- Institut de Pharmacologie et de Biologie Structurale, UMR 5089 CNRS—Université de Toulouse, Toulouse, France
Autophagy is a well-conserved lysosomal degradation pathway that plays key roles in bacterial infections. One of the most studied is probably xenophagy, the selective capture and degradation of intracellular bacteria by lysosomes. However, the impact of autophagy goes beyond xenophagy and involves intensive cross-talks with other host defense mechanisms. In addition, autophagy machinery can have non-canonical functions such as LC3-associated phagocytosis. In this review, we intend to summarize the current knowledge on the many functions of autophagy proteins in cell defenses with a focus on bacteria–macrophage interaction. We also present the strategies developed by pathogens to evade or to exploit this machinery in order to establish a successful infection. Finally, we discuss the opportunities and challenges of autophagy manipulation in improving therapeutics and vaccines against bacterial pathogens.
Introduction
Macroautophagy, hereafter referred to as autophagy, is a lysosomal degradative process that participates in cellular homeostasis by enabling the removal of defective organelles, protein aggregates, or intracellular microorganisms (1). The process is highly regulated by multiple signaling pathways and orchestrated by more than 30 autophagy-related (Atgs) proteins organized in several functional units (2). Upon autophagy activation, Atgs, serine/threonine kinase ULK1, and Beclin-1, in association with Atg14 and type III phosphatidylinositol 3-kinase Vps34, promote the formation of a cup-shaped isolation membrane to engulf the cargo (1). Through concomitant activity of two ubiquitin-like conjugation systems, the covalent linkage of Atg12 with Atg5/Atg16L1 and LC3 lipidation with phosphatidylethanolamime, the isolation membrane elongates into a double-membrane vesicle, called autophagosome. The autophagosome then fuses with lysosomes to form an autolysosome in which the engulfed cargo is degraded. This latter step is mediated by a second Beclin-1 complex, lysosomal-associated membrane protein 1 (LAMP1), and a fusion machinery including SNARE syntaxin-17.
In addition to its role in cellular homeostasis, autophagy is essential to immunity. The autophagy machinery targets intracellular pathogens for degradation, modulates inflammation, and participates in adaptive immune responses (3–5). Here, we review the many functions of autophagy in bacterial infections with a focus on macrophages, the first line of host defenses, and the replicative niche of numerous pathogens.
Autophagy Machinery in Macrophage Antibacterial Defenses
Bacteria induce autophagy mainly via their pathogen-associated molecular patterns (PAMPs) and pathogen-induced damage-associated molecular patterns (DAMPs) (4, 5). Cell surface recognition and cytosolic sensing of these molecules result in signaling cascades that promote rapid and localized autophagy machinery recruitment. Autophagy can further be regulated by several transcriptional factors such as NFkappaB and TFEB to promote expression of different autophagy genes and thus prolong autophagy activation (6, 7). Depending on PAMP/DAMP nature and localization, autophagy can selectively capture bacteria, such event is called xenophagy, damaged organelles, and other signaling platforms activated during the infection (4, 5). Furthermore, Atgs proteins have non-autophagic functions essential for innate immunity against bacteria (Figure 1).
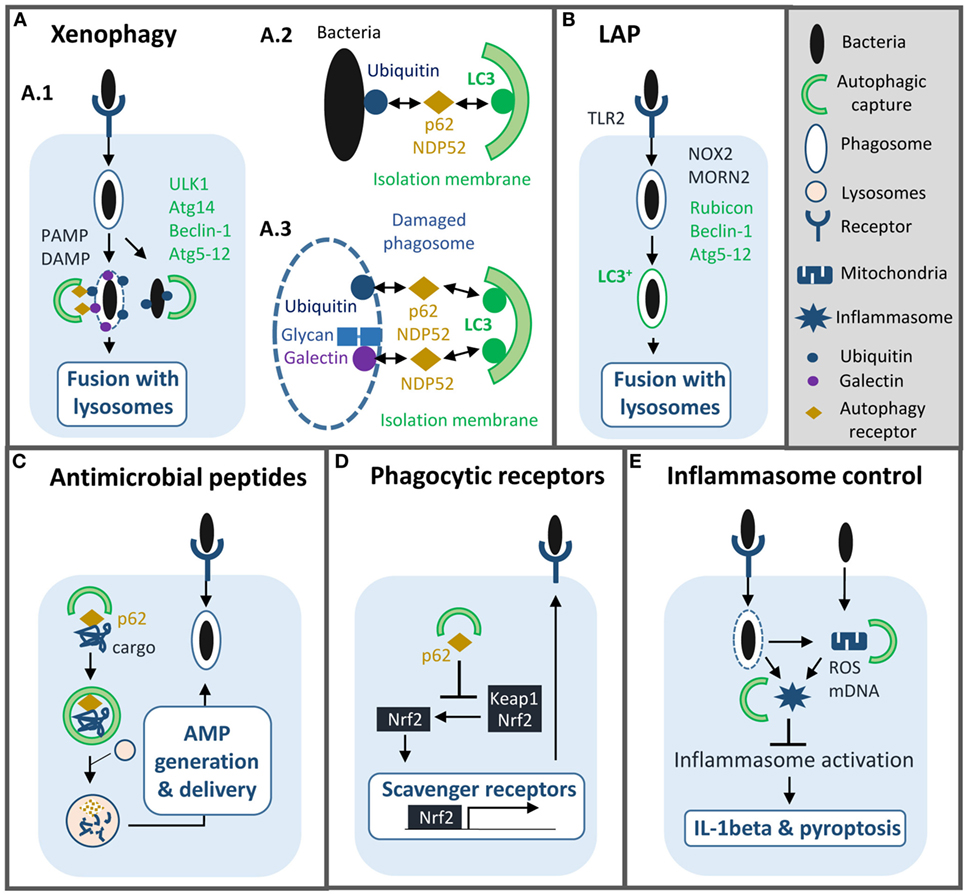
Figure 1. Roles of autophagy machinery in macrophage antibacterial defenses. Autophagy machinery plays several functions in innate immunity to bacterial infection such as: (A) xenophagy: selective capture and lysosomal degradation of cytosolic and vacuolar pathogens. Xenophagy requires formation of an autophagosome and depends on ULK1, autophagy-related (Atg)14, Beclin-1, Atg5-12, and autophagy receptor proteins such as p62 (A.1). Bacterial compartment is captured by autophagosome either via ubiquitination (A.2, 3) or host glycan recognition by galectins (A.3). (B) LC3-associated phagocytosis (LAP): LC3 is conjugated onto the membrane of phagosome containing bacteria to promote fusion with lysosome. LAP depends on Rubicon, Beclin-1, and Atg5-12. (C) Induction of autophagy enables production and delivery of antimicrobial peptides to bacterial compartments. (D) Autophagy machinery reduces the expression of scavenger receptors to limit phagocytosis of some intracellular pathogens. (E) Autophagy controls inflammation by limiting inflammasome activation. PAMP, pathogen-associated molecular pattern; DAMP, damage-associated molecular pattern; AMP, antimicrobial peptide; ROS, reactive oxygen species; mDNA, mitochondrial DNA.
