- Medical Microbiology and Molecular Biology Laboratory, Interdisciplinary Biotechnology Unit, Aligarh Muslim University, Aligarh, India
Urinary tract infection (UTI) is one the common infections caused by the recalcitrant nature of biofilms, developed after the pathogen has adhered to the inner lining of the urinary tract. Although significant research has been made in recent years to control these types of infection, but as of yet, no approach has sufficiently been able to reduce the prevalence of UTIs. The main objective of this study was to prevent UTIs through targeting the fimH gene, which is the major virulent factor responsible for biofilm formation. The novelty of this work lies in the use of CRISPRi, a gene specific editing tool to control such types of infections. Accordingly, the system was designed to target fimH gene, responsible for bacterial adherence and this approach was successfully validated by performing microscopic, biofilm and adherence assays.
Introduction
Urinary tract infection (UTI) is considered as one of the most important causes of health issues and morbidity, worldwide, affecting persons of all ages due to its chronic and recurrent nature (1–3). Uropathogenic Escherichia coli (UPEC) strains are thought to be the main etiologic agents associated with UTIs. These isolates originate from patients’ intestinal normal flora and lead to UTI when fecal E. coli colonizes the periurethral region (4). Consequently it forms both intra- and extracellular biofilm-like communities within the urinary bladder (1), which thwarts the host immune-mediated clearance and develops resistance against antimicrobial therapy that finally results in persistent infections that are difficult to eradicate.
There are several virulence factors associated with UPEC, e.g., fimbriae 1, hemolysis, serum resistance, hydrophobicity, but type 1 fimbriae accounts for more than 95% of the total E. coli virulence factor, causing UTI (5–7).
Type 1 fimbriae are approximately 0.5–1.5 µm long structures, assembled through a chaperone-usher pathway (5, 8). They are also known as attachment pilus and are prevalent in gram negative bacteria (9, 10). Usually, bacteria utilize these structures to carry adhesions at their tip for attachment in order to obtain nutrients and withstand shear forces (10, 11). In E. coli, these type 1 fimbriae carry 30-kDa highly structural conserve adhesive subunit FimH, which is employed for mannose specific adherence (5, 11). FimH occurs in two alternative conformations depending on interaction of its two structural domains, i.e., lectin domain (LD having mannose binding pocket) and pillin domain (PD connecting FimH with other pillin minor subunits like FimG) (5, 12). LD and PD are connected via a short linker chain and on their close interaction LD exists in twisted and compressed conformation that leads to an opening of mannose binding pocket. Hence, as a result, close interaction between LD and PD leads to much lower affinity for mannose binding than in separated domain conformation (5). The binding of mannose to FimH is allosterically regulated that leads to shuffling between its two alternative conformations (5). Apart from FimH, the structural role of type 1 fimbriae is also governed by other subunits, namely FimA (main pillin subunit), FimG, and FimH (minor subunits) (8, 13–15). The mannose binding domain (LD) of the FimH is mainly responsible for adherence, which is the first step of infection. Hence, through fimbriae, bacteria attach to the substratum in order to withstand shear forces and absorb nutrients. The structural components of type 1 fimbriae are shown in Figure 1.
