
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 07 May 2018
Sec. Alloimmunity and Transplantation
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.00964
 Cesar A. Speck-Hernandez1†
Cesar A. Speck-Hernandez1† Amanda F. Assis2†
Amanda F. Assis2† Rafaela F. Felicio1
Rafaela F. Felicio1 Larissa Cotrim-Sousa2
Larissa Cotrim-Sousa2 Nicole Pezzi1
Nicole Pezzi1 Gabriel S. Lopes3
Gabriel S. Lopes3 Karina F. Bombonato-Prado4
Karina F. Bombonato-Prado4 Silvana Giuliatti5
Silvana Giuliatti5 Geraldo A. Passos2,4*
Geraldo A. Passos2,4*
The function of medullary thymic epithelial cells (mTECs) is associated with thymocyte adhesion, which is crucial for the negative selection of autoreactive thymocytes in the thymus. This process represents the root of central tolerance of self-components and prevents the onset of autoimmune diseases. Since thymic epithelia correspond to an important target of donor T cells during the onset of chronic graft-vs-host-disease, mTEC-thymocyte adhesion may have implications for alloimmunity. The Aire and Fezf2 genes function as transcriptome controllers in mTECs. The central question of this study is whether there is a mutual relationship between mTEC-thymocyte adhesion and the control of the mTEC transcriptome and whether Aire is involved in this process. Here, we show that in vitro mTEC-thymocyte adhesion causes transcriptome changes in mTECs and upregulates the transcriptional expression of Aire and Fezf2, as well as cell adhesion-related genes such as Cd80 or Tcf7, among others. Crispr-Cas9-mediated Aire gene disruption demonstrated that this gene plays a role in the process of mTEC-thymocyte adhesion. Consistent with the nuclear localization signal (NLS) encoded by Aire exon 3, which was targeted, we demonstrate that Aire KO−/− mTECs impair AIRE protein localization in the nucleus. Consequently, the loss of function of Aire reduced the ability of these cells to adhere to thymocytes. Their transcriptomes differed from their wild-type Aire+/+ counterparts, even during thymocyte adhesion. A set of mRNA isoforms that encode proteins involved in cell adhesion were also modulated during this process. This demonstrates that both thymocyte interactions and Aire influence transcriptome profiling of mTEC cells.
Thymic crosstalk is an active process that involves both cell migration and cell–cell adhesion, during which thymocytes interact with thymic epithelial cells (TECs) and receive signals to proceed with their differentiation (1–3). Because the T cell receptor (TCR) is expressed on the surface of early thymocytes that are located in the thymic cortex and successfully express the TCRβ chain, these cells pass the β-selection checkpoint and then rearrange and express the TCRα chain. Subsequently, double-positive (CD4+CD8+) cells, which receive weak TCR signals, receive survival signals and undergo positive selection (PS), subsequently becoming single-positive (SP CD4+ or CD8+) cells. Cortical TECs (cTECs) are in control of the PS of thymocytes (4). The SP cells then migrate to the thymic medulla, and clones expressing self-reactive TCRα/β are eliminated by apoptosis through negative selection (NS), which is closely associated with medullary TECs (mTECs) (4–7). This process involves a specific thymic microenvironment that supports the different stages of T cell development (8). This sequence of events can be traced by using molecular markers, such as for the timing of gene recombination and expression of TCRα/β.
The interaction between TECs and thymocytes, in addition to causing the development and selection of T cells, provides distinct sets of signals that modulate transcriptional gene expression in the different regions of the thymic stroma through which the thymocytes migrate (9). In this context, it is quite appropriate to consider that mTECs represent a unique cell type, as they express an enormous variety of genes and mobilize most of their functional genome (10, 11). The significance of this wide-ranging gene expression is immunological, i.e., it results in self-representation in the thymus though promiscuous gene expression (PGE) (7, 12–15).
The autoimmune regulator (Aire) gene is one of the controllers of PGE in mTECs, modulating the so-called Aire-dependent genes. Most of these are genes that encode the self-peripheral tissue antigens (PTAs) and are regulated through a mechanism that releases stalled RNA Pol II in the chromatin (16). As RNA Pol II is nonspecific regarding its target mRNAs, this could explain the wide range of mRNAs that Aire controls. Moreover, it has been shown that the number of mRNA isoforms that is expressed in mTEC cells is higher than in other cell types and that Aire exerts control in the splicing diversity in these cells (17). The implication of these findings concerns the exposure of developing thymocytes to an increased diversity of PTA splice isoforms and diversity of HLA (MHC)-mediated PTA presentation, enforcing NS against self-antigens in the thymus (17–19). Recently, we showed that in addition to PTAs, Aire controls the expression of mRNAs that encode cell adhesion proteins in mTEC cells and, consequently, their thymocyte adhesion (20).
A second PGE controller has been identified in mTEC cells, Forebrain embryonic zinc-finger 2 (Fezf2), whose encoded protein acts directly on chromatin to modulate the expression of a set of Aire-independent genes (21, 22). The sum total of the Aire-dependent and the Aire-independent genes that are expressed by mTECs is approximately 15,000 genes, representing approximately 62% of the murine functional genome (19). This shows that mTECs are an unusual cell type due to the range of their transcriptome modulation while still maintaining their morpho-functional characteristics. The mTECs are usually identified by flow cytometry and characterized as CD45−EpCAM+ (epithelial cell adhesion molecule) Ly51−, in addition to expressing the cell surface markers MHC II and CD80 (4, 23).
The results obtained with reaggregation thymus organ culture, which is consistent with thymic embryogenesis, show that MHC−CD80−AIRE− immature mTECs precede MHC+CD80+AIRE+ mature mTEC development (23–25). This indicates that immature cells proceed to mature mTECs though an intermediate stage of Aire− cells that begin to express MHC II and CD80, a process that is dependent on RANK-mediated signals from CD4+3−RANKL+ lymphoid cells (4, 23, 26). AIRE+ mTECs are short-lived cells; they are post-mitotic and represent the last stage of mTEC differentiation (4, 23, 26). Aire ultimately induces apoptosis in mature mTECs, contributing to the diffusion of PTAs within the medullary thymic compartment (23, 26).
Accordingly, mTECs represent an unusual cell type because they express most of their functional genome in the late phase of their differentiation without losing their morpho-functional characteristics (7).
However, it currently remains unclear whether the physical contact with thymocytes influences the broad transcriptional gene expression modulation in mTECs and what the specific participation of Aire is in this process.
We used 4- to 5-week-old female C57BL/6 mice weighing 18–22 g for the surgical removal of the thymus and further thymocyte preparation, which were isolated according to a previously described protocol and whose procedure yielded a thymocyte population with a purity of ≥93% as determined by flow cytometry with a phycoerythrin-labeled anti-CD3 antibody (20, 27, 28). These cells were used for further cell adhesion assays. All experimental procedures followed ethical guidelines under strict guidance and approval from the University of São Paulo Ethics Committee for Animal Experimental Research (Approval # 006/2016-1).
We used the Aire wild-type (WT) mTEC line (EpCAM+, Ly51−, UEA-1+) mTEC 3.10 cells as previously described (20, 29, 30).
The WT mTEC 3.10 cell line as well as the mutant mTEC 3.10E6 was used in an mTEC-thymocyte adhesion protocol as previously established (31–35) with several modifications introduced by our group (20). Experiments were performed at least six times (six co-cultures with WT mTEC 3.10 cells and six co-cultures with mutant mTEC 3.10E6) with similar results. Then the adhesion index (AI) was calculated as follows: AI = number of adhered thymocytes/number of mTEC cells. Statistical analysis was performed by two-sided Mann–Whitney test with 95% interval.
An all-in-one Crispr-Cas9 vector encompassing gRNA-Cas9-green fluorescent protein (GFP) was designed and purchased from Sigma Aldrich (St. Louis, MO, USA). This vector encodes a specific gRNA, whose complementary sequence is CCCCTTGCTGGTCCCAAGGCCG that targets the Aire exon 3 on Mus musculus chromosomal location GRCm38:10:78030995–78031017, the Cas9 enzyme, and GFP. The gRNA targeted the CGG PAM motif located immediately upstream of the region within the Aire exon 3 that transcribes mRNA nt 346–348 and amino acid residues 113–124 of the AIRE protein. The vector could, in principle, interact with potential off-target gene sequences. Considering the calculated risk for these sequences, which derives from the algorithm proposed by Hsu et al. (36), the gRNA used in this work presents a low off-target probability (Hsu score = 0.7).