Xenophagy
In macrophages, xenophagy has mainly been characterized during Mycobacterium tuberculosis infection but the mechanism is quite similar in epithelial cells infected with Salmonella typhimurium (8). M. tuberculosis through its ESX-1 secretion system damages the phagosomal membrane and access the cytosol (9). Cytosolic sensor c-GAS recognizes bacterial DNA, which results in ubiquitination of the bacterium or its phagosome by ubiquitin ligases Parkin and Smurf1 (10–12). Subsequently, ubiquitin chains bind to autophagy adaptors, such as p62 and NDP52, which recruit LC3 to deliver M. tuberculosis into an autophagosome. Damaged phagosome can also be targeted by autophagy via the recognition of host glycan present on the phagosomal lumen by cytosolic lectins of the galectin family (8, 13). Additionally, in human macrophages, immunity-related GTPase family M protein participates in xenophagy by promoting mitochondrial reactive oxygen species (ROS) production and recruiting autophagy machinery after PAMP exposure (14, 15).
Ultimately, autophagosome sends bacteria to lysosome for degradation (5, 8). In parallel, autophagy can also generate and deliver antimicrobial peptides to bacterial compartment to enhance killing (16, 17). Several, in vitro, studies have shown that xenophagy reduces intracellular survival of M. tuberculosis, however, its role, in vivo, is unclear (9, 12). Mice with monocyte-derived cells and neutrophils lacking Atg5 are more susceptible to M. tuberculosis, but not those lacking other Atgs such as Beclin-1 or Atg14, suggesting that autophagy is not involved in controlling the infection (9, 18, 19). Nonetheless, separate studies suggest that autophagy may play a role in a latter chronic phase of infection and/or that M. tuberculosis may inhibit the process in vivo (12, 20). As we will discuss below, M. tuberculosis and other pathogens have developed multiple strategies to block autophagy and some of them have been relevant in vivo.
LC3-Associated Phagocytosis (LAP)
Aside from xenophagy, a non-canonical autophagy process named LAP is known to play an important role in antibacterial defenses (21, 22). Upon phagocytosis, particles or pathogens that engage surface receptors, such as toll-like receptors, Fcgamma receptors or Dectin-1, trigger LC3 conjugation directly onto the phagosomal membrane and independently of autophagosome formation (23–25). In contrast to canonical autophagy, this process does not require ULK1 or Beclin-1/Atg14 complexes but instead relies on Beclin-1/Rubicon complex and NADPH oxidase-2 (NOX2) activation (25, 26). Rubicon activates phosphatidylinositol 3-phosphate (PI3P) synthesis, which in turn, stimulates ROS production. PI3P and ROS then promote recruitment of the two ubiquitin-like conjugation systems to trigger LC3 conjugation. Another protein MORN2 has recently been implicated in that pathway, however, its action mechanism is unknown (27).
In several instances, LC3 conjugation onto the phagosome enhances the fusion between phagosome and lysosomes. Consequently, macrophages with defects in LAP pathway are less efficient in controlling intracellular growth of various bacteria such as Legionella pneumophila, Staphylococcus aureus, M. tuberculosis, M. bovis BCG, and Listeria monocytogenes (27, 28). Importantly, in vivo studies have shown that Rubicon-deficient mice have greater bacterial load and are more susceptible to L. monocytogenes infection (28). Another important function of LAP is to assist antigen presentation via MHC class II molecules, thus to bridge innate to adaptive immunity (24, 29). Of note, in some specific contexts, LC3 conjugation delays or does not affect at all phagosome maturation suggesting that LC3 alone is not sufficient to boost fusion with lysosomes (29, 30).
Autophagy machinery can also regulate phagocytosis indirectly by altering surface expression of phagocytic receptors. Lack of Atg protein Atg7 in macrophages results in upregulation of two class A scavenger receptors, MARCO and MSR1 that facilitate phagocytosis of M. bovis BCG and M. tuberculosis (31). Autophagy-deficient cells accumulate p62, which promotes dissociation of transcription factor nuclear erythroid-related factor 2 from Keap1 and then its translocation into the nucleus to mediate expression of these receptors. Notably, in another instance, autophagy deficiency can lead to reduced phagocytosis depending on the nature of the bacteria (32).
