
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 07 February 2018
Sec. Microbial Immunology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.00170
This article is part of the Research Topic Immunity to Human Fungal Pathogens: Mechanisms of Host Recognition, Protection, Pathology, and Fungal Interference View all 37 articles
Fungal bioaerosols are ubiquitous in the environment and human exposure can result in a variety of health effects ranging from systemic, subcutaneous, and cutaneous infections to respiratory morbidity including allergy, asthma, and hypersensitivity pneumonitis. Recent research has focused on the role of microRNAs (miRNAs) following fungal exposure and is overlooked, yet important, group of regulators capable of influencing fungal immune responses through a variety of cellular mechanisms. These small non-coding ribose nucleic acids function to regulate gene expression at the post-transcriptional level and have been shown to participate in multiple disease pathways including cancer, heart disease, apoptosis, as well as immune responses to microbial hazards and occupational allergens. Recent animal model studies have characterized miRNAs following the exposure to inflammatory stimuli. Studies focused on microbial exposure, including bacterial infections, as well as exposure to different allergens have shown miRNAs, such as miR-21, miR-146, miR-132, miR-155, and the let-7 family members, to be involved in immune and inflammatory responses. Interestingly, the few studies have assessed that the miRNA profiles following fungal exposure have identified the same critical miRNAs that have been characterized in other inflammatory-mediated and allergy-induced experimental models. Review of available in vitro, animal and human studies of exposures to Aspergillus fumigatus, Candida albicans, Cryptococcus neoformans, Paracoccidioides brasiliensis, and Stachybotrys chartarum identified several miRNAs that were shared between responses to these species including miR-125 a/b (macrophage polarization/activation), miR-132 [toll-like receptor (TLR)2-mediated signaling], miR-146a (TLR mediated signaling, alternative macrophage activation), and miR-29a/b (natural killer cell function, C-leptin signaling, inhibition of Th1 immune response). Although these datasets provide preliminary insight into the role of miRNAs in fungal exposed models, interpretation of miRNA datasets can be challenging for researchers. To assist in navigating this rapidly evolving field, the aim of this review is to describe miRNAs in the framework of host recognition mechanisms and provide initial insight into the regulatory pathways in response to fungal exposure.
Fungi are ubiquitous eukaryotic microorganisms that can be prevalent in indoor, outdoor, and occupational environments. A small portion of 1.5 million fungal species estimated to exist (1) are primary or opportunistic pathogens, whereas the vast majority is ubiquitous saprophytes that obtain nutrients from organic matter. Fungi are composed of membrane bound organelles that are encased by a rigid cell wall but do not contain chlorophyll. The cell wall is composed of ergosterol, chitin, glucans, such as (1 → 3)-β-d-glucan, and mannose proteins (2). Fungal lifeforms broadly vary from unicellular yeasts to multicellular filamentous hyphae that include the production of mitotic or meiotically produced spores. In some cases, fungi are dimorphic and share both lifecycles. Upon disturbance, fungal spores can be aerosolized and in some occupational environments the airborne concentrations may exceed 1 × 105 spores/m3 (3).
Personal exposure to fungal species has been associated with a broad variety of adverse health effects that range from pulmonary, sinus, and subcutaneous infections to respiratory morbidities that may include hypersensitivity pneumonitis, allergy, and asthma (3). Each of these health effects is dependent on the host’s immune responsiveness and fungal species exposed (4). In specific geographical regions, dimorphic fungi that cause endemic mycoses exist as either a filamentous fungus in the environment or as a pathogenic yeast in the host. In the environment, the filamentous hyphae grow in soil at ambient temperatures and produce infectious spores (5, 6). Soil disturbance can aerosolize spores that can be inhaled by a mammalian host. In a process that is thermo-regulated, the spores can germinate into a pathogenic yeast phase that helps these fungi avoid the hosts’ immune responses. For example, Blastomyces dermatitidis can proliferate on the respiratory mucosa, Histoplasma capsulatum modulates the monocyte phagolysosome compartment, and Coccidioides immitis develops a large spherule containing endospores that is resistant to phagocytosis. These dimorphic fungal species as well as others, including Paracoccidioides brasiliensis (paracoccidioidomycosis) and Talaromyces (Penicillium) marneffei (talaromycosis), affect the lungs, although the latter can also affect the liver and mouth.
By contrast, opportunistic fungal pathogens consist of fungi that are environmentally ubiquitous and affect those who are immunocompromised, especially patients who have received a transplant or undergoing chemotherapy or corticosteroid therapy. Examples of fungi that are commonly implicated in opportunistic infections include, Candida albicans (candidiasis), Pneumocystis jirovecii (Pneumocystis pneumonia), Cryptococcus neoformans/gattii (cryptococcosis), and Aspergillus fumigatus (aspergillosis). Infections can be acquired through the inhalation of conidia or yeast depending on the species and can result in systemic mycoses. With the increase in broad-spectrum antibiotic usage and other medical and therapeutic strategies, invasive opportunistic fungal infections are of particular concern in the hospital setting, as nosocomial infections may be life-threatening for critically ill individuals (7).
The World Health Organization and the Institute of Medicine have published consensus documents that report respiratory morbidities are associated with damp indoor environments (8, 9). Recent epidemiological evidence has further built on these consensus findings and shown exposure to mold in damp indoor environments to be associated with adverse respiratory health effects (10, 11). Following recent natural disasters and flooding events associated with Hurricanes Harvey, Irma, and Maria, water-infiltrated occupational, and residential buildings are environments where mold can grow and proliferate on water damaged building materials. Returning to these environments and disturbing contaminated building materials can result in the aerosolization of fungal spores (12) that can pose a significant health risk especially if the person is immunocompromised. Fungi associated with colonizing wet building materials include, Aspergillus versicolor, Ulocladium chartarum, Chaetomium globosum, and Stachybotrys chartarum that are hydrophilic and require a high water activity for growth and proliferation. Of these hydrophilic fungal contaminants, S. chartarum is the most widely studied and many reports have identified exposure to contribute to negative health effects (12–15).
Due to increased community concern regarding personal exposure to these pathogenic fungi and the potential result of life-threatening health outcomes, it is important to characterize the mechanisms that contribute to the host innate and adaptive immune responses. Previous research has focused on host responses in fungal exposure models by analyzing functional, histological, and immunological endpoints; however, research examining the molecular mechanisms that underlie these responses remains unclear for many clinically relevant fungal species. Although many studies have been published that have explored pulmonary immunological responses to acute and chronic fungal spores exposures, the microRNAs (miRNAs) that regulate these deficiencies have not been fully characterized. In this review, the state-of-knowledge of miRNAs characterized in various animal models, including those that have evaluated fungal exposures, will be reviewed with emphasis placed on the mechanistic insights that these studies have provided in relation to the host response following fungal exposure.