Approximately 5 × 105 WT mTEC 3.10 cells were suspended in 82 µl Amaxa Basic Nucleofector™ reagent for primary mammalian epithelial cells (Lonza, Basel, Switzerland) plus 18 µl Lonza supplement containing 5 µg Crispr-Cas9 vector. This cell suspension was placed in an electroporation cuvette and subjected to a single continuous current electric pulse of 200 V during 30 ms in a BTX Square Electroporator (Holliston, MA, USA). Electrotransfected cells were seeded in a well of a six-well Costar® culture plate (Corning) containing 3 ml RPMI medium supplemented with 10% inactivated fetal bovine serum (FBS) plus antibiotics and cultured for 24 h in a 37°C incubator with a 5% CO2 atmosphere and then trypsinized as mentioned above. Wild-type mTEC 3.10 cells electroporated only in the presence of the Nucleofector® reagent plus Lonza supplement were cultured as above and were considered control cells.
The GFP+ cells were separated though a FACS Aria III flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Individual cells were automatically and consecutively placed in separate wells of a polystyrene flat-bottomed 96-well plate (Corning) containing 100 µl RPMI medium supplemented with 10% FBS plus antibiotics and cultured. Data were analyzed via Beckman Coulter Kaluza software.1 Cell proliferation was observed though a conventional inverted microscope during a period of approximately 3 weeks, exchanging fresh culture medium every other day.
The surviving clones from the complete process, i.e., electroporation, sorting, and single cell culture were numbered, cultured until confluence and transferred to 24-well plate, then to 6-well plate, and finally to 25 cm2 culture bottles. Confluent cells were removed from culture bottles by trypsin treatment, processed by a conventional cryopreservation protocol that includes dimethyl sulfoxide, and stocked in liquid N2 for further analysis.
The genomic DNA (200 µg) from individual GFP+ mTEC clones was used in conventional PCR for the amplification of a 415 bp genomic DNA fragment encompassing the Aire exon 3 (Ensembl acc ENSMUSG00000000731), whose primer (forward F or reverse R) sequences were: F = 5′ CCAATGGGTAGCATCGG 3′ and R = 5′ CTCTTGAGTGTACCTGGGCTG 3′. The Primer3 web tool2 was used to select pairs of oligonucleotide primers with an optimal melting temperature of 60°C.
A 50 µl PCR mixture containing the input genomic DNA, PCR buffer, primers, dNTPs, and Taq enzyme (Gotaq G2 Flexi DNA Polymerase, Promega) was subjected to the following thermal cycling conditions: 30× (95°C 20 s, 54°C 30 s, 72°C 30 s), 1× (72°C 5 min). The PCR reactions were performed in triplicate for each GFP+ clone. Genomic DNA from untransfected mTEC 3.10 cells was used as a negative control.
The PCR products were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). For the identification of mutant clones, the respective PCR products were digested with T7 endonuclease (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions. The digestion fragments were resolved though microfluidic electrophoresis using Agilent DNA nanochips and an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). As the T7 endonuclease cleaves mismatched double-stranded PCR products, the DNA of mutant clones were identified by the presence of two bands of approx. 200 bp cut PCR product, in contrast with WT DNA, which featured just one band of undigested DNA.
The PCR products of Aire mutant clones were processed by Sanger sequencing for further characterization of mutations. Sanger electropherograms were analyzed using the Crisp-ID program3 (37) for exact indel size and location in the Crispr-Cas9 Aire exon 3-targeted region.
FASTA sequences of a mutant clone selected for further analysis in this work, here termed mTEC 3.10E6, were deposited at GenBank4 under accession numbers (acc MG493266 for the Aire mutant allele 1 and acc MG493265 for the Aire mutant allele 2).
We initially used the Uniprot databank5 to recover the primary WT sequence of the AIRE protein under acc Q9Z0E3. Then, the FASTA DNA sequences of the mutant 3.10E6 (allele 1 and allele 2) were translated into proteins by using the Expasy Translate Tool.6
To predict the effect of mutations on the amino acid sequence of the encoded AIRE protein, we used the Provean tool,7 which is a software often used to characterize the functional effects of amino acid variations (substitutions or deletions) on proteins. AIRE primary protein sequences (WT and mutant) were aligned and compared by using the Clustal Omega program.8
Total RNA of Aire WT (mTEC 3.10) or Aire mutant (mTEC 3.10E6) cells was prepared using the mirVana kit® (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. Evaluation of RNA integrity was performed by microfluidic electrophoresis using Agilent RNA 6000 nano chips and an Agilent 2100 bioanalyzer (Agilent Technologies Santa Clara, CA, USA) as previously described by our group (20). Only RNA samples that were free of proteins and phenol and had an RNA Integrity Number ≥9.0 were selected for cDNA synthesis using SuperScript reverse transcriptase enzyme according to the manufacturer’s instructions (Invitrogen Corporation, Carlsbad, CA, USA).
The confirmation of transcriptional expression of focused genes was assayed by RT-qPCR. The expression level of each target gene was normalized to the housekeeping gene Hprt, which is commonly used as a reference. The Primer3 web tool (see text footnote 2) was used to select pairs of oligonucleotide primers spanning an intron/exon junction with an optimal melting temperature of 60°C.
The respective sequences were retrieved from the NCBI GenBank database.9 The forward (F) and reverse (R) primer sequences (presented in the 5′–3′ orientation) were the same as those previously used in our group (20) as follows: Aire (acc NM_001271549.1) F = GAAGCTGTACCCACCTCTGG, R = ATTGAGGAGGGACTCCAGGT, Fezf2 (acc NM_NM_080433.3NM) F = GAACGAGGGGGAGTCAAGAG, R = TCTAGCTCCGGTGTGGACAG, CD80 (acc NM_009855.2) F = CCTGGGAAAAACCCCCAGAA, R = ACAACGATGACGACGACTGT, Nfkbia (acc NM_010907.2) F = AGGACGAGGAGTACGAGCAA, R = CGTGGATGATTGCCAAGTGC, and Tcf7 (acc NM_001313981.1) F = CTGTCCCCTTCCTGCGGATA, R = GTCCAGGTACACCAGATCCC.
Gene expression was quantified using a StepOne Real-Time PCR System (Applied Biosystems, USA). The analyses were performed using the cycle threshold (Ct) method, which allows for quantitative analysis of the expression of a factor using the formula 2−ΔΔCt, in which ΔCt = Ct target gene − Ct of the housekeeping gene Hprt, and ΔΔCt = ΔCt sample − ΔCt. Experiments were performed in six independent replicates and statistical analysis of the data was made through the Mann–Whitney two-sided test with 95% interval.
Western blotting analysis of AIRE protein levels was performed according to a conventional protocol, which was described in a previous work from our group (20), including electrotransfer of the SDS-PAGE of total proteins extracted from Aire WT (mTEC 3.10) or Aire mutant (mTEC 3.10E6) to a polyvinylidene fluoride membrane (BioRad, Hercules, CA, USA), incubation with anti-AIRE-1 primary antibody (C-2 mouse monoclonal IgG1 kappa light chain, Santa Cruz Biotechnology, Dallas, TX, USA) and developing for AIRE protein band visualization.
Immunolocalization of AIRE protein in Aire WT (mTEC 3.10) or Aire mutant (mTEC 3.10E6) cells was performed according to a conventional immunofluorescence protocol, which was described in previous work from our group (20) using a goat anti-mouse AIRE D17 IgG polyclonal primary antibody (Santa Cruz Biotechnology) and Novex™ mouse anti-goat IgG rhodamine red conjugate (Life Technologies Corp., Carlsbad, CA, USA) as a secondary antibody. To visualize the cytoplasmic region, actin filaments were labeled with AlexaFluor® 488-conjugated phaloidin (Life Technologies) according to the manufacturer’s instructions. The nuclei were labeled with DAPI. In this study, cells were observed using an Apo Tome immunofluorescence microscope (Zeiss, Oberkochen, Germany). We counted 15 microscopic fields totalizing approximately 150 each cell type.
Quantitative results of RT-qPCR and adhesion assay (from independent experiments performed six times) were analyzed by Mann–Whitney test with 95% confidence intervals. The IBM SPSS Statistics program10 was used for calculations and graphing of results.
We followed a as protocol previously described (38). Briefly, paired-end (2 × 100 bp) sequencing was performed using an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, USA) using a TruSeq Stranded Total RNA Library Prep Kit (Illumina).
The quality of raw FASTQ sequences was first analyzed through a FASTQC program.11 Then, FASTQ sequences were mapped to the Mus musculus reference genome (mm10) using the STAR 2.5 Spliced Aligner program,12 which output a BAM file containing the sequences and their genomic references and a GTF file with gene annotations used for further determinations of the number of reads per transcript through the HTSeq Count program.13
For each RNA sample analyzed, we recovered a list of genes and their respective number of transcripts that served as input for determinations of the differentially expressed (DE) mRNAs through the DESEq2 package14 within the R platform.15 DESEq2 calculates the fold change (FC) for each mRNA considering a contrast matrix for a given experimental condition. In this study, we defined adhesion as a contrast variable and DE as those mRNAs with a p value <0.05 and false discovery rate (FDR Benjamini–Hochberg correction) and FC ≥1.5.
