- College of veterinary medicine, Nanjing Agricultural University, Nanjing, China
The intestine is home to trillions of microorganisms, and the vast diversity within this gut microbiota exists in a balanced state to protect the intestinal mucosal barrier. Research into the association of the intestinal microbiota with health and disease (including diet, nutrition, obesity, inflammatory bowel disease, and cancer) continues to expand, with the field advancing at a rapid rate. Intestinal stem cells (ISCs) are the fundamental component of the mucosal barrier; they undergo continuous proliferation to replace the epithelium, which is also intimately involved in intestinal diseases. The intestinal microbiota, such as Lactobacillus, communicates with ISCs both directly and indirectly to regulate the proliferation and differentiation of ISCs. Moreover, Salmonella infection significantly decreased the expression of intestinal stem cell markers Lgr5 and Bmi1. However, the detailed interaction of intestinal microbiota and ISCs are still unclear. This review considers the progress of research on the model and niches of ISCs, as well as the complex interplay between the gut microbiota and ISCs, which will be crucial for explaining the mechanisms of intestinal diseases related to imbalances in the intestinal microbiota and ISCs.
Introduction
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is the important cause of gastrointestinal disease (1). Although the precise etiology of IBD remains unclear and controversial, the intestinal microbiota and the integrity of mucosal epithelial function have been demonstrated to play key roles in its pathogenesis (2, 3). Moreover, diets high in fat and protein but low in fruits and vegetables have been demonstrated to be associated with particular compositions of intestinal microbiota that increase the risk of IBD (4). Moreover, a high-fat diet induces change of intestinal stem cells (ISCs) and enhances the risk for intestinal cancer incidence (5). Unlike other stem cells, ISCs coexist with the intestinal microbiota, which may influence the growth status of the epithelium (6). The intestinal mucosal barrier is maintained by ISCs, located at the base part of the intestinal crypts, play a key role in governing the proliferation and differentiation of the intestinal epithelium.
The gastrointestinal tract harbors a diverse community of microorganisms, including bacteria, fungi, and viruses, which are considered as the intestinal microbiota (7). The intestinal microbiota interact closely with ISCs. Although the elucidation of host pathways that regulate ISCs function is progressing, the effects of exogenous factors on ISCs biology are poorly understood. Recent studies demonstrated that ISCs protected itself from butyrate produced by beneficial microbes (8, 9). However, when the intestine tract is damaged, butyrate inhibits the proliferation of ISCs through preventing the intestine tissue from repairing itself after damage or injury (10). However, the detailed mechanism by which the intestinal microbiota regulates ISCs remains unknown. This review will focus on ISCs niches and the regulation of ISCs by the intestinal microbiota.
Progress of Research on ISCs
The intestine is composed of columnar epithelial with glandular invaginations, and the intestinal columnar epithelium is continually shed and replaced by the self-renewal capacity of ISCs. Recently, Lgr5 was identified as an important active ISCs marker of ISCs located at the base of crypt, and Bmi1 was another marker of quiescent ISCs predominantly at the +4 position (11, 12). Detailed cell markers and functions of ISCs were listed in Table 1. Studies have shown that the Lgr + crypt base columnar cells (CBC cells) are rapidly dividing ISCs, which is necessary for gut renewal (13). Conversely, the +4 label-retaining cells (LRCs) are more quiescent, protecting them from the environmental stress. The +4 LRCs are activated during stress of injury, and subsequently produce intestinal progenitor cells to replace the damaged intestinal cells (14). Moreover, CBC cells can also regenerate new +4 LRCs under injury (15). Conversely, the active ISCs could replace the damaged quiescent stem cells under special conditions (15). The intestinal crypt maintains a balance between rapid-cycling, easy-to-damage stem cells, and quiescent +4 LRCs to maintain self-renewal and flexible damage repair (Figure 1).
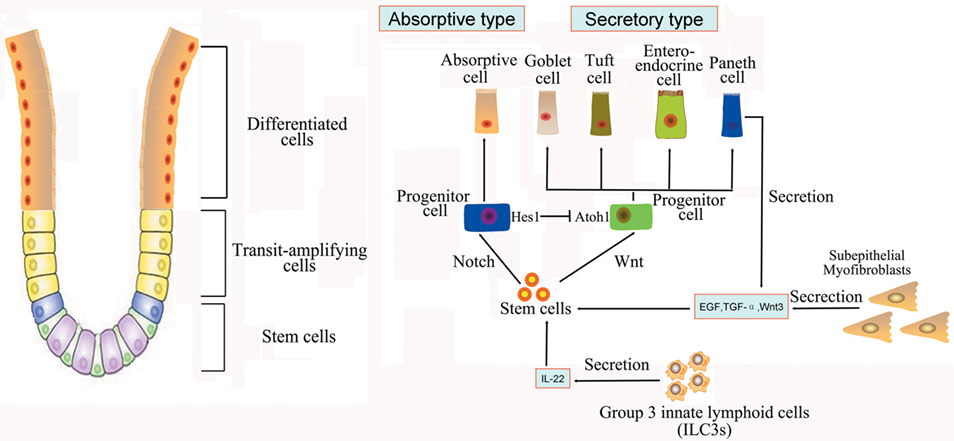
Figure 1. Intestinal stem cells (ISCs) are periodically activated to produce progenitor or transit amplifying cells, which are committed to produce two mature cell lineages: absorptive type (enterocytes) and secretory type (enteroendocrine, goblet, tuft, and Paneth cells). The Paneth cells or subepithelial myofibroblasts could secret epidermal growth factor, TGF-α, Wnt3, and the Notch ligand Dll4, which are essential for the maintenance of ISCs, whereas their maturation depends on Wnt signaling. Innate lymphoid cells (ILC3s) could also activate ISCs regeneration through the secretion of IL-22.