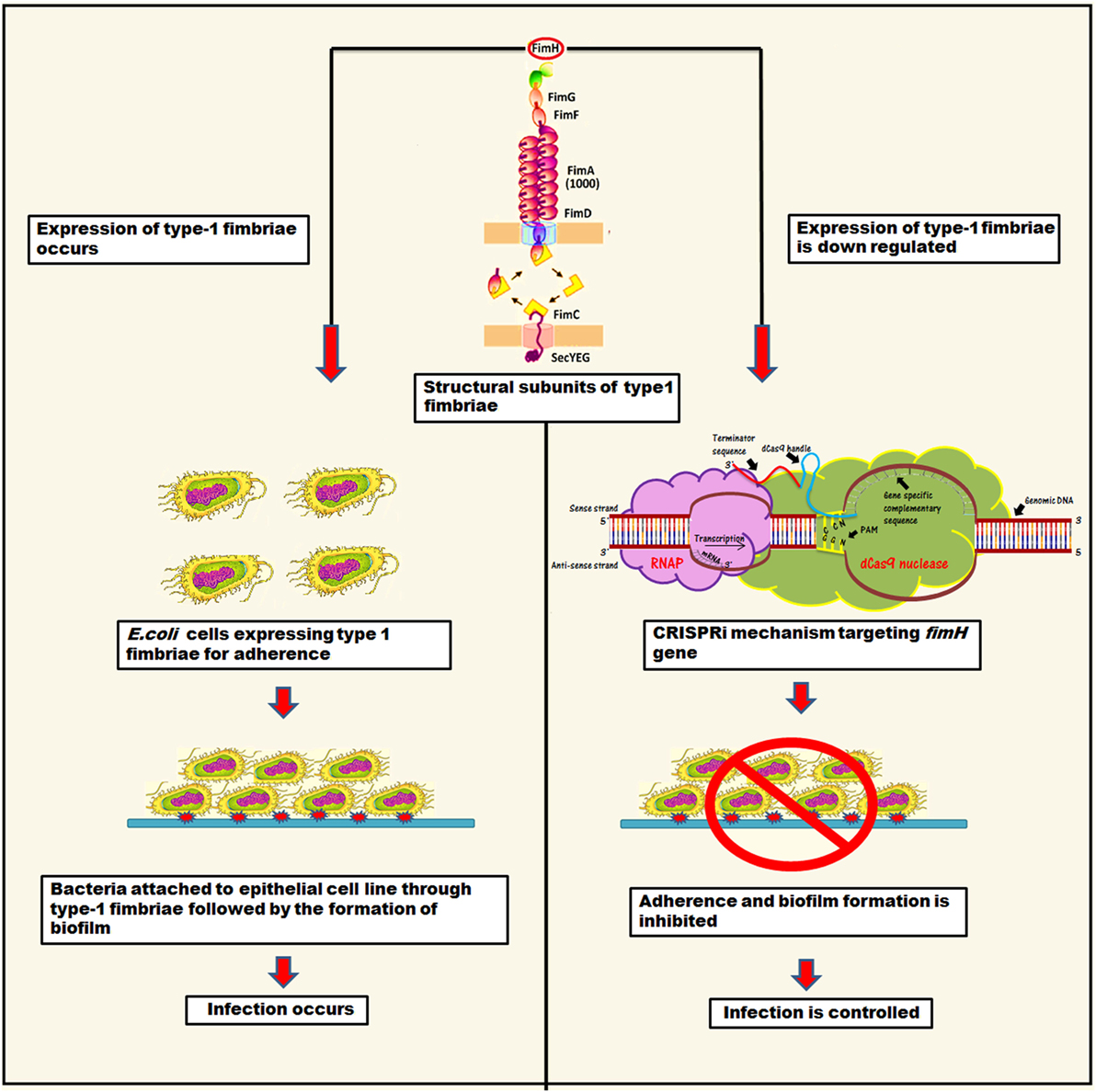
Figure 1. The representation of mechanistic approach of biofilm inhibition through CRISPRi for gene suppression of fimH.
This study was planned to inhibit adhesion property of bacteria by knocking down its fimH gene that ultimately control UTI or other E. coli adherence associated infections. Hence, we designed CRISPRi (CRISPR interference), the gene perturbation technique to inhibit fimH gene expression. It tweaks the gene expression reversibly and accurately by hindering transcriptional machinery through lodging inactive or “dead” Cas9 at specific position (16, 17).
Materials and Methods
Bacterial Strain Collection and Characterization
Strain collection was performed from UTI cases, attending Jawaharlal Nehru Medical College & Hospital (JNMCH), Aligarh Muslim University (AMU) from October 2016 to March 2017. A total of 64 clean catch urine samples were collected from infected patient’s catheters in a wide mouth sterile container from the study subjects. Isolation was performed by a surface streak procedure on Luria Bertani (LB) broth (Himedia Labs, Mumbai, India) using calibrated loops for semiquantitative method and incubated aerobically at 37°C for 24 h, and those cultures which were negative after 24 h incubations, were further incubated for 48 h. A specimen was considered positive for UTI if a single organism was cultured at a concentration of ≥105 cfu/ml. Out of 64 urine samples, 42 samples were found to be positive for UTI. Of 42, 26 samples were characterized to have E. coli isolates, confirmed through BD phoenix-100™ Automated Microbiology system using panel NMIC/ID-55.
For fimH detection a single colony on LB-plate from 26 remaining isolates were suspended in 100 µl of sterilized water and were incubated at 95°C for 10 min followed by centrifugation at 8,000 g for 10 min. Supernatant was used as template to perform colony PCR amplification under the condition, mentioned in supplementary data (Tables S1 and S2 in Supplementary Material) using appropriate primers. Among 26 isolates, 19 were reported to have fimH. Further detailed study was performed on one of the strains (AK-118) of 19 fimH positive strains.
Culture and Growth Conditions
Escherichia coli clinical strain, AK-118 was used in this study. The strain was isolated from UTI patient, admitted in JNMCH AMU, Aligarh, India. The bacteria were cultured in LB broth (Himedia labs, Mumbai, India). The plasmids pdCas9 and pgRNA used in study, already mentioned in our earlier study (16). LB media supplemented with proper antibiotic (ampicillin 100 µg/ml and chloramphenicol 25 µg/ml) and inducer [anhydrotetracycline (aTc) 2 µM] was used, wherever needed. E. coli Top10 cells were used for transformation, while the cotransformation was performed in clinical strain. The microorganism were grown without (on agar) or with (in broth) shaking at 220 rpm at 37°C, overnight. Knockdown strain was supplemented with 2 µM aTc with their respective antibiotics, to suppress gene expression.
