
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 06 March 2017
Sec. Molecular Innate Immunity
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.00210
This article is part of the Research Topic Immunoregulatory mechanisms of Interferon View all 30 articles
 Xu Wang1
Xu Wang1 He Wang1
He Wang1 Man-Qing Liu2
Man-Qing Liu2 Jie-Liang Li1
Jie-Liang Li1 Run-Hong Zhou1
Run-Hong Zhou1 Yu Zhou1
Yu Zhou1 Yi-Zhong Wang1
Yi-Zhong Wang1 Wang Zhou2
Wang Zhou2 Wen-Zhe Ho1,3*
Wen-Zhe Ho1,3*
Type III interferons (IFN-λs) have been demonstrated to inhibit a number of viruses, including HIV. Here, we further examined the anti-HIV effect of IFN-λs in macrophages. We found that IFN-λs synergistically enhanced anti-HIV activity of antiretrovirals [azidothymidine (AZT), efavirenz, indinavir, and enfuvirtide] in infected macrophages. Importantly, IFN-λs could suppress HIV infection of macrophages with the drug-resistant strains, including AZT-resistant virus (A012) and reverse transcriptase inhibitor-resistant virus (TC49). Mechanistically, IFN-λs were able to induce the expression of several important anti-HIV cellular factors, including myxovirus resistance 2 (Mx2), a newly identified HIV post-entry inhibitor and tetherin, a restriction factor that blocks HIV release from infected cells. These observations provide additional evidence to support the potential use of IFN-λs as therapeutics agents for the treatment of HIV infection.
Highly active antiretroviral therapy (HAART) has substantially reduced morbidity and mortality in HIV-infected individuals since its introduction in 1996 (1). Although HAART can suppress plasma viral loads to undetectable levels and improve patient life span (2), a substantial fraction of patients fail therapy and/or experience serious side effects from treatment, accompanied by the emergence of drug-resistant viruses (3, 4). More importantly, patients with HIV-1 infection can harbor the virus in latent reservoirs, such as macrophages, one of the key targets of HIV-1 infection. Studies have shown that the intracellular concentrations of antiretrovirals were significantly lower in macrophages than these in T lymphocytes (5, 6). It is known that macrophages play a crucial role in the host defense against HIV-1 infection, as they produce the multiple intracellular HIV restriction factors (7, 8). As HIV-1 latency is the major obstacle in preventing the eradication of the virus, it is necessary to identify agents that can induce intracellular antiviral factors against HIV-1 in macrophages.
Type III interferons (lambda interferons, IFN-λs) or interleukin-28/29 (IL-28/29) display IFN-like activities (9, 10), although they exert their functions through a receptor distinct from type I IFNs (11, 12). IFN-λ subfamily includes three structurally related cytokine members, IFN-λ1 (IL-29), IFN-λ2 (IL-28A), and IFN-λ3 (IL-28B). IFN-λs could be activated by viral infections or activation of toll-like receptors (13, 14). IFN-λs functionally resemble type I IFNs, the activation of which can trigger antiviral activity in vitro (11, 15–18) as well as in vivo (19, 20). However, unlike type I IFNs that have receptors expressed on many cell types, including the cells in brain, the expression pattern of IFN-λ receptors is more limited to specific cell types (17, 21–24). Thus, IFN-λs have fewer side effects than type I IFNs. The clinical importance of IFN-λs as novel antiviral therapeutic agents has recently become apparent. Several studies (12, 25–27) reported that the endogenous IFN-λ system is associated with treatment-induced clearance of hepatitis C virus (HCV). Furthermore, pegylated IFN-λ works as well as pegylated IFN-α for treating chronic hepatitis C (28–31), but with less side effects in several clinical trial studies. While it has been reported that IFN-λs could inhibit HIV replication in macrophages (17, 18) and CD4+ T cells (32), it is unclear whether IFN-λs can inhibit HIV infection with drug-resistant strains. In the present study, we investigated the antiviral effect of IFN-λs on antiretroviral-drug-resistant HIV strains in primary human macrophages. We also determined whether IFN-λs have synergistic effect on anti-HIV activity of antiretroviral drugs in infected macrophages.
Purified human peripheral blood monocytes were purchased from Human Immunology Core at the University of Pennsylvania (Philadelphia, PA, USA). The Core has the Institutional Review Board approval for blood collection from healthy donors. Monocytes were plated in 48-well culture plates (Corning CellBIND Surface, Corning Incorporated, Corning, NY, USA) at a density of 0.25 × 106 cells/well or 96-well culture plates (Corning CellBIND Surface, Corning Incorporated, Corning, NY, USA) at a density of 105 cells/well in the DMEM containing 10% FCS (33, 34). The medium was half-changed every 2 days. Monocytes differentiated to macrophages after in vitro cultured for 5–7 days. We used 7-day-cultured macrophages for experiments of this study.
Based on their differential use of the major HIV receptors (CCR5 and CXCR4), HIV isolates are classified to R5, X4, and R5X4 strains (35). HIV Bal strain (R5 tropic), AZT-resistant HIV A012 G691-6 strain (R5X4 tropic) (36) and the antiretroviral drugs (AZT, efavirenz, indinavir, and enfuvirtide) were obtained from the AIDS Research and Reference Reagent Program at NIH (Bethesda, MD, USA). Reverse transcriptase (RT) inhibitor-resistant HIV TC49 strain (R5 tropic) was kindly provided by Dr. David Katzenstein (Stanford University, Palo Alto, CA, USA). Recombinant human IFN-λ1 and IFN-λ2 were purchased from PeproTech Inc. (Rocky Hill, NJ, USA). Recombinant human IFN-λ3 was purchased from R&D Systems, Inc. (Minneapolis, MN, USA).
