- Department of Biomedical and Molecular Sciences, Queen’s University, Kingston, ON, Canada
Toll-like receptor (TLR)-7 is an endosomal innate immune sensor capable of detecting single-stranded ribonucleic acid. TLR7-mediated induction of type I interferon and other inflammatory cytokine production is important in antiviral immune responses. Furthermore, altered TLR7 expression levels are implicated in various autoimmune disorders, indicating a key role for this receptor in modulating inflammation. This review is focused on the regulation of TLR7 expression and localization compared to that of the other endosomal TLRs: TLR3, 8, and 9. Endosomal TLR localization is a tightly controlled and intricate process with some shared components among various TLRs. However, TLR-specific mechanisms must also be in place in order to regulate the induction of pathogen- and cell-specific responses. It is known that TLR7 is shuttled from the endoplasmic reticulum to the endosome via vesicles from the Golgi. Several chaperone proteins are required for this process, most notably uncoordinated 93 homolog B1 (Caenorhabditis elegans), recently identified to also be involved in the localization of the other endosomal TLRs. Acidification of the endosome and proteolytic cleavage of TLR7 are essential for TLR7 signaling in response to ligand binding. Cleavage of TLR7 has been demonstrated to be accomplished by furin peptidases in addition to cathepsins and asparagine endopeptidases. Moreover, triggering receptor expressed on myeloid cells like 4, a protein associated with antigen presentation and apoptosis in immune cells, has been implicated in the amplification of TLR7 signaling. Understanding these and other molecular mechanisms controlling TLR7 expression and trafficking will give insight into the specific control of TLR7 activity compared to the other endosomal TLRs.
Introduction
Toll-like receptors (TLRs) are a class of pattern recognition receptors that recognize bacterial or viral pathogen-associated molecular patterns (PAMPs), playing a key role in innate immune responses (1, 2). This family includes plasma membrane receptors identified in mice and humans: TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10 (2, 3), nucleic acid-sensing endosomal receptors also identified in mice and humans: TLR3, TLR7, TLR8, and TLR9, and murine-specific endosomal receptors: TLR11, TLR12, and TLR13 (4–7). The TLR family members have been well reviewed as mediators of innate immunity, as well as being extensively linked to autoimmunity (8–14).
Of the endosomal TLRs, TLR3 recognizes dsRNA and TLR9 recognizes dsDNA. Alternatively, TLR7 and the closely related TLR8 respond to purine-rich single-stranded ribonucleic acid (ssRNA) to elicit an immune response to pathogens which are recognized in the endosome (15–17). Thus, localization of these TLRs to the endosome is a requirement for detection of viral and bacterial nucleic acids that are endocytosed. Control of the trafficking pathways of these TLRs to the endosome is a regimented process involving several steps. In the literature, there is a greater emphasis on the regulation of TLR9 localization, while knowledge of the mechanisms controlling TLR7 localization is mostly deduced from TLR9 studies (18–20). The steps involved in endosomal localization of TLR7 are outlined in Figure 1 and are discussed in detail in this review.
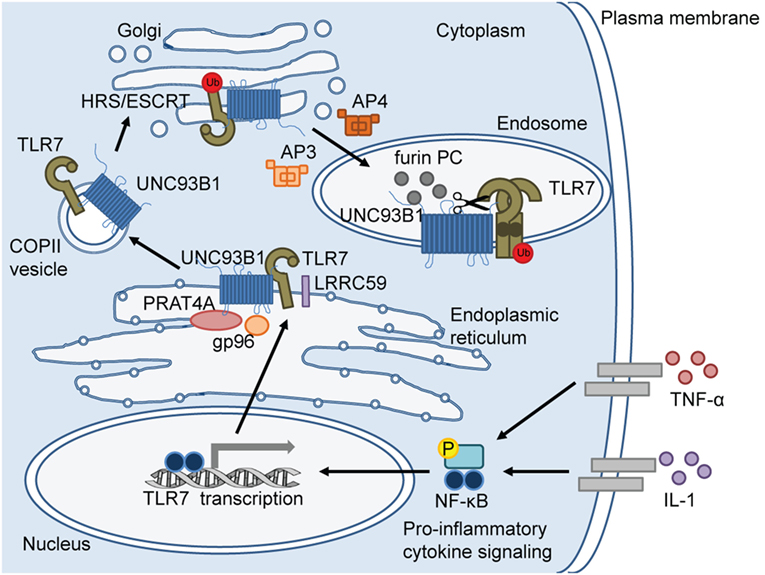
Figure 1. Toll-like receptor (TLR)-7 trafficking from the endoplasmic reticulum (ER) to the endosome. In response to pro-inflammatory cytokine signaling, TLR7 transcription is induced via nuclear factor (NF)-κB. TLR7 protein is synthesized in the ER where ER-resident chaperone proteins, gp96 and protein-associated with TLR4 (PRAT4A), are required to facilitate proper folding of TLR7 before exiting the ER. Upon cellular stimulation (e.g. imiquimod) or infection, leucine-rich repeat (LRR) containing protein 59 (LRRC59) promotes uncoordinated 93 homolog B1 (Caenorhabditis elegans) (UNC93B1)-mediated packaging of TLR7 into coat protein complex II (COPII)-coated vesicles to exit the ER and translocate to the Golgi. In the Golgi, UNC93B1 ubiquitinates TLR7 and HRS/ESCRT machinery sorts ubiquitinated TLR7 for endolysosomal transport. Adaptor protein (AP)-4 and AP-3 deliver TLR7 from the Golgi to lysosome-related organelles, such as “NF-κB endosomes” and “IRF-7 endosomes,” respectively. Once TLR7 reaches the endosome, endosomal acidification occurs for proteolytic cleavage by proteases, including furin proprotein convertases (PC). TLR7 cleavage separates the N-terminal ectodomain (darker circle) from the C-terminal ectodomain, transmembrane domain, and cytosolic region. The N terminal region forms disulfide bonds with the C-terminal region and remains in the endosome for optimal signaling.
TLR7 and TLR8 are often referred to together in the literature due to their high degree of homology and similarity in function. However, in comparison to TLR7, the mechanisms controlling TLR8 localization are less understood, likely due to a lack of appropriate murine model systems for studying TLR8. For example, the commonly used synthetic RNA analog resiquimod (R848) triggers human TLR7 and TLR8 signaling and murine TLR7 signaling, but not murine TLR8 (15, 21). Moreover, many of the studies delineating the mechanisms behind TLR7 localization have been performed in murine models and few studies have focused solely on human cells. Therefore, throughout this review, the studies discussed were performed in murine models unless otherwise specified. As well, much of the published work has examined TLR7 responsiveness, leaving a void in the literature regarding TLR8-specific mechanisms. Herein, we provide an in-depth commentary on the regulation of TLR7 expression and function and include additional highlights on mechanisms which control TLR8 localization.