The DE mRNAs were hierarchically clustered, and a heat-map was constructed to evaluate the expression profiling. The Euclidean distance and the complete linkage method were used for clustering the samples and mRNAs using the R platform.
We used the RSEM software16 to estimate isoform abundance from the mapped reads as generated from the STAR 2.5 Spliced Aligner program as described above. The STAR output Aligned.to.Transcriptome.out.bam files were used as RSEM input to generate isoform counts for each RNA sample analyzed. Then, we compared isoform expression using the EbSeq package17 within the R platform. DE isoforms were determined by comparing each two conditions considering as significant an adjusted FDR p value < 0.05 and an FC ≥ 1.5. The RNA-sequencing data of this work are available on Gene Expression Omnibus18 under the accession number GSE91015.
The list of the DE mRNAs was analyzed in terms of functional enrichment through the Database for Annotation, Visualization, and Integrated Discovery (DAVID) annotation tool.19 This tool was used for the identification of the main biological processes and pathways represented by DE mRNAs. A functional category was considered significant if it had at least three mRNAs and a score of p < 0.005 with Benjamini–Hochberg correction.
The electroporation and nucleofection process with a vector simultaneously expressing GFP, Casp9, and a gRNA targeting Aire exon 3 allowed a satisfactory number of GFP+ cells to be isolated, in light of the fact that the main objective of this procedure was to obtain at least one mTEC mutant clone. Only mTEC cells transfected with the Crispr-Cas9 vector expressed GFP, which allowed their isolation by flow cytometry (Figure S1 in Supplementary Material). Of the 87 single cells deposited in a 96-well plate, we recovered nine clones for further evaluation of the occurrence of mutations in Aire exon 3. We then amplified a 415 bp PCR product that encompassed Aire exon 3, which was then digested with the enzyme T7 endonuclease. Of these nine clones, we were able to identify two that had their PCR DNA product digested, indicating the occurrence of mutations; one clone was named “3.10E6,” which was selected for this study (Figure S2 in Supplementary Material).
For this study, we selected clone 3.10E6 for further characterization through Sanger sequencing of the Aire exon 3-targeted region. Figure S3 in Supplementary Material shows the respective sequencing electropherograms of mTEC CT1 cells (control untransfected WT mTEC 3.10 cells), 3.10A6 (a Crispr-Cas9 vector-transfected GFP+ clone but whose PCR product of the Aire exon 3 was resistant to the T7 endonuclease), and 3.10E6 (a Crispr-Cas9 vector-transfected GFP+ clone whose PCR product of the Aire exon 3 was digested by the T7 endonuclease).
CRISP-ID program analysis of the Sanger electropherograms showed that no mutations were found in the WT mTEC 3.10 CT1 or clone 3.10A6, whereas clone 3.10E6 was characterized as a carrier of indel mutations affecting both Aire alleles [knockout (KO) compound heterozygosis]. This clone was subsequently selected for further analysis. In Aire allele 1, there were two types of mutations: T>G substitution (mRNA position 351) followed by a nine-bp deletion (GCTGGTCCC, mRNA positions 352–360) that transcribed a 1,647 nt Aire mRNA; in allele 2, there was a single G deletion at mRNA position 352 that transcribed a 1,655 nt Aire mRNA (Figure 1A).
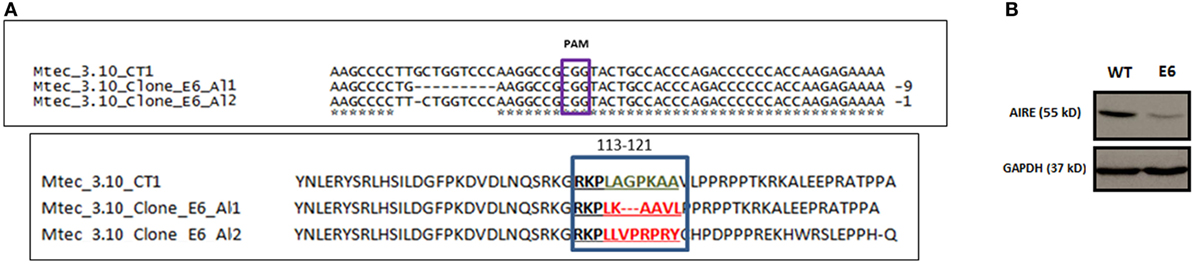
Figure 1. DNA sequence alignment of Mus musculus Aire exon 3 CRISPR-Cas9 target region nearing PAM CGG sequence for wild-type (WT) medullary thymic epithelial cell (mTEC) 3.10 and mTEC 3.10E6 for the characterization of indel size and location (allele 1 and allele 2) and AIRE protein translation from the respective DNA segments (A). Western blot (WB) of SDS-PAGE WT mTEC 3.10 and mTEC 3.10E6 mutant clone cell lysates for detection of Mus musculus AIRE protein expression. WB membrane was probed with an antibody against AIRE protein (upper panel), washed, and then probed with an antibody against GAPDH that was used as an internal load control. This procedure allowed to show that the mTEC 3.10E6 mutant clone express relatively less AIRE protein than its WT mTEC 3.10 counterpart (B).
In silico AIRE protein translation and sequence alignment through the Clustal Omega program (Figure S4 in Supplementary Material) showed that T>G nucleotide substitution in allele 1 resulted in a L118L silent mutation, whereas the nine bp deletion provoked a frameshift leading to deletion of three amino acid residues (A119_P121del) and a consequently shorter 548 amino acid (aa) residue AIRE protein (Figure 1A).
Analysis using the Provean program, which characterizes the functional effects of protein variations from amino acid substitutions or deletions based on calculations of sequence alignments, considers that protein variants featuring a Provean score ≤−2.5 are deleterious. The T>G substitution in allele 1 resulted in no alteration (L118L) in the amino acid sequence (Provean score: −1.46), whereas the deletion of the three amino acid residues (A119_P121del) was deleterious for the AIRE protein nuclear localization signal (NLS) (Provean score: −10.02).
Regarding allele 2, the G nucleotide deletion resulted in a TGA stop codon at mRNA position 352 and, consequently, a 158-aa truncated AIRE protein. For this reason, Provean analysis for allele 2 could not be performed (Table S1 in Supplementary Material).
Moreover, as the anti-AIRE primary antibody used in Western blot (WB) analysis was raised against amino acids 246–545 mapping the C-terminus of AIRE, it could not recognize the truncated AIRE protein from allele 2. The WB depicted in Figure 1B shows the expression of WT AIRE protein in mTEC 3.10 cells and the mutant AIRE in mTEC 3.10E6 clone. Figure S5 in Supplementary Material shows the full WB membrane image.
According to these analyses, clone mTEC 3.10E6 can be considered an Aire compound heterozygous KO that expresses a deleterious AIRE protein from the allele 1. Accordingly, this clone was selected for further study.
To evaluate the extension of the effects of Crispr-Cas9-mediated mutations, we evaluated the relative levels of Aire mRNA transcripts by RT-qPCR and AIRE protein expression by WB analysis. Our results showed that clone 3.10E6 expresses significantly less AIRE protein than its mTEC 3.10 cell line WT counterpart (Figure 1B). Immunolocalization was used to assess whether such a reduction would affect the protein nuclear localization. Compared to the WT mTEC 3.10 cell line, which featured specked red granules of AIRE protein associated with chromatin (50 AIRE+ cells/150 analyzed cells) in immunofluorescence images, the 3.10E6 cells did not exhibit any AIRE protein granules in the nucleus (zero AIRE+ cells/150 analyzed cells) (Figures 2 and 3A).
