- 1Department of Human Oncology, University of Wisconsin, Madison, WI, United States
- 2Department of Biostatistics and Medical Informatics, University of Wisconsin, Madison, WI, United States
- 3Dana-Farber Cancer Institute/Boston Children’s Cancer and Blood Disorder Center, Harvard Medical School, Boston, MA, United States
- 4COG Statistics and Data Center, Department of Biostatistics, University of Florida, Gainesville, FL, United States
- 5Department of Biostatistics, Harvard University, Dana Farber Cancer Institute, Boston, MA, United States
- 6Children’s Hospital of Philadelphia, University of Pennsylvania School of Medicine, Philadelphia, PA, United States
- 7Provenance Biopharmaceuticals, Carlisle, MA, United States
- 8Seattle Children’s Hospital/University, Seattle, WA, United States
- 9University of Washington, Seattle, WA, United States
- 10New York Medical College, Valhalla, NY, United States
- 11Department of Medicine, University of Minnesota, Minneapolis, MN, United States
- 12Levine Children’s Hospital, Charlotte, NC, United States
- 13Department of Medicine, Washington University, St. Louis, MO, United States
- 14Department of Pediatrics, Hematology/Oncology, Moores Cancer Center, University of California San Diego, San Diego, CA, United States
- 15Institute of Stem Cell and Translational Cancer Research, Chang Gung Memorial Hospital, Taoyuan, Taiwan
- 16Department of Pediatrics, University of Wisconsin-Madison, Madison, WI, United States
Killer-cell immunoglobulin-like receptors (KIRs) are a family of glycoproteins expressed primarily on natural killer cells that can regulate their function. Inhibitory KIRs recognize MHC class I molecules (KIR-ligands) as ligands. We have reported associations of KIRs and KIR-ligands for patients in two monoclonal antibody (mAb)-based trials: (1) A Children’s Oncology Group (COG) trial for children with high-risk neuroblastoma randomized to immunotherapy treatment with dinutuximab (anti-GD2 mAb) + GM-CSF + IL-2 + isotretinion or to treatment with isotretinoin alone and (2) An Eastern Cooperative Oncology Group (ECOG) trial for adults with low-tumor burden follicular lymphoma responding to an induction course of rituximab (anti-CD20 mAb) and randomized to treatment with maintenance rituximab or no-maintenance rituximab. In each trial, certain KIR/KIR-ligand genotypes were associated with clinical benefit for patients randomized to immunotherapy treatment (immunotherapy in COG; maintenance rituximab in ECOG) as compared to patients that did not receive the immunotherapy [isotretinoin alone (COG); no-maintenance (ECOG)]. Namely, patients with both KIR3DL1 and its HLA-Bw4 ligand (KIR3DL1+/HLA-Bw4+ genotype) had improved clinical outcomes if randomized to immunotherapy regimens, as compared to patients with the KIR3DL1+/HLA-Bw4+ genotype randomized to the non-immunotherapy regimen. Conversely, patients that did not have the KIR3DL1+/HLA-Bw4+ genotype showed no evidence of a difference in outcome if receiving the immunotherapy vs. no-immunotherapy. For each trial, HLA-Bw4 status was determined by assessing the genotypes of three separate isoforms of HLA-Bw4: (1) HLA-B-Bw4 with threonine at amino acid 80 (B-Bw4-T80); (2) HLA-B-Bw4 with isoleucine at amino acid 80 (HLA-B-Bw4-I80); and (3) HLA-A with a Bw4 epitope (HLA-A-Bw4). Here, we report on associations with clinical outcome for patients with KIR3DL1 and these separate isoforms of HLA-Bw4. Patients randomized to immunotherapy with KIR3DL1+/A-Bw4+ or with KIR3DL1+/B-Bw4-T80+ had better outcome vs. those randomized to no-immunotherapy, whereas for those with KIR3DL1+/B-Bw4-I80+ there was no evidence of a difference based on immunotherapy vs. no-immunotherapy. Additionally, we observed differences within treatment types (either within immunotherapy or no-immunotherapy) that were associated with the genotype status for the different KIR3DL1/HLA-Bw4-isoforms. These studies suggest that specific HLA-Bw4 isoforms may differentially influence response to these mAb-based immunotherapy, further confirming the involvement of KIR-bearing cells in tumor-reactive mAb-based cancer immunotherapy.
Introduction
One modality of cancer immunotherapy utilizes tumor-reactive monoclonal antibodies (mAbs) to elicit a tumor-targeted immune response. Two recently completed clinical trials, in separate disease settings, utilized tumor-reactive mAbs to successfully target and treat the tumors: (1) the combination of dinutuximab with IL-2, GM-CSF, and isotretinoin for patients with high-risk neuroblastoma (1) and (2) rituximab for the treatment of patients with low-tumor burden follicular lymphoma (FL) (2).
Natural killer (NK) cells can contribute to the response to tumor-reactive mAb-based immunotherapeutics through antibody-dependent cellular cytotoxicity (ADCC). The ability of NK cells to elicit ADCC is regulated by activating and inhibiting signaling. Killer-cell immunoglobulin-like receptors (KIRs) are a class of receptors expressed on NK cells that influence such signaling (3, 4). Most inhibitory KIRs interact with HLA class I molecules as their ligands (KIR-ligand) (5). Specifically, KIR2DL1 binds to HLA-C2, KIR2DL2 and KIR2DL3 bind to HLA-C1, and KIR3DL1 recognizes the Bw4 epitope of HLA-A and HLA-B (6, 7). The independent segregation and inheritance of KIRs and KIR-ligands help to shape NK cell function and response to immunotherapeutic agents (8–11). When inhibitory KIRs interact with class I HLA molecules on target cells, NK cell-mediated lysis and ADCC are inhibited. During development, KIR/KIR-ligand interactions lead to self tolerance and NK cells become “licensed NK cells” (12–14). Licensed NK cells have augmented cytotoxicity against class I negative tumors compared to unlicensed NK cells (15, 16).
Killer-cell immunoglobulin-like receptors and KIR-ligands segregate independently: KIR genes are located on chromosome 19; HLA genes (KIR-ligands) are located on chromosome 6. Several studies have shown that genotypic differences of KIR and KIR-ligands can influence clinical outcome of certain cancer immunotherapies (8, 11, 17–19). We recently showed in two clinical trials that KIR3DL1 and its KIR-ligand, HLA-Bw4, appear to influence clinical outcome.