Inflammation Dampening
Although inflammation is central to control bacterial infection, excessive inflammatory responses can lead to host tissue injury, and disease progression. Numerous studies have demonstrated the importance of autophagy in inflammation regulation in infectious and non-infectious settings, the interested readers can refer to recent reviews for a comprehensive view on this subject (3, 5). Here, we will only discuss on the beneficial role of autophagy machinery in modulating inflammation in a context of bacterial infections and macrophages. One key role of autophagy is the down-regulation of inflammasome activation through multiple mechanisms (33, 34). Pseudomonas aeruginosa infection results in mitochondrial damage that leads to NLRC4 inflammasome. Elimination of damaged mitochondria via autophagy, i.e., mitophagy, limits inflammasome activation both in vitro and in vivo (35). Similarly, in a P. aeruginosa septic model, atg7fl/fl mice have an enhanced susceptibility to infection with important neutrophil infiltration and severe lung damage. Loss of Atg7 in alveolar macrophages results in upregulation of IL-1beta and pyroptosis (36). Besides mitophagy, autophagy can also restrain inflammasome activation by capturing inflammasome subunits (37, 38). Lastly, autophagy can prevent non-canonical caspase-11 inflammasome activation by targeting S. typhimurium and thus limiting LPS release into the cytosol (39).
Finally, mechanisms and levels of autophagy dependent greatly on macrophage microenvironment. While IL-4, IL-13, and IL-10 appear to inhibit autophagy, proinflammatory cytokines such as IFNgamma, IL-1beta, and TNFalpha, activate this process (40). Specifically, IFNgamma-mediated xenophagy requires additionally ubiquilin-1 and guanylate-binding proteins 1 and 7 to promote recruitment of autophagy proteins p62 and Atg4B (41, 42). On another hand, the presence of vitamin D in serum enhances significantly macrophage autophagy via expression of cathelicidin antimicrobial peptide (43). Specific T cells can also stimulate autophagic activity in M. tuberculosis-infected human macrophages (44). Finally, microbiota may influence autophagy response too, as recently, probiotic Bacillus amyloliquefaciens has been shown to upregulate autophagy genes in macrophages, which lead to enhanced Escherichia coli killing (45).
Bacterial Pathogens Evade Autophagy
Intracellular bacterial pathogens have developed a wide array of tactics to counterbalance macrophage antibacterial defenses and autophagy is no exception (46). Most of the uncovered strategies are directed against xenophagy and target different steps of the process (Figure 2).
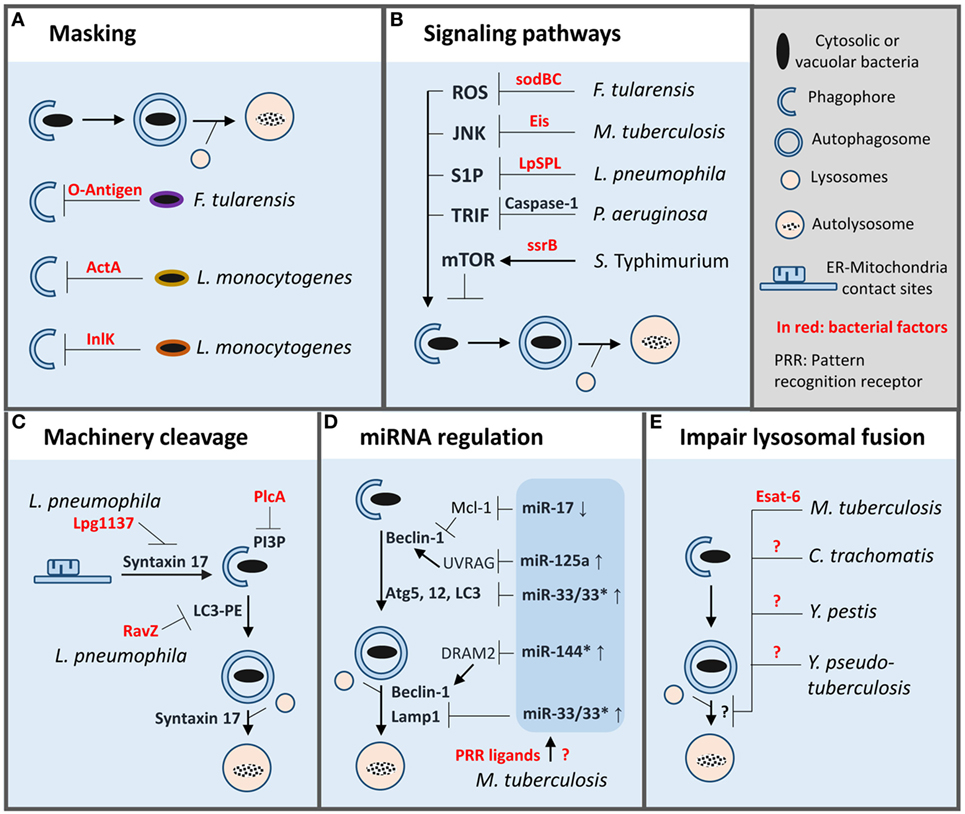
Figure 2. Autophagy evasion strategies adopted by bacterial pathogens inside macrophages. To evade autophagy in macrophages bacterial pathogens have developed a wide array of strategies such as: (A) masking of microbial surface to avoid recognition. (B) Manipulation of macrophage signaling pathways involved in autophagy regulation. (C) Direct cleavage of autophagy proteins or signaling lipid. (D) Limitation of autophagy machinery expression or function by miRNA regulation. (E) Inhibition of autophagosomes/lysosome fusion. PI3P, phosphatidylinositol 3-phosphate; LC3-PE, LC3 lipidated with phosphatidylethanolamine.
Masking Bacterial Surface to Prevent Recognition
Cytosolic L. monocytogenes avoids recognition by autophagy machinery by secreting two virulence factors ActA and InlK (47, 48). These factors promote recruitment of host proteins, actin, and major vault protein, respectively, to form a protective coat that masks the bacteria. Francisella tularensis disguises itself directly by producing a surface polysaccharide, the O-antigen, which prevents cytosolic sensing of the pathogen and thus xenophagy (49). Importantly, cytosolic O-antigen mutants are killed by Atg5-dependent autophagy inside murine macrophages.