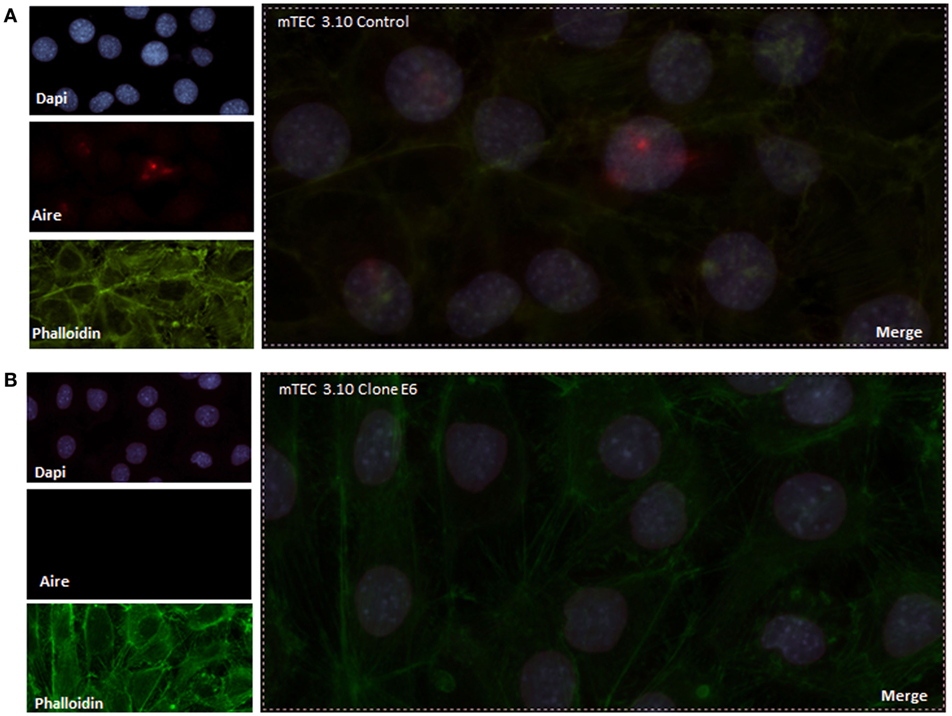
Figure 2. Immunofluorescence of AIRE protein showed that wild-type medullary thymic epithelial cell (mTEC) 3.10 cells exhibited AIRE protein nuclear localization (A) while mTEC 3.10E6 mutant clone did not showed nuclear staining for this protein (B). Cells were labeled with an antibody against Mus musculus AIRE protein (red dots), with phalloidin to show the cytoplasmic region and with DAPI to show nuclei (blue).
Medullary thymic epithelial cell (mTEC)-thymocyte cell adhesion was used to assess the functional consequences of Crispr-Cas9-mediated Aire mutation. Figure 3 shows that clone 3.10E6 had a significantly reduced ability to adhere to thymocytes.
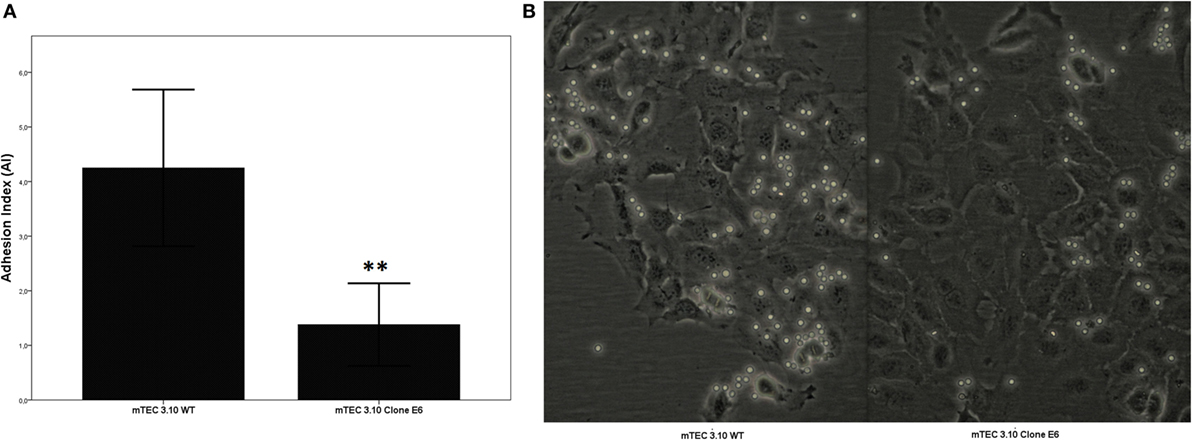
Figure 3. The medullary thymic epithelial cell (mTEC)-thymocyte adhesion assay showed decrease of thymocyte interaction with mTEC 3.10E6 mutant clone in comparison with its wild-type (WT) mTEC 3.10 counterpart. The adhesion degree is plotted as adhesion index (AI), in which values correspond to mean ± SD from three independent experiments and were significantly different between control (WT) vs mutant cells. Mann–Whitney two-sided test with 95% interval (A). Photomicrography of fresh mTEC-thymocyte co-cultures comparing WT mTEC 3.10 vs mutant mTEC 3.10E6 (B).
Figure 4 shows that the WT mTEC 3.10 cell line expresses reduced levels of Aire mRNA transcripts, but comparatively and significantly more than clone 3.10E6. When mTEC 3.10 cells adhere to thymocytes, the transcriptional expression of Aire increases significantly. Similar modulation of Aire expression was observed when clone 3.10E6 cells adhered to thymocytes. However, when comparing the levels of Aire expression between mTEC 3.10 cells and clone 3.10E6 after thymocyte adhesion, we observed that the mutant clone showed reduced Aire expression.
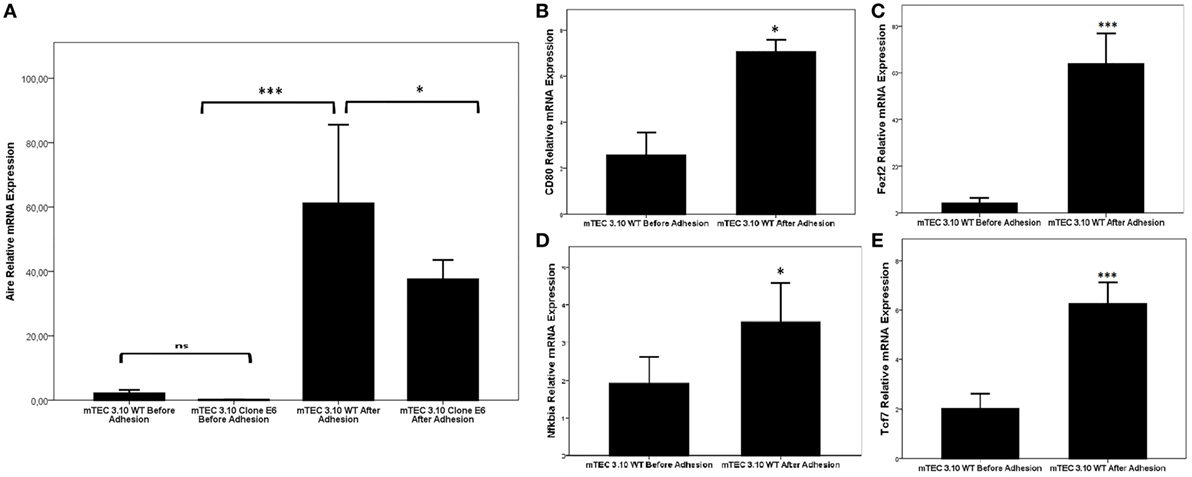
Figure 4. Relative expression of Aire (A), Fezf2 (B), Cd80 (C), Nfkbia (D), and Tcf7 (E) mRNAs in wild-type 3.10 or mutant 3.10E6 medullary thymic epithelial cells (mTECs) before and after thymocyte adhesion. The respective transcriptional expression of these mRNAs were quantified by real-time quantitative-PCR (RT-qPCR). Data were normalized using Hprt mRNA levels and are presented as mean ± SD from six independent experiments. Mann–Whitney two-sided test with 95% interval.
We then evaluated whether thymocyte adhesion itself, independent of Aire, influences specific transcriptional expression in the WT mTEC 3.10 cell line. Figure 4 shows that Aire (Figure 4A), Cd80 (Figure 4B), Fezf2 (Figure 4C), Nfkbia (Figure 4D), and Tcf7 (Figure 4E) were significantly upregulated in mTECs after their adhesion with thymocytes.
After confirming that thymocyte adhesion increases the transcriptional expression of Aire, we used immunolocalization to assess whether such an increase would affect the nuclear localization of the AIRE protein. Figures 5A,B shows that the mTEC 3.10 cell line demonstrated increased nuclear localization of the AIRE protein after adhesion with thymocytes, with granules of AIRE protein associated with chromatin (red dots in the immunofluorescence image). However, clone 3.10E6 cells were negative for AIRE protein localization even after adhesion with thymocytes, showing no red dots in the immunofluorescence images (Figures 5C,D).
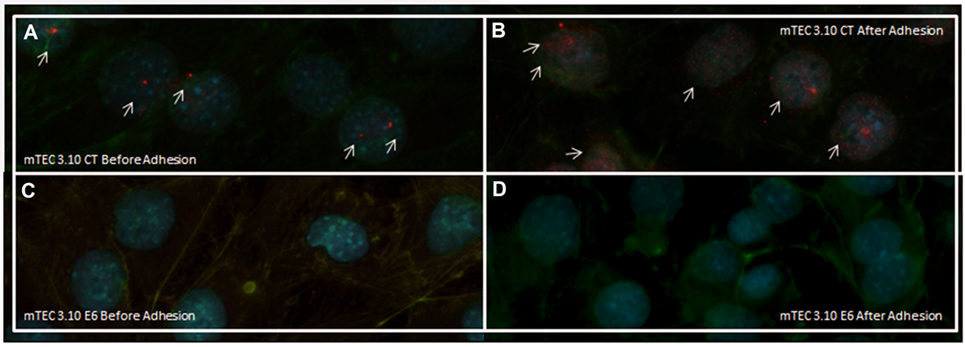
Figure 5. Immunofluorescence of AIRE protein in wild-type (WT) medullary thymic epithelial cell (mTEC) 3.10 cells before and after thymocyte adhesion (A,B) or in mTEC 3.10E6 mutant clone (C,D). Thymocyte adhesion increased AIRE protein nuclear localization in WT mTECs while the mTEC 3.10E6 mutant clone did not exhibited nuclear AIRE protein staining neither before of after thymocyte adhesion. Cells were labeled with an antibody against Mus musculus AIRE protein (red dots), with phalloidin to show the cytoplasmic region and with DAPI to show nuclei (blue).