In a phase III trial (ANBL0032) of high-risk neuroblastoma patients, conducted by the Children’s Oncology Group (COG) (1), patients who inherited the KIR3DL1 gene and the gene for its HLA-Bw4 ligand (KIR3DL1+/Bw4+ genotype) and were treated with an immunotherapy regimen [dinutuximab (anti-GD2), IL-2, GM-CSF, and isotretinoin] had improved event-free survival (EFS) and overall survival as compared to those treated with isotretinoin alone (20, 21). In a separate Eastern Cooperative Oncology Group (ECOG) Phase III clinical trial of low-tumor burden FL (2), patients who were KIR3DL1+/HLA-Bw4+ and treated with a continuous regimen of maintenance rituximab had improved duration of response and % tumor shrinkage compared to KIR3DL1+/HLA-Bw4+ patients who were randomized to not receive maintenance rituximab (22, 23). Conversely, we did not observe improved outcome for patients that were not KIR3DL1+/HLA-Bw4+ when randomized to immunotherapy, in either study (22, 23). Furthermore, in both the COG and ECOG studies, patients who were randomized to the immunotherapy regimen that were KIR3DL1+/HLA-Bw4+ had better outcome compared to patients who were not KIR3DL1+/HLA-Bw4+.
Given these similar associations with outcome for the KIR3DL1/HLA-Bw4 interaction in these two clinical trials, we chose to evaluate these more deeply by evaluating the potential influence of distinct HLA-Bw4 isoforms. Polymorphisms in the α1 helix (positions 77–83) of HLA class I correspond to the sequence site of the Bw4 epitope that is recognized by KIR3DL1 (24). In KIR/KIR-ligand associations, we analyzed in these COG and ECOG trials, individuals were considered positive for HLA-Bw4 if they were found to have at least one of the three isoforms of HLA-Bw4: (1) HLA-B allele with a threonine at amino acid position 80 (B-Bw4-T80), (2) HLA-B allele with an isoleucine at amino acid position 80 (B-Bw4-I80), or (3) HLA-A with a Bw4 epitope (A-Bw4). Patients were negative for HLA-Bw4 if they did not have any of these three isoforms. These polymorphisms of this Bw4 epitope can impact KIR3DL1 recognition (25–29). As such, we describe the impact of the genotype status of B-Bw4-T80, B-Bw4-I80, and A-Bw4, together with the genotype status of KIR3DL1, on the clinical outcome, based on a clinical outcome parameter that measured the duration of response to the treatment regimen (EFS in COG; duration of response in ECOG).
Materials and Methods
Patients
COG ANBL0032 Patients
The phase III neuroblastoma clinical trial (ANBL0032; Clinicaltrials.gov # NCT00026312) evaluated the efficacy of isotretinoin alone as compared to an immunotherapeutic regimen consisting of dinutuximab (anti-GD2), aldesleukin (IL-2), sargramostim (GM-CSF), and isotretinoin (1). Of the 226 patients randomized, 174 patients (immunotherapy: n = 88; isotretinoin: n = 86) had DNA available, allowing evaluation of KIR/KIR-ligand genotype association with updated clinical outcome (>5-year follow-up if no event). All analyses in this study were conducted utilizing an intent-to-treat approach. All patients signed IRB approved consent forms enabling lab-based immune correlative analyses, and the genotyping done at UW-Madison was approved by the UW-IRB.
ECOG E4402 Patients
The Phase III ECOG clinical trial (E4402; ClinicalTrials.gov #NCT00075946) evaluated the efficacy of single agent, rituximab therapy for adults with low-tumor burden FL. Clinical results from this study have been reported elsewhere (2). A total of 408 patients with FL were entered, with 289 patients responding and randomized to no-maintenance or maintenance rituximab regimens. Disease measurements were obtained every 13 weeks (2). Of the 289 randomized patients from this trial, 213 patients had evaluable DNA and clinical data for this study, and 159 of them were randomized to no-maintenance (n = 80) or maintenance rituximab (n = 79) treatment. Of these 79 patients treated with maintenance rituximab, 75 patients had clinical data available for duration of response. All patients signed IRB approved consent forms enabling lab-based immune correlative analyses, and the genotyping done at UW-Madison was approved by the UW-IRB.
Genotyping
KIR3DL1 gene status was determined by a SYBR green real-time PCR reaction (30, 31). The genotype for HLA-Bw4, which includes three known HLA-Bw4 epitopes (B-Bw4-T80, B-Bw4-I80, and A-Bw4) were determined by PCR-SSP reactions using the KIR HLA Ligand SSP typing kit (product number 104.201-12u from Olerup, West Chester, PA, USA) with GoTaq DNA polymerase (M8295, Promega, WI, USA). All genotyping was conducted in a blinded manner, whereby individuals who determined the genotype of the patients did not have access to the clinical outcome data.
Statistical Methods
The goal of these analyses was to evaluate the association of KIR3DL1 in combination with each HLA-Bw4 isoform (B-Bw4-T80, B-Bw4-I80, and A-Bw4) on response to therapy (EFS or duration of response). For the COG trial, EFS time was defined as the time from study enrollment until the first occurrence of relapse, progressive disease, secondary cancer, or death or until the last contact with the patient if none of these events occurred (censored). For the ECOG trial, the duration of response was defined as the time from randomization (following an initial response to the induction rituximab treatment) to documented disease progression (2).
Cox proportional hazards regression models and log-rank tests were used to compare EFS/duration of response curves by treatment and genotype combinations. The proportional hazards assumption was tested, and when the assumption was not met, adjustments were made by incorporating time-dependent covariates into the model. For both trials, only randomized patients were included in the analyses. Statistical analyses were performed using SAS v9.4 (SAS Institute, Cary, NC, USA).
Results
HLA-Bw4 Isoforms, Together with KIR3DL1, Differentially Influence the Impact of mAb-Based Immunotherapy on Clinical Outcome of Neuroblastoma Patients
In our analyses of associations of KIR/KIR-ligand genotypic influence on clinical response in the neuroblastoma study (ANBL0032), we reported on differences in clinical outcome for those KIR3DL1+/Bw4+ (immunotherapy n = 58; isotretinoin n = 61) and those not KIR3DL1+/Bw4+ (immunotherapy n = 30; isotretinoin n = 25), and differences in response were observed dependent upon treatment type (20, 21). Since not all of the isoforms of HLA-Bw4 may interact with KIR3DL1 to the same degree, we further assessed patients with different HLA-Bw4 isoforms in this setting.