The Establishment of ISCs Models
Studies using intestinal organoids are advancing our understanding of the role of the epithelium in intestinal physiology and pathophysiology. Intestinal organoids are composed only of epithelial cell types and are thus useful for understanding intestinal epithelial cell function in the absence of other cell types. For example, studies in mouse intestinal organoids have confirmed the importance of Paneth cells in the epithelial barrier at the intestinal organoid level. Paneth cells synthesize and secrete substantial quantities of antimicrobial peptides to modulate the homeostatic balance with colonizing microbiota and innate immune protection from enteric pathogens (34). Unexpectedly, epidermal growth factor (EGF), delta-like 1/4, and Wnt-3 are also expressed in Paneth cells, suggesting that this cell type provides ISCs niche signaling (35).
The new culture condition established by Wang et al. could produce highly homogeneous population of ISCs, avoiding heterogeneous cell populations in 3D organoids in vivo (36). Current culture method for ISCs relies on a 3D culture system using Matrige, which is not approved by FDA for clinical use. A new 2D culture system for expansion of ISCs as an intestinal epithelial monolayer and cultured on a thin coat of type I collagen or laminin instead of Matrigel was established (37). Moreover, current models of intestinal organoids lack enteric nerves and immune cells, and one goal is to add these components to develop a more complex in vitro intestinal model using coculture approaches. Long-term culture of the human small intestine epithelium requires the presence of subepithelial myofibroblasts, even when exposed to Wnt3a-containing media (38). Most recently, reports have demonstrated the successful regeneration of the human small intestine from collagenase-digested “organoid units” that contain Lgr5 + ISCs and mesenchyme. Group 3 innate lymphoid cells (ILC3s) have also been shown to be important for maintaining ISCs proliferation (39) (Figure 2).
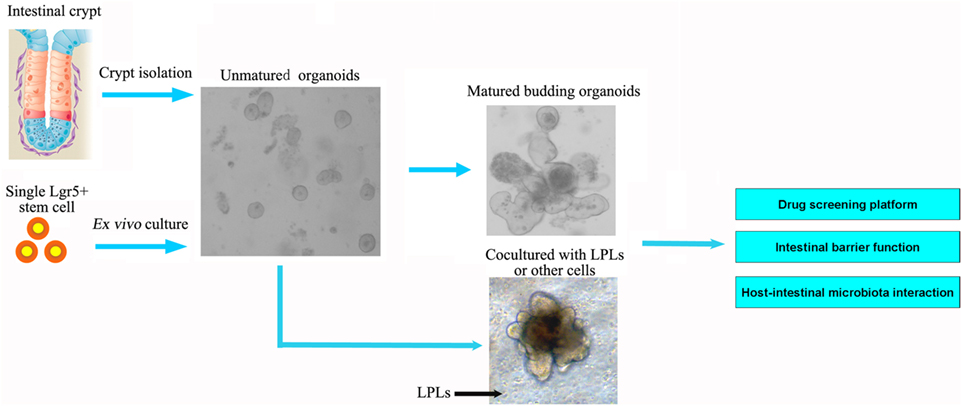
Figure 2. Intestinal organoids, the ex vivo culture systems, are ideal intestinal structure models to explore the interaction between intestinal microbiota and epithelial cells, as well as for drug screening and intestinal barrier function. The organoids could be established from the isolated crypt from the intestine or single intestinal stem cell. The current models of intestinal organoids lack enteric nerves and immune cells. In future, the dendritic cells or lamina propria lymphocytes (LPLs) could be cocultured in vitro to improve the model further.
The intestinal organoids containing ISCs is a promising model to explore the interaction between intestinal microbiota and intestinal mucosa. Salmonella could infect the intestinal organoids, and the infection also significantly decreased the expression of intestinal stem cell marker Lgr5 and Bmi 1 (40, 41). Intestinal organoids could not only be infected by rotavirus, and infected organoids are also capable of producing infectious rotavirus particles (42, 43). The human noroviruses (HuNoVs) have been successfully cultivated in enterocytes in stem cell-derived, non-transformed human intestinal enteroid monolayer cultures, and the Replication of HuNoV Replication occurred in a bile dose-dependent manner (44).
Research Regarding the ISCs Niche
The activity of ISCs is tightly regulated by several niche-signaling pathways to balance the intestinal homeostasis under physiological and pathological stimulation. The Wnt signaling has been identified as the most indispensable pathways for ISCs. Recently, study demonstrated that Wnt3 produced specifically by Paneth cells shapes the concentration gradient in the intestinal crypt, and the ISCs membrane constitutes a reservoir for Wnt proteins (45). R-spondin-1 cooperates with Wnt3 pathway activation through the modulation of Rnf43 (46). Furthermore, ASCL2 regulated downstream of the Wnt pathway has also been shown to be ISCs-specific genes (47).
Besides Wnt signaling, the Notch pathway is another indispensable pathway in regulating the proliferation and differentiation of ISCs (48). Notch1 and Notch2 are two indispensable receptors for maintaining the normal proliferation and differentiation of ISCs in the intestine (49, 50). Loss of the ligands Dll1 and Dll4 could induce the silencing of Notch activation in intestinal epithelial cells (51). Activation of Notch pathway could stimulate ISCs differentiation into absorptive cell lineages, while cis-inhibition of Notch directs ISCs toward secretory lineage cells, such as goblet cells, enteroendocrine cells, Paneth cells, or tuft cells (48). Recent in vitro and in vivo models have suggested that Notch suppression reduces the ratio of BMI1+/LGR5+ ISCs, while Notch stimulation increases this ratio. Furthermore, Notch signaling can activate asymmetric division after intestinal inflammation (52).