Regulation of Signaling Pathways Involved in Autophagy Initiation
In macrophages, S. typhimurium prevents autophagy by activating mTOR, a master repressor of autophagy (50, 51). Two mechanisms seem to be at play, first, the degradation of the energy sensor, Sirt1/AMPK complex, which negatively regulates mTOR resulting in autophagy activation and, second, the recruitment of non-receptor tyrosine kinase focal adhesion kinase (FAK), an activator of Akt/mTOR pathway. SsrB, a response regulator of a two-component system involved in the regulation of SPI2 encoded virulence factors, promotes AMPK down-regulation (50). Notably, in vivo, macrophages deficient in FAK are more efficient in clearing S. typhimurium infection than their wild-type counterpart (51).
Conversely, some pathogens can inhibit signaling pathways that promote autophagy. M. tuberculosis secretes an N-acetyltransferase, Eis, to target JNK-dependent autophagy; however, it does not seem to be sufficient to affect intracellular growth (52, 53). In addition, M. tuberculosis limits LAP by inhibiting NADPH oxidase recruitment onto its phagosome, which favors pathogen intracellular growth in vitro and in vivo (54). Interestingly, antioxidant enzymes SodB and SodC of F. tularensis prevent ROS-induced xenophagy and possibly LAP (55). Further, vacuolar L. pneumophila translocates a sphingosine-1 phosphate lyase into the cytosol to alter sphingosine metabolism implicated in autophagy activation (56). Lastly, some pathogens can take advantage of negative feedback regulatory circuits present in macrophages to target autophagy. P. aeruginosa infection leads to NLRC4-dependent caspase-1 activation which results in cleavage of TRIF, an important mediator of TLR4-induced autophagy (57). Importantly, in vivo, preventing TRIF cleavage restores autophagy and bacterial clearance.
Cleavage of Autophagy Machinery
L. pneumophila can also block autophagy directly by secreting two specific proteases. First, Lpg1137 targets ER-mitochondria contact sites to cleave syntaxin-17, a key SNARE protein implicated in autophagosome formation (58, 59). On another hand, RavZ, a cysteine protease, cleaves phosphatidylethanolamine-conjugated LC3 to produce a permanent unlipidated form of LC3 that is inadequate for the formation of autophagosomes (60). Another example is L. monocytogenes, which secretes phospholipase PlcA to degrade PI3P, a key lipid produced by Beclin-1 complex, and involved in LC3 lipidation (48, 61).
Regulation of miRNA (miR) Targeting Autophagy Machinery
Besides post-translational modification, autophagy can also be modulated at a post-transcriptional level via expression of various small non-coding RNAs (miR). M. tuberculosis thwarts autophagy by inducing expression of miR-33 which down-regulates expression of several autophagy proteins along the pathway (Atg5, Atg12, LC3, and LAMP1) (62). Remarkably, mice with hematopoietic miR-33 deficiency have a superior capacity to control M. tuberculosis infection. Another miR induced by M. tuberculosis, is miR-125a, which targets UVRAG in complex with Beclin-1 to inhibit autophagy and thus promote pathogen intracellular survival inside macrophages (63). Interestingly, miR-33 and miR-125a can be induced by PRR ligands isolated from M. tuberculosis (62, 63). Expression of these miR may be a normal negative feedback loop present in host cells to prevent excessive autophagy, which M. tuberculosis exploits to its advantage. Additionally, M. tuberculosis upregulates miR-144* to reduce expression of DRAM2, a recently discovered autophagy protein, that promotes activation of a second Beclin-1 complex involved in autophagosome maturation (64). Finally, M. tuberculosis can down-regulate miR-17, which targets Mcl-1, an inhibitor of Beclin-1 complex involved in autophagy initiation (65).
Blockade of Autophagosome–Lysosome Fusion
As mentioned above, M. tuberculosis can block autophagosome maturation via miR-144*, although, the bacterial factor(s) involved in that process remain to be identified (65). Separate studies have shown that Esat-6, an important M. tuberculosis virulence factor, plays also a role in blocking autophagosome maturation in macrophages as well as in dendritic cells (66, 67). The action mechanism of Esat-6 is unknown but might be mediated, in part, by miR modulation (68). Several other bacterial pathogens can impair autophagosome maturation for survival or growth in macrophages such as Chlamydia trachomatis, Yersinia pestis, Y. pseudotuberculosis, but like for M. tuberculosis the underlying molecular mechanisms are unknown (69–71). Furthermore, it is unclear whether these mechanisms applied to both xenophagy and LAP.
Autophagy Machinery Favors Bacterial Pathogens
Nutrient and Membrane Acquisition
While some pathogens neutralize autophagy to replicate intracellularly, others exploit autophagy or some autophagy proteins to acquire nutrients and remodel their vacuoles. Y. pseudotuberculosis inhibits autophagosome maturation; however, it relies on autophagosome formation for its growth in macrophages (71). An early study has shown that exogenous autophagy activation promotes the development of Coxiella-replicative vacuole (72). In macrophages and epithelial cells, C. burnetti resides in an large acidified compartment, which fuses with mature autophagosomes via Cig2, a type IV secretion system effector (73, 74). Interestingly, in vivo, Cig2 reduces host tolerance to C. burnetti infection without affecting bacterial load (74).
Brucella abortus subverts ULK1 and Beclin-1 complexes to remodel its ER-derived vacuole into a compartment with autophagic properties (75). Nonetheless, this conversion is independent of the two ubiquitin-like conjugation systems, Atg5-12 and LC3. This selective manipulation of autophagy proteins is important for Brucella lifecycle and cell-to-cell spreading. Similarly, M. marinum, a species closely related to M. tuberculosis, uses autophagy machinery to promote bacterial egress from its natural host, Dictyostelium, a macrophage-like unicellular organism (76). How Brucella and M. marinum manipulate autophagy proteins to favor transmission is still unknown.