To better understand the KIR/KIR-ligand genotypic influence on clinical outcome, we evaluated the effect of Bw4 epitope on either an HLA-A or HLA-B allele. In this study, patients who were KIR3DL1+/A-Bw4+ had a trend toward improved EFS if they were treated with immunotherapy as compared to those treated with isotretinoin alone (p = 0.06; Figure 1A) (Table S1 in Supplementary Material). In contrast, we did not find a significant difference in EFS for patients receiving the immunotherapy vs. those randomized to not receive the immunotherapy (i.e., isotretinoin alone) in the patients that were not KIR3DL1+/A-Bw4+ (p = 0.35; Figure 1A).
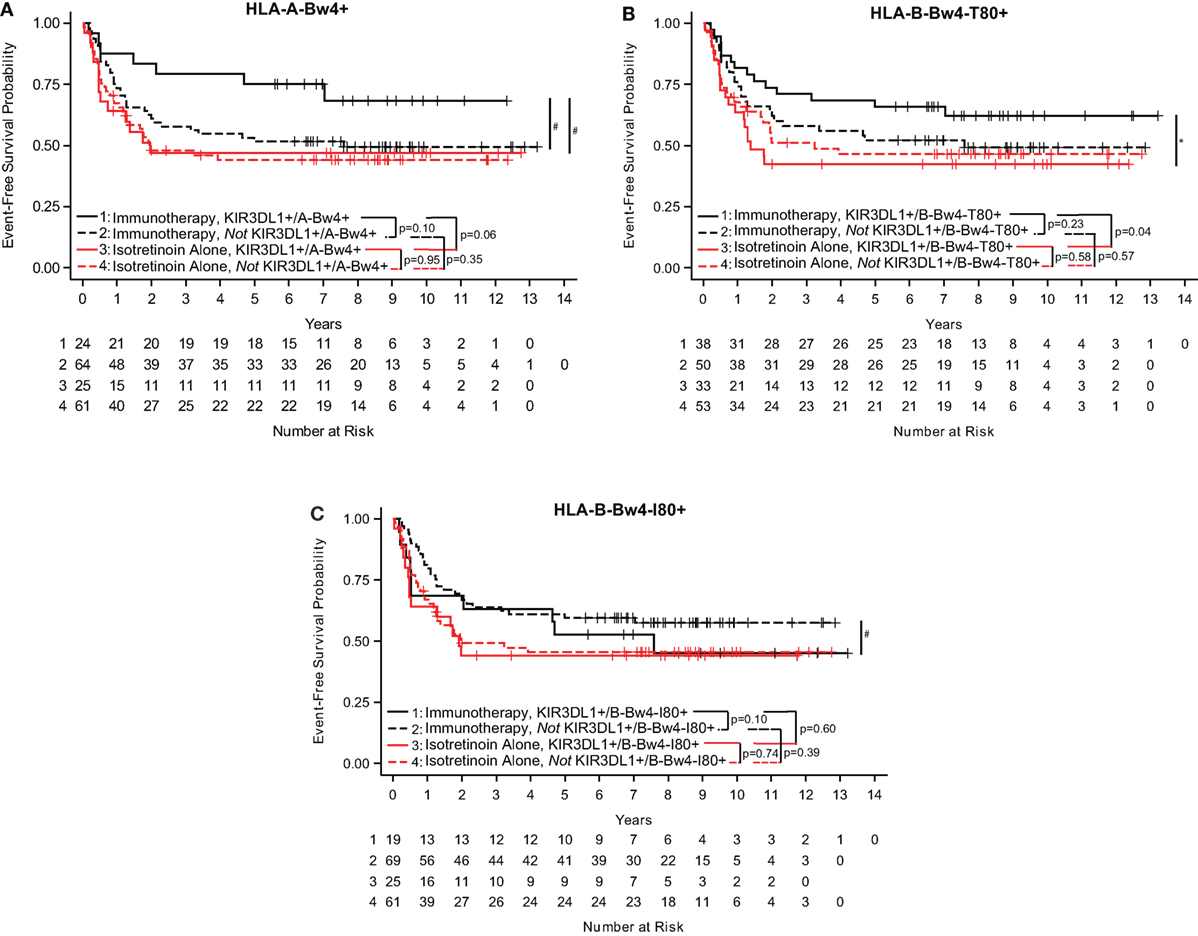
Figure 1. KIR3DL1/HLA-Bw4 isoforms differentially influence event-free survival (EFS) in neuroblastoma patients. In neuroblastoma patients treated with immunotherapy (black lines) or isotretinoin alone (red lines), genotype statuses of KIR3DL1/A-Bw4 (A), KIR3DL1/B-Bw4-T80 (B), and KIR3DL1/B-Bw4-I80 (C) had differential effects on EFS. Those with the genes present are represented by solid lines, and those without the genes present are represented by dotted lines. The number of patients at risk for EFS at a given time point are provided below the x-axis. *p < 0.5 and #p < 0.10.
We found that B-Bw4-T80 and B-Bw4-I80 differentially influenced EFS in these neuroblastoma patients (Table S1 in Supplementary Material). Similar to results in Figure 1A, patients who were KIR3DL1+/B-Bw4-T80+ showed significantly improved EFS if they received immunotherapy compared with isotretinoin alone (p = 0.04; Figure 1B), whereas those that were not KIR3DL1+/B-Bw4-T80+ showed no difference in EFS for patients receiving the immunotherapy vs. those randomized to receive isotretinoin alone (p = 0.57; Figure 1B). However, for B-Bw4-I80+, the results were converse. Patients who were KIR3DL1+/B-Bw4-I80+ showed no sign of improved EFS if they received immunotherapy compared with isotretinoin alone (p = 0.60; Figure 1C). Furthermore, and in contrast to results in Figures 1A,B, while not significant, there appears to be improved EFS for patients receiving the immunotherapy vs. isotretinoin alone in the patients who were not KIR3DL1+/B-Bw4-I80+ (p = 0.10; Figure 1C).
These findings suggest that the different isoforms of HLA-Bw4 differentially influence the impact of anti-GD2-based immunotherapy on EFS for high-risk neuroblastoma patients.
