double domain swapping in bovine seminal RNase: formation of distinct N- and C-swapped tetramers and multimers with increasing biological activities. PLoS One (2012) 7:e46804. doi:10.1371/journal.pone.0046804
222. Tamburrini M, Scala G, Verde C, Ruocco MR, Parente A, Venuta S, et al. Immunosuppressive activity of bovine seminal RNase on T-cell proliferation. Eur J Biochem (1990) 190:145–8. doi:10.1111/j.1432-1033.1990.tb15557.x
223. Souček J, Marinov I, Beneš J, Hilgert I, Matoušek J, Raines RT. Immunosuppressive activity of bovine seminal ribonuclease and its mode of action. Immunobiology (1996) 195:271–85. doi:10.1016/S0171-2985(96)80045-3
224. Maes P, Damart D, Rommens C, Montreuil J, Spik G, Tartar A. The complete amino acid sequence of bovine milk angiogenin. FEBS Lett (1988) 241:41–5. doi:10.1016/0014-5793(88)81027-5
225. Harris P, Johannessen KM, Smolenski G, Callaghan M, Broadhurst MK, Kim K, et al. Characterisation of the anti-microbial activity of bovine milk ribonuclease4 and ribonuclease5 (angiogenin). Int Dairy J (2010) 20:400–7. doi:10.1016/j.idairyj.2009.12.018
226. Gupta SK, Haigh BJ, Seyfert HM, Griffin FJ, Wheeler TT. Bovine milk RNases modulate pro-inflammatory responses induced by nucleic acids in cultured immune and epithelial cells. Dev Comp Immunol (2017) 68:87–97. doi:10.1016/j.dci.2016.11.015
227. Nittoh T, Hirakata M, Mue S, Ohuchi K. Identification of cDNA encoding rat eosinophil cationic protein/eosinophil-associated ribonuclease. Biochim Biophys Acta (1997) 1351:42–6. doi:10.1016/S0167-4781(97)00024-9
228. Dubois J-YF, Jekel PA, Mulder PPMFA, Bussink AP, Catzeflis FM, Carsana A, et al. Pancreatic-type ribonuclease 1 gene duplications in rat species. J Mol Evol (2002) 55:522–33. doi:10.1007/s00239-002-2347-8
229. Zhao W, Kote-Jarai Z, van Santen Y, Hofsteenge J, Beintema JJ. Ribonucleases from rat and bovine liver: purification, specificity and structural characterization. Biochim Biophys Acta (1998) 1384:55–65. doi:10.1016/S0167-4838(97)00213-6
230. Larson KA, Olson EV, Madden BJ, Gleich GJ, Lee NA, Lee JJ. Two highly homologous ribonuclease genes expressed in mouse eosinophils identify a larger subgroup of the mammalian ribonuclease superfamily. Proc Natl Acad Sci U S A (1996) 93:12370–5. doi:10.1073/pnas.93.22.12370
231. Shamri R, Xenakis JJ, Spencer LA. Eosinophils in innate immunity: an evolving story Revital. Cell Tissue Res (2011) 343:57–83. doi:10.1007/s00441-010-1049-6
232. Zhang J, Dyer KD, Rosenberg HF. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc Natl Acad Sci U S A (2000) 97:4701–6. doi:10.1073/pnas.080071397
233. Kumar RK, Foster PS. Modeling allergic asthma in mice: pitfalls and opportunities. Am J Respir Cell Mol Biol (2002) 27:267–72. doi:10.1165/rcmb.F248
234. Malm-Erjefält M, Persson CG, Erjefält JS. Degranulation status of airway tissue eosinophils in mouse models of allergic airway inflammation. Am J Respir Cell Mol Biol (2001) 24:352–9. doi:10.1165/ajrcmb.24.3.4357
235. Clark K, Simson L, Newcombe N, Koskinen AML, Mattes J, Lee NA, et al. Eosinophil degranulation in the allergic lung of mice primarily occurs in the airway lumen. J Leukoc Biol (2004) 75:1001–9. doi:10.1189/jlb.0803391
236. Nitto T, Lin C, Dyer KD, Wagner RA, Rosenberg HF. Characterization of a ribonuclease gene and encoded protein from the reptile, Iguana iguana. Gene (2005) 352:36–44. doi:10.1016/j.gene.2005.03.002
237. Nitta K, Ozaki K, Ishikawa M, Furusawa S, Hosono M, Kawauchi H, et al. Inhibition of cell proliferation by Rana catesbeiana and Rana japonica lectins belonging to the ribonuclease superfamily. Cancer Res (1994) 54:920–7.