MicroRNAs are an important group of regulators capable of influencing gene expression through different mechanisms (16–20). Consisting of short, single stranded noncoding ribose nucleic acids (RNAs), miRNAs bind to target messenger RNA (mRNA) to downregulate gene expression post-transcriptionally through RNA silencing or RNA degradation (21, 22). Depending on the complementarity of base pairing, gene expression is repressed, as observed in humans and animals, or mRNA is cleaved, as observed in plants (23–25). More recently, studies have shown that miRNA can also activate the translation of certain target mRNA (17, 18, 26). Providing insight into how altered miRNA profiles affect upstream processes can be methodologically challenging. For example, a single miRNA can regulate from one to multiple genes, whereas studies have also shown that multiple miRNAs can regulate the same gene (27–30). Several miRNAs, as well as miRNA families, have been extensively studied and have been characterized in models of cancer, heart disease, aging, apoptosis, and immune responses to inflammatory stimuli (19, 20, 31–34).
The let-7 family is the most abundant pulmonary miRNAs and has been identified in cancer, diabetes, and aging studies (35–39). The let-7 family members have been shown to target interleukin (IL)-13 in in vivo and in vitro models, although the regulatory in vivo mechanisms of let-7 are complex (20, 40). miR-21 is another widely studied miRNA and has been shown to participate in the inflammatory response elicited by different stimuli, including doxycycline-induced allergic airway inflammation (41), as well as viral, bacterial and protozoan infections (42–44). One of the most upregulated miRNAs in human patients with allergic eosinophilic esophagitis is miR-21 (45, 46), which correlates with studies that reported miR-21 and miR-223 as regulators of eosinophilic development in an ex vivo model of bone-derived eosinophils (47, 48). miR-375 has also been reported to be downregulated in epithelial cells derived from patients with eosinophilic esophagitis, as well as in IL-13 stimulated epithelial cells indicating the role of miR-375 as a regulator of IL-13-mediated responses (21).
In rodent models exposed to house dust mite allergen, increased miR-126, miR-106a, and miR-145 expression have been shown to contribute to allergic inflammation (49–51). Studies involving airborne pollutants, such as cigarette smoke, reported a downregulation in let-7c, let-7f, miR-34b, miR-34c, and miR-222, all of which contributed to pulmonary inflammation in rodent models (52–54). Research examining aberrant miRNA profiles in asthmatic hosts has also revealed novel miRNAs that contribute to allergic airway disease. Examination of CD4+ T cells isolated from the bronchoalveolar lavage fluid from asthmatic human patients revealed that miR-19a had the highest expression (55), which promoted a Th2-mediated cell response, a known response contributing to allergic asthma. In another study, miR-221 and miR-485-3p were upregulated in peripheral blood from pediatric asthmatic human patients compared with controls, suggesting that these miRNAs contribute to the development of asthma (56). In a chemical allergen model examining the murine miRNA profile following dermal exposure to toluene 2,4-diisocyanate, miR-21, miR-22, miR-27b, miR-31, miR-126, miR-155, miR-210, and miR-301a expression were increased (57). While this study identified miRNAs that are known to participate in the immune response associated with asthma (miR-21, miR-31, miR-126, and miR-155), new miRNAs were proposed as potential biomarkers for allergic sensitization to toluene 2,4-diisocyanate (miR-22, miR-27b, miR-301a, and miR-210).
MicroRNAs critically influence the development and responses of the immune system, but the contributing biological mechanisms are poorly characterized (22, 58–60). Overexpression of the miR-17-92 cluster and miR-181 enhanced B-cell proliferation, while miR-150 regulated B-cell differentiation (61–64). When overexpressed, miR-181 has been shown to decrease T-cell numbers (61), but enhance T-cell receptor signaling (65). When T cells are activated, the miRNA expression profiles are altered (66–68). T-cell activation has additionally been found to induce the miR-17~92 family members (69), as well as the gene that encodes miR-155 (70). miR-155 has also been reported to regulate antigen presentation (71) and to negatively regulate toll-like receptor (TLR) and cytokine signaling (72). The miR-17-92 cluster promotes Th1 type immune responses along with inhibiting regulatory T-cell differentiation (69). Rodriquez et al. showed that miR-155 is required for normal functioning of B and T lymphocytes as well as dendritic cells (73).
In human macrophages, miR-155 targets and subsequently decreases IL-13Rα1, modulating the IL-13 pathway and the switching between classic and alternatively activated macrophages (74). Macrophage polarization is transcriptionally controlled by either miR-146b or miR-34a, directing an M1 macrophage polarization, whereas miR-18a/miR-34a, miR-130b, or miR-125-5p dictates an M2 macrophage phenotype (75). miR-21 has also been reported to direct macrophage polarization from an M1 phenotype toward an M2 phenotype (75). Alveolar macrophages isolated from a fibrotic mouse model showed significantly increased miR-let-7c levels compared with control and that overexpression of this miRNA regulated macrophage polarization (76). Expression of miR-124 and miR-223 in macrophages has also been reported to contribute to macrophage polarization (77, 78).
Located on the surface of sentinel cells, such as macrophages, TLRs play a critical role in the innate immune system by recognizing pathogen-associated molecular patterns expressed on pathogens and signaling for the production of cytokine to elicit an immune response. These TLRs participate in macrophage activation and have been shown to induce miR-155, miR-146, miR-147, miR-9, and miR-21 (79, 80). An upregulation of miR-21 has been observed in both primary human airway epithelial cells (41) and in an IL-13 transgenic mouse model with the latter study identifying that the observed miR-21 upregulation was through an IL-13Rα1-dependent mechanism (81). This increase in miR-21 was also associated with inhibited Th1 cytokine signaling (41). Using an ovalbumin-induced miR-21 deficient mouse model, Th1 cytokines were found to be increased (82), supporting the contribution of miR-21 in Th2 type immune responses. One study confirmed that miR-21 expression inhibited murine pulmonary inflammation by suppressing TLR2 signaling (81). When secreted from tumor cells, miR-21 and miR-29a have also been reported to interact with TLRs, specifically TLR7 and TLR8, respectively (83). Upon lipopolysaccharide (LPS) stimulation, miR-146a/b was shown to be induced and predicted to negatively regulate TLR and cytokine signaling (72).
Macrophage surface activation is induced in the presence of overexpressed miR-125b (84) and in an eosinophilic rhinosinusitis animal model, miR-125b is increased resulting in increased interferon gamma and a Th1 type immune response (85). miR-19a has also been shown to be critical in regulating Th1 type responses through the production of interferon gamma following antigen stimulation in a mouse model (69). Upregulation of miR-19a also caused increased inflammation and promoted a Th2 type response (55). miR-19a is a member of the miR-17–92 cluster, which has been reported to be upregulated during T-cell activation (69, 86). Th17 cell differentiation has also been shown to be regulated by the miR-106-363 cluster (87) and in an experimental autoimmune encephalomyelitis model, Th17 cell-mediated inflammation was shown to be induced by both miR-326 and miR-21 (88, 89).
In summary, multiple studies have characterized the role of miRNAs on immune processes in a variety of diverse animal models of inflammation, but few studies have evaluated the miRNA profiles following fungal exposures (Figure 1). Investigation of these miRNA profiles could provide insight into the immune mechanisms and regulatory pathways involved in the host response to fungal exposure.
