- 1Molecular Infection and Functional Biology Laboratory, Kusuma School of Biological Sciences, Indian Institute of Technology-Delhi, New Delhi, India
- 2School of Life Sciences, Jawaharlal Nehru University, New Delhi, India
- 3Inflammation Biology and Cell Signaling Laboratory, National Institute of Pathology, New Delhi, India
- 4Jamia Hamdard, Institute of Molecular Medicine, New Delhi, India
- 5Dr Reddy’s Institute of Life Sciences, University of Hyderabad Campus, Hyderabad, India
A commentary on
Mycobacterium tuberculosis (M.tb), an obligate slow-growing human pathogen, resides within the macrophage after phagocytosis and develops strategies to escape immune surveillance. It can cause active disease or can persist in a latent stage depending on the host immune responses. The mycobacterial cell wall consists of complex layers of arabinogalactan, peptidoglycan, and unusually long branched mycolic acids that are covalently linked with each other. The cell wall of mycobacteria, containing high proportion of lipids, has 15 times less density of pores in comparison to the outer membrane of Gram-negative bacteria (1). This low density of pores might cause more difficulty in absorption of nutrients and could contribute to slow growth of mycobacteria. The other reasons for slow growth are higher GC content of the promoters, differential orientation of the genes in relation to the direction of replication, low RNA/DNA ratio in growing mycobacteria, and presence of a single ribosomal RNA operon present apart from the oriC. Proteins involved in the formation of the substrate-specific energy-dependent transporters ABC transport systems (ATP-binding cassette) are coded by only 2.5% of the M.tb genome that is very less compared to 5% in case of the Escherichia coli genome (2).
During infection, M.tb targets several host pathways such as induction of glycolytic flux (3), endoplasmic reticulum stress (4, 5), disruption of mitochondrial membrane (6), inhibition of apoptosis (7), induction of necrosis (3), phagosome maturation, suppressing host signaling pathways (8), and regulate autophagy to survive within host cell (9). Inside the granuloma, both the mycobacteria and the macrophages survive under stress conditions because of limitation of nutrients. To persist under such unfavorable conditions, both bacteria and macrophages have to conserve their energy by decreasing metabolic rate to allocate available resources toward the production of dedicated stress management proteins. Stress granules formation is a major adaptive defense mechanism through translation repression for stress survival of host cell infected with mycobacteria (4, 5).
Intracellular mycobacteria are found in different vacuolar compartments in distinct physiological state, gene expression, and survival (10, 11). It has been shown that mycobacterial infection activated phagocytes to secrete different cytokines after triggering several host receptors such as type C lectins such as DC-SIGN (12, 13), NOD/NACHT receptors (14), mannose receptors (15), and toll-like receptor 2 (16). Mincle receptor [macrophage inducible Ca2+-dependent (C-type) lectin] is a calcium-dependent lectin that is a receptor for mycobacterial cord factor, trehalose-6,6’-dimycolate (TDM). Mincle expression on neutrophils is required for TDM infiltration that binds to both the sugar portion of the glycolipid and the hydrocarbon tail (17).
We have reported several key proteins of M.tb that may be functionally important for pathogenesis and survival. Prominent among these are M.tb PE/PPE proteins that have multiple role in terms of providing antigenic variation to the pathogen, acting as a molecular switch toward virulence and altering Th1/Th2 host immune response for survival (18–20), immune quorum sensing (21), etc. Interaction of M.tb virulence factor RipA with chaperone MoxR1 was required for transport through TAT secretion system (22). Inhibition of M.tb chaperonic proteins such as PpiA and PpiB can derail protein folding machinery in M.tb (23) and reticence intracellular bacterial survival through alteration of host cytokine profile (24). PpiB also regulates formation of biofilm and can contribute to drug tolerance. Several M.tb proteins, such as DATIN, modulate host cytokine profile by interacting with TLR-2 (25), Rv2626c induce the production of pro-inflammatory cytokines through NF-κB (26). Rv2430c induces strong B-cell response (27), while Rv2608 induces different humoral and T-cell response in various categories of TB patients (28). Inhibitors of these proteins can help boost host immune system within host and provide an unfavorable environment for M.tb to survive. M.tb ORF Rv1475c encoded aconitase is an iron binding protein that has conserved residues of the iron-responsive class of proteins and binds to iron-responsive elements in case of iron depletion (29). It is one of the several M.tb proteins identified in 30-day infected guinea pig lungs indicating its role in host–pathogen interaction (30).
