
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 31 July 2017
Sec. Molecular Innate Immunity
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.00855
Growing evidence has indicated that the innate immune system can be regulated by microRNAs (miRNAs). However, the mechanism underlying miRNA-mediated simultaneous activation of multiple immune pathways remains unknown. To address this issue, the role of host miR-12 in shrimp (Marsupenaeus japonicus) antiviral immune responses was characterized in the present study. The results indicated that miR-12 participated in virus infection, host phagocytosis, and apoptosis in defense against white spot syndrome virus invasion. miR-12 could simultaneously trigger phagocytosis, apoptosis, and antiviral immunity through the synchronous downregulation of the expression of shrimp genes [PTEN (phosphatase and tensin homolog) and BI-1(transmembrane BAX inhibitor motif containing 6)] and the viral gene (wsv024). Further analysis showed that miR-12 could synchronously mediate the 5′–3′ exonucleolytic degradation of its target mRNAs, and this degradation terminated in the vicinity of the 3′ untranslated region sequence complementary to the seed sequence of miR-12. Therefore, the present study showed novel aspects of the miRNA-mediated simultaneous regulation of multiple immune pathways.
During virus infection, multiple immune pathways are employed by hosts to defend against virus invasion (1). As the first line of the immune systems, innate immunity plays an essential role in immune responses against virus infection in which phagocytosis and apoptosis are two important components (1). Studies have shown that phagocytosis is required for host antiviral immunity through the direct rapid engulfment of virions and apoptotic cells (2–4). Phagocytosis can be divided into multiple stages, including recognition between particles and cell receptors, actin polymerization to engulf exogenous substrates and transfer of a newly formed phagosome into a mature phagolysosome through the by removal of the actin coat (5). In invertebrates, Rab6 protein, which directly interacts with actin, is essential for the correct conformation of actin, thus showing a critical effect on phagocytosis against virus infection (4). During virus infection, a series of signaling pathways can be triggered in the host, leading to apoptosis of infected cells (6, 7). The apoptotic cells attract specialized phagocytes and are subsequently devoured by phagocytes (8, 9). Avian influenza virus encodes NS1, a viral protein with a four-amino acid sequence at its carboxyl terminus termed the PDZ-binding motif. Recent reports have shown that this PDZ-binding motif can protect virus-infected cells from apoptosis by directly disrupting the proapoptotic function of scribble, thereby inhibiting infected cell elimination by immune cells (10). To protect the host from virus invasion, animals have developed various signaling pathways. However, whether an individual molecule can synchronously trigger various antiviral pathways remains unknown. As key regulatory elements of gene expression, an individual microRNA (miRNA) possess multiple target genes involved in different signaling pathways (11, 12), implying that a miRNA may simultaneously trigger multiple antiviral pathways.
MicroRNAs, typically ~22 nucleotides in length, are members of an extensive type of small non-coding RNAs, which exert physiological functions through direct binding to the 3′ untranslated regions (3′UTR) of mRNAs, resulting in mRNA degradation or destabilization (13). In splenic macrophages under hypersplenism, miR-615-3p is highly expressed and targets the ligand-dependent nuclear receptor corepressor (LCoR), an inhibitor of phagocytosis-promoting protein peroxisome proliferator-activated receptor gamma (PPARγ), thus upregulating PPARγ and eventually enhancing the phagocytic capacity of splenic macrophages (14). For the regulation of apoptosis, miR-101 is downregulated in human hepatocellular carcinoma and can markedly inhibit the expression of the antiapoptotic factor Mcl-1 (BCL2 family apoptosis regulator), leading to apoptosis of hepatocellular carcinoma cells (15). Currently, the miRNA-mediated regulation of apoptosis and phagocytosis represent important mechanisms in the host to defend against virus invasion. In shrimp, the host miR-7 directly targets the viral early gene wsv477 of white spot syndrome virus (WSSV) leading to the repression of WSSV infection (16). Previous studies have reported that WSSV infection leads to the accumulation of miR-965, which diminishes the ATG5 expression, thus enhancing host antiviral phagocytosis (12). Currently, miRNA-mediated gene expression regulation is generally a one-to-one type of regulation (i.e., one miRNA to one mRNA). However, the miRNA-mediated regulation mechanism of multiple target gene expressions has not been extensively investigated.
To elucidate whether a single miRNA can simultaneously trigger multiple antiviral pathways by targeting different genes, shrimp miR-12, which has been implicated in antiviral phagocytosis and apoptosis in shrimp (17), was characterized in the present study. In recent years, WSSV has attracted increasing attention as a model for virus–host interactions in vivo (18). In the present study, the results indicated that miR-12 could simultaneously trigger antiviral phagocytosis and apoptosis and inhibit the virus proliferation by synchronously targeting the host genes PTEN and BI-1 and the viral gene wsv024.
Shrimp Marsupenaeus japonicus (10–12 cm in length) were cultured at 20°C in tanks with seawater. For each treatment, 20 randomly selected individuals were raised in an 80 l aquarium. To ensure that the shrimp were virus-free prior to experiments, the shrimp hemocytes were subjected to PCR detection using WSSV-specific primers (5′-TATTGTCTCTCCTGACGTAC-3′ and 5′-CACATTCTTCACGAGTCTAC-3′). The virus-free shrimp were intramuscularly injected with 0.1 ml of WSSV solution (105 copies/ml) using a syringe with a 29-gauge needle. The WSSV solution was obtained from WSSV-infected shrimp (18). At various times postinfection, the shrimp hemocytes were collected for further analysis.
Total miRNAs were extracted from shrimp hemocytes using a mirVana miRNA isolation kit (Ambion, USA) following the manufacturer’s instructions. The RNAs were separated on a denaturing 15% polyacrylamide gel containing 7 M urea in 1× TBE buffer (90 mM Tris–boric, 2 mM EDTA, pH 8.0) and subsequently transferred to a Hybond-N+ nylon membrane. After UV cross-linking, the membrane was prehybridized in DIG (digoxigenin) Easy Hyb granule buffer (Roche, Basel, Switzerland) for 0.5 h at 42°C and then hybridized with DIG-labeled miR-12 (5′-ACCAGTACCTGATGTAATACTCA-3′) or U6 (5′-GGGCCATGCTAATCTTC TCTGTATCGTT-3′) probe at 42°C overnight. The DIG labeling and detection were performed using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Germany) according to the manufacturer’s instructions.
To overexpress or knock down the expression of miR-12 in shrimp, miR-12 (5′-T GAGTATTACATCAGGTACTGGT-3′) synthesized using the in vitro transcription T7 kit (TaKaRa, Japan) or the synthesized anti-miR-12 oligonucleotide (AMO-miR-12) (5′-ACCAGTACCTGATGTAATACTTCA-3′) was injected into shrimp at 30 μg/shrimp. As controls, the sequence of miR-12 or AMO-miR-12 was randomly scrambled, generating miR-12-scrambled (5′-ACTCATAATGTAGTCCAT GACCA-3′) or AMO-miR-12-scrambled (5′-TGGTCATGGACTACATTATGAGT-3′). All synthesized miRNAs were dissolved in miRNA solution (50 mM Tris–HCl, 100 mM NaCl, pH 7.5) and quantified using NanoDrop ND-100 spectrophotometer. At different times after injection, three shrimp were randomly selected from each treatment, and the selected shrimp hemocytes were collected and mixed for later use. The above assays were biologically repeated three times.
To evaluate the proliferation of WSSV in shrimp, the quantitative real-time PCR was performed using WSSV-specific primers (5′-TTGGTTTCATGCCCGAGATT-3′ and 5′-CCTTGGTCAGCCCCTTGA-3′) and a TaqMan fluorogenic probe (5′-FAM-TGCTGCCGTCTCCAATAMRA-3′). The PCR mixture (10 µl) consisted of 5 µl Premix Ex Taq (TaKaRa, Japan), 200 ng DNA template, 0.2 µl of 10 µM primers, and 0.2 µl of 10 µM TaqMan fluorogenic probe at a final concentration of 0.2 µM. The DNA template was extracted from the shrimp hemocytes using an SQ tissue DNA isolation kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s instructions. A plasmid containing a 1,400-bp DNA fragment from the WSSV genome was used as the reference plasmid as previously described (19). The PCR conditions were 95°C for 1 min, followed by 45 cycles at 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s.
