- 1University of Bordeaux, CNRS, ImmunoConcept, UMR 5164, Bordeaux, France
- 2University of Bordeaux, INRA, Mycoplasmal and Chlamydial Infections in Humans, EA 3671, Bordeaux, France
- 3Department of Hepato-Gastroenterology, Bordeaux Hospital University Center, Pessac, France
- 4Department of Rheumatology, Bordeaux Hospital University Center, Bordeaux, France
- 5Department of Parasitology and Mycology, Bordeaux Hospital University Center, Bordeaux, France
- 6University of Bordeaux, INSERM, Cardio-Thoracic Research Centre of Bordeaux, U1045, Bordeaux, France
Resident microbiota do not just shape host immunity, they can also contribute to host protection against pathogens and infectious diseases. Previous reviews of the protective roles of the microbiota have focused exclusively on colonization resistance localized within a microenvironment. This review shows that the protection against pathogens also involves the mitigation of pathogenic impact without eliminating the pathogens (i.e., “disease tolerance”) and the containment of microorganisms to prevent pathogenic spread. Protective microorganisms can have an impact beyond their niche, interfering with the entry, establishment, growth, and spread of pathogenic microorganisms. More fundamentally, we propose a series of conceptual clarifications in support of the idea of a “co-immunity,” where an organism is protected by both its own immune system and components of its microbiota.
Introduction
The immune system is never at rest, nor is it ever isolated from its environment. It is in constant interactions with myriads of microbes, which can be pathogenic, commensal, or mutualistic (1–4). Recent work has shed light on the impact of commensal and mutualistic microbes on the development, induction, training, and functioning of the immune system [reviewed in Ref. (5–7)]. It is becoming increasingly clear that the microbiota, i.e., the microorganisms living persistently on and in a host, play a decisive role in shaping an effective host immune system and, vice versa.
Resident microorganisms do not just induce host immunity but can also directly inhibit pathogens (8–14). This phenomenon has been documented across many species—vertebrates, invertebrates, and plants (15, 16). The ability of commensals and mutualists to interfere with pathogen colonization and growth is known as “colonization resistance” (17–19). While the concept of colonization resistance is not new (Box 1), its underlying mechanisms are just being understood. Two main modes of protection have been proposed: resident microorganisms can protect the host either through direct microbe–microbe competition, which involves niche competition or direct antagonism, or by the indirect induction or priming of host metabolism or immunity, thanks to which the host is better protected against pathogen infections.
Box 1. History of colonization resistance.
Since the beginning of the antibiotics era in the 1950s, it became increasingly clear that indigenous microbial flora—generally assumed as the anaerobic autochthonous symbionts—can be a source of resistance against pathogens. Resistance conferred by the indigenous population explains the side effects of antibiotics (e.g., microbial overgrowth and superinfections) as well as the effectiveness of using antibiotics to prepare animal models for pathogen infection (20–22). The main mechanisms were thought to be competition between microbes, either over substrates or through bactericide molecule production (23, 24). When van der Waaij and colleagues (25) coined the term “colonization resistance,” they cited as possible mechanisms an indirect stimulation of gut movement in addition to direct antagonism between microbes, but rejected the involvement of the immune system. Yet, with the discovery of immunoglobulins (Ig) in the gut lumen and mucosa (26, 27), evidence for indirect, host-mediated mechanisms began to emerge: microbes were thought to assist the immune system by preventing Ig degradation or by significantly lowering pathogen load for Ig effectiveness (28, 29). In the meantime, proposals of direct mechanisms expanded (including competition over substrates and adhesion sites, release of antibacterial substances, prevention of translocation, and alteration of microenvironments to favor growth) (30–33). The 1980s and 1990s saw an explosion of research on colonization resistance and gut immune responses against pathogens, as well as the impact of bacterial flora on immune development (34). But only after the 2000s did breakthrough studies decisively demonstrate microbe-induced innate and adaptive immune responses, e.g., defensins (35), IgA specific to the mucosa (36), angiogenins (37), lectins (38), and modulated T-cell populations (39).
Colonization resistance constitutes an expanded sense of traditional protective immunity, as these mechanisms can in fact be considered part of the host’s defensive repertoire. Of course, the protective effects of microbiota components are context-dependent: some microbes can help protect the host in certain circumstances, but are detrimental in others. In light of recent studies, our main claim is that microorganisms, broadly construed, can exert a protective role, and that a key challenge is now to characterize the different ways this protection can occur.
There are two important gaps in the current literature on colonization resistance. First, current understanding of microbe-induced protection is generally limited to colonization resistance against pathogen establishment and growth. Yet the protection against pathogens also involves the mitigation of pathogenic impact without eliminating the pathogens, also called “disease tolerance” (40), and the containment of microorganisms to prevent pathogenic spread.
Second, the protective effects reviewed under colonization resistance are localized to the immediate vicinity of the protective microbes. Of date, little is known about the protective effects of microorganisms against pathogens in distal organs and tissue sites, even though it is established that microorganisms in one organ (mainly the gut) can influence immune responses in other organs (41). The relative lack of research on long-reaching microbiota-mediated protection in part arises from the assumption that microorganisms, once established within their niches, do not spread or move. Another reason could be the assumption that protective long-reaching effects are too weak to be clinically relevant, and thus, not significant enough for therapeutic purposes. Both assumptions, however, are questionable.
In this review, we fill in the two gaps presented above with a systematic classification of protective mechanisms that include a broader range of defensive strategies differing in range (local, systemic, and long-reaching), mode (direct microbe-to-microbe and indirect host mediated), and effect (resistance, containment, and disease tolerance) (Figure 1). Addressing the first gap, we expand the effects of protection beyond colonization resistance to include containment of microorganisms and suppression of pathogenic impacts. We address the second gap by organizing existing evidence of the long-reaching of protective microbes.
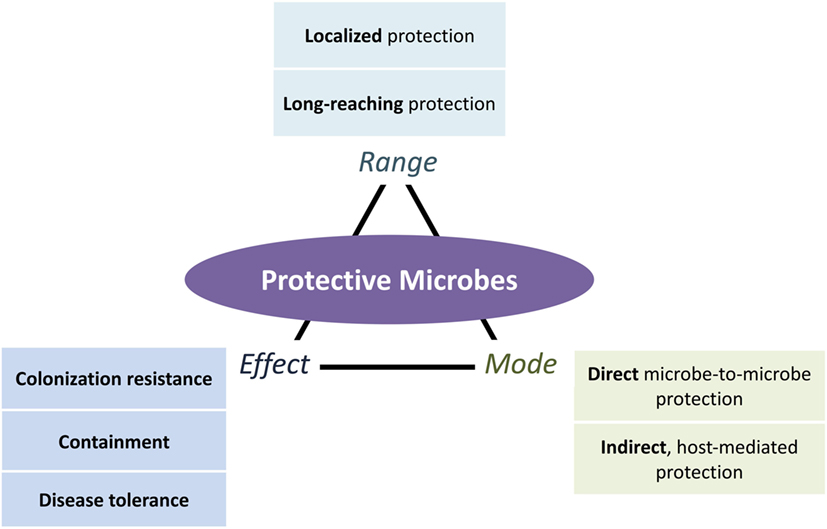
Figure 1. An expanded view of microbe-conferred protection. Microorganisms can protect the host in different ways, depending on the mode of protection (direct ecological and indirect host-mediated), the effects of protection (colonization resistance against pathogenic establishment and growth, containment of pathogens and their effects, and disease tolerance of pathogens while suppressing their negative effects), and the range of protection (localized or long-reaching, with the latter further divided into protection that is systemic or from one locale to another locale). All three aspects of protection can occur in combination. Long-reaching protection, for instance, involves both direct (ecological) and indirect (host-mediated) modes of protection that result in colonization resistance, containment, or disease tolerance effects.
Taking into account space constrains, our examples will focus only on vertebrates, with most of our examples coming from mammalian studies, especially humans and murine models [for example, in invertebrates, see Ref. (15)]. Because many of the terms used in this review have often been employed loosely in the literature and/or have been understood differently by different authors, here we explain in which sense we use them (Box 2). Finally, we offer important conceptual clarifications about the different ways components of the microbiota can exert protective effects on their host, and propose the concept of “co-immunity1” to describe the fact that host protection is in general the emergent and dynamic product of two influences, that of the host and that of some microorganisms (see Concluding Remarks and Perspectives).
Box 2. Definitions.
• Immune protection against pathogens: although immune responses can be very diverse, ranging from protection to development and repair (42), our focus in this article is on protection against pathogens. Consequently, our analysis excludes protection against immune diseases such as allergies, diabetes, and susceptibility to xenobiotics. We distinguish between direct, host-independent protection and indirect, host-mediated protection.
• Microbiota: when talking about the “microbiota,” we broadly consider all the resident microorganisms (microbes, but also “macrobes” such as helminthes) living in or on a host, regardless of the nature of their ecological interaction (parasitic, commensal, and mutualistic), size or taxonomy (parasites such as helminthes, fungi, bacteria, phages, or viruses). We also consider the synergistic and context-dependent effects of microorganisms.
• Co-immunity: a form of immune defense associating components of several organisms.
• Colonization resistance: host-dependent or independent resistance to pathogens that is induced by the microbiota. Current examples only concern the local inhibition of pathogen viability, establishment, or growth, but here we expand this concept to include long-reaching effects as well.
• Containment: controlled localization of microbes within a particular location inside the host body.
• Disease tolerance: limitation of host’s tissue damages induced by pathogens, without direct pathogen elimination.
Localized Protection
In recent years, a general consensus on the mechanisms of colonization resistance has emerged (6, 9, 10, 18, 19, 43–46). Resident microorganisms can inhibit pathogenic viability and growth by antagonizing pathogens or “starving” them of limited resources, two well-recognized mechanisms of microbial ecological competition (47–49). In this section, we review expanded evidence that microorganisms confer local protection directly through microbe-to-microbe interactions (see Direct, Host-Independent Protection) and indirectly through the host immune system (see Indirect, Host-Mediated Protection) through colonization resistance, containment, and disease tolerance.
Direct, Host-Independent Protection
Colonization Resistance: Beyond Niche Competition and Antagonism
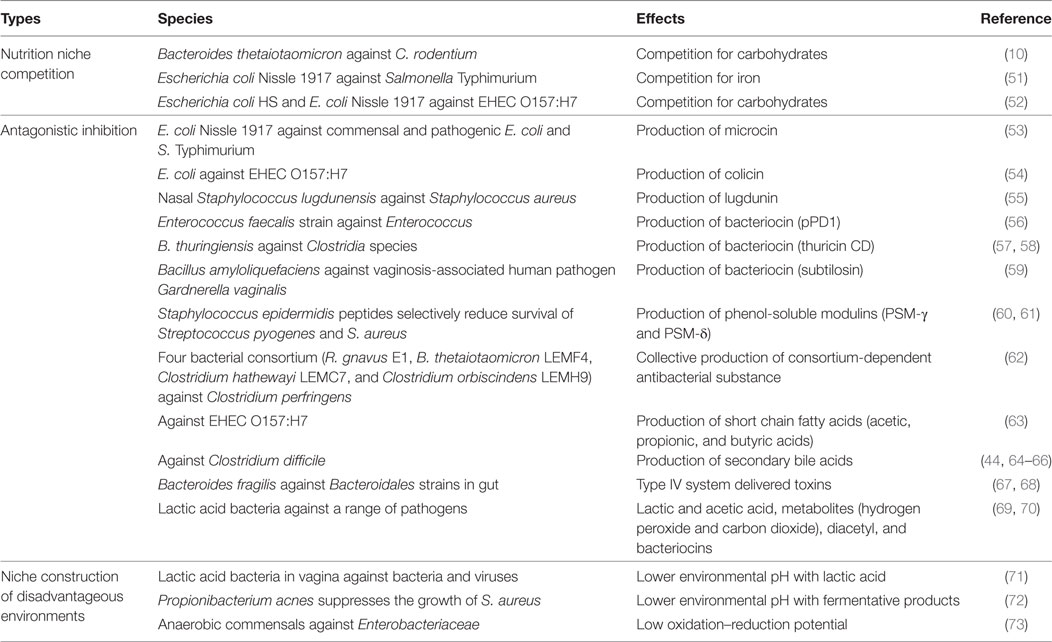
Recent reviews have focused on a few aspects of colonization resistance. Protective microorganisms can antagonize pathogens through contact-dependent inhibition or the release of antimicrobial molecules. They can also outcompete pathogens for limited resources such as trace metals, nutrients, receptor donors, or adhesion sites, or construct environments hostile to pathogens, for instance, by lowering environmental pH (see summary of recent findings in Table 1). A recent finding, for instance, is that bacteria of the Clostridiales order outcompete Listeria monocytogenes for nutrition in the small intestine and likely antagonize the pathogen in other ways in the large intestine, providing the host a first line of defense from systemic infection (50).