238. Hsu CH, Chang CF, Liao YD, Wu SH, Chen C. Solution structure and base specificity of cytotoxic RC-RNase 2 from Rana catesbeiana. Arch Biochem Biophys (2015) 584:70–8. doi:10.1016/j.abb.2015.08.010
239. Liao YD, Huang HC, Leu YJ, Wei CW, Tang PC, Wang SC. Purification and cloning of cytotoxic ribonucleases from Rana catesbeiana (bullfrog). Nucleic Acids Res (2000) 28:4097–104. doi:10.1093/nar/28.21.4097
240. Benito A, Vilanova M, Ribó M. Intracellular routing of cytotoxic pancreatic-type ribonucleases. Curr Pharm Biotechnol (2008) 9:169–79. doi:10.2174/138920108784567281
241. Tao F, Fan M, Zhao W, Lin Q, Ma R. A novel cationic ribonuclease with antimicrobial activity from Rana dybowskii. Biochem Genet (2011) 49:369–84. doi:10.1007/s10528-010-9414-4
242. Ardelt W, Mikulski SM, Shogen K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. Homology to pancreatic ribonucleases. J Biol Chem (1991) 266:245–51.
243. Ardelt W, Ardelt B, Darzynkiewicz Z. Ribonucleases as potential modalities in anticancer therapy. Eur J Pharmacol (2009) 625:181–9. doi:10.1016/j.ejphar.2009.06.067
244. Fagagnini A, Pica A, Fasoli S, Montioli R, Donadelli M, Cordani M, et al. Onconase dimerization through 3D domain swapping: structural investigations and increase in the apoptotic effect in cancer cells. Biochem J (2017) 474:3767–81. doi:10.1042/BCJ20170541
245. Saxena SK, Gravell M, Wu YN, Mikulski SM, Shogen K, Ardelt W, et al. Inhibition of HIV-1 production and selective degradation of viral RNA by an amphibian ribonuclease. J Biol Chem (1996) 271:20783–8. doi:10.1074/jbc.271.34.20783
246. Suhasini AN, Sirdeshmukh R. Transfer RNA cleavages by onconase reveal unusual cleavage sites. J Biol Chem (2006) 281:12201–9. doi:10.1074/jbc.M504488200
247. Pizzo E, Merlino A, Turano M, Russo Krauss I, Coscia F, Zanfardino A, et al. A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish. Biochem J (2011) 433:345–55. doi:10.1042/BJ20100892
248. Pizzo E, Varcamonti M, Di Maro A, Zanfardino A, Giancola C, D’Alessio G. Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon. FEBS J (2008) 275:1283–95. doi:10.1111/j.1742-4658.2008.06289.x
249. Monti DM, Yu W, Pizzo E, Shima K, Hu MG, Di Malta C, et al. Characterization of the angiogenic activity of zebrafish ribonucleases. FEBS J (2009) 276:4077–90. doi:10.1111/j.1742-4658.2009.07115.x
250. Geng R, Liu H, Wang W. Differential expression of six Rnase2 and three Rnase3 paralogs identified in blunt snout bream in response to aeromonas hydrophila infection. Genes (Basel) (2018) 9. doi:10.3390/genes9020095
251. Ferguson BJ, Newland SA, Gibbs SE, Tourlomousis P, Fernandes dos Santos P, Patel MN, et al. The Schistosoma mansoni T2 ribonuclease omega-1 modulates inflammasome-dependent IL-1β secretion in macrophages. Int J Parasitol (2015) 45:809–13. doi:10.1016/j.ijpara.2015.08.005
252. Smirnoff P, Roiz L, Angelkovitch B, Schwartz B, Shoseyov O. A recombinant human RNASET2 glycoprotein with antitumorigenic and antiangiogenic characteristics. Cancer (2006) 107:2760–9. doi:10.1002/cncr.22327
253. McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature (1990) 347:757–60. doi:10.1038/347757a0
254. Ramanauskas K, Igić B. The evolutionary history of plant T2/S-type ribonucleases. PeerJ (2017) 5:e3790. doi:10.7717/peerj.3790
255. Kubo K-I, Entani T, Takara A, Wang N, Fields AM, Hua Z, et al. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science (2010) 330:796–9. doi:10.1126/science.1195243
256. Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA (2008) 14:2095–103. doi:10.1261/rna.1232808
257. Schifano JM, Cruz JW, Vvedenskaya IO, Edifor R, Ouyang M, Husson RN, et al. TRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res (2016) 44:1256–70. doi:10.1093/nar/gkv1370
258. Chao Y, Li L, Girodat D, Förstner KU, Said N, Corcoran C, et al. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell (2017) 65:39–51. doi:10.1016/j.molcel.2016.11.002
259. Masuda H, Inouye M. Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins (Basel) (2017) 9:1–23. doi:10.3390/toxins9040140
260. Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J (1990) 9:1383–9.