Cloning and Construction of Knockdown Strain
The detailed protocol for cloning has already been mentioned in our earlier study (16). The complementary sequences, i.e., 20 bp region immediately following to 5′-CCN-3 [protospacer adjacent motif (PAM)] of fimH gene was commercially synthesized as a primers with additional 35 nt region of dCas9 handle (Table S1 in Supplementary Material). It was finally inserted through inverse PCR (Table S2 in Supplementary Material) in pgRNA (ampicillin resistant). The final PCR products were cleaned and ligated to give new sgRNA expressing plasmid pgRNA-FM to be ready for cotransformation along with pdCas9 (chloramphenicol resistant) in AK-118. The new knockdown strain was named as AK-FM1 for the record. The conditions used for PCR in this study were given in Table S2 in Supplementary Material.
RT-PCR and mRNA Quantification
Extraction of total RNA was performed by Trizol method from respective strains grown in presence of inducer (2 µM aTc) and proper antibiotic concentration till log phase. Total RNA was treated with RNase-free DNase in order to remove DNA contamination and subsequently cDNA was prepared by high capacity cDNA Reverse Transcription Kits (Applied Biosystems, USA) according to manufacturer’s instruction. Quantification of mRNA was performed through comparative Ct method using SYBR green PCR master mix, along with 150 ng of cDNA sample with appropriate primers. The cycle was carried out at 95°C for 10 min, 95°C for 15 s, 60°C for 30 s, and finally 72°C for 30 s. The standard graphs for respective transcripts were observed using 16s rRNA as endogenous gene control.
Mannose Sensitive Hemaggultination Assay on Guinea Pig Erythrocytes for Type 1 Fimbriae Expression
In our study, hemaggultination assay was performed on guinea pig erythrocytes with 1% mannose for type 1 fimbriae detection as described in previous study with the slight modification (18). A consent from institutional animal ethical committee of Interdisciplinary Biotechnology Unit (IBU), AMU (held on June 5, 2017, at IBU, AMU), was taken to perform this experiment. Bacteria were grown overnight in LB at 37°C for full fimbriation. The bacterial suspensions were adjusted to McFarland standard (0.5–0.6) in PBS or PBS/mannose 1% and serially diluted from 1:2 to 1:128 with same buffer in 96-well microtiter plate. 0.5% erythrocytes (in PBS or PBS/mannose 1%) was added to wells. Suspensions of bacteria and erythrocyte were incubated overnight at 4°C. All assays were conducted in triplicate. An E. coli ATCC strain (type 1 fimbriae positive) was used as a positive control.
Morphological Studies on fimH Knockdown
Morphological study was performed using transmission electron microscopy using the standard TEM protocol to investigate the presence and absence of type 1 Fimbriae. Untreated and treated bacterial cultures were suspended using a centrifuge followed by washing with PBS (pH 7.4). Secondary fixation was done using gluteraldehyde (2.5%) and osmium tetroxide (1%) for 2–3 h at 4°C. Further Samples were dehydrated by ethanol and finally embedded in araldite CY212 (Taab, Aldermaston, UK) resin to make cell-pellet blocks. Ultrathin sections of cells stained with uranyl acetate and lead citrate were observed under the TEM (Model: JEM 2100, Jeol, Tokyo, Japan).
Crystal Violet (CV) Assay
Biofilm formation was estimated by CV assay as used in our earlier study (19). Microtiter plates carrying secondary culture of treated and untreated bacterial cells were incubated at 37°C for 24 h, then medium was decanted and the planktonic cells were removed by gentle rinsing with sterile water. The adhered biofilms were stained for 15 min at room temperature with 200 µl of 0.1% CV dye and the bound dye was released from the cells by adding 98% ethanol and keeping the plates on shaker for 5 min. Biofilm quantification was done by measuring the optical density (OD) at 630 nm using a Bio-Rad iMark™ Microplate Reader, India.
Cell Viability Assay (XTT Assay)
To assess metabolic activity and cellular viability, XTT assay was carried out as described earlier (19). XTT (1 mg/ml) was dissolved in filter sterilized PBS solution and menadione (0.4 mM) was freshly prepared in acetone. Fresh mixture of 20:1 volume of XTT and menadione was used. After 24 h of growth, biofilms were washed with 200 µl of PBS solution followed by the addition of XTT-menadione and PBS solution to a volume of 42 and 158 µl in each well, respectively, and kept at 37°C in dark for 4 h. After 4 h, the intensity of orange color formazan compound was measured, that quantify the ability of metabolically active sessile cells to reduce tetrazolium salt (2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide), using microtiter plate reader at 490 nm.