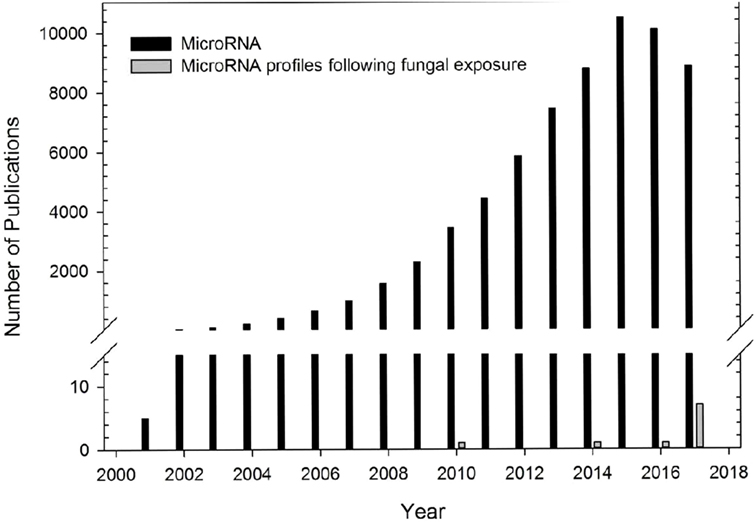
Figure 1. Number of microRNA (miRNA) publications. Number of publications examining miRNA profiles from different diseased models (black bars) or following fungal exposure (gray bars). Data obtained from PubMed search using term “microRNA” for publications using different diseased models. One study in 1972 was not included. Number of publications using a fungal exposed model were obtained from studies included in this review.
Several research studies have focused on the miRNA profiles following acute or chronic fungal exposures (Figure 1). Table 1 shows 10 studies that have preliminarily characterized differentially expressed miRNAs following exposure to five clinically relevant fungal species including A. fumigatus (22, 90, 91), C. albicans (22, 92–94), C. neoformans (95), P. brasiliensis (96, 97), and S. chartarum (98). The paucity of research investigating the regulation of miRNAs on pulmonary and systemic responses to fungal exposure highlights the need for research examining the role miRNAs play in the immunological mechanisms associated with endemic, opportunistic, and environmental fungal exposures.
Paracoccidioidomycosis, caused by the dimorphic fungus P. brasiliensis, is a public health burden in Latin America (117). This fungus can be isolated in the form of yeast from infected individuals and armadillos, and has also been sporadically isolated from soil, dog food, and bat feces (118, 119). The disease begins with the inhalation of spores into the lungs that germinate into yeast and cause a primary lung infection or disseminate systemically resulting in oral and cutaneous lesions. To date, two studies have evaluated differentially expressed miRNAs following P. brasiliensis exposure in a murine model and in a human model. Turini Gonzales Marioto et al. (97) evaluated the miRNA profiles in mice intravenously administered P. brasiliensis and showed that the most upregulated miRNAs at 28 days included miR-126a-5p, miR-340-5p, miR-30b-5p, miR-19b-3p, miR-221-3p, miR-20a-5p, miR-130a-3p, and miR-301a-3p, whereas after 56 days, miRNAs from the let-7 family, as well as miR-26b-5p, and miR-369-3p were the greatest upregulated miRNAs (97). The only miRNA that was upregulated at both time points was miR-466k (Table 1). This study identified differentially expressed miRNAs that are known to contribute to the immune response through T cell function and proliferation, as well as monocyte and erythrocyte differentiation. The contribution of miR-466k on the immune response is unknown; however, this miRNA has been identified in prostate cancer and graft rejection studies (120, 121). Another study examined the miRNA profile in the serum of human patients infected with P. brasiliensis and found that of the 752 miRNAs analyzed, 8 were differentially expressed (96). The upregulated miRNAs included miR-132-3p, miR-604, miR-186-5p, miR-29b-3p, miR-125b-5p, miR-376c-3p, and miR-30b-5p, where the only downregulated miRNA was miR-423-3p (Table 1). These miRNAs are known to mediate macrophage polarization or are involved in TLR2 signaling, indicative of a Th1 immune response. Interestingly, both studies reported an upregulation in miR-30b-5p, suggesting a possible biomarker for P. brasiliensis infection.
Aspergillus fumigatus is a commonly encountered pathogenic fungal species and is often found in the soil, occupational environments [i.e., biowaste containment facilities (122, 123)] or indoor environments [i.e., hospitals (124, 125)]. Inhalation of A. fumigatus unicellular spores can result in varying degrees of infection, known as aspergillosis, depending on the preexisting conditions of the host. miR-132 has been shown to be induced in human monocytes and dendritic cells following stimulation with A. fumigatus compared with control, LPS (90). These datasets suggest a Th2-mediated response, which is further supported by other recent animal models of inhalation exposure to A. fumigatus (91).
Candidiasis, an infection caused by several endogenous Candida species, results in varying symptoms depending on the site of infection (126, 127). Candidiasis is among the most common opportunistic fungal infections localized in the gastrointestinal tract (thrush), occluded regions of the hands, feet, and groin, or can develop into invasive candidiasis and disseminate systemically in the blood (candidemia), heart, brain, eyes, and bones. Invasive candidiasis is the most common type of fungal infection in critically ill patients, with approximately 46,000 healthcare-associated cases occurring each year in the United States (126–128). Although Candida infections are typically resolved by antifungal therapy, some Candida species are resistant or are becoming resistant, such as C. auris (129). In murine macrophages stimulated by 106 cells/mL heat killed C. albicans, miR-146a and miR-155, as well as miR-455 and miR-125a were upregulated (92), indicative of the involvement of these miRNAs in macrophage polarization.
Similar to the findings of Monk et al. (92), miR-146a was also shown to be upregulated in human monocytic THP-1 cells exposed to C. neoformans, inhibiting nuclear factor-κB activation and the release of inflammatory cytokines (95). C. neoformans is one of two pathogenic Cryptococcus species, and along with C. gatti, typically live in the soil surrounding trees and are capable of causing infection following inhalation. Exposure to Cryptococcus species usually causes adverse respiratory health effects; however, it can affect other parts of the body such as the brain, known as cryptococcal meningitis (130, 131).
In a model of mixed fungal exposure, Dix et al. co-cultured human monocyte-derived dendritic cells with A. fumigatus, C. albicans, and LPS and showed that differentially expressed miRNAs were increased after 6 and 12 h, with a stronger regulation observed after 12 h (22). Twenty six miRNAs were identified to be differently expressed in response to the exposure. The authors also showed that strongly modified miRNAs after exposure to fungi clustered separately from the strongly modified miRNAs exposed to LPS. This clustering pattern suggests that examination of miRNA profiles could distinguish between fungal and bacterial exposure. For example, miR-132 and miR-212-5p were specific to fungal exposure at 6 h time point, whereas miR-132, miR-212, and miR-129-5p were specific to fungal exposure at 12 h time point.