Comparative analyses were made for various combinations of the study variables [Aire WT (before or after thymocyte adhesion) vs Aire KO (before or after thymocyte adhesion)]. Initially, we performed comparisons between the WT mTEC 3.10 cell line and the mutant 3.10E6 transcriptome through Euclidean distance heat-maps to evaluate how similar the transcriptomes were between the samples as a whole. Figure S4 in Supplementary Material shows that the transcriptomes of the WT mTEC 3.10 cell line and mutant clone 3.10E6 were significantly different, as the two types of samples were positioned in separate clusters, even considering the conditions before and after adhesion with thymocytes.
Next, the samples and DE mRNAs were hierarchically clustered, and a heat-map was constructed to evaluate the individual expression profiles. Figure 6A shows the expression profiling of 902 DE mRNAs between the WT mTEC 3.10 cell line and mutant clone mTEC 3.10E6. Figure 6B shows the top-10 mRNAs (Timp3, Ddit4, Fabp5, Scd1, Serpin2, Lgals3bp, Tnc, Pvrl3, Tcf7, and Nfkbia), whose biological functions, identified through the DAVID databank, are associated with cell adhesion, that featured significant differences in their expression levels.
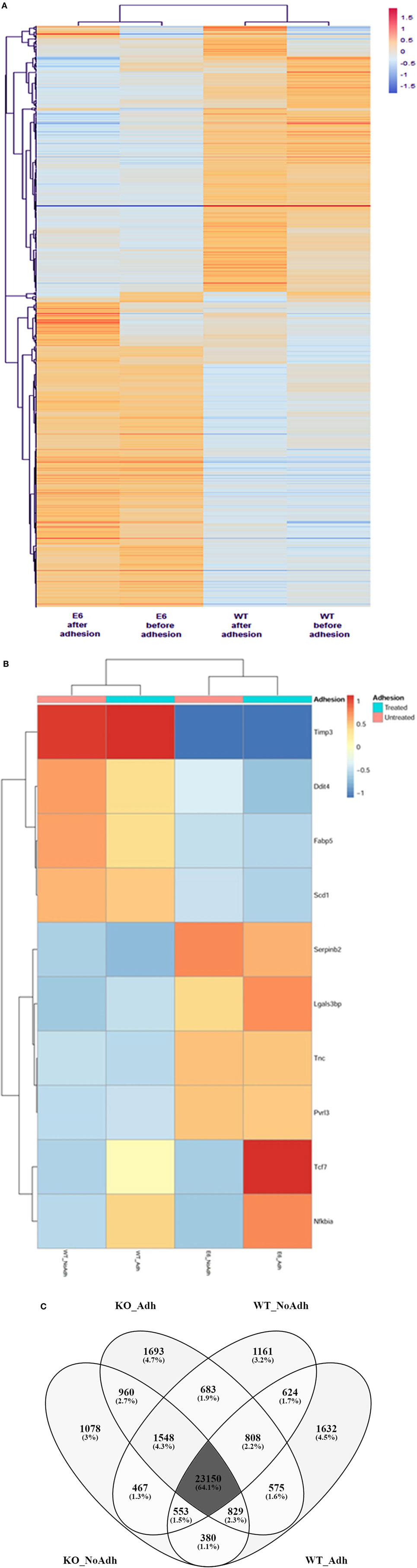
Figure 6. The mRNA transcriptome profile of wild-type (WT) medullary thymic epithelial cell (mTEC) 3.10 or mTEC 3.10E6 mutant clone as evaluated by RNA-Seq. The expression profiling of 902 differentially expressed mRNAs comparing WT mTEC 3.10 cells with mTEC 3.10 mutant clone, before and after thymocyte adhesion (A). The top 10 mRNAs presenting higher expression differences considering the thymocyte adhesion as a variable. Dendrograms and heat-maps were constructed using R platform. Red = upregulation, blue = downregulation [fold change ≥ 1.5 and false discovery rate (FDR) < 0.05] (B). Venn diagram showing the abundance of tissue-specific isoforms when comparing WT mTEC 3.10 cells before (WT_NoAdh) and after (WT_Adh) adhesion and mTEC 3.10E6 mutant clone before (KO_NoAdh) and after (KO_NoAdh) thymocyte adhesion (transcripts per million > 1.0) (C).
Moreover, we evaluated the effects of Aire or thymocyte adhesion on the modulation of mRNA isoforms in mTECs under the different conditions studied. Figure 6C indicates a Venn diagram that shows the number of different isoforms (isoform abundance) and the respective percentages relative to the total isoforms. The different experimental conditions share most of the detected isoforms, but each of the conditions also features populations of specific transcripts.
The different experimental conditions and DE mRNA isoforms were then hierarchically clustered, and heat-maps were constructed to visualize the individual expression profiles. This allowed us to evaluate the influence that Aire has on the modulation of mRNA isoforms that encode proteins involved in cell adhesion and, ultimately, the influence that thymocyte adhesion has on the transcriptomes of mTECs. The top-four DE mRNAs that featured greater isoform abundance involved with cell adhesion were Cd44, Timp3, Tnc, and Tcf7.
In this study, we used mTEC-thymocyte adhesion assays and generated Crispr-Cas9-mediated indel mutations to evaluate the hypothesis that Aire and thymocyte interactions function synergistically in the regulation of cell adhesion-related genes and their isoforms and in comprehensive mTEC mRNA transcriptome modulation.
We have previously observed that Aire controls the expression of genes involved in cell adhesion in mTECs, and its partial inhibition through a small interfering RNA (anti Aire siRNA) system disturbs mTEC-thymocyte adhesion. A reduction in Aire in mTECs was sufficient to decrease the expression of genes, such as Cd80, Vcam, Icam4, Col2a1, Ccl3, and Lama1, impairing adhesion with thymocytes (20). This demonstrated that variations in Aire expression affect mTEC-thymocyte adhesion, which is an essential property for the induction of immunological tolerance.
However, two questions remained: (1) do deleterious mutations in Aire and (2) adhesion to thymocytes affect the mTEC transcriptome, including cell adhesion genes?
More than 100 mutations throughout the human Aire gene sequence, affecting all 14 of its exons, have been described in association with clinical manifestations of autoimmune APECED syndrome (39). Nevertheless, the consequences of Aire mutation in mTECs regarding their adhesion to thymocytes are unknown.
The existing Aire KO mouse model (Aire−/−), which involves a deletion of Aire exon 2 and results in the production of a truncated protein with loss of the SAND and PHD-1 and PHD-2 domains (40, 41), could hypothetically be used to approach these questions.
However, as freshly isolated mTEC cells from the thymus (primary cells) are not easily maintained in culture, and considering that one of our objectives was to make use of the in vitro mTEC-thymocyte adhesion assay, the mTEC cells of these classical Aire KO mice would not be adequate for the present study. For these reasons, we used the mTEC 3.10 cell line (CD45−, EpCam+, Lyn51−, UEA1+), which has been used in previous studies from our laboratory (20, 42).
We therefore used four main experimental and/or bioinformatics approaches to evaluate the hypothesis of this work: (1) the Crispr-Cas9 system to generate mutations within Aire exon 3, (2) the prediction of loss-of-function of the mutant AIRE protein, (3) mTEC-thymocyte adhesion assays, and (4) mRNA transcriptome profiling, including mRNA isoforms.
Interest in Aire exon 3 developed because it encodes the NLS domain of the AIRE protein, a domain involved in translocating that protein from the cytoplasm to the nucleus of mTECs (43–46). Mutations in this region affect not only the transport of the protein to the nucleus but also disrupt its transcriptional behavior (47, 48). The location of the NLS domain involves the amino acid residues at positions 110–114 and 131–133 of the AIRE protein (45). Indel-like mutations in this region may result in truncation in the AIRE protein, causing a loss of the adjacent SAND or PHD1–2 domains (14, 49) (Figure 1A).
Analyses performed with Provean software predicted that clone 3.10E6 showed partial expression of the mutant AIRE protein (Figure 1B). This was confirmed by evaluating mRNA expression levels by RT-qPCR (Figure 4). Regarding the effects of these mutations on Aire mRNA expression levels, it should be noted that in the murine Aire exon 2 KO model, it is still possible to detect mRNA levels comparable to those in the thymi of normal mice (40, 41). Similar to the murine KO model, detection of Aire mRNA levels in clone 3.10E6 could be explained by the fact that the mutations occurred in the Aire exon 3 CDS region and not at splicing sites or promoter sequences.