Adherence Study on Human Intestinal Mucus Visualized through Confocal Microscopy
Confocal microscopy was performed to visualize the adherence property of bacteria with the human intestinal mucus. For that a resected human intestine tissue was taken as a source of intestinal mucus, which was approved by the Institutional ethical committee of IBU, AMU held on June 3, 2017, at IBU (AMU) also an informed written consent was taken from patient which remained confidential as per the protocol, established in our institution. The healthy part of large intestine was taken from the patient of diverticulitis and processed within 20 min of sample collected on ice. Mucus was isolated from the sample after washing it with PBS containing 0.01% gelatin by gently scraping through rubber spatula and collected in HEPES-HANKS buffer (10 mmol/L HEPES; pH 7.4). The collected mucus was centrifuged (13,000 g, 10 min) to remove cell debris and bacteria and stored at −20°C for further use after measuring protein content.
As described elsewhere (20), in adhesion assay mucus was diluted to the concentration 0.5 mg/ml with HEPES-HANKS buffer and 100 µg of it was immobilized directly on confocal dishes and incubates at 4°C overnight. The overnight grown bacteria after washing with PBS and adjustment of OD 0.5 at 600 nm, were stained with DAPI (355 nm wavelength for excitation and 460 nm wavelength for emission) to a final concentration of 0.2 µg/ml (DAPI) by incubating it for 30 min on mild shaking at room temperature. Finally the bacterial cells were washed three times with PBS and added to a volume of 100 µl into confocal dishes. Bacteria were allowed to adhere at 37°C for 1 h and wells were washed with HEPES-HANKS buffer to remove non adherent bacteria. The adherent bacterial cells were observed by Zeiss LSM 780 (Germany) confocal laser scanning microscope equipped with argon and He-Ne laser.
Fluorescence Spectroscopic Studies of Adherent Bacterial Cells with Human Intestinal Mucus
To perform fluorescence spectroscopic studies the above mentioned adherent bacteria were released and lysed with 1% SDS-0.1 M NaOH by incubation at 60°C. The florescence quenching of released stained bacteria (DAPI stained) were determined by using Shimadzu RF-5301PC spectrofluorometer (Shimadzu Corporation, Kyoto, Japan) equipped with a thermostatically controlled cell holder. The filters used were 355 nm for excitation and 460 nm for emission.
Statistical Analysis
The results were reported as mean ± SD. All experiments were conducted in triplicate. Further each experiment was compared with control for analyzing Student’s t-test, two-tailed hypothesis (*p < 0.05, t-test, two sided) (**p < 0.005, t-test, two sided).
Results
CRISPRi-Mediated fimH Gene Inhibition
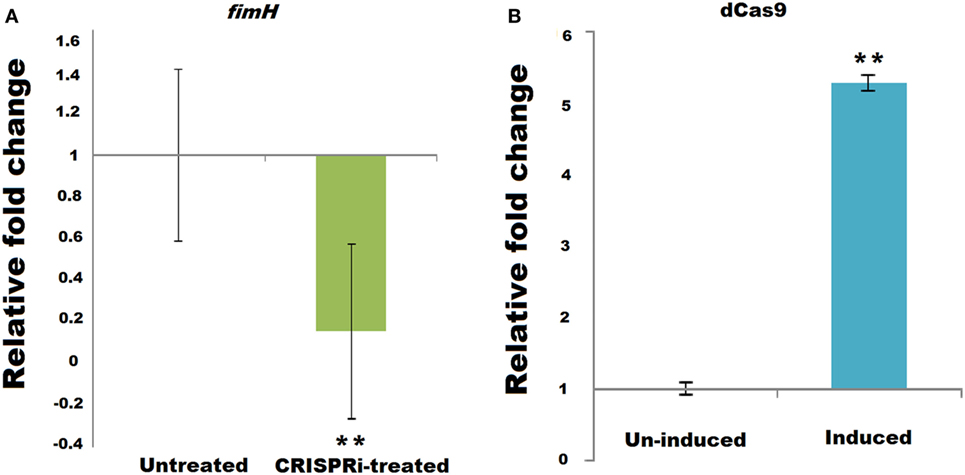
Successful implication of CRISPRi was assayed through fimH mRNA quantification using relative qRT-PCR, taking 16s rRNA gene as endogenos control. Figure 2A corresponds to the relative qRT-PCR data of fimH which showed the downregulation of fimH gene in treated condition. Taking value of relative fold change in untreated condition as 1 ± 0.414, the value of relative fold change in treated condition was observed to be 0.1560 ± 0.419, i.e., 84.39% of downregulation was observed with P-value < 0.005. Figure 2B corresponds to relative qRT-PCR data of dCas9 nuclease in strain AK-118 and proved its successful expression in this strain under induced condition with relative fold change values of 5.2986 ± 0.099 as compare to uninduced condition values 1 ± 0.080 (p-value < 0.005).
