
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol. , 09 March 2017
Sec. Microbial Immunology
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.00256
This article is part of the Research Topic Spirochetes and immune evasion: infection, persistence and clearance View all 15 articles
Leptospirosis is the most widespread zoonosis in the world and a neglected tropical disease estimated to cause severe infection in more than one million people worldwide every year that can be combated by effective immunization. However, no significant progress has been made on the leptospirosis vaccine since the advent of bacterins over 100 years. Although protective against lethal infection, particularly in animals, bacterin-induced immunity is considered short term, serovar restricted, and the vaccine can cause serious side effects. The urgent need for a new vaccine has motivated several research groups to evaluate the protective immune response induced by recombinant vaccines. Significant protection has been reported with several promising outer membrane proteins, including LipL32 and the leptospiral immunoglobulin-like proteins. However, efficacy was variable and failed to induce a cross-protective response or sterile immunity among vaccinated animals. As hundreds of draft genomes of all known Leptospira species are now available, this should aid novel target discovery through reverse vaccinology (RV) and pangenomic studies. The identification of surface-exposed vaccine candidates that are highly conserved among infectious Leptospira spp. is a requirement for the development of a cross-protective universal vaccine. However, the lack of immune correlates is a major drawback to the application of RV to Leptospira genomes. In addition, as the protective immune response against leptospirosis is not fully understood, the rational use of adjuvants tends to be a process of trial and error. In this perspective, we discuss current advances, the pitfalls, and possible solutions for the development of a universal leptospirosis vaccine.
Following the discovery of leptospirosis, it was primarily associated with rural populations (1). This disease is caused by pathogenic Leptospira spp. and can be transmitted by direct contact via infected animals or by indirect contact as leptospires can survive outside the host. Agricultural workers, mineworkers, veterinarians, or individuals that came into direct contact with infected animals or contaminated environments were the main at-risk groups. However, toward the end of the 20th century, there were reports of leptospirosis among the homeless in major cities in the USA (2) and in urban slum communities in developing countries (3). The WHO estimated that the global incidence of leptospirosis more than doubled from approximately 500,000 cases in 1999 (4), to over a million cases in 2015 (5). Urban leptospirosis is now endemic in urban slums due to the lack of sanitation, rodent infestation, extreme poverty, and limited access to public health services in these communities.
Severe leptospirosis or Weil’s disease (jaundice, acute renal failure, and bleeding) has a case fatality rate of >10%. However, leptospirosis-associated pulmonary hemorrhage syndrome (LPHS) is being increasingly reported in developing countries (6) and the fatality rate is >50% (7). Clinical diagnosis of leptospirosis is difficult due to its similarity with other hemorrhagic diseases, and laboratory diagnostic tests are inadequate in these settings (8). There remains an urgent need for point-of-care rapid diagnostic tests. Vaccination of at-risk populations remains the most viable strategy for the control of leptospirosis. Classical, inactivated, vaccines have been available for over 100 years and are used routinely for agricultural and companion animals, reviewed in Ref. (9). Some countries have approved their use in human at-risk populations, although due to the severe side-effects and perceived short-term immunity and lack of cross-protection, they have not been adopted by the global community (10).
Of the 22 known Leptospira spp., 15 are infectious and can cause disease with varying degrees of severity. The pathogenesis of leptospirosis is a multifactorial process that is poorly understood, see, e.g., Ref. (11). Serological classification of leptospires indicates the existence of at least 250 serovars distributed in 18 serogroups (12). All this genetic and phenotypic diversity of pathogenic Leptospira spp. is a major drawback for vaccine development. The idea of a universal vaccine capable of protecting against all infectious Leptospira spp. and serovars would appear to be farfetched. However, some progress has been made with other pathogens such as influenza (13, 14), dengue (15), and others (16, 17). This perspective focuses on current advances, limitations, possible solutions, and looks forward to the possibility of a universal leptospirosis vaccine.
LipL32 is the immunodominant protein in pathogenic Leptospira spp. (18), there are over 38,000 copies per cell (19), and it can comprise up to 75% of the protein content of the outer membrane (OM) (20). However, there is a doubt as to its cellular localization; the latest report suggests it may occupy a subcellular location on the inner leaflet of the OM (21). LipL32 is not required for virulence; an Leptospira interrogans lipL32 knockout mutant could still infect hamsters (22). The biological function of LipL32 remains unknown, yet it is remarkable that such an abundant protein can be removed from the leptospiral OM with little or no effect on growth rate or OM structure. This is an example of the redundancy encoded in the Leptospira genome, as seen with other proteins, e.g., putative adhesins (23). There are over 20 publications on LipL32 and vaccine development. However, when rigorous statistical analysis is applied [e.g., Fisher’s exact test (24)], only five demonstrated significant protection against leptospirosis, reviewed in Ref. (10, 25). In addition, problems with reproducibility, survival in the control groups, high challenge doses (septic shock or leptospirosis), and the subcellular location of LipL32 have complicated its candidacy for inclusion in a universal vaccine formulation.
The leptospiral immunoglobulin-like (Lig) protein family includes LigA, LigB, and LigC and is only found in pathogenic Leptospira spp. (26, 27). While LigA and LigB are highly conserved (28), only LigB is present in all pathogenic Leptospira spp. (29). LigA and LigB are virulence determinants that are upregulated during infection (30), play a role in host cell adherence (31), prevent blood clotting (32, 33), and inhibit complement (34, 35). However, as seen for LipL32, an L. interrogans ligB knockout mutant remained virulent in the hamster model (36). Nevertheless, the Lig proteins are the standout vaccine candidates to date, with high, reproducible, levels of protection in animal models of acute leptospirosis in over 15 scientific reports, although not all withstood rigorous statistical analysis (10). The C-terminal (non-identical) region of LigA is an accepted vaccine candidate, having been evaluated in subunit (37–41), DNA (42), encapsulated (43), lipidated (44), and carbon nanotube vaccine preparations (45). However, when evaluated in a hamster colonization model, LigA failed to prevent infection (46). There is less evidence in support of LigB, the N-terminal conserved (repeat) region conferred significant protection as a subunit vaccine preparation (47) and a DNA vaccine (48) in the hamster model. Our group found that the same LigB polypeptide (LigBrep) not only protected hamsters but also induced sterile immunity in survivors (manuscript submitted).