Critical to antifungal innate immunity, Dectin-1 is a surface receptor that recognizes (1,3)-β-D-glucan found on the cell wall of germinating conidia (91, 132). To date, several studies have attempted to explore the regulatory mechanisms involving miRNAs that underlie Dectin-1 associated immune responses. In a murine model of subchronic A. fumigatus inhalation exposure, Croston et al. showed that significantly downregulated miR-29a-3p was predicted to regulate C-type lectin domain family 7 member A, the gene that codes for Dectin-1 (91). A recent study found that following exposure to C. albicans, Dectin-1 is required for the upregulation of miR-155 in murine macrophages (93). Along with an increase in miR-30-5p and miR-210-3p in THP-1 cells treated with β-glucan isolated from C. albicans, Du et al. found that miR-146a was increased upon Dectin-1 stimulation and negatively regulated the resultant inflammatory response (94). Results from the Croston et al. study using a murine model of subchronic A. fumigatus inhalation exposure also determined that significantly downregulated miR-23b-3p and miR-145a-5p was predicted to regulate the mannose receptor gene, Mrc1 (data not reported).
In order to elucidate the influence of germination on the ensuing immune response, Croston et al. utilized an acoustical generator system to deliver dry fungal spores to mice housed in a multi-animal nose-only inhalation chamber (91). This murine inhalation model reproduces exposures that could be encountered in contaminated indoor or occupational environments (133). Furthermore, this study included a heat inactivated conidia group that was used as a biological control to examine the influence of germination on the miRNA profiles. Along with a downregulation of miR-29a-3p, miR-23b-3p, a miRNA predicted to target SMAD2, as well as genes involved in IL-13 and IL-33 responses, was also downregulated following subchronic exposure to a dry aerosol containing viable A. fumigatus conidia (91). Furthermore, out of 415 miRNAs detected, approximately 50% were altered in mice exposed to viable versus heat inactivated conidia 48 h post fungal exposure. Taken together, these results demonstrate that A. fumigatus germination is an important variable that can lead to the induction of allergic inflammation in the lungs, potentially through an IL-13/IL-33-driven mechanism.
Stachybotrys chartarum is a hydrophilic fungal species prevalent in water infiltrated occupational and residential environments. Exposure to S. chartarum is currently of heightened public health interest following recent natural disasters, such as floods and hurricanes, that can lead to the contamination of indoor building materials. Following the consensus reports published by the IOM and WHO, the immunological mechanisms that contribute to the host response to fungal contaminant exposure require further elucidation. Recent studies have attempted to characterize critical interactions that influence these pulmonary immunological responses. Croston et al. found that miR-21a was the only miRNA upregulated in murine whole lung homogenate 48 h following subchronic exposure to S. chartarum (98) (Figure 2). Although miR-21a is known to promote a Th2 phenotype, a more dominant Th1 phenotype was evident. Since then, miR-706 was also discovered to be upregulated at the same time point. Interestingly, out of 468 miRNA evaluated, only 2 were upregulated with no downregulated miRNAs. These preliminary results suggest that miRNA regulation mechanisms induced by S. chartarum vary from A. fumigatus in these studies using the same exposure system and the pulmonary immunological responses to this species require further evaluation.
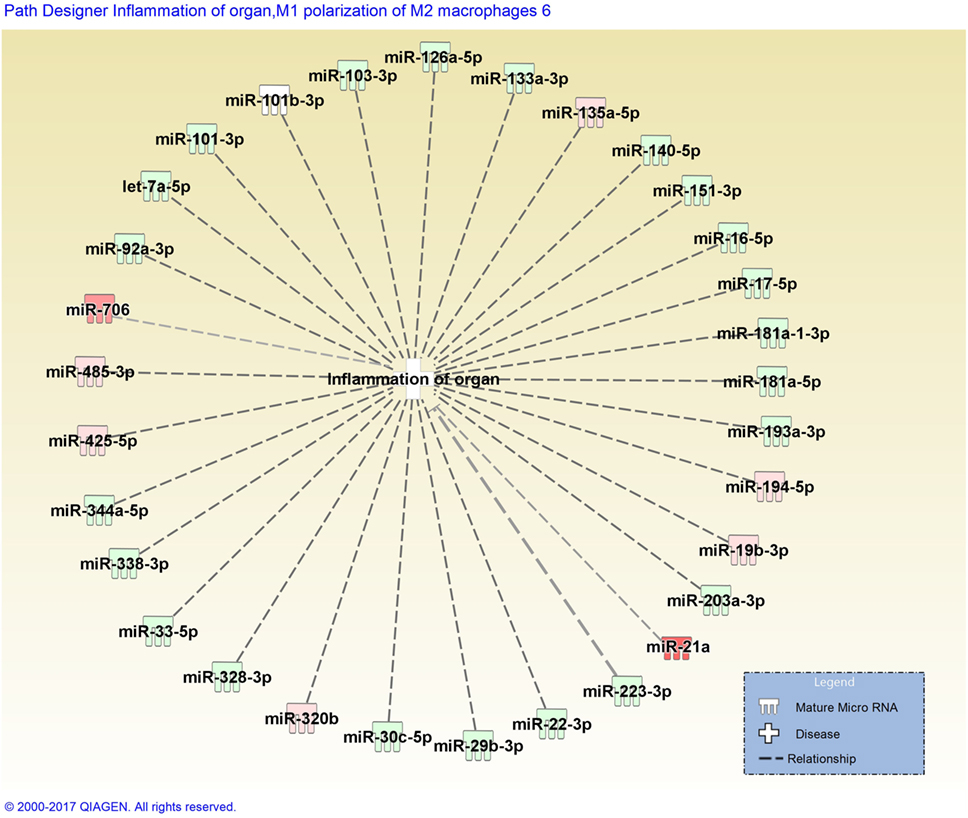
Figure 2. Disease network map generated by Ingenuity Pathway Analysis depicting miRNAs involved in the inflammation of organs. miRNAs are color-coded (red or green for up- and downregulation, respectively) for the expression of miRNAs in Stachybotrys chartarum exposed versus control at 13 weeks, 48 h post-exposure. Gray dotted lines represent predicted regulated relationship.
Figure 2 depicts a disease pathway generated by Ingenuity Pathway Analysis (IPA) that includes predicted miRNAs involved in the inflammation of organs. Once miRNA data are uploaded into IPA, the integrated knowledge base predicts associations between miRNAs from the dataset and different disease pathways or biological functions. These predictions are primarily based on previously publish datasets derived from a broad diversity of animal models. The miRNA dataset included in Figure 2 was obtained from murine whole lung homogenate 48 h following subchronic exposure to S. chartarum (98). When analyzing the top diseases and biological functions of the miRNAs included in the dataset, a handful were predicted to be involved in an inflammatory response, specifically in organ inflammation, illustrated in Figure 2. The miRNAs are color-coded depending on the respective expression level (red or green for up- and downregulation, respectively). The absence of confirmed associations between miRNAs, evidenced by gray dotted lines, supports the lack of miRNA profile studies following fungal exposure.