For infection with the resistant HIV strains, 7-day-cultured macrophages (105 cells/well in 96-well plates) were incubated with or without IFN-λ1, λ2, or λ3 (100 ng/ml each) and/or anti-HIV drugs: azidothymidine (AZT) 10−11M; efavirenz 10−10M; indinavir 10−15M, and enfuvirtide 10−8M for 24 h. Cells were then infected with different strains of HIV (6 ng p24/well) for 2 h. After washed three times with plain DMEM, cells were cultured with fresh 10% DMEM containing IFN-λs and/or antiretroviral drugs. For HIV Bal infection, culture supernatant was harvested at day 8 postinfection for RT and p24 assays. Infected and untreated cells served as controls. HIV Gag gene expression in infected cells was also examined at day 8 postinfection. For anti-HIV drug-resistant virus (A012 G691-6 or TC49) infection, culture supernatant was harvested for HIV p24 protein by ELISA at days 3, 5, 7, and 10 postinfection. The cell cultures were replaced with the fresh media supplemented with IFN-λ1, λ2, or λ3 and/or the antiretrovirals every 2–3 days. The culture supernatant collected at day 10 postinfection was also subjected to RT assay.
HIV RT activity was determined based on the technique (37) with modifications (38, 39). For HIV p24 assay, the cultured supernatant was analyzed ELISA as described in the protocol provided by the manufacturer (Chiron Corp., Emeryville, CA, USA).
RNA was extracted from cell cultures with Tri-Reagent (Molecular Research Center, Cincinnati, OH, USA) as previously described (40, 41). Total RNA (1 μg) was subjected to RTusing the RT system (Promega, Madison, WI, USA) for 1 h at 42°C. The reaction was terminated by incubating the reaction mixture at 99°C for 5 min, and the mixture was then kept at 4°C. The resulting cDNA was then used as a template for real-time PCR quantification. Real-time PCR was performed with 1/10 of the cDNA with the iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) as previously described (41–43). The oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA) and sequences will be available upon request. For the Gag gene expression, the specific oligonucleotide primers are listed as follows: Gag gene primer: 5′-ATAATCCACCTATCCC-AGTAGGAGAAA-3′ (SK38) and 5′TTTGGTCCTTGTCTTATGTCCAGAATGC-3′ (SK39) (44). For the tetherin gene expression, the specific oligonucleotide primers are listed as follows: 5′-AAGAAAGTGGAGGAGCTTTGAGG-3′ (Sense) and 5′-CCTGGTTTTCTCTTCTCAGT-CG-3′ (anti-sense). For the Mx2 gene expression, the specific oligonucleotide primers are listed as follows: 5′-CAGCCACCACCAGGA AACA-3′ (Sense) and 5′-TTCTGCTCGTACTGGCTGTACAG-3′ (anti-sense). The data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH, primers are 5′-GGTGGTCTCCTCTGACTTC AACA-3′ for sense and 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ for anti-sense, respectively) and presented as the change in induction relative to that of untreated control cells.
Cultured macrophages (2.5 × 105 cells/well in 48-well plate) were incubated with or without IFN-λ 1, 2, or 3 (100 ng/ml) for 24 h. Cells were then harvested, washed twice with phosphate-buffered saline containing 1% fetal bovine serum, incubated with PE-conjugated anti-human tetherin (CD317; BioLegend, San Diego, CA, USA) on ice in dark for 30 min. Unstained or isotope-matched mouse immunoglobulin G1-stained cells were included as a negative control. Stained cells were acquired by fluorescence-activated cell sorting (FACSCalibur; BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo software (Tree Star Inc., Ashland, OR, USA).
The expression of the Mx2 and tetherin were evaluated by immunoblot analysis. Following incubation with polyclonal antibodies to Mx2 (Novus, Littleton, CO, USA) or polyclonal rabbit anti-BST-2 (tetherin) serum (AIDS Research and Reference Program, Bethesda, MD, USA) and extensive washing in PBS containing 0.05% Tween-20, the membranes were incubated with horseradish peroxidase-conjugated IgG (Pierce, Chester, UK) for 1 h at room temperature. The membranes were further washed in PBS. The immunoblots were visualized by enhanced chemiluminescence detection (Amersham, Bucks, UK).
For comparison of the mean of two groups, statistical significance was assessed by Student’s t-test. One-way ANOVA were used for comparison of result between the different groups (multiple comparisons). All graphs were generated and statistical analyses were performed with GraphPad InStat Statistical Software (GraphPad Software Inc., San Diego, CA, USA), and the data are presented as mean ± SD. Statistical significance was defined as p < 0.05.
We first determined the effect of IFN-λs and/or the antiretrovirals (AZT, efavirenz, indinavir, and enfuvirtide) on HIV Bal infection of macrophages. IFN-λs (1, 2, or 3) or the antiretrovirals (AZT, efavirenz, indinavir, and enfuvirtide) significantly inhibited the expression of HIV p24 antigen (Figure 1A) and Gag gene (Figure 1B) in macrophages. IFN-λs (1, 2, or 3) also enhanced the anti-HIV (Bal) effect of AZT (Figure 2A), efavirenz (Figure 2B), indinavir (Figure 2C), and enfuvirtide (Figure 2D).