HLA-Bw4 Isoforms, Together with KIR3DL1, Differentially Influence the Impact of mAb-Based Immunotherapy on Clinical Outcome of FL Patients
The ECOG E4402 Phase III clinical trial sought to optimize the rituximab treatment regimen for low-tumor burden FL patients (2). As such, different from the design of the neuroblastoma COG trial described above where one treatment arm was treated with immunotherapy and the other was not, in E4402 all patients were initially treated with rituximab. In E4402, all FL patients received induction rituximab, consisting of four weekly rituximab treatments. After 13 weeks, those patients who achieved ≥50% tumor shrinkage were randomized to two separate treatment regimens: (1) “maintenance” rituximab was given every 13 weeks or (2) “no-maintenance” where rituximab was given only upon disease progression (2). Thus, for the parameter of disease progression, the no-maintenance group received no rituximab between randomization and disease progression. Similar to the COG findings regarding the genotype status of KIR3DL1/Bw4, in this ECOG study, we also found that those KIR3DL1+/Bw4+ (maintenance n = 49; no-maintenance n = 53) had different clinical outcome than those not KIR3DL1+/Bw4+ (maintenance n = 27; no-maintenance n = 26), which was also influenced by the treatment arm.
Analyses of the three separate HLA-Bw4 isoforms suggest that the isoforms of HLA-Bw4 differently influenced the impact of maintenance rituximab. FL patients who were KIR3DL1+/A-Bw4+ that were treated with maintenance rituximab had a longer duration of response (0 of 23 progressed, Figure 2A) as compared to patients who were not KIR3DL1+/A-Bw4+ [13 out of 53 progressed (p = 0.008, Figure 2A) (Table S1 in Supplementary Material)]. Separately, patients who were KIR3DL1+/B-Bw4-T80+ also showed significantly prolonged duration of response if they received maintenance as compared with no-maintenance rituximab (p = 0.007; Figure 2B). In addition, those patients whowere not KIR3DL1+/B-Bw4-T80+ had a trend toward improved duration of response if treated with maintenance as compared with no-maintenance rituximab (p = 0.07; Figure 2B) (Table S1 in Supplementary Material). However, patients who were KIR3DL1+/B-Bw4-I80+ did not show prolonged duration of response if they received maintenance as compared with no-maintenance rituximab (p = 0.40; Figure 2C). Similar to the trends for improved EFS observed in neuroblastoma patients treated with immunotherapy (Figure 1C), those FL patients who were not KIR3DL1+/B-Bw4-I80+ had improved duration of response if treated with maintenance rituximab as compared to no-maintenance (p = 0.002; Figure 2C) (Table S1 in Supplementary Material).
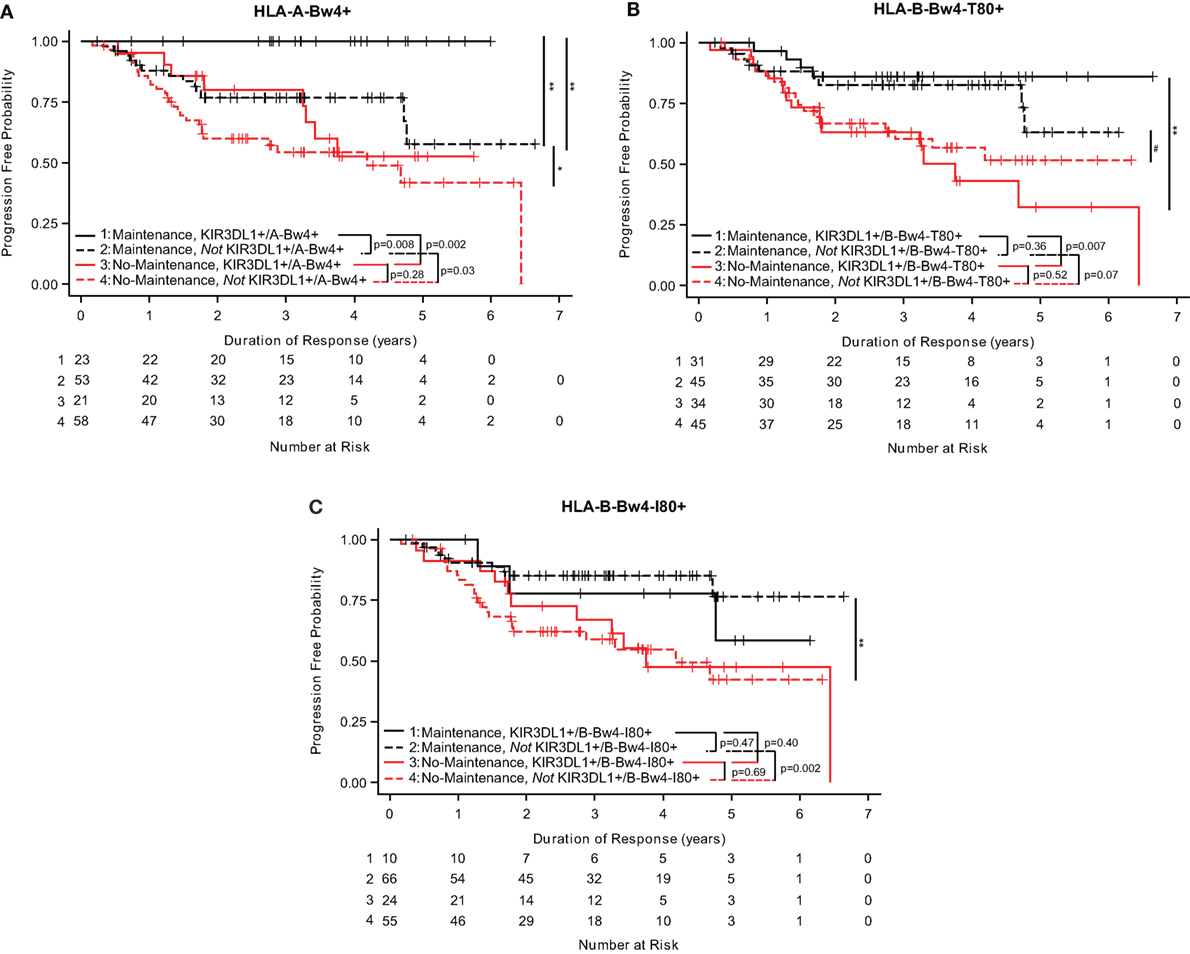
Figure 2. KIR3DL1/HLA-Bw4 isoforms differentially influence duration of response in follicular lymphoma (FL) patients. In FL patients treated with maintenance rituximab (red lines) or no-maintenance rituximab (black lines), genotype statuses of KIR3DL1/A-Bw4 (A), KIR3DL1/B-Bw4-T80 (B), and KIR3DL1/B-Bw4-I80 (C) had differential effects on duration of response. Those with the genes present are represented by solid lines, and those without the genes present are represented by dotted lines. The number of patients at risk for duration of response at a given time point are provided below the x-axis. **p < 0.01, *p < 0.5, and #p < 0.10.