BMP signaling is also required for the maintenance of ISCs replication and the terminal differentiation of intestinal cells. Both epithelial and mesenchymal cells could produce BMP ligands. BMP signaling can suppress Wnt signaling to ensure the appropriate balance of epithelial stem cell self-renewal (53). BMP signaling controls the terminal differentiation of the intestinal secretory cell lineage (51). However, BMP2 inhibits epithelial cell growth in the colon by promoting apoptosis and differentiation (52). Several other signal pathways, such as EGF and Hippo signaling pathway also take part in the formation of ISCs niches (54).
Intestinal Mucosa Injury with Intestinal Microbiota Dysbiosis
In the physiological state, the intestinal microbiota either has direct bactericidal effects or inhibits the adherence and invasion of pathogens to the gut mucosa (55). However, intestinal microbiota dysbiosis may facilitate the adhesion of pathogens that may be associated with irritable bowel syndrome (IBS) symptoms (56). The exact cause of IBS is also unknown and is thought to be multifactorial. Variation in the gut microbiota is thought to be complicit in the low-grade intestinal inflammation associated with this syndrome (57). Alterations in the microbiota composition in IBS patients may aggravate the development of IBS symptoms (58). The relative abundance of the Firmicutes population may be greatly reduced in IBD patients, which is of particular interest because these bacteria are known producers of important short-chain fatty acids, which have potent anti-inflammatory properties (59). The defects in ISCs differentiation to Paneth cells or goblet cells are always observed in IBD, which results in luminal microbe invasion of the mucosa and inflammation (60).
Inoculation of UC patients with feces from healthy population, who were extensively screened for parasites and bacterial pathogens, relieved the severe and recurrent inflammatory symptoms within 1 week, and induce complete reversal of inflammatory symptoms in 4 months after the fecal transfer in UC patients, which indicated that infusion of healthy donor human intestinal flora can reverse UC (61). The promise of intestinal stem cell biology lies in the ability of these remarkable cells to give rise to more differentiated intestinal cell types that can repair damaged or diseased tissues. Several therapeutic approaches, including intestinal organoids, are currently being explored as a possible treatment for intestinal disease (62–64). Moreover, recent studies also demonstrated that gut–microbe interactions are also involved in determination of ISCs activity through Janus kinase–signal transducers and activators of transcription (JAK–STAT) pathway, which indicated modulation intestinal microbiota could also stimulate the ISCs proliferation to treat intestinal injury (65–67).
Effects of the Intestinal Microbiota on ISCs
Regulation of the Intestinal Microbiota by Wnt and Notch Signals through Pattern Recognition Receptors (PRRs)
The intestinal epithelium recognizes bacterial components through PRRs and communicates with the resident luminal bacteria (68). The innate immune system senses the pathogenic invasion or epithelium injury via toll-like receptors (TLRs) and nod-like receptors and provide immediate responses (69). Recent evidence suggests the existence of cross talk among the Wnt and Notch pathways, TLR signaling, and the microbiota (70). Previous study demonstrated that, in alveolar epithelial cells, the Wnt/β-catenin is a negative feedback loop to repress TLR-triggered inflammatory responses (71). TLR signaling has been shown to alter intestinal homeostasis and to affect the proliferation and apoptosis rates in the crypt (72). It has been shown that ISCs also express TLR4, and the direct activation of TLR4 on ISCs, especially Lgr5-positive ISCs, regulates their ability to proliferate in intestinal crypts. TLR4 suppressed Wnt signaling, decreased activation of the Wnt receptor LRP6, and blocked the protective effect of the Wnt3a ligand (73). Epithelial differentiation into goblet cells was increased upon inhibiting the Notch signaling in the intestine, and the Notch signaling could also be modulated by TLR4 (74, 75).
Within the intestinal crypt, Lgr5+ stem cells constitutively express much higher levels of the cytosolic innate immune sensor Nod2 than do Paneth cells (76). Stimulation of Nod2 by its bona fide agonist, muramyl dipeptide, a peptidoglycan motif common to all bacteria, triggers stem cell survival, which leads to strong cytoprotection against oxidative stress-mediated cell death. Thus, gut epithelium restitution is Nod2 dependent and triggered by the presence of microbiota-derived molecules (76).
The Effect of Reactive Oxygen Species (ROS) Produced by the Intestinal Microbiota on the Regulation of ISCs
The interaction between gut and intestinal microbiota is critical for the ISCs proliferation and differentiation, as well as the modulation of epithelium regeneration. However, the detailed mechanism of ISCs activation after microbe-induced issue damage is currently unknown. ROS, traditionally viewed as toxic, is now clearly recognized as a key modulator in all kinds of biological processes. Recent study has demonstrated that intestinal epithelia contacted by enteric commensal bacteria rapidly generate ROS, and physiologically generated ROS acts as signaling molecules to mediate increased cellular proliferation and motility and to modulate innate immune signaling (77, 78) (Figure 3). However, high levels of ROS during enteric infections likely act indiscriminately against both commensals and pathogens (65, 79). S. Typhimurium infects and damages the intestinal epithelial, then promotes migration of neutrophils that produce ROS, which facilitate conversion of S2O32− generated by commensal bacteria, into S4O62− (80, 81). Commensal Lactobacilli stimulates ROS production via Nox rather than Duox, thereby activating ISCs in Drosophila and mice under physiological status (82, 83). Recent studies demonstrated that redox homeostasis is critical in the regulation of stem cell differentiation and ROS specifically modulate the stem cell self-renewal (84). Many molecules involved in ROS-regulated stem cell self-renewal were redox sensors, which were found to be modified at redox-active cysteine residues (78). The Wnt and Notch signaling pathways can also be affected by ROS, which then influence the proliferation of ISCs (85). However, we are still not clear whether ROS act as direct inducers of ISCs signaling or simply cause epithelial damage that signals to ISCs.