Expression Patterns of TLR7
The distribution of TLR7 expression among various cell types is differentially regulated. Constitutive expression of TLR7 is predominant in human and murine plasmacytoid dendritic cells (pDCs) and B cells compared to other circulating immune cells (22–24). Low levels of TLR7 have also been observed in non-immune cells such as hepatocytes, epithelial cells, and keratinocytes (25–28). Unlike TLR7, TLR8 is more strongly expressed in myeloid cells and to a lesser degree in pDCs (22, 29). Under certain circumstances, TLR7 expression is inducible in cells expressing low to undetectable basal TLR7 levels, including immune cells such as macrophages and myeloid DCs, and non-immune cells such as hepatocytes and keratinocytes (25, 28, 30–32). For example, interferon (IFN)-γ stimulation of human macrophages resulted in the induction of TLR7 mRNA expression and consequently enhanced TLR7 responsiveness (16). As well, infection with viruses such as hepatitis C virus (HCV), human immunodeficiency virus (HIV), and influenza A virus (IAV) also result in upregulation of TLR7 expression in hepatocytes, circulating immune cells, and primary macrophages, respectively (25, 30, 33). Under these circumstances, it is likely that the cytokines induced by virus infection are responsible for triggering the induction of TLR7 expression. Indeed, influenza virus infection of human primary macrophages results in upregulation of TLR7 mRNA in a type I IFN-dependent manner (33). Furthermore, augmented TLR7 expression is observed in cancer and autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus (SLE) (31, 34, 35). Other cytokines and PAMPs released during inflammation as a result of infection or disease likely play a role in the regulation of constitutive and inducible TLR7 expression.
Given that the expression levels of TLR7 are relatively well documented under both normal and pathogenic conditions, it is surprising that the molecular mechanisms involved in the signaling pathways controlling TLR7 induction during virus infection or autoimmune conditions have not been well-described. As well, the regulation of constitutive TLR7 expression is relatively understudied. In a study by Lee et al., the TLR7 putative promoter sequence was cloned and characterized (36). Through computer analysis of the human TLR7 gene sequence (accession number: NT_011757), binding sites for the following transcription factors were identified: nuclear factor (NF)-κB, C/EBPα, C/EBPβ, GR, IFN regulatory factor (IRF)-1, MIG1, NF-1, Oct-1, RAP1, RSRFC4, RxR-β, SRF, SP1, and SRY (36). Using a human liver cell line as a model system, TLR7 expression induced by tumor necrosis factor (TNF)-α and interleukin (IL)-1 treatment was shown to require NF-κB activation but not that of IRF-7 (36). Further investigation of the signaling pathways regulating TLR7 expression is warranted in both immune and non-immune cells, particularly in inflammatory conditions.
Antiviral Functions of TLR7
Immune responses to ssRNA viruses include production of TLR7-mediated type I IFNs, a family of key antiviral cytokines that induce a variety of genes collectively known as IFN-inducible genes. Viruses have adapted a number of evasive strategies to avoid and/or shut down type I IFN responses, and there are a variety of innate viral sensors responsible for induction of type I IFN; these include melanoma differentiation-associated protein 5 (MDA5), laboratory of genetic and physiology 2 (LGP2), mitochondrial antiviral-signaling protein (MAVS), stimulator of IFN genes (STING), and retinoic acid inducible gene (RIG)-I (37). The role of these molecules during virus infection has been reviewed in detail elsewhere (37, 38). It is interesting to note that these sensors often work in tandem with each other and/or TLR7 to respond to infection. For example, it has been shown that TLR7 ligation induces expression of the cytoplasmic viral RNA-sensing helicase, RIG-I, in order to clear virus infection, thereby linking TLR7 and RIG-I function (39). Specifically, pDCs generally express low RIG-I but upon stimulation with the TLR7 agonist imiquimod, RIG-I is upregulated and thus this allows for cooperative RIG-I- and TLR7-mediated viral clearance (39).
TLR7 is well-established to bind viral ssRNA; specific examples include vesicular stomatitis virus (VSV), IAV, HIV-1, hepatitis B virus, and HCV (25, 40–43) or synthetic guanine-rich RNA sequence analogs such as R848, gardiquimod, loxoribine, and most notably the human papillomavirus treatment, imiquimod (15, 44, 45). Upon virus infection or agonist stimulation, ssRNA enters the endosome via autophagy or receptor-mediated endocytosis, as discussed in detail further below (46, 47). Dimeric TLR7 then interacts with the ssRNA which subsequently initiates signal transduction (48, 49). Signal transduction of TLR1-10 has been thoroughly reviewed in the literature (1–3, 8, 50–53). Similar to all TLRs, TLR7 contains a Toll/IL-1 receptor domain that associates with myeloid differentiation primary response gene 88 (MyD88) for signal transduction, aside from TLR3 which signals via the MyD88-independent pathway (20, 54). Unlike TLR8 and TLR9, which exist as preformed dimers (55–57), TLR7 dimerizes upon ligand binding in the endosome to initiate TLR7-mediated MyD88 signal transduction (58). This results in the subsequent activation of mitogen-activated protein kinase (MAPK) cascades, NF-κB activation (53, 59), as well as IRF-7 (60, 61) and IRF-5 (62, 63) activation via IL-1 receptor-associated kinases (IRAK)-1/2/4 and TNF receptor-associated factor-3/6 (61, 64, 65). Signaling in human immune cells by TLR7 has been documented to trigger production of pro-inflammatory cytokines including TNF-α, IL-6, IL-1β, IL-12, and IFN-α (61, 66–69). Although the specific signaling pathways triggered by TLR7 activation have been described (53, 60, 61), the underlying mechanisms controlling TLR7 trafficking to the endosomal compartment are now coming into the limelight.
TLR7 Endosomal Localization
Like the other endosomal TLRs, TLR7 can only bind and respond to its ligand in the endosome to prevent recognition of self-genomic information (70). TLR7 is directed to the endosome by an intricate shuttling process mediated by several chaperone proteins as illustrated in Figure 1. Furthermore, proteolytic cleavage must occur for ligand recognition (71, 72). Details regarding TLR7 localization compared to that of the other endosomal TLRs are discussed below.
Exit of TLR7 from the Endoplasmic Reticulum (ER)
TLR7 is initially produced as an inactive protein in the ER and must be correctly folded prior to exiting the ER. Two ER-resident chaperone proteins, the heat shock protein gp96 and protein-associated with TLR4 (PRAT4A), are involved in the folding of TLR7 as well as other TLRs, with the exception of TLR3 (Figure 1) (73–75). In the absence of functional gp96, macrophages, and B cells are unable to respond to TLR7 ligation (73, 74). As well, in PRAT4A-deficient macrophages, TLR7 responsiveness was completely impaired (75). A different ER-resident chaperone protein, leucine-rich repeat (LRR) containing protein 59 (LRRC59), was demonstrated to mediate TLR8 localization as well as TLR3 and TLR9 in human cells (76). Other experiments from the same study also imply that TLR8 and TLR9 endosomal localization requires LRRC59, as the absence of this protein resulted in decreased TLR8 and TLR9 signaling; however, TLR8 and TLR9 localization was not directly measured (76). LRRC59 could also be required for TLR7 signaling, however, such studies are necessary for confirmation. On the other hand, it is interesting to speculate that regulation of TLR7 localization diverges from the other TLRs and evidence to support this is outlined in the sections below.
One of the more researched ER-resident chaperone proteins, uncoordinated 93 homolog B1 (Caenorhabditis elegans) (UNC93B1), has a clearly defined role in the export of nucleotide-sensing TLRs from the ER to the Golgi and endosomes (77–81). UNC93B1 interacts with the TLRs to mediate packaging into coat protein complex II (COPII)-coated vesicles. The COPII complex is composed of conserved coat proteins and is involved in promoting anterograde protein transport from the ER to the Golgi; thus TLR-containing COPII vesicles bud from the ER for transport to the Golgi (19, 79). UNC93B1 is thought to be involved in ubiquitination of TLR9 and potentially TLR7 in a post-ER compartment to facilitate transfer and interactions with other chaperone proteins (82). Once in the Golgi, HRS/endosomal sorting complex required for transport (HRS/ESCRT) machinery sorts ubiquitinated TLR7 for endolysosomal transport through the non-canonical ESCRT pathway to maintain receptor stability within the endosome (Figure 1) (70, 82).