The concept of the ecological niche is central to colonization resistance. Protective microorganisms are thought to “defend” their nutritional niche by killing incoming pathogens or outcompeting them (74). Pathogens, on the other hand, are thought to overcome colonization resistance by creating or exploiting new spatial-temporal niches (19). However, a niche is not just a nutritional environment, but any environmental feature relevant for colony survival and growth. Protective microbes can also inhibit pathogenic establishment and growth by disrupting pathogenic biofilms, bacterial collectives that undergo regular developmental “life cycles” and protected by synthesized extracellular polysaccharides matrices. Proteases released by Staphylococcus epidermidis can degrade the matrices of pathogenic Staphylococcus aureus biofilms (75, 76). Lactic acid bacteria can disrupt matrix synthesis by interfering with pathogen virulence genes, for instance, by decreasing the expression of Streptococcus mutans genes involved in matrix glucan production (77) or modulating S. aureus gene expressions related to the production of intercellular adhesion polysaccharides (78).
Protective microorganisms can also influence quorum sensing, the cell-to-cell communication system that allows bacteria to perceive information about bacterial population density and to regulate collectively virulence factor production and biofilm development (79, 80). Soluble molecules released by probiotics can interfere with the S. aureus accessory gene regulator (agr) quorum sensing system, which regulates the switch between biofilm and free-floating lifestyles (81). Subtilosin, a protein secreted by Bacillus subtilis, also interferes with the quorum sensing of Gardnerella vaginalis, preventing biofilm formation (82). Biosurfactants are well-known anti-adhesion, anti-biofilm agents that can also disrupt cell-to-cell signaling (83, 84). Cell-bound biosurfactants of lactic acid bacteria disrupt the biofilms of multi-drug-resistant Acinetobacter baumannii, Escherichia coli, and S. aureus (85) as well as Serratia marcescens strains (86).
Beyond Colonization Resistance: Containment and Disease Tolerance
Colonization resistance is not the only way microorganisms can protect the host from pathogens, as not all types of protection act by inhibiting pathogenic colonies establishment and growth. The role of resident microorganisms in barring entry and dispersal of alien populations is oftentimes ignored (87). Furthermore, inhibition of quorum sensing can affect pathogenicity, the effects of pathogens, without inhibiting bacterial viability, which, as a therapeutic option can avoid the selection of drug-resistant bacteria (88). We thus propose looking beyond colonization resistance to adopt a community ecology point of view that better understands the multiple ways resident communities can prevent or disrupt pathogen invasion.
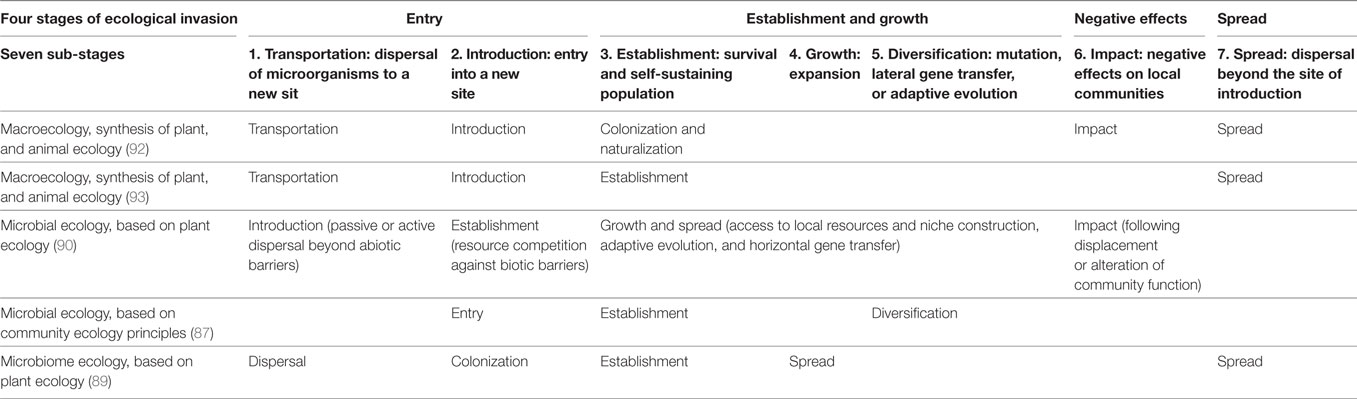
In invasion ecology, residents can disrupt alien invasion by intervening with alien entry and diversification, by inhibiting its establishment or growth, by mitigating its negative impacts on local communities, or by preventing alien dispersal into other vulnerable environments (89–91) (see summary of stages of invasion in Table 2). A more comprehensive picture of resistance against pathogens must take into consideration the multiple stages of invasion and go beyond competition as the only way residents can fight against invaders.
We thus distinguish three major microbial obstacles against invading pathogens (Table 2; Figure 2): colonization resistance exclusively refers to the prevention of pathogen establishment of a persistent colony and population growth, containment is the prevention of pathogen spread into another body site, sometimes leading to systemic infection, and finally, disease tolerance is the limitation of pathogenic impact on host tissues without killing the pathogens.
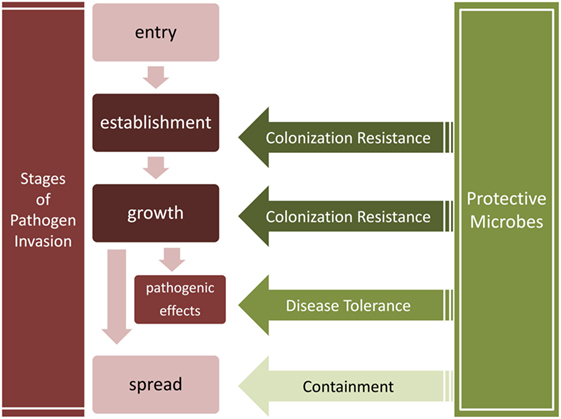
Figure 2. Stages of pathogen invasion and the obstacles presented by resident communities. Colonization resistance, containment, and disease tolerance present obstacles to different stages of the invasion process. Colonization resistance disrupts the establishment and growth of pathogens, the containment of pathogens prevents their spread into other tissues, and disease tolerance suppresses the negative effects of pathogens without decreasing their load.
Containment is the controlled localization of microbes and their effects within a particular location inside the host body. One example is the intestinal lumen. The epithelium barrier and gut mucosal immune system prevents the translocation of microorganisms from the lumen into the host. Mucosal-associated microorganisms have long been postulated as reinforcers of gut barriers, both directly and indirectly through the host (94–97). For instance, in various organisms, bacteriophages that adhere to mucus glycoproteins can prevent translocation of bacteria, thus providing a host-independent protection of mucosal surfaces against bacterial infections (98). Saccharomyces boulardii also prevents Salmonella liver translocation by directly binding to the pathogens (99).
Secreted factors can interfere with the translocation of gut pathogens into and beyond the epithelia. Supernatants of Enterococcus mundtii and Lactobacillus plantarum inhibit the invasion of L. monocytogenes into epithelial cells (100). A secreted, non-bacteriocin component from Escherichia coli Nissle 1917 also reduces the efficiency of Salmonella enterica serovar Typhimurium epithelial invasion and blocks invasion by many pathogens without eliminating them (101). Pathogen translocation can also be prevented by interference with pathogen adhesion to the epithelium (102, 103). It is well known that resident commensals, especially lactic acid bacteria, compete with pathogens for adhesion sites, for instance, the competition between Lactobacillus reuteri and enteropathogenic E. coli (104). Short chain fatty acids are products of anaerobic intestinal microbiota fermentation of dietary fibers. Butyrate inhibits the virulence genes that enable intestinal S. enterica subspecies to enter the epithelium, specifically pathogenicity island 1 (105). Acetate from Bifidobacteria can enhance epithelial cellular defense functions, inhibiting the translocation of Enterohemorrhagic E. coli (EHEC) shiga toxin into the bloodstream (106).
Disease tolerance, on the other hand, is the limitation of pathogen-induced damage of host tissues without elimination of the pathogen. Various types of processes are associated with disease tolerance: the neutralization or inhibition of pathogenic toxins, the healing process of the host, and the regulation of destructive inflammation and maintenance of metabolic homeostasis (see Box 3). While the latter two are mediated through the host, the first can be host-independent.
Box 3. The concept of disease tolerance.
Disease tolerance should be distinguished from “tolerance” or “immune tolerance,” a low or regulated immune reaction toward an antigen (40). It is instead the limitation of pathogenic damage to host tissues. The concept of disease tolerance has long been recognized by plant ecologists as an alternative mode of protection from pathogens or pests besides resistance mechanisms, yet only relatively recently applied to animals (107–110). Even though disease tolerance is currently seen as an indirect process mediated by the immune system to control tissue damage (40), it could be directly regulated by microbes as well. The toleration of pathogens could be a cost-effective alternative when preservation of pathogen fitness does not strongly decrease host fitness (e.g., when pathogen virulence is low), especially since the side effects of pro-inflammatory immune responses are sometimes more damaging to the host than the infections themselves.
An example of host-independent disease tolerance is the ability of resident microbiota to inhibit gene expressions of toxins. Soluble molecules from Bacteroides thetaiotaomicron and the bacteria-produced short chain fatty acid butyrate, for instance, can inhibit EHEC shiga toxin gene expressions and pathways (111, 112). The quorum sensing molecule AI-2 of Ruminococcus obeum also downregulates the Vibrio cholera toxin operon (113). Secreted organic acids can mitigate pathogen toxicity in various ways: lactic acid from Streptococcus thermophilus inhibits Clostridium difficile toxin A gene expression (114) while acetic acid secreted by a Bifidobacteria strain inhibits EHEC shiga toxin production in part by lowering environmental pH (115). Remarkably, the growth of shiga-producing E. coli was not inhibited by the B. breve strain even though toxin production was halted.
The damaging effects of pathogens can also be diminished by the degradation of released toxins. Proteases secreted by a probiotic Bacillus clausii strain acts against toxins from C. difficile and Bacillus cereus (116). Another example is a Saccharomyces boulardii protease that inhibits C. difficile toxin A and B (117). Bifidobacteria and Lactobacilli strains mitigate C. difficile cytotoxic effects by inactivating their secreted toxins (118, 119).
In sum, colonization resistance, containment, and disease tolerance are diverse effects of protective microbes against the spread, establishment and growth, and impact of pathogen invasion. Broadening the concept of protection to include the effects of disease tolerance and containment allows us to consider pathogenicity in a wider context, taking into account the overall ecological process of pathogen invasion (Figure 3). The detailed mechanisms underlying these three types of effects—colonization resistance, containment, and disease tolerance—may overlap. For instance, competition over adherence sites prevents the growth of biofilms that enable the establishment, growth, and spread of many pathogens (Figure 4).
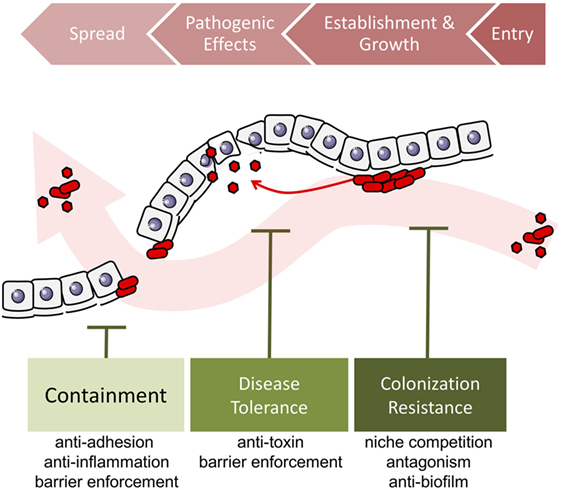
Figure 3. Localized microbe-conferred protection against pathogen invasion. Alien pathogens invade the host by entering a host site, establishing a growing colony, exerting negative effects on the host. When conditions are ripe, they can spread to a different host site, in this case, through the epithelium or endothelium barrier. Protective microorganisms can challenge pathogen invasion at any of the four stages, by disrupting entry, preventing or destroying colony establishment and growth, by suppressing pathogenic effects, or by preventing spread into other tissues.