To label WSSV virions with FITC, the purified virions were incubated in 1 mg/ml FITC (Sigma, USA) solution (dissolved in 0.1 M NaHCO3, pH 9.0) for 1 h at room temperature, followed by washing with 0.1 M NaHCO3. The shrimp hemocytes were rinsed with cold PBS (50 mM Tris–HCl, 100 mM NaCl, pH 7.5). To perform the phagocytosis assay, the FITC-labeled WSSV virions were mixed with shrimp hemocytes at a ratio of 50:1 (WSSV copies: cell numbers) and subsequently incubated at 28°C for 30 min. The hemocytes were rinsed with PBS to remove unphagocytosed FITC-labeled WSSV virions. After centrifugation at 200 × g for 10 min, the hemocytes were resuspended in 1% paraformaldehyde (Sigma, USA). Finally, the hemocytes were analyzed using flow cytometry (Beckman Coulter, USA). For each sample, 10,000–20,000 hemocytes were assessed. The experiments were biologically repeated three times.
The detection of shrimp hemocyte apoptosis using annexin V (Invitrogen, USA) was conducted following the manufacturer’s instructions. Shrimp hemocytes were washed with cold PBS. Then the hemocytes were incubated in 1× annexin binding buffer at 1 × 106 cells/ml, followed by the addition of 5 µl of Alexa Fluor 488-annexin V and 1 µl of 100 µg/ml propidium iodide (PI) in the dark. After incubation for 15 min, 400 µl of 1× annexin binding buffer was added to each sample to stop the reaction. The sample was analyzed using flow cytometry.
Shrimp hemocytes were collected and separated on poly-l-lysine-coated glass slides (Sigma), followed by standing for 10 min at 4°C. The hemocytes were fixed in 4% paraformaldehyde for 25 min at 4°C. The fixed hemocytes were washed with cold PBS and then subjected to the permeabilization with 0.2% Triton X-100 for 5 min. Next, the hemocytes were equilibrated in 100 µl of equilibration buffer at 4°C for 10 min. The equilibrated hemocytes were counterstained with PI after incubation with rTdT mix in a humid environment for 1 h. Subsequently, 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) was added to the slide to stop the reaction. The slide was covered with antifade solution (Invitrogen) to prevent signal quenching.
The apoptosis of shrimp hemocytes was detected using a Caspase-Glo3/7 assay (Promega, USA) according to the manufacturer’s instructions. Briefly, shrimp hemocytes were harvested and washed with PBS. Subsequently the hemocytes were incubated with Caspase-Glo 3/7 reagent at room temperature for 2 h. The caspase 3/7 activity was measured using a Synergy 2 Multi-Mode microplate reader (BioTek).
To predict the target genes of miR-12, four algorithms including TargetScan, miRanda, Pictar, and miRInspector were used to predict the targeted sites in the 3′UTRs of the WSSV genes and the shrimp genes as previously described (20).
To explore the direct interaction between the predicted targets and miR-12, the 3′UTR of a predicted target gene was cloned into the pIZ/V5-His vector (Invitrogen, USA), generating a recombinant plasmid containing EGFP and the 3′UTR of a predicted target. As a control, the 3′UTR sequence matching the seed sequence of miR-12 was scrambled to produce the mutant construct. All the recombinant plasmids were confirmed through sequencing.
Insect High Five cells (Invitrogen) were cultured in Express Five serum-free medium (Invitrogen) containing l-glutamine (Invitrogen) at 27°C. At 70% confluence, the cells were co-transfected with 2 µg of EGFP, EGFP-target gene 3′UTR or EGFP-Δtarget gene 3′UTR and 100 pM of miR-12 or miR-12-scrambled using Cellfectin transfection reagent (Invitrogen) according to the manufacturer’s instructions. The miRNAs were synthesized at Shanghai GenePharma Co., Ltd. (Shanghai, China). At 48 h after co-transfection, the fluorescence intensity of cells was assessed using a Flex Station II microplate reader (Molecular Devices, USA) at 490/510 nm excitation/emission (Ex/Em). The experiments were biologically repeated three times.
The mRNA levels of wsv024, PTEN, PARP, and BI-1 were quantified with real-time PCR using sequence-specific primers (wsv024, 5′-CATCCTGTTAGAGT TTCCTGTTTC-3′ and 5′-ATATTACTGCCATGTTATCTGTTGC-3′; PTEN, 5′-CCAAAATAACCACAACAACAC-3′ and 5′-CACTTCCTGCTCTCCCTT-3′; PARP1, 5′-GAAGAATTACAACTGCGTCCTG-3′ and 5′-GTTCCTCGAAATGGG CTATG-3′; BI-1, 5′-TGCGGCTTCATTGTTTACG-3′ and 5′-CACGGTTCTTCCTCTTGTTCTG-3′; and β-actin, 5′-CGAGCACGGCATCGTTACTA-3′ and 5′-TTGTAG AAAGTGTGATGCCAGATCT-3′). Shrimp hemocytes were collected and subjected to total RNA extraction using the RNAprep Pure Cell/Bacteria kit (Tiangen Biotech Co. Ltd., Beijing, China). The cDNA was synthesized using the PrimeScript™ 1st strand cDNA synthesis kit (Takara, Japan). Quantitative real-time PCR was performed in a total volume of 25 µl containing 5 µl of SYBR® Premix Ex Taq, 0.5 µl of 10 µM forward and reverse primers, and 100 ng of cDNA template. The PCR conditions were 95°C for 1 min, followed by 40 cycles at 95°C for 15 s and 60°C for 45 s.
According to the gene sequence, siRNAs specifically targeting these genes were synthesized, generating the corresponding siRNAs (wsv024-siRNA, 5′-CGAUGAG UACUUGUCUAGCGUUUAA-3′; PTEN-siRNA, 5′-UAGAGUAGCAGAUGUU UGAAGUGUA-3′; and I-1-siRNA, 5′-GCAAACUGGAGAAAGUGCUUUCUGA-3′). As controls, the siRNA sequences were randomly scrambled, producing the corresponding scrambled siRNAs (wsv024-siRNA-scrambled, 5′-CGAAGUACUUG UCUAGCGUUUGUAA-3′; PTEN-siRNA-scrambled, 5′-UAGGAUGACGUAUUU AAGUGAGGUA-3′; BI-siRNA-scrambled, 5′-GCAGGUCAAGAGUGAUUUCCA AUGA-3′). All siRNAs were synthesized using an in vitro transcription T7 kit (TaKaRa, Japan) according to the manufacturer’s instructions. The synthesized siRNAs were quantified using spectrophotometry and dissolved in siRNA buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl).
The shrimp were injected with siRNA (4 nM) or siRNA-scrambled (4 nM) and WSSV (104 copies/shrimp). At different times after injection, the hemocytes of three randomly selected shrimp from each treatment were collected for later use.
The proteins were analyzed in a 12% SDS-PAGE gel and then transferred onto a nitrocellulose membrane (Bio-Rad, USA). The membrane was blocked in blocking buffer [3% bovine serum albumin in TBST (250 mM NaCl, 10 mM Tris–HCl, 0.5 ml/l Tween 20, pH 7.4)] for 2 h at 4°C. Subsequently the membrane was incubated with a primary antibody (anti-wsv024 IgG, anti-BI-1 IgG, anti-PTEN IgG, or anti-actin IgG) for 2 h at 4°C. After two washes with TBST, the membrane was incubated with HRP (horseradish peroxidase)-conjugated anti-mouse IgG (Sigma, USA) for 1 h at 4°C, followed by chemiluminescence detection with ECL substrate (Thermo Scientific, USA). All of the primary antibodies were prepared in our laboratory.