In murine models, different mutations of the UNC93B1 gene, Unc93b1, highlight the role of this protein in TLR shuttling. The UNC93B1 H412R mutation, also called the 3D (or triple D) mutation, causes inefficient processing of TLR3, TLR7, and TLR9 and results in enhanced susceptibility to virus infection (19, 77–79, 81). Similarly, the “loss of endosomal TLR response” (Letr) mutation, a 54-amino acid deletion in exon 4 of murine Unc93b1, leads to increased viral load of IAV (83), likely due to compromised TLR7 trafficking out of the ER. Interestingly, in pDCs, macrophages, B cells, and conventional DCs, the D34A mutation of UNC93B1 leads to increased cytokine production upon TLR7 activation, while cytokine production upon TLR9 activation was decreased and that of TLR3 was unaffected (84, 85). Taken together, these studies suggest that modulation of TLR-UNC93B1 binding capacity may fine-tune specific endosomal TLR responses.
UNC93B1 mRNA expression and protein levels are upregulated upon IFN-α, -β, or -γ stimulation of murine macrophages (86), indicating that antiviral IFN production may, in turn, enhance endosomal TLR7 localization and downstream signaling. However, the direct effect of type I IFN signaling on TLR7 localization has not been well documented. Furthermore, while in the ER, TLR7 competes with TLR9 for interaction with UNC93B1, and TLR9 generally prevails (84, 85, 87). Thus, IFN-mediated upregulation of UNC93B1 represents a potential mechanism to enhance TLR7 trafficking to endosome by increasing the likelihood of TLR7-UNC93B1 interaction over that of TLR9. Additionally, stimulation with imiquimod, as well as with TLR9 or TLR4 agonists, triggers UNC93B1-dependent TLR7 endosomal localization in murine myeloid cells (79). Furthermore, modeling in the human embryonic kidney (HEK)-293 cell line demonstrated that stimulation via other TLRs, in particular that of TLR8, as well as non-specific induction of endocytosis, enhances LRRC59 binding to UNC93B1 (76), indicating that endocytic events, such as virus invasion, may augment endosomal TLR localization to enhance the antiviral response.
Packaging TLR7 in the Endosome
Compared to other chaperone proteins, UNC93B1 appears to be unique in that it also leaves the ER to accompany the TLR in transit to the endosome. Other proteins are required for TLR-UNC93B1 transit to the endosome, these include the adaptor protein (AP) family (AP-1–4), which function to sort membrane proteins (88). Interactions between the APs, UNC93B1, and TLRs appear to be complex, with evidence for direct interaction between APs and UNC93B1 (19, 81) and APs with TLRs (19). Differential requirements for AP-1, AP-2, AP-3, and AP-4 have been proposed for the individual endosomal TLRs. Specifically, AP-4 binds TLR7 to sort into COPII vesicles for transport to the Golgi in macrophages (Figure 1) (19). TLR7 directly translocates from the Golgi to the endosome, accompanied by both UNC93B1 and AP-4 (19). Unlike TLR7, TLR9 is first shuttled from the Golgi to the plasma membrane, then to the endosome in an AP-2-dependent manner (19). Type I IFN induction in response to TLR7 ligation was found to depend on AP-3 for shuttling to specialized lysosome-related organelles (LRO) (89). These LRO, called “IRF-7 endosomes,” are lysosome-associated membrane protein (LAMP)-1+/LAMP2+ and signal via IRF7 to produce type I IFN (89). In contrast, pro-inflammatory cytokine production occurs upon TLR ligation in vesicle-associated membrane protein-3+/LAMP2− LRO that signal by NF-κB, called “NF-κB endosomes” (89). AP-3 deficiency abrogated TLR7-mediated type I IFN production in pDCs, while NF-κB-induced cytokine production was maintained, indicating a specific role for AP-3 in shuttling TLR7 to IRF-7 endosomes (89). Interestingly, Lee et al. demonstrated a direct interaction between UNC93B1 and AP-2, but not AP-1, 3, or 4, as well as a direct interaction between TLR7 and AP-4, but not AP-1, 2, or 3 (19). This suggests that other sorting proteins may be required to facilitate TLR7 travel to IRF-7 endosomes in tandem with AP-3. Furthermore, requirements for the targeting of TLR7 to NF-κB endosomes have not been elucidated and thus represent a focus for future research.
TLR7 Cleavage
Restriction of nucleic acid-sensing TLRs to the endosome, as well as the dependence on cleavage for activation both help prevent recognition of self-DNA/RNA (90, 91). Like all endosomal TLRs, prior to gaining the capacity to bind and respond to ligands, TLR7 must be proteolytically cleaved (92). In general, endosomal TLRs are cleaved by a common mechanism initiated by decreased endosomal pH, required for enzymatic activity. The cleavage events are a complex process and are thought to involve a combination of asparagine endopeptidase (AEP) and/or multiple cathepsins (71, 72, 90, 92). The majority of studies focusing on the regulation of cleavage events used a TLR9-expressing murine macrophage cell line (RAW cells) as a model system. Indeed, the initial role for cathepsins in TLR9 was suggested by Ewald et al.; however, at the time, this group was unable to experimentally block cathepsin activation efficiently, and they concluded that a combination of proteases were likely responsible (90). Later, the same group demonstrated that in RAW cells and murine conventional DCs, TLR cleavage was a two-step event. The initial cleavage event could be performed by either AEP or cathepsin, indicating a redundant role for these proteins (72). The second cleavage step for optimal signaling responses is referred to as TLR trimming and appears to rely exclusively on cathepsin activity (72). In the same report, TLR7 cleavage was shown to require both AEP and cathepsin activity (72). The need for AEP-dependent TLR7 cleavage was further highlighted by inefficient TLR7 signaling in DCs, which was linked to poor induction of adaptive immune responses during IAV infection of AEP-deficient mice (92).
Interestingly, Hipp et al. found that the furin-like proprotein convertases (PC), part of the family of serine endoproteases including furin, PC5/6, and PC7, cleave human TLR7 in the endosome for the generation of competent TLR7 signaling responses (Figure 1) (93). This study demonstrated a novel mechanism for cleavage of human TLR7 compared to the numerous studies focused on murine endosomal TLR7 cleavage by cathepsins and AEP (71, 72, 90, 92). Likewise, furin-like proteases have also been implicated in TLR8 proteolysis in primary human monocytes and macrophages (94). Thus, the use of furin-like proteases may be specific to TLR7 and TLR8 and may represent an alternative mechanism regulating TLR7 cleavage, in addition to AEP and cathepsins. The dependence for TLR7 cleavage on AEP, cathepsins, or furins could be dictated by cell type and differences in human and murine cell models.