Using the classical approach to vaccine candidate discovery, approximately 30 leptospiral, non-LipL32, non-Lig, proteins have been evaluated (10, 25). Of these, 10 proteins conferred significant protection against challenge with Leptospira spp. when the data were reanalyzed using, when necessary, a more rigorous statistical analysis (Fisher’s exact test) (10). The first report of protein-based protection came from studies of recombinant OmpL1 and LipL41 in the hamster model (49), and although only 1/3 experiments demonstrated significant protection, this provided the initial impetus for further research into protein-based vaccine candidates against leptospirosis. In an evaluation of three putative OMPs (Lp1454, Lp1118, and MceII), the subunit formulations failed to protect hamsters (50); however, when combined and encapsulated in liposomes, they conferred significant protection against challenge (51, 52). The putative lipoprotein LemA, identified using a reverse vaccinology (RV) approach (53), significantly protected immunized hamsters when administered as a DNA vaccine and protection increased using a prime-boost strategy (lemA/LemA) (54). In the most extensive study to date, 238 proteins identified using RV were evaluated as vaccine candidates (55). A hamster colonization model was used to evaluate pools of recombinant proteins (5 proteins/pool) and >70% were immunogenic. However, none of the recombinant protein pools conferred protection against colonization.
Cytoplasmic proteins, inner membrane proteins, and OM lipoproteins that are not exposed on the surface (i.e., those attached to the inner leaflet of the OM) are likely to be ineffective recombinant vaccines. Antibodies induced by subsurface proteins would not be able to bind to infecting leptospires making the vaccine ineffective. Therefore, vaccine candidates should be surface exposed on the leptospiral cell. Equally important are the potential roles in pathogenicity and the immunogenicity of these proteins. Furthermore, it is doubtful that a protein-based vaccine candidate would be capable of inducing a protective immune response if the protein components of the vaccine lacked one or more of these characteristics.
Lipoproteins attached to the outer leaflet of the OM and transmembrane β-barrel proteins spanning the OM (OMPs) should be fully or partially surface exposed (Figure 1). The localization of LipL32 is still unresolved; there is experimental data for both surface (56–59) and subsurface locations (21). Leptospiral genomes encode OMPs such as LptD, BamA-like, TonB-dependent receptors, and several other porins that play crucial roles in bacterial survival and potential role in pathogenicity. These proteins are ideal targets and should be evaluated as potential vaccine candidates. RV was developed to identify surface-related proteins in the genome of pathogens using bioinformatics (60). RV has been used to analyze Leptospira genomes and there are several reports in the literature that have used in silico genome mining toward the identification of leptospiral vaccine candidates, reviewed in Ref. (61).
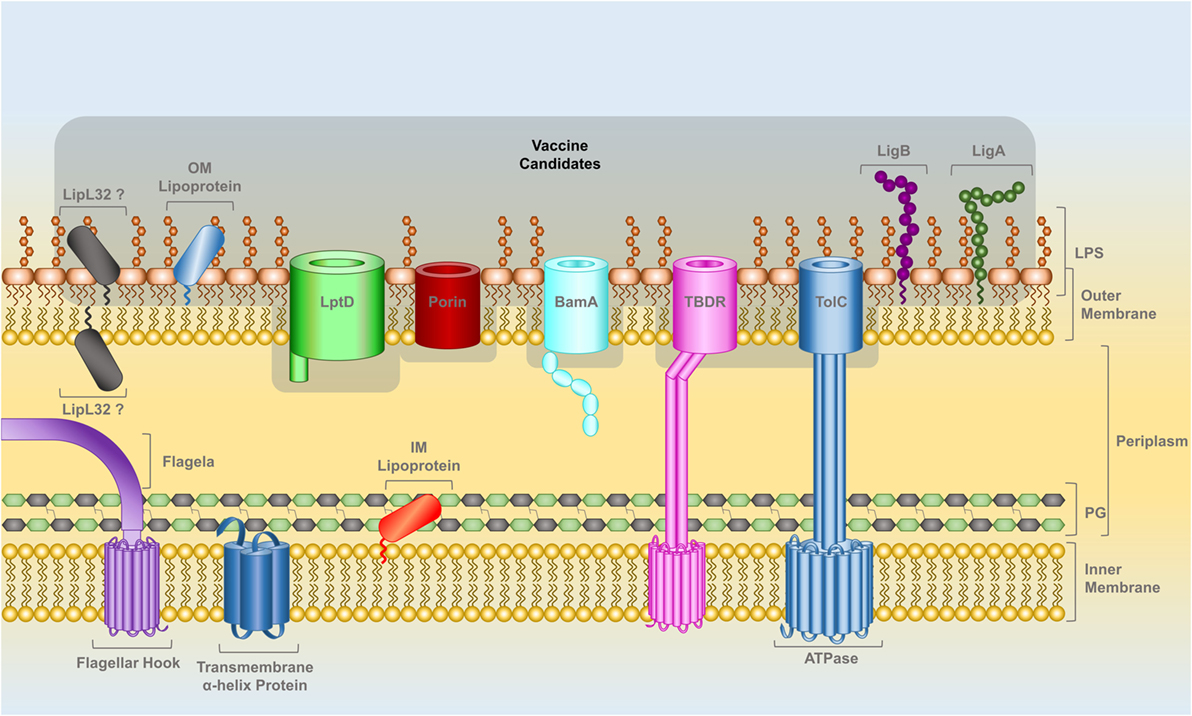
Figure 1. The cell wall of Leptospira spp., a diderm bacteria with inner and outer lipid membranes, is the target for the identification of potential vaccine candidates. The outer leaflet of the outer membrane (OM) is composed of lipopolysaccharide. Lipoproteins can be attached by a lipid anchor to one of the leaflets of either membranes. The IM is spanned by alpha-helix transmembrane proteins while beta-barrel transmembrane proteins span the OM. Leptospiral motility is provided by two flagella that are attached to the IM and are located within the periplasmic space (PS). A peptidoglycan layer is also present in the PS. OM lipoproteins, such as LigA and LigB, as well as OM beta-barrel proteins, such as LptD, BamA, TolC-, TonB-dependent receptors, and other porins have at least a portion of their structure exposed on leptospiral surface and are prospective vaccine candidates, highlighted in gray. The localization of the lipoprotein LipL32 in the OM is controversial; the latest reports indicate that it has a subsurface location (see text).