The 3′UTRs of wsv024, PTEN, and BI-1 mRNAs were amplified with specific primers (wsv024, 5′-TAATACGACTCACTATAGGTCTTCGTTAAAATCAGTTTTACCT-3′ and 5′-GTGCTAATAAACCAACGTCTTTC-3′; PTEN, 5′-TAATACGACTCACTATAGGGCGCACGAAGCTTTGGC-3′ and 5′-TCCTGTACACTACAAATAATAAGCCTC-3′; BI-1-F, 5′-TAATACGACTCACTATAGGGATGCCACA GATGTAATTTAGAAG-3′ and 5′-AATAGGATAATCACATGAAATTCTG-3′), which contained T7 promoter sequence at their 5′-ends of the forward primers, using a T7 transcription kit (TaKaRa, Japan) according to the manufacturer’s instructions. To get the Ago1 complex of shrimp, shrimp muscles were homogenized in lysis buffer [20 mM Tris–HCl, 150 mM NaCl, 1.5 mM MgCl2, 0.25% NP-40 (Non-idet P-40), and 1 mM PMSF (phenylmethanesulfonyl fluoride), pH 7.5] for 15 min on ice and subsequently centrifuged at 10,000 × g for 10 min at 4°C. The cell supernatants were incubated with the shrimp Ago1-specific antibody, prepared in our laboratory, for 1 h at 4°C, followed by mixing with protein-G-coupled agarose beads for 4 h at 4°C. After washes with cold PBS, the beads were resuspended in reaction buffer [100 mM KOAc, 40 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 5 mM MgCl2, 2 mM Dithiothreitol, 0.35% Triton X-100, and 1 mM PMSF, pH 7.6]. To investigate the miRNA-mediated degradation of target mRNAs, the co-immunoprecipitated products of shrimp Ago1, 3′UTR (wsv024, PTEN, and/or BI-1) (100 ng), and miR-12 were mixed at different concentrations and supplemented with 2 µl of 10 mM ATP and 2 µl of 2 mM GTP at a total volume of 20 µl. After incubation at 30°C for different times, the RNA samples were separated using 1% agarose gel electrophoresis and then transferred to a nylon membrane (GE Healthcare, USA). The RNAs were detected using a 5′ DIG (digoxigenin)-labeled probe (wsv024, 5′-TCACGGTTATGGCCAGCGCTACGC-3′; PTEN, 5′-GAACACAGAGCAGAG GGGATGATAG-3′; and BI-1, 5′-TCTCAAACTCATCTATATCATTC-3′).
The mRNA 3′UTR of the miR-12 target gene was incubated with miR-12 (20 nM) and Ago1 complex at 30°C for 2 h. Subsequently, the degraded fragments were separated using 1% agarose electrophoresis. After recovery from the agarose gel, the RNAs were reverse transcribed into cDNAs using the PrimeScript™ II 1st strand cDNA synthesis kit with random 6 primer (TaKaRa, Japan). Then, the single-stranded cDNAs were transformed into double-stranded cDNAs using the second strand cDNA synthesis kit (Beyotime Biotechnology, China). The cDNAs were cloned into the pEASY®-Blunt simple cloning vector (Transgen Biotech, China) and subjected to sequencing.
All numerical data presented in the present study were analyzed using one-way analysis of variance to calculate the means and SDs of triplicate assays.
To explore the role of host miR-12 in virus–host interactions, the expression level of miR-12 in WSSV-challenged shrimp was characterized. Northern blot analysis indicated that miR-12 was significantly upregulated in shrimp in response to WSSV infection, suggesting that miR-12 played important roles in host–virus interactions (Figure 1A).
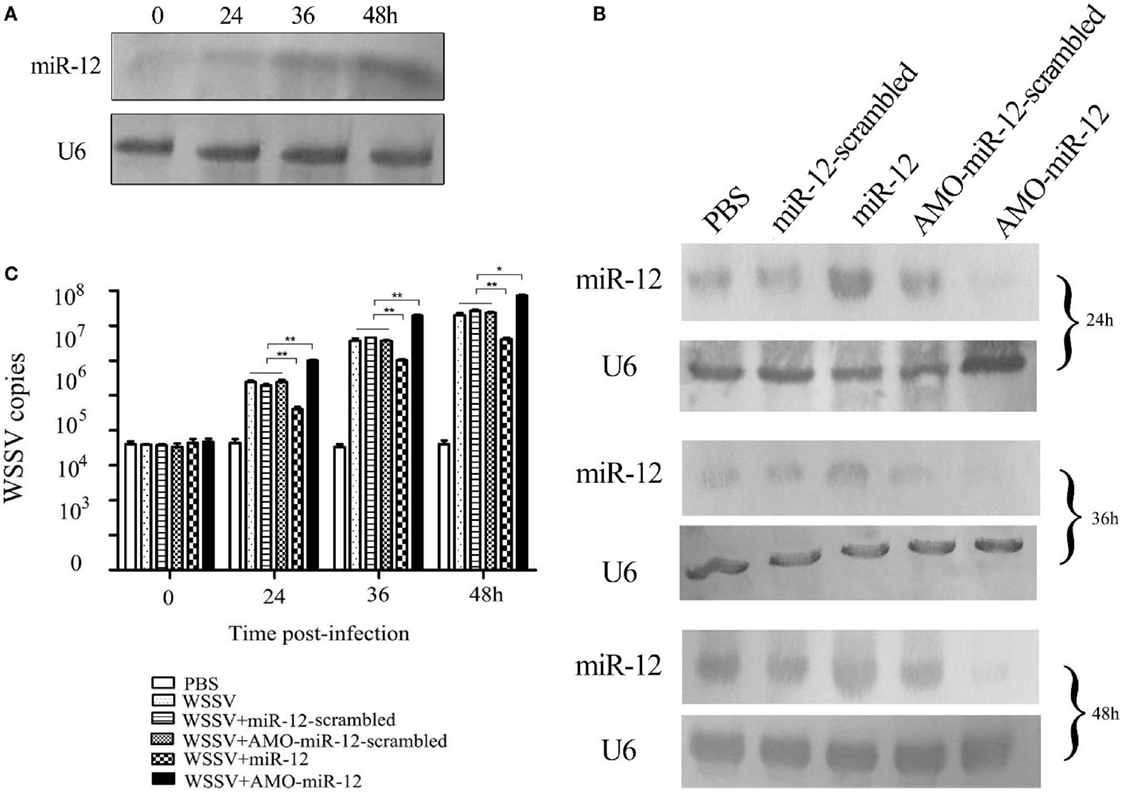
Figure 1. The antiviral activity of miR-12 in shrimp. (A) The expression profile of miR-12 in shrimp in response to virus infection. The shrimp were infected with white spot syndrome virus (WSSV). At different times postinfection, shrimp hemocytes were subjected to northern blot analysis to detect miR-12. U6 was used as a control. The numbers represent the time points postinfection. (B) The overexpression or silencing of miR-12 in shrimp. The virus-free shrimp were injected with miR-12 or AMO-miR-12, followed by detection of miR-12 expression using northern blot analysis. As controls, miR-12-scrambled and AMO-miR-12-scrambled were also injected. The probes are indicated on the left. U6 was used as a control. The numbers indicate the times after treatments. (C) The influence of miR-12 overexpression or silencing on virus infection. The WSSV copies of miR-12-overexpressed or miR-12-silenced shrimp were evaluated at various times postinfection using quantitative real-time PCR. The treatments are indicated on the right. In all panels, the statistical significance between treatments is indicated with asterisks (*p < 0.05; **p < 0.01).
To assess the roles of miR-12 in virus–host interactions, miR-12 expression was overexpressed or knocked down in shrimp, followed by the evaluation of virus infection. Northern blot analysis indicated that miR-12 expression in shrimp hemocytes was significantly upregulated from 24 to 48 h after the miR-12 injection compared with the controls (Figure 1B). The results showed that miR-12 overexpression led to significant decreases in WSSV copies in shrimp compared with the controls (WSSV, WSSV + miR-12-scrambled and WSSV + AMO-miR-12-scrambled) (Figure 1C). However, when miR-12 expression was inhibited by AMO-miR-12 (Figure 1B), the WSSV copies were dramatically increased compared with the controls (Figure 1C), indicating that miR-12 played a negative role in the virus infection. The results revealed that the miR-12 expression level in PBS-treated shrimp was identical to those of miR-12-scrambled and AMO-miR-12-scrambled-treated shrimp (Figure 1B), showing that there was no miR-12 or AMO-miR-12 toxicity in shrimp.
The above data implied that miR-12 possesses antiviral activity in shrimp.
To reveal the mechanism of miR-12-mediated antiviral immunity in shrimp, the WSSV genes targeted by miR-12 were characterized. The prediction results obtained using TargetScan, miRanda, Pictar and miRInspector algorithms showed that wsv024 was a potential target of miR-12 (Figure S1A in Supplementary Material).
To assess the interaction between miR-12 and wsv024, a recombinant plasmid containing EGFP and wsv024 3′UTR (pIZ/EGFP-wsv024-3′UTR) was co-transfected with miR-12 into insect High Five cells (Figure S1B in Supplementary Material). The results indicated that the fluorescence intensity of the insect cells co-transfected with miR-12 and pIZ/EGFP-wsv024-3′UTR was remarkably decreased compared with the controls (Figure 2A), showing that miR-12 could directly target the wsv024 gene.