With regards to the specific amino acid sequences for cleavage sites, TLR7 is susceptible to proteolysis between two LRR domains (LRR14–15) which correspond to residues 450–479 containing asparagine 478, required for AEP-mediated cleavage (92). The role of cleavage in TLR7 localization versus signaling was demonstrated using a model of murine fibroblasts and bone marrow-derived DCs (BMDC) transfected with wild type or mutated TLR7. Mutation of asparagine 478 to glutamine (N478Q) did not hinder TLR7 localization; however, downstream signaling in response to imiquimod was inhibited (92). This cleavage site separates the N-terminal (ectodomain) from the C-terminal (ectodomain, transmembrane, and cytosolic regions) that remains in the endosome for downstream signaling; it is suggested that the TLR7 N-terminus is cleaved but linked to the C-terminus by disulfide bonds and is required for endosomal localization and ligand binding in both murine DCs and human cell lines (THP-1; HEK-293T) (Figure 1) (95, 96). Crystallography studies of human TLR8 and studies in primary human myeloid cells also demonstrate N- and C-terminal association after cleavage (57, 94). Taken together, this information combined with that on the enzymatic cleavage highlights the complexity of how processing of TLR7, as well as TLR8, is regulated.
Ligand Delivery to TLR7
Like the regulation of TLR7 trafficking, ligand delivery to TLR7 is also an intricately regulated process. Upon infection, agonist stimulation, or damage/apoptosis to neighboring cells, ligands must enter the endosome to trigger TLR7 activation. Receptor-mediated endocytosis can deliver virus or purified TLR7 ligands to endosomal TLR7 (48, 49, 97–99). In autoimmunity, delivery of self-ligand to the endosome relies on proteins such as high mobility group box 1 protein and the cathelicidin, LL37 (100). Thus, TLR7 responses are regulated during inflammatory disease conditions by release or overexpression of these molecules; for example, increased LL37 expression is associated with exacerbated TLR7-mediated cytokine production during autoimmune disease (101). As well, B cells bearing B cell receptors (BCR) specific for DNA, and pDCs bearing the Fc receptor FcγRIIa (CD32), were shown to internalize self-DNA for delivery to TLR9 in SLE (102, 103). This sequential engagement of BCR and TLR9 was recapitulated for TLR7 where autoreactive BCRs were shown to bind and internalize ssRNA for delivery to TLR7 in a model of SLE (104).
While viral ssRNA may reach the endosome as a result of endocytosis, murine pDCs have also been shown to utilize autophagy, a homeostatic process whereby damaged or unnecessary cellular material from the cytoplasm is degraded in lysosomes, to serve an endosomal delivery platform for viral RNA-TLR7 ligation (46, 105, 106). Autophagosomes are formed by a sequential series of steps, each involving autophagy-related gene (ATG) proteins; the process is initiated by phagophore formation, a double-membrane vacuole that expands into an autophagosome (107). Once the autophagosome fuses with the endosome or lysosome, autolysosomes are formed for degradation of cellular material (108). A role for autophagy and TLR7 ligand delivery during viral infection is prominent in murine pDCs since these cells exhibit TLR7-dependent type I IFN production in response to virus infection (47). Lee et al. demonstrated that murine ATG5-deficient pDCs infected with VSV were unable to produce IFN-α (47). A few years later, two other groups demonstrated that autophagy was required for TLR7-mediated IFN-α production in primary human pDCs infected with HIV-1 or paramyxovirus simian virus 5 (109, 110). Of note, autophagy has also been implicated in ssRNA delivery to TLR7 in murine B cells in SLE (111). Preference between endocytosis and autophagy for ligand delivery during virus infection may depend on several factors, such as the mode of breaching the cell (endocytosis vs membrane fusion), replication strategies, and expression of viral inhibitory factors. In addition to the role for autophagosome-mediated delivery of TLR7 ligands to the endosome, a role for TLR7 signaling in the induction of autophagy has also been investigated (46). Upon examination of TLRs 1 through 9, ligation of TLR7 demonstrated the most potent induction of autophagy in murine macrophages (46). Furthermore, similar to TLR4, TLR7 induces Beclin 1 (mammalian homolog of yeast ATG6) interaction with MyD88 for enhanced autophagosome formation in murine macrophages (46, 112, 113). These studies provide evidence for a potential positive feedback mechanism between TLR7 signaling and autophagy-mediated viral recognition.
Modulation of TLR7 Signaling
Investigations into the roles for the triggering receptor expressed on myeloid cells (TREM) receptor family and associated ligands have identified these proteins as novel constituents involved in modulating TLR signaling and responsiveness (114–117). Recently, TREM like 4 (TREML4) has been characterized to amplify TLR7 signaling in murine and human cells (Figure 1) (118). Little is known regarding TREML4 functions; it is predominantly found on splenic macrophages as well as CD8+ DCs, and it has been shown to bind apoptotic cells and enhance antigen presentation to T cells (119, 120). In Treml4-deficient BMDC, decreased TLR7 endosomal localization and TLR7-MyD88 interaction was observed with a concomitant decrease in activation of TLR7-mediated signaling and cytokine production (118). Knockdown of Treml4 in human macrophages also resulted in decreased cytokine induction in response to gardiquimod (118). Together, this indicates that upon TLR7 ligation TREML4 may function to stabilize the TLR7-MyD88 interaction, thus promoting signal transduction and resulting cytokine production. Interestingly, a role for TREML4 was also suggested for TLR9 and TLR13 signaling (118), however, the direct impact of TREML4 deficiency on TLR9 or TLR13 association with MyD88 or endosomal localization was not investigated. The cellular localization patterns of TREML4 have not been identified and whether TREML4 is recruited to the endosomes with TLR7 or if it directly interacts with TLR7 remains unknown. Interestingly, stimulation of splenic macrophages with gardiquimod or CpG DNA, a TLR9 agonist, resulted in enhancement of TREML4 mRNA expression (118), although stimulation of splenic DCs with TLR3, TLR4, or TLR9 agonists did not impact TREML4 expression in another study (119). This suggests that TREML4 may play a role in a positive feedback loop for TLR7 signaling and may also have differential effects depending on the cell type. These studies signify a novel mechanism for TLR7 signal amplification in the endosome and this represents an enticing new avenue for investigation to delineate signaling requirements for endosomal TLRs.
Another mechanism to modulate TLR signaling occurs upon multiple subsequent exposures to ligand, which results in abrogation of TLR responsiveness. This is best characterized with TLR4 stimulation using lipopolysaccharide (LPS), whereby LPS (endotoxin) tolerance is characterized by reduced TNF-α and IL-6 production upon repeated LPS stimulation in comparison to a single exposure to LPS (121). Interestingly, TLR7 tolerance has been observed in murine macrophages repeatedly exposed to TLR7 agonists; repeated exposure to imiquimod induced a refractory state whereby cells did not respond to subsequent TLR7 stimulation, due to elevated MyD88 pathway inhibitors: IRAK-M and Src homology 2 domain-containing inositol-5-phosphatase-1, and impaired activation of NF-κB, MAPK p38, and c-Jun N-terminal kinase (122, 123). TLR “hetero-tolerance,” accomplished by exposure to one TLR agonist resulting in decreased responsiveness to secondary stimulation by an agonist for a different TLR, also serves as a general mechanism to control TLR responses. For example, in vivo intravenous administration of LPS in human subjects followed by ex vivo leukocyte challenge with TLR7 ligand (S-27609) led to suppressed secretion of TNF-α, IL-6, IL-1β, and IL-10 compared to S-27609 stimulation alone (124). Moreover, TLR7 ligand stimulation reduces subsequent responses to other TLR agonists. For example, human and murine myeloid cells cultured with R848 or loxoribine exhibited decreased TLR4-responsiveness (125–128). Furthermore, pretreatment of human macrophages with R848 inhibits subsequent induction of TLR2, TLR4, TLR5, and TLR7 signaling (125). Interestingly, costimulation of murine macrophages with the TLR7 agonist, gardiquimod, and the TLR9 agonist, CpG DNA, resulted in a suppressor of cytokine signaling-1-mediated reduction of TNF-α and IL-6 (129). These studies demonstrate multiple mechanisms that regulate TLR responsiveness and further indicate that TLR7 activity can modulate responses to other TLR molecules and vice versa.