Recently, dozens of leptospiral proteins have been described as adhesins, reviewed in Ref. (11), and blocking the adhesion of leptospires is thought to impair their virulence. Similarly, several proteins have been described as host complement activation inhibitors, suggesting that leptospires evade the complement system, reviewed in Ref. (62). In many studies (63–67), the surface localization of the leptospiral antigens were determined by in vitro approaches including proteinase K digestion and a surface immuno-fluorescence assay (IFA) (68). These approaches have contributed to the controversy surrounding the localization of proteins such as LipL32. Another example is that of LIC13166, this protein was originally demonstrated to be an OMP exposed on the surface of the leptospiral cell by surface biotinylation, membrane affinity, and surface-IFA experiments (68). However, in a recent publication, it was shown that LIC13166 is, in fact, a flagellar protein, renamed FcpA, which is located in the periplasm (69). The subcellular location of adhesins, complement binding proteins, and virulence factors described in knockout experiments should be properly investigated; otherwise the biological relevance of these findings will remain unclear. We are currently developing an alternative method to improve the identification of surface leptospiral proteins while maintaining the integrity of the leptospiral OM.
The perceived lack of cross-protection following immunization with a bacterin is another factor that has limited their widespread use. There are, however, several reports of bacterins conferring cross-protection against species-related serovars. An evaluation of bacterins reported 100% cross protection between L. interrogans serovars Canicola, Copenhageni, and Leptospira borgpetersenii serovar Ballum but not serovar Mozdok (70). A multivalent bacterin containing serovars from four different serogroups demonstrated cross-protection in a canine model of leptospirosis (71). Another study of two bacterins based on different serovars, but belonging to the same serogroup and species, reported species-related cross-protection, although 50% of the control group survived (72). It is likely that the protective antigens in these studies were proteins, as leptospiral lipopolysaccharide (LPS) does not induce cross-protection, even among species-related serovars (73). Rather, protein-enriched samples were responsible for cross-protection against species-related serovars in a gerbil model of lethal leptospirosis. This was further supported by a study using a live vaccine based on an LPS defective mutant. Species-related cross-protection was demonstrated, although the vaccine could not prevent colonization by a non-related serovar (74). Several studies of individual proteins have claimed to show cross-protection. An adenovirus construct containing lipL32 conferred cross-protection against a species-related serovar, although >50% of the control groups survived (75). A treatment based on anti-LipL32 monoclonal antibodies protected hamsters challenged with a species-related serovar (76). Prime-boost strategies using LemA and LigBrep conferred cross-protection against a species-related serovar, albeit in one-off experiments (54, 77).
At least one strain for every known Leptospira spp. has been sequenced and new isolates are continually being sequenced and their genomes released on GenBank or other public databases, see, e.g., Ref. (29, 78–83), thereby providing a panoramic view of Leptospira pathogenomics, permitting the identification of orthologs and protein sequence similarity among infectious species. This has significantly contributed to the identification and selection of conserved vaccine candidates based on a simple in silico sequence analysis (Figure 2). Protein sequences are usually highly conserved among the same species regardless of the serovar or serogroup, while they can differ considerably when comparing the same serovar in different species. While, serological classification is unquestionably important for epidemiology and bacterin-based vaccine studies, it is of limited use for recombinant vaccine development. This is a problem associated with a leptospiral bacterin vaccine, the immune response is primarily directed against the leptospiral LPS and while it protects against infection by closely related serovars or serovars from the same Leptospira spp., leptospiral LPS does not stimulate memory B-cells (10). As there is no clear definition of cross-protection in the field of leptospirosis, this is a major drawback to vaccine candidate discovery and evaluation. Ideally, recombinant vaccine-induced cross-protection should be defined as cross-species protection rather than cross-serovar protection. A universal vaccine should therefore protect against all 15 infectious Leptospira spp. regardless of serovar. However, if this is not a viable option, it should be possible to identify the main circulating species and develop a region-specific recombinant vaccine rather than a universal vaccine. This could potentially allow the characterization of the protective immune response and establish standard protocols for the evaluation of cross-protection of recombinant vaccine candidates (Figure 2).

Figure 2. A schematic representation of the development pipeline for a universal vaccine against leptospirosis. The basic research on Leptospira microbiology and molecular biology contributes to vaccine development. Leptospira mutagenesis is an example of basic research that has and will continue to further our understanding of pathogenesis and identification of virulence factors. Genomic and pangenomic studies are of central importance to the development of a universal leptospirosis vaccines, permitting the identification of potential vaccine candidates and the analysis of protein sequences among different Leptospira spp. RV has not been fully explored in leptospirosis and needs to be more thoroughly exploited. Once potential vaccine candidates are identified, an in vitro validation is required, particularly to confirm the localization of antigens on the surface of the leptospiral cell. At this stage, a prospective vaccine candidate can be assessed for immunogenicity. The lack of well-defined correlates of immunity for leptospirosis represents one for the major limitations for leptospirosis vaccine development and remains to be resolved. Therefore, surface-related, conserved (among infectious Leptospira spp.), and immunogenic leptospiral antigens must be evaluated in vaccine challenge experiments using animal models. Cross-protection, defined as cross-species rather than cross-serovar protection should be evaluated. In addition, as the protective immune response is not fully understood, continued research in this field is necessary. Finally, the long-term goal of this pipeline is to identify experimental vaccine preparations for evaluation in clinical trials.