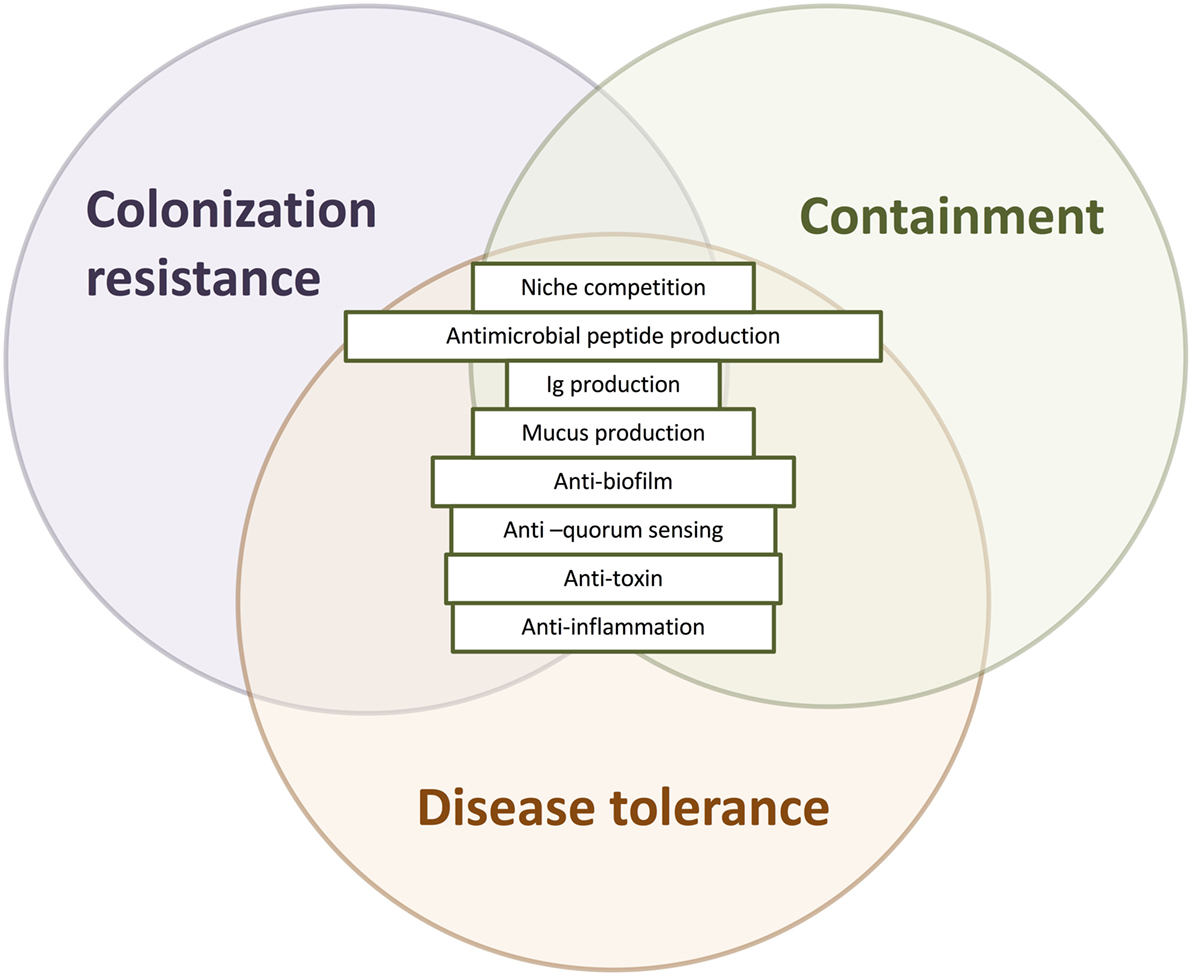
Figure 4. The overlapping mechanisms of colonization resistance, containment, and disease tolerance. Colonization resistance, containment, and disease tolerance are distinct effects on invading pathogens, disrupting their growth and establishment, their spread, and their negative effects, respectively. Nevertheless, the mechanisms underlying these effects can overlap, deployed at different times and stages with different outcomes. Antimicrobial peptides, for instance, prevent the translocation of microorganisms as part of the mucus barrier and disrupt the establishment and growth of pathogens. Another example is the disruption of a pathogenic biofilm, which may destroy the establishment of the colony as well as prevent it from adhering and translocating gut lumen.
The examples reviewed so far are direct microbe-to-microbe effects. We now turn to indirect forms of protection that involve host systems.
Indirect, Host-Mediated Protection
Local microbiota-induced protection of the host can also be indirect, that is, mediated by host immunity and metabolism. We will focus on protection that involves the host immune system. Microorganisms induce proper development of crucial components of the host immune system, such as mucosa-associated lymphoid tissues (120–122). Here, we will more precisely focus on microbe-induced host immunity that specifically acts against pathogens (4, 123), guarding against both harmful (pathogens) and potentially harmful (“pathobionts”) microorganisms.
Colonization Resistance
Examples of host-mediated protection, especially colonization resistance, by gut microbiota have already been extensively reviewed (see Box 1). Indirect colonization resistance occurs when resident microorganisms induce host reactions that act against pathogens.
Resident microbiota sampled by pattern recognition receptors can activate downstream secretion of antimicrobial peptides that protect the host by killing or inactivating bacteria, fungi, and viruses, primarily by destroying cellular membranes (124, 125). These include C-type lectin Reg3-γ (38), α and β defensins (35, 126), and the ribonuclease angiogenin (127). The effects are often pathogen- and host-specific (128–130).
Gut microbes induce the development of lymphoid tissues that sample and secrete Ig A into the gut lumen as well as regulate secretory IgA (SIgA) secretion levels (131). SIgA can help trap and exclude pathogens (“immune exclusion”), but when bound to certain microorganisms, can also selectively promote commensal biofilms that confer colonization resistance against pathogens (“immune inclusion”) (132–136). Specificity to microorganisms could be enhanced through a positive feedback loop mediated by SIgA retrotransportation (137–139).
Microbes can also induce protective pro- or anti-inflammatory immune activities by altering the balance between host T-cell subsets (2, 140–142). By tilting the balance toward pro-inflammatory pathways, commensals help generate host-mediated attacks against pathogens. Segmented filamentous bacteria, for example, induce inflammatory Th17 immunity (143), leading to a generally protective, although context-dependent immune state (144). Protozoan Tritrichomonas musculis protects from gut S. Typhimurium infection through promotion of inflammatory Th1 and Th17 type immunity (145).
In the skin, microbiota could be considered an “endogenous adjuvant” of the skin immune system, exerting its influence via the release of products (such as antimicrobial peptides or metabolites) and/or via the modulation of innate and adaptive immunity without invoking inflammatory responses (146–148).
In the respiratory tract, local manipulation of the microorganism composition may have profound consequences on the capacity of the host to mount protective responses (149). For instance, intranasal inoculation of mice with live or heat-inactivated Lactobacillus spp. and non-pathogenic Listeria spp. protects against secondary lethal infection with the virulent pneumonia virus by suppressing virus-induced pro-inflammatory cytokines and viral load (150).
Beyond Colonization Resistance: Containment and Disease Tolerance
Microbes can also indirectly protect the host by containing pathogens within the gut and lung lumen, as breaching of the epithelial layer triggers systemic inflammation. New developments shed light on the intimate relation between microorganisms and barrier functions. The gut epithelium barrier is fortified by maternal microorganisms before birth (151). After exposure to environmental microbes, microorganisms strengthen and protect the barrier (152). In vitro treatment of Saccharomyces boulardii, for instance, prevents Bacillus anthracis toxins from destroying the integrity of epithelium cells and the tight junctions between them (153). In antibiotics-treated mice, translocation of bacteria and an increase in inflammatory responses can be observed (154, 155).
The separation of host and microorganism with a mucus layer is in part regulated by resident microorganisms. The mucus layer effectively separates luminal components, especially microorganisms, from the epithelium. The constant outgrowth of mucus pushes microorganisms out and provides a medium that concentrates antimicrobial elements such as antimicrobial peptides and SIgA. In the small intestine, Muc2 mucin, the backbone of mucus in mice, requires bacterial cleavage of a proteolytic enzyme to detach mucin from secreting goblet cells; in the colon, “sentinel” goblet cells activated by bacterial components quickly respond by extruding and releasing an explosion of Muc2 (156, 157). Helminthes such as Trichuris muris can restore the Nod2-deficient abnormal goblet cells of mice through TH2-mediated immunity, thereby inhibiting the colonization of pro-inflammatory Bacteroides species (158).
Localized protection is also achieved by disease tolerance. In the gut, Bacteroides fragilis affects systemic T-cell responses through the action of the bacterium-derived polysaccharide A, which protects against pathobiont H. hepaticus colitis via the production of anti-inflammatory IL-10 by CD4+ T cells and the promotion of regulatory T cells (159). Moreover, this protection has no effect on pathogen fitness or on population load in the intestine. SIgA can neutralize intracellular toxins and viruses, as well as directly suppress bacterial virulence mechanisms (133, 135).
In sum, resident microbiota can protect the host by locally inhibiting the establishment and growth of pathogens, mitigating pathogen-induced damage to the host, and preventing the spread of microbes to other sites. Local protection is ensured by mechanisms that are directly targeted against microorganisms or indirectly through the host immune system.
Long-Reaching Protection
We now turn to protective microbes that help the host from a distance, by either inducing systemic protection (that is, a protection realized in the entire organism) or protection at a particular remote site. In host-microbiota symbiosis, the habitat of the microbial communities is a changing and responsive living being. Since host microbial communities are connected by metacommunity dispersal and through the internal sensors and networks of the host, protective microbes can have potentially far-reaching and integrated consequences. A more comprehensive picture of protective microbes requires a global understanding of the ecological and physiological—e.g., metabolic, immune, and neuroendocrine—interactions.
However, our understanding of long-reaching protection is at its infancy. Whereas pathogens can migrate through the blood from one organ to another or travel across microenvironments within an organ system (160–162), there is little evidence that commensals can do the same. Here we review examples of long-reaching protection (see Figure 5).
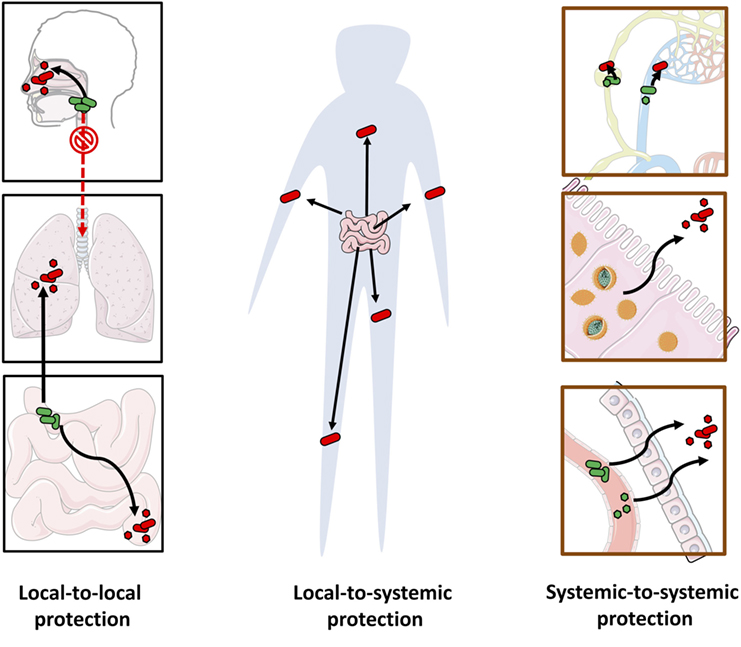
Figure 5. Long-reaching microbe-conferred protection against pathogens. The multiple pathways of long-reaching protection include: local-to-local protection against pathogens at a distal site by protective microorganisms at another site (e.g., gut to lung, upper respiratory tract to lower respiratory tract, and small intestine to large intestine) (left), local-to-systemic protection from one site to pathogens across the body (e.g., systemic protection from gut microbiota) (center) and systemic-to-systemic protection by microorganisms that are distributed systemically (e.g., protective viruses or bacteria that circulate through the blood stream or the lymphatic system, or reside in multiple locations) (right).