There are several proteins present in mycobacteria which help in its survival inside host by slowing down growth at the level of replication (31), transcription (32), and translation (33, 34) (Figure 1). A recent report (35) has described different mycobacterial strategies against host immune responses such as manipulation of the TLR responses, host cytokine responses, antigen presentation by MHCs, inhibition of phagolysosomal fusion, and resistance to reactive nitrogen intermediates. The role of toxin antitoxins systems in mycobacterial growth regulation in unfavorable conditions and role of Clp proteases in reactivation of latent bacilli have been described in detail. It has been shown that arrest of protein synthesis induces formation of persisters (36) that may have similar metabolic and physiological state as the dormant bacteria (37). The persisters are drug tolerant non-grower bacteria, genetically similar sibling of drug susceptible bacteria but physiologically resistant (persistent) against various bacterial drugs (38). Comparative genomic analyses revealed genes associated with survival, virulence, antibiotic resistance, and biofilm formation (39). Many of these genes can act alone or in combination with other genes and thus inhibitors against such genes can prove vital in targeting the virulence and survival of M.tb. Drug re-purposing is an emerging strategy where drugs already in clinical use or approved by US FDA for treatment of mental illness, diabetes, malaria, etc. are being tested against some of the pathogen targets described above. Targeting those host cellular pathways that are also commonly utilized by M.tb for its survival is yet another mode of developing new drugs.
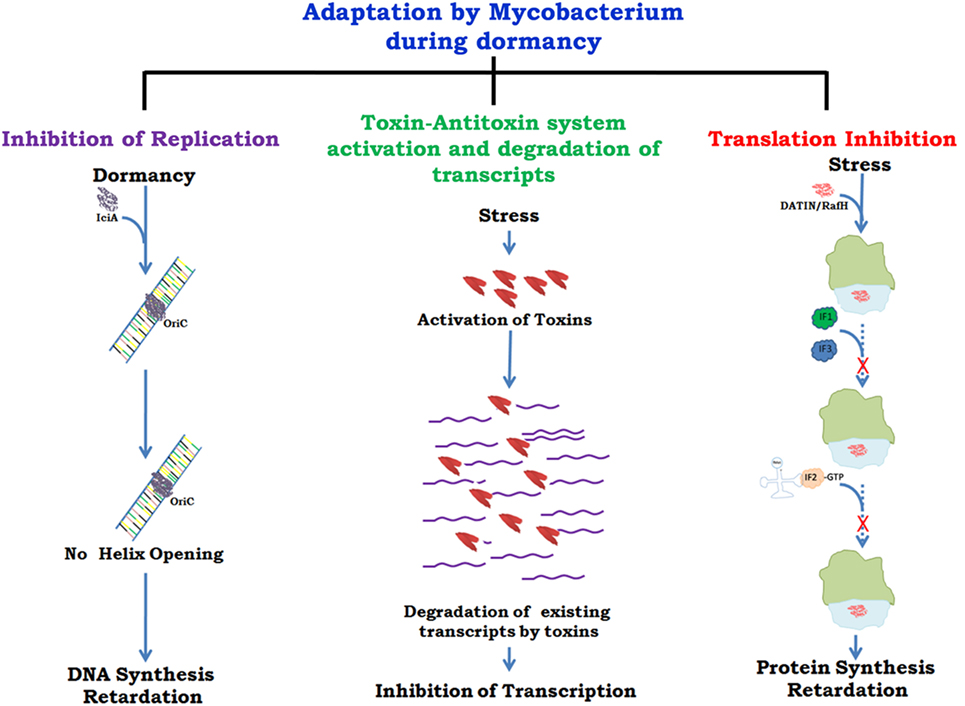
Figure 1. Growth regulation by Mycobacterium for adaptation to stress/dormancy. Mycobacterium tuberculosis (M.tb) IciA (inhibitor of chromosome initiation) binds to the A + T rich oriC region of the M.tb genome and inhibits helix opening resulting in the arrest of chromosomal DNA replication (31). Activated toxin–antitoxin (TA) systems cleave mRNA to shut down metabolic activity (32). Peddireddy et al. (35) have also described the role of TA systems in M.tb and Mycobacterium smegmatis to remain in non-replicating phase that help bacteria in antibiotic tolerance. Highly expressed protein DATIN/RafH of Mycobacterium inhibits translation by binding with the ribosome under conditions of stress (33, 34). Confirmed and putative roles are indicated with continuous and dashed arrows, respectively.
Author Contributions
SH and AK conceived the idea behind this commentary; AK and MR wrote the draft; and NE and SH finalized the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
SH and NE thank the DBT, Ministry of S&T, Government of India, for a Centre of Excellence Grant (BT/PR12817/COE/34/23/2015). SH is a JC Bose National Fellow of the Department of Science and Technology (S&T), Ministry of S&T, Government of India, and a Robert Koch Fellow of the Robert Koch Institute, Berlin, Germany.