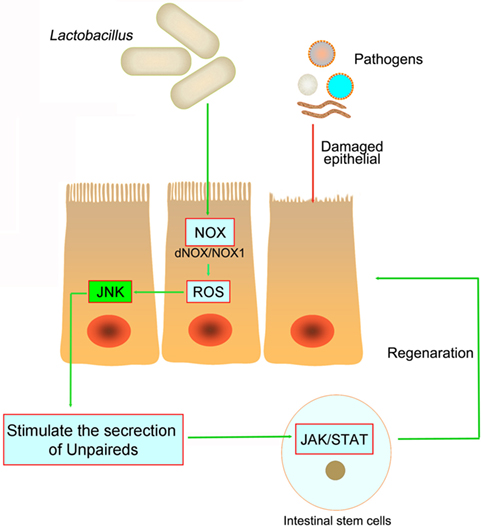
Figure 3. The pathogens in the intestine damage the intestinal epithelial and cause inflammation. However, both in Drosophila and mice, Lactobacillus could stimulate the intestinal epithelial to produce reactive oxygen species (ROS), and then stimulate JNK signaling and increase unpaired expression, thereby stimulating intestinal stem cells proliferation and epithelial turnover or regeneration.
The Effect of the Intestinal Microbiota on Paneth Cells
Paneth cells, located at the base of the crypts in the small intestine, are highly specialized secretory cells. As an important source of antimicrobial peptides, Paneth cells have an antimicrobial function and regulate the intestinal microbiome by secreting bactericidal proteins such as α-defensins and lysozyme (34). Salmonella infection could stimulate the expansion of the Paneth cells population with increased expression of MyD88 in Paneth cells, which is sufficient to limit Salmonella penetration across the mucosal barrier (86, 87).
Recent study also demonstrated that Paneth cells are important for the differentiation and proliferation of ISCs, which are interspersed between Paneth cells (35). Paneth cells secret EGF, TGF-α, Wnt3, and the Notch ligand Dll4, which are essential for the maintenance of ISCs, whereas their maturation depends on Wnt signaling (88). Coculturing of ISCs with Paneth cells could markedly stimulate the formation of intestinal organoids in vitro (35). Crypt cells do not grow ex vivo after the inducible deletion of transcription factor Math1 (Atoh1). However, the complete loss of Paneth cells did not damage the intestinal crypt structure and maintain the physiological proliferation of ISCs in vivo, which implied the underlying mucosal cells may act as a potential ISCs niche (89).
Paneth cells are an initial source of IL-1β signaling during early infection with pathogens, causing gut inflammation. However, intestinal inflammation can be controlled by treatment with Lactobacillus plantarum via the reversal of IL-1β signaling (90). Moreover, the release of antimicrobial products by Paneth cells was controlled by IFN-γ (91). Recently, Paneth cells could augment stem cell function in response to caloric restriction. Calorie intake could regulate the mTORC1 pathway in Paneth cells and affect the ISCs niche (92). Paneth cells are able to directly sense commensal gut bacteria. The role of Paneth cells as critical mediators of microbe-ISC interactions is critical for ISCs, and this topic deserves further exploration.
The Relationship between Crypt-Specific Core Microbiota (CSCM) and ISCs
The microbiota provides continuous stimulation to the intestinal epithelial and affects the ISCs differentiation and proliferation. Recent studies further demonstrated that several signals induced by intestine–microbe interactions are involved in the determination of ISCs activity (93). A particular CSCM was found to survive in the colonic crypt environment (93). The CSCM could prevent the proliferation intestinal pathogens and provide optimal signaling to ISCs (Figure 4). Several particular species, such as Acinetobacter genus, may be evolutionarily selected because they provide advantage to the intestinal epithelial, probably through the expression of particular microbe-associated molecular patterns or the production of specific metabolites to maintain crypt homeostasis (6).
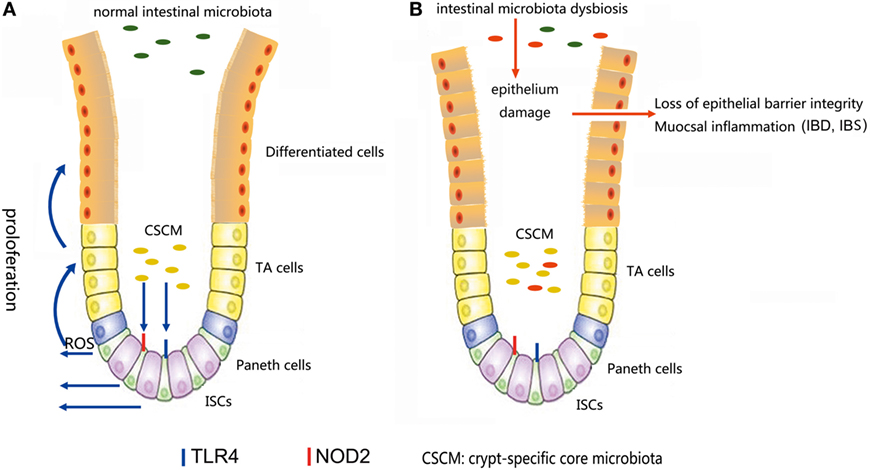
Figure 4. (A) In a normal intestinal microbiota environment, the microbiota, especially the crypt-specific core microbiota (CSCM), survive in the crypt, where they can stimulate the proliferation of intestinal stem cells (ISCs) and induce their differentiation to repair the intestinal epithelium via the TLR4, NOD2, or reactive oxygen species (ROS) signaling pathway to maintain the integrity of the mucosal barrier. (B) Under the pressure of microbiota dysbiosis, the composition of the microbiota and the CSCM will change and affect the stem cell niches, thus hindering the physiological processes regulated by stem cells, inducing epithelial damage and causing mucosal inflammation.