261. Masaki H, Ogawa T. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie (2002) 84:433–8. doi:10.1016/S0300-9084(02)01425-6
262. Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem (2005) 280:42744–9. doi:10.1074/jbc.M510356200
263. Cuthbert BJ, Burley KH, Goulding CW. Introducing the new bacterial branch of the RNase A superfamily. RNA Biol (2018) 15:9–12. doi:10.1080/15476286.2017.1387710
264. Zaccone P, Burton O, Miller N, Jones FM, Dunne DW, Cooke A. Schistosoma mansoni egg antigens induce Treg that participate in diabetes prevention in NOD mice. Eur J Immunol (2009) 39:1098–107. doi:10.1002/eji.200838871
265. Burton OT, Gibbs S, Miller N, Jones FM, Wen L, Dunne DW, et al. Importance of TLR2 in the direct response of T lymphocytes to Schistosoma mansoni antigens. Eur J Immunol (2010) 40:2221–9. doi:10.1002/eji.200939998
266. Durdevic Z, Schaefer M. TRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress? Bioessays (2013) 35:323–7. doi:10.1002/bies.201200158
267. Weller PF, Spencer LA. Functions of tissue-resident eosinophils. Nat Rev Immunol (2017) 17:746–60. doi:10.1038/nri.2017.95
268. Haigis MC, Raines RT. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J Cell Sci (2003) 116:313–24. doi:10.1242/jcs.00214
269. Leland PA, Raines RT. Cancer chemotherapy – ribonucleases to the rescue. Chem Biol (2001) 8:405–13. doi:10.1016/S1074-5521(01)00030-8
270. Lomax JE, Bianchetti CM, Chang A, Phillips GN, Fox BG, Raines RT. Functional evolution of ribonuclease inhibitor: insights from birds and reptiles. J Mol Biol (2014) 426:3041–56. doi:10.1016/j.jmb.2014.06.007
271. Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: structure and function. Prog Nucleic Acid Res Mol Biol (2005) 80:349–74. doi:10.1016/S0079-6603(05)80009-1
272. Thomas SP, Kim E, Kim J-S, Raines RT. Knockout of the ribonuclease inhibitor gene leaves human cells vulnerable to secretory ribonucleases. Biochemistry (2016) 55:6359–62. doi:10.1021/acs.biochem.6b01003
273. Hoang TT, Raines RT. Molecular basis for the autonomous promotion of cell proliferation by angiogenin. Nucleic Acids Res (2017) 45:818–31. doi:10.1093/nar/gkw1192
274. Zasloff M. Antimicrobial RNases of human skin. J Invest Dermatol (2009) 129:2091–3. doi:10.1038/jid.2009.216
275. Spencer JD, Schwaderer AL, Eichler T, Wang H, Kline J, Justice SS, et al. An endogenous ribonuclease inhibitor regulates the antimicrobial activity of ribonuclease 7 in the human urinary tract. Kidney Int (2014) 85:1179–91. doi:10.1038/ki.2013.395
276. Mazzeo C, Canas JA, Zafra MP, Rojas Marco A, Fernandez-Nieto M, Sanz V, et al. Exosome secretion by eosinophils: a possible role in asthma pathogenesis. J Allergy Clin Immunol (2015) 135:1603–13. doi:10.1016/j.jaci.2014.11.026
277. Xu R, Greening DW, Rai A, Ji H, Simpson RJ. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods (2015) 87:11–25. doi:10.1016/j.ymeth.2015.04.008