Some pathogens have a more complex relationship with autophagy in a sense that they can evade, as outlined above, or exploit autophagy depending on the infection stages or the type of autophagy. At early stage, vacuolar Legionella interacts with autophagy pathway and seems to rely on autophagosome for survival in permissive macrophages (77). Similarly, cytosolic F. tularensis triggers an Atg5-independent autophagy pathway to acquire nutrients and replicate while avoiding xenophagy (78). The molecular differences between these two autophagy-related pathways, one that favors bacterial growth and the other promotes killing, definitely call for further investigation. Finally, immediately after entering macrophages, L. monocytogenes manipulates LAP to form spacious non-acidified Listeria-containing phagosomes (SLAPs) which are believed to participate in persistent infection (79, 80). Low expression of virulent factor, Listeriolysin O, appears to play a role in SLAP formation whereas high-expression triggers bacterial escape into the cytosol (80).
Inflammation Dampening
One example is Vibrio parahaemolyticus, which limits NLRC4 inflammasome-mediated IL-1beta production by activating macrophage autophagy via a type III secretion effector, VopQ (81). Other bacteria seem to benefit from the autophagy machinery in a more passive way. For example, Atg16L1-deficient mice clear more efficiently uropathogenic E. coli (UPEC) and Citrobacter rodentium than wild-type controls (82–84). The improved protection against C. rodentium is associated with an enhanced immune response dependent on monocytes; however, it does not rely on a cell-intrinsic role of Atg16L1 in myeloid cells (84). In contrast, loss of Atg16L1 in macrophages is responsible for the phenotype observed during UPEC infection (83). Atg16L1-deficient macrophages, infected with UPEC, activate more NLRP3/Caspase-1 inflammasome, and IL-1beta production. Importantly, in vivo, IL-1beta neutralization reduces the capacity of Atg16L1-deficient mice to control UPEC infection. Testing other autophagy proteins will be essential to confirm the role of autophagy entire pathway in that process.
Autophagy in Host-Directed Therapy (HDT) and Vaccine
Host-Directed Therapy
In the past few years, a number of studies have highlighted the potential of targeting autophagy for the control of bacterial infections. FDA-approved drugs with proautophagy activity, such as statin, gefitinib, carbamazepine, and metformin, have been shown to limit M. tuberculosis growth in mice model of infection (85–88). Similarly, rapamycin enhances clearance of P. aeruginosa and B. cepacia, in vivo, in addition to reducing lung inflammation (89, 90). However, these drugs can module several other cellular functions, so it is unclear whether their action, in vivo, is mediated by autophagy. Another difficulty is the seemingly opposing effects of a same drug depending on the infection settings. Indeed, while rapamycin induces M. tuberculosis killing in macrophages, it promotes bacterial survival in endothelial cells and in HIV-coinfected macrophages (91–93). The only compound tested, in vivo that specifically activates autophagy is a small cell permeant peptide, TAT-Beclin-1 (94). This designed peptide has been shown to improve outcome of chikungunya and West Nile virus infections in mice, thus it would be a good candidate to test in the context of bacterial infection. However, one has to bear in mind that some pathogens use autophagy to thrive in host cells and other can either evade or exploit this pathway according to the infection stage. Therefore, a detailed knowledge of the role(s) of autophagy and its molecular mechanisms for each bacterial pathogen is mandatory to develop more specific autophagy modulators, as well as, the availability of relevant preclinical models to evaluate the efficacy of these compounds.
Vaccine
Manipulation of autophagy in antigen-presenting cells including macrophages holds also great promise in vaccine development. Autophagy plays an important role in antigen processing and MHC presentation (95). Studies already indicate that boosting autophagy can enhance antigen presentation and vaccine efficacy. For instance, mice immunized with BCG-infected dendritic cells treated with mTOR inhibitor, rapamycin, have greater Th1-mediated protection after being challenged with M. tuberculosis (96). Recently, BCGΔureC::hly, a live vaccine with improved immunogenicity and in clinical trial phase II, has been shown to enhance xenophagy (97). However, the importance, in vivo, of phagocyte autophagy in vaccine enhanced-efficacy is unclear as both cited strategies trigger other cellular pathways.
Conclusion
Macrophage autophagy is central to host defenses against bacterial infections, sending intracellular pathogens to lysosomes for degradation while controlling inflammation to limit host damages. Since novel functions for autophagy proteins have emerged in various physiological and pathological situations, it is likely that further contributions of autophagy machinery to macrophage biology will be unveiled in the context of bacterial infection (98, 99). On the other hand, pathogens have found multiple subterfuges to manipulate this machinery in order to persist or proliferate, still, their action mechanisms and significance in vivo are far from being thoroughly understood. A more comprehensive and integrated view of bacteria-autophagy interplay will definitely help in designing more specific HDT and vaccine based on autophagy modulation.
Author Contributions
IV and AB reviewed the relevant literature and wrote the manuscript. IV prepared and AB revised the figures. IV and AB have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitation. The work in the authors’ laboratories was supported by EU FP7 Marie Curie Career Integration Grant 293416 (IV), EU Horizon 2020 TBVAC2020 under grant agreement No. 643381 (IV), and Horizon 2020 COST action TRANSAUTOPHAGY (CA15138).