The ISCs Development under Germ-free Conditions
Mammalian intestines are similar in structure and function with Drosophila, but the gut microbiota of Drosophila is composed of only 5–10 bacterial species, thus making it an ideal model to explore the intestine–microbe interactions (94, 95). Comparative studies of germ-free and conventionally reared Drosophila demonstrated that intestinal microbiota alters intestinal homeostasis, intestinal metabolism, and digestive enzymes expression (95, 96). Furthermore, the intestinal microbiota affects the ISCs activity by the modulation of nutrition metabolism. The damaging effects of a lack of microbiota are more evident in old flies, as the intestinal mucosa of germ-free flies was less over-proliferation and misdifferentiation than conventionally reared flies (97).
In Drosophila larvae, the intestinal microbiota promotes growth in nutrient-scarce conditions, and the intestinal peptidase expression and proteolytic activity were recovered after re-association with L. plantarum alone, thus leading to increased digestive abilities (98). Lactobacilli modulate ISCs and stimulate gut epithelium proliferation via the Nox-mediated generation of ROS (79, 83). The intestinal microbiota can become imbalanced after an injury, and probiotics can help the gut flora to recover and restore balance in the gut (99). Unlike the normal microbiota, Pseudomonas entomophila and P. aeruginosa cause the epithelial cell loss in the midgut and induce apoptosis in intestinal epithelial cells, resulting in ISCs over-proliferation (100).
The intestinal microbiota promotes substantial changes in intestinal morphology, including villus structure, crypt depth, stem cell proliferation, and maturation of mucosa-associated lymphoid tissues. In the absence of bacteria, intestinal crypts are less deep and contain fewer proliferating stem cells (101). During the suckling period in mice, extensive dynamic epigenetic changes are observed in ISCs, and the postnatal DNA methylation increased at 3′ CpG islands (CGIs) are responsible for intestinal maturation. Moreover, the DNA methyltransferases, Dnmt1 is a critical regulator of postnatal epigenetic changes in ISCs. However, the postnatal 3′ CGI methylation and associated gene activation in ISCs are significantly altered by germ-free conditions (102).
Future Possibilities
Most recently, IL-22 was shown to be important for maintaining the proliferation of ISCs. Previous studies have demonstrated that Paneth cells or intestinal subepithelial myofibroblasts can secrete Wnt and R-spondin-1 to regulate ISCs function and induce epithelium regeneration. However, the most recent reports have shown that the immune system can support the intestinal epithelium, activating ISCs to promote regeneration through IL-22 secretion by ILC3s (39). Furthermore, the intestinal microbiota can modulate the mucosal immune response, including the secretion of IL-22. However, whether the intestinal microbiota can stimulate ISCs proliferation through IL-22 secretion remains unknown. This phenomenon opens up the possibility of exploring the interaction between the intestinal microbiota and ISCs.
Intestinal organoids containing ISCs transplantation is a promising therapy method for cure intestinal inflammation. Single Lgr5 + ISC-derived colonoids transplantation accelerated the recovery of epithelial barrier function and reversal of inflammation in the DSS colitis model (103). The regulation effects of intestinal microbiota on ISCs need to be further explored for future application of intestinal microbiota to prevent intestinal inflammation. Several challenges, such as the difference between rodent model and human systems, efficient protocol to enrich ISCs, ensuring the safety and efficacy of ISCs-based products, need to be solved in the further.
Author Contributions
QH and LY contributed equally, responsible for the literature search, figures, study design, and data collection; LH is responsible for data analysis of table design; QY is responsible for the conception and design of the study, drafting the article, and final approval of the version to be submitted.
Conflict of Interest Statement
There are no potential conflicts (financial, professional, or personal) that are relevant to the manuscript.
Funding
This study was supported by National Basic Research Program of China (973 program 2013CB127302), the Fundamental Research Funds for the Central Universities (KJQN201613), National Natural Science Foundation of China (31502024), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
1. Matricon J, Barnich N, Ardid D. Immunopathogenesis of inflammatory bowel disease. Self Nonself (2010) 1(4):299–309. doi:10.4161/self.1.4.13560
2. Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol (2014) 20(5):1192–210. doi:10.3748/wjg.v20.i5.1192
3. Kerman DH, Deshpande AR. Gut microbiota and inflammatory bowel disease: the role of antibiotics in disease management. Postgrad Med (2014) 126(4):7–19. doi:10.3810/pgm.2014.07.2779
4. Graf D, Di Cagno R, Fak F, Flint HJ, Nyman M, Saarela M, et al. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis (2015) 26:26164. doi:10.3402/mehd.v26.26164
5. Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature (2016) 531(7592):53–8. doi:10.1038/nature17173
6. Nigro G, Sansonetti PJ. Microbiota and gut stem cells cross-talks: a new view of epithelial homeostasis. Curr Stem Cell Rep (2015) 1:48–52. doi:10.1007/s40778-014-0005-x
7. Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet (2012) 13:151–70. doi:10.1146/annurev-genom-090711-163814
8. Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, et al. The colonic crypt protects stem cells from microbiota-derived metabolites (vol 165, pg 1708, 2016). Cell (2016) 167(4):1137–1137. doi:10.1016/j.cell.2016.10.034
9. Uchiyama K, Sakiyama T, Hasebe T, Musch MW, Miyoshi H, Nakagawa Y, et al. Butyrate and bioactive proteolytic form of Wnt-5a regulate colonic epithelial proliferation and spatial development. Sci Rep (2016) 6:32094. doi:10.1038/srep32094
10. Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell (2016) 165(7):1708–20. doi:10.1016/j.cell.2016.05.018
11. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature (2007) 449(7165):1003–7. doi:10.1038/nature06196
12. de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev (2014) 28(4):305–16. doi:10.1101/gad.235473.113
13. Bloemendaal AL, Buchs NC, George BD, Guy RJ. Intestinal stem cells and intestinal homeostasis in health and in inflammation: a review. Surgery (2016) 159(5):1237–48. doi:10.1016/j.surg.2016.01.014
14. Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature (2011) 478(7368):255–9. doi:10.1038/nature10408
15. Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science (2010) 327(5965):542–5. doi:10.1126/science.1180794
16. Bastide P, Darido C, Pannequiri J, Kist R, Robine S, Marty-Double C, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol (2007) 178(4):635–48. doi:10.1083/jcb.200704152
17. Mori-Akiyama Y, Van den Born M, Van Es J, Hamilton SR, Clevers H, De Crombrugghe B. Sox9 is required for the differentiation of Paneth cells and regulates intestinal epithelial cell proliferation. Gastroenterology (2007) 132(4):A381–381. doi:10.1053/j.gastro.2007.05.020
18. Jubb AM, Chalasani S, Frantz GD, Smits R, Grabsch HI, Kavi V, et al. Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is upregulated in intestinal neoplasia. Oncogene (2006) 25(24):3445–57. doi:10.1038/sj.onc.1209382
19. Schuijers J, Junker JP, Mokry M, Hatzis P, Koo BK, Sasselli V, et al. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell (2015) 16(2):158–70. doi:10.1016/j.stem.2014.12.006
20. Cambuli FM, Rezza A, Nadjar J, Plateroti M. Brief report: musashi1-eGFP mice, a new tool for differential isolation of the intestinal stem cell populations. Stem Cells (2013) 31(10):2273–8. doi:10.1002/stem.1428
21. Nikpour P, Mowla SJ, Forouzandeh-Moghaddam M, Ziaee SA. The stem cell self-renewal gene, Musashi 1, is highly expressed in tumor and non-tumor samples of human bladder. Indian J Cancer (2013) 50(3):214–8. doi:10.4103/0019-509X.118735
22. Carmon KS, Gong X, Lin QS, Thomas A, Liu QY. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A (2011) 108(28):11452–7. doi:10.1073/pnas.1106083108
23. Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep (2011) 12(10):1055–61. doi:10.1038/embor.2011.175
24. Mulzer SR, Brash JL. Identification of plasma-proteins adsorbed to hemodialyzers during clinical use. J Biomed Mater Res (1989) 23(12):1483–504. doi:10.1002/jbm.820231210
25. Van der Flier LG, Haegebarth A, Stange DE, Van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology (2009) 137(1):15–7. doi:10.1053/j.gastro.2009.05.035
26. Takeda N, Jain R, LeBoeuf MR, Wang QH, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science (2011) 334(6061):1420–4. doi:10.1126/science.1213214
27. Katoh H, Yamashita K, Waraya M, Margalit O, Ooki A, Tamaki H, et al. Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia (2012) 14(7):559. doi:10.1593/neo.12330
28. Geserick C, Tejera A, Gonzalez-Suarez E, Klatt P, Blasco MA. Expression of mTert in primary murine cells links the growth-promoting effects of telomerase to transforming growth factor-beta signaling. Oncogene (2006) 25(31):4310–9. doi:10.1038/sj.onc.1209465
29. Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk MEG, Henderson DE, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A (2011) 108(1):179–84. doi:10.1073/pnas.1013004108
30. Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet (2008) 40(7):915–20. doi:10.1038/Ng.165
31. Yu TX, Chen X, Zhang W, Colon D, Shi JD, Napier D, et al. Regulation of the potential marker for intestinal cells, Bmi1, by beta-catenin and the zinc finger protein KLF4 implications for colon cancer. J Biol Chem (2012) 287(6):3760–8. doi:10.1074/jbc.M111.316349
32. May R, Sureban SM, Hoang N, Riehl TE, Lightfoot SA, Ramanujam R, et al. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells (2009) 27(10):2571–9. doi:10.1002/stem.193
33. Sureban SM, May R, Lightfoot SA, Hoskins AB, Lerner M, Brackett DJ, et al. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res (2011) 71(6):2328–38. doi:10.1158/0008-5472.CAN-10-2738
34. Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol (2013) 75:289–311. doi:10.1146/annurev-physiol-030212-183744
35. Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature (2011) 469(7330):415–8. doi:10.1038/nature09637
36. Wang X, Yamamoto Y, Wilson LH, Zhang T, Howitt BE, Farrow MA, et al. Cloning and variation of ground state intestinal stem cells. Nature (2015) 522(7555):173. doi:10.1038/nature14484
37. Scott A, Rouch JD, Jabaji Z, Khalil HA, Solorzano S, Lewis M, et al. Long-term renewable human intestinal epithelial stem cells as monolayers: a potential for clinical use. J Pediatr Surg (2016) 51(6):995–1000. doi:10.1016/j.jpedsurg.2016.02.074
38. Lahar N, Lei NY, Wang J, Jabaji Z, Tung SC, Joshi V, et al. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One (2011) 6(11):e26898. doi:10.1371/journal.pone.0026898
39. Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature (2015) 528(7583):560–4. doi:10.1038/nature16460