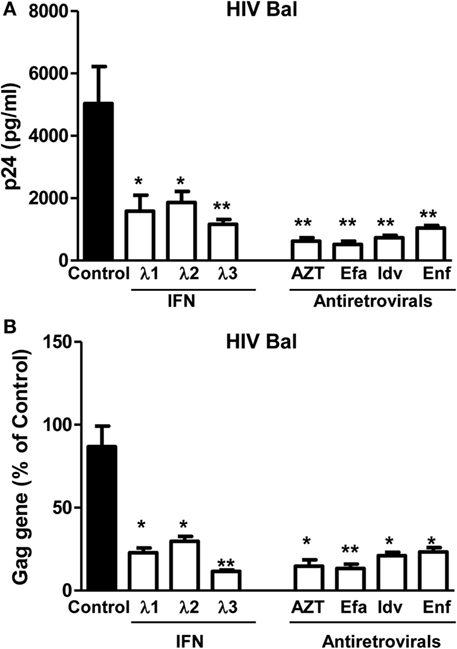
Figure 1. Effect of IFN-λs on HIV (Bal strain) infection of macrophages. Seven-day-cultured macrophages were incubated in the presence or absence of IFN-λs (1, 2, or 3; 100 ng/ml each) or classic antiretrovirals at indicated concentrations (AZT: 10−11M; efavirenz: 10−10M; indinavir: 10−15M; and emfuviride: 10−8M) for 24 h and then infected with HIV Bal strain. HIV-1 p24 production and Gag gene expression was determined at day 8 postinfection. (A) The cell culture supernatant was subjected to ELISA assay to detect HIV p24. (B) Total RNA from cells was subjected to HIV Gag gene expression by real-time RT-PCR. The data are expressed as RNA levels relative (percent) to the control (without treatment, which is defined as 100%). The results shown are the mean ± SD from three independent experiments with triplicate wells (*p < 0.05, **p < 0.01, IFN-λ, or antiretroviral vs. control; Efa, efavirenz; Idv, indinavir; Enf, enfuvirtide).
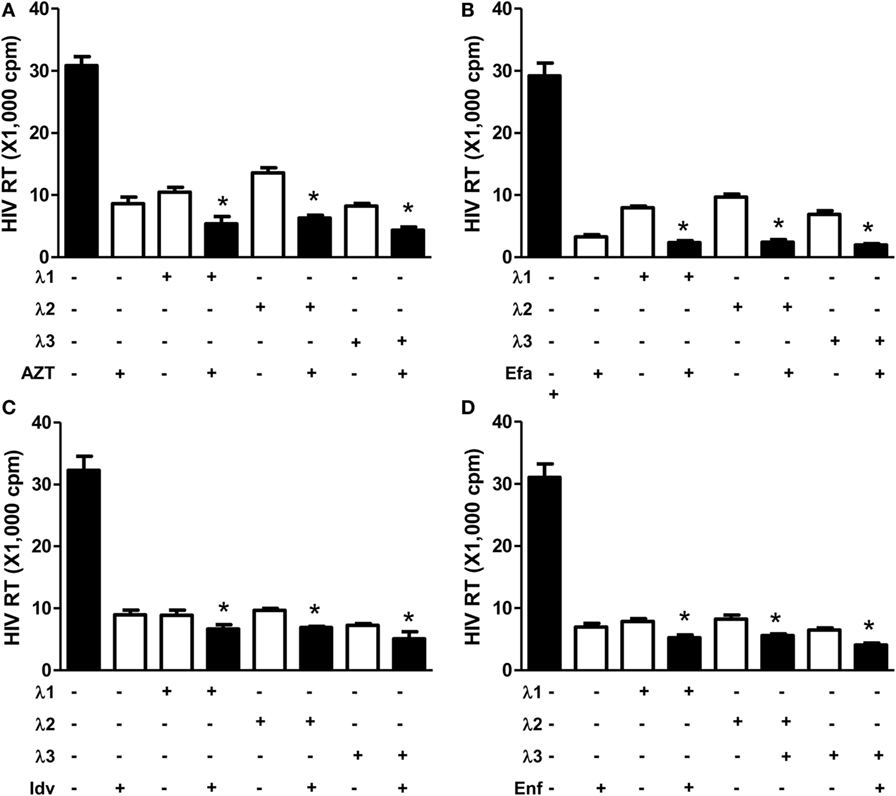
Figure 2. Effect of IFN-λs and/or antiretrovirals on HIV Bal infection of macrophages. Seven-day-cultured macrophages were incubated in the presence or absence of IFN-λs (1, 2, or 3; 100 ng/ml each) or classic antiretrovirals (A) AZT: 10−11M; (B) efavirenz: 10−10M; (C) indinavir: 10−15M; and (D) emfuviride: 10−8M) for 24 h and then infected with HIV Bal strain. HIV RT activity was determined at day 8 postinfection. The data shown are the mean ± SD from three independent experiments with triplicate wells (*p < 0.05, **p < 0.01, IFN-λ+ antiretrovirals vs. antiretrovirals only; Efa, efavirenz; Idv, indinavir; Enf, enfuvirtide).
We next examined whether IFN-λs (1, 2, or 3) can inhibit drug-resistant HIV infection of macrophages. While AZT had little effect on AZT-resistant HIV strain (A012) infection (Figures 3A,C IFN-λs 1, 2, or 3) potently suppressed infection of macrophages by the AZT-resistant HIV strain (A012) (Figures 3A,C). Similarly, IFN-λ 1, 2, or 3) could suppress RT inhibitor-resistant HIV (TC49) infection of macrophages. In contrast, the RT inhibitors (efavirenz) could not inhibit TC49 strain infection of macrophage (Figures 3B,D).