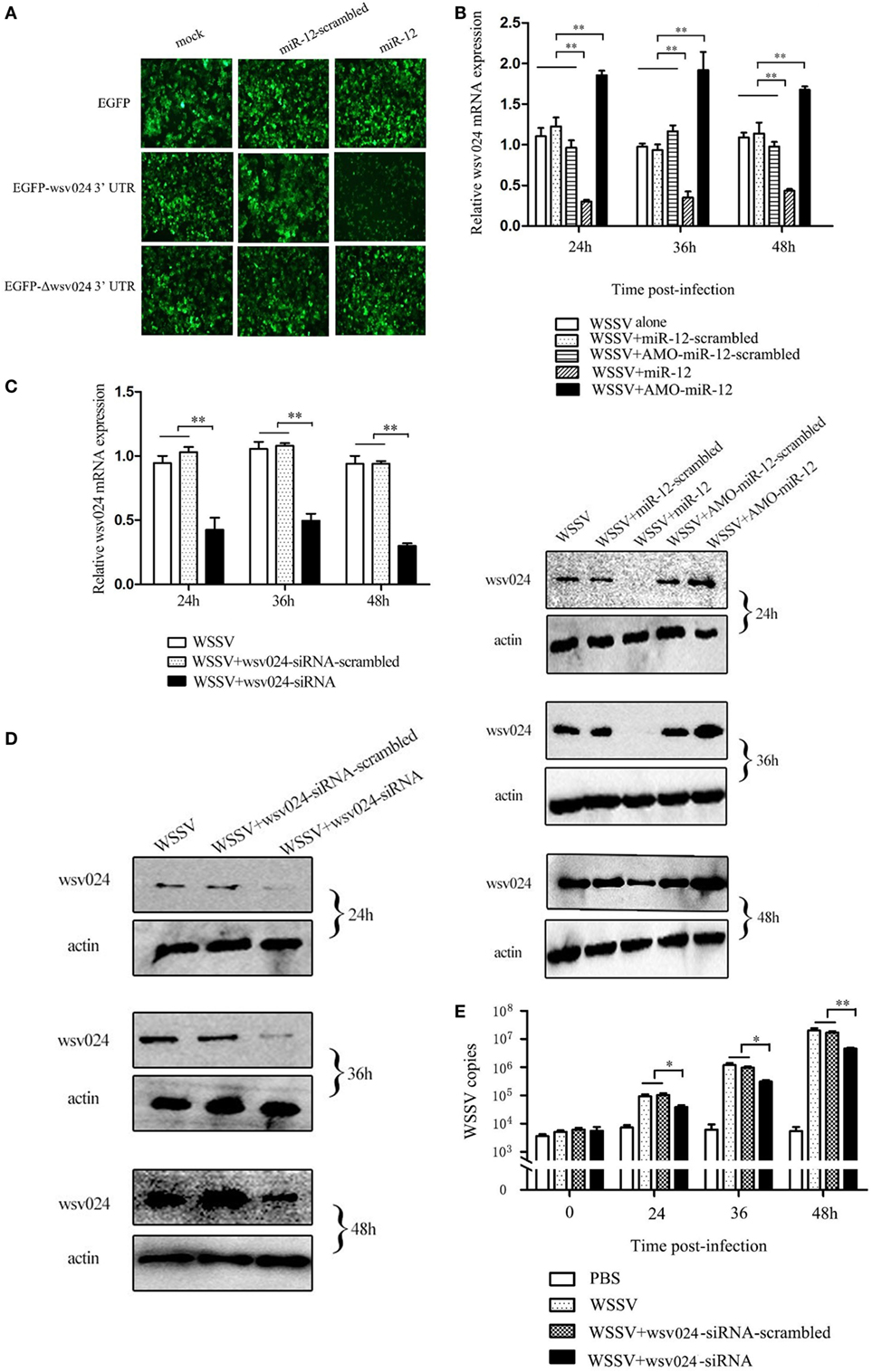
Figure 2. The miR-12-mediated suppression of white spot syndrome virus (WSSV) infection in shrimp by targeting viral wsv024 gene. (A) The direct interaction between miR-12 and wsv024. Insect High Five cells were co-transfected with miR-12 or miR-12-mimic-scrambled with the recombinant plasmids, including EGFP-wsv024-3′UTR, EGFP-Δwsv024-3′UTR, and EGFP. At 48 h after co-transfection, fluorescent images were obtained. (B) The effect of the silencing or overexpression of miR-12 on wsv024 expression in WSSV-infected shrimp. The shrimp were injected with miR-12 and WSSV, AMO-miR-12 and WSSV or WSSV alone. As controls, miR-12-scrambled and AMO-miR-12-scrambled were also injected. At different times after injection, shrimp hemocytes were subjected to quantitative real-time PCR (up) and western blotting (down) to evaluate the wsv024 expression level. The numbers indicated the times postinfection. Shrimp β-actin was used as a control. (C) The knockdown of wsv024 gene expression in shrimp. Shrimp were co-injected with WSSV and wsv024-siRNA. As controls, co-injections of WSSV + wsv024-siRNA-scrambled or WSSV alone were included in the assays. At different times postinfection, the wsv024 mRNA level was determined using quantitative real-time PCR. (D) Western blot analysis of wsv024 silencing in shrimp. The numbers indicate the time points postinfection. (E) The effects of wsv024 silencing on virus infection. At different times postinfection, the WSSV copies in the wsv024-siRNA-treated shrimp were examined using quantitative real-time PCR. The statistically significant differences between treatments are represented with asterisks (*p < 0.05; **p < 0.01).
To evaluate the interaction between miR-12 and wsv024 in vivo, miR-12 expression was upregulated or knocked down in WSSV-infected shrimp, followed by the detection of wsv024 expression levels. The results indicated the miR-12 overexpression decreased wsv024 transcript levels in contrast with the controls (WSSV + miR-12-scrambled and WSSV alone) (Figure 2B). When the expression of endogenous miR-12 was reduced by AMO-miR-12, the wsv024 gene expression was significantly upregulated compared with the controls (WSSV + AMO-miR-12-scrambled and WSSV alone) (Figure 2B). These data showed that miR-12 interacts with wsv024 in vivo.
To evaluate the influence of the interaction between miR-12 and wsv024 on virus infection, the expression of wsv024 was knocked down using wsv024-specific siRNA (wsv024-siRNA) in shrimp, followed by an assessment of WSSV proliferation. The results indicated that the expression of wsv024 was efficiently silenced by wsv024-siRNA compared with the controls (Figures 2C,D). In addition, wsv024 silencing resulted in remarkable inhibition of WSSV proliferation in shrimp (Figure 2E), indicating that wsv024 plays a positive role in WSSV infection.
Taken together, these findings showed that miR-12 functioned in the antiviral immunity of shrimp by targeting the WSSV wsv024 gene.
In a previous study, we revealed that shrimp miR-12 was implicated in antiviral phagocytosis and antiviral apoptosis in shrimp (17). To investigate the mechanism of antiviral phagocytosis and antiviral apoptosis mediated through miR-12, the shrimp genes targeted by miR-12 were predicted. As predicted, PARP1 [poly (ADP-ribose) polymerase 1], PTEN (phosphatase and tensin homolog), and BI-1 (transmembrane BAX inhibitor motif containing 6) genes were potential targets of miR-12 (Figure S2A in Supplementary Material). BI-1 and PARP1 are inhibitors of apoptosis (21, 22), while PTEN is a negative regulator of phagocytosis (23). In this context, the role of miR-12 in the antiviral phagocytosis of shrimp was characterized.
As shown in Figure 3A, when miR-12 was overexpressed, the phagocytic activity of shrimp hemocytes was significantly enhanced. However, miR-12 silencing remarkably decreased shrimp hemocyte phagocytic activity (Figure 3A). These results revealed that miR-12 exerted a positive effect on the antiviral phagocytosis of shrimp to fight against virus infection.