These findings suggest that the different isoforms of HLA-Bw4 differentially influence the impact of rituximab maintenance treatment for these low-tumor burden FL.
Discussion
In both of these clinical trials, in separate disease settings, tumor-reactive mAbs were used to treat the tumors. In the analysis of KIR/KIR-ligand genotypes in each of these studies, we found similar associations with outcomes based upon the influence of KIR3DL1/HLA-Bw4. Specifically, those patients who had both KIR3DL1 and HLA-Bw4 had improved clinical outcomes if they were treated with either the COG immunotherapy regimen or the maintenance rituximab regimen in ECOG as compared to those who did not receive these same immunotherapeutic regimens (20, 21). Here, we report on the analyses of the specific HLA-Bw4 isoforms in both trials. In the ECOG trial of FL patients, patients with a KIR3DL1+/A-Bw4+ genotype or a KIR3DL1+/B-Bw4-T80+ genotype showed improved outcome when randomized to the maintenance regimen rather than to the no-maintenance regimen. In contrast, patients with a KIR3DL1+/B-Bw4-I80+ genotype showed no evidence of improved outcome when randomized to the maintenance treatment vs. no-maintenance regimen. We also observed similar trends for these same analyses in the COG trial of neuroblastoma patients.
Although other mechanisms, such as antibody-dependent cellular phagocytosis and complement-dependent cellular cytotoxicity (32, 33), could also contribute to the anti-tumor efficacy of tumor antigen-specific monoclonal antibodies, we hypothesize that the anti-tumor effect of rituximab and dinutuximab in these FL and neuroblastoma patients, respectively, is primarily through ADCC. NK cells are major contributors to ADCC, and their activity is regulated via the interactions between KIRs/KIR-ligands (34). As such, we hypothesize that the KIR/KIR-ligand genotypes could influence the degree that patients respond to antibody-based immunotherapies. Besides NK cells, KIRs are also expressed by a subset of T cells as well as NKT cells (35, 36). Therefore, it is possible that these other cell types may also be influenced by KIR/KIR-ligand genotypes.
Besides inherited genetic differences in KIR and KIR-ligand genotypes, other individual genetic differences, such as polymorphisms in Fc gamma receptors (FCGRs), may influence patient outcome to immunotherapy. FCGR polymorphisms can alter the affinity of FCGRs for the Fc portion of antibodies (mAbs or endogenous antibodies) (37). For example, in a separate study of patients with metastatic renal cell carcinoma treated with high-dose IL-2, we found that patients with a “higher affinity” FCGR genotype had improved clinical outcome as compared to those patients with a “lower affinity” FCGR genotype (38). In our analysis of those same metastatic renal cell carcinoma patients for KIR/KIR-ligand genotype influence on outcome, we did not observe differences in clinical outcome associated with KIR3DL1 and HLA-Bw4 genotype status (39). The influence of FCGR polymorphisms on clinical outcome to rituximab is variable (40–42). For the FL patients analyzed here from this ECOG study, Kenkre and colleagues reported no association of FCGR genotype polymorphisms with patient outcome (43). In addition, some groups have found associations of FCGR genotype with clinical outcome for patients treated with anti-GD2 immunotherapy (8, 44, 45). For the neuroblastoma patients from this COG trial, FCGR genotype associations with clinical outcome are still under investigation. In addition, it has been reported that the influence from KIR/KIR-ligand interactions on NK cells may be affected by the affinity of the Fc portion of different therapeutic mAb used (46), the rituximab used in this ECOG trial and the dinutuximab used in this COG trial have similar human IgG1 Fc components, which may also help account for why we observed similar influences from HLA-Bw4 epitopes in these two separate studies where two different therapeutic mAbs were used.
These clinical data are consistent with the B-Bw4-I80 isoform functioning somewhat differently than the B-Bw4-T80 or A-Bw4 isoforms, and potentially making the tumor cells less responsive to the potential benefit of the anti-GD2 or anti-CD20 mAb-based immunotherapy. In vitro analyses have shown that a subset of HLA-Bw4 alleles (those with an B-Bw4-I80 isoform) show relative protection from lysis by NK cells (47, 48). The data presented here are consistent with these in vitro results; mAb-based immunotherapy may provide more benefit for patients with weaker NK cell inhibition from B-Bw4-T80 or A-Bw4, than for patients with stronger NK inhibition from B-Bw4-I80.
Given that patients assessed in either trial could be positive for more than one of the HLA-Bw4 epitopes, we did consider whether the HLA-Bw4 epitopes were in linkage disequilibrium. We found that A-Bw4 was not in linkage disequilibrium with either B-Bw4-I80 or B-Bw4-T80 (Table S2 in Supplementary Material). Thus, the influence that each of these HLA-Bw4 epitopes had on the length of patient response in either trial is presumably not due to linkage disequilibrium with each other.
We also considered whether the interaction of KIR3DL1 with these three different HLA-Bw4 isoforms showed any association of outcome among patients randomized to receive the immunotherapy regimens. Within the COG study, we observed a trend for improved outcome for those KIR3DL1+/HLA-A-Bw4+ vs. those not KIR3DL1+/HLA-A-Bw4+ (Figure 1A), and we also observed a trend in the opposite direction for HLA-Bw4-I80, namely, there was a trend for improved outcome for those not KIR3DL1+/HLA-B-Bw4-I80+ vs. those who were KIR3DL1+/HLA-B-Bw4-I80+ (Figure 1C). Although only a trend, this difference in Figure 1A and Figure 1C is consistent with differential function of HLA-A-Bw4 and HLA-B-Bw4-I80. No significant differences or trends were noted when we evaluated among the FL patients randomized to receive the maintenance rituximab regimen (Figures 2A–C).