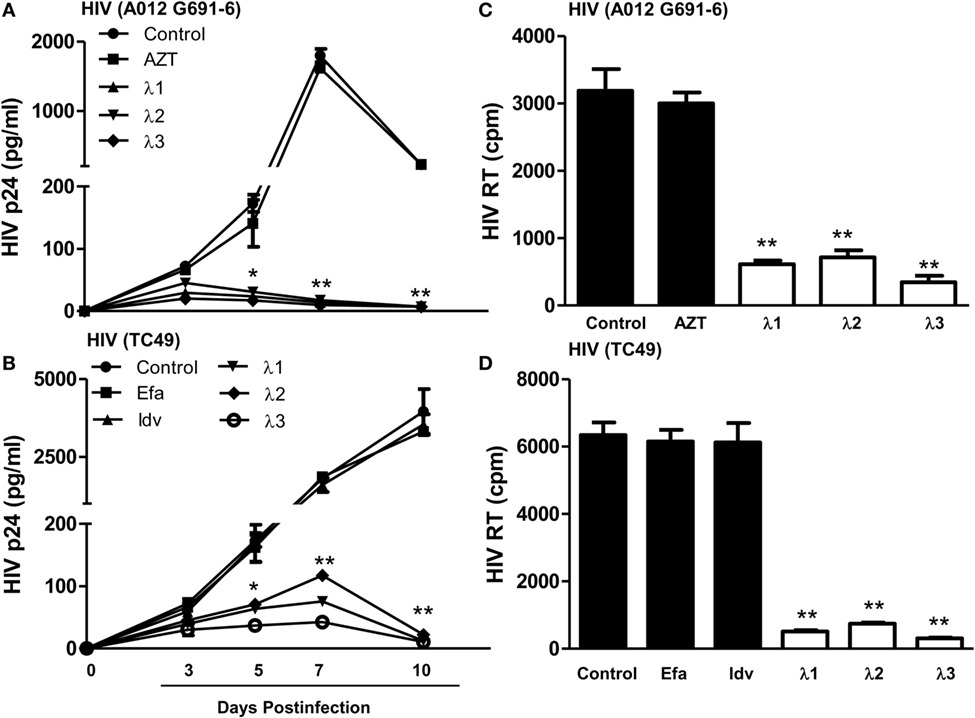
Figure 3. Effect of IFN-λs on drug-resistant HIV infection of macrophages. Seven-day-cultured macrophages were incubated in the presence or absence of IFN-λs (1, 2, or 3; 100 ng/ml each) for 24 h and then infected for 6 h with two drug-resistant viruses (A012 G691-6 or TC49). The HIV p24 antigen (A,C) was detected at indicated time points post HIV infection using a commercially available ELISA kit, and HIV RT activity (B,D) was assayed from the culture supernatant at day 10 postinfection. The data shown are the mean ± SD from three independent experiments with triplicate wells (*p < 0.05, **p < 0.01, IFN-λ vs. control; Efa, Efavirenz, 10−10M; Idv, Indinavir, 10−15M; AZT 10−11M).
Tetherin, an important IFN-α inducible cellular restriction factor, has been shown to inhibit HIV infection of host cells by preventing release of virus from an infected cell (45, 46). Thus, we examined whether IFN-λ treatment of macrophages can induce the tetherin expression. As shown in Figure 4, IFN-λ treatment of macrophages significantly increased the tetherin expression at both messenger RNA (mRNA) (Figure 4A) and protein (Figures 4B,C) levels.
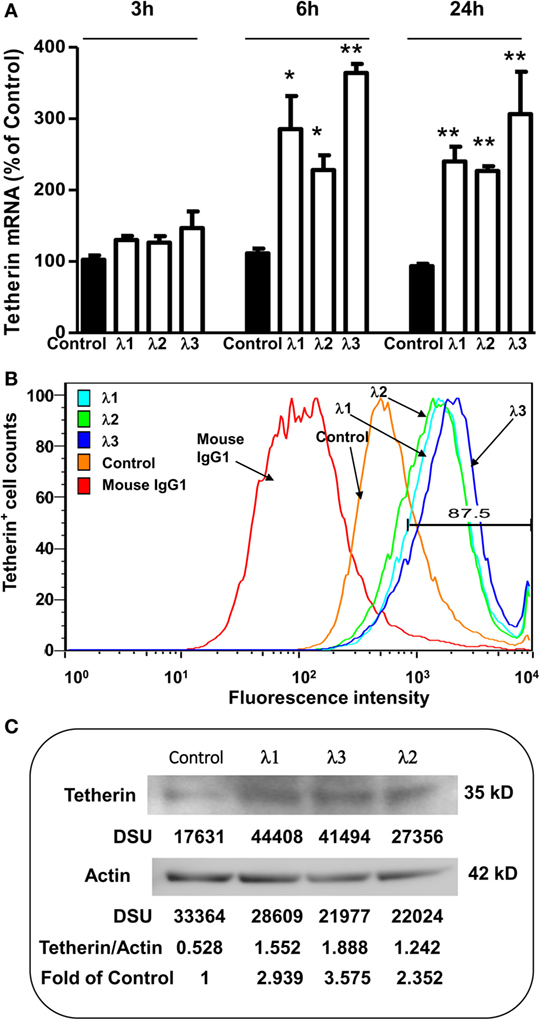
Figure 4. Effect of IFN-λs on tetherin expression. (A) Tetherin messenger RNA (mRNA) expression. Seven-day-cultured macrophages were incubated in the presence or absence of IFN-λs (1, 2, or 3; 100 ng/ml) for 3, 6, or 24 h. Total RNA was extracted from cells and then the real-time RT-PCR was performed to determine the induced mRNA expression of tetherin and GAPDH. The data are expressed as mRNA levels for tetherin relative (fold) to the control (without IFN-λ treatment, which is defined as 1). The results shown are the mean ± SD from three independent experiments with triplicate wells (**p < 0.01; *p < 0.05; IFN-λ vs. control). (B) Tetherin protein expression (flow cytometry). Seven day-cultured macrophages were treated with or without IFN-λs (1, 2, or 3; 100 ng/ml each) for 24 h. Cells were stained with fluorescence-conjugated anti-human tetherin (CD317) antibody and analyzed for tetherin expression by flow cytometry. The isotope control is staining with isotope-matched antibody (immunoglobulin G1). A representative histogram graph was shown. (C) Tetherin protein expression (Western blot). Seven day-cultured macrophages were treated with or without IFN-λ (1, 2, or 3; 100 ng/ml each) for 24 h. Total protein exacted from macrophages was subjected to Western blot assay using antibody against tetherin and actin. The inserts below the panels show the signal intensities [density scan unit (DSU)] of protein bands of the representative blot, expressed as densitometry scanning units. The results shown are representative of three independent experiments.