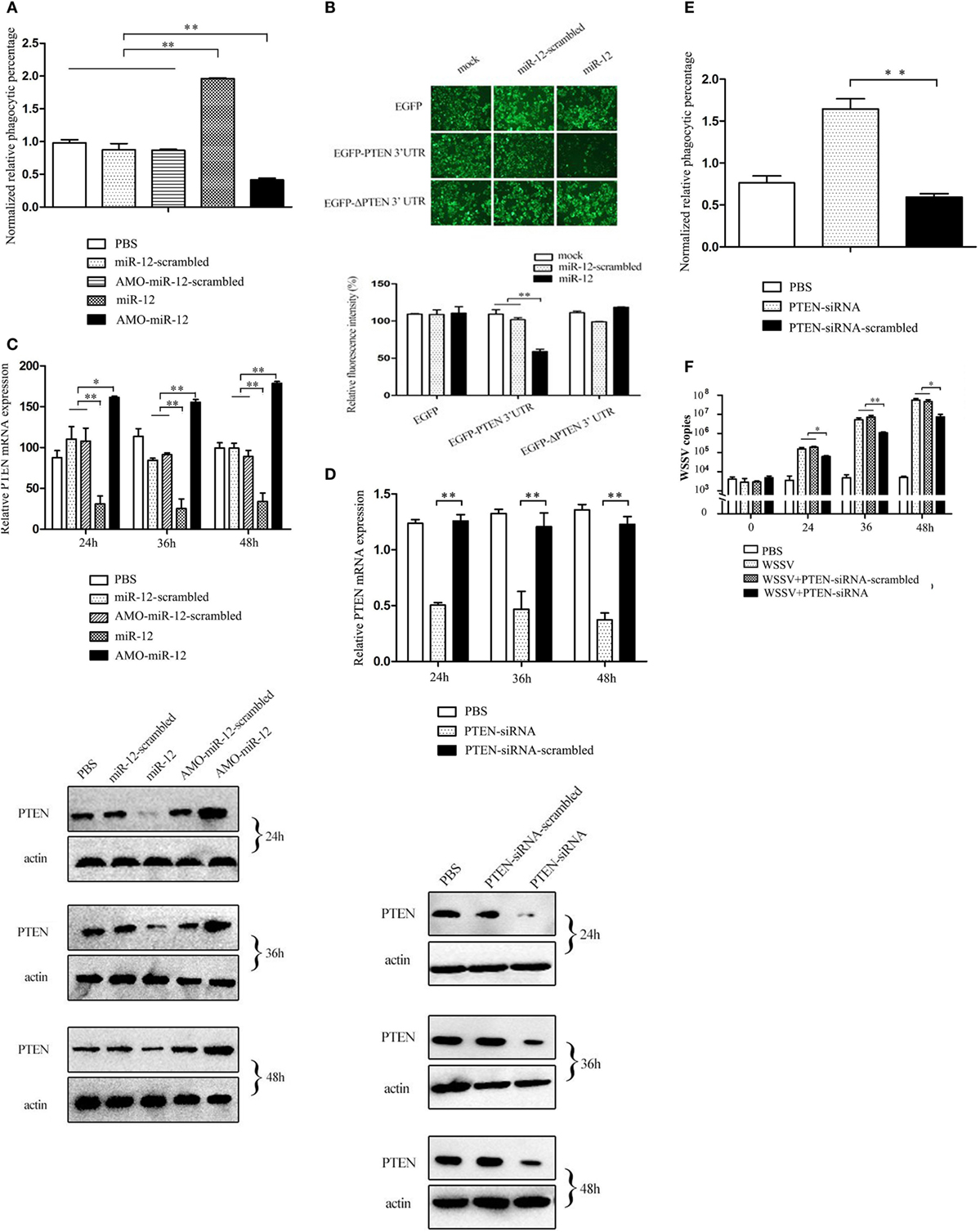
Figure 3. The promotion of shrimp antiviral phagocytosis mediated by miR-12 via targeting the shrimp PTEN gene. (A) The role of miR-12 in phagocytosis. At 36 h after injection, the phagocytic activity of shrimp hemocytes was assessed. The treatments are indicated on the right. (B) The direct interaction between miR-12 and PTEN. The insect cells were co-transfected with miR-12 and various constructs [EGFP, EGFP-PTEN 3′ untranslated region (3′UTR), or EGFP-ΔPTEN 3′UTR]. As a control, miR-12-scrambled was included in the transfection. At 36 h after transfection, the fluorescence intensities of the cells were examined. (C) The interaction between miR-12 and PTEN in vivo. Shrimp were injected with miR-12 or AMO-miR-12 to overexpress or silence miR-12. At different times after injection, the transcript level and protein level of PTEN in shrimp hemocytes were determined using quantitative real-time PCR (up) and Western blotting (down), respectively. In Western blots, the numbers represent the times postinfection. Shrimp β-actin was used as a control. (D) Knockdown of PTEN in shrimp. Shrimp were injected with PTEN-siRNA, followed by an assessment of PTEN expression using quantitative real-time PCR (up) and western blotting (down). PTEN-siRNA-scrambled was used as a control. (E) The role of PTEN in the antiviral phagocytosis of shrimp. Shrimp were injected with PTEN-siRNA or PTEN-siRNA-scrambled. At 36 h post-transfection, the phagocytic activity of shrimp against white spot syndrome virus (WSSV) was examined. (F) The effects of PTEN silencing on WSSV infection. The WSSV copy number in the PTEN-silenced shrimp was evaluated using quantitative real-time PCR at various times postinfection. In all panels, the data are presented as the means ± SD of three independent experiments. The significant differences between treatments are indicated with asterisks (*p < 0.05; **p < 0.01).
To reveal the mechanism of miR-12-mediated antiviral phagocytosis in shrimp, the interaction between miR-12 and its potential target PTEN gene was explored. Recombinant plasmids containing EGFP and PTEN 3′UTR or ΔPTEN 3′UTR were co-transfected with miR-12 or miR-12-scrambled into insect cells (Figure S2B in Supplementary Material). The results indicated that the fluorescence intensity of cells co-transfected with miR-12 and pIZ/EGFP-PTEN-3′UTR remarkably decreased compared with the controls (Figure 3B), showing that miR-12 directly interacted with PTEN. To evaluate the interaction between miR-12 and PTEN in vivo, miR-12 was overexpressed or knocked down, followed by the assessment of PTEN mRNA and protein levels. The data revealed that miR-12 overexpression led to a significant reduction in PTEN transcript and protein levels, while miR-12 silencing significantly increased the PTEN mRNA and protein levels (Figure 3C). These results showed that miR-12 could target PTEN.
To characterize the role of PTEN in phagocytosis, the expression of PTEN was knocked down, followed by an assessment of the antiviral phagocytic activity of shrimp hemocytes. The results showed that the injection of PTEN-siRNA resulted in a significant reduction of the PTEN transcript level (Figure 3D), indicating that PTEN expression was silenced by PTEN-siRNA. Western blots yielded similar results (Figure 3D). When PTEN was silenced, the shrimp phagocytic activity against WSSV was dramatically strengthened compared with controls (Figure 3E), indicating that PTEN was required in shrimp phagocytosis. Further results showed that PTEN silencing led to the remarkable suppression of WSSV replication in contrast with the controls (Figure 3F). These data revealed that PTEN exerted negative effects on the antiviral phagocytosis of shrimp.
The above findings showed that miR-12 could enhance the antiviral phagocytosis of shrimp by directly targeting PTEN, a phagocytosis negative regulation gene.
To evaluate the effects of miR-12 on apoptosis, miR-12 was overexpressed or knocked down in shrimp, and then the apoptotic activity of shrimp hemocytes was assessed. The results of annexin V analysis showed more annexin V-positive hemocytes in WSSV-infected shrimp than in PBS-treated shrimp (Figure 4A). The miR-12 overexpression augmented the percentage of apoptotic hemocytes in contrast with that of shrimp injected with miR-12-scrambled and WSSV or WSSV alone (Figure 4A), suggesting that miR-12 played a positive role in host cell apoptosis. Contrarily, miR-12 silencing diminished the abundance of the annexin V-positive hemocytes (Figure 4A). TUNEL assays and the detection of caspase 3/7 of shrimp hemocytes yielded results similar to those obtained in the annexin V analysis (Figures 4B,C). These results showed that miR-12 was involved in the positive regulation of apoptosis in shrimp.