References
1. Niederweis M. Mycobacterial porins – new channel proteins in unique outer membranes. Mol Microbiol (2003) 49:1167–77. doi:10.1046/j.1365-2958.2003.03662.x
2. Braibant M, Gilot P, Content J. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol Rev (2000) 24:449–67. doi:10.1111/j.1574-6976.2000.tb00550.x
3. Mehrotra P, Jamwal SV, Saquib N, Sinha N, Siddiqui Z, Manivel V, et al. Pathogenicity of Mycobacterium tuberculosis is expressed by regulating metabolic thresholds of the host macrophage. PLoS Pathog (2014) 10(7):e1004265. doi:10.1371/journal.ppat.1004265
4. Seimon TA, Kim MJ, Blumenthal A, Koo J, Ehrt S, Wainwright H, et al. Induction of ER stress in macrophages of tuberculosis granulomas. PLoS One (2010) 5(9):e12772. doi:10.1371/journal.pone.0012772
5. Lim YJ, Choi JA, Choi HH, Cho SN, Kim HJ, Jo EK, et al. Endoplasmic reticulum stress pathway-mediated apoptosis in macrophages contributes to the survival of Mycobacterium tuberculosis. PLoS One (2011) 6(12):e28531. doi:10.1371/journal.pone.0028531
6. Chen M, Gan H, Remold HG. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J Immunol (2006) 176:3707–16. doi:10.4049/jimmunol.176.6.3707
7. Keane J, Remold HG, Kornfeld H. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J Immunol (2000) 164:2016–20. doi:10.4049/jimmunol.164.4.2016
8. Koul A, Herget T, Klebl B, Ullrich A. Interplay between mycobacteria and host signalling pathways. Nat Rev Microbiol (2004) 2:189–202. doi:10.1038/nrmicro840
9. Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell (2010) 140:731–43. doi:10.1016/j.cell.2010.02.012
10. Rohde KH, Veiga DF, Caldwell S, Balazsi G, Russell DG. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog (2012) 8:e1002769. doi:10.1371/journal.ppat.1002769
11. Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, Bhave D, et al. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog (2014) 10(1):e1003902. doi:10.1371/journal.ppat.1003902
12. Geijtenbeek TB, Van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, Appelmelk B, et al. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med (2003) 197:7–17. doi:10.1084/jem.20021229
13. Tailleux L, Schwartz O, Herrmann JL, Pivert E, Jackson M, Amara A, et al. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med (2003) 197:121–7. doi:10.1084/jem.20021468
14. Ferwerda G, Girardin SE, Kullberg B-J, Le Bourhis L, de Jong DJ, Langenberg DML, et al. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog (2005) 1(3):e34. doi:10.1371/journal.ppat.0010034
15. Kang PB, Azad AK, Torrelles JB, Kaufman TM, Beharka A, Tibesar E, et al. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J Exp Med (2005) 202:987–99. doi:10.1084/jem.20051239
16. Udgata A, Qureshi R, Mukhopadhyay S. Transduction of functionally contrasting signals by two mycobacterial PPE proteins downstream of TLR2 receptors. J Immunol (2016) 197(5):1776–87. doi:10.4049/jimmunol.1501816
17. Feinberg H, Jégouzo SA, Rowntree TJ, Guan Y, Brash MA, Taylor ME, et al. Mechanism for recognition of an unusual mycobacterial glycolipid by the macrophage receptor mincle. J Biol Chem (2013) 288(40):28457–65. doi:10.1074/jbc.M113.497149
18. Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie (2012) 94(1):110–6. doi:10.1016/j.biochi.2011.09.026
19. Mohareer K, Tundup S, Hasnain SE. Transcriptional regulation of Mycobacterium tuberculosis PE/PPE genes: a molecular switch to virulence? J Mol Microbiol Biotechnol (2011) 21(3–4):97–109. doi:10.1159/000329489
20. Khubaib M, Sheikh JA, Pandey S, Srikanth B, Bhuwan M, Khan N, et al. Mycobacterium tuberculosis co-operonic PE32/PPE65 proteins alter host immune responses by hampering Th1 response. Front Microbiol (2016) 7:719. doi:10.3389/fmicb.2016.00719
21. Tundup S, Mohareer K, Hasnain SE. Mycobacterium tuberculosis PE25/PPE41 protein complex induces necrosis in macrophages: role in virulence and disease reactivation? FEBS Open Bio (2014) 4:822–8. doi:10.1016/j.fob.2014.09.001