Several adjuvants and delivery systems have been used to enhance the immune response against leptospiral antigens. Aluminum hydroxide (alhydrogel) and Freund’s adjuvant are by far the most common, although others including flagellin (84), CpGs (85), nanostructures (45), liposomes (43, 51, 52), xanthan (85) have been investigated. While Freund’s adjuvant cannot be used in humans due to its high reactogenicity (86), it is the most potent commercially available adjuvant (87), is useful for the primary screening of vaccine antigens, and has been used successfully in vaccine formulations against leptospirosis (39, 41). To date, only partial protection has been demonstrated with vaccines using alhydrogel, the most widely used adjuvant in human vaccines. Recently, other adjuvants have become commercially available and have been approved for use in the formulation of human vaccines, comprising the adjuvants MF59 (squalene), AS01 [monophosphoryl lipid A (MPL), QS21], AS03 (α-tocopherol, squalene, and polysorbate 80), AS04 (MPL combined with alhydrogel), and virosomes (liposome/VLPs) (88, 89). These prospective adjuvants have not yet been evaluated as adjuvants for leptospirosis vaccines.
Rational modulation of the immune response is difficult to achieve for leptospirosis vaccines as little is known about the protective immune response that should be induced by a leptospirosis vaccine. Humoral immunity is believed to be responsible for protection; anti-LPS antibodies are protective in animal models and can be passively transferred between animals (90). As predominantly extracellular organisms, leptospires are most likely cleared from the bloodstream by phagocytosis followed by opsonization. However, at least in some hosts, e.g., cattle, induction of cellular immunity is equally important (90). Until recently, there were no published reports of correlation between antibody titer, induced by leptospiral recombinant vaccines and protection against challenge. However, an oral immunization strategy based on LigA found that survival was dependent on a minimum antibody titer being reached in a 2-week period following immunization (44), and if this can be reproduced, it will be an extremely important finding. The lack of immune correlates is a major limitation in target discovery using RV as they are essential for the in vitro screening of potential vaccine candidates, see, e.g., the bactericidal assay for Neisseria meningitidis (91) and the opsonophagocytosis assay for Staphylococcus aureus (92).
The recommended animal model for acute leptospirosis is the Syrian hamster; this model replicates the human symptoms of the disease, including kidney failure, LPHS, and kidney, liver, and lung tissue damage, which result in death (93). Furthermore, the hamster model is the recommended model for potency testing of bacterin vaccines (94). The acute model is dependent on a virulent challenge strain and the lack thereof has had a major impact on protection studies. However, to date, no well-established correlates of immunity have been identified and, therefore, vaccinated hamsters must be challenged with a virulent Leptospira strain to demonstrate protection. Due to significant variation among the hamster models of acute leptospirosis, we recommend that the research community adopt a standardized model (see Supplementary Material). An alternative to the lethal model is the hamster colonization model, and this is the model of choice when evaluating vaccine candidates for agricultural animals including cattle, swine, and horses (55, 95). Unlike the acute model, the primary endpoint in this model is kidney colonization.
A major limitation of the hamster model is the lack of commercial reagents for characterization of the immune response, e.g., induction of cytokines and chemokines cannot be measured directly. Alternate models include the guinea pig and the gerbil, although there are few commercially available reagents for these models. Due to the wide range of commercially available reagents, the mouse model is attractive, reviewed in Ref. (96). Wild-type mice are naturally resistant to leptospirosis, although colonization is possible with some serovars (9, 97). Lethal leptospirosis has been demonstrated in C3H/HeJ (41), SCID, and Rag1 knockout mice (98). Maintenance host models of chronic infection have been developed using the Wistar strain of Rattus norvegicus (9, 99).
Alternatives to whole-cell inactivated leptospiral vaccines have so far failed to live up to their initial promise, and the concept of a universal leptospiral vaccine remains just that, a concept. Several reviews have highlighted the modest numbers (~30) of leptospiral proteins that have been tested using various vaccine strategies, including subunit, DNA vaccines, prime-boost, encapsulated, and live avirulent strains. Of these, less than a handful has been successful. However, the availability of multiple genome sequences, combined with advances in bioinformatics (e.g., RV) and the characterization of surface-exposed virulence factors, will improve the discovery of potential vaccine candidates. The next challenge is to develop in vitro assays based on correlates of immunity for the high-throughput screening of these vaccine candidates. While there are several animal models of leptospirosis, their standardization is necessary for the critical interpretation of protection data. Cross-protection is a priority for a universal vaccine and will require the identification of vaccine candidates that are conserved among the infectious Leptospira spp. Our poor understanding of the (protective) immune response has hindered the intelligent selection of adjuvants for use in vaccine formulations. Finally, while the field is moving in the right direction, a universal vaccine for leptospirosis remains a long-term goal.
AG, JS, and AM wrote the manuscript. AG and JS created the figures, and all the authors contributed to and revised the manuscript.
AM is an inventor on several patents for the use of Leptospira proteins as vaccines and diagnostics. The other authors declare no conflict of interest.
The work of our group was supported by grants 481133/2011-9, 483052/2012-4, and 311852/2012-2 from the Brazilian National Council for Scientific and Technological Development (CNPq).
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00256/full#supplementary-material.