Future Perspectives
Elucidating the mechanisms controlling TLR7 localization is critical to further our understanding of key elements that dictate TLR7 ligand binding and downstream signaling. TLR7 and TLR8 appear to be redundant, and although there is a paucity of information on the regulation of TLR8 localization, subtle differences do exist between TLR7 and TLR8 regarding the regulation of endosomal localization and function. Thus, understanding precisely how TLR8 is differentially regulated from TLR7 is a key area for future research. Moreover, gaining a better understanding of the differential requirements for localization of each of the different endosomal TLRs will facilitate identification of mechanisms controlling ligand delivery. Ultimately, in-depth analysis of the complex processes regulating the localization and function of TLR7 will garner further information on fine-tuning immune responses in virus infection, cancer, and autoimmune diseases for the development of novel therapeutics.
Author Contributions
CP, NO, and KG contributed to the conceptualization, discussion, and writing of this review. CP designed and composed the figure.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; 342168). CP was supported by Ontario Graduate Scholarship, Dr. Robert John Wilson Graduate Fellowship, and NSERC-PGS D.
References
1. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol (2003) 21(1):335–76. doi:10.1146/annurev.immunol.21.120601.141126
2. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol (2015) 33:257–90. doi:10.1146/annurev-immunol-032414-112240
3. Leifer CA, Medvedev AE. Molecular mechanisms of regulation of toll-like receptor signaling. J Leukoc Biol (2016) 100(5):927–41. doi:10.1189/jlb.2MR0316-117RR
4. Ewald SE, Barton GM. Nucleic acid sensing toll-like receptors in autoimmunity. Curr Opin Immunol (2011) 23(1):3–9. doi:10.1016/j.coi.2010.11.006
5. Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science (2005) 308(5728):1626–9. doi:10.1126/science.1109893
6. Koblansky AA, Jankovic D, Oh H, Hieny S, Sungnak W, Mathur R, et al. Recognition of profilin by toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity (2013) 38(1):119–30. doi:10.1016/j.immuni.2012.09.016
7. Wang J, Chai J, Wang H. Structure of the mouse toll-like receptor 13 ectodomain in complex with a conserved sequence from bacterial 23S ribosomal RNA. FEBS J (2016) 283(9):1631–5. doi:10.1111/febs.13628
8. Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol (2005) 17(1):1–14. doi:10.1093/intimm/dxh186
9. Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol (2006) 6(11):823–35. doi:10.1038/nri1957
10. Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol Rev (2007) 220(1):251–69. doi:10.1111/j.1600-065X.2007.00572.x
11. von Landenberg P, Bauer S. Nucleic acid recognizing toll-like receptors and autoimmunity. Curr Opin Immunol (2007) 19(6):606–10. doi:10.1016/j.coi.2007.10.004
12. Krieg AM. The toll of too much TLR7. Immunity (2007) 27(5):695–7. doi:10.1016/j.immuni.2007.11.001
13. Alexoudi I, Kapsimali V, Vaiopoulos A, Kanakis M, Vaiopoulos G. Toll-like receptors pathways implication in common autoimmune diseases and therapeutic perspectives. G Ital Dermatol Venereol (2015) 150(2):255–60.
14. Pelka K, Shibata T, Miyake K, Latz E. Nucleic acid-sensing TLRs and autoimmunity: novel insights from structural and cell biology. Immunol Rev (2016) 269(1):60–75. doi:10.1111/imr.12375
15. Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88–dependent signaling pathway. Nat Immunol (2002) 3(2):196–200. doi:10.1038/ni758
16. Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, et al. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J Immunol (2008) 180(4):2117–24. doi:10.4049/jimmunol.180.4.2117
17. Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, et al. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol (2009) 10(6):587–94. doi:10.1038/ni.1733
18. Chuang T-H, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw (2000) 11(3):372–8.
19. Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife (2013) 2:e00291. doi:10.7554/eLife.00291
20. Bao M, Liu Y-J. Regulation of TLR7/9 signaling in plasmacytoid dendritic cells. Protein Cell (2013) 4(1):40–52. doi:10.1007/s13238-012-2104-8
21. Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol (2002) 3(6):499. doi:10.1038/ni0602-499
22. Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T, et al. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol (2002) 168(9):4531–7. doi:10.4049/jimmunol.168.9.4531
23. Edwards AD, Diebold SS, Slack E, Tomizawa H, Hemmi H, Kaisho T, et al. Toll-like receptor expression in murine DC subsets: lack of TLR7 expression by CD8α+ DC correlates with unresponsiveness to imidazoquinolines. Eur J Immunol (2003) 33(4):827–33. doi:10.1002/eji.200323797
24. Barr TA, Brown S, Ryan G, Zhao J, Gray D. TLR-mediated stimulation of APC: distinct cytokine responses of B cells and dendritic cells. Eur J Immunol (2007) 37(11):3040–53. doi:10.1002/eji.200636483
25. Lee J, Wu CC, Lee KJ, Chuang T-H, Katakura K, Liu Y-T, et al. Activation of anti-hepatitis C virus responses via toll-like receptor 7. Proc Natl Acad Sci U S A (2006) 103(6):1828–33. doi:10.1073/pnas.0510801103
26. Tengroth L, Millrud CR, Kvarnhammar AM, Georén SK, Latif L, Cardell L-O. Functional effects of toll-like receptor (TLR) 3, 7, 9, RIG-I and MDA-5 stimulation in nasal epithelial cells. PLoS One (2014) 9(6):e98239. doi:10.1371/journal.pone.0098239
27. Schaefer TM, Desouza K, Fahey JV, Beagley KW, Wira CR. Toll-like receptor (TLR) expression and TLR-mediated cytokine/chemokine production by human uterine epithelial cells. Immunology (2004) 112(3):428–36. doi:10.1111/j.1365-2567.2004.01898.x
28. Li ZJ, Sohn KC, Choi DK, Shi G, Hong D, Lee HE, et al. Roles of TLR7 in activation of NF-kappaB signaling of keratinocytes by imiquimod. PLoS One (2013) 8(10):e77159. doi:10.1371/journal.pone.0077159
29. Crozat K, Vivier E, Dalod M. Crosstalk between components of the innate immune system: promoting anti-microbial defenses and avoiding immunopathologies. Immunol Rev (2009) 227(1):129–49. doi:10.1111/j.1600-065X.2008.00736.x
30. Lester RT, Yao XD, Ball TB, McKinnon LR, Kaul R, Wachihi C, et al. Toll-like receptor expression and responsiveness are increased in viraemic HIV-1 infection. AIDS (2008) 22(6):685–94. doi:10.1097/QAD.0b013e3282f4de35
31. Roelofs MF, Joosten LA, Abdollahi-Roodsaz S, van Lieshout AW, Sprong T, van den Hoogen FH, et al. The expression of toll-like receptors 3 and 7 in rheumatoid arthritis synovium is increased and costimulation of toll-like receptors 3, 4, and 7/8 results in synergistic cytokine production by dendritic cells. Arthritis Rheum (2005) 52(8):2313–22. doi:10.1002/art.21278