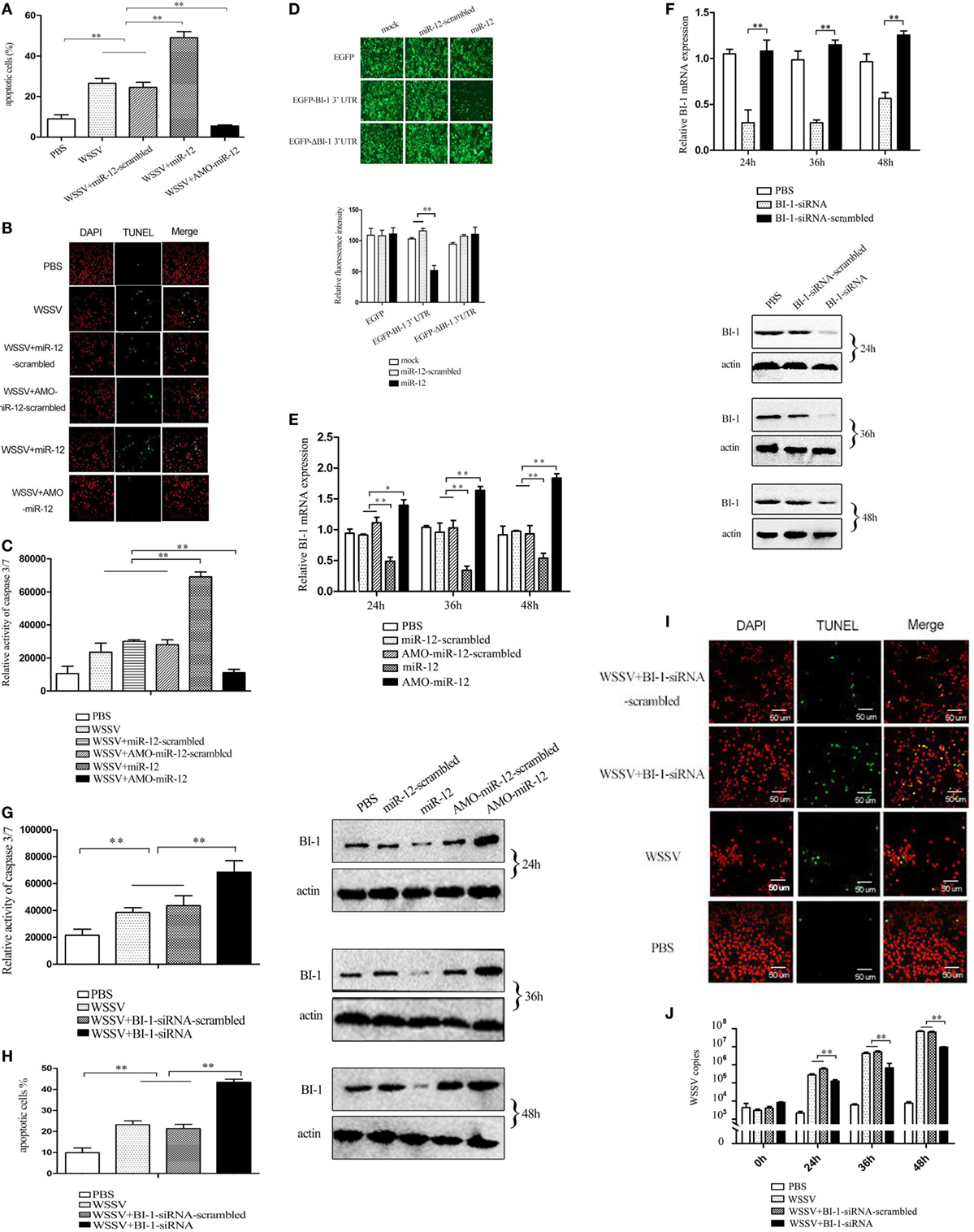
Figure 4. The enhancement of the antiviral apoptotic activity of shrimp mediated by miR-12 through targeting the shrimp BI-1 gene. (A) The effects of miR-12 on apoptosis. Shrimp were injected with miR-12 + white spot syndrome virus (WSSV) or AMO-miR-12 + WSSV. At 36 h after injection, the shrimp hemocytes were subjected to annexin V analysis. (B) The evaluation of the apoptotic activity of shrimp hemocytes using TUNEL assay. (C) The detection of caspase 3/7 activity in shrimp hemocytes. (D) The direct interaction between miR-12 and BI-1 in vitro. Insect High Five cells were simultaneously transfected with miR-12 and different recombinant plasmids. At 48 h after transfection, the fluorescence intensities of the cells were detected. (E) The interaction between miR-12 and BI-1 in vivo. Shrimp were injected with synthesized miR-12 or AMO-miR-12 to overexpress or knockdown the expression of miR-12. At different times after injection, the BI-1 expression level was assessed using quantitative real-time PCR (up) and western blotting (down). In the western blots, the numbers indicate the times postinfection. Shrimp β-actin was used as a control. (F) The silencing of BI-1 expression in shrimp. Shrimp were injected with BI-1-siRNA. BI-1-siRNA-scrambled was used as a control. At different times after injection, the expression level of BI-1 was examined using quantitative real-time PCR (up) and western blotting (down). (G) The influence of BI-1 silencing on the caspase 3/7 activity in shrimp. Shrimp were co-injected with BI-1-siRNA and WSSV. At 36 h after injection, the caspase 3/7 activity in shrimp hemocytes was evaluated. (H) The detection of apoptosis of BI-1-silenced shrimp hemocytes with annexin V. (I) The evaluation of the apoptosis of BI-1-silenced shrimp hemocytes using TUNEL. Scale bar, 50 µm. (J) The impact of BI-1 silencing on virus infection. The expression of BI-1 in WSSV-infected shrimp was knocked down using BI-1-siRNA. At different times postinfection, the WSSV copy number in shrimp hemocytes was evaluated using quantitative real-time PCR. In all panels, the data are representative of three independent experiments (*p < 0.05; **p < 0.01).
As predicted, the PARP1 and BI-1 genes encoding inhibitors of apoptosis were potential targets of miR-12 (Figure S2A in Supplementary Material). To explore the underlying mechanism of miR-12-mediated antiviral apoptosis, the interactions between miR-12 and PARP1 and BI-1 were characterized.
Recombinant pIZ/EGFP-BI-1-3′UTR and pIZ/EGFP-ΔBI-1-3′UTR were constructed (Figure S3 in Supplementary Material), and the recombinant plasmids were co-transfected with miR-12 or miR-12-scrambled into insect cells. The results indicated that the fluorescence intensity of the cells co-transfected with pIZ/EGFP-BI-1-3′UTR and miR-12 was significantly decreased compared with the controls (Figure 4D), showing that miR-12 directly interacted with BI-1. To determine whether miR-12 interacted with BI-1 in vivo, miR-12 was overexpressed or silenced in shrimp, followed by an assessment of the BI-1 expression level. The results showed that miR-12 overexpression significantly reduced BI-1 expression levels compared with the controls, while knocking down miR-12 expression by injection with AMO-miR-12 significantly upregulated BI-1 expression in shrimp (Figure 4E). These data indicated that miR-12 interacted with BI-1 in vivo.
To investigate the interaction between PARP1 and miR-12, the synthesized miR-12 and the pIZ/EGFP-PARP1-3′UTR construct were co-transfected into insect cells. The results showed no significant difference in the fluorescence intensity between various treatments (Figure S4 in Supplementary Material). The in vivo assays revealed that the PARP1 expression level did not change in shrimp when miR-12 was overexpressed or silenced (Figure S4 in Supplementary Material). These results indicated that miR-12 did not interact with PARP1. Therefore, miR-12 could promote apoptosis by targeting the BI-1 gene.
To further reveal the role of BI-1 in apoptosis, BI-1 expression was knocked down by BI-1-siRNA, followed by evaluation of apoptosis. The results showed that the BI-1 expression in shrimp was silenced using BI-1-siRNA (Figure 4F), indicating the specificity of BI-1-siRNA. BI-1 silencing significantly increased caspase3/7 activity in shrimp hemocytes compared with the controls (Figure 4G), showing that BI-1 exerted an inhibitory effect on apoptosis. The data from the annexin V and TUNEL assays yielded results similar to that of caspase 3/7 activity detection (Figures 4H,I). These findings revealed that BI-1 was an inhibitor of apoptosis.
To explore the role of BI-1 in virus infection, the expression of BI-1 was silenced by BI-1-siRNA, and then the WSSV copy number in shrimp was assessed. The results revealed that BI-1 silencing significantly suppressed the proliferation of WSSV compared with the controls (Figure 4J). These data indicated that BI-1 exerted a positive effect on WSSV infection in shrimp.