40. Zhang Y-G, Wu S, Xia Y, Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep (2014) 2(9):e12147. doi:10.14814/phy2.12147
41. Forbester JL, Goulding D, Vallier L, Hannan N, Hale C, Pickard D, et al. Interaction of Salmonella enterica serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun (2015) 83(7):2926–34. doi:10.1128/Iai.00161-15
42. Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio (2012) 3(4):e159–112. doi:10.1128/mBio.00159-12
43. Yin YB, Bijvelds M, Dang W, Xu L, van der Eijk AA, Knipping K, et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res (2015) 123:120–31. doi:10.1016/j.antiviral.2015.09.010
44. Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science (2016) 353(6306):1387–93. doi:10.1126/science.aaf5211
45. Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DV, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature (2016) 530(7590):340–3. doi:10.1038/nature16937
46. Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature (2012) 488(7413):665. doi:10.1038/nature11308
47. Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol (2012) 14(1):106–14. doi:10.1038/ncb2384
48. Tantikanjana T, Nasrallah JB. Ligand-mediated cis-inhibition of receptor signaling in the self-incompatibility response of the Brassicaceae. Plant Physiol (2015) 169(2):1141–54. doi:10.1104/pp.15.00572
49. Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol (2005) 6(3):314–22. doi:10.1038/ni1164
50. Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell (2010) 6(3):251–64. doi:10.1016/j.stem.2010.02.001
51. Shimizu H, Okamoto R, Ito G, Fujii S, Nakata T, Suzuki K, et al. Distinct expression patterns of Notch ligands, Dll1 and Dll4, in normal and inflamed mice intestine. PeerJ (2014) 2:e370. doi:10.7717/peerj.370
52. Srinivasan T, Than EB, Bu PC, Tung KL, Chen KY, Augenlicht L, et al. Notch signalling regulates asymmetric division and inter-conversion between lgr5 and bmi1 expressing intestinal stem cells. Sci Rep (2016) 6:26069. doi:10.1038/srep26069
53. He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet (2004) 36(10):1117–21. doi:10.1038/ng1430
54. Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development (2016) 143(20):3639–49. doi:10.1242/dev.133132
55. Kellow JE, Azpiroz F, Delvaux M, Gebhart GF, Mertz HR, Quigley EM, et al. Applied principles of neurogastroenterology: physiology/motility sensation. Gastroenterology (2006) 130(5):1412–20. doi:10.1053/j.gastro.2005.08.061
56. Rinttila T, Lyra A, Krogius-Kurikka L, Palva A. Real-time PCR analysis of enteric pathogens from fecal samples of irritable bowel syndrome subjects. Gut Pathog (2011) 3(1):6. doi:10.1186/1757-4749-3-6
57. Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol (2013) 6(4):295–308. doi:10.1177/1756283X13482996
58. Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol (2011) 60(Pt 6):817–27. doi:10.1099/jmm.0.028126-0
59. Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol (2000) 66(4):1654–61. doi:10.1128/AEM.66.4.1654-1661.2000
60. Gersemann M, Stange EF, Wehkamp J. From intestinal stem cells to inflammatory bowel diseases. World J Gastroenterol (2011) 17(27):3198–203. doi:10.3748/wjg.v17.i27.3198
61. Borody TJ, Warren EF, Leis SM, Surace R, Ashman O, Siarakas S. Bacteriotherapy using fecal flora – toying with human motions. J Clin Gastroenterol (2004) 38(6):475–83. doi:10.1097/01.mcg.0000128988.13808.dc
62. van Deen WK, Oikonomopoulos A, Hommes DW. Stem cell therapy in inflammatory bowel disease: which, when and how? Curr Opin Gastroenterol (2013) 29(4):384–90. doi:10.1097/MOG.0b013e328361f763
63. Qu B, Xin GR, Zhao LX, Xing H, Lian LY, Jiang HY, et al. Testing stem cell therapy in a rat model of inflammatory bowel disease: role of bone marrow stem cells and stem cell factor in mucosal regeneration. PLoS One (2014) 9(10):e107891. doi:10.1371/journal.pone.0107891
64. Irhimeh MR, Cooney J. Management of inflammatory bowel disease using stem cell therapy. Curr Stem Cell Res Ther (2016) 11(1):72–7. doi:10.2174/1574888x10666150728121738
65. Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe (2009) 5(2):200–11. doi:10.1016/j.chom.2009.01.003
66. Cronin SJF, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science (2009) 325(5938):340–3. doi:10.1126/science.1173164
67. Lee WJ. Bacterial-modulated host immunity and stem cell activation for gut homeostasis. Genes Dev (2009) 23(19):2260–5. doi:10.1101/gad.1858709
68. Moossavi S, Zhang H, Sun J, Rezaei N. Host-microbiota interaction and intestinal stem cells in chronic inflammation and colorectal cancer. Expert Rev Clin Immunol (2013) 9(5):409–22. doi:10.1586/eci.13.27
69. Fukata M, Vamadevan AS, Abreu MT. Toll-like receptors (TLRs) and nod-like receptors (NLRs) in inflammatory disorders. Semin Immunol (2009) 21(4):242–53. doi:10.1016/j.smim.2009.06.005
70. Bansal K, Trinath J, Chakravortty D, Patil SA, Balaji KN. Pathogen-specific TLR2 protein activation programs macrophages to induce Wnt-beta-catenin signaling. J Biol Chem (2011) 286(42):37032–44. doi:10.1074/jbc.M111.260414
71. Li Y, Shi J, Yang JL, Ma Y, Cheng L, Zeng J, et al. A Wnt/beta-catenin negative feedback loop represses TLR-triggered inflammatory responses in alveolar epithelial cells. Mol Immunol (2014) 59(2):128–35. doi:10.1016/j.molimm.2014.02.002
72. Hedayat M, Takeda K, Rezaei N. Prophylactic and therapeutic implications of toll-like receptor ligands. Med Res Rev (2012) 32(2):294–325. doi:10.1002/med.20214
73. Yi H, Patel AK, Sodhi CP, Hackam DJ, Hackam AS. Novel role for the innate immune receptor toll-like receptor 4 (TLR4) in the regulation of the Wnt signaling pathway and photoreceptor apoptosis. PLoS One (2012) 7(5):e36560. doi:10.1371/journal.pone.0036560