The interaction of KIR3DL1 with the Bw4 epitope is dependent not only on the architecture of Bw4 but also on the sequence of the bound peptide (25, 28, 49–51). Additionally, the differences we observed between A-Bw4, B-Bw4-T80, and B-Bw4-I80 may be due to the different inhibition strength for KIR3DL1 from these isoforms. For instance, HLA-A*32:01, HLA-B*51:01, and HLA-B*58:01 strongly inhibit target cells from lysis by KIR3DL1+ NK cells, yet HLA-B*15:13 and HLA-B*27:05 have weaker inhibitory effects, despite all being HLA-Bw4 alleles (6, 25, 26, 48, 52–54). In addition, depending on the KIR3DL1 allele, expression of KIR3DL1 can vary; different HLA-A-Bw4 alleles have differential affinity for KIR3DL1 that is attributed to high vs. low expression of KIR3DL1 (55). Furthermore, the specific Bw4 allele, as well as the KIR3DL1 allele, the strength of KIR3DL1/HLA-Bw4 interaction and the binding avidity can vary (29). For example, Saunders et al. recently showed that HLA-A*24:02 acts as a poor ligand for KIR3DL1, and the strength of its interaction with KIR3DL1 differed depending on the allele of KIR3DL1 (29). The genotyping methodology employed for analyzing the many patients in these two clinical trials reported here was not able to address these more subtle allele-specific or peptide-related issues.
Another possible cause of the differences observed in these HLA-Bw4 isoforms may be due to genetic polymorphisms of KIR3DL1 (26, 28, 29, 56–60). More than 100 alleles of KIR3DL1 have been described. Phylogenetically, these alleles span three lineages based on the polymorphism of the three extracellular domains (D0–D1–D2) (53, 61). In both of these clinical studies analyzed, we did not determine the allelic differences of the KIR genes, but rather we determined their presence or absence. Thus, we cannot assess how different KIR3DL1 alleles may affect the interactions between different isoforms of HLA-Bw4. We did, however, assess if KIR3DL1 allelic status could influence the interactions of KIR3DL1 with HLA-Bw4 and with the separate HLA-Bw4 isoforms. KIR3DL1 and KIR3DS1 are alleles, thus individuals can have 2, 1, or 0 copies of KIR3DL1 (2 copies: KIR3DL1/KIR3DL1, 1 copy: KIR3DL1/KIR3DS1, or 0 copies: KIR3DS1/KIRDS1). Although KIR3DS1 has not been shown to utilize HLA-Bw4 as a ligand in vitro, whether KIR3DS1 may still interact with HLA-Bw4 in vivo is controversial (62–65). We assessed whether the allelic status of KIR3DL1/KIR3DS1 together with HLA-Bw4 (and HLA-Bw4 isoforms) influenced patient response. We found that there was no evidence of an association with outcome in either the COG or the ECOG study that could be linked to the allelic status of KIR3DL1/KIR3DLS1 (data not shown), nor was there evidence of an association of clinical outcome linked to KIR3DL1/KIR3DS1 status together with the HLA-Bw4 ligand isoforms (data not shown). Rather, the mere presence of KIR3DL1 together with its ligand, HLA-Bw4, seemed to influence patients’ response to immunotherapy in both clinical trials. These observations will require validation in a separate study.
In conclusion, this work sheds further light on the role of KIR receptors on NK cells in the antitumor response to immunotherapeutic mAbs. We demonstrate that the KIR3DL1/HLA-Bw4 axis influences response to tumor-targeted mAbs in two separate clinical trials and that the presence of the B-Bw4-T80 isoform or the A-Bw4 isoform is associated with improved response to mAb-based immunotherapy, while the presence of the B-Bw4-I80 isoform is not.
Ethics Statement
This study was carried out in accordance with the recommendations of University of Wisconsin Health Sciences Institutional Review Board with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the University of Wisconsin Health Sciences Institutional Review Board.
Author Contributions
Each author made substantial contributions to the conception and/or design of this research, including the acquisition, analysis, and/or interpretation of data for the work; the drafting of this manuscript, including critical revisions important intellectual content, were shared duties by all authors; each author submitted final approval of this manuscript as submitted to be published; each author is in agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved (AE, WW, PR, LC, KK, EM, YS, JH, WL, AN, FH, MH, JM, JP, MO, JM, AG, BK, AY, and PS).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the COG and ECOG for allowing us to assess the role of the genotypic influences of KIR and KIR-ligands on the clinical outcome from the treatments administered in each of these trials. In addition, we thank the medical and nursing staff who participated in the care of patients in each study, and especially all patients participating in each study.
Funding
This research was supported by Hyundai Hope on Wheels Grant; Midwest Athletes Against Childhood Cancer; Stand Up 2 Cancer; The St. Baldrick’s Foundation; University of Wisconsin-Madison Carbone Cancer Center; Forward Lymphoma Fund; by the ECOG-ACRIN Cancer Research Group (Robert L. Comis, MD, and Mitchell D. Schnall, MD, PhD, Group Co-Chairs); and supported in part by Public Health Service Grants CA014520, CA021115, CA023318, CA066636, CA180820, CA180794, CA021076, CA180799, CA180816, CA166105, and CA197078, from the National Cancer Institute; the National Institutes of Health, the Department of Health and Human Services. Its content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00675/full#supplementary-material.