As an IFN-α-inducible cellular factor, Mx2 has recently been identified to inhibit HIV at post-entry level (47–49). Mx2 could abolish capsid-dependent nuclear import of subviral complexes (41–43). We thus examined whether IFN-λs can induce Mx2 expression in macrophages. As shown in Figure 5, IFN-λ treatment of macrophages significantly upregulated the Mx2 expression at both mRNA (Figure 5A) and protein (Figure 5B) levels.
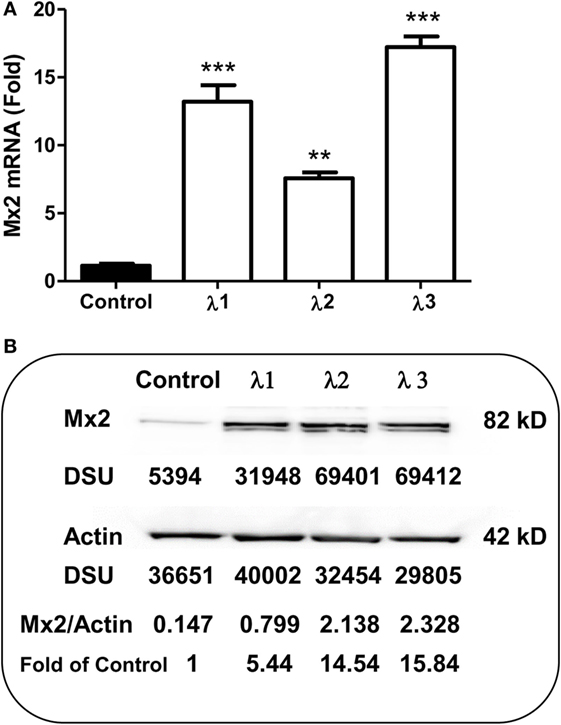
Figure 5. Effect of IFN-λs on Mx2 expression. (A) Mx2 messenger RNA (mRNA) expression. Seven-day-cultured macrophages were incubated in the presence or absence of IFN-λs (1, 2, or 3; 100 ng/ml each) for 24 h. Total RNA extracted from cells was subjected to the real-time RT-PCR was performed for Mx2 mRNA and GAPDH. The data are expressed as Mx2 mRNA levels relative (fold) to the control (without IFN-λ treatment, which is defined as 1). The results shown are the mean ± SD from three independent experiments with triplicate wells (**p < 0.01; *p < 0.05; IFN-λ vs. control). (B) Mx2 protein expression (Western blot). Seven day-cultured macrophages were treated with or without IFN-λ (1, 2, or 3; 100 ng/ml each) for 24 h. Total protein exacted from macrophages was subjected to Western blot assay using antibody against Mx2 and actin. The inserts below the panels show the signal intensities [density scan unit (DSU)] of protein bands of the representative blot, expressed as densitometry scanning units. The results shown are representative of three independent experiments.
To find new antiretroviral agents remains to be an important area of anti-HIV studies. Our earlier studies showed that IFN-λs could inhibit in vitro HIV infection/replication (17, 18, 50). IFN-λs are a class of recently identified members of IFN family, including three IFN-λ (lambda) molecules called IFN-λ1, IFN-λ2, and IFN-λ3 (also called IL-29, IL-28A, and IL-28B, respectively) (51). IFN-λs bind to their own distinctive receptor complex, IL-10Rβ and IL-28Rα, which activates janus kinase/signal transducers and activators of the transcription (JAK/STAT) signaling pathway, resulting in the phosphorylation of STAT proteins and forming of IFN-stimulated gene factor 3 complex (11, 15, 52).
While the IL-10Rβ shows a broad expression pattern (53), expression of the IFN-λ receptor subunit IL-28Rα is much more restricted (11, 54, 55). Earlier analysis of the expression pattern of IL-28Rα in human tissues showed that IL-28Rα mRNA levels were highest in the lung, heart, liver, and prostate, while low mRNA levels were detected in the central nervous system, bone marrow, testis, uterus, and skeletal muscle (22, 54, 55). A few immune cells express IL-28Rα especially at the mRNA level (e.g., B cells, macrophages, and plasmacytoid DCs), but conflicting protein expression data are reported in the literature (17, 22, 54–57). Although some evidence indicates that IFN-λ receptor expression on peripheral leukocytes is not functional (22), other evidence shows clear antiviral innate defense in some of these cells, and IFN-λ signals stimulate monocytes and macrophages to produce IL-6, IL-8, and IL-10 (58).