Systemic Protection
The direct, host-independent systemic effects of microorganisms are poorly illustrated in the literature, and as far as we know, there are no mammalian examples where microorganisms can confer direct systemic protection across microhabitats. Nevertheless, the killing capacities of some microorganisms present promising therapeutic options as “live antibiotics.” For example, the predatory proteobacteria Bdellovibrio bacteriovorus, when injected in a zebrafish model infected with S. flexneri, preys and eliminates the human pathogen in the whole animal (163).
Indirectly, however, the evidence is much more promising as microorganisms have far-reaching immunomodulatory effects on mucosal and systemic immunity (164, 165). Microbiota modulation of myeloid cells preconditions neutrophils induces macrophage killing capacities and calibrates macrophages and dendritic cell responsiveness to infections (166). Microbial-regulated hematopoiesis can fight against systemic infections such as L. monocytogenes (167). In a mice model, injection of chitin from the fungal commensal S. cerevisiae increases resistance to systemic infection with C. albicans by inducing monocyte activity via a fine modulation of TNF-α and IL-6 (168). Without the microbiota, mononuclear phagocytes fail to prime NK cells that normally trigger systemic antiviral immune responses in non-mucosal lymphoid organs (169). Recent studies show that maternal microbes shape offspring immunity before birth. Bacterial products in mothers induce genetic expression of antimicrobial peptides in offspring epithelium and shape the components of their innate immune systems (170) while dampening their adaptive immune responses (171). In fact, the success of vaccination (the injection of microbe-derived products in one site that protect against infections in another site) shows that long-distance immune protection is possible. For instance, the intranasal administration of a vaccine against HSV-1 induces production of specific antibodies in the genital tract (172).
While current evidence is still not clear-cut, it is nevertheless important to conceptually distinguish between various origins of systemic protection. In local-to-systemic protection, microorganisms at a single location, such as the gut microbiota, can confer protection through systemic immune responses or leave the local site and disseminate to other regions. In systemic-to-systemic protection, the protective microorganisms are already distributed systemically, exerting system-wide localized effects. In particular, we discuss how microbiota, their fragments, or by-products can reach distal sites through the host circulatory systems.
Local to Systemic Protection
Microbiota at one location can induce systemic colonization resistance in the host. Composition of the gut microbiota modulates the severity of malaria, where a fecal transplant from resistant mice transfers the resistance to germ-free mice by elevating humoral immune responses. The resistant gut microbiota are characterized by an increased abundance of Lactobacillus and Bifidobacterium (173). In humans, gut microbe E. coli O86:B7 induces antibodies that target plasmodium sporozoites, conferring a protective cross-response against malaria transmission (174).
Systemic innate and adaptive immune responses to Toxoplasma gondii infection in humans rely on the indirect stimulation of dendritic cells by normal gut microflora (175). Several studies have shown that gut microbiota also have the potential to reduce systemic viral infection and disease (176). A clinical trial observed a positive effect of a Lactobacillus strain as an immune adjuvant for live-attenuated H3N2 influenza vaccine in healthy adults, with higher seroconversion rates in patients treated by probiotics (177), suggesting a non-specific immune response activation. In HIV infection, higher abundance of the gut Lactobacillales order in patients is positively associated with CD4+ T cell count and negatively associated with viral load, indicating that bacteria from Lactobacillales could in some direct or indirect way modulate the infectivity or pathology of HIV infection (178).
Finally, as a basal stimulant, bacteria can prime activation threshold of antiviral innate immunity against systemic viruses (179). Antibiotic-treated mice before lymphocytic choriomeningitis (LCMV) infection elicited an impaired innate and adaptive immune response to LCMV infection, and an increased mortality after influenza infection (179).
Systemic to Systemic Protection
Microbiota or its components that are normally distributed across the host system can also induce system-wide colonization resistance in the host. Recently, the idea that viruses can be mutualistic, and in particular can offer protection against pathogens, has gained popularity (180–182). A healthy human is infected by more than 10 permanent chronic systemic viruses, and this number may in fact be far higher (182). Two cases show that viruses induce in their hosts a higher basal immunity that explains normal immunity as well as responses to novel microbes. First, baseline activities can be upregulated and maintained by viruses in latent states, conferring protection against subsequent infections (176, 183). In mice, gammaherpesvirus 68 or murine cytomegalovirus (CMV) (which are models for the human pathogens Epstein–Barr virus and CMV, respectively) in dormancy confers symbiotic protection against bacterial infection to L. monocytogenes and Y. pestis in an antigen-independent way, involving interferon production and macrophages systemic activation (184, 185).
In a clinical study, HIV patients co-infected with non-pathogen GB flavivirus showed reduced mortality rate, suggesting the possibility of an inhibition of HIV replication due to this flavivirus (186). HIV replication was reproducibly inhibited in cultures of peripheral-blood mononuclear cells by GBV-C coinfection (187). Patients with GBV-C infection present an increased proportion of naive T cells and a reduction in T-cell activation and proliferation that could increase disease tolerance and finally the survival among HIV infected individuals co-infected with GBV-C (188).
Circulations and Protective Effects
It is generally assumed that microorganisms are pathogenic when spread throughout the system while non-pathogens are contained within the gut and lung lumens and outside of skin surfaces. Indeed, bacteremia and endotoxemia, i.e., the spreading of bacteria or bacterial structural components through blood circulation, respectively, are usually pathological. For instance, intra-uterine infection during pregnancy in mice can be caused by pathogens that do not belong to vaginal microflora, but to oral microorganisms via hematogenous transmission (189).
However, non-pathological, physiological blood circulation of microorganisms or microbial fragments occurs in healthy subjects (190–192). Substantial numbers of live gut species have also recently been found living in the mesenteric lymph nodes and systemic lymphoid organs under normal situations (164). In this section, we thus consider the possibility that circulation of bacteria or their metabolic compounds can constitute a long-reaching route of protection. Even though a long-reaching seeding through the bloodstream and lymphatic systems has never been demonstrated, it is a possible route protective microbes could take.
The involvement of circulating long-reaching mechanisms in host protection is only a hypothesis at this stage, but investigating this potential long-reaching influence undoubtedly constitutes a very promising avenue for future research, paving the way to targeted experiments.
Bacteremia and Endotoxemia
Bacteremia is the dissemination of bacteria into the circulatory system. Under normal conditions, Gram-negative bacteria in the gut can disseminate systemically, inducing system-wide production of IgG that provide cross-protection against Gram-negative bacteria infections such as E. coli and Salmonella (193).
Recent data suggest a long-distance disease tolerance by challenge-induced translocation of gut microbiota to multiple distal sites. In mice, infection with the respiratory pathogen B. thailandensis (intranasally) or with the pathogen S. Typhimurium (orally) leads to skeletal muscle wasting. Gut commensal E. coli O21:H+ antagonizes muscle wasting during these infections, with no changes in inflammation profile but sustained insulin-like growth factor 1 signaling in skeletal muscle. This protection was associated with the translocation of E. coli O21:H+ from the intestine to white adipose tissue and occurred without a negative impact on B. thailandensis or S. Typhimurium fitness. This beneficial effect promoted both the health of the host and fitness of E. coli O21:H+ (194).
Endotoxemia occurs when bacterial by-products circulate into the bloodstream. It has long been known that circulating lipopolysaccharides, a membrane component of Gram-negative bacteria, has potentially beneficial immunomodulatory effects (195). Lipopolysaccharides endotoxemia is increased by nutritional factors (196) and modulated by changes in gut microbiota (197). The mechanisms of host-mediated modulation of bacteremia and endotoxemia (primarily of lipopolysaccharides) are currently being explored under the context of high-fat intake or obesity, which weaken gut integrity leading to increased penetration of gut microbes or their products into the circulation. The uptake and transportation of lipopolysaccharides is active, reaching from the gut to distant tissues like adipose tissue. After lipoprotein binding and transportation to the lymph and across endothelial barriers, they interact with macrophages and trigger the secretion of pro-inflammatory cytokines (198).
Microbiota-derived peptidoglycan translocated from the gut to neutrophils in the bone marrow systemically primes the innate immune system, enhancing the elimination of two major pathogens, S. pneumoniae and S. aureus via the pattern recognition receptor nucleotide-binding, oligomerization domain-containing protein-1 (199).
Finally, another active long-reaching route is the gut–liver axis, where influx of microbial molecules derived from genetic inflammasome deficiency-associated dysbiosis, passing in portal circulation, can trigger liver inflammation through TLR4 and TLR9 activation. In the case of preexisting lipid accumulation in hepatocytes, this mechanism could lead to non-alcoholic steatohepatitis (200).
Metabolic Compounds
Naïve mass spectrometry-based metabolomics studies comparing blood metabolic profiles between germ-free mice and conventional animals show a drastically different blood metabolic profile, with a drug-like metabolic response of the host to metabolites (i.e., degradation of xenobiotic molecules by host’s enzymes) generated by the microbiome (201).
Gut microbiota produce and regulate multiple compounds that can reach distant organs via systemic circulation, and influence host physiology (202). More precisely, metabolic compounds produced or transformed by gut microbiota may modulate host immunity in distant sites. In particular, short chain fatty acids, especially butyrate, seem to exert broad anti-inflammatory activities by affecting immune cell migration, adhesion, cytokine expression, as well as cellular proliferation, activation, and apoptosis through the activation of signaling pathways (NF-κB) and inhibition of histone deacetylase (203). Moreover, epithelial permeability can be modulated by the microbiota: blood-brain barrier permeability is increased in germ-free mice and reintroduction of a healthy microbiota, of short chain fatty acids producing bacteria or direct short chain fatty acids administration can reverse this effect, up-regulating tight junction proteins expression (204). This mechanism could be involved at the host-environment interface, modulating pathogen susceptibility.
Fungi could also play a crucial role in long-reaching immune modulation. Digestive Candida-produced prostaglandin E2, an active lipid compound with hormone-like effects, can reach the lungs through the bloodstream, act on lung macrophages and promote allergic inflammation, thus suggesting a hypothetical long-reaching route that could be protective (205).
Local to Local Protection
Some studies suggest that local microbiota could modulate distant local conditions, which may confer long-term protection to the host. On the one hand, few studies have highlighted a long-reaching protective role of specific local microbiota to specific local sites, and with poor mechanistic understanding. On the other hand, many papers have brought out various mechanisms that could involve distant modulations of immune conditions, but without a protective effect. Time has perhaps come to combine these approaches, to determine how the microbiota located in a given organ could have a protective effect on another organ (i.e., a local-to-local protection, but without a demonstrated systemic effect). Even though in some cases, later evidence might show that a local-to-local effect is actually a local-to-systemic effect, the conceptual distinction between a highly targeted mechanism versus a system-wide mechanism can guide research questions about the underlying mechanisms and the scope of protective effects. We explore two possible routes of long-reaching protection: the host-mediated gut–lung axis and direct interactions down the respiratory and gastrointestinal tracts.
Indirect, Immune-Mediated Protection along the Gut–Lung Axis
To the best of our knowledge, the only explicit long-reaching protective effects from one local site to another described in the literature are the immune-mediated relation between the gut microbiota and lung infections (203, 206). Klebsiella pneumoniae lung infection has been extensively studied as a model of pulmonary infection. Germ-free mice infected with K. pneumoniae are drastically susceptible to bacterial infection in an IL-10-dependent manner, with an increase of bacterial growth and dissemination. Interestingly, activating microbial pattern recognition receptors helps fight against K. pneumoniae infection in the lungs (207, 208). Another example is tuberculosis. Helicobacter pylori infection is suspected to modify the clinical outcomes of M. tuberculosis infection, with the presence of H. pylori associated with a lower rate of tuberculosis infection (209), but increased tuberculosis severity (210). In this case, a microbe at one location (H. pylori in the stomach) could modulate long-distance immunity at yet another location (in the lungs) in response to infection.
Antibiotic-treated mice exhibit impaired innate and adaptive mucosal immune responses to influenza infection, with increased damages and host mortality (179). A decrease in the number of gut commensals via treatment with a broad-spectrum antibiotic resulted in blunted T-cell and B-cell responses to an intranasal infection with the A/PR8 strain of influenza (211). Activation of Toll-like receptors restored the immune response in antibiotic-treated mice through inflammasome-mediated induction of interleukins such as IL-1β and IL-18. The authors suggest that either the microbial products can diffuse systemically, or activation of the inflammasome does not need to occur at the site of infection.
In an E. coli pneumonia model, antibiotic depletion of commensals in mice also causes a drastic bacterial burden both in lungs and blood, with a significant increase of mortality. Lipopolysaccharides supplementation during antibiotherapy reversed these effects, suggesting a distal action of commensal microbes through toll-like receptors (212). Deleterious effects of antibiotic depletion have been shown in a mouse model of Streptococcus pneumoniae infection, reversed by fecal microbiota transplantations that enhance macrophage functions in the primary alveolar (213).