The above findings suggested that miR-12 could trigger the antiviral apoptosis of shrimp by downregulating expression of the BI-1 gene, which encodes an inhibitor of apoptosis.
The above data showed that shrimp miR-12 promoted antiviral phagocytosis and antiviral apoptosis and inhibited virus infection by targeting the shrimp genes PTEN and BI-1 and the viral gene wsv024 in shrimp. To further explore the mechanism of the miR-12-mediated simultaneous triggering of multiple antiviral pathways in shrimp, the miR-12-mediated degradation of three different target genes was evaluated. The results of time-course experiments showed that the amount of degraded fragments of three target mRNAs increased with extended reaction time (Figure 5A), indicating that miR-12 mediated the degradation of its target mRNAs in the Ago1 complex. Additional data revealed that miR-12 mediated the degradation of three target mRNAs in a miRNA-concentration-dependent manner (Figure 5B).
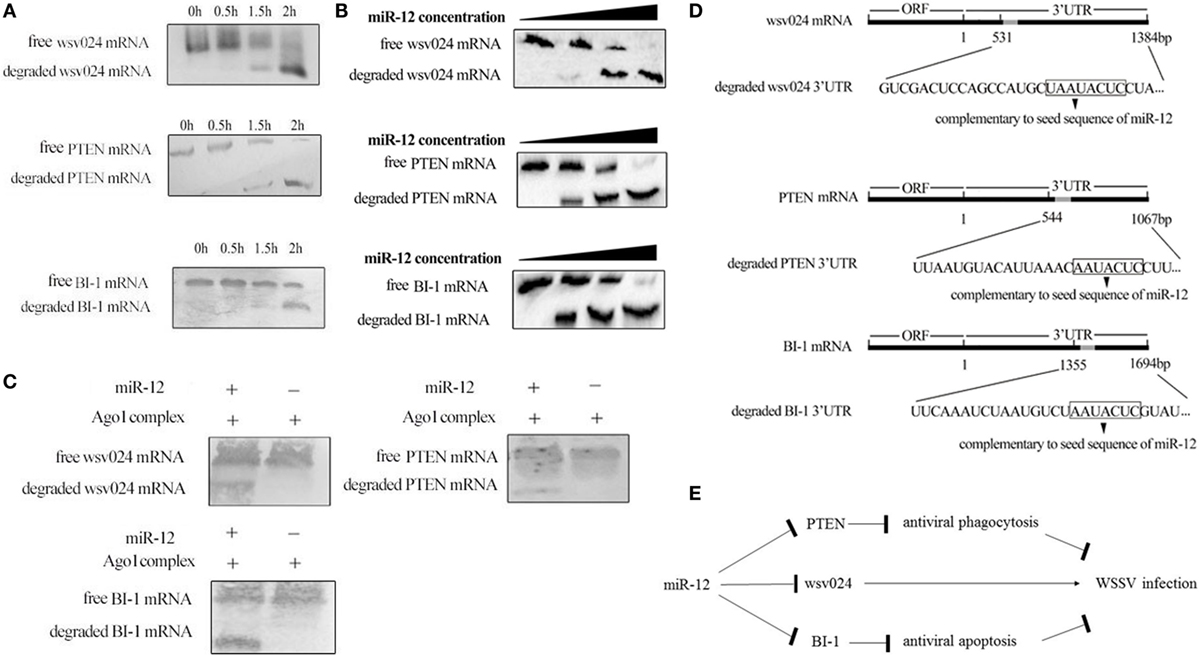
Figure 5. The underlying mechanism of miR-12 synchronously triggering multiple antiviral pathways. (A) Time-course degradation of miR-12 target mRNAs in the shrimp Ago1 complex. The 3′ untranslated region (3′UTR) of wsv024, PTEN, or BI-1 mRNA, miR-12, and the Ago1 complex were incubated for various times (0, 0.5, 1, and 2 h). Subsequently, wsv024, PTEN, and BI-1 mRNAs were detected using northern blotting. (B) The effects of the miR-12 concentration on the degradation of target mRNAs. The 3′UTR of wsv024, PTEN, or BI-1 mRNA and the Ago1 complex were incubated with miR-12 at different concentrations for 2 h, followed by Northern blot analysis to detect the mRNAs. (C) The miR-12-mediated synchronous degradation of target mRNAs. The 3′UTRs of wsv024, PTEN, and BI-1 mRNAs were mixed in equal amounts, followed by incubation with miR-12 and the Ago1 complex for 2 h. The mRNA degradation was monitored using northern blotting. (D) The sequencing of degraded target mRNA (wsv024, PTEN, and BI-1) 3′UTRs mediated by miR-12. (E) Proposed model for the host miR-12-mediated triggering of multiple antiviral pathways.
To evaluate whether miR-12 mediated the synchronous degradation of multiple target mRNAs, the 3′UTRs of PTEN, BI-1, and wsv024 mRNAs were mixed at equivalent amounts and then incubated with miR-12 and the Ago1 complex. Northern blots indicated that the three mRNAs were simultaneously degraded (Figure 5C). These data showed that miR-12 could mediate the synchronous degradation of multiple target genes. To confirm the miR-12-mediated degradation of target mRNAs, the degraded 3′UTR fragments of wsv024, PTEN, and BI-1 mRNAs were sequenced. The results indicated that miR-12 could mediate the 5′–3′ exonucleolytic degradation of its target mRNAs, and the miR-12-mediated degradation terminated at a vicinity of the 3′UTR sequence complementary to the seed sequence of miR-12 (Figure 5D).
Taken together, the findings showed that miR-12 simultaneously triggered phagocytosis, apoptosis, and antiviral immunity through synchronous targeting of shrimp genes (PTEN and BI-1) and a viral gene (wsv024) (Figure 5E).
Virus infection is the major origin of many diseases. To protect the host from virus invasion, the host gradually evolves a series of antiviral mechanisms in response to virus attack. When a virus invades its host or has been initiated to replicate in target cells, multiple antiviral immune pathways of the host are triggered to ensure the clearance of pathogens (24, 25). Owing to the diversity of antiviral immune mechanisms, various trigger factors may be released to initiate multiple antiviral pathways in the host. Toll-like receptors 3, 7, and 8 accumulate when pathogens invade, thus promoting the host antiviral responses through the induction of specific genes, such as interferon regulatory factors (IRFs) and nuclear factor of kappa light polypeptide gene enhancer in B cells (NF-κB) (24). Double-stranded RNA (dsRNA) represents a molecular intermediate during virus replication within infected cells (25). The existence of dsRNA in the cytoplasm can induce the release of RNA helicases, such as RIG-I (DExD/H-box helicase 58), which are used to detect cytoplasmic dsRNA and trigger the production of antiviral molecules, including type I interferons. However, whether an individual molecule can synchronously trigger different host antiviral pathways remains unclear. In the present study, the results indicated that shrimp miR-12 triggered shrimp antiviral apoptosis and phagocytosis through interactions with host BI-1 and PTEN genes and simultaneously inhibited WSSV replication by targeting the viral wsv024 gene. Therefore, the present study revealed that multiple host antiviral pathways could share the same trigger factor (i.e., miRNA). Due to its multiple target genes, a single miRNA revealed the synchronous regulation mechanism of multiple antiviral immune pathways.