Expression of Type 1 Fimbriae
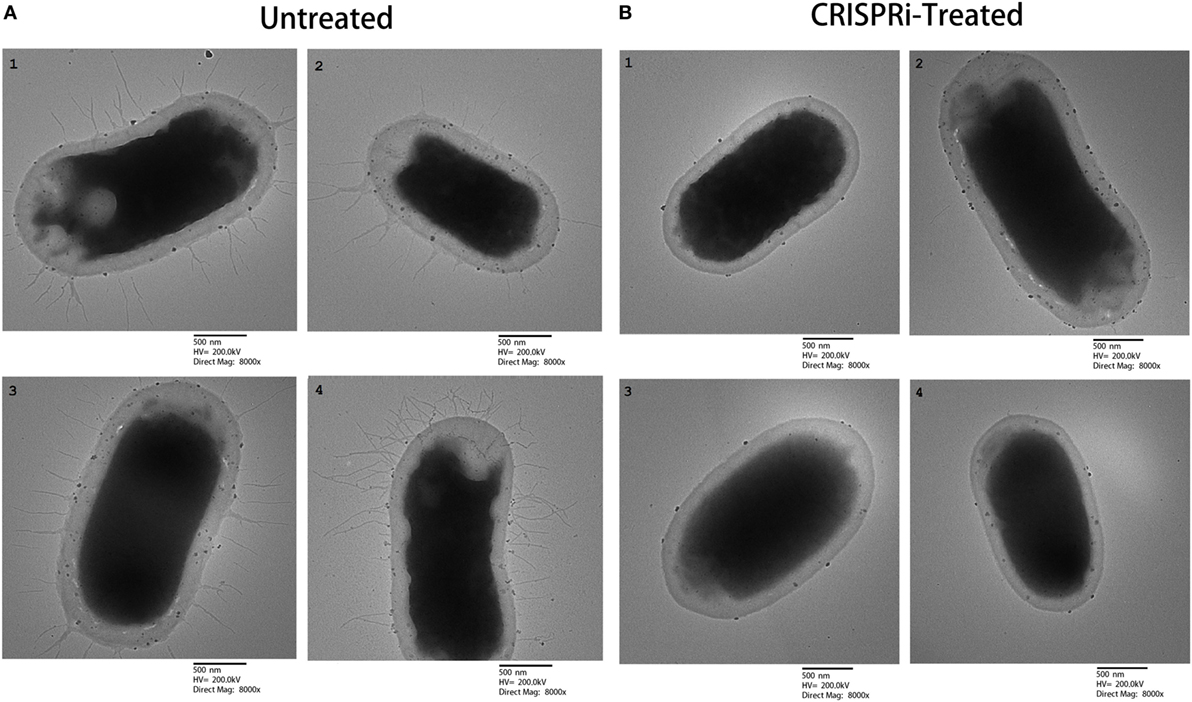
The effect of fimH gene suppression for type 1 fimbriae expression was assayed by Mannose sensitive hemaggultination assay on guinea pig erythrocytes and TEM studies. In Figure 3, row A and row B corresponds to AK-FM1 under treated and untreated conditions, respectively. In row A, a least agglutination was observed in first two wells followed by formation of blood button in the center of the well on serial dilution, while in row B, agglutination of guinea pig erythrocytes were clearly seen in almost all wells. Figure 4 shows the TEM images of AK-FM1 single bacterium in untreated (Figure 4A) and treated (Figure 4B) conditions taken from four different visual fields. In control the fimbriae appendages were present on bacterial surface due to the expression of type 1 fimbriae, while in treated cells nearly smooth bacterial surface was observed.
Quantification of Biofilm and Cell Viability
The biofilm forming tendency of AK-FM1 after CRISPRi treatment was preliminary investigated by CV assay as shown in Figure 5A. The observed value of CV assay in treated sample was 0.445 ± 0.012 (p-value < 0.005) as compared to untreated sample value 0.834 ± 0.006 (p-value < 0.005). The downregulation of fimH gene in knockdown strain, AK-FM1 (treated) showed the dramatic decrease in biofilm formation as quantified by CV assay. The metabolic activity and cell viability of AK-FM1 in treated condition was recorded to be 74.06% with the values of 1.69 ± 0.040 and 2.29 ± 0.126 (p-value < 0.005) in treated and untreated conditions, respectively, as quantified by the converted amount of XTT in XTT reduction assay (Figure 5B).
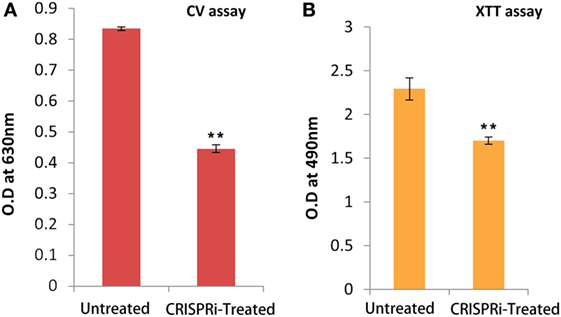
Figure 5. (A) Quantification of biofilm through crystal violet (CV) assay. (B) Cell viability assay (XTT assay). The data represent an average of triplicate experiments ± SD (*p < 0.05, t-test, two sided) (**p < 0.005, t-test, two sided).