Studies from different investigators have demonstrated that IFN-λs have the ability to inhibit the replication of a number of viruses, including HCV and hepatitis B virus (16, 59), cytomegalovirus (60), Apeu virus (61), herpes simplex virus type 2 (HSV-2) (19), encephalomyocarditis virus (11), vesicular stomatitis virus (60), West Nile virus (62), and dengue virus (63). IFN-λs also had antiviral effect in vivo (19, 64, 65). Recent in vivo studies with mice showed that IFN-λs had the ability to reduce hepatic viral titer of HSV-2 and completely blocked HSV-2 replication in vaginal mucosa (19). IFN-λs contribute to innate immunity of mice against influenza A virus (66, 67). In addition, their role in direct antiviral effects in vivo has also been demonstrated in IL-28RA and STAT1 knockout animals, where a significant increase in influenza A virus replication was observed (66–68). Others and we have shown that IFN-λs could inhibit HIV infection of CD4+ T cells (32) and macrophages (17, 18, 50). Mechanistically, IFN-λ1 and IFN-λ2 were able to induce the intracellular expression of type I IFN, CC chemokines (the ligands for CCR5), and APOBEC3G/3F, the cellular HIV restriction factors (17). In addition, we demonstrated that IFN-λ3 could induce multiple antiviral cellular factors (ISG56, MxA, OAS-1) (18). We also showed that all three IFN-λs could induce the expression of pattern recognition receptors in macrophages (50). The in vivo production of IFN-λ1 also was monitored in HIV-infected patients. Tian et al. found that the plasma IFN-λ1 levels were increased along with the depletion of CD4+ T cells in HIV-1-infected patients, but the elevated IFN-λ1 showed limited repression of viral production (32).
In the present study, we further examined the anti-HIV activity of IFN-λs. We showed that all three IFN-λs not only inhibited drug-resistant virus replication (Figure 3) but also enhanced the anti-HIV effect of commonly used antiretrovirals (Figures 1 and 2), including zidovudine (AZT, a nucleoside RT inhibitor), efavirenz (a non-nucleoside RT inhibitor), indinavir (protease inhibitor), and enfuvirtide (HIV fusion inhibitor). In addition to the reported mechanisms involved in IFN-λ-mediated HIV inhibition (17, 18, 50): the induction of extracellular factors, e.g., CC chemokines that block HIV entry into macrophages, and the activation of intracellular innate immunity, e.g., the induction of type I IFNs and APOBEC3G/F, we demonstrated that IFN-λ treatment of macrophages induced the expression of tetherin, a cellular factor that can block HIV infection by preventing virus release from infected cells (Figure 4). In addition, IFN-λs also enhanced the expression of Mx2 (Figure 5), a newly identified HIV post-entry inhibitor that can abolish capsid-dependent nuclear import of subviral complexes (41–43). These anti-HIV cellular factors are the contributors for IFN-λ-mediated anti-HIV activity. These findings in conjunction with our previous observations (17, 18, 50) indicate that IFN-λs are attractive alternative for HIV treatment, as it would be extremely difficult for HIV to develop resistance to IFN-λs that can suppress the virus at various steps of its replication. However, further studies are necessary to determine the impact of IFN-λs on drug-resistant HIV strains in ex vivo and in vivo systems. These additional studies shall explore the clinical potential for developing IFN-λs-based therapy for HIV/AIDS.
In this in vitro study, we obtained primary human monocytes from the Immunology Core at the University of Pennsylvania School of Medicine. The Core has the Institutional Review Board approval for blood collection from healthy donors. Anyone who obtains human cells from the Core is considered as secondary use of de-identified human specimens, which does not subject to human subject review by both NIH and IRB.
XW and W-ZH designed the study. XW, M-QL, YZ, R-HZ, and Y-ZW performed the experiments. J-LL supplied reagents needed for this study. XW, M-QL, and YZ analyzed and interpreted the data and wrote the manuscript. HW, WZ, and W-ZH reviewed and revised the manuscript. All the authors have read, reviewed, and edited the manuscript and agreed for submission to this journal.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Institutes of Health grants DA41302, DA22177, DA40329, and MH109385.
1. Cole SR, Li R, Anastos K, Detels R, Young M, Chmiel JS, et al. Accounting for leadtime in cohort studies: evaluating when to initiate HIV therapies. Stat Med (2004) 23:3351–63. doi:10.1002/sim.1579
2. Qian K, Morris-Natschke SL, Lee KH. HIV entry inhibitors and their potential in HIV therapy. Med Res Rev (2009) 29:369–93. doi:10.1002/med.20138
3. Cheung PK, Wynhoven B, Harrigan PR. 2004: which HIV-1 drug resistance mutations are common in clinical practice? AIDS Rev (2004) 6:107–16.