Gut microbiota also play a role in expanding and maintaining viral-specific memory T-cell populations in the lungs. In a mouse model of MCMV-associated lung disease, the frequency of virus-specific CD8+ T cells in the MCMV-infected lungs of germ-free mice was restored by fecal bacteria from specific pathogen-free mice, likely through cross-activity between gut microbiota peptides and epitopes of MCMV-specific memory T cells (214).
Furthermore, mice challenged with pulmonary staphylococcal infection but lacking segmented filamentous bacteria in their gut microbiota showed more severe infection with higher bacterial load and mortality, associated with diminished lung concentration of Th17 immune effector cells. Reintroduction of segmented filamentous bacteria was sufficient to restore protective effect (215). Antibiotics also significantly decreased lung Th17 cell numbers during pulmonary acute fungal infection, restored by segmented filamentous bacteria colonization (216).
Direct, Ecological Protection through Digestive Flows and Air Circulations
Even though direct and host-mediated causes of microbial protection are oftentimes difficult to distinguish and intertwined at a local scale, the direct versus indirect distinction can help identify ecological versus physiological routes of influence at a global scale. Current evidence indicates that long-distance protection is mostly mediated by the host, but ecological routes are also available when we consider the way fluids and air flow through the host. Many organs are interconnected through airways and liquid channels. We hypothesize microbe-induced protection of downstream conditions in the respiratory tract and the gastrointestinal tract.
The respiratory tract ecosystem consists of the upper and lower respiratory tracts, with the latter further divided into trachea, bronchi, and bronchioles. The gastrointestinal tract ecosystem is divided into the stomach, small intestine, and large intestine, with secretions that come from associated organs such as the gall bladder, liver, and pancreas. It is possible that microbial communities in one patch of the lung and gut meta-communities can protect the host by influencing the entry and growth of pathogens in a downstream patch.
Bronchoalveolar lavage fluid samples of the lungs reveal a substantial microbiota community with multi-kingdom interactions due to air-borne fungi (217). The oral microbiota are the physiological source of lower respiratory tract microorganisms, predominately acquired through microaspiration and especially during sleep (218). It is also the source of disease-inducing pathogens in the lungs, especially pneumonia and cystic fibrosis. The sampled microorganisms of a healthy lung consist primarily of transitory populations determined by a steady balance of incoming migration and outgoing expulsion through physiological clearance or immune responses. In disease states, such as cystic fibrosis, regional growth conditions support the settled colonization and expansion of microbial communities (219, 220). The respiratory tract ecosystem is thus an excellent example of dominance in dispersal processes over processes of establishment and growth, similar to the metacommunity principles governing islands (220–224).
Microbial communities in the upper respiratory tract (e.g., oral cavity) are likely to confer resistance against the colonization and growth of pathogens in the lower respiratory tract. There is an association between oral dysbiosis and lower respiratory tract infections such as pneumonia. The absence of normal residents in the upper respiratory tract may thus contribute to the overwhelming migration of residential or opportunistic pathogens to the lower respiratory tract (225). Pathogen “blooms” in the lower respiratory tract generally occur under two conditions: when there is a positive influx of microorganisms over those eliminated or when altered local conditions favor pathogenic growth [see adapted island biogeography model in Ref. (223), and review in Ref. (224)]. We thus suspect that a loss of colonization resistance of upper respiratory tract residential microbiota may be the reason for lower respiratory tract microbial establishment and growth, resulting in disease.
Another possible downstream protective effect is in the gastrointestinal tract. The habitat of the gastrointestinal tract is similar to a river ecosystem (226). The intestine receives inputs from the source (nutrients and acids from the stomach) and associated organs (liver, gall bladder, and pancreas) and the flow slows down at the colon, which is dominated by greater bacterial loads and the accumulated nutrients and metabolites from upstream.
Secondary bile acids inhibit the germination and growth of C. difficile in the colon. Bile acids are released and mostly reabsorbed upstream in the small intestine, decreasing greatly in concentration further downstream (227, 228). However, small intestinal microbiota can inhibit re-absorption by deconjugating bile acids, thus promoting their excretion down and out of the large intestine (229). Downstream secondary bile acids in the colon can thus depend on upstream commensals. Indeed, antibiotic treatment that decreases colonization resistance against C. difficile also decreases bile acid amounts in fecal matter (64), which are both restored by fecal matter transfer (65). Upstream commensals can thus possibly protect the host from downstream pathogenic growth by releasing metabolite inhibitors.
Microbes can protect the host by regulating the migration flows between the lung “island” communities and by influencing “downstream” communities in the gut. The analogy between lung and gut ecosystems to island and river ecosystems, respectively, point to new perspectives. These are clearly local-to-local effects, although not necessarily organ-to-organ effects (they concern “subparts” of organs rather than organs per se). Yet they point to the possibility of organ-to-organ protective effects, and offer insights about how these effects could be investigated in the future. It is worth investigating whether migrations between other sites, for instance, the upper respiratory tract and the stomach (218), or direct dispersal between oral and vaginal microbiota via sexual behavior, also exhibit upstream-to-downstream colonization resistance against infections.
Concluding Remarks and Perspectives
Previous studies have focused on the local colonization resistance conferred by microorganisms at various sites within the host. Here we examined protective effects beyond colonization resistance through both direct and indirect modes of protection. We highlighted how common types of ecological interactions give rise to the resistance and tolerance against harmful microorganisms as well as the containment of microorganisms, and how the innate and adaptive immune systems are activated by protective microbes, leading to resistance, tolerance, or containment toward pathobionts and pathogens. A better understanding of the range, mode, and effects of microbiota-mediated protection is crucial for therapeutic designs (Box 4). Although evidence for long-reaching microbe-conferred protection is scarce, we examined possible ways protective microbes can reach beyond their local sites, in part by investigating how pathogens spread their influence across the host.
Box 4. Clinical perspectives about microbiota-mediated immunity.
Clinical trials can be designed in two ways to test for the therapeutic effects of protective microorganisms: either look for yet unknown properties of well-known in vivo probiotics, or test the clinical effects of candidate microorganisms found in in vitro or animal models. Such trials are already underway. Based on in vitro studies of isolated oral commensals (230), a trial is designed to test the efficacy of oral probiotics to prevent ventilator-associated pneumonia (231). A pilot study has been conducted to test the tolerance of a nasal probiotic spray against upper respiratory tract infections (232). Microbe-conferred protection is important for host health and, as we have reviewed, can vary in range, mode, and effects. However, such protection may come at a cost for the host. Careful screening of not just protective effects but also of potentially harmful properties and long-term side effects are crucial, as well-known probiotics can exhibit genotoxicity (233) as well as inflammatory diseases and tumors (145).
Why is there so little evidence for long-reaching microbial protection? First, evidence for long-reaching protection is limited in part because microbiome research is still in its infancy. Therefore, despite growing interest in “protective microbes,” only very few detailed studies are available. Second, many protective mechanisms are localized actions. Classical dose-dependent toxicological effects (for instance, bacteriocin production) are unlikely to be involved in long-reaching mechanisms, as dilution volume is considerably higher in comparison with the production of local compounds. Third, technical limitations likely result in the underestimation of long-reaching protective microbes. Long-reaching effects rely probably more on low or even very low concentration mechanisms, with threshold effects (i.e., quorum-sensing molecules) that are extremely difficult to identify with classical shotgun metabolomics studies, thus requiring targeted studies. Another difficulty is the transitory release of long-reaching microbes and signals. For instance, bacteremia or fungaemia are brief events that are difficult to detect. Last, there are very few within-host ecological analyses in animal models. Yet the processes that regulate lung microbiota, for instance, show that we cannot understand respiratory disease without a dynamic ecological point of view.
We suggest a form of “co-immunity” between the host and microbiota. The host is protected against pathogens both by its own immune system and by the direct or indirect action of microorganisms, not only at a local scale but also between local communities and systemically.
It is generally assumed that immunity is the immunity of one organism, and that this immunity is bounded by the conventional frontiers of that organism (e.g., its skin). Yet it is now well-established that several immune phenomena transcend the boundaries of organisms (234). We suggest considering all these phenomena as instances of a more general process that we dub “co-immunity” (see also Box 2). We define co-immunity as a form of immune defense associating components of several organisms (it is, therefore, a “multi-organism” or “cross-organism” immunity). Co-immunity can include the protection of the young by maternal antibodies (235), microbiota-mediated immunity (as detailed in the present review), but also population-level phenomena such as social immunity in insects (236), and “herd immunity” (in particular through vaccination) (237, 238) in humans and cattle.
Rohwer and colleagues use the term “non-host derived immunity” to describe bacteriophage-mediated immunity (98). But this term also applies to immunity mediated by other components of the microbiota (not just bacteriophages) and, even more importantly, all these correspond to subcases of the wider category of what we call here “co-immunity.” This broad perspective helps us realize that the idea that immunity can transcend traditional boundaries of organisms is not as surprising as it might seem at first sight and is in fact certainly a widespread phenomenon in nature.
At the same time, the concept of co-immunity opens up many fascinating questions. One key question is to determine in which circumstances conflicts may arise between different components, and whether these conflicts are regulated. A microorganism that was protective at one moment can become detrimental to the host at another moment, for instance, in immunocompromised states (239). Similarly, maternal antibodies are generally protective for the infant, but they can inhibit the infant’s response to vaccination (240). Overall, a crucial advantage of the concept of co-immunity is that it reminds us that immunity should always be understood as a multi-actor and dynamic phenomenon.
Author Contributions
LC and TB contributed equally to this paper. LC, TB, MT, LD, TS, and TP constituted all together the bibliography. LC, TB, and TP wrote the paper, which was substantially revised by MT, LD, and TS. LC was the primary author of the figures in this paper, with input and discussion from the other authors, especially TB and TP. TB was the main contributor to the clinical aspects. TP designed the project and managed the group.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Samuel Alizon, Silvia Bulgheresi, Nadine Cerf-Bensussan, Gérard Eberl, lvo Gomperts Boneca, Akiko Iwasaki, Bruno Lemaitre, Sarkis Mazmanian, Carmen Lía Murall, Philippe Sansonetti, Tz-Ching Yeh, and the ImmunoConcept lab for comments and suggestions. Parts of the figures were adapted with changes from Servier Medical Art (http://smart.servier.com/), licensed under Creative Commons Attribution 3.0 Unported.
Funding
TP and LC have received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme—grant agreement no. 637647—IDEM.
Footnote
- ^Derek Skillings helped dubbed the term, noting its similarity to “community.”
References
2. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol (2009) 9:313–23. doi:10.1038/nri2515
3. Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science (2001) 292:1115–8. doi:10.1126/science.1058709
4. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336:1268–73. doi:10.1126/science.1223490
5. Shi Y, Mu L. An expanding stage for commensal microbes in host immune regulation. Cell Mol Immunol (2017) 14:339–48. doi:10.1038/cmi.2016.64
6. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell (2014) 157:121–41. doi:10.1016/j.cell.2014.03.011
7. Sommer F, Bäckhed F. The gut microbiota – masters of host development and physiology. Nat Rev Microbiol (2013) 11:227–38. doi:10.1038/nrmicro2974
8. Abt MC, Pamer EG. Commensal bacteria mediated defenses against pathogens. Curr Opin Immunol (2014) 29:16–22. doi:10.1016/j.coi.2014.03.003
9. Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol (2013) 13:790–801. doi:10.1038/nri3535
10. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol (2013) 14:685–90. doi:10.1038/ni.2608
11. McKenney PT, Pamer EG. From hype to hope: the gut microbiota in enteric infectious disease. Cell (2015) 163:1326–32. doi:10.1016/j.cell.2015.11.032
12. Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol (2015) 194:4081–7. doi:10.4049/jimmunol.1403169
13. Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunology (2017) 6:e128. doi:10.1038/cti.2017.2
14. Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature (2008) 455:804–7. doi:10.1038/nature07250
15. Masson F, Lemaitre B. Symbiosis: protection from within. Elife (2017) 6:e24111. doi:10.7554/eLife.24111
16. Oldroyd GED. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol (2013) 11:252–63. doi:10.1038/nrmicro2990
17. Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science (2016) 352:535–8. doi:10.1126/science.aad9382
18. Lawley TD, Walker AW. Intestinal colonization resistance. Immunology (2013) 138:1–11. doi:10.1111/j.1365-2567.2012.03616.x
19. Stecher B, Berry D, Loy A. Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol Rev (2013) 37:793–829. doi:10.1111/1574-6976.12024
20. Ashburner FM, Mushin R. Experimental intestinal coliform infections in mice. J Hyg (Lond) (1962) 60:175–81. doi:10.1017/S0022172400039449
21. Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med (1954) 86:132–7. doi:10.3181/00379727-86-21030
22. Freter R. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J Infect Dis (1955) 97:57–65. doi:10.1093/infdis/97.1.57
23. Freter R. Experimental enteric Shigella and Vibrio infections in mice and guinea pigs. J Exp Med (1956) 104:411–8. doi:10.1084/jem.104.3.411
24. Mushin R, Dubos R. Colonization of the mouse intestine with Escherichia coli. J Exp Med (1965) 122:745–57. doi:10.1084/jem.122.4.745
25. van der Waaij D, Vries JMB, der Wees JECL. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) (1971) 69:405–11. doi:10.1017/S0022172400021653
26. Benveniste J, Lespinats G, Adam C, Salomon J-C. Immunoglobulins in intact, immunized, and contaminated axenic mice: study of serum IgA. J Immunol (1971) 107:1647–55.