1. Adler B. History of leptospirosis and Leptospira. Curr Top Microbiol Immunol (2015) 387:1–9. doi:10.1007/978-3-662-45059-8_1
2. Vinetz JM, Glass GE, Flexner CE, Mueller P, Kaslow DC. Sporadic urban leptospirosis. Ann Intern Med (1996) 125(10):794–8. doi:10.7326/0003-4819-125-10-199611150-00002
3. McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis (2005) 18(5):376–86. doi:10.1097/01.qco.0000178824.05715.2c
5. Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis (2015) 9(9):e0003898. doi:10.1371/journal.pntd.0003898
6. Croda J, Neto AN, Brasil RA, Pagliari C, Nicodemo AC, Duarte MI. Leptospirosis pulmonary haemorrhage syndrome is associated with linear deposition of immunoglobulin and complement on the alveolar surface. Clin Microbiol Infect (2010) 16(6):593–9. doi:10.1111/j.1469-0691.2009.02916.x
7. Gouveia EL, Metcalfe J, de Carvalho AL, Aires TS, Villasboas-Bisneto JC, Queirroz A, et al. Leptospirosis-associated severe pulmonary hemorrhagic syndrome, Salvador, Brazil. Emerg Infect Dis (2008) 14(3):505–8. doi:10.3201/eid1403.071064
8. Haake DA, Levett PN. Leptospirosis in humans. Curr Top Microbiol Immunol (2015) 387:65–97. doi:10.1007/978-3-662-45059-8_5
9. Ellis WA. Animal leptospirosis. Curr Top Microbiol Immunol (2015) 387:99–137. doi:10.1007/978-3-662-45059-8_6
10. Adler B. Vaccines against leptospirosis. Curr Top Microbiol Immunol (2015) 387:251–72. doi:10.1007/978-3-662-45059-8_10
11. Murray GL. The molecular basis of leptospiral pathogenesis. Curr Top Microbiol Immunol (2015) 387:139–85. doi:10.1007/978-3-662-45059-8_7
12. Levett PN. Systematics of leptospiraceae. Curr Top Microbiol Immunol (2015) 387:11–20. doi:10.1007/978-3-662-45059-8_2
13. Sridhar S. Heterosubtypic T-cell immunity to influenza in humans: challenges for universal T-cell influenza vaccines. Front Immunol (2016) 7:195. doi:10.3389/fimmu.2016.00195
14. Cho A, Wrammert J. Implications of broadly neutralizing antibodies in the development of a universal influenza vaccine. Curr Opin Virol (2016) 17:110–5. doi:10.1016/j.coviro.2016.03.002
15. Chokephaibulkit K, Perng GC. Challenges for the formulation of a universal vaccine against dengue. Exp Biol Med (Maywood) (2013) 238(5):566–78. doi:10.1177/1535370212473703
16. Pier GB. Will there ever be a universal Staphylococcus aureus vaccine? Hum Vaccin Immunother (2013) 9(9):1865–76. doi:10.4161/hv.25182
17. Guan Q, Wang X, Wang X, Teng D, Mao R, Zhang Y, et al. Recombinant outer membrane protein A induces a protective immune response against Escherichia coli infection in mice. Appl Microbiol Biotechnol (2015) 99(13):5451–60. doi:10.1007/s00253-014-6339-6
18. Haake DA, Chao G, Zuerner RL, Barnett JK, Barnett D, Mazel M, et al. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun (2000) 68(4):2276–85. doi:10.1128/IAI.68.4.2276-2285.2000
19. Malmstrom J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature (2009) 460(7256):762–5. doi:10.1038/nature08184
20. Cullen PA, Cordwell SJ, Bulach DM, Haake DA, Adler B. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect Immun (2002) 70(5):2311–8. doi:10.1128/IAI.70.5.2311-2318.2002
21. Pinne M, Haake DA. LipL32 is a subsurface lipoprotein of Leptospira interrogans: presentation of new data and reevaluation of previous studies. PLoS One (2013) 8(1):e51025. doi:10.1371/journal.pone.0051025
22. Murray GL, Srikram A, Hoke DE, Wunder EA Jr, Henry R, Lo M, et al. Major surface protein LipL32 is not required for either acute or chronic infection with Leptospira interrogans. Infect Immun (2009) 77(3):952–8. doi:10.1128/IAI.01370-08
23. Adler B, Lo M, Seemann T, Murray GL. Pathogenesis of leptospirosis: the influence of genomics. Vet Microbiol (2011) 153(1–2):73–81. doi:10.1016/j.vetmic.2011.02.055
25. Dellagostin OA, Grassmann AA, Hartwig DD, Felix SR, da Silva EF, McBride AJ. Recombinant vaccines against leptospirosis. Hum Vaccin (2011) 7(11):1215–24. doi:10.4161/hv.7.11.17944
26. Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol (2003) 49(4):929–45. doi:10.1046/j.1365-2958.2003.03619.x
27. Cerqueira GM, McBride AJ, Picardeau M, Ribeiro SG, Moreira AN, Morel V, et al. Distribution of the leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira species and application of ligB to typing leptospiral isolates. J Med Microbiol (2009) 58(Pt 9):1173–81. doi:10.1099/jmm.0.009175-0
28. McBride AJ, Cerqueira GM, Suchard MA, Moreira AN, Zuerner RL, Reis MG, et al. Genetic diversity of the leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira spp. Infect Genet Evol (2009) 9(2):196–205. doi:10.1016/j.meegid.2008.10.012
29. Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, Berg DE, et al. What makes a bacterial species pathogenic? Comparative genomic analysis of the genus Leptospira. PLoS Negl Trop Dis (2016) 10(2):e0004403. doi:10.1371/journal.pntd.0004403
30. Matsunaga J, Sanchez Y, Xu X, Haake DA. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect Immun (2005) 73(1):70–8. doi:10.1128/IAI.73.1.70-78.2005
31. Choy HA, Kelley MM, Chen TL, Moller AK, Matsunaga J, Haake DA. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect Immun (2007) 75(5):2441–50. doi:10.1128/IAI.01635-06
32. Choy HA, Kelley MM, Croda J, Matsunaga J, Babbitt JT, Ko AI, et al. The multifunctional LigB adhesin binds homeostatic proteins with potential roles in cutaneous infection by pathogenic Leptospira interrogans. PLoS One (2011) 6(2):e16879. doi:10.1371/journal.pone.0016879