To date, only a handful of studies have examined the altered miRNA profiles following fungal exposure; therefore, more researches are required to fully understand the mechanistic influence miRNAs have on the immune response. With the increased interest in studying miRNAs, methodological approaches are becoming more advanced by using next-generation sequencing methods that examine miRNA profiles in more depth and at a higher precision compared with miRNA arrays. Once the more influential miRNAs are identified, strategies can be developed in order to manipulate the host response. With the use of transgenic or knockout animal models, the functionality of miRNAs or genes can be elucidated; however, the manipulation of the genome may in fact alter normal miRNA production or function, contributing to the phenotype of the disease (134). Targeting miRNAs that are upregulated or replacing the expression of miRNAs that are downregulated are potential strategies that could be tested in animal models as a new therapeutic strategy to treat fungal infections and diseases. This targeting strategy could be completed through the use of an anti-miR or a miRNA mimic (135), and may allow for the manipulation of a group of genes or proteins that participate in the progression of the infection or disease. Ultimately, these targeting strategies will help bridge the knowledge gap between the identification of miRNAs and the host responses to fungal exposure, potentially leading to advanced therapeutics to combat adverse effects resulting from exposure to pathogenic fungi.
In this review, the identification and influence of miRNAs on the host immune responses following fungal exposure were examined. Compared with existing datasets examining miRNA profiles in allergic and inflammatory models, some common differentially expressed miRNAs were identified in fungal exposed models. Influential miRNAs altered in different disease models, such as miR-132, functions to maintain a normal hematopoietic output during an immune response and regulates genes at the beginning of an immune response to regain homeostasis of the immune system. Other common miRNAs identified in multiple inflammatory disease models, including miR-155 and miR-146a, regulates critical genes involved in the host defense system through opposing mechanisms. For example, miR-146a is known to decrease cytokine production and inhibits Th1 cells following an inflammatory stimulus, as well as induces alternative activation of macrophages, whereas miR-155 stimulates both Th1 and Th17 immune responses and induces classical activation of macrophages. Taken together, these miRNAs act in concert to defend the host from infection. In addition to common miRNAs identified in multiple diseased models, the miRNAs that were observed to be differentially expressed specifically in fungal exposed models could potentially serve as biomarkers for fungal exposures.
Recent discoveries in miRNA biology have heightened the research community’s interest in examining the altered genetic profiles in different disease models; however, only a few studies have examined miRNA profiles following fungal exposure. As such, the description of the immune responses to the corresponding miRNAs listed in Table 1 was not all compiled from fungal exposure studies due to the lack of research examining the influence of miRNAs on the immune responses following fungal exposure. Although advancements made in this field have helped elucidate mechanisms underlying host responses to a variety of infections and diseases, further examination of miRNA profiles, specifically in fungal exposed models, is required in order to provide greater mechanistic insight into the immunological response to clinically and environmentally relevant fungal species.
TC and BG designed the manuscript; TC drafted the manuscript and prepared figures/tables; TC, AL, DB, and BG revised and edited the manuscript; TC, AL, DB, and BG approved the final version of the manuscript and agree to be accountable for the content of the work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention. The author declares no conflict of interest.
Clec7a, C-type lectin domain family 7 member A; IL, interleukin; LPS, lipopolysaccharide; RNA, ribose nucleic acid; RNAi, RNA interference; mRNA, messenger RNA; miRNA, microRNA; TLRs, Toll-like receptors.
1. Hawksworth DL. The magnitude of fungal diversity: the 1.5 million species estimate revised. Mycol Res (2001) 105:1422–32. doi:10.1017/S0953756201004725
2. Cannon R, Firth N. Fungi and fungal infections of the oral cavity. In: Lamont R, Burne R, Lantz M, LeBlanc D, editors. Oral Microbiology and Immunology Washington, DC: ASM Press (2006). p. 333–48.
3. Eduard W. Fungal spores: a critical review of the toxicological and epidemiological evidence as a basis for occupational exposure limit setting. Crit Rev Toxicol (2009) 39(10):799–864. doi:10.3109/10408440903307333
4. Benedict K, Park BJ. Invasive fungal infections after natural disasters. Emerg Infect Dis (2014) 20(3):349–55. doi:10.3201/eid2003.131230
5. Klein BS, Tebbets B. Dimorphism and virulence in fungi. Curr Opin Microbiol (2007) 10(4):314–9. doi:10.1016/j.mib.2007.04.002
6. Gauthier GM. Dimorphism in fungal pathogens of mammals, plants, and insects. PLoS Pathog (2015) 11(2):e1004608. doi:10.1371/journal.ppat.1004608
7. Bajwa SJ, Kulshrestha A. Fungal infections in intensive care unit: challenges in diagnosis and management. Ann Med Health Sci Res (2013) 3(2):238–44. doi:10.4103/2141-9248.113669
9. WHO. WHO Guidelines for Indoor Air Quality: Dampness and Mould. Geneva: WHO Regional Office for Europe (2009).
10. Mendell MJ, Mirer AG, Cheung K, Tong M, Douwes J. Respiratory and allergic health effects of dampness, mold, and dampness-related agents: a review of the epidemiologic evidence. Environ Health Perspect (2011) 119(6):748–56. doi:10.1289/ehp.1002410
11. Quansah R, Jaakkola MS, Hugg TT, Heikkinen SA, Jaakkola JJ. Residential dampness and molds and the risk of developing asthma: a systematic review and meta-analysis. PLoS One (2012) 7(11):e47526. doi:10.1371/journal.pone.0047526
12. Kuhn DM, Ghannoum MA. Indoor mold, toxigenic fungi, and Stachybotrys chartarum: infectious disease perspective. Clin Microbiol Rev (2003) 16(1):144–72. doi:10.1128/CMR.16.1.144-172.2003
13. Johanning E, Biagini R, Hull D, Morey P, Jarvis B, Landsbergis P. Health and immunology study following exposure to toxigenic fungi (Stachybotrys chartarum) in a water-damaged office environment. Int Arch Occup Environ Health (1996) 68(4):207–18. doi:10.1007/s004200050052
14. Andersson MA, Nikulin M, Koljalg U, Andersson MC, Rainey F, Reijula K, et al. Bacteria, molds, and toxins in water-damaged building materials. Appl Environ Microbiol (1997) 63(2):387–93.