32. Yin C, Zhang T, Qiao L, Du J, Li S, Zhao H, et al. TLR7-expressing cells comprise an interfollicular epidermal stem cell population in murine epidermis. Sci Rep (2014) 4:5831. doi:10.1038/srep05831
33. Miettinen M, Sareneva T, Julkunen I, Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun (2001) 2(6):349. doi:10.1038/sj.gene.6363791
34. Sheyhidin I, Nabi G, Hasim A, Zhang R-P, Ainiwaer J, Ma H, et al. Overexpression of TLR3, TLR4, TLR7 and TLR9 in esophageal squamous cell carcinoma. World J Gastroenterol (2011) 17(32):3745. doi:10.3748/wjg.v17.i32.3745
35. Komatsuda A, Wakui H, Iwamoto K, Ozawa M, Togashi M, Masai R, et al. Up-regulated expression of toll-like receptors mRNAs in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin Exp Immunol (2008) 152(3):482–7. doi:10.1111/j.1365-2249.2008.03646.x
36. Lee J, Hayashi M, Lo J-F, Fearns C, Chu W-M, Luo Y, et al. Nuclear factor κB (NF-κB) activation primes cells to a pro-inflammatory polarized response to a toll-like receptor 7 (TLR7) agonist. Biochem J (2009) 421(2):301–10. doi:10.1042/BJ20090013
37. Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses (2011) 3(6):920–40. doi:10.3390/v3060920
38. Felix J, Savvides SN. Mechanisms of immunomodulation by mammalian and viral decoy receptors: insights from structures. Nat Rev Immunol (2017) 17(2):112–29. doi:10.1038/nri.2016.134
39. Szabo A, Magyarics Z, Pazmandi K, Gopcsa L, Rajnavolgyi E, Bacsi A. TLR ligands upregulate RIG-I expression in human plasmacytoid dendritic cells in a type I IFN-independent manner. Immunol Cell Biol (2014) 92(8):671–8. doi:10.1038/icb.2014.38
40. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science (2004) 303(5663):1526–9. doi:10.1126/science.1093620
41. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa CR. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (2004) 303(5663):1529–31. doi:10.1126/science.1093616
42. Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, et al. Recognition of single-stranded RNA viruses by toll-like receptor 7. Proc Natl Acad Sci U S A (2004) 101(15):5598–603. doi:10.1073/pnas.0400937101
43. Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol (2005) 79(11):7269–72. doi:10.1128/JVI.79.11.7269-7272.2005
44. Lee J, Chuang T-H, Redecke V, She L, Pitha PM, Carson DA, et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of toll-like receptor 7. Proc Natl Acad Sci U S A (2003) 100(11):6646–51. doi:10.1073/pnas.0631696100
45. Gosu V, Basith S, Kwon O-P, Choi S. Therapeutic applications of nucleic acids and their analogues in toll-like receptor signaling. Molecules (2012) 17(11):13503–29. doi:10.3390/molecules171113503
46. Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J (2008) 27(7):1110–21. doi:10.1038/emboj.2008.31
47. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science (2007) 315(5817):1398–401. doi:10.1126/science.1136880
48. Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity (2010) 32(3):305–15. doi:10.1016/j.immuni.2010.03.012
49. Perry AK, Chen G, Zheng D, Tang H, Cheng G. The host type I interferon response to viral and bacterial infections. Cell Res (2005) 15(6):407–22. doi:10.1038/sj.cr.7290309
50. Kopp EB, Medzhitov R. The toll-receptor family and control of innate immunity. Curr Opin Immunol (1999) 11(1):13–8. doi:10.1016/S0952-7915(99)80003-X
51. Doyle SL, O’Neill LA. Toll-like receptors: from the discovery of NFκB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol (2006) 72(9):1102–13. doi:10.1016/j.bcp.2006.07.010
52. Kaisho T, Akira S. Toll-like receptor function and signaling. J Allergy ClinImmunol (2006) 117(5):979–87. doi:10.1016/j.jaci.2006.02.023
53. Kawai T, Akira S. Signaling to NF-κB by toll-like receptors. Trends Mol Med (2007) 13(11):460–9. doi:10.1016/j.molmed.2007.09.002
54. Kenny EF, O’Neill LA. Signalling adaptors used by toll-like receptors: an update. Cytokine (2008) 43(3):342–9. doi:10.1016/j.cyto.2008.07.010
55. Zhu J, Brownlie R, Liu Q, Babiuk LA, Potter A, Mutwiri GK. Characterization of bovine toll-like receptor 8: ligand specificity, signaling essential sites and dimerization. Mol Immunol (2009) 46(5):978–90. doi:10.1016/j.molimm.2008.09.024
56. Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DC, et al. Ligand-induced conformational changes allosterically activate toll-like receptor 9. Nat Immunol (2007) 8(7):772–9. doi:10.1038/ni1479
57. Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T. Structural reorganization of the toll-like receptor 8 dimer induced by agonistic ligands. Science (2013) 339(6126):1426–9. doi:10.1126/science.1229159
58. Manavalan B, Basith S, Choi S. Similar structures but different roles–an updated perspective on TLR structures. Front Physiol (2011) 2:41. doi:10.3389/fphys.2011.00041
59. Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, et al. IκB kinase-α is critical for interferon-α production induced by toll-like receptors 7 and 9. Nature (2006) 440(7086):949–53. doi:10.1038/nature04641
60. Honda K, Yanai H, Mizutani T, Negishi H, Shimada N, Suzuki N, et al. Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in toll-like receptor signaling. Proc Natl Acad Sci U S A (2004) 101(43):15416–21. doi:10.1073/pnas.0406933101
61. Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, et al. Interferon-α induction through toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol (2004) 5(10):1061–8. doi:10.1038/ni1118
62. Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, et al. Integral role of IRF-5 in the gene induction programme activated by toll-like receptors. Nature (2005) 434(7030):243–9. doi:10.1038/nature03308
63. Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem (2005) 280(17):17005–12. doi:10.1074/jbc.M412584200
64. Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, et al. Critical role of TRAF3 in the toll-like receptor-dependent and-independent antiviral response. Nature (2006) 439(7073):208–11. doi:10.1038/nature04374
65. Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu L-C, Wang GG, et al. Specificity in toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature (2006) 439(7073):204–7. doi:10.1038/nature04369
66. Ito T, Amakawa R, Kaisho T, Hemmi H, Tajima K, Uehira K, et al. Interferon-α and interleukin-12 are induced differentially by toll-like receptor 7 ligands in human blood dendritic cell subsets. J Exp Med (2002) 195(11):1507–12. doi:10.1084/jem.20020207
67. Testerman T, Gerster J, Imbertson L, Reiter M, Miller R, Gibson S, et al. Cytokine induction by the immunomodulators imiquimod and S-27609. J Leukoc Biol (1995) 58(3):365–72.
68. Reiter MJ, Testerman TL, Miller RL, Weeks CE, Tomai MA. Cytokine induction in mice by the immunomodulator imiquimod. J Leukoc Biol (1994) 55(2):234–40.