33. Hsieh CL, Chang E, Tseng A, Ptak C, Wu LC, Su CL, et al. Leptospira immunoglobulin-like protein B (LigB) binds to both the C-terminal 23 amino acids of fibrinogen alphaC domain and factor XIII: insight into the mechanism of LigB-mediated blockage of fibrinogen alpha chain cross-linking. PLoS Negl Trop Dis (2016) 10(9):e0004974. doi:10.1371/journal.pntd.0004974
34. Choy HA. Multiple activities of LigB potentiate virulence of Leptospira interrogans: inhibition of alternative and classical pathways of complement. PLoS One (2012) 7(7):e41566. doi:10.1371/journal.pone.0041566
35. Castiblanco-Valencia MM, Fraga TR, Pagotto AH, Serrano SM, Abreu PA, Barbosa AS, et al. Plasmin cleaves fibrinogen and the human complement proteins C3b and C5 in the presence of Leptospira interrogans proteins: a new role of LigA and LigB in invasion and complement immune evasion. Immunobiology (2016) 221(5):679–89. doi:10.1016/j.imbio.2016.01.001
36. Croda J, Figueira CP, Wunder EA Jr, Santos CS, Reis MG, Ko AI, et al. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun (2008) 76(12):5826–33. doi:10.1128/IAI.00989-08
37. Hartwig DD, Bacelo KL, Oliveira PD, Oliveira TL, Seixas FK, Amaral MG, et al. Mannosylated LigANI produced in Pichia pastoris protects hamsters against leptospirosis. Curr Microbiol (2014) 68(4):524–30. doi:10.1007/s00284-013-0505-4
38. Coutinho ML, Choy HA, Kelley MM, Matsunaga J, Babbitt JT, Lewis MS, et al. A LigA three-domain region protects hamsters from lethal infection by Leptospira interrogans. PLoS Negl Trop Dis (2011) 5(12):e1422. doi:10.1371/journal.pntd.0001422
39. Silva EF, Medeiros MA, McBride AJ, Matsunaga J, Esteves GS, Ramos JG, et al. The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine (2007) 25(33):6277–86. doi:10.1016/j.vaccine.2007.05.053
40. Palaniappan RU, McDonough SP, Divers TJ, Chen CS, Pan MJ, Matsumoto M, et al. Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospira interrogans serovar Pomona infection. Infect Immun (2006) 74(3):1745–50. doi:10.1128/IAI.74.3.1745-1750.2006
41. Koizumi N, Watanabe H. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine (2004) 22(11–12):1545–52. doi:10.1016/j.vaccine.2003.10.007
42. Faisal SM, Yan W, Chen CS, Palaniappan RU, McDonough SP, Chang YF. Evaluation of protective immunity of Leptospira immunoglobulin like protein A (LigA) DNA vaccine against challenge in hamsters. Vaccine (2008) 26(2):277–87. doi:10.1016/j.vaccine.2007.10.029
43. Faisal SM, Yan W, McDonough SP, Chang YF. Leptospira immunoglobulin-like protein A variable region (LigAvar) incorporated in liposomes and PLGA microspheres produces a robust immune response correlating to protective immunity. Vaccine (2009) 27(3):378–87. doi:10.1016/j.vaccine.2008.10.089
44. Lourdault K, Wang LC, Vieira A, Matsunaga J, Melo R, Lewis MS, et al. Oral immunization with Escherichia coli expressing a lipidated form of LigA protects hamsters against challenge with Leptospira interrogans serovar Copenhageni. Infect Immun (2014) 82(2):893–902. doi:10.1128/IAI.01533-13
45. Oliveira TL, Bacelo KL, Schuch RA, Seixas FK, Collares T, Rodrigues OE, et al. Immune response in hamsters immunised with a recombinant fragment of LigA from Leptospira interrogans, associated with carrier molecules. Mem Inst Oswaldo Cruz (2016) 111(11):712–6. doi:10.1590/0074-02760160214
46. Lucas DS, Cullen PA, Lo M, Srikram A, Sermswan RW, Adler B. Recombinant LipL32 and LigA from Leptospira are unable to stimulate protective immunity against leptospirosis in the hamster model. Vaccine (2011) 29(18):3413–8. doi:10.1016/j.vaccine.2011.02.084
47. Yan W, Faisal SM, McDonough SP, Divers TJ, Barr SC, Chang CF, et al. Immunogenicity and protective efficacy of recombinant Leptospira immunoglobulin-like protein B (rLigB) in a hamster challenge model. Microbes Infect (2009) 11(2):230–7. doi:10.1016/j.micinf.2008.11.008
48. Forster KM, Hartwig DD, Seixas FK, Bacelo KL, Amaral M, Hartleben CP, et al. A conserved region of leptospiral immunoglobulin-like A and B proteins as a DNA vaccine elicits a prophylactic immune response against leptospirosis. Clin Vaccine Immunol (2013) 20(5):725–31. doi:10.1128/CVI.00601-12
49. Haake DA, Mazel MK, McCoy AM, Milward F, Chao G, Matsunaga J, et al. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect Immun (1999) 67(12):6572–82.
50. Chang YF, Chen CS, Palaniappan RU, He H, McDonough SP, Barr SC, et al. Immunogenicity of the recombinant leptospiral putative outer membrane proteins as vaccine candidates. Vaccine (2007) 25(48):8190–7. doi:10.1016/j.vaccine.2007.09.020
51. Faisal SM, Yan W, McDonough SP, Chang CF, Pan MJ, Chang YF. Leptosome-entrapped leptospiral antigens conferred significant higher levels of protection than those entrapped with PC-liposomes in a hamster model. Vaccine (2009) 27(47):6537–45. doi:10.1016/j.vaccine.2009.08.051
52. Faisal SM, Yan W, McDonough SP, Mohammed HO, Divers TJ, Chang YF. Immune response and prophylactic efficacy of smegmosomes in a hamster model of leptospirosis. Vaccine (2009) 27(44):6129–36. doi:10.1016/j.vaccine.2009.08.029
53. Hartwig DD, Seixas FK, Cerqueira GM, McBride AJ, Dellagostin OA. Characterization of the immunogenic and antigenic potential of putative lipoproteins from Leptospira interrogans. Curr Microbiol (2011) 62(4):1337–41. doi:10.1007/s00284-010-9865-1