The results of the present study revealed that shrimp miR-12 synchronously initiated multiple antiviral pathways by directly targeting the virus wsv024 gene and the host PTEN and BI-1 genes. During phagocytosis, PTEN abrogates ITAM (immunoreceptor tyrosine-based activation motif)-dependent signaling events in vivo and controls RAC activities to inhibit phagocytic capacities, showing negative regulation on antiviral phagocytosis (26). In fungi, PTEN directly activates the actin depolymerization factor CFL1 (cofilin-1) during PGE2-mediated inhibition of phagocytosis (23). BI-1, an inhibitor of the apoptotic activator BAX (BCL2 associated X), can suppress apoptosis (27). In the present study, the results indicated that the viral wsv024 gene was required for WSSV infection. Therefore, the miR-12 target genes wsv024, PTEN, and BI-1 played negative roles in the antiviral immunity of shrimp. The results of the present study demonstrated that miR-12 mediated the synchronous degradation of three target genes (wsv024, PTEN, and BI-1) in the shrimp Ago1 complex, demonstrating the simultaneous regulation of multiple antiviral immune pathways. As previously reported, miRNAs mediate target gene silencing through the induction of mRNA degradation or transcriptional repression, and target mRNA degradation affords a dominating contribution to silencing (28–30). In plants, miRNAs directly induce the endonucleolytic cleavage of full complementary mRNAs in the middle of the matching regions of their target genes (31). However, in animals, the majority of target mRNAs are incompletely complementary. In animals, miRNAs can guide their targets to the cellular 5′- to 3′-exonucleolytic digestion of mRNAs (32–34), consistent with the findings of the present study. In the case of 5′- to 3′-exonucleolytic degradation, mRNAs are initially deadenylated by the CAF1 (CCR4-NOT transcription complex subunit 8)-CCR4 (C–C motif chemokine receptor 4)-NOT deadenylase complex, followed by decapping via the enzyme DCP2 (decapping mRNA 2) (35). DCP2 typically exerts its decapping function with the aid of other co-factors, such as DCP1 (decapping mRNA 1), EDC4 (enhancer of mRNA decapping 4), and the DEAD-box protein RCK (also known as Me31B), for full activity or stability. However, the mechanism of the miRNA-mediated deadenylation of mRNAs remains unclear. In this context, the present study contributed novel insights to the miRNA-mediated simultaneous regulation of multiple antiviral pathways in invertebrates.
XZ conceived the project. LS performed the experiments and wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, AT, and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
This work was financially supported through grants from the National Natural Science Foundation of China (31430089) and the National Program on Key Basic Research Projects (2015CB755903).
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00855/full#supplementary-material.
1. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell (2006) 124(4):783–801. doi:10.1016/j.cell.2006.02.015
2. Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol (2007) 178(4):2448–57. doi:10.4049/jimmunol.178.4.2448
3. Chung KM, Thompson BS, Fremont DH, Diamond MS. Antibody recognition of cell surface-associated NS1 triggers Fc-γ receptor-mediated phagocytosis and clearance of West Nile virus-infected cells. J Virol (2007) 81(17):9551–5. doi:10.1128/JVI.00879-07
4. Ye T, Tang W, Zhang X. Involvement of Rab6 in the regulation of phagocytosis against virus infection in invertebrates. J Proteome Res (2012) 11(11):4834–46. doi:10.1021/pr300274k
5. Flannagan RS, Jaumouillé V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol (2012) 7(4):61–98. doi:10.1146/annurev-pathol-011811-132445
6. Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, et al. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J Virol (2008) 82(21):10375–85. doi:10.1128/JVI.00395-08
7. Wang L, Zhi B, Wu W, Zhang X. Requirement for shrimp caspase in apoptosis against virus infection. Dev Comp Immunol (2008) 32(6):706–15. doi:10.1016/j.dci.2007.10.010
8. Fujimoto I, Pan J, Takizawa T, Nakanishi Y. Virus clearance through apoptosis-dependent phagocytosis of influenza A virus-infected cells by macrophages. J Virol (2000) 74(7):3399–403. doi:10.1128/JVI.74.7.3399-3403.2000
9. Wang W, Zhang X. Comparison of antiviral efficiency of immune responses in shrimp. Fish Shellfish Immunol (2008) 25(5):522–7. doi:10.1016/j.fsi.2008.07.016
10. Liu H, Golebiewski L, Dow EC, Krug RM, Javier RT, Rice AP. The ESEV PDZ binding-motif of the avian influenza A virus NS1 protein protects infected cells from apoptosis through directly targeting scribble. J Virol (2010) 84(21):11164–74. doi:10.1128/JVI.01278-10
11. Smith JL, Grey FE, Uhrlaub JL, Nikolich-Zugich J, Hirsch AJ. Induction of the cellular microRNA, Hs_154, by West Nile virus contributes to virus-mediated apoptosis through repression of antiapoptotic factors. J Virol (2012) 86(9):5278–87. doi:10.1128/JVI.06883-11
12. Le S, Li C, Zhang X. The role of shrimp miR-965 in virus infection. Fish Shellfish Immunol (2016) 54:427–34. doi:10.1016/j.fsi.2016.04.129
13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi:10.1016/S0092-8674(04)00045-5
14. Jiang A, Zhang S, Li Z, Liang R, Ren S, Li J, et al. miR-615-3p promotes the phagocytic capacity of splenic macrophages by targeting ligand-dependent nuclear receptor corepressor in cirrhosis-related portal hypertension. Exp Biol Med (2011) 236(6):672–80. doi:10.1258/ebm.2011.010349
15. Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res (2009) 69(3):1135–42. doi:10.1158/0008-5472.CAN-08-2886
16. Huang T, Zhang X. Functional analysis of a crustacean microRNA in host-virus interactions. J Virol (2012) 86(23):12997–3004. doi:10.1128/JVI.01702-12
17. Yang G, Yang L, Zhao Z, Wang J, Zhang X. Signature miRNAs involved in the innate immunity of invertebrates. PLoS One (2012) 7(6):e39015. doi:10.1371/journal.pone.0039015
18. He Y, Zhang X. Comprehensive characterization of viral miRNAs involved in white spot syndrome virus (WSSV) infection. RNA Biol (2012) 9(7):1019–29. doi:10.4161/rna.20741
19. Liu W, Han F, Zhang X. Ran GTPase regulates hemocytic phagocytosis of shrimp by interaction with myosin. J Proteome Res (2009) 8(3):1198–206. doi:10.1021/pr800840x
20. Huang T, Xu D, Zhang X. Characterization of host microRNAs that respond to DNA virus infection in a crustacean. BMC Genomics (2012) 13(1):159. doi:10.1186/1471-2164-13-159
21. Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, et al. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol (2012) 19(7):685–92. doi:10.1038/nsmb.2335
22. Hay T, Jenkins H, Sansom OJ, Martin NM, Smith GC, Clarke AR. Efficient deletion of normal Brca2-deficient intestinal epithelium by poly(ADP-ribose) polymerase inhibition models potential prophylactic therapy. Cancer Res (2005) 65(22):10145–8. doi:10.1158/0008-5472.CAN-05-1186
23. Serezani CH, Kane S, Medeiros AI, Cornett AM, Kim SH, Marques MM, et al. PTEN activates the actin depolymerization factor cofilin-1 during PGE2-mediated inhibition of fungal phagocytosis. Sci Signal (2012) 5(210):ra12. doi:10.1126/scisignal.2002448
24. Takeda K, Akira S. TLR signaling pathways. Semin Immunol (2004) 16(1):3–9. doi:10.1016/j.smim.2003.10.003
25. Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet (2011) 48(12):856–9. doi:10.1136/jmedgenet-2011-100156
26. Kim JS, Peng X, De PK, Geahlen RL, Durden DL. PTEN controls immunoreceptor (immunoreceptor tyrosine-based activation motif) signaling and the activation of Rac. Blood (2002) 99(2):694–7. doi:10.1182/blood.V99.2.694
27. Reimers K, Choi CY, Bucan V, Vogt PM. The Bax inhibitor-1 (BI-1) family in apoptosis and tumorigenesis. Curr Mol Med (2008) 8(2):148–56. doi:10.2174/156652408783769562
28. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell (2009) 136(4):642–55. doi:10.1016/j.cell.2009.01.035
29. Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell (2008) 29(1):1–7. doi:10.1016/j.molcel.2007.12.010
30. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature (2010) 466(7308):835–40. doi:10.1038/nature09267
31. Yekta S, Shih I, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science (2004) 304(5670):594–6. doi:10.1126/science.1097434
32. Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA (2005) 11(11):1640–7. doi:10.1261/rna.2191905
33. Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev (2006) 20(14):1885–98. doi:10.1101/gad.1424106
34. Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A (2006) 103(11):4034–9. doi:10.1073/pnas.0510928103
Keywords: microRNA, antiviral immunity, phagocytosis, apoptosis, degradation of target microRNA
Citation: Shu L and Zhang X (2017) Shrimp miR-12 Suppresses White Spot Syndrome Virus Infection by Synchronously Triggering Antiviral Phagocytosis and Apoptosis Pathways. Front. Immunol. 8:855. doi: 10.3389/fimmu.2017.00855
Received: 07 March 2017; Accepted: 06 July 2017;
Published: 31 July 2017
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Philippe Georgel, University of Strasbourg, FranceCopyright: © 2017 Shu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobo Zhang, enhiMDgxMkB6anUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.