References
1. Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med (2010) 363(14):1324–34. doi:10.1056/NEJMoa0911123
2. Kahl BS, Hong F, Williams ME, Gascoyne RD, Wagner LI, Krauss JC, et al. Rituximab extended schedule or re-treatment trial for low-tumor burden follicular lymphoma: Eastern Cooperative Oncology Group Protocol e4402. J Clin Oncol (2014) 32(28):3096–102. doi:10.1200/JCO.2014.56.5853
3. Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol (2002) 38(14):1007–21. doi:10.1016/S0161-5890(02)00030-5
4. Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol (2002) 20:217–51. doi:10.1146/annurev.immunol.20.092501.134942
5. Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, et al. Gene map of the extended human MHC. Nat Rev Genet (2004) 5(12):889–99. doi:10.1038/nrg1489
6. Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med (1994) 180(2):537–43. doi:10.1084/jem.180.2.537
7. Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, di Donato C, et al. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med (1996) 184(2):505–18. doi:10.1084/jem.184.2.505
8. Delgado DC, Hank JA, Kolesar J, Lorentzen D, Gan J, Seo S, et al. Genotypes of NK cell KIR receptors, their ligands, and Fcgamma receptors in the response of neuroblastoma patients to Hu14.18-IL2 immunotherapy. Cancer Res (2010) 70(23):9554–61. doi:10.1158/0008-5472.CAN-10-2211
9. Du J, Lopez-Verges S, Pitcher BN, Johnson J, Jung SH, Zhou L, et al. CALGB 150905 (alliance): rituximab broadens the anti-lymphoma response by activating unlicensed NK cells. Cancer Immunol Res (2014) 2(9):878–89. doi:10.1158/2326-6066.CIR-13-0158
10. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol (2008) 180(6):3969–79. doi:10.4049/jimmunol.180.6.3969
11. Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest (2012) 122(9):3260–70. doi:10.1172/JCI62749
12. Jonsson AH, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol (2009) 101:27–79. doi:10.1016/S0065-2776(08)01002-X
13. Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature (2005) 436(7051):709–13. doi:10.1038/nature03847
14. Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol Rev (2006) 214:143–54. doi:10.1111/j.1600-065X.2006.00458.x
15. Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science (1991) 253(5016):199–202. doi:10.1126/science.1853205
16. Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell (2010) 142(6):847–56. doi:10.1016/j.cell.2010.08.031
17. Cheung NK, Cheung IY, Kushner BH, Ostrovnaya I, Chamberlain E, Kramer K, et al. Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte-macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. J Clin Oncol (2012) 30(26):3264–70. doi:10.1200/JCO.2011.41.3807
18. Forlenza CJ, Boudreau JE, Zheng J, Le Luduec JB, Chamberlain E, Heller G, et al. KIR3DL1 allelic polymorphism and HLA-B epitopes modulate response to anti-GD2 monoclonal antibody in patients with neuroblastoma. J Clin Oncol (2016) 34(21):2443–51. doi:10.1200/JCO.2015.64.9558
19. Venstrom JM, Zheng J, Noor N, Danis KE, Yeh AW, Cheung IY, et al. KIR and HLA genotypes are associated with disease progression and survival following autologous hematopoietic stem cell transplantation for high-risk neuroblastoma. Clin Cancer Res (2009) 15(23):7330–4. doi:10.1158/1078-0432.CCR-09-1720
20. Erbe A, Wang W, Carmichael L, Kim K, Reville P, London W, et al. Impact of KIR/KIR ligand genotype for neuroblastoma patients in a phase 3 COG immunotherapy trial. AACR Annual Meeting (Abstract). Washington, DC: Cancer Research (2017).
21. Reville P, Erbe A, Wang W, Carmichael L, Kim K, London W, et al. KIR/KIR-ligand genotypes influence clinical response to dinutuximab-based immunotherapy in high-risk neuroblastoma patients. Frontiers in Cancer Immunotherapy Conference (Abstract). New York, NY: The New York Academy of Sciences (2017).
22. Erbe A, Wang W, Grzywacz B, Ranheim E, Hank J, Kim K, et al. Rituximab response in follicular lymphoma: contributions from KIR 2DS1 and HLA-C. Society for Immunotherapy of Cancer Annual Conference (abstract): Journal for ImmunoTherapy of Cancer. Maryland: National Harbor (2013).
23. Grzywacz B, Erbe A, Wang W, Ranheim E, Hank J, Kim K, et al. Specific KIR and HLA genotypes affect outcomes of single-agent anti-CD20 immunotherapy of follicular lymphoma. ASH Annual Meeting (Abstract): Blood. New Orleans, LA (2013).
24. Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol (2002) 20:853–85. doi:10.1146/annurev.immunol.20.100301.064812
25. Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol (1997) 158(11):5237–41.
26. O’Connor GM, Vivian JP, Widjaja JM, Bridgeman JS, Gostick E, Lafont BA, et al. Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J Immunol (2014) 192(6):2875–84. doi:10.4049/jimmunol.1303142
27. Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol (2008) 181(9):6293–300. doi:10.4049/jimmunol.181.9.6293
28. Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, et al. Cutting edge: allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol (2007) 178(1):33–7. doi:10.4049/jimmunol.178.1.33
29. Saunders PM, Pymm P, Pietra G, Hughes VA, Hitchen C, O’Connor GM, et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med (2016) 213(5):791–807. doi:10.1084/jem.20152023
30. Alves LG, Rajalingam R, Canavez F. A novel real-time PCR method for KIR genotyping. Tissue Antigens (2009) 73(2):188–91. doi:10.1111/j.1399-0039.2008.01184.x
31. Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens (2007) 70(5):415–22. doi:10.1111/j.1399-0039.2007.00923.x
32. Sondel PM, Hank JA. Antibody-directed, effector cell-mediated tumor destruction. Hematol Oncol Clin North Am (2001) 15(4):703–21. doi:10.1016/S0889-8588(05)70243-4
33. Weiskopf K, Weissman IL. Macrophages are critical effectors of antibody therapies for cancer. MAbs (2015) 7(2):303–10. doi:10.1080/19420862.2015.1011450
34. Wang W, Erbe AK, Hank JA, Morris ZS, Sondel PM. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol (2015) 6:368. doi:10.3389/fimmu.2015.00368
35. Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat Rev Immunol (2004) 4(3):190–8. doi:10.1038/nri1306
36. Patterson S, Chaidos A, Neville DC, Poggi A, Butters TD, Roberts IA, et al. Human invariant NKT cells display alloreactivity instructed by invariant TCR-CD1d interaction and killer Ig receptors. J Immunol (2008) 181(5):3268–76. doi:10.4049/jimmunol.181.5.3268
37. Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol (2013) 6:1. doi:10.1186/1756-8722-6-1
38. Erbe AK, Wang W, Goldberg J, Gallenberger M, Kim K, Carmichael L, et al. FCGR polymorphisms influence response to IL2 in metastatic renal cell carcinoma. Clin Cancer Res (2017) 23(9):2159–68. doi:10.1158/1078-0432.CCR-16-1874
39. Wang W, Erbe AK, Gallenberger M, Kim K, Carmichael L, Hess D, et al. Killer immunoglobulin-like receptor (KIR) and KIR-ligand genotype do not correlate with clinical outcome of renal cell carcinoma patients receiving high-dose IL2. Cancer Immunol Immunother (2016) 65(12):1523–32. doi:10.1007/s00262-016-1904-8
40. Carlotti E, Palumbo GA, Oldani E, Tibullo D, Salmoiraghi S, Rossi A, et al. FcgammaRIIIA and FcgammaRIIA polymorphisms do not predict clinical outcome of follicular non-Hodgkin’s lymphoma patients treated with sequential CHOP and rituximab. Haematologica (2007) 92(8):1127–30. doi:10.3324/haematol.11288
41. Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood (2002) 99(3):754–8. doi:10.1182/blood.V99.3.754
42. Weng WK, Weng WK, Levy R. Immunoglobulin G Fc receptor polymorphisms do not correlate with response to chemotherapy or clinical course in patients with follicular lymphoma. Leuk Lymphoma (2009) 50(9):1494–500. doi:10.1080/10428190903128660
43. Kenkre VP, Hong F, Cerhan JR, Lewis M, Sullivan L, Williams ME, et al. Fc gamma receptor 3A and 2A polymorphisms do not predict response to rituximab in follicular lymphoma. Clin Cancer Res (2016) 22(4):821–6. doi:10.1158/1078-0432.CCR-15-1848
44. Cheung NK, Sowers R, Vickers AJ, Cheung IY, Kushner BH, Gorlick R. FCGR2A polymorphism is correlated with clinical outcome after immunotherapy of neuroblastoma with anti-GD2 antibody and granulocyte macrophage colony-stimulating factor. J Clin Oncol (2006) 24(18):2885–90. doi:10.1200/JCO.2005.04.6011
45. Lode HN, Troschke-Meurer S, Valteau-Couanet D, Garaventa A, Gray J, Castel V, et al. Correlation of killer-cell Ig like receptor (KIR) haplotypes and Fcγ-receptor polymorphisms with survival of high-risk relapsed/refractory neuroblastoma patients treated by long-term infusion of anti-GD2 antibody ch14.18/CHO. ASCO Annual Meeting (abstract): Journal of Clinical Oncology. Chicago, IL (2016).