74. Sodhi CP, Shi XH, Richardson WM, Grant ZS, Shapiro RA, Prindle T Jr, et al. Toll-like receptor-4 inhibits enterocyte proliferation via impaired beta-catenin signaling in necrotizing enterocolitis. Gastroenterology (2010) 138(1):185–96. doi:10.1053/j.gastro.2009.09.045
75. Sodhi CP, Neal MD, Siggers R, Sho S, Ma CR, Branca MF, et al. Intestinal epithelial toll-like receptor 4 regulates goblet cell development and is required for necrotizing enterocolitis in mice. Gastroenterology (2012) 143(3):708–U234. doi:10.1053/j.gastro.2012.05.053
76. Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe (2014) 15(6):792–8. doi:10.1016/j.chom.2014.05.003
77. Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem (2012) 19(10):1519–29. doi:10.2174/092986712799828283
78. Wang K, Zhang T, Dong Q, Nice EC, Huang C, Wei Y. Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis (2013) 4:e537. doi:10.1038/cddis.2013.50
79. Patel PH, Maldera JA, Edgar BA. Stimulating cROSstalk between commensal bacteria and intestinal stem cells. EMBO J (2013) 32(23):3009–10. doi:10.1038/emboj.2013.244
80. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol (2013) 14(7):685–90. doi:10.1038/ni.2608
81. Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature (2016) 535(7610):85–93. doi:10.1038/nature18849
82. Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal (2012) 24(5):981–90. doi:10.1016/j.cellsig.2012.01.008
83. Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J (2013) 32(23):3017–28. doi:10.1038/emboj.2013.224
84. Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J (2007) 26(21):4457–66. doi:10.1038/sj.emboj.7601867
85. Caliceti C, Nigro P, Rizzo P, Ferrari R. ROS, Notch, and Wnt signaling pathways: crosstalk between three major regulators of cardiovascular biology. Biomed Res Int (2014) 2014:318714. doi:10.1155/2014/318714
86. Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A (2008) 105(52):20858–63. doi:10.1073/pnas.0808723105
87. Rodriguez NRM, Eloi MD, Huynh A, Dominguez T, Lam AHC, Carcamo-Molina D, et al. Expansion of Paneth cell population in response to enteric Salmonella enterica serovar Typhimurium infection. Infect Immun (2012) 80(1):266–75. doi:10.1128/Iai.05638-11
88. van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol (2005) 7(4):381–6. doi:10.1038/ncb1240
89. Durand A, Donahue B, Peignon G, Letourneur F, Cagnard N, Slomianny C, et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A (2012) 109(23):8965–70. doi:10.1073/pnas.1201652109
90. Hirao LA, Grishina I, Bourry O, Hu WK, Somrit M, Sankaran-Walters S, et al. Early mucosal sensing of SIV infection by Paneth cells induces IL-1beta production and initiates gut epithelial disruption. PLoS Pathog (2014) 10(8):e1004311. doi:10.1371/journal.ppat.1004311
91. Farin HF, Karthaus WR, Kujala P, Rakhshandehroo M, Schwank G, Vries RG, et al. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-gamma. J Exp Med (2014) 211(7):1393–405. doi:10.1084/jem.20130753
92. Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature (2012) 486(7404):490–5. doi:10.1038/nature11163
93. Pedron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, et al. A crypt-specific core microbiota resides in the mouse colon. MBio (2012) 3(3):e116–112. doi:10.1128/mBio.00116-12
94. Wong CN, Ng P, Douglas AE. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol (2011) 13(7):1889–900. doi:10.1111/j.1462-2920.2011.02511.x
95. Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio (2014) 5(3):e1117–1114. doi:10.1128/mBio.01117-14
96. Erkosar B, Defaye A, Bozonnet N, Puthier D, Royet J, Leulier F. Drosophila microbiota modulates host metabolic gene expression via IMD/NF-kappaB signaling. PLoS One (2014) 9(4):e94729. doi:10.1371/journal.pone.0094729
97. Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, et al. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun (2015) 6:6312. doi:10.1038/ncomms7312
98. Bonfini A, Liu X, Buchon N. From pathogens to microbiota: how Drosophila intestinal stem cells react to gut microbes. Dev Comp Immunol (2016) 64:22–38. doi:10.1016/j.dci.2016.02.008
99. Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science (2016) 351(6275):854–7. doi:10.1126/science.aad8588
100. Apidianakis Y, Pitsouli C, Perrimon N, Rahme L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci U S A (2009) 106(49):20883–8. doi:10.1073/pnas.0911797106
101. Sommer F, Backhed F. The gut microbiota – masters of host development and physiology. Nat Rev Microbiol (2013) 11(4):227–38. doi:10.1038/nrmicro2974
102. Yu DH, Gadkari M, Zhou Q, Yu SY, Gao N, Guan YT, et al. Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol (2015) 16:211. doi:10.1186/s13059-015-0763-5
Keywords: intestinal stem cells, intestinal microbiota, niche, Paneth cell, intestinal epithelium
Citation: Hou Q, Ye L, Huang L and Yu Q (2017) The Research Progress on Intestinal Stem Cells and Its Relationship with Intestinal Microbiota. Front. Immunol. 8:599. doi: 10.3389/fimmu.2017.00599
Received: 28 March 2017; Accepted: 08 May 2017;
Published: 23 May 2017
Edited by:
Kuldeep Dhama, Indian Veterinary Research Institute (IVRI), IndiaReviewed by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranRuchi Tiwari, DUVASU Mathura UP, India
Copyright: © 2017 Hou, Ye, Huang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghua Yu, eXVxaW5naHVhMTk4MUBuamF1LmVkdS5jbg==
†These authors have contributed equally to this work and are co-first authors.
 Qihang Hou†
Qihang Hou† Lulu Huang
Lulu Huang Qinghua Yu
Qinghua Yu