69. Imbertson LM, Couture AM, Gibson SJ, Smith RM, Miller RL, Reiter MJ, et al. Cytokine induction in hairless mouse and rat skin after topical application of the immune response modifiers imiquimod and S-28463. J Invest Dermatol (1998) 110(5):734–9. doi:10.1046/j.1523-1747.1998.00174.x
70. Lee BL, Barton GM. Trafficking of endosomal toll-like receptors. Trends Cell Biol (2014) 24(6):360–9. doi:10.1016/j.tcb.2013.12.002
71. Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, et al. Critical role for asparagine endopeptidase in endocytic toll-like receptor signaling in dendritic cells. Immunity (2009) 31(5):737–48. doi:10.1016/j.immuni.2009.09.013
72. Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med (2011) 208(4):643–51. doi:10.1084/jem.20100682
73. Liu B, Yang Y, Qiu Z, Staron M, Hong F, Li Y, et al. Folding of toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun (2010) 1:79. doi:10.1038/ncomms1070
74. Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrançois L, et al. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity (2007) 26(2):215–26. doi:10.1016/j.immuni.2006.12.005
75. Takahashi K, Shibata T, Akashi-Takamura S, Kiyokawa T, Wakabayashi Y, Tanimura N, et al. A protein associated with toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J Exp Med (2007) 204(12):2963–76. doi:10.1084/jem.20071132
76. Tatematsu M, Funami K, Ishii N, Seya T, Obuse C, Matsumoto M. LRRC59 regulates trafficking of nucleic acid–sensing TLRs from the endoplasmic reticulum via association with UNC93B1. J Immunol (2015) 195(10):4933–42. doi:10.4049/jimmunol.1501305
77. Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via toll-like receptors 3, 7 and 9. Nat Immunol (2006) 7(2):156–64. doi:10.1038/ni1297
78. Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim Y-M. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol (2007) 177(2):265–75. doi:10.1083/jcb.200612056
79. Kim Y-M, Brinkmann MM, Paquet M-E, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature (2008) 452(7184):234–8. doi:10.1038/nature06726
80. Itoh H, Tatematsu M, Watanabe A, Iwano K, Funami K, Seya T, et al. UNC93B1 physically associates with human TLR8 and regulates TLR8-mediated signaling. PLoS One (2011) 6(12):e28500. doi:10.1371/journal.pone.0028500
81. Pelka K, Phulphagar K, Zimmermann J, Stahl R, Schmid-Burgk JL, Schmidt T, et al. Cutting edge: the UNC93B1 tyrosine-based motif regulates trafficking and TLR responses via separate mechanisms. J Immunol (2014) 193(7):3257–61. doi:10.4049/jimmunol.1301886
82. Chiang C-Y, Engel A, Opaluch AM, Ramos I, Maestre AM, Secundino I, et al. Cofactors required for TLR7-and TLR9-dependent innate immune responses. Cell Host Microbe (2012) 11(3):306–18. doi:10.1016/j.chom.2012.02.002
83. Lafferty EI, Flaczyk A, Angers I, Homer R, d’Hennezel E, Malo D, et al. An ENU-induced splicing mutation reveals a role for Unc93b1 in early immune cell activation following influenza A H1N1 infection. Genes Immun (2014) 15(5):320–32. doi:10.1038/gene.2014.22
84. Fukui R, Saitoh S-I, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, et al. Unc93B1 biases toll-like receptor responses to nucleic acid in dendritic cells toward DNA-but against RNA-sensing. J Exp Med (2009) 206(6):1339–50. doi:10.1084/jem.20082316
85. Fukui R, Saitoh S-I, Kanno A, Onji M, Shibata T, Ito A, et al. Unc93B1 restricts systemic lethal inflammation by orchestrating toll-like receptor 7 and 9 trafficking. Immunity (2011) 35(1):69–81. doi:10.1016/j.immuni.2011.05.010
86. Panchanathan R, Liu H, Choubey D. Expression of murine Unc93b1 is up-regulated by interferon and estrogen signaling: implications for sex bias in the development of autoimmunity. Int Immunol (2013) 25(9):521–9. doi:10.1093/intimm/dxt015
87. Wang J, Shao Y, Bennett TA, Shankar RA, Wightman PD, Reddy LG. The functional effects of physical interactions among toll-like receptors 7, 8, and 9. J Biol Chem (2006) 281(49):37427–34. doi:10.1074/jbc.M605311200
88. Nakatsu F, Ohno H. Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct Funct (2003) 28(5):419–29. doi:10.1247/csf.28.419
89. Sasai M, Linehan MM, Iwasaki A. Bifurcation of toll-like receptor 9 signaling by adaptor protein 3. Science (2010) 329(5998):1530–4. doi:10.1126/science.1187029
90. Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi G-P, Chapman HA, et al. The ectodomain of toll-like receptor 9 is cleaved to generate a functional receptor. Nature (2008) 456(7222):658–62. doi:10.1038/nature07405
91. Barton GM, Kagan JC, Medzhitov R. Intracellular localization of toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol (2006) 7(1):49–56. doi:10.1038/ni1280
92. Maschalidi S, Hässler S, Blanc F, Sepulveda FE, Tohme M, Chignard M, et al. Asparagine endopeptidase controls anti-influenza virus immune responses through TLR7 activation. PLoS Pathog (2012) 8(8):e1002841. doi:10.1371/journal.ppat.1002841
93. Hipp MM, Shepherd D, Gileadi U, Aichinger MC, Kessler BM, Edelmann MJ, et al. Processing of human toll-like receptor 7 by furin-like proprotein convertases is required for its accumulation and activity in endosomes. Immunity (2013) 39(4):711–21. doi:10.1016/j.immuni.2013.09.004
94. Ishii N, Funami K, Tatematsu M, Seya T, Matsumoto M. Endosomal localization of TLR8 confers distinctive proteolytic processing on human myeloid cells. J Immunol (2014) 193(10):5118–28. doi:10.4049/jimmunol.1401375
95. Kanno A, Yamamoto C, Onji M, Fukui R, Saitoh S-I, Motoi Y, et al. Essential role for toll-like receptor 7 (TLR7)-unique cysteines in an intramolecular disulfide bond, proteolytic cleavage and RNA sensing. Int Immunol (2013) 25(7):413–22. doi:10.1093/intimm/dxt007
96. Hipp MM, Shepherd D, Booth S, Waithe D, Reis e Sousa C, Cerundolo V. The processed amino-terminal fragment of human TLR7 acts as a chaperone to direct human TLR7 into endosomes. J Immunol (2015) 194(11):5417–25. doi:10.4049/jimmunol.1402703
97. Brencicova E, Diebold SS. Nucleic acids and endosomal pattern recognition: how to tell friend from foe? Front Cell Infect Microbiol (2013) 3:37. doi:10.3389/fcimb.2013.00037
98. Xagorari A, Chlichlia K. Toll-like receptors and viruses: induction of innate antiviral immune responses. Open Microbiol J (2008) 2:49–59. doi:10.2174/1874285800802010049
99. Wagner H, Bauer S. All is not toll: new pathways in DNA recognition. J Exp Med (2006) 203(2):265–8. doi:10.1084/jem.20052191