References
1. Ktistakis NT, Tooze SA. Digesting the expanding mechanisms of autophagy. Trends Cell Biol (2016) 26(8):624–35. doi:10.1016/j.tcb.2016.03.006
2. Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev (2010) 90(4):1383–435. doi:10.1152/physrev.00030.2009
3. Cadwell K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat Rev Immunol (2016) 16(11):661–75. doi:10.1038/nri.2016.100
4. Shibutani ST, Saitoh T, Nowag H, Munz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol (2015) 16(10):1014–24. doi:10.1038/ni.3273
5. Deretic V, Kimura T, Timmins G, Moseley P, Chauhan S, Mandell M. Immunologic manifestations of autophagy. J Clin Invest (2015) 125(1):75–84. doi:10.1172/JCI73945
6. van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense [corrected]. Cell Host Microbe (2014) 15(6):753–67. doi:10.1016/j.chom.2014.05.005
7. Pastore N, Brady OA, Diab HI, Martina JA, Sun L, Huynh T, et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy (2016) 12(8):1240–58. doi:10.1080/15548627.2016.1179405
8. Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell (2014) 54(2):224–33. doi:10.1016/j.molcel.2014.03.009
9. Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell (2012) 150(4):803–15. doi:10.1016/j.cell.2012.06.040
10. Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe (2015) 17(6):811–9. doi:10.1016/j.chom.2015.05.004
11. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature (2013) 501(7468):512–6. doi:10.1038/nature12566
12. Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, et al. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe (2017) 21(1):59–72. doi:10.1016/j.chom.2016.11.002
13. Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, et al. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell (2016) 39(1):13–27. doi:10.1016/j.devcel.2016.08.003
14. Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol (2010) 12(12):1154–65. doi:10.1038/ncb2119
15. Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell (2015) 58(3):507–21. doi:10.1016/j.molcel.2015.03.020
16. Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity (2010) 32(3):329–41. doi:10.1016/j.immuni.2010.02.009
17. Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A (2007) 104(14):6031–6. doi:10.1073/pnas.0700036104
18. Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A (2012) 109(46):E3168–76. doi:10.1073/pnas.1210500109
19. Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature (2015) 528(7583):565–9. doi:10.1038/nature16451
20. Saini NK, Baena A, Ng TW, Venkataswamy MM, Kennedy SC, Kunnath-Velayudhan S, et al. Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE_PGRS47. Nat Microbiol (2016) 1(9):16133. doi:10.1038/nmicrobiol.2016.133
21. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J (2017) 36(13):1811–36. doi:10.15252/embj.201796697
22. Mitchell G, Isberg RR. Innate immunity to intracellular pathogens: balancing microbial elimination and inflammation. Cell Host Microbe (2017) 22(2):166–75. doi:10.1016/j.chom.2017.07.005
23. Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature (2007) 450(7173):1253–7. doi:10.1038/nature06421
24. Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem (2012) 287(41):34149–56. doi:10.1074/jbc.M112.382812
25. Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A (2009) 106(15):6226–31. doi:10.1073/pnas.0811045106
26. Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol (2015) 17(7):893–906. doi:10.1038/ncb3192
27. Abnave P, Mottola G, Gimenez G, Boucherit N, Trouplin V, Torre C, et al. Screening in planarians identifies MORN2 as a key component in LC3-associated phagocytosis and resistance to bacterial infection. Cell Host Microbe (2014) 16(3):338–50. doi:10.1016/j.chom.2014.08.002
28. Yang CS, Lee JS, Rodgers M, Min CK, Lee JY, Kim HJ, et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe (2012) 11(3):264–76. doi:10.1016/j.chom.2012.01.018
29. Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol (2013) 203(5):757–66. doi:10.1083/jcb.201308173
30. Cemma M, Grinstein S, Brumell JH. Autophagy proteins are not universally required for phagosome maturation. Autophagy (2016) 12(9):1440–6. doi:10.1080/15548627.2016.1191724
31. Bonilla DL, Bhattacharya A, Sha Y, Xu Y, Xiang Q, Kan A, et al. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity (2013) 39(3):537–47. doi:10.1016/j.immuni.2013.08.026
32. Jacquel A, Obba S, Boyer L, Dufies M, Robert G, Gounon P, et al. Autophagy is required for CSF-1-induced macrophagic differentiation and acquisition of phagocytic functions. Blood (2012) 119(19):4527–31. doi:10.1182/blood-2011-11-392167
33. Harris J, Lang T, Thomas JPW, Sukkar MB, Nabar NR, Kehrl JH. Autophagy and inflammasomes. Mol Immunol (2017) 86:10–5. doi:10.1016/j.molimm.2017.02.013
34. Saitoh T, Akira S. Regulation of inflammasomes by autophagy. J Allergy Clin Immunol (2016) 138(1):28–36. doi:10.1016/j.jaci.2016.05.009
35. Jabir MS, Hopkins L, Ritchie ND, Ullah I, Bayes HK, Li D, et al. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy (2015) 11(1):166–82. doi:10.4161/15548627.2014.981915
36. Pu Q, Gan C, Li R, Li Y, Tan S, Li X, et al. Atg7 deficiency intensifies inflammasome activation and pyroptosis in Pseudomonas sepsis. J Immunol (2017) 198(8):3205–13. doi:10.4049/jimmunol.1601196
37. Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol (2012) 13(3):255–63. doi:10.1038/ni.2215
38. Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol (2015) 210(6):973–89. doi:10.1083/jcb.201503023
39. Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature (2014) 509(7500):366–70. doi:10.1038/nature13157