15. Pestka JJ, Yike I, Dearborn DG, Ward MD, Harkema JR. Stachybotrys chartarum, trichothecene mycotoxins, and damp building-related illness: new insights into a public health enigma. Toxicol Sci (2008) 104(1):4–26. doi:10.1093/toxsci/kfm284
16. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi:10.1016/S0092-8674(04)00045-5
17. Li L-C, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A (2006) 103(46):17337–42. doi:10.1073/pnas.0607015103
18. Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol (2007) 3(3):166–73. doi:10.1038/nchembio860
19. Place RF, Li L-C, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A (2008) 105(5):1608–13. doi:10.1073/pnas.0707594105
20. Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol (2013) 132(1):3–13; quiz 14. doi:10.1016/j.jaci.2013.04.039
21. Lu TX, Lim E-J, Wen T, Plassard AJ, Martin LJ, Aronow BJ, et al. miR-375 is down-regulated in epithelial cells after IL-13 stimulation and regulates an IL-13 induced epithelial transcriptome. Mucosal Immunol (2012) 5(4):388–96. doi:10.1038/mi.2012.16
22. Dix A, Czakai K, Leonhardt I, Schäferhoff K, Bonin M, Guthke R, et al. Specific and novel microRNAs are regulated as response to fungal infection in human dendritic cells. Front Microbiol (2017) 8:270. doi:10.3389/fmicb.2017.00270
23. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell (2003) 115(7):787–98. doi:10.1016/S0092-8674(03)01018-3
24. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell (2005) 120(1):15–20. doi:10.1016/j.cell.2004.12.035
25. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell (2009) 136(4):642–55. doi:10.1016/j.cell.2009.01.035
26. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science (2007) 318(5858):1931–4. doi:10.1126/science.1149460
27. Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet (2005) 37(5):495–500. doi:10.1038/ng1536
28. Rajewsky N. MicroRNA target predictions in animals. Nat Genet (2006) 38:S8–13. doi:10.1038/ng1798
29. Bartel DP. MicroRNA target recognition and regulatory functions. Cell (2009) 136(2):215–33. doi:10.1016/j.cell.2009.01.002
30. Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res (2009) 19(1):92–105. doi:10.1101/gr.082701.108
31. van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest (2007) 117(9):2369–76. doi:10.1172/JCI33099
32. Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C, et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell (2010) 9(2):291–6. doi:10.1111/j.1474-9726.2010.00549.x
33. Leopold JA, Maron BA. Molecular mechanisms of pulmonary vascular remodeling in pulmonary arterial hypertension. Int J Mol Sci (2016) 17(5):761. doi:10.3390/ijms17050761
34. Posch W, Heimdörfer D, Wilflingseder D, Lass-Flörl C. Invasive candidiasis: future directions in non-culture based diagnosis. Expert Rev Anti Infect Ther (2017) 15:829–38. doi:10.1080/14787210.2017.1370373
35. Frost RJA, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A (2011) 108(52):21075–80. doi:10.1073/pnas.1118922109
36. Jen-Liang S, Pai-Sheng C, Gunnar J, Min-Liang K. Function and regulation of let-7 family microRNAs. Microrna (2012) 1(1):34–9. doi:10.2174/2211536611201010034
37. Jun-Hao ET, Gupta RR, Shyh-Chang N. Lin28 and let-7 in the metabolic physiology of aging. Trends Endocrinol Metab (2016) 27(3):132–41. doi:10.1016/j.tem.2015.12.006
38. Brennan E, Wang B, McClelland A, Mohan M, Marai M, Beuscart O, et al. Protective effect of let-7 miRNA family in regulating inflammation in diabetes-associated atherosclerosis. Diabetes (2017) 66(8):2266. doi:10.2337/db16-1405
39. Pal AS, Kasinski AL. Animal models to study microRNA function. Adv Cancer Res (2017) 135:53–118. doi:10.1016/bs.acr.2017.06.006
40. Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem (2010) 285(39):30139–49. doi:10.1074/jbc.M110.145698
41. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol (2009) 182(8):4994–5002. doi:10.4049/jimmunol.0803560
42. Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog (2009) 5(12):e1000681. doi:10.1371/journal.ppat.1000681
43. Eulalio A, Schulte L, Vogel J. The mammalian microRNA response to bacterial infections. RNA Biol (2012) 9(6):742–50. doi:10.4161/rna.20018
44. Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, et al. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog (2013) 9(4):e1003248. doi:10.1371/journal.ppat.1003248
45. Lu S, Mukkada VA, Mangray S, Cleveland K, Shillingford N, Schorl C, et al. MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS One (2012) 7(7):e40676. doi:10.1371/journal.pone.0040676
46. Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol (2012) 129(4):1064–75.e9. doi:10.1016/j.jaci.2012.01.060
47. Lu TX, Lim E-J, Besse JA, Itskovich S, Plassard AJ, Fulkerson PC, et al. miR-223 deficiency increases eosinophil progenitor proliferation. J Immunol (2013) 190(4):1576–82. doi:10.4049/jimmunol.1202897
48. Lu TX, Lim E-J, Itskovich S, Besse JA, Plassard AJ, Mingler MK, et al. Targeted ablation of miR-21 decreases murine eosinophil progenitor cell growth. PLoS One (2013) 8(3):e59397. doi:10.1371/journal.pone.0059397
49. Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of T(H)2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A (2009) 106(44):18704–9. doi:10.1073/pnas.0905063106
50. Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol (2011) 128(1):160–167.e4. doi:10.1016/j.jaci.2011.04.005
51. Sharma A, Kumar M, Ahmad T, Mabalirajan U, Aich J, Agrawal A, et al. Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. J Appl Physiol (1985) (2012) 113(3):459–64. doi:10.1152/japplphysiol.00001.2012
52. Izzotti A, Bagnasco M, Cartiglia C, Longobardi M, Balansky RM, Merello A, et al. Chemoprevention of genome, transcriptome, and proteome alterations induced by cigarette smoke in rat lung. Eur J Cancer (2005) 41(13):1864–74. doi:10.1016/j.ejca.2005.04.011
53. Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J (2009) 23(3):806–12. doi:10.1096/fj.08-121384
54. Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J (2009) 23(9):3243–50. doi:10.1096/fj.09-135251
55. Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol (2014) 15(12):1162–70. doi:10.1038/ni.3026
56. Liu F, Qin HB, Xu B, Zhou H, Zhao DY. Profiling of miRNAs in pediatric asthma: upregulation of miRNA-221 and miRNA-485-3p. Mol Med Rep (2012) 6(5):1178–82. doi:10.3892/mmr.2012.1030
57. Anderson SE, Beezhold K, Lukomska E, Richardson J, Long C, Anderson K, et al. Expression kinetics of miRNA involved in dermal toluene 2,4-diisocyanate sensitization. J Immunotoxicol (2014) 11(3):250–9. doi:10.3109/1547691X.2013.835891
58. Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev (2009) 8(6):520–4. doi:10.1016/j.autrev.2009.01.008
59. Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell (2009) 136(1):26–36. doi:10.1016/j.cell.2008.12.027
60. Gracias DT, Katsikis PD. MicroRNAs: key components of immune regulation. In: Pulendran B, Katsikis PD, Schoenberger SP, editors. Crossroads between Innate and Adaptive Immunity III. New York, NY: Springer (2011). p. 15–26.
61. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science (2004) 303(5654):83–6. doi:10.1126/science.1091903
62. Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. miR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell (2007) 131(1):146–59. doi:10.1016/j.cell.2007.07.021
63. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with elevated miR-17-92 expression in lymphocytes. Nat Immunol (2008) 9(4):405–14. doi:10.1038/ni1575
64. Lu L-F, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology (2009) 127(3):291–8. doi:10.1111/j.1365-2567.2009.03092.x
65. Li Q-J, Chau J, Ebert PJR, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell (2007) 129(1):147–61. doi:10.1016/j.cell.2007.03.008
66. Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai T-H, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol (2005) 6(8):R71–71. doi:10.1186/gb-2005-6-8-r71
67. Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, et al. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One (2007) 2(10):e1020. doi:10.1371/journal.pone.0001020
68. Jeker LT, Bluestone JA. MicroRNA regulation of T-cell differentiation and function. Immunol Rev (2013) 253(1):65–81. doi:10.1111/imr.12061
69. Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood (2011) 118(20):5487–97. doi:10.1182/blood-2011-05-355644
70. Haasch D, Chen Y-W, Reilly RM, Grace Chiou X, Koterski S, Smith ML, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol (2002) 217(1):78–86. doi:10.1016/S0008-8749(02)00506-3
71. Pedersen I, David M. MicroRNAs in the immune response. Cytokine (2008) 43(3):391–4. doi:10.1016/j.cyto.2008.07.016
72. Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A (2006) 103(33):12481–6. doi:10.1073/pnas.0605298103
73. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (2007) 316(5824):608–11. doi:10.1126/science.1139253
74. Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor α1 (IL13Rα1). J Biol Chem (2011) 286(3):1786–94. doi:10.1074/jbc.M110.169367
75. Roy S. miRNA in macrophage development and function. Antioxid Redox Signal (2016) 25(15):795–804. doi:10.1089/ars.2016.6728
76. Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol (2013) 190(12):6542–9. doi:10.4049/jimmunol.1202496
77. Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, et al. A novel regulator of macrophage activation clinical perspective miR-223 in obesity-associated adipose tissue inflammation. Circulation (2012) 125(23):2892–903. doi:10.1161/CIRCULATIONAHA.111.087817
78. Veremeyko T, Siddiqui S, Sotnikov I, Yung A, Ponomarev ED. IL-4/IL-13-dependent and independent expression of miR-124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PLoS One (2013) 8(12):e81774. doi:10.1371/journal.pone.0081774
79. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol (2010) 10(2):111–22. doi:10.1038/nri2708
80. O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of toll-like receptor signalling. Nat Rev Immunol (2011) 11(3):163–75. doi:10.1038/nri2957
81. Case SR, Martin RJ, Jiang D, Minor MN, Chu HW. MicroRNA-21 inhibits toll-like receptor 2 agonist-induced lung inflammation in mice. Exp Lung Res (2011) 37(8):500–8. doi:10.3109/01902148.2011.596895
82. Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol (2011) 187(6):3362–73. doi:10.4049/jimmunol.1101235
83. Chen X, Liang H, Zhang J, Zen K, Zhang CY. MicroRNAs are ligands of toll-like receptors. RNA (2013) 19(6):737–9. doi:10.1261/rna.036319.112
84. Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O’Connell RM, et al. MicroRNA-125b potentiates macrophage activation. J Immunol (2011) 187(10):5062–8. doi:10.4049/jimmunol.1102001
85. Zhang XH, Zhang YN, Li HB, Hu CY, Wang N, Cao PP, et al. Overexpression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med (2012) 185(2):140–51. doi:10.1164/rccm.201103-0456OC
86. Podshivalova K, Salomon DR. MicroRNA regulation of T lymphocyte immunity: modulation of molecular networks responsible for T cell activation, differentiation and development. Crit Rev Immunol (2013) 33(5):435–76. doi:10.1615/CritRevImmunol.2013006858
87. Kastle M, Bartel S, Geillinger-Kastle K, Irmler M, Beckers J, Ryffel B, et al. MicroRNA cluster 106a~363 is involved in T helper 17 cell differentiation. Immunology (2017) 152:402–13. doi:10.1111/imm.12775
88. Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med (2008) 205(9):1993–2004. doi:10.1084/jem.20081062
89. Murugaiyan G, da Cunha AP, Ajay AK, Joller N, Garo LP, Kumaradevan S, et al. MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J Clin Invest (2015) 125(3):1069–80. doi:10.1172/JCI74347
90. Das Gupta M, Fliesser M, Springer J, Breitschopf T, Schlossnagel H, Schmitt AL, et al. Aspergillus fumigatus induces microRNA-132 in human monocytes and dendritic cells. Int J Med Microbiol (2014) 304(5–6):592–6. doi:10.1016/j.ijmm.2014.04.005
91. Croston TL, Nayak AP, Lemons AR, Goldsmith WT, Gu JK, Germolec DR, et al. Influence of Aspergillus fumigatus conidia viability on murine pulmonary microRNA and mRNA expression following subchronic inhalation exposure. Clin Exp Allergy (2016) 46(10):1315–27. doi:10.1111/cea.12783
92. Monk CE, Hutvagner G, Arthur JSC. Regulation of miRNA transcription in macrophages in response to Candida albicans. PLoS One (2010) 5(10):e13669. doi:10.1371/journal.pone.0013669
93. Agustinho DP, de Oliveira MA, Tavares AH, Derengowski L, Stolz V, Guilhelmelli F, et al. Dectin-1 is required for miR155 upregulation in murine macrophages in response to Candida albicans. Virulence (2017) 8(1):41–52. doi:10.1080/21505594.2016.1200215
94. Du L, Chen X, Duan Z, Liu C, Zeng R, Chen Q, et al. miR-146a negatively regulates dectin-1-induced inflammatory responses. Oncotarget (2017) 8(23):37355–66. doi:10.18632/oncotarget.16958
95. Chen H, Jin Y, Chen H, Liao N, Wang Y, Chen J. MicroRNA-mediated inflammatory responses induced by Cryptococcus neoformans are dependent on the NF-kappaB pathway in human monocytes. Int J Mol Med (2017) 39(6):1525–32. doi:10.3892/ijmm.2017.2951
96. De Lacorte Singulani J, De Fátima Da Silva J, Gullo FP, Costa MC, Fusco-Almeida AM, Enguita FJ, et al. Preliminary evaluation of circulating microRNAs as potential biomarkers in paracoccidioidomycosis. Biomed Rep (2017) 6(3):353–7. doi:10.3892/br.2017.849
97. Turini Gonzales Marioto D, Navarro Dos Santos Ferraro AC, Goulart de Andrade F, Barros Oliveira M, Itano EN, Petrofeza S, et al. Study of differential expression of miRNAs in lung tissue of mice submitted to experimental infection by Paracoccidioides brasiliensis. Med Mycol (2017) 55:774–84. doi:10.1093/mmy/myw135
98. Croston TL, Nayak AP, Lemons AR, Goldsmith WT, Germolec DM, Beezhold DH, et al. Pulmonary immune response following subchronic Stachybotrys chartarum exposure. J Allergy ClinImmunol (2017) 139(2):AB75. doi:10.1016/j.jaci.2016.12.291
99. Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy ClinImmunol (2011) 128(5):1077–85.e1–10. doi:10.1016/j.jaci.2011.04.034
100. Liu G, Abraham E. MicroRNAs in immune response and macrophage polarization. Arterioscler Thromb Vasc Biol (2013) 33(2):170–7. doi:10.1161/ATVBAHA.112.300068