Effect on Bacterial Adherence
The bacterial adherence level on human intestinal mucus after CRISPRi-mediated fimH gene suppression was visualized through confocal microscopy. Figures 6A,B correspond to confocal microscopy images of untreated and treated samples of AK-FM1 taken from four different visual fields, respectively. The difference in the intensity of blue color due to DAPI stained adherent bacterial cells in untreated and treated samples, indicated the noticeable reduction in the adherence property of a bacteria after CRISPRi treatment. The same result came out from fluorescence spectroscopic studies where the quenching was observed in treated samples due to decrease in number of bacterial cells adhered to human intestinal mucus (Figure 7).
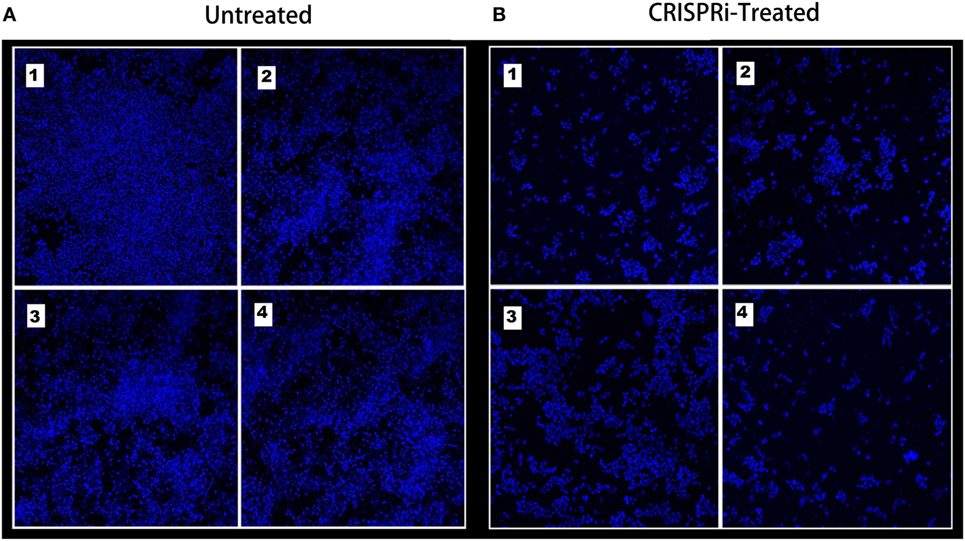
Figure 6. Adherence assay through confocal microscopy, images are taken from four different visual fields at 63× magnification. (A) Untreated and (B) CRISPRi treated.
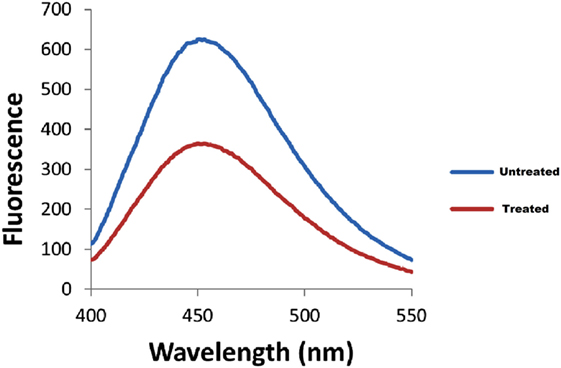
Figure 7. Fluorescence spectra of adherent bacterial cells. Quenching is observed in treated samples.
Discussion
The main cause of UTI is adherence of bacterial aggregates on the inner lining of urinary tract or inner part of catheters, consequently leading to the formation of resistant biofilms, which are difficult to be eradicated. About 10–15% of patients develop UTI on short term catheterization mainly due to the bacterial growth and biofilm formation in its inner part (4). Among all bacterial strains, UPEC strains are responsible for about 50 and 80% of hospital and community acquired UTI infections, respectively (21). Such a high percentage of UPEC strains in these infections are due to the presence of different virulence factors like adhesions, hydrophobicity, hemolysin, serum resistance, drug resistance, gelatinase production, etc. Among all adhesions, type 1 fimbriae is thought to be the most important virulence factor that may result in worsening of UTIs (21). In UTI, FimH is a major detriment of type 1 fimbriae for adherence, because of the presence of high tropism for urinary tract receptors. It was further explained in detail that FimH contains mannose binding pocket that recognizes mannose containing glycoprotein receptors present on the host cell surface. In case of UTI, the apical surface of urinary bladders bears integral membrane uroplakin 1a that acts as a main receptor for FimH (22). Hence, the epidemiological typing of UPEC was performed through single-nucleotide polymorphism analysis of FimH (23, 24). Keeping this in mind, we have designed a CRISPRi based gene editing study against fimH gene to inhibit bacterial adherence and biofilm formation in UTI cases.