To evaluate the effects of the mutations generated by the Crispr-Cas9 system on AIRE protein levels, we used a specific monoclonal antibody that recognizes the region of the protein from amino acid residues 246–545, i.e., the SAND and PHD1–2 domains. WB analysis (Figure 1B) showed that there was a reduction in AIRE protein expression in clone 3.10E6. This may be explained by the fact that one of its alleles encodes a truncated AIRE protein and that the other allele, although it encodes a nonfunctional protein, shows no change in the domains recognized by the primary anti-AIRE antibody used.
Several patients with APECED have mutations in the CARD or NLS domain, and the AIRE protein ceases to be located in the nucleus and is instead deposited in the cytoplasm (47, 49). This observation became of interest for the present work, since the potential consequences of the mutations induced by the Crispr-Cas9 system on AIRE protein function could be due to possible changes in its capacity for intracellular translocation rather than through direct association with DNA or other proteins.
AIRE is a protein that exerts its function in the nucleus, and its nuclear localization occurs due to its NLS domain (45). Mutations generated by the Crispr-Cas9 system occurred within the NLS coding region, which most likely caused perturbations in the nuclear AIRE translocation. This was confirmed through immunofluorescence analysis (Figure 2). Clone 3.10E6, although showing expression of the mutant AIRE protein, does not show localization to the nucleus of mTEC cells. Accordingly, clone 3.10E6 can be considered a KO since the mutant AIRE protein from allele 1, although expressed, had its function altered and its translocation from the cytoplasm to the nucleus impaired. The absence of labeling in the cytoplasm is likely due to insufficient expression of the AIRE protein. Mutations of this type have been described previously in patients with APECED and are, respectively, associated with different symptoms of the disease (39).
Disturbance of Aire expression both in vitro and in vivo leads to changes in regulation of the expression of a large set of genes that are associated with NS, autoantigen processing and other biological processes, and autoantigens themselves (20, 40, 41, 50). In addition, genes encoding adhesion molecules are also perturbed by the reduction of Aire expression (20). This led us to evaluate whether Aire mutations could affect cell adhesion. Accordingly, we determined whether the mutations induced by Crispr-Cas9 could disrupt the adhesive capacity of mTEC cells with thymocytes, which was demonstrated by means of adhesion assays (Figure 3).
There have been no investigations to date of the link between Aire, cell adhesion, and transcriptome profiles of mTECs. However, we find such a relationship plausible since a group of thymocytes expressing RANKL can interact with mTEC cells, induce Aire expression (51, 52) and consequently modulate cell adhesion genes (Figure 4) and the transcriptomes of these cells (Figure 6). Since we observed that mTEC-thymocyte adhesion stimulates the expression of Aire in mTECs, and considering that Aire controls more than 3,300 genes in these cells (11), we next evaluated their transcriptomes. This question was addressed by sequencing the transcriptomes (RNA-Seq) of WT mTECs and 3.10E6 clones from both groups, both before and after thymocyte adhesion.
The mTEC-thymocyte adhesion itself functions as a stimulus for the transcription of mRNAs that encode molecules such as the transcription factor TCF7, a protein that was initially identified in thymocytes and participates in their differentiation in the thymus (53, 54). The Tcf7 gene has only recently been associated with mTEC cells. It has been shown that this protein is part of an essential transcriptional complex along with other transcription factors, such as Irf4, Irf8, Tbx21, and Ctcfl, that regulates the expression of Aire in these cells (55). We also showed that mTEC-thymocyte adhesion causes increased expression of mRNA transcripts encoding NFKB alpha inhibitor (Nfkbia), which is the main inhibitor of the Nfkb pathway.
The expression of Aire in mTECs is activated by the RANK–RANKL pathway (51, 52) and is also dependent on the binding of NFKB to an enhancer located in a non-coding region between the Aire and Dnmt3L genes (56). Defects in the Nfkb-mediated signaling pathway also lead to a decrease in Aire expression in mTECs (57).
We asked whether the mTEC-thymocyte adhesion that activates the expression of Tcf7 would activate the Nfkb pathway and consequently Aire (Figure 4). The increase in Aire mRNA expression by mTEC-thymocyte adhesion was found to intensify the AIRE protein localization to the nucleus of WT mTEC 3.10 cells but, as expected, not in clone 3.10E6 cells (Figure 5).
In fact, the transcriptomes of WT mTECs differ from those of clone 3.10E6 cells. This finding has a significant physiological correlation, as the transcriptomes of mTEChi cells are different from mTEClo cells, which have low Aire expression (11).
The mTEC 3.10 cells are characterized by a CD80lo or MHC-IIlo phenotype (20) that, under proper stimulation, initiates Aire expression. With altered expression of Aire, either by means of anti-Aire siRNA (20) or by means of Crispr-Cas9 KO (this study), these cells exhibit marked dysregulation of genes involved with NS. Among the set of mRNAs with dysregulated expression, we have identified several that control one or more of the following processes: cell adhesion, migration, positive regulation of gene expression, signal transduction, or positive regulation of RNA Pol II function.
We have found that several important mRNAs associated with cell adhesion, such as those that encode proteins belonging to the claudins, integrins, selectins, or extracellular matrix protein families, were DE in clone 3.10E6 cells (Figures 4A,B). For example, the mRNAs that encode the Timp3, Cd24a, or Fabp5 proteins, which are important for the occurrence of mTEC-thymocyte interactions, were repressed in clone 3.10E6 cells. Conversely, Tnc and Lgals3bp mRNAs were induced in this clone. This set of results suggests that thymocyte adhesion and Aire, although functioning in different ways, have synergistic activity. Cell adhesion stimulates transcriptional expression in mTEC cells, including of Aire itself, which in turn regulates a downstream cascade of mRNAs that encode cell adhesion molecules.
Evidence suggests that Aire plays a role in the processing of mRNAs and therefore contributes to the expression of tissue-specific isoforms in mTECs (17–19). We quantified these isoforms and verified that their abundance was different, but not significantly so, between each of the situations studied. In addition, mTEC-thymocyte adhesion does not appear to affect the abundance of transcripts (Figure 6C). These results corroborate those of St-Pierre et al. (11), who found no differences in the complexity of splicing between mTEChi, mTEClo, and cTEC populations, and Danan-Gotthold et al. (58), who observed equivalent representation of isoforms when comparing WT vs Aire KO mTECs for most tissue-specific transcripts.
However, when analyzing these results in a comparative way, we observed the occurrence of specific transcripts for each of the experimental groups studied. This led us to investigate the influence of Aire or mTEC-thymocyte adhesion on isoform modulation. When comparing WT mTEC 3.10 cells with clone 3.10E6 cells (both before and after thymocyte adhesion), it was evident that Aire modulates mRNA isoforms that encode cell adhesion molecules.
The occurrence of isoforms in mTECs has an important immunological consequence. This increases the variability of PTAs and consequently increases the chance of negatively selecting autoreactive thymocyte clones in the thymus (17–19, 58). In this work, we have shown that Aire can control the relative abundance of isoforms of mRNAs that encode adhesion molecules. The immunological consequences of this might be associated with the maintenance of different mTEC clones, each exhibiting a different degree of thymocyte adhesion.
As discussed in a previous study from our group (13), healthy individuals also maintain self-reactive T cell clones, and the development of central tolerance is a particularly delicate process during which NS must be sufficiently stringent to avoid the escape of self-reactive T cell clones into the periphery (7).
The results from this work increase our knowledge of the biological function of Aire at two levels: (1) the existence of synergy between Aire and thymocyte adhesion on the transcriptomes of mTECs and (2) the role played by Aire in modulating the expression of mRNAs and their isoforms that encode cell adhesion molecules.
This opens new perspectives for further studies concerning the molecular mechanisms controlling gene expression in mTEC cells upon interaction with thymocytes, as well as the fine-tuning of NS. Finally, considering that mTEC cells are an important target in donor T cell alloimmunity, there is significant implication of these results for further studies of transcriptional expression in the thymus during onset of graft-vs-host-disease (59–61).
Local Animal Ethical Committee of the Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto Campus, Brazil (permit Number 006/2016-1).