4. Wainberg MA, Turner D. Resistance issues with new nucleoside/nucleotide backbone options. J Acquir Immune Defic Syndr (2004) 37(Suppl 1):S36–43. doi:10.1097/01.qai.0000137005.63376.6e
5. Gavegnano C, Detorio MA, Bassit L, Hurwitz SJ, North TW, Schinazi RF. Cellular pharmacology and potency of HIV-1 nucleoside analogs in primary human macrophages. Antimicrob Agents Chemother (2013) 57:1262–9. doi:10.1128/AAC.02012-12
6. Jin X, McGrath MS, Xu H. Inhibition of HIV expression and integration in macrophages by methylglyoxal-bis-guanylhydrazone. J Virol (2015) 89:11176–89. doi:10.1128/JVI.01692-15
7. Kuhl BD, Cheng V, Wainberg MA, Liang C. Tetherin and its viral antagonists. J Neuroimmune Pharmacol (2011) 6:188–201. doi:10.1007/s11481-010-9256-1
8. Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, et al. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood (2012) 120:778–88. doi:10.1182/blood-2012-01-407395
9. Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld JC. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem (2004) 279:32269–74. doi:10.1074/jbc.M404789200
10. Meager A, Visvalingam K, Dilger P, Bryan D, Wadhwa M. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine (2005) 31:109–18. doi:10.1016/j.cyto.2005.04.003
11. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol (2003) 4:69–77. doi:10.1038/ni875
12. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet (2009) 41:1100–4. doi:10.1038/ng.447
13. Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol (2008) 180:2474–85. doi:10.4049/jimmunol.180.4.2474
14. Pagliaccetti NE, Robek MD. Interferon-lambda in HCV infection and therapy. Viruses (2010) 2:1589–602. doi:10.3390/v2081589
15. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol (2002) 4:63–8. doi:10.1038/ni873
16. Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol (2005) 79:3851–4. doi:10.1128/JVI.79.6.3851-3854.2005
17. Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, et al. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol (2009) 83:3834–42. doi:10.1128/JVI.01773-08
18. Liu MQ, Zhou DJ, Wang X, Zhou W, Ye L, Li JL, et al. IFN-lambda3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PLoS One (2012) 7:e35902. doi:10.1371/journal.pone.0035902
19. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol (2006) 80:4501–9. doi:10.1128/JVI.80.9.4501-4509.2006
20. Ank N, West H, Paludan SR. IFN-lambda: novel antiviral cytokines. J Interferon Cytokine Res (2006) 26:373–9. doi:10.1089/jir.2006.26.373
21. Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog (2008) 4:e1000017. doi:10.1371/journal.ppat.1000017
22. Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, et al. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun (2009) 10:702–14. doi:10.1038/gene.2009.72
23. Zhou L, Wang X, Wang YJ, Zhou Y, Hu S, Ye L, et al. Activation of toll-like receptor-3 induces interferon-lambda expression in human neuronal cells. Neuroscience (2009) 159:629–37. doi:10.1016/j.neuroscience.2008.12.036
24. Li J, Hu S, Zhou L, Ye L, Wang X, Ho J, et al. Interferon lambda inhibits herpes simplex virus type I infection of human astrocytes and neurons. Glia (2011) 59:58–67. doi:10.1002/glia.21076
25. Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature (2009) 461:399–401. doi:10.1038/nature08309
26. Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet (2009) 41:1105–9. doi:10.1038/ng.449
27. Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature (2009) 461:798–801. doi:10.1038/nature08463
28. Vierling JM, Lataillade M, Gane EJ, Lüth S, Serfaty L, Taliani G, et al. Sustained virologic response (SVR12) in HCV genotype 1 patients receiving peginterferon lambda in combination with ribavirin and either daclatasvir or asunaprevir: interim results from the D-LITE study. Hepatol Int (2013) 7(suppl 1):S438–9. doi:10.1007/s12072-013-9429-0
29. Andersen H, Meyer J, Freeman J, Doyle SE, Klucher K, Miller DM, et al. Peginterferon lambda-1a, a new therapeutic for hepatitis C infection, from bench to clinic. J Clin Transl Hepatol (2013) 1:116–24. doi:10.14218/JCTH.2013.00014
30. Fredlund P, Hillson J, Gray T, Shemanski L, Dimitrova D, Srinivasan S. Peginterferon lambda-1a is associated with a low incidence of autoimmune thyroid disease in chronic hepatitis C. J Interferon Cytokine Res (2015) 35:841–3. doi:10.1089/jir.2014.0233
31. Flisiak R, Kawazoe S, Znoyko O, Assy N, Gadano A, Kao JH, et al. Peginterferon lambda-1a/ribavirin with daclatasvir or peginterferon alfa-2a/ribavirin with telaprevir for chronic hepatitis C genotype 1b. J Interferon Cytokine Res (2016) 36(11):635–43. doi:10.1089/jir.2015.0173
32. Tian RR, Guo HX, Wei JF, Yang CK, He SH, Wang JH. IFN-lambda inhibits HIV-1 integration and post-transcriptional events in vitro, but there is only limited in vivo repression of viral production. Antiviral Res (2012) 95:57–65. doi:10.1016/j.antiviral.2012.04.011
33. Liang H, Wang X, Chen H, Song L, Ye L, Wang SH, et al. Methamphetamine enhances HIV infection of macrophages. Am J Pathol (2008) 172:1617–24. doi:10.2353/ajpath.2008.070971
34. Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood (2009) 113:671–4. doi:10.1182/blood-2008-09-175000
35. Berger EA, Doms RW, Fenyo EM, Korber BT, Littman DR, Moore JP, et al. A new classification for HIV-1. Nature (1998) 391:240. doi:10.1038/35091
36. Larder BA, Darby G, Richman DD. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science (1989) 243:1731–4. doi:10.1126/science.2467383
37. Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, et al. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol (1988) 62:139–47.
38. Ho WZ, Lioy J, Song L, Cutilli JR, Polin RA, Douglas SD. Infection of cord blood monocyte-derived macrophages with human immunodeficiency virus type 1. J Virol (1992) 66:573–9.