CRISPRi is a technique of genetic perturbation at transcriptional level that enable gene repression or activation. Although there are some potential limitations with the use of this technique for, e.g., requirement of PAM sequence within the target site, off-targeting, etc., but this can be solved by using different Cas9 homologs with longer PAM (25). Moreover, this technique has shown its advantage over other gene manipulating techniques because of its successful delivery through cell-mediated transfer in bacterial system (26). Our study was planned to inhibit fimH, the gene responsible for mannose receptor adhesion and type 1 fimbriae polymerization initiation. This is a lead study or a proof of concept for future implementation of this technology/approach to inhibit biofilm formation as it can be said that mixing this CRISPRi modified strain with natural population of E. coli (or preexisting biofilm) would hinder new biofilm formation and adherence of bacteria that will ultimately control the infection. The novelty of using CRISPRi in this study is that, in future this could be a potential approach to control infections like UPEC in clinical settings by directly delivering the CRISPRi modified cells at the site of infection through natural conjugation.
The experimental setup was established by designing CRISPRi to knockdown fimH. The effect of fimH downregulation on type 1 fimbriae was then checked through mannose-sensitive hemagglutination assay and TEM microscopic studies. The possible reason behind these observations is that, being an important component, FimH adhesion regulates the synthesis of type 1 fimbriae via chaperone usher pathway (27). It stimulates and initiates the polymerization of type 1 fimbriae through interacting FimD (28). For quantification of biofilm formation in these strains, CV assay was performed, where noticeable reduction was observed because of the inhibition of type 1 fimbriae that guides the first stage of biofilm formation. Finally, to check the adherence level of fimH knockdown strain, a study was performed on human intestinal mucus and the adhered bacteria were visualized through confocal microscopy. The dense layer of bacteria adhered on mucus was observed in untreated sample as compared to treated. The density of cells was reduced to very low level due to the absence of type 1 fimbriae expressions that abrogate the adherence property of bacteria to a large extent. These results highlight a new era where genomic manipulations are no more a hindrance to experiments and moreover they pave the way toward the major discoveries in science, with applications in all areas of biotechnology and human therapeutics.
Conclusion
We conclude the role of CRISPRi in reducing the fimbriae of UPEC by suppressing the expression of fimH. The study has opened the new vistas to treat UTI by inhibiting biofilm formation of UPEC. Therefore, this approach has been proved as one of the potential strategies to control biofilm-mediated UTI. Hence, we further propose this approach to be validated and implemented in vivo infection model as to proof its use in UTI treatment therapy.
Ethics Statement
Approved by the Institutional ethical committee of Interdisciplinary Biotechnology Unit (IBU), Aligarh Muslim University (AMU) held on 03-06-2017 at IBU (AMU) also an informed written consent was taken from patient who remained confidential as per the protocol, established in our institution.
Author Contributions
AK designed experiments, analyzed data, and wrote the manuscript. AZ performed experiments and wrote the manuscript. NA performed experiments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CF and handling editor declared their shared affiliation.
Acknowledgments
Amit Kumar is acknowledged for the refinement of figures. AZ is thankful to Department of biotechnology for the support of fellowship. DST-PURSE Phase-II Grant is highly acknowledged.
Funding
This work was supported by internal funds of Interdisciplinary Biotechnology Unit.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/article/10.3389/fimmu.2017.01552/full#supplementary-material.
References
1. Blango MG, Mulvey MA. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother (2010) 54:1855–63. doi:10.1128/AAC.00014-10
2. Kunin CM. Urinary tract infections in females. Clin Infect Dis (1994) 18:1–12. doi:10.1093/clinids/18.1.1
3. Soto SM. Importance of biofilms in urinary tract infections: new therapeutic approaches. Adv Biol (2014) 2014:13. doi:10.1155/2014/543974
4. Mittal S, Sharma M, Chaudhary U. Biofilm and multidrug resistance in uropathogenic Escherichia coli. Pathog Glob Health (2015) 109:26–9. doi:10.1179/2047773215Y.0000000001
5. Tchesnokova V, Aprikian P, Kisiela D, Gowey S, Korotkova N, Thomas W, et al. Type 1 fimbrial adhesin FimH elicits an immune response that enhances cell adhesion of Escherichia coli. Infect Immun (2011) 79:3895–904. doi:10.1128/IAI.05169-11
6. Johnson JR, Stamm WE. Diagnosis and treatment of acute urinary tract infections. Infect Dis Clin North Am (1987) 1:773–91.
7. Warren JW. Clinical Presentations and Epidemiology of Urinary Tract Infections. Urinary Tract Infections: Molecular Pathogenesis and Clinical Management. Washington, DC: ASM Press (1996). p. 3–27.
8. Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science (2008) 320:376–9. doi:10.1126/science.1154994
9. Berne C, Ducret A, Hardy GG, Brun YV. Adhesions involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol Spectr (2015) 3(4). doi:10.1128/microbiolspec.MB-0018-2015
10. Tripathi P, Beaussart A, Alsteens D, Dupres V, Claes I, von Ossowski I, et al. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano (2013) 7:3685–97. doi:10.1021/nn400705u
11. Weissman SJ, Chattopadhyay S, Aprikian P, Obata Yasuoka M, Yarova Yarovaya Y, Stapleton A, et al. Clonal analysis reveals high rate of structural mutations in fimbrial adhesions of extra intestinal pathogenic Escherichia coli. Mol Microbiol (2006) 59:975–88. doi:10.1111/j.1365-2958.2005.04985.x
12. Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, et al. Structural basis for mechanical force regulation of the adhesion FimH via finger trap-like β sheet twisting. Cell (2014) 141:645–55. doi:10.1016/j.cell.2010.03.038
13. Munera D, Palomino C, Fernández LÁ. Specific residues in the N terminal domain of FimH stimulate type 1 fimbriae assembly in Escherichia coli following the initial binding of the adhesin to FimD usher. Mol Microbiol (2008) 69:911–25. doi:10.1111/j.1365-2958.2008.06325.x
14. Antão EM, Wieler LH, Ewers C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog (2009) 1:22. doi:10.1186/1757-4749-1-22
15. Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol (2015) 13:343–59. doi:10.1038/nrmicro3456
16. Zuberi A, Misba L, Khan AU. CRISPR Interference (CRISPRi) inhibition of luxS gene expression in E. coli: an approach to inhibit biofilm. Front Cell Infect Microbiol (2017) 7:214. doi:10.3389/fcimb.2017.00214
17. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell (2013) 152:1173–83. doi:10.1016/j.cell.2013.02.022
18. Borowsky L, Corção G, Cardoso M. Mannanoligosaccharide agglutination by Salmonella enterica strains isolated from carrier pigs. Braz J Microbiol (2009) 40:458–64. doi:10.1590/S1517-83822009000300007
19. Misba L, Kulshrestha S, Khan AU. Antibiofilm action of a toluidine blue O-silver nanoparticle conjugate on Streptococcus mutans: a mechanism of type I photodynamic therapy. Biofouling (2016) 32:313–28. doi:10.1080/08927014.2016.114189
20. Vesterlund S, Paltta J, Karp M, Ouwehand AC. Measurement of bacterial adhesion—in vitro evaluation of different methods. J Microbiol Methods (2005) 28:225–33. doi:10.1016/j.mimet.2004.09.013
21. Hojati Z, Zamanzad B, Hashemzadeh M, Molaie R, Gholipour A. The FimH gene in uropathogenic Escherichia coli strains isolated from patients with urinary tract infection. Jundishapur J Microbiol (2015) 8(2):e17520. doi:10.5812/jjm.17520
22. Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol (2008) 85:11–9. doi:10.1016/j.yexmp.2008.03.007
23. Dias RC, Moreira BM, Riley LW. Use of fimH single-nucleotide polymorphisms for strain typing of clinical isolates of Escherichia coli for epidemiologic investigation. J Clin Microbiol (2010) 48:483–8. doi:10.1128/JCM.01858-09
24. Tartof SY, Solberg OD, Riley LW. Genotypic analyses of uropathogenic Escherichia coli based on fimH single nucleotide polymorphisms (SNPs). J Med Microbiol (2007) 56:1363–9. doi:10.1099/jmm.0.47262-0
25. Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc (2013) 8:2180. doi:10.1038/nprot.2013.132
26. Ji W, Lee D, Wong E, Dadlani P, Dinh D, Huang V, et al. Specific gene repression by CRISPRi system transferred through bacterial conjugation. ACS Synth Biol (2014) 3:929–31. doi:10.1021/sb500036q
27. Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc Natl Acad Sci U S A (2000) 97:9240–5. doi:10.1073/pnas.160070497
Keywords: biofilm, CRISPRi, fimH, bacteria, immunity, gene, fimbriae
Citation: Zuberi A, Ahmad N and Khan AU (2017) CRISPRi Induced Suppression of Fimbriae Gene (fimH) of a Uropathogenic Escherichia coli: An Approach to Inhibit Microbial Biofilms. Front. Immunol. 8:1552. doi: 10.3389/fimmu.2017.01552
Received: 28 August 2017; Accepted: 31 October 2017;
Published: 13 November 2017
Edited by:
Michael Harrison Hsieh, Children’s National Health System, United StatesReviewed by:
Christina Ching, Nationwide Children’s Hospital, United StatesParameth Thiennimitr, Chiang Mai University, Thailand
Catherine Forster, Children’s National Health System, United States
Copyright: © 2017 Zuberi, Ahmad and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asad U. Khan, YXNhZC5rJiN4MDAwNDA7cmVkaWZmbWFpbC5jb20=
 Azna Zuberi
Azna Zuberi Nayeem Ahmad
Nayeem Ahmad Asad U. Khan
Asad U. Khan