CS-H: conceived and performed all the experiments, transfected mTEC cells with Crispr-Cas9 vector, analyzed the mutant mTEC clone, collected, analyzed, and interpreted all data. AA: extracted and analyzed the total RNA samples, analyzed and interpreted the mTEC transcriptome by bioinformatics. RF: transfected mTEC cells with Crispr-Cas9 vector. LC-S: performed the cell adhesion assay. NP and GL: prepared the cell lysate and performed the WB experiments. KB-P: performed the immunofluorescence assays. SG: analyzed and interpreted the mTEC transcriptome by bioinformatics. GP: conceived the study, designed the experiments, decided the Crispr-Cas9 system, interpreted and organized all data, interpreted transcriptome data, and wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by the following agencies, Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, São Paulo, Brazil, Project # 13/17481-1 to GAP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, Brazil, Projects # 306315/2013-0 and 305787/2017-9 to GP), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, Brazil, Project # 8888105/2014-01 to GP). We thank Drs. W. Savino and Daniella Arêas Mendes-da-Cruz (Laboratory on Thymus Research, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Rio de Janeiro, RJ, Brazil) who ceded the mTEC 3.10 cells, for their help and discussion, Mrs. Denise B. Ferraz (Ribeirão Preto Medical School), Mr. Roger R. Fernandes, Mrs. Tania M. Sousa, and Mrs. Aline Ap. Ferrarese Tiballi (School of Dentistry of Ribeirão Preto) for their technical assistance.
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.00964/full#supplementary-material.
Figure S1. Flow cytometry for sorting separation of green-fluorescent-positive (GFP+) mTEC 3.10 cells transfected (or not) with the CRISPR-Cas9 vector.
Figure S2. T7 endonuclease assay. The PCR product of Aire gene exon 3 from two wild-type mTEC 3.10 cell samples (mTEC CT1 and mTEC CT2) were used as controls whose 415 bp PCR amplicon was not digested by the T7 enzyme. The PCR amplicon from two mutant clones (mTEC 3.10E6 and mTEC 3.10G10) were digested by the T7 enzyme resulting, as expected, in two DNA fragments with approximately 200 bp. The mTEC 3.10G10 mutant clone was not included in this study. Agilent Bioanalyzer model 2100 microfluidic electrophoresis.
Figure S3. Electropherograms obtained from the Sanger sequencing of the Aire exon 3, partial sequence of the 415 bp PCR amplicon. Amplicons of mTEC CT1 and mTEC CT2 cell samples are Aire wild-type for both alleles, whereas mTEC 3.10E6 is a compound heterozygous (red rectangle). Position of the PAM sequence is indicated.
Figure S4. Euclidean distances comparing the global differences of the transcriptome as evaluated by RNA-Seq of wild-type mTEC 3.10 (before and after thymocyte adhesion) vs mTEC 3.10E6 mutant clone (before and after thymocyte adhesion).
Figure S5. Image of full Western blot (WB) membrane of SDS-PAGE wild-type mTEC 3.10 and mTEC 3.10E6 mutant clone cell lysates for detection of Mus musculus AIRE protein. WB membrane was probed with an antibody against AIRE protein (upper panel), washed and then probed with an antibody against GAPDH that was used as an internal load control.
Table S1. Molecular characterization of the Aire exon 3, mTEC 3.10E6 mutant clone through Sanger DNA sequencing and Provean protein sequence analysis. GenBank NCBI accession numbers: Aire mutant allele 1 (MG493266), Aire mutant allele 2 (MG493265).
1. Petrie HT, Zúñiga-Pflücker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol (2007) 25:649–79. doi:10.1146/annurev.immunol.23.021704.115715
2. Love PE, Brandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol (2011) 11:469–77. doi:10.1038/nri2989
3. Hu Z, Lancaster JN, Ehrlich LI. The contribution of chemokines and migration to the induction of central tolerance in the thymus. Front Immunol (2015) 6:398. doi:10.3389/fimmu.2015.00398
4. Abramson J, Anderson G. Thymic epithelial cells. Annu Rev Immunol (2017) 35:85–118. doi:10.1146/annurev-immunol-051116-052320
5. Klein L, Kyewski B, Allen PM, Hogguist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol (2014) 14:377–91. doi:10.1038/nri3667
6. Akiyama T, Tateishi R, Akiyama N, Yoshinaga R, Kobayashi TJ. Positive and negative regulatory mechanisms for fine-tuning cellularity and functions of medullary thymic epithelial cells. Front Immunol (2015) 6:461. doi:10.3389/fimmu.2015.00461
7. Passos GA, Speck-Hernandez CA, Assis AF, Mendes-da-Cruz DA. Update on Aire and negative selection. Immunology (2018) 153:1. doi:10.1111/imm.12831
8. Lind EF, Prockop SE, Porritt HE, Petrie HT. Mapping precursor movement through the postnatal thymus reveals specific microenvironments supporting defined stages of early lymphoid development. J Exp Med (2001) 194:127–34. doi:10.1084/jem.194.2.127
9. Griffith AV, Fallahi M, Nakase H, Gosink M, Young B, Petrie HT. Spatial mapping of thymic stromal microenvironments reveals unique features influencing T lymphoid differentiation. Immunity (2009) 31:999–1009. doi:10.1016/j.immuni.2009.09.024
10. Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, et al. Population and single-cell genomics reveal the Aire dependency relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res (2014) 24:1918–31. doi:10.1101/gr.171645.113
11. St-Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C. Differential features of AIRE-induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J Immunol (2015) 195:498–506. doi:10.4049/jimmunol.1500558
12. Ucar O, Rattay K. Promiscuous gene expression in the thymus: a matter of epigenetics, miRNA, and more? Front Immunol (2015) 6:93. doi:10.3389/fimmu.2015.00093
13. Passos GA, Mendes-Da-Cruz DA, Oliveira EH. The thymic orchestration involving Aire, miRNAs and cell–cell interactions during the induction of central tolerance. Front Immunol (2015) 6:352. doi:10.3389/fimmu.2015.00352
14. Anderson MS, Su MA. AIRE expands: new roles in immune tolerance and beyond. Nat Rev Immunol (2016) 16:247–58. doi:10.1038/nri.2016.9
15. Abramson J, Goldfarb Y. AIRE: from promiscuous molecular partnerships to promiscuous gene expression. Eur J Immunol (2016) 46:22–33. doi:10.1002/eji.201545792
16. Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A (2012) 109:535–40. doi:10.1073/pnas.1119351109
17. Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell (2010) 140:123–35. doi:10.1016/j.cell.2009.12.030
18. Keane P, Ceredig R, Seoighe C. Promiscuous mRNA splicing under the control of AIRE in medullary thymic epithelial cells. Bioinformatics (2015) 31:986–90. doi:10.1093/bioinformatics/btu785
19. Alvarez I, Collado JA, Colobran R, Carrascal M, Ciudad MT, Canals F, et al. Central T cell tolerance: identification of tissue-restricted autoantigens in the thymus HLA-DR peptidome. J Autoimmun (2015) 60:12–9. doi:10.1016/j.jaut.2015.03.004
20. Pezzi N, Assis AF, Cotrim-Sousa LC, Lopes GS, Mosella MS, Lima DS, et al. Aire knockdown in medullary thymic epithelial cells affects Aire protein, deregulates cell adhesion genes and decreases thymocyte interaction. Mol Immunol (2016) 77:157–73. doi:10.1016/j.molimm.2016.08.003
21. Klein L. Aire gets company for immune tolerance. Cell (2015) 163:794–5. doi:10.1016/j.cell.2015.10.057
22. Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, et al. Fezf2 Orchestrates a thymic program of self-antigen expression for immune tolerance. Cell (2015) 163:975–87. doi:10.1016/j.cell.2015.10.013
23. Lopes N, Sergé A, Ferrier P, Irla M. Thymic crosstalk coordinates medulla organization and T-cell tolerance induction. Front Immunol (2015) 6:365. doi:10.3389/fimmu.2015.00365
24. Gäbler J, Arnold J, Kyewski B. Promiscuous gene expression and the developmental dynamics of medullary thymic epithelial cells. Eur J Immunol (2007) 37:3363–72. doi:10.1002/eji.200737131
25. Rossi SW, Kim MY, Leibbrandt A, Parnell SM, Jenkinson WE, Glanville SH, et al. RANK signals from CD4(+)3(-) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med (2007) 204:1267–72. doi:10.1084/jem.20062497
26. Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med (2007) 204(11):2521–8. doi:10.1084/jem.20070795
27. Donate PB, Fornari TA, Macedo C, Cunha TM, Nascimento DC, Sakamoto-Hojo ET, et al. T cell post-transcriptional miRNA-mRNA interaction networks identify targets associated with susceptibility/resistance to collagen-induced arthritis. PLoS One (2013) 8(1):e54803. doi:10.1371/journal.pone.0054803
28. Fornari TA, Donate PB, Assis AF, Macedo C, Sakamoto-Hojo ET, Donadi EA, et al. Comprehensive survey of miRNA-mRNA interactions reveals that Ccr7 and Cd247 (CD3 zeta) are posttranscriptionally controlled in pancreas infiltrating T lymphocytes of non-obese diabetic (NOD) mice. PLoS One (2015) 10(11):e0142688. doi:10.1371/journal.pone.0142688
29. Hirokawa K, Utsuyama M, Moriizumi E, Handa S. Analysis of the thymic microenvironment by monoclonal antibodies with special reference to thymic nurse cells. Thymus (1986) 8:349–60.