39. Li Y, Wang X, Tian S, Guo CJ, Douglas SD, Ho WZ. Methadone enhances human immunodeficiency virus infection of human immune cells. J Infect Dis (2002) 185:118–22. doi:10.1086/338011
40. Wang X, Douglas SD, Metzger DS, Guo CJ, Li Y, O’Brien CP, et al. Alcohol potentiates HIV-1 infection of human blood mononuclear phagocytes. Alcohol Clin Exp Res (2002) 26:1880–6. doi:10.1097/01.ALC.0000042148.50808.04
41. Zhou Y, Wang X, Liu M, Hu Q, Song L, Ye L, et al. A critical function of toll-like receptor-3 in the induction of anti-human immunodeficiency virus activities in macrophages. Immunology (2010) 131:40–9. doi:10.1111/j.1365-2567.2010.03270.x
42. Ye L, Wang X, Wang S, Wang Y, Song L, Hou W, et al. CD56+ T cells inhibit hepatitis C virus replication in human hepatocytes. Hepatology (2009) 49:753–62. doi:10.1002/hep.22715
43. Wang X, Ye L, Zhou Y, Liu MQ, Zhou DJ, Ho WZ. Inhibition of anti-HIV microRNA expression: a mechanism for opioid-mediated enhancement of HIV infection of monocytes. Am J Pathol (2011) 178:41–7. doi:10.1016/j.ajpath.2010.11.042
44. Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature (2003) 424:94–8. doi:10.1038/nature01707
45. Kawai S, Azuma Y, Fujii E, Furugaki K, Ozaki S, Matsumoto T, et al. Interferon-alpha enhances CD317 expression and the antitumor activity of anti-CD317 monoclonal antibody in renal cell carcinoma xenograft models. Cancer Sci (2008) 99:2461–6. doi:10.1111/j.1349-7006.2008.00968.x
46. Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature (2008) 451:425–30. doi:10.1038/nature06553
47. Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature (2013) 502:559–62. doi:10.1038/nature12542
48. Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature (2013) 502:563–6. doi:10.1038/nature12653
49. Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe (2013) 14:398–410. doi:10.1016/j.chom.2013.08.015
50. Wang Y, Li J, Wang X, Zhou Y, Zhang T, Ho W. Comparison of antiviral activity of lambda-interferons against HIV replication in macrophages. J Interferon Cytokine Res (2015) 35:213–21. doi:10.1089/jir.2014.0064
52. Li M, Liu X, Zhou Y, Su SB. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol (2009) 86:23–32. doi:10.1189/jlb.1208761
53. Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity (2001) 15:35–46. doi:10.1016/S1074-7613(01)00169-8
54. Steen HC, Gamero AM. Interferon-lambda as a potential therapeutic agent in cancer treatment. J Interferon Cytokine Res (2010) 30:597–602. doi:10.1089/jir.2010.0058
55. Lasfar A, Abushahba W, Balan M, Cohen-Solal KA. Interferon lambda: a new sword in cancer immunotherapy. Clin Dev Immunol (2011) 2011:349575. doi:10.1155/2011/349575
56. Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M, et al. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun (2007) 8:254–61. doi:10.1038/sj.gene.6364348
57. Guenterberg KD, Grignol VP, Raig ET, Zimmerer JM, Chan AN, Blaskovits FM, et al. Interleukin-29 binds to melanoma cells inducing Jak-STAT signal transduction and apoptosis. Mol Cancer Ther (2010) 9:510–20. doi:10.1158/1535-7163.MCT-09-0461
58. Jordan WJ, Eskdale J, Boniotto M, Rodia M, Kellner D, Gallagher G. Modulation of the human cytokine response by interferon lambda-1 (IFN-lambda1/IL-29). Genes Immun (2007) 8:13–20. doi:10.1038/sj.gene.6364348
59. Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, Macdonald MR, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology (2006) 131:1887–98. doi:10.1053/j.gastro.2006.09.052
60. Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, et al. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol (2005) 289:G960–8. doi:10.1152/ajpgi.00126.2005
61. Almeida GM, De Oliveira DB, Magalhaes CL, Bonjardim CA, Ferreira PC, Kroon EG. Antiviral activity of type I interferons and interleukins 29 and 28a (type III interferons) against Apeu virus. Antiviral Res (2008) 80:302–8. doi:10.1016/j.antiviral.2008.06.016
62. Ma D, Jiang D, Qing M, Weidner JM, Qu X, Guo H, et al. Antiviral effect of interferon lambda against West Nile virus. Antiviral Res (2009) 83:53–60. doi:10.1016/j.antiviral.2009.03.006
63. Palma-Ocampo HK, Flores-Alonso JC, Vallejo-Ruiz V, Reyes-Leyva J, Flores-Mendoza L, Herrera-Camacho I, et al. Interferon lambda inhibits dengue virus replication in epithelial cells. Virol J (2015) 12:150. doi:10.1186/s12985-015-0383-4
64. Ank N, Paludan SR. Type III IFNs: new layers of complexity in innate antiviral immunity. Biofactors (2009) 35:82–7. doi:10.1002/biof.19
65. Muzammil, Jayanthi D, Faizuddin M, Noor Ahamadi HM. Association of interferon lambda-1 with herpes simplex viruses-1 and -2, Epstein-Barr virus, and human cytomegalovirus in chronic periodontitis. J Investig Clin Dent (2015). doi:10.1111/jicd.12200
66. Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, et al. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog (2008) 4:e1000151. doi:10.1371/journal.ppat.1000151
67. Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol (2010) 84:5670–7. doi:10.1128/JVI.00272-10
Keywords: IFN-λ, drug-resistant HIV, antiretrovirals, Mx2, tetherin
Citation: Wang X, Wang H, Liu M-Q, Li J-L, Zhou R-H, Zhou Y, Wang Y-Z, Zhou W and Ho W-Z (2017) IFN-λ Inhibits Drug-Resistant HIV Infection of Macrophages. Front. Immunol. 8:210. doi: 10.3389/fimmu.2017.00210
Received: 29 September 2016; Accepted: 15 February 2017;
Published: 06 March 2017
Edited by:
Jorg Hermann Fritz, McGill University, CanadaReviewed by:
Taruna Madan, National Institute for Research in Reproductive Health, IndiaCopyright: © 2017 Wang, Wang, Liu, Li, Zhou, Zhou, Wang, Zhou and Ho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Zhe Ho, d2VuemhlaG9AdGVtcGxlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.