41. Sakowski ET, Koster S, Portal Celhay C, Park HS, Shrestha E, Hetzenecker SE, et al. Ubiquilin 1 promotes IFN-gamma-induced xenophagy of Mycobacterium tuberculosis. PLoS Pathog (2015) 11(7):e1005076. doi:10.1371/journal.ppat.1005076
42. Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science (2011) 332(6030):717–21. doi:10.1126/science.1201711
43. Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe (2009) 6(3):231–43. doi:10.1016/j.chom.2009.08.004
44. Petruccioli E, Romagnoli A, Corazzari M, Coccia EM, Butera O, Delogu G, et al. Specific T cells restore the autophagic flux inhibited by Mycobacterium tuberculosis in human primary macrophages. J Infect Dis (2012) 205(9):1425–35. doi:10.1093/infdis/jis226
45. Wu Y, Wang Y, Zou H, Wang B, Sun Q, Fu A, et al. Probiotic Bacillus amyloliquefaciens SC06 induces autophagy to protect against pathogens in macrophages. Front Microbiol (2017) 8:469. doi:10.3389/fmicb.2017.00469
46. Mitchell G, Chen C, Portnoy DA. Strategies used by bacteria to grow in macrophages. Microbiol Spectr (2016) 4(3):1–22. doi:10.1128/microbiolspec.MCHD-0012-2015
47. Dortet L, Mostowy S, Samba-Louaka A, Gouin E, Nahori MA, Wiemer EA, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog (2011) 7(8):e1002168. doi:10.1371/journal.ppat.1002168
48. Mitchell G, Ge L, Huang Q, Chen C, Kianian S, Roberts MF, et al. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect Immun (2015) 83(5):2175–84. doi:10.1128/IAI.00110-15
49. Case ED, Chong A, Wehrly TD, Hansen B, Child R, Hwang S, et al. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell Microbiol (2014) 16(6):862–77. doi:10.1111/cmi.12246
50. Ganesan R, Hos NJ, Gutierrez S, Fischer J, Stepek JM, Daglidu E, et al. Salmonella typhimurium disrupts Sirt1/AMPK checkpoint control of mTOR to impair autophagy. PLoS Pathog (2017) 13(2):e1006227. doi:10.1371/journal.ppat.1006227
51. Owen KA, Meyer CB, Bouton AH, Casanova JE. Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLoS Pathog (2014) 10(6):e1004159. doi:10.1371/journal.ppat.1004159
52. Shin DM, Jeon BY, Lee HM, Jin HS, Yuk JM, Song CH, et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog (2010) 6(12):e1001230. doi:10.1371/journal.ppat.1001230
53. Kim KH, An DR, Song J, Yoon JY, Kim HS, Yoon HJ, et al. Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc Natl Acad Sci U S A (2012) 109(20):7729–34. doi:10.1073/pnas.1120251109
54. Koster S, Upadhyay S, Chandra P, Papavinasasundaram K, Yang G, Hassan A, et al. Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proc Natl Acad Sci U S A (2017) 114(41):E8711–20. doi:10.1073/pnas.1707792114
55. Rabadi SM, Sanchez BC, Varanat M, Ma Z, Catlett SV, Melendez JA, et al. Antioxidant defenses of Francisella tularensis modulate macrophage function and production of proinflammatory cytokines. J Biol Chem (2016) 291(10):5009–21. doi:10.1074/jbc.M115.681478
56. Rolando M, Escoll P, Nora T, Botti J, Boitez V, Bedia C, et al. Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy. Proc Natl Acad Sci U S A (2016) 113(7):1901–6. doi:10.1073/pnas.1522067113
57. Jabir MS, Ritchie ND, Li D, Bayes HK, Tourlomousis P, Puleston D, et al. Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and beta-interferon production during Pseudomonas aeruginosa infection. Cell Host Microbe (2014) 15(2):214–27. doi:10.1016/j.chom.2014.01.010
58. Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, et al. Autophagosomes form at ER-mitochondria contact sites. Nature (2013) 495(7441):389–93. doi:10.1038/nature11910
59. Arasaki K, Mikami Y, Shames SR, Inoue H, Wakana Y, Tagaya M. Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat Commun (2017) 8:15406. doi:10.1038/ncomms15406
60. Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science (2012) 338(6110):1072–6. doi:10.1126/science.1227026
61. Tattoli I, Sorbara MT, Yang C, Tooze SA, Philpott DJ, Girardin SE. Listeria phospholipases subvert host autophagic defenses by stalling pre-autophagosomal structures. EMBO J (2013) 32(23):3066–78. doi:10.1038/emboj.2013.234
62. Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S, et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol (2016) 17(6):677–86. doi:10.1038/ni.3434
63. Kim JK, Yuk JM, Kim SY, Kim TS, Jin HS, Yang CS, et al. MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J Immunol (2015) 194(11):5355–65. doi:10.4049/jimmunol.1402557
64. Kim JK, Lee HM, Park KS, Shin DM, Kim TS, Kim YS, et al. miR144* inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2. Autophagy (2017) 13(2):423–41. doi:10.1080/15548627.2016.1241922
65. Kumar R, Sahu SK, Kumar M, Jana K, Gupta P, Gupta UD, et al. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell Microbiol (2016) 18(5):679–91. doi:10.1111/cmi.12540
66. Chandra P, Ghanwat S, Matta SK, Yadav SS, Mehta M, Siddiqui Z, et al. Mycobacterium tuberculosis inhibits RAB7 recruitment to selectively modulate autophagy flux in macrophages. Sci Rep (2015) 5:16320. doi:10.1038/srep16320
67. Romagnoli A, Etna MP, Giacomini E, Pardini M, Remoli ME, Corazzari M, et al. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy (2012) 8(9):1357–70. doi:10.4161/auto.20881
68. Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe (2015) 17(3):345–56. doi:10.1016/j.chom.2015.01.007
69. Yasir M, Pachikara ND, Bao X, Pan Z, Fan H. Regulation of chlamydial infection by host autophagy and vacuolar ATPase-bearing organelles. Infect Immun (2011) 79(10):4019–28. doi:10.1128/IAI.05308-11
70. Pujol C, Klein KA, Romanov GA, Palmer LE, Cirota C, Zhao Z, et al. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect Immun (2009) 77(6):2251–61. doi:10.1128/IAI.00068-09