46. Terszowski G, Klein C, Stern M. KIR/HLA interactions negatively affect rituximab- but not GA101 (obinutuzumab)-induced antibody-dependent cellular cytotoxicity. J Immunol (2014) 192(12):5618–24. doi:10.4049/jimmunol.1400288
47. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet (2007) 39(6):733–40. doi:10.1038/ng2035
48. Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med (1994) 180(4):1235–42. doi:10.1084/jem.180.4.1235
49. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med (1995) 181(3):1133–44. doi:10.1084/jem.181.3.1133
50. Saunders PM, Vivian JP, Baschuk N, Beddoe T, Widjaja J, O’Connor GM, et al. The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope. J Immunol (2015) 194(2):781–9. doi:10.4049/jimmunol.1402542
51. Vivian JP, Duncan RC, Berry R, O’Connor GM, Reid HH, Beddoe T, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature (2011) 479(7373):401–5. doi:10.1038/nature10517
52. Luque I, Solana R, Galiani MD, Gonzalez R, Garcia F, Lopez de Castro JA, et al. Threonine 80 on HLA-B27 confers protection against lysis by a group of natural killer clones. Eur J Immunol (1996) 26(8):1974–7. doi:10.1002/eji.1830260845
53. Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res (2009) 19(5):757–69. doi:10.1101/gr.085738.108
54. Rojo S, Wagtmann N, Long EO. Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol (1997) 27(2):568–71. doi:10.1002/eji.1830270231
55. Foley BA, De Santis D, Van Beelen E, Lathbury LJ, Christiansen FT, Witt CS. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood (2008) 112(2):435–43. doi:10.1182/blood-2008-01-132902
56. Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol (2005) 175(8):5222–9. doi:10.4049/jimmunol.175.8.5222
57. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med (2006) 203(3):633–45. doi:10.1084/jem.20051884
58. Hughes AL. Natural selection and the diversification of vertebrate immune effectors. Immunol Rev (2002) 190:161–8. doi:10.1034/j.1600-065X.2002.19012.x
59. Selvakumar A, Steffens U, Dupont B. Polymorphism and domain variability of human killer cell inhibitory receptors. Immunol Rev (1997) 155:183–96. doi:10.1111/j.1600-065X.1997.tb00951.x
60. Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol (2002) 168(5):2307–15. doi:10.4049/jimmunol.168.5.2307
61. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res (2015) 43(Database issue):D423–31. doi:10.1093/nar/gku1161
62. Morvan M, Willem C, Gagne K, Kerdudou N, David G, Sebille V, et al. Phenotypic and functional analyses of KIR3DL1+ and KIR3DS1+ NK cell subsets demonstrate differential regulation by Bw4 molecules and induced KIR3DS1 expression on stimulated NK cells. J Immunol (2009) 182(11):6727–35. doi:10.4049/jimmunol.0900212
63. Jiang Y, Chen O, Cui C, Zhao B, Han X, Zhang Z, et al. KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors. BMC Infect Dis (2013) 13:405. doi:10.1186/1471-2334-13-405
64. Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet (2002) 31(4):429–34. doi:10.1038/ng934
Keywords: KIR, KIR-ligand, HLA-Bw4, HLA, MHC class I, natural killer cells, cancer immunotherapy
Citation: Erbe AK, Wang W, Reville PK, Carmichael L, Kim K, Mendonca EA, Song Y, Hank JA, London WB, Naranjo A, Hong F, Hogarty MD, Maris JM, Park JR, Ozkaynak MF, Miller JS, Gilman AL, Kahl B, Yu AL and Sondel PM (2017) HLA-Bw4-I-80 Isoform Differentially Influences Clinical Outcome As Compared to HLA-Bw4-T-80 and HLA-A-Bw4 Isoforms in Rituximab or Dinutuximab-Based Cancer Immunotherapy. Front. Immunol. 8:675. doi: 10.3389/fimmu.2017.00675
Received: 01 March 2017; Accepted: 24 May 2017;
Published: 12 June 2017
Edited by:
Gianfranco Pittari, Hamad Medical Corporation, QatarReviewed by:
Daniel Olive, Institut national de la santé et de la recherche médicale (INSERM), FranceRaquel Tarazona, University of Extremadura, Spain
Alessandro Moretta, Università di Genova, Italy
Copyright: © 2017 Erbe, Wang, Reville, Carmichael, Kim, Mendonca, Song, Hank, London, Naranjo, Hong, Hogarty, Maris, Park, Ozkaynak, Miller, Gilman, Kahl, Yu and Sondel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul M. Sondel, cG1zb25kZWxAaHVtb25jLndpc2MuZWR1
†Co-first authors.
 Amy K. Erbe
Amy K. Erbe Wei Wang
Wei Wang Patrick K. Reville
Patrick K. Reville Lakeesha Carmichael2
Lakeesha Carmichael2 Jacquelyn A. Hank
Jacquelyn A. Hank Arlene Naranjo
Arlene Naranjo Michael D. Hogarty
Michael D. Hogarty M. F. Ozkaynak
M. F. Ozkaynak Jeffrey S. Miller
Jeffrey S. Miller Paul M. Sondel
Paul M. Sondel