101. Essandoh K, Li Y, Huo J, Fan G-C. miRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock (2016) 46(2):122–31. doi:10.1097/SHK.0000000000000604
102. Rivera A, Barr T, Rais M, Engelmann F, Messaoudi I. MicroRNAs regulate host immune response and pathogenesis during influenza infection in rhesus macaques. Viral Immunol (2016) 29(4):212–27. doi:10.1089/vim.2015.0074
103. Zhang N, Bevan MJ. Dicer controls CD8(+) T-cell activation, migration, and survival. Proc Natl Acad Sci U S A (2010) 107(50):21629–34. doi:10.1073/pnas.1016299107
104. Nahid MA, Yao B, Dominguez-Gutierrez PR, Kesavalu L, Satoh M, Chan EK. Regulation of TLR2-mediated tolerance and cross-tolerance through IRAK4 modulation by miR-132 and miR-212. J Immunol (2013) 190(3):1250–63. doi:10.4049/jimmunol.1103060
105. Testa U, Pelosi E, Castelli G, Labbaye C. miR-146 and miR-155: two key modulators of immune response and tumor development. Noncoding RNA (2017) 3(3):22. doi:10.3390/ncrna3030022
106. Suman S, Jones-Reed DZZ, Schmidt ML, Clark GJ, Klinge C, Barve S, et al. Alteration of miR-186 expression modifies inflammatory markers in normal epithelial and prostate cancer cell models. FASEB J (2017) 31(1 Suppl):757.716. doi:10.1096/fasebj.31.1_supplement.757.16
107. Liu S-Q, Jiang S, Li C, Zhang B, Li Q-J. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros family zinc finger 4 to promote T(H)17-mediated inflammation. J Biol Chem (2014) 289(18):12446–56. doi:10.1074/jbc.M114.550723
108. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol (2007) 9(7):775–87. doi:10.1038/ncb1613
109. Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res (2016) 35(1):103. doi:10.1186/s13046-016-0375-2
110. Quinn SR, O’Neill LA. A trio of microRNAs that control toll-like receptor signalling. Int Immunol (2011) 23(7):421–5. doi:10.1093/intimm/dxr034
111. Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol (2016) 16(5):279–94. doi:10.1038/nri.2016.40
112. Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-[gamma]. Nat Immunol (2011) 12(9):861–9. doi:10.1038/ni.2073
113. Huang L, Liu Y, Wang L, Chen R, Ge W, Lin Z, et al. Down-regulation of miR-301a suppresses pro-inflammatory cytokines in toll-like receptor-triggered macrophages. Immunology (2013) 140(3):314–22. doi:10.1111/imm.12139
114. Vegh P, Magee DA, Nalpas NC, Bryan K, McCabe MS, Browne JA, et al. MicroRNA profiling of the bovine alveolar macrophage response to Mycobacterium bovis infection suggests pathogen survival is enhanced by microRNA regulation of endocytosis and lysosome trafficking. Tuberculosis (2015) 95(1):60–7. doi:10.1016/j.tube.2014.10.011
115. Warg LA, Oakes JL, Burton R, Neidermyer AJ, Rutledge HR, Groshong S, et al. The role of the E2F1 transcription factor in the innate immune response to systemic LPS. Am J Physiol Lung Cell Mol Physiol (2012) 303(5):L391–400. doi:10.1152/ajplung.00369.2011
116. Wong JJL, Ritchie W, Gao D, Lau KA, Gonzalez M, Choudhary A, et al. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J Hematol Oncol (2014) 7:42. doi:10.1186/1756-8722-7-42
117. Goldani LZ, Wirth F. Animal models and antifungal agents in paracoccidioidomycosis: an overview. Mycopathologia (2017) 182(7):633–43. doi:10.1007/s11046-017-0130-z
118. Terçarioli GR, Bagagli E, Reis GM, Theodoro RC, Bosco Sde M, Macoris SA, et al. Ecological study of Paracoccidioides brasiliensis in soil: growth ability, conidia production and molecular detection. BMC Microbiol (2007) 7(1):92. doi:10.1186/1471-2180-7-92
119. Bagagli E, Theodoro RC, Bosco SM, McEwen JG. Paracoccidioides brasiliensis: phylogenetic and ecological aspects. Mycopathologia (2008) 165(4–5):197–207. doi:10.1007/s11046-007-9050-7
120. Seo M, Choi J-S, Rho CR, Joo C-K, Lee SK. MicroRNA miR-466 inhibits lymphangiogenesis by targeting prospero-related homeobox 1 in the alkali burn corneal injury model. J Biomed Sci (2015) 22(1):3. doi:10.1186/s12929-014-0104-0
121. Colden M, Dar AA, Saini S, Dahiya PV, Shahryari V, Yamamura S, et al. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis (2017) 8(1):e2572. doi:10.1038/cddis.2017.15
122. Wéry N. Bioaerosols from composting facilities—a review. Front Cell Infect Microbiol (2014) 4:42. doi:10.3389/fcimb.2014.00042
123. Pearson C, Littlewood E, Douglas P, Robertson S, Gant TW, Hansell AL. Exposures and health outcomes in relation to bioaerosol emissions from composting facilities: a systematic review of occupational and community studies. J Toxicol Environ Health B Crit Rev (2015) 18(1):43–69. doi:10.1080/10937404.2015.1009961
124. Symoens F, Burnod J, Lebeau B, Viviani MA, Piens MA, Tortorano AM, et al. Hospital-acquired Aspergillus fumigatus infection: can molecular typing methods identify an environmental source? J Hosp Infect (2002) 52(1):60–7. doi:10.1053/jhin.2002.1263
125. Kauffman CA, Nicolasora NP. Epidemiology of invasive pulmonary aspergillosis. In: Comarú Pasqualotto A, editor. Aspergillosis: From Diagnosis to Prevention. Dordrecht, Netherlands: Springer (2010). p. 329–44.
126. CDC. Fungal Diseases: Candidiasis. (2017). Available from: https://www.cdc.gov/fungal/diseases/candidiasis/index.html
127. CDC. Fungal Diseases: Invasive Candidiasis Statistics. (2017). Available from: https://www.cdc.gov/fungal/diseases/candidiasis/invasive/statistics.html
128. Epelbaum O, Chasan R. Candidemia in the intensive care unit. Clin Chest Med (2017) 38(3):493–509. doi:10.1016/j.ccm.2017.04.010
129. CDC. Fungal Diseases: General Information about Candida auris. (2016). Available from: https://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-qanda.html
130. CDC. Fungal Diseases: C. gattii Infection. (2015). Available from: https://www.cdc.gov/fungal/diseases/cryptococcosis-gattii/index.html
131. CDC. Fungal Diseases: C. neoformans Infection. (2015). Available from https://www.cdc.gov/fungal/diseases/cryptococcosis-neoformans/index.html
132. Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol (2006) 6(1):33–43. doi:10.1038/nri1745
133. Buskirk AD, Green BJ, Lemons AR, Nayak AP, Goldsmith WT, Kashon ML, et al. A murine inhalation model to characterize pulmonary exposure to dry Aspergillus fumigatus conidia. PLoS One (2014) 9(10):e109855. doi:10.1371/journal.pone.0109855
134. Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol (2014) 51(4):759–74. doi:10.1177/0300985813502820
Keywords: fungal exposure, microRNA, fungi, immune response, inflammatory response
Citation: Croston TL, Lemons AR, Beezhold DH and Green BJ (2018) MicroRNA Regulation of Host Immune Responses following Fungal Exposure. Front. Immunol. 9:170. doi: 10.3389/fimmu.2018.00170
Received: 30 October 2017; Accepted: 19 January 2018;
Published: 07 February 2018
Edited by:
Amariliz Rivera, New Jersey Medical School, United StatesReviewed by:
Agostinho Carvalho, University of Minho, PortugalCopyright: © 2018 Croston, Lemons, Beezhold and Green. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tara L. Croston, eHp1OUBjZGMuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.