100. Lee CC, Avalos AM, Ploegh HL. Accessory molecules for toll-like receptors and their function. Nat Rev Immunol (2012) 12(3):168–79. doi:10.1038/nri3151
101. Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, et al. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med (2009) 206(9):1983–94. doi:10.1084/jem.20090480
102. Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin–IgG complexes activate B cells by dual engagement of IgM and toll-like receptors. Nature (2002) 416(6881):603–7. doi:10.1038/416603a
103. Viglianti GA, Lau CM, Hanley TM, Miko BA, Shlomchik MJ, Marshak-Rothstein A. Activation of autoreactive B cells by CpG dsDNA. Immunity (2003) 19(6):837–47. doi:10.1016/S1074-7613(03)00323-6
104. Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/toll-like receptor 7 engagement. J Exp Med (2005) 202(9):1171–7. doi:10.1084/jem.20050630
105. Kroemer G, Jäättelä M. Lysosomes and autophagy in cell death control. Nat Rev Cancer (2005) 5(11):886–97. doi:10.1038/nrc1738
106. Weindel CG, Richey LJ, Mehta AJ, Shah M, Huber BT. Autophagy in dendritic cells and B cells is critical for the inflammatory state of TLR7-mediated autoimmunity. J Immunol (2017) 198(3):1081–92. doi:10.4049/jimmunol.1601307
107. Yordy B, Tal MC, Hayashi K, Arojo O, Iwasaki A. Autophagy and selective deployment of Atg proteins in antiviral defense. Int Immunol (2013) 25(1):1–10. doi:10.1093/intimm/dxs101
108. Eskelinen E-L. Maturation of autophagic vacuoles in mammalian cells. Autophagy (2005) 1(1):1–10. doi:10.4161/auto.1.1.1270
109. Zhou D, Kang KH, Spector SA. Production of interferon α by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J Infect Dis (2012) 205(8):1258–67. doi:10.1093/infdis/jis187
110. Manuse MJ, Briggs CM, Parks GD. Replication-independent activation of human plasmacytoid dendritic cells by the paramyxovirus SV5 Requires TLR7 and autophagy pathways. Virology (2010) 405(2):383–9. doi:10.1016/j.virol.2010.06.023
111. Weindel CG, Richey LJ, Bolland S, Mehta AJ, Kearney JF, Huber BT. B cell autophagy mediates TLR7-dependent autoimmunity and inflammation. Autophagy (2015) 11(7):1010–24. doi:10.1080/15548627.2015.1052206
112. Xu Y, Jagannath C, Liu X-D, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity (2007) 27(1):135–44. doi:10.1016/j.immuni.2007.05.022
113. Shi C-S, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem (2008) 283(48):33175–82. doi:10.1074/jbc.M804478200
114. Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol (2000) 164(10):4991–5. doi:10.4049/jimmunol.164.10.4991
115. Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol (2006) 177(4):2051–5. doi:10.4049/jimmunol.177.4.2051
116. Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol (2006) 177(6):3520–4. doi:10.4049/jimmunol.177.6.3520
117. Ornatowska M, Azim AC, Wang X, Christman JW, Xiao L, Joo M, et al. Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. Am J Physiol Lung Cell Mol Physiol (2007) 293(6):L1377–84. doi:10.1152/ajplung.00140.2007
118. Ramirez-Ortiz ZG, Prasad A, Griffith JW, Pendergraft WF III, Cowley GS, Root DE, et al. The receptor TREML4 amplifies TLR7-mediated signaling during antiviral responses and autoimmunity. Nat Immunol (2015) 16(5):495–504. doi:10.1038/ni.3143
119. Hemmi H, Idoyaga J, Suda K, Suda N, Kennedy K, Noda M, et al. A new Trem family member, Treml4, binds to dead cells and is a DAP12 linked marker for subsets of mouse macrophages and dendritic cells. J Immunol (2009) 182(3):1278. doi:10.4049/jimmunol.182.3.1278
120. Hemmi H, Zaidi N, Wang B, Matos I, Fiorese C, Lubkin A, et al. Treml4, an Ig superfamily member, mediates presentation of several antigens to T cells in vivo, including protective immunity to HER2 protein. J Immunol (2012) 188(3):1147–55. doi:10.4049/jimmunol.1102541
121. West MA, Heagy W. Endotoxin tolerance: a review. Crit Care Med (2002) 30(1):S64–73. doi:10.1097/00003246-200201001-00009
122. Hassan F, Islam S, Tumurkhuu G, Dagvadorj J, Naiki Y, Komatsu T, et al. Involvement of interleukin-1 receptor-associated kinase (IRAK)-M in toll-like receptor (TLR) 7-mediated tolerance in RAW 264.7 macrophage-like cells. Cell Immunol (2009) 256(1):99–103. doi:10.1016/j.cellimm.2009.01.013
123. Hayashi T, Gray CS, Chan M, Tawatao RI, Ronacher L, McGargill MA, et al. Prevention of autoimmune disease by induction of tolerance to toll-like receptor 7. Proc Natl Acad Sci U S A (2009) 106(8):2764–9. doi:10.1073/pnas.0813037106
124. de Vos AF, Pater JM, van den Pangaart PS, de Kruif MD, van’t Veer C, van der Poll T. In vivo lipopolysaccharide exposure of human blood leukocytes induces cross-tolerance to multiple TLR ligands. J Immunol (2009) 183(1):533–42. doi:10.4049/jimmunol.0802189
125. Nahid MA, Benso LM, Shin JD, Mehmet H, Hicks A, Ramadas RA. TLR4, TLR7/8 agonist-induced miR-146a promotes macrophage tolerance to MyD88-dependent TLR agonists. J Leukoc Biol (2016) 100(2):339–49. doi:10.1189/jlb.2A0515-197R
126. Tsukada K, Kitazawa T, Fukushima A, Okugawa S, Yanagimoto S, Tatsuno K, et al. Macrophage tolerance induced by stimulation with toll-like receptor 7/8 ligands. Immunol Lett (2007) 111(1):51–6. doi:10.1016/j.imlet.2007.05.001
127. Sato S, Takeuchi O, Fujita T, Tomizawa H, Takeda K, Akira S. A variety of microbial components induce tolerance to lipopolysaccharide by differentially affecting MyD88-dependent and-independent pathways. Int Immunol (2002) 14(7):783–91. doi:10.1093/intimm/dxf046
128. Broad A, Kirby JA, Jones DE. Toll-like receptor interactions: tolerance of MyD88-dependent cytokines but enhancement of MyD88-independent interferon-β production. Immunology (2007) 120(1):103–11. doi:10.1111/j.1365-2567.2006.02485.x
Keywords: toll-like receptor 7, single-stranded ribonucleic acid, endosomal trafficking, uncoordinated 93 homolog B1 (Caenorhabditis elegans), furin peptidases, asparagine endopeptidases
Citation: Petes C, Odoardi N and Gee K (2017) The Toll for Trafficking: Toll-Like Receptor 7 Delivery to the Endosome. Front. Immunol. 8:1075. doi: 10.3389/fimmu.2017.01075
Received: 05 June 2017; Accepted: 18 August 2017;
Published: 04 September 2017
Edited by:
Janos G. Filep, Université de Montréal, CanadaReviewed by:
Maura Rossetti, University of California, Los Angeles, United StatesSandra Stephanie Diebold, King’s College London, United Kingdom
Copyright: © 2017 Petes, Odoardi and Gee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katrina Gee, a2dlZUBxdWVlbnN1LmNh
 Carlene Petes
Carlene Petes Natalya Odoardi
Natalya Odoardi Katrina Gee
Katrina Gee