54. Hartwig DD, Forster KM, Oliveira TL, Amaral M, McBride AJ, Dellagostin OA. A prime-boost strategy using the novel vaccine candidate, LemA, protects hamsters against leptospirosis. Clin Vaccine Immunol (2013) 20(5):747–52. doi:10.1128/CVI.00034-13
55. Murray GL, Lo M, Bulach DM, Srikram A, Seemann T, Quinsey NS, et al. Evaluation of 238 antigens of Leptospira borgpetersenii serovar Hardjo for protection against kidney colonisation. Vaccine (2013) 31(3):495–9. doi:10.1016/j.vaccine.2012.11.028
56. Cullen PA, Xu X, Matsunaga J, Sanchez Y, Ko AI, Haake DA, et al. Surfaceome of Leptospira spp. Infect Immun (2005) 73(8):4853–63. doi:10.1128/IAI.73.8.4853-4863.2005
57. Haake DA, Matsunaga J. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect Immun (2002) 70(9):4936–45. doi:10.1128/IAI.70.9.4936-4945.2002
58. Oliveira R, de Morais ZM, Goncales AP, Romero EC, Vasconcellos SA, Nascimento AL. Characterization of novel OmpA-like protein of Leptospira interrogans that binds extracellular matrix molecules and plasminogen. PLoS One (2011) 6(7):e21962. doi:10.1371/journal.pone.0021962
59. Fernandes CP, Seixas FK, Coutinho ML, Vasconcellos FA, Seyffert N, Croda J, et al. Monoclonal antibodies against LipL32, the major outer membrane protein of pathogenic Leptospira: production, characterization, and testing in diagnostic applications. Hybridoma (Larchmt) (2007) 26(1):35–41. doi:10.1089/hyb.2006.033
60. Seib KL, Zhao X, Rappuoli R. Developing vaccines in the era of genomics: a decade of reverse vaccinology. Clin Microbiol Infect (2012) 18(Suppl 5):109–16. doi:10.1111/j.1469-0691.2012.03939.x
61. Dellagostin OA, Grassmann AA, Rizzi C, Schuch RA, Jorge S, Oliveira TL, et al. Reverse vaccinology: an approach for identifying leptospiral vaccine candidates. Int J Mol Sci (2017) 18(1):158–73. doi:10.3390/ijms18010158
62. Fraga TR, Isaac L, Barbosa AS. Complement evasion by pathogenic Leptospira. Front Immunol (2016) 7:623. doi:10.3389/fimmu.2016.00623
63. Domingos RF, Fernandes LG, Romero EC, de Morais ZM, Vasconcellos SA, Nascimento AL. Novel Leptospira interrogans protein Lsa32 is expressed during infection and binds laminin and plasminogen. Microbiology (2015) 161(Pt 4):851–64. doi:10.1099/mic.0.000041
64. Evangelista KV, Hahn B, Wunder EA Jr, Ko AI, Haake DA, Coburn J. Identification of cell-binding adhesins of Leptospira interrogans. PLoS Negl Trop Dis (2014) 8(10):e3215. doi:10.1371/journal.pntd.0003215
65. Figueredo J, Siqueira G, Souza G, Heinemann M, Vasconcellos S, Chapola E, et al. Characterization of two new putative adhesins of Leptospira interrogans. Microbiology (2016) 163:37–51. doi:10.1099/mic.0.000411
66. Robbins GT, Hahn BL, Evangelista KV, Padmore L, Aranda PS, Coburn J. Evaluation of cell binding activities of Leptospira ECM adhesins. PLoS Negl Trop Dis (2015) 9(4):e0003712. doi:10.1371/journal.pntd.0003712
67. Silva LP, Fernandes LG, Vieira ML, de Souza GO, Heinemann MB, Vasconcellos SA, et al. Evaluation of two novel leptospiral proteins for their interaction with human host components. Pathog Dis (2016) 74(5):ftw040. doi:10.1093/femspd/ftw040
68. Pinne M, Haake DA. A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS One (2009) 4(6):e6071. doi:10.1371/journal.pone.0006071
69. Wunder EA Jr, Figueira CP, Benaroudj N, Hu B, Tong BA, Trajtenberg F, et al. A novel flagellar sheath protein, FcpA, determines filament coiling, translational motility and virulence for the Leptospira spirochete. Mol Microbiol (2016) 101(3):457–70. doi:10.1111/mmi.13403
70. Rosario LA, Arencibia DF, Suarez YE, Infante JF, Valdes BY, Batista N. Cross-protection among unrelated Leptospira pathogens serovars: an unfinished story. Adv Clin Exp Med (2012) 21(5):581–9.
71. Klaasen HL, van der Veen M, Molkenboer MJ, Sutton D. A novel tetravalent Leptospira bacterin protects against infection and shedding following challenge in dogs. Vet Rec (2013) 172(7):181. doi:10.1136/vr.101100
72. Dib CC, Goncales AP, de Morais ZM, de Souza GO, Miraglia F, Abreu PA, et al. Cross-protection between experimental anti-leptospirosis bacterins. Braz J Microbiol (2014) 45(3):1083–8. doi:10.1590/S1517-83822014000300042
73. Sonrier C, Branger C, Michel V, Ruvoen-Clouet N, Ganiere JP, Andre-Fontaine G. Evidence of cross-protection within Leptospira interrogans in an experimental model. Vaccine (2000) 19(1):86–94. doi:10.1016/S0264-410X(00)00129-8
74. Srikram A, Zhang K, Bartpho T, Lo M, Hoke DE, Sermswan RW, et al. Cross-protective immunity against leptospirosis elicited by a live, attenuated lipopolysaccharide mutant. J Infect Dis (2011) 203(6):870–9. doi:10.1093/infdis/jiq127
75. Branger C, Sonrier C, Chatrenet B, Klonjkowski B, Ruvoen-Clouet N, Aubert A, et al. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect Immun (2001) 69(11):6831–8. doi:10.1128/IAI.69.11.6831-6838.2001
76. Maneewatch S, Sakolvaree Y, Saengjaruk P, Srimanote P, Tapchaisri P, Tongtawe P, et al. Monoclonal antibodies to LipL32 protect against heterologous Leptospira spp. challenge. Hybridoma (Larchmt) (2008) 27(6):453–65. doi:10.1089/hyb.2008.0056
77. Forster KM, Hartwig DD, Oliveira TL, Bacelo KL, Schuch R, Amaral MG, et al. DNA prime-protein boost based vaccination with a conserved region of leptospiral immunoglobulin-like A and B proteins enhances protection against leptospirosis. Mem Inst Oswaldo Cruz (2015) 110(8):989–95. doi:10.1590/0074-02760150222