278. Mateescu B, Kowal EJK, van Balkom BWM, Bartel S, Bhattacharyya SN, Buzás EI, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J Extracell Vesicles (2017) 6:1286095. doi:10.1080/20013078.2017.1286095
279. Folkman J, Merler E, Abernathy C, Williams G. Isolation of a tumor factor responsible for angiogenesis. J Exp Med (1971) 133:275–88. doi:10.1084/jem.133.2.275
280. Xu H, Liu Y, Meng F, He B, Han N, Li G, et al. Multiple bursts of pancreatic ribonuclease gene duplication in insect-eating bats. Gene (2013) 526:112–7. doi:10.1016/j.gene.2013.04.035
281. Rucksaken R, Pairojkul C, Pinlaor P, Khuntikeo N, Roytrakul S, Selmi C, et al. Plasma autoantibodies against heat shock protein 70, enolase 1 and ribonuclease/angiogenin inhibitor 1 as potential biomarkers for cholangiocarcinoma. PLoS One (2014) 9:e103259. doi:10.1371/journal.pone.0103259
282. Pizzo E, Sarcinelli C, Sheng J, Fusco S, Formiggini F, Netti P, et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci (2013) 126:4308–19. doi:10.1242/jcs.134551
283. Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A (2018) 4:1–6. doi:10.1073/pnas.1800038115
284. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet (2011) 12:861–74. doi:10.1038/nrg3074
285. Tafech A, Bassett T, Sparanese D, Lee CH. Destroying RNA as a therapeutic approach. Curr Med Chem (2006) 13:863–81. doi:10.2174/092986706776361021
286. Krauss J, Arndt MAE, Dübel S, Rybak SM. Antibody-targeted RNase fusion proteins (immunoRNases) for cancer therapy. Curr Pharm Biotechnol (2008) 9:231–4. doi:10.2174/138920108784567317
287. Newton DL, Rybak SM. Antibody targeted therapeutics for lymphoma: new focus on the CD22 antigen and RNA Introduction: putting them together. Expert Opin Biol Ther (2001) 1:995–1003. doi:10.1517/14712598.1.6.995
288. Weber T, Mavratzas A, Kiesgen S, Haase S, Botticher B, Exner E, et al. A humanized anti-CD22-onconase antibody-drug conjugate mediates highly potent destruction of targeted tumor cells. J Immunol Res (2015) 2015:561814. doi:10.1155/2015/561814
289. Jordaan S, Akinrinmade O, Nachreiner T, Cremer C, Naran K, Chetty S, et al. Updates in the development of ImmunoRNases for the selective killing of tumor cells. Biomedicines (2018) 6:28. doi:10.3390/biomedicines6010028
Keywords: ribonucleases, innate immunity, RNA, extracellular, inflammation, infection
Citation: Lu L, Li J, Moussaoui M and Boix E (2018) Immune Modulation by Human Secreted RNases at the Extracellular Space. Front. Immunol. 9:1012. doi: 10.3389/fimmu.2018.01012
Received: 01 February 2018; Accepted: 23 April 2018;
Published: 16 May 2018
Edited by:
Tobias Schuerholz, Universitätsmedizin Rostock, GermanyReviewed by:
Lukas Martin, Uniklinik RWTH Aachen, GermanyCaroline Jefferies, Cedars-Sinai Medical Center, United States
Copyright: © 2018 Lu, Li, Moussaoui and Boix. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammed Moussaoui, bW9oYW1tZWQubW91c3Nhb3VpQHVhYi5lcw==;
Ester Boix, ZXN0ZXIuYm9peEB1YWIuZXM=
 Lu Lu
Lu Lu Jiarui Li
Jiarui Li Mohammed Moussaoui
Mohammed Moussaoui Ester Boix
Ester Boix