30. Nihei OK, Campos de Carvalho AC, Spray DC, Savino W, Alves LA. A novel form of cellular communication among thymic epithelial cells: intercellular calcium wave propagation. Am J Physiol Cell Physiol (2003) 285:C1304–13. doi:10.1152/ajpcell.00568.2002
31. Ribeiro-Carvalho MM, Farias-de-Oliveira DA, Villa-Verde DM, Savino W. Triiodothyronine modulates extracellular matrix-mediated interactions bet-ween thymocytes and thymic microenvironmental cells. Neuroimmuno-modulation (2002–2003) 10:142–52. doi:10.1159/000067175
32. Savino W, Mendes-Da-Cruz DA, Smaniotto S, Silva-Monteiro E, Villa-Verde DM. Molecular mechanisms governing thymocyte migration: combined role of chemokines and extracellular matrix. J Leukoc Biol (2004) 75:951–61. doi:10.1189/jlb.1003455
33. Savino W. Neuroendocrine control of T cell development in mammals: role of growth hormone in modulating thymocyte migration. Exp Physiol (2007) 92:813–7. doi:10.1113/expphysiol.2007.038422
34. Ocampo JS, de Brito JM, Corrêa-de-Santana E, Borojevic R, Villa-Verde DM, Savino W. Laminin-211 controls thymocyte–thymic epithelial cell interactions. Cell Immunol (2008) 254:1–9. doi:10.1016/j.cellimm.2008.06.005
35. Linhares-Lacerda L, Ribeiro-Alves M, Nogueira AC, Mendes-da-Cruz DA, Magalhães DA, Dardenne M, et al. RNA interference-mediated knockdown of CD49e (α5 integrin chain) in human thymic epithelial cells modulates the expression of multiple genes and decreases thymocyte adhesion. BMC Genomics (2010) 11:S2. doi:10.1186/1471-2164-11-S5-S2
36. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol (2013) 31:827–32. doi:10.1038/nbt.2647
37. Dehairs J, Talebi A, Cherifi Y, Swinnen JV. CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci Rep (2016) 6:28973. doi:10.1038/srep28973
38. St-Pierre C, Brochu S, Vanegas JR, Dumont-Lagacé M, Lemieux S, Perreault C. Transcriptome sequencing of neonatal thymic epithelial cells. Sci Rep (2013) 3:1860. doi:10.1038/srep01860
39. Bruserud Ø, Oftedal BE, Wolff AB, Husebye ES. AIRE-mutations and autoimmune disease. Curr Opin Immunol (2016) 43:8–15. doi:10.1016/j.coi.2016.07.003
40. Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow with in the thymus by the Aire protein. Science (2002) 298(5597):1395–401. doi:10.1126/science.1075958
41. Yano M, Kuroda N, Han H, Meguro-Horike M, Nishikawa Y, Kiyonari H, et al. Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J Exp Med (2008) 205:2827–38. doi:10.1084/jem.20080046
42. Macedo C, Evangelista AF, Marques MM, Octacílio-Silva S, Donadi EA, Sakamoto-Hojo ET, et al. Autoimmune regulator (Aire) controls the expression of microRNAs in medullary thymic epithelial cells. Immunobiology (2013) 218(4):554–60. doi:10.1016/j.imbio.2012.06.013
43. Björses P, Pelto-Huikko M, Kaukonen J, Aaltonen J, Peltonen L, Ulmanen I. Localization of the APECED protein in distinct nuclear structures. Hum Mol Genet (1999) 8:259–66. doi:10.1093/hmg/8.2.259
44. Pitkänen J, Vähämurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. Characterization of nuclear targeting and transcriptional activation domain. J Biol Chem (2001) 276:19597–602. doi:10.1074/jbc.M008322200
45. Ilmarinen T, Melén K, Kangas H, Julkunen I, Ulmanen I, Eskelin P. The monopartite nuclear localization signal of autoimmune regulator mediates its nuclear import and interaction with multiple importin alpha molecules. FEBS J (2006) 273:315–24. doi:10.1111/j.1742-4658.2005.05065.x
46. Perniola R, Musco G. The biophysical and biochemical properties of the autoimmune regulator (AIRE) protein. Biochim Biophys Acta (2014) 1842:326–37. doi:10.1016/j.bbadis.2013.11.020
47. Björses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, et al. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am J Hum Genet (2000) 66:378–92. doi:10.1086/302765
48. Ramsey C, Bukrinsky A, Peltonen L. Systematic mutagenesis of the functional domains of AIRE reveals their role in intracellular targeting. Hum Mol Genet (2002) 11:3299–308. doi:10.1093/hmg/11.26.3299
49. Oftedal BE, Hellesen A, Erichsen MM, Bratland E, Vardi A, Perheentupa J, et al. Dominant mutations in the autoimmune regulator AIRE are associated with common organ-specific autoimmune diseases. Immunity (2015) 42:1185–96. doi:10.1016/j.immuni.2015.04.021
50. Nishijima H, Kajimoto T, Matsuoka Y, Mouri Y, Morimoto J, Matsumoto M, et al. Paradoxical development of polymyositis-like autoimmunity through augmented expression of autoimmune regulator (AIRE). J Autoimmun (2018) 86:75–92. doi:10.1016/j.jaut.2017.09.006
51. Akiyama T, Shimo Y, Yanai H, Qin J, Ohshima D, Maruyama Y, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity (2008) 29:423–37. doi:10.1016/j.immuni.2008.06.015
52. Akiyama N, Takizawa N, Miyauchi M, Yanai H, Tateishi R, Shinzawa M, et al. Identification of embryonic precursor cells that differentiate into thymic epithelial cells expressing autoimmune regulator. J Exp Med (2016) 213:1441–58. doi:10.1084/jem.20151780
53. Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature (1995) 374:70–4. doi:10.1038/374070a0
54. Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity (1998) 8:11–20. doi:10.1016/S1074-7613(00)80454-9
55. Herzig Y, Nevo S, Bornstein C, Brezis MR, Ben-Hur S, Shkedy A, et al. Transcriptional programs that control expression of the autoimmune regulator gene Aire. Nat Immunol (2017) 18:161–72. doi:10.1038/ni.3638
56. Haljasorg U, Bichele R, Saare M, Guha M, Maslovskaja J, Kõnd K, et al. A highly conserved NF-κB-responsive enhancer is critical for thymic expression of Aire in mice. Eur J Immunol (2015) 45:3246–56. doi:10.1002/eji.201545928
57. Fletcher AL, Seach N, Reiseger JJ, Lowen TE, Hammett MV, Scott HS, et al. Reduced thymic Aire expression and abnormal NF-kappa B2 signaling in a model of systemic autoimmunity. J Immunol (2009) 182:2690–9. doi:10.4049/jimmunol.0801752
58. Danan-Gotthold M, Guyon C, Giraud M, Levanon EY, Abramson J. Extensive RNA editing and splicing increase immune self-representation diversity in medullary thymic epithelial cells. Genome Biol (2016) 17:219. doi:10.1186/s13059-016-1079-9
59. Dertschnig S, Nusspaumer G, Ivanek R, Hauri-Hohl MM, Holländer GA, Krenger W. Epithelial cytoprotection sustains ectopic expression of tissue-restricted antigens in the thymus during murine acute GVHD. Blood (2013) 122:837–41. doi:10.1182/blood-2012-12-474759
60. Dertschnig S, Hauri-Hohl MM, Vollmer M, Holländer GA, Krenger W. Impaired thymic expression of tissue-restricted antigens licenses the de novo generation of autoreactive CD4+ T cells in acute GVHD. Blood (2015) 125:2720–3. doi:10.1182/blood-2014-08-597245
Keywords: Aire gene, cell adhesion, transcriptome, medullary thymic epithelial cells, immune tolerance
Citation: Speck-Hernandez CA, Assis AF, Felicio RF, Cotrim-Sousa L, Pezzi N, Lopes GS, Bombonato-Prado KF, Giuliatti S and Passos GA (2018) Aire Disruption Influences the Medullary Thymic Epithelial Cell Transcriptome and Interaction With Thymocytes. Front. Immunol. 9:964. doi: 10.3389/fimmu.2018.00964
Received: 03 December 2017; Accepted: 18 April 2018;
Published: 07 May 2018
Edited by:
Antoine Toubert, Paris Diderot University, FranceReviewed by:
Michael Uhlin, Karolinska Institute (KI), SwedenCopyright: © 2018 Speck-Hernandez, Assis, Felicio, Cotrim-Sousa, Pezzi, Lopes, Bombonato-Prado, Giuliatti and Passos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geraldo A. Passos, cGFzc29zQHVzcC5icg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.