27. Fubara ES, Freter R. Availability of locally synthesized and systemic antibodies in the intestine. Infect Immun (1972) 6:965–81.
28. Freter R. Interactions between mechanisms controlling the intestinal microflora. Am J Clin Nutr (1974) 27:1409–16.
29. Rolfe RD. Interactions among microorganisms of the indigenous intestinal flora and their influence on the host. Rev Infect Dis (1984) 6:S73–9. doi:10.1093/clinids/6.Supplement_1.S73
30. Berg RD, Owens WE. Inhibition of translocation of viable Escherichia coli from the gastrointestinal tract of mice by bacterial antagonism. Infect Immun (1979) 25:820–7.
31. Freter R, Abrams GD. Function of various intestinal bacteria in converting germfree mice to the normal state. Infect Immun (1972) 6:119–26.
32. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol (1977) 31:107–33. doi:10.1146/annurev.mi.31.100177.000543
34. Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr (1999) 69:1046s–51s.
35. Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol (2000) 1:113–8. doi:10.1038/77783
36. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science (2000) 288:2222–6. doi:10.1126/science.288.5474.2222
37. Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A (2002) 99:15451–5. doi:10.1073/pnas.202604299
38. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science (2006) 313:1126–30. doi:10.1126/science.1127119
39. Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity (2006) 24:677–88. doi:10.1016/j.immuni.2006.06.002
40. Soares MP, Teixeira L, Moita LF. Disease tolerance and immunity in host protection against infection. Nat Rev Immunol (2017) 17:83–96. doi:10.1038/nri.2016.136
41. Tomkovich S, Jobin C. Microbiota and host immune responses: a love-hate relationship. Immunology (2016) 147:1–10. doi:10.1111/imm.12538
42. Laurent P, Jolivel V, Manicki P, Chiu L, Contin-Bordes C, Truchetet M-E, et al. Immune-mediated repair: a matter of plasticity. Front Immunol (2017) 8:454. doi:10.3389/fimmu.2017.00454
43. Brugiroux S, Beutler M, Pfann C, Garzetti D, Ruscheweyh H-J, Ring D, et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol (2016) 2:16215. doi:10.1038/nmicrobiol.2016.215
44. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature (2015) 517:205–8. doi:10.1038/nature13828
45. Spees AM, Lopez CA, Kingsbury DD, Winter SE, Baumler AJ. Colonization resistance: battle of the bugs or ménage a trois with the host? PLoS Pathog (2013) 9:e1003730. doi:10.1371/journal.ppat.1003730
46. Stecher B, Hardt W-D. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol (2011) 14:82–91. doi:10.1016/j.mib.2010.10.003
47. Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol (2010) 8:15–25. doi:10.1038/nrmicro2259
48. Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol (2008) 62:375–401. doi:10.1146/annurev.micro.030608.101423
49. Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol (2013) 21:230–7. doi:10.1016/j.tim.2013.02.003
50. Becattini S, Littmann ER, Carter RA, Kim SG, Morjaria SM, Ling L, et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med (2017) 214:1973–89. doi:10.1084/jem.20170495
51. Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, et al. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe (2013) 14:26–37. doi:10.1016/j.chom.2013.06.007
52. Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One (2013) 8:e53957. doi:10.1371/journal.pone.0053957
53. Sassone-Corsi M, Nuccio S-P, Liu H, Hernandez D, Vu CT, Takahashi AA, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature (2016) 540:280–3. doi:10.1038/nature20557
54. Schamberger GP, Diez-Gonzalez F. Selection of recently isolated colicinogenic Escherichia coli strains inhibitory to Escherichia coli O157:H7. J Food Prot (2002) 65:1381–7. doi:10.4315/0362-028X-65.9.1381
55. Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature (2016) 535:511–6. doi:10.1038/nature18634
56. Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, et al. Bacteriocin production augments niche competition by enterococci in the mammalian GI tract. Nature (2015) 526:719–22. doi:10.1038/nature15524
57. Rea MC, Sit CS, Clayton E, O’Connor PM, Whittal RM, Zheng J, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A (2010) 107:9352–7. doi:10.1073/pnas.0913554107
58. Rea MC, Dobson A, O’Sullivan O, Crispie F, Fouhy F, Cotter PD, et al. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A (2011) 108:4639–44. doi:10.1073/pnas.1001224107
59. Sutyak Noll K, Prichard MN, Khaykin A, Sinko PJ, Chikindas ML. The natural antimicrobial peptide subtilosin acts synergistically with glycerol monolaurate, lauric arginate, and ε-poly-l-lysine against bacterial vaginosis-associated pathogens but not human Lactobacilli. Antimicrob Agents Chemother (2012) 56:1756–61. doi:10.1128/AAC.05861-11
60. Cogen AL, Yamasaki K, Muto J, Sanchez KM, Crotty Alexander L, Tanios J, et al. Staphylococcus epidermidis antimicrobial delta-toxin (phenol-soluble modulin-gamma) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLoS One (2010) 5:e8557. doi:10.1371/journal.pone.0008557
61. Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol (2010) 130:192–200. doi:10.1038/jid.2009.243
62. Crost EH, Pujol A, Ladiré M, Dabard J, Raibaud P, Carlier JP, et al. Production of an antibacterial substance in the digestive tract involved in colonization-resistance against Clostridium perfringens. Anaerobe (2010) 16:597–603. doi:10.1016/j.anaerobe.2010.06.009
63. Shin R, Suzuki M, Morishita Y. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J Med Microbiol (2002) 51:201–6. doi:10.1099/0022-1317-51-3-201
64. Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere (2016) 1:e00045-15. doi:10.1128/mSphere.00045-15
65. Weingarden AR, Dosa PI, DeWinter E, Steer CJ, Shaughnessy MK, Johnson JR, et al. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PLoS One (2016) 11:e0147210. doi:10.1371/journal.pone.0147210
66. Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology (2014) 146:1547–53. doi:10.1053/j.gastro.2014.01.059
67. Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A (2016) 113:3627–32. doi:10.1073/pnas.1522510113
68. Wexler AG, Bao Y, Whitney JC, Bobay L-M, Xavier JB, Schofield WB, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A (2016) 113:3639–44. doi:10.1073/pnas.1525637113
69. Annuk H, Shchepetova J, Kullisaar T, Songisepp E, Zilmer M, Mikelsaar M. Characterization of intestinal Lactobacilli as putative probiotic candidates. J Appl Microbiol (2003) 94:403–12. doi:10.1046/j.1365-2672.2003.01847.x
70. Mishra C, Lambert J. Production of anti-microbial substances by probiotics. Asia Pac J Clin Nutr (1996) 5:20–4.
71. Danielsson D, Teigen PK, Moi H. The genital econiche: focus on microbiota and bacterial vaginosis. Ann N Y Acad Sci (2011) 1230:48–58. doi:10.1111/j.1749-6632.2011.06041.x
72. Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One (2013) 8:e55380. doi:10.1371/journal.pone.0055380
73. Altier C. Genetic and environmental control of Salmonella invasion. J Microbiol (2005) 43:85–92.
74. Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun (1983) 39:686–703.
75. Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature (2010) 465:346–9. doi:10.1038/nature09074
76. Vandecandelaere I, Depuydt P, Nelis HJ, Coenye T. Protease production by Staphylococcus epidermidis and its effect on Staphylococcus aureus biofilms. Pathog Dis (2014) 70:321–31. doi:10.1111/2049-632X.12133
77. Wu C-C, Lin C-T, Wu C-Y, Peng W-S, Lee M-J, Tsai Y-C. Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol Oral Microbiol (2015) 30:16–26. doi:10.1111/omi.12063
78. Melo TA, dos Santos TF, de Almeida ME, Junior LAGF, Andrade EF, Rezende RP, et al. Inhibition of Staphylococcus aureus biofilm by Lactobacillus isolated from fine cocoa. BMC Microbiol (2016) 16:250. doi:10.1186/s12866-016-0871-8
79. Bassler BL, Losick R. Bacterially speaking. Cell (2006) 125:237–46. doi:10.1016/j.cell.2006.04.001
80. Davis MF, Baron P, Price LB, Williams DL, Jeyaseelan S, Hambleton IR, et al. Dry collection and culture methods for recovery of methicillin-susceptible and methicillin-resistant Staphylococcus aureus strains from indoor home environments. Appl Environ Microbiol (2012) 78:2474–6. doi:10.1128/AEM.06886-11
81. Cotar A, Ionescu B, Pelinescu D, Voidarou C, Lazar V, Bezirtzoglou E, et al. Current solutions for the interception of quorum sensing in Staphylococcus aureus. Curr Org Chem (2013) 17:97–104. doi:10.2174/1385272811317020004
82. Algburi A, Zehm S, Netrebov V, Bren AB, Chistyakov V, Chikindas ML. Subtilosin prevents biofilm formation by inhibiting bacterial quorum sensing. Probiotics Antimicrob Proteins (2017) 9:81–90. doi:10.1007/s12602-016-9242-x
83. Irie Y, O’Toole GA, Yuk MH. Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. FEMS Microbiol Lett (2005) 250:237–43. doi:10.1016/j.femsle.2005.07.012
84. Valle J, Da Re S, Henry N, Fontaine T, Balestrino D, Latour-Lambert P, et al. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc Natl Acad Sci U S A (2006) 103:12558–63. doi:10.1073/pnas.0605399103
85. Sambanthamoorthy K, Feng X, Patel R, Patel S, Paranavitana C. Antimicrobial and antibiofilm potential of biosurfactants isolated from Lactobacilli against multi-drug-resistant pathogens. BMC Microbiol (2014) 14:197. doi:10.1186/1471-2180-14-197
86. Shokouhfard M, Kermanshahi RK, Shahandashti RV, Feizabadi MM, Teimourian S. The inhibitory effect of a Lactobacillus acidophilus derived biosurfactant on biofilm producer Serratia marcescens. Iran J Basic Med Sci (2015) 18:1001–7.
87. Kinnunen M, Dechesne A, Proctor C, Hammes F, Johnson D, Quintela-Baluja M, et al. A conceptual framework for invasion in microbial communities. ISME J (2016) 10:2773–9. doi:10.1038/ismej.2016.75
88. Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol (2006) 296:149–61. doi:10.1016/j.ijmm.2006.02.005
89. Murall CL, Abbate JL, Puelma Touzel M, Allen-Vercoe E, Alizon S, Froissart R, et al. Invasions of host-associated microbiome networks. In: Bohan DA, Dumbrell AJ, Massol F, editors. Networks of Invasion: Empirical Evidence and Case Studies. Oxford: Elsevier (2017). p. 201–81.