22. Bhuwan M, Arora N, Sharma A, Khubaib M, Pandey S, Chaudhuri TK, et al. Interaction of Mycobacterium tuberculosis virulence factor ripa with chaperone MoxR1 is required for transport through the TAT secretion system. MBio (2016) 7(2):e02259. doi:10.1128/mBio.02259-15
23. Pandey S, Sharma A, Tripathi D, Kumar A, Khubaib M, Bhuwan M, et al. Mycobacterium tuberculosis peptidyl-prolyl isomerases also exhibit chaperone like activity in-vitro and in-vivo. PLoS One (2016) 11(3):e0150288. doi:10.1371/journal.pone.0150288
24. Pandey S, Tripathi D, Khubaib M, Kumar A, Sheikh JA, Sumanlatha G, et al. Mycobacterium tuberculosis peptidyl-prolyl isomerases are immunogenic, alter cytokine profile and aid in intracellular survival. Front Cell Infect Microbiol (2017) 7:38. doi:10.3389/fcimb.2017.00038
25. Kumar A, Lewin A, Rani PS, Qureshi IA, Devi S, Majid M, et al. Dormancy associated translation inhibitor (DATIN/Rv0079) of Mycobacterium tuberculosis interacts with TLR2 and induces proinflammatory cytokine expression. Cytokine (2013) 64:258–64. doi:10.1016/j.cyto.2013.06.310
26. Bashir N, Kounsar F, Mukhopadhyay S, Hasnain SE. Mycobacterium tuberculosis conserved hypothetical protein rRv2626c modulates macrophage effector functions. Immunology (2010) 130(1):34–45. doi:10.1111/j.1365-2567.2009.03196.x
27. Choudhary RK, Mukhopadhyay S, Chakhaiyar P, Sharma N, Murthy KJ, Katoch VM, et al. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect Immun (2003) 71(11):6338–43. doi:10.1128/IAI.71.11.6338-6343.2003
28. Chakhaiyar P, Nagalakshmi Y, Aruna B, Murthy KJ, Katoch VM, Hasnain SE. Regions of high antigenicity within the hypothetical PPE major polymorphic tandem repeat open-reading frame, Rv2608, show a differential humoral response and a low T cell response in various categories of patients with tuberculosis. J Infect Dis (2004) 190(7):1237–44. doi:10.1086/423938
29. Banerjee S, Nandyala AK, Raviprasad P, Ahmed N, Hasnain SE. Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J Bacteriol (2007) 189:4046–52. doi:10.1128/JB.00026-07
30. Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS One (2010) 5(11):e13938. doi:10.1371/journal.pone.0013938
31. Kumar S, Farhana A, Hasnain SE. In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein. PLoS One (2009) 4:e4139. doi:10.1371/journal.pone.0004139
32. Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet (2009) 5:e1000767. doi:10.1371/journal.pgen.1000767
33. Kumar A, Majid M, Kunisch R, Rani PS, Qureshi IA, Lewin A, et al. Mycobacterium tuberculosis DosR regulon gene Rv0079 encodes a putative, ‘dormancy associated translation inhibitor (DATIN)’. PLoS One (2012) 7:e38709. doi:10.1371/journal.pone.0038709
34. Trauner A, Lougheed KE, Bennett MH, Hingley-Wilson SM, Williams HD. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J Biol Chem (2012) 287:24053–63. doi:10.1074/jbc.M112.364851
35. Peddireddy V, Doddam S, Ahmed N. Mycobacterial dormancy systems and host responses in tuberculosis. Front Immunol (2017) 8:84. doi:10.3389/fimmu.2017.00084
36. Kwan BW, Valenta JA, Benedik MJ, Wood TK. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother (2013) 57:1468–73. doi:10.1128/AAC.02135-12
37. Wood TK, Knabel SJ, Kwan BW. Bacterial persister cell formation and dormancy. Appl Environ Microbiol (2013) 79(23):7116. doi:10.1128/AEM.02636-13
38. Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers? Nat Rev Microbiol (2013) 11:587–91. doi:10.1038/nrmicro3076
Keywords: Mycobacterium tuberculosis, stress adaptation, virulence factors, host pathways, persisters
Citation: Kumar A, Rani M, Ehtesham NZ and Hasnain SE (2017) Commentary: Modification of Host Responses by Mycobacteria. Front. Immunol. 8:466. doi: 10.3389/fimmu.2017.00466
Received: 31 January 2017; Accepted: 05 April 2017;
Published: 28 April 2017
Edited by:
Hridayesh Prakash, University of Hyderabad, IndiaReviewed by:
Elsa Anes, Universidade de Lisboa, PortugalCopyright: © 2017 Kumar, Rani, Ehtesham and Hasnain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seyed E. Hasnain, c2V5ZWRoYXNuYWluQGdtYWlsLmNvbQ==, c2VoQGJpb3NjaG9vbC5paXRkLmFjLmlu
 Ashutosh Kumar1
Ashutosh Kumar1 Nasreen Z. Ehtesham
Nasreen Z. Ehtesham Seyed E. Hasnain
Seyed E. Hasnain