78. Varni V, Koval A, Nagel A, Ruybal P, Caimi K, Amadio AF. First genome sequence of Leptospira interrogans serovar Pomona, isolated from a bovine abortion. Genome Announc (2016) 4(3):e00345–16. doi:10.1128/genomeA.00345-16
79. Nally JE, Bayles DO, Hurley D, Fanning S, McMahon BJ, Arent Z. Complete genome sequence of Leptospira alstonii serovar Room22 strain GWTS #1. Genome Announc (2016) 4(6):e01230–16. doi:10.1128/genomeA.01230-16
80. Llanes A, Restrepo CM, Rajeev S. Whole genome sequencing allows better understanding of the evolutionary history of Leptospira interrogans serovar Hardjo. PLoS One (2016) 11(7):e0159387. doi:10.1371/journal.pone.0159387
81. Lehmann JS, Corey VC, Ricaldi JN, Vinetz JM, Winzeler EA, Matthias MA. Whole genome shotgun sequencing shows selection on Leptospira regulatory proteins during in vitro culture attenuation. Am J Trop Med Hyg (2016) 94(2):302–13. doi:10.4269/ajtmh.15-0401
82. Kremer FS, Eslabao MR, Jorge S, Oliveira NR, Labonde J, Santos MN, et al. Draft genome of the Leptospira interrogans strains, Acegua, RCA, Prea, and Capivara, obtained from wildlife maintenance hosts and infected domestic animals. Mem Inst Oswaldo Cruz (2016) 111(4):280–3. doi:10.1590/0074-02760160010
83. Kremer FS, Eslabao MR, Provisor M, Woloski RD, Ramires OV, Moreno LZ, et al. Draft genome sequences of Leptospira santarosai strains U160, U164, and U233, isolated from asymptomatic cattle. Genome Announc (2015) 3(4):e00910–15. doi:10.1128/genomeA.00910-15
84. Monaris D, Sbrogio-Almeida ME, Dib CC, Canhamero TA, Souza GO, Vasconcellos SA, et al. Protective immunity and reduced renal colonization induced by vaccines containing recombinant Leptospira interrogans outer membrane proteins and flagellin adjuvant. Clin Vaccine Immunol (2015) 22(8):965–73. doi:10.1128/CVI.00285-15
85. Bacelo KL, Hartwig DD, Seixas FK, Schuch R, Moreira Ada S, Amaral M, et al. Xanthan gum as an adjuvant in a subunit vaccine preparation against leptospirosis. Biomed Res Int (2014) 2014:636491. doi:10.1155/2014/636491
86. Guy B. The perfect mix: recent progress in adjuvant research. Nat Rev Microbiol (2007) 5(7):505–17. doi:10.1038/nrmicro1681
87. Azmi F, Ahmad Fuaad AA, Skwarczynski M, Toth I. Recent progress in adjuvant discovery for peptide-based subunit vaccines. Hum Vaccin Immunother (2014) 10(3):778–96. doi:10.4161/hv.27332
88. Stassijns J, Bollaerts K, Baay M, Verstraeten T. A systematic review and meta-analysis on the safety of newly adjuvanted vaccines among children. Vaccine (2016) 34(6):714–22. doi:10.1016/j.vaccine.2015.12.024
89. Liang F, Lore K. Local innate immune responses in the vaccine adjuvant-injected muscle. Clin Transl Immunology (2016) 5(4):e74. doi:10.1038/cti.2016.19
90. Zuerner RL. Host response to Leptospira infection. Curr Top Microbiol Immunol (2015) 387:223–50. doi:10.1007/978-3-662-45059-8_9
91. Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science (2000) 287(5459):1816–20. doi:10.1126/science.287.5459.1816
92. Etz H, Minh DB, Henics T, Dryla A, Winkler B, Triska C, et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc Natl Acad Sci U S A (2002) 99(10):6573–8. doi:10.1073/pnas.092569199
93. Freudenstein H, Hein B. Potency of leptospiral vaccines and protection against chronic infection in golden hamsters. Comp Immunol Microbiol Infect Dis (1991) 14(3):229–34. doi:10.1016/0147-9571(91)90003-V
94. Srinivas GB, Walker A, Rippke B. USDA regulatory guidelines and practices for veterinary Leptospira vaccine potency testing. Biologicals (2013) 41(5):298–302. doi:10.1016/j.biologicals.2013.06.005
95. Zuerner RL, Alt DP, Palmer MV. Development of chronic and acute golden Syrian hamster infection models with Leptospira borgpetersenii serovar Hardjo. Vet Pathol (2012) 49(2):403–11. doi:10.1177/0300985811409252
96. Gomes-Solecki M, Santecchia I, Werts C. Animal models of leptospirosis: of mice and hamsters. Front Immunol (2017) 8(58). doi:10.3389/fimmu.2017.00058
97. Santos CS, Macedo JO, Bandeira M, Chagas-Junior AD, McBride AJ, McBride FW, et al. Different outcomes of experimental leptospiral infection in mouse strains with distinct genotypes. J Med Microbiol (2010) 59(Pt 9):1101–6. doi:10.1099/jmm.0.021089-0
98. Bandeira M, Santos CS, de Azevedo EC, Soares LM, Macedo JO, Marchi S, et al. Attenuated nephritis in inducible nitric oxide synthase knockout C57BL/6 mice and pulmonary hemorrhage in CB17 SCID and recombination activating gene 1 knockout C57BL/6 mice infected with Leptospira interrogans. Infect Immun (2011) 79(7):2936–40. doi:10.1128/IAI.05099-11
Keywords: Leptospira, reverse vaccinology, genome mining, vaccine discovery, vaccine candidate, recombinant vaccine, subunit vaccine, animal model
Citation: Grassmann AA, Souza JD and McBride AJA (2017) A Universal Vaccine against Leptospirosis: Are We Going in the Right Direction? Front. Immunol. 8:256. doi: 10.3389/fimmu.2017.00256
Received: 05 December 2016; Accepted: 21 February 2017;
Published: 09 March 2017
Edited by:
Monica E. Embers, Tulane University, USAReviewed by:
Arup Sarkar, Trident Academy of Creative Technology, IndiaCopyright: © 2017 Grassmann, Souza and McBride. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan John Alexander McBride, YWxhbi5tY2JyaWRlQHVmcGVsLmVkdS5icg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.