90. Mallon CA, van Elsas JD, Salles JF. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol (2015) 23:719–29. doi:10.1016/j.tim.2015.07.013
91. Levine JM, Adler PB, Yelenik SG. A meta-analysis of biotic resistance to exotic plant invasions: biotic resistance to plant invasion. Ecol Lett (2004) 7:975–89. doi:10.1111/j.1461-0248.2004.00657.x
92. Catford JA, Jansson R, Nilsson C. Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers Distrib (2009) 15:22–40. doi:10.1111/j.1472-4642.2008.00521.x
93. Blackburn TM, Pyšek P, Bacher S, Carlton JT, Duncan RP, Jarošík V, et al. A proposed unified framework for biological invasions. Trends Ecol Evol (2011) 26:333–9. doi:10.1016/j.tree.2011.03.023
94. Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol (2012) 30:149–73. doi:10.1146/annurev-immunol-020711-075001
95. Bron PA, Kleerebezem M, Brummer R-J, Cani PD, Mercenier A, MacDonald TT, et al. Can probiotics modulate human disease by impacting intestinal barrier function? Br J Nutr (2017) 117:93–107. doi:10.1017/S0007114516004037
96. Gill N, Wlodarska M, Finlay BB. Roadblocks in the gut: barriers to enteric infection. Cell Microbiol (2011) 13:660–9. doi:10.1111/j.1462-5822.2011.01578.x
97. Wootton L. Phage biology: a new barrier at mucosal surfaces. Nat Rev Microbiol (2013) 11:430–1. doi:10.1038/nrmicro3064
98. Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A (2013) 110:10771–6. doi:10.1073/pnas.1305923110
99. Martins FS, Vieira AT, Elian SDA, Arantes RME, Tiago FCP, Sousa LP, et al. Inhibition of tissue inflammation and bacterial translocation as one of the protective mechanisms of Saccharomyces boulardii against Salmonella infection in mice. Microbes Infect (2013) 15:270–9. doi:10.1016/j.micinf.2012.12.007
100. Botes M, Loos B, van Reenen CA, Dicks LMT. Adhesion of the probiotic strains Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 to Caco-2 cells under conditions simulating the intestinal tract, and in the presence of antibiotics and anti-inflammatory medicaments. Arch Microbiol (2008) 190:573–84. doi:10.1007/s00203-008-0408-0
101. Altenhoefer A, Oswald S, Sonnenborn U, Enders C, Schulze J, Hacker J, et al. The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol Med Microbiol (2004) 40:223–9. doi:10.1016/S0928-8244(03)00368-7
102. Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol (1999) 276:G941–50.
103. Tuomola EM, Ouwehand AC, Salminen SJ. The effect of probiotic bacteria on the adhesion of pathogens to human intestinal mucus. FEMS Immunol Med Microbiol (1999) 26:137–42. doi:10.1111/j.1574-695X.1999.tb01381.x
104. Walsham ADS, MacKenzie DA, Cook V, Wemyss-Holden S, Hews CL, Juge N, et al. Lactobacillus reuteri inhibition of enteropathogenic Escherichia coli adherence to human intestinal epithelium. Front Microbiol (2016) 7:244. doi:10.3389/fmicb.2016.00244
105. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, et al. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol (2006) 72:946–9. doi:10.1128/AEM.72.1.946-949.2006
106. Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature (2011) 469:543–9. doi:10.1038/nature09646
107. Ayres JS. Cooperative microbial tolerance behaviors in host-microbiota mutualism. Cell (2016) 165:1323–31. doi:10.1016/j.cell.2016.05.049
108. Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science (2012) 335:936–41. doi:10.1126/science.1214935
109. Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol (2008) 8:889–95. doi:10.1038/nri2432
110. Råberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science (2007) 318:812–4. doi:10.1126/science.1148526
111. de Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. Human microbiota-secreted factors inhibit Shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun (2009) 77:783–90. doi:10.1128/IAI.01048-08
112. Carey CM, Kostrzynska M, Ojha S, Thompson S. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J Microbiol Methods (2008) 73:125–32. doi:10.1016/j.mimet.2008.01.014
113. Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature (2014) 515:423–6. doi:10.1038/nature13738
114. Kolling GL, Wu M, Warren CA, Durmaz E, Klaenhammer TR, Timko MP, et al. Lactic acid production by Streptococcus thermophilus alters Clostridium difficile infection and in vitro Toxin A production. Gut Microbes (2012) 3:523–9. doi:10.4161/gmic.21757
115. Asahara T, Shimizu K, Nomoto K, Hamabata T, Ozawa A, Takeda Y. Probiotic Bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect Immun (2004) 72:2240–7. doi:10.1128/IAI.72.4.2240-2247.2004
116. Ripert G, Racedo SM, Elie A-M, Jacquot C, Bressollier P, Urdaci MC. Secreted compounds of the probiotic Bacillus clausii strain o/c inhibit the cytotoxic effects induced by Clostridium difficile and Bacillus cereus toxins. Antimicrob Agents Chemother (2016) 60:3445–54. doi:10.1128/AAC.02815-15
117. Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun (1999) 67:302–7.
118. Valdés-Varela L, Hernández-Barranco AM, Ruas-Madiedo P, Gueimonde M. Effect of Bifidobacterium upon Clostridium difficile growth and toxicity when co-cultured in different prebiotic substrates. Front Microbiol (2016) 7:738. doi:10.3389/fmicb.2016.00738
119. Valdés-Varela L, Alonso-Guervos M, García-Suárez O, Gueimonde M, Ruas-Madiedo P. Screening of Bifidobacteria and Lactobacilli able to antagonize the cytotoxic effect of Clostridium difficile upon intestinal epithelial HT29 monolayer. Front Microbiol (2016) 7:577. doi:10.3389/fmicb.2016.00577
120. Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature (2008) 456:507–10. doi:10.1038/nature07450
121. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell (2005) 122:107–18. doi:10.1016/j.cell.2005.05.007
122. Pradeu T. A mixed self: the role of symbiosis in development. Biol Theory (2011) 6:80–8. doi:10.1007/s13752-011-0011-5
123. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity (2017) 46:562–76. doi:10.1016/j.immuni.2017.04.008
124. Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol (2011) 9:356–68. doi:10.1038/nrmicro2546
125. Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol (2012) 12:503–16. doi:10.1038/nri3228
126. Nagy I, Pivarcsi A, Kis K, Koreck A, Bodai L, McDowell A, et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect (2006) 8:2195–205. doi:10.1016/j.micinf.2006.04.001
127. Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nature Immunology (2003) 4:269–73. doi:10.1038/ni888
128. Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG, et al. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A (2013) 110:E3730–8. doi:10.1073/pnas.1304960110
129. Salzman NH. Paneth cell defensins and the regulation of the microbiome: détente at mucosal surfaces. Gut Microbes (2010) 1:401–6. doi:10.4161/gmic.1.6.14076
130. Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A (2008) 105:20858–63. doi:10.1073/pnas.0808723105
131. Macpherson AJ, Köller Y, McCoy KD. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol (2015) 36:460–70. doi:10.1016/j.it.2015.06.006
132. Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev (2012) 245:132–46. doi:10.1111/j.1600-065X.2011.01072.x
133. Strugnell RA, Wijburg OLC. The role of secretory antibodies in infection immunity. Nat Rev Microbiol (2010) 8:656–67. doi:10.1038/nrmicro2384
134. Gutzeit C, Magri G, Cerutti A. Intestinal IgA production and its role in host-microbe interaction. Immunol Rev (2014) 260:76–85. doi:10.1111/imr.12189
135. Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol (2011) 4:603–11. doi:10.1038/mi.2011.41
136. Everett ML, Palestrant D, Miller SE, Bollinger RR, Parker W. Immune exclusion and immune inclusion: a new model of host-bacterial interactions in the gut. Clin Appl Immunol Rev (2004) 4:321–32. doi:10.1016/j.cair.2004.03.001
137. Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S, et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity (2015) 43:527–40. doi:10.1016/j.immuni.2015.08.011
138. Zagato E, Mazzini E, Rescigno M. The variegated aspects of Immunoglobulin A. Immunol Lett (2016) 178:45–9. doi:10.1016/j.imlet.2016.04.009
139. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell (2014) 158:1000–10. doi:10.1016/j.cell.2014.08.006
140. Ivanov II, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol (2011) 14:106–14. doi:10.1016/j.mib.2010.12.003
141. Molloy M, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol (2012) 24:58–66. doi:10.1016/j.smim.2011.11.008
142. Pang IK, Iwasaki A. Control of antiviral immunity by pattern recognition and the microbiome. Immunol Rev (2012) 245:209–26. doi:10.1111/j.1600-065X.2011.01073.x
143. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139:485–98. doi:10.1016/j.cell.2009.09.033
144. Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol (2010) 10:735–44. doi:10.1038/nri2850
145. Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, et al. Host-protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell (2016) 167:444–56.e14. doi:10.1016/j.cell.2016.08.076
146. Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science (2014) 346:954–9. doi:10.1126/science.1260144
147. Belkaid Y, Tamoutounour S. The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol (2016) 16:353–66. doi:10.1038/nri.2016.48
148. Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence? Br J Dermatol (2008) 158:442–55. doi:10.1111/j.1365-2133.2008.08437.x
149. Man WH, de Steenhuijsen Piters WAA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol (2017) 15:259–70. doi:10.1038/nrmicro.2017.14
150. Gabryszewski SJ, Bachar O, Dyer KD, Percopo CM, Killoran KE, Domachowske JB, et al. Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J Immunol (2011) 186:1151–61. doi:10.4049/jimmunol.1001751
151. Stinson LF, Payne MS, Keelan JA. Planting the seed: origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota. Crit Rev Microbiol (2016) 43:352–69. doi:10.1080/1040841X.2016.1211088
152. Sharma R, Young C, Neu J. Molecular modulation of intestinal epithelial barrier: contribution of microbiota. J Biomed Biotechnol (2010) 2010:15. doi:10.1155/2010/305879
153. Pontier-Bres R, Rampal P, Peyron J-F, Munro P, Lemichez E, Czerucka D. The Saccharomyces boulardii CNCM I-745 strain shows protective effects against the B. anthracis LT toxin. Toxins (Basel) (2015) 7:4455–67. doi:10.3390/toxins7114455
154. Frantz AL, Bruno MEC, Rogier EW, Tuna H, Cohen DA, Bondada S, et al. Multifactorial patterns of gene expression in colonic epithelial cells predict disease phenotypes in experimental colitis. Inflamm Bowel Dis (2012) 18:2138–48. doi:10.1002/ibd.22923
155. Knoop KA, McDonald KG, Kulkarni DH, Newberry RD. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut (2016) 65:1100–9. doi:10.1136/gutjnl-2014-309059
156. Birchenough GMH, Johansson ME, Gustafsson JK, Bergström JH, Hansson GC. New developments in goblet cell mucus secretion and function. Mucosal Immunol (2015) 8:712–9. doi:10.1038/mi.2015.32
157. Birchenough GMH, Nyström EEL, Johansson MEV, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science (2016) 352:1535–42. doi:10.1126/science.aaf7419
158. Ramanan D, Bowcutt R, Lee SC, Tang MS, Kurtz ZD, Ding Y, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science (2016) 352:608–12. doi:10.1126/science.aaf3229
159. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature (2008) 453:620–5. doi:10.1038/nature07008
160. Budden KF, Gellatly SL, Wood DLA, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol (2017) 15:55–63. doi:10.1038/nrmicro.2016.142
161. El-Solh AA. Association between pneumonia and oral care in nursing home residents. Lung (2011) 189:173–80. doi:10.1007/s00408-011-9297-0
162. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature (2014) 513:59–64. doi:10.1038/nature13568
163. Willis AR, Moore C, Mazon-Moya M, Krokowski S, Lambert C, Till R, et al. Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr Biol (2016) 26:3343–51. doi:10.1016/j.cub.2016.09.067
164. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell (2017) 168:928–43.e11. doi:10.1016/j.cell.2017.01.022
165. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol (2004) 4:478–85. doi:10.1038/nri1373
166. Gorjifard S, Goldszmid RS. Microbiota-myeloid cell crosstalk beyond the gut. J Leukoc Biol (2016) 100:865–79. doi:10.1189/jlb.3RI0516-222R
167. Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe (2014) 15:374–81. doi:10.1016/j.chom.2014.02.006
168. Rizzetto L, Ifrim DC, Moretti S, Tocci N, Cheng S-C, Quintin J, et al. Fungal chitin induces trained immunity in human monocytes during cross-talk of the host with Saccharomyces cerevisiae. J Biol Chem (2016) 291:7961–72. doi:10.1074/jbc.M115.699645
169. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity (2012) 37:171–86. doi:10.1016/j.immuni.2012.05.020
170. de Agüero MG, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science (2016) 351:1296–302. doi:10.1126/science.aad2571
171. Koch MA, Reiner GL, Lugo KA, Kreuk LSM, Stanbery AG, Ansaldo E, et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell (2016) 165:827–41. doi:10.1016/j.cell.2016.04.055
172. Gallichan WS, Woolstencroft RN, Guarasci T, McCluskie MJ, Davis HL, Rosenthal KL. Intranasal immunization with CpG oligodeoxynucleotides as an adjuvant dramatically increases IgA and protection against herpes simplex virus-2 in the genital tract. J Immunol (2001) 166:3451–7. doi:10.4049/jimmunol.166.5.3451
173. Villarino NF, LeCleir GR, Denny JE, Dearth SP, Harding CL, Sloan SS, et al. Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci U S A (2016) 113:2235–40. doi:10.1073/pnas.1504887113
174. Yilmaz B, Portugal S, Tran TM, Gozzelino R, Ramos S, Gomes J, et al. Gut microbiota elicits a protective immune response against malaria transmission. Cell (2014) 159:1277–89. doi:10.1016/j.cell.2014.10.053
175. Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe (2009) 6:187–96. doi:10.1016/j.chom.2009.06.005
176. Pfeiffer JK, Sonnenburg JL. The intestinal microbiota and viral susceptibility. Front Microbiol (2011) 2:92. doi:10.3389/fmicb.2011.00092
177. Davidson LE, Fiorino A-M, Snydman DR, Hibberd PL. Lactobacillus GG as an immune adjuvant for live-attenuated influenza vaccine in healthy adults: a randomized double-blind placebo-controlled trial. Eur J Clin Nutr (2011) 65:501–7. doi:10.1038/ejcn.2010.289
178. Pérez-Santiago J, Gianella S, Massanella M, Spina CA, Karris MY, Var SR, et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS (2013) 27:1921–31. doi:10.1097/QAD.0b013e3283611816
179. Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity (2012) 37:158–70. doi:10.1016/j.immuni.2012.04.011
180. Pradeu T. Mutualistic viruses and the heteronomy of life. Stud Hist Philos Biol Biomed Sci (2016) 59:80–8. doi:10.1016/j.shpsc.2016.02.007
181. Roossinck MJ. The good viruses: viral mutualistic symbioses. Nat Rev Microbiol (2011) 9:99–108. doi:10.1038/nrmicro2491
182. Virgin HW. The virome in mammalian physiology and disease. Cell (2014) 157:142–50. doi:10.1016/j.cell.2014.02.032
183. Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell (2009) 138:30–50. doi:10.1016/j.cell.2009.06.036
184. Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature (2007) 447:326–9. doi:10.1038/nature05762
185. Robinson CM, Pfeiffer JK. Viruses and the microbiota. Annu Rev Virol (2014) 1:55–69. doi:10.1146/annurev-virology-031413-085550
186. Tillmann HL, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, Wilber JC, et al. Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med (2001) 345:715–24. doi:10.1056/NEJMoa010398
187. Xiang J, Wünschmann S, Diekema DJ, Klinzman D, Patrick KD, George SL, et al. Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med (2001) 345:707–14. doi:10.1056/NEJMoa003364
188. Stapleton JT, Chaloner K, Martenson JA, Zhang J, Klinzman D, Xiang J, et al. GB virus C infection is associated with altered lymphocyte subset distribution and reduced T cell activation and proliferation in HIV-infected individuals. PLoS One (2012) 7:e50563. doi:10.1371/journal.pone.0050563
189. Fardini Y, Chung P, Dumm R, Joshi N, Han YW. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun (2010) 78:1789–96. doi:10.1128/IAI.01395-09
190. Donnet-Hughes A, Perez PF, Doré J, Leclerc M, Levenez F, Benyacoub J, et al. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc Nutr Soc (2010) 69:407–15. doi:10.1017/S0029665110001898
191. Nikkari S, McLaughlin IJ, Bi W, Dodge DE, Relman DA. Does blood of healthy subjects contain bacterial ribosomal DNA? J Clin Microbiol (2001) 39:1956–9. doi:10.1128/JCM.39.5.1956-1959.2001
192. Tomás I, Diz P, Tobías A, Scully C, Donos N. Periodontal health status and bacteraemia from daily oral activities: systematic review/meta-analysis. J Clin Periodontol (2012) 39:213–28. doi:10.1111/j.1600-051X.2011.01784.x
193. Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, et al. Gut microbiota-induced Immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity (2016) 44:647–58. doi:10.1016/j.immuni.2016.02.006
194. Schieber AMP, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, et al. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science (2015) 350:558–63. doi:10.1126/science.aac6468
195. Bocci V. The neglected organ: bacterial flora has a crucial immunostimulatory role. Perspect Biol Med (1992) 35:251–60. doi:10.1353/pbm.1992.0004
196. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes (2007) 56:1761–72. doi:10.2337/db06-1491
197. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes (2008) 57:1470–81. doi:10.2337/db07-1403
198. Hersoug L-G, Møller P, Loft S. Gut microbiota-derived lipopolysaccharide uptake and trafficking to adipose tissue: implications for inflammation and obesity. Obes Rev (2016) 17:297–312. doi:10.1111/obr.12370
199. Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med (2010) 16:228–31. doi:10.1038/nm.2087
200. Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature (2012) 482:179–85. doi:10.1038/nature10809
201. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A (2009) 106:3698–703. doi:10.1073/pnas.0812874106
202. Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol (2014) 28:1221–38. doi:10.1210/me.2014-1108
203. Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol (2015) 6:1085. doi:10.3389/fmicb.2015.01085
204. Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med (2014) 6:263ra158. doi:10.1126/scitranslmed.3009759
205. Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol (2014) 14:405. doi:10.1038/nri3684
206. Denny JE, Powell WL, Schmidt NW. Local and long-distance calling: conversations between the gut microbiota and intra- and extra-gastrointestinal tract infections. Front Cell Infect Microbiol (2016) 6:41. doi:10.3389/fcimb.2016.00041
207. Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, et al. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol (2012) 188:1411–20. doi:10.4049/jimmunol.1101682
208. Clarke TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via Nod-like receptor ligands. Infect Immun (2014) 82:4596–606. doi:10.1128/IAI.02212-14
209. Perry S, de Jong BC, Solnick JV, de la Luz Sanchez M, Yang S, Lin PL, et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One (2010) 5:e8804. doi:10.1371/journal.pone.0008804
210. Majlessi L, Sayes F, Bureau J-F, Pawlik A, Michel V, Jouvion G, et al. Colonization with Helicobacter is concomitant with modified gut microbiota and drastic failure of the immune control of Mycobacterium tuberculosis. Mucosal Immunol (2017) 10:1178–89. doi:10.1038/mi.2016.140
211. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A (2011) 108:5354–9. doi:10.1073/pnas.1019378108
212. Chen L-W, Chen P-H, Hsu C-M. Commensal microflora contribute to host defense against Escherichia coli pneumonia through toll-like receptors. Shock (2011) 36:67–75. doi:10.1097/SHK.0b013e3182184ee7
213. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJTH, de Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut (2016) 65:575–83. doi:10.1136/gutjnl-2015-309728
214. Tanaka K, Sawamura S, Satoh T, Kobayashi K, Noda S. Role of the indigenous microbiota in maintaining the virus-specific CD8 memory T cells in the lung of mice infected with murine cytomegalovirus. J Immunol (2007) 178:5209–16. doi:10.4049/jimmunol.178.8.5209
215. Gauguet S, D’Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, et al. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun (2015) 83:4003–14. doi:10.1128/IAI.00037-15
216. McAleer JP, Nguyen NLH, Chen K, Kumar P, Ricks DM, Binnie M, et al. Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J Immunol (2016) 197:97–107. doi:10.4049/jimmunol.1502566
217. Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One (2012) 7:e36313. doi:10.1371/journal.pone.0036313
218. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio (2015) 6:e00037. doi:10.1128/mBio.00037-15
219. Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, et al. Application of a neutral community model to assess structuring of the human lung microbiome. MBio (2015) 6:e02284-14. doi:10.1128/mBio.02284-14
220. Dickson RP, Erb-Downward JR, Huffnagle GB. Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol (2015) 309:L1047–55. doi:10.1152/ajplung.00279.2015
221. Whiteson KL, Bailey B, Bergkessel M, Conrad D, Delhaes L, Felts B, et al. The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis. Parallels from island biogeography. Am J Respir Crit Care Med (2014) 189:1309–15. doi:10.1164/rccm.201312-2129PP
222. Dickson RP, Erb-Downward JR, Freeman CM, Walker N, Scales BS, Beck JM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS One (2014) 9:e97214. doi:10.1371/journal.pone.0097214
223. Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet (2014) 384:691–702. doi:10.1016/S0140-6736(14)61136-3
224. Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol (2016) 1:16113. doi:10.1038/nmicrobiol.2016.113
225. de Steenhuijsen Piters WAA, Sanders EAM, Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc Lond B Biol Sci (2015) 370(1675):20140294. doi:10.1098/rstb.2014.0294
226. Buddington R. Using probiotics and prebiotics to manage the gastrointestinal tract ecosystem. In: Charalampopoulos D, Rastall RA, editors. Prebiotics and Probiotics Science and Technology. New York: Springer (2009). p. 1–31.
227. Wright ML, Pendarvis K, Nanduri B, Edelmann MJ, Jenkins HN, Reddy JS, et al. The effect of oxygen on bile resistance in Listeria monocytogenes. J Proteomics Bioinform (2016) 9:107–19. doi:10.4172/jpb.1000396
228. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol (2015) 14:20–32. doi:10.1038/nrmicro3552
229. Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol (2013) 14:676–84. doi:10.1038/ni.2640
230. Santagati M, Scillato M, Patanè F, Aiello C, Stefani S. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol Med Microbiol (2012) 65:23–31. doi:10.1111/j.1574-695X.2012.00928.x
231. Probiotics to Prevent Severe Pneumonia and Endotracheal Colonization Trial (PROSPECT). ClinicalTrials.gov Identifier: NCT02462590 (2016). Available from: https://clinicaltrials.gov/ct2/show/NCT02462590
232. Santagati M, Scillato M, Muscaridola N, Metoldo V, La Mantia I, Stefani S. Colonization, safety, and tolerability study of the Streptococcus salivarius 24SMBc nasal spray for its application in upper respiratory tract infections. Eur J Clin Microbiol Infect Dis (2015) 34:2075–80. doi:10.1007/s10096-015-2454-2
233. Olier M, Marcq I, Salvador-Cartier C, Secher T, Dobrindt U, Boury M, et al. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes (2012) 3:501–9. doi:10.4161/gmic.21737
234. Pradeu T. The Limits of the Self: Immunology and Biological Identity. New York: Oxford University Press (2012).
235. Van de Perre P. Transfer of antibody via mother’s milk. Vaccine (2003) 21:3374–6. doi:10.1016/S0264-410X(03)00336-0
236. Cremer S, Armitage SAO, Schmid-Hempel P. Social immunity. Curr Biol (2007) 17:R693–702. doi:10.1016/j.cub.2007.06.008
237. Anderson RM. The concept of herd immunity and the design of community-based immunization programmes. Vaccine (1992) 10:928–35. doi:10.1016/0264-410X(92)90327-G
238. Fine PEM. Herd immunity: history, theory, practice. Epidemiol Rev (1993) 15:265–302. doi:10.1093/oxfordjournals.epirev.a036121
239. Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe (2010) 7:265–76. doi:10.1016/j.chom.2010.03.004
Keywords: host–microbiota symbiosis, colonization resistance, microbial ecology, disease tolerance, pathogens, containment, infectious diseases
Citation: Chiu L, Bazin T, Truchetet M-E, Schaeverbeke T, Delhaes L and Pradeu T (2017) Protective Microbiota: From Localized to Long-Reaching Co-Immunity. Front. Immunol. 8:1678. doi: 10.3389/fimmu.2017.01678
Received: 13 July 2017; Accepted: 15 November 2017;
Published: 07 December 2017
Edited by:
Mats Bemark, University of Gothenburg, SwedenReviewed by:
Siegfried Hapfelmeier, University of Bern, SwitzerlandWim Van Den Broeck, Ghent University, Belgium
Copyright: © 2017 Chiu, Bazin, Truchetet, Schaeverbeke, Delhaes and Pradeu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Pradeu, dGhvbWFzLnByYWRldUB1LWJvcmRlYXV4LmZy
†Co-first authors.
 Lynn Chiu
Lynn Chiu Thomas Bazin
Thomas Bazin Marie-Elise Truchetet
Marie-Elise Truchetet Thierry Schaeverbeke2,4
Thierry Schaeverbeke2,4 Thomas Pradeu
Thomas Pradeu