71. Moreau K, Lacas-Gervais S, Fujita N, Sebbane F, Yoshimori T, Simonet M, et al. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol (2010) 12(8):1108–23. doi:10.1111/j.1462-5822.2010.01456.x
72. Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol (2005) 7(7):981–93. doi:10.1111/j.1462-5822.2005.00527.x
73. Winchell CG, Steele S, Kawula T, Voth DE. Dining in: intracellular bacterial pathogen interplay with autophagy. Curr Opin Microbiol (2016) 29:9–14. doi:10.1016/j.mib.2015.09.004
74. Kohler LJ, Reed Sh C, Sarraf SA, Arteaga DD, Newton HJ, Roy CR. Effector protein Cig2 decreases host tolerance of infection by directing constitutive fusion of autophagosomes with the Coxiella-containing vacuole. MBio (2016) 7(4):e1127–1116. doi:10.1128/mBio.01127-16
75. Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe (2012) 11(1):33–45. doi:10.1016/j.chom.2011.12.002
76. Gerstenmaier L, Pilla R, Herrmann L, Herrmann H, Prado M, Villafano GJ, et al. The autophagic machinery ensures nonlytic transmission of mycobacteria. Proc Natl Acad Sci U S A (2015) 112(7):E687–92. doi:10.1073/pnas.1423318112
77. Amer AO, Byrne BG, Swanson MS. Macrophages rapidly transfer pathogens from lipid raft vacuoles to autophagosomes. Autophagy (2005) 1(1):53–8. doi:10.4161/auto.1.1.1589
78. Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog (2013) 9(8):e1003562. doi:10.1371/journal.ppat.1003562
79. Lam GY, Cemma M, Muise AM, Higgins DE, Brumell JH. Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy (2013) 9(7):985–95. doi:10.4161/auto.24406
80. Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature (2008) 451(7176):350–4. doi:10.1038/nature06479
81. Higa N, Toma C, Koizumi Y, Nakasone N, Nohara T, Masumoto J, et al. Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS Pathog (2013) 9(1):e1003142. doi:10.1371/journal.ppat.1003142
82. Wang C, Mendonsa GR, Symington JW, Zhang Q, Cadwell K, Virgin HW, et al. Atg16L1 deficiency confers protection from uropathogenic Escherichia coli infection in vivo. Proc Natl Acad Sci U S A (2012) 109(27):11008–13. doi:10.1073/pnas.1203952109
83. Symington JW, Wang C, Twentyman J, Owusu-Boaitey N, Schwendener R, Nunez G, et al. ATG16L1 deficiency in macrophages drives clearance of uropathogenic E. coli in an IL-1beta-dependent manner. Mucosal Immunol (2015) 8(6):1388–99. doi:10.1038/mi.2015.7
84. Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe (2013) 14(2):216–24. doi:10.1016/j.chom.2013.07.013
85. Parihar SP, Guler R, Khutlang R, Lang DM, Hurdayal R, Mhlanga MM, et al. Statin therapy reduces the Mycobacterium tuberculosis burden in human macrophages and in mice by enhancing autophagy and phagosome maturation. J Infect Dis (2014) 209(5):754–63. doi:10.1093/infdis/jit550
86. Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, et al. Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog (2014) 10(2):e1003946. doi:10.1371/journal.ppat.1003946
87. Schiebler M, Brown K, Hegyi K, Newton SM, Renna M, Hepburn L, et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol Med (2015) 7(2):127–39. doi:10.15252/emmm.201404137
88. Singhal A, Jie L, Kumar P, Hong GS, Leow MK-S, Paleja B, et al. Metformin as adjunct antituberculosis therapy. Sci Transl Med (2014) 6(263):263ra159. doi:10.1126/scitranslmed.3009885
89. Junkins RD, Shen A, Rosen K, McCormick C, Lin TJ. Autophagy enhances bacterial clearance during P. aeruginosa lung infection. PLoS One (2013) 8(8):e72263. doi:10.1371/journal.pone.0072263
90. Abdulrahman BA, Khweek AA, Akhter A, Caution K, Kotrange S, Abdelaziz DH, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy (2011) 7(11):1359–70. doi:10.4161/auto.7.11.17660
91. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell (2004) 119(6):753–66. doi:10.1016/j.cell.2004.11.038
92. Lerner TR, de Souza Carvalho-Wodarz C, Repnik U, Russell MR, Borel S, Diedrich CR, et al. Lymphatic endothelial cells are a replicative niche for Mycobacterium tuberculosis. J Clin Invest (2016) 126(3):1093–108. doi:10.1172/JCI83379
93. Andersson AM, Andersson B, Lorell C, Raffetseder J, Larsson M, Blomgran R. Autophagy induction targeting mTORC1 enhances Mycobacterium tuberculosis replication in HIV co-infected human macrophages. Sci Rep (2016) 6:28171. doi:10.1038/srep28171
94. Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature (2013) 494(7436):201–6. doi:10.1038/nature11866
95. Munz C. Autophagy proteins in antigen processing for presentation on MHC molecules. Immunol Rev (2016) 272(1):17–27. doi:10.1111/imr.12422
96. Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med (2009) 15(3):267–76. doi:10.1038/nm.1928
97. Saiga H, Nieuwenhuizen N, Gengenbacher M, Koehler AB, Schuerer S, Moura-Alves P, et al. The recombinant BCG DeltaureC::hly vaccine targets the AIM2 inflammasome to induce autophagy and inflammation. J Infect Dis (2015) 211(11):1831–41. doi:10.1093/infdis/jiu675
98. Kenific CM, Wittmann T, Debnath J. Autophagy in adhesion and migration. J Cell Sci (2016) 129(20):3685–93. doi:10.1242/jcs.188490
Keywords: autophagy, macrophage, bacteria, pathogen, phagocytosis, xenophagy, inflammation
Citation: Bah A and Vergne I (2017) Macrophage Autophagy and Bacterial Infections. Front. Immunol. 8:1483. doi: 10.3389/fimmu.2017.01483
Received: 13 September 2017; Accepted: 23 October 2017;
Published: 06 November 2017
Edited by:
Luciana Balboa, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Roberta Olmo Pinheiro, Oswaldo Cruz Foundation, BrazilElsa Anes, Universidade de Lisboa, Portugal
Copyright: © 2017 Bah and Vergne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabelle Vergne, aXNhYmVsbGUudmVyZ25lQGlwYnMuZnI=
 Aïcha Bah
Aïcha Bah Isabelle Vergne
Isabelle Vergne